Abstract
The aim of this study was to assess potential candidate gene regions and corresponding universal primer pairs as secondary DNA barcodes for the fungal kingdom, additional to ITS rDNA as primary barcode. Amplification efficiencies of 14 (partially) universal primer pairs targeting eight genetic markers were tested across > 1 500 species (1 931 strains or specimens) and the outcomes of almost twenty thousand (19 577) polymerase chain reactions were evaluated. We tested several well-known primer pairs that amplify: i) sections of the nuclear ribosomal RNA gene large subunit (D1–D2 domains of 26/28S); ii) the complete internal transcribed spacer region (ITS1/2); iii) partial β -tubulin II (TUB2); iv) γ-actin (ACT); v) translation elongation factor 1-α (TEF1α); and vi) the second largest subunit of RNA-polymerase II (partial RPB2, section 5–6). Their PCR efficiencies were compared with novel candidate primers corresponding to: i) the fungal-specific translation elongation factor 3 (TEF3); ii) a small ribosomal protein necessary for t-RNA docking; iii) the 60S L10 (L1) RP; iv) DNA topoisomerase I (TOPI); v) phosphoglycerate kinase (PGK); vi) hypothetical protein LNS2; and vii) alternative sections of TEF1α. Results showed that several gene sections are accessible to universal primers (or primers universal for phyla) yielding a single PCR-product. Barcode gap and multi-dimensional scaling analyses revealed that some of the tested candidate markers have universal properties providing adequate infra- and inter-specific variation that make them attractive barcodes for species identification. Among these gene sections, a novel high fidelity primer pair for TEF1α, already widely used as a phylogenetic marker in mycology, has potential as a supplementary DNA barcode with superior resolution to ITS. Both TOPI and PGK show promise for the Ascomycota, while TOPI and LNS2 are attractive for the Pucciniomycotina, for which universal primers for ribosomal subunits often fail.
Keywords: DNA barcoding, ITS supplement, molecular taxonomy, phylogeny, species identification, universal primers
INTRODUCTION
Identification and classification of eukaryotes increasingly depends on DNA sequences of standardised genetic markers, a concept known as DNA barcoding (Hebert et al. 2003a, b, Hebert & Gregory 2005, Meyer & Paulay 2005, Schindel & Miller 2005, Schoch et al. 2012). An intense debate is ongoing concerning whether the identification of organisms of unresolved alpha taxonomy is amenable to DNA barcoding, because in silico-based identification requires gene sequences that accurately reflect natural classifications (Eberhardt 2010, Schlick-Steiner et al. 2010). If this prior condition is not adequately fulfilled a posteriori nesting of ‘unknowns’ among known fungal taxa is impossible. DNA barcoding has evolved, despite its unresolved theoretical and taxonomic issues, as a standard procedure in organism identification among various disciplines of modern biology (Tautz et al. 2003, Shokralla et al. 2014, Stockinger et al. 2014, Stoeckle & Thaler 2014).
The milestone paper on amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics by White et al. (1990, now > 15 000 citations), described primers that allowed simple, rapid PCR-based amplification and Sanger sequencing of sections of the fungal rDNA operon. Inspired by the efforts to resolve bacterial taxonomy using 16S sequences summarised by Woese (1987) and further elaborated by Weisburg et al. (1991) and Stackebrandt & Goebel (1994), the White et al. paper generated an explosion of phylogenetic research that now dominates fungal taxonomy. The modern concept of fungal DNA barcoding does not differ substantially from approaches proposed more than two decades ago. Sanger DNA sequencing of nuclear rDNA domains remains the most widely accepted approach in molecular mycology to classify and to identify unknown fungal specimens or cultures. The first comprehensive database of fungal DNA barcodes was established for yeasts (Kurtzman & Robnett 1998, Fell et al. 2000, Scorzetti et al. 2002) and revolutionised the approach to species recognition in that group. Where once a single taxonomist could identify only a handful of yeast strains in a week because of the plethora of physiological tests required, now it is possible to process hundreds. The result has been a rapid increase in the number of recognised yeast species, and a wealth of ecological data (Kurtzman et al. 2015).
The broad acceptance of DNA barcoding today relies on public repositories such as the INSDC (http://www.insdc.org/), which accessions hundreds of thousands of sequence entries (Schoch et al. 2012, 2014). In a concerted action with the Consortium for the Barcode Of Life (CBOL), the Fungal Barcoding Consortium (Schoch et al. 2012) ratified the ITS as the universal DNA barcode for the fungal kingdom using the same gene section proposed by White et al. (1990) more than 20 years earlier. This study (Schoch et al. 2012) was widely accepted by the community and already has been highly cited > 850 times.
Abandoning dual nomenclature of pleomorphic fungi in favour of a single name system in the nomenclatural code was a radical change (Gams & Jaklitsch 2011, Taylor 2011), partly enabled by the possibility to unequivocally demonstrate phylogenetic relationships between asexual and sexual states that were formerly classified and named separately in parallel systems under the provisions of the former Article 59 (Guadet et al. 1989, Bruns et al. 1991, Berbee & Taylor 1992, Reynolds & Taylor 1993). This change to the botanical code was driven by DNA data, ITS and other rDNA sequences in particular, and demonstrated the impact of DNA sequencing on our understanding of genetic diversity, species identification, delimitation and nomenclatural changes in mycology.
Despite the strength and impact of rDNA ITS as the sanctioned universal fungal DNA barcode, its resolution of higher taxonomic level relationships is inferior to many protein-coding genes such as RPB1, RPB2 or TUB2 (Nilsson et al. 2006, Seifert 2009, Begerow et al. 2010, Schoch et al. 2014). However, as a barcode ITS outperforms alternative loci because of its highly robust PCR amplification fidelity (> 90 % success rates), a Probability of Correct Identification (PCI) of about 70 %, and applicability to a wide range of sample conditions. Although many alternatives to ITS were considered by the mycological community, its sanctioning as the primary barcode marker rested on its practicality and reliability, not on the highly desired ‘resolution power’ (Schoch et al. 2012, 2014). Many mycologists would prefer one or several universal, but phylogenetically informative loci as barcodes, with higher species resolution power than is feasible with ITS. The ideal genetic marker would have high inter- and low intra-species sequence divergence, i.e. a discrete barcode gap (Schoch et al. 2012, Samerpitak et al. 2015), and accurately reflect higher-level taxonomic affiliations. It could then serve as a substitute for ITS, or as a supplementary, secondary or tertiary marker in concert with ITS as the primary barcode.
Molecular taxonomists thus continue to search for genes conserved enough to allow reliable priming but sufficiently variable to yield highly resolved and well-supported phylograms and useful barcode gaps (Schmitt et al. 2009, Feau et al. 2011, Lewis et al. 2011, Robert et al. 2011, Walker et al. 2012, Capella-Gutierrez et al. 2014). Although the concepts of phylogenetic markers and DNA barcodes differ in principle, they overlap in application. Their ideal characteristics include adequate species resolution, ease of amplification, absence of extreme length variation, the presence of only single copies, and low intra-species variability. Numerous attempts were made to identify loci with suitable primary barcode characteristics. These include efforts targeting: i) Cox1 (or CO1; Seifert et al. 2007, Dentinger et al. 2011, Robideau et al. 2011), the primary barcode for animals ratified by CBOL (Hebert et al. 2003a, b, Schindel & Miller 2005); ii) the AFTOL (http://aftol.org/about.php) genes (e.g. RPB1, RPB2, nucLSU, nucSSU, mtSSU, TEF1α and mtATP6), partially used by James et al. (2006) and evaluated for their barcoding potential by Schoch et al. (2012); iii) non-universal regions such as ND6 (hypothetical protein), CAL (Calmodulin), ACT (Gamma Actin) or TUB2 (Beta Tubulin 2) (Carbone & Kohn 1999, Aveskamp et al. 2009, Lee & Young 2009, Verkley et al. 2014); iv) the minichromosome maintenance complex MCM7 (a DNA helicase) and Tsr1 (a pre-mRNA processing protein homolog), the first loci extracted from genome-based computational predictions (Aguileta et al. 2008, Schmitt et al. 2009) and FG1093 and MS204, selected from a screen of 25 single copy protein coding genes (Walker et al. 2012).
The relatively small number of barcode markers now available strongly reflect the past ‘poor man’s approach’ of data-driven gene selection (Eberhardt 2010, Capella-Gutierrez et al. 2014). As emphasised by Eberhardt (2010), group or clade-specific questions continue to require different, more specialised species identification solutions (Balajee et al. 2009, Lumbsch & Leavitt 2011, Gao & Zhang 2013, Heinrichs et al. 2012). This persists because attempts to identify alternatives or substitutes for rDNA sequences that meet the requirements of a single ‘primary barcode’ have not yet succeeded. Discovery of the ‘golden bullet barcode’ seems more unlikely than ever. Inconsistencies between the results of different computational analysis and their identified ‘best’ genes (Tautz et al. 2003, Avise 2004, Feau et al. 2011, Lewis et al. 2011, Robert et al. 2011, Capella-Gutierrez et al. 2014) remain discouraging. Most mycologists agree that a compromise solution of combining ITS with secondary or tertiary, ‘group-specific’ DNA barcode(s) is the most realistic solution. This would combine the universal primer fidelity and high taxon coverage possible with ITS, with equally robust primers for secondary barcodes specific to the group or taxon of interest, enhancing precision of species identification. To take an analogy from medical diagnostics, this is an iterative diagnostic process, where the results of a primary analysis (ITS sequencing) can be used to determine what secondary analyses should be performed (Irinyi et al. 2015a, b).
In adopting additional barcodes, the obvious absence of complete reference data is a serious problem, and was one of the main arguments presented against considering Cox1 a primary barcode for fungi (other problems eventually emerged, see e.g. Gilmore et al. 2009). To be truly effective for specialised and non-specialised applications, such as phytopathology (plant diseases control and quarantine), clinical applications (diagnostics, disease control, epidemiology) or environmental studies (ecology, conservation, metagenomics), formally adopted barcodes will need to be highly predictive, and thus comprehensive reference datasets will need to be developed. Ideally, such data should leave almost no margin for error, reduce false-positives even in the absence of a perfect match, and allow for additive compensation via the primary barcode (ITS) when unknowns are likely to occur and reference data for rare taxa is scarce, as is the case in fungal ecology (Tedersoo et al. 2008, Buée et al. 2009, Pawlowski et al. 2012).
The selection process for novel high fidelity ‘phylogenetic markers’ and ‘DNA barcodes’ is mostly determined by laboratory practicalities. In silico analyses of complete genomes (Capella-Gutierrez et al. 2014) to identify combinations of genes most informative to establish phylogenetic relationships are conceptually similar. Current marker selection procedures are biased towards ranking single genes, rather than combinations of genes, reducing resolution power. The question arises which approach is optimal for identifying potential secondary or tertiary barcodes and how selection procedures, formerly achieved by ‘trial-and-error’, can be optimised to avoid arbitrary selections?
At present, the growth of the number of complete fungal genome sequences and the steady increase of comparative genomic tools allow an unprecedented view on variability among genes and species, on sequence homology and gene synteny. The detection of orthologs and paralogs is crucial for inference of robust molecular classifications. Understanding should go beyond phylogenetics and determination of species limits (Grigoriev et al. 2014, Nagy et al. 2014).
A key concept in the present study was the in silico selection process for alternative candidate barcodes, following strategies described by Robert et al. (2011) and Lewis et al. (2011) to search for gene regions in complete fungal genomes under several optimality criteria. Lewis et al. (2011) inferred suitable candidate genes using translated protein sequences (Pfam domains), whereas Robert et al. (2011) focused entirely on nucleic acids. Both studies identified gene sections and identified novel primer pairs that functioned in silico for the genomes available at the time, either as universal fungal primers, or as primers targeting phyla or classes. In our study, the primers were rigorously tested and later distributed for independent verification to contributing laboratories. Departing from the study of Robert et al. (2011), we tested and compared amplification efficiency of two nuclear ribosomal regions (ITS and LSU, D1–D2 domains of 26/28S), the 5’ primed end of β -tubulin2 (TUB2) and γ-actin (ACT), the section ‘6–7’ of the second largest subunit of the RNA-polymerase II gene (RPB2), the commonly used intermediate section of translation elongation factor 1-α (TEF1α) (Sasikumar et al. 2012) corresponding to the section 983–1567 bp (in the rust Puccinia graminis), two novel universal high fidelity TEF1α primer alternatives covering approximately the same region, three sections of the newly identified candidate region and fungus-specific gene, translation elongation factor 3 (TEF3) (Ypma-Wong et al. 1992, Belfield et al. 1995, Belfield & Tuite 2006, Greganova et al. 2011), and one of the small ribosomal proteins required for tRNA transfer, the 60S L10 (L1) (Brodersen & Nissen 2005). From the study of Lewis et al. (2011) based on Pfam domains, we tested ITS against the seven genes that could theoretically amplify a wide range of fungal taxa with a single primer pair, namely, phosphoglycerate kinase (PGK, PF00162), DNA topoisomerase I (TOPI, PF02919), Lipin/Ned1/Smp2 (LNS2, PF08235), Indole-3-glycerol phosphate synthase (IGPS, PF00218), Phosphoribosylaminoimidazole carboxylase PurE domain (PurE, PF00731), Peptide methionine sulphoxide reductase (Msr, PF01625) and Vacuolar ATPase (vATP, PF01992). Overall, universal primer performance was recorded for almost 20 000 individual PCR reactions, and selected sequence data were evaluated using a ‘distance’ – rather than a ‘character’ – matrix approach to accommodate a puristic barcoding concept, since the ‘barcoding-gap’ is traditionally calculated from Kimura-2-parametric distance model (K2P) (Kimura 1980). Our taxon selection covers several major lineages of the fungal kingdom and represents hundreds of species of economic, phytopathological and clinical importance among the Agaricomycotina, Pezizomycotina, Pucciniomycotina, Saccharomycotina and Ustilaginomycotina.
MATERIAL AND METHODS
Fungal cultures and specimens
For the barcode primer search by the complete genome approach of Robert et al. (2011), axenic cultures preserved and deposited in several biological resource centres, private collections and fungaria were used as sources for DNA. Tested fungal cultures are circumscribed according to their higher level taxonomic affiliation, main taxa (species level), quantity per ‘dataset’ source and corresponding collaborator in Table 1. A detailed taxon list for each ‘dataset’ is available upon request. Cultures were either activated from lyophilised or cryopreserved material and inoculated on various media, such as oatmeal (OA) and 2 % malt extract (MEA, Oxoid) agars, prepared according to Crous et al. (2009). Alternatively, preserved cultures were directly used for DNA isolation from lyophilised or cryopreserved stocks. The selected ‘datasets’ covered several important taxa within the Agaricomycotina, Pezizomycotina, Pucciniomycotina, Saccharomycotina and Ustilaginomycotina. In total 1 931 fungal strains and fungarium specimens representing > 1 500 fungal species (exact taxonomic status for some strains was not precisely determined ‘spp.’) were selected for benchmarking newly designed against published primers. For the barcode primer search by the Pfam domain approach of Lewis et al. (2011), fungal cultures were obtained from a number of laboratories at the Eastern Cereal and Oilseed Research Centre, ECORC, of Agriculture and Agri-Food Canada, CEF, Ottawa. They were supplied as fungal cultures from which DNA was extracted or as already purified DNA samples.
