Abstract
The marine pennate diatom Phaeodactylum tricornutum has become a model for diatom biology, due to its ease of culture and accessibility to reverse genetics approaches. While several features underlying the molecular mechanisms of cell division have been described, morphological analyses are less advanced than they are in other diatoms. We therefore examined cell ultrastructure changes prior to and during cytokinesis. Following chloroplast division, cleavage furrows are formed at both longitudinal ends of the cell and are accompanied by significant vesicle transport. Although neither spindle nor microtubules were observed, the nucleus appeared to be split by the furrow after duplication of the Golgi apparatus. Finally, centripetal cytokinesis was completed by fusion of the furrows. Additionally, F-actin formed a ring structure and its diameter became smaller, accompanying the ingrowing furrows. To further analyse vesicular transport during cytokinesis, we generated transgenic cells expressing yellow fluorescent protein (YFP) fusions with putative diatom orthologs of small GTPase Sec4 and t-SNARE protein SyntaxinA. Time-lapse observations revealed that SyntaxinA-YFP localization expands from both cell tips toward the center, whereas Sec4-YFP was found in the Golgi and subsequently relocalizes to the future division plane. This work provides fundamental new information about cell replication processes in P. tricornutum.
Key words: Diatom, cytokinesis, actin, Sec4, Syntaxin, contractile ring.
Introduction
Diatoms are unicellular heterokont protists believed to have arisen via serial secondary endosymbiotic events and characterized by peculiar structural features such as silica cell walls composed of two unequal, overlapping halves called the frustule (Moustafa et al., 2009, Round et al., 1990). They have global significance, being responsible for one fifth of global primary production (Nelson et al. 1995), and they are thought to be major players in the biological pump since dead diatoms with heavy siliceous walls transfer carbon, nitrogen, and silicon to the ocean interior as a result of gravitational settling (Armbrust et al., 2004, Bowler et al., 2010, Street-Perrott and Barker, 2008).
Diatoms have been studied for more than a hundred years to understand their unique cellular structures, such as the spindle apparatus, ornamented silica cell walls, and silica deposition vesicles (SDVs), in which the silica frustules are synthesized (Cohn et al., 1989, Tesson and Hildebrand, 2010a, Van de Meene and Pickett-Heaps, 2002, Van de Meene and Pickett-Heaps, 2004, Zurzolo and Bowler, 2001). Intensive cytological research has revealed that diatoms possess two different types of microtubule-organizing centers (MTOCs), named the microtubule center and the polar complex (reviewed by De Martino et al., 2009, Pickett-Heaps and Tippit, 1978, Pickett-Heaps et al., 1982). Actin filaments are known to be involved in meso- and micro-scale morphogenesis and also in cell motility (Cohn et al., 1989, Poulsen et al., 1999, Tesson and Hildebrand, 2010b). For example, it has been suggested that the SDVs may be restrained by circumferential actin bands that control valve shape (Pickett-Heaps, 1998, Tesson and Hildebrand, 2010a, Van de Meene and Pickett-Heaps, 2002, Van de Meene and Pickett-Heaps, 2004).
During cytokinesis, cells are cleaved longitudinally by ingrowing furrows from both ends and the cleavage plane grows parallel to and inbetween the two halves of overlapping silica valves (Pickett-Heaps et al., 1975, Schmid and Schulz, 1979). The centripetal mode of cytokinesis seen in diatoms is seemingly similar to what is observed in animal and yeast systems, by contrast with the centrifugal cytokinesis observed in plant cells. However, studies of diatom cytokinesis are quite sparse and are limited to large sized diatoms (De Martino et al. 2009).
In animal and yeast cells, it is well known that an actomyosin-based ring promotes the concentric cell division that accompanies the addition of new membranes (Barr and Gruneberg, 2007, Pollard, 2010). By contrast, plant cells possess a phragmoplast, which facilitates centrifugal formation of the cell plate and plasma membrane expansion by vesicle fusion, leading to cell division (reviewed by Barr and Gruneberg, 2007, Jürgens, 2005, Smith, 2001). Although actin filaments participate in the partition process of plant cells, higher plants have lost an actomyosin-based cleavage furrow (Jürgens 2005). In spite of such distinct systems for cytokinesis, plant-like targeted membrane addition is a widely conserved mechanism and is also observed during furrow formation in animal and yeast cytokinesis (Albertson et al., 2005, Finger and White, 2002). Indeed, there are several common proteins participating in membrane trafficking among animals, yeasts, and plants. For example, the Rab-family small GTPase Sec4 is shared by yeast and animals and is involved in vesicle transport from the Golgi apparatus to the plasma membrane during cytokinesis (Barr and Gruneberg, 2007, Jantsch-Plunger and Glotzer, 1999, Walworth et al., 1989). Additionally, homologs of the t-SNARE proteins, such as Syntaxins in animals, Sso in yeast, and KNOLLE in plants, mediate selective membrane fusion events in the plane of cell division in diverse organisms (Aalto et al., 1993, Bennett et al., 1992, Lukowitz et al., 1996). While homologs of both Sec4 and SyntaxinA appear to be encoded in the genomes of the centric diatom Thalassiosira pseudonana and the pennate diatom P. tricornutum (Armbrust et al., 2004, Bowler et al., 2008), little is known about diatom membrane trafficking related to cytokinesis itself.
The marine pennate diatom Phaeodactylum tricornutum has become a major model species for the diatoms and is well suited to study cytokinesis and membrane trafficking. Unlike most diatoms it is generally only poorly silicified, which facilitates analysis by transmission electron microscopy (Borowitzka and Volcani, 1978, De Martino et al., 2007, De Martino et al., 2011, Francius et al., 2008, Lewin et al., 1958, Tesson et al., 2009a, Tesson et al., 2009b). Furthermore, a non-chemical synchronization method has been established (Huysman et al. 2010), and gene manipulation tools are well developed for this species (Daboussi et al., 2014, De Riso et al., 2009, Siaut et al., 2007). These unique advantages of P. tricornutum are powerful allies for cytological studies, even though its small cell size is less optimal for observations. Furthermore, recent studies using P. tricornutum have uncovered important aspects underpinning the regulation of diatom cell division. For instance, a light-induced diatom specific cyclin 2 has been proposed to control the onset of cell division 15 min after light illumination, and diatom cyclin-dependent kinase A2 functions as a mitotic regulator and distributes to the future dividing plane after chloroplast replication (Huysman et al., 2013, Huysman et al., 2015).
In this study we have performed an ultrastructural examination of the global morphological transitions occurring during the cell cycle in P. tricornutum, including nuclear, Golgi and mitochondria replication. We then utilized transgenic lines expressing yellow fluorescent protein (YFP) fusions to Sec4 and SyntaxinA (SytA) to observe membrane trafficking in vivo. Transformants expressing YFP fused to carbamoyl phosphate synthase III (unCPS), which is known to localize to mitochondria (Supplementary figure 2 in Allen et al. 2011), was additionally used for observations of mitochondria. Results with these organellar markers were compared with electron micrographs and with observations of actin fibers during the cell cycle using fluorescent phalloidin. Using these tools, we report here the sequential process of cytokinesis, including organelle division, actin dynamics and membrane trafficking in this diatom.
Results
Sequential Observation of the Cell Division Process
The doubling time of P. tricornutum cells is generally one to one and a half days in liquid culture. In synchronized cultures, DNA synthesis begins around 4 hours after the onset of illumination, and divided chloroplasts begin to appear shortly afterwards following constriction at their center (Huysman et al. 2010). Cell division typically occurs at the end of the day, around 12 hours after the onset of illumination.
