Abstract
We investigated the phylogenetic diversity of 144 Colletotrichum isolates associated with symptomatic and asymptomatic tissues of Camellia sinensis and other Camellia spp. from seven provinces in China (Fujian, Guizhou, Henan, Jiangxi, Sichuan, Yunnan, Zhejiang), and seven isolates obtained from other countries, including Indonesia, UK, and the USA. Based on multi-locus (ACT, ApMat, CAL, GAPDH, GS, ITS, TUB2) phylogenetic analyses and phenotypic characters, 11 species were distinguished, including nine well-characterised species (C. alienum, C. boninense, C. camelliae, C. cliviae, C. fioriniae, C. fructicola, C. gloeosporioides, C. karstii, C. sia-mense), and two novel species (C. henanense and C. jiangxiense). Of these, C. camelliae proved to be the most dominant and probably host specific taxon occurring on Camellia. An epitype is also designated for the latter species in this study. Colletotrichum jiangxiense is shown to be phylogenetically closely related to the coffee berry pathogen C. kahawae subsp. kahawae. Pathogenicity tests and the pairwise homoplasy index test suggest that C. jiangxiense and C. kahawae subsp. kahawae are two independent species. This study represents the first report of C. alienum and C. cliviae occurring on Camellia sinensis. In addition, our study demonstrated that the combined use of the loci ApMat and GS in a phylogenetic analysis is able to resolve all currently accepted species in the C. gloeosporioides species complex.
Keywords: Camellia, Colletotrichum, morphology, phylogeny, tea plants
INTRODUCTION
Camellia, a genus of flowering plants in the family Theaceae, is cultivated in eastern and southern Asia, from the Himalayas east to Japan and Indonesia. Many species of Camellia (Ca.) are of major commercial importance. For example, leaves of Ca. sinensis are processed to produce tea, a popular beverage, while Ca. japonica, Ca. oleifera, and Ca. sasanqua and their hybrids are cultivated as ornamentals. Camellia production is affected by a large number of diseases, of which anthracnose, caused by species of the genus Colletotrichum, is one of the most important (Copes & Thomson 2008, Farr & Rossman 2014, Guo et al. 2014). Several Colletotrichum species have been reported from Camellia, e.g. C. boninense (Damm et al. 2012b), C. camelliae (Thompson & Johnston 1953, Tai 1979, Alfieri et al. 1984), C. carveri (Cash 1952), C. coccodes (Thaung 2008), C. gloeosporioides (Alfieri et al. 1984, Shivas 1989, Lu et al. 2000, Chen 2003, Guo et al. 2014), C. pseudomajus (Liu et al. 2014), C. queenslandicum (Simmonds 1966; syn. C. gloeosporioides var. minor, Weir et al. 2012), and Glomerella major (Tunstall 1934).
The genus Colletotrichum was also considered as one of the dominant endophytic genera in Camellia plants (Lu et al. 2007, Dai et al. 2008, Osono 2008, Fang et al. 2013). Colletotrichum acutatum and C. gloeosporioides were recognised as frequently occurring endophytic species in Ca. japonica based on morphological characteristics (Osono 2008). Fang et al. (2013) also found that C. gloeosporioides was one of the dominant endophytic species in Ca. sinensis based on ITS sequence data. Other reports of endophytic isolates of Colletotrichum on Camellia were, however, only identified to genus level.
Because of the commercial yield losses experienced in tea plantations due to Colletotrichum infections, as well as the limited knowledge of their identity and endophytic growth in Camellia plants, accurate identification of the causal organisms is of extreme importance. Most of the recent taxonomic treatments have primarily focused on the study of different Colletotrichum species complexes, for example C. acutatum (Damm et al. 2012a), C. boninense (Damm et al. 2012b), C. caudatum (Crouch 2014), C. destructivum (Damm et al. 2014), C. gigasporum (Liu et al. 2014), C. gloeosporioides (Weir et al. 2012), C. graminicola (Crouch et al. 2009), and C. orbiculare (Damm et al. 2013). Robust identification of Colletotrichum species relies on multi-locus sequence data (Cai et al. 2009, Cannon et al. 2012, Weir et al. 2012, Damm et al. 2013, Liu et al. 2013a, Crouch 2014). However, previous phylogenetic studies have rarely included isolates from Camellia. Thus far only a few strains of C. boninense, C. fioriniae, C. lupini, and Glomerella cingulata ‘f. sp. camelliae’ from Camellia were included in multi-locus phylogenies (Damm et al. 2012a,b, Weir et al. 2012, Sharma et al. 2014). In contrast, most of the studies that focused on the identification of Colletotrichum species associated with Camellia were only based on host, morphology or ITS sequence data (Tai 1979, Alfieri et al. 1984, Copes & Thomson 2008, Thaung 2008, Fang et al. 2013, Guo et al. 2014). Published reports of C. acutatum and C. gloeosporioides on Camellia should therefore be interpreted with care. Furthermore, although C. camelliae is regarded as the causal agent of brown blight disease of tea, the taxonomic and phylogenetic status of this pathogen remains unresolved (Weir et al. 2012).
The aim of the present study was thus to investigate the taxonomic and phylogenetic diversity of Colletotrichum spp. associated with Ca. sinensis and other Camellia spp. based on sequence data of six loci (ACT, CAL, GAPDH, GS, ITS, TUB2). A further aim was to test the usefulness of the ApMat locus in resolving taxa in the C. gloeosporioides complex (Crouch et al. 2009, Rojas et al. 2010, Silva et al. 2012b, Doyle et al. 2013, Sharma et al. 2013a, 2014) in combination with the other loci listed above.
MATERIALS AND METHODS
Collection and isolates
Diseased and healthy leaves of tea plants (Ca. sinensis) and other Camellia spp. were collected from seven provinces in China (Fujian, Guizhou, Henan, Jiangxi, Sichuan, Yunnan, and Zhejiang). Plant pathogenic fungi were isolated from leaf spots using both single spore and tissue isolation methods. Single spore isolation following the protocol of Choi et al. (1999) was adopted for collections with visible foliar sporulation, while tissue isolation was used for sterile isolates. Fungal endophytes were isolated by cutting four fragments (4 mm2) per leaf from the apex, base and lateral sides, surface sterilised with 70 % ethanol for 1 min, 0.5 % NaClO for 3 min, 70 % ethanol for 1 min, rinsed in sterile water, and then transferred to quarter-strength potato dextrose agar (1/4 PDA; 9.75 g Difco PDA, 15 g Difco agar and 1 L distilled water). After 3–21 d, mycelial transfers were made from the colony periphery onto PDA. Colletotrichum colonies were primarily identified based on cultural characteristics on PDA, morphology of the spores, and ITS sequence data.
Type specimens of new species from this study were deposited in the Mycological Herbarium, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), and ex-type living cultures deposited in the China General Microbiological Culture Collection centre (CGMCC). A further seven isolates from Camellia originating from other countries including Indonesia, UK, and the USA used in this study were obtained from the culture collection of the International Collection of Microorganisms from Plants, Landcare Research, Auckland, New Zealand (ICMP) and the CBS-KNAW Fungal Biodiversity Centre, Utrecht, the Netherlands (CBS).
Morphological analysis
Agar plugs (5-mm-diam) were taken from the periphery of actively growing cultures and transferred to the centre of 9-cm-diam Petri dishes containing PDA or synthetic nutrient-poor agar medium (SNA; Nirenberg 1976) amended with double-autoclaved stems of Anthriscus sylvestris placed onto the agar surface. Cultures were incubated at room temperature (c. 25 °C) for 7 d. Colony characters and pigment production on PDA were noted after 7 d. Colony colours were rated according to Rayner (1970). Colony diameters were measured after 7 and 10 d.
Conidia were taken from acervuli on PDA and mounted in clear lactic acid. Cultures were examined periodically for the development of ascomata. Ascospores were described from ascomata crushed in lactic acid. If a fungus was not sporulating on PDA, morphological characters were described from SNA or from inoculated stems of Anthriscus sylvestris. Hyphal appressoria were observed on the reverse side of colonies grown on SNA plates. At least 30 measurements per structure were noted and observed with a Nikon Eclipse 80i microscope using differential interference contrast (DIC) illumination. Descriptions and illustrations of taxonomic novelties were deposited in MycoBank (www.MycoBank.org; Crous et al. 2004).
DNA extraction, PCR amplification and sequencing
Total genomic DNA was extracted from axenic cultures with a modified CTAB protocol as described in Guo et al. (2000). Seven loci including the 5.8S nuclear ribosomal gene with the two flanking internal transcribed spacers (ITS), an intron of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a partial sequence of the actin (ACT), beta-tubulin (TUB2), glutamine synthetase (GS), calmodulin (CAL) and Apn2-Mat1-2 intergenic spacer and partial mating type (Mat1-2) gene (ApMat) were amplified and sequenced using the primer pairs ITS1 + ITS4 (White et al. 1990), GDF1 + GDR1 (Guerber et al. 2003), ACT-512F + ACT-783R (Carbone & Kohn 1999), T1 + Bt-2b (Glass & Donaldson 1995, O’Donnell & Cigelnik 1997), GSF1 + GSR1 (Stephenson et al. 1997), CL1C + CL2C (Weir et al. 2012), and AMF1 + AMR1 (Silva et al. 2012b), respectively. PCR amplification protocols were performed as described by Liu et al. (2012), but the denaturing temperatures were adjusted to 52 °C for ITS, GAPDH, ACT, GS, CAL, and ApMat, and 55 °C for TUB2. Purification and sequencing of PCR amplicons were carried out by the SinoGenoMax Company, Beijing, China. DNA sequences generated with forward and reverse primers were used to obtain consensus sequences using MEGA v. 5.1 (Tamura et al. 2011). All novel sequences were deposited in NCBIs GenBank database (www.ncbi.nlm.nih.gov/; KJ954359–KJ955371, KM360143–KM360146, KM610172–KM610185, Table 1, 2), and the alignments and trees in TreeBASE (www.treebase.org/treebase-web/home.html; study S16761).
Table 1.
Strains of the C. gloeosporioides s.l. species studied in this paper with details about host and location, and GenBank accessions of the sequences generated.
| Species | Accession numbera | Host | Locality | GenBank accessions |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | ACT | TUB2 | CAL | GS | ApMat | ||||
| C. aenigma | ICMP 18608* | Persea americana | Israel | JX010244 | JX010044 | JX009443 | JX010389 | JX009683 | JX010078 | KM360143 |
| ICMP 18686 | Pyrus pyrifolia | Japan | JX010243 | JX009913 | JX009519 | JX010390 | JX009684 | JX010079 | ||
| C. aeschynomenes | ICMP 17673, ATCC 201874* | Aeschynomene virginica | USA | JX010176 | JX009930 | JX009483 | JX010392 | JX009721 | JX010081 | KM360145 |
| C. alatae | CBS 304.67, ICMP 17919* | Dioscorea alata | India | JX010190 | JX009990 | JX009471 | JX010383 | JX009738 | JX010065 | KC888932 |
| ICMP 18122 | Dioscorea alata | Nigeria | JX010191 | JX010011 | JX009470 | JX010449 | JX009739 | JX010136 | ||
| C. alienum | ICMP 12071* | Malus domestica | New Zealand | JX010251 | JX010028 | JX009572 | JX010411 | JX009654 | JX010101 | KM360144 |
| ICMP 18621 | Persea americana | New Zealand | JX010246 | JX009959 | JX009552 | JX010386 | JX009657 | JX010075 | ||
| IMI 313842, ICMP 18691 | Persea americana | Australia | JX010217 | JX010018 | JX009580 | JX010385 | JX009664 | JX010074 | ||
| LC3114, LF322 | Ca. sinensis, endophyte | China | KJ955131 | KJ954832 | KJ954411 | KJ955279 | KJ954684 | KJ954982 | KJ954545 | |
| C. aotearoa | ICMP 17324 | Kunzea ericoides | New Zealan | JX010198 | JX009991 | JX009538 | JX010418 | JX009619 | JX010109 | |
| ICMP 18532 | Vitex lucens | New Zealand | JX010220 | JX009906 | JX009544 | JX010421 | JX009614 | JX010108 | ||
| ICMP 18537* | Coprosma sp. | New Zealand | JX010205 | JX010005 | JX009564 | JX010420 | JX009611 | JX010113 | KC888930 | |
| C. asianum | GM595, MTCC 11680 | Mangifera indica | India | JQ894679 | JQ894623 | JQ894545 | JQ894601 | KC790789 | JQ894554 | |
| ICMP 18580, CBS 130418* | Coffea arabica | Thailand | FJ972612 | JX010053 | JX009584 | JX010406 | FJ917506 | JX010096 | FR718814 | |
| IMI 313839, ICMP 18696 | Mangifera indica | Australia | JX010192 | JX009915 | JX009576 | JX010384 | JX009723 | JX010073 | ||
| C. boninense | MAFF 305972, CBS 123755* | Crinum asiaticum var. sinicum | Japan | JQ005153 | JQ005240 | JQ005501 | JQ005588 | JQ005674 | ||
| C. camelliae | CBS 125502 | Camellia sp., pathogen | unknown | KJ955077 | KJ954778 | KJ954359 | KJ954630 | KJ954928 | ||
| ICMP 10643, LF897, LC3667 | Camellia × williamsii | UK | JX010224 | JX009908 | JX009540 | JX010436 | JX009630 | JX010119 | KJ954625 | |
| ICMP 10646, LF898, LC3668 | Ca. sasanqua | USA | JX010225 | JX009993 | JX009563 | JX010437 | JX009629 | JX010117 | KJ954626 | |
| ICMP 18542, LF899, LC3669 | Ca. sasanqua | USA | JX010223 | JX009994 | JX009488 | JX010429 | JX009628 | JX010118 | KJ954627 | |
| CGMCC 3.14924, LC1363 | Ca. sinensis, pathogen | China | KJ955080 | KJ954781 | KJ954362 | KJ955229 | KJ954633 | KJ954931 | KJ954496 | |
| CGMCC 3.14925, LC1364* | Ca. sinensis, pathogen | China | KJ955081 | KJ954782 | KJ954363 | KJ955230 | KJ954634 | KJ954932 | KJ954497 | |
| CGMCC 3.14926, LC1365 | Ca. sinensis, pathogen | China | KJ955082 | KJ954783 | KJ954364 | KJ955231 | KJ954635 | KJ954933 | KJ954498 | |
| LC2944, LF152 | Camellia sp., pathogen | China | KJ955090 | KJ954791 | KJ954372 | KJ955239 | KJ954643 | KJ954941 | KJ954506 | |
| LC2962, LF170 | Camellia sp., pathogen | China | KJ955091 | KJ954792 | KJ954373 | KJ955240 | KJ954644 | KJ954942 | KJ954507 | |
