Aid-based RCC1 conditional knockout cells are established to examine the functional domains of RCC1. Based on an analysis using RCC1-deficient cells, two pathways are proposed for nuclear localization of RCC1: NLS-dependent and NLS-independent pathways.
Abstract
RCC1, a guanine nucleotide exchange factor of the small GTPase Ran, plays various roles throughout the cell cycle. However, the functions of RCC1 in biological processes in vivo are still unclear. In particular, although RCC1 has multifunctional domains, the biological significance of each domain is unclear. To examine each domain of RCC1, we established an RCC1 conditional knockout chicken DT40 cell line and introduced various RCC1 mutants into the knockout cells. We found that nuclear reformation did not occur properly in RCC1-deficient cells and examined whether specific RCC1 mutants could rescue this phenotype. Surprisingly, we found that neither the nuclear localization signal nor the chromatin-binding domain of RCC1 is essential for its function. However, codisruption of these domains resulted in defective nuclear reformation, which was rescued by artificial nuclear localization of RCC1. Our data indicate that chromatin association of RCC1 during mitosis is crucial for its proper nuclear localization in the next interphase. Moreover, proper nuclear localization of RCC1 in interphase is essential for its function through its nucleotide exchange activity.
INTRODUCTION
The small GTPase Ran regulates multiple cellular processes, including nucleocytoplasmic transport, mitotic spindle assembly, and nuclear envelope assembly (Clarke and Zhang, 2008). The GTPase cycle of Ran is driven by the guanine nucleotide exchange factor (GEF) regulator of chromosome condensation 1 (RCC1) and the GTPase-activating protein RanGAP1 (Clarke and Zhang, 2008). Throughout the cell cycle, RCC1 is associated with chromatin, whereas RanGAP1 localizes in the cytoplasm. The distinct subcellular localization of these Ran-specific regulators results in accumulation of Ran-GTP within interphase nuclei or near mitotic chromosomes, whereas Ran-GDP accumulates within interphase cytoplasm or distal to mitotic chromosomes. Therefore proper localization of RCC1 is crucial for asymmetric Ran-GTP distribution.
RCC1 is a 45-kDa nuclear protein composed of a short, flexible N-terminal domain (NTD) and a major catalytic domain (Renault et al., 1998). The NTD contains a lysine-rich region, which functions as a nuclear localization signal (NLS). This NLS mediates the nuclear import of RCC1 by the importin α3/β pathway in a Ran-GTP–dependent manner (Nemergut and Macara, 2000; Talcott and Moore, 2000; Quensel et al., 2004). Because RCC1 is the sole Ran-GTP producer, it is unlikely that its localization relies exclusively on self-produced Ran-GTP. In fact, the N-terminal–truncated mutant of RCC1 can also localize to the nucleus, suggesting the existence of additional nuclear localization mechanisms (Seino et al., 1992; Nemergut and Macara, 2000; Moore et al., 2002). It is possible that the chromatin-binding feature of RCC1 may contribute to its nuclear localization.
Although RCC1 is associated with chromatin throughout the cell cycle, the regions of RCC1 involved in this association remain unclear. Previous studies suggest that the NTD contributes to chromatin binding of RCC1. In addition, binding of NTD to chromatin DNA requires α-methylation of N-terminal serine 2 by the N-terminal RCC1 methyltransferase (Chen et al., 2007; Tooley et al., 2010). On the other hand, structural analysis of the RCC1–nucleosome complex indicates that the major catalytic domain of RCC1 binds directly with histones H2A/H2B and nucleosomal DNA (Makde et al., 2010). However, the biological significance of this site remains elusive.
In this study, we used RCC1-deficient DT40 cells to examine the functional role of each RCC1 domain, including the chromatin-binding domain. We found that the histone/DNA-binding site of the catalytic domain largely contributed to chromatin association of RCC1, whereas the NTD was not involved in chromatin binding. Surprisingly, neither the NTD nor the histone/DNA-binding domain was indispensable for RCC1 function. However, codisruption of both of these domains resulted in cell death, and this phenotype was rescued by artificial nuclear localization of RCC1. Furthermore, we found that although the transiently expressed, NTD-truncated mutant RCC1 localized in the cytoplasm during interphase, it loaded onto the chromatin just after nuclear envelope breakdown (NEB) and remained in the nuclei until the next interphase. These data indicate that chromatin association of RCC1 during mitosis is crucial for its nuclear localization in the following interphase, and we conclude that this is essential for the nuclear accumulation of Ran-GTP.
