Abstract
The class Kinetoplastea encompasses both free-living and parasitic species from a wide range of hosts. Several representatives of this group are responsible for severe human diseases and for economic losses in agriculture and livestock. While this group encompasses over 30 genera, most of the available information has been derived from the vertebrate pathogenic genera Leishmaniaand Trypanosoma. Recent studies of the previously neglected groups of Kinetoplastea indicated that the actual diversity is much higher than previously thought. This article discusses the known segment of kinetoplastid diversity and how gene-directed Sanger sequencing and next-generation sequencing methods can help to deepen our knowledge of these interesting protists.
Keywords: Kinetoplastea, metagenomics, metabarcoding, taxonomy, Trypanosomatida
Overview of kinetoplastid classification and diversity
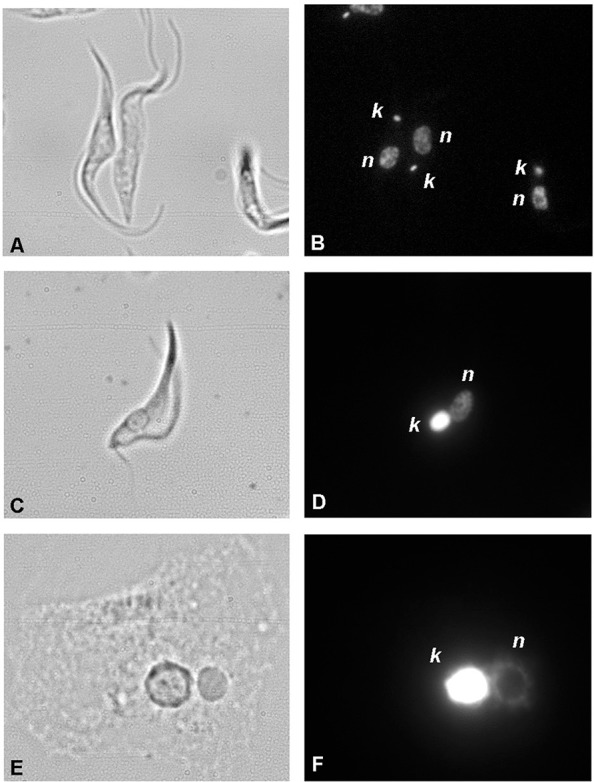
Kinetoplastid protists belonging to the phylum Euglenozoa (Cavalier-Smith 1981) are characterised by the presence of a kinetoplast, which is the apomorphy for the group and which is easily identifiable as a large mass of mitochondrial DNA (kDNA) (Vickerman & Preston 1976, Adl et al. 2012). The distribution of kDNA within the mitochondrion has three patterns: compacted and lying close to the flagellar pocket (termed eukinetoplast), dispersed throughout the mitochondrial lumen in several identical clusters (termed polykinetoplast), or unevenly dispersed as a diffuse mass (termed pankinetoplast) (Fig. 1) (Lukeš et al. 2002, Moreira et al. 2004). The lifestyle (parasitic vs. free-living, monoxenous vs. dixenous, intracellular vs. extracellular, and others), disease manifestation, and morphological traits have historically been used to classify these organisms (Lukeš et al. 2014, Votýpka et al. 2015a).
Fig. 1. : images of the main patterns of kinetoplast DNA arrangement. Eukinetoplast of Trypanosomabrucei (A, B), pankinetoplast of Trypanoplasma borreli (C, D) and polykinetoplast (E, F) of Perkinsela sp. (A, C, E) bright field (B, D, F) DAPI staining. k: kDNA; n: nucleolus.
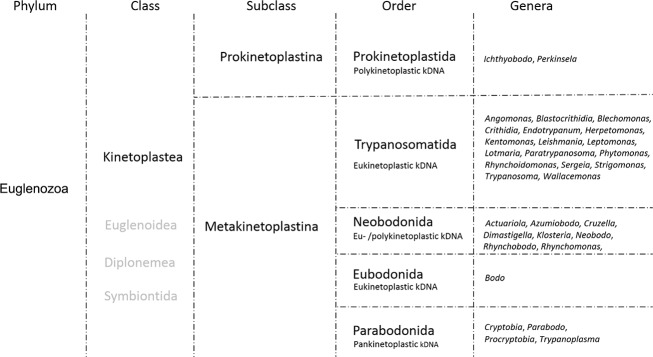
Recently, 18S (small subunit) rRNA-based phylogenetic analyses have led to extensive changes in the classification of kinetoplastid flagellates. The class Kinetoplastea, hierarchically equivalent to the formerly accepted order Kinetoplastida, is now divided into two subclasses: Prokinetoplastina and Metakinetoplastina (Moreira et al. 2004, Adl et al. 2012). The latter brings together four orders, of which the Trypanosomatida contains the majority of catalogued species. Notably, the National Center for Biotechnology Information database still uses the former version of classification, i.e., order Kinetoplastida, encompassing the families Bodonidae, Ichthyobodonidae and Trypanosomatidae (Fig. 2).
Fig. 2. : updated taxonomy of kinetoplastids. The phylum Euglenozoa (Cavalier-Smith 1981) encompasses five classes, among which the class Kinetoplastea is subdivided into two subclasses. The bulk of the diversity described is within the Metakinetoplastina that is further subdivided into four orders. The order Kinetoplastida encompasses representatives responsible for human diseases and contains the largest number of described genera and species. This organogram compiles taxonomic data from Moreira et al. (2004) and Adl et al. (2012). It should be pointed out that the National Center for Biotechnology Information database still uses the formerly accepted classification, i.e., order Kinetoplastida encompassing three families: Bodonidae, Ichthyobodonidae and Trypanosomatidae.
The order Trypanosomatida encompasses parasitic species responsible for economic losses in agriculture and livestock and for severe human diseases. The order Trypanosomatida is composed of a single family, Trypanosomatidae, which covers a diverse group of strictly parasitic uniflagellated protists with either monoxenous or dixenous life cycles. Regarding the latter, Chagas disease, leishmaniases and African sleeping sickness are diseases caused by Trypanosoma cruzi,Leishmania spp. and two subspecies of Trypanosoma brucei (T. b. rhodesiense and T. b. gambiense), respectively, and these diseases affect millions of people worldwide (Vickerman 1994, Stuart et al. 2008). In addition to humans, a wide range of domestic and wild animals can be infected by T. b. brucei, Trypanosoma congolense and Trypanosoma vivax, which are responsible for a complex of animal trypanosomiases in Africa that are collectively called nagana. T. b. evansi causes a globally distributed disease calledsurra in domestic and wild animals found in Asia, Africa, South America, and Europe (Carnes et al. 2015), and several other species can occasionally cause atypical human trypanosomiases (Truc et al. 2013). Moreover, new clades of potentially pathogenic trypanosomes are emerging in phylogenetic trees, further expanding the landscape of African trypanosomes (Votýpka et al. 2015b). Interestingly, some Trypanosoma or Leishmaniaspecies are nonpathogenic to mammals and can infect hosts such as lizards, fish, snakes and frogs (Simpson 1986, Simpson et al. 2006, Viola et al. 2009, Zídková et al. 2012, Grybchuk-Ieremenko et al. 2014, Stoco et al. 2014, Ferreira et al. 2015). Moreover, severalPhytomonas species can cause damage to economically important fruits and plants such as coffee, corn, coconut, oil palm, and cassava, although the phytopathology is not well established (Dollet 1984, Camargo 1999, Jaskowska et al. 2015).
