Abstract
The oocyte-to-embryo transition entails genome activation and a dramatic reprogramming of gene expression that is required for continued development. Superimposed on genome activation and reprogramming is development of a transcriptionally repressive state at the level of chromatin structure. Inducing global histone hyperacetylation relieves this repression and histone deacetylases 1 and 2 (HDAC1 and HDAC2) are involved in establishing the repressive state. Because SIN3A is an HDAC1/2-containing complex, we investigated whether it is involved in reprogramming gene expression during the course of genome activation. We find that Sin3a mRNA is recruited during maturation and that inhibiting its recruitment not only inhibits development beyond the 2-cell stage but also compromises the fidelity of reprogramming gene expression. The SIN3A that is synthesized during oocyte maturation reaches a maximum level in the mid-1-cell embryo and is essentially absent by the mid-2-cell stage. Overexpressing SIN3A in 1-cell embryos has no obvious effect on pre- and postimplantation development. These results provide a mechanism by which reprogramming can occur using a maternally inherited transcription machinery, namely, recruitment of mRNAs encoding transcription factors and chromatin remodelers, such as SIN3A.
Keywords: gene expression, histone modifications, preimplantation embryo, reprogramming gene expression, zygotic gene activation
INTRODUCTION
Full-grown mouse oocytes are transcriptionally quiescent. Following fertilization, a low level of transcription initiates in 1-cell embryos with genomewide transcription of intra- and intergenic regions [1]. This minor wave of genome activation has minimal promoter requirements for transcription, and the transcripts are nonfunctional because they are inefficiently spliced and polyadenylated. The major wave of genome activation occurs during the 2-cell stage and entails a major change in the transcriptome [2–5] that likely is critical for further development and conversion of a highly differentiated oocyte into totipotent blastomeres.
Superimposed on genome activation is development of a chromatin-mediated transcriptionally repressive state. Evidence for this transition comes from work with plasmid-borne reporter genes as well as endogenous genes. For example, 1-cell embryos do not require an enhancer for efficient expression, whereas an enhancer is required for efficient expression in 2-cell embryos [6, 7]. The requirement for an enhancer reflects formation of a transcriptionally repressive state in 2-cell embryos. Consistent with a role of enhancers to relieve chromatin-mediated repression of transcription is the observation that increasing histone acetylation relieves this requirement. Although these conclusions were drawn using viral promoters, similar conclusions were reached using expression the endogenous Eif1a gene [8, 9].
Histone deacetylase 1 (HDAC1) is the major HDAC involved in the formation of the transcriptional repressive state that develops during the 2-cell stage because knockdown of HDAC1, but not HDAC2, in preimplantation embryos results in hyperacetylation of histone H4 and prevents the normal decrease of endogenous genes, for example, Eif1a [10]. Class I HDACs such as HDAC1 and 2 typically reside in complexes, for example, NuRD, NODE, COREST, and SIN3A [11–13]. We focused our attention on SIN3A, which is a scaffolding protein that interacts with HDAC1/2, because Sin3a is essential for mouse development; Sin3a embryonic null embryos perish around Embryonic Day 6.5 (E6.5) [14, 15]. Because Sin3a null embryos were generated from crossing Sin3a heterozygous mice, the role of maternal SIN3A in early preimplantation development, and in particular, the role in development of the chromatin-mediated transcriptionally repressive state was not known.
We report here that SIN3A is encoded by a dormant maternal mRNA that is recruited during oocyte maturation and the newly synthesized protein is essentially degraded by the early 2-cell stage in a proteasome-dependent manner. Inhibiting the maturation-associated increase in SIN3A protein using a combined small interfering RNA (siRNA)/morpholino approach markedly inhibits development beyond the 2-cell stage following in vitro fertilization (IVF). Microarray analysis reveals that a subset of normally expressed genes is expressed at higher levels in SIN3A-knockdown embryos and that another subset of genes not expressed in 2-cell embryos become expressed. Interestingly, no significant increase in global histone acetylation is observed in 1-cell embryos. Last, overexpressing SIN3A in 2-cell embryos does not inhibit the incidence of development to the blastocyst or implantation.
MATERIALS AND METHODS
Oocyte and Embryo Collection, and Embryo Culture and Transfer
Germinal vesicle (GV)-intact oocytes were collected from 6-wk-old CF-1 mice primed with eCG for 44 h before isolation as previously described [16]; 10 μM milrinone was present in the collection medium to prevent resumption of meiosis [17]. Metaphase I (MI) oocytes were collected 7 h after transferring full-grown oocytes to milrinone-free Chatot Ziomek Brinster medium [18]. Metaphase II (MII) eggs were collected from eCG-primed 6-wk-old CF-1 female mice 13–16 h following hCG administration as previously described [19]. MII eggs were also obtained following in vitro maturation (IVM) for 16–18 h after transferring full-grown oocytes to milrinone-free Chatot Ziomek Brinster medium. Mid-1-cell, late 1-cell, early 2-cell, mid-2-cell, 8-cell, and blastocyst stage embryos were collected from eCG-primed 6-wk-old CF-1 female mice mated to B6D2F1/J males (Jackson Laboratory) by flushing either the oviduct or uterus 20–21, 30–32, 36, 44, 68, and 94–96 h post-hCG as previously described [20].
Synchronization of 1- and 2-cell embryos was performed as follows. One-cell embryos generated from IVF were cultured in KSOM medium and examined for the appearance of pronuclei at 1 h intervals or for the first cleavage at 1 h intervals starting at 19 h post-IVF. Embryos that formed pronuclei or underwent the first cleavage within the previous hour were collected and cultured separately. One-cell embryos used for global histone modification analysis were fixed 6 h after pronucleus formation. Two-cell embryos used for microarray analysis were collected and frozen 12 h postcleavage. For global transcriptional analysis, 2-cell embryos were added to 2 mM 5-ethynyl uridine (EU) in KSOM medium 12 h postcleavage.
In some experiments, mid-1-cell embryos were cultured in KSOM medium [21, 22] containing either 20 μM N-(benzyloxycarbonyl)leucinylleucinylleucinal (MG132) (Sigma) or dimethyl sulfoxide (DMSO) (Sigma) at 37°C under an atmosphere of 5% O2, 5% CO2, and 90% N2. After 5 and 10 h of culture, 40 1-cell embryos were collected from the MG132 group and the DMSO control group at each of the two time points for immunoblot analysis.
