Abstract
Muscle denervation resulting from injury, disease or aging results in impaired motor function. Restoring neuromuscular communication requires axonal regrowth and endplate reinnervation. Muscle activity inhibits the reinnervation of denervated muscle. The mechanism by which muscle activity regulates muscle reinnervation is poorly understood. Dach2 and Hdac9 are activity-regulated transcriptional co-repressors that are highly expressed in innervated muscle and suppressed following muscle denervation. Dach2 and Hdac9 control the expression of endplate-associated genes such as those encoding nicotinic acetylcholine receptors (nAChRs). Here we tested the idea that Dach2 and Hdac9 mediate the effects of muscle activity on muscle reinnervation. Dach2 and Hdac9 were found to act in a collaborative fashion to inhibit reinnervation of denervated mouse skeletal muscle and appear to act, at least in part, by inhibiting denervation-dependent induction of Myog and Gdf5 gene expression. Although Dach2 and Hdac9 inhibit Myog and Gdf5 mRNA expression, Myog does not regulate Gdf5 transcription. Thus, Myog and Gdf5 appear to stimulate muscle reinnervation through parallel pathways. These studies suggest that manipulating the Dach2-Hdac9 signaling system, and Gdf5 in particular, might be a good approach for enhancing motor function in instances where neuromuscular communication has been disrupted.
KEY WORDS: Muscle, Nerve, Regeneration, Denervation, Neuromuscular junction, Myogenin, Dach2, Hdac9, Gdf5, Brachypodia, Motor nerve
Summary: Dach2 and Hdac9 work in a collaborative fashion to inhibit the reinnervation of denervated mouse skeletal muscle by suppressing the denervation-dependent expression of Myog and Gdf5.
INTRODUCTION
Communication between nerve and muscle takes place at the neuromuscular junction (NMJ) where presynaptic motor axons release neurotransmitter that is bound by receptors concentrated in the muscle's endplate region. This communication is often disrupted in people suffering from neuromuscular diseases and age-related sarcopenia, and results in reduced motor function. Therefore, devising strategies for restoring neuromuscular communication might help people with these conditions. Indeed, miR206 was found to promote regeneration of neuromuscular synapses and this appeared to slow disease progression and increase survival in a mouse model of amyotrophic lateral sclerosis (ALS) (Williams et al., 2009).
Although mechanisms controlling muscle reinnervation are poorly understood, it is clear that muscle activity plays an inhibitory role (Jansen et al., 1973; Cangiano et al., 1980; Hennig, 1987). Dach2 and Hdac9 are transcriptional co-repressors highly expressed in innervated muscle and suppressed following muscle denervation (Méjat et al., 2005; Tang and Goldman, 2006; Tang et al., 2009). They mediate activity-dependent suppression of Myog, a muscle-specific transcription factor that is induced in denervated muscle (Buonanno et al., 1992) and an activator of genes encoding endplate-associated proteins, like nicotinic acetylcholine receptors (nAChRs) and Musk (Méjat et al., 2005; Tang and Goldman, 2006; Tang et al., 2006). Thus, Dach2 and Hdac9 are candidates for mediating the inhibitory effects of muscle activity on muscle reinnervation.
In order for muscle activity to regulate muscle reinnervation it is expected to affect the expression of secreted factors that impact motor axons and their ability to reform neuromuscular connections. Previous studies have suggested that secreted factors like Igf, Bdnf, Gdnf, Nt-3 and Nt-4 (also known as Ntf3 and Ntf5, respectively) are released from muscle and might be involved in motor neuron survival, axonal sprouting and maturation (Smith et al., 1985; Rassendren et al., 1992; Caroni, 1993; Koliatsos et al., 1993; Funakoshi et al., 1995; Wang et al., 1995; Gould et al., 2008); however, none have been shown to regulate the reinnervation of endplates.
Here we report that Dach2 and Hdac9 collaborate to inhibit muscle reinnervation of pre-existing endplates. We show that Myog is regulated in a Dach2/Hdac9-dependent fashion and that loss of Myog expression results in delayed muscle reinnervation. Importantly, we found that Dach2-Hdac9 signaling also inhibits Gdf5 expression and that Gdf5 is a muscle tissue-derived factor that appears to act in a retrograde fashion to stimulate endplate reinnervation.
RESULTS
Dach2 and Hdac9 inhibit reinnervation of denervated muscle endplates
To investigate if Dach2 and Hdac9 controlled muscle reinnervation, we first used a nerve transfer model where the distal end of the tibial nerve innervating the soleus muscle is severed and transplanted to the soleus's periphery, and then allowed to re-grow over the muscle surface where it can form ectopic endplates and also reconnect with old endplates (Payne and Brushart, 1997). These experiments were performed on wild-type (Wt), Dach2−/−, Hdac9−/− and Dach2−/−/Hdac9−/− mice. Unlike the predominantly slow fiber composition of rat soleus muscle, the mouse soleus muscle is mixed with a slightly higher percentage of fast fibers over slow fibers (Wigston and English, 1992). Upon dissection of soleus muscles at 6 weeks post nerve transfer, Dach2−/−/Hdac9−/− muscles appeared to display less relative atrophy than those from Wt, Dach2−/− and Hdac9−/− animals (Fig. 1A, Fig. S1A). Quantification of muscle mass confirmed this observation (Fig. 1B). By contrast, when soleus muscles were not allowed to reinnervate in Wt and Dach2−/−/Hdac9−/− mice, a similar amount of denervation-dependent atrophy was observed (Fig. 1C). This suggested that the reduced atrophy noted in Dach2−/−/Hdac9−/− muscles after nerve transfer might have resulted from an enhanced rate of muscle reinnervation.
Fig. 1.
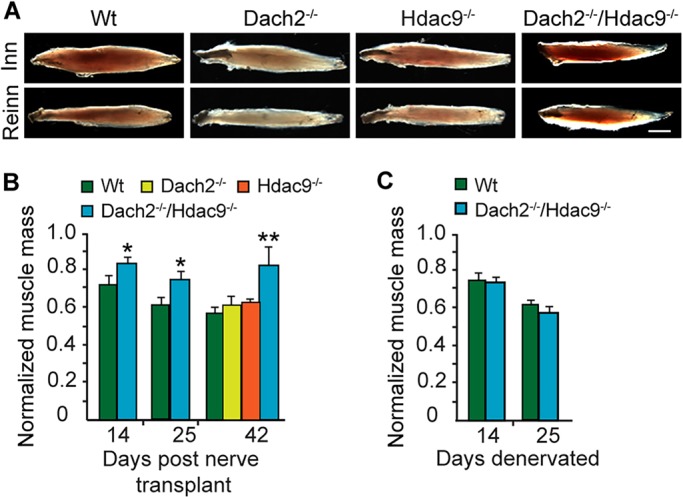
Recovery of soleus muscle mass and function reflects muscle reinnervation in Dach2−/−/Hdac9−/− mice. (A) Representative images of dissected soleus muscles from Wt, Dach2−/−, Hdac9−/− and Dach2−/−/Hdac9−/− mice at 6 weeks post muscle denervation and nerve transplant. Scale bar: 2 mm. (B) Quantification of reinnervated soleus muscle mass normalized to the contralateral innervated control. Error bars are s.d.; n=3 for 14 and 25 days and n=5 for 42 days post nerve transplant. *P<0.05; **P<0.01 relative to Wt. (C) Quantification of denervated soleus muscle mass normalized to the contralateral innervated control. Error bars are s.e.m.; n=3.
Encouraged by the above results, we quantified the number of reinnervated endplates in soleus muscles of Wt, Dach2−/−, Hdac9−/− and Dach2−/−/Hdac9−/− mice after muscle denervation with subsequent nerve transfer. For this analysis βIII tubulin immunofluorescence was used to visualize regenerating nerves and presynaptic terminals, and fluorescent α-bungarotoxin (α-Btx) was used to visualize muscle endplates (Fig. 2A). Depending on the extent of βIII tubulin and α-Btx overlap, endplates were scored as completely reinnervated (100% overlap), partially reinnervated (∼10-80% overlap) or denervated (no overlap). Within the first couple of days of muscle denervation, Wt and Dach2−/−/Hdac9−/− mice exhibited a similar rate of motor nerve degeneration with ∼90% of the muscle endplates exhibiting no association with βIII tubulin immunofluorescence and a complete loss of SV2 presynaptic terminal staining (data not shown). When denervated soleus muscles were evaluated for endplate reinnervation at 6 weeks post nerve transfer, we found significant differences at the original pre-existing endplates, which were more completely innervated in Dach2−/−/Hdac9−/− mice compared with Wt, Dach2−/−, or Hdac9−/− mice (Fig. 2B,C). The enhanced reinnervation of Dach2−/−/Hdac9−/− muscle was reflected by a stronger muscle twitch and tetanic force when stimulating the transplanted nerve (Fig. 2D). Consistent with these observations, we also found a tendency for an increase in the total number of ectopic αBtx+ plaques formed in Dach2−/−/Hdac9−/− muscles when they were evaluated 14 days post nerve transfer (Wt=433±132 versus Dach2−/−/Hdac9−/−=580±161). Over time it became increasingly difficult to visualize ectopic endplates as the transplanted nerve preferentially migrated toward the original endplates.
