Abstract
Peroxynitrite is formed in biological systems when nitric oxide and superoxide rapidly interact at near equimolar ratio. Peroxynitrite, though not a free radical by chemical nature, is a powerful oxidant which reacts with proteins, DNA and lipids. These reactions trigger a wide array of cellular responses ranging from subtle modulations of cell signaling to overwhelming oxidative injury, committing cells to necrosis or apoptosis. The present review outlines the various peroxynitrite-induced DNA modifications with special mention to the formation of 8-nitroguanine and 8-oxoguanine as well as the induction of DNA single strand breakage. Low concentrations of peroxynitrite cause apoptotic death, whereas higher concentrations cause necrosis with cellular energetics (ATP and NAD+) serving as control between the two modes of cell death. DNA damage induced by peroxynitrite triggers the activation of DNA repair systems. A DNA nick sensing enzyme, poly(ADP-ribose) polymerase-1 (PARP-1) becomes activated upon detecting DNA breakage and it cleaves NAD+ into nicotinamide and ADP-ribose and polymerizes the latter on nuclear acceptor proteins. Over-activation of PARP induced by peroxynitrite consumes NAD+ and consequently ATP decreases, culminating in cell dysfunction, apoptosis or necrosis. This mechanism has been implicated in the pathogenesis of various diseases like diabetes, cardiovascular diseases and neurodegenerative diseases. In this review, we have discussed the cytotoxic effects (apoptosis and necrosis) of peroxynitrite in the etiology of the mentioned diseases, focusing on the role of PARP in DNA repair in presence of peroxynitrite.
Keywords: Peroxynitrite, DNA, Poly(ADP-ribose) polymerase-1, Diabetes, Cardiovascular diseases, Neurodegenerative diseases
Introduction
The two major pathways involved in the nitric oxide (NO) induced DNA damage involve a reaction of NO with molecular oxygen, yielding N2O3 with subsequent nitrosation of secondary amines and the formation of N-nitrosoamines or nitrosation of primary amines and nucleic acid bases [1] and the production of peroxynitrite through the reaction of NO with superoxide radical (O•−2). Peroxynitrite (ONOO−) is a potent oxidant and nitrating agent with a short half life (~10 ms) [2, 3]. The peroxynitrite formation sites are considered to be spatially associated with the sources of superoxide (such as the plasma membrane NAD(P)H oxidases and the mitochondrial respiratory complexes). Since NO is a relatively stable and highly diffusible free radical while superoxide is much short lived and has restricted diffusion rates across biological membranes. The rate of peroxynitrite production in vivo has been estimated to be as high as 50–100 μM per min. Inspite of the short half-life of peroxynitrite at physiological pH, its ability to cross cell membranes [4] influences the surrounding target cells within one to two cell diameters (~5–20 μm).
The oxidant reactivity of peroxynitrite is highly pH-dependent and both peroxynitrite anion (ONOO−) and peroxynitrous acid (ONOOH) can participate directly in one- and two-electron oxidation reactions with biological macromolecules (Fig. 1), where many of them have transition metal centres and thiols. The one of the fundamental reactions of ONOO− in biological systems is the fast reaction with carbon dioxide (in equilibrium with physiological levels of bicarbonate anion), which leads to the formation of carbonate (CO•−3) and nitrogen dioxide (•NO2) radicals (yield ~35 %), which are one-electron oxidants [5]. Nitrogen dioxide can undergo diffusion-controlled radical–radical termination reactions with biological macromolecules, while in nitrated compounds alternatively ONOOH can undergo homolytic fission to generate one-electron oxidants, hydroxyl (•OH) and •NO2 radicals (with a 30 % yield). Nonetheless, this reaction is slow compared with the other reactions of ONOO− and ONOOH in biological systems and therefore considered as a modest component of the in vivo reactivity of peroxynitrite. The proton-catalysed decomposition of ONOO− to form •OH and •NO2 radicals becomes relevant in hydrophobic phases and results in the initiation of lipid peroxidation processes [5, 6].
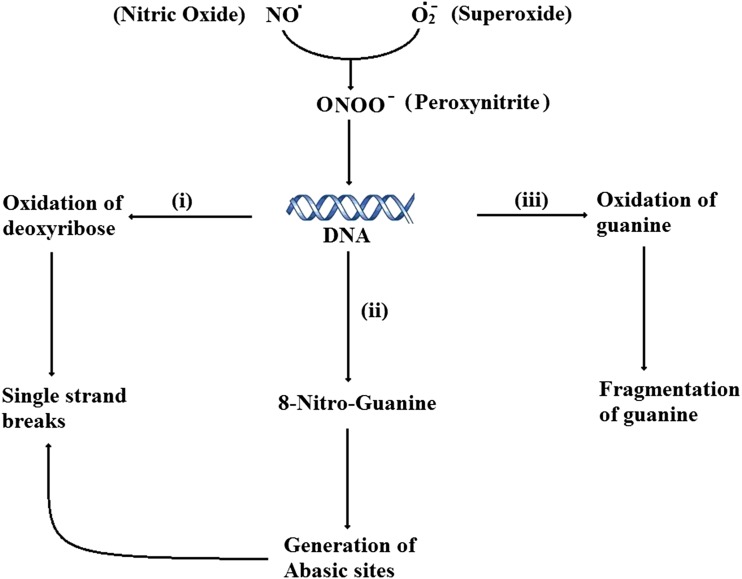
Fig. 1.
Effects of peroxynitrite on DNA. The superoxide (O•−2) and nitric oxide (NO) reacts to form peroxynitrite, which attacks DNA resulting in (i) oxidation of deoxyribose, (ii) formation of 8-Nitro-Guanine, ultimately result in single strand breaks, (iii) oxidation of guanine owing its fragmentation
Various biological macromolecules including proteins, DNA and unsaturated fatty-acid-containing phospholipids are oxidized and/or nitrated by peroxynitrite derived radicals. In fact, tyrosine nitration, dimerization and hydroxylation caused by peroxynitrite to form 3-nitrotyrosine, 3,3′-dityrosine and 3,4′-dihydrophenylalanine, respectively, are totally dependent on free-radical pathways [7]. Thiols can be oxidized through one-electron reactions by peroxynitrite-derived radicals and initiate radical-dependent chain reactions to produce higher oxidation states of sulphur, including sulphinic and sulphonic acid derivatives [8, 9]. In DNA, purine nucleotides are susceptible to oxidation and adduct formation [10–13], with 8-oxo and 8-nitroguanine existing as two of the major products. Peroxynitrite can also cause deoxyribose oxidation and strand breaks [14].
Peroxynitrite induced oxidative and nitrosative changes in lipids result in peroxidation [5] and the formation of nitrito-, nitro-, nitrosoperoxo- and/or nitrated lipid oxidation adducts (malondialdehyde, conjugated diene and lipid hydroperoxide formation) [15–17]. Peroxynitrite also causes the oxidation of arachidonic acid and leads to the formation of F2-isoprostanes through the oxidation of low-density lipoprotein [18]. The nitration of fatty acids may lead to the secondary inhibition of protein function via thiol-based modifications [19].
Exposure of DNA to peroxynitrite has been shown to lead to the formation of 8-hydroxydeoxyguanosine [20] and 8-nitroguanine from guanosine [21].
Peroxynitrite-Induced DNA Damage
The DNA damage caused by peroxynitrite is mostly oxidative. DNA treatment with peroxynitrite usually leads to much more damage than treatment with an equivalent dose of nitric oxide. Besides the higher levels of damage present in DNA after ONOO− treatment, the scale of damage also tends to be much more complex. This gives the assumption that peroxynitrite is intrinsically much more reactive than nitric oxide. Ischiropoulos and co-workers were among the first to observe the formation of peroxynitrite from activated macrophages [22]. They detected significant concentration of peroxynitrite (as high as 0.11 nmol/106 cells min−1) using the nitration of 4-hydroxyphenylacetate in the media as a marker of peroxynitrite activity. Another group led by Lewis performed a kinetic analysis of the fate of NO• synthesized by activated macrophages using end product measurements of nitrite and nitrate (N2O3 hydrolysis forms nitrite while ONOO− decay leads to nitrate formation) [23]. Their results showed that approximately half of the released nitric oxide forms N2O3 while the remainder combines with O•−2 to form ONOO−. Interestingly, from this study, it comes into view that that ONOO− formation occurs partially extracellularly and not exclusively inside the macrophage. This can be assumed from the observation that adding SOD to the media significantly reduced ONOO− formation. If peroxynitrite would have been formed only within the cell, it would have not been influenced by the extracellular addition of superoxide dismutase.
Modification of Bases in DNA
The reaction of peroxynitrite with purine bases, such as guanine and adenine, resulted in the production of a strong yellow color, whereas pyrimidine bases, such as thymine, cytosine, 5-methylcytosine, and uracil, did not [21] because guanine (G) has the lowest reduction potential among the four normal DNA bases (Eº = 1.29 V [24] ), and its reaction with the peroxynitrite-derived HO• (Eº = 1.9–2.1 V) [25] and CO•−3 (Eº = 1.5 V) [26] radicals is thermodynamically favorable. Furthermore, the bimolecular rate constants for reaction of these radicals with G (7.8 × 109 M−1 s−1 for HO•) [27] and (7 × 107 M−1 s−1 for CO•−3) [28] dictate extremely fast kinetics for the initial oxidation event. The yellow coloured compound formed by the reaction between guanine and peroxynitrite has been identified by HPLC as 8-nitroguanine (first nitration product) and its formation was most favorable at pH 8 and increased dose-dependently with peroxynitrite concentration, but was not dependent on the concentration of guanine. It was proposed that either heterolytic cleavage of peroxynitrite to form a nitronium ion (NO2+) or a high energy intermediate (ONOOH) derived from trans-peroxynitrite (pKa 7.9) [29] could be involved in the formation of 8-nitroguanine [21]. The reaction of 2′-deoxyguanosine with peroxynitrite yields several compounds, two of which were identified as 4,5-dihydro-5-hydroxy-4-(nitrosooxy)-2′-deoxyguanosine (nox-dG) and 8-nitroguanine [30]. The 8-nitroguanine could be formed either from guanine (depurination of dG) by peroxynitrite or depurination of 8-nitro-2′-deoxyguanosine generated with peroxynitrite. The reaction of various deoxyribonucleosides with peroxynitrite was also shown to yield 2-thiobarbituric acid (TBA)-reactive substances dose-dependently [31, 32]. Yermilov et al. have shown the formation of 8-nitroguanine dose-dependently in calf thymus DNA when incubated with low concentrations of peroxynitrite [33]. Among peroxynitrite, nitrous acid, tetra-nitromethane, and NO-releasing compounds, only peroxynitrite was responsible for the formation of 8-nitroguanine. This reaction was inhibited by antioxidants like urate, ascorbate, N-acetylcysteine and desferrioxamine [33]. Studies have shown that 8-Nitroguanine was depurinated rapidly from DNA incubated at physiological conditions (t1/2 = ~4 h), suggesting its formation in DNA is potentially mutagenic because depurination yields apurinic sites, which can induce G:C → T:A transversions [33]. Spencer et al. have shown increased levels of both oxidized and deaminated base products, including 5-hydroxyhydantoin, 5-(hydroxymethyl)uracil, thymine glycol, 4,6-diamino-5-formamidepyrimidine (FAPy-adenine), 2,6-diamino-5-formamidepyrimidine (FAPy-guanine), 8-oxoadenine, 8-oxoguanine, hypoxanthine and xanthine in addition to 8-nitroguanine [34]. Among these, 8-nitroguanine and xanthine were formed in 100–1,000 times greater concentrations than other modified bases.
During the peroxynitrite-induced oxidation of guanine, two main oxidation products namely, 2,5-diamino-4H-imidazol-4-one (Iz) [30] and 8-oxoG [14] were formed. Iz has previously been identified as a product of Type I G photooxidation which undergoes hydrolysis to form 2,2,4-triamino-5(2H)-oxazolone (Oz) at physiologic pH [35] (Fig. 2). Oz is expected to accumulate in tissues under oxidative stress in which Iz is initially formed. Niles et al. have isolated and identified two highly oxidized products namely spiroiminodihydantoin, Sp and guanidinohydantoin, Gh [36]. 8-oxoG was detected in genomic DNA isolated from tissues not under chronically elevated levels of oxidative stress ranging from 4 to 11 in 107 bases [37]. Uppu et al. have showed that 103-fold excess of G over 8-oxoG protected only half of the 8-oxoG from reacting with peroxynitrite implying that 8-oxoG is inherently more reactive than G with peroxynitrite [38].
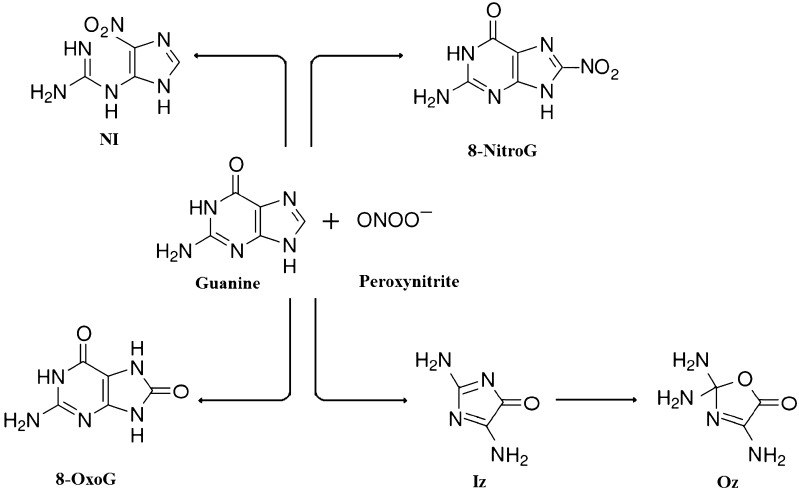
Fig. 2.