To assess the effect of overexpressing SIN3A on development, embryo transfer experiments were conducted as follows. Gfp−/− virgin CF-1 females were mated to Gfp+/− males. The resulting Gfp+/− embryos were collected 18 h after fertilization, and embryos with two distinct pronuclei were microinjected at 19–21 h after fertilization with Sin3a cRNA; controls were injected with buffer. The embryos were cultured for 96 h (to E4.5) in KSOM medium as described above at which time the number of blastocysts was scored and GFP expression assessed. To determine the incidence of implantation of Sin3a cRNA-injected embryos compared with control embryos, blastocyst stage embryos were transferred to pseudopregnant female mice on Postcoital Day 3.5 using the Non-Surgical Embryo Transfer Device (Paratechs) according to the manufacturer's protocol. Each female received 8–10 embryos, half of which were injected with Sin3a (Gfp+/− or Gfp−/−) and the other half injected with (buffer) control (Gfp−/− or Gfp−/+). Thus, each female received four to five GFP-positive (or negative) Sin3a cRNA-injected embryos and four to five GFP-negative (or positive) control embryos. The females were killed 7 days after embryo transfer (E10.5), and the presence of GFP expression in the implanted embryos was assessed. All the animal experiments were approved by the University of Pennsylvania Institutional Animal Use and Care Committee and were consistent with the National Institutes of Health guidelines.
Microinjection
Full-grown GV-intact oocytes isolated from cumulus cell-enclosed oocytes (CEOs) were microinjected with 5 pl of a solution containing 5 μM siRNA and 1 mM morpholino that target Sin3a mRNA; oocytes were injected while being cultured in bicarbonate-free minimum essential medium (MEM) containing 10 mM HEPES, pH 7.2, 10 μM milrinone, and 20% fetal bovine serum [23]. The cRNA for Sin3a-T7 and GFP were injected at 580 ng/μl. The Sin3a (s73784; Ambion) and control Luciferase siRNA (D-001100-01-05; Dharmacon) were both injected at a concentration of 5 μM. The concentration of the Sin3a (5′-CCTGGTCATCCAAACGTCGCTTCAT-3′; GeneTools) and standard control morpholinos (GeneTools) was 1 mM.
IVM and IVF
For IVM and IVF, CEOs from 24-day-old B6SJLF1/J females primed with eCG 44 h before isolation were collected in MEM supplemented with 10 μM milrinone and 20% fetal bovine serum as previously described [24]. The cumulus cells were gently stripped from the oocytes using a mouth-operated pipette. The medium used for oocyte maturation was MEM supplemented with 0.23 mM pyruvate and 20% fetal bovine serum. The denuded oocytes were cultured in this medium under drops of mineral oil at 37°C in 5% O2, 5% CO2, and 90% N2 for 13−14 h in the presence of 10–15 CEOs as previously described [25].
Fertilization of eggs in vitro was performed as previously described [25]. The fertilization medium was Tyrode medium supplemented with 4 mg/ml of bovine serum albumin [26]. The MII eggs were washed in 4 drops of fertilization medium and placed into 50 μl drops overlaid with oil. The eggs were then inseminated using sperm (5 × 105 sperm/ml) capacitated for 3 h at 37°C in 5% O2, 5% CO2, and 90% N2. Sperm adhering to the eggs were removed using a fine-bore pipette, and the inseminated eggs were then transferred to and cultured in KSOM medium under oil at 37°C in 5% O2, 5% CO2, and 90% N2. MII eggs were activated by SrCl2 treatment as previously described [27].
Immunocytochemistry
Oocyte, MI, egg, or embryo samples were collected and fixed in 2.5% paraformaldehyde for 20 min at room temperature within 2 days. The samples were permeabilized for 15 min in PBS containing 0.1% Triton X-100, washed, and blocked for 30 min with PBS containing 0.1% bovine serum albumin, 0.01% Tween-20. The samples were next incubated with a rabbit anti-SIN3A antibody (MBL International) at 1:100 in blocking buffer overnight at 4°C, followed by three 15-min washes in blocking solution (PBS containing 0.1% bovine serum albumin and 0.01% Tween-20). After the washes, the samples were incubated for 1 h with the anti-rabbit cy5-conjugated secondary antibody (Jackson ImmunoResearch) diluted 1:100 in blocking solution. In some cases 1-cell embryos were first permeabilized for 15 min in PBS containing 0.1% Triton X-100, washed, and then fixed in 2.5% paraformaldehyde for 20 min at room temperature. The samples were then processed as described above.
Polyclonal antibodies against histone H3 acetylated on K18 (1:100, 39756; Active Motif), histone H4 acetylated on K5 (1:50, 06-759; Millipore), histone H4 acetylated on K8 (1:100, 06-760; Millipore), histone H4 acetylated on K12 (1:100, 06-761; Millipore), histone H4 acetylated on K16 (1:100, 06-762; Millipore) were used to assess histone modifications. Trichostatin A (TSA) (Sigma), a histone deacetylase inhibitor, was used to induce histone hyperacetylation of early 1-cell embryos by treating the cells with 50 nM TSA for 5 h. After three 15-min washes with blocking solution, the samples were incubated with 1 μM SYTOX Green (Molecular Probes) to stain DNA. The cells were then mounted under a coverslip in VECTASHIELD medium (Vector Laboratories). A Leica TCS SP laser-scanning confocal microscope captured the images and detected the fluorescence intensity of the samples. For each experiment, all the samples were processed in parallel. For SIN3A, the laser power was adjusted so that the signal intensity was below saturation for the developmental stage that showed the highest signal intensity, and all the images were then scanned at that laser power. The images were processed, and the fluorescence intensity was quantified using ImageJ software (National Institutes of Health).
Immunoblot Analysis
Protein extracts from 40 oocytes, eggs, or embryos were solubilized in Laemmli sample buffer [28], resolved by SDS-PAGE (7.5% gel), and transferred to a polyvinylidene fluoride membrane. The membrane was blocked in 2% Amersham ECL prime blocking reagent (GE Healthcare Life Sciences) for 1 h and incubated at 4°C overnight with the primary antibody in blocking solution. The membrane was then washed four times with PBS with 0.1% Tween-20, incubated with a secondary antibody conjugated with horseradish peroxidase for 1 h in blocking solution, and washed four times with PBS containing 0.1% Tween-20. The signal was detected with the Amersham ECL Select Western blot detection reagent (GE Healthcare Life Sciences) according to the manufacturer's instructions. The SIN3A primary antibody (BMP004; MBL International) was diluted 1:2000 in blocking solution. The TUBA primary antibody (T6074; Sigma) was diluted 1:5000 in blocking solution. The Amersham ECL secondary antibody (NA934V and NA931V; GE Healthcare Life Sciences) was diluted 1:75 000 in blocking solution.
RNA Extraction, RT-PCR, and Real-Time PCR
Total RNA from 20 embryos was extracted using the Arcturus PicoPure RNA Isolation Kit (Life Technologies) according to the manufacturer's instructions using added Gfp RNA as an external standard. The reverse transcription reaction using random hexamers was performed as previously described [10]. The cDNA was then quantified by quantitative real-time PCR (qRT-PCR) using the ABI Taqman Assay-on-demand probe/primer sets for Sin3a and GFP as previously described [29]. For each qRT-PCR, one embryo equivalent of cDNA was used with a minimum of three replicates as well as a minus reverse transcription and minus template control. Quantification was normalized to GFP.
