Abstract
Translocation of cell-penetrating peptides is often promoted by increased content of arginine or other guanidinum groups. However, relatively little research has considered the role of these functional groups on antimicrobial peptide activity. This study compared the activity of three histone-derived antimicrobial peptides—buforin II, DesHDAP1, and parasin— with variants that contain only lysine or arginine cationic residues. These peptides operate via different mechanisms as parasin causes membrane permeabilization while buforin II and DesHDAP1 translocate into bacteria. For all peptides, antibacterial activity increased with increased arginine content. Higher arginine content increased permeabilization for parasin while it improved translocation for buforin II and DesHDAP1. These observations provide insight into the relative importance of arginine and lysine in these antimicrobial peptides.
Keywords: histone-derived antimicrobial peptide, buforin, parasin, membrane translocation, membrane permeabilization, arginine
1. Introduction
Bacterial resistance to antimicrobial drugs has become stronger, but there have been few recent advances in antimicrobials that combat this resistance.1-5 Antimicrobial peptides (AMPs) are effective antibacterial agents, which also can have antiviral, antifungal and antitumor activities.1, 2 Many antimicrobial peptides use a lytic mechanism, disrupting the bacterial membrane.2-6 However, other AMPs act by translocating through the membrane and affecting intracellular processes.7-10 These translocating antimicrobial peptides may be particularly apt to avoid antimicrobial resistance.3-5
In nature, antimicrobial peptides are often cationic. Because cationic residues attract the peptide to the anionic bacterial membrane, changing the arginine or lysine content can affect the antimicrobial activity. Despite their identical charge, arginine residues are more prevalent in naturally occurring antimicrobial peptides than lysine, implying that the guanidinium group in arginine may be preferable for activity than the amine group found in lysine.6, 11 Based on the observed prevalence of arginine residues, a few researchers have attempted to enhance antimicrobial activity of peptides and synthetic peptoids by increasing the relative composition of arginine residues or guanidinium moieties. In many studies, increased arginine/guanidinium vs. lysine/amine composition seems to enhance activity,12-18 although this trend is not universally observed.12, 19-22 The fact that several studies have noted the importance of guanidinium groups in promoting the translocation of cell-penetrating peptides (CPPs),14, 15, 23-25 supports the importance of arginine/guanidinium composition for AMPs with a translocation-based mechanism. There is less evidence supporting the importance of these groups in the activity of AMPs with a membrane permeabilizing mechanism, although recent results have shown increased activity of defensin variants containing increased arginine composition.18
This study systematically considers the role of arginine composition on three histone-derived peptides (HDAPs), buforin II, DesHDAP1 and parasin (Table 1), by observing how mutating their cationic residues to all arginine or all lysine affects their antibacterial activity and mechanism. Histones and many of their fragments have been shown to have antibacterial properties.8, 9, 26 BF2, a naturally occurring peptide, and DesHDAP1, a designed peptide, have been shown to kill microbes by crossing the bacterial membrane and interacting with DNA without significant membrane permeabilization.8-10, 27-29 Conversely, the naturally occurring peptide parasin kills cells through a lytic mechanism.30 Our results show that increasing the arginine composition of these peptides enhances their activity regardless of their mechanism. These results provide support for the general strategy of enhancing the membrane activity of AMPs through increased arginine content.
Table 1.
Primary structure of peptides analyzed in this study with abbreviated names. Cationic residues are shown in boldface with mutated residues underlined.
| Peptide | Primary Structure |
|---|---|
| Buforin II (21 amino acids) | |
| Wildtype (BF2) | TRSSRAGLQWPVGRVHRLLRK |
| Lysine Mutant (BF2K) | TKSSKAGLQWPVGKVHKLLKK |
| Arginine Mutant (BF2R) | TRSSRAGLQWPVGRVHRLLRR |
| DesHDAP1 (19 amino acids) | |
| Wildtype (Des1) | ARDNKKTRIWPRHLQLAVRN |
| Lysine Mutant (Des1K) | AKDNKKTKIWPKHLQLAVKN |
| Arginine Mutant (Des1R) | ARDNRRTRIWPRHLQLAVRN |
| Parasin (20 amino acids) | |
| Wildtype (Parasin) | KGRGKQGGKVRAKWKTRSS |
| Lysine Mutant (ParasinK) | KGKGKQGGKVKAKWKTKSS |
| Arginine Mutant (ParasinR) | RGRGRQGGRVRARWRTRSS |
2. Materials and Methods
2.1 Materials
All peptides used in this study were synthesized by NeoBioSci (Cambridge, MA) and obtained at >95% purity. BF2 and parasin contained F10W and A14W mutations, respectively, for peptide membrane insertion and translocation studies. POPC, POPE and DNS-PE lipids were obtained from Avanti Polar Lipids (Alabaster, AL) and other chemicals were obtained from Sigma unless otherwise noted.