RESULTS
Establishment of efficient RCC1-conditional knockout cell lines
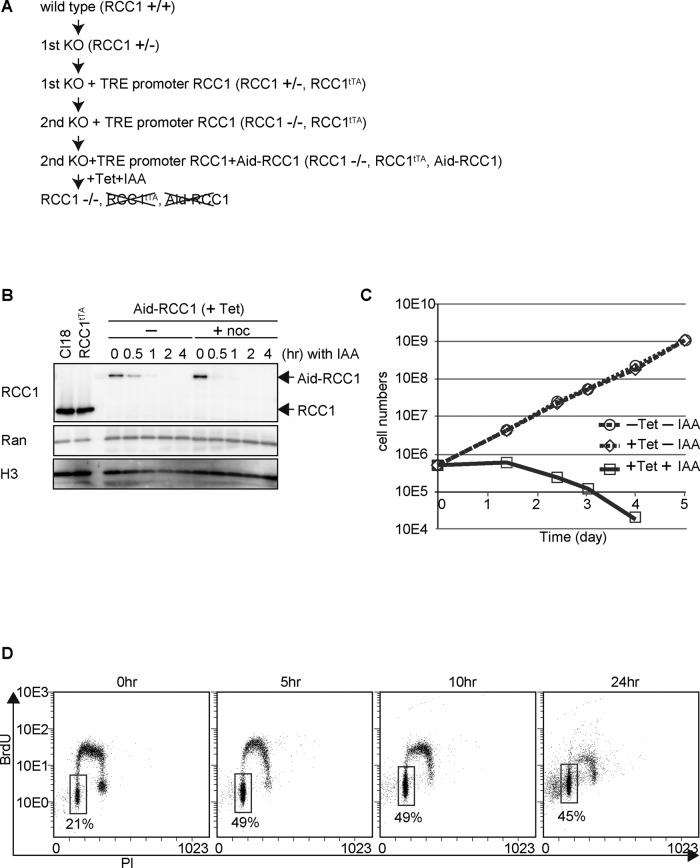
To analyze the function of RCC1 in vivo, we first created an RCC1-deficient DT40 chicken cell line (Figure 1A). Because RCC1 is essential for cell viability, a conditional knockout cell line was generated by expressing RCC1 from cDNA, under the control of a tetracycline-responsive promoter (TRE), after disruption of the endogenous RCC1 alleles (Figure 1A and Supplemental Figure S1A). Establishment of the tetracycline (TET)-induced knockout cell lines (RCC1−/−, Rcc1tTA) was confirmed by Southern hybridization (Supplemental Figure S1, A and B). Although RCC1 protein expression was gradually lost after TET addition, its levels reached <1% after 3 d (Supplemental Figure S1C). Consistent with the slow RCC1 depletion response, cells stopped growing and died within 3–5 d after TET addition (Supplemental Figure S1, D and E). However, evaluation of substantial RCC1 function at each cell cycle stage required its rapid depletion. For this, an auxin-induced-degradation (Aid) system (Nishimura et al., 2009) was introduced in the TET-induced knockout cell line to generate a highly efficient conditional knockout cell line (Figure 1A). The TET-responsive RCC1 was replaced with Aid-tagged RCC1, which could be degraded by the addition of indole-3-acetic acid (IAA; a natural auxin) in the presence of TET. As expected, Aid-RCC1 expression was completely lost 1 h after the addition of IAA (Figure 1B). This rapid degradation occurred both in the presence and in the absence of nocodazole (a spindle poison), and the amount of Ran was unchanged by the addition of IAA (Figure 1B). These observations indicate that this degradation system specifically affects RCC1 expression throughout the cell cycle. