Abstract
Objective
HIV-1 persists indefinitely in memory CD4+ T cells and other long-lived cellular reservoirs despite antiretroviral therapy (ART). Our group had previously demonstrated that HIV-1 can establish a productive infection in renal epithelial cells and that the kidney represents a separate compartment for HIV-1 replication. Here, to better understand the viruses in this unique site, we genetically characterized and compared the viruses in blood and urine specimens from twenty-four HIV-1 infected subjects with detectable viremia.
Design and Methods
Blood and urine samples were obtained from 35 HIV-1 positive subjects. Single-genome amplification was performed on HIV-1 env RNA and DNA isolated from urine supernatants and urine derived cell pellets respectively, as well as from plasma and PBMC from the same individuals. Neighbor-joining trees were constructed under the Kimura 2-parameter mode.
Results
We amplified and sequenced the full-length HIV-1 envelope (env) gene from twelve of the twenty-four individuals, indicating that fifty percent (50%) of the viremic HIV-1 positive patients had viral RNA in their urine. Phylogenetic analysis of the env sequences from four subjects with more than fifteen urine-derived env sequences showed that the majority of the sequences from urine formed distinct cluster(s) independent of those PBMC and plasma-derived sequences, consistent with viral compartmentalization in the urine.
Conclusions
Our results suggest the presence of a distinct HIV compartment in the genitourinary tract.
Keywords: HIV-1, Urine, Genitourinary tract, Compartmentalization, Reservoir
INTRODUCTION
HIV-1 can infect and persist in different organs and tissues. The infection of different cell types results in the generation of distinct viral populations that replicate within their unique environments and are subject to diverse selective pressure [1]. Some organs can maintain distinct viral compartments that can persist and serve as reservoirs for HIV-1 despite ART [2]. Compartments, defined as anatomic environments, restrict HIV-1 trafficking and gene flow, favor viral evolution and divergence from the viral population in the peripheral blood [3]. HIV-1 compartmentalization has been documented in the central nervous system (CNS), lymph nodes, breast milk and genital tract [4–6]. Understanding HIV-1 dynamics in different compartments and reservoirs is crucial for designing effective strategies to cure infected individuals. In our previous studies we have shown that HIV-1 infects renal epithelial cells in vitro and in vivo and that the kidney represents a separate compartment for HIV-1 replication in patients with HIV associated nephropathy (HIVAN) [7–9]. In a recent examination of renal biopsies from ART suppressed HIV positive patients undergoing kidney transplantation, HIV-1 was detected in graft renal tubular cells and/or podocytes in 68% of patients despite the absence of detectable plasma viremia [10], underscoring the importance of renal epithelial cells as a unique target for HIV-1. Understanding the long-term consequences of this non-hematologic reservoir is challenging as renal biopsies pose a risk to patients and repeated biopsies are rarely performed. We therefore amplified and characterized HIV-1 env sequences in urine specimens from HIV-1 positive patients with normal kidney function (non-HIVAN), to determine if viruses in urine represent a separate compartment that might reflect the renal reservoir.
MATERIALS AND METHODS
Study subjects and samples processing
Blood and overnight urine samples were obtained from 35 HIV-1 positive subjects. All subjects gave informed consent, and sample collections were performed with institutional review board approval (Pro00008576). Large volumes of urine ranging from 35 to 630 mL (Supplementary table 1) were collected overnight in 100 ml urine containers and kept at 4°C until processed the following morning. EDTA anticoagulated blood samples were processed within 2 hours from collection to isolate plasma and PBMC by Ficoll gradient centrifugation. Urine samples were spun at 1500 rpm for 10 minutes to separate urine supernatants from urinary cells. Supernatants were then filtered through a 0.45 μm filter unit to remove cellular debris followed by 2 hours of ultracentrifugation to pellet HIV virions. Pelleted viruses were then resuspended in 400 μl of 1X PBS and either immediately subjected to RNA extraction or stored at −80°C. Urinary cell pellets were stored at −20°C until DNA extraction.
