Abstract
Nanopores are being hailed as a potential next-generation DNA sequencer that could provide cheap, high-throughput DNA analysis. In this review we present a detailed summary of the various sensing techniques being investigated for use in DNA sequencing and mapping applications. A crucial impasse to the success of nanopores as a reliable DNA analysis tool is the fast and stochastic nature of DNA translocation. We discuss the incorporation of biological motors to step DNA through a pore base-by-base, as well as the many experimental modifications attempted for the purpose of slowing and controlling DNA transport.
Keywords: single-molecule, DNA sequencing, nanopores
Nanopores have recently emerged as a label-free platform for interrogating sequence and structure in nucleic acids. The concept of DNA sequencing using nanopores is as follows: as a DNA strand is driven through a nanopore by some means in a single-file manner, the steric occupation of each nucleobase (A, T, C, and G), or each short string of nucleobases, should correlate to a distinct measureable signal. Since the advent of nanopore-based DNA sensing in the 1990s, the prized objective of many research groups has been to use this principle in order to afford low-cost DNA sequencing. This goal was fueled by the $1000 human genome project initiated by the National Human Genome Research Institute in 2004, a program that has consistently been supporting the development of nanopore-based DNA sequencers [1].
Single-molecule methods are challenging because they rely on interpreting some indirect measured signal into a molecular property (e.g., position, speed, force, conformation, stiffness). When considering the parameters that one needs to control and interpret for nanopore-based DNA sequencing, it is not surprising that nanopores are among the most difficult techniques to interpret. DNA sequencing using nanopores requires the ability to move a DNA strand at intervals of ~1 nm and to read sequence information at each position. However, controlling the stepwise motion of a highly entropic polymer coil through a nanopore to a level that permits sequence readout is challenging. The straightforward approach of applying voltage across the pore to drive DNA translocation results in high velocities and large velocity fluctuations, i.e., motion is fast and noisy. Use of an enzyme motor to regulate DNA motion through a pore greatly reduces DNA transport speeds and affords sequence information, although the use of enzymes poses limitations such as time-dependent stochasticity, uncontrollable forward–reverse motion, and enzyme malfunction. Married to all of the above challenges in DNA motion control is yet another layer of complication: the need for a nanopore signal that reports on the DNA sequence. Several nanopore signal detection schemes have been proposed, some that show molecular recognition/discrimination, and others that demonstrate DNA base recognition. However, even the highest-resolution nanopore devices lack the spatial resolution required for single base recognition, instead relying on deconvolving simultaneous signals from strings of bases. This increased number of sequence permutations increases the required signal resolution and necessitates the use of complex algorithms for deciphering sequence, all of which can compromise the DNA readout accuracy. In the current spotlight, Oxford Nanopore Technologies (ONT) has already begun to commercialize a miniature DNA sequence detector based on enzyme-driven motion of DNA molecules through a nanopore array, although to date the details of the process are intentionally obscured and the quality of the readout raises concerns about the suitability of this product for DNA sequencing applications [67]. At the same time, other companies are emerging with alternative nanopore approaches to DNA sequencing. In the midst of this exciting period in nanopore research, we focus this review on the various types of approaches for nanopore-based DNA sequence detection, as well as on the various efforts to regulate DNA motion through these detectors.
DNA detection schemes
Ion current sensors
The most common detection method for DNA sequencing applications is to monitor transient changes in ionic current through a nanopore. When two wells of electrolytic fluid are separated with an ultrathin membrane that contains a nanoscale aperture, a steady transmembrane current can be established by applying a small potential bias across the membrane. If a dilute solution (<1 μM) of nucleic acid molecules is present in the chamber that is held at a lower potential, stochastic threading of individual nucleic acid molecules can be observed as transient spikes in the current signal. Based on the magnitude and duration of the ion current pulses, basic structural features of biomolecules can be probed in great detail without labeling or amplification of sample. For example, nucleic acids that contain two homopolymeric segments can be resolved based on their different current amplitudes [2]. To obtain greater detail in time resolution for nanopore experiments, data acquisition amplifiers with bandwidths on the order of 1 MHz can be used [17, 81], although the cost of this increased time resolution is a higher current noise which can compromise the signal quality [93, 100, 106]. In nanopore experiments that involve ionic current detection, several sources of noise need to be recognized and reduced. A common way to eliminate high-frequency capacitive noise is to apply a low-pass filter to the raw signal, although cutting the bandwidth of a measurement deteriorates the time resolution of an experiment. In addition, when a voltage is applied across the nanopore a steep increase in low-frequency noise is observed. This region of the noise spectrum is called 1/f noise, which is due to transient factors that influence the current flux (e.g., surface charge fluctuations, hydrophobic pockets on the pore surface) [33, 74, 76, 92].
Tunneling-based electronic readout
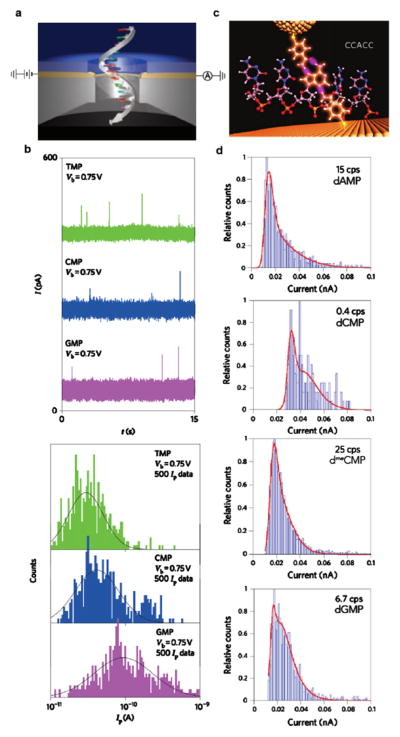
Another avenue for sequencing DNA aims to harvest differences in the electronic structure of the DNA nucleobases in order to record differences in transverse electrical current through DNA bases [16, 35, 39, 47, 104, 123]. The envisioned device would contain a metallic electrode nanogap across the nanopore. As a DNA molecule translocates, a process that is driven by an electrochemical bias across the pore generated by a different pair of electrodes, tunneling currents across the nanogap are recorded at high bandwidth (see figure 2(a)). In figure 2(b), a coarse proof-of-principle is shown in which transient events of nucleotide monophosphate residence within the gap trigger upward current spikes [104]. Only three of the four DNA bases have been distinguished in this work, although broad overlapping distributions of tunneling current amplitudes are observed, likely due to a number of sources including a molecule’s possible orientations in the nanogap. In order to generate a recognition ability at the nanogap for nucleotides, research groups have developed functionalized gold electrodes that are capable of forming hydrogen bonds with DNA [15, 30]. By coating a gold surface and scanning tunneling microscope’s probe with the reagent 4-mercaptobenzamide, the Lindsay group recently were able to probe short oligomers of different sequence in solution that diffused into the tunneling gap (see figure 2(c)) [16, 35]. Combining the amplitude, duration, and frequency of tunneling current bursts shows that individual DNA nucleotides could be distinguished with the exception of dTMP, which exhibited no tunneling activity due to its strong binding affinity to the functionalized gold surface (see figure 2(d)). Notably, deoxy-5-methylcytosine monophosphate (dmeCMP) produced lower current amplitudes than dCMP, which suggests an opportunity for label-free epigenetics studies of DNA fragments. Tunneling-based sensing of this nature has also demonstrated the ability to recognize amino acids and short peptides [72, 121]. However, combining this recognition-based technology with a nanopore to drive/read DNA sequence has yet to be demonstrated due to the device’s increasing complexity.
Figure 2.
Nucleic acid base detection by transverse tunneling current read-out. (a) A representation of ssDNA translocating through a gold tunneling junction sandwiched inside a solid-state membrane. (b) Top: sample tunneling current traces recorded for the detection of the RNA monomers TMP (green), CMP (blue), and GMP (purple) at a trans-electrode bias of 0.75 V. Bottom: histograms of the measured tunneling current Ip of 500 data points for each nucleotide. When fitting the distributions to Gaussian functions, the peaks indicate signature mean currents for each of the three bases [104]. (c) An illustration depicting the binding of a short oligomer CCACC between a gold probe and surface, both functionalized with the reagent 4-mercaptobenzamide. (d) Current histograms that show differentiation between the DNA nucleotides dAMP, dCMP, dmeCMP, and dGMP due to distinction in both tunneling current amplitude and the current spikes per second. The red solid lines are double Gaussian fits in the logarithm of the current, implying two binding conformations for each nucleotide [35].
