Summary
The colonization of the land by plants, sometime before 470 million years ago, was accompanied by the evolution tissue systems [1, 2, 3]. Specialized structures with diverse functions—from nutrient acquisition to reproduction—derived from single cells in the outermost layer (epidermis) were important sources of morphological innovation at this time [2, 4, 5]. In extant plants, these structures may be unicellular extensions, such as root hairs or rhizoids [6, 7, 8, 9], or multicellular structures, such as asexual propagules or secretory hairs (papillae) [10, 11, 12]. Here, we show that a ROOTHAIR DEFECTIVE SIX-LIKE (RSL) class I basic helix-loop-helix transcription factor positively regulates the development of the unicellular and multicellular structures that develop from individual cells that expand out of the epidermal plane of the liverwort Marchantia polymorpha; mutants that lack MpRSL1 function do not develop rhizoids, slime papillae, mucilage papillae, or gemmae. Furthermore, we discovered that RSL class I genes are also required for the development of multicellular axillary hairs on the gametophyte of the moss Physcomitrella patens. Because class I RSL proteins also control the development of rhizoids in mosses and root hairs in angiosperms [13, 14], these data demonstrate that the function of RSL class I genes was to control the development of structures derived from single epidermal cells in the common ancestor of the land plants. Class I RSL genes therefore controlled the generation of adaptive morphological diversity as plants colonized the land from the water.
Highlights
-
•
Class I RSL genes control the development of structures derived from single cells
-
•
Class I RSL function is conserved among land plants
-
•
Class I RSL controlled epidermal development in the land plant common ancestor
-
•
These genes controlled the development of the first plant rooting systems
The colonization of the land by plants was accompanied by the evolution of specialized cells and structures that develop from single cells. Proust et al. show that RSL class I basic helix-loop-helix transcription factors controlled the development of specialized structures from single epidermal cells in the last common ancestor of the land plants.
Results and Discussion
Mutations Define a Gene Required for the Formation of Structures that Develop from Single Epidermal Cells
Unicellular rhizoids [15, 16], multicellular slime papilla [17], mucilage papillae, and gemmae (asexual propagules) [10, 18] develop from single epidermal cells that grow out of the plane of the epidermis and differentiate in the haploid phase of the life cycle in the liverwort M. polymorpha. To identify genes that control the development of structures derived from single epidermal cells that expand out from the epidermal surface, we identified two rhizoidless mutants, ST46-1 and ST44-8, from a population of T-DNA-transformed plants (Figures 1A, 1B, and S1A). In wild-type gemmae, rhizoids develop after approximately 24 hr from rhizoid precursor cells that are larger and paler than the surrounding epidermal cells (Figure 1E) [15]. Rhizoid precursors did not form in the gemma epidermis of either mutant ST46-1 or ST44-8 (Figures 1F and S1C), and rhizoids did not develop at any stage in the life cycle (Figures 1B, 1J, S1A, and S1D). The absence of rhizoids in ST46-1 and ST44-8 suggests that these lines are defective in genes required for rhizoid development. To determine whether the gene defective in ST46-1 and ST44-8 also controls the development of other structures derived from individual epidermal cells, we compared the development of multicellular gemmae, mucilage papillae, and slime papillae in wild-type and mutants. Both gemmae and mucilage papillae develop from individual epidermal cells located on the floor of wild-type gemma cups (Figures 1M and 1U) [10, 18]. The gemma precursor cell expands out of the plane of the epidermis and then undergoes a transverse division producing a proximal and a distal cell. The proximal cell develops as a uniseriate stalk, whereas the distal cell forms the disc-shaped multicellular gemma (Figure 1U). During mucilage papilla development, an epidermal cell swells out of the plane of the epidermal plane before dividing to form a proximal cell in the plane of the epidermis and a distal cell that swells and secretes mucilage [18]. Neither gemmae nor mucilage papillae developed in either mutant ST46-1 or ST44-8; mutant gemma cups were completely empty (Figures 1N and S1B). Whereas the epidermal surface of the floor of wild-type gemma cups was covered with cellular outgrowths (Figures 1Q and 1U), these surfaces were smooth and flat in mutants; cells did not break the plane of the epidermis (Figures 1R and 1V). This demonstrates that the activity of the wild-type gene is required for the early development of gemma and mucilage papilla precursor cells in the epidermis of the gemma cup. Slime papillae develop from single cells in the dorsal epidermis near the meristem of wild-type gemmae, where they are visible as projections from the dorsal gemma epidermis (Figure 1Y) [17]. These papillae did not develop in either of ST46-1 or ST44-8 (Figure 1Y). These data demonstrate that the wild-type function of the genes defective in the ST46-1 and ST44-8 mutants is required for the development of structures—unicellular rhizoids and multicellular mucilage papillae, slime papillae, and gemmae—that are derived from individual epidermal cells in M. polymorpha.