DNA Replication Assay
Inseminated MII eggs were cultured in KSOM medium containing 10 μM bromodeoxyuridine (BrdU) 3 h postinsemination or soon after first cleavage. One-cell embryos were fixed 19 h postinsemination, and the 2-cell embryos were fixed 34 h postinsemination. The cells were then processed using the aforementioned immunofluorescence protocol with the addition of a denaturing step and neutralizing step after permeabilization. The samples were denatured for 30 min in 2 N HCl and neutralized for 20 min in 100 mM Tris-HCl, pH 8.5, at room temperature. The samples were incubated overnight at 4°C with the mouse monoclonal anti-BrdUTP antibody (11170376001; Roche) at 1:50 in blocking solution. The samples were incubated with the secondary fluorescein isothiocyanate anti-mouse IgG1 antibody (1144-02; Southern Biotech) at 1:100 in blocking solution for 1 h and then mounted in VECTASHIELD medium containing 2 μM TO-PRO-3 (Life Technologies).
Global Transcriptional Assay
Click-iT RNA Imaging kit (Invitrogen) was used to assay global transcription following the manufacturer's instructions. Briefly, early 2-cell embryos were cultured with 2 mM EU in KSOM medium for 1 h before fixation in 2.5% for 20 min at room temperature. To assay transcription in 1-cell embryos, the cells were incubated in KSOM medium containing 2 mM EU from the time the first pronucleus formed until 17 h postinsemination.
After washing and membrane permeabilization, incorporated EU was detected using the Click-iT detection molecule. The samples were mounted in VECTASHIELD medium containing 2 μM TO-PRO-3 to visualize the DNA. DNA and EU were visualized using a Leica TCS SP laser-scanning confocal microscopy. The intensity of the fluorescence was quantified using ImageJ software as previously described [30].
Plasmid DNA Constructs
Firefly luciferase reporter constructs under the control of the Sin3a 3′-untranslated region (3′-UTR) were generated as previously described [31]. The entire 1-kb Sin3a 3′-UTR with a poly (A) site was amplified using the forward primer 5′-GATATCTAGACTGCAGAGCCAGAGCAGGTAGC-3′ and reverse primer 5′-GCGCCGAATTCACTTATTTCCTTAAGAATCAAGCT-3′. Amplified Sin3a 3′-UTR were digested by XbaI and EcoRI and subcloned downstream of the coding sequence of the pIVT-Luc vector.
To generate a T7 C-terminal-tagged Sin3a cRNA, mouse Sin3a coding sequence was amplified from mouse cDNA clone 6837144 (Thermo Scientific Open Biosystems) by PCR using the forward primer 5′-gaggGCATGCcATGAAGaGgaGacTGGAcGACCA-3′ and the reverse primer 5′-gcgGTCGACCCGGCCACGCGTAGGGGCTTTGAATACTGTGCCGTA-3′. After enzymatic digestion by SphI and SalI-HF, the amplified Sin3a coding sequence was subcloned into the pIVT-T7 vector to make pIVT-Sin3a-T7.
In Vitro Transcription
The DNA sequence-verified pIVT-Sin3a-T7 construct was linearized by SfoI digestion. Capped cRNAs were made using in vitro transcription with T7 mMESSAGE mMachine (Ambion) according to the manufacturer's instructions. Following in vitro transcription, template plasmid DNAs were digested by adding RNase-free DNase, and the synthesized cRNA were purified by MEGAclear Kit (Ambion), precipitated, and redissolved in RNase-free water.
The DNA sequence-verified pIVT-firefly Luc/Sin3a 3′-UTR construct was linearized by EcoRI digestion. The product was then processed as described above. For both constructs, a single band of the correct size was observed following electrophoresis in an agarose gel containing 1% formaldehyde. Synthesized cRNA was aliquoted and stored at −80°C. For microinjection controls, polyadenylated Renilla luciferase cRNA was generated as previously described [31].
Luciferase Assay
Luciferase assays following injection of cRNA encoding the different reporters were performed as previously described [31].
Microarrays Analysis
Total RNA was extracted from 20 oocytes/embryos as described above and amplified with an Ovation Pico WTA system V2 (NuGEN). The material was then fragmented and labeled with the Encore Biotin Module V2 (NuGEN). Four independent biological replicates were hybridized to GeneChip Mouse 2.0 ST microarrays (Affymetrix).
The microarray datasets from the MoGene-2_ (GPL16570) platform were processed using the Oligo package [32] from the Bioconductor framework [33]. Raw data was background corrected, normalized, and summarized using the robust multiarray average procedure.
Differential expression analysis was done using a robust linear model with empirical Bayes from the limma package [34]. We compared the following conditions: α-amanitin versus control 2-cell embryo; Sin3a KD (knockdown) 2-cell embryo versus control 2-cell embryo; control 2-cell embryo versus GV oocyte; and Sin3a KD 2-cell embryo versus GV oocyte. The P-values were calculated using a moderate t-statistic. The calculated P-values were adjusted for multiple testing using the false discovery rate procedure. Genes were marked as differentially expressed if they had a minimal absolute fold-change >1.5 and the calculated P-value was <0.05. A list of all differentially expressed genes for which a gene was differentially expressed for a least one comparison group is found in Supplemental Table S1 (Supplemental Data are available online at www.biolreprod.org).
Platform annotations were downloaded from the Gene Expression Omnibus repository, and all genes that mapped to differentially expressed Affymetrix probesets (defined as false discovery rate <0.05 and absolute logarithmic fold-change >0.58, in any of the comparisons), were kept for further analysis. The 29 680 Affymetrix probesets, which could be annotated, were used for the differential expression analysis, and functional analysis of differentially expressed gene sets was done using the pantherdb database [35].
Statistical Analysis
One-way ANOVA were used to evaluate the differences between groups using GraphPad Prism 6. A level of P < 0.05 was considered to be significant.