Table 1.
Overview on datasets and selected taxa.
| Dataset | Family | Main taxa | Quantity | Source, Collaborator |
|---|---|---|---|---|
| Penicillium I | Trichocomaceae | Penicillium spp. | 96 | CBS, Houbraken |
| Penicillium II | Trichocomaceae | Penicillium spp., Talaromyces spp. | 96 | CBS, Yilmaz |
| Scedosporium I | Microascaceae | Scedosporium apiospermum | 30 | Inst. Pasteur, Hermoso |
| Scedosporium II | Microascaceae | Scedosporium spp. | 22 | Westmead Hospital, Meyer & Irinyi |
| Onygenales | Arthrodermataceae and Ajellomycetaceae | Trichophyton, Epidermophyton, Arthroderma, Microsporum, Chrysosporium spp. | 180 | CBS, Dukik, Moreno, De Hoog, Stielow |
| Ascomycetous yeast I | Saccharomycetaceae | Candida albicans, C. catenulata, C. dubliniensis, C. glabrata, Kodamaea spp., Clavispora spp. | 207 | Westmead Hospital, Meyer & Irinyi |
| Ascomycetous yeast II | Saccharomycetaceae | Candida albicans, C. tropicalis, C. orthopsilosis, C. metapsilosis | 96 | CBS, Groenewald, Boekhout |
| Ascomycetous yeast III | Saccharomycetaceae | Debaromyces, Millerozyma spp. | 24 | CIRM-Levures, INRA, Casaregola, Mallet & Jacques |
| Ascomycetous yeast IV | Saccharomycetaceae | Debaromyces, Millerozyma, Saccharomyces spp. | 83 | Microbiology Perugia, Cardinali & Roscini |
| Colletotrichum | Glomerellaceae | Colletotrichum spp. | 50 | Senkenberg Museum Goerlitz, Damm |
| Hypocreales | Nectriaceae | Cylindrocarpon, Fusarium, Nectria spp. | 96 | CBS, Lombard |
| Fusarium | Nectriaceae | Fusarium fujikuroi sensu stricto | 96 | CBS, Al-Hatmi, Van Diepeningen |
| Ochroconis | Sympoventuriaceae | Ochroconis spp. | 50 | CBS, Samerpitak |
| Ceratocystis | Ceratocystidaceae | Ceratocystis spp. | 21 | FABI, Wingfield, De Beer, Barnes & Duong |
| Coniothyrium | Montagnulaceae | Coniothyrium spp. | 96 | CBS, Verkley & Stielow |
| Mycosphaerellaceae | Mycosphaerellaceae | Mycosphaerella, Septoria spp. | 45 | CBS, Quaedvlieg |
| Teratosphaeriaceae | Teratosphaeriaceae | Teratosphaeria spp. sensu lato | 90 | CBS, Egidi, Binder & Quaedvlieg |
| Polyporaceae | Polyporaceae | Polyporus, Trametes spp. | 20 | INRA, Chaduli, Lomascolo, Welti & Lesage-Meessen |
| Russula | Russulaceae | Russula spp. | 47 | Museum für Naturkunde Stuttgart, Eberhardt |
| Gasteromycetes | Sclerodermataceae, Phallaceae and Diplocystaceae | Pisolithus, Phallus and Astraeus spp. | 32 | Royal Botanic Garden Madrid, Martin |
| Basidiomycetous yeast I | Tremellaceae | Cryptococcus, Rhodotorula spp. | 98 | Westmead Hospital, Meyer & Irinyi |
| Basidiomycetous yeast II | Tremellaceae sensu lato | Cryptococcus, Kwoniella, Bullera spp. | 174 | DSMZ & Ruhr University Bochum, Yrukov & Begerow |
| Multi-Phyla | Chytridiaceae, Mortierellaceae, Nectriaceae, | Rhizoclosmatium, Mortierella spp., Fusarium spp., Drechslera spp., Cadophora, | 182 | Agriculture and Agri-Food Canada (AAFC), Ottawa |
| Pleosporaceae, Pucciniaceae, Rhizophydiaceae, | Pyrenophora spp., Ulocladium, Epicoccum, Dendryphion, Alternaria, Puccinia spp., | |||
| Tilletiaceae, Wallemiaceae and 35 more | Rhizophydium spp., Tilletia spp., Wallemia spp. and 40 more |
Standard molecular procedures
For the primers inferred from the complete genome through the approach by Robert et al. (2011), total genomic DNA was extracted from living cultures, cells preserved in liquid nitrogen or in lyophilisation and from dried fungarium specimens (Agaricomycotina only) using different variations of one-by-one or high-throughput 96-well plate DNA extraction techniques, routinely used by the respective collaborating laboratories (Ferrer et al. 2001, Ivanova et al. 2006, Yurkov et al. 2012, 2015, Feng et al. 2013, Verkley et al. 2014). DNA extraction and PCR protocols differed between participating laboratories.
Primers and amplification conditions used varied between laboratories, an example of the ones used at the CBS is provided. PCR reactions for amplification of the ITS barcode employed primers ITS5/ITS1/ITS1F and ITS4 were performed under standard or semi-nested conditions in 12.5 μL reactions (the CBS-KNAW barcoding lab protocol) containing 2.5 μL purified DNA, 1.25 μL PCR buffer (Takara, Japan, incl. 2.5 mM MgCl2), 1 μL dNTPs (1 mM stock; Takara, Japan), 0.6 μL v/v DMSO (Sigma, Netherlands), forward-reverse primer 0.25 μL each (10 mM stock), 0.06 μL (5 U) Takara HS Taq polymerase, 7.19 μL MilliQ water (White et al. 1990, Stielow et al. 2012, Yurkov et al. 2012). PCR conditions for amplifying partial LSU rDNA using primers LR0R and LR5 differed only by their annealing temperature (55 °C instead of 60 °C) and increased cycle extension time (90 s per cycle). Amplification of partial γ-actin (ACT), covering the more variable 5’-end including two small introns, and partial β -tubulin 2 (TUB2), covering the variable 5’-end with up to four small introns, followed the protocol of Aveskamp et al. (2009) and Carbone & Kohn (1999) using the primers Btub2Fd and Btub4Rd, and ACT-512F, ACT-783R, respectively. TEF1α and RPB2 were amplified following the protocols of Rehner & Buckley (2005) and Liu et al. (1999), respectively (Table 2). PCR products were directly purified using FastAP thermosensitive alkaline phosphatase and shrimp alkaline phosphatase (Fermentas, Thermo Fisher Scientific). Cycle-sequencing reactions were set up using ABI BigDye Terminator v. 3.1 Cycle Sequencing kit (Thermo Fisher Scientific), with the manufacturers’ protocol modified by using a quarter of the recommended volumes, followed by bidirectional sequencing with a 3730xl DNA Analyser (Thermo Fisher Scientific). Sequences were archived, bidirectional reads assembled and manually corrected for sequencing artefacts using BioloMICS software v. 8.0 (www.bio-aware.com) (Vu et al. 2012). Edited sequences were exported to and aligned with MAFFT v. 7.0 (Katoh et al. 2005) and further corrected for indels and SNPs (single nucleotide polymorphisms) by replacing respective positions with ambiguity code letters.
Table 2.
Benchmarking primers used to assess performance and versatility of newly designed primers.
| Locus | Primer | Oligo nucleotides (5’-3’) | Reference |
|---|---|---|---|
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | Ward & Adams (1998) |
| ITS4 | TCCTCCGCTTATTGATATGC | White et al. (1990) | |
| ITS1 | TCC GTA GGT GAA CCT GCG G | White et al. (1990) | |
| ITS1-F | CTT GGT CAT TTA GAG GAA GTA A | Gardes & Bruns (1993) | |
| LSU | LROR | ACCCGCTGAACTTAAGC | Vilgalys & Hester (1990) |
| LR5 | TCCTGAGGGAAACTTCG | ||
| TUB2 | Btub2Fd | GTBCACCTYCARACCGGYCARTG | Woudenberg et al. (2009) |
| Btub4Rd | CCRGAYTGRCCRAARACRAAGTTGTC | ||
| Bsens | ATCACWCACTCICTIGGTGGTGG | Lesage-Meessen et al. (2011) | |
| Brev | CATGAAGAARTGIAGACGIGGG | ||
| ACT | ACT512f | ATGTGCAAGGCCGGTTTCG | Carbone & Kohn (1999) |
| ACT783r | TACGAGTCCTTCTGGCCCAT | ||
| CA5R | GTGAACAATGGATGGACCAGATTCGTCG | Daniel et al. (2001), Daniel & Meyer (2003) | |
| CA14 | AACTGGGATGACATGGAGAAGATCTGGC | ||
| RPB2 | fRPB2-5f | GAY GAY MGW GAT CAY TTY GG | Liu et al. (1999), Binder & Hibbett (Clark University website) |
| fRPB2-7cF | ATG GGY AAR CAA GCY ATG GG | ||
| fRPB2-7cR | CCC ATR GCT TGY TTR CCC AT | ||
| TEF1α | EF1-983F | GCY CCY GGH CAY CGT GAY TTY AT | Rehner & Buckley (2005) |
| EF1-1567R | ACH GTR CCR ATA CCA CCR ATC TT |
For the primers inferred from the complete proteome through the Pfam approach by Lewis et al. (2011), genomic DNA from fungal cultures was extracted using the OmniPrep™ Genomic DNA Extraction Kit (G-Biosciences, St. Louis, Missouri). Fungal tissue was ground into a fine powder in a mortar and pestle with liquid nitrogen and stored at -80 °C. Approximately 50 mg of ground tissue was placed in a 1.7 mL microfuge tube, resuspended in 250 μL of lysis buffer and vortexed for several seconds. An additional 250 μL of lysis buffer was added to the resuspension and the tube was incubated for 15 min at 55–60 °C without the addition of Proteinase K. The samples were cooled to room temperature and 200 μL of chloroform was added to the tube. The tube was mixed by inversion several times and then centrifuged for 10 min at 14 000 ×g. The upper aqueous phase was removed to a new microfuge tube and 100 μL of precipitation solution was added. If no white precipitate was produced an additional 50 μL of precipitation solution was added to the tube. The white precipitate was pelleted by centrifugation and the supernatant was moved to a fresh tube. The genomic DNA was precipitated by the addition of 500 μL of isopropanol to the supernatant and inversion of the tube several times. The genomic DNA was pelleted by centrifugation and washed with 700 μL of 70 % ethanol. The ethanol was decanted and the pellet was air dried for 15 s prior to resuspension in 50 μL of TE Buffer. DNA concentration was determined using the Qubit® 2.0 Fluorometer (Life Technologies, Burlington, Canada) and working solutions were prepared at a concentration of 0.05 ng/μL. PCR was carried out on 0.1 ng of genomic DNA in a total volume of 20 μL using 0.2 mM dNTPs, 0.5 μM of each primer, 1× of Titanium Taq Buffer and Titanium Taq DNA Polymerase (Clontech) using an Eppendorf GradientS thermal cycler. For the ITS primers, an initial denaturation at 95 °C for 3 min was followed by 40 cycles at the following conditions: 30 s at 95 °C, 45 s at 58 °C and 2 min at 72 °C. A final extension at 72 °C for 8 min completed the PCR. For the remaining primers, Touchdown PCR was performed where an initial denaturation at 95 °C for 5 min was followed by 10 cycles of 45 s at 95 °C, 45 s starting at 68 °C and dropping by 1 °C per cycle until a temperature of 58 °C was reached and a 1 min extension at 72 °C. The initial 10 cycles were then followed by 35 cycles of 45 s at 95 °C, 45 s at 58 °C and 1 min at 72 °C. A final extension at 72 °C for 5 min completed the PCR.
In silico selection of gene and protein regions and initial seed primers
Two different genome-mining strategies were employed to identify potential new barcode markers and different strategies were used to develop them further and for validation.
For the DNA-based approach, complete genome sequences of 74 fungi were downloaded from public repositories such as Broad, Genoscope and NCBI in 2011, covering most major lineages of the fungal kingdom. The initial approach to find ideal gene regions was described in Robert et al. (2011) as the ‘Ideal Locus Method’ (ILM). An ideal locus is a gene or gene region that would provide a phylogram possibly close to a ‘whole-genome phylogram’ (a phylogeny inferred from single copy orthologous genes). A ‘Best Pair of Primers Method’ (BPPM) was devised to identify short conserved sections at < 1 Kb apart that could serve as kernels to construct forward and reverse primers for species or selected taxonomic groups. Suitability as barcode candidate was assessed by calculating Pearson coefficients against the super matrix, built from overlapping orthologs (Kuramae et al. 2007).
PCR primers are normally 18–24 bp long, but in practice only the 3’ end fit is critical for PCR success. Hence, an alternative ‘Short Primer Method’ (SPM) was developed as follows: i) listing all possible 12 bp primers as ‘Length 12’ (maximum to limit calculations); ii) searching for these 12-mer primers in all available fungal genomes, which is considerably faster than searching for primer pairs; iii) maintaining primers with sufficient quality in terms of PCR applicability and wide distribution. Factors in the equation are: F1 = number of most frequent nucleotides / size of the sequence; F2 = comparison of the sequence with itself, with an offset of three nucleotides, this factor is high for repetitive triplets (e.g. ACCACCACC…n); F3 = comparison of the sequence with its reverse complement, with an offset greater than half the sequence size (avoidance of hairpins). All factors (Fn) take a value of 0.25 for a random sequence, and a value of 1.0 for a poor sequence. Thus, the factors F2 and F3 were rescaled with the following equation: F = (F – 0.25) / 0.75, the final equation is given by: Quality = 1.0 – max (F1, F2, F3); iv) using these best primers to search for possible primer pairs; v) using the best primer pairs to extract the intervening DNA sequence. The target length of the intermediate sequence was set to 200 to 1 000 nucleotides, thresholds chosen to ensure a minimum variability and allow ease of amplification and Sanger sequencing. The formal DNA standard recommends amplicons of about 700 bp, reflecting the technical constraints of the Sanger sequencing prevalent when the standard was implemented; vi) comparing the in silico sequences and building distance matrices and trees to compare with a reference matrix and tree. This last step ensures that the selected DNA region produces a relatively coherent phylogeny compared to the reference matrix and that it would also be suitable for discriminating closely related species.
The in silico design, execution and publication of the resulting primers based on identification of protein families (see Lewis et al. 2011) was done concurrently with the DNA-based method described above. These primers were extensively tested only at the Agriculture & Agri-Food Canada laboratory on smaller fungal test sets by Levesque, Seifert, Hambleton, Lewis and McCormick, with additional testing of primers done at the CBS-KNAW Fungal Biodiversity Centre. The in silico pipeline for identifying suitable protein sequences for primer design is briefly described below.
The approach was inspired by the CARMA algorithm for taxonomic identification of metagenomic sequences (Krause et al. 2008). CARMA matches short environmental gene fragments to Pfam protein families and constructs a taxonomic profile for the sample. The objective is to assign translated input nucleotide sequences to Pfam accessions that can identify single copy gene regions in source organisms and then design degenerate primers from the alignment of these putatively orthologous sequences. This requires pre-processing the Pfam dataset, translating and assigning DNA sequences to Pfam groups, and adding sequences to the reference Pfam alignments. The CARMA pipeline was adapted for use in this project, although the original intent and our application are quite different.