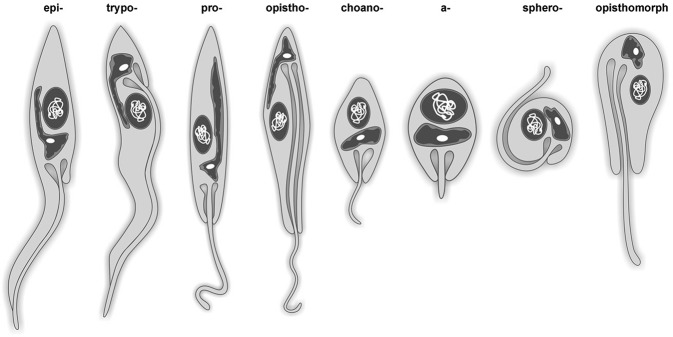
The most comprehensive and up-to-date catalogue of trypanosomatid genera and species was published 25 years ago and described known species, their synonymies, hosts, and distribution (Podlipaev 1990). Since then, substantial progress has been made in systematics and taxonomy primarily due to the introduction of molecular approaches. For a long time, trypanosomatid taxonomy was based solely on morphology and life cycles (Hoare & Wallace 1966, Vickerman 1976, McGhee & Cosgrove 1980), yet both parameters have a range of limitations, with morphology requiring the examiner to have a high level of proficiency (Fig. 3).
Fig. 3. : schematic representation of the main morphological forms present in trypanosomatids. The main typical morphotypes observed are represented; the dash should be replaced by the word “mastigote”.
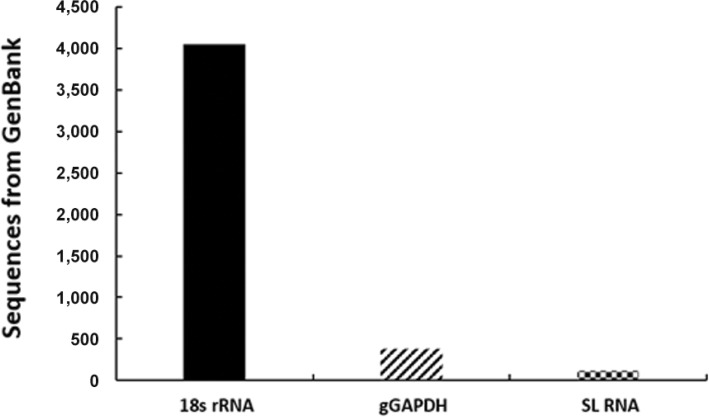
However, during the last decade, the traditional taxonomy has been integrated with DNA sequencing data. The 18S rRNA gene, glycosomal glyceraldehyde phosphate dehydrogenase (gGAPDH) and spliced leader (SL) RNA gene repeats are the most commonly used markers for molecular phylogenetic reconstructions of kinetoplastid flagellates (Maslov et al. 1996, Croan et al. 1997, Lukeš et al. 1997, Hollar et al. 1998, Yurchenko et al. 2000,Merzlyak et al. 2001, Hamilton et al. 2004, Teixeira et al. 2011, Borghesan et al. 2013) (Fig. 4). Using these molecular markers, species identification can be made by direct comparison with available DNA databases. However, if the match with the reference sequence is not full, the identification depends on accurate interpretation of molecular phylogenetic reconstructions and a rather arbitrary decision regarding whether the difference is intraspecific, interspecific or intergeneric. Commonly, the reference sequences are not correctly reassembled to updated taxonomic reclassifications, thus creating another challenging task of correctly comparing new isolates to previously described species.
Fig. 4. : comparison on the number of available kinetoplastid sequences from GenBank database. The most abundant sequences are: 18S (small subunit) rRNA gene, glycosomal glyceraldehyde phosphate dehydrogenase (gGAPDH) and spliced leader (SL) RNA. Mitochondrial cytochrome b, internal transcribed spacer one (ITS1) or two (ITS2) of rRNA, glucose 6 phosphate isomerase and the 70 kilodaltons heat shock protein also possess a considerable number of sequences, yet concentrated on Leishmaniaand Trypanosoma.
In this sense, it is not surprising that our knowledge about the apparently extensive diversity of this group of protists remains fragmented. Moreover, a taxonomic bias towards vertebrate pathogenic species exists; this bias improves our knowledge of their nutritional requirements, therefore favouring their isolation and cultivation in vitro, leaving a vast segment of the free-living species diversity unexplored. Indeed, the order Trypanosomatida has more described genera and species than the sum of the other four orders (Fig. 2).
Insect and plant trypanosomatids, although not usually pathogenic to humans, have been widely used in basic research as model organisms to unveil aspects of cellular biology, biochemistry and genetics and in the search for antitrypanosomatid drugs (Hoare & Wallace 1966, Vickerman & Preston 1976, McGhee & Cosgrove 1980, Camargo 1999). Another possible explanation for the great expansion of the known trypanosomatid diversity not correlated with that of other groups of Kinetoplastea may be morphological uniformity and, hence, wide occurrence of cryptic species (Von der Heyden & Cavalier-Smith 2005). Exploring the diversity of the entire Kinetoplastea class is thus relevant for (i) filling the gaps in the tree of life, which would help to reconstruct more robust phylogenetic and evolutionary histories, (ii) comprehension of protistan synecology (i.e., the composition of their communities) and (iii) diversity inventory and conservation for future generations.
The primary aim of this article is to discuss achievements and potentials to screen kinetoplastid diversity directly within the hosts and in the environment using modern molecular approaches.
Kinetoplastid diversity screen in the metagenomics era
Thus far, diversity and taxonomy studies have been based on polymerase chain reaction (PCR) amplification of molecular markers followed by DNA sequencing. This field is facing a dynamic and tremendous revolution. Over the past decade, the development of generations of sequencing technologies has resulted in an almost exponential increase in throughput and accuracy. Despite being relatively new, current sequencing techniques and associated bioinformatics analyses are now highly accurate and reasonably priced, with whole-genome sequencing of eukaryotes becoming a standard approach.
Complete genomic data of reference organisms are the best sources of information for diversity and phylogenetic studies. However, free-living protist genome projects encompass only a small fraction of completed and ongoing eukaryotic genome projects (Dawson & Fritz-Laylin 2009, del Campo et al. 2014), and the primary impediment to sequencing genomes is the scarcity of representative free-living protists in stable, axenic cultures (Dawson & Fritz-Laylin 2009). From 2,213 fully sequenced eukaryotic genomes, 59 belong to kinetoplastid protists, the majority of which pertain to the genera Leishmania (n = 24) andTrypanosoma (n = 16); these genera are over-represented due to their medical importance. The other genera with available genomic data are as follows: Crithidia (n = 3), Leptomonas (n = 2),Trypanoplasma (n = 1), Strigomonas (n = 4),Angomonas (n = 3), Lotmaria (n = 1),Herpetomonas (n = 1), Endotrypanum (n = 1),Bodo (n = 1) and Phytomonas (n = 3) (ncbi.nlm.nih.gov/genomes, sanger.ac.uk, tritrypdb.org). High-quality, well-annotated genomes are available for trypanosomatids. Additionally, molecular tools, such as gene knockouts, ectopic gene expression, RNAi and CRISPR, have been developed to improve genome annotation and to determine gene function and localisation (Dean et al. 2015). New bioinformatics tools for reanalysis of genome databases allow further identification of “partial” genes that can be categorised as C-terminal extensions, gene joining, tandemly repeated paralogs and wrong chromosomal assignments (Pawar et al. 2014).
The microeukaryotic diversity that resides in ecological niches such as animal microbiotas (for instance, insect gut or salivary glands), lakes, oceans and soil remains poorly understood (Foster et al. 2012, Weinstock 2013). Furthermore, any existing relationships among these species remain largely undiscovered. Due to the reduction in costs, labour intensity and time, new generation sequencing has the potential to reveal both the diversity and/or ecological and metabolic functions in virtually any environment. A recent salient example is the qualitative and quantitative new insights into this problem achieved by the Tara Oceans project, which not only massively extended the known eukaryotic diversity in the world oceans (de Vargas et al. 2015), but also explored a wealth of putative interactions among them (Lima-Mendez et al. 2015). However, because DNA sequencing from environmental samples generates a large amount of information, correctly and clearly formulated questions are of major importance.