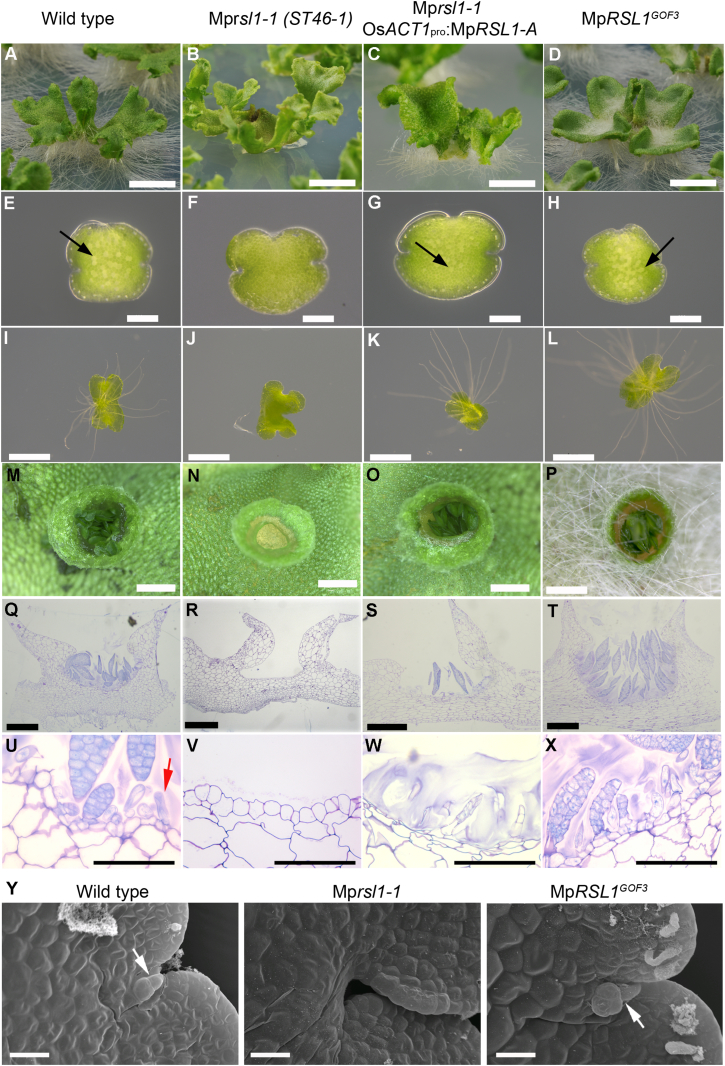
Figure 1.
MpRSL1 Positively Regulates the Initiation of Rhizoids, Gemmae, Mucilage Papillae, and Slime Papillae in M. polymorpha
(A–X) Four genotypes are presented, one in each column: (first column) wild-type, (second column) Mprsl1-1 mutants, (third column) Mprsl1-1 OsACT1pro:MpRSL1-A, and (fourth column) MpRSL1GOF3. Six different stages are presented in rows.
(A–D) Rhizoids on 28-day-old thalli grown from gemmae. Scale bars represent 1 cm.
(E–H) Zero-day-old gemmae. Arrows highlight the position of pale rhizoid precursor cells. Scale bars represent 200 μm.
(I–L) Rhizoids developing on 4-day-old gemmae. Scale bars represent 1 mm.
(M–P) Surface view of gemma cups of 1-month-old plant. Scale bars represent 1 mm.
(Q–T) Toluidine-blue-stained longitudinal sections of gemma cups in a 1-month-old plant. Scale bars represent 500 μm.
(U–X) Toluidine-blue-stained longitudinal sections of gemma cups in a 1-month-old plant. The red arrow indicates a mucilage papilla; the black arrow indicates a gemma primordium. Scale bars represent 100 μm.
(Y) Wild-type, Mprsl1-1, and MpRSL1GOF3 1-day-old gemmae. Arrows indicate slime papillae. Scale bars represent 20 μm.
See also Figure S1.