Fig. 2.
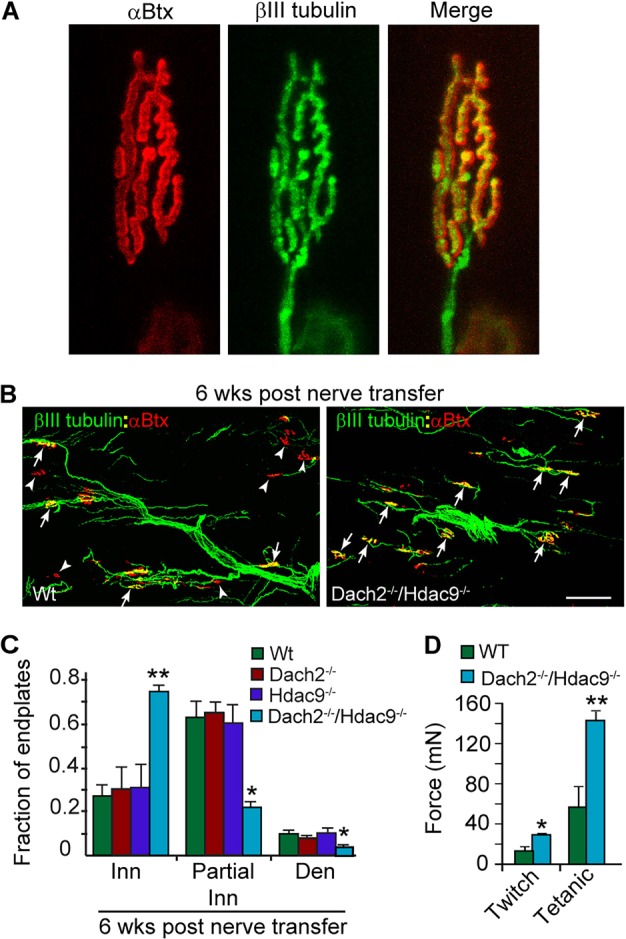
Dach2 and Hdac9 inhibit reinnervation of endplates in denervated soleus muscle following nerve transfer. (A) βIII tubulin staining of motor nerve terminals overlaps precisely with αBtx stained endplates. (B) Representative images and (C) quantification of reinnervated endplates at 6 weeks post nerve transfer. βIII tubulin+ regenerating tibial nerve branches are green and αBtx+ endplates are red. Arrows point to fully innervated endplates. Arrowheads point to denervated endplates. Scale bar: 50 µm. Error bars are s.e.m.; n=4 for Wt and Dach2−/−; n=3 for Hdac9−/− and Dach2−/−/Hdac9−/−. *P<0.05; **P<0.01 relative to Wt. (D) Soleus force measurements following indirect stimulation through the transplanted tibial nerve. Error bars are s.e.m.; n=6 for Wt and n=5 for Dach2−/−/Hdac9−/−. *P<0.05; **P<0.01 relative to Wt. Inn, innervated; Den, denervated.
When our analysis of presynaptic nerve terminal overlap with endplates was applied to innervated muscle, we found no significant differences between Wt and Dach2−/−/Hdac9−/− mice (Fig. 3B). Similarly, endplate surface areas and muscle fiber widths were unaffected by Dach2 and Hdac9 deletion (Fig. S1B), and no significant endplate fragmentation was noted. However, consistent with previous studies suggesting that Dach2 and Hdac9 mediate activity-dependent suppression of Myog and genes encoding nAChR proteins (Méjat et al., 2005; Tang and Goldman, 2006), we found their combined deletion in innervated and denervated muscle generally increased the expression of these genes more so than their individual deletion (Fig. S1C,D). Taken together, these data suggested that Dach2 and Hdac9 collaborate to inhibit muscle reinnervation.
Fig. 3.
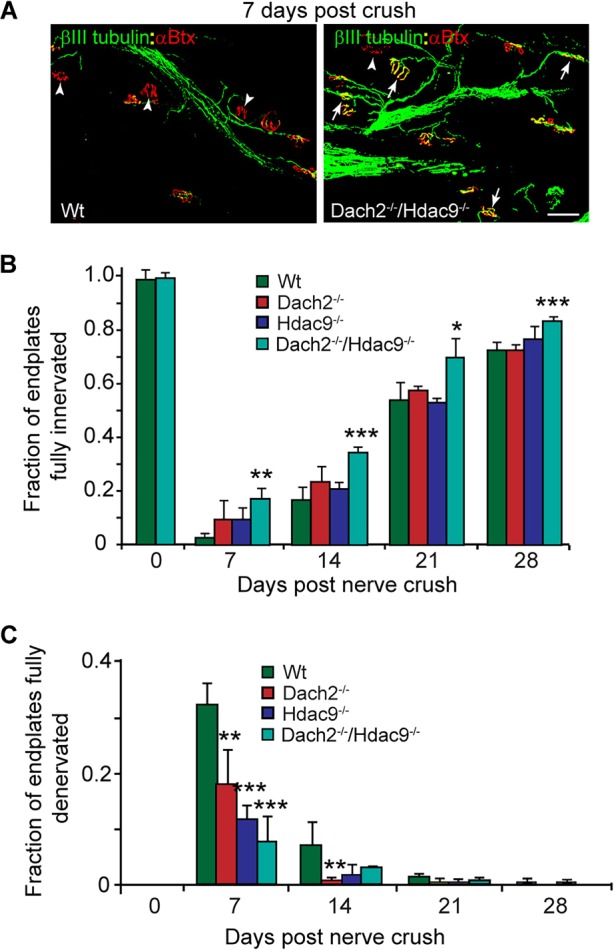
Dach2 and Hdac9 inhibit reinnervation of endplates in denervated soleus muscle following nerve crush. (A) Representative images and (B,C) quantification of innervated and denervated NMJs at various times post nerve crush. βIII tubulin+ regenerating tibial nerve branches are green and αBtx+ endplates are red. Arrows point to fully innervated endplates. Arrowheads point to denervated endplates. Scale bar: 50 µm. Error bars are s.e.m.; n=3. *P<0.05; **P<0.01; ***P<0.001 relative to Wt.
In the above model of muscle reinnervation, the interpretation of the results could have been influenced by the precise location of the nerve transplant as well as the degree of muscle damage induced during transplantation. Therefore, we next tested if Dach2 and Hdac9 also inhibited endplate reinnervation using a nerve crush model in which the tibial nerve branch to the soleus muscle is specifically directed to original endplates by regenerating through its perineural sheath. As in the nerve transplant experiments, we found a significant enhancement of endplate reinnervation in Dach2−/−/Hdac9−/− mice compared with Wt, Dach2−/−, or Hdac9−/− mice (Fig. 3A-C). This enhancement was not restricted to the soleus muscle, as denervated Dach2−/−/Hdac9−/− EDL muscle also exhibited a greater degree of reinnervation compared with Wt muscle at 14 days post peroneal nerve crush (Fig. S1F). The quantitative differences noted in muscle reinnervation when nerves were allowed to grow through their perineural sheath versus across the muscle surface might reflect differences in these two environments that are further amplified in mutant muscle.
Dach2 and Hdac9 act on muscle to inhibit endplate reinnervation
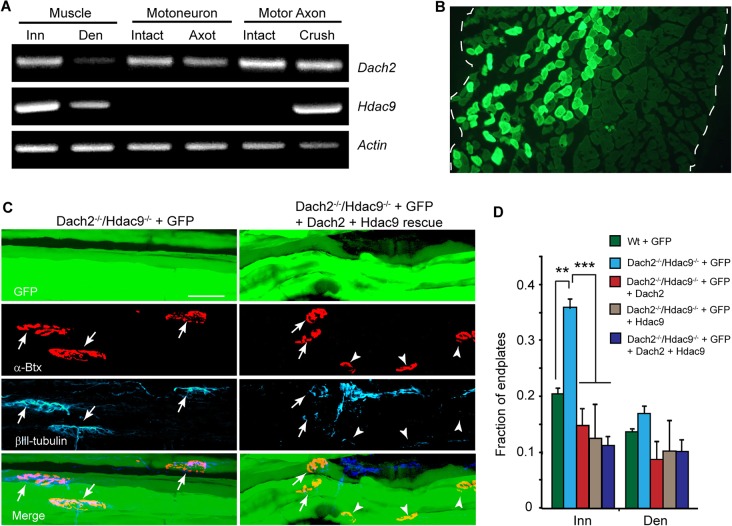
In addition to their expression in muscle, we found that Dach2 was expressed in motor neurons and cells associated with the motor nerves, whereas Hdac9 was induced in cells associated with injured motor nerves (Fig. 4A). This raised the possibility that Dach2 and Hdac9 might act on both muscle and nerve to regulate muscle reinnervation. To test if Dach2 and Hdac9 acted on muscle to regulate endplate reinnervation, we electroporated Dach2−/−/Hdac9−/− soleus muscle with sCMV expression vectors, allowing forced expression of Dach2 and Hdac9 individually or in combination (Fig. 4C,D). Electroporations also included sCMV:GFP so electroporated fibers could be identified. Previous studies have shown that soleus muscle electroporation results in expression that is restricted to electroporated muscle fibers (Kong et al., 2004; Sadasivam et al., 2005). Consistent with these studies, we find GFP confined to muscle and no indication of electroporation of Schwann cells or nerve (Fig. S1G). We also note that in contrast to denervated muscle, there are no macrophages in innervated muscle so they too cannot be electroporated (Fig. S1H). We generally observe ∼30% of the soleus fibers are electroporated at the site of DNA injection (Fig. 4B). At 12 days post nerve crush, the number of fully innervated and denervated endplates on GFP+ electroporated fibers was quantified. Consistent with the idea that Dach2 and Hdac9 act on muscle gene expression programs to regulate endplate reinnervation, forced expression of Dach2 and/or Hdac9 in denervated Dach2−/−/Hdac9−/− soleus muscle suppressed reinnervation (Fig. 4C,D). The incomplete inhibition of endplate reinnervation by Dach2 and Hdac9 overexpression in muscle might indicate insufficient expression levels, the contribution of other cell types, or the involvement of additional inhibitory pathways. Nonetheless, these data suggest Dach2 and Hdac9 act in a redundant fashion to suppress muscle reinnervation.