| LC2998, LF206 | Ca. sinensis, pathogen | China | KJ955094 | KJ954795 | KJ954376 | KJ955243 | KJ954647 | KJ954945 | KJ954510 | |
| LC2999, LF207 | Ca. sinensis, pathogen | China | KJ955095 | KJ954796 | KJ954377 | KJ955244 | KJ954648 | KJ954946 | KJ954511 | |
| LC3000, LF208 | Ca. sinensis, pathogen | China | KJ955096 | KJ954797 | KJ954378 | KJ955245 | KJ954649 | KJ954947 | ||
| LC3001, LF209 | Ca. sinensis, pathogen | China | KJ955097 | KJ954798 | KJ954379 | KJ955246 | KJ954650 | KJ954948 | KJ954512 | |
| LC3002, LF210 | Ca. sinensis, pathogen | China | KJ955098 | KJ954799 | KJ954380 | KJ955247 | KJ954651 | KJ954949 | KJ954513 | |
| LC3004, LF212 | Ca. sinensis, pathogen | China | KJ955099 | KJ954800 | KJ954381 | KJ955248 | KJ954652 | KJ954950 | KJ954514 | |
| LC3005, LF213 | Ca. sinensis, pathogen | China | KJ955100 | KJ954801 | KJ954382 | KJ955249 | KJ954653 | KJ954951 | KJ954515 | |
| LC3006, LF214 | Ca. sinensis, pathogen | China | KJ955101 | KJ954802 | KJ954383 | KJ955250 | KJ954654 | KJ954952 | KJ954516 | |
| LC3007, LF215 | Ca. sinensis, pathogen | China | KJ955102 | KJ954803 | KJ954384 | KJ955251 | KJ954655 | KJ954953 | KJ954517 | |
| LC3008, LF216 | Ca. sinensis, pathogen | China | KJ955103 | KJ954804 | KJ954385 | KJ955252 | KJ954656 | KJ954954 | KJ954518 | |
| LC3014, LF222 | Ca. sinensis, pathogen | China | KJ955104 | KJ954805 | KJ954386 | KJ955253 | KJ954657 | KJ954955 | KJ954519 | |
| LC3015, LF223 | Ca. sinensis, pathogen | China | KJ955105 | KJ954806 | KJ954387 | KJ954658 | KJ954956 | KJ954520 | ||
| LC3017, LF225 | Ca. sinensis, pathogen | China | KJ955106 | KJ954807 | KJ954388 | KJ955254 | KJ954659 | KJ954957 | KJ954521 | |
| LC3018, LF226 | Ca. sinensis, pathogen | China | KJ955107 | KJ954808 | KJ954389 | KJ955255 | KJ954660 | KJ954958 | KJ954522 | |
| LC3019, LF227 | Ca. sinensis, pathogen | China | KJ955108 | KJ954809 | KJ954390 | KJ955256 | KJ954661 | KJ954959 | KJ954523 | |
| LC3054, LF262 | Ca. sinensis, pathogen | China | KJ955110 | KJ954811 | KJ954391 | KJ955258 | KJ954663 | KJ954961 | KJ954525 | |
| LC3057, LF265 | Ca. sinensis, pathogen | China | KJ955111 | KJ954812 | KJ954392 | KJ955259 | KJ954664 | KJ954962 | KJ954526 | |
| LC3070, LF278 | Ca. sinensis, pathogen | China | KJ955112 | KJ954813 | KJ954393 | KJ955260 | KJ954665 | KJ954963 | KJ954527 | |
| LC3071, LF279 | Ca. sinensis, pathogen | China | KJ955113 | KJ954814 | KJ955261 | KJ954666 | KJ954964 | KJ954528 | ||
| LC3076, LF284 | Ca. sinensis, endophyte | China | KJ955114 | KJ954815 | KJ954394 | KJ955262 | KJ954667 | KJ954965 | KJ954529 | |
| LC3089, LF297 | Ca. sinensis, endophyte | China | KJ955115 | KJ954816 | KJ954395 | KJ955263 | KJ954668 | KJ954966 | KJ954530 | |
| LC3091, LF299 | Ca. sinensis, endophyte | China | KJ955116 | KJ954817 | KJ954396 | KJ955264 | KJ954669 | KJ954967 | KJ954531 | |
| LC3092, LF300 | Ca. sinensis, endophyte | China | KJ955117 | KJ954818 | KJ954397 | KJ955265 | KJ954670 | KJ954968 | KJ954532 | |
| LC3095, LF303 | Ca. sinensis, endophyte | China | KJ955118 | KJ954819 | KJ954398 | KJ955266 | KJ954671 | KJ954969 | KJ954533 | |
| LC3096, LF304 | Ca. sinensis, endophyte | China | KJ955119 | KJ954820 | KJ954399 | KJ955267 | KJ954672 | KJ954970 | KJ954534 | |
| LC3100, LF308 | Ca. sinensis, endophyte | China | KJ955120 | KJ954821 | KJ954400 | KJ955268 | KJ954673 | KJ954971 | KJ954535 | |
| LC3101, LF309 | Ca. sinensis, endophyte | China | KJ955121 | KJ954822 | KJ954401 | KJ955269 | KJ954674 | KJ954972 | KJ954536 | |
| LC3102, LF310 | Ca. sinensis, endophyte | China | KJ955122 | KJ954823 | KJ954402 | KJ955270 | KJ954675 | KJ954973 | KJ954537 | |
| LC3103, LF311 | Ca. sinensis, endophyte | China | KJ955123 | KJ954824 | KJ954403 | KJ955271 | KJ954676 | KJ954974 | KJ954538 | |
| LC3107, LF315 | Ca. sinensis, endophyte | China | KJ955124 | KJ954825 | KJ954404 | KJ955272 | KJ954677 | KJ954975 | KJ954539 | |
| LC3109, LF317 | Ca. sinensis, endophyte | China | KJ955126 | KJ954827 | KJ954406 | KJ955274 | KJ954679 | KJ954977 | KJ954540 | |
| LC3111, LF319 | Ca. sinensis, endophyte | China | KJ955128 | KJ954829 | KJ954408 | KJ955276 | KJ954681 | KJ954979 | KJ954542 | |
| LC3112, LF320 | Ca. sinensis, endophyte | China | KJ955129 | KJ954830 | KJ954409 | KJ955277 | KJ954682 | KJ954980 | KJ954543 | |
| LC3113, LF321 | Ca. sinensis, endophyte | China | KJ955130 | KJ954831 | KJ954410 | KJ955278 | KJ954683 | KJ954981 | KJ954544 | |
| LC3116, LF324 | Ca. sinensis, endophyte | China | KJ955132 | KJ954833 | KJ954412 | KJ955280 | KJ954685 | KJ954983 | KJ954546 | |
| LC3117, LF325 | Ca. sinensis, endophyte | China | KJ955133 | KJ954834 | KJ954413 | KJ955281 | KJ954686 | KJ954984 | KJ954547 | |
| LC3123, LF331 | Ca. sinensis, endophyte | China | KJ955134 | KJ954835 | KJ954414 | KJ955282 | KJ954687 | KJ954985 | KJ954548 | |
| LC3128, LF336 | Ca. sinensis, pathogen | China | KJ955135 | KJ954836 | KJ954415 | KJ955283 | KJ954688 | KJ954986 | KJ954549 | |
| LC3129, LF337 | Ca. sinensis, pathogen | China | KJ955136 | KJ954837 | KJ954416 | KJ955284 | KJ954689 | KJ954987 | KJ954550 | |
| LC3130, LF338 | Ca. sinensis, pathogen | China | KJ955137 | KJ954838 | KJ954417 | KJ955285 | KJ954690 | KJ954988 | KJ954551 | |
| LC3131, LF339 | Ca. sinensis, pathogen | China | KJ955138 | KJ954839 | KJ955286 | KJ954691 | KJ954989 | KJ954552 | ||
| LC3142, LF350 | Ca. sinensis, pathogen | China | KJ955139 | KJ954840 | KJ954418 | KJ955287 | KJ954692 | KJ954990 | KJ954553 | |
| LC3143, LF351 | Ca. sinensis, pathogen | China | KJ955140 | KJ954841 | KJ954419 | KJ955288 | KJ954693 | KJ954991 | KJ954554 | |
| LC3147, LF355 | Ca. sinensis, pathogen | China | KJ955141 | KJ954842 | KJ954420 | KJ955289 | KJ954694 | KJ954992 | KJ954555 | |
| LC3148, LF356 | Ca. sinensis, pathogen | China | KJ955142 | KJ954843 | KJ954421 | KJ955290 | KJ954695 | KJ954993 | KJ954556 | |
| LC3158, LF367 | Ca. sinensis, endophyte | China | KJ955144 | KJ954845 | KJ954423 | KJ955292 | KJ954697 | KJ954995 | KJ954558 | |
| LC3173, LF383 | Ca. sinensis, endophyte | China | KJ955147 | KJ954848 | KJ954425 | KJ955295 | KJ954998 | KJ954560 | ||
| LC3269, LF491 | Ca. sinensis, pathogen | China | KJ955150 | KJ954851 | KJ955297 | KJ954702 | KJ955001 | KJ954562 | ||
| LC3270, LF492 | Ca. sinensis, pathogen | China | KJ955151 | KJ954852 | KJ954428 | KJ955298 | KJ954703 | KJ955002 | KJ954563 | |
| LC3274, LF496 | Ca. sinensis, pathogen | China | KJ955153 | KJ954854 | KJ954430 | KJ955300 | KJ954705 | KJ955004 | KJ954564 | |
| LC3279, LF501 | Ca. sinensis, pathogen | China | KJ955154 | KJ954855 | KJ954431 | KJ955301 | KJ954706 | KJ955005 | KJ954565 | |
| LC3282, LF504 | Ca. sinensis, pathogen | China | KJ955155 | KJ954856 | KJ954432 | KJ955302 | KJ954707 | KJ955006 | KJ954566 | |
| LC3319, LF541 | Ca. sinensis, pathogen | China | KJ955160 | KJ954861 | KJ954436 | KJ955307 | KJ954712 | KJ954571 | ||
| LC3322, LF544 | Ca. sinensis, pathogen | China | KJ955161 | KJ954862 | KJ954437 | KJ955308 | KJ954713 | KJ955011 | KJ954572 | |
| LC3323, LF545 | Ca. sinensis, pathogen | China | KJ955162 | KJ954863 | KJ955309 | KJ954714 | KJ955012 | KJ954573 | ||
| LC3328, LF550 | Ca. sinensis, pathogen | China | KJ955163 | KJ954864 | KJ955310 | KJ954715 | KJ955013 | KJ954574 | ||
| LC3330, LF552 | Ca. sinensis, pathogen | China | KJ955164 | KJ954865 | KJ954438 | KJ955311 | KJ954716 | KJ955014 | KJ954575 | |
| LC3335, LF557 | Ca. sinensis, pathogen | China | KJ955165 | KJ954866 | KJ954439 | KJ955312 | KJ954717 | KJ955015 | KJ954576 | |
| LC3350, LF572 | Ca. sinensis, pathogen | China | KJ955166 | KJ954867 | KJ954440 | KJ955313 | KJ954718 | KJ955016 | KJ954577 | |
| LC3352, LF574 | Ca. sinensis, pathogen | China | KJ955167 | KJ954868 | KJ954441 | KJ955314 | KJ954719 | KJ955017 | KJ954578 | |
| LC3355, LF577 | Ca. sinensis, pathogen | China | KJ955168 | KJ954869 | KJ954442 | KJ955315 | KJ954720 | KJ955018 | KJ954579 | |
| LC3367, LF589 | Ca. sinensis, pathogen | China | KJ955170 | KJ954871 | KJ954444 | KJ955317 | KJ954722 | KJ955020 | ||
| LC3374, LF596 | Ca. sinensis, pathogen | China | KJ955173 | KJ954874 | KJ954447 | KJ955320 | KJ954725 | KJ955023 | KJ954582 | |
| LC3379, LF601 | Ca. sinensis, pathogen | China | KJ955174 | KJ954875 | KJ954448 | KJ955321 | KJ954726 | KJ955024 | KJ954583 | |
| LC3385, LF607 | Ca. sinensis, pathogen | China | KJ955178 | KJ954879 | KJ954451 | KJ955325 | KJ954730 | KJ955028 | KJ954586 | |
| LC3387, LF609 | Ca. sinensis, pathogen | China | KJ955179 | KJ954880 | KJ954452 | KJ955326 | KJ954731 | KJ955029 | KJ954587 | |
| LC3389, LF611 | Ca. sinensis, pathogen | China | KJ955180 | KJ954881 | KJ954453 | KJ955327 | KJ954732 | KJ955030 | KJ954588 | |
| LC3395, LF617 | Ca. sinensis, pathogen | China | KJ955181 | KJ954882 | KJ954454 | KJ955328 | KJ954733 | KJ955031 | KJ954589 | |
| LC3398, LF620 | Ca. sinensis, pathogen | China | KJ955182 | KJ954883 | KJ954455 | KJ955329 | KJ954734 | KJ955032 | KJ954590 | |
| LC3401, LF623 | Ca. sinensis, pathogen | China | KJ955183 | KJ954884 | KJ954456 | KJ955330 | KJ954735 | KJ955033 | KJ954591 | |
| LC3403, LF625 | Ca. sinensis, pathogen | China | KJ955185 | KJ954886 | KJ954458 | KJ955332 | KJ954737 | KJ955035 | KJ954593 | |
| LC3408, LF630 | Ca. sinensis, pathogen | China | KJ955186 | KJ954887 | KJ954459 | KJ955333 | KJ954738 | KJ955036 | KJ954594 | |
| LC3469, LF694 | Ca. sinensis, pathogen | China | KJ955204 | KJ954905 | KJ954474 | KJ955350 | KJ954755 | KJ955054 | KJ954610 | |
| LC3488, LF715 | Ca. sinensis, pathogen | China | KJ955206 | KJ954907 | KJ954476 | KJ955352 | KJ954757 | KJ955056 | KJ954612 | |
| LC3492, LF720 | Ca. sinensis, pathogen | China | KJ955208 | KJ954909 | KJ954478 | KJ955354 | KJ954759 | KJ955058 | KJ954614 | |
| LC3506, LF734 | Ca. sinensis, pathogen | China | KJ955209 | KJ954910 | KJ954479 | KJ955355 | KJ954760 | KJ955059 | KJ954615 | |
| LC3513, LF741 | Camellia sp., pathogen | China | KJ955210 | KJ954911 | KJ955356 | KJ954761 | KJ955060 | KJ954616 | ||
| LC3514, LF742 | Camellia sp., pathogen | China | KJ955211 | KJ954912 | KJ954480 | KJ955357 | KJ954762 | KJ955061 | KJ954617 | |
| LC3515, LF743 | Camellia sp., pathogen | China | KJ955212 | KJ954913 | KJ954481 | KJ955358 | KJ954763 | KJ955062 | KJ954618 | |
| LC3516, LF744 | Camellia sp., pathogen | China | KJ955213 | KJ954914 | KJ955359 | KJ954764 | KJ955063 | KJ954619 | ||
| LC3561, LF789 | Ca. sinensis, pathogen | China | KJ955217 | KJ954918 | KJ954485 | KJ955363 | KJ954768 | KJ955067 | KJ954621 | |
| LC3562, LF790 | Ca. sinensis, pathogen | China | KJ955218 | KJ954919 | KJ954486 | KJ954769 | KJ955068 | KJ954622 | ||
| C. clidemiae | ICMP 18658* | Clidemia hirta | USA, Hawaii | JX010265 | JX009989 | JX009537 | JX010438 | JX009645 | JX010129 | KC888929 |
| ICMP 18706 | Vitis sp. | USA | JX010274 | JX009909 | JX009476 | JX010439 | JX009639 | JX010128 | ||
| C. cordylinicola | LC0886, ICMP 18579* | Cordyline fruticosa | Thailand | JX010226 | JX009975 | HM470235 | JX010440 | HM470238 | JX010122 | JQ899274 |
| C. dianesei | CMM4083, MFLU 1300058* | Mangifera indica | Brazil | KC329779 | KC517194 | KC517298 | KC517254 | KC517209 | KC430894 | |
| CMM4088, MFLU 1300059 | Mangifera indica | Brazil | KC329781 | KC517162 | KC517300 | KC517255 | KC517210 | KC430900 | ||
| CMM4089, MFLU 1300060 | Mangifera indica | Brazil | KC329783 | KC517163 | KC517302 | KC517256 | KC517211 | KC430879 | ||
| C. endophytica | MFLUCC 130417, LC1216 | Pennisetum purpureum | Thailand | KC633853 | KC832853 | KC692467 | KC810017 | |||
| MFLUCC 130418, LC0324* | Pennisetum purpureum | Thailand | KC633854 | KC832854 | KF306258 | KC810018 | ||||
| MFLUCC 130419, LC0327 | Pennisetum purpureum | Thailand | KC633855 | KC832846 | KC692468 | KC810016 | ||||
| C. fructicola | CBS 125395, ICMP 18645 | Theobroma cacao | Panama | JX010172 | JX009992 | JX009543 | JX010408 | JX009666 | JX010098 | |
| CBS 238.49, ICMP 17921 | Ficus edulis | Germany | JX010181 | JX009923 | JX009495 | JX010400 | JX009671 | JX010090 | ||
| GM567, MTCC 11679 | Mangifera indica | India | JQ894676 | JQ894630 | JQ894543 | JQ894600 | KC790787 | JQ894576 | ||
| ICMP 18581, CBS 130416* | Coffea arabica | Thailand | JX010165 | JX010033 | FJ907426 | JX010405 | FJ917508 | JX010095 | JQ807838 | |
| ICMP 18646, CBS 125397, MTCC 10906 | Tetragastris panamensis | Panama | JX010173 | JX010032 | JX009581 | JX010409 | JX009674 | JX010099 | ||
| LC2923, LF130 | Ca. sinensis, pathogen | China | KJ955083 | KJ954784 | KJ954365 | KJ955232 | KJ954636 | KJ954934 | KJ954499 | |
| LC2924, LF131 | Ca. sinensis, pathogen | China | KJ955084 | KJ954785 | KJ954366 | KJ955233 | KJ954637 | KJ954935 | KJ954500 | |
| LC2925, LF132 | Ca. sinensis, pathogen | China | KJ955085 | KJ954786 | KJ954367 | KJ955234 | KJ954638 | KJ954936 | KJ954501 | |
| LC2926, LF133 | Ca. sinensis, pathogen | China | KJ955086 | KJ954787 | KJ954368 | KJ955235 | KJ954639 | KJ954937 | KJ954502 | |
| LC3155, LF364 | Ca. sinensis, endophyte | China | KJ955143 | KJ954844 | KJ954422 | KJ955291 | KJ954696 | KJ954994 | KJ954557 | |
| LC3167, LF376 | Ca. sinensis, endophyte | China | KJ955145 | KJ954846 | KJ955293 | KJ954698 | KJ954996 | KJ954559 | ||
| LC3284, LF506 | Ca. sinensis, pathogen | China | KJ955156 | KJ954857 | KJ954433 | KJ955303 | KJ954708 | KJ955007 | KJ954567 | |
| LC3288, LF510 | Ca. sinensis, pathogen | China | KJ955157 | KJ954858 | KJ955304 | KJ954709 | KJ955008 | KJ954568 | ||
| LC3315, LF537 | Ca. sinensis, pathogen | China | KJ955159 | KJ954860 | KJ954435 | KJ955306 | KJ954711 | KJ955010 | KJ954570 | |
| LC3368, LF590 | Ca. sinensis, pathogen | China | KJ955171 | KJ954872 | KJ954445 | KJ955318 | KJ954723 | KJ955021 | KJ954580 | |