The concept of DNA metabarcoding relies upon the identification of species present in environmental samples directly, without the need for microscopic observation or cultivation. This method is performed by direct extraction of DNA and PCR amplification of a selected gene (fragment) used to barcode the targeted group of eukaryotes (Pompanon et al. 2011, Epp et al. 2012, Taberlet et al. 2012, Aylagas et al. 2014, Pompanon & Samadi 2015). The metabarcoding approach aims to answer the following question: who is out there? In contrast, metagenomics aspires to functionally analyse the whole DNA present in a given sample from the perspective of the following question: how does the organismal assembly function? The two approaches have thus far been used by the research community somewhat indistinctly, although a distinction is advisable (Mendoza et al. 2015).
The utilisation of DNA sequences of short standardised gene fragments for quick and accurate determination of the species is called DNA barcoding. Because no consensus of a single marker able to distinguish and classify all the species on the planet exists, group-specific markers have been proposed (Pawlowski et al. 2012) (also see BOLD; boldsystems.org/). The regions of the mitochondrial gene encoding cytochrome c oxidase subunit 1 (CO1) and mitoribosomal RNAs are used for animals (Hebert et al. 2003), while two large subunits of the chloroplast RuBisCO and maturase K genes are used for plants, 16S rRNA for bacteria, internal transcribed spacer region 1 for fungi, and some other genes for less studied groups (Pawlowski et al. 2012). Although CO1 was shown to be insufficient for species delimitations for many microorganisms (Begerow et al. 2010, Pawlowski et al. 2012, Lebonah et al. 2014), it is applicable to a number of eukaryotic groups including trypanosomatids (Chantangsi et al. 2007,Nassonova et al. 2010, Stern et al. 2010, Kher et al. 2011, KA Morelli et al., unpublished observations). However, a consensual barcoding approach for kinetoplastids does not exist, although barcoding by means of 18S rRNA and gGAPDH is applied frequently.
The majority of the microeukaryotic diversity remains undiscovered primarily due to the methodological approaches used to assess it. While prokaryotic diversity studies are based mainly on 16S rRNA sequencing of their communities, for historical reasons, protistan diversity described without the establishment of axenic cultures and/or microscopic observation was considered incomplete and insufficient during the genomic era (Votýpka et al. 2015a). The identification of a kinetoplastid species has been traditionally based on its introduction into an axenic culture, with the culture-dependent approach considered critical for species validation. However, although establishment in culture is not feasible in many cases, the metabarcoding approach is not yet widely used even in studies of protistan diversity (Stoeck et al. 2005, Von der Heyden et al. 2005, Sauvadet et al. 2010, McCarthy et al. 2011, Bates et al. 2013, Glaser et al. 2014). Other hurdles include the low number of reference genomes in databases available for comparison and difficulties in establishing universally accepted markers (Sturm et al. 2008), as discussed above. In many cases, culture establishment is prevented by our limited knowledge of kinetoplastid metabolism and nutritional requirements, which is improving at a very slow pace even in well-studied groups (Škodová-Sveráková et al. 2015). Consequently, we are confined only to the cultivable fraction of protist diversity. Direct microscopic observation of environmental samples provides substantial morphological and ecological data related to eukaryotic communities in vivo. However, these data are hard to compare with the existing formally recognised species primarily due to high morphological variability (Dawson & Fritz-Laylin 2009, Votýpka et al. 2015a). Culture-independent approaches to assess diversity, such as single-cell sequencing methodology, which was recently successfully applied to protists (Kolísko et al. 2014), should help address these questions. Overall, the exploration of protistan diversity in general and kinetoplastid diversity in particular, appears significantly restrained by established and rather rigid traditional approaches.
Genes used for molecular phylogeny of kinetoplastids
The SL RNA gene has been repeatedly used to explore trypanosomatid diversity using either parasites isolated in culture or direct insect gut contents, allowing many new trypanosomatid taxa to be described (Westenberger et al. 2004, Maslov et al. 2007, 2010, Yurchenko et al. 2008, 2009, Votýpka et al. 2010, 2012, 2013, 2014, Wilfert et al. 2011). This gene is absent from host genomes and from nonkinetoplastid microorganisms that could occur within such samples (Westenberger et al. 2004). The SL RNA gene consists of regions with different levels of variability (exon, intron, and intergenic spacer variability), which makes this gene suitable for both inter and intraspecific comparisons. Additionally, differences in the product amplification length among trypanosomatid species often allow the detection of mixed infections by standard agarose gel electrophoresis.
Species discrimination using the SL RNA gene is based on a 90% sequence similarity threshold (Westenberger et al. 2004). Although this criterion is arbitrary, it has withstood scrutiny and has provided a simple operational rule necessary for broad-scale studies. Hence, this criterion is an integral part of taxonomic studies of insect trypanosomatids (Kostygov et al. 2014).
Meanwhile, using the SL RNA gene in diversity studies has several disadvantages, particularly for PCR-based approaches. First, universal primers for this marker are not suitable for its amplification in some trypanosomatids (Podlipaev et al. 2004), making its use for metabarcoding analysis of the entire Kinetoplastea class questionable. Thus, SL RNA-based mapped diversity may be narrower than the actual diversity. Second, the very short conserved region of the gene (the exon and intron together are approximately 100 bp in length) does not provide sufficient data for deeper phylogenetic analysis. Third, different SL RNA gene classes varying in size and sequence have been described in a few species (Lamontagne & Papadopoulou 1999), yet this finding was not confirmed by whole-genome analyses (Berriman et al. 2005, Thomas et al. 2005). Fourth, the size differences of SL RNA gene repeats lead to competitive amplification favouring shorter PCR products. Hence, in the case of mixed infections, some species with longer repeats may remain undetected; this particular issue can be effectively addressed using new generation sequencing.
Due to these disadvantages, several research groups adopted a more habitual marker in diversity studies, the 18S rRNA gene, which can be amplified either from environmental samples or from cultured materials. The usage of different kinetoplastid-specific primers allows either the nearly complete gene or its most variable part to be obtained (Maslov et al. 1996, Kostygov & Frolov 2007,Votýpka et al. 2015b). Thus far, the 18S rRNA gene has been successfully used in diversity studies not only for insect trypanosomatids (Votýpka et al. 2010, 20,2012, Týč et al. 2013), but also for fish trypanosomes and trypanoplasms (Grybchuk-Ieremenko et al. 2014, Losev et al. 2015), as well as for flagellates from deep-sea samples (Sauvadet et al. 2010, Scheckenback et al. 2010, Pawlowski et al. 2011, Salani et al. 2012d, de Vargas et al. 2015). A few reports used the 18S rRNA gene to scrutinise lake sediments (van Hannen et al. 1999) and soil (Glaser et al. 2014). No generally accepted criterion of species discrimination exists based on this gene most likely due to its unpredictable variability in different groups of eukaryotes. For example, the observed multiple closely related haplotypes of this gene in trypanosomes parasitising fishes suggest that some intraspecific variability of this marker exists within the given group (Grybchuk-Ieremenko et al. 2014).