Fig. 4.
Dach2/Hdac9 inhibits muscle reinnervation. (A) RT-PCR analysis of Dach2, Hdac9 and γ-Actin mRNA in innervated (Inn) and denervated (Den) muscle; intact or axotomized (Axot) motor neurons and intact or crushed motor nerves and associated cells (Crush). (B) Representative image of soleus muscle cross section electroporated with CMV:GFP expression vector and stained for GFP immunofluorescence. Dashed line marks the muscle borders. (C) Representative images and (D) quantification of reinnervated endplates at 12 days post nerve crush in Wt and Dach2−/−/Hdac9−/− soleus muscles that were electroporated with GFP or Dach2, Hdac9 and GFP expression vectors. GFP marks electroporated muscle fibers green, αBtx marks endplates red and βIII tubulin marks regenerating axons cyan. Arrows point to fully and partially innervated endplates, whereas arrowheads point to denervated endplates. Error bars are s.d.; n=3 for Wt+GFP; n=4 for Dach2−/−/Hdac9−/−+GFP, n=3 for Dach2−/−/Hdac9−/− + GFP + Hdac9 and Dach2−/−/Hdac9−/− + GFP + Dach2 + Hdac9. **P<0.01; ***P<0.001 relative to Dach2−/−/Hdac9−/− + GFP. Scale bar: 50 µm.
Myog stimulates muscle reinnervation
Our data suggested that Dach2 and Hdac9 might act by repressing the expression of genes contributing to enhanced reinnervation of denervated muscle. Previous studies have shown Dach2 and Hdac9 repress Myog expression and that Myog regulated the expression of genes that are associated with endplate formation, like those encoding nAChRs and Musk (Méjat et al., 2005; Macpherson et al., 2006; Tang and Goldman, 2006; Tang et al., 2006, 2009). Interestingly, miR206, a gene previously shown to participate in endplate reinnervation (Williams et al., 2009) is also regulated in a Dach2/Hdac9-dependent manner (Fig. S1E).
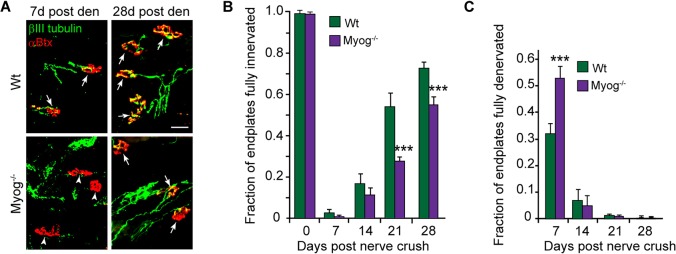
Because Myog is a transcription factor that has the potential to regulate gene expression programs impacting synapse regeneration, we investigated whether Myog deletion in adult muscle repressed endplate reinnervation. In innervated muscle, expression of nAChR-encoding genes is repressed (Goldman et al., 1988; Goldman and Staple, 1989; Buonanno et al., 1992) and Myog deletion modestly reduced this basal expression (Fig. S2A). Importantly, endplate surface areas and muscle fiber widths were unaffected by Myog deletion (Fig. S2B) and no significant endplate fragmentation was noted. By contrast, Myog deletion prior to nerve crush resulted in significant delays in the rate of endplate reinnervation (Fig. 5) and this was reversed by forced expression of Myog (Fig. S2C). Thus, Myog is under control of the Dach2-Hdac9 signaling system and its induction following soleus muscle denervation enhances endplate reinnervation.
Fig. 5.
Myog stimulates endplate reinnervation. (A) Representative images and (B,C) quantification of innervated and denervated endplates in Wt and Myog−/− soleus muscle at different times post nerve crush. βIII tubulin+ regenerating motor nerve branches are green and αBtx+ endplates are red. Arrows point to partially or fully innervated endplates and arrowheads point to denervated endplates. Scale bar: 50 µm. Error bars are s.d.; n=3 for 7 days and n=4 for 14, 21 and 28 days post nerve crush. ***P<0.001 relative to Wt.
Gdf5 is induced in denervated muscle in a Dach2/Hdac9-dependent manner
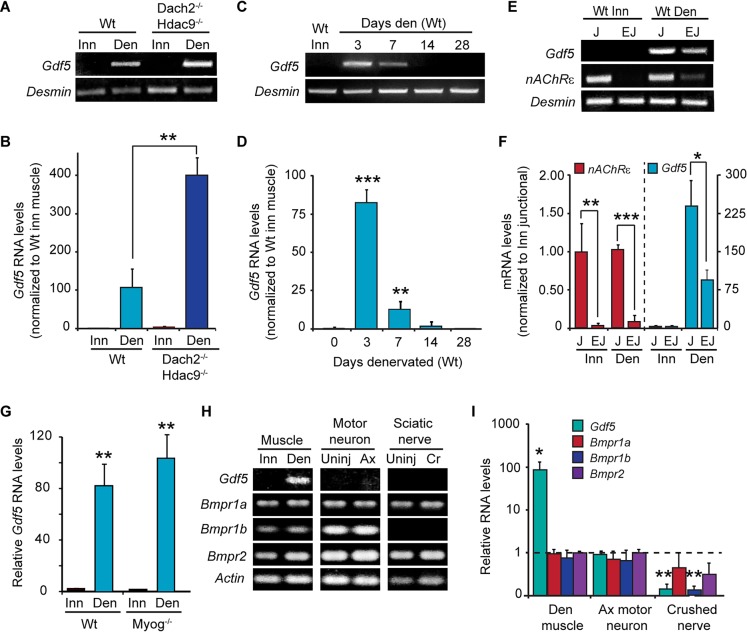
Our data are consistent with the idea that Dach2 and Hdac9 suppress the expression of genes that stimulate muscle reinnervation in adult muscle. Interestingly, even the reduced levels of Dach2 and Hdac9 found in denervated muscle are sufficient to inhibit muscle reinnervation (Figs 2, 3). Based on these observations we hypothesized that genes mediating the effects of Dach2 and Hdac9 would be induced following muscle denervation and further increased in denervated muscle of Dach2−/−/Hdac9−/− mice. Furthermore, because the products of these genes stimulate muscle reinnervation, we suspected they would encode secreted proteins that could act on motor neurons. Previously identified candidate genes included Gdnf, Bdnf, Cntf, Nt-3 and Fgfbp1 (Caroni, 1993; Koliatsos et al., 1993; Funakoshi et al., 1995; Griesbeck et al., 1995; Wang et al., 1995; Gould et al., 2008; Williams et al., 2009); however, none of these met our criterion of being super-induced in 3-day denervated muscle from Dach2−/−/Hdac9−/− mice (Fig. S3A). Therefore, we developed an RNAseq-based screen to identify genes that fit the above criterion. For this analysis, we compared the transcriptomes from Wt innervated muscle, Wt denervated muscle and Dach2−/−/Hdac9−/− denervated muscle. Candidate genes were identified by focusing on differentially expressed genes with an abundance greater than five fragments per kilobase of exon per million fragments mapped (FPKM), induced at least two-fold after Wt muscle denervation, super-induced in denervated muscle of Dach2−/−/Hdac9−/− mice and encoded a secreted protein (http://spd.cbi.pku.edu.cn/). This analysis identified Serpina3m, Serpina3n and Gdf5 (Fig. S3B). Of these, the most highly induced gene was Gdf5, a member of the bone morphogenetic protein (Bmp) family (Nishitoh et al., 1996). Although Bmps have not been previously associated with mammalian NMJs, they have been demonstrated to serve as retrograde signals that mediate activity-dependent remodeling of the Drosophila NMJ (Berke et al., 2013; Piccioli and Littleton, 2014). Because of its high induction in Dach2−/−/Hdac9−/− denervated muscle and this evolutionary connection, we decided to investigate if Gdf5 was a muscle tissue-derived factor that could regulate muscle reinnervation in an activity-dependent manner.