Formation of major products from the reaction of peroxynitrite and guanine residue. The reaction between peroxynitrite (ONOO−) and guanine (G) residue forms 8-nitroguanine (8-nitroG), 8-oxoguanine (8-oxoG), 5-guanidino-4-nitroimidazole (NI) and 2,5-diamino-4H-imidazol-4-one (Iz) and it undergoes hydrolysis to form 2,2,4-triamino-5(2H)-oxazolone (Oz) at physiologic pH
There is substantial controversy existing regarding whether synthesized peroxynitrite reacts with guanine to form 8-oxoguanine in DNA. On one hand, Yermilov et al., Douki and Cadet, and Uppu et al. reported no significant increase in 8-oxoguanine in calf thymus DNA treated with synthesized peroxynitrite, compared to non-treated DNA or DNA treated with decomposed peroxynitrite [30, 33, 38]. While on the other hand, Inoue and Kawanishi, Fiala et al. and Spencer et al. showed considerable increase in 8-oxoguanine in calf thymus DNA treated with peroxynitrite [34, 39, 40]. The formation of 8-oxoguanine in peroxynitrite-treated DNA was inhibited by hydroxyl radical scavengers like ethanol and sodium formate [39]. Kennedy and co-workers also reported that low doses of peroxynitrite in pUC19 plasmid formed 8-oxoguanine dose-dependently, where its level being decreased with a high dose of peroxynitrite [14]. A similar decrease in 8-oxoguanine formation at high concentrations of peroxynitrite was also reported [30, 33].
Some possible reasons for this discrepancy could be that (i) 8-oxoguanine formation is mediated by contaminants (e.g., hydrogen peroxide and metal ions), the concentrations of which may fluctuate in different preparations of peroxynitrite; (ii) 8-oxoguanine may be formed artificially during isolation, hydrolysis, and analyses of DNA (e.g., conversion of 8-nitroguanine to 8-oxoguanine) and (iii) under certain circumstances, especially with high concentrations of peroxynitrite, the 8-oxoguanine produced may be further oxidized into the ring cleavage product by peroxynitrite [14, 38, 39].
Human skin epidermal keratinocytes exposure to preformed peroxynitrite or SIN-1 led to extensive DNA base modification [34]. Large increases in xanthine, hypoxanthine and 8-nitroguanine were observed, while only small increases in some oxidized bases including 8-oxoguanine and FAPy-guanine were found in the DNA from keratinocytes [34]. A variety of DNA base modifications have also been depicted in immunostimulated macrophages which produce oxy-radicals and NO, as well as peroxynitrite. Levels of xanthine, 5-(hydroxymethyl)uracil, and 8-oxoguanine were found elevated in the DNA of macrophages activated with lipopolysaccharide and interferon-γ, which were indicative of both oxidative and deaminative DNA damage [41, 42]. Both xanthine and 8-oxoguanine formation was inhibited by an NO synthase inhibitor, suggesting that NO plays a role in both deamination and oxidation reactions [42].
Kaneko et al. showed that 8-nitroguanosine induced oxidative damage and formation of abasic sites in DNA in AS52 cells [43]. Formation of 8-nitroguanosine has been considered as mutagenic lesion because of its rapid removal from DNA strand by depurination to form mutagenic abasic sites, that can possibly induce G-to-T transversions [44].
Single Strand Breaks in DNA
Over the past few years, several studies have shown the induction of single strand breakage in DNA upon exposure to peroxynitrite or to NO and superoxide parallel in different systems. In vitro, peroxynitrite can induce DNA strand breaks in plasmid DNA such as pBR322, PM2, CMV and pUC19, converting the supercoiled form to relaxed or linear forms, which can be separated by agarose gel electrophoresis [31, 32, 41]. A significant induction of single strand breakage was observed in pBR322 plasmid treated with low peroxynitrite, whereas much higher concentrations of peroxynitrite [31] or the presence of a catalyst such as manganese porphyrin [45] were required to induce double strand breakage. DNA breakage caused by peroxynitrite or SIN-1 (donor of peroxynitrite) was observed at almost every nucleotide with a dominance at guanine residues [39]. Peroxynitrite stimulated considerably more single strand breaks at acidic pH than at neutral or alkaline pH, suggesting that hydroxy radical-like intermediate(s) (ONOOH•) or peroxynitrous acid (ONOOH) are responsible for the damage [32]. Salgo et al. reported that benzoate and dimethyl sulfoxide amplified the breakage by reacting with peroxynitrite to form NO2, which is responsible for the increased DNA damage indicating that the free hydroxyl radical is not involved in the damage [46].
Manganese porphyrins have been shown to catalyze the peroxynitrite-induced DNA single strand breakage in an in vitro system [45]. SIN-1 induced strand breakage was inhibited by superoxide dismutase [41] as well as NO-trapping agents such as oxyhemoglobin and carboxy-PTIO [47], suggesting that simultaneous generation of NO and superoxide is necessary to cause strand breaks, supporting the assumption that these radicals react with each other to form peroxynitrite or other oxidant(s). Most of the above mentioned studies were not performed in intact cells. However, DNA single strand breakage has also been reported in intact cells exposed to peroxynitrite [10, 48, 49], indicating that extracellular peroxynitrite has the ability to enter the cells and reach the nucleus. More notably, indirect evidence supports the observation that endogenously produced peroxynitrite can also cause DNA single strand breakage during immunostimulation of various cells in addition to DNA base modifications [34, 50]. For example, the generation of DNA single strand breaks in immunostimulated macrophages parallels the production of peroxynitrite [50].
The mechanism for the formation of strand breaks by peroxynitrite includes its reaction with 2′-deoxyribose and deoxynucleosides to form malondialdehyde and base-propenals, respectively [2, 31, 32]. The initial reaction could involve hydrogen abstraction and O2 attack at either deoxyribose C4′ or C5′ by hydroxyl radical-like intermediate(s) (ONOOH•) or peroxynitrous acid (ONOOH). After the modification at C4′, the strand breakage can be induced either by C3′–(phosphate-O) cleavage or C3′–C4′ plus C1′–(ring-O) bond cleavages [51]. The strand breakage could also be induced by cleavage between C4′ and C5′, following the damage at C5′ [52]. Studies by Habib et al. and Dixit et al. have shown the single breaks resulted from peroxynitrite upon exposure to DNA. Due to this exposure, neo-epitopes were generated in the modified DNA owing to its antigenicity [53, 54].
Biological Effects of Peroxynitrite-Induced DNA Damage
Apoptosis Induced by Peroxynitrite
Once the level of cellular damage imposed by peroxynitrite supercedes any possibility of repair, the cell eventually dies through one of the two main pathways of cell demise, necrosis or apoptosis. Necrosis is associated with loss of cellular ATP, leading to membrane disruption, release of noxious cellular debris, and the development of secondary inflammation. On the contrary, apoptosis occurs in a well-strategized sequence of morphological events characterized by nuclear and cytoplasmic condensation with blebbing of the plasma membrane. The dying cell ultimately breaks up into membrane-enclosed particles termed apoptotic bodies, which are rapidly ingested and degraded by professional phagocytes or neighboring cells, without inducing any inflammatory response. Apoptosis is organized by the proteolytic activation of cysteine proteases known as caspases, that requires preserved ATP levels to proceed properly, and which may be triggered either by the activation of death receptors (extrinsic pathway) or by the permeabilization of the outer membrane of mitochondria (intrinsic pathway) [55, 56].
Several studies have shown that NO and peroxynitrite either cause acute cell death (necrosis) or delayed cell death (apoptosis) in various cell types [2, 57–59]. Sustained low level exposure of NO or peroxynitrite cause apoptosis, whereas cell necrosis results from sudden exposure to high concentrations of peroxynitrite or NO. Therefore, necrosis induced by peroxynitrite has been suggested to play an important role in inflammation.
The cellular mechanisms of peroxynitrite-induced apoptosis have not been clearly explained which is demonstrated in CHO-Em9 cells defective in their ability to repair DNA single strand breaks and unrepaired single strand breaks lead to the formation of double strand breaks ultimately resulting in cell death [60]. There are number of regulatory factors such as p53 and CPP32 which appears to determine the fate of the cell [61, 62] in relation to NO or peroxynitrite induced DNA damage.
It is noteworthy that different types of DNA injuries can cause cell cycle arrest in different phases which consecutively can affect the fate of the cell [61, 62]. It is quite clear that the NO-induced apoptosis associated with proteolytic cleavage of PARP [63], involves mechanisms that vary from the apoptosis triggered by potent DNA single strand breaking agents, such as peroxynitrite and hydroxyl radical.
Poly(ADP-ribose) Polymerase PARP Activation and Acute Cell Injury
The post-translational modification of nuclear proteins that results in the poly-adenosine diphosphate (ADP) ribosylation is called as PARylation. PAR polymerase (PARP) enzymes initiate the reaction by converting the substrate nicotinamide adenine dinucleotide (NAD+) to ADP-ribose and nicotinamide, and then catalyze ADP-ribose polymerization on nuclear acceptor proteins [64, 65]. The major acceptors of poly(ADP-ribose) are PARP itself (automodification domain), topoisomerase I and II, DNA polymerases α and β, and DNA ligase 2. The ADP-ribosylation reduced the catalytic activities of these enzymes [66]. Chromatin relaxation is also caused by the presence of ADP-ribose on histones (mainly histone H1) [66]. PAR turnover is regulated by PAR glycohydrolase (PARG), which catalyzes the degradation of poly(ADP ribose) into free ADP-ribose (ADPr) and adenosine monophosphate [67].
So far, about 17 isoforms of PARP with different structural domains and functions have been identified: PARP-1, PARP-2, PARP-3, PARP-4 (Vault-PARP), PARP-5 (Tankyrases-1 and 2), PARP-6, PARP-7 (tiPARP), PARP-8, PARP-9 (BAL1), PARP-10, PARP-11, PARP-12, PARP-13 (ZAP), PARP-14 (CoaSt6), PARP-15, and PARP-16 [68, 69]. PARP-1 is the most abundant and best characterized isoform of the PARP enzyme family. This nuclear enzyme is a highly conserved protein of 116 kDa. This protein is composed of three primary functional domains: an N-terminal DNA-binding domain (46 kDa), including a nuclear localization signal, a central auto modification domain (16 kDa), and a C-terminal catalytic domain (55 kDa). The DNA binding domain contains two zinc-finger motifs that help in binding to both single and double-stranded DNA breaks [70].
Poly ADP-ribosylation has been associated in the regulation of several physiological cellular functions such as DNA repair, gene transcription, cell cycle progression, cell death, chromatin function and genomic stability [71]. PARP-1 becomes hyperactivated in response to free radicals, reactive oxygen species and peroxynitrite resulting in the depletion of NAD+ and adenosine triphosphate (ATP) and ultimately cell death and organ dysfunction. Also, PARP-1 upregulates the expression of pro-inflammatory genes by transcription factors activation. Hence, PARP-1 plays important roles in the pathogenesis of many diseases such as stroke, myocardial infarction, circulatory shock, diabetes, neurodegenerative disorders including Parkinson’s and Alzheimer’s diseases, autoimmune diseases, allergy, asthma, colitis and other inflammatory disorders [72, 73].
Peroxynitrite has been identified as a pathophysiologically relevant trigger of PARP activation [72, 74] and it can also induce pathophysiological alterations independently from PARP as well. These alterations are numerous and include protein modifications (of which the most studied is tyrosine nitration), DNA modifications, alterations in cellular signal transduction pathways, leading to changes in inflammatory responses and promotion of cell death via apoptotic and necrotic routes [75, 76]. The pathway of cell injury involving the pronounced activation of PARP can rapidly deplete the intracellular concentration of its substrate, NAD+, retarding the rate of glycolysis, electron transport and, therefore, ATP formation resulting in acute cell dysfunction and cell death. It develops rapidly and is apparently distinguishable from apoptosis (Fig. 3). It has been suggested that there is an additive or synergistic relationship involving the PARP-dependent and the PARP-independent pathophysiological actions [76–78].
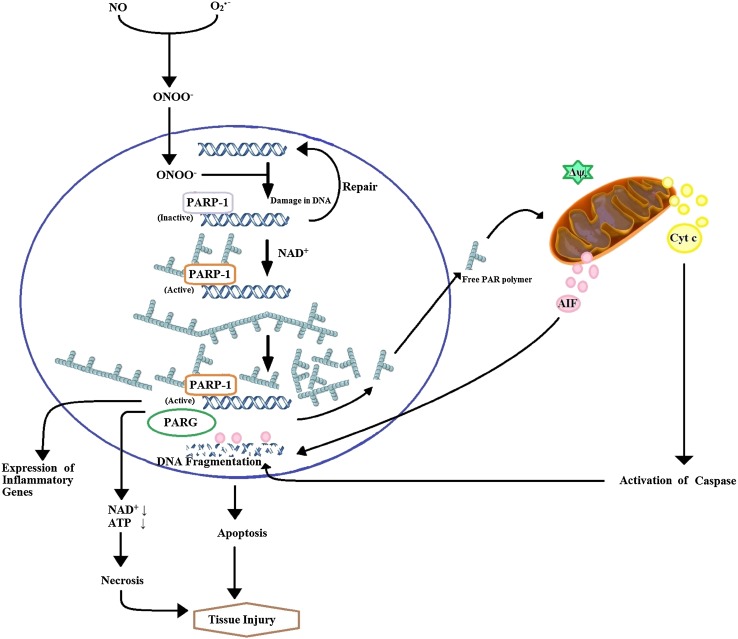
Fig. 3.
Peroxynitrite mediated downstream caspase dependent and independent apoptotic pathways
PARP activation can contribute to the development of disease through two main mechanisms: by driving the cell into an energetically deficit state of dysfunction and by catalyzing the activation of proinflammatory pathways. In the first pathway, PARP-1 functions as a DNA damage sensor and signaling molecule, binding to both single and double-stranded DNA breaks. PARP-1 forms homodimers on binding to damaged DNA and catalyzes the cleavage of NAD+ into nicotinamide and ADP-ribose to form long branches of ADP-ribose polymers on target proteins such as histones and PARP-1 itself, resulting in cellular energetic depletion, mitochondrial dysfunction and ultimately necrosis [72, 74]. But in second pathway, various transcription factors, DNA replication factors and signaling molecules have also been shown to become poly(ADP-ribosylated) by PARP-1, but a PARP-mediated activation of the pluripotent transcription factor nuclear factor-κB (NF-κB) appears to be of crucial importance [72, 79]. Notably, recent studies have suggested that PARP-1 activity can be modulated by several endogenous factors and PARP-1 can also modulate important signaling pathways [78].
