Abstract
Reprogrammed glucose metabolism is an emerging hallmark of cancer cells, which show a unique metabolic phenotype known as the Warburg effect. Lactate dehydrogenase A (LDHA), a key enzyme in the glycolytic process, executes the final step by conversion of lactate into pyruvate. However, little is known about the roles of LDHA in human glioblastoma (GBM). In this study, we aimed to determine the effects of LDHA and elucidate related underlying mechanisms. Data derived from Oncomine database showed that LDHA is commonly upregulated in GBM tissues in comparison with corresponding normal controls. Silencing of LDHA expression resulted in reduced glycolysis, decreased cell growth, increased cell apoptosis, and attenuated invasive ability. In the presence of 2-deoxyglucose, a glycolysis inhibitor, the oncogenic activities of LDHA were completely blocked. These findings provide evidence of the cellular functions of LDHA in the progression of GBM and suggest that LDHA might act as a potential therapeutic target for GBM treatment.
Keywords: apoptosis, growth, invasion, lactate dehydrogenase A, Warburg effect
Introduction
Glioblastoma (GBM) is the most common malignant brain tumor, with high morbidity and mortality 1. Despite considerable improvements in surgical resection, radiotherapy as well as immune and gene therapy, GBM remains a challenging disease to treat 2,3. Therefore, it is very important to understand the molecular mechanisms underlying the aggressive phenotype of GBM cells.
Tumor cells differ from their normal counterparts through uncontrolled cell division and show a greater requirement for energy for rapid proliferation 4. Reprogrammed glucose metabolism is pervasively involved in the pathogenesis of cancer cells, which prefer glycolysis to oxidative phosphorylation even in the presence of oxygen 5. This is known as the Warburg effect, which is characterized by increased glucose consumption and lactate production. Lactate dehydrogenase A (LDHA), which executes the last step of anaerobic glycolysis, is critical to this highly glycolytic phenotype. In GBM, tumor cell-derived lactate dehydrogenase induces Natural killer group 2, member D ligands on myeloid cells, and ultimately subverts antitumor immune responses 6. In the isocitrate dehydrogenase mutant GBM cells, LDHA is highly methylated and underexpressed 7. Knockdown of LDHA with a subsequent decrease in lactate concentration leads to reduced levels of thrombospondin 1 and transforming growth factor-β2, which is crucial to the migration in glioma cells 8. Meanwhile, the oncogenic roles of deregulated LDHA in tumors have been reported in many previous studies, including colorectal cancer 9, gastric cancer 10, prostate cancer 11, liver cancer 12, and so on 13–15. However, the potential cellular functions and related mechanisms of LDHA in GBM cells remain largely unknown.
In this study, we aimed to investigate the expression pattern and roles of suppression of LDHA expression in GBM cells. We found that upregulated LDHA expression contributes toward tumor growth and invasion and this effect is dependent on an altered Warburg effect.
Materials and methods
Cell culture and reagent
Human GBM cell lines, A172 and U87, were all purchased from the American Type Culture Collection (ATCC, Manassas, Virginia, USA). A172 cells were maintained in Dulbecco’s modified Eagle medium (Gibco, Grand Island, New York, USA), U87 cells were grown in Eagle’s minimum essential medium (ATCC) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% antibiotics (penicillin and streptomycin) at 37°C in a humidified incubator at 5% CO2. 2-deoxyglucose (2-DG) was purchased from Sigma (Shanghai, China) and diluted to a preferable concentration before use.
Oncomine gene expression analysis
LDHA gene expression was analyzed using microarray gene expression datasets derived from the Oncomine database (https://www.oncomine.org/resource/login.html). To determine the differential expression of LDHA between GBM and their normal counterparts, a combined filter was used to display the corresponding datasets. Briefly, the Cancer Type was defined as Glioblastoma, Data Type was mRNA, and Analysis Type was Cancer versus Normal Analysis.
Transfection
Small interfering RNAs (siRNAs)-mediated interference was used in this study. Specific siRNAs targeting LDHA were cited from a previous report 16 and synthesized by Stanta Cruz (Santa Cruz Biotechnology, Carlsbad, California, USA). For transfection, A172 or U87 cells were transfected with 50 μM siRNAs using the lipofectamine 2000 transfection reagent kit according to the manufacturer’s instructions (Invitrogen, Carlsbad, California, USA). The interference efficiency was evaluated by western blotting.