RESULTS
Sin3a Is a Dormant Maternal mRNA That Is Recruited for Translation During Oocyte Maturation and Following Fertilization
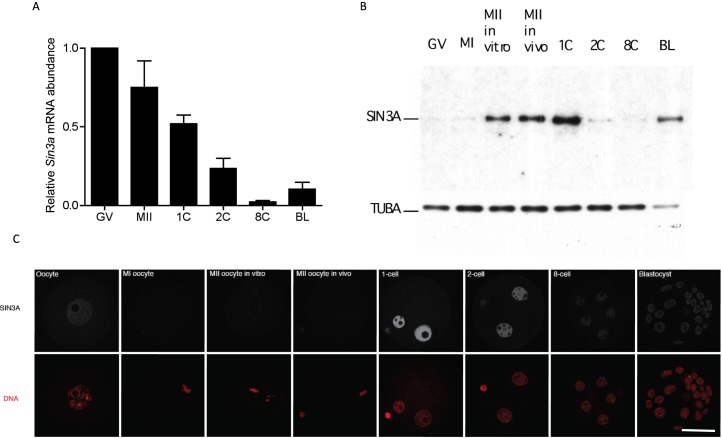
Oocyte maturation triggers degradation of many maternal mRNAs [36], and not surprisingly, the relative abundance of Sin3a mRNA decreased during maturation reaching a low point at the 8-cell stage before increasing, likely due to zygotic expression (Fig. 1A). Analysis of the relative amount of SIN3A protein by immunoblotting, however, revealed only a small amount of SIN3A protein in GV oocytes with a dramatic increase between MI and MII and a further increase following fertilization (Fig. 1B). Striking is a dramatic loss of SIN3A protein by the 2-cell stage. Consistent with Sin3a transcript abundance is the observation that the amount of SIN3A protein appeared lowest in 8-cell embryos and then increased by the blastocyst stage. Immunocytochemical detection of SIN3A protein, which was nuclear, was consistent with the immunoblotting results (Fig. 1C). The failure to detect SIN3A in MI and MII eggs is attributed to the >25-fold dilution into the cytoplasm following GV breakdown and the pronounced staining in the pronuclei due to accumulation of the newly synthesized protein. In addition, SIN3A localization in male and female pronuclei was not altered when the 1-cell embryos were permeabilized prior to fixation (Supplemental Fig. S1), suggesting that SIN3A is chromatin-associated in both pronuclei.
FIG. 1.
Developmental expression profile of Sin3a/SIN3A. A) Sin3a mRNA levels were measured by qRT-PCR at the indicated stages. Data were normalized against the detected levels of exogenously added GFP and expressed relative to the value obtained for mid-1-cell embryos. The experiment was conducted three times, and the data are expressed as mean ± SEM. GV, full-grown GV-intact oocytes; MII, metaphase II-arrested egg; 1C, 2C, and 8C refer to 1-cell, 2-cell, and 8-cell stages, respectively; BL, blastocyst. B) The relative amount of SIN3A was measured by immunoblot analysis. MII in vitro, oocytes were matured in vitro to MII; MII in vivo, MII eggs were collected following maturation in vivo. The TUBA signal was used to normalize total protein loading. The experiment was performed three times, and similar results were obtained in each case; shown is a representative example. C) Immunocytochemical analysis of SIN3A expression during preimplantation development. The experiment was conducted twice, and at least 15 oocytes/embryos were analyzed for each experiment. Bar = 50 μm.
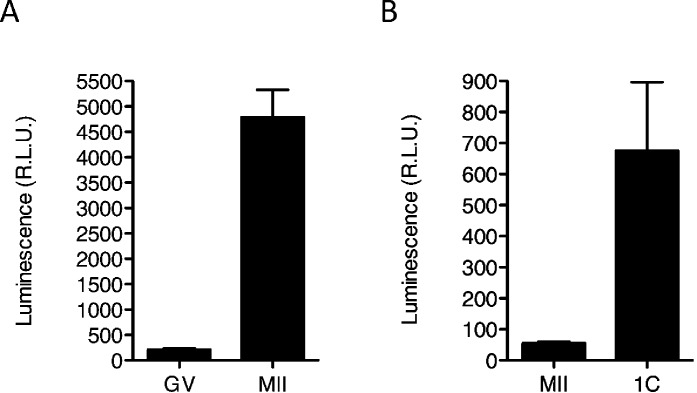
Further support that Sin3a mRNA is recruited during maturation is the observation that there was a pronounced increase in luciferase activity following maturation of GV oocytes injected with a cRNA encoding luciferase fused to the Sin3a 3′-UTR (Fig. 2A). In addition, injection of the cRNA into MII eggs that were then activated revealed an increase in luciferase activity (Fig. 2B). This increase in luciferase activity is consistent with the additional increase in the amount of SIN3A protein detected by immunoblotting between the MII egg and 1-cell stage (Fig. 1B).
FIG. 2.
Sin3a 3′-UTR contains elements that drive translational recruitment during oocyte maturation and following activation. Full-grown oocytes (A) or MII eggs (B) were microinjected with the luciferase reporter cRNAs. Firefly luciferase reporter activities were normalized to the coinjected Renilla luciferase. The data are expressed as mean ± SEM, and at least 10 oocytes/embryos were analyzed for each group.
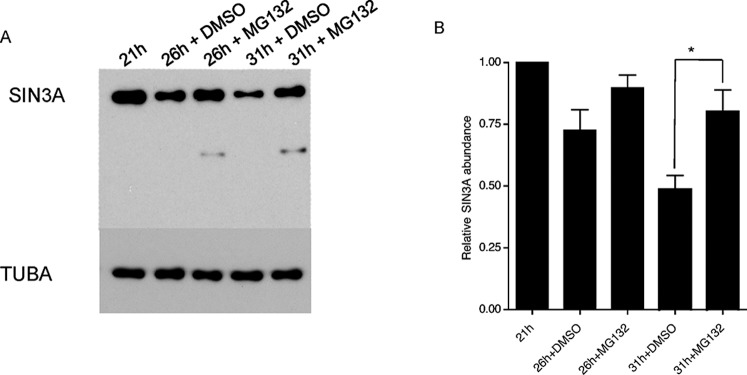
To define better the time course for degradation of newly synthesized SIN3A protein, an immunoblot analysis was conducted using 1-cell and 2-cell embryos (Fig. 3). The results of these experiments indicated that ∼65% of SIN3A is degraded by the late 1-cell stage, ∼85% by the early 2-cell stage, and it being virtually absent by the mid-2-cell stage. SIN3A degradation was presumably mediated by the proteasome because MG132, a proteasome inhibitor, substantially inhibited SIN3A degradation (Fig. 4).
FIG. 3.
Time course for SIN3A degradation. A) Immunoblot analysis for SIN3A was conducted at the indicated developmental stages. The TUBA signal was used to normalize total protein loading. The experiment was performed three times, and similar results were obtained in each case; shown is a representative example. B) Quantification of the relative amount of SIN3A shown in A. The data are expressed as mean ± SEM.
FIG. 4.
SIN3A degradation is proteasome-dependent. A) Mid-1-cell embryos were isolated and cultured in vitro in the presence of the proteasome inhibitor MG132 or DMSO, the vehicle. SIN3A abundance was measured by immunoblot analysis at the indicated times. The TUBA signal was used to normalize total protein loading. The experiment was performed three times, and a representative example is shown. B) Quantification of the data shown in A. The data are expressed as the mean ± SEM, and the SIN3A signal is relative to the mid-1-cell embryo. The times refer to the number of hours post-hCG injection. *P < 0.05.
Inhibiting the Maturation-Associated Increase in SIN3A Perturbs Histone Acetylation in 1-Cell Embryos
The results described above suggest that the function of newly synthesized SIN3A is restricted mainly to the 1-cell stage. Given this restricted window, we focused our efforts on ascertaining the effect of inhibiting the maturation-associated increase in SIN3A on histone acetylation in 1-cell embryos. We used a combined siRNA/morpholino approach that effectively inhibited the maturation-associated increase in SIN3A (Fig. 5). We analyzed 1-cell embryos produced following maturation and insemination in vitro rather than activating the in-vitro matured, microinjected oocytes because of the known differences in histone modifications in the male and female pronuclei [37].