| LC3370, LF592 | Ca. sinensis, pathogen | China | KJ955172 | KJ954873 | KJ954446 | KJ955319 | KJ954724 | KJ955022 | KJ954581 | |
| LC3384, LF606 | Ca. sinensis, pathogen | China | KJ955177 | KJ954878 | KJ954450 | KJ955324 | KJ954729 | KJ955027 | KJ954585 | |
| LC3402, LF624 | Ca. sinensis, pathogen | China | KJ955184 | KJ954885 | KJ954457 | KJ955331 | KJ954736 | KJ955034 | KJ954592 | |
| LC3417, LF639 | Ca. sinensis, endophyte | China | KJ955188 | KJ954889 | KJ954461 | KJ955335 | KJ954740 | KJ955038 | KJ954595 | |
| LC3425, LF647 | Ca. sinensis, endophyte | China | KJ955190 | KJ954891 | KJ954463 | KJ955337 | KJ954741 | KJ955040 | KJ954596 | |
| LC3427, LF649 | Ca. sinensis, endophyte | China | KJ955191 | KJ954892 | KJ954464 | KJ955338 | KJ954742 | KJ955041 | KJ954597 | |
| LC3430, LF652 | Ca. sinensis, endophyte | China | KJ955192 | KJ954893 | KJ954465 | KJ955339 | KJ954743 | KJ955042 | KJ954598 | |
| LC3433, LF655 | Ca. sinensis, endophyte | China | KJ955193 | KJ954894 | KJ954466 | KJ955340 | KJ954744 | KJ955043 | KJ954599 | |
| LC3434, LF656 | Ca. sinensis, endophyte | China | KJ955194 | KJ954895 | KJ954467 | KJ955341 | KJ954745 | KJ955044 | KJ954600 | |
| LC3447, LF670 | Ca. sinensis, endophyte | China | KJ955195 | KJ954896 | KJ955342 | KJ954746 | KJ955045 | KJ954601 | ||
| LC3451, LF674 | Ca. sinensis, endophyte | China | KJ955196 | KJ954897 | KJ955343 | KJ954747 | KJ955046 | KJ954602 | ||
| LC3457, LF681 | Ca. sinensis, endophyte | China | KJ955197 | KJ954898 | KJ954468 | KJ955344 | KJ954748 | KJ955047 | KJ954603 | |
| LC3461, LF685 | Ca. sinensis, pathogen | China | KJ955199 | KJ954900 | KJ955346 | KJ954750 | KJ955049 | KJ954605 | ||
| LC3462, LF686 | Ca. sinensis, pathogen | China | KJ955200 | KJ954901 | KJ954470 | KJ955347 | KJ954751 | KJ955050 | KJ954606 | |
| LC3464, LF689 | Ca. sinensis, pathogen | China | KJ955202 | KJ954903 | KJ954472 | KJ954753 | KJ955052 | KJ954608 | ||
| LC3465, LF690 | Ca. sinensis, pathogen | China | KJ955203 | KJ954904 | KJ954473 | KJ955349 | KJ954754 | KJ955053 | KJ954609 | |
| LC3471, LF696 | Ca. sinensis, pathogen | China | KJ955205 | KJ954906 | KJ954475 | KJ955351 | KJ954756 | KJ955055 | KJ954611 | |
| LC3489, LF716 | Ca. sinensis, endophyte | China | KJ955207 | KJ954908 | KJ954477 | KJ955353 | KJ954758 | KJ955057 | KJ954613 | |
| LC3545, LF773 | Ca. sinensis, endophyte | China | KJ955214 | KJ954915 | KJ954482 | KJ955360 | KJ954765 | KJ955064 | KJ954620 | |
| LC3569, LF797 | Ca. sinensis, pathogen | China | KJ955219 | KJ954920 | KJ954487 | KJ955364 | KJ954770 | KJ955069 | KJ954623 | |
| LC3666, LF896, ICMP 18656 | Ca. sinensis, pathogen | Indonesia | KJ955221 | KJ954922 | KJ954489 | KJ955366 | KJ954772 | KJ955071 | KJ954624 | |
| LC3670, LF900, ICMP 10642 | Camellia sp., pathogen | UK | KJ955225 | KJ954926 | KJ954492 | KJ955370 | KJ954776 | KJ955075 | KJ954628 | |
| C. fructivorum | Coll1092, BPI 884114, CBS 133135 | Rhexia virginica | USA | JX145133 | JX145184 | |||||
| Coll1414, BPI 884103, CBS 133125* | Vaccinium macrocarpon | USA | JX145145 | JX145196 | ||||||
| C. gloeosporioides | IMI 356878, ICMP 17821, CBS 112999* | Citrus sinensis | Italy | JX010152 | JX010056 | JX009531 | JX010445 | JX009731 | JX010085 | JQ807843 |
| LC3110, LF318 | Ca. sinensis, endophyte | China | KJ955127 | KJ954828 | KJ954407 | KJ955275 | KJ954680 | KJ954978 | KJ954541 | |
| LC3312, LF534 | Ca. sinensis, pathogen | China | KJ955158 | KJ954859 | KJ954434 | KJ955305 | KJ954710 | KJ955009 | KJ954569 | |
| LC3382, LF604 | Ca. sinensis, pathogen | China | KJ955176 | KJ954877 | KJ954450 | KJ955323 | KJ954728 | KJ955026 | KJ954584 | |
| LC3686, LF916 | Ca. sinensis, pathogen | China | KJ955226 | KJ954927 | KJ954493 | KJ955371 | KJ954777 | KJ955076 | KJ954629 | |
| C. grevilleae | CBS 132879, CPC 15481* | Grevillea sp. | Italy | KC297078 | KC297010 | KC296941 | KC297102 | KC296963 | KC297033 | |
| C. henanense | LC3030, CGMCC 3.17354, LF238* | Ca. sinensis, pathogen | China | KJ955109 | KJ954810 | KM023257 | KJ955257 | KJ954662 | KJ954960 | KJ954524 |
| LC2820, LF24 | Cirsium japonicum, pathogen | China | KM610182 | KM610178 | KM610172 | KM610184 | KM610176 | KM610180 | KM610174 | |
| LC2821, LF25 | Cirsium japonicum, pathogen | China | KM610183 | KM610179 | KM610173 | KM610185 | KM610177 | KM610181 | KM610175 | |
| C. horii | ICMP 17968 | Diospyros kaki | China | JX010212 | GQ329682 | JX009547 | JX010378 | JX009605 | JX010068 | |
| NBRC 7478, ICMP 10492, MTCC 10841* | Diospyros kaki | Japan | GQ329690 | GQ329681 | JX009438 | JX010450 | JX009604 | JX010137 | JQ807840 | |
| C. jiangxiense | LC3266, CGMCC 3.17361, LF488 | Ca. sinensis, pathogen | China | KJ955149 | KJ954850 | KJ954427 | KJ954701 | KJ955000 | KJ954561 | |
| LC3460, CGMCC 3.17362, LF684 | Ca. sinensis, endophyte | China | KJ955198 | KJ954899 | KJ954469 | KJ955345 | KJ954749 | KJ955048 | KJ954604 | |
| LC3463, CGMCC 3.17363, LF687* | Ca. sinensis, pathogen | China | KJ955201 | KJ954902 | KJ954471 | KJ955348 | KJ954752 | KJ955051 | KJ954607 | |
| C. kahawae subsp. ciggaro | ICMP 12952 | Persea americana | New Zealand | JX010214 | JX009971 | JX009431 | JX010426 | JX009648 | JX010126 | |
| ICMP 18534 | Kunzea ericoides | New Zealand | JX010227 | JX009904 | JX009473 | JX010427 | JX009634 | JX010116 | HE655657 | |
| ICMP 18539* | Olea europaea | Australia | JX010230 | JX009966 | JX009523 | JX010434 | JX009635 | JX010132 | ||
| C. kahawae subsp. kahawae | IMI 319418, ICMP 17816* | Coffea arabica | Kenya | JX010231 | JX010012 | JX009452 | JX010444 | JX009642 | JX010130 | JQ894579 |
| CBS 982.69, ICMP 17915 | Coffea arabica | Angola | JX010234 | JX010040 | JX009474 | JX010435 | JX009638 | JX010125 | ||
| IMI 361501, ICMP 17905 | Coffea arabica | Cameroon | JX010232 | JX010046 | JX009561 | JX010431 | JX009644 | JX010127 | ||
| C. melanocaulon | Coll126, BPI 884101, CBS 133123 | Vaccinium macrocarpon | USA | JX145142 | JX145193 | JX145309 | ||||
| Coll131, BPI 884113, CBS 133251* | Vaccinium macrocarpon | USA | JX145144 | JX145195 | JX145313 | |||||
| C. musae | CBS 116870, ICMP 19119, MTCC 11349* | Musa sp. | USA | JX010146 | JX010050 | JX009433 | HQ596280 | JX009742 | JX010103 | KC888926 |
| IMI 52264, ICMP 17817 | Musa sapientum | Kenya | JX010142 | JX010015 | JX009432 | JX010395 | JX009689 | JX010084 | ||
| C. nupharicola | CBS 469.96, ICMP 17938 | Nuphar lutea subsp. polysepala | USA | JX010189 | JX009936 | JX009486 | JX010397 | JX009661 | JX010087 | |
| CBS 470.96, ICMP 18187* | Nuphar lutea subsp. polysepala | USA | JX010187 | JX009972 | JX009437 | JX010398 | JX009663 | JX010088 | JX145319 | |
| CBS 472.96, ICMP 17940 | Nymphaea ordorata | USA | JX010188 | JX010031 | JX009582 | JX010399 | JX009662 | JX010089 | ||
| C. proteae | CBS 132882, CPC 14859* | Protea sp. | South Africa | KC297079 | KC297009 | KC296940 | KC297101 | KC296960 | KC297032 | |
| CBS 134301, CPC 14860 | Protea sp. | South Africa | KC842385 | KC842379 | KC842373 | KC842387 | KC842375 | KC842387 | ||
| C. psidii | CBS 145.29, ICMP 19120* | Psidium sp. | Italy | JX010219 | JX009967 | JX009515 | JX010443 | JX009743 | JX010133 | KC888931 |
| C. queenslandicum | ICMP 1778* | Carica papaya | Australia | JX010276 | JX009934 | JX009447 | JX010414 | JX009691 | JX010104 | KC888928 |
| ICMP 18705 | Coffea sp. | Fiji | JX010185 | JX010036 | JX009490 | JX010412 | JX009694 | JX010102 | ||
| C. rhexiae | Coll1026, BPI 884112, CBS 133134* | Rhexia virginica | USA | JX145128 | JX145179 | JX145290 | ||||
| Coll877, BPI 884110, CBS 133132 | Vaccinium macrocarpon | USA | JX145157 | JX145209 | JX145302 | |||||
| C. salsolae | ICMP 19051* | Salsola tragus | Hungary | JX010242 | JX009916 | JX009562 | JX010403 | JX009696 | JX010093 | KC888925 |
| C. siamense | DAR 76934, ICMP 18574 | Pistacia vera | Australia | JX010270 | JX010002 | JX009535 | JX010391 | JX009707 | JX010080 | |
| GM018, MTCC 11672 | Mangifera indica | India | JQ894653 | JQ894624 | JQ894533 | JQ894594 | KC790778 | |||
| GM057, MTCC 11590 | Mangifera indica | India | JQ894658 | JQ894620 | JQ894534 | JQ894590 | KC790780 | JQ894551 | ||
| GM172, MTCC 11591 | Mangifera indica | India | JQ894662 | JQ894621 | JQ894535 | JQ894591 | KC790781 | JQ894562 | ||
| GM385 | Mangifera indica | India | JQ894668 | JQ894626 | JQ894536 | JQ894596 | KC790782 | JQ894568 | ||
| GM390, MTCC 11677 | Mangifera indica | India | JQ894670 | JQ894627 | JQ894537 | JQ894597 | KC790783 | JQ894570 | ||
| GM473, MTCC 11589 | Mangifera indica | India | JQ894673 | JQ894622 | JQ894539 | JQ894592 | KC790785 | JQ894553 | ||
| GM529, MTCC 11592 | Mangifera indica | India | JQ894675 | JQ894629 | JQ894540 | JQ894599 | KC790786 | JQ894575 | ||
| GZAAS 5.09538 | Murraya sp. | China | JQ247632 | JQ247608 | JQ247656 | JQ247645 | JQ247597 | JQ247620 | ||
| ICMP 12567 | Persea americana | Australia | JX010250 | JX009940 | JX009541 | JX010387 | JX009697 | JX010076 | ||
| ICMP 18121 | Dioscorea rotundata | Nigeria | JX010245 | JX009942 | JX009460 | JX010402 | JX009715 | JX010092 | ||
| ICMP 18578, CBS 130417* | Coffea arabica | Thailand | JX010171 | JX009924 | FJ907423 | JX010404 | FJ917505 | JX010094 | JQ899289 | |
| LC0148 | Camellia sp., pathogen | China | KJ955078 | KJ954779 | KJ954360 | KJ955227 | KJ954631 | KJ954929 | KJ954494 | |
| LC0149 | Camellia sp., pathogen | China | KJ955079 | KJ954780 | KJ954361 | KJ955228 | KJ954632 | KJ954930 | KJ954495 | |
| LC2931, CGMCC 3.17353, LF139 | Camellia sp., pathogen | China | KJ955087 | KJ954788 | KJ954369 | KJ955236 | KJ954640 | KJ954938 | KJ954503 | |
| LC2940, LF148 | Camellia sp., pathogen | China | KJ955088 | KJ954789 | KJ954370 | KJ955237 | KJ954641 | KJ954939 | KJ954504 | |
| LC2941, LF149 | Camellia sp., pathogen | China | KJ955089 | KJ954790 | KJ954371 | KJ955238 | KJ954642 | KJ954940 | KJ954505 | |
| LC2969, LF177 | Camellia oleifera, pathogen | China | KJ955092 | KJ954793 | KJ954374 | KJ955241 | KJ954645 | KJ954943 | KJ954508 | |
| LC2974, LF182 | Camellia sp., endophyte | China | KJ955093 | KJ954794 | KJ954375 | KJ955242 | KJ954646 | KJ954944 | KJ954509 | |
| LC3409, LF631 | Ca. sinensis, pathogen | China | KJ955187 | KJ954888 | KJ954460 | KJ955334 | KJ954739 | KJ955037 | ||
| MTCC 9660 | Mangifera indica | India | JQ894649 | JQ894619 | JQ894532 | JQ894589 | KC790790 | JQ894548 | ||
| NK24, MTCC 11599 | Mangifera indica | India | JQ894681 | JQ894632 | JQ894546 | JQ894602 | KC790791 | JQ894582 | ||
| NK28, MTCC 11593 | Mangifera indica | India | JQ894687 | JQ894633 | JQ894547 | JQ894603 | KC790792 | |||
| C. siamense (syn. C. hymenocallidis) | CBS 125378, ICMP 18642, LC0043 | Hymenocallis americana | China | JX010278 | JX010019 | JX009441 | JX010410 | JX009709 | JX010100 | JQ899283 |
| C. siamense (syn. C. jasmini-sambac) | CBS 130420, ICMP 19118 | Jasminum sambac | Vietnam | HM131511 | HM131497 | HM131507 | JX010415 | JX009713 | JX010105 | JQ807841 |
| C. siamense (syn. C. murrayae) | GZAAS 5.09506 | Murraya sp. | China | JQ247633 | JQ247609 | JQ247657 | JQ247644 | JQ247596 | JQ247621 | |
| C. temperatum | Coll1103, BPI 884098, CBS 133120 | Vaccinium macrocarpon | USA | JX145135 | JX145186 | JX145297 | ||||
| Coll883, BPI 884100, CBS 133122* | Vaccinium macrocarpon | USA | JX145159 | JX145211 | JX145298 | |||||
| C. theobromicola | MTCC 11350, CBS 124945, ICMP 18649* | Theobroma cacao | Panama | JX010294 | JX010006 | JX009444 | JX010447 | JX009591 | JX010139 | KC790726 |
| C. theobromicola (syn. C. fragariae) | CBS 142.31, ICMP 17927, MTCC 10325 | Fragaria × ananassa | USA | JX010286 | JX010024 | JX009516 | JX010373 | JX009592 | JX010064 | JQ807844 |
| C. ti | ICMP 4832* | Cordyline sp. | New Zealand | JX010269 | JX009952 | JX009520 | JX010442 | JX009649 | JX010123 | KM360146 |
| ICMP 5285 | Cordyline australis | New Zealand | JX010267 | JX009910 | JX009553 | JX010441 | JX009650 | JX010124 | ||
| C. tropicale | CBS 124949, ICMP 18653, MTCC 11371* | Theobroma cacao | Panama | JX010264 | JX010007 | JX009489 | JX010407 | JX009719 | JX010097 | KC790728 |
| MAFF 239933, ICMP 18672 | Litchi chinensis | Japan | JX010275 | JX010020 | JX009480 | JX010396 | JX009722 | JX010086 | ||
| C. viniferum | GZAAS 5.08601, yg1* | Vitis vinifera cv. Shuijing | China | JN412804 | JN412798 | JN412795 | JQ309639 | JN412787 | ||
| GZAAS 5.08608, yg4 | Vitis vinifera cv. Hongti | China | JN412802 | JN412800 | JN412793 | JN412782 | JN412784 | |||
| C. xanthorrhoeae | BRIP 45094, ICMP 17903, CBS 127831* | Xanthorrhoea preissii | Australia | JX010261 | JX009927 | JX009478 | JX010448 | JX009653 | JX010138 | KC790689 |
| IMI 350817a, ICMP 17820 | Xanthorrhoea sp. | Australia | JX010260 | JX010008 | JX009479 | JX009652 | ||||
a AS, CGMCC: China General Microbiological Culture Collection; ATCC: American Type Culture Collection; BPI: U.S. National Fungus Collections, USA; BRIP: Plant Pathology Herbarium, Department of Employment, Economic, Development and Innovation, Queensland, Australia; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; CPC: Working collection of Pedro W. Crous, housed at CBS, The Netherlands; DAR: Plant pathology Herbarium, Australia; GZAAS: Guizhou Academy of Agricultural Sciences Herbarium, China; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; IMI: Culture collection of CABI Europe UK Centre, Egham, UK; LC: Working collection of Lei Cai, housed at CAS, China; LF: Working collection of Fang Liu, housed at CAS, China; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Japan; MFLUCC: Mae Fah Luang University Culture Collection, ChiangRai, Thailand; MTCC: Microbial type culture collection and gene bank, India; NBRC: NITE Biological Resource Centre, Japan.
* = ex-type culture. Strains/sequences studied in this paper are in bold font.
Table 2.
