The role of the Nck adaptor protein in balancing linear versus branched actin nucleation in comet tails is evaluated. Nck recruits both linear and branched nucleation-promoting factors, both of which are necessary for the formation of actin comets. The findings highlight a novel role for Nck in pathogen-like actin motility.
Abstract
The Nck adaptor protein recruits cytosolic effectors such as N-WASP that induce localized actin polymerization. Experimental aggregation of Nck SH3 domains at the membrane induces actin comet tails—dynamic, elongated filamentous actin structures similar to those that drive the movement of microbial pathogens such as vaccinia virus. Here we show that experimental manipulation of the balance between unbranched/branched nucleation altered the morphology and dynamics of Nck-induced actin comets. Inhibition of linear, formin-based nucleation with the small-molecule inhibitor SMIFH2 or overexpression of the formin FH1 domain resulted in formation of predominantly circular-shaped actin structures with low mobility (actin blobs). These results indicate that formin-based linear actin polymerization is critical for the formation and maintenance of Nck-dependent actin comet tails. Consistent with this, aggregation of an exclusively branched nucleation-promoting factor (the VCA domain of N-WASP), with density and turnover similar to those of N-WASP in Nck comets, did not reconstitute dynamic, elongated actin comets. Furthermore, enhancement of branched Arp2/3-mediated nucleation by N-WASP overexpression caused loss of the typical actin comet tail shape induced by Nck aggregation. Thus the ratio of linear to dendritic nucleation activity may serve to distinguish the properties of actin structures induced by various viral and bacterial pathogens.
INTRODUCTION
Actin-based cell motility is an important and well-studied physiological process. At its core is the polymerization of actin monomers into filaments (Pollard et al., 2000). Polymerization and organization of these filaments into different cellular structures is a highly coordinated process regulated by many proteins (dos Remedios et al., 2003; Disanza et al., 2005). The force generated by growing actin barbed ends extends the plasma membrane into ruffles, lamellipodial and filopodial protrusions (Campellone and Welch, 2010). It also propels intracellular vesicles and pathogens that infect the host cell (Stevens et al., 2006; Bhavsar et al., 2007).
Polymerization of G-actin monomers into F-actin occurs in a polarized manner. Actin monomers tend to add to the barbed (growing/plus) end of an existing filament. Formation of a primer (dimer or trimer) initiates actin filament assembly. This process, termed nucleation, is kinetically unfavorable in vitro. The actin-related proteins Arp 2 and Arp 3 (Arp2/3) complex and formin family proteins are two of the three major types of actin nucleators in cells (Mullins et al., 1998; Amann and Pollard, 2001; Pruyne et al., 2002; Sagot et al., 2002). Arp2/3 creates new actin branches at the sides of preexisting filaments. It is activated by the C-terminal verprolin-homology, cofilin-homology, and acidic domains (VCAs) of class I nucleation-promoting factors (NPFs; Hufner et al., 2001; Hitchcock-DeGregori, 2003). The VCA domain binds G-actin and the Arp2/3 protein complex, inducing a conformational change that primes Arp2/3 for activity. Formin family proteins catalyze nucleation, increase elongation rate, and prevent capping of the actin barbed ends (Krause and Gautreau, 2014). The highly conserved C-terminal formin homology (FH) domains FH1 and FH2 increase the elongation rate compared with the elongation of free barbed ends (Kovar and Pollard, 2004; Romero et al., 2004). Formins bind to Src homology 3 (SH3)–containing proteins through the FH1 proline-rich domain, which also recruits profilin–actin complexes (Paul and Pollard, 2009) for addition of actin monomers onto the elongating barbed ends. The FH2 domain has a donut-shaped structure that caps and processively moves with the growing actin ends and adds actin monomers to the barbed ends of actin filaments.
Both formins and class I NPFs such as neural Wiskott–Aldrich syndrome protein (N-WASP) are activated and spatiotemporally controlled through interaction with specific regulatory proteins that bind to their N-termini (Campellone and Welch, 2010; Burianek and Soderling, 2013). Many formins are autoinhibited by intramolecular interaction between their N- and C-termini. Activation and recruitment of formins to the membrane are mainly achieved through binding of Rho GTPases. However other factors contribute to the regulation of the activity of specific formins. SH3 domain–containing proteins such as Src family kinases (Young and Copeland, 2010) and the adaptor protein mDia-interacting protein (DIP; known as SH3 protein interacting with Nck 90 kDa [SPIN90], Nck-interacting protein with SH3 domain [NCKIPSD], and WASP-interacting SH3 protein [WISH]; Eisenmann et al., 2007) interact with the proline-rich FH1 domain, suggesting that these interactions might contribute to the regulation of formin activity. N-WASP is a class I NPF, and therefore it contains a C-terminal catalytic VCA domain (Campellone and Welch, 2010; Burianek and Soderling, 2013). Similarly to the formins, it is maintained in the autoinhibited state in which the VCA domain is caged and therefore inactive. To stimulate N-WASP, signaling pathways target multiple cellular factors that interact with the N-terminus of N-WASP. For example, binding of the Rho family GTPase Cdc42 and phosphatidylinositol (4,5)-bisphosphate (PIP2) induces conformational changes that free the VCA domain. Another activation route that relieves N-WASP autoinhibition is the cooperative binding of PIP2 and the adaptor protein Nck (Rohatgi et al., 2001). WASP-interacting protein (WIP) binds to both Nck and N-WASP and is essential in stimulating N-WASP/Arp2/3–dependent actin polymerization (Ditlev et al., 2012; Donnelly et al., 2013).
Nck comprises one SH2 and three SH3 domains. It has a pivotal role in pTyr signaling from the cell surface through N-WASP and to the actin cytoskeleton (Stein et al., 1998; Li et al., 2001; Rao, 2005; Jones et al., 2006; Lettau et al., 2009, 2014; New et al., 2013). The pathogen vaccinia virus targets the Nck adaptor of the host cell (Hayward et al., 2006; Haglund and Welch, 2011) by mimicking host phosphotyrosine motif. Vaccinia introduces the viral A36 protein into the membrane of the infected cell. A36 undergoes tyrosine phosphorylation by Src and Abl kinases at Y112 creating a binding site for the Nck SH2 domain (Dodding and Way, 2009). By recruiting Nck, vaccinia localizes and activates N-WASP in the host cell. This results in production of actin comet tails beneath the surface of the virus (Roberts and Smith, 2008). Other host proteins, including the adaptor Grb2, small G protein Cdc42, and the Rho guanine-nucleotide-exchange factor (GEF) intersectin-1 have been shown to contribute to formation of vaccinia comet tails (Scaplehorn et al., 2002; Weisswange et al., 2009; Humphries et al., 2014).
We show here that whereas the clustering of Nck SH3 domains at the membrane produces mostly elongated dynamic actin structures similar to those induced by vaccinia virus (Rivera et al., 2004; Ditlev et al., 2012), the clustering of N-WASP VVCA (referred to henceforth as VCA) domains produces slow-moving actin “blobs.” We investigated why Nck SH3 clustering and VCA clustering result in formation of such different actin structures. First, we test whether the density or turnover of the N-WASP VCA domain differentiates elongated dynamic actin assemblies from actin blobs. We show that clustering of VCA at low density, or when turnover at the membrane is allowed, does not reproduce typical actin comets. Second, we demonstrate that both branched and linear actin polymerization are necessary for the assembly of dynamic actin comet tails. Interfering with either branched or linear nucleation decreased the velocity and altered the morphology of actin structures induced by aggregation of Nck SH3 domains. Our results suggest that the activity of linear NPFs such as formins is critical for the formation of Nck-dependent actin comets and the maintenance of their phenotype. We propose that Nck serves as an integrator of linear and branched nucleation in Nck-induced comet tails.
RESULTS
Membrane clustering of Nck SH3 and VCA produces distinct actin structures
Aggregating the SH3 domains of the Nck adaptor at the membrane induces the formation of dynamic actin comet tails (Figure 1A; Rivera et al., 2004, 2009; Ditlev et al., 2012). Nck SH3 domains fused to a CD7 transmembrane segment and an extracellular CD16 domain were aggregated using primary anti-CD16 monoclonal antibodies and secondary anti-mouse immunoglobulin G (IgG). Fluorescently labeled Nck SH3 aggregates and actin were visualized using time-lapse confocal microscopy. This assay mimics the recruitment and increased local concentration of full-length Nck that occurs during biological processes such as axon growth cone guidance during Drosophila eye development (Rao, 2005), formation of the immunological synapse (Lettau et al., 2009), and actin rearrangements in kidney podocytes (Jones et al., 2006). Actin comet tails formed beneath multiple mCherry-tagged Nck SH3 clusters (Figure 1B). Nck is an activator of NWASP-Arp2/3–mediated actin assembly in mammalian cells (Rohatgi et al., 2001; Kempiak et al., 2005), and so clustering and activation of N-WASP is believed to be the critical step in Nck SH3–induced actin comets. To test this, we directly aggregated mCherry-tagged N-WASP VCA domains at the membrane (Figure 1C). Compared to clustering of Nck SH3 domains, this bypasses WIP-dependent N-WASP recruitment and activation steps, as well as signaling to the actin cytoskeleton through Nck SH3–binding proteins other than N-WASP. To our surprise, we found the actin structures induced by aggregation of Nck SH3 and VCA (Figure 1, B and D) have distinctly different morphologies (Figure 1E) and velocity (Figure 1F and Supplemental Movie S1) distributions in cells.