MpRSL1 Controls the Morphogenesis of Structures Derived from Individual Epidermal Cells
To identify DNA sequence flanking each of the T-DNA insertion sites in ST46-1 and ST44-8 mutants, we carried out thermal asymmetric interlaced (TAIL) PCR using a combination of random primers and nested primers specific to the T-DNA on isolated genomic DNA [19]. The DNA sequence flanking these insertions demonstrated that the T-DNAs had inserted into intron one and exon one, respectively, of a protein-coding gene (Figures 2A and 2D; Data S1 and S2). The presence of an “RSL” motif—QLQVKVLMNDEYWP—at the carboxyl end of a basic helix-loop-helix (bHLH) domain suggested that the gene encodes a member of the RSL class I bHLH proteins (Figure 2B) [20]. To test this hypothesis, we constructed trees (Figures 2C and S3) using an LG maximum likelihood model [21] and approximate likelihood ratio statistics [22]. The topology of the gene tree demonstrated that MpRSL1 is a ROOTHAIR DEFECTIVE SIX-LIKE (RSL) class I basic helix-loop-helix protein (Figure 2C). Members of the RSL class I proteins are required for the development of rhizoids in the moss P. patens and root hairs in the angiosperm Arabidopsis thaliana (Figure 2C) [13, 14]. We designated the protein encoded by the gene disrupted by a T-DNA insertion in the ST46-1 and ST44-8 mutants as M. polymorpha ROOTHAIR SIX-LIKE1 (MpRSL1).
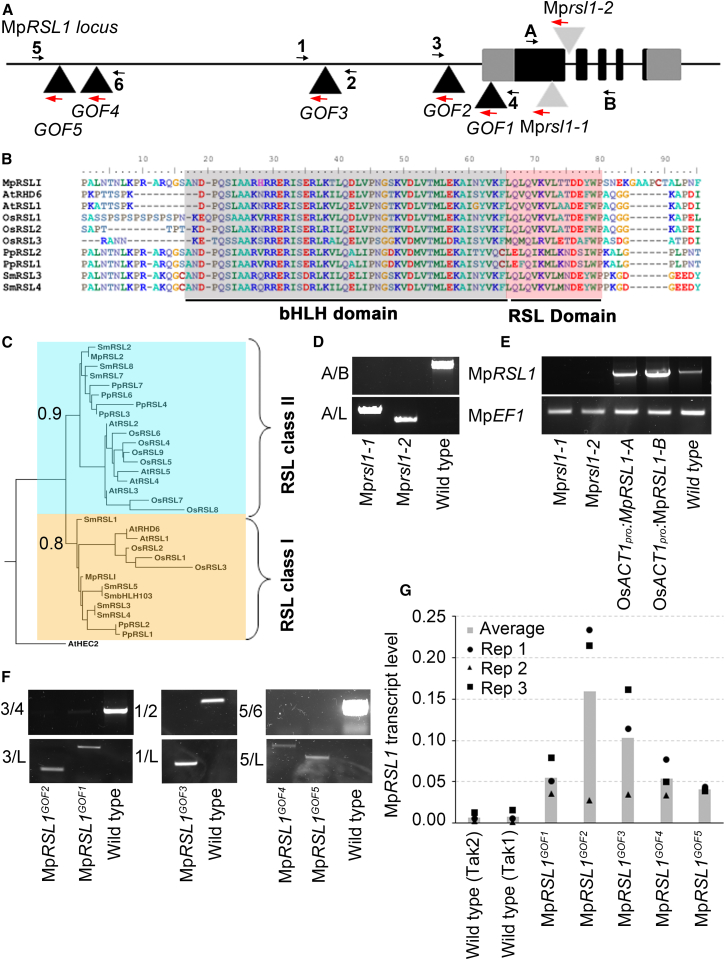
Figure 2.
Molecular Characterization of Mprsl1 Loss-of-Function and MpRSL1GOF Mutations
(A) Schematic of MpRSL1 gene structure. Black and gray boxes represent exons of CDS and UTR of MpRSL1, respectively. Black and light gray triangles indicate the T-DNA insertion site in each MpRSL1GOF and Mprsl1 loss-of-function line, respectively.
(B) Alignment of land plant RSL class I proteins. bHLH and RSL domains are highlighted in gray and red, respectively.
(C) A maximum-likelihood tree of RSL class I (yellow box) and RSL class II proteins (blue box) rooted with AtHEC2. Bootstrap values are indicated at the nodes.
(D) Amplification of Mprsl1-1 and Mprsl1-2 T-DNA insertion site from genomic DNA. A and B are MpRSL1 locus-specific primers, and L is a T-DNA left-border-specific primer.
(E) RT-PCR analysis of the presence or absence of MpRSL1 full-length cDNA in the Mprsl1-1 and Mprsl1-2 loss-of-function mutants, two Mprsl1-1 loss of function complemented with OsACTpro:MpRSL1 lines, and wild-type. MpEF1 is a reference gene.