The main evidence on the role of peroxynitrite and PARP in the development of disease is based on in vitro studies (including studies in human primary cells and cell lines), as well as on various animal models of disease [78, 80–86]. Evidences were accumulating on the pathophysiological pathways associated with the production of peroxynitrite and the activation of PARP specifically in human disease. The relationship between these two pathophysiological pathways is close and diverse, well supported by in vitro data and preclinical animal studies using various pharmacological inhibitors. The pathological activation of PARP requires the development of DNA single-strand breaks in vivo [87]. Theoretically, breaks in the DNA can be brought about by a range of oxidant and free radical species, including hydroxyl radical and peroxynitrite (but not by superoxide or nitric oxide alone). From numerous studies, however, it is quite clear that peroxynitrite is the key pathophysiological species that can trigger such DNA injury in vivo. Peroxynitrite can cross cell membranes, enter the nucleus and cause breaks in the strands of the DNA [74–76]. Presently, no other species is known with the capacity of travelling within and between cells and having the ability to break the DNA [76]. In vitro studies clearly demonstrate that exogenously applied or endogenously produced peroxynitrite induces DNA strand breakage and PARP activation, and peroxynitrite-induced cell death can be attenuated by pharmacological inhibition of PARP or specifically by genetic inactivation of PARP isoform [49, 88].
Besides, above mentioned roles, a new role of PARP has been discovered in regulating the mitochondria-to-nucleus translocation of apoptosis-inducing factor (AIF). AIF is a 67-kDa mitochondrial flavoprotein that is involved in cellular metabolism and apoptotic pathways [89]. It maintains mitochondrial structure [90] and plays a necessary role in oxidative phosphorylation [91] under physiological conditions where it acts as a key mediator of caspase-independent cell death under pathological conditions.
The translocation of AIF from the mitochondria to the nucleus appears to be the central event, leading to induction of chromatin condensation and large-scale (≈ 50 kb) DNA fragmentation and ultimately cell death [92]. AIF plays a pivotal role in mediating PAR polymerase-1 (PARP-1)-dependent cell death. Activation of PARP induces cell death by a mechanism that differs from apoptosis, necrosis and autophagy [93, 94]. This process involves translocation of AIF from mitochondria to the nucleus [95]. Harraz et al. coined the term “parthanatos” to PARP-1-mediated cell death in order to differentiate this process from other forms of cell death [96]. PARP-1 does not cause apoptotic body formation (unlike apoptosis), cell swelling (unlike necrosis) and also does not involve autophagic vacuole formation and lysosomal degradation like autophagy. Parthanatos results in phosphatidylserine flipping onto the outer plasma membrane, dissipation of the mitochondrial membrane potential, chromatin condensation and large DNA fragmentation [97]. Reduction in AIF nuclear translocation has been shown by pharmacological inhibition of PARP activity [98].
Peroxynitrite-Induced DNA Damage: Aspect in Inflammation-Linked Cancer
Several risk factors that are recognized for human cancers at various sites include chronic infection by bacteria, parasites, or viruses and tissue inflammation such as gastritis, hepatitis and colitis [99, 100]. Nitric oxide and other oxygen radicals generated in infected and inflamed tissues have been suggested as one of the contributing factors to the multistage process of carcinogenesis by damaging DNA and tissues. Since peroxynitrite is currently considered to be a major compound responsible for tissue damage caused by inflammation, it may also play an essential role in carcinogenesis [101].
Peroxynitrite causes numerous types of modifications in DNA and induces strand breakage. The transversions comprising of G:C → T:A are very common in a variety of genes from all types of human cancers [102] and these transversions account for 30 % of all p53 gene mutations in lung cancer and are also high in liver and breast cancer. It has been suggested that G:C → T:A transversions are mostly caused by polycyclic aromatic hydrocarbons such as benzo(a)pyrene present in tobacco smoke. Nonetheless, peroxynitrite also caused G:C → T:A transversions. NO (formed endogenously by NO synthase in the lung) and superoxide may react to form peroxynitrite in the lungs of smokers and play an essential role in smoking-related diseases like lung cancer. Similarly, the reaction between catechol–estrogens and nitric oxide can generate oxidants which are analogous to peroxynitrite [103]. NO is produced by constitutive and inducible types of NO synthases in human breast tissues [104, 105] which may react with superoxide generated from catechol–estrogens to produce peroxynitrite and can induce the G:C → T:A transversions observed commonly in breast tumors.
There are other numerous sites where inflammation, peroxynitrite production and DNA injury have been linked to carcinogenesis which include Helicobacter pylori infection, gastritis and gastric cancer [106–109], as well as ulcerative colitis and colon cancer [110–112].
Role in Diseases
Various diseases such as diabetes, atherosclerosis, ischemic heart diseases, stroke, autoimmune and neurodegenerative diseases are resulted from the disruption of mitochondrial functions. Peroxynitrite may be either directly produced within the mitochondria or may reach mitochondria from extra mitochondrial compartments. Consecutively, peroxynitrite nitrates and inhibits Mn-SOD [113] and therefore preventing the breakdown of locally produced superoxide, which further increases the formation of peroxynitrite. The peroxynitrite toxicity in mitochondria results from both direct oxidative reactions and free radical-mediated damage [114, 115], secondary to peroxynitrite reacting with CO2, giving rise to CO•−3 and NO•2 radicals. The latter reaction is largely favored within mitochondria, which are the main organelles where CO2 is produced during decarboxylation reactions [114, 115]. Peroxynitrite further reduces energy metabolism by inhibiting the tricarboxylic acid cycle enzyme aconitase, located in the mitochondrial matrix, through oxidative disruption of the 4Fe–4S center of the enzyme [116, 117], as well as mitochondrial creatine kinase, which is present in the intermembrane space [118].
Diabetes
Diabetes is one of the major chronic diseases with an estimated worldwide dominance of 170 million in 2002 and is expected to double by 2030 according to the World Health Organization [119]. In this disorder, system’s ability to regulate blood glucose level in a physiological (5 mM) range is altered, either by a reduced production of insulin as a result of pancreatic β-cell destruction (Type 1) or by insulin resistance (Type 2). Primary Diabetes (Type 1 diabetes) is caused by the autoimmune destruction of insulin producing β-cells of the pancreatic islet resulting in prolonged periods of hyperglycemia via reduced uptake of glucose and relative increase in glucagon secretion and gluconeogenesis. However, the resulting long-term effects are far more dangerous than acute hyperglycemia itself. The endothelial cells are damaged in the vessels by chronic hyperglycemia and in fact most of the diabetic chronic complications result from pathological alterations of the micro and macrovascular system, leading to increased risk of blindness, infarction of brain and heart, kidney failure and impaired wound healing [120–122].
Atherosclerosis is the most common macrovascular complication of diabetes which increases the risk for myocardial infarction, stroke and peripheral artery disease, the latter being the principal cause of limb amputation in civilized countries whereas microvascular complications consist of retinopathy and nephropathy which are the leading causes of blindness and renal failure respectively [123]. Diabetic cardiomyopathy is a complication resulting from diabetes and is characterized by complex changes in the mechanical, structural, biochemical and electrical properties of the heart, which may underlie the development of an early diastolic and a late systolic dysfunction, or both, and increased incidence of cardiac arrhythmias in diabetic patients [124].
Considerable evidences support the pathogenetic role of endogenous peroxynitrite formation in diabetic cardiovascular complications both in experimental animals and in humans [125–127]. Peroxynitrite has been reported to attack different biological macromolecules, leading to compromised cardiovascular function in diabetes through numerous mechanisms. One of these pathways involves DNA strand breakage and consequent activation of the nuclear enzyme PARP. This PARP activation (specifically PARP-1 isoform) is a crucial process in the development of diabetic cardiovascular dysfunction both in diabetic animals and humans [128–130] and may also contribute to the progress of other diabetic complications such as neuropathy, nephropathy and retinopathy [84]. The kidney biopsies of patients having diabetic nephropathy and of diabetic animals have shown increased peroxynitrite formation and oxidative stress [131, 132]. There is an elevation of superoxide anion and peroxynitrite formation in retinal endothelial cells maintained in high glucose [133] and in retinas of diabetic animals [133, 134] and these alterations could be attenuated with various antioxidants like NOS inhibitors or peroxynitrite scavengers [133–135]. Also, increased formation of peroxynitrite has been documented in both experimental [135] and clinical diabetic neuropathy [136]. A novel mechanism has been identified in which PARP activation induces various pathways leading to diabetic complications [137]. According to this concept, hyperglycemia induced superoxide generation from the mitochondria (directly or indirectly via generation of peroxynitrite) induces DNA strand breaks and PARP activation which, in turn, induces the poly ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The resulting metabolic alterations activate NFκB, polyol pathway and aldose reductase leading to increased oxidative stress [137]. Studies have shown that inhibition of PARPs diminished the concomitant NAD+ loss and prevented PAR formation [138]. Similarly, isolated β-cells from Parp1 knockout animals were protected from cell death induced by ROS [139]. In streptozotocin-induced diabetes, Parp1 knockout mice were protected from β-cell loss and subsequent type 1 diabetes. Gene dosage appeared to be essential as Parp1+/− animals were partially protected [140].
Animals were also protected after altering the application of streptozotocin from a single high dose to multiple low-dose injections, if PARP enzymes were inhibited or the Parp1 gene was disrupted [141]. Hence, inhibiting PARP activity directly suppresses islet cell loss and therefore prevents the onset of type 1 diabetes. The increased cellular damage also induces DNA strand breaks, which in turn activates PARP. PARP inhibitors significantly improved diastolic dysfunction of the heart and loss of endothelium-dependent vasodilation [129]. The administration of PARP inhibitor PJ34 even one week after onset of diabetes was effective in spite of persistent hyperglycemia [129]. Rats with streptozotocin-induced diabetes showed increased heart dysfunction and a larger infarcted area after myocardial ischemia–reperfusion injury as well as higher mortality rates during reperfusion. Blocking PARP activity with the inhibitor INO-1001 improved myocardial function in both groups. In diabetic rats, INO-1001 significantly lowered the mortality during the experiment [142]. The increase in nuclear AIF after ischemia–reperfusion was prevented in both control and diabetic groups. These potent PARP inhibitors reduce the clinical complications usually seen in diabetes and therefore considered to be therapeutically good candidates. But PARP inhibition has been shown to promote cancer formation under genotoxic stress [143]. Likewise, because many of the cellular functions are regulated by poly ADP-ribosylation, very specific inhibitors against few isoforms of PARP have to be designed [144].
Cardiovascular System Diseases
The one of the major causes of death in developed countries are diseases of cardiovascular system. Therefore, improvement in treatment will have a great impact on mortality rates and average life span. After challenge with oxidants like peroxynitrite and hydrogen peroxide, PARP activation was detected in cardiomyoblasts, but not with NO-donors (S-nitroso-N-acetyl-dl-penicillamine, diethyltriamine NONOate) [145]. All these chemicals cause reduction in mitochondrial respiration. Furthermore, hypoxia and re-oxygenation stimulated PAR formation. Unsurprisingly, PARP activity was suppressed by addition of 3AB and nicotinamide in all cases. In Parp-1 knockout mouse fibroblasts, NO-donors diminished mitochondrial respiration, whereas peroxynitrite and hydrogen peroxide did not. In brief, PARP activation depends on DNA damage induced by peroxynitrite or hydrogen peroxide. In this case, mitochondrial respiratory failure can be rescued by PARP inhibitors, but not if reduction of mitochondrial respiration is achieved by NO-donors, which do not activate PARPs.
Thiemermann and co-workers showed that PARP inhibitors (3AB, nicotinamide and 5-ISO, 1,5-isoquinolinediol) improved heart functionality and reduced infarct size after occlusion and reperfusion of the left coronary artery in ischemia–reperfusion of rabbit hearts whereas structurally similar compounds like 3-AB (3-aminobenzoic acid) and nicotinic acid, which did not inhibit PARPs, were not effective [146]. Similarly, Zingarelli and colleagues reported similar effects in rats, and additionally elevated necrosis, neutrophil infiltration and nitrotyrosine formation with loss of ATP. 3AB diminished neutrophil activation and partially preserved myocardial ATP levels [147]. The application of 3AB also proved effective in reducing infarct size and contractile dysfunction in different animal model (pigs) [148].
In the Parp1 knockout mouse model, it was shown that INO-1001 attenuated heart remodeling after ischemia–reperfusion injury (hypertrophy and formation of collagen in the hearts) in addition to preserving functionality and translocation of AIF into the nucleus [149]. In a cell culture-model of cardiomyoblasts, hypoxia followed by re-oxygenation led to oxidative stress, PARP activation and a drop in energy-metabolite levels (NAD+ and ATP) [150]. Therefore, necrosis was the common pathway of death and AIF dependent apoptosis was increased. The administering of PJ34 attenuated the decline in levels of NAD+ and ATP, possibly due to reduced PAR formation. A shifting took place from caspase-independent to caspase-dependent apoptosis. Besides cell survival, PARP inhibition also modulates the way in which cells die, changing it from the pro-inflammatory necrosis to the less harmful apoptosis. Pacher and co-workers showed that inhibition of poly(ADP-ribose) polymerases is also effective in a chronic heart-failure model [151]. They ligated the left anterior descending coronary artery in rats and tested functionality and stress parameters. The levels of nitrotyrosine and PAR were increased in controls and the performance of the left ventricle was reduced and relaxation was impaired (measured ex vivo). PJ34 facilitated the reduction of PAR amount (as expected) but had no significant impact on nitrotyrosine formation. Hence, it was not the early damage (nitrotyrosine level), but the activity of PARPs (polymer amount) that was suppressed.
In conclusion, PARP inhibition possibly does not reduce initial damage, but diminishes the ongoing formation of reactive compounds like NO• and subsequently peroxynitrite. This results in preservation of energy metabolites and blocks the translocation of AIF from the mitochondria into the nucleus. As a result, necrosis is reduced as well as overall cell death. Inflammation (neutrophil activation) is suppressed and the resulting morphological changes like hypertrophy are reduced. These all collectively lead to an improved functionality of the heart and a reduction in infarct size [86, 152, 153].
Neurodegenerative Diseases
Neurodegenerative diseases consist of different conditions which primarily affect the neurons in the human brain and spinal cord leading to either functional loss (ataxia) or sensory dysfunction (dementia). Peroxynitrite formation has been associated to neurodegenerative diseases like Alzheimer’s disease, Parkinson’s disease, Amyotrophic lateral sclerosis (ALS) and other brain pathologies like Huntington’s disease, multiple sclerosis (MS) [154–157]. The immunoreactivity of nitrotyrosine was established in early stages of all of these diseases in human autopsy samples as well as in experimental animal models. Elevated levels of nitrite, nitrate and free nitrotyrosine have been found to be present in the cerebral spinal fluid (CSF) and proposed to be useful marker of neurodegeneration [154–157]. When peroxynitrite is formed in diseased brain, it may exert its toxic effects through multiple mechanisms, including lipid peroxidation, mitochondrial damage, protein nitration and oxidation, depletion of antioxidant reserves (especially glutathione), activation or inhibition of various signaling pathways, and DNA damage followed by the activation of the nuclear enzyme PARP [155–158].