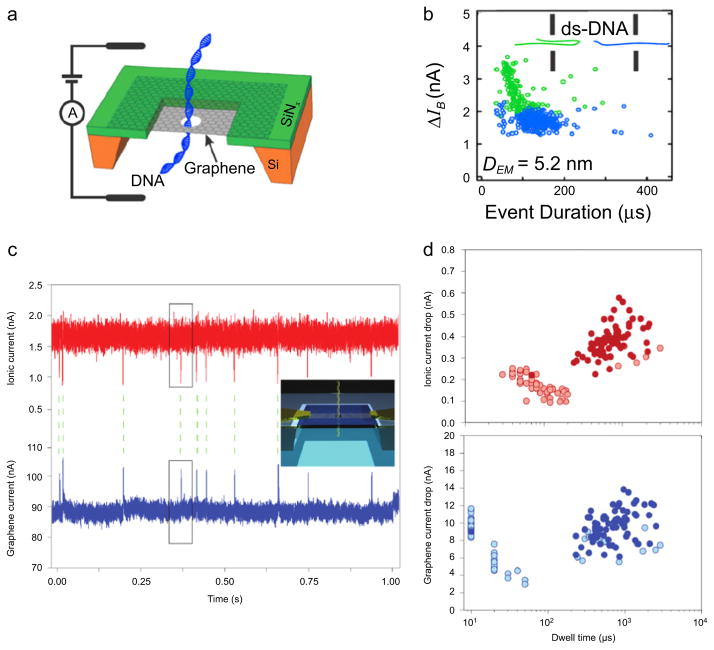
Graphene, a robust material with 2D geometry and good electronic properties, has recently sparked interest as a base material for nanopore-based DNA sequencing applications [22]. Concepts for graphene-based sequencing include the detection of fluctuations in transverse tunneling current [7, 75, 77], ion current [83, 116], or nanoribbon conductance [27, 70, 82], yet none have been experimentally realized to date. Several groups have reported on ion transport-based detection of DNA translocation through pores in single- and multi-layer graphene (see figures 3(a) and (b)) [25, 26], titania-coated graphene [66], monolayer-coated graphene [86], and alumina-coated graphene [108]. In addition, similar experiments through other 2D materials such as molybdenum sulfide [57] and hexagonal boron nitride nanopores [58, 122] have been demonstrated. These experimental works have collectively pinpointed to a problem with graphene as a membrane/pore material: graphene’s hydrophobicity reduces its compatibility with processes that require a dynamic aqueous biomolecular environment, namely, ion/DNA transport. It is therefore key that graphene’s surface is modified with the appropriate agent in order to reduce undesirable DNA sticking and ion current signal fluctuations.
Figure 3.
Graphene nanopores for ionic current and electric field detection of DNA. (a) A schematic of a dsDNA molecule translocating through a graphene nanopore. (b) A sample scatter plot of current blockade ΔIB versus the event duration for 10 kbp DNA transport through a 5.2 nm pore (160 mV applied bias, 3 M KCl, pH 10). A pore of this size permits two types of translocation events: unfolded, linear events (blue dots) and folded events (green dots). This heterogeneity of events can be eliminated by raising the solution pH or fabricating pores of smaller diameter [26]. (c) Graphene nanoribbon (GNR) for field-effect sensing of translocating DNA molecules through a nearby nanopore. By monitoring both the transmembrane ion current (red trace) and GNR transistor current (blue trace), individual translocations of circular pNEB DNA (2713 bp) are detected by and correlated between the two signals. Inset: an illustration of DNA translocation a nanopore embedded inside a GNR. (d) Scatter plots of ionic current drop and graphene current drop versus dwell time. Solid-colored circles are translocation events (n = 125) that were correlated between the ion and graphene current signals (i.e., 56% were successfully correlated). The majority of uncorrelated events occur at faster time scales, perhaps due to DNA collisions [103].
In recent simulations, the Aksimentiev group demonstrated that the surface charge applied to a graphene sheet containing a small nanopore can drastically alter the translocation velocity of a ssDNA [88]. By repeatedly alternating the polarity of the graphene surface charge, a DNA molecule occupying the pore has a stop and go motion, which if refined properly could prove useful for step-wise single base identification by ion or tunneling current readout. The ability to incorporate electronically-active 2D materials as high-resolution sensors with nanopores as DNA transport agents is attractive, and has led to the development of devices that sense a field effect due to DNA presence near the material.
Field-effect sensors
Another proposed approach for base readout utilizes a field-effect induced conductance change upon DNA occupation in a pore-containing 2D nanoribbon. Graphene nanoribbons (GNRs) on the order of <200 nm in width have a high electronic conductance as compared with the ionic current through a ~2 nm pore, which is ~3 orders of magnitude lower [78, 82, 103]. By combining chemical vapor deposition and electron-beam lithography, narrow GNRs can be fabricated onto thin silicon nitride (SiN) membranes. Due to the high mobility of electrons and holes in graphene, electronic conductance through GNRs is sensitive to subtle variations in local electric potential fields, which renders the device a sensitive field-effect transistor (FET). As a DNA molecule translocates through a fabricated nanopore inside or adjacent to a GNR, single-base sensitive sequencing is potentially viable if a differentiable field is generated upon the presence of different bases in the sensing region of the device. As shown in figures 3(c) and (d), the Radenovic group has recently demonstrated DNA detection via simultaneous ion current and GNR electronic current measurements [103], although no indication about base recognition or single-base resolution has been shown. Results from this work were similar to an earlier work that utilized a Si nanowire/nanopore as a FET-based detector [119]. In this work the use of symmetric salt conditions (1 M KCl) led to no observable events in the nanowire signal, while application of a 100-fold salt gradient such that the FET device was in the lower ionic strength compartment produced measurable translocation events. Although promising advances have been made in field-effect detection of DNA, many refinements and challenges to overcome will still be required towards achieving single-nucleotide resolution.
Sequence conversion/detection
To bypass difficulties associated with ionic current readout resolution, many groups pursued sequencing by the optical detection of fluorescently labeled DNA molecules. McNally et al reported on the use of circular DNA conversion to expand each nucleobase of a ssDNA by 24-fold into a 2-bit binary-encoded sequence [6, 64]. This sequence expansion relies on a multistep circular biochemical assay, and results in an amplified ssDNA that can be used for nanopore-based detection. Once converted, the expanded sequence template is hybridized to two fluorescently-labeled 12 mer molecular beacons, and the hybrid is unzipped using a 2 nm solid-state nanopore while monitoring the fluorescence color using an ultrafast electron-multiplying CCD camera. As the beacons are unzipped sequentially at the pore, the 2-bit information gathered from the sequence of color bursts is translated into the sequence. Ultimately, the major advantage of this approach is enzyme-free readout, which in principle speeds up the rate of sequence readout to the 1 ms regime from its 10–100 ms native rate. However, possible flaws to this method are the converted sequence will be too long to allow readout of multi-kb length DNA due to reduced yield of expanded DNA production and inefficient capture at the pore. In addition, point deletion/insertion errors in the 2-bit converted sequence can lead to sub-base frame shifts, which would have to be dealt with using non-standard algorithms.
A similar technique for sequence conversion is being developed by Stratos Genomics. Sequencing By Xpansion involves enzymatic conversion of a DNA sequence into a longer surrogate molecule called an xpandomer, a molecule that is ~50 times longer than the original DNA molecule. Once the sequence is completely encoded into the xpandomer, it is then translocated through a nanopore device. Two major advantages of this method are electrical detection (compact and fast) and a potential for increased resolution afforded by the expansion.