Strains of Colletotrichum excluded from the C. gloeosporioides species complex. Details are provided about host and location, and GenBank accessions of the sequences generated.
| Species | Association numbera | Host | Locality | GenBank accessions |
|||
|---|---|---|---|---|---|---|---|
| ITS | GAPDH | ACT | TUB2 | ||||
| C. acutatum | CBS 112996, ATCC 56816* | Carica papaya | Australia | JQ005776 | JQ948677 | JQ005839 | JQ005860 |
| CBS 979.69 | Coffea arabica | Kenya | JQ948400 | JQ948731 | JQ949721 | JQ950051 | |
| C. boninense | CBS 123755, MAFF 305972* | Crinum asiaticum var. sinicum | Japan | JQ005153 | JQ005240 | JQ005501 | JQ005588 |
| CBS 128526, ICMP 18591 | Dacrycarpus dacrydioides | New Zealand | JQ005162 | JQ005249 | JQ005510 | JQ005596 | |
| CBS 128547, ICMP 10338 | Camellia sp. | New Zealand | JQ005159 | JQ005246 | JQ005507 | JQ005593 | |
| LC3422, CGMCC 3.14356, | Camellia sinensis, endophyte | China | KJ955189 | KJ954890 | KJ954462 | KJ955336 | |
| LF644 | |||||||
| C. brasiliense | CBS 128501, ICMP 18607* | Passiflora edulis | Brazil | JQ005235 | JQ005322 | JQ005583 | JQ005669 |
| CBS 128528, ICMP 18606 | Passiflora edulis | Brazil | JQ005234 | JQ005321 | JQ005582 | JQ005668 | |
| C. cliviae | CBS 125375* | Clivia miniata | China | JX519223 | JX546611 | JX519240 | JX519249 |
| LC3546, CGMCC 3.17358, | Camellia sinensis, endophyte | China | KJ955215 | KJ954916 | KJ954483 | KJ955361 | |
| LF774 | |||||||
| C. coccodes | CBS 369.75* | Solanum tuberosum | Netherlands | HM171679 | HM171673 | HM171667 | JX546873 |
| C. colombiense | CBS 129817 | Passiflora edulis | Colombia | JQ005173 | JQ005260 | JQ005521 | JQ005607 |
| CBS 129818* | Passiflora edulis | Colombia | JQ005174 | JQ005261 | JQ005522 | JQ005608 | |
| C. constrictum | CBS 128504, ICMP 12941* | Citrus limon | New Zealand | JQ005238 | JQ005325 | JQ005586 | JQ005672 |
| C. dracaenophilum | CBS 118199* | Dracaena sanderana | China | JX519222 | JX546707 | JX519238 | JX519247 |
| C. fioriniae | CBS 119293 | Vaccinium corymbosum | New Zealand | JQ948314 | JQ948644 | JQ949635 | JQ949965 |
| CBS 128517* | Fiorinia externa | USA | JQ948292 | JQ948622 | JQ949613 | JQ949943 | |
| CBS 129948 | Tulipa sp. | UK | JQ948344 | JQ948674 | JQ949665 | JQ949995 | |
| LC3381, CGMCC 3.17357, LF603 | Camellia sinensis, pathogen | China | KJ955175 | KJ954876 | KJ954449 | KJ955322 | |
| C. karstii | CBS 129824 | Musa sp. | Colombia | JQ005215 | JQ005302 | JQ005563 | JQ005649 |
| CBS 132134, CORCG6, | Vanda sp. | China | HM585409 | HM585391 | HM581995 | HM585428 | |
| CGMCC 3.14194* | |||||||
| LC3108, LF316 | Camellia sinensis, endophyte | China | KJ955125 | KJ954826 | KJ954405 | KJ955273 | |
| LC3168, LF377 | Camellia sinensis, endophyte | China | KJ955146 | KJ954847 | KJ954424 | KJ955294 | |
| LC3210, LF421 | Camellia sinensis, endophyte | China | KJ955148 | KJ954849 | KJ954426 | KJ955296 | |
| LC3272, LF494 | Camellia sinensis, pathogen | China | KJ955152 | KJ954853 | KJ954429 | KJ955299 | |
| LC3357, LF579 | Camellia sinensis, pathogen | China | KJ955169 | KJ954870 | KJ954443 | KJ955316 | |
| LC3560, LF788 | Camellia sinensis, pathogen | China | KJ955216 | KJ954917 | KJ954484 | KJ955362 | |
| LC3570, CGMCC 3.17359, | Camellia sinensis, pathogen | China | KJ955220 | KJ954921 | KJ954488 | KJ955365 | |
| LF798 | |||||||
| MAFF 305973, ICMP 18598 | Passiflora edulis | Japan | JQ005194 | JQ005281 | JQ005542 | JQ005628 | |
| C. orchidophilum | CBS 632.80* | Dendrobium sp. | USA | JQ948151 | JQ948481 | JQ949472 | JQ949802 |
| C. phormii | CBS 118194* | Phormium sp. | Germany | JQ948446 | JQ948777 | JQ949767 | JQ950097 |
| CBS 199.35 | Phormium sp. | UK | JQ948447 | JQ948778 | JQ949768 | JQ950098 | |
| C. rusci | CBS 119206* | Ruscus sp. | Italy | GU227818 | GU228210 | GU227916 | GU228112 |
| C. spaethianum | CBS 167.49* | Funkia sieboldiana | Germany | GU227807 | GU228199 | GU227905 | GU228101 |
| C. walleri | CBS 125472* | Coffea sp. | Vietnam | JQ948275 | JQ948605 | JQ949596 | JQ949926 |
| C. yunnanense | AS 3.9167, CBS 132135* | Buxus sp. | China | JX546804 | JX546706 | JX519239 | JX519248 |
| Monilochaetes infuscans | CBS 869.96* | Ipomoea batatas | South Africa | JQ005780 | JX546612 | JQ005843 | JQ005864 |
a AS, CGMCC: China General Microbiological Culture Collection; ATCC: American Type Culture Collection; CBS: Culture collection of the Centraalbureau voor Schimmelcultures, Fungal Biodiversity Centre, Utrecht, The Netherlands; ICMP: International Collection of Microorganisms from Plants, Auckland, New Zealand; LC: Working collection of Lei Cai, housed at CAS, China; LF: Working collection of Fang Liu, housed at CAS, China; MAFF: Ministry of Agriculture, Forestry and Fisheries, Tsukuba, Japan.
* = ex-type culture. Strains/sequences studied in this paper are in bold font.
Phylogenetic analyses
Multiple sequence alignments were generated using MAFFT v. 7 (Katoh & Standley 2013), and if necessary, manually edited in MEGA v. 5.1. Bayesian analyses were performed on concatenated alignments using MrBayes v. 3.2.2 (Ronquist et al. 2012) as described by Crous et al. (2006) using nucleotide substitution models that were selected by MrModeltest v. 2.3 (Nylander 2004), with critical values for the topological convergence diagnostic set to 0.01. Maximum likelihood (ML) analyses were implemented using the CIPRES Science Gateway v. 3.3 (www.phylo.org), and the RAxML-HPC BlackBox was selected with default parameters. Six loci (ACT, CAL, GAPDH, GS, ITS, and TUB2) were concatenated for the multi-locus analysis of C. gloeosporioides s.l., while four loci (ACT, GAPDH, ITS, TUB2) were used for the multi-locus analysis of other Colletotrichum species. Due to the lack of available ApMat gene sequences of most of the recently identified Colletotrichum isolates, the ApMat locus could not be included in the concatenated alignment. Therefore, a single ApMat phylogeny was generated including sequences of 136 C. gloeosporioides s.l. isolates obtained from Camellia in this study, and 181 reference sequences that were retrieved from NCBI-GenBank. An additional phylogeny using a concatenated ApMat and GS sequence alignment was constructed which included 126 C. gloeosporioides s.l. isolates from Camellia and 33 reference isolates.
Genealogical concordance phylogenetic species recognition analysis
Phylogenetically related but ambiguous species were analysed using the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) model by performing a pairwise homoplasy index (PHI) test as described by Quaedvlieg et al. (2014). The PHI test was performed in SplitsTree4 (Huson 1998, Huson & Bryant 2006) in order to determine the recombination level within phylogenetically closely related species using a 6-locus concatenated dataset (ACT, CAL, GAPDH, GS, ITS, and TUB2). If the pairwise homoplasy index results were below a 0.05 threshold (Φw < 0.05), it was indicative for significant recombination present in the dataset. The relationship between closely related species was visualised by constructing a splits graph.
Pathogenicity
Koch’s postulates were conducted as described in Cai et al. (2009). Six Colletotrichum isolates were selected for pathogenicity tests: C. camelliae CGMCC 3.14925, C. henanense CGMCC 3.17354, C. jiangxiense CGMCC 3.17362 and CGMCC 3.17363, C. kahawae subsp. kahawae IMI 319418 and IMI 363578. Healthy leaves of intact 2-yr-old tea plants were washed with sterilised water, and then inoculated using the wound/drop and non-wound/drop inoculation methods. Plants inoculated with sterile water were used as control. The inoculated samples were incubated at room temperature in normal light regimes in the greenhouse for 14 d.
RESULTS
Isolates
In total, 144 Colletotrichum isolates were obtained from Camellia tissues from the main tea growing regions in China. Of these, 102 isolates were isolated from diseased tissues, and 42 from asymptomatic tissues (Table 1, 2).
Phylogenetic analyses of the combined datasets
Based on the BLAST search results of the NCBI database with the ITS sequences, all Colletotrichum isolates in this study were preliminarily allocated to species complexes: 141 iso-lates belonged to the C. gloeosporioides species complex, eight isolates belonged to the C. boninense species complex, one isolate belonged to C. acutatum species complex, and one isolate was identified as C. cliviae.
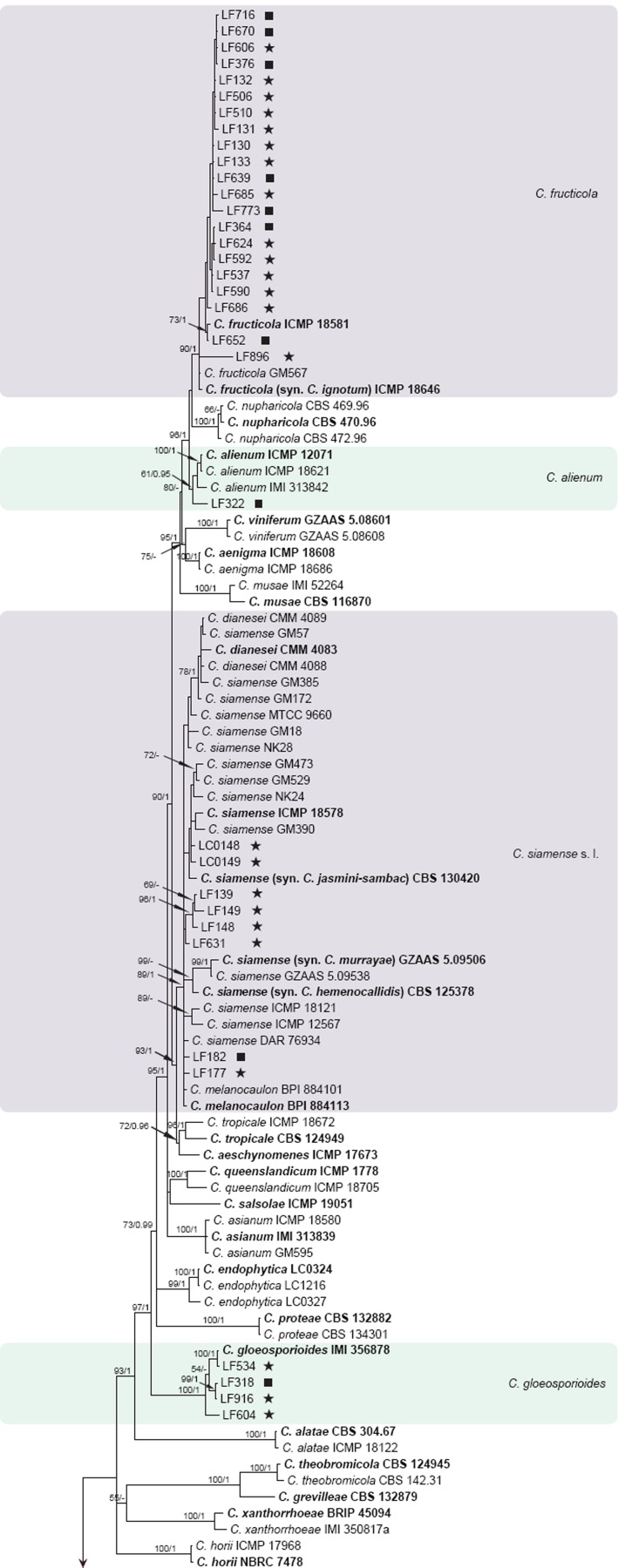
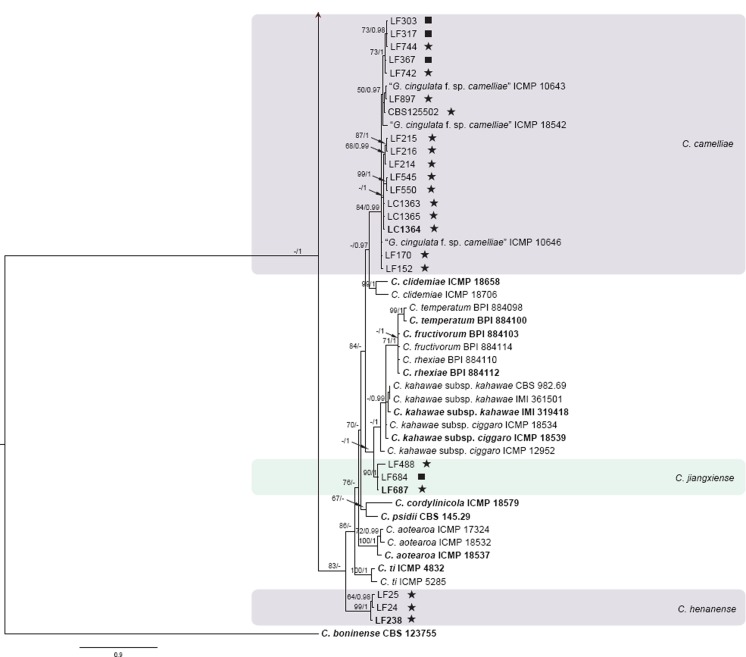
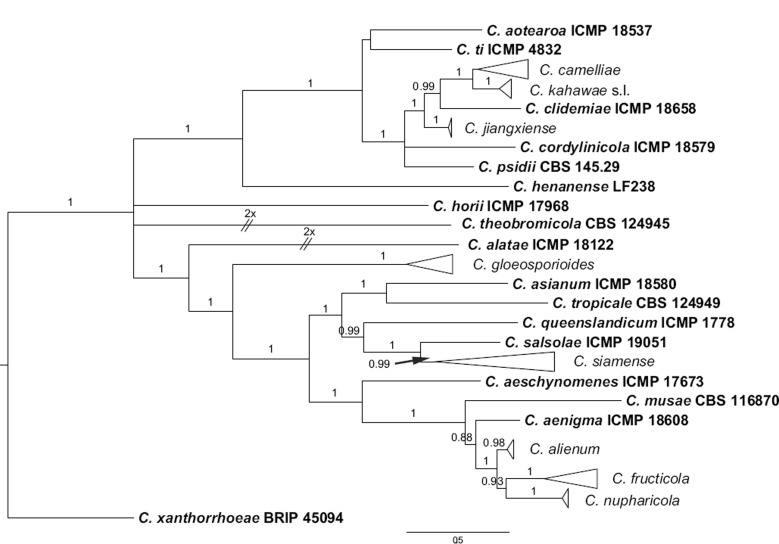
The 6-locus (ACT, CAL, GAPDH, GS, ITS, TUB2) phylogenetic analysis of the C. gloeosporioides species complex included 229 isolates from Camellia and other hosts, with C. boninense (CBS 123755) as the outgroup (see Fig. 1 for a version of this phylogeny with selected identical isolates removed; the complete alignment and tree, as Fig. S1, is available from TreeBASE). The dataset comprised 3 522 characters including the alignment gaps. For the Bayesian inference, a GTR+I+G model with inverse gamma-distributed rate was selected for ACT, HKY+G with gamma-distributed rates for CAL and ITS, GTR+G with gamma-distributed rates for GAPDH, GS, and TUB2. The maximum likelihood tree confirmed the tree topology and posterior probabilities of the Bayesian consensus tree. Isolates from Camellia in the C. gloeosporioides complex clustered in seven clades (data present in TreeBASE as Fig. S1): one Camellia isolate clustered with the ex-type isolate of C. alienum, 32 isolates clustered with C. fructicola, four isolates clustered with C. gloeosporioides, 91 isolates clustered with C. camelliae (syn. Glomerella cingulata ‘f. sp. camelliae’), and eight isolates clustered with the ex-type isolates of C. siamense, C. dianesei, and C. melanocaulon in one clade. Three Camellia isolates formed a distinct clade (posterior probability = 1), most closely related to C. kahawae s.l. A simplified tree was subsequently generated by removing 87 isolates of C. camelliae and C. fructicola (Fig. 1).
Fig. 1.
Fifty percent majority rule consensus tree from a Bayesian analysis based on a 6-gene combined dataset (ACT, CAL, GAPDH, GS, ITS, TUB2) showing phylogenetic affinities of a reduced set of Colletotrichum isolates from Camellia isolated in this study with species of the C. gloeosporioides species complex. The RAxML bootstrap support values (ML > 50) and Bayesian posterior probabilities (PP > 0.95) are displayed at the nodes (ML/PP). The tree was rooted to C. boninense (CBS 123755). The scale bar indicates 0.9 expected changes per site. Ex-type cultures are emphasised in bold, and include the taxonomic name as originally described. Coloured blocks are used to indicate clades containing Chinese isolates from Camellia; stars indicate pathogens, squares indicate endophytes.
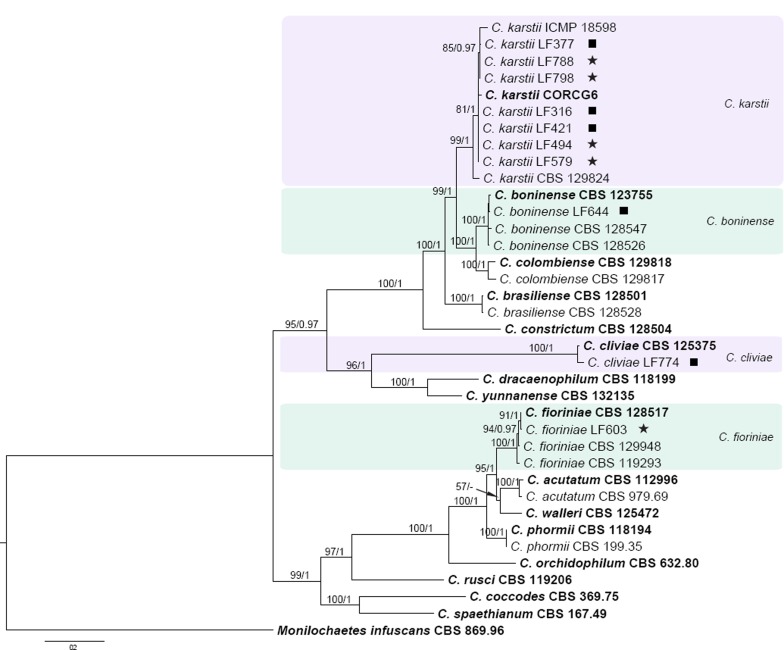
Fig. 2 shows the identity of the Camellia isolates that fell outside of the C. gloeosporioides species complex. The concatenated alignment (ACT, GAPDH, ITS, TUB2) contained 37 isolates, with Monilochaetes infuscans (CBS 869.96) as outgroup. The dataset comprised 1 559 characters including the alignment gaps. For the Bayesian inference, a HKY+G model with gamma-distributed rate was selected for ACT, HKY+I+G with inverse gamma-distributed rate for GAPDH, GTR+I+G with inverse gamma-distributed rates for ITS and TUB2. The maximum likelihood tree confirmed the tree topology and posterior probabilities of the Bayesian consensus tree. Seven Camellia isolates clustered with the ex-type isolate of C. karstii, one isolate clustered with C. boninense, one isolate clustered with C. fioriniae and one isolate clustered with C. cliviae.