Alzheimer’s Disease (AD)
It is a progressive neurodegenerative disorder which is characterized by the formation of neuritic plaques rich in β-amyloid (Aβ) peptide, neurofibrillary tangles rich in hyperphosphorylated tau protein, gliosis and a neuroinflammatory response involving astrocytes and microglia, certainly leading to progressive global cognitive decline, and accounting for the vast majority of age-related dementia [159]. This disease is known to cause loss of functional neurons from several regions of the brain, although the precise mechanisms of cell loss are still unclear, but several proposed mechanisms ultimately converge on disrupted calcium signaling [160].
It appears that increased oxidative stress is an early event in the process of neurodegeneration associated with Alzheimer’s disease [161–166] and both neuronal and glial NOS may play a role in the pathogenesis of AD and peroxynitrite formation. Expression of nNOS was increased in neurons with neurofibrillary tangles in the hippocampus and enthorinal cortex of AD patients as well as in reactive astrocytes near amyloid plaques [167, 168].
Neuropathological studies have revealed fragmentation of nuclear DNA (by use of in situ end-labelling, ISEL) in a much higher proportion of neurons, oligodendrocytes, astrocytes and microglia in the brains of patients with Alzheimer’s disease than in age-matched controls [169–172]. This led to the interpretation that the loss of neurons is due to apoptosis, probably initiated by Aβ-protein or other inducers of oxidative stress. The extent of apoptosis which is morphologically evident in Alzheimer’s disease is very limited and difficult to reconcile with the high density of ISEL-positive cells. It remains unconfirmed that the fragmentation of nuclear DNA in Alzheimer’s disease denotes apoptosis, particularly in the context of non-mitotic cells.
The connection between DNA damage and oxidative stress is known to activate DNA repair proteins, including poly(ADPribose) polymerase (PARP). PARP is stimulated in response to free radical-mediated injury to DNA after brain ischaemia and reperfusion [173]. Its overactivation in injured cells can cause massive consumption of NAD+ and finally resulting in cell death due to energy depletion [173–175]. Love and co-workers have shown that increased expression of PARP and intranuclear accumulation of its end-product poly(ADP-ribose) in neurons in brains from patients with Alzheimer’s disease than of control brains. So, there may be the possibility that overactivation of PARP may contribute to neuronal loss in Alzheimer’s disease [176].
Parkinson Disease (PD)
It is the second most common neurodegenerative disease of adult onset, characterized by progressive loss of dopaminergic neurons within substantia nigra pars reticulata (SNr), resulting in reduced dopamine levels and a loss of dopaminergic neurotransmission in the striatum, which interferes with the function of the basal ganglia critical to motor function and coordination [177, 178]. The common symptoms are hypokinesia, rigidity, resting tremor and postural instability. The characteristics of the disease are Lewy body inclusions in the dopaminergic cells and cell loss in the substantia nigra (SN). Similar characteristics are accompanied by Lewy bodies in the cortical neurons and consequent dementia in diffuse Lewy body disease (DLBD). Both oxidative stress and excitotoxic injury play a significant role in the degeneration mechanism of the dopaminergic neurons in the SN in PD [179], resulting in intraneuronal calcium level increase and can lead to the activation of PARP to repair DNA damage. The excessive activation of PARP leads to massive consumption of NAD+ and subsequently of ATP resulting in cell death due to energy depletion [180]. PARP is needed as co-activator for the nuclear translocation and transcriptional activation of NF-κB in the nuclei of the dopaminergic neurons in PD [181], which has been involved in the regulation of genes involved in immune and inflammatory responses, cell survival, apoptosis, development, differentiation, cell growth and neoplastic transformation [182]. This ultimately leads to apoptosis by its influence on p53 gene expression [183].
Soós and co-workers using immunohistochemical methods confirmed that the expression of PARP is increased together with the nuclear translocation of NF-κB in the dopaminergic cells in the SN in PD and DLBD patients as compared with age-matched control patients with other neurodegenerative diseases and with normal controls [184]. This significant increase in PARP expression in the nuclei of a subset of dopaminergic neurons in the SN in PD and DLBD can reflect both oxidative damage and an increased intracellular calcium level leading to neuronal death by over-activation of PARP.
Besides PARP, peroxynitrite may also induce nitration of tyrosine hydroxylase, the initial and rate-limiting enzyme in the biosynthesis of dopamine, leading to inhibition of enzyme activity and consequent failure in the synthesis of dopamine [185]. The nitration of tyrosine residues in tyrosine hydroxylase paralleled the decline in dopamine levels in mouse striatum following 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) administration [185]. Tyrosine residues are also crucial for the substrate specificity of monoamine oxidase B (MAO B) resulting in impaired dopamine catabolism once nitrated [186]. Peroxynitrite also involved in the loss of intracellular glutathione from substantia nigra (an early event in PD) by inactivating glutathione reductase, the enzyme responsible for the regeneration of glutathione from its oxidized form [187, 188], and to induce apoptosis in dopaminergic neurons in PD [189]. Some recent evidences suggested that mitochondrial complex I inhibition may be the central cause of sporadic PD and these derangements in complex I lead to α-synuclein aggregation, which contributes to the demise of dopamine neurons [190]. Accumulation and aggregation of α-synuclein may further assist the death of dopamine neurons through impairments in protein handling and detoxification [190].
Amyotrophic Lateral Sclerosis (ALS)
It is also known as Lou Gehrig’s disease or motor neuron disease and most common adult-onset progressive fatal neurodegenerative disease, which is characterized by rapid, progressive degeneration of both upper and lower motor neurons in the motor cortex, brain stem, and spinal cord. This finally leads to progressive weakness, paralysis and premature death within 3–5 years after the onset of symptoms [191, 192]. Patients are cognitively intact and therefore completely aware of their progressive disability. About ~10 % have familial inheritance, usually with an autosomal dominant pattern, while most ALS cases are sporadic with no genetic basis. Various mechanisms like neuroinflammation, oxidative stress (in sporadic ALS, sALS), autoimmunity, defect in neuronal glutamate transport, neurofilament accumulation, exogenous factors (viruses, toxins), mitochondrial dysfunction and mutations in the SOD gene were implicated to play a role, but the pathogenesis of ALS is incompletely understood [164, 192–195].
Protein and lipid oxidation markers are elevated in postmortem tissues of sALS [196–199]. Additionally, the level of 8-hydroxy-2′-deoxyguanosine (8OH2′dG), a marker of oxidative injury of DNA, increases with disease progression [197] and correlates with disease severity [200]. These alterations were originally supposed to be mostly confined to neurons. But, recent studies using spinal cords of sALS patients or ALS animal models exhibit that markers of oxidative stress have been localized not only to motor neurons but also astrocytes, microglia, and macrophages [198–203].
DNA damage as a result of oxidation, alkylation or ionizing radiation is associated with activation of PARP [204–207]. When PARP is activated by DNA strand breaks, it subsequently transfers branched chains of ADP-ribose to a variety of nuclear proteins, including PARP itself [208]. It can also be activated by inositol 1,4,5,-triphosphate-Ca2+ mobilization without DNA damage [209]. Ca2+ promotes PARP activation and Ca2+ acts as an activating factor in PARP-mediated cell killing [210, 211]. Even though PARP facilitates DNA repair [212], excessive PARP activation can cause massive consumption of its substrate NAD+ and results in cell death due to energy depletion [205, 208, 210, 213–215]. Recently, studies have shown that FUS (fused-in-sarcoma, member of the heterogeneous nuclear ribonucleoprotein family of proteins that bind thousands of pre-mRNAs and can regulate their splicing) is a component of the PARP-1 dependent response to oxidative chromosomal DNA damage and defect in this response might contribute to ALS [216].
Protein nitration may also play a role in ALS pathogenesis, acting directly by inhibiting the function of specific proteins and indirectly interfering with protein degradation pathways and phosphorylation cascades [217]. Peroxynitrite can participate in the pathogenesis of ALS, because it can activate spinal cord astrocytes, which normally provide excellent trophic support to motor neurons, to assume a reactive phenotype that induces the death of motor neurons [218]. Reactive astrocytes are common characteristic of neurodegeneration. The transformation of astrocytes into reactive astrocytes is the reason behind progressive nature of ALS and cause the relentless death of neighboring motor neurons.
The involvement of PARP has been emphasized as a pathogenic mechanism in MPTP-induced Parkinson’s disease [219–221], Alzheimer’s disease [222] and ischemic brain injury [174, 222]. Moreover, PARP inhibitors have been proposed as potential therapies in neurodegenerative diseases in which oxidative stress is suspected to play an important pathogenic role [207, 219–226].
Conclusions
Peroxynitrite is a charged strong oxidant capable of modifying various types of biological macromolecules to produce variety of products. It primarily acts as a trigger for multiple forms of DNA damage (Base modifications, single strand breakage, and apoptotic double strand breakages). It is known to induce damage in the DNA bases predominantly at guanine and 8-oxoG nucleobases through thermodynamically and kinetically favorable oxidation reactions. Peroxynitrite induced DNA damage activates PARP and it catalyzes ADP-ribose polymerization on nuclear acceptor proteins by converting the substrate NAD+ to ADP-ribose and nicotinamide, triggering caspase-independent cell death through Apoptosis Inducing Factor. It helps in upregulation of pro-inflammatory genes expression by activating transcription factors. As a result, activated PARP plays an important role in pathogenesis of numerous diseases like diabetes, cardiovascular diseases and neurodegenerative diseases. Several strategies were suggested for the inhibition of peroxynitrite-induced damage. One of them involves targeting of cytotoxic pathways stimulated by peroxynitrite which is now becoming a feasible approach to alleviate disease signs in numerous disorders. Controlled blocking of the activity of PARP could provide a possible direction to overcome such oxidative and nitrosative damages. But excess of DNA damage results in over stimulation of PARP, leading to an energy deficit state and ultimately necrsosis. So, there is a need for exploration of more beneficial approaches in future.
Acknowledgments
One of the authors B. U. I. is thankful to UGC-MANF for financial support as senior research fellow. Assistance from the Institution (AMU) as well as infrastructural support from DST-FIST to the department is also duly acknowledged.
References
- 1.Zingarelli B, Day BJ, Crapo J, Salzman AL, Szabó C. The potential involvement of peroxynitrite in the pathogenesis of endotoxic shock. Br J Pharmacol. 1997;120:259–267. doi: 10.1038/sj.bjp.0700872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Radi R, Peluffo G, Alvarez MN, Naviliat M, Cayota A. Unraveling peroxynitrite formation in biological systems. Free Radic Biol Med. 2001;30:463–488. doi: 10.1016/S0891-5849(00)00373-7. [DOI] [PubMed] [Google Scholar]
- 4.Denicola A, Souza JM, Radi R. Diffusion of peroxynitrite across erythrocyte membranes. Proc Natl Acad Sci USA. 1998;95:3566–3571. doi: 10.1073/pnas.95.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls: the cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 6.Bartesaghi S, Valez V, Trujillo M, Peluffo G, Romero N, Zhang H, Kalyanaraman B, Radi R. Mechanistic studies of peroxynitrite-mediated tyrosine nitration in membranes using the hydrophobic probe N-t-BOC-l-tyrosine tert-butyl ester. Biochemistry. 2006;45:6813–6825. doi: 10.1021/bi060363x. [DOI] [PubMed] [Google Scholar]
- 7.Radi R. NO, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quijano C, Alvarez B, Gatti R, Augusto O, Radi R. Pathways of peroxynitrite oxidation of thiol groups. Biochem J. 1997;322:167–173. doi: 10.1042/bj3220167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonini MG, Augusto O. Carbon dioxide stimulates the production of thiyl, sulfinyl, and disulfide radical anion from thiol oxidation by peroxynitrite. J Biol Chem. 2001;276:9749–9754. doi: 10.1074/jbc.M008456200. [DOI] [PubMed] [Google Scholar]
- 10.Salgo MG, Bermudez E, Squadrito GL, Pryor WA. Peroxynitrite causes DNA damage and oxidation of thiols in rat thymocytes. Arch Biochem Biophys. 1995;322:500–505. doi: 10.1006/abbi.1995.1493. [DOI] [PubMed] [Google Scholar]
- 11.Szabó C, Ohshima H. DNA damage induced by peroxynitrite: subsequent biological effects. Nitric Oxide. 1997;1:373–385. doi: 10.1006/niox.1997.0143. [DOI] [PubMed] [Google Scholar]
- 12.Burney S, Niles JC, Dedon PC, Tannenbaum SR. DNA damage in deoxynucleosides and oligonucleotides treated with peroxynitrite. Chem Res Toxicol. 1999;12:513–520. doi: 10.1021/tx980254m. [DOI] [PubMed] [Google Scholar]
- 13.Niles JC, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced oxidation and nitration products of guanine and 8-oxoguanine: structures and mechanisms of product formation. Nitric Oxide. 2006;14:109–121. doi: 10.1016/j.niox.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy LJ, Moore K, Jr, Caulfield JL, Tannenbaum SR, Dedon PC. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem Res Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- 15.Villa LM, Salas E, Darley-Usmar VM, Radomski MW, Moncada S. Peroxynitrite induces both vasodilatation and impaired vascular relaxation in the isolated perfused rat heart. Proc Natl Acad Sci USA. 1994;91:12383–12387. doi: 10.1073/pnas.91.26.12383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubbo H, Radi R, Trujillo M, Telleri R, Kalyanaraman B, Barnes S, Kirk M, Freeman BA. Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives. J Biol Chem. 1994;269:26066–26075. [PubMed] [Google Scholar]
- 17.Violi F, Marino R, Milite MT, Loffredo L. NO and its role in lipid peroxidation. Diabetes Metab Res Rev. 1999;15:283–288. doi: 10.1002/(SICI)1520-7560(199907/08)15:4<283::AID-DMRR42>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Moore KP, Darley-Usmar V, Morrow J, Roberts LJ. Formation of F2-isoprostanes during oxidation of human low-density lipoprotein and plasma by peroxynitrite. Circ Res. 1995;77:335–341. doi: 10.1161/01.RES.77.2.335. [DOI] [PubMed] [Google Scholar]
- 19.Wright MM, Schopfer FJ, Baker PR, Vidyasagar V, Powell P, Chumley P, Iles KE, Freeman BA, Agarwal A. Fatty acid transduction of nitric oxide signaling: nitrolinoleic acid potently activates endothelial heme oxygenase 1 expression. Proc Natl Acad Sci USA. 2006;103:4299–4304. doi: 10.1073/pnas.0506541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faulkner KM, Liochev SI, Fridowich I. Stable Mn(III) prophyrins mimic cuperoxide dismutase in vitro and substitute for it in vivo. J Biol Chem. 1994;269:23471–23476. [PubMed] [Google Scholar]
- 21.Yermilov V, Rubio J, Becchi M, Friesen MD, Pignatelli B, Ohshima H. Formation of 8-nitroguanine by the reaction of guanine with peroxynitrite in vitro. Carcinogenesis. 1995;16:2045–2050. doi: 10.1093/carcin/16.9.2045. [DOI] [PubMed] [Google Scholar]
- 22.Ischiropoulos H, Zhu L, Beckman JS. Peroxynitrite formation from macrophage-derived nitric oxide. Arch Biochem Biophys. 1992;298:446–451. doi: 10.1016/0003-9861(92)90433-W. [DOI] [PubMed] [Google Scholar]
- 23.Lewis RS, Tamir S, Tannenbaum SR, Deen WM. Kinetic analysis of the fate of nitric oxide synthesized by macrophages in vitro. J Biol Chem. 1995;270:29350–29355. doi: 10.1074/jbc.270.10.5057. [DOI] [PubMed] [Google Scholar]
- 24.Steenken S, Jovanovic SV. How easily oxidizable is DNA? One electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J Am Chem Soc. 1997;119:617–618. doi: 10.1021/ja962255b. [DOI] [Google Scholar]
- 25.Stanbury DM. Reduction potentials involving inorganic free radicals in aqueous solution. Adv Inorg Chem. 1989;33:69–138. doi: 10.1016/S0898-8838(08)60194-4. [DOI] [Google Scholar]
- 26.Huie RE, Clifton CL, Neta P. Electron transfer reaction rates and equilibria of the carbonate and sulfate radical anions. Radiat Phys Chem. 1991;38:477–481. [Google Scholar]
- 27.Steenken S. Purine-bases, nucleosides and nucleotides-aqueous solution redox chemistry and transformation reactions of their radical cations and e − and & ·OH adducts. Chem Rev. 1989;89:503–520. doi: 10.1021/cr00093a003. [DOI] [Google Scholar]
- 28.Shafirovich V, Dourandin A, Huang WD, Geacintov NE. The carbonate radical is a site-selective oxidizing agent of guanine in double-stranded oligonucleotides. J Biol Chem. 2001;276:24621–24626. doi: 10.1074/jbc.M101131200. [DOI] [PubMed] [Google Scholar]
- 29.Koppenol WH, Moreno JJ, Pryor WA, Ischiropoulos H, Beckman JS. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem Res Toxicol. 1992;5:834–842. doi: 10.1021/tx00030a017. [DOI] [PubMed] [Google Scholar]
- 30.Douki T, Cadet J. Peroxynitrite mediated oxidation of purine bases of nucleosides and isolated DNA. Free Radicals Res. 1996;24:369–380. doi: 10.3109/10715769609088035. [DOI] [PubMed] [Google Scholar]
- 31.Rubio J, Yermilov V, Ohshima H. DNA damage induced by peroxynitrite: Formation of 8-nitroguanine and base propenals. In The Biology of Nitric Oxide (Mon cada, S., Stamler J, Gross S, and Higgs EA, Eds.) Portland Press proceedings, London; 1996. p. 34 part 5.