(F) Amplification of MpRSL1GOF1 -MpRSL1GOF5 T-DNA insertion site with MpRSL1 promoter-specific primers flanking the insertion site (top) and with an MpRSL1 promoter-specific primer and a T-DNA left-border-specific primer (bottom). Amplification was performed on genomic DNA. Primers 1–6 are MpRLS1 promoter-specific primers. L is a T-DNA left-border-specific primer.
(G) Analysis by qRT-PCR of MpRSL1 expression in the different MpRSL1GOF mutants. Histograms represent the average MpRSL1 transcript levels (n = 3) normalized with the geometric mean of MpEF1 and MpCUL transcript levels. The three biological replicates are represented with circle, triangle, and square for replicate 1, replicate 2, and replicate 3, respectively.
To determine whether the ST46-1 and ST44-8 mutant alleles resulted from loss of MpRSL1 function, we compared the levels of MpRSL1 mRNA in each mutant background with wild-type. MpRSL1 mRNA was not detectable in mRNA isolated from either mutant ST46-1 or ST44-8 thalli, whereas the transcript was detected in wild-type thalli (Figure 2E). This suggests that the mutant phenotype in the ST46-1 and ST44-8 lines was due to loss of MpRSL1 function. Therefore ST46-1 and ST44-8 were designated Mprsl1-1 and Mprsl1-2, respectively. To verify that loss of MpRSL1 function is responsible for the defects in rhizoid development in these mutants, we transformed Mprsl1-1 thalli with the MpRSL1 cDNA under the transcriptional control of the rice ACTIN1 promoter (OsACT1pro). MpRSL1 mRNA was detected in two independent Mprsl1-1 lines transformed with OsACT1pro:MpRSL1, indicating that the MpRSL1 transgene was expressed in the transformed lines (Figure 2E). Rhizoids, mucilage papillae, and gemmae developed in both transformed lines, indicating that expression of MpRSL1 is sufficient to restore rhizoid (Figures 1C, 1G, and 1K), mucilage papilla, and gemma development in Mprsl1-1 mutant plants (Figures 1O, 1S, and 1W). These results verify that MpRSL1 is required for the initiation and the development of unicellular rhizoids, mucilage papillae, and gemmae in M. polymorpha.
To test the hypothesis that RSL class I genes positively regulate the development of M. polymorpha rhizoids, we screened T-DNA mutants for putative gain-of-function mutations in the MpRSL1 gene. Rhizoids do not develop on the dorsal surface of the wild-type thallus (Figure 1A). We identified five mutants in which rhizoids developed on the dorsal surface of the thallus (Figures 1D and S1E). The dorsal rhizoid phenotype segregated 1:1 after crossing each of the mutants to wild-type (Table S1), indicating that the dorsal rhizoid phenotype was caused by a single gene mutation in each line. 100% of the offspring from crosses between each of these mutants developed rhizoids on the dorsal epidermis, indicating that the mutation is in the same gene in each mutant line (Table S2). Furthermore, 100% of the mutants produced from crosses to wild-type were hygromycin resistant (Table S1). This indicates that a T-DNA was closely linked to the rhizoid mutation in each of these mutants. To identify the genomic DNA sequences flanking each of the T-DNAs in these five dorsal rhizoid mutants, we carried out TAIL PCR using a combination of random primers and nested primers in the T-DNA [19]. The flanking sequences demonstrated that T-DNAs were inserted 1.6 kb, 2.6 kb, 6 kb, 14.5 kb, and 15 kb upstream (5′) of the start codon of the MpRSL1 gene in these mutants (Figures 2A and 2F; Data S1 and S2). Steady-state levels of MpRSL1 mRNA were higher in each mutant than in wild-type (Figure 2G). These data demonstrate that the insertion of the T-DNA in the 5′ region of the gene results in higher steady-state levels of MpRSL1 mRNA. These mutant lines were therefore designated MpRSL1GOF1, MpRSL1GOF2, MpRSL1GOF3, MpRSL1GOF4, and MpRSL1GOF5, respectively (where GOF indicates that these are gain-of-function mutations). The demonstration that supernumerary rhizoids develop on MpRSL1 gain-of-function mutants and rhizoids do not develop on Mprsl1 loss-of-function mutants indicates that MpRSL1 positively regulates rhizoid development. These data also demonstrate that MpRSL1 is sufficient for rhizoid development. Ectopic slime papillae (Figure 1Y), mucilage papillae, and gemmae (Figures 1P, 1T, and 1X) did not develop in the MpRSL1 gain-of-function mutants. This suggests that whereas MpRSL1 function is sufficient for rhizoid development, it is most likely not sufficient for slime papilla, mucilage papilla, or gemma development.