Viral RNA Extraction and cDNA Synthesis
Viral RNA was extracted from 400 μl of concentrated urine or plasma by using the EZ1 virus Mini Kit v2.0 (Qiagen). RNA was eluted in a final volume of 60 μl, 20 μl of which were immediately subjected to cDNA synthesis. Reverse transcription of RNA to single-stranded cDNA was performed with Super-Script III reverse transcriptase following manufacturer’s instructions (Invitrogen Life Technologies). Briefly, each cDNA reaction included 1X reverse transcription (RT) buffer, 0.5 mM each deoxynucleoside triphosphate, 5 mM DTT, 2 units/μl RNaseOUT (RNase inhibitor), 10 units/μl Super-Script III reverse transcriptase, and 0.25 μM antisense primer 1.R3.B3R 5’-ACTACTTGAAGCACTCAAGGCAAGCTTTATTG- 3’. The mixture was incubated at 50°C for 60 min, followed by an increase in temperature to 55°C for an additional 60 min. The reaction was then heat-inactivated at 70°C for 15 min and then treated with RNaseH at 37°C for 20 min. The newly synthesized cDNA was used immediately or kept frozen at −80°C.
Viral DNA Extraction
Viral DNA was extracted from 5 × 106 PBMC by using the QIAamp mini kit and from the urine cell pellet by using the QIAamp micro kit (Qiagen) following the manufacturer’s instructions, and eluted in 50 μl of water.
Single Genome Amplification
cDNA was serially diluted and 1 μl of each dilution was distributed among wells of replicate 96-well plates so as to identify a dilution where PCR positive wells constituted less than 30% of the total number of reactions. At this dilution, most positive wells contain amplicons derived from a single cDNA molecule. This was confirmed in every positive reaction by direct sequencing of the amplicon and inspection of the sequence for double peaks, which would be evidence of priming from more than one original template or the introduction of PCR errors in early cycles. Any sequence with evidence of mixed bases was excluded from further analysis. First-round PCR primers included sense primer Env5out 5’-TAGAGCCCTGGAAGCATCCAGGAAG- 3’ and antisense primer Env3out 5’-TTGCTACTTGTGATTGCTCCATGT- 3’, which generated a ~3-kb product. PCR was performed in 96-well PCR reaction plates with the following PCR parameters: 1 cycle of 94°C for 2 min; 35 cycles of a denaturing step of 94°C for 15 s, an annealing step of 55°C for 30 s, an extension step of 68°C for 4 min, followed by a final extension of 68°C for 10 min. 2 μl from first-round PCR product were added to a second-round PCR that included the sense primer Env5in 5’-CACCTTAGGCATCTCCTATGGCAGGAAGAAG-3’ and antisense primer Env3in 5’-GTCTCGAGATACTGCTCCCACCC-3’. The second-round PCR was carried out under the same conditions used for first-round PCR but for a total of 45 cycles. Amplicons were separated on 1% agarose gels and positive bands were excised from the gel and PCR products were purified using the QIAquick gel extraction kit (Qiagen). All PCR procedures were carried out under PCR clean room conditions with procedural safeguards against sample contamination. Amplifications were performed using the Eppendorf Mastercycler pro.
Sequencing and sequence alignments
The env gene amplicons were sequenced by using the primer walking method. Individual sequence fragments for each amplicon were assembled and edited by using the Sequencher program 5.0 (Gene Codes). Inspection of individual chromatograms allowed for the identification of amplicons derived from single versus multiple templates. The absence of mixed bases at each nucleotide position throughout the entire env gene was taken as evidence of amplification from a single viral RNA/cDNA template. All alignments and phylogenetic trees were made with MEGA6 [25]. Neighbor-joining trees were constructed under the Kimura 2-parameter mode and the reliability of topologies was estimated by performing bootstrap analysis with 1000 replicates.
Statistics
Non-parametric Spearman correlation analysis between number of urinary sequences and plasma viral load was performed using Graph-Pad Prism 6.0 software (GraphPad Software, Inc., San Diego, Ca).
RESULTS
Fifty percent of viremic HIV-1 positive subjects have viral RNA in their urine
Blood and urine specimens were simultaneously collected from a total of 35 HIV-1 positive subjects (Supplementary table 1), 24 with detectable viremia and 11 with undetectable viral load. We performed single-genome amplification [2] on HIV-1 env RNA and DNA isolated from urine supernatants and urine derived cell pellets respectively, as well as from plasma and PBMC from the same individuals. We were able to amplify the full-length HIV-1 env gene in urine supernatants from 12 out of the 24 viremic subjects analyzed in this study and obtained multiple env sequences from the majority of them (table 1 and Supplementary figure 1). On the other hand, no envelope sequence could be derived from urine samples in the 11 suppressed subjects, suggesting that ART is effective in suppressing viral replication in the urine compartment.
Table 1.