Sequence mapping
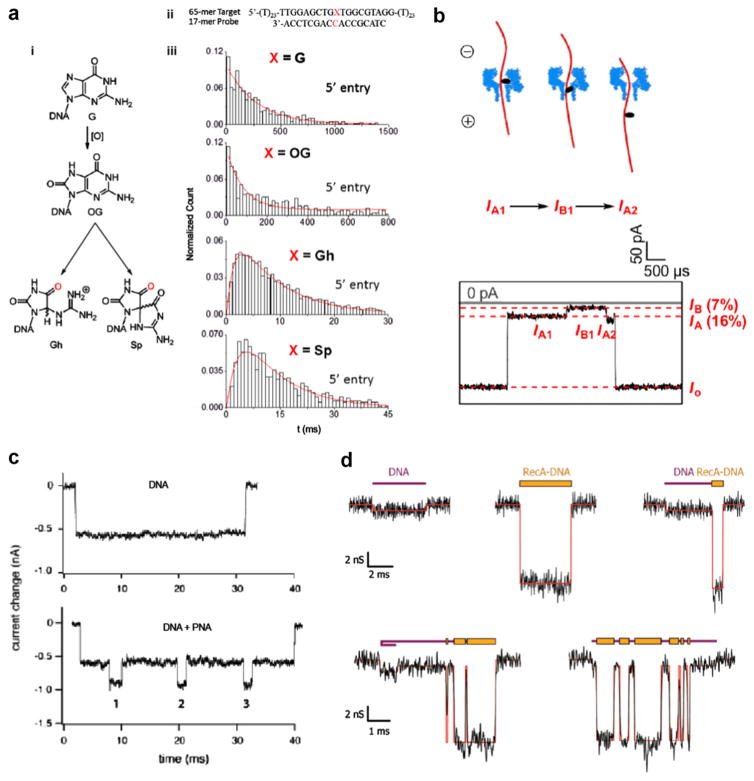
Mapping DNA damage through the use of nanopores could prove groundbreaking since DNA base lesions have been associated with age-related diseases such as cancer and Alzheimer’s disease [21, 59, 62]. One of the most common base lesions in DNA sequence occurs in the form of the mutation of guanine, 8-oxo-7, 8-dihydroguanine (OG), and its further oxidized products spiroiminodihydantoin (Sp) and guanidinohydantoin (Gh). The differentiation of these base lesions has been achieved by immobilizing biotinylated oligonucleotides containing a single guanine modification inside of an αHL pore with a streptavidin molecule as an anchor [84], as well as unzipping DNA duplexes containing single oxidized guanines (see figure 4(a)) [40]. The subtle shifts in current blockade and unzipping duration revealed structural information, which correlated specifically to each oxidized lesion of guanine. Another prevalent form of DNA damage in the human genome are abasic (AP) sites, which, if unrepaired, can lead to DNA mutagenesis, polymerase stalling, DNA strand breaks, and cytotoxicity [11, 117]. Protein nanopore sensors have proven capable of detecting AP lesions by binding 2-aminomethyl-18-crown-6 (18c6) as a label molecule, which causes a noticeably deeper current blockade during pore translocation (see figure 4(b)) [4], as well as when immobilizing DNA strands containing AP sites using a streptavidin–biotin–DNA complex [44]. In a recent publication, the Burrows group pinpointed the precise location of a furan group, a type of AP lesion, in a DNA duplex by detecting a different current blockade when this site was positioned at the latch or central constriction site of an α-hemolysin (αHL) pore while immobilized [41].
Figure 4.
Mapping of DNA damage and DNA–protein complexes using nanopores. (a) Using αHL pores to unzip DNA duplexes containing a single oxidized guanine. (i) The molecular structure of guanine (G) and it’s oxidatively-damaged products 8-oxo-8,8-dihydroguanine (OG), guanidinohydantoid (Gh), and spiroiminodihydantoid (Sp). (ii) The hybridized DNA sequence used experimentally to detect a single guanine base lesion. (iii) The unzipping duration histograms for all the guanine base modifications. Two different distribution shapes emerge, one shape for G and OG and another for Gh and Sp, which indicates two distinct unzipping processes. The G and OG distributions fit to a single exponential decay, which points to a single-step unzipping process, whereas the Gh and Sp distributions fit to a more complex double-exponential function, which indicates a two-step unzipping process [40, 85]. (b) Detection of an abasic site in DNA by the selective attachment of 2-aminomethyl-18-crown-6, which causes a noticeable increase in current blockade when translocating an αHL pore. This approach can be expanded to detect multiple abasic sites in a single DNA strand [4]. (c) The binding of sequence-specific peptide nucleic acids (PNA) to a DNA molecule makes it possible to map these regions in a DNA strand by monitoring the current signal while a single DNA electrophoreses through the pore [90]. (d) The non-specific binding of the protein RecA to DNA results in a larger current blockade in regions where it coats the bare DNA. The five sample current traces show events with two distinct blockade levels caused by alternating regions of DNA and RecA–DNA complex translocating a 30 nm nanopore. RecA is a DNA-stiffening protein that has shown promise in DNA-tagging for sequence mapping (Nabsys) [45].
Peptide nucleic acids (PNAs), are charge-neutral oligomers that contain nucleobases as a side-chain. Due to their strong and specific affinity to ssDNA and double-stranded DNA (dsDNA) sequences, PNA probes have great potential as sensors for genomic profiling, as recently demonstrated by Singer et al [6, 91]. The number of occurrences of a particular HIV gene and their relative locations has been detected via the higher current blockade magnitude of the DNA/PNA complex as compared to just bare DNA (see figure 4(c)) [90]. Extension of this technique to map and multiplex a number of sequences in a particular DNA sample could be performed, although it would require specific current signature identifiers for a target sequence.
DNA coating using proteins can also be of use for sequence-based information. Detailed studies of how the binding of proteins such as RecA to DNA affect its translocation time, current blockade amplitude [45, 94], (see figure 4(d)) and electrophoretic force [29, 95] in a nanopore have been carried out. However, no sequence information has been revealed using this approach due to the inherent non-specificity of RecA binding. In order to map specific sequences in a genome, Nabsys has suggested the binding of labeled oligomers that are complements to the ssDNA genome, which is then completely hybridized to dsDNA and coated in RecA for control of DNA coiling and secondary structure [102]. By mapping a genome with enough unique oligomers, this sequencing by hybridization technique was able to differentiate between multiple strains of viral genomes with great accuracy [101].
Two common epigenetic markers that are commonplace in eukaryotic DNA are methylated and hydroxymethylated cytosine, mC and hmC, respectively. These epigenetic modifications play an active role in regulating gene expression [8, 12], DNA–histone interactions [43, 71], and thus are often good markers of various diseases and cancers [23, 80]. The importance of these epigenetic markers in DNA samples is illustrated by the coining of mC and hmC as the fifth and sixth DNA bases, respectively. Furthermore, there is a particular challenge in discriminating between different types of cytosine modifications. Differentiation of mC and hmC is not possible using standard bisulfite sequencing techniques, which only distinguishes C from mC and hmC [99]. The Timp group demonstrated that they were able to discriminate DNA containing varying degrees of mC in solid-state nanopores by using pores <2 nm and applying high voltages across the pore [68]. When quantifying the amount of DNA in the trans chamber by qPCR, they discovered that the amount of methylation determined what threshold voltage was required for DNA translocation. The Bayley group identified C, mC, and hmC bases by immobilizing a ssDNA molecule of known sequence inside of an αHL pore using a biotin–streptavidin linkage [110]. Wallace et al established that each nucleobase had an easily distinguishable current blockade by mapping different sequences with C, mC, and hmC at various locations along the immobilized strand. Wanunu et al investigated differences in transport properties between identical dsDNA sequences with native and chemically-modified cytosines. Although differences were observed, complementary atomic force microscopy and melting temperature analysis measurements pointed to changes in DNA flexibility of the DNA with modified cytosines: DNA with the hydrophobic base mC was the least flexible, whereas hmC-DNA was the most flexible, giving rise to longer dwell times [112]. Using proteins that bind specifically to methylated DNA, Shim et al distinguished dsDNA that contained strictly C or mC sites by detecting drastic increases in both the ionic current blockade and dwell time of the methylated DNA–protein complexes as compared to bare unmethylated DNA [89]. Recent work by the Akeson and Gundlach groups has also proved that differences in cytosine methylation (i.e., C, mC, and hmC) could be detected by ratcheting DNA sequences with known methylation locations base-by-base through an MspA pore using the phi29 DNA polymerase (DNAP) and analyzing the current blockade levels, as described below [50, 87].
DNA motion control
Kasianowicz et al first realized the concept of polymer translocation through a nanopore experimentally in 1996 using a lipid-embedded αHL channel to detect single ssDNA molecules [42]. This proof-of-principle finding spurred many other groups to study the possibility of nanopores being able to discriminate between the different bases that comprise both ssDNA and ssRNA [2]. In 2000, Meller et al accurately detected the difference between poly(dA) and poly (dC) based upon the characteristic time required for each to translocate through αHL [65], a feature that was correctly attributed to purine stacking, later shown by the same group [55]. However, the regularity with which homopolymers translocate through nanopores is not similar for random DNA sequences.
For all methods of data acquisition used in DNA sequencing, one common difficulty that limits accuracy is the positional uncertainty of DNA in a nanopore. DNA transport through a nanopore is inherently a drift-diffusion process that includes a combination of directed and random motion, which contributes to anomalous DNA velocity in any translocation event [60]. As shown in figure 6(a), the random, diffusive motion of a DNA molecule can easily cause base insertions and deletions, causing incorrect sequence reads. In order to control and reduce the translocation speed of DNA to a rate at which single-nucleotide resolution is feasible (1–100 ms/nt), two major approaches have been attempted: (i) enzyme-mediated transport by incorporation of a biological motor and (ii) voltage-driven transport controlled by adjustment of pore geometry and experimental conditions.