Western blotting
Treated A172 or U87 cells were harvested and washed twice with cold PBS and lysed in radioimmunoprecipitation Assay buffer [50 mM Tris, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 1% nadeoxycholate (pH 7.4)] (Beyotime, Shanghai, China) with a fresh protease inhibitor. Protein lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred electrophoretically to a nitrocellulose membrane (Amersham Biosciences, Piscataway, New Jersey, USA). The blots were blocked with 5% bovine serum albumin, followed by incubation with primary antibodies against LDHA (Cat#ab47010; Abcam, Los Angeles, USA). Blots were then incubated with horseradish peroxidase-conjugated secondary antibodies (Abmart, Shanghai, China) and visualized using the enhanced chemiluminescent Plus kit (Millipore, Boston, Massachusetts, USA). And β-actin (Cat#ab6276; Abcam) was set as an internal control.
Colony formation assay
A total of 3000 LDHA-silenced or si-Ctrl GBM cells were seeded in a six-well plate in triplicate at 24 h after transfection and cultured in medium supplemented with 10% FBS. The culture medium was replaced every 2 days. After incubation for 10–14 days, colonies formed were fixed with methanol and visualized by staining with 0.1% crystal violet. After staining, the number of fixed colonies in each group was counted. Three independent experiments were conducted and each experiment had three duplicates.
Cell apoptosis assay
The caspase-3/7 activity assay kit was used to detect cell apoptosis. Briefly, LDHA-silenced or si-Ctrl GBM cells were seeded in 96-well plates at a density of 10 000 cells per well. After serum deprivation for 48 h, cell number and caspase-3/7 activity were measured in the same sample using the BCA kit (Thermo Scientific, Chicago, Illinois, USA) and Apo-ONE Caspase-3/7 assay (Promega, Beijing, China), respectively. Caspase-3/7 activity was estimated as the ratio of Apo-ONE/BCA signals. Three independent experiments were conducted and each experiment had six duplicates.
Cell invasion assays
The invasive potential of A172 or U87 cells was measured using a Transwell model (Corning, New York, USA) according to the manufacturer’s instructions. Briefly, a density of 20 000 cells was seeded in the upper chamber of matrigel-coated filters (BD Bioscience, San Diego, California, USA); the lower chamber was filled with 600 μl of Dulbecco’s modified Eagle medium or Eagle’s minimum essential medium containing 2% FBS. After incubation for 48 h, the invaded cells were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. The number of invaded cells was counted under a light microscope in three random fields. Three independent experiments were conducted and each experiment had three duplicates.
Measurement of glucose and lactate
A Glucose Colorimetric Assay Kit II (K686-100; BioVision, San Francisco, California, USA) and a Lactate Assay Kit (K607-100; BioVision) were used to estimate the intracellular glucose utilization and lactate concentration in A172 or U87 cells. GBM cells were cultured in fresh phenol red-free medium. The glucose utilization and lactate production were estimated using a standard glucose calibration curve prepared under the same condition according to the manufacturer’s instructions. Three independent experiments were conducted and each experiment had six duplicates.
Statistical analysis
All of the results reported were derived from at least three independent experiments. Data were presented as the means±SD. The SPSS software program (SPSS, Chicago, Illinois, USA) was used for statistical analysis. Graphical representations were performed using GraphPad Prism 5 (GraphPad Software, Inc., San Diego, California, USA) software. Comparison of results from experimental groups versus control groups using the Student t-test was performed using a two-sided Student’s t-test. P values less than 0.05 were considered statistically significant.
Results
LDHA expression is upregulated in GBM tissues
To investigate the expression pattern of LDHA in GBM, we determined the differential mRNA expression of LDHA between GBM tissues and normal brain tissues by analysis of the Oncomine microarray gene expression datasets. The expression of LDHA was significantly increased in GBM tissues compared with the corresponding normal brain tissues in the Lee brain dataset (Fig. 1a, P=1.95E−4), the Shai brain dataset (Fig. 1b, P=1.78E−8), the Sun brain dataset (Fig. 1c, P=3.53E−4), and the Liang brain dataset (Fig. 1d, P=0.013). Meanwhile, data derived from the TCGA database indicated that LDHA expression was also significantly higher in GBM tissues than normal brain tissues (Fig. 1e, P=9.77E−7). These observations highlighted a deregulated expression of LDHA in GBM.