Fig. 2.
Fifty percent majority rule consensus tree from a Bayesian analysis based on a 4-gene combined dataset (ITS, GAPDH, ACT, TUB2) showing phylogenetic affinities of Colletotrichum isolates from Camellia with members of the Colletotrichum species outside of the C. gloeosporioides species complex. The RAxML bootstrap support values (ML > 50) and Bayesian posterior probabilities (PP > 0.95) are displayed at the nodes (ML/PP). The tree was rooted to Monilochaetes infuscans (CBS 869.96). The scale bar indicates 0.2 expected changes per site. Ex-type cultures are emphasised in bold. Coloured blocks are used to indicate clades containing Chinese isolates from Camellia; stars indicate pathogens, squares indicate endophytes.
The pathogenic and endophytic isolates of Colletotrichum studied here were labelled with stars and squares, respectively, on the multi-locus phylogenetic trees (Fig. 1, 2). Isolates from symptomatic Camellia leaves belong to eight clades, representing C. camelliae, C. fioriniae, C. fructicola, C. gloeosporioides, C. henanense, C. jiangxiense, C. karstii, and C. siamense. Isolates from asymptomatic tissues belong to nine clades representing C. alienum, C. boninense, C. camelliae, C. cliviae, C. fructicola, C. gloeosporioides, C. henanense, C. karstii, and C. siamense.
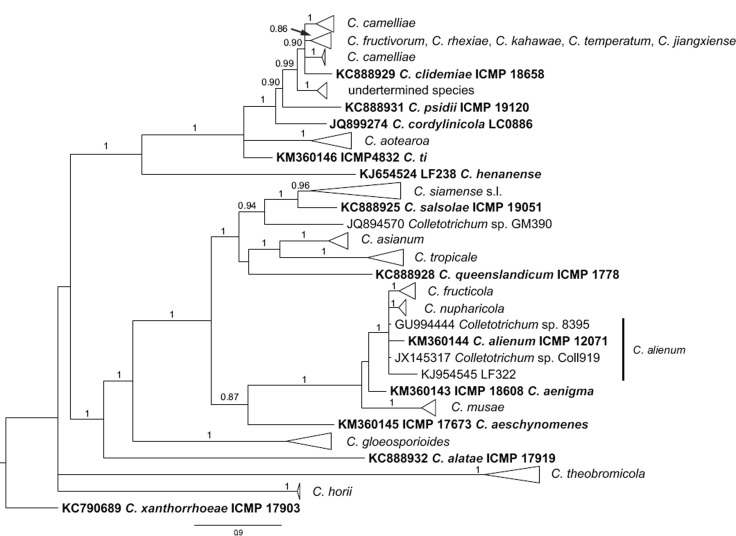
ApMat-based phylogenetic analysis
The phylogenetic analysis of the C. gloeosporioides species complex using the ApMat locus included 317 isolates from Camellia and other hosts (rooted with C. xanthorrhoeae), and 785 characters with alignment gaps were involved in the dataset. All isolates included in this analysis were separated into 15 main clades and 12 single-isolate lineages (see Fig. 3 for a cartoon version of this phylogeny; the complete alignment and tree, as Fig. S2, is available from TreeBASE). One of the clades is represented by an assemblage of more than one species, including C. fructivorum, C. jiangxiense, C. kahawae, C. rhexiae, and C. temperatum (Fig. 3, S2). Of these five species, C. fructivorum, C. rhexiae, and C. temperatum formed monophyletic species clades. However, strains from C. jiangxiense and C. kahawae were intermingled in one clade and the two species could not be differentiated from each other. The C. camelliae isolates were separated into two distinct clades, while the other species formed monophyletic clades.
Fig. 3.
Collapsed cartoon of the 50 % majority rule consensus tree from a Bayesian analysis based on the ApMat dataset showing phylogenetic affinities of Colletotrichum isolates from Camellia with members of the C. gloeosporioides species complex. Bayesian posterior probabilities values are displayed at the node. The tree was rooted to C. xanthorrhoeae (ICMP 17903). The scale bar indicates 0.6 expected changes per site. Ex-type cultures are emphasised in bold, and include the taxonomic name as originally described.
ApMat & GS-based phylogenetic analysis
Colletotrichum jiangxiense and C. kahawae subsp. kahawae cannot be separated on the basis of the ApMat locus. They are mainly distinguished from one another based on the GS gene (see also notes under C. jiangxiense); the two species formed distinct clades in the GS gene phylogeny (not shown). The potential of the concatenated ApMat and GS genes to serve as a barcode for the C. gloeosporioides species complex was demonstrated by re-constructing a phylogenetic tree using the sequences listed in Table 1 (Fig. 4). All species of the C. gloeosporioides species complex included in the analysis could be delimited clearly based on the concatenated ApMat & GS gene tree.
Fig. 4.
Collapsed cartoon of the 50 % majority rule consensus tree from a Bayesian analysis based on the combined ApMat and GS alignment showing phylogenetic affinities of Colletotrichum isolates from Camellia with species of the C. gloeosporioides species complex. Bayesian posterior probabilities values are displayed at the node. The tree was rooted to C. xanthorrhoeae (ICMP 17903). The scale bar indicates 0.5 expected changes per site. Ex-type cultures are emphasised in bold. Extremely long branches were halved in length (indicated with 2× above two diagonal lines) to better fit the tree to the page.
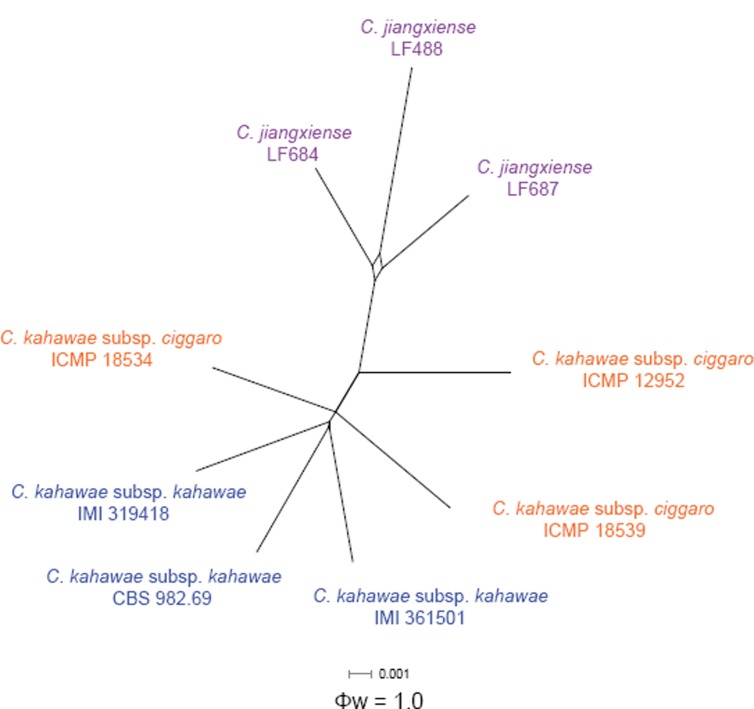
Pairwise homoplasy index (PHI) test
A pairwise homoplasy index (PHI) test using a 6-gene dataset (ACT, CAL, GAPDH, GS, ITS, TUB2) was further performed to determine the recombination level between C. jiangxiense and its phylogenetically closely related species, C. kahawae subsp. ciggaro and C. kahawae subsp. kahawae. Based on the result no significant recombination events could be detected between C. kahawae s.l. and C. jiangxiense (Φw = 1) (Fig. 5).
Fig. 5.
The result of the pairwise homoplasy index (PHI) test of closely related species using both LogDet transformation and splits decomposition. PHI test results (Φw) < 0.05 indicate significant recombination within the dataset.
Pathogenicity
The tea plant leaves inoculated with a conidial suspension of Colletotrichum isolates from symptomatic tea leaves (C. camelliae CGMCC 3.14925, C. henanense CGMCC 3.17354, C. jiangxiense CGMCC 3.17363) developed typically brown lesions around the leaf wounds after 14 d (Fig. 6). The inoculated Colletotrichum isolates could be re-isolated from the periphery of these lesions, thereby fulfilling Koch’s postulates. Leaves of the control plants were inoculated with sterile water, and leaves inoculated with isolates of C. kahawae subsp. kahawae did not develop any symptoms after 14 d past inoculation (Fig. 6).
Fig. 6.
Pathogenicity test of selected isolates on tea plant leaves after 14 d. a. C. jiangxiense (CGMCC 3.17363); b, c. C. henanense (CGMCC 3.17354); d. C. kahawae subsp. kahawae (IMI 363578); e. C. camelliae (CGMCC 3.14925); f. control.
Taxonomy
Based on the multi-locus phylogenies (Fig. 1–4 and Fig. S1, S2 in TreeBASE), the 151 Colletotrichum isolates from Camellia sinensis and other Camellia spp. belonged to 11 species, including two species that proved to be new to science.
Colletotrichum alienum B. Weir & P.R. Johnst, Stud. Mycol. 73: 139. 2012
Description and illustrations — See Weir et al. (2012) and Liu et al. (2013b).
Material examined. CHINA, Jiangxi Province, Ganzhou, Yangling National Forest Park, on living leaf of Ca. sinensis, Apr. 2013, F. Liu, culture CGMCC 3.17355 = LC3114 = LF322.
Notes — Colletotrichum alienum was previously only known from Australia, New Zealand, Portugal, and South Africa (Weir et al. 2012, Liu et al. 2013b). In the present study, one endophytic isolate CGMCC 3.17355 from a tea leaf clustered together with the ex-type culture of C. alienum (ICMP 12071) in the multi-locus phylogenetic tree (Fig. 1); this is the first reported occurrence of C. alienum on Ca. sinensis and in China.
Both conidia and ascospores of the tea isolate (CGMCC 3.17355) are slightly shorter than that of the ex-type (ICMP 12071) of C. alienum (conidia 14.5 × 4.6 μm vs 16.5 × 5 μm, ascospores 16.3 × 4.4 μm vs 18.1 × 4.6 μm; Weir et al. 2012).
Colletotrichum boninense Moriwaki, Toy. Sato & Tsukib., Mycoscience 44: 48. 2003
Description and illustrations — See Moriwaki et al. (2003) and Damm et al. (2012b).
Material examined. CHINA, Jiangxi Province, Ganzhou, Fengshan Mountain, on living leaf of Ca. sinensis, Sept. 2013, Y. Zhang, culture CGMCC 3.14356 = LC3422 = LF644.
Notes — The endophytic isolate (LF644) from a tea leaf eva-luated in this study was identified as C. boninense based on the multi-locus phylogenetic analyses (Fig. 2). This species was previously reported on Camellia sp. from New Zealand (Damm et al. 2012b).
Conidia of the tea isolate (CGMCC 3.14356) on PDA are wider, and the L/W ratio is smaller than that of the ex-type culture (CBS 123755) of C. boninense on Anthriscus stem and SNA (CGMCC 3.14356: 10–15 × 6.5–8 μm, mean = 13.7 × 7.3 μm, L/W ratio = 1.9 vs CBS 123755: on Anthriscus stem (9–)12–14.5(–16.5) × (4–)5.5–6.5 μm, av = 13.2 × 5.8 μm, L/W ratio = 2.3, on SNA (8.5–)11–14.5(–17.5) × (4–)5–6(–6.5) μm, av = 12.8 × 5.4 μm, L/W ratio = 2.4). Conidia of CBS 123755 often contain two large polar guttules, which were absent in the tea isolate.
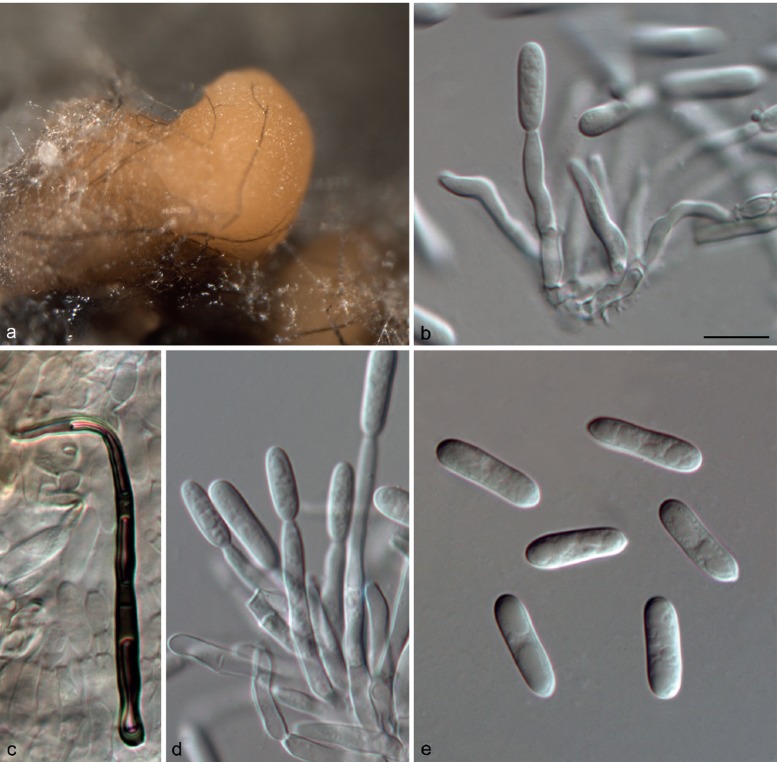
Colletotrichum camelliae Massee, Bull. Misc. Inform. Kew 1899: 91. 1899. — Fig. 7
Fig. 7.
Colletotrichum camelliae (CGMCC 3.14925). a. Symptom on tea leaf; b, c. forward and reverse view of culture 7 d after inoculation; d. conidiophores; e, f, i. conidia; g, h. appressoria (b–f, i from PDA; g, h from SNA). — Scale bar: d–i = 10 μm.
= Glomerella cingulata ‘f. sp. camelliae’ Dickens & R.T.A. Cook, Pl. Pathol. 38: 85. 1989.
On PDA: Colonies 69–71 mm diam in 7 d, > 90 mm diam in 10 d, flat with entire edge, aerial mycelium white, cottony, sparse; reverse white at first, then grey to black at the centre. Conidiomata not observed, conidiophores formed directly on aerial mycelium, hyaline, septate. Conidiogenous cells hyaline, cylindrical, 16–42 × 1.5–4.5 μm. Conidia hyaline, smooth-walled, guttulate, cylindrical with obtuse ends, sometimes narrowed at the centre or towards the base, 9–25 × 3.5–7.5 μm, av ± SD = 15.5 ± 3.3 × 5.0 ± 0.9 μm, L/W ratio = 3.1. Appressoria irregularly shaped, clavate, crenate, lobed, brown to dark brown, solitary, branched, catenate, with age sometimes complex chlamydospore-like structures develop, 6.5–13.5 × 5.0–10.5 μm, av ± SD = 10.0 ± 1.8 × 7.5 ± 1.3, L/W ratio = 1.3.
Materials examined. CHINA, Fujian Province, Zhangzhou, on Ca. sinensis, Nov. 2012, L. Cai, culture LF214; Guizhou Province, Huishui District, on Ca. sinensis, 11 Nov. 2010, P. Tan (HMAS 243126 epitype designated here MBT178292, culture ex-epitype CGMCC 3.14925 = LC1364); ibid., HMAS 243127, culture CGMCC 3.14924 = LC1363; ibid., HMAS 243128, culture CGMCC 3.14926 = LC1365; Jiangxi Province, Yangling National Forest Park, on Ca. sinensis, Apr. 2013, F. Liu, culture LC3095 = LF303; ibid., culture LC3109 = LF317. – Sri Lanka, on leaves of Camellia sp., 8 Apr. 1899, J.C. Willis, K(M) 173540 holotype. – USA, South Carolina, on Ca. sasanqua, 1982, unknown collector, culture LC3668 = LF898 = ICMP 10646.
Notes — To our knowledge, the earliest known record of tea anthracnose was described in 1899 by Massee (in Willis 1899) from living leaves of Ca. sinensis from Sri Lanka. The holotype sample is preserved in K(M) 173540 and labelled C. camelliae (Fig. 8). Although it was subsequently synonymised with C. gloeosporioides (von Arx 1957), the name C. camelliae is still widely used in fungaria, websites, trade and semi-popular literature as the causal agent of the brown blight disease of tea plants (Weir et al. 2012). In 1989, Glomerella cingulata ‘f. sp. camelliae’ was proposed as the causal agent of disease on ornamental Ca. saluenensis hybrids, but without distinguishable morphological characteristics compared to G. cingulata (Dickens & Cook 1989). Weir et al. (2012) revealed G. cingulata ‘f. sp. camelliae’ to belong to the C. gloeosporioides complex. However, due to the lack of an ex-type culture of C. camelliae, the genetic relationship between C. camelliae and G. cingulata ‘f. sp. camelliae’ remained unresolved.
Fig. 8.
Holotype of C. camelliae (K (M) 173540). a. Label of the specimen; b. tea leaf with C. camelliae colonisation from above and below; c–g. conidia. — Scale bars: c–g = 10 μm.
We evaluated the holotype specimen of C. camelliae from K, but very few morphological characters could be observed on this old specimen, and DNA extraction was unsuccessful. Conidia on the holotype specimen are hyaline and cylindrical (Fig. 8), 14.5–20 × 4–6 μm, av ± SD = 17.2 ± 1.2 × 4.9 ± 0.4 μm. Conidial dimensions of isolates in this study on PDA (9–25 × 3.5–7.5 μm, av ± SD = 15.5 ± 3.3 × 5.0 ± 0.9 μm) are in accordance with the holotype specimen.