MpRSL1 Is Expressed in Epidermal Cells that Form Specialized Structures
We predicted that MpRSL1 would be expressed in epidermal cells that swell and break the plane of the epidermis during morphogenesis. We measured steady-state levels of MpRSL1 mRNA in isolated cells and tissues and found expression in rhizoids (Figures 3A and 3B). MpRSL1 transcript was also detected in gemmae and ventral thallus, where rhizoids develop. In contrast, no significant MpRSL1 mRNA transcript was detected in the cells of the mature dorsal thallus where epidermal cells that break the epidermal plane do not develop. To determine when MpRSL1 is first expressed in cells that break the epidermal plane during their development, we carried out semi-qRT-PCR analysis on mRNA isolated from single cells of the epidermis inside the gemma cup where mucilage papillae and gemmae develop using laser capture microdissection (Figure S3). RNA was isolated from epidermal cells that had expanded out of the plane of the epidermis to form buds (GCEB) inside the gemma cup, epidermal cells from the inside of the of the gemma cup that were not forming outgrowths (GCEP), and dorsal thallus epidermal cells (DTEP) in which growth out of the epidermal plane does not occur (Figures 3C and S3A–S3C). MpRSL1 mRNA was detectable in the bud cells (GCEB), was less abundant in the epidermal cells that do not break the plane of the epidermis (GCEP), and was not detectable in the cells of the dorsal thallus epidermis from which epidermis-derived structures do not develop (DTEP) (Figures 3D and S3). This indicates that MpRSL1 is expressed in cells that swell out of the epidermal plane during the formation of rhizoids, mucilage papillae, and gemmae. Taken together, these data indicate that MpRSl1 is expressed in the cells that expand out of the epidermis and subsequently form unicellular and multicellular structures in M. polymorpha.
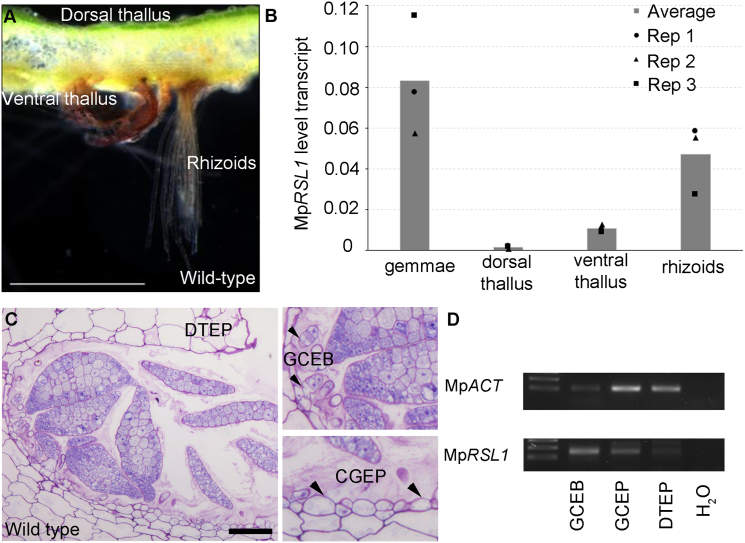
Figure 3.
MpRSL1 Is Expressed in Rhizoid and Gemma Precursor Cells
(A and B) Expression of MpRSL1 in different tissues of 1-month-old wild-type gemmalings.
(A) Cross-section of 1-month-old wild-type thallus showing dorsal and ventral thallus and rhizoid.
(B) Analysis of MpRSL1 level transcript by qRT-PCR. Histograms represent the mean transcript levels (n = 3) normalized with the geometric mean of MpEF1 and MpCUL transcript level in different organs. The three biological replicates are represented with circle, triangle, and square for replicate 1, replicate 2, and replicate 3, respectively.
(C and D) Expression of MpRSL1 in gemmae primordia.
(C) Toluidine-blue-stained cross-section of a gemma cup showing gemma buds (GCEB), gemma cup epidermis cells (GCEP), and thallus epidermis cells (DTEP). The two small pictures are magnification of the first picture. The scale bar represents 200 μm.
(D) Analysis by semi-qRT-PCR of MpRSL1 level transcript in gemma buds (GCEB), gemma cup epidermis cells (GCEP), and thallus epidermis cells (DTEP). MpACT was used as reference gene.