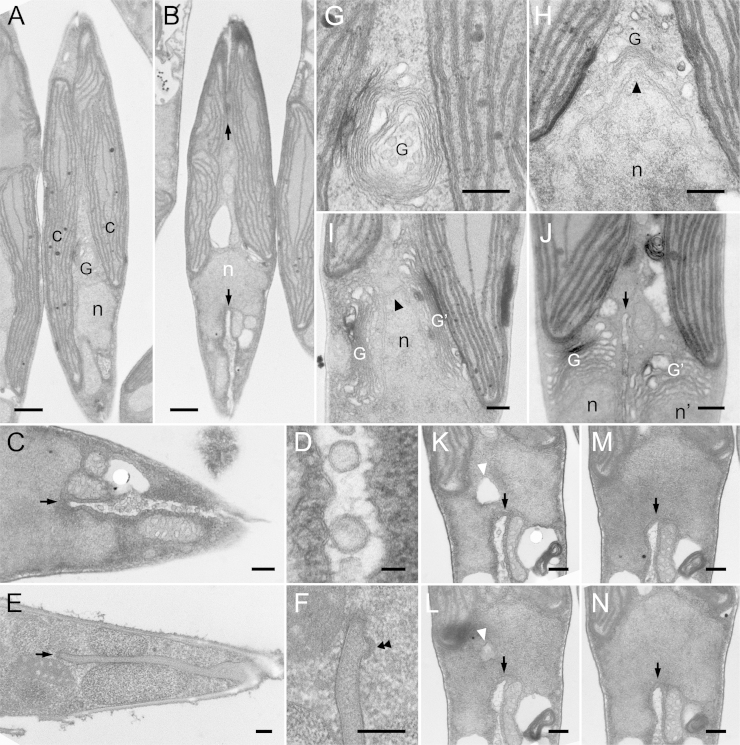
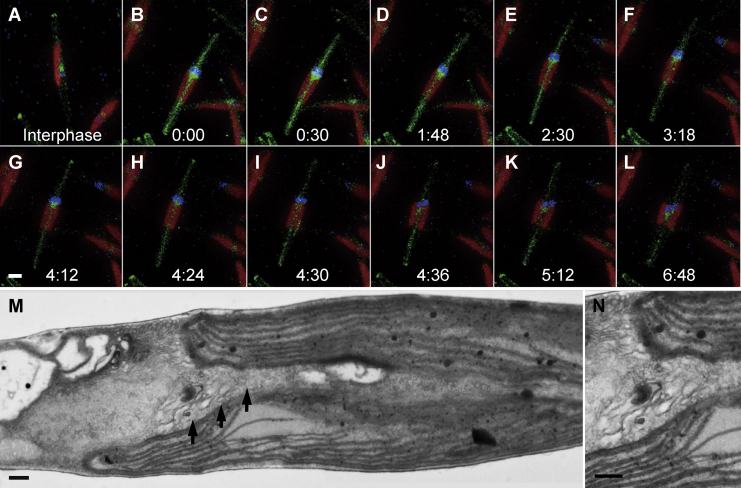
Chloroplast replication is the first major observable event of the serial cell replication process (Fig. 1A). After completion of chloroplast replication, cleavage furrows begin to be formed from both tips of the fusiform cell around 8 to 10 hours after illumination (Fig. 1B, arrows). Coalescence of cleavage furrows is completed only after organelle replication has been completed (see below). Detailed observations clarified several notable characteristics of the cleavage furrow, including numerous vesicles that were observed inside them (Fig. 1C). These vesicles have been observed previously in the pennate diatom Diatoma vulgare and were denoted cleavage vesicles (Pickett-Heaps et al. 1975). They are surrounded by double membranes whose electron density is quite similar to that of the newly formed plasma membrane (furrow membrane; Fig. 1D). However, in cryo-fixed cells, no cleavage vesicles were apparent (Fig. 1E and F). In addition, the ingrowing edge is lined with dense filaments, particularly the corners of the cleavage furrow, which are covered with radial fibrils (Fig. 1F, double arrowheads).
Figure 1.
Electron micrographs of cytokinesis in P. tricornutum. A and B. Whole cell images during cytokinesis. C-F. A part of cleavage furrows. G-J. Duplication of Golgi apparatus. K-N. Serial sections of nucleus. A. Completion of chloroplast duplication before initiation of cytokinesis. A single nucleus and Golgi apparatus are observed. B. Initial phase of cytokinesis. Arrows indicate the both edges of the cleavage furrows. Interestingly, nucleus is invaginated and seemly bisected by the cleavage furrow. C. Vesicles in the cleavage furrow. Front portion of the cleavage furrow is indicated by arrow. D. Magnified image of C. Vesicular membrane is similar to the new plasma membrane. E. Cleavage furrow in cryo-fixed cell. Front portion is indicated by arrow. F. Magnified image of the front portion of the cleavage furrow in E. Electron-dense materials are localized under the membrane of cleavage furrow (double arrowheads). G. Magnified Golgi apparatus in A. H. Curved Golgi apparatus covering the protrusion of nucleus (arrowhead). I. Duplicated Golgi apparatuses located with both sides of nuclear protrusion. J. Cleavage furrow (arrow) separating each set of nucleus and Golgi apparatus. K. A large vesicle in mid part of nucleus (white arrowhead). It seemed that the two parts of the nucleus were connected only with a narrow portion between the furrow and the large vesicle. L. The large vesicle (white arrowhead) became unclear and the connecting portion extended. M. The large vesicle completely disappeared and furrow only passes through nucleus unilaterally (arrow). N. Nucleus maintaining both protruded and penetrated portions. Spindle apparatus and microtubules were not observed in any sections. c: chloroplast, G: Golgi apparatus, n: nucleus. Scale bar: 0.5 μm (A and B), 0.2 μm (C and E-N) and 20 nm (D).
Duplication of Golgi Apparatus and Nuclear Division
Following the onset of furrow formation, the Golgi apparatus and nucleus initiate morphological changes. A part of the nucleus begins to protrude towards the center of the cell, and the Golgi bends into a heart shape, enclosing the protruding portion of the nucleus (Fig. 1H, arrowhead). The Golgi apparatus duplicates and then re-positions at both sides of the protruding nucleus (Fig. 1I). This repositioning is likely to be a preparation for equal segregation to daughter cells and, indeed, the cleavage furrow is formed inbetween the duplicated Golgi apparatus about 12 hours after illumination (Fig. 1J).
The next step, nuclear division, should be accompanied by the appearance of two MTOCs and partial nuclear envelope breakdown, as described previously in other diatoms (Manton et al., 1969, Tippit and Pickett-Heaps, 1977, Tippit et al., 1980). However, we have never been able to observe neither the mitotic apparatus nor microtubules in P. tricornutum, in spite of extensive efforts. Unlike previous observations, we observed that the nucleus, which indeed maintained a clear nuclear membrane, was penetrated and bisected by the ingrowing furrow from the opposite side with respect to the Golgi apparatus (arrows in Fig. 1B and K to N). Large vesicles were occasionally observed next to the penetrated nucleus (white arrowhead in Fig. 1K and L). Serial sections indicate that the nucleus is likely to be segmented by fusion of the ingrowing furrow and large vesicle (Fig. 1K to N). Following karyokinesis, the cleavage furrow assigns one Golgi apparatus and one nucleus to each daughter cell (Fig. 1J).
Spatial Dynamics of the Mitochondria
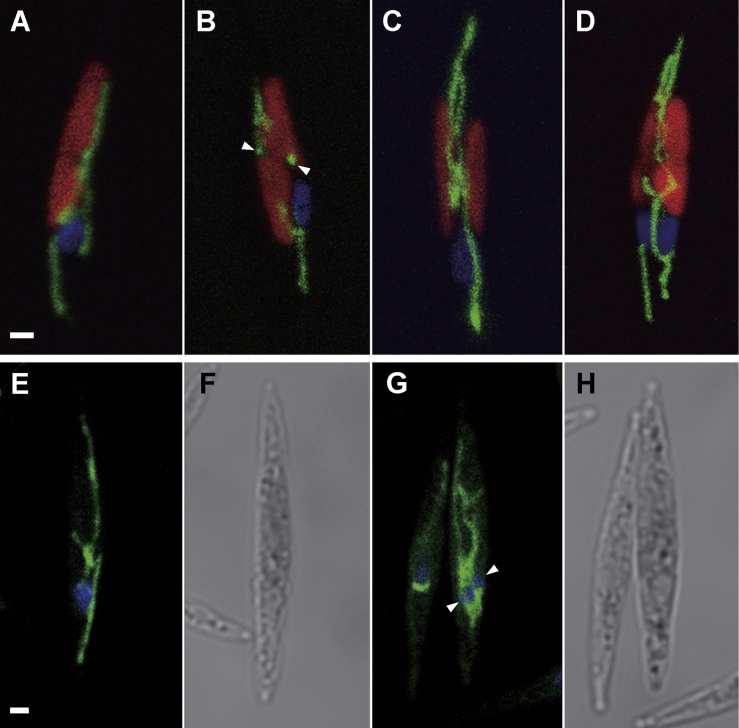
It is already known that changes in mitochondria shape play significant roles in various processes of physiology, such as calcium signaling and reactive oxygen species production (Da Silva et al. 2014). In order to grasp the whole picture of mitochondrial dynamics with respect to the nucleus during the cell cycle, we utilized a transgenic line expressing both unCPS-YFP and CFP-histone H4 (H4) fusion genes. In interphase, branches of elongated, branched mitochondria reach both extremities of fusiform cells (Fig. 2A), and are also positioned at both sides of the chloroplast constriction (arrowheads in Fig. 2B). After chloroplast duplication, elongated mitochondria were located along the future division plane (Fig. 2C), such that each daughter cell contained approximately equal portions of mitochondria (Fig. 2D). To confirm that unCPS-YFP-derived fluorescence originated from mitochondria, we stained cells with MitoTracker Orange, which indicated a strikingly similar elongated branched morphology (Fig. 2E-H).