Several efforts to obtain a fresh culture from tea plants from Sri Lanka, the original location from where C. camelliae was reported, proved to be unsuccessful. However, we collected many anthracnose diseased samples in the tea fields from different provinces in China. Leaf lesions were dark brown and circular at first, then enlarged to become more irregular, with many of the lesions coalescing; raised black circular masses were found at the centre of lesions, bordered by a discoloured margin (Fig. 7a). Isolates from these samples clustered together with authentic isolates of G. cingulata ‘f. sp. camelliae’ (cited by Dickens & Cook 1989) in the 6-gene and ApMat phylogenetic trees (Fig. 1 and Fig. S2 in TreeBASE). Inoculations using conidial suspensions were performed on tea plants under controlled environmental conditions to test whether this fungus was the causal agent of tea anthracnose disease. The inoculations resulted in leaf infection of Ca. sinensis consistent with the original natural infections. Re-isolation and re-sequencing confirmed that the culture was identical to the one used for inoculation. No symptoms were produced in the negative control plants. A pathogenicity test with isolates of G. cingulata ‘f. sp. camelliae’ from ornamental Camellia on detached tea (Ca. sinensis) leaves was performed by Weir et al. (2012) and the isolates proved to be highly virulent. The Colletotrichum isolates from tea brown blight symptoms from India, showing affinities to G. cingulata ‘f. sp. camelliae’, were also pathogenic to detached tea leaves (Sharma et al. 2014). All the tests and analyses demonstrated that the isolates collected from typical brown blight symptoms on tea in the field and those from ornamental varieties are the same species. Since C. camelliae was published earlier than G. cingulata ‘f. sp. camelliae’ (1899 vs 1989), and there is no nomenclatural priority for formae speciales (Art. 4, http://www.iapt-taxon.org/nomen/main.php?page=art4), the name C. camelliae is adopted for the anthracnose pathogen of tea and is epitypified in this study, and G. cingulata ‘f. sp. camelliae’ is synonymised with C. camelliae.
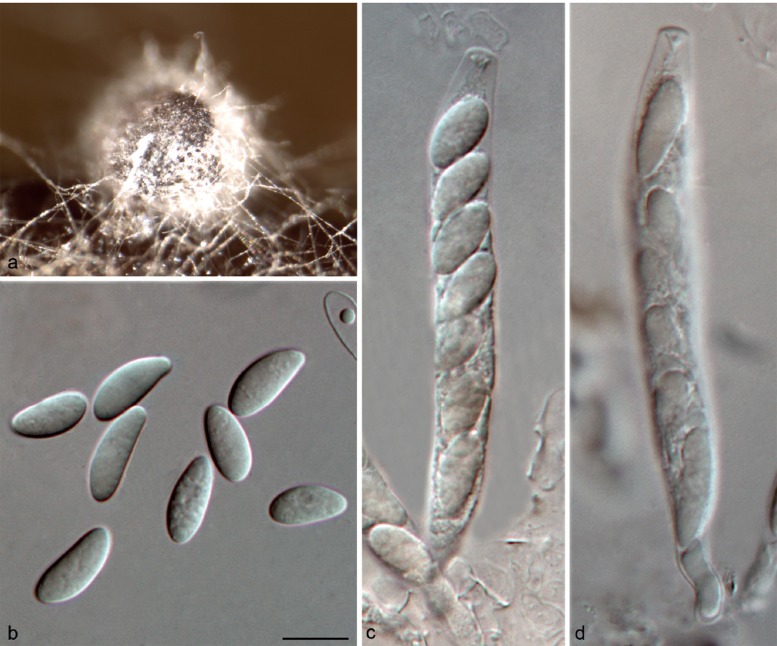
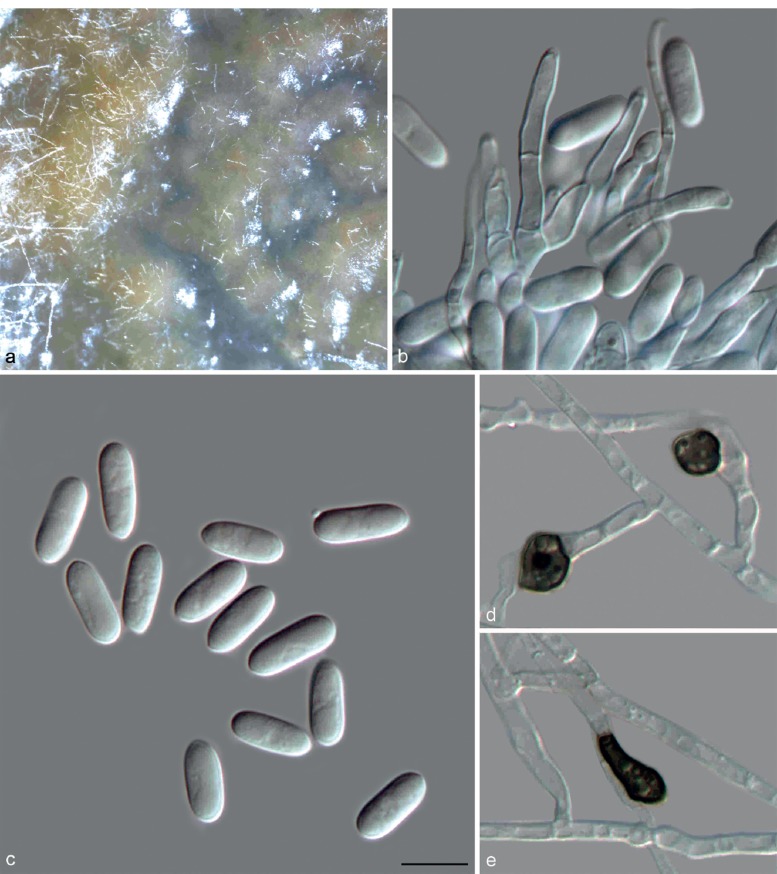
Colletotrichum cliviae Y.L. Yang et al., Fung. Diversity 39: 133. 2009 — Fig. 9
Fig. 9.
Colletotrichum cliviae on Anthriscus stem (CGMCC 3.17358). a. Ascomata; b. ascospores; c, d. asci and ascospores. — Scale bar: b = 10 μm, scale bar of b applies to b–d.
On PDA: Colonies 65–69 mm diam in 7 d, > 90 mm diam in 10 d, flat with entire edge. Cultures on PDA and SNA are sterile, but a sexual morph developed on Anthriscus stem. Ascomata glo-bose, brown to black, covered by sparse and white aerial mycelium, outer wall composed of flattened angular cells. Asci cylindrical, 62–92 × 8–12 μm, 8-spored. Ascospores uni- or biseriately arranged, hyaline, aseptate, smooth-walled, allantoid, ellipsoidal or ovoid with rounded ends, 11–16.5 × 4–6.5 μm, av ± SD = 13.8 ± 1.6 × 5.8 ± 0.5 μm, L/W ratio = 2.4. No asexual morph was observed in this study. Yang et al. (2009) provided a description of the asexual morph of this species.
Material examined. CHINA, Guangxi Province, Guilin, on living leaf of Ca. sinensis, Sept. 2013, T.W. Hou, culture CGMCC 3.17358 = LC3546 = LF774.
Notes — Colletotrichum cliviae was reported to cause anthracnose diseases on Clivia miniata, Arundina graminifolia and Cymbidium hookerianum in China (Yang et al. 2009, 2011). The host range was recently extended to include Cattleya, Calamus thwaitesii, Phaseolus, and Saccharum (Sharma et al. 2013b). In the present study, a single isolate (CGMCC 3.17358) of Colletotrichum from a healthy tea leaf proved to belong to C. cliviae, but the asexual morph was not observed. Conversely, this is the first report of a sexual morph of C. cliviae, and the first report of this species on Ca. sinensis.
Colletotrichum fioriniae (Marcelino & Gouli) R.G. Shivas & Y.P. Tan, Fung. Diversity 39: 117. 2009
Basionym. Colletotrichum acutatum var. fioriniae Marcelino & Gouli, Myco-logia 100: 362. 2008.
Description and illustration — See Damm et al. (2012a).
Materials examined. CHINA, Jiangxi Province, Ganzhou, Yangling National Forest Park, on Ca. sinensis, Apr. 2013, F. Liu, culture CGMCC 3.17357 = LC3381 = LF603.
Notes — Colletotrichum fioriniae was previously reported from Ca. reticulata in Kunming, Yunnan Province and from Ca. sinensis in Fujian Province in China (Damm et al. 2012a, Liu 2013).
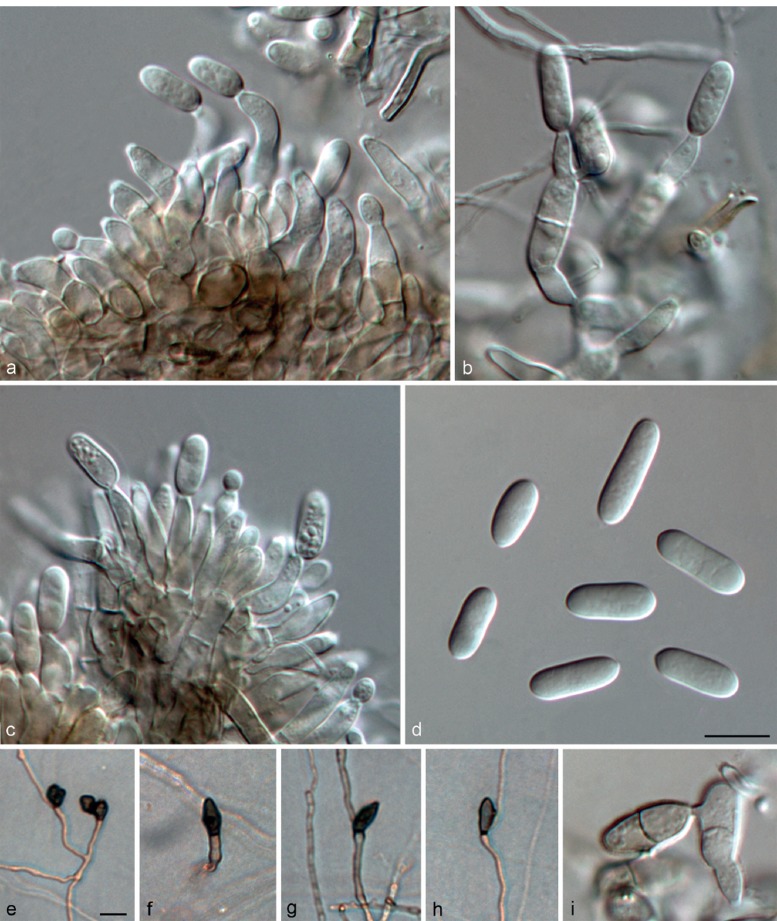
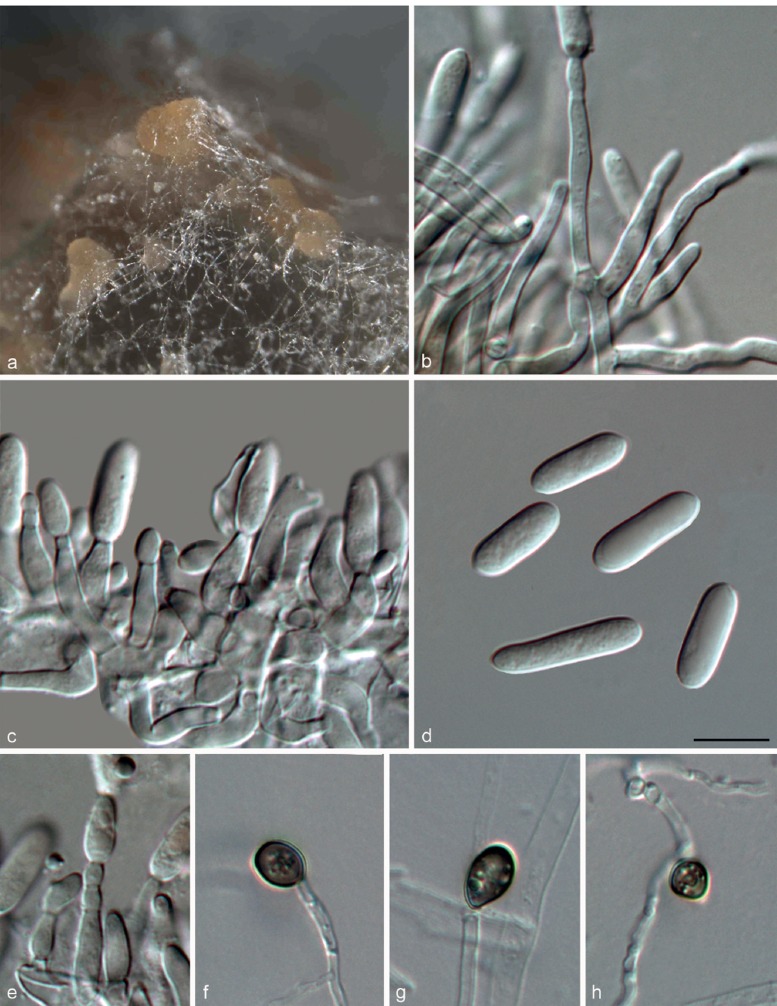
Colletotrichum fructicola Prihast., L. Cai & K.D. Hyde, Fung. Diversity 39: 158. 2009 — Fig. 10
Fig. 10.
Colletotrichum fructicola on PDA (a, b, d, e from LC2923; c from LC3451). a. Acervulus; b, d. conidiophores; c. seta; e. conidia. — Scale bar: b = 10 μm, scale bar of b applies to b–e.
On PDA: Colonies 74–79 mm diam in 7 d, > 90 mm diam in 10 d, flat with entire edge, aerial mycelium dense, cottony, grey to dark grey in the centre, white at the margin; reverse greyish green with white halo. Chlamydospores not observed. Conidiomata acervular, only one seta was observed, brown, smooth-walled, 1-septate, 64 μm long, base inflated, 4 μm diam, tip more or less acute. Conidiophores hyaline, septate, branched. Conidiogenous cells hyaline, cylindrical or ampulliform, 7.5–18.5 μm, apex 1–3 μm diam. Conidia hyaline, aseptate, smooth-walled, cylindrical, both ends rounded, 11.5–17.5 × 3–5.5 μm, av ± SD = 14.9 ± 1.3 × 4.4 ± 0.4 μm, L/W ratio = 3.4. Appressoria not observed.
Materials examined. CHINA, Guangxi Province, Guilin, on Ca. sinensis, Sept. 2013, T.W. Hou, culture LC3545 = LF773; ibid., culture LC3489 = LF716; Hangzhou, on Ca. sinensis, Oct. 2013, F. Liu, culture LC3569 = LF797; on Ca. sinensis, Sept. 2012, L. Cai, culture CGMCC 3.17352 = LC2923 = LF130; Jiangxi Province, Ganzhou, Fengshan Mountain, on Ca. sinensis, Sept. 2013, Y. Zhang, culture LC3462 = LF686; ibid., culture LC3451 = LF674; Yangling National Forest Park, on Ca. sinensis, Apr. 2013, F. Liu, culture LC3284 = LF506. – Indonesia, on Ca. sinensis, Jan. 1979, H. Semangun, culture LC3666 = LF896 = ICMP 18656. UK, on a shipment of Camellia flowers from New Zealand, on Camellia sp., 1982, staff of Ministry of Agriculture, Fisheries & Food, culture LC3670 = LF900 = ICMP 10642.
Notes — This study supplements the morphological characteristics of setae of C. fructicola that were not observed in the previous studies. Colletotrichum fructicola was reported to cause anthracnose diseases on several varieties of Ca. sinensis in many regions in Fujian Province, China (Liu 2013). In the present study, the species was found to be widely distributed throughout China, although there appears to be some variation in sequence data among isolates from Ca. sinensis. Conidia of the tea isolates (LC2923, av = 14.9 × 4.4 μm and LC3451, av = 15.03 × 4.35 μm) are longer than that of the ex-type (MFLU 090228, av = 11.53 × 3.55) of C. fructicola.
Colletotrichum gloeosporioides (Penz.) Penz. & Sacc., Atti Reale Ist. Veneto Sci. Lett. Arti., ser. 6, 2: 670. 1884 — Fig. 11
Fig. 11.
Colletotrichum gloeosporioides (LC3686). a. Acervulus; b. conidiophores; c. conidia; d, e. appressoria (a–c from PDA; d, e from SNA). — Scale bar: c = 10 μm, scale bar of c applies to b–e.
Basionym. Vermicularia gloeosporioides Penz., Michelia 2: 450. 1882.
On PDA: Colonies 56–58 mm diam in 7 d, > 90 mm diam in 10 d, flat with erose edge, scattered acervuli with orange conidial ooze near centre, fuscous black pigment near the edge; reverse honey with fuscous black near the edge. Chlamydospores not observed. Conidiomata acervular, conidiophores formed on a cushion of roundish and medium brown cells. Setae not observed. Conidiophores hyaline to pale brown, septate, branched. Conidiogenous cells hyaline, cylindrical to ampulliform, 5.5–17.5 μm, apex 1–2 μm diam. Conidia hyaline, aseptate, smooth-walled, cylindrical, both ends bluntly rounded, 11–15.5 × 4.5–6 μm, av ± SD = 13.5 ± 1.2 × 5.5 ± 0.3 μm, L/W ratio = 2.5. Appressoria medium to dark brown, aseptate, solitary or in groups, variable in shape, circular, clavate, ellipsoidal or irregular in outline, crenate or slightly lobed at edge, 7.5–13.5 × 5–9.5 μm, av ± SD = 9.5 ± 1.4 × 6.5 ± 0.9 μm, L/W ratio = 1.5.
Materials examined. CHINA, Jiangxi Province, on Ca. sinensis, Sept. 2013, Y.H. Gao, culture CGMCC 3.17360 = LC3686 = LF916; Ganzhou, Yangling National Forest Park, on Ca. sinensis, Apr. 2013, F. Liu, culture LC3110 = LF318; ibid., culture LC3312 = LF534; ibid., culture LC3382 = LF604.
Notes — Colletotrichum gloeosporioides is listed as a pathogen of Camellia in Australia, Brazil, China, Hong Kong, Japan, and the USA (Farr & Rossman 2014). However, many of these reports probably refer to this species in its broader sense as a species complex and need to be further verified (Watson 1950, Shivas 1989, Osono 2008, Guo et al. 2014). For example, the anthracnose pathogen C. gloeosporioides was recently detected in 30–60 % of the Ca. sinensis fields in the Yellow Mountain region in China during 2011 to 2012 (Guo et al. 2014), the identification of which, however, was solely based on morphology and NCBI BLAST searches with ITS sequences, and was not based on the presently accepted classification system in Colletotrichum (Cannon et al. 2012). Colletotrichum gloeosporioides was also considered to be one of the dominant endophytic taxa of Camellia in the study of Fang et al. (2013) based on ITS analysis, the identification of which needs to be verified by multi-locus analysis. In our investigation, four isolates of C. gloeosporioides were associated with Camellia, confirming this species to occur on this host. However, C. gloeosporioides is not the dominant Colletotrichum species on Camellia spp. at the localities where we sampled.