See also Figure S3.
RSL Class I Function Is Conserved in Land Plants
To determine whether RSL class I genes control the development of cells that break the plane of the epidermis in other taxa, we characterized the development of epidermis-derived structures in the Pprsl1 Pprsl2 double mutant of the moss P. patens. We previously showed that rhizoid development was defective in Pprsl1 Pprsl2 double mutants [13, 14]. To determine whether these genes control the development of other structures derived from individual epidermal cells, we compared the development of the multicellular hairs that develop in the axils of leaves [23] on wild-type and Pprsl1 Pprsl2 double mutants (Figures 4A and 4B). Whereas wild-type developed between one and two axillary hairs per node (1.45 ± 0.57 hairs per axil), Pprsl1 Pprsl2 double mutants developed 0.47 (±0.3) hairs per axil (Student’s t test indicates that these means are different at p < 0.0001). This demonstrates that RSL class I genes control the development of structures—rhizoids and axillary hairs—derived from single cells that break the epidermal plane during development in mosses.
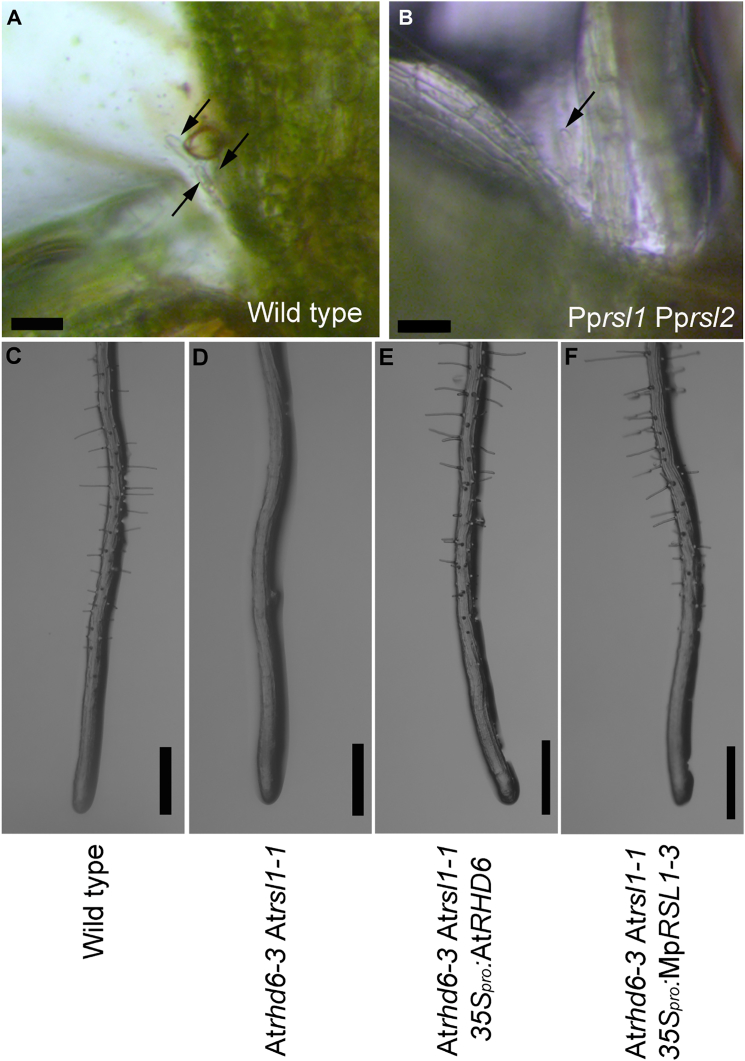
Figure 4.
The Function of RSL Class I Proteins in the Formation of Structures that Develop from Individual Epidermal Cells Has Been Conserved Among Land Plants
(A and B) Papillae on P. patens gametophores from 1-month-old colonies. Arrows highlight axillary papillae. The scale bar represents 50 μm.
(A) More than one slime papilla develops at each node on wild-type gametophores (arrows).
(B) One or no slime papilla develops at each node of Pprsl1 Pprsl2 double mutant gametophores. The arrow identifies the single papilla that develops at a node of the Pprsl1 Pprsl2 double mutant.
(C–F) Five-day-old seedlings of A. thaliana. The scale bar represents 500 μm.
(C) Wild-type (with root hairs).
(D) Atrhd6-3 Atrsl1-1 double mutant (root hairless).
(E and F) Atrhd6-3 Atrsl1-1 double mutant transformed with 35Spro:AtRHD6 (with root hairs) (E) and Atrhd6-3 Atrsl1-1 double mutant transformed with 35Spro:MpRSL1 (F).