FIG. 5.
Combined Sin3a siRNA and morpholino inhibit maturation-associated increase in the amount of SIN3A. A) Full-grown oocytes microinjected with control or Sin3a morpholino (MO) and siRNA were cultured for 1 h in medium containing 2.5 μM milrinone and then cultured in inhibitor-free medium for maturation. MII eggs were collected 16 h after maturation and used for immunoblot analysis to detect SIN3A protein levels. TUBA was used to normalize total protein loading. The experiment was performed three times, and a representative example is shown. B) Quantification of the relative amount of SIN3A shown in A. The data are expressed as mean ± SEM.
Because SIN3A is part of the SIN3A-HDAC complex, we assessed whether inhibiting the increase in SIN3A that occurs during maturation affects histone acetylation in 1-cell embryos as assessed by immunocytochemistry (Fig. 6). We analyzed H3K18ac, H4K5ac, H4K8ac, H4K12ac, and H4K16ac because these marks are affected when HDAC function is lost (e.g., [10]) and because these acetylated histones are located within three distinct regions of genes; H3K18ac is mainly located in the region surrounding the transcriptional start site, whereas the others are enriched in the promoter and transcribed regions of active genes [38]. Although TSA, an HDAC inhibitor, results in a modest hyperacetylation of all of these marks, surprisingly no hyperacetylation was observed when the maturation-associated increase in SIN3A was inhibited, and if anything, hypoacetylation was observed, for example, H3K18ac, H4K8ac, and H4K12ac.
FIG. 6.
Effect of inhibiting the maturation-associated increase in SIN3A on histone acetylation in 1-cell embryos. A) The indicated histone were assayed by immunocytochemistry for their acetylation status using maternal SIN3A-depleted, control 1-cell embryos (Ctrl), and 1-cell embryos that were incubated with TSA (an HDAC inhibitor) to generate the maximum increase in histone acetylation. The experiment was performed twice, and at least a total of 10 embryos were analyzed for each sample. Bar = 50 μm. B) Quantification of the data shown in A. The data are expressed relative to the control 1-cell embryos and are expressed as mean ± SEM. *P < 0.05.
Inhibiting the Maturation-Associated Increase in SIN3A Inhibits Development Beyond the 2-Cell Stage
We next assessed the effect of inhibiting the maturation-associated increase in SIN3A on preimplantation development and found that whereas control-injected embryos developed beyond the 2-cell stage, development arrested at the 2-cell stage for the experimental group (Fig. 7). The low incidence of development in the control group likely reflects the deleterious effects on development when denuded oocytes, which have been microinjected, are then matured and inseminated in vitro. It is unlikely that arrest at the 1-cell and 2-cell stages was due to failure to replicate DNA because a similar fraction of both experimental and control embryos displayed BrdU incorporation with comparable signal intensity in both groups (Supplemental Fig. S2).
FIG. 7.
Inhibiting the maturation-associated increase in SIN3A leads to arrest at the 2-cell stage. After inhibiting the maturation-associated increase in SIN3A, the maternal SIN3A-depleted and control MII eggs were in vitro fertilized (IVF) and embryo development was assessed at 24, 48, and 72 h after fertilization; hpf, hours postfertilization. The experiment was performed three times and the data pooled. A total of 56 experimental and 43 control embryos were analyzed.
Inhibiting zygotic genome activation with α-amanitin leads to developmental arrest at the 2-cell stage in mice [39]. Transcription, as assessed by EU incorporation, was reduced in 2-cell embryos derived from eggs in which the maturation-associated increase in SIN3A was inhibited (Fig. 8) relative to controls. The ∼50% decrease, however, was not sufficient to cause arrest at the 2-cell stage because transcription must be reduced by at least 70% to result in such an arrest [40]. Inhibiting the maturation-associated increase in SIN3A had no effect on transcription in either the male or female pronucleus of 1-cell embryos (Supplemental Fig. S3). Although transcription in 1-cell embryos is not required for cleavage to the 2-cell stage, recent results have now shown that it is required for development beyond the 2-cell stage [1].
FIG. 8.
Transcription is reduced in 2-cell embryo when maternal SIN3A is depleted. A) The experiment was performed four times and shown are representative images. At least 10 embryos were analyzed for each treatment group. Bar = 25 μm. B) Quantification of the images shown in A. The data are expressed as mean ± SEM. *P < 0.05.
Reprogramming of Gene Expression in SIN3A-Depleted Embryos
A major reprogramming of gene expression occurs during the mid- to late 2-cell stage in mouse [2–5], and inappropriate reprogramming is associated with 2-cell arrest (e.g., [41]). The observed 2-cell arrest when the maturation-associated increase in SIN3A was inhibited suggested that reprogramming of gene expression was compromised. Accordingly, we conducted microarray analyses with the expectation that a subset of zygotically expressed genes, that is, genes whose expression is inhibited by α-amanitin, would be perturbed.
Hierarchical cluster analysis revealed that as anticipated the transcriptome of GV oocytes and α-amanitin-treated 2-cell embryos was similar and differed from the transcriptome of control and experimental 2-cell embryos (Fig. 9). Furthermore, finding that the transcriptomes of the control and α-amanitin-treated 2-cell embryos was consistent with perturbed expression of a subset of zygotically expressed genes.
FIG. 9.
Heat map of all samples from different treatment groups constructed using hierarchical clustering. Replicate sample numbers are indicated at the bottom of the figure. GV, full-grown GV-intact oocytes; a-am, 1-cell embryos incubated in α-amanitin to the 2-cell stage; 2C, 2-cell embryos derived from oocytes injected with control siRNA and morpholino matured and fertilized in vitro; KD, 2-cell embryos derived from oocytes injected with Sin3a siRNA and morpholino matured and fertilized in vitro. Colors correspond to relative RNA abundance (on the log2 scale) for the detected genes each of which is represented by one horizontal bar. The numbers correspond to each replicate within each group.
A chromatin-based transcriptionally repressive state develops during the 2-cell stage, and this repression is relieved by histone hyperacetylation. Accordingly, we first filtered the dataset for α-amanitin-sensitive genes whose expression was higher in SIN3A-depleted embryos (Supplemental Table S2); the criterion used was that expression in control 2-cell embryos was at least 1.5-fold higher than in GV oocytes. Expression of 145 zygotically expressed genes was higher in 2-cell embryos when the maturation-associated increase in SIN3A was inhibited. These genes included not only protein-coding genes but also ribosomal genes, small nuclear RNAs, and noncoding RNAs.