Fig. 1.
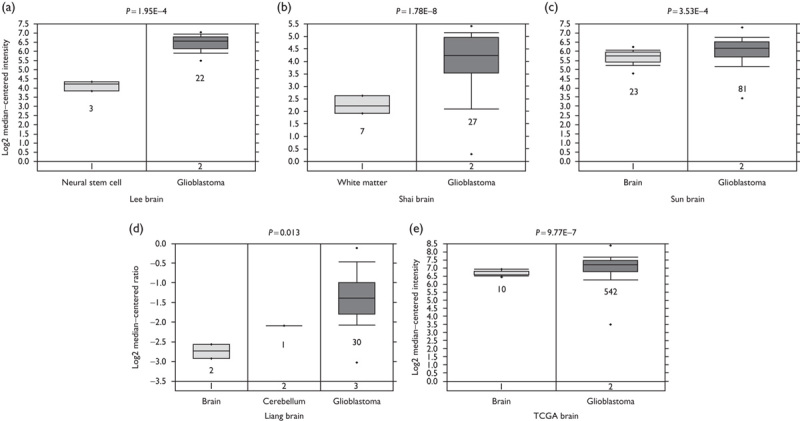
LDHA expression is upregulated in GBM tissues. The mRNA expression of LDHA in Oncomine datasets including Lee brain (a), Shai brain (b), Sun brain (c), and Liang brain (d), as well as TCGA brain (e) is upregulated in GBM tissues compared with the normal control tissues. GBM, glioblastoma; LDHA, lactate dehydrogenase A.
Silencing of LDHA compromises tumor growth and invasion in GBM cells
To further determine the potential oncogenic roles of LDHA in GBM, we suppressed LDHA expression in two GBM cell lines: A172 and U87. As shown in Fig. 2a, treatment of LDHA siRNAs markedly reduced LDAH protein expression relative to the control siRNAs in both A172 and U87 cells. After LDHA was silenced, we found that the acidification of culture medium was markedly decreased (Fig. 2b). Accordingly, the lactate level in the culture medium of si-LDHA cells was also reduced nearly 50% compared with that of si-Ctrl cells (Fig. 2c). Expectedly, silencing of LDHA also resulted in ∼25–30% reduction in glucose utilization (Fig. 2d), indicating that the LDHA level is closely associated with the Warburg effect. We next investigated the cellular functions of LDHA in A172 and U87 cells in vitro. Colony formation assay showed a significant decrease in the colony number of LDHA-silenced A172 (206±17 vs. 314±28) and U87 cells (184±25 vs. 277±14) compared with their si-Ctrl cells (Fig. 2e). Meanwhile, we found that LDHA-silenced cells showed enhanced caspase-3/7 activity compared with si-Ctrl cells in both A172 (4146±220 vs. 5351±491) and U87 cells (3420±271 vs. 4908±183) (Fig. 2f). Using the Transwell model, we observed that the invaded A172 cells in the LDHA-silenced group (235±26) were significantly less than those in the si-Ctrl group (404±23); a similar result was also found in U87 cells (Fig. 2g). Collectively, these results suggest that upregulated LDHA contributes toward tumor progression in GBM.
Fig. 2.