FIGURE 1:
Membrane clustering of Nck SH3 and VCA induces formation of dissimilar actin structures. (A, C) Schematic of the CD16/7-mCherry-Nck-SH3 and CD16/7-mCherry-VCA transmembrane fusion proteins. Both were aggregated by sequential application of primary mouse anti-CD16 and secondary anti-mouse antibodies. N-WASP recruitment to Nck SH3 required WIP (not shown in the schematic). (B) Aggregation of the Nck SH3 domains induces formation of actin comet tails. Staining: CD16/7-mCherry-Nck (red), actin (green). (D) Aggregation of the VCA domain of N-WASP induces formation of blob-like actin structures. Staining: CD16/7-mCherry-VCA (red), actin (green). Scale bars, 5 μm. (E) Morphology comparison of Nck- and VCA-induced actin structures. The bars represent the average percentage of particles per cell with circularity <0.6. Nck: three cells, 540 aggregates; VCA: three cells, 336 aggregates. Error bars are mean ± SEM. *p < 0.01. (F) Velocity comparison of Nck- and VCA-induced actin structures and vaccinia actin comet tails. Distributions of mean velocities (μm/s) are shown with boxplots. On each box, the red mark is the median, the black mark is the mean, and the lower edge is the 25th and the upper edge the 75th percentile. The whiskers are the most extreme data points, and outliers are shown by red plus signs. Nck: seven cells, 654 aggregates; VCA: seven cells, 945 aggregates.
We compared the morphology of actin particles by determining a circularity parameter (scale of 0 to 1), with elongated objects such as comet tails having low values and circular-shaped objects such as blobs approaching circularity values of 1 (see Materials and Methods). We quantified the percentage of particles per cell with circularity <0.6 (elongated objects). Nck SH3 clustering produces 37% (Figure 1E), whereas VCA induced only 16% of elongated actin structures. We compared the velocity of actin assemblies by tracking actin particles over time and plotting the distribution of average velocity per cell. Nck SH3 actin aggregates have higher mean and median velocities than VCA aggregates (Figure 1F). The population of fairly motile (v > 0.06 μm/s) actin structures is much greater for Nck SH3 aggregates (23%) than for VCA aggregates (4%), as calculated based on the data shown in Figure 1F. By these quantitative parameters, Nck SH3–induced actin structures are similar to the comets induced by vaccinia virus (Figure 1B and Supplemental Figure S6B). Nck SH3 and vaccinia comets have comparable circularity (Supplemental Figure S6E) and a subset of highly motile actin particles (Supplemental Figure S6F and Supplemental Movies S1 and S6).
The dramatic differences in shape and dynamic behavior of actin structures induced by clustering of Nck SH3 and VCA led us to investigate the molecular mechanisms that might underlie these differences.
Does VCA density differentiate Nck SH3– and VCA-induced actin structures?
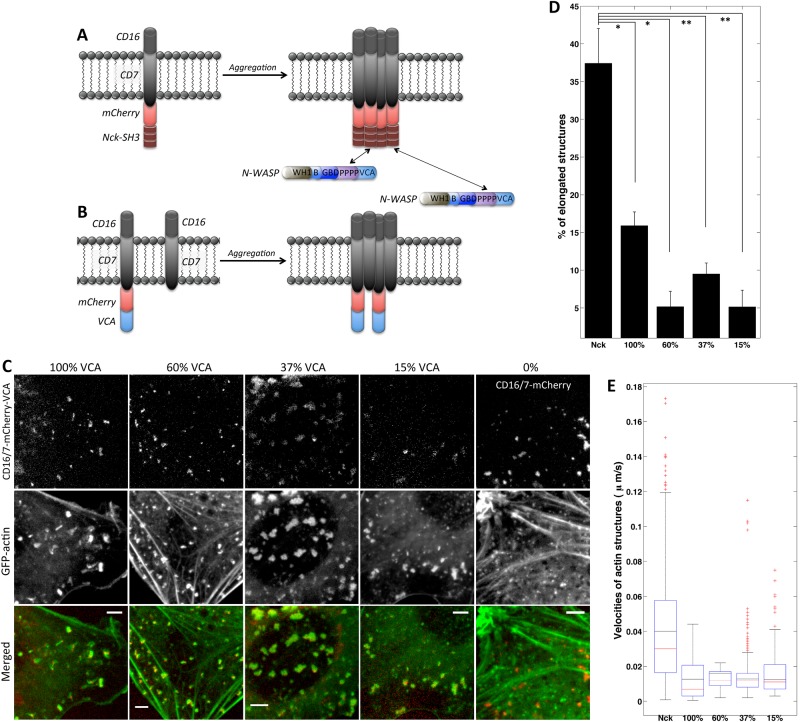
We first explored whether the density of VCA domains in membrane clusters might explain differences between the Nck SH3–and VCA-induced actin structures. Dilution of functional A36 viral protein, which stimulates N-WASP/Arp2/3–mediated actin assembly, resulted in formation of longer and faster vaccinia actin comets (Humphries et al., 2012). The density of VCA domains in the CD16/7-mCherry-VCA aggregates is 100% because each CD16/7-mCherry membrane protein has VCA covalently linked at the C-terminus (Figure 1C). The VCA density in Nck SH3 clusters is always <100% (Figure 2A). This is because Nck SH3 domains have multiple binding partners besides N-WASP (Kitamura et al., 1996; Quilliam et al., 1996; Wunderlich et al., 1999; Zhao et al., 2000; Schmidt and Dikic, 2005; Antoku et al., 2008; Ramesh and Geha, 2009) and N-WASP can dissociate (Weisswange et al., 2009; Siton et al., 2011; Smith et al., 2013) from Nck SH3 domains after a new actin branch has been formed. In addition, at equilibrium, only 38% of Nck molecules are predicted to be bound by N-WASP (based on estimates of Nck/N-WASP affinity and N-WASP abundance), and experimental and computational modeling data strongly suggest that the Nck:VCA stoichiometry in Nck comets is 2:1 (Ditlev et al., 2012).
FIGURE 2:
Decreasing VCA density in membrane clusters is not sufficient for comet tail formation. (A) Schematic of CD16/7-mCherry-Nck aggregation and binding by endogenous N-WASP protein. (B) Schematic of CD16/7-mCherry-VCA coaggregation with empty CD16/7 fusion proteins resulting in membrane VCA cluster of decreased density (comparable to density of VCA in Nck aggregates). (C) Aggregation of VCA diluted with Empty fusion proteins. Staining in merged images: GFP-actin (green), CD16/7-mCherry-VCA (red) in 100, 60, 37, and 15% VCA, and CD16/7-mCherry (red) in 0% VCA. Scale bars, 5 μm. (D) Morphology comparison of Nck- and VCA-induced and actin structures induced with diluted VCA clusters. Nck: three cells, 540 aggregates; 100% VCA: three cells, 336 aggregates; 60% VCA: three cells, 619 aggregates; 37% VCA: three cells, 425 aggregates; 15% VCA: three cells, 811 aggregates. Error bars are mean ± SEM. *p < 0.01, **p < 0.05. (E) Velocity comparison of Nck- and VCA-induced and actin structures induced with diluted VCA clusters. Nck: seven cells, 654 aggregates; 100% VCA: six cells, 872 aggregates; 60% VCA: three cells, 597 aggregates; 37% VCA: three cells, 804 aggregates; 15% VCA: three cells, 496 aggregates.
To test whether VCA-domain density differentiates Nck SH3– and VCA-induced actin assemblies, we experimentally lowered the density of VCA molecules in CD16/7-mCherry-VCA clusters. CD16/7-mCherry-VCA proteins were coclustered with CD16/7 proteins lacking VCA (“Empty”; Figure 2B). NIH3T3 cells were transfected with different ratios of VCA and Empty constructs so that VCA expression was 100, 60, 37, 15, and 0% of the combined VCA and Empty protein amount (Supplemental Figure S1). CD16/7 fusion proteins were aggregated, the cells were imaged (Figure 2C and Supplemental Movie S2), and actin particle morphology and velocity (Figure 2, D and E) were analyzed. Our prediction was that membrane clusters with 60, 37, or 15% VCA density would induce more comet-like actin structures. However at 60 and 37% VCA in the clusters, fairly typical actin blobs formed (Figures 1D and 2C). At densities <37%, VCA still could induce actin polymerization, even though the actin structures appeared to be smaller. Of note, decreasing VCA density in membrane clusters did not in any case result in formation of elongated dynamic actin structures similar to those induced by Nck SH3 (Figure 1B) aggregation. These results are inconsistent with the hypothesis that the lower density of recruited VCA in Nck SH3 clusters is responsible for the phenotypic differences between Nck SH3–induced (Figure 1B) and VCA-induced (Figure 1D) actin assemblies. Comet tail morphology and dynamics cannot be reproduced solely by decreasing VCA density in membrane clusters.
Does VCA turnover differentiate Nck SH3– and VCA-induced actin structures?