We first confirmed the RNAseq data using RT-PCR and qPCR. This analysis revealed ∼80-fold induction of Gdf5 mRNA at 3 days post denervation which was further enhanced when Dach2 and Hdac9 were deleted (Fig. 6A,B). Electroporation of denervated muscle with either a combination of sCMV:GFP, sCMV:Dach2 and sCMV:Hdac9 expression vectors or an sCMV:GFP vector alone, also suggested Dach2/Hdac9 inhibit denervation-dependent Gdf5 mRNA expression (Fig. S3C). Gdf5 mRNA levels peaked within 3 days post denervation of soleus muscle and then decreased over the next 10 days (Fig. 6C,D). This timeframe corresponds to when most endplates begin to reinnervate, as only ∼30% of the endplates are fully denervated at 7 days post nerve crush and this is reduced to less than 10% at 14 days post nerve crush (Fig. 3C).
Fig. 6.
Gdf5 gene expression is regulated by muscle activity in a Dach2/Hdac9-dependent manner. (A) RT-PCR and (B) qPCR show Gdf5 mRNA levels in innervated (Inn) and denervated (Den) soleus muscle from Wt and Dach2−/−/Hdac9−/− mice. Error bars are s.d.; n=3. **P<0.01. (C) RT-PCR and (D) qPCR shows changes in Gdf5 mRNA at various times post muscle denervation. Error bars are s.d.; n=3. **P<0.01; ***P<0.001. (E) RT-PCR and (F) qPCR show Gdf5 and nAChRε mRNA expression in junctional and extrajunctional regions of innervated and denervated soleus muscle. mRNA values are relative to innervated nAChRε mRNA (Y-axis on the left) and to innervated Gdf5 mRNA (Y-axis on the right). Error bars are s.d.; n=4. *P<0.05; **P<0.01; ***P<0.001. (G) qPCR show Gdf5 mRNA levels in innervated (Inn) and denervated (Den) soleus muscle from Wt and Myog−/− mice. Error bars are s.d.; n=3 for Wt and n=2 for Myog−/−. (H) RT-PCR and (I) qPCR shows expression levels of the indicated mRNAs in innervated and denervated soleus muscle, uninjured and axotomized (Ax) motor neurons and cells associated with uninjured or crushed (Cr) sciatic nerve. For qPCR values were normalized to γ-Actin for muscle and Gapdh for motor neuron and sciatic nerve. Error bars are s.d.; n=3. *P<0.05; **P<0.01 relative to uninjured for each tissue.
Using nAChRε (also known as Chrne) mRNA as a positive control for an RNA localized to NMJs (Goldman and Staple, 1989), we found Gdf5 mRNA is ∼3-fold more abundant in junctional than extrajunctional regions of denervated muscle (Fig. 6E,F), which might suggest a gradient of Gdf5 expression emanating from endplates. Surprisingly, denervation-dependent Gdf5 induction was independent of Myog expression (Fig. 6G). When comparing nerve and muscle components, we found Gdf5 predominantly in denervated muscle, whereas Gdf5 receptors (Bmpr1a, 1b and 2) are in muscle, motor nerve and sciatic nerve-associated cells (Fig. 6H,I). However, motor neurons appeared to preferentially express high levels of the preferred Gdf5 receptor complex Bmpr1b/Bmpr2 (Fig. 6H) (Nishitoh et al., 1996). These data are consistent with the idea that Gdf5 is a factor produced by muscle tissue that can act on both muscle and nerve.
Gdf5 stimulates muscle reinnervation
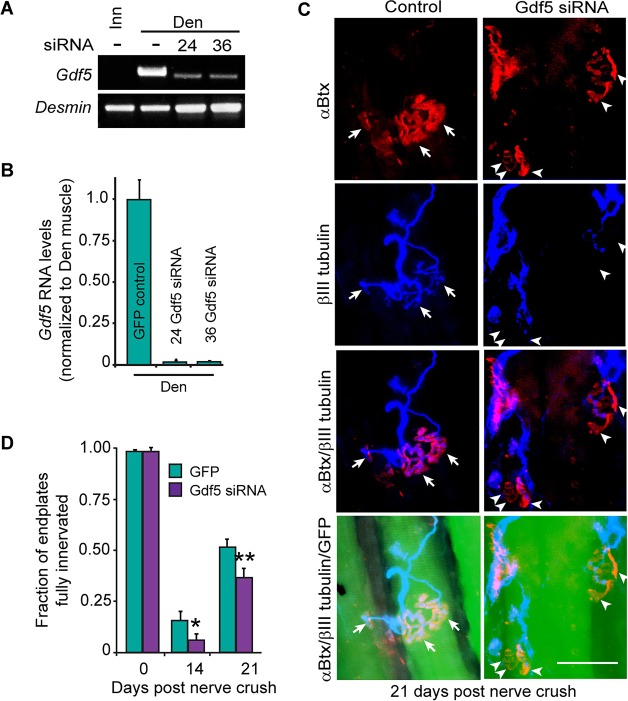
We next investigated if Gdf5 knockdown in denervated muscle affected muscle reinnervation. For this analysis we first electroporated muscle with a CMV:GFP expression vector and one of two different Gdf5-targeting siRNAs. We then crushed the tibial nerve branch leading to the soleus muscle. Dissection of GFP+ fibers and qPCR for Gdf5 mRNA indicated robust Gdf5 knockdown (Fig. 7A,B). Although Gdf5 knockdown had no effect on NMJs in innervated muscle (Fig. 7D), nerve crush experiments showed that muscle fibers with Gdf5 knockdown (GFP+ fibers) did not reinnervate endplates as well as Wt muscle (Fig. 7C,D). A control siRNA targeting E. coli lacZ mRNA had no effect on muscle reinnervation (Fig. 9A).
Fig. 7.
Gdf5 knockdown inhibits muscle reinnervation. (A) RT-PCR and (B) qPCR shows siRNAs 24 and 36 suppress Gdf5 mRNA levels. Error bars are s.d.; n=3 for control, n=1 for siRNA 24 and n=2 for siRNA 36. (C) Representative images of reinnervated endplates at 21 days post nerve crush in Wt soleus muscle electroporated with sCMV:GFP or a combination of sCMV:GFP and Gdf5-targeting siRNA24. αBtx marks endplates red; βIII tubulin marks regenerating axons cyan; and GFP identifies electroporated fibers. Arrows point to innervated endplates; arrowheads point to denervated endplates. Scale bar: 50 µm. (D) Quantification of regenerated neuromuscular synapses with and without Gdf5 knockdown. Error bars are s.d.; n=4 for GFP-treated 14 days post nerve crush and n=3 for 21 days post nerve crush. *P<0.05; **P<0.01 relative to GFP control.
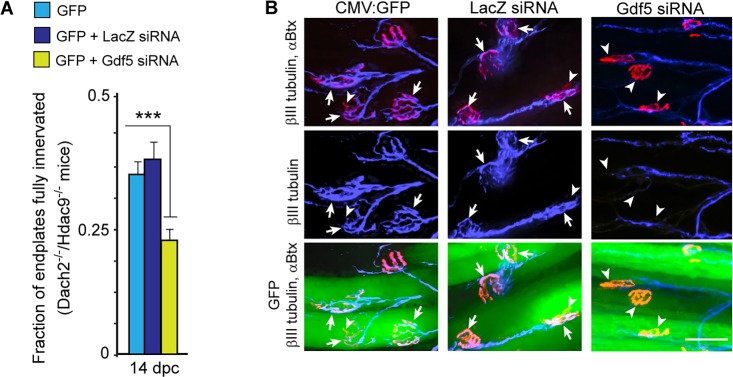
Fig. 9.
Gdf5 knockdown inhibits endplate reinnervation in Dach2−/−/Hdac9−/− mice. (A) Quantification of the percentage of fully innervated endplates at 14 days post nerve crush (dpc) in Dach2−/−/Hdac9−/− mice whose denervated soleus muscles were electroporated with a CMV:GFP expression vector and a siRNA targeting either E. coli lacZ mRNA or mouse Gdf5 mRNA. Error bars are s.d.; n=3. ***P<0.001. (B) Representative images of CMV:GFP and siRNA electroporated Dach2−/−/Hdac9−/− muscle fibers. GFP (green) identifies electroporated fibers, αBtx (red) identifies muscle endplates and βIII tubulin (cyan) identifies motor nerves. Arrowheads point to partially innervated and denervated endplates, whereas arrows point to fully innervated endplates. Scale bar: 50 µm.