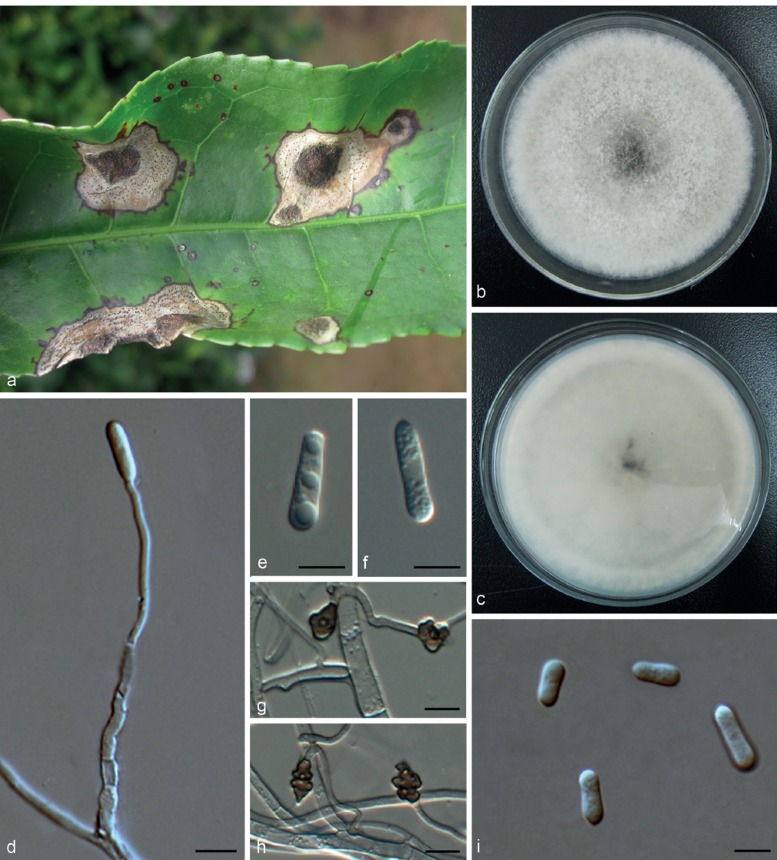
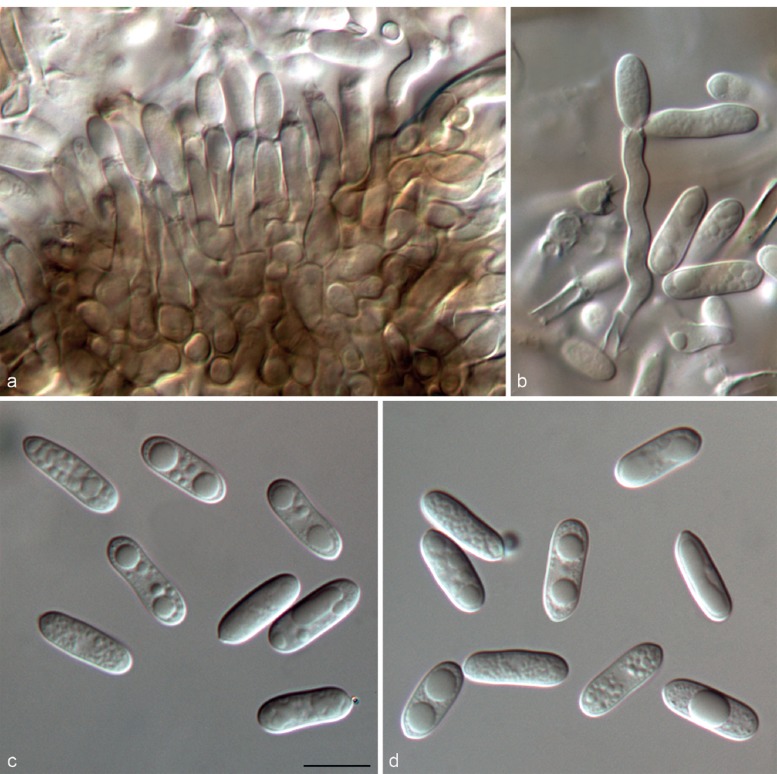
Colletotrichum henanense F. Liu & L. Cai, sp. nov. — MycoBank MB809160; Fig. 12
Fig. 12.
Colletotrichum henanense (CGMCC 3.17354). a–c. Conidiophores; d, i. conidia; e–h. appressoria (a–d, i from PDA; e–h from SNA). — Scale bars: d, e = 10 μm, scale bar of d applies to a–d, i; scale bar of e applies to e–h.
Etymology. Named after the collection site, Henan province, China.
On PDA: Colonies 53–59 mm diam in 7 d, > 90 mm diam in 10 d, aerial mycelium pale olivaceous-grey to olivaceous-grey; reverse sulphur-yellow to straw with pale olivaceous-grey to iron-grey in the centre. Chlamydospores not observed. Conidiomata acervular, conidiophores formed on a cushion of roundish and medium brown cells. Setae not observed. Conidiophores hyaline to pale brown, septate, branched. Conidiogenous cells hyaline to pale brown, cylindrical to ovoid or ampulliform, 5.5–12.5 μm, apex 1–2 μm diam. Conidia hya-line, usually aseptate, sometimes becoming 1-septate with age, smooth-walled, cylindrical, both ends obtusely rounded, contents sometimes with guttulae, 8–17 × 3–5.5 μm, av ± SD = 12.5 ± 1.8 × 4.5 ± 0.6 μm, L/W ratio = 2.8. Appressoria single or in small groups, medium brown, outline mostly clavate or elliptical, rarely lobate, 7–14.5 × 5–9 μm, av ± SD = 11.2 ± 3.7 × 6.7 ± 2 μm, L/W ratio = 1.7.
Materials examined. CHINA, Henan Province, Xinyang, on Ca. sinensis, 23 Sept. 2012, M. Zhang & R. Zang (holotype HMAS 245381, culture ex-type CGMCC 3.17354 = LC3030 = LF238 = CSBX001); Beijing, Water Great Wall, on Cirsium japonicum, 2010, L. Cai, culture LC2820 = LF24; ibid., culture LC2821 = LF25.
Notes — The isolates of C. henanense isolated from tea plants and Cirsium japonicum formed a distinct clade that could be clearly distinguished from other species in the C. gloeo-sporioides species complex (Fig. 1). A BLASTn search of NCBI GenBank with the ITS sequence of CGMCC 3.17354 showed 99 % similarity to quite a number of sequences from isolates previously identified as C. gloeosporioides in other studies. The closest match in a BLASTn search in GenBank with the GAPDH sequence of CGMCC 3.17354 was GenBank JX009967 (99 % identity, 3 bp differences), the sequence generated from an authentic isolate of C. psidii CBS 145.29 (Weir et al. 2012), and with 98 % identity (5–6 bp differences) to some sequences of C. aotearoa, C. ti, and Glomerella cingulata ‘f. sp. camelliae’ isolates (Weir et al. 2012). The top 10 closest matches with the TUB2 sequence (with 97 % identity, 20–23 bp differences) were the isolates of C. aotearoa and C. kahawae subsp. ciggaro analysed in the study of Weir et al. (2012).
Colletotrichum jiangxiense F. Liu & L. Cai, sp. nov. — MycoBank MB809161; Fig. 13
Fig. 13.
Colletotrichum jiangxiense on PDA (CGMCC 3.17363). a, b. Conidiophores; c, d. conidia. — Scale bar: c = 10 μm, scale bar of c applies to a–d.
Etymology. Named after the collection site, Jiangxi Province, China.
On PDA: Colonies 50–53 mm diam in 7 d, > 90 mm diam in 10 d, flat with entire edge, aerial mycelium dense, cottony, white to grey, numerous small acervuli with orange conidial masses near the margin; reverse olivaceous with pale orange near the margin. Appressoria-like structures pale brown to brown, circular, ellipsoidal or irregular. Conidiomata acervular, conidiophores formed on a cushion of roundish and medium brown cells. Setae not observed. Conidiophores hyaline to pale brown, branched. Conidiogenous cells hyaline to pale brown, cylindrical, 11.5–20 μm, apex 1–2.5 μm diam. Conidia hyaline, aseptate, smooth-walled, cylindrical, both ends bluntly rounded, or one end bluntly rounded and one end acutely rounded, 13–19 × 4–6 μm, av ± SD = 15.2 ± 1.0 × 5.2 ± 0.4 μm, L/W ratio = 2.9. Appressoria not observed.
Materials examined. CHINA, Jiangxi Province, Ganzhou, Fengshan Mountain, on Ca. sinensis, Sept. 2013, Y. Zhang (holotype HMAS 245382, culture ex-type CGMCC 3.17363 = LC3463 = LF687); ibid., culture CGMCC 3.17362 = LC3460 = LF684; Yangling National Forest Park, on Ca. sinensis, Apr. 2013, F. Liu, culture CGMCC 3.17361 = LC3266 = LF488.
Notes — Based on multi-locus sequence data (ACT, CAL, GAPDH, GS, ITS, TUB2), C. jiangxiense is phylogenetically closely related to the devastating coffee berry pathogen C. kahawae subsp. kahawae, and up to four other taxa, namely C. kahawae subsp. ciggaro, C. temperatum, C. fructivorum, and C. rhexiae (Fig. 1). All of the C. jiangxiense isolates differ from both C. kahawae subsp. kahawae and C. kahawae subsp. ciggaro by 1 bp change in CAL, 2 bp changes in ITS, and 17 bp changes and 1 bp indel in GS. Additionally, the 22 bp deletion in the GS sequence used to distinguish C. kahawae subsp. ciggaro from C. kahawae subsp. kahawae (Weir et al. 2012) is also lacking in the sequences of the C. jiangxiense isolates. Phylogenetic analyses based on single genes (except GS) could not clearly separate C. jiangxiense from the above listed species (results not shown). Comparisons of morphological and ecological characteristics were also made between these species. Conidia of the tea isolate (CGMCC 3.17363, av = 15.2 × 5.2 μm) are shorter than that of the ex-type culture (ICMP 18539, av = 17.8 × 5.1) of C. kahawae subsp. ciggaro. Colletotrichum kahawae subsp. kahawae is host-specific to Coffea and was confirmed causing no disease symptoms on Camellia sinensis by cross infection experiments (Fig. 6). In conclusion, the pathogenicity test, PHI test (Φw = 1) and phylogenetic analyses all suggested that C. jiangxiense is distinct from C. ka- hawae s.l.
The closest match in a BLASTn search with the ITS sequences of CGMCC 3.17363 was GenBank JN715848 (with 100 % iden-tity) from isolate R046 from a fruit of Rubus glaucus in Colombia, which was identified as C. kahawae subsp. ciggaro (Afanador-Kafuri et al. unpubl. data). Closest matches with the TUB2 sequence were GenBank KC297083 and KC297082 (with 100 % identity) from isolate CBS 115194 and CBS 112984 from Banksia sp., both of which are C. kahawae subsp. ciggaro (Liu et al. 2013b). The GAPDH blast result showed that the sequence of CGMCC 3.17363 was identical to those of the C. kahawae subsp. ciggaro isolates ICMP 18534 (GenBank JX009904) and ICMP 18544 (GenBank JX009920) (Weir et al. 2012), while CGMCC 3.17363 could be distinguished from ICMP 18534 in the multi-locus tree (Fig. 1).
Colletotrichum karstii Y.L. Yang et al., Cryptog. Mycol. 32: 241. 2011
Description and illustrations — See Yang et al. (2011) and Damm et al. (2012b).
Materials examined. CHINA, Hangzhou, on Ca. sinensis, Oct. 2013, F. Liu, culture CGMCC 3.17359 = LC3570 = LF798; on Ca. sinensis, Oct. 2013, F. Liu, culture LC3560 = LF788.
Notes — Colletotrichum karstii is a common and geographically diverse species, occurring on various host plants. It was previously reported to be pathogenic to Ca. sinensis in China (Liu 2013) and Camellia in Italy (Schena et al. 2013). Comparing it to the available TUB2 sequences from Camellia in Schena et al. (2013), 4 bp differences were detected between the Italian C. karstii and the Chinese isolates.
Colletotrichum siamense Prihast., L. Cai & K.D. Hyde, Fung. Diversity 39: 98. 2009 — Fig. 14
Fig. 14.
Colletotrichum siamense on PDA (CGMCC 3.17353). a. Acervulus; b, c, e. conidiophores; d. conidia; f–h. appressoria. — Scale bar: d = 10 μm, scale bar of d applies to b–h.
On PDA: Colonies 79 mm diam in 7 d, > 90 mm diam in 10 d, aerial mycelium white, cottony, sparse, surface of colony with numerous small acervuli with orange conidial ooze; reverse pale yellowish. Chlamydospores not observed. Conidiomata acervular, conidiophores formed on a cushion of roundish and medium brown cells. Setae not observed. Conidiophores hyaline, branched. Conidiogenous cells hyaline, cylindrical to ampulliform, 6.5–16 μm, apex 1–2 μm diam. Conidia hyaline, aseptate, smooth-walled, cylindrical, both ends bluntly rounded, 12–15.5 × 4–5.5 μm, mean ± SD = 13.8 ± 0.9 × 4.7 ± 0.35 μm, L/W ratio = 2.9. Appressoria medium brown, aseptate, solitary, circular, clavate or ellipsoidal, 5.5–9.5 × 5–7.5 μm, mean ± SD = 7.5 ± 1.32 × 5.8 ± 0.7 μm, L/W ratio = 1.3.
Materials examined. CHINA, Sichuan Province, Chengdu Botanical Garden, on Ca. oleifera, Oct. 2012, F. Liu, culture LC2969 = LF177; on Camellia sp., Oct. 2012, F. Liu, culture LC2974 = LF182; ibid., culture CGMCC 3.17353 = LC2931 = LF139; Yunnan Province, Pu’er, on Camellia sp., 2010, D.M. Hu, culture LC0149 = PE007-2.
Notes — Conidiogenous cells of C. siamense were not well-illustrated in the original publication (Prihastuti et al. 2009), but are illustrated here based on our isolate from Camellia (Fig. 14). Colletotrichum melanocaulon was proposed as a novel species closely related to C. siamense based on the sequence data of ITS, TUB2, DNA lyase (APN2) and an intergenic spacer between the 3’ end of the DNA lyase and the mating type locus MAT1-2 (apn2mat/IGS) (Doyle et al. 2013). Since ACT, CAL, GAPDH and GS gene sequences of C. melanocaulon were unavailable, only ITS and TUB2 sequences of the ex-type culture (BPI 884101) were included in our genetic analysis. Another recently published new species C. dianesei (Lima et al. 2013), phylogenetically related to C. siamense, was also included in the study. The multi-locus phylogenetic analysis result showed that both C. melanocaulon and C. dianesei clustered together with the ex-type isolate of C. siamense (CBS 18578), and its synonyms C. murrayae (GZAAS 5.09506), C. jasmini-sambac (CBS 130420) and C. hymenocallidis (CBS 125378) (Fig. 1). As the ex-type of these species and isolates from tea plants formed a robust clade with high posterior probability (1, Fig. 1, and 0.96, Fig. 3), we suspect C. melanocaulon and C. dianesei to be synonyms of C. siamense. Further studies are needed to confirm if these taxa are synonymous, or if C. siamense is a species complex (Sharma et al. 2013a).
DISCUSSION
Colletotrichum species on Camellia
In this study, pathogenic and endophytic Colletotrichum isolates associated with Ca. sinensis and other Camellia spp. were allocated to different species complexes and further assigned to 11 species, including nine known and two new species. Furthermore, this study also represents the first report of C. alie-num, C. cliviae, C. jiangxiense, and C. henanense from tea plants. Six species were isolated from both symptomatic and asymptomatic leaves tissues, namely C. camelliae, C. fructicola, C. gloeosporioides, C. jiangxiense, C. karstii, and C. sia-mense. This indicates that they could switch their lifestyle from endophytic to plant pathogenic in nature, and provides additional support for the hypothesis that endophytes can be latent pathogens (Photita et al. 2001, Romero et al. 2001). Some Colletotrichum species were collected only once from this host; C. fioriniae and C. henanense were obtained from symptomatic tea leaves, while C. alienum, C. boninense and C. cliviae were only encountered as endophytes in tea plants. Previous pathogenicity tests showed that C. fructicola isolates from symptomless tissues could cause disease on Citrus fruits (Huang et al. 2013). Consequently, we hypothesise that endophytic species in Camellia could also be potential latent pathogens. Further investigations are therefore required to clarify the ecological relationships of the pathogenic and endophytic Colletotrichum species on Camellia.
Based on this study, C. camelliae is the dominant Colletotrichum species on Camellia in China and is probably host-specific to Camellia. These findings make C. camelliae an appropriate model for addressing questions of population structure and dispersal at broad geographical and landscape level. Knowledge of molecular demographic parameters, such as rates of gene flow, levels of species divergence and migration patterns between populations will elucidate the biogeographic history, and the evolutionary and adaptive mechanisms. Information on the genetic structure of the populations can also assist in the development of disease management strategies (Rampersad et al. 2013). Additional collections from Camellia growing regions across the world would therefore aid us to characterise the population structure of this important pathogen and to confirm whether this species is indeed the dominant Colletotrichum species globally.
Colletotrichum acutatum and C. gloeosporioides were previously reported as the dominant endophytic species in Camellia based on morphological characteristics or ITS sequence data (Osono 2008, Fang et al. 2013). However, we did not isolate any C. acutatum s.str. isolates in our study, and only a single isolate of C. fioriniae, belonging to the C. acutatum species complex, was obtained from symptomatic tissue. In addition, although the majority of strains from Camellia in this study belong to the C. gloeosporioides species complex, only four of them are C. gloeosporioides s.str., including three pathogenic and one endophytic isolates. This indicates that many of the previous identifications of Colletotrichum species on Camellia were probably incorrect.