Data were processed in two stages. The first stage translates input sequences and assigns them to Pfam accessions; the second stage prepares alignments and attempts primer design for the selected set of taxa. Adding additional or updating data for existing organisms requires repetition of the first stage. Restricting the analysis to a taxonomic subset of the data requires that the final stage be repeated for the selected organisms. The pipeline requires up-to-date versions of the Pfam-A and NCBI taxonomy datasets; if the Pfam dataset is updated, the first stage of processing must be repeated. The input datasets are placed in a single folder for processing, with each organism in a unique directory named by ‘genus_species_strain’. The pipeline first processes source nucleotide sequences to identify Pfam domains within each sequence and creates clusters of conserved domains. Sequences are then translated into protein sequences using the orientation and frame from a translated BLAST search against the Pfam sequences. Each translated sequence is then processed to identify the region corresponding to the matched protein family and the protein sequence is written to a file. This functionality is as originally implemented in the CARMA pipeline. The second stage of the pipeline involves screening the Pfam matches for a user-defined subset of organisms to identify families that contain a single match for each of the selected organisms. Such families are considered to represent conserved, single copy gene regions. Next, these putatively orthologous sequences are added to the corresponding Pfam multiple sequence alignment. Adding the sequences to an existing curated alignment results in a superior alignment to de novo sequence alignment. Original sequences are then removed from the reference alignments so that only targeted organisms remain and the resulting alignment is screened for conserved blocks for primer design. Conserved regions flanking variable sites are identified and only alignments with two or more conserved blocks are selected so that pairs of forward and reverse primers can be sought in the separate blocks. The resulting primers were further screened for protein families containing primer pairs with additional desirable characteristics, such as optimum amplicon length.
Final primer design
Two different genome and proteome mining strategies were employed to identify the initial potential new barcode markers (Lewis et al. 2011, Robert et al. 2011) and different strategies were also used to develop them further and for validation.
The selected gene regions by the complete genome approach of Robert et al. (2011) corresponding to a total of 54 alignments (available on request) were manually re-annotated using n- and x-BLAST and alignments resulting in identical annotations were realigned and redundant alignments discarded. These seed primers are given in Table 3. To design PCR primers, non-redundant alignments were imported into BioEdit v. 7.0.52 and visually screened for the most conserved sites to identify suitable primer binding sites. Priority was given to stable 3’ prime ends with at least eight nucleotide binding sites and the least possible amount of degeneracy. For several primers, ‘N’s were replaced with an inosine, ‘I’s, to improve binding stability and to reduce the quantity of primers in the actual synthesized wobble pool. Individual primer pairs are discussed in the results section.
Table 3.
Initial seed primers from ‘nucleic acid based’ computational predictions (Robert et al. 2011).
| Alignment group | Species in alignment (Al) | Primers (forward-reverse) | Functional annotation |
|---|---|---|---|
| 1 | 51 | acaagcgtttct-catcaagttcca | Hypothetical protein |
| 2 | 41 | acatggagaaga-catcaaggagaa | Gamma actin |
| 3 | 44 | accttcttgatg-atgttcttgatg | Translation elongation factor 1α |
| 3b | 46 | caagaacatgat-catcaagaaggt | Translation elongation factor 1α |
| 4 | 38 | agtacttgtagg-cttggccttgta | 60S ribosomal protein L15b |
| 5 | 51 | ggaacttgatgg-agaaacgcttgt | 60S ribosomal protein L15b |
| 6 | 45 | ggtatcaccatc-caacaagatgga | Translation elongation factor 1α |
| 7 | 45 | tccatcttgttg-gatggtgatacc | Translation elongation factor 1α |
| 8 | 36 | gtccatcttgtt-tacttgaaggaa | Translation elongation factor 1α |
| 9 | 56 | gttcttggagtc-gtccatcttgtt | Translation elongation factor 1α |
| 10 | 56 | aacaagatggac-ctccaagaacga | Translation elongation factor 1α |
| 11 | 41 | aacaagatggac-tcaccactgaag | Translation elongation factor 1α |
| 11b | 42 | tggtatctccaa-aacgtcaagaac | Translation elongation factor 1α |
| 12 | 58 | caacaagatgga-ctccaagaacga | Translation elongation factor 1α |
| 13 | 42 | cacttcttcatg-atggacgagatg | 60S ribosomal protein L15b |
| 14 | 37 | catatgcttgtc-gactcgtcatct | Putative CD box sno-RNA protein,putative protein S6 kinase |
| 15 | 43 | cttcagtggtga-ccatcttgttga | Translation elongation factor 1α |
| 16 | 51 | gaacttgatggt-agaaacgcttgt | Hypothetical protein |
| 17 | 36 | gttccttcaagt-caacaagatgga | Hypothetical protein |
| 18 | 36 | tccatcttgttg-acttgaaggaac | Translation elongation factor 1α |
| 19 | 48 | tcttgacgttga-gtccatcttgtt | Translation elongation factor 1α |
| 20 | 48 | ttcttgacgttg-gtccatcttgtt | Translation elongation factor 1α |
| 21 | 39 | tcgttcttggag-tcttgatgaagt | Translation elongation factor 1α |
| 21b | 53 | ttcttggagtca-tccatcttgttg | Translation elongation factor 1α |
| 22 | 45 | ttcttgacgttg-catgttcttgat | Translation elongation factor 1α |
| 23 | 37 | ttccttcaagta-caacaagatgga | Translation elongation factor 1α |
| 24 | 45 | ttcatcaagaac-tcaacgtcaaga | Translation elongation factor 1α |
| 25 | 41 | ttcagtggtgac-atcatgttcttg | Translation elongation factor 1α |
| 26 | 39 | tccttgatttcg-cttcatgacctt | Translation elongation factor 1α |
| 27 | 37 | tcaagaaggtcg-ctccaagaacga | Translation elongation factor 1α |
| 28 | 46 | tcaagaacatga-caacgtcaagaa | Translation elongation factor 1α |
| 29 | 40 | caacaagatgga-aagttcatcaag | Translation elongation factor 1α |
| 30 | 47 | gttcttgacgtt-gtccatcttgtt | Translation elongation factor 1α |
| 31 | 48 | aacaagatggac-caacgtcaagaa | Translation elongation factor 1α |
| 32 | 39 | gacttgatgaac-ccatcttgttga | Translation elongation factor 1α |
| 33 | 45 | catcaagaacat-tcaacgtcaaga | Translation elongation factor 1α |
| 34 | 52 | catcaagaacat-gactccaagaac | Translation elongation factor 1α |
| 35 | 40 | catcaagaaggt-gactccaagaac | Translation elongation factor 1α |
| 36 | 40 | atcaagaacatg-tcaccactgaag | Translation elongation factor 1α |
| 37 | 51 | caagcgtttctc-ccatcaagttcc | Translation elongation factor 1α |
| 38 | 39 | atctccaaggat-ctccaagaacga | Translation elongation factor 1α |
| 39 | 52 | atcaagaacatg-gactccaagaac | Translation elongation factor 1α |
| 40 | 45 | atcaagaacatg-tcaacgtcaaga | Translation elongation factor 1α |
| 41 | 40 | acttgatgaact-tccatcttgttg | Translation elongation factor 1α |
| 42 | 37 | acttcatcaaga-tcaacgtcaaga | Translation elongation factor 1α |
| 43 | 40 | acaagatggaca-gactccaagaac | Translation elongation factor 1α |
| 44 | 38 | aacaagatggac-aagttcatcaag | Translation elongation factor 1α |
| 45 | 39 | aaggtcatgaag-cgaaatcaagga | Translation elongation factor 2 |
| 46 | 42 | catctcgtccat-catgaagaagtg | Beta tubulin 2 |
| 47 | 45 | gttcttgaactt-cttcatcttcca | Translation elongation factor 3 |
| 48 | 39 | tccttgatttcg-ccatcttggaga | Translation elongation factor 3 |
| 49 | 45 | tggaagatgaag-aagttcaagaac | Translation elongation factor 3 |
| 50 | 42 | tggaagatgaag-tgtcaagaccaa | Translation elongation factor 3 |
| 51 | 42 | ttggtcttgaca-cttcatcttcca | Translation elongation factor 3 |
| 52 | 51 | tggaacttgatg-gagaaacgcttg | 60S ribosomal protein L10 (L1) |
| 53 | 35 | ggtatcaccatc-ctccaagaacga | Translation elongation factor 1α |
| 54 | 40 | ttcatcaagaac-tcaccactgaag | Translation elongation factor 1α |
Seed primer sequences highlighted bold, were those qualifying for the last and final cross-laboratory trial.
For the Pfam domain approach by Lewis et al. (2011), a nucleotide search was first conducted within NCBI for Interpro names to help with primer validation, using keywords found within the paper (e.g., PF01625, PGK, IPR001576) and NCBI was searched for more recent accessions using trimmed amino acid sequences for each of the inferred protein regions as queries. The downloaded nucleotide sequences were added to the original data from Lewis et al. (2011). A ClustalW alignment was performed individually on the supplied trimmed sequences and NCBI downloaded sequences. For LNS2, the alignments were primarily from basidiomycete sequences as the objective was to design primers that would have a higher success for this group. The alignments were handled individually to ensure that all possible primers for each gene could be identified. Within each gene alignment, frequency tables were developed for conserved regions. All areas of the protein region that contained, at a minimum, 3 conserved nucleotides (> 85 %) at the 3’ end, were examined for the potential of becoming a primer site. In developing the primers, all base positions were examined visually. Any base with a frequency > 80 % was maintained in the primer. Primers in which there was no base with a frequency > 80 %, were resolved into degenerate primers, such that the minimum frequency > 80 % was maintained. Close to 500 primer pair combinations were tested. The overall number of forward primers designed, of reverse primers designed, and number of primer pairs tested, were 26, 23 and 125 for PGK; 16, 13 and 74 for TOP1; 20, 28 and 49 for LNS2; 24, 13 and 62 for IGPS; 17, 11 and 64 for PurE; 12, 9 and 27 for Msr; and 12, 8 and 32 for vATP, respectively.
Laboratory trials of gene-based primers (phases I and II)
For the primers designed by the complete genome approach from Robert et al. (2011), annealing temperatures were set at 42 °C and slowly ramped up to 52 °C. This allowed us to identify the most universally applicable primer pairs (one-by-one proof-of-principle) and those yielding nothing, or single or multiple fragments. The general PCR cycler setup was: 7 min 95 °C, 1 min 95 °C, 1 min 42–52 °C (+ 0.5 °C/cycle until 52 °C is reached), 2 min 72 °C, the latter three steps repeated 40 times, final elongation at 10 min 72 °C and cooling at 10 °C. The laboratory work was conducted in two initial phases. Phase I included a small set of genomic DNA extracts roughly covering several major fungal lineages (Table 4) representing strains with completely sequenced genomes, thus the same strains used for the in silico work described above (Robert et al. 2011). This trial aimed to identify well-performing primer sets and to assess their versatility, i.e. their ability to amplify the targeted gene from various genera. Primer pairs that yielded no product were discarded. Phase II was conducted with 48 DNA extracts (Table 5) to decrease taxon bias and increase the reliability and consistency of the entire experiment. Only primers that successfully amplified, either highly accurately and/or slightly inaccurately (defined as a single PCR product not exactly the calculated size), were further tested employing the PCR conditions described above. Amplifications that resulted in a single, clear fragment, without any further reaction optimisation, were tested under widely used successful PCR conditions, defined as: 5 min 95 °C, 1 min 95 °C, 1 min 48 or 50 °C, 2 min 72 °C, the latter three steps repeated 40 times, final elongation at 10 min 72 °C and cooling at 10 °C. This PCR protocol is generally applicable to those primers listed in Table 6. Only those pairs yielding single fragments under these conditions were kept for the final phase. Phase III included in depth testing of the best candidate primers, described in the Results section, with large sets of DNA extracts covering multiple species representing economically important genera such as Penicillium sensu stricto (s.str.), Fusarium s.str., or higher level ranks covering several genera within orders such as the Onygenales (Table 1). The selected taxa (number of strains or specimens) for this phase represent those covered by the consortium of participating laboratories. Benchmarking of the best newly designed candidate primers was conducted against commonly used and well-recognised primer pairs (Table 2). All reagents for phases I and II were standardised with enzymes and dNTPs from Takara (Japan), oligonucleotides synthesized by Integrated DNA Technologies (The Netherlands) and PCR reactions ran on SensoQuest PCR cyclers (Germany) as described above under ‘standard molecular procedures’. Reagents for phase III varied among laboratories and institutions according to their internal protocols, and served as a robust verification of the tested primers (detailed protocols can be requested from the respective ‘collaborator’ as indicated in Table 1).
Table 4.
Initial test cultures used for primary laboratory trial I for primers inferred from ‘nucleic acid based’ computational predictions (Robert et al. 2011).
| CBS number | Taxon |
|---|---|
| CBS 513.88 | Aspergillus niger |
| CBS 818.72 | Aspergillus oryzae var. brunneus |
| CBS 1954 | Candida parapsilosis var. parapsilosis |
| CBS 115846 | Cryphonectria parasitica |
| CBS 123668 | Fusarium oxysporum f.sp. lycopersici |
| CBS 445.79 | Laccaria bicolor |
| CBS 277.49 | Mucor circinelloides f. lusitanicus |
| FGSC 9596 | Nectria haematococca |
| CBS 708.71 | Neurospora crassa |
| FGSC 10004 | Phycomyces blakesleeanus |
| CBS 405.96 | Schizophyllum commune |
| CBS 142.95 | Trichoderma atroviride |
| CBS 109036 | Trichophyton equinum |
| CBS 127170 | Verticillium dahliae |
| CBS 115943 | Zymoseptoria tritici |
Table 5.