Characteristics of study participants with chronic HIV-1 Infection in which HIV-1 RNA was amplified from urine supernatants.
| Subject ID | Ethnicity | Gender | Plasma viral load (copies/mL) | CD4 count (cells/mm3) | Urine Volume (mL) | # of Urine SGA |
|---|---|---|---|---|---|---|
| 264 | Black/African American | F | 3,600.00 | 26 | 400 | 5 |
| 265 | Black/African American | M | 23,400 | 151 | 400 | 2 |
| 266 | Asian | M | 180,000 | 495 | 550 | 1 |
| 267* | White | F | 4460 | 466 | 35 | 17 |
| 269* | Black/African American | F | 211,000 | 316 | 430 | 28 |
| 270 | Black/African American | F | 140,000 | 233 | 600 | 3 |
| 271 | Black/African American | M | 1,100.00 | 1119 | 200 | 2 |
| 275 | Black/African American | M | 158,000 | 252 | 485 | 2 |
| 276 | Black/African American | F | 46,500 | 425 | 83 | 2 |
| 280* | Black/African American | F | 220,000 | 47 | 320 | 31 |
| 290* | Black/African American | M | 490,000 | 320 | 550 | 26 |
| 291 | White | M | 81,700 | 306 | 530 | 2 |
HIV-1 variants in urine are compartmentalized from the viral population in blood and clonally amplified
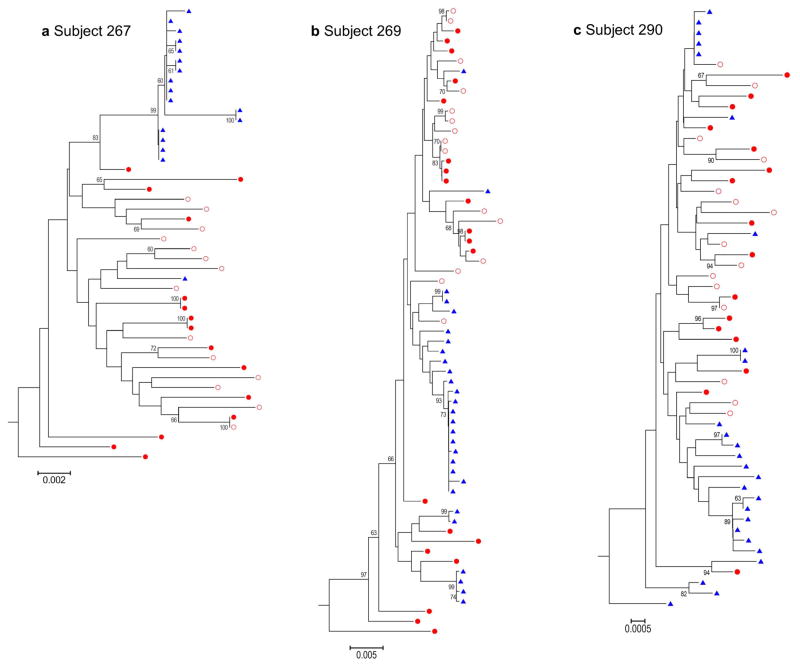
To determine whether the viral population in the urine is different from the viruses in the blood, we performed phylogenetic tree analysis of sequences obtained from 4 of the subjects from which we were able to amplify multiple HIV-1 env sequences (>15). As shown in figure 1(a–c), the majority of the urine derived env sequences form independent clusters among the PBMC and plasma derived sequences, suggesting that viruses found in the urine were generated from a separate compartment in the genitourinary tract. In the one male patient (subject 290) the lower genital tract (i.e. seminal tract) could have contributed to the urine sequences. However, the presence of urine derived sequence in the three female patients precludes the male lower genitourinary tract as the only source of virus in the urine. We observed a higher frequency of identical or nearly identical sequences in urine than in blood. Because of the nature of the SGA approach, this cannot be the result of PCR resampling, since each amplicon was generated from a single template, therefore it suggests clonal amplification of virus in this compartment. To assess whether viruses derived from the urinary compartment had a different co-receptor tropism from plasma viruses, we determined co-receptor usage of viruses by using the Geno2pheno [coreceptor] algorithm with a 10% false positive rate [13]. No difference in co-receptor usage was observed between blood and urine derived sequences.
Figure 1. Analysis of HIV-1 RNA in urine reveals viral compartmentalization and the presence of HIV genomes with identical sequences.
(a – c) The full-length env sequences were obtained by SGA from PBMC DNA, plasma viral RNA and urine viral RNA from three different subjects. Sequences were aligned by using Clustal W, and neighbor-joining trees were rooted on the HXB2 sequence (subgroup B). Blue triangles: urine RNA; Full red circles: PBMCs; Empty red circles: Plasma RNA.