2.2 Circular dichroism spectroscopy
Circular dichroism (CD) spectra of each peptide were collected on an Olis DSM 1000 (Bogart, GA) using a 1-mm path-length quartz cell. Peptides were 25 μM solutions in 1:1 TFE:phosphate buffer (10 mM sodium phosphate, 45 mM NaCl, 1 mM EDTA, pH 7.4), a solution previously shown to mimic membrane environments for these peptide structures.9, 31 For each sample, three scans from 250 to 195 nm with 60 s integration at each wavelength were averaged at 37 °C.
2.3 Bacterial preparation
For radial diffusion and propidium iodide assays, 50 mL of Tryptic Soy Broth (TSB) was inoculated with 50 μL of overnight E. coli bacterial culture in TSB and incubated for 3 hours at 37 °C while shaking. The culture was then centrifuged at 4 °C for 10 minutes at 880×g and resuspended in 10 mM sodium phosphate buffer (pH 7.4). This sample was pelleted under the same conditions and resuspended again. Data is presented for E. coli strain Carolina #25922, although analogous trends also were observed for Top10 E. coli (Invitrogen).
2.4 Radial diffusion assay
Radial diffusion assays were based on the protocols presented previously.32, 33 10 mL of melted underlay (1% TSB, 1% agarose w/v, 10 mM sodium phosphate, pH 7.4) was vortexed with 4×106 CFU of bacteria resuspended in phosphate buffer and poured into a petri dish. Wells were made using a pipette attached to a bleach trap, and each well was filled with 2.0 μL of 3×10−4 M of peptide solution or water. After a three hour incubation at 37 °C, the plates were covered with 10 mL of overlay agar (6% w/v TSB, 1% w/v agarose) and incubated overnight at 37 °C. Following incubation, the diameter of bacterial clearance was measured. Measurements were taken for at least three separate overnight cultures for each peptide, each with multiple wells.
2.5 Peptide membrane insertion
To form vesicles, lipids dissolved in chloroform at a 75:25 ratio of POPC:POPG were dried and reconstituted in a 1:1 mixture by volume of HEPES buffer (10 mM HEPES, 45 mM NaCl, 1 mM EDTA, pH 7.4). The lipids were subjected to five freeze-thaw cycles in liquid nitrogen and then were extruded twenty-one times through a Whatman 0.1μm Nucleopore filter (Whatman) at 25°C using an extruder (Avanti Polar Lipids). The sample was then returned to its original volume in 10 mM HEPES buffer. The concentration of vesicles was determined using a phosphorous assay.34
Three solutions were then prepared for titration with lipid: two containing 5×10−6 M of peptide in 10 mM HEPES buffer and one containing 0.015 mL tryptophan in HEPES buffer. Vesicles were added to one of the peptide solutions and the tryptophan solution and allowed to incubate for 5 minutes before taking a fluorescence emission spectrum from 300-450 nm of all three samples with an excitation of 280 nm. Vesicle additions were continued until peptide samples showed no additional blue shift. Each spectrum was collected as the average of 10 scans using a Varian Cary Eclipse fluorescence spectrophotometer.
The fluorescence emission for the peptide titrated with vesicles was then adjusted at each collected wavelength using the control peptide solution without added lipid and the tryptophan solution with added vesicles (Equation 1). The peptide alone corrects for any loss of fluorescence due to adsorption to the glass cuvette, as observed previously for BF2,35 while the tryptophan solution corrects for light scattering caused by vesicles.36 For each addition of lipid, the peak of the adjusted peptide fluorescence was measured by fitting the symmetric portion of the spectra data to a Gaussian curve using KaliedaGraph (Synergy Software). Peak wavelengths reported for each peptide were averaged over three titrations performed with at least two independently produced vesicle samples.
| Equation 1. |
2.6 Propidium iodide (PI) uptake assay
The PI uptake assay was used to determine the relative membrane permeabilization caused by each peptide. The change in fluorescence intensity of PI (1 mg/mL) mixed with 3.75×108 CFU E. coli culture in 10 mM phosphate buffer upon addition of 2×10−6 M peptide was measured using a Varian Cary Eclipse fluorescence spectrophotometer with excitation and emission of 535 nm and 617 nm, respectively. The permeabilizing ability of each peptide was calculated as the relative fluorescence increase due to propidium iodide•DNA binding five minutes after the addition of peptide to bacteria (Equation 2). Each peptide was tested using bacteria from at least two different overnight cultures.
| Equation 2. |
2.7 Translocation assay
The translocation assay was performed analogous manner to previous HDAP studies.27, 37-39 To form experimental vesicles, lipids dissolved in chloroform at a 75:20:5 ratio of POPC:POPG:DNS-PE were dried and reconsituted in a 1:1 volume mixture by volume of 10 mM HEPES buffer and 0.4 mM trypsin in 10 mM HEPES buffer solution for a final concentration of 0.2 mM trypsin. For control vesicles, Bowman-Birk Inhibitor was added to the reconstitution mixture for a final concentration of 2.0 mM. Extrusion and quantification was performed as described above for membrane insertion studies. Samples of control vesicles used for quantification were centrifuged for 10 minutes at 14,000 × g in Nanosep 10K centrifugal devices with omega membrane (Pall Corporation) to remove phosphorous salts.