Initial test cultures used for secondary laboratory trial II for primers inferred from ‘nucleic acid based’ computational predictions (Robert et al. 2011).
| CBS number | Taxon |
|---|---|
| CBS 674.68 | Ajellomyces dermatitidis |
| CBS 118699 | Alternaria brassicicola |
| CBS 131.61 | Aspergillus flavus var. flavus |
| CBS 126972 | Aspergillus nidulans |
| FGSC 1144 | Aspergillus niger |
| FGSC 1156 | Aspergillus terreus |
| CBS 136.29 | Bipolaris maydis |
| CBS 114389 | Blastomyces dermatitidis |
| CBS 8758 | Candida albicans var. albicans |
| CBS 113850 | Coccidioides immitis |
| CBS 113843 | Coccidioides posadasii |
| CBS 126970 | Coprinus cinereus |
| CBS 8710 | Filobasidiella neoformans |
| FGSC 9075 | Fusarium graminarum |
| FGSC 9935 | Fusarium oxysporum f.sp. lycopersici |
| CBS 123670 | Fusarium verticillioides |
| FGSC 1089 | Gibberella fujikuroi |
| CBS 287.54 | Histoplasma capsulatum |
| CBS 2605 | Lodderomyces elongisporus |
| CBS 113480 | Microsporum canis |
| CBS 180.27 | Neurospora tetrasperma |
| CBS 223.38 | Neurospora tetrasperma |
| CBS 101191 | Neurospora tetrasperma |
| CBS 372.73 | Paracoccidioides brasiliensis |
| CBS 127171 | Parastagonospora nodorum |
| FGSC 9002 | Phanerochaete chrysosporium |
| CBS 6054 | Pichia stipitis |
| CBS 126969 | Podospora pauciseta |
| CBS 120258 | Pseudocercospora fijiensis |
| CBS 117146 | Pyrenophora tritici-repentis |
| CBS 128304 | Pyricularia grisea |
| CBS 658.66 | Pyricularia grisea |
| CBS 126971 | Rhizopus orysae |
| CBS 124811 | Schizophyllum commune |
| CBS 7116 | Schizosaccharomyces japonicus |
| CBS 484 | Sporidiobolus pararoseus |
| CBS 208.27 | Sporobolomyces roseus |
| CBS 375.48 | Talaromyces stipitatus |
| CBS 693.94 | Trichoderma atroviride |
| CBS 392.92 | Trichoderma reesei |
| CBS 383.78 | Trichoderma reesei |
| CBS 127.97 | Trichophyton equinum var. equinum |
| CBS 668.78 | Uncinocarpus reesii |
| CBS 127172 | Ustilago maydis |
| CBS 127169 | Verticillium alboatrum |
| CBS 599 | Yarrowia lipolytica |
| CBS 732 | Zygosaccharomyces rouxii |
Table 6.
Super primers and best candidate primers inferred from ‘nucleic acid based’ computational predictions (Robert et al. 2011).
| Locus | Original primer name | Final primer name | Sequence (5’-3’) |
|---|---|---|---|
| TEF1α | Al33_54_73_F1_forward | EF1-1018F | GAYTTCATCAAGAACATGAT |
| Al33_879_859_R2_reverse | EF1-1620R | GACGTTGAADCCRACRTTGTC | |
| Al_34_EF1_300_F1_forward | EF1-1002F | TTCATCAAGAACATGAT | |
| AL34_EF1_1050_R_Tail_reverse | EF1-1688R | GCTATCATCACAATGGACGTTCTTGGAG | |
| Al33_129_148_F2_forward | Al33_alternative_f | GARTTYGARGCYGGTATCTC | |
| Al28_EF1_400_f | EF1_alterantive_3f | TTYGARGCYGGTATCTC | |
| Al28_EF1_900_R | EF1_alternative_3r | GAVACRTTCTTGACGTTGAA | |
| 60S L10 (L1) | AlGr52_412-433_f1 | 60S-908R | CTTVAVYTGGAACTTGATGGT |
| Algr52_1102_1084_R1 | 60S-506F | GHGACAAGCGTTTCTCNGG | |
| TEF3 | Al50+51_EF3_2900_f | EF3-3185F | TCYGGWGGHTGGAAGATGAAG |
| Al50+51_EF3_3300_R | EF3-3538R | YTTGGTCTTGACACCNTC | |
| Al47_EF3_1650_forward | EF3-3188F | GGHGGHTGGAAGATGAAG | |
| A47_EF3_2451_R1_reverse | EF3-3984R | TCRTAVSWGTTCTTGAACTT | |
| Al49_ EF3_44_63_F1_forward | EF3-3186F | CYGGHGGHTGGAAGATGAAG | |
| Al49_ EF3_846_829_r1_reverse | EF3-3984R2 | TCRTAVSWGTTCTTGAAC |
For the primers designed from Pfam domains (Lewis et al. 2011), initial PCR analysis of potential barcoding primers was performed on DNA from the ascomycetes Cadophora fastigiata and Pyrenophora teres f. teres using a standard PCR protocol and gradient annealing temperatures, ranging from 56–70 °C, to determine an optimum annealing temperature for further testing. Touchdown PCR (Don et al. 1991) was used to increase primer specificity. The final Touchdown temperatures of 68–58 °C was selected on the basis of the PCR giving the best combination of product concentration and single fragment size produced for two of the three initial ascomycetes tested. For IGPS and vATP, none of the primer pairs passed this first panel. Primer pairs that resulted in acceptable sequences for this first panel were carried forward and tested against a panel of eight organisms: Rhizophydium littoreum (Chytridiomycetes), Mortierella vinacea (Zygomycetes), the basidiomycetes Tilletia indica and Puccinia graminis, and the ascomycetes Pyrenophora teres f. teres, Debaryomyces hansenii, Penicillium verrucosum and Fusarium graminearum. Only primers for PGK, TOP1 and LNS2 were tested further with additional isolates and with tagging of M13 primers to improve sequencing success (Table 7). Throughout the process of primer development, newly acquired long sequences, as well as newly mined results of full length genes from GenBank (performed on a monthly basis) were used to improved primers for shorter fragments.
Table 7.
Super primers inferred from ‘protein-based’ computational predictions (Lewis et al. 2011).
| Locus | Original primer name | Final primer name | Sequence (5’ to 3’) |
|---|---|---|---|
| PGK | PF00162.1120.M13-8-F | PGK_480-F | TGTAAAACGACGGCCAGTACGATATCCGAGTCGACTTCAAYGTCCC |
| PF00162.2081.M13-8-R | PGK_480-R | CAGGAAACAGCTATGACTCGAAGACACCRGGGGGACCGTTCCA | |
| PF00162.1433.M13-8-F | PGK_483-F | TGTAAAACGACGGCCAGTACGATGAGAACYTGCGHTTCCACRYYGAGGAGGARGG | |
| PF00162.1793.M13-8-R | PGK_483-R | CAGGAAACAGCTATGACCTTCTTGAAGGTGAARGCCAT | |
| PF00162A.675.1-F | PGK_511-F | GTYGSTGCYYTGCCMACCATCAA | |
| PF00162A.1915.1-R | PGK_511-R | ATCTTGTCRGMRACCTTRGCACC | |
| PF00162.1127.1-F | PGK_533-F | GTYGAYTTCAAYGTYCC | |
| PF00162.2081.1-R | PGK_533-R | ACACCDGGDGGRCCGTTCCA | |
| TOP1 | PF02919.2708.M13.1-F | TOP1_501-F | TGTAAAACGACGGCCAGTACGATACTGCCAAGGTTTTCCGTACHTACAACGC |
| PF02919.3469.M13.8-R | TOP1_501-R | CAGGAAACAGCTATGACCCAGTCCTCGTCAACWGACTTRATRGCCCA | |
| LNS2 | PF08235.1463.8-F | LNS2_468-F | GGCCATGTGCTGAACATGATCGGHCGWGAYTGGAC |
| PF08235.1821.8-R | LNS2_468-R | CGGTTGCCRAAKCCRGCATAGAAKGG |
Bases in bold: M13 primers with an ACGAT spacer for the forward primers.
Sequence and data analysis (phase III)
To assure standardised laboratory procedures, a working agreement defining experimental conditions was distributed prior to primer testing in phase III, which defined the basis for recording amplification efficiency. To assemble a data matrix of binary variables, successful amplification was scored as ‘true’ and unsuccessful amplification as ‘false’. Recovery of multiple fragments was also classified as ‘false’. Single fragments of unexpected sizes were assumed to represent length variations (mostly false negatives), a common phenomenon when comparing intron-containing sequences, were classified as ‘true’. This phenomenon does not always reciprocally correlate with an incorrect amplification of targeted gene section or, subsequently, poor downstream (sequencing) success.
All data were analysed and visualised with R statistical software (R Core Team 2014) with the ‘lattice’, ‘vcd’ and ‘MASS’ libraries, as well as related packages; the source code is available from Sarkar (2008). Primer amplification efficiencies were visualised as ‘barchart’ and ‘mosaicplot’. Gene maps with primer locations were created with the Qiagen CLC genomics suite (http://www.clcbio.com/products/clc-genomics-workbench/) primers designed by the complete genome approach by Robert et al. (2011) and with Geneious v. R6 (Biomatters http://www.geneious.com/) for the primers designed from Pfam domains (Lewis et al. 2011).
For sequences generated from primers designed by the complete genome approach by Robert et al. (2011), sequences were stored as bidirectional reads and edited as described above, using BioloMICS (Vu et al. 2012) at CBS. Various software packages, differing between laboratories, were employed to assemble consensus sequences. Sequences were visually corrected for sequencing artefacts and quality controlled within individually aligned datasets. Quality controlled data was uploaded to the BioloMICS database and pairwise distances for selected (symmetric/orthogonal matrices only) datasets calculated using the optimistic reverse pairwise alignment algorithm. Symmetrical datasets were assembled to include sequence data for at least: complete ITS1-5.8S-ITS2, LSU, TEF1α (either from one of the newly designed TEF1α primer pairs, see Results), or the TEF1α AFTOL primer set EF1-983F / EF1-1567R, Table 2, spanning almost the same section), TEF3, and the section spanning the 60S ribosomal protein L10 (L1). The global dataset comprised 502 strains or specimens representing a fully symmetrical distance matrix with 5 gene partitions. Each dataset was analysed individually to create a single locus similarity matrix to generate an overview UPGMA tree. A second analysis used all loci in a concatenated matrix, followed by a multi-dimensional scaling analysis (MDS), visualised in n-dimensional space. All six similarity matrices (5 loci + 1 concatenated) were compared, to obtain pairwise cophenetic coefficients of correlation (Mantel test) between each similarity matrix (Smouse et al. 1986). A correlation super matrix was subsequently created, which was numerically rescaled and another final MDS performed; the results were plotted in an n-dimensional space to compare the ‘global’ between-loci performance. Eigenvalues for each axis were computed to assign weights to dimensions. Two examples, the Onygenales and Fusarium datasets were analysed individually as described above, with identical loci sampling, except for the Onygenales which was supplemented with ‘γ-Actin’ sequence data to provide examples on ‘local gene optima’.
For sequences generated from primers designed by the Pfam approach of Lewis et al. (2011), bidirectional reads were edited manually using Geneious to generate consensus sequences. Sequences were aligned through MAFFT within Geneious and processed through R to compare intra- and inter-species pairwise distances (Schoch et al. 2014). TUB2 and TEF1 sequences of Penicillium and Fusarium strains, respectively, that were already available were added to the dataset as comparison for species resolution. The aligned sequences of each marker were analysed individually with the ‘ape’ package (Paradis et al. 2004) for R (R Core Team 2014) to generate raw pairwise distances for each marker and each pair of strains. The values were separated as intra- or inter-species distances and the function ggplot2 (Wickham 2009) was used with R to generate a barcode gap plot for each gene by overlaying the distribution of the intra- and inter-species distances.
RESULTS
In silico selection of gene regions and manual design of initial seed-primers (gene-based primers)
The distance super matrix, underlying the multiple alignments used to generate the reference topology, was correlated with all individual distance matrices using Pearson coefficient. One third of the genes (29.8 %) produced a phylogeny highly correlated with the super matrix (Robert et al. 2011). Seventy percent of the gene matrices correlated at > 0.70 with the super matrix, and only 25 genes had no or a very low correlation. For the 531-gene matrix, maximum correlations were found for 190 concatenated individual genes. The results from the computational inference indicated that a single or a very small number of genes were sufficient to reflect the reference topology. Unfortunately, none of the alignment sections used later for the design of initial seed-primers scored high among candidates that reflected the ideal reference topology (see Robert et al. 2011). However, key criteria for in silico selection of gene regions that would successfully qualify as secondary barcodes were universally met, including the conservation of primer sites among distant taxa (e.g. Ascomycota and Basidiomycota), with predicted amplicons no longer than 1 000 bp, a technical constraint for Sanger DNA sequencing and a requirement of the barcode standard.
Qualification of gene regions was restricted to the 54 alignments that served for seeding novel primers and manual redesign, described above, and reflected a compromise between score against the super-matrix and PCR versatility (1 Kbp cut-off). However, the alignments are essentially the results of the in silico searches described as BPPM and SPM above, and non-redundant alignments were selected. Manually inspected alignments (available upon request) and the identified conserved sites yielded a large number of primers (Table 3), which were tested in trial I and reduced to the best performing candidates (Table 6) for trial II. A more rigorous test of primers successful in trial II was undertaken in trial III (Table 6), using sets of extracts with increased species sampling of specific taxonomic groups. Because of time and resource constraints, some of the putative primers were not tested and should be tested in future studies, e.g. those corresponding to TEF2 (Table 3; Al45, Al48).