We also asked whether a subset of genes that are not zygotically expressed in 2-cell embryo were now expressed in SIN3A-depleted embryos (Supplemental Table S2). We identified 98 genes that displayed no difference in expression between α-amanitin-treated and control 2-cell embryos but whose expression was increased in SIN3A-depleted embryos when compared to control 2-cell embryos. Similar to the situation for zygotically expressed genes, these genes included protein coding genes, ribosomal genes, small nuclear RNAs, and noncoding RNAs but also small nucleolar RNAs, and microRNAs. We also selected four genes from each class to confirm by qRT-PCR that their expression was increased in the SIN3A-depleted 2-cell embryos and found this to be the case for seven of them (Supplemental Table S3).
Mapping the genes whose expression was increased when the maturation-associated increase in SIN3A was inhibited to chromosomes revealed that although there are observable clusters, these clusters fall within gene clusters on the chromosomes, that is, gene-dense regions (Fig. 10). Thus, their positional enrichment is not unexpected. Nevertheless, some chromosomes were enriched for these genes (e.g., chromosome 12), whereas others were not (e.g., chromosome 1) (Supplemental Table S4).
FIG. 10.

Karyogram of genes up-regulated in 2-cell embryos depleted of maternal SIN3A (left panel) and density of genes (right panel). The comparison showed that most of the SIN3A sensitive genes emanated from gene dense regions.
Exogenously Expressing SIN3A Beyond the 1-Cell Stage Does Not Impair Preimplantation Development
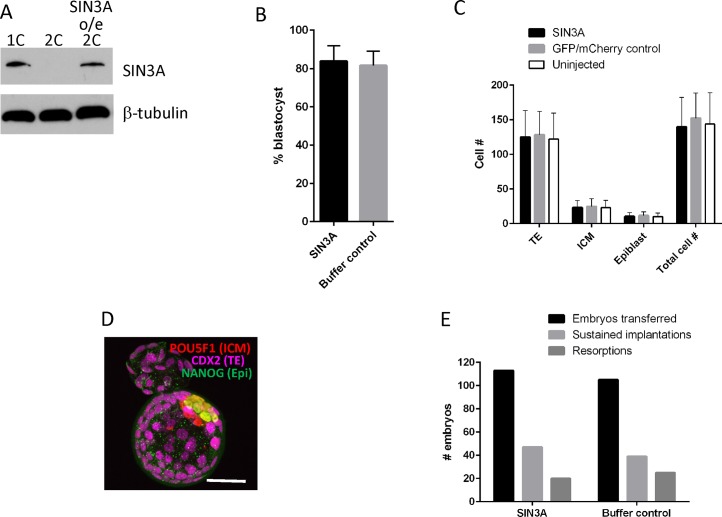
The highly restricted window for the presence of SIN3A—it accumulates during maturation and then is essentially absent by the early 2-cell stage—contrasts to other proteins encoded by dormant maternal mRNAs that remain present for much longer periods of time during preimplantation development, (see, e.g., [31]). This transient pattern of expression suggests that SIN3A function could be restricted to this short window and that the decrease in the amount of SIN3A is required for development beyond the 2-cell stage. To test whether such is the case, we identified conditions in which we increased the amount of SIN3A in 2-cell embryos to an amount comparable to that observed following oocyte maturation by injecting 1-cell embryos with a cRNA encoding Sin3a (Fig. 11A).
FIG. 11.
Overexpressing SIN3A does not affect pre- and postimplantation development. A) Immunoblot analysis of SIN3A demonstrating that SIN3A is expressed at a similar level in 2-cell embryos (SIN3A o/e 2C) when compared to 1-cell embryos (1C). As shown in Figure 1, SIN3A is essentially absent in control 2-cell embryos (2C). B) Effect of overexpressing SIN3A on development to the blastocyst stage. Control embryos were injected with buffer. The experiment was conducted seven times, and at least 261 embryos were analyzed in each group. The data are expressed as mean ± SEM. C) Effect of overexpressing SIN3A on cell numbers in blastocysts. The experiment was conducted four times, and at least 32 embryos were analyzed in each group. The data are expressed as mean ± SEM. D) Immunocytochemical detection of lineage markers in blastocysts derived from embryos overexpressing SIN3A. Bar = 50 μm. E) Incidence of postimplantation development following blastocyst transfer of control and SIN3A-overexpressing embryos. The experiment was conducted six times.
The control group for these experiments was buffer-injected 1-cell embryos. The rationale for using buffer-injected as a control rather than injecting a cRNA that encodes for some protein, which is the typical control, is that the encoded protein is typically stable, whereas SIN3A is not. In addition, we noted that development to the blastocyst stage depended on what control cRNA was injected (Supplemental Fig. S4). The results of these experiments demonstrated that maintaining elevated levels of SIN3A for several hours beyond that time when it has normally decreased to undetectable levels had no effect on the incidence of development to the blastocyst stage and total cell numbers in blastocysts and their types (trophectoderm, inner cell mass, epiblast) (Fig. 11B–D). In addition, embryo transfer experiments did not detect any differences in the incidence of implantation and resorption (Fig. 11E). Thus, maintaining elevated amounts of SIN3A, albeit for only a limited duration (Supplemental Fig. S5), has little, if any, effect on development.
DISCUSSION
The results described here suggest that inhibiting recruitment of maternal Sin3a mRNA during oocyte maturation relieves repression of a subset of genes during formation of the global transcriptionally repressive that develops during the course of genome activation. This finding is consistent with the observation that SIN3A is one of several HDAC-containing complexes [11–13] and inducing genomewide hyperacetylation relieves repression of the chromatin-based transcriptionally repressive state that starts to develop during the 2-cell stage [8, 42, 43]. From this perspective, recruitment of maternal Sin3a mRNA during oocyte maturation contributes to the first major wave of reprogramming of gene expression during preimplantation development in mouse. The molecular basis for how reprogramming occurs using a maternally inherited transcription machinery remains poorly understood. Although posttranslational modifications of existing components of the transcription machinery present in oocytes could contribute to reprograming, recruitment of dormant maternal mRNAs encoding chromatin remodelers, such as SIN3A, and transcription factors provides an elegant and effective solution.
Examination of microarray data and the nucleotide sequence of Sin3a mRNA is consistent with Sin3a being a dormant maternal mRNA that is recruited during maturation and following fertilization. Microarray data [5] reveal an increase in the relative abundance of Sin3a mRNA between the GV oocyte and 1-cell stages that likely reflects elongation of the poly (A) tail during maturation that leads to more efficient oligo (dT) priming when generating the cDNA libraries. Examination of the 3′-UTR of Sin3a mRNA reveals a well-defined cytoplasmic polyadenylation element (U5AU) [44] within ∼200 nucleotides of the polyadenylation signal sequence. In addition, a poly (U13) tract directly preceding the polyadenylation signal sequence could also contribute to the increase in SIN3A observed following fertilization [45]. Microarray data also indicate that Sin3b transcript is present in oocytes. Given that this transcript does not exhibit properties that suggest it a dormant maternal mRNA that is recruited during maturation, for example, no apparent cytoplasmic polyadenylation element, there is every expectation that SIN3B protein is present in oocytes. Nevertheless, such maternal SIN3B is not sufficient to compensate for loss of the maturation-associated increase in SIN3A that is essential for preimplantation development.
A striking observation is that the increase in SIN3A protein is very transient, essentially restricting its function to the 1-cell stage. The presence of a significant amount of Sin3a mRNA in 2-cell embryos (when compared to oocytes) makes it unlikely that the decrease in the amount of Sin3a mRNA that occurs between the MII and 2-cells is responsible for the virtual absence of SIN3A protein in 2-cell embryos. Rather, the newly synthesized SIN3A appears to be intrinsically unstable and subject to proteasome-dependent degradation.
The transient nature of SIN3A expression led us to focus our efforts on the 1-cell stage. We used MII eggs fertilized in vitro, which is technically more demanding than using parthogenetically activated MII eggs, because of the known differences in modified histone composition between the male and female pronuclei [37] and differences in nuclear concentration of specific chromatin-associated proteins, for example, PG7/STELLA [46]. We did not observe any difference in the pronuclear concentration of SIN3A, regardless of whether the 1-cell embryos were permeabilized before fixation or not. In addition, we did not observe any pronuclear differences in the extent of acetylation of the modified histones that were examined.
Inhibiting the maturation-associated increase in SIN3A led to histone hypoacetylation, not hyperacetylation. It should be noted that nuclear localization of HDAC1 and HDAC2 is not altered in Sin3a−/− oocytes (Ma and Schultz, unpublished results). A possible explanation for this observation is that whereas deletion of HDAC1/2 leads to histone hyperacetylation [10, 47], HDAC1/2 would still be present and could function in an unregulated fashion no longer targeting specific nucleosomes. Alternatively, the HDACs would be available to associate with other HDAC1/2-containing complexes, and should the activity of these complexes be greater than that of SIN3A, this difference in activity could lead to the observed histone hypoacetylation. Although transcription in 1-cell embryos is not affected when the maturation-associated increase in SIN3A is inhibited, the observed hypoacetylation in 1-cell embryos could contribute to the decrease in global transcription in 2-cell embryos. For example, H4K8ac is linked with transcriptional activation and enriched in the promoter and transcribed regions of active genes [38], and H3K18ac may be involved in POL2R recruitment and transcription initiation [48].
Inhibiting the maturation-associated increase in SIN3A results in few embryos developing beyond the 2-cell stage. Such an arrest is the hallmark of failure to activate the genome, in particular, a failure to reprogram correctly. For example, deleting maternal Brg1 results in arrest at the 2-cell stage and a failure to reprogram faithfully, and expressing BRG1 in Brg1−/− zygotes overcomes the 2-cell arrest [41]. The combination of misexpression of a subset of genes normally activated during the course of genome activation and reprogramming and genes that are not normally expressed in 2-cell embryos is a likely source for the observed developmental arrest. Consistent with SIN3A being one of several HDAC-containing complexes is the observation that only a subset of genes normally activated during the course of genome activation are affected.
Analysis of the misexpressed genes was essentially uninformative, but one interesting correlation emerged when using the molecular signatures database (MSigDB.v5) [49], namely, about 40 of the genes up-regulated when the maturation-associated increase in SIN3A was inhibited 9% to ∼20% are genes marked by H3K27me3, EED, or SUZ12 in embryonic stem cells. Further examination of these genes indicates they are marked with a bivalent promoter, that is, promoters enriched both in H3K27me3 and H3K4me3 [50–52]. Any mechanistic linkage between reduced amounts of SIN3A and expression of these genes remains unknown, and the small amounts of biological material essentially preclude chromatin immunoprecipitation experiments to determine whether the promoters of these genes bear these histone marks.
We were unable to maintain elevated amounts of SIN3A following injecting a Sin3a cRNA into 1-cell embryos and therefore not able to assess whether the rapid turnover of SIN3A is essential for continued development. Identifying amino sequences responsible for rapid SIN3A degradation may permit such an analysis. These overexpression experiments, however, highlight the need to be careful about what constitutes the control in the experiment because certain cRNAs led to decreased development (e.g., mCherry cRNA) whereas others do not (e.g., Luc cRNA).
In summary, the results presented here document a critical role of recruitment of maternal Sin3a mRNA during maturation in reprogramming gene expression during the course of genome activation. Although the precise mechanism as to why expression of certain genes is affected whereas others are unaffected is not known, this strategy of recruiting dormant maternal mRNAs that encode proteins central to a process that should be minimal in the oocyte but then required by the 1-cell stage likely provided a selective pressure for this sophisticated mechanism of regulation of gene expression to evolve in oocytes.
ACKNOWLEDGMENT
The authors thank Paula Stein for performing the experiments described in Supplemental Table S3.
Footnotes
Current address: Embrapa Recursos Geneticos e Biotecnologia, Cenargen PBI sala 07B, PqEB Final W5, Asa Norte, Brasilia DF, Brazil 70770-901.
Current address: JH TECHNOLOGIES, INC., 225 Hammond Avenue, Fremont, CA 94539.
Supported by grants from NIH (HD022681 to R.M.S. and HD-000849 to M.M.). O.D. was supported by 5TL1CA133837 and R.J. by T32 GM008216. V.F. was supported through the European Commission Seventh Framework Program (Integra-Life; Grant 315997), and Croatian Ministry of Science, Education and Sports Grant 119-0982913-1211 awarded to Kristian Vlahovicek.