For both control and experimental samples in the assay, 200 μL samples of 0.250 mM vesicles in 10 mM HEPES buffer were placed in a microcentrifuge vial. For experimental vesicles, the 200 μL sample also included 4.0 mM Bowman Birk inhibitor. Vesicle solutions were heated to 37 °C, gently vortexed and added to the wells of a 96 well plate containing 3×10−6 M peptide. Fluorescence measurements were begun immediately and measured for 25 minutes on a Spectra Max M3 plate reader with excitation and emission of 280 nm and 525 nm, respectively.
Translocation was quantitated by dividing the average control relative fluorescence in the last minute by the average experimental relative fluorescence in the last minute (Equation 3), with relative fluorescence equaling the FRET divided by the FRET at t=0. Thus, a translocation ratio of 1 would imply no translocation, and ratios greater than one would indicate translocation. Translocation was only calculated for trials when the relative control fluorescence did not drop below 0.85, since in an ideal trial all of the vesicle-encapsulated trypsin would be inhibited, giving a relative control fluorescence of 1. The translocation of each peptide was averaged over at least three trials using at least two independently prepared sets of vesicle solutions.
| Equation 3. |
3. Results and Discussion
3.1 Changing cationic residues between arginine and lysine does not significantly affect peptide structure
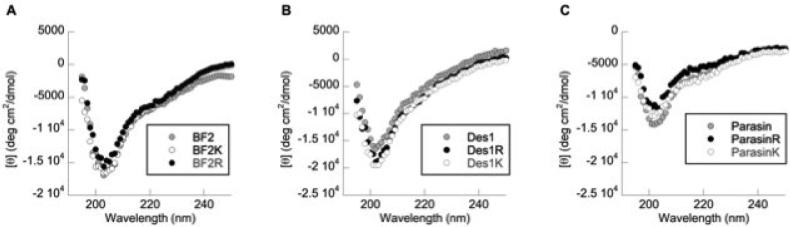
The secondary structures of the native HDAPs were compared to their lysine and arginine mutants using circular dichroism (CD) spectroscopy in 1:1 TFE:phosphate solutions to provide a membrane-mimetic environment.27 CD spectra of the wild type peptides correspond with previous measurements (Fig. 1).27, 28, 30, 35 The spectra of both BF2 and DesHDAP1 are consistent with a partially helical structure. Parasin similarly shows a mixed structure with somewhat increased α-helical character relative to the other peptides. Changing the identity of the basic residues had no appreciable change in the secondary structure of the mutants in comparison to the wildtype peptides (Fig. 1).
Figure 1.
Circular dichroism spectra of 25 μM wildtype and mutant variants of (a) BF2, (b) DesHDAP1 and (c) parasin in 1:1 TFE:phosphate buffer.
3.2 Increasing arginine composition of peptides increases antimicrobial activity
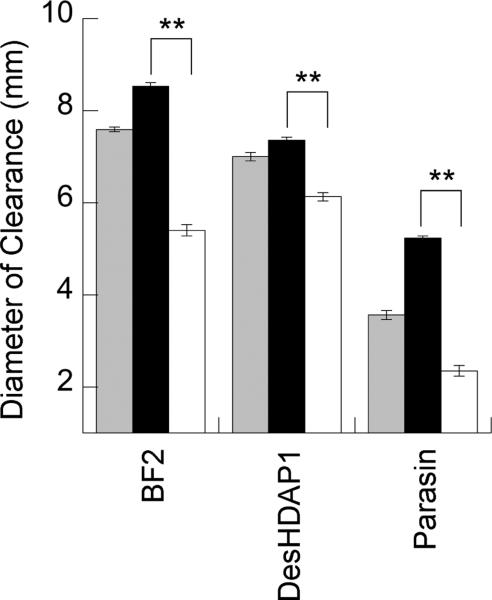
The antimicrobial activity of BF2, DesHDAP1, parasin and their mutants against E. coli was measured using a radial diffusion assay (Fig. 2), where an increased diameter of bacterial clearance indicated increased antimicrobial activity. For all three peptides, arginine mutants had significantly increased antimicrobial activity compared to the lysine mutants. The region of bacterial clearance was only partial for both parasin and parasinK, with an inner fully cleared circle surrounded by a dense ring of bacteria and a larger outer ring of partially cleared (less dense) E. coli. A similar observation was made for parasin in previous studies.31 ParasinR, on the other hand, completely cleared the radius, consistent with its increased activity.