Apart from the Colletotrichum species found in this study, Camellia spp. could also be infected or colonised by a few other species, i.e. C. lupini (Damm et al. 2012a), C. acutatum, C. car-veri, C. coccodes, and C. queenslandicum (syn. C. gloeosporioides var. minor, Weir et al. 2012) (Farr & Rossman 2014). These reports (except C. lupini), however, need to be verified based on the presently accepted classification system in Colletotrichum.
Combined use of ApMat and GS in the C. gloeosporioides species complex
The Apn2-Mat1 locus was introduced for differentiation of Colletotrichum species in the C. graminicola species complex by Crouch et al. (2009), while Rojas et al. (2010) applied it to the C. gloeosporioides species complex. Following this, a new marker in the intergenic region of APN2 and MAT1-2-1 was specifically designed to improve the systematics of the C. gloeosporioides species complex (Silva et al. 2012b), and the locus was renamed as ApMat, which has subsequently been used in molecular phylogenetic analyses of this group (Sharma et al. 2013a, 2014, Vieira et al. 2014).
In the study of Silva et al. (2012a), the ApMat locus proved to be the most informative marker compared to other standard markers, and could resolve species in the C. gloeosporioides species complex and provide a similar amount of information and support as the concatenated tree based on seven loci (ApMat, Apn25L, MAT5L, MAT1-2-1, ITS, β-tub2, GS). However, it is noteworthy that the sample size in their study was rather limited, including only 22 isolates belonging to six divergent species from Coffea. Subsequently, the ApMat marker was employed to analyse species in the C. gloeosporioides complex that are associated with Mangifera indica using a larger sample size, in which 39 Colletotrichum isolates were separated into nine lineages, namely C. fragariae, C. fructicola, C. jasmini-sambac, C. melanocaulon and five unnamed lineages (Sharma et al. 2013a). In that study, only 15 of the Colletotrichum isolates used in the ApMat gene analysis were also included in a multi-locus phylogenetic tree (ACT, CAL, CHS, GAPDH, ITS, TUB2) where they were separated into four clades corresponding to C. theobromicola, C. asianum, C. siamense and C. fructicola. However, no comparison was made between the results of the single-locus ApMat and the multi-locus phylogenetic analysis.
In order to determine if the ApMat sequences provide adequate phylogenetic information compared to that of a multi-locus dataset, we constructed both single-locus ApMat and combined 6-marker (ACT, CAL, GAPDH, GS, ITS, TUB2) trees using the same Colletotrichum isolates associated with Camellia collected in this study. All ApMat reference sequences used in Sharma et al. (2013a) were incorporated in our ApMat analysis, except for GenBank KC888927 from C. alienum isolate ICMP 12071 (incorrect sequence deposited by the original author). The ApMat sequence of isolate ICMP 12071 was re-sequenced and submitted to GenBank as GenBank KM360144 in this study. Our study demonstrated that 22 species (C. aenigma, C. aeschynomenes, C. alatae, C. alienum, C. asianum, C. aotearoa, C. camelliae, C. clidemiae, C. cordylinicola, C. fructicola, C. gloeosporioides, C. henanense, C. horii, C. musae, C. nupharicola, C. psidii, C. queenslandicum, C. salsolae, C. siamense, C. theobromicola, C. ti, and C. tropicale) could be clearly delimitated with ApMat (Fig. 3 and Fig. S2 in TreeBASE). Although C. fructivorum, C. jiangxiense, C. kahawae subsp. kahawae, C. rhexiae, and C. temperatum clustered together in one big clade, the species C. fructivorum, C. rhexiae, and C. temperatum could be delimitated by forming three small subclades with high posterior probabilities (Fig. S2 in TreeBASE). However, C. jiangxiense and C. kahawae subsp. kahawae could not be distinguished from each other. Furthermore, isolates of C. camelliae were separated into two subclades (Fig. 3 and Fig. S2 in TreeBASE). Although C. jiangxiense could be distinguished from C. kahawae s.l. by the GS marker, the other species in the C. gloeosporioides species complex could not be delimited very well, e.g. C. camelliae, C. fructicola, C. siamense, and C. queenslandicum (data not shown). This study demonstrates that the ApMat marker provides superior phylogenetic information compared to other used loci and can distinguish most species in the C. gloeo-sporioides species complex. A further phylogenetic analysis using the concatenated ApMat and GS alignment showed that all species could be delimited, including C. jiangxiense and C. kahawae subsp. kahawae. We therefore recommend a combination of ApMat and GS as an effective way of identifying species in the C. gloeosporioides species complex.
In the present study we mainly focused on the taxonomy and biodiversity of Colletotrichum species associated with tea plants in China as plant pathogens and/or endophytes. Further attention should be given to surveys from different geographical regions to help resolve the life cycles and ecology of these species, especially of C. camelliae. Because of the important commercial value of tea plantations, appropriate disease management strategies in tea plantations should also be developed to control infection by Colletotrichum species.
Acknowledgments
We thank Dianming Hu, Qian Chen, Yu Zhang, Yahui Gao and Nan Zhou from CAS, Tianwen Hou from Guilin Forestry Bureau, Zongxiu Luo from Chinese Academy of Agricultural Sciences for their help in sample collections. We also thank Ping Tan in performing a part of the pathogenicity testing. Drs Eric H.C. McKenzie, Jo Anne Crouch and Gunjan Sharma are thanked for inspiration and useful discussions on the tea pathogen C. camellia and G. cingulata ‘f. sp. camelliae’. Dr William Quaedvlieg is thanked for his technical assistance with performing the pairwise homoplasy index tests. Dr Johannes Z. Groenewald is thanked for commenting on the manuscript and helpful suggestions. This study was financially supported by the external Cooperation Program of CAS (GJHZ1310), the NSFC (31322001 & 31400017), and Project for Fundamental Research on Science and Technology, MOST (2014FY120100). Yong Wang acknowledged Guizhou Province (Grant 20113045) for supporting his investigation on foliar pathogens.
REFERENCES
- Alfieri SA, Langdon KR, Wehlburg C, et al. 1984. Index of plant diseases in Florida. Florida Department of Agriculture & Consumer Services, Division of Plant Industry 11: 1–389. [Google Scholar]
- Arx JA von. 1957. Die Arten der Gattung Colletotrichum Cda. Phytopathologische Zeitschrift 29: 413–468. [Google Scholar]
- Cai L, Hyde KD, Taylor PWJ, et al. 2009. A polyphasic approach for studying Colletotrichum. Fungal Diversity 39: 183–204. [Google Scholar]
- Cannon PF, Damm U, Johnston PR, et al. 2012. Colletotrichum – current status and future directions. Studies in Mycology 73: 181–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone I, Kohn LM. 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556. [Google Scholar]
- Cash EK. 1952. A record of the fungi named by J.B. Ellis, Part I. Division of Mycology and Disease Survey, Bureau of Plant Industry, Soils, and Agricultural Engineering, Agricultural Research Administration, USDA: 2: 1–165. [Google Scholar]
- Chen MM. 2003. Forest fungi phytogeography: Forest fungi phytogeography of China, North America, and Siberia and international quarantine of tree pathogens. Pacific Mushroom Research and Education Center, Sacramento, California. [Google Scholar]
- Choi YW, Hyde KD, Ho W. 1999. Single spore isolation of fungi. Fungal Diversity 3: 29–38. [Google Scholar]
- Copes WE, Thomson JL. 2008. Survival analysis to determine the length of the incubation period of Camellia twig blight caused by Colletotrichum gloeosporioides. Plant Disease 92: 1177–1182. [DOI] [PubMed] [Google Scholar]
- Crouch JA. 2014. Colletotrichum caudatum s.l. is a species complex. IMA Fungus 5: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch JA, Clarke BB, White JF, et al. 2009. Systematic analysis of the falcate-spored graminicolous Colletotrichum and a description of six new species from warm-season grasses. Mycologia 101: 717–732. [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, et al. 2004. MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Crous PW, Slippers B, Wingfield MJ, et al. 2006. Phylogenetic lineages in the Botryosphaeriaceae. Studies in Mycology 55: 235–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai QL, Xu YP, Lin QQ, et al. 2008. Distribution and establishment of Colletotrichum sp. as an endophyte in tea plants (Camellia sinensis). Scientia Silvae Sinicae 44: 84–89. [Google Scholar]
- Damm U, Cannon PF, Liu F, et al. 2013. The Colletotrichum orbiculare species complex: Important pathogens of field crops and weeds. Fungal Diversity 61: 29–59. [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012a. The Colletotrichum acutatum species complex. Studies in Mycology 73: 37–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, Cannon PF, Woudenberg JHC, et al. 2012b. The Colletotrichum boninense species complex. Studies in Mycology 73: 1–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damm U, O’Connell R, Groenewald J, et al. 2014. The Colletotrichum destructivum species complex-hemibiotrophic pathogens of forage and field crops. Studies in Mycology 79: 49–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens JSW, Cook RTA. 1989. Glomerella cingulata on Camellia. Plant Pathology 38: 75–85. [Google Scholar]
- Doyle VP, Oudemans PV, Rehner SA, et al. 2013. Habitat and host indicate lineage identity in Colletotrichum gloeosporioides s.l. from wild and agricultural landscapes in North America. PloS ONE 8: e62394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang WP, Yang LC, Zhu XJ, et al. 2013. Seasonal and habitat dependent variations in culturable endophytes of Camellia sinensis. Journal of Plant Pathology & Microbiology 4: 169. [Google Scholar]
- Farr DF, Rossman AY. 2014. Fungal databases. Systematic Mycology and Microbiology Laboratory, ARS, USDA. Retrieved May 18, 2014, from http://nt.ars-grin.gov/fungaldatabases/. [Google Scholar]
- Glass NL, Donaldson GC. 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerber JC, Liu B, Correll JC, et al. 2003. Characterization of diversity in Colletotrichum acutatum sensu lato by sequence analysis of two gene introns, mtDNA and intron RFLPs, and mating compatibility. Mycologia 95: 872–895. [PubMed] [Google Scholar]
- Guo LD, Hyde KD, Liew ECY. 2000. Identification of endophytic fungi from Livistona chinensis based on morphology and rDNA sequences. New Phytologist 147: 617–630. [DOI] [PubMed] [Google Scholar]
- Guo M, Pan YM, Dai YL, et al. 2014. First report of brown blight disease caused by Colletotrichum gloeosporioides on Camellia sinensis in Anhui Province, China. Plant Disease 98: 284. [DOI] [PubMed] [Google Scholar]
- Huang F, Chen Q, Hou X, et al. 2013. Colletotrichum species associated with cultivated citrus in China. Fungal Diversity 61: 61–74. [Google Scholar]
- Huson DH. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14: 68–73. [DOI] [PubMed] [Google Scholar]
- Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution 23: 254–267. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima NB, Batista MVdA, Morais MA de, Jr, et al. 2013. Five Colletotrichum species are responsible for mango anthracnose in northeastern Brazil. Fungal Diversity 61: 75–88. [Google Scholar]
- Liu F, Cai L, Crous PW, et al. 2013a. Circumscription of the anthracnose pathogens Colletotrichum lindemuthianum and C .nigrum. Mycologia 105: 844–860. [DOI] [PubMed] [Google Scholar]
- Liu F, Cai L, Crous PW, et al. 2014. The Colletotrichum gigasporum species complex. Persoonia 33: 83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Damm U, Cai L, et al. 2013b. Species of the Colletotrichum gloeosporioides complex associated with anthracnose diseases of Proteaceae. Fungal Diversity 61: 89–105. [Google Scholar]
- Liu F, Hu DM, Cai L. 2012. Conlarium duplumascospora gen. et. sp. nov. and Jobellisia guangdongensis sp. nov. from freshwater habitats in China. Mycologia 104: 1178–1186. [DOI] [PubMed] [Google Scholar]
- Liu W. 2013. Anthracnose pathogens identification and the genetic diversity of tea plant. PhD thesis, Fujian Agriculture and Forestry University, China. [Google Scholar]
- Lu B, Hyde KD, Ho WH, et al. 2000. Checklist of Hong Kong fungi. Fungal Diversity Press, Hong Kong. [Google Scholar]
- Lu DS, Wang JP, Wu XQ, et al. 2007. The species and distribution of endophytic fungi in tea trees. Journal of Henan Agricultural Sciences 10: 54–56. [Google Scholar]
- Moriwaki J, Sato T, Tsukiboshi T. 2003. Morphological and molecular characterization of Colletotrichum boninense sp. nov. from Japan. Mycoscience 44: 47–53. [Google Scholar]
- Nirenberg H. 1976. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Sektion Liseola. Mitteilungen aus der Biologischen Bundesanstalt für Land- und Forstwirtschaft Berlin-Dahlem 169: 1–117. [Google Scholar]
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. [Google Scholar]
- O’Donnell K, Cigelnik E. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. [DOI] [PubMed] [Google Scholar]
- Osono T. 2008. Endophytic and epiphytic phyllosphere fungi of Camellia japonica: seasonal and leaf age-dependent variations. Mycologia 100: 387–391. [DOI] [PubMed] [Google Scholar]
- Photita W, Lumyong S, Lumyong P, et al. 2001. Endophytic fungi of wild banana (Musa acuminata) at Doi Suthep Pui National Park, Thailand: Mycologial Research 105: 1508–1513. [Google Scholar]
- Prihastuti H, Cai L, Chen H, et al. 2009. Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Diversity 39: 89–109. [Google Scholar]
- Quaedvlieg W, Binder M, Groenewald JZ, et al. 2014. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 33: 1–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampersad SN, Perez-Brito D, Torres-Calzada C, et al. 2013. Genetic structure and demographic history of Colletotrichum gloeosporioides sensu lato and C. truncatum isolates from Trinidad and Mexico. BMC Evolutionary Biology 13:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner RW. 1970. A mycological colour chart. Commonwealth Mycological Institute, Kew. [Google Scholar]
- Rojas EI, Rehner SA, Samuels GJ, et al. 2010. Colletotrichum gloeosporioides s.l. associated with Theobroma cacao and other plants in Panamá: multilocus phylogenies distinguish host-associated pathogens from asymptomatic endophytes. Mycologia 102: 1318–1338. [DOI] [PubMed] [Google Scholar]
- Romero A, Carrión G, Rico-Gray V. 2001. Fungal latent pathogens and endophytes from leaves of Parthenium hysterophorus (Asteraceae). Fungal Diversity 7: 81–87. [Google Scholar]
- Ronquist F, Teslenko M, Mark P van der, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena L, Mosca S, Cacciola SO, et al. 2013. Species of the Colletotrichum gloeosporioides and C. boninense complexes associated with olive anthracnose. Plant Pathology 63: 437–446. [Google Scholar]
- Sharma G, Kumar N, Weir BS, et al. 2013a. The ApMat marker can resolve Colletotrichum species: a case study with Mangifera indica. Fungal Diversity 61: 117–138. [Google Scholar]
- Sharma G, Pinnaka AK, Shenoy BD. 2013b. ITS-based diversity of Colletotrichum from India. Current Research in Environmental & Applied Mycology 3: 194–220. [Google Scholar]
- Sharma G, Pinnaka AK, Shenoy BD. 2014. Resolving the Colletotrichum siamense species complex using ApMat marker. Fungal Diversity. doi: 10.1007/s13225-014-0312-7. [Google Scholar]
- Shivas RG. 1989. Fungal and bacterial diseases of plants in Western Australia. Journal of the Royal Society of Western Australia 72: 1–62. [Google Scholar]
- Silva DN, Talhinhas P, Cai L, et al. 2012a. Host-jump drives rapid and recent ecological speciation of the emergent fungal pathogen Colletotrichum kahawae. Molecular Ecology 21: 2655–2670. [DOI] [PubMed] [Google Scholar]
- Silva DN, Talhinhas P, Várzea V, et al. 2012b. Application of the Apn2/MAT locus to improve the systematics of the Colletotrichum gloeosporioides complex: an example from coffee (Coffea spp.) hosts. Mycologia 104: 396–409. [DOI] [PubMed] [Google Scholar]
- Simmonds JH. 1966. Host index of plant diseases in Queensland: 111. Queensland Department of Primary Industries, Brisbane. [Google Scholar]
- Stephenson SA, Green JR, Manners JM, et al. 1997. Cloning and characterisation of glutamine synthetase from Colletotrichum gloeosporioides and demonstration of elevated expression during pathogenesis on Stylosanthes guianensis. Current Genetics 31: 447–454. [DOI] [PubMed] [Google Scholar]
- Tai FL. 1979. Sylloge Fungorum Sinicorum. Science Press, Academica Sinica, Peking. [Google Scholar]
- Tamura K, Peterson D, Peterson N, et al. 2011. MEGA 5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaung MM. 2008. Biodiversity survey of coelomycetes in Burma. Australasian Mycologist 27: 74–110. [Google Scholar]
- Thompson A, Johnston A. 1953. A host list of plant diseases in Malaya. Mycological Papers 52: 1–38. [Google Scholar]
- Tunstall A. 1934. A new species of Glomerella on Camellia theae. Transactions of the British Mycological Society 19: 331–336. [Google Scholar]
- Vieira WA, Michereff SJ, Morais MA de, Jr, et al. 2014. Endophytic species of Colletotrichum associated with mango in northeastern Brazil. Fungal Diversity 67: 181–202. [Google Scholar]
- Watson AJ. 1950. Fungi associated with camellia flowers. Plant Disease Reporter 34: 186–187. [Google Scholar]
- Weir BS, Johnston PR, Damm U. 2012. The Colletotrichum gloeosporioides species complex. Studies in Mycology 73: 115–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee J, et al. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In:Innis MA, Gelfand DH, Sninsky JJ, et al. (eds), PCR protocols: a guide to methods and applications: 315–322. Academic Press, San Diego, California, USA. [Google Scholar]
- Willis JC. 1899. DCLII – Tea and coffee diseases. Bulletin of Miscellaneous Information Royal Botanical Gardens Kew; 1899: 89–94. [Google Scholar]
- Yang YL, Cai L, Yu ZN, et al. 2011. Colletotrichum species on Orchidaceae in southwest China. Cryptogamie, Mycologie 32: 229–253. [Google Scholar]
- Yang YL, Liu ZY, Cai L, et al. 2009. Colletotrichum anthracnose of Amaryllidaceae. Fungal Diversity 39: 123–146. [Google Scholar]