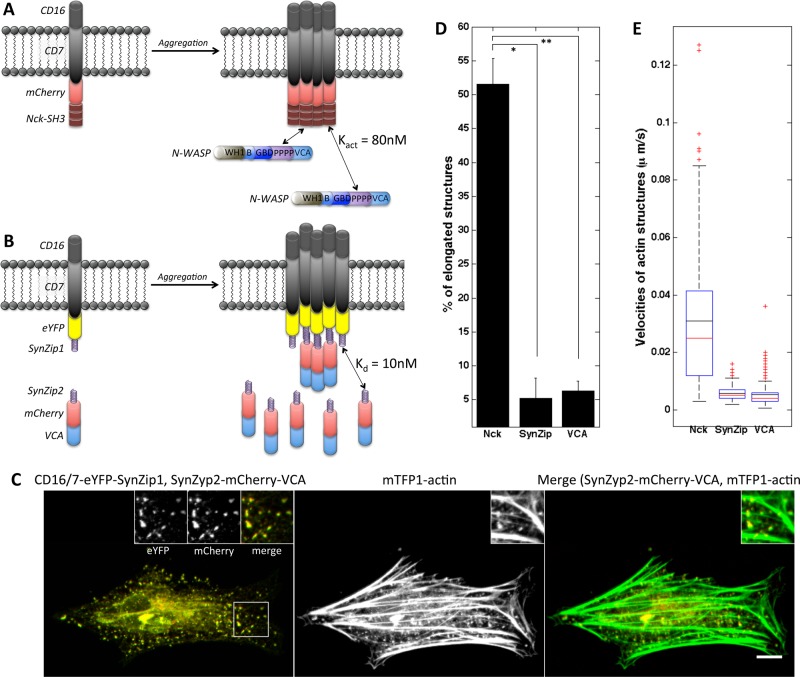
In CD16/7-mCherry-Nck clusters, endogenous N-WASP protein has the ability to dissociate from the membrane aggregates and undergo turnover. After photobleaching, green fluorescent protein (GFP)–N-WASP recovers in the head of the vaccinia-induced comet with a half-time of 1–3 s (Weisswange et al., 2009; Donnelly et al., 2013; Humphries et al., 2014). In vitro, Nck SH3 domains activate N-WASP with Kact ≈ 80 nM (Rohatgi et al., 2001), suggesting that they interact with modest affinity, and thus complexes turn over relatively rapidly. By contrast, in CD16/7-mCherry-VCA clusters, the C-terminal VCA domain of N-WASP is covalently attached to the transmembrane protein (Figure 1C). As expected, mCherry fluorescence of CD16/7-mCherry-VCA clusters does not recover after being bleached (Supplemental Figure S2A and Supplemental Movie S3A).
To test whether turnover of the VCA domain is critical for producing actin comets, we generated VCA membrane clusters where turnover is possible. To allow interaction between CD16/7 membrane fusion proteins and mCherry-VCA domains, we used a coiled-coil motif interaction interface that consists of the parallel coiled-coil pair SYNZIP1:SYNZIP2 (Reinke et al., 2010). These 47–amino acid (aa)-long peptides form tight heterospecific complexes (Kd ≤ 10 nM) and display minimal self-association (Thompson et al., 2012). CD16/7 was tagged with enhanced yellow fluorescent protein (eYFP) and fused to SYNZIP1 (Figure 3B). SYNZIP2 was attached to the N-terminus of the mCherry-VCA (Figure 3B). Expression of CD16/7-eYFP-SYNZIP1 and SYNZIP2-mCherry-NWASP-VCA in NIH3T3 cells was verified by Western blotting (Supplemental Figure S2B). We confirmed that clusters of membrane-embedded SYNZIP1 do in fact recruit cytosolic SYNZIP2-mCherry-VCA protein (Supplemental Figure S2C). Turnover of SYNZIP2-mCherry-VCA in the clusters was shown by performing fluorescent recovery after photobleaching (FRAP) of mCherry (Supplemental Figure S2A and Supplemental Movie S3A; half-time, ∼40 s). Clusters of SYNZIP2-mCherry-VCA and actin aggregates did not form without inducing antibody-mediated aggregation of CD16/7-eYFP-SYNZIP1 (Supplemental Figure S2C). Clustering of CD16/7-eYFP-SYNZIP1 resulted in localized actin recruitment (Figure 3C). However, morphology and velocity analysis (Figure 3, D and E) of the actin structures revealed that they do not exhibit the dynamic behavior and morphological features of comet tails. Thus, even though VCA turnover is likely to be important for comet tail behavior (Smith et al., 2013), it is not sufficient to reproduce the phenotype of elongated dynamic Nck SH3–induced actin assemblies and cannot explain the differences between VCA- and Nck SH3–induced actin structures.
FIGURE 3:
Allowing VCA turnover in membrane clusters does not result in formation of comet tails. (A) Schematic of CD16/7-mCherry-Nck aggregation and binding by endogenous N-WASP protein. (B) Schematic of VCA clusters with turnover through SynZip-mediated interaction. CD16/7-eYFP-SynZip1 aggregation results in recruitment of cytosolic SynZip2-mCherry-VCA fusion proteins. (C) Actin recruitment and shape of actin structures induced by clustering of SynZip2-mCherry-VCA through interaction with CD16/7-eYFP-SynZip1. Left, merged staining of SynZip2-mCherry-VCA (red) and CD16/7-eYFP-SynZip1 (green). Insets, magnified views of CD16/7-eYFP-SynZip1 (left inset), SynZip2-mCherry-VCA (middle inset), and a composite of the two (right inset). Middle, actin staining alone. Right, merged staining of SynZip2-mCherry-VCA (red) and mTFP1-actin (green). Scale bar, 10μm. (D) Morphology analysis of actin structures induced by VCA clustered through SynZip binding. Nck SH3 clustering: three cells, 124 aggregates; VCA clustering though SynZip interaction: eight cells, 152 aggregates; VCA clustering: three cells, 233 aggregates. Error bars are mean ± SEM. *p < 0.001, **p < 0.01. (E) Velocity of actin structures induced by VCA clustered through SynZip binding. Nck SH3 clustering: three cells, 121 aggregates; VCA clustering though SynZip interaction: four cells, 174 aggregates; VCA clustering: three cells, 246 aggregates.
Inhibition of formin FH2 domain disrupts Nck SH3–induced actin comets
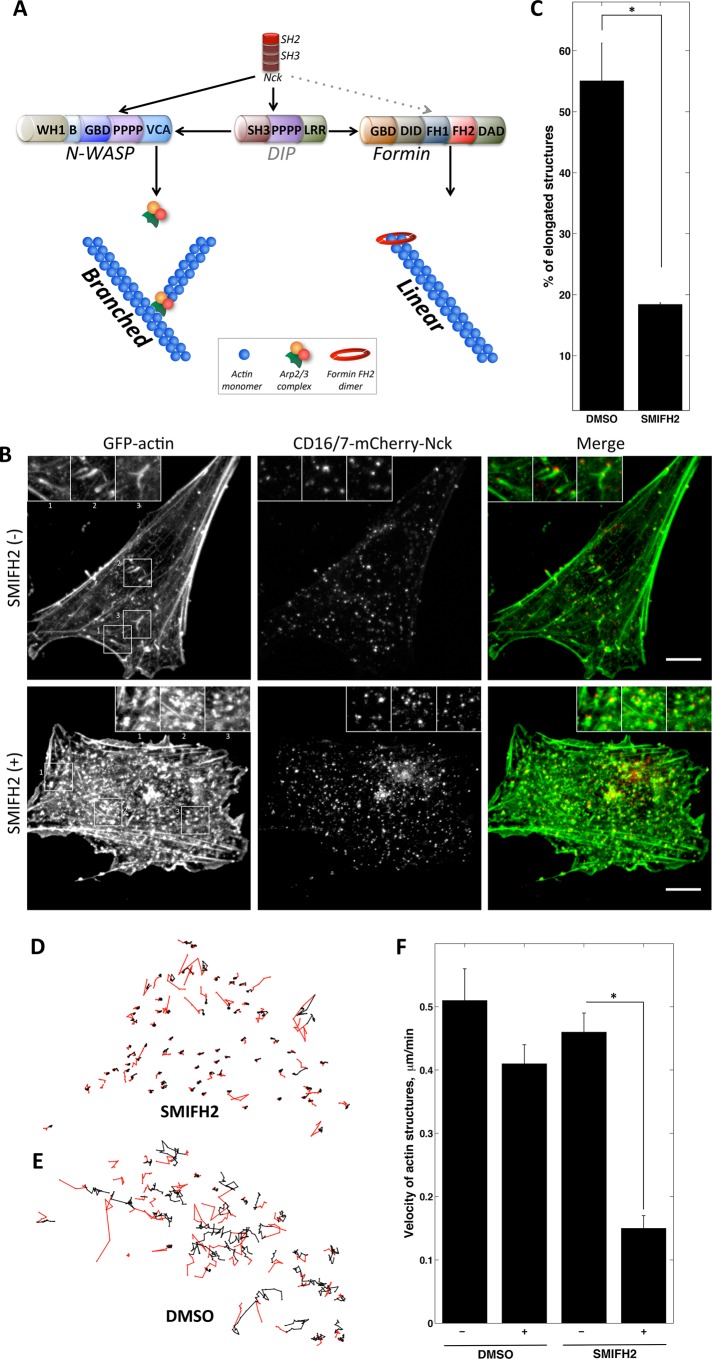
The actin architecture of the baculovirus comet tail was recently visualized by electron microscopy (Mueller et al., 2014). There are multiple actin branches that stem from the center axis of the comet. There are also long, parallel, unbranched actin filaments in the center of these comets. In addition, in the branched actin network, filament fragmentation and subsequent elongation often take place. These facts suggest that a linear elongation mechanism should be an important contributor to the structure and behavior of actin comet tails. Formins catalyze actin polymerization by adding monomers to barbed ends and thus elongate existing actin filaments. We hypothesized that formin-mediated barbed-end elongation activity would be higher in the Nck SH3–induced comets than in VCA-induced actin structures (Figure 4A). Nck SH3 domains can potentially recruit formin through the adaptor DIP (Lim et al., 2001; Eisenmann et al., 2007) or possibly by directly binding the proline-rich FH1 domain of formin (Supplemental Figure S3).