Figure 2.
Radius of clearance observed for E. coli in radial diffusion assays with histone-derived peptides (gray) and their arginine (black) and lysine (white) mutants. The average region of clearance noted for parasin and its mutants for E. coli are reported as the inner radial clearance that contained no bacteria. ** denotes p≤0.001 for comparisons between arginine and lysine mutants of a peptide. Uncertainty shown as standard error.
Interestingly, a correlation between α-helical structure and activity does not emerge in this study as enhanced activity of arginine mutants (Fig. 2) is not accompanied by notably different structures in our CD data (Fig. 1). In the case of BF2 and DesHDAP1, this may be related to the presence of a helix breaking proline residue in the middle of the peptide. Previous results have shown the importance of these proline residues for BF2 and DesHDAP1 activity,9, 10, 27, 37 and computer simulations have implied that the membrane entry of BF2 is enhanced by its ability to deform its N-terminal structure.40 Future studies could consider whether deformation may play a similar role in DesHDAP1 and parasin.
3.3 Increased arginine content enhances peptide embedding into membranes
In order to explain the increased antimicrobial activity of peptides containing arginine instead of lysine, we considered the ability of our peptides to embed into lipid membranes by monitoring their blue shift in the presence of vesicles. For these experiments, we titrated peptides with 3:1 POPC:POPG vesicles until obtaining a maximum blue shift, shown in Table 2. Of the three peptides, only DesHDAP1 showed a statistically significant trend in comparing arginine and lysine mutants, and the shift for these peptides was relatively small albeit consistent with somewhat increased membrane embedding. No discernable effect was observed for parasin mutants, and any potential trend between BF2 arginine and lysine mutants was too small to reliably interpret. While quantitatively small, these observations led us to further consider whether arginine/lysine mutants altered other membrane-related mechanistic properties in these three peptides.
Table 2.
Final average peak wavelength of peptide tryptophan emission spectra after addition of 3:1 POPC:POPG vesicles. Starred values are significantly different between arginine and lysine mutants with p≤0.01.
| Peptide | Wildtype final peak wavelength (nm) | Arginine mutant final peak wavelength (nm) | Lysine mutant final peak wavelength (nm) |
|---|---|---|---|
| BF2 | 349.8±0.5 | 351.0±1 | 351.8±0.2 |
| DesHDAP1 | 354.2±0.6 | 353.5±0.5* | 355.5±0.2* |
| Parasin | 355.7±0.4 | 355.8±0.2 | 355.2±0.6 |
3.4 Changes in antimicrobial mechanism with lysine and arginine mutations
3.4.1 Arginine promotes membrane permeabilization more than lysine
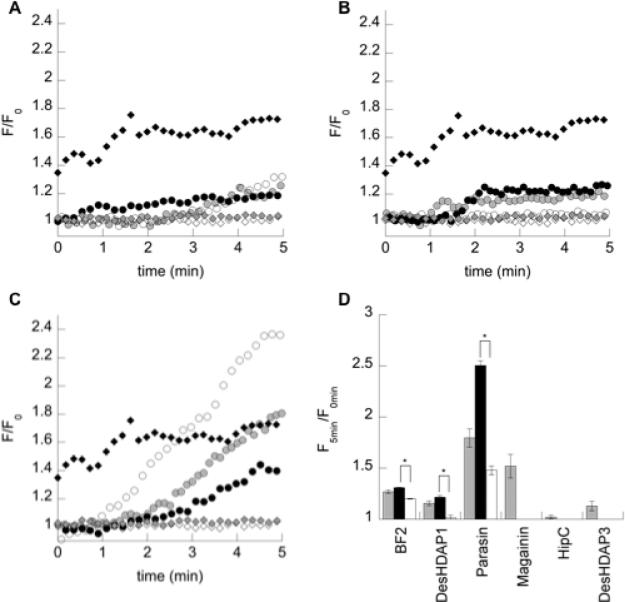
To analyze one potential mechanism of bacterial death based on membrane interactions, the ability of the HDAPs and their mutants to permeabilize the bacterial membrane of E. coli was determined using a PI uptake assay (Fig. 3). In this assay, increased membrane permeabilization allows PI to bind intracellular DNA, giving an increased fluorescence. For all three peptides, arginine mutants caused significantly greater membrane permeabilization than lysine mutants. This was particularly notable in parasin, which primarily functions through causing membrane permeabilization.30 However, even though wild type BF2 and DesHDAP1 cause relatively low levels of permeabilization compared to a prototypical lytic peptide, such as magainin, their lysine mutations still showed significantly decreased permeabilization similar to that of two non-permeabilizing peptides considered in previous work (HipC and DesHDAP3).37, 39 Relatively smaller changes in permeabilization for the arginine mutations in BF2 and DesHDAP1 compared to wild type may be attributed to a difference of only one (BF2) or two (DesHDAP1) residues in those mutants versus wildtype.