- 32.Yermilov V, Yoshie Y, Rubio J, Ohshima H. Effects of carbon dioxide/bicarbonate on induction of DNA single-strand breaks and formation of 8-nitroguanine, 8-oxo-guanine and base-propenal mediated by peroxynitrite. FEBS Lett. 1996;399:67–70. doi: 10.1016/S0014-5793(96)01288-4. [DOI] [PubMed] [Google Scholar]
- 33.Yermilov V, Rubio J, Ohshima H. Formation of 8-nitroguanine in DNA treated with peroxynitrite in vitro and its rapid removal from DNA by depurination. FEBS Lett. 1995;376:207–210. doi: 10.1016/0014-5793(95)01281-6. [DOI] [PubMed] [Google Scholar]
- 34.Spencer JP, Wong J, Jenner A, Aruoma OI, Cross CE, Halliwell B. Base modification and strand breakage in isolated calf thymus DNA and in DNA from human skin epidermal keratinocytes exposed to peroxynitrite or 3-morpholinosydnonimine. Chem Res Toxicol. 1996;9:1152–1158. doi: 10.1021/tx960084i. [DOI] [PubMed] [Google Scholar]
- 35.Cadet J, Berger M, Buchko GW, Joshi PC, Raoul S, Ravanat J-L. 2,2-Diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-D-erythro-pentofuranosyl)amino-5-(2H) oxazolone: a novel and predominant radical oxidation product of 3ʹ, 5ʹ-di-O-acetyl-2ʹ-deoxyguanosine. J Am Chem Soc. 1994;116:7403–7404. doi: 10.1021/ja00095a052. [DOI] [Google Scholar]
- 36.Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin and guanidinohydantoin are the dominant products of 8-oxoguanosine oxidation at low fluxes of peroxynitrite: mechanistic studies with O-18. Chem Res Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- 37.Helbock HJ, Beckman KB, Shigenaga MK, Walter PB, Woodall AA, Yeo HC, Ames BN. DNA oxidation matters: the HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc Natl Acad Sci USA. 1998;95:288–293. doi: 10.1073/pnas.95.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uppu RM, Cueto R, Squadrito GL, Salgo MG, Pryor WA. Competitive reactions of peroxynitrite with 2ʹ-deoxyguanosine and 7,8-dihydro-8-oxo-2ʹ-deoxyguanosine (8-oxodG): relevance to the formation of 8-oxodG in DNA exposed to peroxynitrite. Free Radic Biol Med. 1996;21:407–411. doi: 10.1016/0891-5849(96)00220-1. [DOI] [PubMed] [Google Scholar]
- 39.Inoue S, Kawanishi S. Oxidative DNA damage induced by simultaneous generation of nitric oxide and superoxide. FEBS Lett. 1995;371:86–88. doi: 10.1016/0014-5793(95)00873-8. [DOI] [PubMed] [Google Scholar]
- 40.Fiala ES, Sodum RS, Bhattacharya M, Li H. (-)-Epigallocatechin gallate, a polyphenolic tea antioxidant, inhibits peroxynitrite-mediated formation of 8-oxo-deoxyguanosine and 3-nitrotyrosine. Experientia. 1996;52:922–926. doi: 10.1007/BF01938881. [DOI] [PubMed] [Google Scholar]
- 41.Epe B, Ballmaier D, Roussyn I, Briviba K, Sies H. DNA damage by peroxynitrite characterized with DNA repair enzymes. Nucleic Acids Res. 1996;24:4105–4110. doi: 10.1093/nar/24.21.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeRojas-Walker T, Tamir S, Ji H, Wishnok JS, Tannenbaum SR. Nitric oxide induces oxidative damage in addition to deamination in macrophage DNA. Chem Res Toxicol. 1995;8:473–477. doi: 10.1021/tx00045a020. [DOI] [PubMed] [Google Scholar]
- 43.Kaneko K, Akuta T, Sawa T, Kim HW, Fujii S, Okamoto T, Nakayama H, Ohigashi H, Murakami A, Akaike T. Mutagenicity of 8-nitroguanosine, a product of nitrative nucleoside modification by reactive nitrogen oxides, in mammalian cells. Cancer Lett. 2008;262:239–247. doi: 10.1016/j.canlet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki N, Yasui M, Geacintov NE, Shafirovich V, Shibutani S. Miscoding events during DNA synthesis past the nitration-damaged base 8-nitroguanine. Biochemistry. 2005;44:9238–9245. doi: 10.1021/bi050276p. [DOI] [PubMed] [Google Scholar]
- 45.Groves JT, Marla SS. Peroxynitrite-induced DNA strand scission mediated by a manganese porphyrin. J Am Chem Soc. 1995;117:9578–9579. doi: 10.1021/ja00142a032. [DOI] [Google Scholar]
- 46.Salgo MG, Stone K, Squadrito GL, Battista JR, Pryor WA. Peroxynitrite causes DNA nicks in plasmid pBR322. Biochem Biophys Res Commun. 1995;210:1025–1030. doi: 10.1006/bbrc.1995.1759. [DOI] [PubMed] [Google Scholar]
- 47.Yoshie Y, Ohshima H. Nitric oxide synergistically enhances DNA strand breakage induced by polyhydroxyaromatic compounds, but inhibits that induced by the Fenton reaction. Arch Biochem Biophys. 1997;342:13–21. doi: 10.1006/abbi.1997.0100. [DOI] [PubMed] [Google Scholar]
- 48.Kennedy M, Szabó C, Salzman AL. Activation of poly(ADP-ribose) synthetase mediates hyperpermeability induced by peroxynitrite in human intestinal epithelial cells. Crit Care Med. 1997;5(Suppl):A68. [Google Scholar]
- 49.Szabó C, Zingarelli B, O’Connor M, Salzman AL. DNA strand breakage, activation of poly-ADP ribosyl synthetase, and cellular energy depletion are involved in the cytotoxicity in macrophages and smooth muscle cells exposed to peroxynitrite. Proc Natl Acad Sci USA. 1996;93:1753–1758. doi: 10.1073/pnas.93.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zingarelli B, O’Connor M, Wong H, Salzman AL, Szabó C. Peroxynitrite-mediated DNA strand breakage activates poly-ADP ribosyl synthetase and causes cellular energy depletion in macrophages stimulated with bacterial lipopolysaccharide. J Immunol. 1996;156:350–358. [PubMed] [Google Scholar]
- 51.Burger RM, Drlica K, Birdsall B. The DNA cleavage pathway of iron bleomycin. Strand scission precedes deoxyribose 3-phosphate bond cleavage. J Biol Chem. 1994;269:25978–25985. [PubMed] [Google Scholar]
- 52.Shulte-Frohlinde D, von Sonntag C. Radiolysis of DNA and model systems in the presence of oxygen. In: Sies H, editor. Oxidative stress. London: Academic Press; 1985. pp. 11–40. [Google Scholar]
- 53.Habib S, Moinuddin, Ali R. Acquired antigenicity of DNA after modification with peroxynitrite. Int J Biol Macromol. 2005;35:221–225. doi: 10.1016/j.ijbiomac.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 54.Dixit K, Moinuddin, Ali A. Immunological studies on peroxynitrite modified human DNA. Life Sci. 2005;77:2626–2642. doi: 10.1016/j.lfs.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 55.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/S0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 56.Bouchier-Hayes L, Lartigue L, Newmeyer DD. Mitochondria: pharmacological manipulation of cell death. J Clin Invest. 2005;115:2640–2647. doi: 10.1172/JCI26274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beckman JS, Carson M, Smith CD, Koppenol WH. ALS, SOD and peroxynitrite. Nature. 1993;364:584. doi: 10.1038/364584a0. [DOI] [PubMed] [Google Scholar]
- 58.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol Cell Physiol. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 59.Beckman JS, Estevez AG, Crow JP, Barbeito L. Superoxide dismutase and the death of motoneurons in ALS. Trends Neurosci. 2001;24:S15–S20. doi: 10.1016/S0166-2236(00)01981-0. [DOI] [PubMed] [Google Scholar]
- 60.Tamir S, Burney S, Tannenbaum SR. DNA damage by nitric oxide. Chem Res Toxicol. 1996;9:821–827. doi: 10.1021/tx9600311. [DOI] [PubMed] [Google Scholar]
- 61.Kröncke KD, Fehsel K, Kolb-Bachofen V. Nitric oxide: cytotoxicity versus cytoprotection—how, why, when, where? Nitric Oxide Biol Chem. 1997;1:107–120. doi: 10.1006/niox.1997.0118. [DOI] [PubMed] [Google Scholar]
- 62.Burney S, Tamir S, Gal A, Tannenbaum SR. A mechanistic analysis of nitric oxide induced cytotoxicity. Nitric Oxide Biol Chem. 1997;1:130–144. doi: 10.1006/niox.1996.0114. [DOI] [PubMed] [Google Scholar]
- 63.Messmer UK, Reimer DM, Reed JC, Brune B. Nitric oxide induced poly(ADP-ribose) polymerase cleavage in RAS 264.7 macrophage apoptosis is blocked by Bcl-2. FEBS Lett. 1996;384:162–166. doi: 10.1016/0014-5793(96)00311-0. [DOI] [PubMed] [Google Scholar]
- 64.Riquelme PT, Burzio LO, Koide SS. ADP ribosylation of rat liver lysine-rich histone in vitro. J Biol Chem. 1979;254:3018–3028. [PubMed] [Google Scholar]
- 65.Suzuki H, Quesada P, Farina B, Leone E. In vitro poly(ADP-ribosyl)ation of seminal ribonuclease. J Biol Chem. 1986;261:6048–6055. [PubMed] [Google Scholar]
- 66.Lautier D, Lagueux J, Thiboldeau J, Menard L, Poirier GG. Molecular and biochemical features of poly(ADP-ribose)metabolism. Mol Cell Biochem. 1993;122:171–193. doi: 10.1007/BF01076101. [DOI] [PubMed] [Google Scholar]
- 67.Min W, Wang ZQ. Poly (ADP-ribose) glycohydrolase (PARG) and its therapeutic potential. Front Biosci (Landmark Ed). 2009;14:1619–1626. doi: 10.2741/3329. [DOI] [PubMed] [Google Scholar]
- 68.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs) BMC Genom. 2005;6:139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 70.Dantzer F, Amé JC, Schreiber V, Nakamura J, Ménissier-de Murcia J, de Murcia G. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods Enzymol. 2006;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- 71.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 72.Jagtap P, Szabó C. Poly(ADP-ribose) polymerase and the therapeutic effects of its inhibitors. Nat Rev Drug Discov. 2005;4:421–440. doi: 10.1038/nrd1718. [DOI] [PubMed] [Google Scholar]
- 73.Beneke S. Poly(ADP-ribose) polymerase activity in different pathologies-the link to inflammation and infarction. Exp Gerontol. 2008;43:605–614. doi: 10.1016/j.exger.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 74.Virág L, Szabó C. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol Rev. 2002;54:375–429. doi: 10.1124/pr.54.3.375. [DOI] [PubMed] [Google Scholar]
- 75.Szabó C, Ischiropoulos H, Radi R. Peroxynitrite: biochemistry, pathophysiology and development of therapeutics. Nat Rev Drug Discov. 2007;6:662–680. doi: 10.1038/nrd2222. [DOI] [PubMed] [Google Scholar]
- 76.Pacher P, Beckman JS, Liaudet L. Nitric oxide and peroxynitrite in health and disease. Physiol Rev. 2007;87:315–424. doi: 10.1152/physrev.00029.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Szabó C. Multiple pathways of peroxynitrite cytotoxicity. Toxicol Lett. 2003;140–141:105–112. doi: 10.1016/S0378-4274(02)00507-6. [DOI] [PubMed] [Google Scholar]
- 78.Szabó C, Pacher P, Swanson RA. Novel modulators of poly(ADPribose) polymerase. Trends Pharmacol Sci. 2006;27:626–630. doi: 10.1016/j.tips.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Aguilar-Quesada R, Muñoz-Gámez JA, Martín-Oliva D, Peralta-Leal A, Quiles-Pérez R, Rodríguez-Vargas JM, Ruiz de Almodóvar M, Conde C, Ruiz-Extremera A, Oliver FJ. Modulation of transcription by PARP-1: consequences in carcinogenesis and inflammation. Curr Med Chem. 2007;14:1179–1187. doi: 10.2174/092986707780597998. [DOI] [PubMed] [Google Scholar]
- 80.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 81.Chiarugi A. Poly(ADP-ribosyl)ation and stroke. Pharmacol Res. 2005;52:15–24. doi: 10.1016/j.phrs.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 82.Komjáti K, Besson VC, Szabó C. Poly(ADP-ribose) polymerase inhibitors as potential therapeutic agents in stroke and neurotrauma. Curr Drug Targets CNS Neurol Disord. 2005;4:179–194. doi: 10.2174/1568007053544138. [DOI] [PubMed] [Google Scholar]
- 83.Pacher P, Schulz R, Liaudet L, Szabó C. Nitrosative stress and pharmacological modulation of heart failure. Trends Pharmacol Sci. 2005;26:302–310. doi: 10.1016/j.tips.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Obrosova IG, Julius UA. Role for poly(ADP-ribose) polymerase activation in diabetic nephropathy, neuropathy and retinopathy. Curr Vasc Pharmacol. 2005;3:267–283. doi: 10.2174/1570161054368634. [DOI] [PubMed] [Google Scholar]