FIGURE 4:
Formin FH2 domain inhibition decreases mobility and disrupts comet tail shape of Nck SH3–induced actin structures. (A) Potential role of Nck as integrator of linear and branched elongation in actin comet tails. Schematic of branched and linear elongation molecular pathways that may be initiated by Nck SH3 clustering. Red, donut-shaped structure at the barbed end of the unbranched filament is a dimer formed by FH2 domains of formin. Gray dotted line represents potential but not demonstrated interaction. Black lines represent known interactions of DIP with Nck, N-WASP, Arp2/3, and formin. (B) Nck SH3-induced actin aggregates in fixed, DMSO-treated NIH3T3 cells (top) or those treated with SMIFH2 inhibitor (bottom). Scale bars, 10 μm. (C) Morphology analysis of Nck SH3–induced actin aggregates in DMSO-treated (control) and SMIFH2-treated fixed NIH3T3 cells. DMSO treated: three cells, 197 aggregates; SMIFH2 treated: three cells, 413 aggregates). Error bars are mean ± SEM. *p < 0.05. (D, E) Tracks of Nck SH3–induced actin structures in NIH3T3 cells treated with SMIFH2 inhibitor (D) or DMSO-treated control (E). Part of the track before treatment is colored in red; after treatment, in black. Frame rate, 153 s, 20 frames. (F) Velocity of Nck SH3–induced actin aggregates before and after DMSO and SMIFH2 treatment. DMSO: Vbefore = 0.51 μm/min and Vafter = 0.41 μm/min. SMIFH2: Vbefore = 0.46 μm/min and Vafter = 0.15 μm/min. DMSO treated: three cells, 92 tracks; SMIFH2 treated: three cells, 148 tracks. Error bars are mean ± SEM. *p < 0.01.
To test whether inhibition of formin-mediated actin assembly would affect Nck SH3-induced actin structures, we treated NIH3T3 cells with a small-molecule inhibitor of formin homology 2 domains (SMIFH2) (Rizvi et al., 2009). We aggregated mCherry-Nck SH3 in both SMIFH2-treated and control cells. Then, we fixed cells and visualized mCherry membrane proteins and GFP-stained actin (Figure 4B). SMIFH2 treatment had a dramatic effect on Nck SH3–associated actin (Figure 4B). The amount of elongated actin assemblies associated with Nck SH3 clusters was significantly lower in treated cells (18%) than in dimethyl sulfoxide (DMSO)–treated control cells (55%; Figure 4C).
To observe the effect of the formin inhibitor on Nck SH3–induced actin in live cells, we subjected cells expressing CD16/7-mCherry-Nck and GFP-actin to the aggregation protocol and then imaged them every 2.5 min (Supplemental Movie S4A). The inhibitor was added after five frames of acquisition. For control cells, DMSO was added at the same time point during the acquisition. We tracked all of the Nck SH3–induced actin particles (Figure 4, D and E) and analyzed their velocity before and after the treatment (Figure 4F). After addition of SMIFH2, the velocity of Nck SH3–induced actin aggregates drops on average by 37% (Figure 4, D and F), and 72% of actin aggregates moved more than twofold more slowly after drug treatment. The velocity of actin particles in the control cells did not change significantly (Figure 4, E and F, and Supplemental Movie S4A). We observed a similar decrease in velocity due to SMIFH2 treatment when we imaged Nck SH3–induced actin aggregates every 30 s (Supplemental Movie S4B).
To test whether inhibition of formin-mediated actin assembly would affect vaccinia actin comets, we inhibited the FH2 domain in HeLa cells that were infected with vaccinia virus. After treatment with the SMIFH2 inhibitor (or DMSO alone as control), the cells were fixed, and then immunostained virus and phalloidin-stained actin were visualized (Supplemental Figure S4A). SMIFH2 treatment had a dramatic effect on vaccinia-associated actin structures, in accord with the findings by Alvarez and Agaisse (2013). The amount of elongated actin assemblies associated with the virus was significantly lower in treated cells (5%) than in DMSO-treated control cells (40%; Supplemental Figure S4B).
These results suggest that formin-mediated barbed-end assembly is important for the elongated morphology and rapid motility characteristic of actin comets induced by Nck SH3 aggregation.
Formin FH1 domain overexpression disrupts Nck SH3–induced actin comets
The FH1 domain of formin interacts with the actin-binding protein profilin. Profilin-actin complexes are important for pathogenic Listeria motility (Grenklo et al., 2003). Recruitment of profilin-actin complexes by FH1 feeds actin monomers to the FH2 domain at the growing barbed ends (Vavylonis et al., 2006; Paul and Pollard, 2008; Truong et al., 2014). The FH1 domain is rich in polyproline repeats and also interacts with SH3 domain–containing proteins such as DIP and Src (Young and Copeland, 2010). Therefore we decided to use the formin FH1 domain as a competitive inhibitor of the endogenous formin activity on locally induced Nck SH3 actin structures. If formins are important for the formation of actin comets, we expect FH1 domain overexpression to have a greater effect on actin structures induced by Nck SH3 than on those induced by VCA.
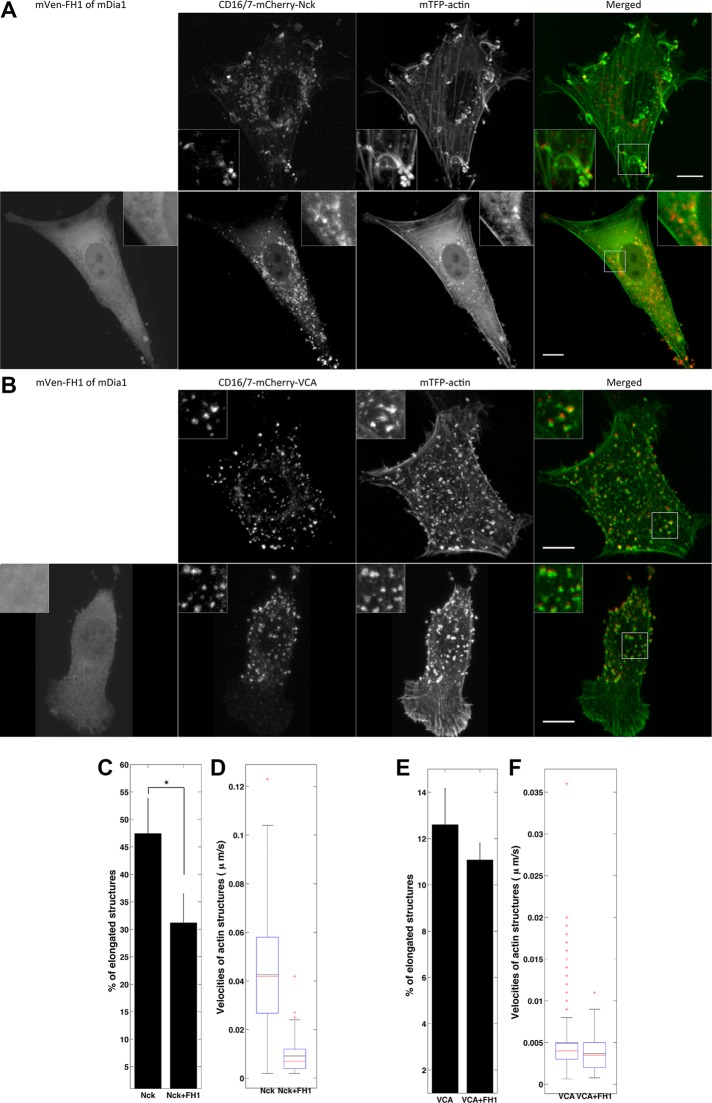
NIH3T3 cells were cotransfected with aggregatable VCA or Nck SH3 and fluorescently labeled actin with or without the FH1 domain of mammalian Diaphanous 1 formin (mDia1). Compared to control, overexpression of the FH1 domain (Supplemental Figure S5) reduced the number of actin assemblies induced by Nck SH3 aggregation that had low circularity (31 vs. 47%; Figure 5, A and C), whereas the morphology of VCA-induced actin structures was not significantly affected by FH1 overexpression (11 vs. 13%; Figure 5, B and E). Velocity analysis revealed that Nck SH3–induced structures moved more slowly (Supplemental Movie S5A) in the cells expressing FH1 (Figure 5D), whereas the velocity of the N-WASP–VCA actin structures (Supplemental Movie S5B) was not affected by the presence of the FH1 domain (Figure 5F).