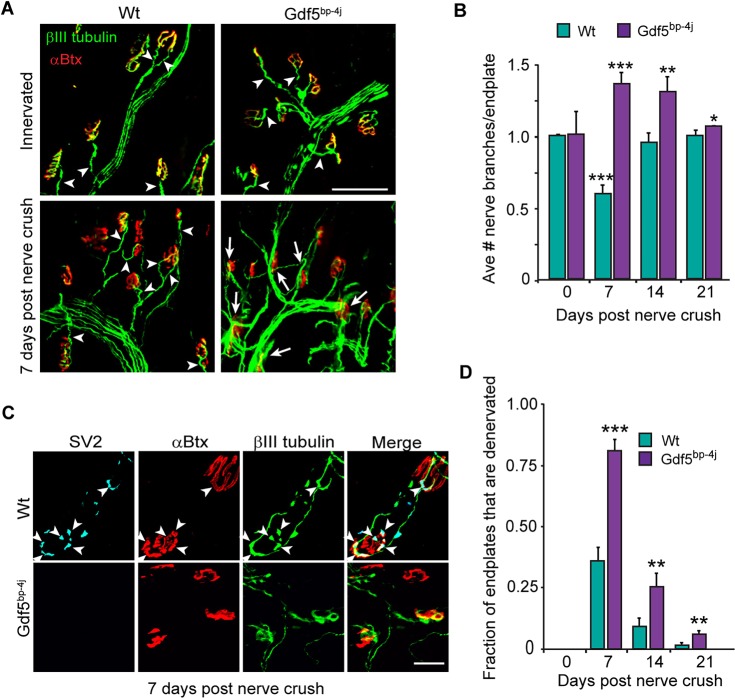
To further investigate a role for Gdf5 in muscle reinnervation and confirm the above results, we obtained Gdf5bp-4j mice from the Jackson Laboratory (stock 014183) that harbor a Gdf5 mutation that causes brachypodism (shortened digits and limbs). Inspection of the soleus muscle in these animals revealed thinner muscle fibers and smaller endplates that despite their diminished size, maintained their appropriate proportionality and were fully innervated (Fig. S4A,B). Although Gdf5bp-4j mice exhibited a trend towards decreased nAChRα (also known as Chrna1) and increased nAChRε gene expression these were not statistically significant changes (Fig. S4C). At 7 days post tibial nerve crush, the regenerating Gdf5bp-4j tibial nerve appeared to exhibit excessive branching, which was quantified by counting the number of βIII tubulin+ nerve branches that contacted or crossed over an individual endplate (Fig. 8A,B). Because of the excessive branching noted in the regenerating tibial nerve of Gdf5bp-4j mice, we were concerned that βIII tubulin staining might have difficulty in distinguishing nerves growing over endplates from those forming NMJs. Therefore, we used SV2 immunofluorescence to quantify synapse regeneration at 7 days post nerve crush and found more fully denervated endplates in Gdf5bp-4j mice (Fig. 8C,D; Fig. S4D-F). Although many endplates had nerve branches growing over them in the Gdf5bp-4j mice at 7 days post nerve crush (Fig. 8A,B), most (∼80%) were scored as denervated as they lacked differentiated SV2-positive nerve terminals (Fig. 8C,D). By contrast, most nerve branches contacting endplates in Wt mice at 7 days post nerve crush exhibited some SV2 staining (Fig. 8C,D). It is noteworthy that even at 7 days post nerve crush in Wt muscle, βIII tubulin and SV2 staining gave comparable results (∼35% fully denervated endplates; Fig. 3C, Fig. 8D). At later time points βIII tubulin staining proved reliable in labeling nerve terminals in Gdf5bp-4j mice and demonstrated that the lag in muscle reinnnervation in Gdf5bp-4j mice persisted out to 21 days post nerve crush (Fig. 8D). Importantly, Gdf5bp-4j motor nerve terminals degenerated at a similar rate as that observed in Wt and Dach2−/−/Hdac9−/− mice. We electroporated Gdf5bp-4j soleus muscle with either CMV:GFP alone or a combination of CMV:GFP and CMV:Gdf5 expression vectors and found that forced expression of Gdf5 in muscle rescued the poor reinnervation phenotype of Gdf5bp-4j mice (Fig. S4G). This analysis is consistent with the idea that denervation-dependent Gdf5 expression in muscle enhances muscle reinnervation and synapse formation.
Fig. 8.
Gdf5 stimulates muscle reinnervation. (A) Representative images showing increased axonal branching over the soleus muscle in Gdf5bp-4j mice at 7 days post nerve crush. βIII tubulin+ regenerating motor nerve branches are green and αBtx+ endplates are red. Arrowheads point to a single axonal branch innervating endplates, whereas arrows point to multiple axonal branches coursing over the endplate. Scale bar: 100 µm. (B) Quantification of the number of regenerating axonal branches/endplate in Wt and Gdf5bp-4j mice. Error bars are s.d.; n=3. *P<0.05; **P<0.01; ***P<0.001 relative to Wt. (C) Representative images showing reduced differentiation of axon terminals at endplates in Gdf5bp-4j mice at 7 days post nerve crush compared with Wt mice. SV2+ differentiated axon terminals are cyan, βIII tubulin+ regenerating motor nerves are green and αBtx+ endplates are red. Arrowheads point to SV2+ differentiated axon terminals that are also βIII tubulin+ and innervating αBtx+ endplates. (D) Quantification of denervated endplates in Wt and Gdf5bp-4j mice. Error bars are s.d.; n=3 for 7 and 14 days post nerve crush; n=4 for 21 days post nerve crush. **P<0.01; ***P<0.001 relative to Wt.
Our analysis of muscle reinnervation in Dach2−/−/Hdac9−/− mice revealed enhanced rates of muscle reinnervation (Figs 2, 3) that was correlated with Gdf5 mRNA super-induction (Fig. 6A,B). To directly test if Gdf5 mRNA induction contributed to the enhanced muscle reinnervation noted in Dach2−/−/Hdac9−/− mice, we electroporated denervated Dach2−/−/Hdac9−/− muscle with either a control siRNA targeting E. coli lacZ mRNA or an experimental siRNA targeting mouse Gdf5 mRNA. Fourteen days post muscle denervation, muscles were sectioned and fully innervated endplates were quantified. This analysis showed that knockdown of Gdf5 reduced the number of fully innervated endplates by ∼40% (Fig. 9A,B). Thus, Gdf5 contributes to both the normal reinnervation of endplates in Wt mice and the enhanced reinnervation of endplates noted in Dach2−/−/Hdac9−/− mice.
Identification of the Gdfbp-4j mutation causing brachypodism and reduced endplate reinnervation
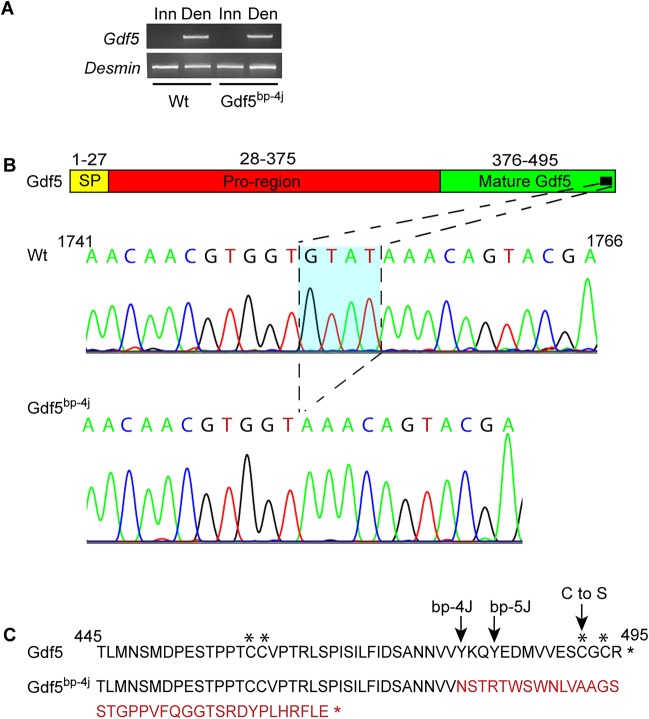
The Gdf5bp-4j mouse harbors a spontaneous recessive mutation that maps to the brachypodism locus on chromosome 2 and complementation tests with other brachypodism alleles proved this mutation is an allele of Gdf5 (Jackson Laboratory, stock 014183). However, the nature of this mutation remained unknown. RT-PCR confirmed normal denervation-dependent Gdf5 mRNA expression in Gdf5bp-4j mouse muscle (Fig. 10A). To identify the Gdf5bp-4j mutation, we sequenced Gdf5 cDNA prepared from denervated Wt and Gdf5bp-4j muscle. This analysis identified a 4 bp deletion in the Gdf5 mRNA from Gdf5bp-4j mice that causes a reading frame shift in the carboxy-terminal region (Fig. 10B,C). Amino acid changes in this region have previously been implicated in causing brachypodism in the Gdf5bp-5j mouse (T484N) (Jackson Laboratory, stock 021333) and in humans (C498S) (Everman et al., 2002). These data suggest the possibility that human mutations causing brachypodism might also inhibit muscle reinnervation.
Fig. 10.
Identification of Gdf5 gene mutation in Gdf5bp-4j mice. (A) RT-PCR shows normal denervation-dependent induction of Gdf5 mRNA in the Gdf5bp-4j mouse soleus muscle. (B) Diagram of Gdf5 protein sequence and DNA sequence of the Gdf5 carboxy-terminal region cDNA prepared from denervated Wt and Gdf5bp-4j muscle. A four nucleotide deletion is identified that alters the Gdf5 reading frame (C) in Gdf5bp-4j mice (amino acids in red). Mutations known to cause brachypodism in this region of the Gdf5 protein are indicated with arrows above the protein sequence. Cysteines that contribute to intrachain disulfide bonds are indicated with asterisks above the protein sequence.