Figure 2.
Mitochondrial dynamics during cell division. A-D. Doubly transgenic lines expressing unCPS-YFP and CFP-H4. Green, blue and red fluorescence correspond to mitochondrion, nucleus and chloroplast, respectively. E and G. Mitochondrion and nucleus stained with MitoTracker Orange and DAPI. F and H. DIC images of same cells in E and G respectively. A. Branched mitochondrion is stretched to the tip of the cell along the chloroplast. B. Two spots of mitochondrial signals beside the chloroplast constrictions (arrowheads). C. Extended mitochondria located in the future dividing plane prior to karyokinesis. D. Two identical cells with elongated mitochondria just after cell division. E. Branched mitochondrion is extended throughout the cell. DIC image of the cell is shown in F. G. Signals from mitochondria accumulate between two daughter nuclei (arrowheads). DIC image is shown in H. Scale bar: 1 μm.
Actin Dynamics During the Cell Cycle
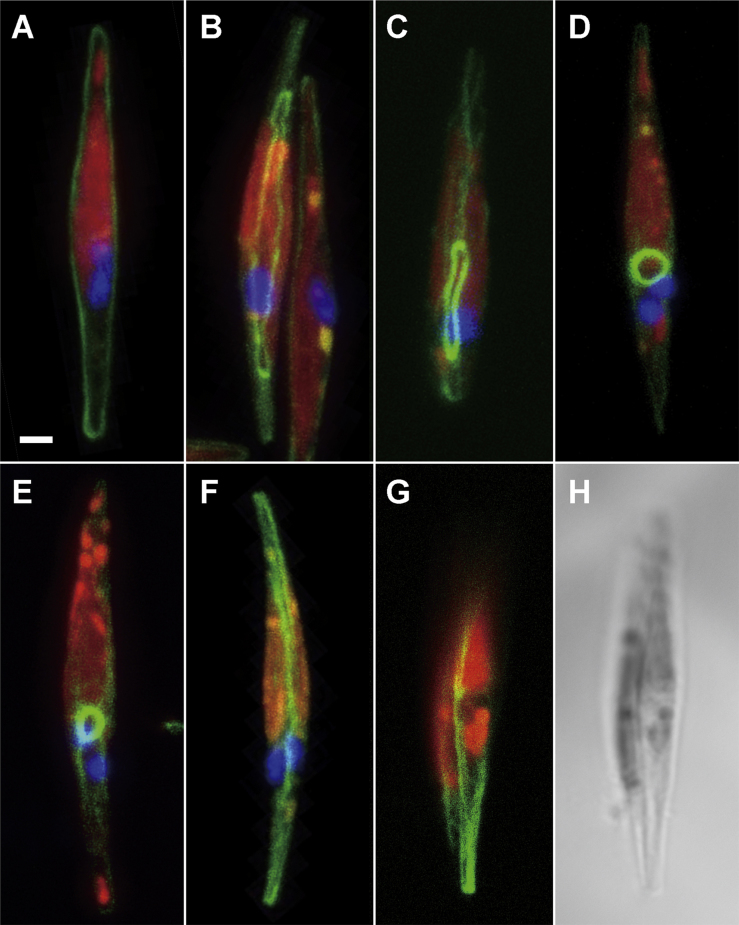
Actin is a major component of contractile rings in mammalian and yeast cytokinesis. To elucidate the contracting mechanism in diatom cytokinesis, we investigated actin distribution during the cell cycle by labelling F-actin with phalloidin. Actin bundles in the peripheral regions were always observed during interphase (Fig. 3A), and ring structures then appeared following chloroplast division (Fig. 3B). The oblique view displayed a clear contractile actin ring involving the nucleus (Fig. 3B and Supplementary Material Movie S1). Figure 3C shows the advanced phase with a smaller actin ring. During the appearance of ring structures inbetween the two daughter chloroplasts, peripheral actin remains, albeit at low concentrations. However, as the ring begins to contract four branches emerge from the ring, extending towards the peripheral regions of the cell. The fibers are primarily assembled on the future dividing plane around the actin ring (Supplementary Material Movie S2). The diameter of the contractile ring becomes smaller after karyokinesis, while some actin fibers are still maintained to cover the whole cell (Fig. 3D, E and Supplementary Material Movie S3). During the appearance of the ring structure, weak actin signals along the periphery of the cell are maintained and four actin bundles radially branched from the actin ring (Fig. 3E). Eventually, the actin contractile ring disappears, and a strong accumulation of actin fibers emerges in the cytokinetic face (Fig. 3F). Finally, cell division is completed and two daughter cells are surrounded by F-actin bundles just below the plasma membrane (Fig. 3G, H).
Figure 3.
Actin dynamics throughout the cell cycle. A-G. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively. A. Interphase cell surrounded by actin bundle under the plasma membrane. B. Actin ring appeared after the duplication of chloroplast. C. Contracted actin ring with fibrous actin in the cell. D. Smaller contractile ring after karyokinesis. The ring positioned between two daughter nuclei. E. Oblique view of the cell at a similar stage as in D. Actin fibers connected with the contractile ring. F. Final stage of cytokinesis. Actin accumulated in the cytokinetic face. G. Completion of cell division. Two daughter cells were surrounded by actin bundles. H. Bright field image in same frame with G. Cell walls for both daughter cells had been completely generated. Scale bar: 1 μm.