See also Figure S4.
We previously demonstrated that RSL class I genes control root hairs in A. thaliana [13]. To determine whether the molecular function of RSL class I genes has diverged since evolving from the RSL class I gene in the common ancestor of extant land plants, we tested whether the MpRSL1 gene could program the development of root hairs in A. thaliana mutants plants devoid of RSL class I gene function (Atrhd6 Atrsl1 double mutants). Whereas root hairs do not develop in Atrhd6 Atrsl1 double mutants (Figures 4C and 4D), root hairs develop on Atrhd6 Atrsl1 double mutants transformed with MpRSL1 (Figures 4E and S4). These plants are identical to plants transformed with the RSL class I gene from A. thaliana, AtRHD6 (Figure 4F). Whereas 81% of the amino acids are identical in the bHLH-RSL domains of MpRSL1 and AtRHD6, there is no conservation elsewhere in the protein. This demonstrates that, despite sequence divergence, the molecular function of RSL class I proteins has not diverged since M. polymorpha and A. thaliana last shared a common ancestor more than 470 million years ago.
A diversity of structures in land plants—unicellular rhizoids and root hairs to multicellular organs such as gemmae—develop from single cells in the epidermis [6, 15, 16, 17, 18]. Early in development, individual cells change their growth polarity and expand perpendicular to the plane of the epidermal surface to form balloon-shaped swellings. Tip growth may initiate from these outgrowths during the differentiation of rhizoids and root hairs. Alternatively, the swollen cell may divide once or many times to form multicellular structures and organs, such as gemmae and slime papillae. We discovered that the RSL class I mechanism is required for the development of a variety of cells that develop from cells that break the epidermal plane before undergoing morphogenesis in liverworts, mosses, and angiosperms. The conserved role of class I RSL genes in development of structures developing from single epidermal cells that break the plane of the epidermis suggests that these genes controlled this process in the common ancestor of all land plants. Given that many epidermal structures develop from single epidermal cells that expand beyond the surface plane of the epidermis prior to differentiation and organogenesis, our data demonstrate that RSL class I genes played an important role in the generation of cellular and organ diversity in the newly evolved epidermis early in land plant evolution. Many of these structures were key adaptations to life on land—protecting the plant and plant parts from desiccation and providing access to inorganic nutrients [1, 3, 24]. Therefore, the evolution of the RSL class I mechanism allowed the generation of morphological diversity that facilitated adaptation of green plants to life in the terrestrial environment as plants colonized the land from the water.
Author Contributions
H. Proust performed the experiments and analyzed the data with L.D. S.H. isolated lines with gain-of-function and loss-of-function mutations and identified insertions sites; V.A.S.J. and G.M. isolated lines with gain-of-function mutations and identified insertions sites. H. Proust, G.M., and S.K. isolated and sequenced mRNA and analyzed the transcriptomes and the genome of M. polymorpha. K.I. and T.K. provided MpRSL1 sequences at the start of the project. H. Prescott initiated the transformation and establishment of the Marchantia system in the lab. H. Proust and L.D. designed the experiments and wrote the paper. L.D. conceived and supervised the project.
Acknowledgments
This research was funded by the EVO500 ERC AdG award to L.D. and a JSPS Fellowship for Overseas Researchers (short-term: PE12565) award to H. Proust held with T.K. We are grateful to Dr. Chulmin Kim, Dr. Denis Saint-Marcoux, Dr. Clémence Bonnot, Bruno Catarino, and Sandy Hetherington for helpful discussions. We are grateful to Bruno Catarino for his help with the phylogenetic analysis. We are grateful to Dr. Denis Saint-Marcoux for his help for the laser microdissection experiments. We would like to thank Dr. Keke Yi for providing the Arabidopsis thaliana line transformed with the 35S:AtRHD6 construct.
Published: December 24, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, four figures, two tables, and two data files and can be found with this article online at http://dx.doi.org/10.1016/j.cub.2015.11.042.
Accession Numbers
The accession numbers for the MpRSL1 and MpRSL2 sequences reported in this paper are Genbank: KT633827 and Genbank: KT633828, respectively.