- 85.Kauppinen TM, Swanson RA. The role of poly(ADP-ribose) polymerase-1 in CNS disease. Neuroscience. 2007;145:1267–1272. doi: 10.1016/j.neuroscience.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 86.Pacher P, Szabó C. Role of poly(ADP-ribose) polymerase 1 (PARP-1) in cardiovascular diseases: the therapeutic potential of PARP inhibitors. Cardiovasc Drug Rev. 2007;25:235–260. doi: 10.1111/j.1527-3466.2007.00018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Szabó C, Dawson VL. Role of poly(ADP-ribose) synthetase in inflammation and ischaemia-reperfusion. Trends Pharmacol Sci. 1998;19:287–298. doi: 10.1016/S0165-6147(98)01193-6. [DOI] [PubMed] [Google Scholar]
- 88.Szabó C, Virag L, Cuzzocrea S, Scott GS, Hake P, O’Connor MP, Zingarelli B, Salzman A, Kun E. Protection against peroxynitrite induced fibroblast injury and arthritis development by inhibition of poly(ADP-ribose) synthase. Proc Natl Acad Sci USA. 1998;95:3867–3872. doi: 10.1073/pnas.95.7.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boujrad H, Gubkina O, Robert N, Krantic S, Susin SA. AIF-mediated programmed necrosis: a highly regulated way to die. Cell Cycle. 2007;6:2612–2619. doi: 10.4161/cc.6.21.4842. [DOI] [PubMed] [Google Scholar]
- 90.Cheung EC, Joza N, Steenaart NA, McClellan KA, Neuspiel M, McNamara S, MacLaurin JG, Rippstein P, Park DS, Shore GC, McBride HM, Penninger JM, Slack RS. Dissociating the dual roles of apoptosis-inducing factor in maintaining mitochondrial structure and apoptosis. EMBO J. 2006;25:4061–4073. doi: 10.1038/sj.emboj.7601276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joza N, Oudit GY, Brown D, Bénit P, Kassiri Z, Vahsen N, Benoit L, Patel MM, Nowikovsky K, Vassault A, Backx PH, Wada T, Kroemer G, Rustin P, Penninger JM. Muscle-specific loss of apoptosis-inducing factor leads to mitochondrial dysfunction, skeletal muscle atrophy, and dilated cardiomyopathy. Mol Cell Biol. 2005;25:10261–10272. doi: 10.1128/MCB.25.23.10261-10272.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, Mangion J, Jacotot E, Costantini P, Loeffler M, Larochette N, Goodlett DR, Aebersold R, Siderovski DP, Penninger JM, Kroemer G. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 93.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: a key event in parthanatos. Exp Neurol. 2009;218:193–202. doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann N Y Acad Sci. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297:259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 96.Harraz MM, Dawson TM, Dawson VL. Advances in neuronal cell death 2007. Stroke. 2008;39:286–288. doi: 10.1161/STROKEAHA.107.511857. [DOI] [PubMed] [Google Scholar]
- 97.Delettre C, Yuste VJ, Moubarak RS, Bras M, Lesbordes-Brion JC, Petres S, Bellalou J, Susin SA. AIFsh, a novel apoptosis-inducing factor (AIF) pro-apoptotic isoform with potential pathological relevance in human cancer. J Biol Chem. 2006;281:6413–6427. doi: 10.1074/jbc.M509884200. [DOI] [PubMed] [Google Scholar]
- 98.Culmsee C, Zhu C, Landshamer S, Becattini B, Wagner E, Pellecchia M, Blomgren K, Plesnila N. Apoptosis-inducing factor triggered by poly(ADP-ribose) polymerase and Bid mediates neuronal cell death after oxygen-glucose deprivation and focal cerebral ischemia. J Neurosci. 2005;25:10262–10272. doi: 10.1523/JNEUROSCI.2818-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gordon LI, Weitzman SA. Review: inflammation and cancer. Cancer J. 1993;6:257–261. doi: 10.1002/1097-0142(19930101)71:1<257::AID-CNCR2820710139>3.0.CO;2-B. [DOI] [Google Scholar]
- 100.Ohshima H, Bartsch H. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat Res. 1994;305:253–264. doi: 10.1016/0027-5107(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 101.Ohshima H, Tatemichi M, Sawa T. Chemical basis of inflammation-induced carcinogenesis. Arch Biochem Biophys. 2003;417:3–11. doi: 10.1016/S0003-9861(03)00283-2. [DOI] [PubMed] [Google Scholar]
- 102.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 103.Yoshie Y, Ohshima H. Synergistic induction of DNA strand breakage by catecholestrogen and nitric oxide: implications for hormonal carcinogenesis. Free Radic Biol Med. 1998;24:341–348. doi: 10.1016/S0891-5849(97)00269-4. [DOI] [PubMed] [Google Scholar]
- 104.Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, Moncada S. Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tschugguel W, Knogler W, Czerwenka K, Mildner M, Weninger W, Zeillinger R, Huber JC. Presence of endothelial calcium-dependent nitric oxide synthase in breast apocrine metaplasia. Br J Cancer. 1996;74:1423–1426. doi: 10.1038/bjc.1996.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mannick EE, Bravo LE, Zarama G, Realpe JL, Zhang XJ, Ruiz B, Fontham ET, Mera R, Miller MJ, Correa P. Inducible nitric oxide synthase, nitrotyrosine, and apoptosis in Helicobacter pylori gastritis: effect of antibiotics and antioxidants. Cancer Res. 1996;56:3238–3243. [PubMed] [Google Scholar]
- 107.Tsuji S, Kawano S, Tsujii M, Takei Y, Tanaka M, Sawaoka H, Nagano K, Fusamoto H, Kamada T. Helicobacter pylori extract stimulates inflammatory nitric oxide production. Cancer Lett. 1996;108:195–200. doi: 10.1016/S0304-3835(96)04410-2. [DOI] [PubMed] [Google Scholar]
- 108.Rachmilewitz D, Karmeli F, Eliakim R, Stalnikowicz R, Ackerman Z, Amir G, Stamler JS. Enhanced gastric nitric oxide synthase activity in duodenal ulcer patients. Gut. 1994;35:1394–1397. doi: 10.1136/gut.35.10.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pignatelli B, Bancel B, Estève J, Malaveille C, Calmels S, Correa P, Patricot LM, Laval M, Lyandrat N, Smit F, Ohshima H. Helicobacter pylori infection and oxidative stress in human gastric carcinogenesis: expression of inducible nitric oxide synthase and anti-oxidant enzymes. In: Tahara E, Sugimachi K, Oohara T, editors. Recent advances in gastroenterological carcinogenesis I. Bologna: Monduzzi Editore; 1996. pp. 1221–1225. [Google Scholar]
- 110.Boughton-Smith NK, Evans SM, Hawkey CJ, Cole AT, Balsitis M, Whittle BJ, Moncada S. Nitric oxide synthase activity in ulcerative colitis and Crohn’s disease. Lancet. 1993;342:338–340. doi: 10.1016/0140-6736(93)91476-3. [DOI] [PubMed] [Google Scholar]
- 111.Middleton SJ, Shorthouse M, Hunter JO. Increased nitric oxide synthesis in ulcerative colitis. Lancet. 1993;341:465–466. doi: 10.1016/0140-6736(93)90211-X. [DOI] [PubMed] [Google Scholar]
- 112.Singer II, Kawka DW, Scott S, Weidner JR, Mumford RA, Riehl TE, Stenson WF. Expression of inducible nitric oxide synthase and nitrotyrosine in colonic epithelium in inflammatory bowel disease. Gastroenterology. 1996;111:871–885. doi: 10.1016/S0016-5085(96)70055-0. [DOI] [PubMed] [Google Scholar]
- 113.MacMillan-Crow LA, Thompson JA. Tyrosine modifications and inactivation of active site manganese superoxide dismutase mutant (Y34F) by peroxynitrite. Arch Biochem Biophys. 1999;366:82–88. doi: 10.1006/abbi.1999.1202. [DOI] [PubMed] [Google Scholar]
- 114.Radi R, Cassina A, Hodara R. Nitric oxide and peroxynitrite interactions with mitochondria. Biol Chem. 2002;383:401–409. doi: 10.1515/BC.2002.044. [DOI] [PubMed] [Google Scholar]
- 115.Radi R, Cassina A, Hodara R, Quijano C, Castro L. Peroxynitrite reactions and formation in mitochondria. Free Radic Biol Med. 2002;33:1451–1464. doi: 10.1016/S0891-5849(02)01111-5. [DOI] [PubMed] [Google Scholar]
- 116.Castro L, Rodriguez M, Radi R. Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 117.Han D, Canali R, Garcia J, Aguilera R, Gallaher TK, Cadenas E. Sites and mechanisms of aconitase inactivation by peroxynitrite: modulation by citrate and glutathione. Biochemistry. 2005;44:11986–11996. doi: 10.1021/bi0509393. [DOI] [PubMed] [Google Scholar]
- 118.Stachowiak O, Dolder M, Wallimann T, Richter C. Mitochondrial creatine kinase is a prime target of peroxynitrite-induced modification and inactivation. J Biol Chem. 1998;273:16694–16699. doi: 10.1074/jbc.273.27.16694. [DOI] [PubMed] [Google Scholar]
- 119.Wild S, Roglic G, Green A, Sicree R, King H. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care. 2004;27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 120.Hartge MM, Unger T, Kintscher U. The endothelium and vascular inflammation in diabetes. Diab Vasc Dis Res. 2007;4:84–88. doi: 10.3132/dvdr.2007.025. [DOI] [PubMed] [Google Scholar]
- 121.Hermans MP. Diabetes and the endothelium. Acta Clin Belg. 2007;62:97–101. doi: 10.1179/acb.2007.017. [DOI] [PubMed] [Google Scholar]
- 122.Nicolls MR, Haskins K, Flores SC. Oxidant stress, immune dysregulation, and vascular function in type I diabetes. Antioxid Redox Signal. 2007;9:879–889. doi: 10.1089/ars.2007.1631. [DOI] [PubMed] [Google Scholar]
- 123.Sobel BE, Schneider DJ. Cardiovascular complications in diabetes mellitus. Curr Opin Pharmacol. 2005;5:143–148. doi: 10.1016/j.coph.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 124.Hayat SA, Patel B, Khattar RS, Malik RA. Diabetic cardiomyopathy: mechanisms, diagnosis and treatment. Clin Sci. 2004;107:539–557. doi: 10.1042/CS20040057. [DOI] [PubMed] [Google Scholar]
- 125.Pacher P, Obrosova IG, Mabley JG, Szabó C. Role of nitrosative stress and peroxynitrite in the pathogenesis of diabetic complications. Emerging new therapeutical strategies. Curr Med Chem. 2005;12:267–275. doi: 10.2174/0929867053363207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pacher P, Szabó C. Role of peroxynitrite in the pathogenesis of cardiovascular complications of diabetes. Curr Opin Pharmacol. 2006;6:136–141. doi: 10.1016/j.coph.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Giacco F, Brownlee M. Oxidative stress and diabetic complications. Circ Res. 2010;107:1058–1070. doi: 10.1161/CIRCRESAHA.110.223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Du X, Matsumura T, Edelstein D, Rossetti L, Zsengellér Z, Szabó C, Brownlee M. Inhibition of GAPDH activity by poly(ADP-ribose) polymerase activates three major pathways of hyperglycemic damage in endothelial cells. J Clin Invest. 2003;112:1049–1057. doi: 10.1172/JCI18127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pacher P, Liaudet L, Soriano FG, Mabley JG, Szabó E, Szabó C. The role of poly(ADP-ribose) polymerase activation in the development of myocardial and endothelial dysfunction in diabetes. Diabetes. 2002;51:514–521. doi: 10.2337/diabetes.51.2.514. [DOI] [PubMed] [Google Scholar]
- 130.Szabó C, Zanchi A, Komjáti K, Pacher P, Krolewski AS, Quist WC, LoGerfo FW, Horton ES, Veves A. Poly(ADP-Ribose) polymerase is activated in subjects at risk of developing type 2 diabetes and is associated with impaired vascular reactivity. Circulation. 2002;106:2680–2686. doi: 10.1161/01.CIR.0000038365.78031.9C. [DOI] [PubMed] [Google Scholar]
- 131.Thuraisingham RC, Nott CA, Dodd SM, Yaqoob MM. Increased nitrotyrosine staining in kidneys from patients with diabetic nephropathy. Kidney Int. 2000;57:1968–1972. doi: 10.1046/j.1523-1755.2000.00046.x. [DOI] [PubMed] [Google Scholar]
- 132.Onozato ML, Tojo A, Goto A, Fujita T, Wilcox CS. Oxidative stress and nitric oxide synthase in rat diabetic nephropathy: effects of ACEI and ARB. Kidney Int. 2002;61:186–194. doi: 10.1046/j.1523-1755.2002.00123.x. [DOI] [PubMed] [Google Scholar]
- 133.El-Remessy AB, Behzadian MA, Abou-Mohamed G, Franklin T, Caldwell RW, Caldwell RB. Experimental diabetes causes breakdown of the blood-retina barrier by a mechanism involving tyrosine nitration and increases in expression of vascular endothelial growth factor and urokinase plasminogen activator receptor. Am J Pathol. 2003;162:1995–2004. doi: 10.1016/S0002-9440(10)64332-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kowluru RA. Effect of reinstitution of good glycemic control on retinal oxidative stress and nitrative stress in diabetic rats. Diabetes. 2003;52:818–823. doi: 10.2337/diabetes.52.3.818. [DOI] [PubMed] [Google Scholar]
- 135.Coppey LJ, Gellett JS, Davidson EP, Dunlap JA, Lund DD, Yorek MA. Effect of antioxidant treatment of streptozotocin-induced diabetic rats on endoneurial blood flow, motor nerve conduction velocity, and vascular reactivity of epineurial arterioles of the sciatic nerve. Diabetes. 2001;50:1927–1937. doi: 10.2337/diabetes.50.8.1927. [DOI] [PubMed] [Google Scholar]