Amplification of HIV-1 DNA from cells shed in urine
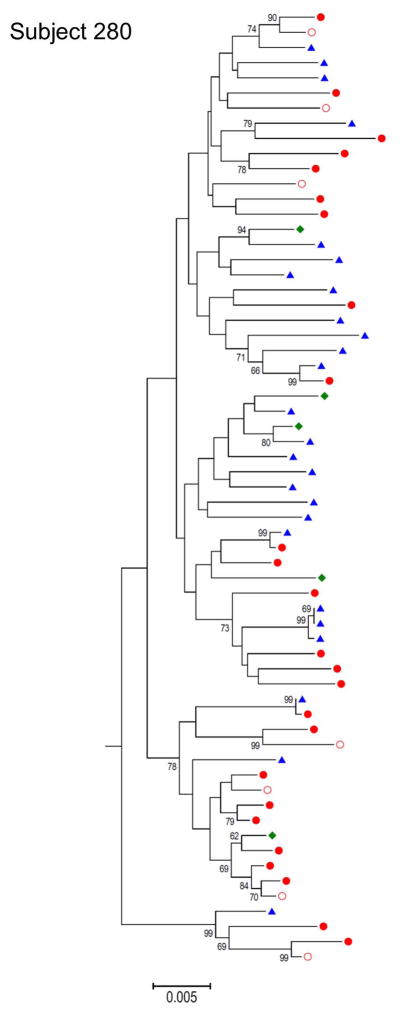
Although we did not have simultaneous renal biopsies from the subjects enrolled in this study to allow a comparison between HIV-1 DNA in the kidney with the virus RNA found in the urine, we amplified HIV-1 env DNA sequences from epithelial cells shed in the urine. As expected, the number of cells found in the urine was variable among the different subjects and often very low. Nevertheless, in three of the twelve subjects positive for HIV RNA in the urine we were able to amplify and sequence HIV-1 env DNA from the urine derived cell pellet (Supplementary figure 1). In figure 2 the phylogenetic tree for one of the three subjects from which we amplified a total of 5 HIV-1 env sequences from the urine derived cell pellet is shown. Interestingly, we found evidence of co-clustering of some of these sequences with some of the urine RNA sequences, while other sequences are interspersed among the PBMC and plasma derived sequences. We observed contaminating blood cells in the urine cell pellet from this subject, which could explain the presence of more urine/blood intermixed sequences in this patient compared to the others. These data suggest that, as it has been described for semen [14], the virus in urine can arise through multiple mechanisms: i) direct import from blood, which results in equilibrated viral populations between urine and blood, as observed in subject 280; ii) autonomous viral replication in the GU tract, which results in compartmentalization and iii) clonal amplification of virus in this compartments, as observed in the other three subjects.
Figure 2. Neighbor-joining tree for subject 280.
from which in addition to PBMC DNA, plasma viral RNA and urine viral RNA, the full-length env sequences were also obtained from urine cell pellet DNA. Blue triangles: urine RNA; Full red circles: PBMCs; Empty red circles: Plasma RNA; Green diamond: urine Cell-associate DNA.