DISCUSSION
The work reported here identifies a Dach2-Hdac9 signaling system that mediates the inhibitory effects of muscle activity on muscle reinnervation and appears to do so, at least in part, by inhibiting the expression of RNAs encoding Myog, miR206 and Gdf5. We found that Gdf5 is specifically expressed in muscle tissue and appears to act in a retrograde fashion to stimulate endplate reinnervation. Our analysis of Gdf5 mRNA expression suggests that Gdf5 is enriched in endplate regions of the denervated muscle fiber. Finally, we identified a 4-nucleotide deletion in the Gdf5 gene that is responsible for both brachypodia and reduced muscle reinnervation in Gdf5bp-5j mice.
The reinnervation of denervated muscle is a multistep process where motor axons must regrow over the muscle surface, recognize and approach denervated endplates, and then grow over this structure so axon terminals are in perfect register with the underlying endplate. Our data suggests that the Dach2-Hdac9-Gdf5 signaling system might participate in many of these events. The observation that Gdf5 mRNA is expressed in a gradient emanating from denervated endplates might indicate that the regenerating motor axons use this gradient to identify old endplates. Furthermore, the reduced differentiation of axon terminals and increased axonal branching in Gdf5bp-4j mice suggests that the Gdf5 signaling gradient might contribute to endplate recognition by the axon, axonal growth arrest and axon terminal differentiation.
Dach2 and Hdac9 are transcriptional corepressors that are highly expressed in innervated muscle and decrease following muscle denervation (Méjat et al., 2005; Tang and Goldman, 2006; Tang et al., 2009). Using two models of nerve injury we show that Dach2 and Hdac9 reduce the rate of muscle reinnervation. These factors appear to act in a collaborative fashion as their individual deletion had only small effects. Remarkably, even in denervated muscle, residual Dach2 and Hdac9 are sufficient to inhibit muscle reinnervation. Our data suggest that Dach2 and Hdac9 mediate their effects, at least in part, by controlling Myog and Gdf5 expression. Both Myog and Gdf5 mRNAs are induced in Wt denervated muscle and super-induced in denervated muscle from Dach2/Hdac9 null mice. Importantly, Myog deleted mice and Gdf5bp-4j mutant mice had a slower rate of soleus muscle reinnervation than Wt mice, and the enhanced reinnervation of endplates noted in Dach2−/−/Hdac9−/− mice is inhibited by Gdf5 knockdown. Although our observations on the effect of Myog and Gdf5 on muscle reinnervation were limited to soleus muscle, these proteins are induced after denervation of other skeletal muscles (Tang and Goldman, 2006; Tang et al., 2009; Sartori et al., 2013); however, the generality of their effect on muscle reinnervation remains to be tested.
The Dach2-Hdac9-Myog signaling system might contribute to endplate reinnervation by regulating endplate-associated genes, like those encoding Musk and nAChRs that are crucial components of the endplate complex. In addition, miR206 is an important regulator of muscle reinnervation (Williams et al., 2009) and our data indicates that miR206 is regulated by the Dach2-Hdac9 signaling system. Interestingly, the miR206 promoter harbors Myog consensus E-box binding sites that control its induction in denervated muscle (Rao et al., 2006; Williams et al., 2009). Based on these regulatory properties and the observations that loss of Myog or miR206 expression results in attenuated rates of reinnervation, we propose that the Dach2-Hdac9 signaling system inhibits muscle reinnervation, in part by suppressing a Myog-miR206 signaling cascade. In addition, we suspect other factors will also impinge on these signaling pathways and two such candidates are Msy3 (also known as Ybx3) and MyoD (also known as Myod1). Msy3 is induced in innervated muscle and, like Dach2 and/or Hdac9, suppresses Myog gene expression (Berghella et al., 2008) and thereby, might inhibit muscle reinnervation. MyoD is an activity-regulated transcription factor that is induced in denervated muscle (Buonanno et al., 1992) and often collaborates with Myog to regulate gene expression (Cao et al., 2006).
Myog induction has previously been noted to regulate denervation-dependent muscle atrophy, at least in part by activating atrophy-related genes (Moresi et al., 2010; Macpherson et al., 2011). In innervated muscle of Dach2−/−/Hdac9−/− mice, we find increased Myog without increased muscle atrophy. This might suggest that Myog is insufficient to drive muscle atrophy on its own and collaborates with other factors induced in denervated muscle, or that the levels of Myog induction noted in Dach2−/−/Hdac9−/− innervated muscle are below the threshold for stimulating muscle atrophy. Indeed, although Myog RNA levels are elevated in Dach2−/−/Hdac9−/− innervated muscle, they are around one-tenth that observed after muscle denervation.
An analysis of the denervated muscle transcriptome for RNAs encoding secreted factors that were induced in denervated muscle and super-induced in denervated muscle from Dach2/Hdac9 null mice identified Gdf5, Serpina3m and Serpina3n. Serpina3m and Serpina3n are members of a large family of secreted serine protease inhibitors (Forsyth et al., 2003) that appear to be expressed at the NMJ and are regulated by muscle denervation (Akaaboune et al., 1993, 1994). Although their function remains untested, they could influence the availability of trophic factors and modify the extracellular environment of the muscle so it is more conducive to muscle reinnervation.
We chose to focus on Gdf5, a member of the Bmp and Tgf-β family of secreted proteins that has been shown to regulate mammalian bone and/or cartilage formation and muscle atrophy (Storm et al., 1994; Jin and Li, 2013; Sartori et al., 2013). Whereas Bmp family members have been shown to regulate activity-dependent formation and plasticity of the Drosophila NMJ (Berke et al., 2013; Piccioli and Littleton, 2014), our study is the first to suggest a role for these proteins at the mammalian NMJ. Although we show that Gdf5 is super-induced in Dach2/Hdac9 null mice and that loss of Gdf5 inhibits muscle reinnervation, we found that denervation-dependent induction of Gdf5 was independent of Myog. This suggests that the Dach2-Hdac9 signaling system participates in the regulation of at least two signaling pathways that converge on endplate reinnervation.
Although our data suggest Gdf5 regulates muscle reinnervation, a number of additional considerations regarding Gdf5 remain to be resolved. First, it is not known if the Gdf5 mRNA gradient is reflected by Gdf5 protein expression and whether a gradient of Gdf5 expression is important to endplate reinnervation. Second, although our data demonstrates that Gdf5 is highly enriched in denervated muscle tissue relative to nerve-associated cells and motor neurons, we cannot currently rule out the possibility that this enrichment occurs in nerve terminal associated Schwann cells, or some other muscle-associated cell type. However, its expression in extrajunctional regions of the muscle does suggest that at least part of the Gdf5 signal is derived from muscle. Third, we cannot unequivocally state that Gdf5 functions in a retrograde manner to promote muscle reinnervation. Our data indicate that receptors for Gdf5 are expressed in both muscle tissue and motor neurons. Given the observation that Gdf5 expression attenuates the magnitude of denervation-induced atrophy (Sartori et al., 2013), it is possible that the delay in reinnervation observed in Gdf5bp-4j mice was secondary to the loss of Gdf5 signaling within the muscle itself. Resolving this issue will require experiments involving tissue-specific knockout of Gdf5 receptors in both muscle and motor nerves.
It is interesting that the Dach2-Hdac9 signaling system regulates a variety of activity-dependent processes like muscle reinnervation, muscle metabolism, muscle fiber type determination and muscle atrophy (Méjat et al., 2005; Tang and Goldman, 2006; Tang et al., 2006, 2009; Moresi et al., 2010; Macpherson et al., 2011). The coordinate activation of these processes in denervated muscle might help promote muscle survival and reinnervation after nerve damage. Thus, this signaling system appears to play a key role in maintaining motor function in adult animals and might represent a crucial target for enhancing motor function when neuromuscular communication is compromised.
In conclusion, our data suggest that a Dach2-Hdac9 signaling system mediates the inhibitory effects of muscle activity on muscle reinnervation. Dach2 and Hdac9 are highly expressed in innervated muscle and reduced by muscle denervation. This reduction correlates with enhanced reinnervation of denervated muscle and we found that forced overexpression of Dach2 and Hdac9 inhibits this reinnervation. Our studies suggest that the Dach2-Hdac9 signaling system inhibits muscle reinnervation by acting on at least two different pathways; one controlled by Myog and another acting via Gdf5. Our studies suggest Gdf5 might represent a secreted factor that can be used to enhance motor function in people suffering from neuromuscular diseases and age-related sarcopenia.
MATERIALS AND METHODS
Mouse lines
Dach2−/− and Hdac9−/− mice were previously described (Zhang et al., 2002; Méjat et al., 2005; Davis et al., 2006). Dach2−/−/Hdac9−/− double knockout mice were generated by breeding Dach2−/− and Hdac9−/− mice with each other. Myog conditional knockout mice (Myogflox/flox;CAGG-CreER) were described previously (Knapp et al., 2006). Tamoxifen-mediated recombination was performed as previously described (Knapp et al., 2006; Macpherson et al., 2011). Gdf5bp-4j mice were obtained from The Jackson Laboratory (Bar Harbor, Maine). All animal care and experimental procedures were carried out in accordance with guidelines and approval of the University Committee on Use and Care of Animals at the University of Michigan. See supplementary materials and methods for details.