Distributions of Two Distinct Proteins Involved in Vesicular Transport
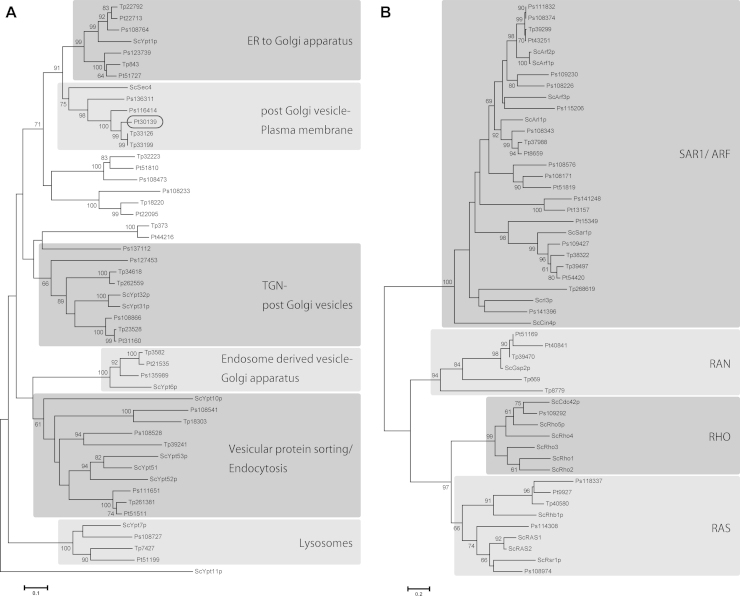
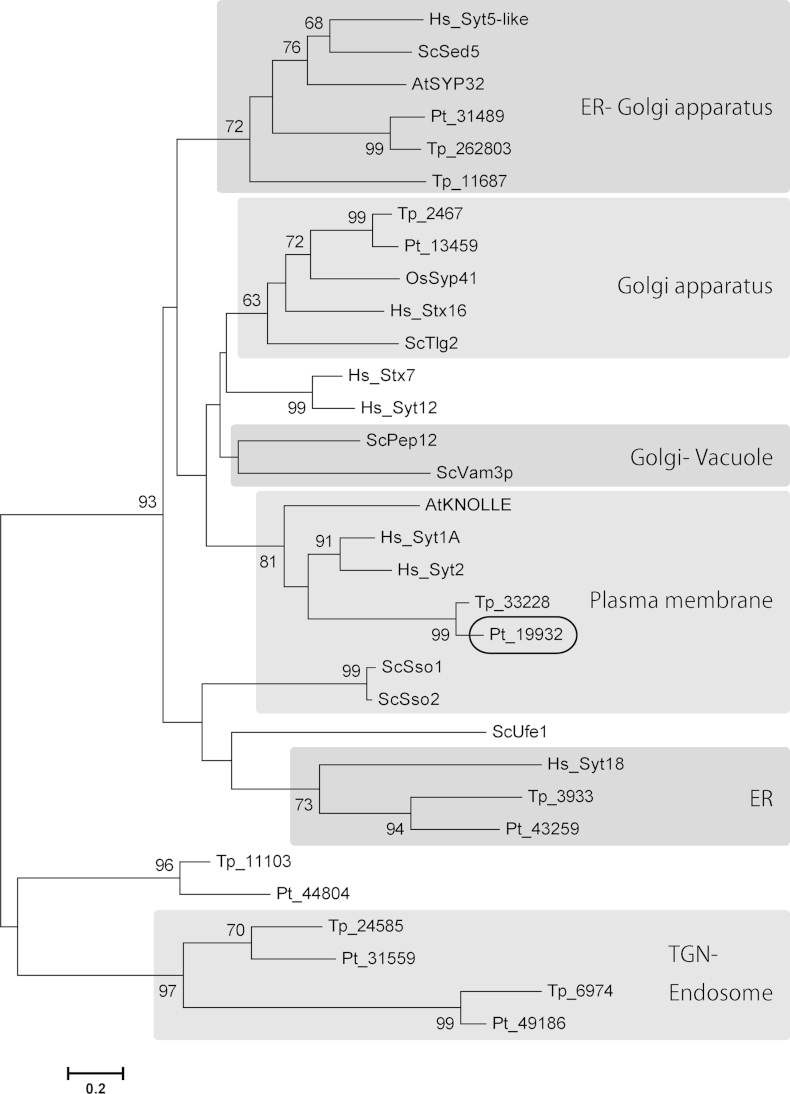
Vesicular transport is essential for cytokinesis and small GTPases and SNARE complexes play crucial roles (Albertson et al., 2005, Barr and Gruneberg, 2007, Jürgens, 2005, Smith, 2001). Small GTPases are hydrolase enzymes for guanosine triphosphate (GTP) and are often called Ras superfamily GTPases. In particular, Rab family proteins, including temperature-sensitive yeast secretion (Sec) proteins, play key regulatory roles in membrane trafficking, e.g., Sec4 is involved in the final stages of the yeast secretory pathway (Schimmöller et al., 1998, Walworth et al., 1989). Proteins of the VAMP (v-SNARE) and syntaxin (t-SNARE) families are located on transport vesicles and their targets, respectively, and the pairing of cognate of v- and t-SNAREs, is believed to provide the specificity of membrane fusion reactions (Hay and Scheller 1997). In order to reveal membrane trafficking during diatom cytokinesis, we first analyzed the diatom genomes to identify genes encoding the key proteins Sec4 and Syntaxin. The P. tricornutum genome was found to encode one copy of Sec4 (Pt30139 in Fig. 4A), whereas T. pseudonana encodes two (Tp33126 and Tp33199), all supported by high bootstrap values (Fig. 4). Additionally, all the other major clades of small GTPases contain diatom homologs with strong support (Fig. 4). On the other hand, both diatom genomes include a single homolog (Pt19932 and Tp33228 in Fig. 5) of human syntaxin 1A and Arabidopsis KNOLLE, and all of them form a monophyletic clade with high support (Fig. 5).
Figure 4.
Neighbor-joining (NJ) trees of Rab family (A) and other small GTPases (B). The 19 small GTPases from P. tricornutum were aligned with selected small GTPases from the centric diatom Thalassiosira pseudonana (Tp), the oomycete Phytophthora sojae (Ps) and, as a reference, yeast Saccharomyces cerevisiae (Sc). Detailed information about sequences used here is listed in Supplementary TableS1. Numbers at branch points indicate bootstrap percentages from NJ analyses with 1000 replicates. Only values greater than 60% are shown. ScSec4 was clustered with Pt_30139 and this monophyletic clade was supported with significant bootstrap value. Diatom T. pseudonana and oomycete P. sojae were almost always clustered together with P. tricornutum as they all belonged to Heterokonta. It is noteworthy that there are three unknown monophyletic clades, which are completely independent from the others. These could be Heterokont-specific Rab family small GTPases.
Figure 5.
Neighbor-joining (NJ) tree of Syntaxins. Each phylum forms a monophyletic clade. The 7 Syntaxins from P. tricornutum were aligned with selected Syntaxins from the centric diatom Thalassiosira pseudonana (Tp), the oomycete Phytophthora sojae (Ps) and, as references, yeast Saccharomyces cerevisiae (Sc), the plants Arabidopsis thaliana (At) and Oryza sativa (Os), and Homo sapiens (Hs). Pt_19932 was clustered with KNOLLE, Sso, Syt1 and Syt2 with significant bootstrap support. There was also one monophyletic clade consisting of only diatom sequences and this might correspond to a diatom-specific Syntaxin.
Based on these results, we generated double fluorescent marker lines; SEC4-YFP/CFP-H4 and SYTA-YFP/CFP-H4, in order to observe membrane trafficking during cytokinesis in vivo (see Methods). Time-lapse observations revealed that SEC4-YFP was constantly accumulating inbetween the chloroplasts and nucleus during the cell cycle (Fig. 6 and Supplementary Material Movie S4). Following chloroplast duplication and prior to nuclear division, the Sec4 signal spreads equally to the future dividing face, and the signal is maintained for more than six hours, up to the completion of cell division (Fig. 6A-L; karyokinesis in Fig. 6G-I). During the earliest times of observation, the signal was localized very strongly in a region between the nucleus and the divided chloroplasts (Fig. 6B-D) that corresponds to where the Golgi was observed by transmission electron microscopy (TEM; Fig. 1A, H and Fig. 6 M). The heart-shaped structure observed by both TEM and fluorescence further suggests that the Sec4 fusion protein co-localizes with the Golgi. The intensity of the fluorescence signal was gradually lost because of bleaching due to the long-term illumination performed for these experiments, but nonetheless the localization of the signals was not affected. Following the time-lapse observations, and by comparison with the TEM images, the collective information indicated that large amounts of vesicles are generated from the Golgi apparatus, and sent to the future dividing plane following chloroplast duplication and prior to nuclear division (Fig. 6 M and N).
Figure 6.
Dynamics of Sec4 distribution. A-L. Images from cells transformed with SEC4-YFP/CFP-H4 constructs. Sec4 (green) and H4 (blue) are displayed together with red chlorophyll autofluorescence. M and N. Micrographs of wild type cell. A: Interphase cell. The Sec4 core signals are presumably in the Golgi apparatus (based on similarity of morphology compared with TEM images). B-L. Time-lapse images for 6 hours and 48 minutes. Numbers in each image indicate the passage of time. Nuclear division occurred in G-I. Two daughter nuclei then started to move separately (J-L) and cell division was completed at the end. M. Ultrastructures just after the duplication of chloroplast and Golgi apparatus. Two Golgi apparatuses secreted a lot of vesicles to the future dividing plane (arrows). N. Magnified image of arrowed part of M. Scale bar: 2 μm (A-L) and 0.2 μm (M and N).
By contrast, the dynamics of SytA localization were quite different. Prior to chloroplast duplication, SYTA-YFP signals were weakly localized in the cytoplasm at the periphery of the cell (Fig. 7A). Following chloroplast division, the signals began to accumulate at both tips of the cell, and the accumulating sites elongated towards the center of the cell (Fig. 7B, C). Finally the fluorescence signal was observed as a continuous line on what is likely to be the cytokinetic face (Fig. 7D), until daughter cell separation. The similarity of SYTA-YFP localization with the extension of the cleavage furrow observed by TEM (Fig. 1) indicates that this protein may selectively localize to the extending cell membrane during cytokinesis.