Assessment of molecular diversity by metagenomic approaches
Comprehensive assessment of the molecular diversity of unicellular eukaryotes retrieved from deep-sea water has been the focus of several studies in the past 15 years. Although prokaryotic communities have been studied extensively, protists have been generally much less explored in aquatic environments, where they thrive even under conditions of high pressure, high toxic product concentrations and high and low temperatures. A study devoted to revealing microeukaryotic diversity in the abyssal sea floor of the Atlantic Ocean used general eukaryotic and kinetoplastid-specific primers to discover members of the generaIchthyobodo, Rhynchobodo andNeobodo (Scheckenbach et al. 2010). In cultivation-independent studies of the South Atlantic, Mediterranean and other sites, kinetoplastid-specific 18S rRNA primers were used to detect Neobodo designis, Rhynchobodo sp. andIchthyobodo. Notably, a particular percentage of identical clones is shared among even geographically distant regions, suggesting global distribution (Von der Heyden & Cavalier-Smith 2005, Salani et al. 2012). Protist community surveys from deep-sea waters from hydrothermal vents in the Pacific Ocean using general 18S rRNA primers revealed the presence of Bodo sp. and Bodo saliens (Brown & Wolfe 2006, Sauvadet et al. 2010). In other hydrothermal areas in the Mid-Atlantic Ridge and the eastern Pacific Ocean, kinetoplastids such as Ichthyobodo necator, Procryptobia sorokini, Rhynchomonas nasuta,Bodosaltans and B. saliens were also abundant (Atkins et al. 2000, López-García et al. 2003). Although these data reveal the ubiquitous distribution of kinetoplastids and their exciting plasticity, which allows them to adapt to extreme environments, no cultured representatives from these environments are available. In spite of these advances, deep-sea kinetoplastid sequences have disproportionally low representation in public databases (Salani et al. 2012). An extensive 18S rRNA metabarcoding study of the sunlit zone of the world oceans by the Tara Oceans initiative revealed a surprisingly highly abundant presence of diplonemids (Lukeš et al. 2015) and a much less conspicuous presence of kinetoplastids (de Vargas et al. 2015). In another 18S rRNA-based survey targeting aquatic microeukaryotes in The Netherlands, sequences related to parasitic trypanosomatids have been described (van Hannen et al. 1999). However, their re-analysis against recently available sequences revealed their high identity with N. designis (KA Morelli, unpublished observations).
A cultivation-independent survey of kinetoplastid diversity in soil employed 18S rRNA primers and revealed an abundance of sequences related to the neobodonid clade, followed by parabodonids and eubodonids (Glaser et al. 2014). While approximately 30% of the obtained sequences have low similarity to databases, whether these sequences are derived from unknown taxa, the so-called rare biosphere, or represent methodological “noise” remains to be established (Glaser et al. 2014). In a study that aimed to investigate the role of free-living protists in contaminated food,Bodo sp. and Parabodo caudatus were frequently detected, along with related sequences with low BLAST scores (Vaerewijck et al. 2008).
Collectively, these data emphasise the need for more comprehensive studies targeting free-living kinetoplastids, the diversity of which remains fractionated, underestimated and, consequently, poorly taxonomically and phylogenetically studied. As a result of the increasing application of 18S rRNA gene-based approaches, new protistan phylotypes are constantly being revealed (López-García et al. 2001, Taib et al. 2013), improving our knowledge of the diversity, distribution and function of eukaryotic microorganisms.
Museums and institutional collections as a basis for diversity screening
In comparison to macroscopic eukaryotes, protist collections are generally unknown to the public certainly because they concern microscopic organisms that are not spectacular or emblematic. These collections are often accumulated in dusty boxes of slides stored on shelves in an obscure corner. However, for protistologists, such collections are gold mines primarily because they contain type material (hapantotypes) deposited since the end of the XIX century by generations of scientists (Votýpka et al. 2015a).
With the beginning of the molecular era in the 1990s, natural history collections evolved to meet the challenges of the current and future interdisciplinary studies. Many institutions developed new collections and information databases (DNA, tissues, cultures, cryobanks, photographs, ethanol-fixed specimens, publication collections, and geographical and ecological information databases), which are of first-rate importance, offering opportunities to conduct integrative studies, including temporal and spatial surveys (Suarez & Tsutsui 2004).
With the worldwide awareness of the dramatic erosion of both macro and microorganismal diversity, the necessity of its inventory and preservation is now a priority. Many museums and academic institutions are engaged in large surveys in diversity host spots [see for example laplaneterevisitee.org/en/ (Bouchet et al. 2008)]. In addition to traditional taxonomy, DNA barcoding approaches are used to describe diversity. Furthermore, recent works have demonstrated the possibility of extracting relevant genetic information from ancient archived specimens such as archaeological remains (Frías et al. 2013), formalin-fixed tissues (Gilbert et al. 2007), and fixed and stained smears (Hayes et al. 2014). For a long time, such material was considered useless for molecular analyses due to DNA degradation. Studies on ancient human remains have changed the widely accepted theory of the origin of Chagas disease. Approximately 9,000-year-old pre-Colombian mummies were shown to be PCR-positive for T. cruzi, indicating that Chagas disease is at least as old as human presence in the Americas (Aufderheide et al. 2004). Another example derived from the museum collections is the rapid extinction of endemic rats on Christmas Island around the year 1900 due to Trypanosoma lewisiintroduced by black rats and their fleas (Wyatt et al. 2008).
The possibility of extracting DNA suitable for amplification from fixed and stained blood smears and other difficult samples opens new avenues for the molecular characterisation of kinetoplastid type specimens deposited in collections. Their potential use in studying kinetoplastid diversity can be illustrated by the recent work on trypanosomes of marine fishes from South Africa and their leech vectors (Hayes et al. 2014).
Trends in metabarcoding of kinetoplastids
Direct sequencing of the environmental DNA, either total or focused on barcoding markers, has been the basis for “blind” diversity screens. After the early studies of diversity through direct DNA sequencing, the overall ratio of cultivable microorganisms has been generally accepted not to exceed 1% of the total diversity on earth (Pace 1997). For protists in particular, less than 10% of the sequences revealed by cultivation-independent molecular surveys were previously known (Šlapeta et al. 2005, Medinger et al. 2010). These approaches revealed not only putatively novel species, but also new kingdoms (Dawson & Pace 2002, Berney et al. 2004, Cavalier-Smith 2004). However, these data are problematic because nothing beyond the molecular signature is known, such as morphological and/or biochemical characteristics of the new organisms, their ecological roles, or in situ abundance. Hence, we can only speculate whether these distinct molecular signatures represent existing unknown microbes or are only methodological artefacts.
Although taxonomic information of an unknown microorganism through DNA sequencing is interesting per se, ideally, this information must be combined with morphological, biochemical and ecological data (Votýpka et al. 2015a). For example, in the order Neobodonida, an undescribed sequence indicated the existence of a novel clade that appeared to consist of free-living organisms from aquatic and terrestrial habitats (López-García et al. 2003, Von der Heyden & Cavalier-Smith 2005). However, no cultured representatives of this clade were available. Later, a diversity survey using combined molecular and culturing approaches succeeded in isolating and culturing an organism that branched within that undescribed neobodonid clade according to its phylogenetic position (Stoeck et al. 2005).
Another issue to consider while screening environmental sequences is whether the infrequent sequences are indeed members of a highly diverse microbial “rare biosphere” or only represent sequencing artefacts. To address this question, tintinnid ciliates, a species-rich group that can be easily distinguished morphologically, were surveyed to assess the accuracy of 18S rRNA pyrosequencing of Mediterranean samples with different patterns of tintinnid diversity. The inferred number of typing units outnumbered tintinnid cells in the samples, which was found to be primarily dependent on the data treatment, suggesting that many undescribed environmental sequences might indeed be artefacts (Bachy et al. 2013).
The intention of this review is to critically evaluate the usefulness of methodological advances for studies of kinetoplastid diversity. The scarcity of protist environmental data is a large obstacle for the perception of true eukaryotic diversity. An analysis of the SILVA SSU database of the eukaryotic phyla (Pruesse et al. 2007) showed that less than 5% of the 18S rRNA sequences originated from protists (Pace 2009). A recent re-evaluation of environmental studies revealed that protists that were previously overlooked constitute the bulk of extant eukaryotic diversity (Pawlowski et al. 2011).