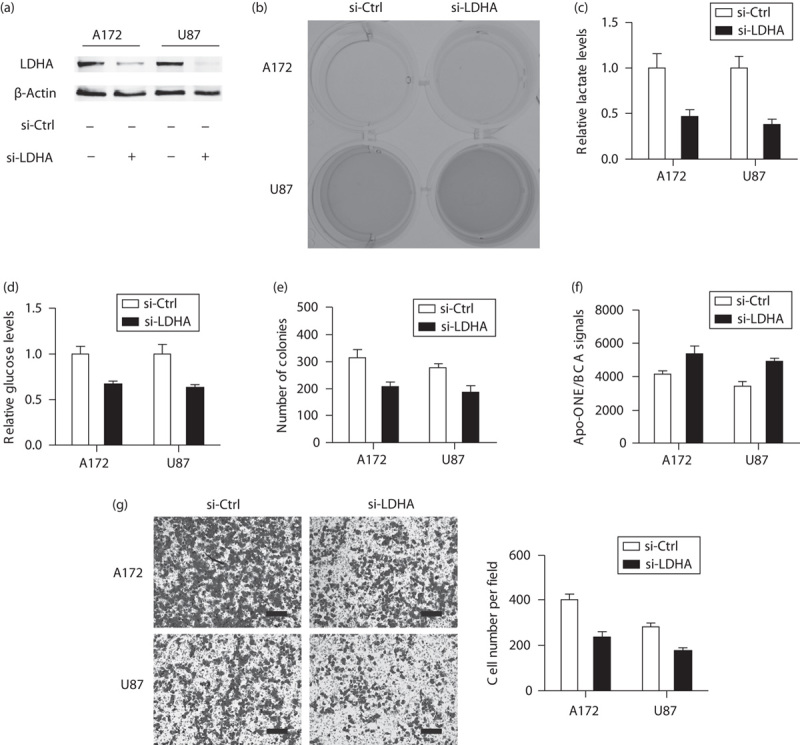
Silencing of LDHA compromises tumor growth and invasion in GBM cells. (a) Interfere efficiency of LDHA in A172 and U87 cells as shown by western blotting. (b) Representative photograph of culture medium in si-LDHA and si-Ctrl cells. Lactate production (c) and glucose utilization (d) in GBM cells were reduced after LDHA was silenced. Cell colony formation assay (e), cell apoptosis assay (f), and Transwell assay (g) were performed to detect cell proliferation ability, caspase-3/7 activity, and cell invasive ability in si-LDHA and si-Ctrl cells, respectively. LDHA-silenced cells showed decreased cell proliferation ability and invasive ability, and enhanced caspase-3/7 activity compared with si-Ctrl cells. Scale bar: 50 μm; si-Ctrl versus si-LDHA; *P<0.05; **P<0.01. GBM, glioblastoma; LDHA, lactate dehydrogenase A.
The oncogenic roles of LDHA are dependent on an enhanced Warburg effect
To elucidate the mechanisms underlying LDHA-mediated oncogenic roles, we observed the implications of silencing of LDHA in the presence of a glycolysis inhibitor 2-DG. As shown in Fig. 3a, no significant difference in acidification of culture medium was found between the si-LDHA and the si-Ctrl group. Consistent with this phenomenon, 2-DG also abolished the effects of LDHA on cell growth (Fig. 3b), cell apoptosis (Fig. 3c), and cell invasion (Fig. 3d), indicating that the oncogenic roles of LDHA were mediated by an altered Warburg effect.
Fig. 3.
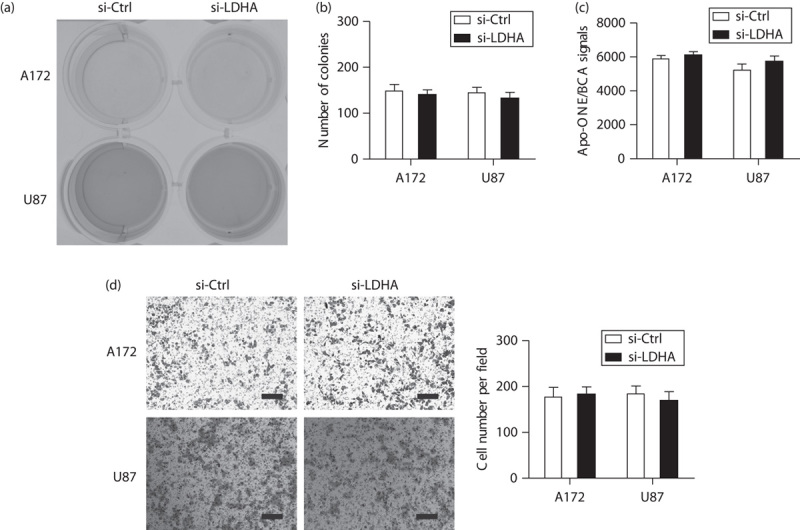
The oncogenic roles of LDHA are dependent on an enhanced Warburg effect. (a) Representative photograph of culture medium in si-LDHA and si-Ctrl cells in the presence of 2-DG. Effects of silencing of LDHA on cell growth as shown by colony formation assay (b), cell apoptosis as measured by caspase-3/7 activity, (c) and cell invasion as shown by the Transwell model (d) in A172 and U87 cells were measured. In the presence of 2-DG, the effects of LDHA on cell growth, cell apoptosis, and cell invasion were completely blocked. Scale bar: 50 μm; si-Ctrl versus si-LDHA; *P<0.05; **P<0.01. 2-DG, 2-deoxyglucose; GBM, glioblastoma; LDHA, lactate dehydrogenase A.