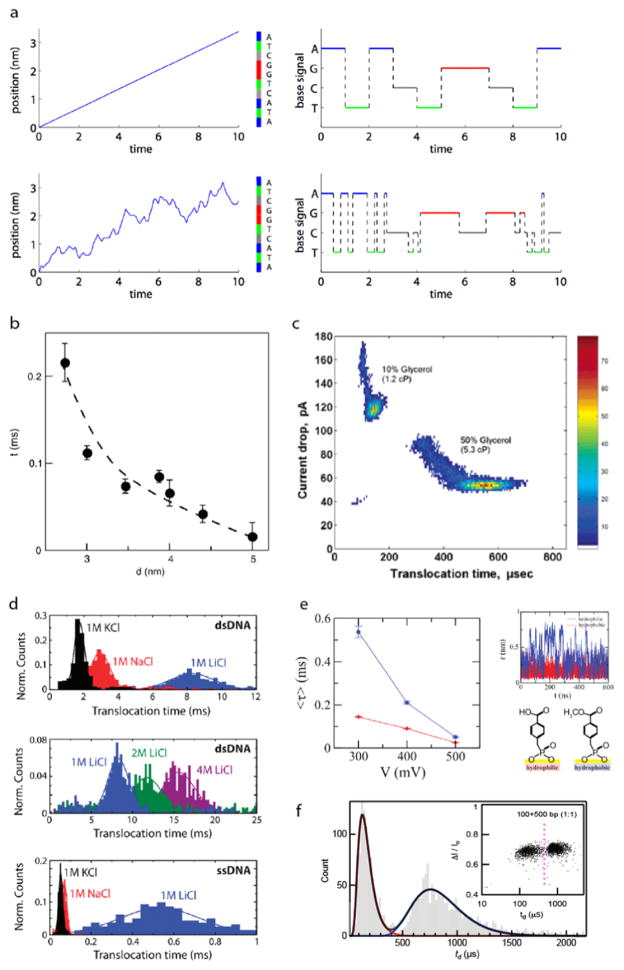
Figure 6.
Slowing down and controlling voltage-driven DNA translocation through solid-state nanopores. (a) Simulating the effect of random velocity fluctuations in DNA sequencing in an ideal single-base resolution nanopore device. Top: when a 10 nt ssDNA translocates the pore with a constant velocity, the current signal shows perfect identification of each base. Bottom: conversely, when realistic, random fluctuations are introduced to the system, the DNA sequence is detected incorrectly with multiple, erroneous base insertions and deletions [60]. (b) Reducing pore diameter has the effect of slowing down DNA transport due to greater DNA–pore interactions [115]. (c) The addition of glycerol to the electrolyte solution has the effect of unfolding DNA molecules and increasing DNA dwell time in the pore. For a 3 kbp DNA, an increase from 10% glycerol to 50% glycerol results in a four-fold increase in dwell time [24]. (d) The salt type and concentration used in electrolyte buffer has an effect on translocation time. The Dekker group observed that NaCl and LiCl resulted in longer dwell times for λ-DNA and further found that increasing the concentration of LiCl to 4 M gave even slower translocation times. An even more drastic increase in dwell time was seen for an M13mp18 ssDNA when switching the salt type to LiCl [46]. (e) Hydrophobic (blue) and hydrophilic (red) monolayer coating of pores effect voltage-driven DNA translocation. Under a moderate applied voltage, the hydrophobic monolayer causes DNA to translocate faster than in the case of a hydrophilic monolayer. By conducting MD simulations, it was seen that slower transport when using hydrophilic monolayer was due to the DNA molecule fluctuating radially inside the pore because of its great affinity to the hydrophilic pore surface [111]. (f) Using a 2.5 nm pore allows for the distinction between 100 bp and 500 bp in a mixture with an accuracy >98% [14].
Enzyme-mediated transport
Two classes of protein motors were initially used to modify DNA transport in a nanopore for the purpose of enhanced base recognition: DNA exonucleases and DNAPs. The binding of the Escherichia coli exonuclease I (ExoI) to ssDNA was observed to slow down translocation in αHL [34], but not in the controlled manner required for precision base readout. Later, ONT demonstrated that an engineered-αHL pore was capable of discriminating between individual dNMPs hydrolyzed by ExoI digestion in solution [19]. Despite successful base recognition, the use of exonucleases for DNA sequencing has two major shortcomings that caused researchers to move away from its use. First, by sequencing in this manner, the original sequence cannot be recovered for multiple reads since it is digested irreversibly into dNMPs. Second, this method requires the precise feeding of individual nucleotides into the pore in the correct order, which is inherently difficult since Brownian forces could cause missed or out of order base reads [79]. Due to these issues, further exploration into using exonucleases for DNA sequencing was set aside for more promising biological motors, including the (DNAP).
The first polymerase widely studied was the Klenow fragment (KF) of E coli DNAP I in conjunction with αHL pores. Benner et al first showed that a DNA–KF complex in the presence of the correct dNTP slowed down DNA transport through αHL by ~2 orders of magnitude [9]. By immobilizing a ssDNA–PEG complex inside an αHL pore, the Ghadiri group demonstrated that individual base synthesis using TopoTaq and KF DNAP could be observed by the detection of nine different current blockade levels, each representing a different ratio of ssDNA to PEG in the αHL barrel constriction [20]. Following these initial results, fruitful studies of KF binding kinetics [118], concentration dependence of dGTP and KF on dwell time [36], and detection of abasic regions in KF–DNA complexes [28] were demonstrated in αHL pores.
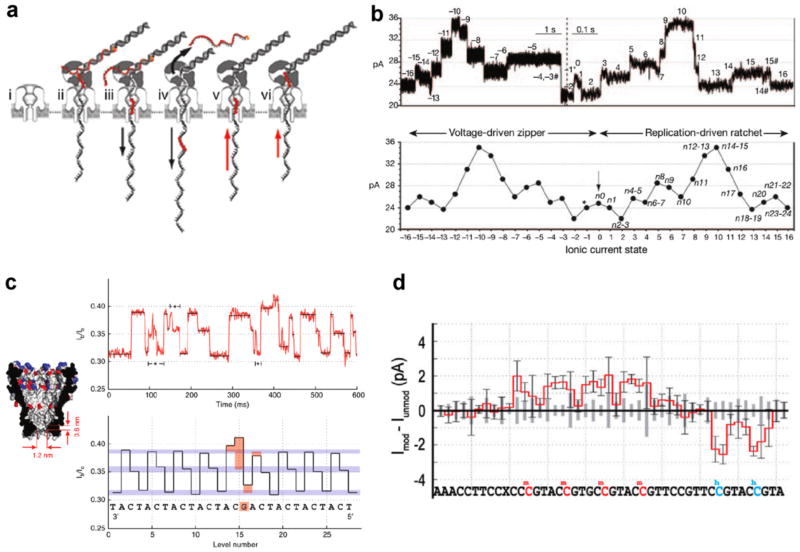
Although these experiments were valuable to understanding polymerase activity, experiments shifting toward real-time base sequencing required the development of a technique capable of eliminating bulk extension or digestion of DNA templates before arrival at the pore mouth. When binding a blocking oligomer, a complementary ssDNA with a non-complementary 3′ tail and acridine residue, adjacent to the primer, Olasagasti et al established that replication by KF and T7, another A-family DNAP, is eradicated in bulk solution [73]. Issues surrounding the stability and processivity of KF and T7 DNAPs led researchers to replace these polymerases with the B-family DNAP phi29 [54], which has been shown to synthesize up to 70 kb before dissociation [10] and retains synthetic activity at applied forces as large as 37 pN [37]. Later, by using αHL or modified MspA pores, the Akeson and Gundlach groups achieved seconds-long, phi29 mediated translocations of ssDNA with sensitivity to both sequence [18, 63] and epigenetic modifications in cytosine (see figure 5) [50, 87]. Despite this breakthrough, there are still a few issues with enzyme-driven translocation of ssDNA through pores for the application of DNA sequencing. The first major shortcoming is that MspA has a constriction region that is about 0.6 nm long, which means the ion current measured at any moment is the average of ~4 nt. This smearing of information requires the application of analysis algorithms that can precisely and repeatedly decipher the differences between the possible 256 permutations of 4 nt reads. By predicting the current levels of each distinct 4 nt sequence, or quadromer, the Gundlach group were recently able to sequence and map portions of the phi X174 genome, with read lengths as long as 4500 bases (and mean read length of ~1300 bases) [51]. Secondly, the use of a DNAP introduces the possibility of skipping or double-reading bases in a DNA strand due to fluctuations in synthesis rate and the inherent proofreading trait of phi29 DNAP. Lastly, the stochastic duration of any current level in a synthesis-driven event can cause inconsistencies in reading homopolymeric regions of a DNA molecule. To minimize the read out errors due to the latter two difficulties, the implementation of alternative protein motors such as helicases should be explored in detail. The latest product released for early access use from ONT, the MinIon, which incorporates a proprietary protein motor in its nanopore sequencing process, showed an accurate base recognition of 10% on average [67]. These less than optimal results suggest that further work is needed before the MinIon can be considered for de novo sequencing applications.