Metabarcoding has become a fundamental approach for diversity assessment in recent years. The possibility of revealing previously unknown microorganisms through metabarcoding and the potential of unveiling their physiology and ecology through metagenomics pose great opportunities and challenges to protistologists.
ACKNOWLEDGEMENTS
To all the members of our laboratories, for helpful and stimulating discussions, and to Dr Marta H Branquinha and André LS Santos (Institute of Microbiology Prof Paulo de Góes, Federal University of Rio de Janeiro), for critical reading of the paper.
Footnotes
Financial support: FP7 (316304), Bioglobe (CZ.1.07/2.3.00/30.0032) (to JL), Praemium Academiae (to JL), CAPES, MCT/CNPq, FAPERJ, FIOCRUZ
VY is supported by the Moravskoslezský Kraj research initiative (MSK.DT1/2014, 00955/RRC/2015) and the Czech Science Foundation (P506/13/24983S), CMD-L receives fellowship from CNPq and FAPERJ, LD was supported by a post-doctoral grant of the LABEX BCDiv, AK worked in the context of state budgetary topic 01201376804 of ZIN RAS. CMD-L, VY, PG and JL contributed equally to this work.
REFERENCES
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick RS, Schoch CL, Smirnov A, Spiegel FW. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins MS, Teske AP, Anderson OR. A survey of flagellate diversity at four deep-sea hydrothermal vents in the eastern Pacific Ocean using structural and molecular approaches. J Eukaryot Microbiol. 2000;47:400–411. doi: 10.1111/j.1550-7408.2000.tb00067.x. [DOI] [PubMed] [Google Scholar]
- Aufderheide AC, Salo W, Madden M, Streitz J, Buikstra J, Guhl F, Arriaza B, Renier C, Wittmers LE, Jr, Fornaciari G, Allison M. A 9,000-year record of Chagas disease. Proc Natl Acad Sci USA. 2004;101:2034–2039. doi: 10.1073/pnas.0307312101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylagas E, Borja A, Rodríguez-Ezpeleta N. Environmental status assessment using DNA metabarcoding: towards a genetics based marine biotic index (gAMBI) PLoS ONE. 2014;9: doi: 10.1371/journal.pone.0090529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachy C, Dolan JR, López-García P, Deschamps P, Moreira D. Accuracy of protist diversity assessments: morphology compared with cloning and direct pyrosequencing of 18S rRNA genes and ITS regions using the conspicuous tintinnid ciliates as a case study. ISME J. 2013;7:244–255. doi: 10.1038/ismej.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates ST, Clemente JC, Flores GE, Walters WA, Parfrey LW, Knight R, Fierer N. Global biogeography of highly diverse protistan communities in soil. ISME J. 2013;7:652–659. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begerow D, Nilsson H, Unterseher M, Maier W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl Microbiol Biotechnol. 2010;87:99–108. doi: 10.1007/s00253-010-2585-4. [DOI] [PubMed] [Google Scholar]
- Berney C, Fahrni J, Pawlowski J. How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol. 2004;2: doi: 10.1186/1741-7007-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Böhme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, Macleod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;5733:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Borghesan TC, Ferreira RC, Takata CS, Campaner M, Borda CC, Paiva F, Milder RV, Teixeira MM, Camargo EP. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist. 2013;164:129–152. doi: 10.1016/j.protis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Bouchet P, Héros V, Lozouet P, Maestrati P. Héros V, Cowe RH, Bouchet P.eds. Tropical deep-sea Benthos 25. Muséum National d’Histoire Naturelle; Paris: 2008. A quarter-century of deep-sea malacological exploration in the South and West Pacific: where do we stand? How far to go? pp. 9–40. [Google Scholar]
- Brown PB, Wolfe GV. Protist genetic diversity in the acidic hydrothermal environments of Lassen Volcanic National Park, USA. J Eukaryot Microbiol. 2006;53:420–431. doi: 10.1111/j.1550-7408.2006.00125.x. [DOI] [PubMed] [Google Scholar]
- Camargo EP. Phytomonas and other trypanosomatid parasites of plants and fruit. Adv Parasitol. 1999;42:29–112. doi: 10.1016/s0065-308x(08)60148-7. [DOI] [PubMed] [Google Scholar]
- Carnes J, Anupama A, Balmer O, Jackson A, Lewis M, Brown R, Cestari I, Desquesnes M, Gendrin C, Gibson W, Hertz-Fowler C, Imamura H, Ivens A, Jansen A, Kořený L, Lai D-H, MacLeod A, Martin D, McDermott SM, Merritt C, Monnerat S, Moon W, Myler P, Phan I, Ramasamy G, Sivam D, Lun Z-R, Lukeš J, Stuart K, Schnaufer A. Genome and phylogenetic analyses of Trypanosoma evansi reveal extensive similarity to T. brucei and multiple independent origins for dyskinetoplasty. PLoS Negl Trop Dis. 2015;9: doi: 10.1371/journal.pntd.0003404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Eukaryote kingdoms: seven or nine? Biosystems. 1981;14:461–481. doi: 10.1016/0303-2647(81)90050-2. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Only six kingdoms of life. Proc Biol Sci. 2004;271:1251–1262. doi: 10.1098/rspb.2004.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantangsi C, Lynn DH, Brandl MT, Cole JC, Hetrick N, Ikonomi P. Barcoding ciliates: a comprehensive study of 75 isolates of the genus Tetrahymena. Int J Syst Evol Microbiol. 2007;57:2412–2425. doi: 10.1099/ijs.0.64865-0. [DOI] [PubMed] [Google Scholar]
- Croan DG, Morrison DA, Ellis JT. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol Biochem Parasitol. 1997;89:149–159. doi: 10.1016/s0166-6851(97)00111-4. [DOI] [PubMed] [Google Scholar]
- Dawson SC, Fritz-Laylin LK. Sequencing free-living protists: the case for metagenomics. Environ Microbiol. 2009;7:1627–1631. doi: 10.1111/j.1462-2920.2009.01965.x. [DOI] [PubMed] [Google Scholar]
- Dawson SC, Pace NR. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci USA. 2002;99:8324–8329. doi: 10.1073/pnas.062169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vargas C, Audic S, Henry N, Decelle J, Mahé F, Logares R, Lara E, Berney C, Le Bescot N, Probert I, Carmichael M, Poulain J, Romac S, Colin S, Aury J-M, Bittner L, Chaffron S, Dunthorn M, Engelen S, Flegontova O, Guidi L, Horák A, Jaillon O, Mendez GL, Lukeš J, Malviya S, Morard R, Mulot M, Scalco E, Siano R, Vincent F, Zingone A, Dimier C, Picheral M, Searson S, Kandels-Lewis S, Acinas SG, Bork P, Bowler C, Gaill F, Gorsky G, Grimsley N, Hingcamp P, Iudicone D, Not F, Ogata H, Pesant S, Raes J, Sieracki M, Speich S, Stemmann L, Sunagawa S, Weissenbach J, Wincker P, Karsenti E. Eukaryotic plankton diversity in the sunlit global ocean. Science. 2015;348: doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- Dean S, Sunter J, Wheeler RJ, Hodkinson I, Gluenz E, Gull K. A toolkit enabling efficient, scalable and reproducible gene tagging in trypanosomatids. Open Biol. 2015;5: doi: 10.1098/rsob.140197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Campo J, Sieracki ME, Molestina R, Keeling P, Massana R, Ruiz-Trillo I. The others: our biased perspective of eukaryotic genomes. Trends Ecol Evol. 2014;29:252–258. doi: 10.1016/j.tree.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollet M. Plant diseases caused by flagellate protozoa (Phytomonas) Annu Rev Phytopathol. 1984;22:115–132. [Google Scholar]