Figure 7.
Localization of SyntaxinA during cell cycle. SytA signals (green) and chlorophyll autofluorescence (red) are shown here. A. SytA signals localized to cytoplasmic periphery of the cell. B. The signals began to accumulate in both tips of the cell following chloroplast duplication. C. The accumulation of SytA-YFP fluorescence extended to the center of the cell. D. The extended signals from both tips ultimately combined to make a contiguous line in the future dividing plane. Scale bar: 1 μm.
Discussion
Intensive observations throughout the cell cycle have enabled us to elucidate the intracellular dynamics of cell replication in P. tricornutum. Following chloroplast division, cleavage furrows start to be formed at both tips of the cell. This early-onset of cytokinesis precedes karyokinesis and Golgi apparatus replication, and is an interesting characteristic of P. tricornutum cell division identified in this study. The nucleus, Golgi apparatus and probably also the mitochondria are all aligned on the future dividing plane prior to their division. Accompanying the centripetal growth of the furrows, cleavage vesicles gather close to them (Fig. 1C and D). Cleavage vesicles have already been described in another pennate diatom, Diatoma vulgare, but in this case it was suggested that ramification of the plasma membrane could be distorted to result in multiple vesicles being apparent inside the cleavage furrow and that the authors were actually observing a ramification of the plasma membrane (Pickett-Heaps et al. 1975). However, in our study, cryo-fixed samples showed neither ramification of the plasma membrane nor cleavage vesicles in P. tricornutum, so cleavage vesicles may be an artefact caused by chemical fixation. It is noteworthy that cryo-fixed cells displayed dense filaments accumulating just below the neo-synthesized plasma membrane at the fronts of the furrows (Fig. 1F), an observation also reported in D. vulgare. Pickett-Heaps et al. (1975) mentioned the possibility that these filaments could be involved in cytokinetic contractile processes.
In our study, we succeeded to detect the actin ‘contractile’ ring and found that its diameter reduced as cell division progressed. This actin contractile ring could be equivalent to the filaments observed by TEM (Fig. 1F), based on the spatial relationships with organelles. While we currently have no idea about the involvement of myosin in these contractile rings, Pelham and Chang (2002) suggested that the rate of actin polymerization could influence the rate of cleavage in fission yeast. We therefore propose that P. tricornutum possesses a contractile ring composed of actin interacting with cleavage furrows, and that actin is likely to be essential for diatom cytokinesis as it is for animal and yeast cytokinesis.
We also succeeded to detect the timing of Golgi apparatus duplication during cell division. Although P. tricornutum cells normally contain a single Golgi apparatus nearby the nucleus, it duplicates into two just prior to karyokinesis (Fig. 1I, J). In animal cells, duplication of the Golgi apparatus is completed after cell division by reconstructing it from fragmented vesicles. However, in P. tricornutum, it appears that the Golgi apparatus is maintained throughout and is seemingly duplicated by medial fission prior to cytokinesis. Such medial fission of the Golgi apparatus has been reported previously in the apicomplexan Toxoplasma gondii (fig. 1 in Pelletier et al. 2002). Further observations are required to confirm whether the two processes are indeed similar, but it is interesting to note that apicomplexans and heterokonts are believed to share the same evolutionary origins (Cavalier-Smith, 1999, Moustafa et al., 2009).
In contrast to most other diatoms, P. tricornutum fusiform cells are only very poorly silicified (Francius et al. 2008). High concentrations of polysaccharides replace silica as a structural component of this novel cell wall (Francius et al., 2008, Tesson et al., 2009b, Willis et al., 2013, Willis et al., 2014). This fact implies that P. tricornutum must rely much more on secretion of polysaccharides instead of a silicified frustule for formation of the cell wall. Indeed, active membrane trafficking was observed during cell division in our study. Syntaxin is a member of the t-SNARE protein family and is known to participate in membrane fusion processes such as exocytosis and cytokinesis (Burgess et al., 1997, Jantsch-Plunger and Glotzer, 1999). Similar syntaxin distribution in ingrowing furrows has already been reported in Caenorhabditis elegans embryos (Jantsch-Plunger and Glotzer 1999), and depletion of syntaxin abolishes cellularization in Drosophila embryos (Loncar and Singer 1995). It is noteworthy that the failed cellularization in syntaxin-depleted Drosophila embryos is caused by inadequate actin recruitment (Burgess et al.1997). In addition, the plant syntaxin KNOLLE also localizes to the plane of cell division and specifically mediates cytokinetic vesicle fusion in the cell plate (Boutté et al., 2010, Lauber et al., 1997). Taken together, these observations indicate that in P. tricornutum SyntaxinA may play a similar role in vesicle fusion specialized to the cleavage furrow (i.e., for neo-synthesis of the plasma membrane). However, in spite of the similarity of P. tricornutum SyntaxinA and A. thaliana KNOLLE localization to the plane of cell division, cytokinesis proceeds differently in the two lineages, centripetally in diatoms and centrifugally in plants (De Martino et al. 2009).
On the other hand, P. tricornutum Sec4 fluorescence always accumulated between the chloroplasts and nucleus at a location very likely to be the Golgi apparatus (Fig. 1A and 6A). Since cleavage furrows never reach the middle of the cell before karyokinesis, it is clear that Sec4 distribution is irrespective of the formation of cleavage furrows, in contrast to SyntaxinA, although Sec4 becomes distributed throughout the plane of future cell division. While we could not follow the movement of fluorescence from the area corresponding to the Golgi to the future dividing plane, TEM observations indicate that the Golgi apparatus releases large amounts of vesicles to the future dividing plane (Fig. 6 M). The yeast Sec4 protein is known to locate on the cytoplasmic face of both the plasma membrane and the secretory vesicles during transit to the cell surface (Goud et al. 1988), so Sec4 in P. tricornutum may similarly be involved in post-Golgi secretory vesicle formation during cytokinesis.
In contrast to homogeneous distribution of Sec4 in the future division plane before karyokinesis (Fig. 6A-L), SyntaxinA localization spread to the division plane in accordance with development of the cleavage furrow (Fig. 7). As observed in yeast, animal and plant cells (Assaad et al., 2001, Hay and Scheller, 1997, Schimmöller et al., 1998), SyntaxinA could be present on newly-formed plasma membranes as a t-SNARE protein, and Sec4 may facilitate SNARE complex formation for vesicle docking to the future division plane also in P. tricornutum.
During karyokinesis, one or two large vesicles without any components inside appear next to the nucleus on the line of cytokinesis at the time when the furrows are in-growing (Fig. 1B, white arrowhead in Fig. 1K). These large vesicles appear to associate with the cleavage furrow coincident with the progress of karyokinesis. While our other observations suggest that diatom cytokinesis is quite similar to animal cytokinesis, the coexistence of the actin contractile ring with cleavage furrows and large vesicles on the cytokinetic plane implies that P. tricornutum may utilize both centrifugal vesicle fusion and centripetal furrow formation for cellularization. Furthermore, in contrast to plant cells, wall formation does not coincide with cellularization in diatom cells although they are covered with silica cell walls, which are formed in the SDV after the completion of cell separation.
Besides diatoms, members of the Heterokonta are known to utilize diverse cytokinetic processes. For example, brown algae have centripetal development of a plasma membrane based on an actin plate and vesicle fusion (Karyophyllis et al., 2000, Markey and Wilce, 1975, Nagasato and Moromura, 2002), oomycetes generate a dividing force using an actin-based system without a contractile ring during zoospore morphogenesis (Heath and Harold 1992), and a raphidophyte has been found to display actin contractile rings like in animal cells (Yamagishi and Kawai 2012). These observations indicate that members of the Heterokonta may have evolved a range of cytokinetic systems with identical components.
The observations reported here provide fundamental information about diatom cell replication processes. P. tricornutum is highly suited for these kinds of studies because of the absence of silica in the cell wall, which has impeded similar studies in other diatoms, as well as gene manipulation tools, synchronization protocols, and the presence of single organelles in each cell. It is to be hoped that with these tools progress in diatom cell division research will proceed faster than has been the case in previous decades.
Methods
Culture conditions and synchronization: Phaeodactylum tricornutum Bohlin (Pt1 8.6; CCMP 2561) cells were grown in f/2 medium (Guillard 1975) without silicate made from sterilized seawater. Cultures were grown at 19 °C in 12-h light/12-h dark (80-100 μmol photon m-2 s-1) with shaking at 100 rpm and maintained in exponential phase.