Figure 5.
Enzyme-mediated transport of DNA through protein nanopores. (a) A step-by-step schematic describing sequencing by phi29 synthesis of a DNA sequence containing an a basic region. (i) An αHL pore embedded in a lipid bilayer before DNA capture. (ii) The capture of a DNA–polymerase complex by the protruding electric field. (iii) The electrophoretic force starts the unzipping of the blocking oligomer, which prevented DNA digestion and synthesis in the bulk solution. (iv) Once completely unzipped, the blocking oligo diffuses away from the pore. (v) DNA synthesis by phi29 reverses the direction of DNA motion against the applied bias. (vi) Once the abasic region of the DNA template reaches the polymerase, synthesis concludes and the DNA–polymerase complex disassociates from the pore mouth. (b) Top: a sample current trace of a single DNA molecule being unzipped by the electrophoretic force, then synthesized by the phi29 DNAP. Bottom: when averaging multiple current traces, multiple current levels are discerned that represent an averaged combination of the bases lying within the pore constriction [18]. (c) Using phi29 DNAP to ratchet a repeating CAT DNA sequence with a single substitution of G through a modified MspA nanopore. Left: the narrower and shorter constriction of MspA yield greater spatial resolution for base recognition. Right: despite the inherent stochastic motion of the DNA sequence, the raw and analyzed current traces detect an increase in fractional current blockade at the location of the guanine substitution [63]. (d) By driving DNA through a modified MspA nanopore by polymerase catalysis, the epigenetic modifications methylcytosine (mC) and hydroxymethylcytosine (hmC) can be distinguished from cytosine (C), despite not having single base resolution [50].
Voltage-driven transport
Various approaches have been taken to slow down and regulate the otherwise irregular DNA velocity profile during voltage-driven translocation. The Golovchenko group simulated the difficulties surrounding random ssDNA motion through an ideal base-sequencing nanopore, as shown in figure 6(a) [60]. When completely eliminating random stop and go motion of DNA, perfect base recognition is achievable in their simulations. However, when a realistic diffusive motion of DNA is incorporated into the simulation, multiple insertions and deletions of base reads are seen. Due to issues with scalability, stability, limited pore geometries, and incompatibility with different detection schemes, research expanded from protein pores to include the use of solid-state nanopores. First formed by ion-beam sculpting [52] and fabricated more recently by TEM drilling [97, 98] or dielectric breakdown [13], solid-state nanopores can resolve the above limitations, but at the cost of reduced spatial resolution due to the required thickness needed for membrane stability. Li et al first demonstrated that dsDNA could translocate these synthetic pores [52], sparking many subsequent studies of dsDNA transport dynamics. Due to the greater stiffness of dsDNA, however, translocation velocity increased since the entropic barrier for threading through the pore decreased. This fueled many endeavors into modifying experimental conditions to reduce this transport speed and to minimize motion stochasticity due to DNA diffusion.
An obvious approach to slow DNA is to reduce the applied bias, which increases its dwell time inside the pore [24, 115]. This adjustment, however, has the detrimental effect of decreasing the capture rate of DNA [114] and reducing the current signal-to-noise ratio. Another approach involves slowing down DNA by threading it through a sub-5 nm diameter nanopore, which increases DNA–pore wall interactions (see figure 6(b)) [3, 24, 31, 115]. However, this has resulted in noticeable smearing of the dwell-time distribution widths, a phenomenon that suggests stop–go motion of the DNA due to stalling interactions at the pore. Modest reductions in translocation velocity have been observed for dsDNA when increasing the solution viscosity [24, 53] (see figure 6(c)) or operating at low temperatures (i.e., <20 °C) [24, 115], but these come with the adverse effect of reducing the current signal. As shown in figure 6(d), electrolytic solutions with various molarities and cationic species have been shown to decrease pore dwell time relative to 1 M KCl [46, 105], but have little effect on reducing the average drift velocity of DNA. Chemical modifications of the nanopore surface [5, 38, 96, 111, 113] have shown promise in slowing DNA transport, although an undesirable irregularity in motion has been observed (see figure 6(e)). Aside from using SiNx and SiO2, researchers have also fabricated nanopores in other materials such as aluminum oxide [107] and hafnium oxide [49] and have embedded nanopores formed from DNA origami structures [32, 48] all of which have been shown to reduce translocation velocity moderately. Also, researchers have observed that by applying a pressure difference in opposition to the electrophoretic motion, DNA velocity can be slowed noticeably through nanopores [61, 120].
Recently, Carson et al found using ultrathin SiN nanopores (<10 nm thick) that reducing the nanopore diameter to d ~3 nm results in smooth, well-behaved voltage-driven dsDNA transport [14] that can be modeled as a first-passage time distribution that is derived from the 1D Fokker–Planck equation [53, 56, 69]. When fitting dwell time distributions to this model, the greatest agreement between the data and model occurred in pores was found for diameters in range 2.8–3.0 nm, which suggests ‘smooth’ motion through the pore. In larger pores, broader dwell-time distributions were observed, which was attributed to self-interaction of DNA due to field-driven formation of a DNA loop at the pore mouth. Physically, this signifies that pores of this size confine DNA in a manner that balances the hydrodynamic forces and pore surface interactions optimally, all of which give rise to a greatly reduced DNA axial diffusion. Using pores in this diameter regime, differentiation of 100 bp and 500 bp DNA was achieved with an accuracy of 98% in an equimolar mixture by detecting single current pulses as seen in figure 6(f) [14].
Conclusions
In conclusion, nanopores have tremendous potential for revolutionizing nucleic acid analysis, specifically DNA sequencing. To our knowledge, there is no equivalent nanoscale device to a nanopore that allows localization/transport of DNA sequentially in space, while in addition to detect its identity. However, controlling DNA motion through a nanopore, and readout of its sequence, are independently grand challenges that require further development before this method can be viable as a platform. Further, with the proposed use of emerging synthetic materials (e.g., 2D materials, metals), various approaches to minimize interactions of the DNA with these materials need to be explored. Looking forward, it will be interesting to see breakthroughs in methods to control the motion of DNA through pores and its readout.
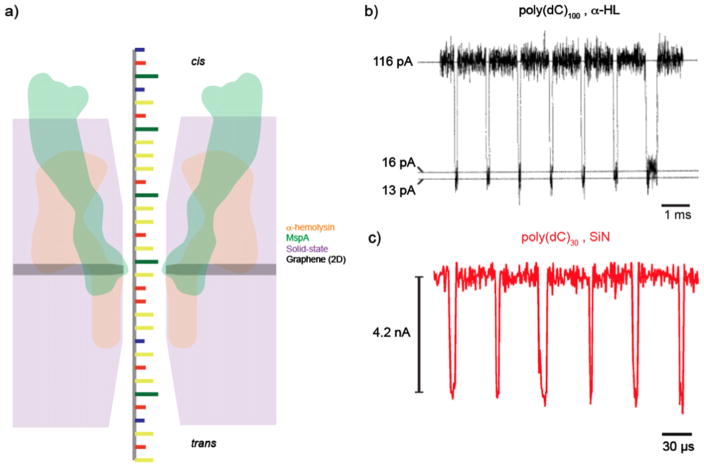
Figure 1.
Ionic current detection of ssDNA in protein and solid-state nanopores. (a) An illustration comparing the relative sizes of α-hemolysin (αHL) (orange), MspA (green), solid-state (purple), and graphene nanopores (gray). The reduced thickness of MspA and graphene pores allows for the greatest spatial resolution, which is the motivation behind their usage towards DNA sequencing applications. (b) A concatenated current trace of 100 mer cytosine homopolymer translocations through an αHL channel in 1.0 M KCl solution when applying a transmembrane voltage of 120 mV. The restriction of the pore diameter and low voltage application allows for transport times on the order of 100 μs [2]. (c) A concatenated current trace of poly(dC) 30 mer translocations through an ultrathin silicon nitride nanopore (diameter of 1.4 nm) in 1.0 M KCl when applying a voltage bias of 1 V. While these small, thin pores can provide an improvement in structural details and signal to noise ratio, the reduced thickness and higher applied voltage causes the average dwell time to be on the order of 10 μs [109].
Acknowledgments
We gratefully acknowledge financial support from the National Institutes of Health, Grant Nos. R21-HG006873 and R01-HG6321.