Supplemental Information
References
- 1.Kenrick P., Crane P.R. The origin and early evolution of plants on land. Nature. 1997;389:33–39. [Google Scholar]
- 2.Ligrone R., Duckett J.G., Renzaglia K.S. Major transitions in the evolution of early land plants: a bryological perspective. Ann. Bot. 2012;109:851–871. doi: 10.1093/aob/mcs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle J.A. Phylogenetic analyses and morphological evolution in land plants. In: Ambrose B.A., Purugganan M., editors. Annual Plant Reviews Volume 45: The Evolution of Plant Form. John Wiley and Sons; 2013. pp. 1–50. [Google Scholar]
- 4.Ramsay N.A., Glover B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005;10:63–70. doi: 10.1016/j.tplants.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Goffinet B., Buck W.R. The evolution of body form in bryophytes. In: Ambrose B.A., Purugganan M., editors. Annual Plant Reviews Volume 45: The Evolution of Plant Form. John Wiley and Sons; 2013. pp. 51–90. [Google Scholar]
- 6.Carol R.J., Dolan L. Building a hair: tip growth in Arabidopsis thaliana root hairs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2002;357:815–821. doi: 10.1098/rstb.2002.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfano F., Russell A., Gambardella R., Duckett G. The actin cytoskeleton of the liverwort Riccia fluitans: effects of cytochalasin B and aluminum ions on rhizoid tip growth. J. Plant Physiol. 1993;142:569–574. [Google Scholar]
- 8.Pressel S., Ligrone R., Duckett J.G. Cellular differentiation in moss protonemata: a morphological and experimental study. Ann. Bot. 2008;102:227–245. doi: 10.1093/aob/mcn080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones V.A., Dolan L. The evolution of root hairs and rhizoids. Ann. Bot. 2012;110:205–212. doi: 10.1093/aob/mcs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barnes C.R., Land W.J.G. Bryological papers. II. The origin of the cupule of Marchantia. Bot. Gaz. 1908;46:401–409. [Google Scholar]
- 11.Duckett J.G., Ligrone R. The formation of catenate foliar gemmae and the origin of oil bodies in the liverwort Odontoschisma denudatum (Mart.) Dum. (Jungermanniales): a light and electron microscope study. Ann. Bot. 1995;76:405–419. [Google Scholar]
- 12.Ligrone R., Duckett J.G., Gambardella R. Serial development of foliar gemmae in Tortula (Pottiales, Musci), an ultrastructural study. Ann. Bot. 1996;78:305–315. [Google Scholar]
- 13.Menand B., Yi K., Jouannic S., Hoffmann L., Ryan E., Linstead P., Schaefer D.G., Dolan L. An ancient mechanism controls the development of cells with a rooting function in land plants. Science. 2007;316:1477–1480. doi: 10.1126/science.1142618. [DOI] [PubMed] [Google Scholar]
- 14.Jang G., Yi K., Pires N.D., Menand B., Dolan L. RSL genes are sufficient for rhizoid system development in early diverging land plants. Development. 2011;138:2273–2281. doi: 10.1242/dev.060582. [DOI] [PubMed] [Google Scholar]
- 15.Tarén N. Factors regulating the initial development of gemmae in Marchantia polymorpha. Bryologist. 1958;61:191–204. [Google Scholar]
- 16.Cao J.-G., Dai X.-L., Zou H.-M., Wang Q.-X. Formation and development of rhizoids of the liverwort Marchantia polymorpha. J. Torrey Bot. Soc. 2014;141:126–134. [Google Scholar]
- 17.Renzaglia K.S., Duff R.J.T., Nickrent D.L., Garbary D.J. Vegetative and reproductive innovations of early land plants: implications for a unified phylogeny. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:769–793. doi: 10.1098/rstb.2000.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vashishta B.R., Sinha A.K., Kumar A. S. Chand; 2011. Botany for Degree Students Part III: Bryophyta. [Google Scholar]
- 19.Ishizaki K., Chiyoda S., Yamato K.T., Kohchi T. Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 2008;49:1084–1091. doi: 10.1093/pcp/pcn085. [DOI] [PubMed] [Google Scholar]
- 20.Pires N., Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol. Biol. Evol. 2010;27:862–874. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le S.Q., Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 22.Anisimova M., Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst. Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 23.Eklund D.M., Thelander M., Landberg K., Ståldal V., Nilsson A., Johansson M., Valsecchi I., Pederson E.R., Kowalczyk M., Ljung K. Homologues of the Arabidopsis thaliana SHI/STY/LRP1 genes control auxin biosynthesis and affect growth and development in the moss Physcomitrella patens. Development. 2010;137:1275–1284. doi: 10.1242/dev.039594. [DOI] [PubMed] [Google Scholar]
- 24.Langdale J.A. Evolution of developmental mechanisms in plants. Curr. Opin. Genet. Dev. 2008;18:368–373. doi: 10.1016/j.gde.2008.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.