Discussion
In the current study, we determined the oncogenic roles of LDHA in GBM cell growth and invasion and its correlation with the Warburg effect. We found that the mRNA expression of LDHA was significantly upregulated in GBM tissues compared with normal tissues and suppression of LDHA repressed cell growth, increased cell apoptosis, and decreased cell invasion.
Upregulated LDHA has been reported in several previous studies. In colon cancer, overexpression of LDHA renders colon cancer cells resistant to 5-fluorouracil and this effect can be reversed by miR-34a, which inhibits LDHA expression 17. In colorectal cancer, elevated LDHA is also negatively regulated by miRNAs, including miR-34a, miR-34c, miR-369-3p, miR-374a, and miR-4524a/b 9. In thyroid cancer, estrogen-related receptor α regulates the expression of LDHA expression and the LDHA/LDHB ratio through interaction with the LDH promoter 18. In Corynebacterium glutamicum, it has been identified that SugR, a global repressor of genes involved in sugar uptake and glycolysis, represses LDHA transcription through binding to the its promoter region 19. Consistently with these findings in tumors, we also observed that LDHA expression is upregulated in several Oncomine datasets and the TCGA database in this study. However, the regulator involved in the expression of LDHA in GBM remains to be further identified.
Apart from its critical roles in glycolysis, LDHA also facilitates tumor progression 11,20,21. In line with this notion, we found that knockdown of LDHA resulted in a reduced Warburg effect as indicated by decreased glucose utilization and lactate production. In this study, the glucose consumption was measured at first 24 h after silencing of LDHA and shown as relative levels on the basis of protein quantification. At this time point, cell proliferation was slightly influenced by silencing of LDHA. Thus, we are more inclined to suggest that reduction in glucose utilization was a result of LDHA suppression on a per-cell basis. Consistently, cell growth arrest, increased cell apoptosis, and decreased cell invasion were induced by silencing of LDHA. Glycolysis not only provides cancer cells with building blocks but also reductive NADPH to protect cancer cells from apoptosis through the pentose phosphate pathway 22,23. A high lactate level induced by an enhanced Warburg effect contributes toward the acidification of the tumor microenvironment, which ultimately favors the activation of prometastatic genes (MMP2, MMP7, and PLAU) and inactivation of antimetastatic genes (MTSS1, TIMP2, and CTSK) 24. Therefore, it is reasonable to envisage the oncogenic roles of LDHA in GBM cell proliferation, apoptosis, and invasion. Expectedly, when glycolysis was inhibited by 2-DG treatment, the oncogenic activities of LDHA were completely abolished.
Conclusion
GBM is one of the most devastating primary malignant brain malignancies. In the past few years, considerable progress was achieved in our understanding of the molecular basis of GBM and these advances contributed toward the development of novel targeted therapies and individualized treatment. To the best of our knowledge, this is the first time that the expression pattern and cellular functions of LDHA in GBM have been determined. We described upregulated LDHA expression in GBM tissues and silencing of LDHA inhibits cell proliferation, invasion, and promotes cell apoptosis through a decreasing Warburg effect. Therefore, our present study provides evidence that LDHA plays crucial roles in the development and progression of human GBM and indicates that LDHA might be a therapeutic target in the treatment of GBM.
Acknowledgements
Conflicts of interest
There are no conflicts of interest.