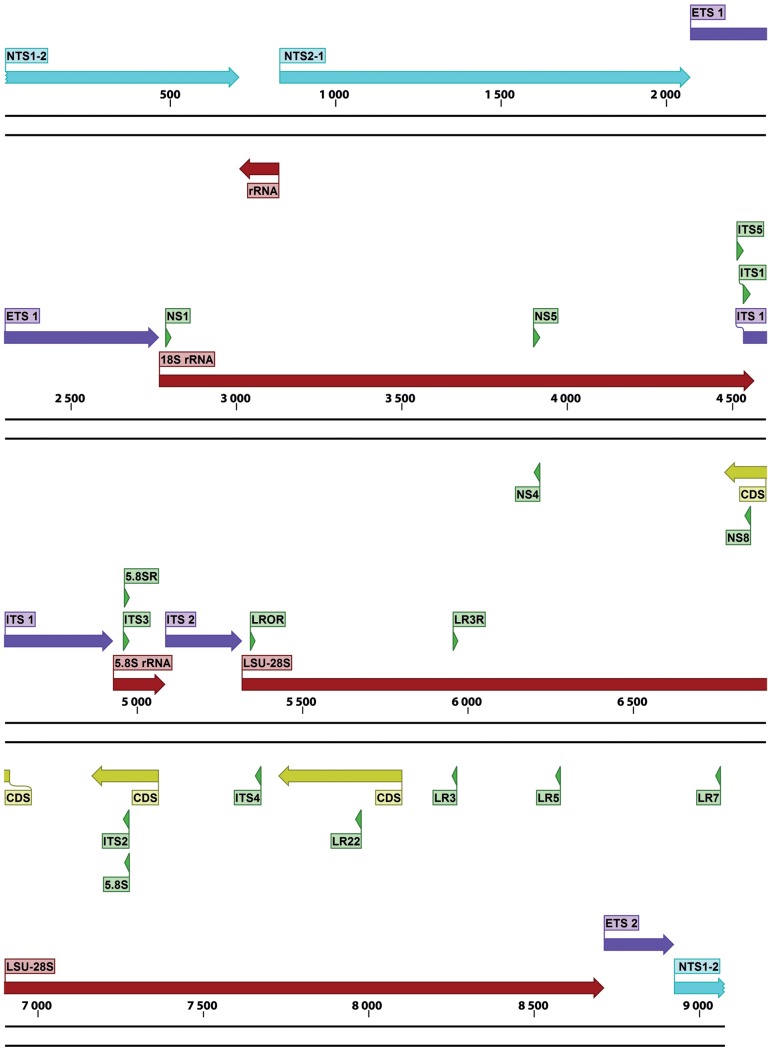
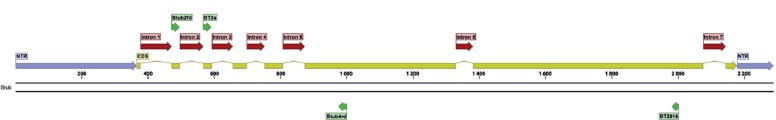
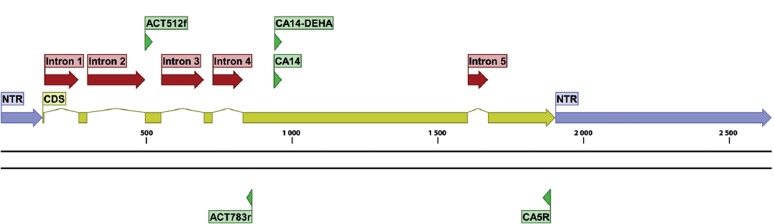
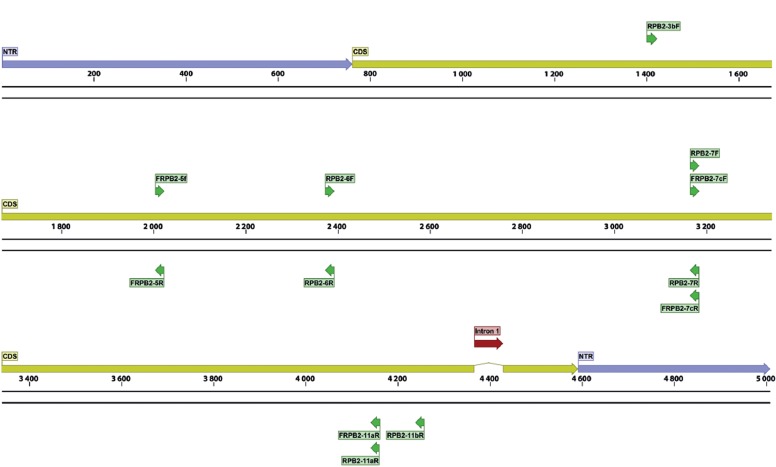
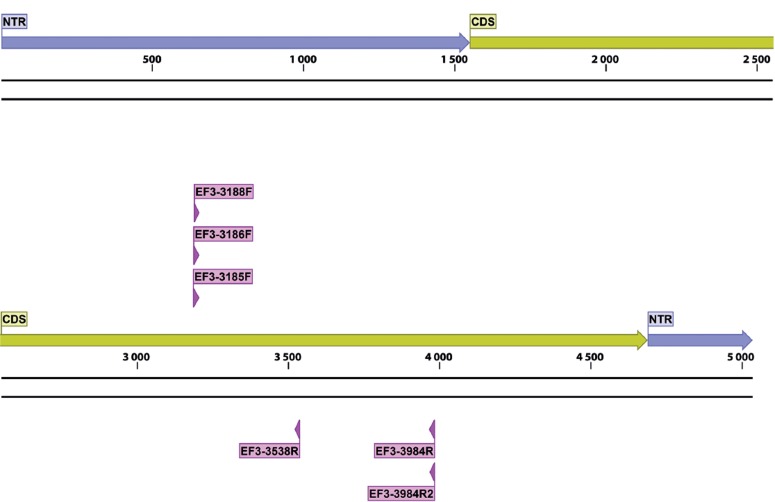
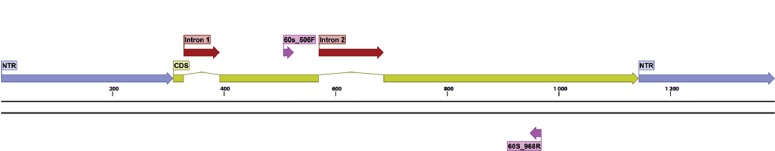
Gene maps, indicating exact primer locations, and exon and intron boundaries for each gene, were designed for the best performing candidates (Table 6) and for some important ‘standard’ markers (Table 2; the latter supplemented with positional information of commonly used primers). These gene maps represent the fungal rDNA operon (Fig. 1), TUB2 (Fig. 2), ACT (Fig. 3), RPB2 (Fig. 4), TEF1 (Fig. 5), TEF3 (Fig. 6) and 60S L10 (L1) (Fig. 7).
Fig. 1.
Structural gene map and positional primer locations for fungal rDNA array.
Fig. 2.
Structural gene map and positional primer locations for fungal β-tubulin 2.
Fig. 3.
Structural gene map and positional primer locations for γ-actin.
Fig. 4.
Structural gene map and positional primer locations for fungal RNA polymerase subunit II.
Fig. 5.
Structural gene map and positional primer locations for fungal translation elongation factor-1α.
Fig. 6.
Structural gene map and positional primer locations for fungal translation elongation factor 3.
Fig. 7.
Structural gene map and positional primer locations for fungal 60S ribosomal protein L 10 (L1).
In silico design of initial seed-primers of protein regions and manual optimization (Pfam based primers)
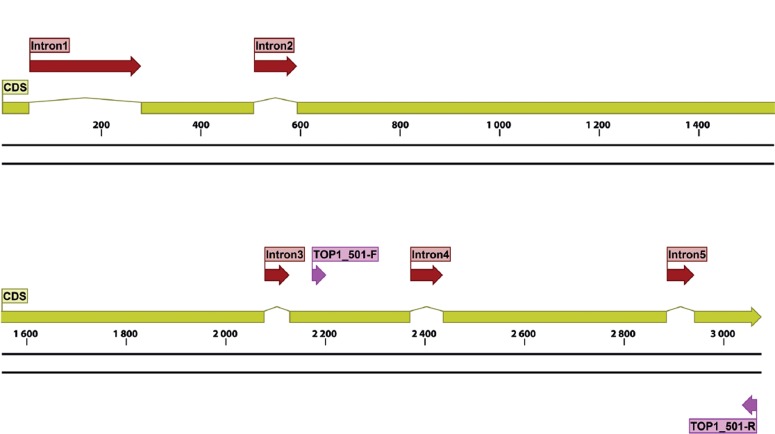
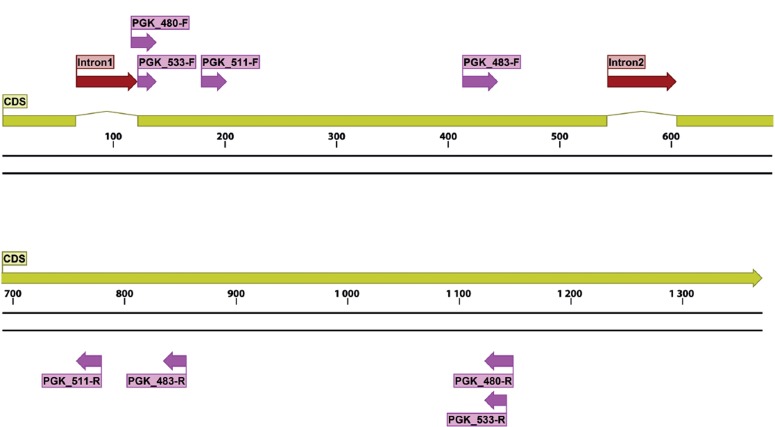
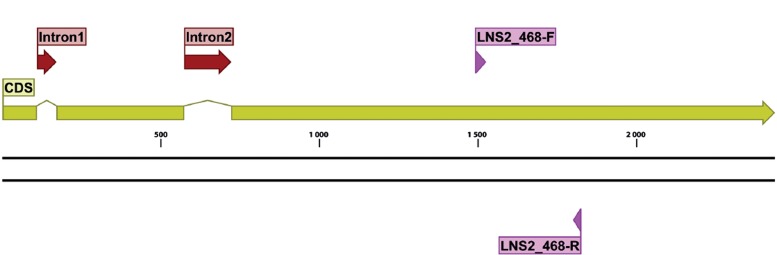
For the seven protein families found by Lewis et al. (2011) that could be potentially amplified with a single primer pair, a total of 127 forward primers and 105 reverse primer pairs were tested, including some modified with M13 sequencing primers, for a total of 433 primer pairs always tested individually. In silico analysis and manual refinement resulted in six primer pairs that passed all quality filters, which were subsequently used as described above (Table 7). Gene maps for TOP1, PGK and LNS2, indicating primer locations, exon and intron boundaries for each gene are shown in Fig. 8, Fig. 9, Fig. 10, respectively.
Fig. 8.
Structural gene map and positional primer locations for fungal TOP1 primarily based on Penicillium chrysogenum genome.
Fig. 9.
Structural gene map and positional primer locations for fungal PGK primarily based on Penicillium chrysogenum genome.
Fig. 10.
Structural gene map and positional primer locations for fungal LNS2 primarily based on Penicillium chrysogenum genome.
Laboratory testing of in silico designed primer sets designed from gene alignments
Laboratory tests were conducted for 71 sets of primers corresponding to nine distinct regions (or 15 alignment groups) listed in Table 3 for their ability to consistently amplify a short standardised gene region resulting in a single PCR fragment.
Trial I
With trial I, we identified pairs of primers that resulted in a single PCR product employing a low annealing temperature (Ta), what required adjustment of PCR parameters and conditions. PCR tests with several primer pairs that contained an inosine (= I), instead of third-base wobbles, resulted in almost no visible amplicons. We excluded all such primer pairs from further testing, including those that did not result in any detectable PCR products, and these are not mentioned further.
Trial II
For trial II, the Ta was unchanged but the set of extracts was expanded to test a broader range of fungi, and a subset of seven primer pairs was selected that gave optimal results during trial I. These primer sets were tested for their reliability and consistently over a broad range of fungal species, with preference for those amplifying the desired marker as single fragment. Some primer sets yielded multiple fragments. Some of the best primer sets, derived from alignment groups 2 (ACT), 33 (TEF1α) and 52 (L1) yielded a strong single signal with almost no secondary fragments. Primers for alignment groups 46 (TUB2) and 49 (TEF3) yielded multiple fragments, but had a strong primary signal. Of our Trial II candidates, the most problematic primers were those for a gene corresponding to a small RNA processing protein, a putative S6 kinase (alignment group 14), which resulted in multiple intense bands.
Tailed primer design was a successful approach, resulting in PCR products from evolutionary diverse fungi for TEF1α, a gene until now difficult to amplify on a universal basis with known standard primers. However, because of our stringent testing procedure, only alignment groups highlighted in bold (Table 3) qualified further for trial III, and these selections were strictly based on universal fidelity among all tested strains in trial II.
Trial III
In the last experimental phase, primer testing was extended to species-specific DNA sets. These extracts were received from 10 collaborating taxonomic experts at CBS (Table 1) in sets of 96 extracts. This more extensive testing evaluated the capacity of sequences yielded by the selected primer pairs for their power to delimit closely related fungal species. The extracts represented relevant groups such as medically or phytopathologically relevant fungi, or were selected because reference datasets were available for comparison, or because the taxa represented complexes of species that were previously shown to be difficult to resolve.
The performance of several primer pairs amplifying TEF3 (alignment groups 47, 49, 50 and 51) and newly designed primers for TEF1α (alignment groups 17, 28, 31, 33) was consistently excellent. During trial III, we sequenced larger numbers of PCR products corresponding to TEF3, the 60S L10 (L1) and TEF1α to test the performance of our primers in standardised Sanger sequencing applications. Our results convinced us that capillary sequencing and production of high quality trace files did not require major modifications for the tested PCR primers or amplification/sequence parameters. For all amplicons assessed by Sanger sequencing, the sequences obtained were identical to those recorded in the original in silico inferred alignments for the respective target species.
The designated primer pairs for TEF2 (alignment groups 45, 48) gave very poor results and were excluded from further testing. The primer pair for alignment group 28 (TEF1α) initially gave good results, but performed poorly in later experiments and was excluded from further testing. However, in common with the ‘untested’ pairs, the primers for TEF2 and TEF1α (alignment group 28) may be amenable to species-specific redesign (Table 6; Al28 = EF1_alternative_3f, EF1_alternative_3r), and thus represent interesting ‘seed’ candidates for further experiments. Best performing primers (Table 6), were distributed to collaborating colleagues for independent testing on taxon sets (Table 1).
Amplification efficiencies
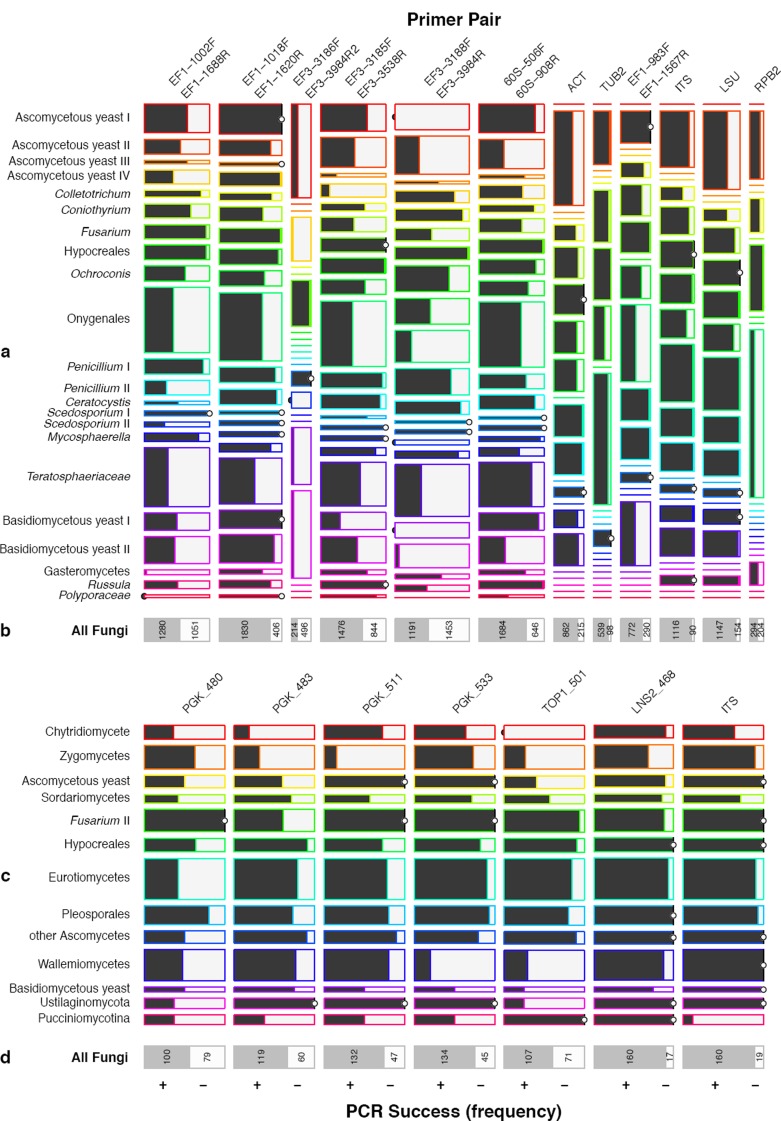
Optimal visualisation of multivariate data matrices, showing relative proportions of ‘within’ and ‘between’ categories (i.e. datasets vs primers), was achieved with the ‘mosaic plot’ function using ‘lattice’ in R (Sarkar 2008), with our categorical variables encoded as typical ‘survival data’. Each plot is sectioned in rows representing datasets and columns representing primer pairs separated by primers designed by the complete genome approach (Fig. 11a, b) and primers designed by the Pfam approach (Fig. 11c, d). Horizontal expansion of boxes indicates the ratio of PCR reactions within a ‘dataset’ or taxonomic group relative to the global quantity (proportion) of all other PCR values (outcome), and vertical expansion of boxes indicates the ratio of individuals within quantity (proportion) of PCR values (outcome) of a specific primer test (e.g. ITS). Horizontal lines indicate value ‘zero’. When a line with a dot is on the left, it equals ‘zero’ positives (= no amplification), and when the line with a dot is on the right, it indicates ‘zero’ negatives (= 100 % amplification). Darkly shaded boxes indicate proportion of amplifications, relative to no amplification, which are shown by pale grey shaded boxes. Mosaic plots outperform other plot types in visualising large quantities of data and provide a global overview on data proportions only. The frequencies of positive and negative amplification for each primer pair are shown in Fig. 11b and d.