FIGURE 5:
Formin FH1 domain overexpression affects the shape and dynamics of actin structures induced by Nck SH3 but not those induced by VCA clustering. (A) Nck SH3–induced actin structures in cells without (top) or with (bottom) overexpression of formin FH1 domain. Merged images show the composites of CD16/7-mCherry-Nck SH3 and mTFP1-actin. (B) VCA-induced actin structures in cells without (top) or with (bottom) overexpression of formin FH1 domain. Merged images show the composites of CD16/7-mCherry-VCA and mTFP1-actin. Scale bars, 10 μm. (C) Morphology analysis of Nck SH3–induced actin structures in the control and in cells overexpressing formin FH1 domain. Control: three cells, 334 aggregates; FH1 overexpressing: three cells, 117 aggregates. Error bars are mean ± SEM. *p < 0.01. (D) Velocity analysis of Nck SH3–induced actin structures in the control and in cells overexpressing formin FH1 domain. Control: three cells, 201 aggregates; FH1 overexpressing: four cells, 86 aggregates. (E) Morphology of VCA-induced actin structures in the control and in cells overexpressing formin FH1 domain. Control: three cells, 420 aggregates; FH1 overexpressing: three cells, 248 aggregates. Error bars are mean ± SEM. (F) Velocity of VCA-induced actin structures in the control and in cells overexpressing formin FH1 domain. Control: three cells, 459 aggregates; FH1 overexpressing: three cells, 278 aggregates.
The results indicate that competitive inhibition of formin in membrane clusters strongly affected the properties of Nck SH3–induced but not VCA-induced actin assemblies, suggesting that formin-based polymerization distinguishes dynamic, elongated actin comets from slow-moving actin blobs.
N-WASP overexpression changes the morphology of Nck SH3–induced actin structures
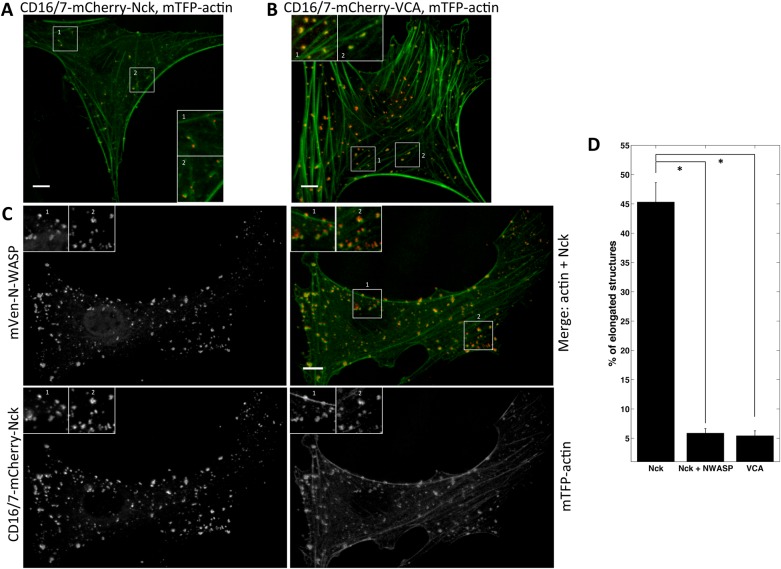
As another approach to probe whether the balance of linear and dendritic nucleation is crucial in Nck SH3-induced actin comets, we overexpressed mVen–N-WASP in cells expressing CD16/7-mCherry-Nck SH3 fusion proteins and fluorescently labeled actin (Figure 4A). We hypothesized that excess N-WASP would occupy more of the Nck SH3 domains in the membrane clusters at the expense of formin, resulting in a higher ratio of branched nucleation in the Nck SH3–induced actin comets. We first subjected NIH3T3 cells expressing CD16/7-mCherry-VCA and CD16/7-mCherry-Nck with or without mVen-N-WASP to antibody-mediated aggregation. Then we fixed and imaged the cells. Compared with control cells (Figure 6A), overexpression of N-WASP had a dramatic effect on the shape of the Nck SH3–induced actin assemblies (Figure 6C). This effect is reflected in a decrease in the number of actin particles with low circularity (45 vs. 6%) due to N-WASP overexpression (Figure 6D). In fact, the amount of elongated actin structures induced by Nck SH3 clustering in N-WASP–overexpressing cells (6%) was as low as in the cells with VCA-induced actin blobs (5%; Figure 6, B and D). This result is consistent with the proposal that shifting the balance in favor of branched versus linear nucleation affects Nck SH3–dependent actin comet tail formation and causes assembly of slow-moving, blob-like actin structures.
FIGURE 6:
N-WASP overexpression changes shape of Nck SH3–induced actin structures. (A–C) Actin structures induced by aggregation of Nck SH3 (A, C) or VCA (B) in NIH3T3 cells (A, B) or NIH3T3 cells overexpressing mVen-N-WASP (C). Cells were fixed. (C) mVen-N-WASP (top left), merge of mTFP1-actin (green) and Nck SH3 clusters (red; top right), CD16/7-mCherry-Nck (bottom left), and mTFP1-actin (bottom right). Scale bars, 10 μm. (D) Morphology analysis of Nck SH3–induced actin structures in the control and in cells overexpressing N-WASP. Nck-SH3: three cells, 200 aggregates; Nck-SH3 plus N-WASP overexpression: three cells, 329 aggregates; VCA: three cells, 250 aggregates. Error bars are mean ± SEM. *p < 0.001.
DISCUSSION
The main goal of these studies was to understand the role of the Nck adaptor protein in determining the morphology and dynamical behavior of actin comet tails. To address this question, we used antibody-mediated aggregation of proteins of interest at the membrane to induce localized actin assembly (Rivera et al., 2004); this gives us the ability to manipulate the signaling inputs leading to actin polymerization in a much more direct way than is possible in the case of pathogen-induced comets. Aggregation of Nck SH3 domains resulted in formation of actin comet tails (Rivera et al., 2004) similar to the vaccinia-induced comets (Frischknecht and Way, 2001; Dodding and Way, 2009). We reasoned that if the role of Nck was merely to recruit and activate the NPF N-WASP to the membrane, then it should be possible to recreate comet tails by clustering just the catalytic VCA domain of N-WASP. Surprisingly, we found that VCA clustering caused formation of actin blob structures, which do not have comet-like morphology and dynamics. Comparing Nck comets and VCA blobs deepened our understanding of mechanisms of formation of phenotypically distinct actin structures and revealed a critical and previously unappreciated role for Nck in the assembly of actin comet tails.
We first tested whether Nck might promote comet tail formation by regulating the density and/or turnover of VCA domains in the membrane clusters. Clustering CD16/7-mCherry-VCA with various ratios of CD16/7-XFP (Empty) transmembrane proteins allowed us to produce clusters in which VCA density could be systematically varied. We found that comet tails were not induced at lower densities of VCA. We sought to mimic cytosolic N-WASP interaction with Nck SH3 clusters through coiled-coil–mediated interaction of VCA with CD16/7 (Kd = 10 nM), which should fairly well simulate N-WASP–Nck binding. Nevertheless, providing VCA turnover in membrane clusters in this manner did not yield production of comet tails. Thus differences in VCA density or turnover are unlikely to explain why aggregation of Nck SH3 domains can induce actin comet tails, whereas aggregation of VCA alone cannot.
Another possible role of Nck SH3 is to promote linear actin nucleation in actin comet tails via recruitment and activation of formins. Accelerating barbed-end elongation is a crucial mechanism supporting growth of filopodial membrane protrusions (Pellegrin and Mellor, 2005; Schirenbeck et al., 2005; Yang and Svitkina, 2011). Similar to filopodia, actin comet tails are narrow, actin-rich structures pushing against the plasma membrane. Extremely long membrane protrusions can be formed in this manner. Indeed, we observe some Nck SH3–induced comets extending away from the cell boundary up to a distance greater than the diameter of a cell. This process is in fact very important for cell-to-cell spread of vaccinia virus (Cudmore et al., 1995; Doceul et al., 2010).
The Agaisse group showed that N-WASP and formin FHOD1 activities are both present in vaccinia comet tails (Alvarez and Agaisse, 2013, 2014). FHOD1 depletion results in less efficient comet tail production and lower velocity of vaccinia comets in host cells. Hence we reasoned that formin-based actin polymerization could be critical for maintenance of comet tail shape and dynamics. By contrast, VCA-induced actin structures are predicted to contain exclusively a branched actin network because clusters of VCA at the membrane should activate predominantly Arp2/3 molecules. Thus differences in the ability to promote linear polymerization might explain phenotypic differences between Nck SH3 comets and VCA blobs.
We hypothesized that Nck engages and controls the balance between the linear and branched nucleation machinery, thereby determining the shape and dynamic behavior of the resulting actin structures. The branched/linear actin nucleation balance in Nck SH3–induced comets could be upset by either inhibiting formin-based nucleation or increasing Arp2/3-based nucleation. We first inhibited FH2-mediated elongation of actin barbed ends (Rizvi et al., 2009), which altered the morphology and decreased the mobility of Nck SH3 actin assemblies. As an alternate method for reducing unbranched actin growth, we overexpressed the proline-rich FH1 domain of the formin mDia1. We found that Nck SH3 clusters produced mostly circular, slow-moving actin structures when FH1 was overexpressed. These results are consistent with an important role for linear actin nucleation in comet tail formation. To shift the balance toward branched nucleation in actin comets, we overexpressed N-WASP, which promotes branched nucleation. Under these conditions, Nck SH3–induced actin assemblies drastically changed their morphology. Thus experimentally shifting the unbranched/branched nucleation balance altered the morphology and dynamics of Nck SH3 actin comets. This is consistent with our idea that the integration of signaling pathways between linear and branched polymerization is critical for the comet tail phenotype.