Figure 3.
(a-c) Representative traces of relative fluorescence increases (F/F0) over time for (a) BF2, (b) DesHDAP1 and (c) parasin wild type (gray circle), arginine (black circle) and lysine (white circle) variants. Data for magainin (black diamond), HipC (white diamond) and DesHDAP3 (gray diamond) from 37, 39 provided for comparison. (d) Relative fluorescence increase of propidium iodide five minutes after the addition of histone-derived peptides (gray) and their arginine (black) and lysine (white) mutants to E. coli cells. Data for wild type magainin, HipC and DesHDAP3 from 37, 39 provided for comparison. * denotes p≤0.01 for comparisons between arginine and lysine mutants of a peptide. Uncertainty shown as standard error.
3.4.2 Arginine improves membrane translocation relative to lysine
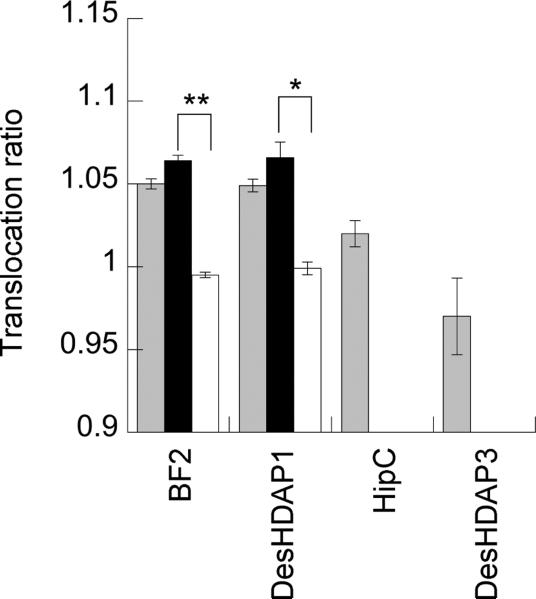
To analyze the translocation ability of BF2, DesHDAP1 and their mutants, vesicles made of 70:25:5 POPC:POPG:DNS-POPE encapsulated with trypsin were mixed with peptide to determine whether peptide crosses the membrane and becomes proteolyzed. In this assay, increased translocation causes peptide cleavage by the encapsulated trypsin, leading to a decreased FRET signal. This FRET signal is compared to the FRET for the same peptide with a control sample of vesicles in which the encapsulated trypsin is inhibited to ensure decreased fluorescence is due to membrane translocation. Parasin and its mutants were not amenable to this assay because they induce relatively high membrane permeabilization, and the assay relies on membrane integrity to maintain trypsin on the inside of vesicles for reliable results. Data for peptides previously observed to show a low level (HipC) and essentially no (DesHDAP3) translocation in lipid vesicle assays and cellular microscopy experiments were shown for comparison (Fig. 4).37, 39
Figure 4.
Translocation ratios (defined in equation 3) for histone-derived peptides (gray) and their arginine (black) and lysine (white) mutants into lipid vesicles. An increased ratio implies increased translocation across cell membranes. Data for HipC and DesHDAP3 from 37, 39 provided for comparison. * denotes p≤0.01 and ** denotes p≤0.001 for comparisons between arginine and lysine mutants of a peptide. Uncertainty shown as standard error.
The BF2K and DesHDAP1K mutants caused a clear decrease in translocation compared to their arginine counterparts (Fig. 4). These peptides show translocation very similar to that of an entirely non-translocating peptide (DeHDAP3), consistent with their decreased antibacterial activity. As seen for permeabilization, the slight increases in translocation for BF2R and DesHDAP1R were not necessarily significant, again potentially stemming from the more conservative nature of these mutations.
4. Conclusions
In this study, we considered the effect of arginine and lysine content on the antimicrobial activity and membrane properties of three HDAPs with differing mechanisms of action. Increasing the composition of arginine versus lysine residues improved antibacterial activity for these peptides. For parasin, a lytic peptide, this increase seems to result from increased membrane permeabilization. This observation highlights the potential for altered arginine content to enhance the activity of membrane permeabilizing AMPs. For BF2 and DesHDAP1, which rely on translocating into the cell as their main antimicrobial mechanism, the presence of arginine residues seems to promote translocation as shown by the dramatic loss of translocation in BF2K and Des1K in comparison to that of the wildtype. In addition, there was some effect on membrane permeabilization as arginine content increased BF2 and DesHDAP1. This leaves open the possibility that the altered antimicrobial activity in the BF2 and DesHDAP1 arginine mutants is not solely based on the increase in translocation. Interestingly, the comparable relative translocation and permeabilization of wild type BF2 and BF2R could not explain the increased antimicrobial activity observed for BF2R. Instead, the increased activity may have resulted from altered binding to its intracellular target. Possible changes in nucleic acid binding for these peptides is the focus on ongoing work in our lab.