Fig. 11.
Mosaic plot, indicating proportions of success rates between datasets (a, b: complete genome approach; c, d: Pfam approach) and within tested primer pairs.
The mosaic plot shows that the overall amplification testing between laboratories to test the primers designed through the complete genome approach (Robert et al. 2011) was strongly biased towards novel primer pairs, revealed by the many horizontal lines in some datasets corresponding to the ACT, TUB2, TEF1α (AFTOL primers), ITS, LSU, and RPB2 pairs (Fig. 11a). Data corresponding to these pairs was mainly obtained from PCR experiments performed at CBS, and lack of data from collaborating laboratories can simply be explained by time and financial constraints. However, the results of these amplification experiments provide an important benchmark, even when inconsistently assessed, and valuable information on the universal performance of certain pairs.
Promising results were obtained for several amplicons. The TEF1α region, with the AFTOL pair (EF1-983F/EF1-1567R), enjoyed relatively consistent amplification for different taxon sets, with exceptions being relatively poor results for the Onygenales and rock-inhabiting Teratosphaeriaceae (80 % average).
Results for sections of the fungal rDNA operon, especially the complete ITS and partial LSU (D1/D2 domains), were high in efficiency for all taxon sets, 92 % and 91 %, respectively (Fig. 11b). These results confirm the outstanding potential of ITS as primary universal fungal DNA barcode; it performs exceptionally well under standard laboratory conditions. No particular distinction was made between the possible forward primer combinations using ITS5/ITS1/ITS1-F. Similarly, LSU had very similar amplification efficiencies with LR0R/LR5, and augments the useful species-level signal ITS with increased phylogenetic information.
The results for RPB2 (fRPB2-5f/fRPB2-7cR) were less encouraging; only five taxon sets were tested for a single primer combination and the global performance of RPB2 can only be partly judged from our data.
Two very commonly known gene sections widely employed in fungal phylogenetics (Fig. 2, 3), ACT and TUB2, had relatively consistent amplification efficiencies among datasets (mostly grey shaded boxes), showing that these pairs qualify well for universal amplification (84 % ACT and 86 % TUB2 on average, respectively). Unfortunately, as is obvious from Table 2, we tested different primer pairs for these genes, and no universal performing set was found (CA5R/CA14 for ascomycetous yeasts II only; Bsens/Brev for Polyporaceae only, amplification success data not shown). The problem is apparent with ascomycetous yeasts group II, where experiments were performed with CA5R and CA14 instead of ACT-512F/ACT-783R, and the entirely negative results of the latter PCRs were excluded from the analysis. A similar issue was observed in tests prior to this study for a Polyporaceae taxon set (data not shown), where only Bsens and Brev resulted in positive amplification (Lesage-Meessen et al. 2011). Surprisingly amplification of TUB2 (using Btub2Fd/Btub4Rd) in ascomycetous yeasts group II always resulted in a single PCR product, but these amplicons always yielded noisy trace files. β -Tubulins are known to be duplicated, particularly in basidiomycetous fungi (Ayliffe et al. 2001, Zhao et al. 2014). The inconsistent PCR efficiencies and taxon-specific primer pairs disqualify ACT and TUB2 as universal barcodes.
Overall amplification efficiencies of primer pairs were inconsistently recorded in different laboratories and our conclusions on universality remain speculative. The most comprehensive results were retrieved for five of our newly designed primer pairs, EF1-1018F/EF1-1620R, EF1-1002F/1688R, 60S-908R/60S-506F, EF3-3185F/EF3-3538R and EF3-3188F/EF3-3984R. Performance of these three gene targets, as visualised in Fig. 11, varied among these pairs, but successful amplification proved that predictions of universal primer binding sites were accurately computed. Of these, the TEF1α primer candidates tested consistently among all labs. The poorest performance (57 % average) occurred with EF1-1002F/1688R, which had low efficiencies for all ascomycetous and basidiomycetous yeasts, and for the Ceratocystis, Onygenales, Penicillium 2, Russula and Teratosphaeriaceae taxon sets. In contrast, the TEF1α pair EF1-1018F/EF1-1620R had the highest fidelity for all taxon sets and among all tested protein coding genes with an average amplification success of 88 % (Fig. 11), almost equal to ITS (92 %) and LSU (91 %). As a second best among our novel primer pairs, we identified a pair corresponding to a gene encoding the 60S ribosomal protein L10 (L1), previously unused for barcoding or phylogenetic purposes. This primer pair had an average success rate of 77 %, with local optima over 95 %, and performed poorly only for some ascomycetous and basidiomycetous yeasts, and the Coniothyrium, Mycosphaerellaceae and Onygenales sets (∼40–60 %, Fig. 11). The novel region TEF3, although a fungal-specific gene, could not be universally amplified with high success rates, with neither the short nor long sections yielding satisfactory results for fidelity (short EF3-3185F/EF3-3538R, 68 % on average; long EF3-3188F/EF3-3984R, 52 % on average). Nevertheless, promising local optima for both TEF3 pairs were observed. The long section could be retrieved (∼70–90 % success) for the Sordariales (Colletotrichum, Ceratocystis, Scedosporium), Hypocreales (Fusarium s.str., Hypocreales s.lat.), the Dothidiomycetes (Mycosphaerellaceae) and Eurotiales (Penicillium 1, 2). The shorter TEF3 section was amplifiable for a broader spectrum of fungi with relatively high success rates, including the Sordariales, Hypocreales, Onygenales, Eurotiales and even within the basidiomycetes (∼70–90 % success). Despite this, failure to retrieve products for other taxa, such as the yeasts sensu lato, strongly decreased the universal efficiency of both TEF3 primers. We did not globally test a third TEF3 pair, EF3-3186F/EF3-3984R2, because of poor performance in the trial III. Nevertheless some taxon specific efficiency, in particular for Fusarium (Hypocreales) and Scedosporium, (Sordariales) were recorded.
For the primers designed by the Pfam domain approach (Lewis et al. 2011), PGK533 primer pair performed the best for this locus, generating fragments of about 1 kb with a success rate about as good as ITS with a wide range of fungi, except for most Basidiomycetes (Fig. 11c, d). The TOP1 primer pair amplified an 800 bp fragment efficiently for most ascomycetes but performed rather poorly for lower fungi and most basidiomycetes, the notable exception being Pucciniomycotina which are very challenging for ITS (Fig. 1c, d). LNS2 generated a short fragment (less than 400 bp) but amplified efficiently in most lower fungi and in all basidiomycetes.
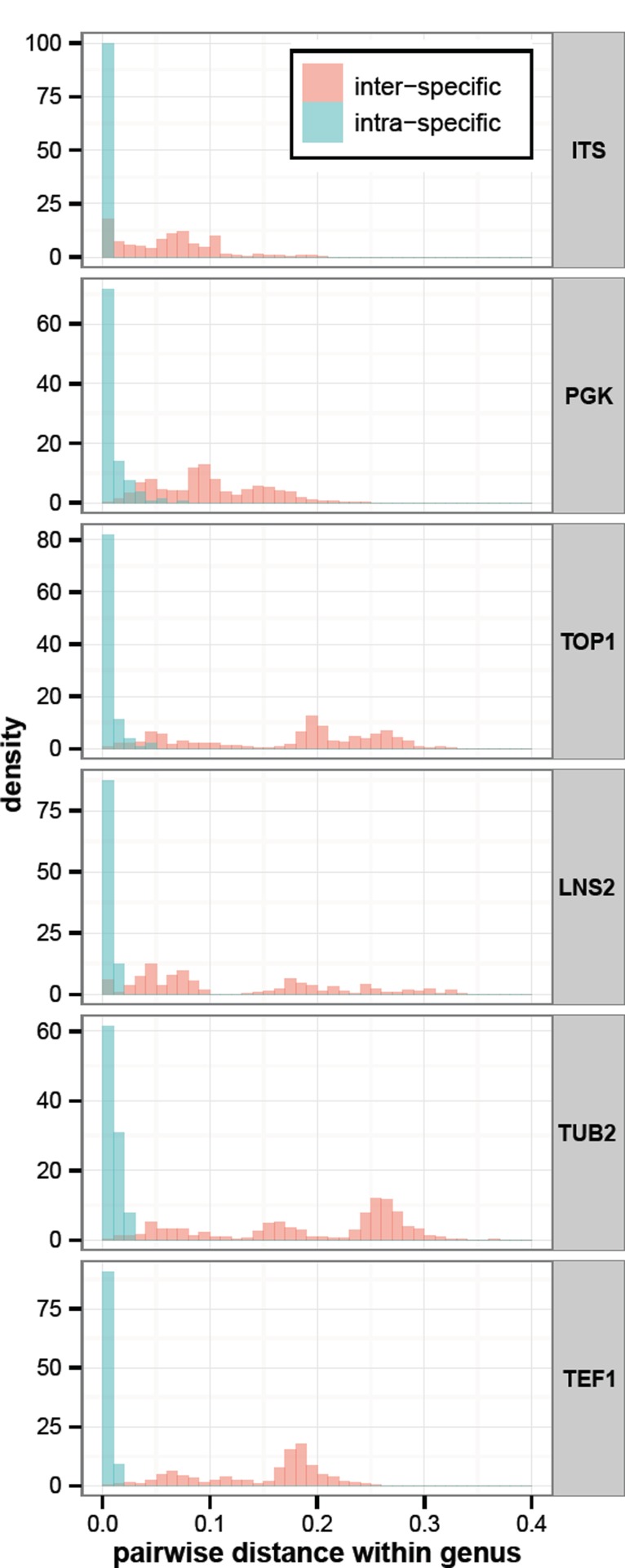
Barcode gap analyses
As expected, many closely related species have a pairwise distance of zero (Fig. 12). PGK, TOP1 and LNS2 all show a higher resolution than ITS. Our data shows that PGK and TOP1 are as good as TUB2 or TEF1α to resolve closely related Penicillium and Fusarium species, respectively (data not shown). There is insufficient data to make such statement for LNS2 as testing for ascomycetes was less of a priority than testing for basidiomycetes.
Fig. 12.
Barcode gap analysis for ITS, PGK, TOP1, LNS2, TUB2 and TEF1.
Multi-dimensional scaling of sequence data
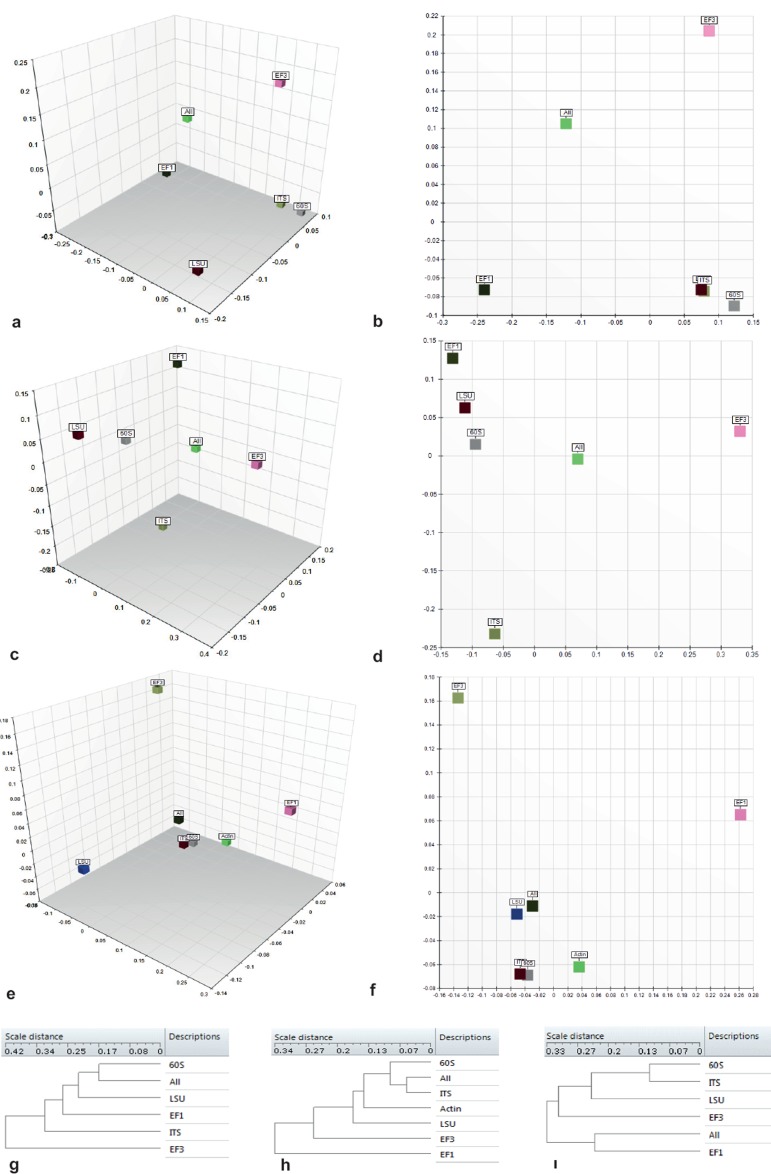
Results of the Mantel test (Smouse et al. 1986), comparing pairwise distance matrices of: i) the ‘global 502 taxa dataset’ (Fig. 13a, b, i); ii) ‘Fusarium’ (Fig. 13c, d, g); and iii) ‘Onygenales’ (Fig. 13e, f, h) by pairwise cophenetic coefficients of correlation, were plotted (numerically rescaled 0–1) in n-dimensional space. Rescaled cophenetic coefficients for each dataset are shown in Table 8. Assessment of global performance employing a comparison of TEF1α distances matrices (with sequences obtained from pair EF1-1018F/EF1-1620R; AFTOL pair EF1-983/EF1-1567 combined), TEF3 (sequences obtained from pair EF3-3185F/EF3-3538R, EF3-3188F/EF3-3984R combined), 60S, ITS and LSU for 502 taxa, showed that the first two axes contributed more than 75 % of the total information and the first three axes over 90 %. While 60S and ITS were rendered closely together and apart from LSU, neither TEF1α nor TEF3 were correlated closely to one of the rDNA matrices (Fig. 13a, b). The most significant result from visualising the cophenetic coefficients of correlation is apparent in Fig. 13a and i. The 3D plot and hierarchical clustering indicated that TEF1α correlated optimally with the overall concatenated (ALL) data matrix. Scoring of individual genes (matrices) is equivalent to distances as obvious from the dendrogram in Fig. 13i. The comparison of the ‘Fusarium’ matrices showed the same trend with the first three axes describing the vast majority of variance. Our results show that optimal correlation of the global dataset (ALL) was achieved with the 60S matrix (Fig. 13c, d, g), and the universal barcode ITS correlated poorly with the concatenated (ALL) object. The impact of the second dimension is obvious, because TEF3 is more distant from the concatenated matrix than ITS in the second but not the first dimension. Therefore, the weighted impact of dimensions causes the performance of TEF3 to appear inferior to ITS, presumably an artefact of distance over true character-based (cladistic/phylogenetic) inference (Felsenstein 2003). This could also be related to comparisons of cophenetic coefficients employing the Mantel test (Harmon & Glor 2010). Visualisation of the results from comparing data matrices obtained from the ‘Onygenales’ dataset contradicted those of the ‘502 taxon dataset’. We observed that the combination of two genes, ITS and TEF3, resulted in the ordination highly resembling the one of the concatenated (ALL) matrix. The impact of the third dimension scaled the LSU matrix more distantly than the two latter genes in Fig. 13e but was less pronounced in Fig. 13f. Second closest, and in addition to the two previous datasets, ACT was inferred as inferior to 60S and ITS when compared to the overall dataset. Both elongation factor matrices, TEF1α and TEF3, were inferior to ITS and were both distant from the overall concatenated matrix (ALL). Hierarchical clustering of individual matrices performance against the overall dataset is shown in Fig. 13h. In general, TEF1α performed the best among all ‘502’ investigated taxa when comparing 5+1 gene datasets, which was biased by local optima. The best candidate genes for Fusarium (60S) and Onygenales (ITS and 60S) rendered one of our new candidates as an optimal ‘local’ barcode for these two sets. The pairwise comparison for each dataset and each gene is given in Table 8 by rescaled (0–1) cophenetic correlation coefficients, with key results reflected by MDS analysis (Fig. 13) above.