- 136.Hoeldtke RD, Bryner KD, McNeill DR, Hobbs GR, Riggs JE, Warehime SS, Christie I, Ganser G, Van Dyke K. Nitrosative stress, uric acid, and peripheral nerve function in early type 1 diabetes. Diabetes. 2002;51:2817–2825. doi: 10.2337/diabetes.51.9.2817. [DOI] [PubMed] [Google Scholar]
- 137.Jagtap PG, Baloglu E, Southan GJ, Mabley JG, Li H, Zhou J, van Duzer J, Salzman AL, Szabó C. Discovery of potent poly(ADP-ribose) polymerase-1 inhibitors from the modification of indeno[1,2-c]isoquinolinone. J Med Chem. 2005;48:5100–5103. doi: 10.1021/jm0502891. [DOI] [PubMed] [Google Scholar]
- 138.Inada C, Yamada K, Takane N, Nonaka K. Poly(ADP-ribose) synthesis induced by nitric oxide in a mouse beta-cell line. Life Sci. 1995;56:1467–1474. doi: 10.1016/0024-3205(95)00109-J. [DOI] [PubMed] [Google Scholar]
- 139.Heller B, Wang ZQ, Wagner EF, Radons J, Bürkle A, Fehsel K, Burkart V, Kolb H. Inactivation of the poly(ADP-ribose) polymerase gene affects oxygen radical and nitric oxide toxicity in islet cells. J Biol Chem. 1995;270:11176–11180. doi: 10.1074/jbc.270.19.11176. [DOI] [PubMed] [Google Scholar]
- 140.Pieper AA, Brat DJ, Krug DK, Watkins CC, Gupta A, Blackshaw S, Verma A, Wang ZQ, Snyder SH. Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:3059–3064. doi: 10.1073/pnas.96.6.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mabley JG, Suarez-Pinzon WL, Haskó G, Salzman AL, Rabinovitch A, Kun E, Szabó C. Inhibition of poly (ADP-ribose) synthetase by gene disruption or inhibition with 5-iodo-6-amino-1,2-benzopyrone protects mice from multiple-low-dose-streptozotocin-induced diabetes. Br J Pharmacol. 2001;133:909–919. doi: 10.1038/sj.bjp.0704156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xiao CY, Chen M, Zsengellér Z, Szabó C. Poly(ADP-ribose) polymerase contributes to the development of myocardial infarction in diabetic rats and regulates the nuclear translocation of apoptosis-inducing factor. J Pharmacol Exp Ther. 2004;310:498–504. doi: 10.1124/jpet.104.066803. [DOI] [PubMed] [Google Scholar]
- 143.Beneke S, Bürkle A. Poly(ADP-ribosyl)ation in mammalian ageing. Nucleic Acids Res. 2007;35:7456–7465. doi: 10.1093/nar/gkm735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pacher P, Szabó C. Role of poly(ADP-ribose) polymerase-1 activation in the pathogenesis of diabetic complications: endothelial dysfunction, as a common underlying theme. Antioxid Redox Signal. 2005;7:1568–1580. doi: 10.1089/ars.2005.7.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gilad E, Zingarelli B, Salzman AL, Szabó C. Protection by inhibition of poly (ADP-ribose) synthetase against oxidant injury in cardiac myoblasts in vitro. J Mol Cell Cardiol. 1997;29:2585–2597. doi: 10.1006/jmcc.1997.0496. [DOI] [PubMed] [Google Scholar]
- 146.Thiemermann C, Bowes J, Myint FP, Vane JR. Inhibition of the activity of poly(ADP ribose) synthetase reduces ischemia-reperfusion injury in the heart and skeletal muscle. Proc Natl Acad Sci USA. 1997;94:679–683. doi: 10.1073/pnas.94.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zingarelli B, Cuzzocrea S, Zsengellér Z, Salzman AL, Szabó C. Protection against myocardial ischemia and reperfusion injury by 3-aminobenzamide, an inhibitor of poly (ADP-ribose) synthetase. Cardiovasc Res. 1997;36:205–215. doi: 10.1016/S0008-6363(97)00137-5. [DOI] [PubMed] [Google Scholar]
- 148.Bowes J, Ruetten H, Martorana PA, Stockhausen H, Thiemermann C. Reduction of myocardial reperfusion injury by an inhibitor of poly (ADP-ribose) synthetase in the pig. Eur J Pharmacol. 1998;359:143–150. doi: 10.1016/S0014-2999(98)00638-4. [DOI] [PubMed] [Google Scholar]
- 149.Xiao CY, Chen M, Zsengellér Z, Li H, Kiss L, Kollai M, Szabó C. Poly(ADP-Ribose) polymerase promotes cardiac remodeling, contractile failure, and translocation of apoptosis-inducing factor in a murine experimental model of aortic banding and heart failure. J Pharmacol Exp Ther. 2005;312:891–898. doi: 10.1124/jpet.104.077164. [DOI] [PubMed] [Google Scholar]
- 150.Fiorillo C, Ponziani V, Giannini L, Cecchi C, Celli A, Nassi N, Lanzilao L, Caporale R, Nassi P. Protective effects of the PARP-1 inhibitor PJ34 in hypoxic-reoxygenated cardiomyoblasts. Cell Mol Life Sci. 2006;63:3061–3071. doi: 10.1007/s00018-006-6345-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pacher P, Liaudet L, Mabley JG, Komjáti K, Szabó C. Pharmacologic inhibition of poly(adenosine diphosphate-ribose) polymerase may represent a novel therapeutic approach in chronic heart failure. J Am Coll Cardiol. 2002;40:1006–1016. doi: 10.1016/S0735-1097(02)02062-4. [DOI] [PubMed] [Google Scholar]
- 152.Csiszar A, Pacher P, Kaley G, Ungvari Z. Role of oxidative and nitrosative stress, longevity genes and poly(ADP-ribose) polymerase in cardiovascular dysfunction associated with aging. Curr Vasc Pharmacol. 2005;3:285–291. doi: 10.2174/1570161054368616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Szabó C. Cardioprotective effects of poly(ADP-ribose) polymerase inhibition. Pharmacol Res. 2005;52:34–43. doi: 10.1016/j.phrs.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 154.Chabrier PE, Demerlé-Pallardy C, Auguet M. Nitric oxide synthases: targets for therapeutic strategies in neurological diseases. Cell Mol Life Sci. 1999;55:1029–1035. doi: 10.1007/s000180050353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Torreilles F, Salman-Tabcheh S, Guerin M, Torreilles J. Neurodegenerative disorders: the role of peroxynitrite. Brain Res. 1999;30:153–163. doi: 10.1016/S0165-0173(99)00014-4. [DOI] [PubMed] [Google Scholar]
- 156.Ischiropoulos H, Beckman JS. Oxidative stress and nitration in neurodegeneration: cause, effect, or association? J Clin Invest. 2003;111:163–169. doi: 10.1172/JCI200317638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sarchielli P, Galli F, Floridi A, Floridi A, Gallai V. Relevance of protein nitration in brain injury: a key pathophysiological mechanism in neurodegenerative, autoimmune, or inflammatory CNS diseases and stroke. Amino Acids. 2003;25:427–436. doi: 10.1007/s00726-003-0028-6. [DOI] [PubMed] [Google Scholar]
- 158.Calabrese V, Scapagnini G, Ravagna A, Bella R, Foresti R, Bates TE, Giuffrida Stella AM, Pennisi G. Nitric oxide synthase is present in the cerebrospinal fluid of patients with active multiple sclerosis and is associated with increases in cerebrospinal fluid protein nitrotyrosine and S-nitrosothiols and with changes in glutathione levels. J Neurosci Res. 2002;70:580–587. doi: 10.1002/jnr.10408. [DOI] [PubMed] [Google Scholar]
- 159.Weksler ME, Gouras G, Relkin NR, Szabo P. The immune system, amyloid-beta peptide, and Alzheimer’s disease. Immunol Rev. 2005;205:244–256. doi: 10.1111/j.0105-2896.2005.00264.x. [DOI] [PubMed] [Google Scholar]
- 160.Mattson MP, Chan SL. Neuronal and glial calcium signaling in Alzheimer’s disease. Cell Calcium. 2003;34:385–397. doi: 10.1016/S0143-4160(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 161.Butterfield DA, Boyd-Kimball D. Amyloid beta-peptide(1-42) contributes to the oxidative stress and neurodegeneration found in Alzheimer disease brain. Brain Pathol. 2004;14:426–432. doi: 10.1111/j.1750-3639.2004.tb00087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Emerit J, Edeas M, Bricaire F. Neurodegenerative diseases and oxidative stress. Biomed Pharmacother. 2004;58:39–46. doi: 10.1016/j.biopha.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 163.Markesbery WR. Oxidative stress hypothesis in Alzheimer’s disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/S0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 164.Mhatre M, Floyd RA, Hensley K. Oxidative stress and neuroinflammation in Alzheimer’s disease and amyotrophic lateral sclerosis: common links and potential therapeutic targets. J Alzheimers Dis. 2004;6:147–157. doi: 10.3233/jad-2004-6206. [DOI] [PubMed] [Google Scholar]
- 165.Moreira PI, Siedlak SL, Aliev G, Zhu X, Cash AD, Smith MA, Perry G. Oxidative stress mechanisms and potential therapeutics in Alzheimer disease. J Neural Transm. 2005;112:921–932. doi: 10.1007/s00702-004-0242-8. [DOI] [PubMed] [Google Scholar]
- 166.Togo T, Katsuse O, Iseki E. Nitric oxide pathways in Alzheimer’s disease and other neurodegenerative dementias. Neurol Res. 2004;26:563–566. doi: 10.1179/016164104225016236. [DOI] [PubMed] [Google Scholar]
- 167.Simic G, Lucassen PJ, Krsnik Z, Kruslin B, Kostovic I, Winblad B, Bogdanovi N. nNOS expression in reactive astrocytes correlates with increased cell death related DNA damage in the hippocampus and entorhinal cortex in Alzheimer’s disease. Exp Neurol. 2000;165:12–26. doi: 10.1006/exnr.2000.7448. [DOI] [PubMed] [Google Scholar]
- 168.Thorns V, Hansen L, Masliah E. nNOS expressing neurons in the entorhinal cortex and hippocampus are affected in patients with Alzheimer’s disease. Exp Neurol. 1998;150:14–20. doi: 10.1006/exnr.1997.6751. [DOI] [PubMed] [Google Scholar]
- 169.Su JH, Anderson AJ, Cummings BJ, Cotman CW. Immunohistochemical evidence for apoptosis in Alzheimer’s disease. NeuroReport. 1994;5:2529–2533. doi: 10.1097/00001756-199412000-00031. [DOI] [PubMed] [Google Scholar]
- 170.Dragunow M, Faull RL, Lawlor P, Beilharz EJ, Singleton K, Walker EB, Mee E. In situ evidence for DNA fragmentation in Huntington’s disease striatum and Alzheimer’s disease temporal lobes. NeuroReport. 1995;6:1053–1057. doi: 10.1097/00001756-199505090-00026. [DOI] [PubMed] [Google Scholar]
- 171.Lassmann H, Bancher C, Breitschopf H, Wegiel J, Bobinski M, Jellinger K, Wisniewski HM. Cell death in Alzheimer’s disease evaluated by DNA fragmentation in situ. Acta Neuropathol. 1995;89:35–41. doi: 10.1007/BF00294257. [DOI] [PubMed] [Google Scholar]
- 172.Troncoso JC, Sukhov RR, Kawas CH, Koliatsos VE. In situ labeling of dying cortical neurons in normal aging and in Alzheimer’s disease: correlations with senile plaques and disease progression. J Neuropathol Exp Neurol. 1996;55:1134–1142. doi: 10.1097/00005072-199611000-00004. [DOI] [PubMed] [Google Scholar]
- 173.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity [see comments]. Science. 1994; 263: 687–689. Comment in: Science. 1994; 265: 722–723. [DOI] [PubMed]
- 174.Endres M, Wang ZQ, Namura S, Waeber C, Moskowitz MA. Ischemic brain injury is mediated by the activation of poly(ADP-ribose)polymerase. J Cereb Blood Flow Metab. 1997;17:1143–1151. doi: 10.1097/00004647-199711000-00002. [DOI] [PubMed] [Google Scholar]
- 175.Ying W, Alano CC, Garnier P, Swanson RA. NAD+ as a metabolic link between DNA damage and cell death. J Neurosci Res. 2005;79:216–223. doi: 10.1002/jnr.20289. [DOI] [PubMed] [Google Scholar]
- 176.Love S, Barber R, Wilcock GK. Neuronal accumulation of poly(ADP-ribose) after brain ischaemia. Neuropathol Appl Neurobiol. 1999;25:98–103. doi: 10.1046/j.1365-2990.1999.00179.x. [DOI] [PubMed] [Google Scholar]
- 177.Hattori N, Mizuno Y. Pathogenetic mechanisms of parkin in Parkinson’s disease. Lancet. 2004;364:722–724. doi: 10.1016/S0140-6736(04)16901-8. [DOI] [PubMed] [Google Scholar]
- 178.Eriksen JL, Wszolek Z, Petrucelli L. Molecular pathogenesis of Parkinson disease. Arch Neurol. 2005;62:353–357. doi: 10.1001/archneur.62.3.353. [DOI] [PubMed] [Google Scholar]
- 179.Jenner P. Oxidative stress in Parkinson’s disease. Ann Neurol. 2003; 53 Suppl 3:S26–36; p no. S36–8. [DOI] [PubMed]
- 180.Kim SH, Henkel JS, Beers DR, Sengun IS, Simpson EP, Goodman JC, Engelhardt JI, Siklós L, Appel SH. PARP expression is increased in astrocytes but decreased in motor neurons in the spinal cord of sporadic ALS patients. J Neuropathol Exp Neurol. 2003;62:88–103. doi: 10.1093/jnen/62.1.88. [DOI] [PubMed] [Google Scholar]