The results presented here are consistent with the importance of arginine residues in the activity of AMPs and provide further evidence for considering increased arginine or guanidinium content in the design of antimicrobial peptides and peptoids. Recent work by Wong and co-workers has implied that the ability of arginine sidechains to form bidentate interactions with lipids can lead to enhanced Gaussian, or “saddle-splay,” membrane curvature, while lysine residues that only form monodentate interactions do not readily induce this curvature on their own.18, 41, 42 The ability to induce increased curvature can enhance the activity of both translocating and membrane permeabilizing peptides.
As well, our observations are interesting in light of other recent work more broadly considering the role of arginine residues in membrane proteins. For example, Moon and Fleming found that arginine residues were more energetically favorable than lysine in the transmembrane regions of OmpLA.43 They also observed that there may be some cooperativity for placing multiple arginine residues into the membrane. These finding appear consistent with the increased membrane effects of arginine mutants in this study, although our systems are very different in nature. In another well-characterized system, mutating the twin-arginine motif of Tat signal peptides to a twin-lysine typically removes their ability to interact with the Tat translocation machinery.44-46 Although Tat systems involve a distinct mechanism of translocation from the peptides considered in this study, these results further emphasize the different role of arginine and lysine residues in membrane protein function. Other studies have compared the structural stability of arginine versus lysine in a four-helix bundle47 and have considered the role of arginine residues in the transmembrane regions of ion channels.48, 49 Together with the results presented here, these studies increasingly highlight the important role that arginine can have in mediating protein•membrane interactions.
Cutrona et al. highlights.
Arginine enhances antimicrobial peptide activity compared to lysine
Both permeabilizing and translocating peptides exhibit arginine effect
Increasing arginine content is a potential strategy for antimicrobial peptide design
Acknowledgements
The authors would like to thank Carla Perez for helpful conversations and technical assistance. Research funded by National Institute of Allergy and Infectious Diseases (NIH-NIAID) award R15AI079685 with instrumentation support from National Science Foundation award CHE-0922860. B.A.K. was supported by National Science Foundation award CHE-1005032. D.E.E. is a Henry Dreyfus Teacher-Scholar. D.M.F. was supported by funds from the Sherman Fairchild Foundation.
Abbreviations
- AMP
antimicrobial peptide
- BF2
buforin II
- CD
circular dichroism
- CPP
cell-penetrating peptide
- HDAP
histone-derived antimicrobial peptide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hoskin DW, Ramamoorthy A. Studies on anticancer activities of antimicrobial peptides. Bba-Biomembranes. 2008;1778:357–375. doi: 10.1016/j.bbamem.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jenssen H, Hamill P, Hancock REW. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–+. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock REW, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 4.Toke O. Antimicrobial peptides: New candidates in the fight against bacterial infections. Biopolymers. 2005;80:717–735. doi: 10.1002/bip.20286. [DOI] [PubMed] [Google Scholar]
- 5.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 6.Yeaman MR, Yount NY. Mechanisms of antimicrobial peptide action and resistance. Pharmacol Rev. 2003;55:27–55. doi: 10.1124/pr.55.1.2. [DOI] [PubMed] [Google Scholar]
- 7.Hale JD, Hancock RE. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev Anti-Infe. 2007;5:951–959. doi: 10.1586/14787210.5.6.951. [DOI] [PubMed] [Google Scholar]
- 8.Park CB, Kim HS, Kim SC. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem Bioph Res Co. 1998;244:253–257. doi: 10.1006/bbrc.1998.8159. [DOI] [PubMed] [Google Scholar]
- 9.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: The proline hinge is responsible for the cell-penetrating ability of buforin II. P Natl Acad Sci USA. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Y, Fleming E, Chen JL, Elmore DE. Effect of proline position on the antimicrobial mechanism of buforin II. Peptides. 2011;32:677–682. doi: 10.1016/j.peptides.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hristova K, Wimley WC. A look at arginine in membranes. J Membr Biol. 2011;239:49–56. doi: 10.1007/s00232-010-9323-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svenson J, Karstad R, Flaten GE, Brandsdal BO, Brandl M, Svendsen JS. Altered Activity and Physicochemical Properties of Short Cationic Antimicrobial Peptides by Incorporation of Arginine Analogues. Mol Pharmaceut. 2009;6:996–1005. doi: 10.1021/mp900057k. [DOI] [PubMed] [Google Scholar]