- Epp LS, Boessenkool S, Bellemain EP. New environmental metabarcodes for analysing soil DNA: potential for studying past and present ecosystems. Mol Ecol. 2012;21:1821–1833. doi: 10.1111/j.1365-294X.2012.05537.x. [DOI] [PubMed] [Google Scholar]
- Ferreira JI, da Costa AP, Ramirez D, Roldan JA, Saraiva D, Founier GFS, Sue A, Zambelli ER, Minervino AH, Verdade VK, Gennari SM, Marcili A. Anuran trypanosomes: phylogenetic evidence for new clades in Brazil. Syst Parasitol. 2015;91:63–70. doi: 10.1007/s11230-015-9558-z. [DOI] [PubMed] [Google Scholar]
- Foster JA, Bunge J, Gilbert JA. Measuring the microbiome: perspectives on advances in DNA-based techniques for exploring microbial life. Brief Bioinform. 2012;13:420–429. doi: 10.1093/bib/bbr080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frías L, Leles D, Araújo A. Studies on protozoa in ancient remains - A Review. Mem Inst Oswaldo Cruz. 2013;108:1–12. doi: 10.1590/S0074-02762013000100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MT, Haselkorn T, Bunce M, Sanchez JJ, Lucas SB, Jewell LD, Van Marck E, Worobey M. The isolation of nucleic acids from fixed, paraffin-embedded tissues-which methods are useful when? PLoS ONE. 2007;2: doi: 10.1371/journal.pone.0000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser K, Kuppardt A, Krohn S, Heidtmann A, Harms H, Chatzinotas A. Primer pairs for the specific environmental detection and T-RFLP analysis of the ubiquitous flagellate taxa Chrysophyceae and Kinetoplastea. J Microbiol Methods. 2014;100:8–16. doi: 10.1016/j.mimet.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Grybchuk-Ieremenko A, Losev A, Kostygov AY, Lukeš J, Yurchenko V. High prevalence of trypanosome co-infections in freshwater fishes. Folia Parasitol. 2014;61:495–504. [PubMed] [Google Scholar]
- Hamilton PB, Stevens JR, Gaunt MW, Gidley J, Gibson WC. Trypanosomes are monophyletic: evidence from genes for glyceraldehyde phosphate dehydrogenase and small subunit ribosomal RNA. Int J Parasitol. 2004;34:1393–1404. doi: 10.1016/j.ijpara.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Hayes PM, Lawton SP, Smit NJ, Gibson WC, Davies AJ. Morphological and molecular characterization of a marine fish trypanosome from South Africa, including its development in a leech vector. Parasit Vectors. 2014;7: doi: 10.1186/1756-3305-7-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoare CA, Wallace FG. Developmental stages of trypanosomatid flagellates: a new terminology. Nature. 1966;212:1385–1386. [Google Scholar]
- Hollar L, Lukeš J, Maslov DA. Monophyly of endosymbiont containing trypanosomatids: phylogeny versus taxonomy. J Eukaryot Microbiol. 1998;45:293–297. doi: 10.1111/j.1550-7408.1998.tb04539.x. [DOI] [PubMed] [Google Scholar]
- Jaskowska E, Butler C, Preston G, Kelly S. Phytomonas: trypanosomatids adapted to plant environment. PLoS Pathogens. 2015;11: doi: 10.1371/journal.ppat.1004484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kher CP, Doerder FP, Cooper J, Ikonomi P, Achilles-Day U, Küpper FC, Lynn DH. Barcoding Tetrahymena: discriminating species and identifying unknowns using the cytochrome c oxidase subunit I (cox-1) barcode. Protist. 2011;162:2–13. doi: 10.1016/j.protis.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Kolísko M, Boscaro V, Burki F, Lynn DH, Keeling PJ. Single-cell transcriptomics for microbial eukaryotes. Curr Biol. 2014;24:R1081–R1082. doi: 10.1016/j.cub.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Kostygov A, Frolov AO. Leptomonas jaculum (Leger, 1902) Woodcock 1914: a leptomonas or a blastocrithidia? Parazitologiia. 2007;41:126–136. [PubMed] [Google Scholar]
- Kostygov AY, Grybchuk-Ieremenko A, Malysheva MN, Frolov AO, Yurchenko V. Molecular revision of the genus Wallaceina. Protist. 2014;165:594–604. doi: 10.1016/j.protis.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Lamontagne J, Papadopoulou B. Developmental regulation of spliced leader RNA gene in Leishmania donovani amastigotes is mediated by specific polyadenylation. J Biol Chem. 1999;274:6602–6609. doi: 10.1074/jbc.274.10.6602. [DOI] [PubMed] [Google Scholar]
- Lebonah DE, Dileep A, Chandrasekhar K, Sreevani S, Sreedevi B, Kumari JP. DNA barcoding on bacteria: A Review. Adv Biol. 20142014:1–9. [Google Scholar]
- Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, Chaffron S, Ignacio-Espinosa JC, Roux S, Vincent F, Bittner L, Darzi Y, Wang J, Audic S, Berline L, Bontempi G, Cabello AM, Coppola L, Cornejo-Castillo FM, d’Ovidio F, Meester L, Ferrera I, Garet-Delmas M-J, Guidi L, Lara E, Pesant S, Royo-Llonch M, Salazar G, Sánchez P, Sebastian M, Souffreau C, Dimier C, Picheral M, Searson S, Kandels-Lewis S, coordinators Tara Oceans, Gorsky G, Not F, Ogata H, Speich S, Stemmann L, Weissenbach J, Wincker P, Acinas SG, Sunagawa S, Bork P, Sullivan MB, Karsenti E, Bowler C, Vargas C, Raes J. Determinants of community structure in the global plankton interactome. 10.1126/science.1262073.Science. 2015;348 doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- López-García P, Philippe H, Gail F, Moreira D. Autochthonous eukaryotic diversity in hydrothermal sediment and experimental microcolonizers at the Mid-Atlantic Ridge. Proc Natl Acad Sci USA. 2003100:697–702. doi: 10.1073/pnas.0235779100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Losev A, Grybchuk-Ieremenko A, Kostygov AY, Lukeš J, Yurchenko V. Host specificity, pathogenicity, and mixed infections of trypanoplasms from freshwater fishes. Parasitol Res. 2015;114:1071–1078. doi: 10.1007/s00436-014-4277-y. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Flegontova O, Horák A. Diplonemids. Curr Biol. 2015;25:R1–R3. doi: 10.1016/j.cub.2015.04.052. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Guilbride DL, Votýpka J, Zíková A, Benne R, Englund PT. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot Cell. 2002;1:495–502. doi: 10.1128/EC.1.4.495-502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš J, Jirků M, Doležel D, Král’ová I, Hollar L, Maslov DA. Analysis of ribosomal RNA genes suggests that trypanosomes are monophyletic. J Mol Evol. 1997;44:521–527. doi: 10.1007/pl00006176. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol. 2014;195:115–122. doi: 10.1016/j.molbiopara.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Lukeš L, Jirku M, Simpson L. Phylogeny of trypanosomes as inferred from the small and large subunit rRNAs: implications for the evolution of parasitism in the trypanosomatid protozoa. Mol Biochem Parasitol. 1996;75:197–205. doi: 10.1016/0166-6851(95)02526-x. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Westenberger SJ, Xu X, Campbell DA, Sturm NR. Discovery and barcoding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J Eukaryot Microbiol. 2007;54:57–65. doi: 10.1111/j.1550-7408.2006.00150.x. [DOI] [PubMed] [Google Scholar]
- Maslov DA, Yurchenko VY, Jirků M, Lukeš J. Two new species of trypanosomatid parasites isolated from Heteroptera in Costa Rica. J Eukaryot Microbiol. 2010;57:177–188. doi: 10.1111/j.1550-7408.2009.00464.x. [DOI] [PubMed] [Google Scholar]