The subversion of signaling to the Arp2/3-mediated branched actin network by pathogens has been actively studied (Haglund and Welch, 2011; Welch and Way, 2013). Although the critical role of Nck in recruiting and activating N-WASP to promote the branched nucleation has been well established in those studies, our results show that Nck performs a balancing act to also promote linear nucleation. We demonstrate that in Nck SH3–induced actin comets, the linear and dendritic polymerization machinery contribute to both the morphology and the dynamic behavior of actin comets. Of interest, it was reported that Rickettsia uses two modes of actin polymerization sequentially: Arp2/3 based, for the early stage of infection, and formin-like, for later comet tail motility (Haglund et al., 2010; Jermy, 2010; Reed et al., 2014). Recent studies of the F-actin composition in Listeria monocytogenes (Jasnin et al., 2013) and baculovirus (Mueller et al., 2014) comet tails suggested the presence of fairly long unbranched and bundled actin filaments.
Enteropathogenic Escherichia coli (EPEC) integrates its translocated intimin receptor (Tir) effector into the plasma membrane; Y474 of Tir is phosphorylated by host kinases Fyn and Abl to generate a binding site for the Nck SH2 domain (Hayward et al., 2006; Bhavsar et al., 2007). By recruiting Nck, EPEC localizes and activates N-WASP in the host cell (Supplemental Figure S6, C and D). This results in production of actin pedestals beneath the surface of the bacteria (Goosney et al., 1999). The morphology and dynamics of EPEC pedestals are similar to those of VCA-induced actin blobs (Supplemental Figure S6, E and F, and Supplemental Movie S6). On the basis of our results, we predict that the ratio of branched to linear actin nucleation activity may distinguish slow-moving actin pedestals from long, dynamic comet tails and that the formin activity in EPEC pedestals is lower (compared with N-WASP activity) than in vaccinia comets. Consistent with this hypothesis, we did not observe a significant effect of SMIFH2 (25 μM) on the dynamics and morphology of EPEC actin pedestals (unpublished data).
In summary, our results point to a role for Nck in maintaining a tight balance between the formin-mediated linear and the Arp2/3-based branched nucleation pathways. The dynamical behavior and elongated morphology of Nck SH3–induced actin comets depend on the presence of both pathways. Manipulations that inhibit linear nucleation or enhance branched nucleation have dramatic effects on actin comets in this system. We expect that the Nck adaptor plays a similar role in integrating the branched and linear nucleation pathways in actin assemblies induced in host cells by microbial pathogens, thus determining the dynamic properties of the resulting actin structures. The dynamics and morphology of other actin-rich structures, such as kidney podocyte foot processes and invadopodia, potentially also depend on the integration of different nucleation pathways through the Nck adaptor protein. Further studies will address the specific mechanism by which Nck engages the formin-based linear nucleation pathway.
MATERIALS AND METHODS
Plasmid construction
Details of constructions in the mammalian expression vector pEBB encoding fusion proteins consisting of the extracellular domain of CD16, the transmembrane domain of CD7, and hemagglutinin (HA) tag have been previously given (Rivera et al., 2004). CD16/7-mCherry-Nck SH3-HA, CD16/7-Nck SH3-HA, and CD16/7-HA (Empty) constructs are described in Ditlev et al. (2012). CD16/7-mCherry-VCA-HA and CD16/7-VCA-HA were cloned by replacing Nck SH3 domains with N-WASP VVCA (rat, aa P388–D501) in CD16/7-mCherry-Nck SH3-HA and CD16/7-Nck SH3-HA, respectively. CD16/7-eYFP-SynZip1 was generated by replacing HA in CD16/7-eYFP-HA with SynZip1 sequence (Reinke et al., 2010). HA-SynZip2-mCherry-VCA was generated by cloning SynZip2-mCherry-VCA (obtained by overlapping PCR; SynZip2 as in Reinke et al., 2010) into parental pEBB vector containing N-terminal HA tag. mVen-FH1 construct was generated by cloning the FH1(aa S543–P747) domain of mDia1 (mouse) into pTriEx4-mVen plasmid. mVen-N-WASP (rat) construct was generated by cloning full-length N-WASP into pTriEx4-mVen plasmid. GFP-actin is described in Gerisch and Muller-Taubenberger (2003) and Rivera et al. (2004). pmTFP1-actin was purchased from Allele Biotechnology (San Diego, CA).
Cell culture, transfections, antibody-mediated aggregation, fixation, and drug treatment
Mouse NIH3T3 cells were cultured in DMEM supplemented with 10% calf serum and 1% penicillin-streptomycin solution (Mediatech). Transient transfections were carried out using the Lipofectamine, Lipofectamine 2000 (Themo Fisher Scientific, Waltham, MA), or FuGene6 (Promega Life Sciences, Madison, WI) transfection reagent according to manufacturer’s protocol. The antibody-mediated aggregation protocol is described in detail elsewhere (Rivera et al., 2004; Ditlev et al., 2012). Briefly, ∼36–48 h after transfection, cells were sequentially incubated with a monoclonal antibody against human CD16 (Santa Cruz Biotechnology, Dallas, TX; 0.75 μg/ml, 20 min, 37°C) and then with an unlabeled goat anti–mouse IgG (Thermo Fisher Scientific; 0.50 μg/ml, 30 min, 37°C). Unbound antibody was removed from the cells by washing with complete medium after each antibody application, and then cells were immediately imaged using confocal microscopy or fixed for imaging at a later time. Fixation was performed with 4% paraformaldehyde in phosphate-buffered saline (PBS), followed by mounting on glass coverslips with Aqua-Poly/Mount (Polysciences, Warrington, PA). The small-molecule inhibitor of FH2 domains SMIFH2 was purchased from Tocris Bioscience (Bristol, UK) and dissolved in DMSO to 25 mM stock. Cells were treated with 100 μM SMIFH2 (or the same amount of DMSO for control) during the antibody-mediated aggregation procedure for later fixed-cell imaging (Figure 4B). For live-cell imaging (Supplemental Movie S4), SMIFH2 (or DMSO for control) was diluted in 100 μl of imaging medium and added to cells at the indicated time.
Western blotting
For Western immunoblotting, cell lysates were obtained from NIH3T3 cells as in Rivera et al. (2009). Protein content was determined by the Bradford assay (Bio-Rad, Hercules, CA), and equal amounts of protein were subjected to SDS–PAGE. After transfer to nitrocellulose membranes, blots were probed with anti-GFP (rabbit polyclonal IgG; Santa Cruz Biotechnology) to detect CD16/7-eYFP-SynZip1, anti-dsRed (rabbit polyclonal; Clontech, Mountain View, CA) to detect HA-SynZip2-mCherry-VCA, and anti-tubulin (mouse monoclonal; Abcam, Cambridge, UK) for loading control (Supplemental Figure S2B).
For the VCA density decrease experiments in Figure 2, the amount CD16/7-mCherry-VCA DNA was 100, 60, 33, 14, and 0% from the total amount of CD16/7-mCherry-VCA (VCA) and CD16/7 (Empty) DNA that was used for transfection. CD16/7 fusion proteins (CD16/7-mCherry-VCA and CD16/7) were detected with anti-CD16 antibody (mouse monoclonal; sc-19620; Santa Cruz Biotechnology). In cells coexpressing VCA and Empty fusion proteins (Supplemental Figure S1, lanes, 3–5), the expression of VCA was estimated to be 60, 37, and 15%, respectively, of the combined VCA and Empty expression.
To estimate overexpression of the formin FH1 domain (Figure 5 and Supplemental Figure S4), cells expressing this domain were compared with cells expressing full-length mDia1. The cells expressing mDia1 were transfected with the same amount of DNA (mVen-mDia1) as was used for transfection of mVen-FH1–expressing cells. Both mVen-FH1 and mVen-mDia1 were detected with anti-GFP (rabbit polyclonal IgG; Santa Cruz Biotechnology); mVen-mDia1 was also detected with anti-mDia1 (mouse monoclonal; 610848; BD Biosciences, Franklin Lakes, NJ). The level of FH1 domain overexpression versus endogenous FH1 was estimated by comparing the Ven-mDia1 band to the Ven-FH1 band (Supplemental Figure S4, IB: GFP).
Microscopy
At 18–24 h after transfection, cells were trypsinized and replated on either 25-mm cover glass for fixation or 35-mm glass-bottomed culture dishes (MatTek [Ashland, MA] or WPI [Sarasota, FL]) for live imaging.
Transfected cells were imaged in phenol red–free DMEM (Thermo Fisher) supplemented with 10% calf serum in glass-bottomed culture dishes and maintained at 37°C using a stage/objective heating (and CO2 regulation for LSM 780) system. Live-cell images were obtained using a spinning microlens confocal system (UltraView; PerkinElmer, Waltham, MA) mounted on an inverted microscope (TE2000; Nikon, Tokyo, Japan) equipped with a charge-coupled device camera (Orca-ER Firewire; Hamamatsu Photonics, Hamamatsu, Japan), MetaMorph (Molecular Devices, Sunnyvale, CA) software, and a 40×/1.25 numerical aperture (NA) oil immersion objective (Figures 1 and 2 and Supplemental Movies S1 and S2) or a laser scanning confocal microscope (LSM 780; Carl Zeiss, Oberkochen, Germany) with a 63×/1.4 NA oil immersion objective using ZEN (Carl Zeiss) software (Figures 3–6, Supplemental Figures S1 and S2, and Supplemental Movies S3–S5) to acquire the time-lapse images. Images were collected as vertical Z-stacks of optical sections. Maximum Z-projections were used for analysis. FRAP of mCherry-VCA (Supplemental Figure S2A and Supplemental Movie S3A) was done on an LSM 780 AxioObserver with C-Apochromat 40×/1.20 W Korr objective. Fluorescence was acquired from single confocal Z-slices of regions of interest (ROIs) with multiple CD16/7 aggregates. mCherry-VCA was bleached (5–10 times) after frame 3 in ROIs smaller than the acquisition ROI. For the FH2 inhibition experiment (Figure 4), cells were treated with 100 μm of SMIFH2 or DMSO (for control); multiple cells (using multistage acquisition mode) from each sample were imaged live every 153 s. The drug (or DMSO) was added between frames 5 and 6.