For synchronization of cell cycle, cultures were placed in prolonged darkness to arrest the cells in G1 (Gap 1) phase (Brzezinski et al., 1990, Huysman et al., 2010). Before starting the dark period for synchronization, cells were cultivated for two weeks in normal conditions as pre-culture.
Phylogenetic analysis: A total of 101 protein sequences (66 amino acids in length) and 32 protein sequences (60 amino acids in length) were used for small GTPase and Syntaxin analyses, respectively (Supplementary Material Table S1). All protein sequences were collected from the National Center for Biotechnology Information (NCBI) and the US Department of Energy Joint Genome Institute (JGI). Reference proteins are listed in Supplementary Material Table S1 and others were collected with protein ID used as sequence codes in the phylogenetic trees. Each data set was aligned by ClustalW enclosed in the free software Molecular Evolutionary Genetics Analysis version 5 (MEGA5, Tamura et al. 2011). Neighbor-joining trees derived by applying the Jones-Taylor-Thornton (JTT) distance matrix calculation method and bootstrap values were calculated with 1000 replications. All the steps were performed with MEGA5.
Vector cloning and biolistic transformation: Homologs of target proteins (Sec4 and SynA) were selected by BLAST protein searches in NCBI and JGI websites and phylogenetic analyses confirmed their putative functions (Figs 5 and 6). The expression vector (pEXP) for P. tricornutum was cloned via the Gateway® cloning System (Invitrogen, Carlsbad, CA USA; Siaut et al. 2007). The full length sequences of SyntaxinA (protein ID: 19932) and Sec4 (protein ID: 30139) of P. tricornutum were amplified by PCR with gene-specific primer sets (SYTA-Fw 5′-CACCATGAACAACCGTTTGG-3′ SYTA-Rv 5′-AAGAGCAATTCATGGCAAGAC-3′; SEC4-Fw 5′- CACCATGTCCCAAGCCAGCAACAG-3′ SEC4-Rv 5′- TTAGCAACACTTTTTGTCTTTGC-) and inserted into the pENTR vectors. PtGENE ENTR clones were recombined into diatom adapted pDEST-CEYFP or pDEST- NEYFP through attL× attR recombination reaction (Invitrogen) to generate the pEXP- gene-YFP vector (pEXP YFP-PtSEC4, and pEXP YFP-PtSYTA). The vector pEXP PtCPS-YFP was kindly provided by Dr. Andrew E. Allen (J. Craig Venter Institute). The pEXP CFP-PtH4 used for labeling histones in the nucleus was the original one constructed by Siaut et al. (2007).
Constructs were integrated into the genome of P. tricornutum by microparticle bombardment using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad, Marnes-la-Coquette, France) with a protocol adapted from Falciatore et al. (1999). We did simultaneous co-transformation with three vectors: pAF6 that confers resistance to phleomycin (PhlR; Falciatore et al. 1999), pEXP CFP-PtH4 (Siaut et al. 2007) to label the nucleus, and a vector containing YFP fused to the gene encoding the protein of interest. To optimize transformation efficiency, a hepta-adaptor (Bio-Rad) was used. The pEXP plasmids (3 μg or 2.2 μg for co-transformation with two or three vectors, respectively) were mixed with pAF6 plasmid (3 μg or 1.6 μg for co-transformation with two or three vectors). Transformants were selected on solid medium (1:1 f/2:agarose) containing 100 μg ml-1 phleomycin (Sigma) for two-three weeks, and individual colonies were then transferred to f/2 liquid medium for screening by epifluorescence microscopy. Growth curve analyses confirmed that none of the transformed lines were affected in their normal growth and cell division (data not shown).
Actin labeling: P. tricornutum cells were fixed with 3.7% formaldehyde in f/2 medium. During fixation, cells in the fixative were settled on the coverslips thinly coated by poly-L-lysine. After 30 min of fixation, the cells on coverslips were washed with mixed solutions, f/2 medium: marine phosphate buffer (mPBS, Van de Meene and Pickett-Heaps 2004) = 2:1, 1:1 and 1:2, and finally washed with mPBS. One unit of FITC-phalloidin (Molecular Probes, Eugene, USA) was used to label actin, and cells were incubated with it on coverslips for 30 min at room temperature. Cells were then stained with 0.5 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) for 10 min and mounted with Vectashield (Vector Laboratories, Inc., USA) after a rinse. Samples were observed using a Leica SP5 confocal microscope (Leica Microsystems, Austria).
Mitochondria labeling: Live P. tricornutum cells were stained in the f/2 culture media with 250 mM MitoTracker Orange (Invitrogen, USA) for 40 min in ordinary culture conditions. After rinsing with f/2 medium, cells were fixed with 3.7% formaldehyde in f/2 medium for 30 min at room temperature. Fixed cells were washed with f/2 medium and incubated with 5% Triton-X100 for 30 min to remove chlorophyll. Following three washes, cells were then stained with DAPI and mounted as described above and observed using a Leica SP5 confocal microscope (Leica Microsystems, Austria).
Time-lapse observations: Synchronized cells in exponential phase were harvested 6 hours after illumination, and highly concentrated cells were placed into a custom handmade chamber. For time-lapse observation, a Leica SP2AOBS confocal microscope (Leica Microsystems, Austria) was used. During the experiments, cells were illuminated by white light to maintain the light period. Chlorophyll autofluorescence was detected at 600-700 nm by excitation with a 488 nm beam. The interval of Z-axis was set with the optimized setting (approximately 0.6 μm thickness) and 10-15 images were captured along the Z-axis every 7 minutes. All frames were squashed using Z-projection at each time point using ImageJ.
Chemical fixation and transmission electron microscopy: Cells were harvested at precise time points after the synchronization and fixed with 2% glutaraldehyde in phosphate buffer (0.2 M) for 1 hour on ice. After washing with phosphate buffer, samples were fixed with 2% osmium tetroxide in phosphate buffer for 1 hour on ice as a post fixation. Dehydration was done with acetone after washing with phosphate buffer and distilled water. Samples were then embedded in Spurr's resin and cut for making pale yellow ultra-thin sections (70-80 nm thickness) with a microtome ULTRACUT (Reichert-Jung, Inc. US). Sections were counterstained with uranyl acetate and lead citrate for inspection with a Philips tecnai 12 electron microscope (FEI the Eindhoven, The Netherlands).
Cryo-fixation and freeze substitution: Cells were collected by gentle centrifugation and added to gold carrier. Cells in the gold carrier were fixed with a high pressure Leica HPM100 machine (Leica Microsystems, Austria). Afterwards, cells were transferred to automatic freeze substitution unit EM ASF2 (Leica Microsystems, Austria) with following protocols; following at least 18 hours at -90 °C in acetone, the acetone was replaced with 2% OsO4 in acetone at -90 °C for 16 hours. The temperature was gradually raised to -20 °C over 23 hours and samples were then kept at -20 °C for at least 13 hours. Temperature was then gradually raised again to 4 °C over 5 hours. Finally the cells were kept at 4 °C and embedded with graded concentrations of Spurr's resin. Following procedures were same with chemical-fixed samples described above.
Acknowledgements
The authors wish to thank Laure Wingertsmann for her technical support. Funding is acknowledged from the ERC “Diatomite” and ANR DiaDomOil projects. We additionally thank the French Government “Investissements d’Avenir” programmes MEMO LIFE (ANR-10-LABX-54) and PSL* Research University (ANR-11-IDEX-0001-02)
Monitoring Editor: Michael Melkonian
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.protis.2015.07.005.
Appendix A. Supplementary data
The following are the supplementary data to this article:
Reconstruction of whole cell just after chloroplast division. Actin ring had just appeared. Two daughter chloroplasts and a single nucleus were surrounded by the ellipse ring structure. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively.
Reconstruction of whole cell before karyokinesis. The actin ring, whose diameter became smaller, located in-between two chloroplasts. Some bundles were also maintained. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively.
Reconstruction of whole cell after karyokinesis. Smaller actin ring with two daughter nuclei. Clear four bundles of actin were generated from the ring. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively.
Reconstructed three-dimensional movie from SEC4-YFP/CFP-H4 expressing cell. Sec4 (green) and H4 (blue) are displayed together with red chlorophyll autofluorescence.