Muscle denervation
Muscle denervations followed previously published protocols (Goldman et al., 1988; Macpherson et al., 2011). To prevent muscle reinnervation after nerve sectioning, the proximal nerve stump was implanted into the hip muscle, which allows for permanent denervation of the lower leg (Carlson and Faulkner, 1988). See supplementary materials and methods for details.
Laser capture microdissection, RNA isolation and real-time PCR
The Arcturus AutoPix system was used to isolate motor neurons from adult spinal cord sections. Sections were stained with Cresyl Violet as previously described (Veldman et al., 2007). RNA was isolated from microdissected samples using a PicoPure RNA Isolation Kit according to manufacturer's directions. For all other tissues, total RNA was isolated using TRIzol (Invitrogen). A Turbo DNase Kit (Ambion) was used to remove residual DNA and total RNA was quantified spectrophotometrically. One microgram of total RNA was used to generate cDNA with random primers and Superscript II reverse transcriptase (Invitrogen). Real-time PCR was performed with SYBR Green mix (1:20,000 SYBR Green) on an iCycler iQ real-time detection system (BioRad) according to the manufacturer's instructions. Each reaction was performed in duplicate or triplicate on at least three samples for each condition in a volume of 25 μl. Real-time PCR results for muscle were normalized to γ-Actin or desmin mRNA whereas those for spinal cord and sciatic nerve were normalized to gapdh mRNA. To assay miR206, poly(A) tails were first added to total RNA (Ambion) and subsequent reverse transcription was performed with Superscript II and a poly(T) adapter [5′-GCGAGCACAGAATTAATACGACTCACTATAGG-(T)12VN-3′]. Following cDNA synthesis real-time qPCR was performed as described above. PCR primers are listed in Table S1.
Immunofluorescence analysis of NMJs and nerves
Standard immunofluorescence protocols were used. Briefly, soleus muscles were fixed for 10 min in 4% paraformaldehyde and cut to 40 to 80 μm-thick longitudinal sections. After blocking, endplates were visualized by staining with Alexa Fluor 594-conjugated α-bungarotoxin (α-Btx; 1:2000; ThermoFisher, B-13423) and rabbit anti-βIII-tubulin antibody (1:1000; BioLegend, 802001) or mouse anti-SV2 antibody (1:50; Developmental Studies Hybridoma Bank, SV2). Washed muscles were incubated in secondary antibodies (see the supplementary materials and methods). Following secondary antibody incubation, slides were washed, coverslipped and examined with either a Zeiss Axiophot fluorescence microscope, Zeiss Axiophot, Axio Observer Z.1 or an Olympus FV1000 laser scanning confocal microscope. Quantification of muscle reinnervation in Wt and knockout mice was done blindly by two different individuals using software associated with the Axio Observer Z.1 or Olympus FV1000 microscopes.
Nerve branching analysis was performed on soleus muscles after tibial nerve crush. For this analysis the number of nerve branches that either contacted or crossed over an individual endplate was counted. Approximately 100 endplates from each muscle were analyzed and the data represent the average number of nerve branches per endplate for three muscles. See supplementary materials and methods for details.
Muscle functional measurements
Soleus muscles were surgically freed from surrounding muscle and connective tissue, the distal tendon was secured to the lever arm of a servomotor (Cambridge Instruments) with 5-0 suture and the hindlimb was securely tied to a fixed post with 4-0 suture at the knee. Soleus muscle maximum isometric twitch and tetanic force measurements were determined by indirect nerve stimulation. See supplementary materials and methods for details.
Plasmids, siRNA and muscle electroporation
Plasmids and siRNAs were injected individually or together into innervated soleus muscles. The molar ratio of stealth siRNA to enhanced green fluorescent protein (EGFP) plasmid was >500-fold so as to ensure all GFP+ fibers harbor siRNA. Nucleic acid uptake was facilitated by electroporation as previously described (Tang et al., 2009). Following electroporation, the tibial nerve to the soleus muscle was crushed as described above. See supplementary materials and methods for details.
RNAseq and DNA sequencing
DNA sequencing was performed by the University of Michigan DNA Sequencing Core. RNA was isolated from innervated and 3 day denervated Wt soleus muscle and denervated Dach2−/−/Hdac9−/− soleus muscle using Direct-zol RNA MiniPrep Kit (Zymo Research) and was treated with DNase (Invitrogen). RNA was submitted to the University of Michigan DNA Sequencing Core for the generation of RNAseq libraries according to the Illumina TruSeq protocol. Two independent libraries were prepared for each sample. Sequencing was performed on an Illumina HiSeq 2000 sequencer. The University of Michigan's Bioinformatics Core analyzed DNA sequencing data. CUFFDIFF2 (http://cole-trapnell-lab.github.io/cufflinks/) was used to determine differentially expressed genes, with a false discovery rate of 5%, fold-changes of at least two and abundance greater than five FPKM. The secreted protein database (SPD) (http://spd.cbi.pku.edu.cn/) was used to annotate differentially expressed genes. RNAseq data was deposited in GEO with accession number GSE58669. See supplementary materials and methods for details.
Statistics
All experiments were performed a minimum of three times. Statistical analysis was performed using two-tailed Student's t-test and, where appropriate, ANOVA with post-hoc Bonferroni t-tests. The level of significance was set a priori at P<0.05.
Acknowledgements
We thank Eric Olson, Graeme Mardon, and William Klein for providing Hdac9−/−, Dach2−/− and Myogflox/flox;CAGG-CreER mice, respectively; The University of Michigan DNA Sequencing Core for RNAseq library preparation and DNA sequencing; Ana Rodrigues Grant and the University of Michigan Bioinformatics Core for bioinformatics support; Carol Davis and Susan Brooks for help with nerve stimulation and muscle force measurements; Robert Thompson for help with laser capture microscopy; Dennis Claflin for assistance with instrumentation for muscle force assays; Curtis Powell for the CMV:Gdf5 construct; and Eli Cornblath for help in quantifying regenerated neuromuscular junctions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
D.G. conceived the study, designed experiments and wrote the manuscript. P.C.D.M. designed and conducted experiments and edited the paper. P.F. conducted experiments.
Funding
This work was supported by NIH Grant [RO1 NS059848] from National Institute of Neurological Disorders and Stroke (D.G.). Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.125674/-/DC1
References
- Akaaboune M., Ma J., Festoff B. W., Greenberg B. D. and Hantaï D. (1993). The influence of denervation on beta-amyloid protein precursor and alpha 1-antichymotrypsin in mouse skeletal muscle. Neuromuscul. Disord. 3, 477-481. 10.1016/0960-8966(93)90100-X [DOI] [PubMed] [Google Scholar]
- Akaaboune M., Ma J., Festoff B. W., Greenberg B. D. and Hantaï D. (1994). Neurotrophic regulation of mouse muscle beta-amyloid protein precursor and alpha 1-antichymotrypsin as revealed by axotomy. J. Neurobiol. 25, 503-514. 10.1002/neu.480250505 [DOI] [PubMed] [Google Scholar]
- Berghella L., De Angelis L., De Buysscher T., Mortazavi A., Biressi S., Forcales S. V., Sirabella D., Cossu G. and Wold B. J. (2008). A highly conserved molecular switch binds MSY-3 to regulate myogenin repression in postnatal muscle. Genes Dev. 22, 2125-2138. 10.1101/gad.468508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke B., Wittnam J., McNeill E., Van Vactor D. L. and Keshishian H. (2013). Retrograde BMP signaling at the synapse: a permissive signal for synapse maturation and activity-dependent plasticity. J. Neurosci. 33, 17937-17950. 10.1523/JNEUROSCI.6075-11.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonanno A., Apone L., Morasso M. I., Beers R., Brenner H. R. and Eftimie R. (1992). The MyoD family of myogenic factors is regulated by electrical activity: isolation and characterization of a mouse Myf-5 cDNA. Nucleic Acids Res. 20, 539-544. 10.1093/nar/20.3.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cangiano A., Lømo T., Lutzemberger L. and Sveen O. (1980). Effects of chronic nerve conduction block on formation of neuromuscular junctions and junctional AChE in the rat. Acta Physiol. Scand. 109, 283-296. 10.1111/j.1748-1716.1980.tb06599.x [DOI] [PubMed] [Google Scholar]
- Cao Y., Kumar R. M., Penn B. H., Berkes C. A., Kooperberg C., Boyer L. A., Young R. A. and Tapscott S. J. (2006). Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25, 502-511. 10.1038/sj.emboj.7600958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson B. M. and Faulkner J. A. (1988). Reinnervation of long-term denervated rat muscle freely grafted into an innervated limb. Exp. Neurol. 102, 50-56. 10.1016/0014-4886(88)90077-5 [DOI] [PubMed] [Google Scholar]