References
- 1.www.genome.gov/12513210
- 2.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–33. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aksimentiev A, Heng JB, Timp G, Schulten K. Microscopic kinetics of DNA translocation through synthetic nanopores. Biophys J. 2004;87:2086–97. doi: 10.1529/biophysj.104.042960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An N, Fleming AM, White HS, Burrows CJ. Crown ether–electrolyte interactions permit nanopore detection of individual DNA abasic sites in single molecules. Proc Natl Acad Sci. 2012;109:11504–9. doi: 10.1073/pnas.1201669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson BN, Muthukumar M, Meller A. pH Tuning of DNA translocation time through organically functionalized nanopores. ACS Nano. 2012;7:1408–14. doi: 10.1021/nn3051677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atas E, Singer A, Meller A. DNA sequencing and bar-coding using solid-state nanopores. Electrophoresis. 2012;33:3437–47. doi: 10.1002/elps.201200266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avdoshenko SM, Nozaki D, Gomes da Rocha C, González JW, Lee MH, Gutierrez R, Cuniberti G. Dynamic and electronic transport properties of DNA translocation through graphene nanopores. Nano Lett. 2013;13:1969–76. doi: 10.1021/nl304735k. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Esteller M, Rountree MR, Bachman KE, Schuebel K, Herman JG. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genetics. 2001;10:687–92. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- 9.Benner S, Chen RJA, Wilson NA, Abu-Shumays R, Hurt N, Lieberman KR, Deamer DW, Dunbar WB, Akeson M. Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore. Nat Nanotechnology. 2007;2:718–24. doi: 10.1038/nnano.2007.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco L, Bernad A, Lázaro JM, Martin G, Garmendia C, Salas M. Highly efficient DNA synthesis by the phage phi 29 DNA polymerase. Symmetrical mode of DNA replication. J Biol Chem. 1989;264:8935–40. [PubMed] [Google Scholar]
- 11.Boiteux S, Guillet M. Abasic sites in DNA: repair and biological consequences in. Saccharomyces cerevisiae DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Brena RM, Huang TH, Plass C. Toward a human epigenome. Nat Genetics. 2006;38:1359–60. doi: 10.1038/ng1206-1359. [DOI] [PubMed] [Google Scholar]
- 13.Briggs K, Kwok H, Tabard-Cossa V. Automated fabrication of 2 nm solid-state nanopores for nucleic acid analysis. Small. 2014;10:2077–86. doi: 10.1002/smll.201303602. [DOI] [PubMed] [Google Scholar]
- 14.Carson S, Wilson J, Aksimentiev A, Wanunu M. Smooth DNA transport through narrowed pore geometry. Biophys J. 2014;107:2381–93. doi: 10.1016/j.bpj.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang S, He J, Kibel A, Lee M, Sankey O, Zhang P, Lindsay S. Tunnelling readout of hydrogen-bonding-based recognition. Nat Nanotechnology. 2009;4:297–301. doi: 10.1038/nnano.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang S, Huang S, He J, Liang F, Zhang P, Li S, Chen X, Sankey O, Lindsay S. Electronic signatures of all four DNA nucleosides in a tunneling gap. Nano Lett. 2010;10:1070–5. doi: 10.1021/nl1001185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Yemenicioglu S, Uddin A, Corgliano E, Theogarajan L. A CMOS Enhanced Solid-State Nanopore Based Single Molecule Detection Platform: IEEE. 2013:164–7. doi: 10.1109/EMBC.2013.6609463. [DOI] [PubMed] [Google Scholar]
- 18.Cherf GM, Lieberman KR, Rashid H, Lam CE, Karplus K, Akeson M. Automated forward and reverse ratcheting of DNA in a nanopore at 5-A precision. Nat Biotechnol. 2012;30:344–8. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnology. 2009;4:265–70. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 20.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–20. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooke MS, Olinski R, Evans MD. Does measurement of oxidative damage to DNA have clinical significance? Clin Chim Acta. 2006;365:30–49. doi: 10.1016/j.cca.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Drndic M. Sequencing with graphene pores. Nat Nanotehnology. 2014;9:743. doi: 10.1038/nnano.2014.232. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Herman JG. Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 24.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–7. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garaj S, Hubbard W, Reina A, Kong J, Branton D, Golovchenko JA. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467:190–3. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garaj S, Liu S, Golovchenko JA, Branton D. Molecule-hugging graphene nanopores. Proc Natl Acad Sci. 2013;110:12192–6. doi: 10.1073/pnas.1220012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Girdhar A, Sathe C, Schulten K, Leburton J-P. Graphene quantum point contact transistor for DNA sensing. Proc Natl Acad Sci. 2013;110:16748–53. doi: 10.1073/pnas.1308885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gyarfas B, Olasagasti F, Benner S, Garalde D, Lieberman KR, Akeson M. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 angstrom resolution. ACS Nano. 2009;3:1457–66. doi: 10.1021/nn900303g. [DOI] [PubMed] [Google Scholar]
- 29.Hall AR, van Dorp S, Lemay SG, Dekker C. Electrophoretic force on a protein-coated DNA molecule in a solid-state nanopore. Nano Lett. 2009;9:4441–5. doi: 10.1021/nl9027318. [DOI] [PubMed] [Google Scholar]
- 30.He J, Lin L, Zhang P, Lindsay S. Identification of DNA basepairing via tunnel-current decay. Nano Lett. 2007;7:3854–8. doi: 10.1021/nl0726205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heng JB, Ho C, Kim T, Timp R, Aksimentiev A, Grinkova YV, Sligar S, Schulten K, Timp G. Sizing DNA using a nanometer-diameter pore. Biophys J. 2004;87:2905–11. doi: 10.1529/biophysj.104.041814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernández-Ainsa S, Bell NA, Thacker VV, Göpfrich K, Misiunas K, Fuentes-Perez ME, Moreno-Herrero F, Keyser UF. DNA origami nanopores for controlling DNA translocation. ACS Nano. 2013;7:6024–60230. doi: 10.1021/nn401759r. [DOI] [PubMed] [Google Scholar]
- 33.Hoogerheide DP, Garaj S, Golovchenko JA. Probing surface charge fluctuations with solid-state nanopores. Phys Rev Lett. 2009;102:256804. doi: 10.1103/PhysRevLett.102.256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornblower B, Coombs A, Whitaker RD, Kolomeisky A, Picone SJ, Meller A, Akeson M. Single-molecule analysis of DNA–protein complexes using nanopores. Nat Methods. 2007;4:315–7. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 35.Huang S, He J, Chang S, Zhang P, Liang F, Li S, Tuchband M, Fuhrmann A, Ros R, Lindsay S. Identifying single bases in a DNA oligomer with electron tunnelling. Nat Nanotechnology. 2010;5:868–73. doi: 10.1038/nnano.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hurt N, Wang HY, Akeson M, Lieberman KR. Specific nucleotide binding and rebinding to individual DNA polymerase complexes captured on a nanopore. J Am Chem Soc. 2009;131:3772–8. doi: 10.1021/ja809663f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibarra B, Chemla YR, Plyasunov S, Smith SB, Lazaro JM, Salas M, Bustamante C. Proofreading dynamics of a processive DNA polymerase. EMBO J. 2009;28:2794–802. doi: 10.1038/emboj.2009.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iqbal SM, Akin D, Bashir R. Solid-state nanopore channels with DNA selectivity. Nat Nanotechnology. 2007;2:243–8. doi: 10.1038/nnano.2007.78. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov AP, Instuli E, McGilvery CM, Baldwin G, McComb DW, del Albrecht TEJB. DNA tunneling detector embedded in a nanopore. Nano Lett. 2011;11:279–85. doi: 10.1021/nl103873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin Q, Fleming AM, Burrows CJ, White HS. Unzipping kinetics of duplex DNA containing oxidized lesions in an alpha-hemolysin nanopore. J Am Chem Soc. 2012;134:11006–11. doi: 10.1021/ja304169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson RP, Fleming AM, Burrows CJ, White HS. Effect of an electrolyte cation on detecting DNA damage with the latch constriction of α-hemolysin. J Phys Chem Lett. 2014;5:3781–6. doi: 10.1021/jz502030e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci USA. 1996;93:13770–3. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–43. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Maitra RD, Pedrotti K, Dunbar WB. Detecting single-abasic residues within a DNA strand immobilized in a biological nanopore using an integrated CMOS sensor. Sensors Actuators B. 2013;177:1075–82. doi: 10.1016/j.snb.2012.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kowalczyk SW, Hall AR, Dekker C. Detection of local protein structures along DNA using solid-state nanopores. Nano Lett. 2010;10:324–8. doi: 10.1021/nl903631m. [DOI] [PubMed] [Google Scholar]