Protein list for the phylogenetic analyses. All protein sequences were collected from the National Center for Biotechnology Information (NCBI). Upper frame was used for the analysis of small GTPase and lower one was for syntaxin analysis.
References
- Aalto M.K., Ronne H., Keranen S. Yeast syntaxins Ssolp and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson R., Riggs B., Sullivan W. Membrane traffic: a driving force in cytokinesis. Trends Cell Biol. 2005;15:92–101. doi: 10.1016/j.tcb.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Allen A.E., Dupont C.L., Oborník M., Horák A., Nunes-Nesi A., McCrow J.P., Zheng H., Johnson D.A., Hu H., Fernie A.R., Bowler C. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature. 2011;473:203–207. doi: 10.1038/nature10074. [DOI] [PubMed] [Google Scholar]
- Armbrust E.V. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Assaad F.F., Huet Y., Mayer U., Jürgens G. The cytokinesis gene KEULE encodes a Sac1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., Gruneberg U. Cytokinesis: placing and making the final cut. Cell. 2007;131:847–860. doi: 10.1016/j.cell.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Bennett M.K., Calakos N., Scheller R.H. Syntaxin: a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255–259. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- Borowitzka M.A., Volcani B.E. The polymorphic diatom Phaeodactylum tricornutum: Ultrastructure of its morphotypes. J Phycol. 1978;14:10–21. [Google Scholar]
- Boutté Y., Frescatada-Rosa M., Men S., Chow C.M., Ebine K., Gustavsson A., Johansson L., Ueda T., Moore I., Jürgens G., Grebe M. Endocytosis restricts Arabidopsis KNOLLE syntaxin to the cell division plane during late cytokinesis. EMBO J. 2010;29:546–558. doi: 10.1038/emboj.2009.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Bowler C., De Martino A., Falciatore A. Diatom cell division in an environmental context. Curr Opin Plant Biol. 2010;13:623–630. doi: 10.1016/j.pbi.2010.09.014. [DOI] [PubMed] [Google Scholar]
- Brzezinski M.A., Olson R.J., Chisholm S.W. Silicon availability and cell-cycle progression in marine diatoms. Marine Ecol Prog Ser. 1990;67:83–96. [Google Scholar]
- Burgess R.W., Deitcher D.L., Schwarz T.L. The synaptic protein syntaxin1 is required for cellularization of Drosophila embryos. J Cell Biol. 1997;138:861–875. doi: 10.1083/jcb.138.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, Dinoflagellate, and Sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cohn S.A., Nash J., Picket-Heaps J.D. The effect of drugs on diatom valve morphogenesis. Protoplasma. 1989;149:130–143. [Google Scholar]
- Daboussi F., Leduc S., Maréchal A., Dubois G., Guyot V., Perez-Michaut C., Amato A., Falciatore A., Juillerat A., Beurdeley M., Voytas D.F., Cavarec L., Duchateau P. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat Commun. 2014;5:3831. doi: 10.1038/ncomms4831. [DOI] [PubMed] [Google Scholar]
- Da Silva A.F., Mariotti F.R., Máximo, Campello S. Mitochondria dynamism: of shape, transport and cell migration. Cell Mol Life Sci. 2014;71:2313–2324. doi: 10.1007/s00018-014-1557-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Martino A., Amato A., Bowler C. Mitosis in diatoms: rediscovering an old model for cell division. BioEssays. 2009;31:874–884. doi: 10.1002/bies.200900007. [DOI] [PubMed] [Google Scholar]
- De Martino A., Meichenin A., Shi J., Pan K., Bowler C. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. J Phycol. 2007;43:992–1009. [Google Scholar]
- De Martino A., Bartual A., Willis A., Meichenin A., Villazan B., Maheswari U., Bowler C. Physiological and molecular evidence that environmental changes elicit morphological interconversion in the model diatom Phaeodactylum tricornutum. Protist. 2011;162:462–481. doi: 10.1016/j.protis.2011.02.002. [DOI] [PubMed] [Google Scholar]
- De Riso V., Raniello R., Maumus F., Rogato A., Bowler C., Falciatore A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009;37:e96. doi: 10.1093/nar/gkp448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falciatore A., Casotti R., Leblanc C., Abrescia C., Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol. 1999;1:239–251. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- Finger F.P., White J.G. Fusion and fission: membrane trafficking in animal cytokinesis. Cell. 2002;108:727–730. doi: 10.1016/s0092-8674(02)00668-2. [DOI] [PubMed] [Google Scholar]
- Francius G., Tesson B., Dague E., Martin-Jézéquel V., Dufrêne Y.F. Nanostructure and nanomechanics of live Phaeodactylum tricornutum morphotypes. Environ Microbiol. 2008;10:1344–1356. doi: 10.1111/j.1462-2920.2007.01551.x. [DOI] [PubMed] [Google Scholar]
- Goud B., Salminen A., Walworth N.C., Novick P.J. A GTP-binding protein required for secretion rapidly associates with secretory vesicles and the plasma membrane in Yeast. Cell. 1988;53:753–768. doi: 10.1016/0092-8674(88)90093-1. [DOI] [PubMed] [Google Scholar]
- Guillard R.R.L. Culture of Phytoplankton for Feeding Marine Invertebrates. In: Smith W.L., Chanley M.H., editors. Culture of Marine Invertebrate Animals. Plenum Publishing Corporation; New York, NY: 1975. pp. 29–60. [Google Scholar]
- Hay J.C., Scheller R.H. SNAREs and NSF in targeted membrane fusion. Curr Opin Cell Biol. 1997;9:505–512. doi: 10.1016/s0955-0674(97)80026-9. [DOI] [PubMed] [Google Scholar]
- Heath I.B., Harold R.L. Actin has multiple roles in the formation and architecture of zoospores of the oomycetes, Saprolegnia ferax and Achlya bisexualis. J Cell Sci. 1992;102:611–627. [Google Scholar]
- Huysman M.J.J., Tanaka A., Bowler C., Vyverman W., De Veylder L. Functional characterization of the diatom cyclin-dependent kinase A2 as a mitotic regulator reveals plant-like properties in a non-green lineage. BMC Plant Biol. 2015;15:86. doi: 10.1186/s12870-015-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysman M.J.J., Martens C., Vandepoele K., Gillard J., Rayko E., Heijde M., Bowler C., Inzé D., Van de Peer Y., De Veylder L., Vyverman W. Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling. Genome Biol. 2010;11:R17. doi: 10.1186/gb-2010-11-2-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysman M.J.J., Fortunato A.E., Matthijs M., Schellenberger Costa B., Vanderhaeghen R., Van den Daele H., Sachse M., Inzé D., Bowler C., Kroth P.G., Wilhelm C., Falciatore A., Vyverman W., De Veylder L. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum) Plant Cell. 2013;25:215–228. doi: 10.1105/tpc.112.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Glotzer M. Depletion of syntaxins in the early Caenorhabditis elegans embryo reveals a role for membrane fusion events in cytokinesis. Curr Biol. 1999;9:738–745. doi: 10.1016/s0960-9822(99)80333-9. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Plant cytokinesis: fission by fusion. Trends Cell Biol. 2005;15:277–283. doi: 10.1016/j.tcb.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Karyophyllis D., Katsaros C., Galatis B. F-actin organization during the cell cycle of Sphacelaria rigidula (Phaeophyceae) Eur J Phycol. 2000;35:25–33. [Google Scholar]
- Lauber M.H., Waizenegger I., Steinmann T., Schwarz H., Mayer U., Hwang I., Lukowitz W., Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin J.C., Lewin R.A., Philpott D.E. Observations on Phaeodactylum tricornutum. J Gen Microbiol. 1958;18:418–426. doi: 10.1099/00221287-18-2-418. [DOI] [PubMed] [Google Scholar]
- Loncar D., Singer S.J. Cell membrane formation during the cellularization of the syncytial blastoderm of Drosophila. Proc Natl Acad Sci USA. 1995;92:2199–2203. doi: 10.1073/pnas.92.6.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W., Mayer U., Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Markey D.R., Wilce R.T. The ultrastructure of reproduction in the brown alga Pylaiella littoralis I. Mitosis and cytokinesis in the plurilocular gametangia. Protoplasma. 1975;85:219–241. doi: 10.1007/BF01567948. [DOI] [PubMed] [Google Scholar]