DISCUSSION
Our study represents the first genetic characterization of HIV-1 RNA and DNA in urine. We found viral RNA in the urine of fifty percent of viremic HIV-1 positive subjects and sequence analysis showed features characteristic of viral compartmentalization distinct from blood. We also showed that in subjects with undetectable plasma viral load, viral RNA is also undetectable in urine, suggesting that ART is effective in suppressing viral replication in the urine compartment. Interestingly, we found that plasma viral load does not strictly correlate with the number of distinct viral sequences derived from urine (Spearman r, 0.3759; P = 0.0703; Supplementary figure 2); in subject 267 (4460 copies/mL) we amplified more than 15 HIV-1 env sequences from a volume of urine as low as 35 ml, while in other subjects with very high viral load (> 1 million copies/mL), none or only few sequences were derived from a large volume of urine (Supplementary table 1). We collected high volumes of urine from the majority of the subjects enrolled in this study in order to increase the chances of amplifying HIV-1 RNA, but interestingly multiple HIV-1 env sequences were derived from as low as 8 ml of urine. These data suggest that the dynamics of virus release in the urine is independent from virus production in the blood and is more likely dependent on the number of cells infected in this separate compartment. Interestingly, in our previously reported in situ hybridization study, we found that ~ 50% of patients had detectable viral RNA and/or DNA in their renal biopsies [26]. It is also possible that, as it has been observed in semen [15, 16], HIV-1 shedding in urine may be intermittent leading to the absence of virus in some samples. Future studies will address this question by analyzing follow-up urine specimens. Notably, in many instances we amplified a certain number of identical env sequences from the same subject in independent PCR reactions. Identical sequences have been previously isolated from plasma of patients both chronically infected [17, 18] or under suppressive ART and were found to be distinct from most of the viruses found in resting CD4+ T cells or activated CD4+ T cells in the circulation, suggesting that those similar viruses come from a different source [19]. Furthermore, in a recent study by Maldarelli F. et al., identical sequences isolated from patients on ART were present also before therapy initiation and were shown to be the result of clonal expansion of infected cells in which HIV-1 had integrated in genes that regulate cell growth and division [20]. These data were confirmed by Wagner TA et al., and suggest that clonal expansion of infected cells can contribute to HIV persistence [20, 21]. Interestingly, three recent publications independently showed that individual renal epithelial cells mediate tubulogenesis and renal epithelial repair during acute renal injury with single cell clones that contribute to segmental tubular regeneration [22–24]. In the setting of HIV-1 infection of renal epithelial cells it is possible that clones of infected renal tubule epithelial cells undergo cell division in an attempt to repair the injured renal tubule giving rise to clonal expansion of HIV-1 infected cells in the tubules. Our previous observation of the unique pattern of HIV-1 infection of renal tubules in HIVAN biopsies, in which the majority of cells lining a renal tubule were infected [26], supports this hypothesis or alternatively cell to cell transfer of HIV and further provides a possible explanation for the presence of identical sequences in the urine. In a previous study by Li et al., HIV-1 DNA and RNA were detected in a small number of urinary cells in 66% of HIV-1 positive subjects [27]. Although in these samples we were able to obtain cell-associated HIV-1 env sequences from the limited urine cells, the data suggest that increasing the number of renal epithelial cells in the urine, as might be found in HIV-1 positive subjects with acute tubular necrosis (ATN), will provide a more definitive comparison between HIV-1 viral RNA and cell associated DNA in the urine. This would also allow determination of preferential integration sites in the genome of urine epithelial cells. In male subjects it is possible that viruses produced in the lower genital tract could be present in urine specimens. In the present study three of the four subjects for which separate phylogenetic trees are shown are female, precluding the male lower genitourinary tract as the only source of virus in the urine. In conclusion, our study demonstrates that the amplification and analysis of urine-derived HIV-1 sequences is a feasible non-invasive approach and allows analysis of a unique genitourinary compartment that could serve as a non-blood viral reservoir to be studied in developing broader strategies to cure HIV-1.
Supplementary Material
Acknowledgments
The authors thank Janet Muller, Amanda Stemke, Deborah Murray, Kathy Ramadanovic and Katherine Frankey for assistance with study participants.
Funding: This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases P01DK056492.
Footnotes
Author contributions: M.B. designed and performed the majority of the experiments including urine processing, single genome amplification for urine and plasma samples and sequence analysis and wrote the manuscript. JHC processed blood samples and performed single genome amplification on PBMC and urine cell pellets. B.B. processed some of the samples and provided input to the initial draft of the manuscript. A.C. provided input on the study design. F.G. provided protocols and input on the study designs, assisted with sequence analysis and interpretation of the data. M.E.K. oversaw the planning and direction of the project including analysis and interpretation of the data and editing of the manuscript.
Competing interests: The authors declare no competing interests.
Data availability: All env sequences determined in this study were deposited in GenBank under accession numbers KR182173 - KR182473.