- 181.Hunot S, Brugg B, Ricard D, Michel PP, Muriel MP, Ruberg M, Faucheux BA, Agid Y, Hirsch EC. Nuclear translocation of NF-kappaB is increased in dopaminergic neurons of patients with parkinson disease. Proc Natl Acad Sci USA. 1997;94:7531–7536. doi: 10.1073/pnas.94.14.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Denk A, Wirth T, Baumann B. NF-kappaB transcription factors: critical regulators of hematopoiesis and neuronal survival. Cytokine Growth Factor Rev. 2000;11:303–320. doi: 10.1016/S1359-6101(00)00009-5. [DOI] [PubMed] [Google Scholar]
- 183.de Erausquin GA, Hyrc K, Dorsey DA, Mamah D, Dokucu M, Mascó DH, Walton T, Dikranian K, Soriano M, García Verdugo JM, Goldberg MP, Dugan LL. Nuclear translocation of nuclear transcription factor-kappa B by alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors leads to transcription of p53 and cell death in dopaminergic neurons. Mol Pharmacol. 2003;63:784–790. doi: 10.1124/mol.63.4.784. [DOI] [PubMed] [Google Scholar]
- 184.Soós J, Engelhardt JI, Siklós L, Havas L, Majtényi K. The expression of PARP, NF-kappa B and parvalbumin is increased in Parkinson disease. NeuroReport. 2004;15:1715–1718. doi: 10.1097/01.wnr.0000136175.51954.ce. [DOI] [PubMed] [Google Scholar]
- 185.Ara J, Przedborski S, Naini AB, Jackson-Lewis V, Trifiletti RR, Horwitz J, Ischiropoulos H. Inactivation of tyrosine hydroxylase by nitration following exposure to peroxynitrite and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) Proc Natl Acad Sci USA. 1998;95:7659–7663. doi: 10.1073/pnas.95.13.7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Geha RM, Rebrin I, Chen K, Shih JC. Substrate and inhibitor specificities for human monoamine oxidase A and B are influenced by a single amino acid. J Biol Chem. 2001;276:9877–9882. doi: 10.1074/jbc.M006972200. [DOI] [PubMed] [Google Scholar]
- 187.Barker JE, Bolaños JP, Land JM, Clark JB, Heales SJ. Glutathione protects astrocytes from peroxynitrite-mediated mitochondrial damage: implications for neuronal/astrocytic trafficking and neurodegeneration. Dev Neurosci. 1996;18:391–396. doi: 10.1159/000111432. [DOI] [PubMed] [Google Scholar]
- 188.Bharath S, Andersen JK. Glutathione depletion in a midbrain-derived immortalized dopaminergic cell line results in limited tyrosine nitration of mitochondrial complex I subunits: implications for Parkinson’s disease. Antioxid Redox Signal. 2005;7:900–910. doi: 10.1089/ars.2005.7.900. [DOI] [PubMed] [Google Scholar]
- 189.Naoi M, Maruyama W. Future of neuroprotection in Parkinson’s disease. Parkinsonism Relat Disord. 2001;8:139–145. doi: 10.1016/S1353-8020(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 190.Dawson TM, Dawson VL. Molecular pathways of neurodegeneration in Parkinson’s disease. Science. 2003;302:819–822. doi: 10.1126/science.1087753. [DOI] [PubMed] [Google Scholar]
- 191.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–1700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 192.Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- 193.Bar-Shai M, Reznick AZ. Peroxynitrite induces an alternative NF-kappaB activation pathway in L8 rat myoblasts. Antioxid Redox Signal. 2006;8:639–652. doi: 10.1089/ars.2006.8.639. [DOI] [PubMed] [Google Scholar]
- 194.Bertram L, Tanzi RE. The genetic epidemiology of neurodegenerative disease. J Clin Invest. 2005;115:1449–1457. doi: 10.1172/JCI24761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Weydt P, Möller T. Neuroinflammation in the pathogenesis of amyotrophic lateral sclerosis. NeuroReport. 2005;16:527–531. doi: 10.1097/00001756-200504250-00001. [DOI] [PubMed] [Google Scholar]
- 196.Beal MF, Ferrante RJ, Browne SE, Matthews RT, Kowall NW, Brown RH., Jr Increased 3-nitrotyrosine in both sporadic and familial amyotrophic lateral sclerosis. Ann Neurol. 1997;42:644–654. doi: 10.1002/ana.410420416. [DOI] [PubMed] [Google Scholar]
- 197.Ferrante RJ, Browne SE, Shinobu LA, Bowling AC, Baik MJ, MacGarvey U, Kowall NW, Brown RH, Jr, Beal MF. Evidence of increased oxidative damage in both sporadic and familial amyotrophic lateral sclerosis. J Neurochem. 1997;69:2064–2074. doi: 10.1046/j.1471-4159.1997.69052064.x. [DOI] [PubMed] [Google Scholar]
- 198.Pedersen WA, Fu W, Keller JN, Markesbery WR, Appel S, Smith RG, Kasarskis E, Mattson MP. Protein modification by the lipid peroxidation product 4-hydroxynonenal in the spinal cords of amyotrophic lateral sclerosis patients. Ann Neurol. 1998;44:819–824. doi: 10.1002/ana.410440518. [DOI] [PubMed] [Google Scholar]
- 199.Shibata N, Nagai R, Uchida K, Horiuchi S, Yamada S, Hirano A, Kawaguchi M, Yamamoto T, Sasaki S, Kobayashi M. Morphological evidence for lipid peroxidation and protein glycoxidation in spinal cords from sporadic amyotrophic lateral sclerosis patients. Brain Res. 2001;917:97–104. doi: 10.1016/S0006-8993(01)02926-2. [DOI] [PubMed] [Google Scholar]
- 200.Bogdanov M, Brown RH, Matson W, Smart R, Hayden D, O’Donnell H, Flint Beal M, Cudkowicz M. Increased oxidative damage to DNA in ALS patients. Free Radic Biol Med. 2000;29:652–658. doi: 10.1016/S0891-5849(00)00349-X. [DOI] [PubMed] [Google Scholar]
- 201.Sasaki S, Warita H, Abe K, Iwata M. Inducible nitric oxide synthase (iNOS) and nitrotyrosine immunoreactivity in the spinal cords of transgenic mice with a G93A mutant SOD1 gene. J Neuropathol Exp Neurol. 2001;60:839–846. doi: 10.1093/jnen/60.9.839. [DOI] [PubMed] [Google Scholar]
- 202.Sasaki S, Shibata N, Komori T, Iwata M. iNOS and nitrotyrosine immunoreactivity in amyotrophic lateral sclerosis. Neurosci Lett. 2000;291:44–48. doi: 10.1016/S0304-3940(00)01370-7. [DOI] [PubMed] [Google Scholar]
- 203.Cha CI, Chung YH, Shin CM, Shin DH, Kim YS, Gurney ME, Lee KW. Immunocytochemical study on the distribution of nitrotyrosine in the brain of the transgenic mice expressing a human Cu/Zn SOD mutation. Brain Res. 2000;853:156–161. doi: 10.1016/S0006-8993(99)02302-1. [DOI] [PubMed] [Google Scholar]
- 204.Schraufstatter IU, Hinshaw DB, Hyslop PA, Spragg RG, Cochrane CG. Oxidant injury of cells. DNA strand-breaks activate polyadenosine diphosphate-ribose polymerase and lead to depletion of nicotinamide adenine dinucleotide. J Clin Invest. 1986;77:1312–1320. doi: 10.1172/JCI112436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Virag L, Salzman AL, Szabó C. Poly(ADP-ribose) synthetase activation mediates mitochondrial injury during oxidant-induced cell death. J Immunol. 1998;161:3753–3759. [PubMed] [Google Scholar]
- 206.Farkas B, Magyarlaki M, Csete B, Nemeth J, Rabloczky G, Bernath S, Literáti Nagy P, Sümegi B. Reduction of acute photodamage in skin by topical application of a novel PARP inhibitor. Biochem Pharmacol. 2002;63:921–932. doi: 10.1016/S0006-2952(01)00929-7. [DOI] [PubMed] [Google Scholar]
- 207.Ying W, Sevigny MB, Chen Y, Swanson RA. Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci USA. 2001;98:12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang J, Pieper A, Snyder SH. Poly(ADP-ribose) synthetase activation: an early indicator of neurotoxic DNA damage. J Neurochem. 1995;65:1411–1414. doi: 10.1046/j.1471-4159.1995.65031411.x. [DOI] [PubMed] [Google Scholar]
- 209.Homburg S, Visochek L, Moran N, Dantzer F, Priel E, Asculai E, Schwartz D, Rotter V, Dekel N, Cohen-Armon M. A fast signal-induced activation of Poly(ADP-ribose) polymerase: a novel downstream target of phospholipase C. J Cell Biol. 2000;150:293–307. doi: 10.1083/jcb.150.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Karczewski JM, Peters JG, Noordhoek J. Prevention of oxidant-induced cell death in Caco-2 colon carcinoma cells after inhibition of poly(ADP-ribose) polymerase and Ca2+ chelation: involvement of a common mechanism. Biochem Pharmacol. 1999;57:19–26. doi: 10.1016/S0006-2952(98)00286-X. [DOI] [PubMed] [Google Scholar]
- 211.Garthwaite G, Goodwin DA, Batchelor AM, Leeming K, Garthwaite J. Nitric oxide toxicity in CNS white matter: an in vitro study using rat optic nerve. Neuroscience. 2002;109:145–155. doi: 10.1016/S0306-4522(01)00447-X. [DOI] [PubMed] [Google Scholar]
- 212.Satoh MS, Lindahl T. Role of poly(ADP-ribose) formation in DNA repair. Nature. 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]
- 213.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase is a mediator of necrotic cell death by ATP depletion. Proc Natl Acad Sci USA. 1999;96:13978–13982. doi: 10.1073/pnas.96.24.13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schraufstatter IU, Hyslop PA, Hinshaw DB, Spragg RG, Sklar LA, Cochrane CG. Hydrogen peroxide-induced injury of cells and its prevention by inhibitors of poly(ADP-ribose) polymerase. Proc Natl Acad Sci USA. 1986;83:4908–4912. doi: 10.1073/pnas.83.13.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wallis RA, Panizzon KL, Henry D, Wasterlain CG. Neuroprotection against nitric oxide injury with inhibitors of ADP-ribosylation. NeuroReport. 1993;5:245–248. doi: 10.1097/00001756-199312000-00015. [DOI] [PubMed] [Google Scholar]
- 216.Rulten SL, Rotheray A, Green RL, Grundy GJ, Moore DA, Gómez-Herreros F, Hafezparast M, Caldecott KW. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Casoni F, Basso M, Massignan T, Gianazza E, Cheroni C, Salmona M, Bendotti C, Bonetto V. Protein nitration in a mouse model of familial amyotrophic lateral sclerosis: possible multifunctional role in the pathogenesis. J Biol Chem. 2005;280:16295–16304. doi: 10.1074/jbc.M413111200. [DOI] [PubMed] [Google Scholar]
- 218.Cassina P, Peluffo H, Pehar M, Martinez-Palma L, Ressia A, Beckman JS, Estevez AG, Barbeito L. Peroxynitrite triggers a phenotypic transformation in spinal cord astrocytes that induces motor neuron apoptosis. J Neurosci Res. 2002;67:21–29. doi: 10.1002/jnr.10107. [DOI] [PubMed] [Google Scholar]
- 219.Mandir AS, Przedborski S, Jackson-Lewis V, Wang ZQ, Simbulan-Rosenthal CM, Smulson ME, Hoffman BE, Guastella DB, Dawson VL, Dawson TM. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc Natl Acad Sci USA. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cosi C, Marien M. Decreases in mouse brain NAD+ and ATP induced by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP): prevention by the poly(ADP-ribose) polymerase inhibitor, benzamide. Brain Res. 1998;809:58–67. doi: 10.1016/S0006-8993(98)00829-4. [DOI] [PubMed] [Google Scholar]
- 221.Cosi C, Marien M. Implication of poly (ADP-ribose) polymerase (PARP) in neurodegeneration and brain energy metabolism. Decreases in mouse brain NAD1 and ATP caused by MPTP are prevented by the PARP inhibitor benzamide. Ann N Y Acad Sci. 1999;890:227–239. doi: 10.1111/j.1749-6632.1999.tb07998.x. [DOI] [PubMed] [Google Scholar]
- 222.Love S, Barber R, Wilcock GK. Increased poly(ADP-ribosyl)ation of nuclear proteins in Alzheimer’s disease. Brain. 1999;122:247–253. doi: 10.1093/brain/122.2.247. [DOI] [PubMed] [Google Scholar]
- 223.Hivert B, Cerruti C, Camu W. Hydrogen peroxide-induced motoneuron apoptosis is prevented by poly ADP ribosyl synthetase inhibitors. NeuroReport. 1998;9:1835–1838. doi: 10.1097/00001756-199806010-00031. [DOI] [PubMed] [Google Scholar]
- 224.Mandir AS, Poitras MF, Berliner AR, Herring WJ, Guastella DB, Feldman A, Poirier GG, Wang ZQ, Dawson TM, Dawson VL. NMDA but not non-NMDA excitotoxicity is mediated by Poly(ADP-ribose) polymerase. J Neurosci. 2000;20:8005–8011. doi: 10.1523/JNEUROSCI.20-21-08005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 226.Fatokun AA, Liu JO, Dawson VL, Dawson TM. Identification through high-throughput screening of 4′-methoxyflavone and 3′,4′-dimethoxyflavone as novel neuroprotective inhibitors of parthanatos. Br J Pharmacol. 2013;169:1263–1278. doi: 10.1111/bph.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]