Fig. 13.
Multi-dimensional scaling of rescaled cophenetic correlation coefficients comparing distance matrices.
Table 8.
Rescaled cophenetic correlation coefficients from Mantel test comparing distance matrices.
| Gene/Dataset | Gene | ||||||
|---|---|---|---|---|---|---|---|
| Onygenales | 60S | Actin | All | TEF1α | TEF3 | ITS | LSU |
|
| |||||||
| 60S | 1 | 0.869922 | 0.945842 | 0.665818 | 0.741716 | 0.878008 | 0.828932 |
| Actin | 0.869922 | 1 | 0.86677 | 0.718523 | 0.709779 | 0.833556 | 0.7986 |
| All | 0.945842 | 0.86677 | 1 | 0.703723 | 0.801272 | 0.947347 | 0.879415 |
| TEF1α | 0.665818 | 0.718523 | 0.703723 | 1 | 0.58968 | 0.656179 | 0.654457 |
| TEF3 | 0.741716 | 0.709779 | 0.801272 | 0.58968 | 1 | 0.739432 | 0.751655 |
| ITS | 0.878008 | 0.833556 | 0.947347 | 0.656179 | 0.739432 | 1 | 0.822984 |
| LSU | 0.828932 | 0.7986 | 0.879415 | 0.654457 | 0.751655 | 0.822984 | 1 |
| Fusarium | 60S | All | TEF1α | TEF3 | ITS | LSU | |
|
|
|||||||
| 60S | 1 | 0.833529 | 0.748277 | 0.564056 | 0.717349 | 0.818429 | |
| All | 0.833529 | 1 | 0.770656 | 0.773935 | 0.766443 | 0.737515 | |
| TEF1α | 0.748277 | 0.770656 | 1 | 0.502797 | 0.616714 | 0.687664 | |
| TEF3 | 0.564056 | 0.773935 | 0.502797 | 1 | 0.523543 | 0.532899 | |
| ITS | 0.717349 | 0.766443 | 0.616714 | 0.523543 | 1 | 0.645249 | |
| LSU | 0.818429 | 0.737515 | 0.687664 | 0.532899 | 0.645249 | 1 | |
| Global | 60S | All | TEF1α | TEF3 | ITS | LSU | |
|
|
|||||||
| 60S | 1 | 0.678678 | 0.621512 | 0.68173 | 0.88979 | 0.748801 | |
| All | 0.678678 | 1 | 0.77138 | 0.754324 | 0.698097 | 0.694224 | |
| TEF1α | 0.621512 | 0.77138 | 1 | 0.572193 | 0.68485 | 0.635149 | |
| TEF3 | 0.68173 | 0.754324 | 0.572193 | 1 | 0.724653 | 0.670825 | |
| ITS | 0.88979 | 0.698097 | 0.68485 | 0.724653 | 1 | 0.778558 | |
| LSU | 0.748801 | 0.694224 | 0.635149 | 0.670825 | 0.778558 | 1 | |
DISCUSSION
DNA-barcoding: a conceptual overview
In the early 1960s some milestone papers by Edward & Cavalli-Sforza (1964), Zuckerkandl & Pauling (1965) and Hennig (1965) gave rise to the novel field now known as molecular phylogenetics (Felsenstein 2003), including some decades later ‘DNA barcoding’. The goal of DNA barcoding is to apply high-throughput DNA sequencing technology to large-scale screening of one or a few reference genes to identify unknown specimens to species, and enhance discovery of new species (Hebert et al. 2003a, b, Stoeckle 2003). Proponents of ‘DNA-based taxonomy’ envisage the development of comprehensive databases of reference sequences, centred on voucher specimens or living cultures that represent all described species, against which sequences from newly sampled individuals can be compared. Given the long history of use of molecular markers for this purpose for identifying microbes (e.g. 16S rDNA for prokaryotes, Avise 2004), there is nothing fundamentally novel about DNA barcoding as applied in microbiology and mycology today, except for the increased scale and proposed standardisation. Standardisation by the selection of one or more reference genes is critical and stimulates large-scale phylogenetic analyses, but whether ‘one gene fits it all’ is still an open debate.
Initial reactions to the coordinated DNA barcoding movement ranged from great enthusiasm, especially among ecologists (Janzen 2004), to condemnation, largely from traditional taxonomists. The proponents emphasized its application to modern meta-barcoding studies, increasing the precision and efficiency of field studies of rare, diverse and difficult to identify taxa across all kingdoms of organisms, while DNA barcoding is not possible without large-scale and well curated sequence databases (Schoch et al. 2014), founded in traditional systematics and referencing authoritatively identified voucher specimens. DNA barcoding should not be confused or confounded with efforts to resolve the ‘tree of life’ or establish large-scale phylogenies. DNA barcodes often have limited phylogenetic resolution, reflecting the use of a single-gene assay in an attempt to identify an entity to species or reveal inconsistencies between molecular variation and current species delimitations. Thus, while barcoding initiatives use similar knowledge and techniques, resolving phylogenies from species to major higher clade levels requires a different strategy for gene selection.
Irrespective of which kingdom of organisms is targeted, the conceptual character and constraints of DNA barcoding always remain identical. We need to separate the two applications: i) molecular diagnostics of individuals relative to described taxa; and ii) DNA-enabled discovery of new species. Both are inherently phylogenetic and rely on a solid taxonomic background, including adequate sampling of variation within species and inclusion of all previously described extant species within a given genus or clade. Accurate species diagnoses rely on careful examination of intra-species variation compared with that observed between species. So a short DNA sequence (barcode) will allow reliable allocation of an individual to a described taxon. Such identifications should be accompanied by a clear statement of statistical support such as branch support in a phylogenetic analysis. This view is in agreement with best taxonomic practice that is acceptance of name changes or subdivisions should be based on multiple lines of evidence. As a strictly designated concept to species identification and discovery, a key theory of DNA barcoding is testing of ‘boundaries’ between taxa established by prior solid taxonomic research (Hebert et al. 2003a, b, Hebert & Gregory 2005, Meyer & Paulay 2005, Schindel & Miller 2005, Schoch et al. 2012).
One fungus, which genes?
In this study, we identified alternative universal gene targets for the fungal kingdom from available fully sequenced genomes (data processed in 2011) using taxonomy-aware in silico pipelines. Two approaches were used to infer potential novel candidates and corresponding primer pairs. Each approach yielded promising, but different candidate barcodes.
The search for the ideal gene, using the genomic approach and alignment templates of Robert et al. (2011) was based on five criteria: i) presence in the majority of investigated genomes; ii) potential ease of PCR amplification and Sanger DNA sequencing; iii) restriction to putative single copy orthologs, constrained to genes harbouring conserved sites < 1 Kbp apart; iv) the potential to accurately delimit fungal species; and eventually v) an inherent phylogenetic signal. Often, the distance between conserved nucleotide stretches was too long, or conserved sites were not present across a sufficient proportion of the analysed genomes. Primers were tested and marker sequences we sequenced were the best compromise between technical feasibility, and a holistic ‘one-gene-fits-all’ barcode locus. We identified known genes and some not yet used for either barcoding or phylogenetic applications, resulting in a long list of candidate primers. While most alignment templates were functionally annotated (e.g. TEF1α), only 14 of 54 were identified as putatively novel candidate regions with no known track record in fungal DNA barcoding or phylogenetics.
Although TEF1α is already well known to provide superior subordinate taxon resolution in some groups of Ascomycetes such as Trichoderma and Fusarium, and it is sufficiently present in public repositories, there was still a need to improve the universality of primers. From the genomic studies, TEF1α qualified as the only universal barcode candidate. Some of the newly identified genes through the Pfam approach, in particular TOP1 and PGK, which were tested more intensively for sequencing and analyses of barcode gaps, show great promise, especially for ascomycetes, where both performed well in genera with particularly narrow barcode gaps in the ITS, i.e. Penicillium and Fusarium.
In general, slowly evolving genes (e.g. TEF1α, RPB2 exons) are more suitable for inference of deep phylogenetic relationships, while genes with higher evolutionary rates (e.g. ITS, TUB2) reflect more recent evolutionary and speciation events. Thus, genes or gene sections reflecting both of these characteristics are most suitable as barcodes if phylogenetic signal is considered important. Fragments of protein-coding genes can potentially meet both requirements, either by incorporating more variable intronic and more conserved exonic sequences, or by providing one level of variability as nucleotide sequences and a more conserved suite for phylogenies as amino acid sequences. Our results show that TEF1α is likely to be one of the most ideal candidates for non-rDNA barcodes because of its balanced ‘trade-off’s’, in particular when it comes to a broader community acceptance of extant users, and versatility among important fungal orders.
Finding primer candidates to amplify a standardised DNA fragment with high phylogenetic information content in distantly related species was difficult with the computational resources available in 2011 when our research was conducted. In our in silico search of available genomes of dikaryotic fungi, only seven primer pairs covered 72 species of the 74 species studied. For ascomycetes, six primer pairs were identified as compatible with 57 species studied and 186 primer pairs with 56 species. The universality of our best-performing primers in silico was experimentally verified, at least for Asco- and Basidiomycota. Some primer candidates were located almost adjacent to each other with only a few bases difference at either the 5’ or 3’ primed ends. No new candidates with high local optima such as the MCM7 and Tsr1 primers (Schmitt et al. 2009) were discovered.
From the genomic scans, the fragment of the translation elongation factor 1α (TEF1α) gene appears to be the most promising candidate as a universal secondary barcode. We remain cautious about fidelity for basal fungal lineages but we believe our primers for TEF1α are the most optimal compromise between universal taxon applicability and fidelity. In our study we have particularly tested fungal taxa of economic or applied importance. We foresee that a high fidelity secondary DNA barcode would be included in a very large portion of ongoing taxonomic studies. Based on our results and anticipating community acceptance, we propose the use of primers EF1-1018F and EF1-1620R (corresponding to TEF1α) as secondary universal DNA barcode primer pair for the fungal kingdom. Given the results of this extensive four-year multi-lab project, we are convinced that fungal DNA barcoding with ITS and TEF1α for the identification of unknown or validation of ‘primary-barcoded’ specimens will increase the resolution and lead to the development of more complete, structured datasets. Global consensus might also speed up the discovery and description of novel taxa in a standardised way.
Another critical question is whether the proposal of a secondary DNA barcode actually is still relevant in a world where next generation sequencing is becoming routine in many laboratories, at a cost that is comparable to Sanger sequencing? The costs for sequencing a complete fungal genome are dropping rapidly. Despite this, the fungal taxonomic community lacks the computational resources and capacities to routinely process complete genomes for specimen identification or for phylogenetic studies, especially in the developing world. Undoubtedly, the rapid increase of publicly available fungal genomes will enhance our understanding of evolutionary processes governing niche adaption, mating or gene family expansion related to pathogenicity and other functional capabilities. Genome plasticity, architecture and high genome sequence identity between sister species will force us to rethink ‘tree thinking’ (Bapteste et al. 2005). Despite this, genome sequencing and its subsequent Big Data explosion have yet to establish a solid basis for molecular species identification, in particular when there is low sequence coverage per taxon, insufficient sampling of species diversity or, reciprocally, when complete genomic sequences may require an updated classification.
Rank inflation, i.e. the tendency to reclassify taxa at higher taxonomic levels (such as a previously recognised family as an order) is currently commonplace in fungal taxonomy, and requires a more standardised approach. Agreement on conserved universal protein coding markers, such as TEF1α, that would allow a more precise nesting of unknown fungi among known higher level taxa will be valuable.
DNA barcoding has great impact on human and animal health diagnostics. We hope that this paper, with its detailed gene maps and large quantity of tested and yet to be tested primers, will serve as a reference for expanded barcoding (e.g. further testing of the fungal specific translation elongation factor 3 in a clinical setting). It seems unlikely that there will be a similar study of this magnitude, employing PCR, Sanger DNA sequencing and testing so many universal primers on a large collection of fungal taxa. Therefore, we believe that the knowledge obtained during this study would convince the community to ratify TEF1α as secondary barcode in order to ensure its rapid application with the improved primer system, but also to promote the research on other target genes for other lineages of fungi.
Acknowledgments
Primer development and testing by partners in the European Consortium of Microbial Resource Centres (EMbaRC) was supported through funding of the European Community’s Seventh Framework Programme (FP7, 2007–2013), Research Infrastructures action, under grant agreement no. FP7-228310. Part of sequencing work in CBS was supported by Fonds Economische Structuurversterking (FES), Dutch Ministry of Education, Culture and Science grant BEK/BPR-2009/137964-U). WM and VR were supported by research grant NH&MRC # APP1031952. Genome mining at CBS and AAFC, and primer development and testing at AAFC, were supported by grants from the A.P. Sloan Foundation Programme on the Microbiology of the Built Environment. We acknowledge the Deanship of Scientific Research (DSR), King Abdulaziz University, under grant No. 1-965/1434 HiCi for technical and financial support. AY was supported by Fundação para a Cieência e a Tecnologia (Portugal), project PTDC/BIA-BIC/4585/2012. MPM was supported by grant CGL2012-359 (Spain).