REFERENCES
- Abe K, Yamamoto R, Franke V, Cao M, Suzuki Y, Suzuki MG, Vlahovicek K, Svoboda P, Schultz RM, Aoki F. The first murine zygotic transcription is promiscuous and uncoupled from splicing and 3′ processing. EMBO J. 2015;34:1523–1537. doi: 10.15252/embj.201490648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Park SJ, Komata M, Inoue F, Yamada K, Nakai K, Ohsugi M, Shirahige K. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 2013;27:2736–2748. doi: 10.1101/gad.227926.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QT, Piotrowska K, Ciemerych MA, Milenkovic L, Scott MP, Davis RW, Zernicka-Goetz M. A genome-wide study of gene activity reveals developmental signaling pathways in the preimplantation mouse embryo. Dev Cell. 2004;6:133–144. doi: 10.1016/s1534-5807(03)00404-0. [DOI] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283:40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
- Majumder S, DePamphilis ML. A unique role for enhancers is revealed during early mouse development. Bioessays. 1995;17:879–889. doi: 10.1002/bies.950171010. [DOI] [PubMed] [Google Scholar]
- Nothias JY, Majumder S, Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development. J Biol Chem. 1995;270:22077–22080. doi: 10.1074/jbc.270.38.22077. [DOI] [PubMed] [Google Scholar]
- Davis W, Jr, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol. 1996;174:190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- Davis W, Jr, Schultz RM. Role of the first round of DNA replication in reprogramming gene expression in the preimplantation mouse embryo. Mol Reprod Dev. 1997;47:430–434. doi: 10.1002/(SICI)1098-2795(199708)47:4<430::AID-MRD9>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110–120. doi: 10.1016/j.ydbio.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int J Dev Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- Kelly RD, Cowley SM. The physiological roles of histone deacetylase (HDAC) 1 and 2: complex co-stars with multiple leading parts. Biochem Soc Trans. 2013;41:741–749. doi: 10.1042/BST20130010. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley SM, Iritani BM, Mendrysa SM, Xu T, Cheng PF, Yada J, Liggitt HD, Eisenman RN. The mSin3A chromatin-modifying complex is essential for embryogenesis and T-cell development. Mol Cell Biol. 2005;25:6990–7004. doi: 10.1128/MCB.25.16.6990-7004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, David G, Zhong S, van der Torre J, Wong WH. Depinho RA. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005;19:1581–1595. doi: 10.1101/gad.1286905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz RM, Montgomery RR, Belanoff JR. Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev Biol. 1983;97:264–273. doi: 10.1016/0012-1606(83)90085-4. [DOI] [PubMed] [Google Scholar]
- Tsafriri A, Chun SY, Zhang R, Hsueh AJ, Conti M. Oocyte maturation involves compartmentalization and opposing changes of cAMP levels in follicular somatic and germ cells: studies using selective phosphodiesterase inhibitors. Dev Biol. 1996;178:393–402. doi: 10.1006/dbio.1996.0226. [DOI] [PubMed] [Google Scholar]
- Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- Endo Y, Schultz RM, Kopf GS. Effects of phorbol esters and a diacylglycerol on mouse eggs: inhibition of fertilization and modification of the zona pellucida. Dev Biol. 1987;119:199–209. doi: 10.1016/0012-1606(87)90221-1. [DOI] [PubMed] [Google Scholar]
- Manejwala F, Kaji E, Schultz RM. Development of activatable adenylate cyclase in the preimplantation mouse embryo and a role for cyclic AMP in blastocoel formation. Cell. 1986;46:95–103. doi: 10.1016/0092-8674(86)90863-9. [DOI] [PubMed] [Google Scholar]
- Erbach GT, Lawitts JA, Papaioannou VE, Biggers JD. Differential growth of the mouse preimplantation embryo in chemically defined media. Biol Reprod. 1994;50:1027–1033. doi: 10.1095/biolreprod50.5.1027. [DOI] [PubMed] [Google Scholar]
- Ho Y, Wigglesworth K, Eppig JJ, Schultz RM. Preimplantation development of mouse embryos in KSOM: augmentation by amino acids and analysis of gene expression. Mol Reprod Dev. 1995;41:232–238. doi: 10.1002/mrd.1080410214. [DOI] [PubMed] [Google Scholar]
- Kurasawa S, Schultz RM, Kopf GS. Egg-induced modifications of the zona pellucida of mouse eggs: effects of microinjected inositol 1,4,5-trisphosphate. Dev Biol. 1989;133:295–304. doi: 10.1016/0012-1606(89)90320-5. [DOI] [PubMed] [Google Scholar]
- Downs SM, Coleman DL, Eppig JJ. Maintenance of murine oocyte meiotic arrest: uptake and metabolism of hypoxanthine and adenosine by cumulus cell-enclosed and denuded oocytes. Dev Biol. 1986;117:174–183. doi: 10.1016/0012-1606(86)90359-3. [DOI] [PubMed] [Google Scholar]
- Schroeder AC, Eppig JJ. The developmental capacity of mouse oocytes that matured spontaneously in vitro is normal. Dev Biol. 1984;102:493–497. doi: 10.1016/0012-1606(84)90215-x. [DOI] [PubMed] [Google Scholar]
- Tateno H, Kamiguchi Y. Evaluation of chromosomal risk following intracytoplasmic sperm injection in the mouse. Biol Reprod. 2007;77:336–342. doi: 10.1095/biolreprod.106.057778. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Wakayama T. Efficient strontium-induced activation of mouse oocytes in standard culture media by chelating calcium. J Reprod Dev. 2007;53:1207–1215. doi: 10.1262/jrd.19067. [DOI] [PubMed] [Google Scholar]
- Laemmli UK, Quittner SF. Maturation of the head of bacteriophage T4. IV. The proteins of the core of the tubular polyheads and in vitro cleavage of the head proteins. Virology. 1974;62:483–499. doi: 10.1016/0042-6822(74)90409-7. [DOI] [PubMed] [Google Scholar]
- Zeng F, Baldwin DA, Schultz R. Transcript profiling during preimplantation mouse development. Dev Biol. 2004;272:483–496. doi: 10.1016/j.ydbio.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Ma J, Flemr M, Strnad H, Svoboda P, Schultz RM. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod. 2013;88:11. doi: 10.1095/biolreprod.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YQ, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol. 2007;302:104–117. doi: 10.1016/j.ydbio.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaujean N. Histone post-translational modifications in preimplantation mouse embryos and their role in nuclear architecture. Mol Reprod Dev. 2014;81:100–112. doi: 10.1002/mrd.22268. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flach G, Johnson MH, Braude PR, Taylor RA, Bolton VN. The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1982;1:681–686. doi: 10.1002/j.1460-2075.1982.tb01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Schultz RM. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol Reprod. 2011;85:409–416. doi: 10.1095/biolreprod.111.090886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev. 2006;20:1744–1754. doi: 10.1101/gad.1435106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Miranda M, DePamphilis ML. Analysis of gene expression in mouse preimplantation embryos demonstrates that the primary role of enhancers is to relieve repression of promoters. EMBO J. 1993;12:1131–1140. doi: 10.1002/j.1460-2075.1993.tb05754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiekowski M, Miranda M, DePamphilis ML. Requirements for promoter activity in mouse oocytes and embryos distinguish paternal pronuclei from maternal and zygotic nuclei. Dev Biol. 1993;159:366–378. doi: 10.1006/dbio.1993.1248. [DOI] [PubMed] [Google Scholar]
- Oh B, Hwang S, McLaughlin J, Solter D, Knowles BB. Timely translation during the mouse oocyte-to-embryo transition. Development. 2000;127:3795–3803. doi: 10.1242/dev.127.17.3795. [DOI] [PubMed] [Google Scholar]
- Fox CA, Wickens M. Poly(A) removal during oocyte maturation: a default reaction selectively prevented by specific sequences in the 3′ UTR of certain maternal mRNAs. Genes Dev. 1990;4:2287–2298. doi: 10.1101/gad.4.12b.2287. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, Tanaka S, Shiota K, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- Ma P, Pan H, Montgomery RL, Olson EN, Schultz RM. Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development. Proc Natl Acad Sci U S A. 2012;109:E481–E489. doi: 10.1073/pnas.1118403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]