- McCarthy CB, Diambra LA, Pomar RVR. Metagenomic analysis of taxa associated with Lutzomyia longipalpis, vector of visceral leishmaniasis using an unbiased high-throughput approach. PLoS Negl Trop Dis. 2011;5: doi: 10.1371/journal.pntd.0001304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee R, Cosgrove WB. Biology and physiology of the lower Trypanosomatidae. Microbiol Rev. 1980;44:140–173. doi: 10.1128/mr.44.1.140-173.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medinger R, Nolte V, Pandey RV, Jost S, Ottenwälder B, Schlötterer C, Boenigk J. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol Ecol. 2010;19:32–40. doi: 10.1111/j.1365-294X.2009.04478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MLZ, Sicheritz-Pontén T, Gilbert MTP. Environmental genes and genomes: understanding the differences and challenges in the approaches and software for their analyses. Brief Bioinform. 2015;16:745–758. doi: 10.1093/bib/bbv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak E, Yurchenko V, Kolesnikov AA, Alexandrov K, Podlipaev SA, Maslov DA. Diversity and phylogeny of insect trypanosomatids based on small subunit rRNA genes: polyphyly of Leptomonas and Blastocrithidia. J Eukaryot Microbiol. 2001;48:161–169. doi: 10.1111/j.1550-7408.2001.tb00298.x. [DOI] [PubMed] [Google Scholar]
- Moreira D, López-García P, Vickerman K. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. Int J Syst Evol Microbiol. 2004;54:1861–1875. doi: 10.1099/ijs.0.63081-0. [DOI] [PubMed] [Google Scholar]
- Nassonova E, Smirnov A, Fahrni J, Pawlowski J. Barcoding amoebae: comparison of SSU, ITS and COI genes as tools for molecular identification of naked lobose amoebae. Protist. 2010;161:102–115. doi: 10.1016/j.protis.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Pace N. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Pace N. Mapping the tree of life: progress and prospects. Microbiol Mol Biol Rev. 2009;73:565–576. doi: 10.1128/MMBR.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawar H, Kulkarni A, Dixit T, Chaphekar D, Patole MS. A bioinformatics approach to reanalyze the genome annotation of kinetoplastid protozoan parasite Leishmania donovani. Genomics. 2014;104:554–561. doi: 10.1016/j.ygeno.2014.09.008. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Audic S, Adl S, Bass D, Belbahri L, Berney C, Bowser SS, Čepička I, Decelle J, Dunthorn M, Fiore-Donno AM, Gile GH, Holzmann M, Jahn R, Jirků M, Keeling PJ, Kostka M, Kudryavtsev A, Lara E, Lukeš J, Mann DG, Mitchell EA, Nitsche F, Romeralo M, Saunders GW, Simpson AG, Smirnov AV, Spouge JL, Stern RF, Stoeck T, Zimmermann J, Schindel D, de Vargas C. CBOL Protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 2012;10: doi: 10.1371/journal.pbio.1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski J, Christen R, Lecroq B, Bachar D, Shahbazkia HR, Amaral-Zettler L, Guillou L. Eukaryotic richness in the abyss: insights from pyrotag sequencing. PLoS ONE. 2011;6: doi: 10.1371/journal.pone.0018169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podlipaev SA. Catalogue of world fauna of Trypanosomatidae (Protozoa) Proc Zool Inst Leningrad. 1990;144:1–178. [Google Scholar]
- Podlipaev SA, Sturm NR, Fiala I, Fernandes O, Westenberger SJ, Dollet M, Campbell DA, Lukeš J. Diversity of insect trypanosomatids assessed from the spliced leader RNA and 5S rRNA genes and intergenic regions. J Eukaryot Microbiol. 2004;51:283–290. doi: 10.1111/j.1550-7408.2004.tb00568.x. [DOI] [PubMed] [Google Scholar]
- Pompanon F, Coissac E, Taberlet P. Metabarcoding, une nouvelle façon d’analyser la biodiversité. Biofutur. 2011;319:30–32. (Fre). [Google Scholar]
- Pompanon F, Samadi S. Next generation sequencing for characterizing diversity: promises and challenges. Genetica. 2015;143:133–138. doi: 10.1007/s10709-015-9816-7. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salani FS, Arndt H, Hausmann K, Nitsche F, Scheckenbach F. Analysis of the community structure of abyssal kinetoplastids revealed similar communities at larger spatial scales. ISME J. 2012;6:713–723. doi: 10.1038/ismej.2011.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvadet AL, Gobet A, Guillou Comparative analysis between protist communities from the deep-sea pelagic ecosystem and specific deep hydrothermal habitats. Environ Microbiol. 2010;12:2946–2964. doi: 10.1111/j.1462-2920.2010.02272.x. [DOI] [PubMed] [Google Scholar]
- Scheckenbach F, Hausmann K, Wylezich C, Weitere M, Arndt H. Large-scale patterns in biodiversity of microbial eukaryotes from the abyssal sea floor. Proc Natl Acad Sci USA. 2010;107:115–120. doi: 10.1073/pnas.0908816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AG, Stevens JR, Lukes J. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 2006;4:168–174. doi: 10.1016/j.pt.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Simpson L. Behavior of the kinetoplast of Leishmania tarentolae upon cell rupture. J Protozool. 1986;15:132–136. doi: 10.1111/j.1550-7408.1968.tb02097.x. [DOI] [PubMed] [Google Scholar]
- Škodová-Sveráková I, Verner Z, Skalický T, Votýpka J, Horváth A, Lukeš J. Lineage-specific activities of a multipotent mitochondrion of trypanosomatid flagellates. Mol Microbiol. 2015;96:55–67. doi: 10.1111/mmi.12920. [DOI] [PubMed] [Google Scholar]
- Šlapeta J, Moreira D, Lopez-Garcia P. The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes. Proc Biol Sci. 2005;272:2073–2081. doi: 10.1098/rspb.2005.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern RF, Horak A, Andrew RL, Coffroth MA, Andersen RA, Küpper FC, Jameson I, Hoppenrath M, Véron B, Kasai F, Brand J, James ER, Keeling PJ. Environmental barcoding reveals massive dinoflagellate diversity in marine environments. PLoS ONE. 2010;5: doi: 10.1371/journal.pone.0013991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoco PH, Wagner G, Talavera-Lopez C, Gerber A, Zaha A, Thompson CE, Bartholomeu DC, Lückemeyer DD, Bahia D, Loreto E, Prestes EB, Lima FM, Rodrigues-Luiz G, Vallejo GA, da Silveira JF, Filho, Schenkman S, Monteiro KM, Tyler KM, de Almeida LG, Ortiz MF, Chiurillo MA, de Moraes MH, Cunha OL, Mendonça R, Neto, Silva R, Teixeira SM, Murta SM, Sincero TC, Mendes TA, Urmenyi TP, Silva VG, DaRocha WD, Andersson B, Romanha AJ, Steindel M, de Vasconcelos AT, Grisard EC. Genome of the avirulent human-infective Trypanosome - Trypanosoma rangeli. PLoS Negl Trop Dis. 2014;8: doi: 10.1371/journal.pntd.0003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, Schwarz MV, Boenigk J, Schweikert M, von der Heyden S, Behnke A. Cellular identity of an 18S rRNA gene sequence clade within the class Kinetoplastea: the novel genus Actuariola gen. nov. (Neobodonida) with description of the type species Actuariola framvarensis sp. nov. Int J Syst Evol Microbiol. 2005;55:2623–2635. doi: 10.1099/ijs.0.63769-0. [DOI] [PubMed] [Google Scholar]
- Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm NR, Martinez LLIT, Thomas S. Kinetoplastid genomics: the thin end of the wedge. Infect Genet Evol. 2008;8:901–906. doi: 10.1016/j.meegid.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez AK, Tsutsui ND. The value of museum collections for research and society. Bioscience. 2004;54:66–73. [Google Scholar]
- Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol. 2012;21:2045–2050. doi: 10.1111/j.1365-294X.2012.05470.x. [DOI] [PubMed] [Google Scholar]
- Taib N, Mangot JF, Domaizon I, Bronner G, Debroas D. Phylogenetic affiliation of SSU rRNA genes generated by massively parallel sequencing: new insights into the freshwater protist diversity. PLoS ONE. 2013;8: doi: 10.1371/journal.pone.0058950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MM, Borghesan TC, Ferreira RC, Santos MA, Takata CS, Campaner M, Nunes VL, Milder RV, de Souza W, Camargo EP. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist. 2011;162:503–524. doi: 10.1016/j.protis.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Thomas S, Westenberger SJ, Campbell DA, Sturm NR. Intragenomic spliced leader RNA array analysis of kinetoplastids reveals unexpected transcribed region diversity in Trypanosoma cruzi. Gene. 2005;352:100–108. doi: 10.1016/j.gene.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Truc P, Büscher P, Cuny G, Gonzatti MI, Jannin J, Joshi P, Juyal P, Lun ZR, Mattioli R, Pays E, Simarro PP, Teixeira MM, Touratier L, Vincendeau P, Desquesnes M. Atypical human infections by animal trypanosomes. PLoS Negl Trop Dis. 2013;7: doi: 10.1371/journal.pntd.0002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Týč J, Votýpka J, Klepetková H, Šuláková H, Jirků M, Lukeš J. Growing diversity of trypanosomatid parasites of flies (Diptera: Brachycera): frequent cosmopolitism and moderate host specificity. Mol Phylogenet Evol. 2013;69:255–264. doi: 10.1016/j.ympev.2013.05.024. [DOI] [PubMed] [Google Scholar]
- Vaerewijck MJ, Sabbe K, Baré J, Houf K. Microscopic and molecular studies of the diversity of free-living protozoa in meat-cutting plants. Appl Environ Microbiol. 2008;74:5741–5749. doi: 10.1128/AEM.00980-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hannen EJ, Zwart G, van Agterveld MP, Gons HJ, Ebert J, Laanbroek HJ. Changes in bacterial and eukaryotic community structure after mass lysis of filamentous cyanobacteria associated with viruses. Appl Environ Microbiol. 1999;65:795–801. doi: 10.1128/aem.65.2.795-801.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. Lumsden WHR, Evans DA.eds. Biology of the Kinetoplastida. Academic Press; London/New York/San Francisco: 1976. The diversity of the kinetoplastid flagellates; pp. 1–34. [Google Scholar]
- Vickerman K. The evolutionary expansion of the trypanosomatid flagellates. Int J Parasitol. 1994;24:1317–1331. doi: 10.1016/0020-7519(94)90198-8. [DOI] [PubMed] [Google Scholar]
- Vickerman K, Preston TM. Lumsden WHR, Evans DA.eds. Biology of the Kinetoplastida. Academic Press: London/New York/San Francisco; 1976. Comparative cell biology of the kinetoplastid flagellates; pp. 35–130. [Google Scholar]
- Viola LB, Almeida RS, Ferreira RC, Campaner M, Takata CSA, Rodrigues AC, Paiva F, Camargo EP, Teixeira MMG. Evolutionary history of trypanosomes from South American Caiman (Caiman yacare) and African crocodiles inferred by phylogenetic analyses using SSU rDNA and gGAPDH genes. Int J Parasitol. 2009;40:345–355. doi: 10.1017/S003118200800512X. [DOI] [PubMed] [Google Scholar]
- Von der Heyden S, Cavalier-Smith T. Culturing and environmental DNA sequencing uncover hidden kinetoplastid diversity and a major marine clade within ancestrally freshwater Bodo designis. Int J Syst Evol Microbiol. 2005;55:2605–2621. doi: 10.1099/ijs.0.63606-0. [DOI] [PubMed] [Google Scholar]
- Votýpka J, d’Avila-Levy CM, Grellier P, Maslov D, Lukeš J, Yurchenko V. Systematics of Trypanosomatidae: criteria for taxonomic (re-) description. Trends Parasitol. 2015a;31:460–469. doi: 10.1016/j.pt.2015.06.015. [DOI] [PubMed] [Google Scholar]
- Votýpka J, Klepetková H, Jirků M, Kment P, Lukeš J. Phylogenetic relationships of trypanosomatids parasitising true bugs (Insecta: Heteroptera) in sub-Saharan Africa. Int J Parasitol. 2012;42:489–500. doi: 10.1016/j.ijpara.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, Grybchuk D, Lukeš J, Yurchenko V. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist. 2014;165:825–838. doi: 10.1016/j.protis.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Votýpka J, Maslov DA, Yurchenko V, Jirků M, Kment P, Lun ZR, Lukeš J. Probing into the diversity of trypanosomatid flagellates parasitizing insect hosts in South-West China reveals both endemism and global dispersal. Mol Phylogenet Evol. 2010;54:243–253. doi: 10.1016/j.ympev.2009.10.014. [DOI] [PubMed] [Google Scholar]
- Votýpka J, Rádrová J, Skalický T, Jirků M, Jirsová D, Mihalca AD, D‘Amico G, Petrželková KJ, Modrý D, Lukeš J. A tsetse and tabanid fly survey of African great apes habitats reveals the presence of a novel trypanosome lineage but the absence of Trypanosoma brucei. 10.1016/j.ijpara.2015.06.005.Int J Parasitol. 2015b doi: 10.1016/j.ijpara.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Votýpka J, Suková E, Kraeva N, Ishemgulova A, Duží I, Lukeš J, Yurchenko V. Diversity of trypanosomatids (Kinetoplastea: Trypanosomatidae) parasitizing fleas (Insecta: Siphonaptera) and description of a new genus Blechomonas gen. n. Protist. 2013;164:763–781. doi: 10.1016/j.protis.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2013;489:250–256. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberger SJ, Sturm NR, Yanega D, Podlipaev SA, Zeledón R, Campbell DA, Maslov DA. Trypanosomatid biodiversity in Costa Rica: genotyping of parasites from Heteroptera using the spliced leader RNA gene. Parasitology. 2004;129:537–547. doi: 10.1017/s003118200400592x. [DOI] [PubMed] [Google Scholar]
- Wilfert L, Longdon B, Ferreira AG, Bayer F, Jiggins FM. Trypanosomatids are common and diverse parasites of Drosophila. Parasitology. 2011;138:858–865. doi: 10.1017/S0031182011000485. [DOI] [PubMed] [Google Scholar]
- Wyatt KB, Campos PF, Gilbert MT, Kolokotronis SO, Hynes WH, DeSalle R, Ball SJ, Daszak P, MacPhee RD, Greenwood AD. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS ONE. 2008;3: doi: 10.1371/journal.pone.0003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko VY, Kolesnikov AA, Lukeš J. Phylogenetic analysis of Trypanosomatina (Protozoa: Kinetoplastida) based on minicircle conserved regions. Folia Parasitol. 2000;47:1–5. doi: 10.14411/fp.2000.001. [DOI] [PubMed] [Google Scholar]
- Yurchenko VY, Lukeš J, Jirků M, Maslov DA. Selective recovery of the cultivation-prone components from mixed trypanosomatid infections: a case of several novel species isolated from Neotropical Heteroptera. Int J Syst Evol Microbiol. 2009;59:893–909. doi: 10.1099/ijs.0.001149-0. [DOI] [PubMed] [Google Scholar]
- Yurchenko VY, Lukeš J, Tesařová M, Jirků M, Maslov DA. Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus Crithidia. Protist. 2008;159:99–114. doi: 10.1016/j.protis.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Zídková L, Cepicka I, Szabová J, Svobodová M. Biodiversity of avian trypanosomes. Infect Genet Evol. 2012;12:102–112. doi: 10.1016/j.meegid.2011.10.022. [DOI] [PubMed] [Google Scholar]