- 13.Andreev K, Bianchi C, Laursen JS, Citterio L, Hein-Kristensen L, Gram L, Kuzmenko I, Olsen CA, Gidalevitz D. Guanidino groups greatly enhance the action of antimicrobial peptidomimetics against bacterial cytoplasmic membranes. Biochim Biophys Acta. 2014;1838:2492–2502. doi: 10.1016/j.bbamem.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bahnsen JS, Franzyk H, Sandberg-Schaal A, Nielsen HM. Antimicrobial and cell-penetrating properties of penetratin analogs: effect of sequence and secondary structure. Biochim Biophys Acta. 2013;1828:223–232. doi: 10.1016/j.bbamem.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Gabriel GJ, Madkour AE, Dabkowski JM, Nelson CF, Nusslein K, Tew GN. Synthetic mimic of antimicrobial peptide with nonmembrane-disrupting antibacterial properties. Biomacromolecules. 2008;9:2980–2983. doi: 10.1021/bm800855t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Locock KE, Michl TD, Valentin JD, Vasilev K, Hayball JD, Qu Y, Traven A, Griesser HJ, Meagher L, Haeussler M. Guanylated polymethacrylates: a class of potent antimicrobial polymers with low hemolytic activity. Biomacromolecules. 2013;14:4021–4031. doi: 10.1021/bm401128r. [DOI] [PubMed] [Google Scholar]
- 17.Vedel L, Bonke G, Foged C, Ziegler H, Franzyk H, Jaroszewski JW, Olsen CA. Antiplasmodial and prehemolytic activities of alpha-peptide-beta-peptoid chimeras. Chembiochem. 2007;8:1781–1784. doi: 10.1002/cbic.200700385. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt NW, Tai KP, Kamdar K, Mishra A, Lai GH, Zhao K, Ouellette AJ, Wong GC. Arginine in alpha-defensins: differential effects on bactericidal activity correspond to geometry of membrane curvature generation and peptide-lipid phase behavior. J Biol Chem. 2012;287:21866–21872. doi: 10.1074/jbc.M112.358721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen PW, Shyu CL, Mao FC. Antibacterial activity of short hydrophobic and basic-rich peptides. Am J Vet Res. 2003;64:1088–1092. doi: 10.2460/ajvr.2003.64.1088. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen LT, de Boer L, Zaat SA, Vogel HJ. Investigating the cationic side chains of the antimicrobial peptide tritrpticin: hydrogen bonding properties govern its membrane-disruptive activities. Biochim Biophys Acta. 2011;1808:2297–2303. doi: 10.1016/j.bbamem.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Schibli DJ, Nguyen LT, Kernaghan SD, Rekdal O, Vogel HJ. Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures. Biophys J. 2006;91:4413–4426. doi: 10.1529/biophysj.106.085837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strom MB, Haug BE, Skar ML, Stensen W, Stiberg T, Svendsen JS. The pharmacophore of short cationic antibacterial peptides. J Med Chem. 2003;46:1567–1570. doi: 10.1021/jm0340039. [DOI] [PubMed] [Google Scholar]
- 23.Nakase I, Okumura S, Katayama S, Hirose H, Pujals S, Yamaguchi H, Arakawa S, Shimizu S, Futaki S. Transformation of an antimicrobial peptide into a plasma membrane-permeable, mitochondria-targeted peptide via the substitution of lysine with arginine. Chem Commun. 2012;48:11097–11099. doi: 10.1039/c2cc35872g. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 25.Stanzl EG, Trantow BM, Vargas JR, Wender PA. Fifteen years of cell-penetrating, guanidinium-rich molecular transporters: basic science, research tools, and clinical applications. Accounts Chem Res. 2013;46:2944–2954. doi: 10.1021/ar4000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouvry-Patat SA, Schey KL. Characterization of antimicrobial histone sequences and posttranslational modifications by mass spectrometry. J Mass Spectrom. 2007;42:664–674. doi: 10.1002/jms.1200. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi S, Takeshima K, Park CB, Kim SC, Matsuzaki K. Interactions of the novel antimicrobial peptide buforin 2 with lipid bilayers: Proline as a translocation promoting factor. Biochemistry-Us. 2000;39:8648–8654. doi: 10.1021/bi0004549. [DOI] [PubMed] [Google Scholar]