Cell culture, transfection, infection, and imaging of vaccinia comets
HeLa cells (American Type Culture Collection, Manassas, VA) were grown in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Life Technologies) at 37°C in a 5% CO2 incubator. Cells were seeded on glass coverslips at 20% confluency, infected with vB5R-GFP vaccinia virus (Ward and Moss, 2001) at 1 plaque-forming unit (pfu)/cell for 1 h, and then washed three times with PBS before addition of fresh medium. At 12 h after infection, cells were washed three times in PBS, fixed in 4% paraformaldehyde in PBS, permeabilized with 0.05% Triton X-100 in PBS, and stained with phalloidin–Alexa Fluor 568 (Life Technologies) to visualize actin tails. For time-lapse video microscopy (Alvarez and Agaisse, 2013), cells were seeded on a 35-mm MatTek glass-bottom dish (MatTek) at a confluency of 30–50%. Cells were transfected with pYFP–N-WASP and actin-CFP the day before infection and infected with WR vaccinia virus (Ward and Moss, 2001) at 10 pfu/cell. Images were captured 10 h after infection every 12 s on a spinning-disk confocal microscope (TE2000E) using a 60× oil objective with Volocity software (PerkinElmer). See Supplemental Figures S4 and S6B and Supplemental Movie S6.
Cell culture, transfection, infection, and imaging of enteropathogenic E. coli pedestals
NIH3T3 cells stably expressing mCherry–β-actin were maintained in subconfluent monolayers in DMEM supplemented with 10% heat-inactivated fetal calf serum, 1× antibiotic/antimycotic (Life Technologies), and 500 μg/ml G418 at 37˚C/5% CO2. Two days before infection, cells were seeded into 35-mm glass-bottom plates and induced to express mCherry-actin with 7.6 mM sodium butyrate 16 h before infection. EPEC cultures were grown overnight in DMEM plus 100 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) at 37˚C/5% CO2 to enhance type 3 secretion. The overnight culture was diluted 1:200 into DMEM plus 3.5% FBS plus 20 mM HEPES for infection at a multiplicity of infection of ∼6. Bacteria were centrifuged onto the cells at 200 × g for 5 min to synchronize the infection. After 3 h of infection, cells were washed twice with PBS, given fresh medium, and imaged 3–5 h postinfection. Live imaging of mCherry fluorescence was performed using a Nikon Eclipse Ti microscope with a Plan Apoλ 100×/1.45 NA objective. Images were captured at 10- to 15-s intervals using an Andor Clara-E camera and NIS Elements software. See Supplemental Figure S6D and Supplemental Movie S6.
Image analysis
Morphometric analysis was done in ImageJ (National Institutes of Health, Bethesda, MD) with a custom-written macro. In brief, each actin-dense structure was roughly outlined manually, including the surrounding area; background was subtracted; and the object was thresholded (20–40%) against the local background region and subjected to circularity measurement in ImageJ. Circularity ranges from 0 (infinitely long polygon or a line) to 1 (perfect circle) and is calculated as 4π(area/perimeter2). For velocity analysis, all actin structures associated with membrane CD16/7 clusters were tracked with the MTrackJ plug-in to ImageJ (www.imagescience.org/meijering/software/mtrackj/). Velocity distributions (3–13 cells/sample) of average velocity per cell are presented as box plots, where the red mark is the median, the black mark is the mean, and the lower edge is the q1 (25th) and the upper edge the q3 (75th) percentile. The whiskers are the most extreme data points, and outliers are shown by red plus signs. Outliers are points larger than q3 + w(q3 – q1) or smaller than q1 – w(q3 – q1), where w = 1.5. The plotted whisker extends to the adjacent value, which is the most extreme data value that is not an outlier.
For the step size analysis (Figure 4F), the average step size (distance between two consecutive positions of the particle) after the treatment (frames 6–20) was divided by the average step size before the treatment (frames 1–5).
Supplementary Material
Acknowledgments
We thank A. Cowan, S. Krueger, and W. Mohler for expert advice in imaging techniques and image analysis. We are grateful to A. Cowan for critically reading and providing insightful comments on the manuscript. This work was supported by National Institutes of Health grants P41GM103313 to L.M.L. and R01 CA82258 to B.J.M.
Abbreviations used:
- Arp2/3
actin-related proteins Arp 2 and Arp 3
- DIP
mDia-interacting protein
- mDia1
mammalian Diaphanous 1 formin
- NCKIPSD
Nck-interacting protein with SH3 domain
- NPF
nucleation-promoting factor
- N-WASP
neural Wiskott–Aldrich syndrome protein
- SH
Src homology
- SPIN90
SH3 protein interacting with Nck 90 kDa
- VCA
verprolin-homology, cofilin-homology, and acidic domains
- WISH
WASP-interacting SH3 protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-11-1555) on November 25, 2015.
REFERENCES
- Alvarez DE, Agaisse H. The formin FHOD1 and the small GTPase Rac1 promote vaccinia virus actin–based motility. J Cell Biol. 2013;202:1075–1090. doi: 10.1083/jcb.201303055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez DE, Agaisse H. A role for the small GTPase Rac1 in vaccinia actin-based motility. Small GTPases. 2014;5, e29038 doi: 10.4161/sgtp.29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann KJ, Pollard TD. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci USA. 2001;98:15009–15013. doi: 10.1073/pnas.211556398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoku S, Saksela K, Rivera GM, Mayer BJ. A crucial role in cell spreading for the interaction of Abl PxxP motifs with Crk and Nck adaptors. J Cell Sci. 2008;121:3071–3082. doi: 10.1242/jcs.031575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhavsar AP, Guttman JA, Finlay BB. Manipulation of host-cell pathways by bacterial pathogens. Nature. 2007;449:827–834. doi: 10.1038/nature06247. [DOI] [PubMed] [Google Scholar]
- Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol. 2013;24:258–266. doi: 10.1016/j.semcdb.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636-638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- Disanza A, Steffen A, Hertzog M, Frittoli E, Rottner K, Scita G. Actin polymerization machinery: the finish line of signaling networks, the starting point of cellular movement. Cell Mol Life Sci. 2005;62:955–970. doi: 10.1007/s00018-004-4472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditlev JA, Michalski PJ, Huber G, Rivera GM, Mohler WA, Loew LM, Mayer BJ. Stoichiometry of Nck-dependent actin polymerization in living cells. J Cell Biol. 2012;197:643–658. doi: 10.1083/jcb.201111113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science. 2010;327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding MP, Way M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe. 2009;6:536–550. doi: 10.1016/j.chom.2009.10.011. [DOI] [PubMed] [Google Scholar]
- Donnelly SK, Weisswange I, Zettl M, Way M. WIP provides an essential link between Nck and N-WASP during Arp2/3-dependent actin polymerization. Curr Biol. 2013;23:999–1006. doi: 10.1016/j.cub.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Remedios CG, Chhabra D, Kekic M, Dedova IV, Tsubakihara M, Berry DA, Nosworthy NJ. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- Eisenmann KM, Harris ES, Kitchen SM, Holman HA, Higgs HN, Alberts AS. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr Biol. 2007;17:579–591. doi: 10.1016/j.cub.2007.03.024. [DOI] [PubMed] [Google Scholar]
- Frischknecht F, Way M. Surfing pathogens and the lessons learned for actin polymerization. Trends Cell Biol. 2001;11:30–38. doi: 10.1016/s0962-8924(00)01871-7. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Muller-Taubenberger A. GFP-fusion proteins as fluorescent reporters to study organelle and cytoskeleton dynamics in chemotaxis and phagocytosis. Methods Enzymol. 2003;361:320–337. doi: 10.1016/s0076-6879(03)61017-7. [DOI] [PubMed] [Google Scholar]
- Goosney DL, de Grado M, Finlay BB. Putting E. coli on a pedestal: a unique system to study signal transduction and the actin cytoskeleton. Trends Cell Biol. 1999;9:11–14. doi: 10.1016/s0962-8924(98)01418-4. [DOI] [PubMed] [Google Scholar]
- Grenklo S, Geese M, Lindberg U, Wehland J, Karlsson R, Sechi AS. A crucial role for profilin-actin in the intracellular motility of Listeria monocytogenes. EMBO Rep. 2003;4:523–529. doi: 10.1038/sj.embor.embor823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, Kovar DR, Welch MD. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–1063. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward RD, Leong JM, Koronakis V, Campellone KG. Exploiting pathogenic Escherichia coli to model transmembrane receptor signalling. Nat Rev Microbiol. 2006;4:358–370. doi: 10.1038/nrmicro1391. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE. Now, swing your partner! 3D-domain switching of WASP activates Arp2/3 complex. Nat Struct Biol. 2003;10:583–584. doi: 10.1038/nsb0803-583. [DOI] [PubMed] [Google Scholar]
- Hufner K, Higgs HN, Pollard TD, Jacobi C, Aepfelbacher M, Linder S. The verprolin-like central (vc) region of Wiskott-Aldrich syndrome protein induces Arp2/3 complex-dependent actin nucleation. J Biol Chem. 2001;276:35761–35767. doi: 10.1074/jbc.M106520200. [DOI] [PubMed] [Google Scholar]