- 46.Kowalczyk SW, Wells DB, Aksimentiev A, Dekker C. Slowing down DNA translocation through a nanopore in lithium chloride. Nano Lett. 2012;12:1038–44. doi: 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagerqvist J, Zwolak M, Di Ventra M. Fast DNA sequencing via transverse electronic transport. Nano Lett. 2006;6:779–82. doi: 10.1021/nl0601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langecker M, Arnaut V, Martin TG, List J, Renner S, Mayer M, Dietz H, Simmel FC. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338:932–6. doi: 10.1126/science.1225624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin J, Henley R, Bell DC, Cohen-Karni T, Rosenstein JK, Wanunu M. Slow DNA transport through nanopores in hafnium oxide membranes. ACS Nano. 2013;7:10121–8. doi: 10.1021/nn404326f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laszlo AH, Derrington IM, Brinkerhoff H, Langford KW, Nova IC, Samson JM, Bartlett JJ, Pavlenok M, Gundlach JH. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc Natl Acad Sci. 2013;110:18904–9. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Laszlo AH, Derrington IM, Ross BC, Brinkerhoff H, Adey A, Nova IC, Craig JM, Langford KW, Samson JM, Daza R. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829–33. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–9. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Talaga DS. The distribution of DNA translocation times in solid-state nanopores. J Phys : Condens Matter. 2010;22:454129. doi: 10.1088/0953-8984/22/45/454129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lieberman KR, Cherf GM, Doody MJ, Olasagasti F, Kolodji Y, Akeson M. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J Am Chem Soc. 2010;132:17961–72. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin J, Kolomeisky A, Meller A. Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel. Phys Rev Lett. 2010;104:158101. doi: 10.1103/PhysRevLett.104.158101. [DOI] [PubMed] [Google Scholar]
- 56.Ling DY, Ling XS. On the distribution of DNA translocation times in solid-state nanopores: an analysis using Schrödinger’s first-passage-time theory. J Phys : Condens Matter. 2013;25:375102. doi: 10.1088/0953-8984/25/37/375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu K, Feng J, Kis A, Radenovic A. Atomically thin molybdenum disulfide nanopores with high sensitivity for DNA translocation. ACS Nano. 2014;8:2504–11. doi: 10.1021/nn406102h. [DOI] [PubMed] [Google Scholar]
- 58.Liu S, et al. Boron nitride nanopores: highly sensitive DNA single-molecule detectors. Adv Mater. 2013;25:4549–54. doi: 10.1002/adma.201301336. [DOI] [PubMed] [Google Scholar]
- 59.Lonkar P, Dedon PC. Reactive species and DNA damage in chronic inflammation: reconciling chemical mechanisms and biological fates. Int J Cancer. 2011;128:1999–2009. doi: 10.1002/ijc.25815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu B, Albertorio F, Hoogerheide DP, Golovchenko JA. Origins and consequences of velocity fluctuations during DNA passage through a nanopore. Biophys J. 2011;101:70–9. doi: 10.1016/j.bpj.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lu B, Hoogerheide DP, Zhao Q, Zhang H, Tang Z, Yu D, Golovchenko JA. Pressure-controlled motion of single polymers through solid-state nanopores. Nano Lett. 2013;13:3048–52. doi: 10.1021/nl402052v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangialasche F, Polidori MC, Monastero R, Ercolani S, Camarda C, Cecchetti R, Mecocci P. Biomarkers of oxidative and nitrosative damage in Alzheimer’s disease and mild cognitive impairment. Ageing Res Rev. 2009;8:285–305. doi: 10.1016/j.arr.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 63.Manrao EA, Derrington IM, Laszlo AH, Langford KW, Hopper MK, Gillgren N, Pavlenok M, Niederweis M, Gundlach JH. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–53. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McNally B, Singer A, Yu ZL, Sun YJ, Weng ZP, Meller A. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010;10:2237–44. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meller A, Nivon L, Brandin E, Golovchenko J, Branton D. Rapid nanopore discrimination between single polynucleotide molecules. Proc Natl Acad Sci USA. 2000;97:1079–84. doi: 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Merchant CA, et al. DNA Translocation through graphene nanopores. Nano Lett. 2010;10:2915–21. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 67.Mikheyev AS, Tin MMY. A first look at the Oxford nanopore MinION sequencer. Mol Ecol Resour. 2014;14:1097–102. doi: 10.1111/1755-0998.12324. [DOI] [PubMed] [Google Scholar]
- 68.Mirsaidov U, Timp W, Zou X, Dimitrov V, Schulten K, Feinberg AP, Timp G. Nanoelectromechanics of methylated DNA in a synthetic nanopore. Biophys J. 2009;96:L32–4. doi: 10.1016/j.bpj.2008.12.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Muthukumar M. Polymer Translocation. Boca Raton, FL: CRC Press; 2011. [Google Scholar]
- 70.Nelson T, Zhang B, Prezhdo OV. Detection of nucleic acids with graphene nanopores: ab initio characterization of a novel sequencing device. Nano Lett. 2010;10:3237–42. doi: 10.1021/nl9035934. [DOI] [PubMed] [Google Scholar]
- 71.Ng H-H, Adrian B. DNA methylation and chromatin modification. Curr Opin Genetics Dev. 1999;9:158–63. doi: 10.1016/s0959-437x(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 72.Ohshiro T, Tsutsui M, Yokota K, Furuhashi M, Taniguchi M, Kawai T. Detection of post-translational modifications in single peptides using electron tunnelling currents. Nat Nanotechnology. 2014;9:835–40. doi: 10.1038/nnano.2014.193. [DOI] [PubMed] [Google Scholar]
- 73.Olasagasti F, Lieberman KR, Benner S, Cherf GM, Dahl JM, Deamer DW, Akeson M. Replication of individual DNA molecules under electronic control using a protein nanopore. Nat Nanotechnology. 2010;5:798–806. doi: 10.1038/nnano.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedone D, Langecker M, Munzer AM, Wei R, Nagel RD, Rant U. Fabrication and electrical characterization of a pore-cavity-pore device. J Phys : Condens Matter. 2010;22:454115. doi: 10.1088/0953-8984/22/45/454115. [DOI] [PubMed] [Google Scholar]
- 75.Postma HWC. Rapid sequencing of individual DNA molecules in graphene nanogaps. Nano Lett. 2010;10:420–5. doi: 10.1021/nl9029237. [DOI] [PubMed] [Google Scholar]
- 76.Powell MR, Vlassiouk I, Martens C, Siwy ZS. Nonequilibrium 1/f noise in rectifying nanopores. Phys Rev Lett. 2009;103:248104. doi: 10.1103/PhysRevLett.103.248104. [DOI] [PubMed] [Google Scholar]
- 77.Prasongkit J, Grigoriev A, Pathak B, Ahuja R, Scheicher RH. Transverse conductance of DNA nucleotides in a graphene nanogap from first principles. Nano Lett. 2011;11:1941–5. doi: 10.1021/nl200147x. [DOI] [PubMed] [Google Scholar]
- 78.Puster M, Rodríguez-Manzo JA, Balan A, Drndić M. Toward sensitive graphene nanoribbon–nanopore devices by preventing electron beam-induced damage. ACS Nano. 2013;7:11283–9. doi: 10.1021/nn405112m. [DOI] [PubMed] [Google Scholar]