- Caroni P. (1993). Activity-sensitive signaling by muscle-derived insulin-like growth factors in the developing and regenerating neuromuscular system. Ann. N. Y. Acad. Sci. 692, 209-222. 10.1111/j.1749-6632.1993.tb26219.x [DOI] [PubMed] [Google Scholar]
- Davis R. J., Pesah Y. I., Harding M., Paylor R. and Mardon G. (2006). MouseDach2 mutants do not exhibit gross defects in eye development or brain function. Genesis 44, 84-92. 10.1002/gene.20188 [DOI] [PubMed] [Google Scholar]
- Everman D. B., Bartels C. F., Yang Y., Yanamandra N., Goodman F. R., Mendoza-Londono J. R., Savarirayan R., White S. M., Graham J. M. Jr, Gale R. P. et al. (2002). The mutational spectrum of brachydactyly type C. Am. J. Med. Genet. 112, 291-296. 10.1002/ajmg.10777 [DOI] [PubMed] [Google Scholar]
- Forsyth S., Horvath A. and Coughlin P. (2003). A review and comparison of the murine alpha1-antitrypsin and alpha1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics 81, 336-345. 10.1016/S0888-7543(02)00041-1 [DOI] [PubMed] [Google Scholar]
- Funakoshi H., Belluardo N., Arenas E., Yamamoto Y., Casabona A., Persson H. and Ibanez C. F. (1995). Muscle-derived neurotrophin-4 as an activity-dependent trophic signal for adult motor neurons. Science 268, 1495-1499. 10.1126/science.7770776 [DOI] [PubMed] [Google Scholar]
- Goldman D. and Staple J. (1989). Spatial and temporal expression of acetylcholine receptor RNAs in innervated and denervated rat soleus muscle. Neuron 3, 219-228. 10.1016/0896-6273(89)90035-4 [DOI] [PubMed] [Google Scholar]
- Goldman D., Brenner H. R. and Heinemann S. (1988). Acetylcholine receptor alpha-, beta-, gamma-, and delta-subunit mRNA levels are regulated by muscle activity. Neuron 1, 329-333. 10.1016/0896-6273(88)90081-5 [DOI] [PubMed] [Google Scholar]
- Gould T. W., Yonemura S., Oppenheim R. W., Ohmori S. and Enomoto H. (2008). The neurotrophic effects of glial cell line-derived neurotrophic factor on spinal motoneurons are restricted to fusimotor subtypes. J. Neurosci. 28, 2131-2146. 10.1523/JNEUROSCI.5185-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O., Parsadanian A. S., Sendtner M. and Thoenen H. (1995). Expression of neurotrophins in skeletal muscle: quantitative comparison and significance for motoneuron survival and maintenance of function. J. Neurosci. 42, 21-33. 10.1002/jnr.490420104 [DOI] [PubMed] [Google Scholar]
- Hennig R. (1987). Late reinnervation of the rat soleus muscle is differentially suppressed by chronic stimulation and by ectopic innervation. Acta Physiol. Scand. 130, 153-160. 10.1111/j.1748-1716.1987.tb08121.x [DOI] [PubMed] [Google Scholar]
- Jansen J. K. S., Lomo T., Nicolaysen K. and Westgaard R. H. (1973). Hyperinnervation of skeletal muscle fibers: dependence on muscle activity. Science 181, 559-561. 10.1126/science.181.4099.559 [DOI] [PubMed] [Google Scholar]
- Jin L. and Li X. (2013). Growth differentiation factor 5 regulation in bone regeneration. Curr. Pharm. Des. 19, 3364-3373. 10.2174/1381612811319190003 [DOI] [PubMed] [Google Scholar]
- Knapp J. R., Davie J. K., Myer A., Meadows E., Olson E. N. and Klein W. H. (2006). Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development 133, 601-610. 10.1242/dev.02249 [DOI] [PubMed] [Google Scholar]
- Koliatsos V. E., Clatterbuck R. E., Winslow J. W., Cayouette M. H. and Prices D. L. (1993). Evidence that brain-derived neurotrophic factor is a trophic factor for motor neurons in vivo. Neuron 10, 359-367. 10.1016/0896-6273(93)90326-M [DOI] [PubMed] [Google Scholar]
- Kong X. C., Barzaghi P. and Ruegg M. A. (2004). Inhibition of synapse assembly in mammalian muscle in vivo by RNA interference. EMBO Rep. 5, 183-188. 10.1038/sj.embor.7400065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson P. C. D., Cieslak D. and Goldman D. (2006). Myogenin-dependent nAChR clustering in aneural myotubes. Mol. Cell. Neurosci. 31, 649-660. 10.1016/j.mcn.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Macpherson P. C. D., Wang X. and Goldman D. (2011). Myogenin regulates denervation-dependent muscle atrophy in mouse soleus muscle. J. Cell. Biochem. 112, 2149-2159. 10.1002/jcb.23136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méjat A., Ramond F., Bassel-Duby R., Khochbin S., Olson E. N. and Schaeffer L. (2005). Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 8, 313-321. 10.1038/nn1408 [DOI] [PubMed] [Google Scholar]
- Moresi V., Williams A. H., Meadows E., Flynn J. M., Potthoff M. J., McAnally J., Shelton J. M., Backs J., Klein W. H., Richardson J. A. et al. (2010). Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 143, 35-45. 10.1016/j.cell.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitoh H., Ichijo H., Kimura M., Matsumoto T., Makishima F., Yamaguchi A., Yamashita H., Enomoto S. and Miyazono K. (1996). Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J. Biol. Chem. 271, 21345-21352. 10.1074/jbc.271.35.21345 [DOI] [PubMed] [Google Scholar]
- Payne S. H. Jr., and Brushart T. M. (1997). Neurotization of the rat soleus muscle: a quantitative analysis of reinnervation. J. Hand Surg. 22, 640-643. 10.1016/S0363-5023(97)80121-9 [DOI] [PubMed] [Google Scholar]
- Piccioli Z. D. and Littleton J. T. (2014). Retrograde BMP signaling modulates rapid activity-dependent synaptic growth via presynaptic LIM kinase regulation of cofilin. J. Neurosci. 34, 4371-4381. 10.1523/JNEUROSCI.4943-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao P. K., Kumar R. M., Farkhondeh M., Baskerville S. and Lodish H. F. (2006). Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA 103, 8721-8726. 10.1073/pnas.0602831103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassendren F. A., Bloch-Gallego E., Tanaka H. and Henderson C. E. (1992). Levels of mRNA coding for motoneuron growth-promoting factors are increased in denervated muscle. Proc. Natl. Acad. Sci. USA 89, 7194-7198. 10.1073/pnas.89.15.7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam G., Willmann R., Lin S., Erb-Vögtli S., Kong X. C., Rüegg M. A. and Fuhrer C. (2005). Src-family kinases stabilize the neuromuscular synapse in vivo via protein interactions, phosphorylation, and cytoskeletal linkage of acetylcholine receptors. J. Neurosci. 25, 10479-10493. 10.1523/JNEUROSCI.2103-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori R., Schirwis E., Blaauw B., Bortolanza S., Zhao J., Enzo E., Stantzou A., Mouisel E., Toniolo L., Ferry A. et al. (2013). BMP signaling controls muscle mass. Nat. Genet. 45, 1309-1318. 10.1038/ng.2772 [DOI] [PubMed] [Google Scholar]
- Smith R. G., McManaman J. and Appel S. H. (1985). Trophic effects of skeletal muscle extracts on ventral spinal cord neurons in vitro: separation of a protein with morphologic activity from proteins with cholinergic activity. J. Cell Biol. 101, 1608-1621. 10.1083/jcb.101.4.1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storm E. E., Huynh T. V., Copeland N. G., Jenkins N. A., Kingsley D. M. and Lee S.-J. (1994). Limb alterations in brachypodism mice due to mutations in a new member of the TGF beta-superfamily. Nature 368, 639-643. 10.1038/368639a0 [DOI] [PubMed] [Google Scholar]
- Tang H. and Goldman D. (2006). Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. USA 103, 16977-16982. 10.1073/pnas.0601565103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Veldman M. B. and Goldman D. (2006). Characterization of a muscle-specific enhancer in human MuSK promoter reveals the essential role of myogenin in controlling activity-dependent gene regulation. J. Biol. Chem. 281, 3943-3953. 10.1074/jbc.M511317200 [DOI] [PubMed] [Google Scholar]
- Tang H., Macpherson P., Marvin M., Meadows E., Klein W. H., Yang X.-J. and Goldman D. (2009). A histone deacetylase 4/myogenin positive feedback loop coordinates denervation-dependent gene induction and suppression. Mol. Biol. Cell 20, 1120-1131. 10.1091/mbc.E08-07-0759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman M. B., Bemben M. A., Thompson R. C. and Goldman D. (2007). Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev. Biol. 312, 596-612. 10.1016/j.ydbio.2007.09.019 [DOI] [PubMed] [Google Scholar]
- Wang T., Xie K. and Lu B. (1995). Neurotrophins promote maturation of developing neuromuscular synapses. J. Neurosci. 15, 4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigston D. J. and English A. W. (1992). Fiber-type proportions in mammalian soleus muscle during postnatal development. J. Neurobiol. 23, 61-70. 10.1002/neu.480230107 [DOI] [PubMed] [Google Scholar]
- Williams A. H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J. L., Bassel-Duby R., Sanes J. R. and Olson E. N. (2009). MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549-1554. 10.1126/science.1181046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A. and Olson E. N. (2002). Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110, 479-488. 10.1016/S0092-8674(02)00861-9 [DOI] [PMC free article] [PubMed] [Google Scholar]