- Manton I., Kowallik K., von Stosch H.A. Observations on the fine structure and development of the spindle at mitosis and meiosis in a marine centric diatom (Lithodesmium undulatum). II. The early meiotic stages in male gametogenesis. J Cell Sci. 1969;5:271–298. doi: 10.1242/jcs.5.1.271. [DOI] [PubMed] [Google Scholar]
- Moustafa A., Beszteri B., Maier U.G., Bowler C., Valentin K., Bhattacharya D. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Nagasato C., Moromura T. Ultrastructual study on mitosis and cytokinesis in Scytosiphon lomentaria zygotes (Scytosiphonales, Phaeophyceae) by freeze-substitution. Protoplasma. 2002;219:140–149. doi: 10.1007/s007090200015. [DOI] [PubMed] [Google Scholar]
- Nelson D.M., Treguer P., Brzezinski M.A., Leynaert A., Queguiner B. Production and dissolution of biogenic silica in the ocean-revised global estimates, comparison with regional data and relationship to biogenic sedimentation. Global Biogeochem Cycl. 1995;9:359–372. [Google Scholar]
- Pelham R.J., Chang F. Actin dynamics in the contractile ring during cytokinesis in fission yeast. Nature. 2002;419:82–86. doi: 10.1038/nature00999. [DOI] [PubMed] [Google Scholar]
- Pelletier L., Stern C.A., Pypaert M., Sheff D., Ngô H.M., Roper N., He C.Y., Hu K., Toomre D., Coppens I., Roos D.S., Joiner K.A., Warren G. Golgi biogenesis in Toxoplasma gondii. Nature. 2002;418:548–552. doi: 10.1038/nature00946. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J.D. Cell division and morphogenesis of the centric diatom Chaetoceros decipiens (Bacillariophyceae) II. Electron microscopy and a new paradigm for tip growth. J Phycol. 1998;34:995–1004. [Google Scholar]
- Pickett-Heaps J.D., Tippit D.H. The diatom spindle in perspective. Cell. 1978;14:455–467. doi: 10.1016/0092-8674(78)90232-5. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., McDonald K.L., Tippit D.H. Cell division in the pennate diatom Diatoma vulgare. Protoplasma. 1975;86:205–242. doi: 10.1007/BF01275633. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J.D., Tippit D.H., Porter K.R. Rethinking mitosis. Cell. 1982;29:729–744. doi: 10.1016/0092-8674(82)90435-4. [DOI] [PubMed] [Google Scholar]
- Pollard T.D. Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen N.C., Spector I., Spurck T.P., Schults T.F., Wetherbee R. Diatom gliding is the result of an actin-myosin motility system. Cell Motil Cytoskeleton. 1999;44:23–33. doi: 10.1002/(SICI)1097-0169(199909)44:1<23::AID-CM2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Round F.E., Crawford R.M., Mann D.G. Cambridge University Press; Cambridge: 1990. The Diatoms, Biology and Morphology of the Genera. 747 p. [Google Scholar]
- Schimmöller F., Simon I., Pfeffer S.R. Rab GTPases, directors of vesicle docking. J Biol Chem. 1998;273:22161–22164. doi: 10.1074/jbc.273.35.22161. [DOI] [PubMed] [Google Scholar]
- Schmid A.M.M., Schulz D. Wall morphogenesis in diatoms: deposition of silica by cytoplasmic vesicles. Protoplasma. 1979;100:267–288. [Google Scholar]
- Siaut M., Heijde M., Mangogna M., Montsant A., Coesel S., Allen A., Manfredonia A., Falciatore A., Bowler C. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum. Gene. 2007;406:23–35. doi: 10.1016/j.gene.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Smith L.G. Plant cell division: building walls in the right places. Nat Rev. 2001;2:33–39. doi: 10.1038/35048050. [DOI] [PubMed] [Google Scholar]
- Street-Perrott F.A., Barker P.A. Biogenic silica: a neglected component of the coupled global continental biogeochemical cycles of carbon and silicon. Earth Surf Process Landforms. 2008;33:1436–1457. [Google Scholar]
- Tamura K., Peterspn D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson B., Hildebrand M. Dynamics of silica cell wall morphogenesis in the diatom Cyclotella cryptica: substructure formation and the role of microfilaments. J Struct Biol. 2010;169:62–74. doi: 10.1016/j.jsb.2009.08.013. [DOI] [PubMed] [Google Scholar]
- Tesson B., Hildebrand M. Extensive and intimate association of the cytoskeleton with forming silica in diatoms: control over patterning on the meso- and micro- scale. PloS ONE. 2010;5:e14300. doi: 10.1371/journal.pone.0014300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson B., Gaillard C., Martin-Jézéquel V. Insights into the polymorphism of the diatom Phaeodactylum tricornutum Bohlin. Bot Mar. 2009;52:104–116. [Google Scholar]
- Tesson B., Genet M.J., Fernandez V., Degand S., Rouxhet P.G., Martin-Jézéquel V. Surface chemical composition of diatoms. Chem Bio Chem. 2009;10:2011–2024. doi: 10.1002/cbic.200800811. [DOI] [PubMed] [Google Scholar]
- Tippit D.H., Pickett-Heaps J.D. Mitosis in the pennate diatom Surirella ovalis. J Cell Biol. 1977;73:705–727. doi: 10.1083/jcb.73.3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippit D.H., Pickett-Heaps J.D., Leslie R. Cell division in two large pennate diatoms Hantzschia and Nitzschia. III. A new proposal for kinetochore function during prometaphase. J Cell Biol. 1980;86:402–416. doi: 10.1083/jcb.86.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Meene A.M.L., Pickett-Heaps J.D. Valve morphogenesis in the centric diatom Proboscia alata Sundstrom. J Phycol. 2002;38:351–363. [Google Scholar]
- Van de Meene A.M.L., Pickett-Heaps J.D. Valve morphogenesis in the centric diatom Rhizosolenia setigera (Bacillariophyceae, Centrales) and its taxonomic implications. Eur J Phycol. 2004;39:93–104. [Google Scholar]
- Walworth N.C., Goud B., Kabcenell A.K., Novick P.J. Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A., Chiovitti A., Dugdale T.M., Wetherbee R. Characterization of the extracellular matrix of Phaeodactylum tricornutum (Bacillariophyceae): Structure, composition, and adhesive characteristics. J Phycol. 2013;49:937–949. doi: 10.1111/jpy.12103. [DOI] [PubMed] [Google Scholar]
- Willis A., Eason-Hubbard M., Hodson O., Maheswari U., Bowler C., Wetherbee R. Adhesion molecules from the diatom Phaeodactylum tricornutum (Bacillariophyceae): Genomic identification by amino-acid profiling and in vivo analysis. J Phycol. 2014;50:837–849. doi: 10.1111/jpy.12214. [DOI] [PubMed] [Google Scholar]
- Yamagishi T., Kawai H. Cytoskeleton organization during the cell cycle in two stramenopile microalgae, Ochromonas danica (Chrysophyceae) and Heterosigma akashiwo (Raphidophyceae), with special reference to F-actin organization and its role in cytokinesis. Protist. 2012;163:686–700. doi: 10.1016/j.protis.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Zurzolo C., Bowler C. Exploring bioinorganic pattern formation in diatoms. A story of polarized trafficking. Plant Physiol. 2001;127:1339–1345. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reconstruction of whole cell just after chloroplast division. Actin ring had just appeared. Two daughter chloroplasts and a single nucleus were surrounded by the ellipse ring structure. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively.
Reconstruction of whole cell before karyokinesis. The actin ring, whose diameter became smaller, located in-between two chloroplasts. Some bundles were also maintained. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively.
Reconstruction of whole cell after karyokinesis. Smaller actin ring with two daughter nuclei. Clear four bundles of actin were generated from the ring. F-actin, nucleus and chloroplast were visualized with FITC-phalloidin (green), DAPI (blue) and chlorophyll autofluorescence (red), respectively.
Reconstructed three-dimensional movie from SEC4-YFP/CFP-H4 expressing cell. Sec4 (green) and H4 (blue) are displayed together with red chlorophyll autofluorescence.
Protein list for the phylogenetic analyses. All protein sequences were collected from the National Center for Biotechnology Information (NCBI). Upper frame was used for the analysis of small GTPase and lower one was for syntaxin analysis.