- 79.Reiner JE, Balijepalli A, Robertson JWF, Drown BS, Burden DL, Kasianowicz JJ. The effects of diffusion on an exonuclease/nanopore-based DNA sequencing engine. J Chem Phys. 2012;137:214903. doi: 10.1063/1.4766363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genetics. 2000;1:11–9. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- 81.Rosenstein JK, Wanunu M, Merchant CA, Drndic M, Shepard KL. Integrated nanopore sensing platform with sub-microsecond temporal resolution. Nat Methods. 2012;9:487–92. doi: 10.1038/nmeth.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Saha KK, Drndić M, Nikolić BK. DNA base-specific modulation of microampere transverse edge currents through a metallic graphene nanoribbon with a nanopore. Nano Lett. 2011;12:50–5. doi: 10.1021/nl202870y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sathe C, Zou X, Leburton J-P, Schulten K. Computational investigation of DNA detection using graphene nanopores. ACS Nano. 2011;5:8842–51. doi: 10.1021/nn202989w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schibel AE, An N, Jin Q, Fleming AM, Burrows CJ, White HS. Nanopore detection of 8-oxo-7, 8-dihydro-2′-deoxyguanosine in immobilized single-stranded DNA via adduct formation to the DNA damage site. J Am Chem Soc. 2010;132:17992–5. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schibel AEP, Fleming AM, Jin Q, An N, Liu J, Blakemore CP, White HS, Burrows CJ. Sequence-specific single-molecule analysis of 8-Oxo-7, 8-dihydroguanine lesions in DNA based on unzipping kinetics of complementary probes in ion channel recordings. J Am Chem Soc. 2011;133:14778–84. doi: 10.1021/ja205653v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider GF, Kowalczyk SW, Calado VE, Pandraud G, Zandbergen HW, Vandersypen LMK, Dekker C. DNA Translocation through graphene nanopores. Nano Lett. 2010;10:3163–7. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 87.Schreiber J, Wescoe ZL, Abu-Shumays R, Vivian JT, Baatar B, Karplus K, Akeson M. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc Natl Acad Sci. 2013;110:18910–5. doi: 10.1073/pnas.1310615110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shankla M, Aksimentiev A. Conformational transitions and stop-and-go nanopore transport of single-stranded DNA on charged graphene. Nat Commun. 2014;5:5171. doi: 10.1038/ncomms6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shim J, et al. Detection and quantification of methylation in DNA using solid-state nanopores. Sci Rep-UK. 2013;3:1389. doi: 10.1038/srep01389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Singer A, Rapireddy S, Ly DH, Meller A. Electronic barcoding of a viral gene at the single-molecule level. Nano Lett. 2012;12:1722–8. doi: 10.1021/nl300372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singer A, Wanunu M, Morrison W, Kuhn H, Frank-Kamenetskii M, Meller A. Nanopore based sequence specific detection of duplex DNA for genomic profiling. Nano Lett. 2010;10:738–42. doi: 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smeets RMM, Dekker NH, Dekker C. Low-frequency noise in solid-state nanopores. Nanotechnology. 2009;20:095501. doi: 10.1088/0957-4484/20/9/095501. [DOI] [PubMed] [Google Scholar]
- 93.Smeets RMM, Keyser UF, Dekker NH, Dekker C. Noise in solid-state nanopores. Proc Natl Acad Sci USA. 2008;105:417–21. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smeets RMM, Kowalczyk SW, Hall AR, Dekker NH, Dekker C. Translocation of reca-coated double-stranded DNA through solid-state nanopores. Nano Lett. 2009;9:3089–95. doi: 10.1021/nl803189k. [DOI] [PubMed] [Google Scholar]
- 95.Spiering A, Getfert S, Sischka A, Reimann P, Anselmetti D. Nanopore translocation dynamics of a single DNA-bound protein. Nano Lett. 2011;11:2978–82. doi: 10.1021/nl201541y. [DOI] [PubMed] [Google Scholar]
- 96.Squires AH, Hersey JS, Grinstaff MW, Meller A. A nanopore–nanofiber mesh biosensor to control DNA translocation. J Am Chem Soc. 2013;135:16304–7. doi: 10.1021/ja408685x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Storm AJ, Chen J, Ling XS, Zandbergen HW, Dekker C. Fabrication of solid-state nanopores with single-nanometre precision. Nat Mater. 2003;2:537–40. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 98.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocation of double-strand DNA through a silicon oxide nanopore. Phys Rev E. 2005;71:051903. doi: 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- 99.Susan JC, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nuc Acid Res. 1994;22:2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tabard-Cossa V, Trivedi D, Wiggin M, Jetha NN, Marziali A. Noise analysis and reduction in solid-state nanopores. Nanotechnology. 2007;18:305505. [Google Scholar]
- 101.Thompson JF, et al. Identification of Highly Similar Microbial Strains and Pathogens 2012 [Google Scholar]
- 102.Thompson JF, Oliver JS. Mapping and sequencing DNA using nanopores and nanodetectors. Electrophoresis. 2012;33:3429–36. doi: 10.1002/elps.201200136. [DOI] [PubMed] [Google Scholar]
- 103.Traversi F, Raillon C, Benameur S, Liu K, Khlybov S, Tosun M, Krasnozhon D, Kis A, Radenovic A. Detecting the translocation of DNA through a nanopore using graphene nanoribbons. Nat Nanotechnology. 2013;8:939–45. doi: 10.1038/nnano.2013.240. [DOI] [PubMed] [Google Scholar]
- 104.Tsutsui M, Taniguchi M, Yokota K, Kawai T. Identifying single nucleotides by tunnelling current. Nat Nanotechnology. 2010;5:286–90. doi: 10.1038/nnano.2010.42. [DOI] [PubMed] [Google Scholar]
- 105.Uplinger J, Thomas B, Rollings R, Fologea D, McNabb D, Li J. K+, Na+ and Mg2+ on DNA translocation in silicon nitride nanopores. Electrophoresis. 2012;33:3448–57. doi: 10.1002/elps.201200165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uram JD, Ke K, Mayer M. Noise and bandwidth of current recordings from submicrometer pores and nanopores. ACS Nano. 2008;2:857–72. doi: 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- 107.Venkatesan BM, Dorvel B, Yemenicioglu S, Watkins N, Petrov I, Bashir R. Highly sensitive, mechanically stable nanopore sensors for DNA analysis. Adv Mater. 2009;21:2771–6. doi: 10.1002/adma.200803786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Venkatesan BM, Estrada D, Banerjee S, Jin X, Dorgan VE, Bae M-H, Aluru NR, Pop E, Bashir R. Stacked graphene–Al2O3 nanopore sensors for sensitive detection of DNA and DNA–protein complexes. ACS Nano. 2011;6:441–50. doi: 10.1021/nn203769e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Venta K, Shemer G, Puster M, Rodríguez-Manzo JA, Balan A, Rosenstein JK, Shepard K, Drndić M. Differentiation of short, single-stranded DNA homopolymers in solid-state nanopores. ACS Nano. 2013;7:4629–36. doi: 10.1021/nn4014388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wallace EVB, Stoddart D, Heron AJ, Mikhailova E, Maglia G, Donohoe TJ, Bayley H. Identification of epigenetic DNA modifications with a protein nanopore. Chem Commun. 2010;46:8195–7. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wang D, Harrer S, Luan B, Stolovitzky G, Peng H, Afzali-Ardakani A. Regulating the transport of DNA through biofriendly nanochannels in a thin solid membrane. Sci Rep-UK. 2014;4:3985. doi: 10.1038/srep03985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wanunu M, Cohen-Karni D, Johnson RR, Fields L, Benner J, Peterman N, Zheng Y, Klein ML, Drndic M. Discrimination of methylcytosine from hydroxymethylcytosine in DNA molecules. J Am Chem Soc. 2011;133:486–92. doi: 10.1021/ja107836t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wanunu M, Meller A. Chemically modified solid-state nanopores. Nano Lett. 2007;7:1580–5. doi: 10.1021/nl070462b. [DOI] [PubMed] [Google Scholar]
- 114.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnology. 2010;5:160–5. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wanunu M, Sutin J, McNally B, Chow A, Meller A. DNA translocation governed by interactions with solid-state nanopores. Biophys J. 2008;95:4716–25. doi: 10.1529/biophysj.108.140475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wells DB, Belkin M, Comer J, Aksimentiev A. Assessing graphene nanopores for sequencing DNA. Nano Lett. 2012;12:4117–23. doi: 10.1021/nl301655d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilson DM, III, Barsky D. The major human abasic endonuclease: formation, consequences and repair of abasic lesions in DNA. Mutation Res DNA Repair. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 118.Wilson NA, Abu-Shumays R, Gyarfas B, Wang H, Lieberman KR, Akeson M, Dunbar WB. Electronic control of DNA polymerase binding and unbinding to single DNA molecules. ACS Nano. 2009;3:995–1003. doi: 10.1021/nn9000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xie P, Xiong Q, Fang Y, Qing Q, Lieber CM. Local electrical potential detection of DNA by nanowire–nanopore sensors. Nat Nanotechnology. 2012;7:119–25. doi: 10.1038/nnano.2011.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang HB, et al. Slowing down DNA translocation through solid-state nanopores by pressure. Small. 2013;9:4112–7. doi: 10.1002/smll.201301263. [DOI] [PubMed] [Google Scholar]
- 121.Zhao Y, Ashcroft B, Zhang P, Liu H, Sen S, Song W, Im J, Gyarfas B, Manna S, Biswas S. Single-molecule spectroscopy of amino acids and peptides by recognition tunnelling. Nat Nanotechnology. 2014;9:466–73. doi: 10.1038/nnano.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou Z, Hu Y, Wang H, Xu Z, Wang W, Bai X, Shan X, Lu X. DNA Translocation through hydrophilic nanopore in hexagonal boron nitride. Sci Rep-UK. 2013;3:3287. doi: 10.1038/srep03287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zwolak M, Di Ventra M. Electronic signature of DNA nucleotides via transverse transport. Nano Lett. 2005;5:421–4. doi: 10.1021/nl048289w. [DOI] [PubMed] [Google Scholar]