- 28.Tsao HS, Spinella SA, Lee AT, Elmore DE. Design of novel histone-derived antimicrobial peptides. Peptides. 2009;30:2168–2173. doi: 10.1016/j.peptides.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 29.Uyterhoeven ET, Butler CH, Ko D, Elmore DE. Investigating the nucleic acid interactions and antimicrobial mechanism of buforin II. Febs Letters. 2008;582:1715–1718. doi: 10.1016/j.febslet.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 30.Koo YS, Kim JM, Park IY, Yu BJ, Jang SA, Kim KS, Park CB, Cho JH, Kim SC. Structure-activity relations of parasin I, a histone H2A-derived antimicrobial peptide. Peptides. 2008;29:1102–1108. doi: 10.1016/j.peptides.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 31.Birkemo GA, Mantzilas D, Luders T, Nes IF, Nissen-Meyer J. Identification and structural analysis of the antimicrobial domain in hipposin, a 51-mer antimicrobial peptide isolated from Atlantic halibut. Bba-Proteins Proteom. 2004;1699:221–227. doi: 10.1016/j.bbapap.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Lehrer RI, Rosenman M, Harwig SSSL, Jackson R, Eisenhauer P. Ultrasensitive Assays for Endogenous Antimicrobial Polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 33.Steinberg DA, Lehrer RI. Designer assays for antimicrobial peptides. Disputing the “one-size-fits-all” theory. Methods Mol. Biol. 1997;78:169–186. doi: 10.1385/0-89603-408-9:169. [DOI] [PubMed] [Google Scholar]
- 34.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. Structure-activity analysis of buforin II, a histone H2A-derived antimicrobial peptide: the proline hinge is responsible for the cell-penetrating ability of buforin II. Proc. Natl. Acad. Sci. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleming E, Maharaj NP, Chen JL, Nelson RB, Elmore DE. Effect of lipid composition on buforin II structure and membrane entry. Proteins. 2008;73:480–491. doi: 10.1002/prot.22074. [DOI] [PubMed] [Google Scholar]
- 36.Ladokhin AS, Jayasinghe S, White SH. How to measure and analyze tryptophane fluorescence in membrane properly, and why bother? Anal Biochem. 2000;285:235–245. doi: 10.1006/abio.2000.4773. [DOI] [PubMed] [Google Scholar]
- 37.Pavia KE, Spinella SA, Elmore DE. Novel histone-derived antimicrobial peptides use different antimicrobial mechanisms. Bba-Biomembranes. 2012;1818:869–876. doi: 10.1016/j.bbamem.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spinella SA, N. RB, Elmore DE. Measuring Peptide Translocation into Large Unilamellar Vesicles. Journal of Visualized Experiments. 2012;59 doi: 10.3791/3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bustillo ME, Fischer AL, LaBouyer MA, Klaips JA, Webb AC, Elmore DE. Modular analysis of hipposin, a histone-derived antimicrobial peptide consisting of membrane translocating and membrane permeabilizing fragments. Biochim Biophys Acta. 2014;1838:2228–2233. doi: 10.1016/j.bbamem.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmore DE. Insights into buforin II membrane translocation from molecular dynamics simulations. Peptides. 2012;38:357–362. doi: 10.1016/j.peptides.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt NW, Lis M, Zhao K, Lai GH, Alexandrova AN, Tew GN, Wong GC. Molecular basis for nanoscopic membrane curvature generation from quantum mechanical models and synthetic transporter sequences. Journal of the American Chemical Society. 2012;134:19207–19216. doi: 10.1021/ja308459j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt NW, Mishra A, Lai GH, Davis M, Sanders LK, Tran D, Garcia A, Tai KP, McCray PB, Ouellette AJ, Selsted ME, Wong GC. Criterion for amino acid composition of defensins and antimicrobial peptides based on geometry of membrane destabilization. Journal of the American Chemical Society. 2011;133:6720–6727. doi: 10.1021/ja200079a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moon CP, Fleming KG. Side-chain hydrophobicity scale derived from transmembrane protein folding into lipid bilayers. P Natl Acad Sci USA. 2011;108:10174–10177. doi: 10.1073/pnas.1103979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alami M, Trescher D, Wu LF, Muller M. Separate analysis of twin-arginine translocation (Tat)-specific membrane binding and translocation in Escherichia coli. J Biol Chem. 2002;277:20499–20503. doi: 10.1074/jbc.M201711200. [DOI] [PubMed] [Google Scholar]
- 45.Cline K. Mechanistic Aspects of Folded Protein Transport by the Twin Arginine Translocase (Tat) J Biol Chem. 2015;290:16530–16538. doi: 10.1074/jbc.R114.626820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cristobal S, de Gier JW, Nielsen H, von Heijne G. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. The EMBO journal. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diez-Garcia F, Chakrabartty A, Gonzalez C, Laurents DV. An Arg-rich putative prebiotic protein is as stable as its Lys-rich variant. Arch Biochem Biophys. 2012;528:118–126. doi: 10.1016/j.abb.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Jiang YX, Lee A, Chen JY, Cadene M, Chait BT, MacKinnon R. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt D, Jiang QX, MacKinnon R. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature. 2006;444:775–779. doi: 10.1038/nature05416. [DOI] [PubMed] [Google Scholar]