- Humphries AC, Dodding MP, Barry DJ, Collinson LM, Durkin CH, Way M. Clathrin potentiates vaccinia-induced actin polymerization to facilitate viral spread. Cell Host Microbe. 2012;12:346–359. doi: 10.1016/j.chom.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Humphries AC, Donnelly SK, Way M. Cdc42 and the Rho GEF intersectin-1 collaborate with Nck to promote N-WASP-dependent actin polymerisation. J Cell Sci. 2014;127:673–685. doi: 10.1242/jcs.141366. [DOI] [PubMed] [Google Scholar]
- Jasnin M, Asano S, Gouin E, Hegerl R, Plitzko JM, Villa E, Cossart P, Baumeister W. Three-dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc Natl Acad Sci USA. 2013;110:20521–20526. doi: 10.1073/pnas.1320155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermy A. Formin’ a comet tail. Nat Rev Microbiol. 2010;8:837. doi: 10.1038/nrmicro2481. [DOI] [PubMed] [Google Scholar]
- Jones N, Blasutig IM, Eremina V, Ruston JM, Bladt F, Li H, Huang H, Larose L, Li SS, Takano T, et al. Nck adaptor proteins link nephrin to the actin cytoskeleton of kidney podocytes. Nature. 2006;440:818–823. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- Kempiak SJ, Yamaguchi H, Sarmiento C, Sidani M, Ghosh M, Eddy RJ, Desmarais V, Way M, Condeelis J, Segall JE. A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin. J Biol Chem. 2005;280:5836–5842. doi: 10.1074/jbc.M410713200. [DOI] [PubMed] [Google Scholar]
- Kitamura T, Kitamura Y, Yonezawa K, Totty NF, Gout I, Hara K, Waterfield MD, Sakaue M, Ogawa W, Kasuga M. Molecular cloning of p125Nap1, a protein that associates with an SH3 domain of Nck. Biochem Biophys Res Commun. 1996;219:509–514. doi: 10.1006/bbrc.1996.0264. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101:14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- Lettau M, Kliche S, Kabelitz D, Janssen O. The adapter proteins ADAP and Nck cooperate in T cell adhesion. Mol Immunol. 2014;60:72–79. doi: 10.1016/j.molimm.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Lettau M, Pieper J, Janssen O. Nck adapter proteins: functional versatility in T cells. Cell Commun Signal. 2009;7:1. doi: 10.1186/1478-811X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Fan J, Woodley DT. Nck/Dock: An adapter between cell surface receptors and the actin cytoskeleton. Oncogene. 2001;20:6403–6417. doi: 10.1038/sj.onc.1204782. [DOI] [PubMed] [Google Scholar]
- Lim CS, Park ES, Kim DJ, Song YH, Eom SH, Chun JS, Kim JH, Kim JK, Park D, Song WK. SPIN90 (SH3 protein interacting with Nck, 90 kDa), an adaptor protein that is developmentally regulated during cardiac myocyte differentiation. J Biol Chem. 2001;276:12871–12878. doi: 10.1074/jbc.M009411200. [DOI] [PubMed] [Google Scholar]
- Mueller J, Pfanzelter J, Winkler C, Narita A, Le Clainche C, Nemethova M, Carlier MF, Maeda Y, Welch MD, Ohkawa T, et al. Electron tomography and simulation of baculovirus actin comet tails support a tethered filament model of pathogen propulsion. PLoS Biol. 2014;12:e1001765. doi: 10.1371/journal.pbio.1001765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: Nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New LA, Keyvani Chahi A, Jones N. Direct regulation of nephrin tyrosine phosphorylation by Nck adaptor proteins. J Biol Chem. 2013;288:1500–1510. doi: 10.1074/jbc.M112.439463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18:9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66:606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrin S, Mellor H. The Rho family GTPase Rif induces filopodia through mDia2. Curr Biol. 2005;15:129–133. doi: 10.1016/j.cub.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Quilliam LA, Lambert QT, Mickelson-Young LA, Westwick JK, Sparks AB, Kay BK, Jenkins NA, Gilbert DJ, Copeland NG, Der CJ. Isolation of a NCK-associated kinase, PRK2, an SH3-binding protein and potential effector of Rho protein signaling. J Biol Chem. 1996;271:28772–28776. doi: 10.1074/jbc.271.46.28772. [DOI] [PubMed] [Google Scholar]
- Ramesh N, Geha R. Recent advances in the biology of WASP and WIP. Immunol Res. 2009;44:99–111. doi: 10.1007/s12026-008-8086-1. [DOI] [PubMed] [Google Scholar]
- Rao Y. Dissecting Nck/Dock signaling pathways in Drosophila visual system. Int J Biol Sci. 2005;1:80–86. doi: 10.7150/ijbs.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed Shawna CO, Lamason Rebecca L, Risca Viviana I, Abernathy E, Welch Matthew D. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke AW, Grant RA, Keating AE. A synthetic coiled-coil interactome provides heterospecific modules for molecular engineering. J Am Chem Soc. 2010;132:6025–6031. doi: 10.1021/ja907617a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera GM, Briceno CA, Takeshima F, Snapper SB, Mayer BJ. Inducible clustering of membrane-targeted SH3 domains of the adaptor protein Nck triggers localized actin polymerization. Curr Biol. 2004;14:11–22. doi: 10.1016/j.cub.2003.12.033. [DOI] [PubMed] [Google Scholar]
- Rivera GM, Vasilescu D, Papayannopoulos V, Lim WA, Mayer BJ. A reciprocal interdependence between Nck and PI(4,5)P(2) promotes localized N-WASp-mediated actin polymerization in living cells. Mol Cell. 2009;36:525–535. doi: 10.1016/j.molcel.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi SA, Neidt EM, Cui J, Feiger Z, Skau CT, Gardel ML, Kozmin SA, Kovar DR. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem Biol. 2009;16:1158–1168. doi: 10.1016/j.chembiol.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KL, Smith GL. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008;16:472–479. doi: 10.1016/j.tim.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Nollau P, Ho HY, Kirschner MW, Mayer BJ. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J Biol Chem. 2001;276:26448–26452. doi: 10.1074/jbc.M103856200. [DOI] [PubMed] [Google Scholar]
- Romero S, Le Clainche C, Didry D, Egile C, Pantaloni D, Carlier M-F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and Profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- Scaplehorn N, Holmström A, Moreau V, Frischknecht F, Reckmann I, Way M. Grb2 and Nck act cooperatively to promote actin-based motility of vaccinia virus. Curr Biol. 2002;12:740–745. doi: 10.1016/s0960-9822(02)00812-6. [DOI] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol. 2005;7:619–625. doi: 10.1038/ncb1266. [DOI] [PubMed] [Google Scholar]
- Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- Siton O, Ideses Y, Albeck S, Unger T, Bershadsky AD, Gov NS, Bernheim-Groswasser A. Cortactin releases the brakes in actin-based motility by enhancing WASP-VCA detachment from Arp2/3 branches. Curr Biol. 2011;21:2092–2097. doi: 10.1016/j.cub.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Correa IR, Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife. 2013;2:e01008. doi: 10.7554/eLife.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E, Huynh-Do U, Lane AA, Cerretti DP, Daniel TO. Nck recruitment to Eph receptor, EphB1/ELK, couples ligand activation to c-Jun kinase. J Biol Chem. 1998;273:1303–1308. doi: 10.1074/jbc.273.3.1303. [DOI] [PubMed] [Google Scholar]
- Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat Rev Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- Thompson KE, Bashor CJ, Lim WA, Keating AE. SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains. ACS Synthetic Biol. 2012;1:118–129. doi: 10.1021/sb200015u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D, Copeland JW, Brumell JH. Bacterial subversion of host cytoskeletal machinery: Hijacking formins and the Arp2/3 complex. BioEssays. 2014;36:687–696. doi: 10.1002/bies.201400038. [DOI] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BM, Moss B. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J Virol. 2001;75:4802–4813. doi: 10.1128/JVI.75.10.4802-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisswange I, Newsome TP, Schleich S, Way M. The rate of N-WASP exchange limits the extent of ARP2/3-complex-dependent actin-based motility. Nature. 2009;458:87–91. doi: 10.1038/nature07773. [DOI] [PubMed] [Google Scholar]
- Welch MD, Way M. Arp2/3-mediated actin-based motility: a tail of pathogen abuse. Cell Host Microbe. 2013;14:242–255. doi: 10.1016/j.chom.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich L, Farago A, Buday L. Characterization of interactions of Nck with Sos and dynamin. Cell Signal. 1999;11:25–29. doi: 10.1016/s0898-6568(98)00027-8. [DOI] [PubMed] [Google Scholar]
- Yang C, Svitkina T. Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh Migr. 2011;5:402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young KG, Copeland JW. Formins in cell signaling. Biochim Biophys Acta. 2010;1803:183–190. doi: 10.1016/j.bbamcr.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Zhao ZS, Manser E, Lim L. Interaction between PAK and nck: a template for Nck targets and role of PAK autophosphorylation. Mol Cell Biol. 2000;20:3906–3917. doi: 10.1128/mcb.20.11.3906-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.