References
- 1.Nickle DC, Jensen MA, Shriner D, Brodie SJ, Frenkel LM, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–5546. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nickle DC1, Shriner D, Mittler JE, Frenkel LM, Mullins JI. Importance and detection of virus reservoirs and compartments of HIV infection. Curr Opin Microbiol. 2003;6:410–416. doi: 10.1016/s1369-5274(03)00096-1. [DOI] [PubMed] [Google Scholar]
- 3.Karris MA, Smith DM. Tissue-specific HIV-1 infection: why it matters. Future Virol. 2011;6:869–882. doi: 10.2217/fvl.11.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haggerty S, Stevenson M. Predominance of distinct viral genotypes in brain and lymph node compartments of HIV-1-infected individuals. Viral Immunol. 1991;4:123–131. doi: 10.1089/vim.1991.4.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong JK, Ignacio CC, Torriani F, Havlir D, Fitch NJ, Richman DD. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becquart P, Chomont N, Roques P, Ayouba A, Kazatchkine MD, Bélec L, et al. Compartmentalization of HIV-1 between breast milk and blood of HIV-infected mothers. Virology. 2002;300:109–117. doi: 10.1006/viro.2002.1537. [DOI] [PubMed] [Google Scholar]
- 7.Blasi M, Balakumaran B, Chen P, Negri DR, Cara A, Chen BK, et al. Renal epithelial cells produce and spread HIV-1 via T-cell contact. AIDS. 2014;28:2345–2353. doi: 10.1097/QAD.0000000000000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marras D, Bruggeman LA, Gao F, Tanji N, Mansukhani MM, Cara A, et al. Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat Med. 2002;8:522–526. doi: 10.1038/nm0502-522. [DOI] [PubMed] [Google Scholar]
- 9.Chen P, Chen BK, Mosoian A, Hays T, Ross MJ, Klotman PE, et al. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J Am Soc Nephrol. 2011;22:496–507. doi: 10.1681/ASN.2010040379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Canaud G, Dejucq-Rainsford N, Avettand-Fenoël V, Viard JP, Anglicheau D, Bienaimé F, et al. The kidney as a reservoir for HIV-1 after renal transplantation. J Am Soc Nephrol. 2014;25:407–19. doi: 10.1681/ASN.2013050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, Keele BF, et al. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J Virol. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci US A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lengauer T, Sander O, Sierra S, Thielen A, Kaiser R. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol. 2007;25:1407–1410. doi: 10.1038/nbt1371. [DOI] [PubMed] [Google Scholar]
- 14.Anderson JA, Ping LH, Dibben O, Jabara CB, Arney L, Kincer L, et al. HIV-1 Populations in Semen Arise through Multiple Mechanisms. PLoS pathog. 2010;6(8) doi: 10.1371/journal.ppat.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta P, Leroux C, Patterson BK, Kingsley L, Rinaldo C, Ding M, et al. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. J Infect Dis. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- 16.Bujan L, Daudin M, Matsuda T, Righi L, Thauvin L, Berges L, et al. Factors of intermittent HIV-1 excretion in semen and efficiency of sperm processing in obtaining spermatozoa without HIV-1 genomes. AIDS. 2004;26:757–66. doi: 10.1097/00002030-200403260-00006. [DOI] [PubMed] [Google Scholar]
- 17.Kirchherr JL, Hamilton J, Lu X, Gnanakaran S, Muldoon M, Daniels M, et al. Identification of amino acid substitutions associated with neutralization phenotype in the human immunodeficiency virus type-1 subtype C gp120. Virology. 2011;409:163–174. doi: 10.1016/j.virol.2010.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci US A. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wagner TA, McLaughlin S, Garg K, Cheung CY, Larsen BB, Styrchak S, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345:570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kusaba T, Lalli M, Kramann R, Kobayashi A, Humphreys BD. Differentiated kidney epithelial cells repair injured proximal tubule. Proc Natl Acad Sci US A. 2014;111:1527–1532. doi: 10.1073/pnas.1310653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinkevich Y, Montoro DT, Contreras-Trujillo H, Harari-Steinberg O, Newman AM, Tsai JM, et al. In vivo clonal analysis reveals lineage-restricted progenitor characteristics in mammalian kidney development, maintenance, and regeneration. Cell Rep. 2014;7:1270–1283. doi: 10.1016/j.celrep.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger K, Bangen JM, Hammerich L, Liedtke C, Floege J, Smeets B. Moeller Origin of regenerating tubular cells after acute kidney injury. Proc Natl Acad Sci US A. 2014;111:1533–1538. doi: 10.1073/pnas.1316177111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruggeman LA1, Ross MD, Tanji N, Cara A, Dikman S, Gordon RE, et al. Renal epithelium is a previously unrecognized site of HIV-1 infection. J Am Soc Nephrol. 2000;11:2079–2087. doi: 10.1681/ASN.V11112079. [DOI] [PubMed] [Google Scholar]
- 27.Li JJ, Huang YQ, Poiesz BJ, Zaumetzger-Abbot L, Friedman-Kien AE. Detection of human immunodeficiency virus type 1 (HIV-1) in urine cell pellets from HIV-1-seropositive individuals. J Clin Microbiol. 1992;30(5):1051–5. doi: 10.1128/jcm.30.5.1051-1055.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.