References
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA 2013; 310:1842–1850. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005; 352:987–996. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Jiang T. Understanding high grade glioma: molecular mechanism, therapy and comprehensive management. Cancer Lett 2013; 331:139–146. [DOI] [PubMed] [Google Scholar]
- 4.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2012; 2:881–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer cell 2012; 21:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crane CA, Austgen K, Haberthur K, Hofmann C, Moyes KW, Avanesyan L, et al. Immune evasion mediated by tumor-derived lactate dehydrogenase induction of NKG2D ligands on myeloid cells in glioblastoma patients. Proc Natl Acad Sci USA 2014; 111:12823–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesnelong C, Chaumeil MM, Blough MD, Al-Najjar M, Stechishin OD, Chan JA, et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol 2014; 16:686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seliger C, Leukel P, Moeckel S, Jachnik B, Lottaz C, Kreutz M, et al. Lactate-modulated induction of THBS-1 activates transforming growth factor (TGF)-beta2 and migration of glioma cells in vitro. PLoS One 2013; 8:e78935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, Wang H, Liu A, Fang C, Hao J, Wang Z. Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 2015; 6:19456–19468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X, Yang Z, Chen Z, Chen R, Zhao D, Zhou Y, Qiao L. Effects of the suppression of lactate dehydrogenase A on the growth and invasion of human gastric cancer cells. Oncol Rep 2015; 33:157–162. [DOI] [PubMed] [Google Scholar]
- 11.Xian ZY, Liu JM, Chen QK, Chen HZ, Ye CJ, Xue J, et al. Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumour Biol 2015; 36:8093–8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheng SL, Liu JJ, Dai YH, Sun XG, Xiong XP, Huang G. Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. FEBS J 2012; 279:3898–3910. [DOI] [PubMed] [Google Scholar]
- 13.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell 2006; 9:425–434. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Hanai J, Ren JG, Kats L, Burgess K, Bhargava P, et al. Targeting lactate dehydrogenase – a inhibits tumorigenesis and tumor progression in mouse models of lung cancer and impacts tumor-initiating cells. Cell Metab 2014; 19:795–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao YH, Zhou M, Liu H, Ding Y, Khong HT, Yu D, et al. Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene 2009; 28:3689–3701. [DOI] [PubMed] [Google Scholar]
- 16.Arseneault R, Chien A, Newington JT, Rappon T, Harris R, Cumming RC. Attenuation of LDHA expression in cancer cells leads to redox-dependent alterations in cytoskeletal structure and cell migration. Cancer Lett 2013; 338:255–266. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Zhao H, Zhou X, Song L. Inhibition of lactate dehydrogenase A by microRNA-34a resensitizes colon cancer cells to 5-fluorouracil. Mol Med Rep 2015; 11:577–582. [DOI] [PubMed] [Google Scholar]
- 18.Mirebeau-Prunier D, Le Pennec S, Jacques C, Fontaine JF, Gueguen N, Boutet-Bouzamondo N, et al. Estrogen-related receptor alpha modulates lactate dehydrogenase activity in thyroid tumors. PLoS One 2013; 8:e58683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyoda K, Teramoto H, Inui M, Yukawa H. Molecular mechanism of SugR-mediated sugar-dependent expression of the ldhA gene encoding L-lactate dehydrogenase in Corynebacterium glutamicum. Appl Microbiol Biotechnol 2009; 83:315–327. [DOI] [PubMed] [Google Scholar]
- 20.Shi M, Cui J, Du J, Wei D, Jia Z, Zhang J, et al. A novel KLF4/LDHA signaling pathway regulates aerobic glycolysis in and progression of pancreatic cancer. Clin Cancer Res 2014; 20:4370–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He TL, Zhang YJ, Jiang H, Li XH, Zhu H, Zheng KL. The c-Myc-LDHA axis positively regulates aerobic glycolysis and promotes tumor progression in pancreatic cancer. Med Oncol 2015; 32:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dang CV, Le A, Gao P. Links between metabolism and cancer. Genes Dev 2012; 26:877–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finley LW, Zhang J, Ye J, Ward PS, Thompson CB. SnapShot: cancer metabolism pathways. Cell Metab 2013; 17:466–466.e2. [DOI] [PubMed] [Google Scholar]
- 24.Han T, Kang D, Ji D, Wang X, Zhan W, Fu M, et al. How does cancer cell metabolism affect tumor migration and invasion? Cell Adh Migr 2013; 7:395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]