REFERENCES
- Aguileta G, Marthey S, Chiapello H, et al. 2008. Assessing the performance of single-copy genes for recovering robust phylogenies. Systematic Biology 57: 613–627. [DOI] [PubMed] [Google Scholar]
- Aveskamp M, Verkley GJM, De Gruyter J, et al. 2009. DNA phylogeny reveals polyphyly of Phoma section Peyronellea and multiple taxonomic novelties. Mycologia 101: 363–382. [DOI] [PubMed] [Google Scholar]
- Avise JC. 2004. Molecular markers, natural history, and evolution, 2nd ed Sunderland (Massachusetts), Sinauer Associates. [Google Scholar]
- Ayliffe MA, Dodds PN, Lawrence GJ. 2001. Characterisation of a β -tubulin gene from Melampsora lini and comparison of fungal β -tubulin genes. Mycological Research 105: 818–826. [Google Scholar]
- Balajee SA, Borman AM, Brandt ME, et al. 2009. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: Where are we and where should we go from here? Journal of Clinical Microbiology 47: 877–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, Susko E, Leigh J, et al. 2005. Do orthologous gene phylogenies really support tree-thinking. BMC Evolutionary Biology 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begerow D, Nilsson H, Unterseher M, et al. 2010. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Applied Microbiology and Biotechnology 87: 99–108. [DOI] [PubMed] [Google Scholar]
- Belfield GP, Ross-Smith NJ, Tuite MF. 1995. Translation elongation factor-3 (EF-3): an evolving eukaryotic ribosomal protein? Journal of Molecular Evolution 41: 376–387. [PubMed] [Google Scholar]
- Belfield GP, Tuite MF. 2006. Translation elongation factor 3: a fungus-specific translation factor? Molecular Microbiology 9: 411–418. [DOI] [PubMed] [Google Scholar]
- Berbee ML, Taylor JW. 1992. 18S ribosomal RNA gene sequence characters place the human pathogen Sporothrix schenckii in the genus Ophiostoma. Experimental Mycology 16: 87–91. [Google Scholar]
- Brodersen DE, Nissen P. 2005. The social life of ribosomal proteins. FEBS Journal 272: 2098–2108. [DOI] [PubMed] [Google Scholar]
- Bruns TD, White TJ, Taylor JW. 1991. Fungal molecular systematics. Annual Review of Ecology and Systematics 22: 525–564. [Google Scholar]
- Buée M, Reich M, Murat C, et al. 2009. 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytologist 184: 449–456. [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Kauff F, Gabaldón T. 2014. A phylogenomics approach for selecting robust sets of phylogenetic markers. Nucleic Acids Research 42: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Crous PW, Verkley GJM, Groenewald JZ, et al. 2009. Fungal biodiversity. CBS Laboratory Manuals Series 1. Centraalbureau voor Schimmelcultures, Utrecht, Netherlands. [Google Scholar]
- Daniel HM, Meyer W. 2003. Evaluation of ribosomal RNA and actin gene sequences for the identification of ascomycetous yeast. International Journal of Food Microbiology 86: 61–78. [DOI] [PubMed] [Google Scholar]
- Daniel HM, Sorrell TC, Meyer W. 2001. Partial sequence analysis of the actin gene and its potential for studying the phylogeny of Candida species and their teleomorphs. International Journal of Systematic and Evolutionary Microbiology 51: 1593–1606. [DOI] [PubMed] [Google Scholar]
- Dentinger BTM, Didukh MY, Moncalvo JM. 2011. Comparing COI and ITS as DNA barcode markers for mushrooms and allies (Agaricomycotina). PLoS One 6:e25081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don RH, Cox PT, Wainwright BJ, et al. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Research 19: 4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt U. 2010. A constructive step towards selecting a DNA barcode for fungi. New Phytologist 187: 265–268. [DOI] [PubMed] [Google Scholar]
- Edward AWF, Cavalli-Sforza LL. 1964. Reconstruction of evolutionary trees. In: Heywood VH, McNeill J. (eds), Phenetic and phylogenetic classification: 67–76. Systematic Association, Publ. No. 6, London, UK. [Google Scholar]
- Feau N, Decourcelle T, Husson C, et al. 2011. Finding single copy genes out of sequenced genomes for multilocus phylogenetics in non-model fungi. PlosOne e18803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fell JW, Boekhout T, Fonseca A, et al. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology 50: 1351–1371. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 2003. Inferring Phylogenies. 2nd ed Sinauer Association. [Google Scholar]
- Feng P, Klaassen CHW, Meis JF, et al. 2013. Identification and typing of isolates of Cyphellophora and relatives by use of amplified fragment length polymorphism and rolling circle amplification. Journal of Clinical Microbiology 51: 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer C, Colom F, Frases S, et al. 2001. Detection and identification of fungal pathogens by PCR and ITS2 and 5.8S ribososmal DNA typing in ocular infections. Journal of Clinical Microbiology 38: 2873–2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gams W, Jaklitsch W. 2011. Fungal nomenclature 3. A critical response to the ‘Amsterdam declaration’. Mycotaxon 116: 501–512. [Google Scholar]
- Gao R, Zhang G. 2013. Potential of DNA barcoding for detecting quarantine fungi. Phytopathology 103: 1103–1107. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. 1993. ITS primers with enhanced specificity for basidiomycetes – application for the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Gilmore SR, Gräfenhan T, Louis-Seize G, et al. 2009. Multiple copies of cytochrome oxidase 1 in species of the fungal genus Fusarium. Molecular Ecology Resources 9: 90–98. [DOI] [PubMed] [Google Scholar]
- Greganova E, Altmann M, Buetikofer P. 2011. Unique modifications of translation elongation factors. FEBS Journal 278: 2613–2624. [DOI] [PubMed] [Google Scholar]
- Grigoriev , IV, Nikitin R, Haridas S, et al. 2014. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research 42: D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadet J, Julien J, Lafay JF, et al. 1989. Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Molecular Biology and Evolution 6: 227–242. [DOI] [PubMed] [Google Scholar]
- Harmon LJ, Glor RE. 2010. Poor statistical performance of the Mantel test in phylogenetic comparative analyses. Evolution 64: 2173–2178. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, et al. 2003a. Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Gregory TR. 2005. The promise of DNA barcoding for taxonomy. Systematic Biology 54: 852–859. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, DeWaard JR. 2003b. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society of London B: Biological Sciences 270 (suppl.1): S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs G, De Hoog GS, Haase G. 2012. Barcode identifier – a practical tool for reliable species assignment of medically important black yeast species. Journal of Clinical Microbiology 50: 3023–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig W. 1965. Phylogenetic systematics. Annual Review of Entomology 10: 97–116. [Google Scholar]
- Irinyi L, Lackner M, De Hoog S, et al. 2015a. DNA barcoding of fungi causing infections in humans and animals. Fungal Biology (in press, doi:10.1016/j.funbio.2015.04.007). [DOI] [PubMed] [Google Scholar]
- Irinyi L, Serena C, Garcia-Hermoso D, et al. 2015b. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database – the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Medical Mycology 53: 313–337. [DOI] [PubMed] [Google Scholar]
- Ivanova NV, DeWaard J, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Molecular Ecology Notes 6: 998–1002. [Google Scholar]
- James TY, Kauff F, Schoch CL, et al. 2006. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- Janzen DH. 2004. Now is the time. Philosophical Transactions of the Royal Society of London B 359: 731–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, et al. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research 33: 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M.1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Krause L, Diaz NN, Goesmann A, et al. 2008. Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Research 36: 2230–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramae E, Robert V, Echavarri-Erasun C, et al. 2007. Cophenetic correlation analysis as a strategy to select phylogenetically informative proteins: an example from the fungal kingdom. BMC Evolutionary Biology 7: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzman CP, Quintilla Mateo R, Kolecka A, et al. 2015. Yeast systematics and phylogeny and their use as predictors of biotechnologically important metabolic pathways. FEMS Yeast Research (in press). doi: 10.1093/femsyr/fov050. E-pub: 2015 June 30. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. 1998, Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73: 331–371. [DOI] [PubMed] [Google Scholar]
- Lee J, Young PW. 2009. The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytologist 1: 200–211. [DOI] [PubMed] [Google Scholar]
- Lesage-Meessen L, Haon M, Uzan E, et al. 2011. Phylogeographic relationships in the polypore fungus Pycnoporus inferred from molecular data. FEMS Microbiology Letters 325: 32–48. [DOI] [PubMed] [Google Scholar]
- Lewis CT, Bilkhu S, Robert V, et al. 2011. Identification of fungal DNA barcode targets and PCR primers based on Pfam protein families and taxonomic hierarchy. The Open and Applied Informatics Journal 5: 30–44. [Google Scholar]
- Liu YJ, Whelen S, Hall DB. 1999. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808. [DOI] [PubMed] [Google Scholar]
- Lumbsch TH, Leavitt SD. 2011. Goodbye morphology? A paradigm shift in the delimitation of species in lichenized fungi. Fungal Diversity 50: 59–72. [Google Scholar]
- Meyer CP, Paulay G. 2005. DNA barcoding: error rates based on comprehensive sampling. PLoS Biology 3: e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy LG, Ohm RA, Kovács GM, et al. 2014. Latent homology and convergent regulatory evolution underlies the repeated emergence of yeasts. Nature Communications 5: 4471. [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Ryberg M, Kristiansson E, et al. 2006. Taxonomic reliability of DNA sequences in public sequence databases: A fungal perspective. PLoS One 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20: 289–290. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Audic S, Adl S, et al. 2012. CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant and fungal kingdoms. PLoS Biology 10: e1001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Rehner SA, Buckley E. 2005. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–89. [DOI] [PubMed] [Google Scholar]
- Reynolds DR, Taylor JW. (eds). 1993. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. Wallingford, CAB International. [Google Scholar]
- Robert V, Szoke S, Eberhardt U, et al. 2011. The quest for a general and reliable fungal DNA barcode. The Open and Applied Informatics Journal 5: 45–61. [Google Scholar]
- Robideau GP, De Cock AWAM, Coffey MD, et al. 2011. DNA barcoding of Oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Molecular Ecology Resources 11: 1002–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samerpitak K, Gerrits van den Ende AHG, Stielow JB, et al. 2015. Barcoding and species recognition of opportunistic pathogens in Ochroconis and Verruconis. Fungal Biology (in press). [DOI] [PubMed] [Google Scholar]
- Sarkar D. 2008. Lattice: multivariate data visualization with R. Use R! Series, Springer, Berlin. [Google Scholar]
- Sasikumar AN, Perez WB, Goss Kinzy T. 2012. The many roles of eukaryotic elongation factor 1 complex. Wiley Interdisciplinary Reviews: RNA 3: 543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindel DE, Miller SE. 2005. DNA barcoding a useful tool for taxonomists. Nature 435: 17. [DOI] [PubMed] [Google Scholar]
- Schlick-Steiner BC, Steiner FM, Seifert B, et al. 2010. Integrative taxonomy: A multisource approach to exploring biodiversity. Annual Review of Entomology 55: 421–438. [DOI] [PubMed] [Google Scholar]
- Schmitt I, Crespo A, Divakar PK, et al. 2009. New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Robbertse B, Robert V, et al. 2014. Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database (Oxford) doi: 10.1093/database/bau061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzetti G, Fell JW, Fonseca A, et al. 2002, Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research 4: 495–517. [DOI] [PubMed] [Google Scholar]
- Seifert KA. 2009. Progress towards DNA barcoding in fungi. Molecular Ecology Resources 9: 83–89. [DOI] [PubMed] [Google Scholar]
- Seifert KA, Samson RA, DeWaard JR, et al. 2007. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proceedings of the National Academy of Sciences 104: 3901–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokralla S, Gibson JF, Nikbakht H, et al. 2014. Next-generation DNA barcoding: using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Molecular Ecology Resources 14: 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smouse PE, Long JC, Sokal RR. 1986. Multiple regression and correlation extensions of the mantel test of matrix correspondence. Systematic Zoology 4: 627–632. [Google Scholar]
- Stackebrandt E, Goebel BM. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. International Journal of Systematic Bacteriology 44: 846–849. [Google Scholar]
- Stielow B, Hensel G, Strobelt D, et al. 2012. Hoffmannoscypha, a novel genus of brightly coloured, cupulate Pyronemataceae closely related to Tricharina and Geopora. Mycological Progress 12: 675–686. [Google Scholar]
- Stockinger H, Peyret-Guzzon M, Koegel S, et al. 2014. The largest subunit of RNA Polymerase II as a new marker gene to study assemblages of arbuscular mycorrhizal fungi in the field. PlosOne 9: e107783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckle M. 2003. Taxonomy, DNA, and the Bar Code of Life. Bioscience 53: 796–797. [Google Scholar]
- Stoeckle MY, Thaler DS. 2014. DNA barcoding works in practice but not in (neutral) theory. PlosOne 9: e100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Acrtander P, Minelli A, et al. 2003. A plea for DNA taxonomy. Trends in Ecology and Evolution 18: 70–74. [Google Scholar]
- Taylor JW. 2011. One fungus = one name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus 2: 113–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Jairus T, Horton BM, et al. 2008. Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytologist 180: 479–490. [DOI] [PubMed] [Google Scholar]
- Verkley GJM, Dukik K, Renfurm R, et al. 2014. Novel genera of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 32: 25–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TD, Eberhardt U, Szöke S, et al. 2012. A laboratory information management system for DNA barcoding workflows. Integrative Biology 4: 744–755. [DOI] [PubMed] [Google Scholar]
- Walker DM, Castlebury LA, Rossman AY, et al. 2012. New molecular markers for fungal phylogenetics: Two genes for species-level systematics in the Sordariomycetes (Ascomycota). Molecular Phylogenetics and Evolution 64: 500–512. [DOI] [PubMed] [Google Scholar]
- Ward E, Adams MJ. 1998. Analysis of ribosomal DNA sequences of Polymyxa species and related fungi and the development of genus- and species-specific PCR primers. Mycological Research 102: 965–974. [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, et al. 1991. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology 173: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, Inc., New York, USA. [Google Scholar]
- Wickham W. 2009. ggplot2: Elegant graphics for data analysis. Springer; New York. [Google Scholar]
- Woese CR. 1987. Bacterial evolution. Microbiological Reviews 51: 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woudenberg JHC, Aveskamp MM, De Gruyter J, et al. 2009. Multiple Didymella teleomorphs are linked to the Phoma clematidina morphotype. Persoonia 22: 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ypma-Wong MF, Fonzi WA, Syphred PS. 1992. Fungus-specific translation elongation factor 3 gene present in Pneumocystis carinii. Infection and Immunity 60: 4140–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov A, Guerreiro MA, Sharma L, et al. 2015. Correction: Multigene assessment of the species boundaries and sexual status of the basidiomycetous yeasts Cryptococcus flavescens and C. terrestris (Tremellales). PLoS ONE 10, 4: e0126996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkov A, Krueger D, Begerow D, et al. 2012, Basidiomycetous yeasts from Boletales fruiting bodies and their interactions with the mycoparasite Sepedonium chrysospermum and the host fungus Paxillus. Microbial Ecology 63: 295–303. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Liu H, Luo Y, et al. 2014. Molecular evolution and functional divergence of tubulin superfamily in the fungal tree of life. Scientific Reports 4: 6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L. 1965. Molecules as documents of evolutionary history. Journal of Theoretical Biology 8: 357–366. [DOI] [PubMed] [Google Scholar]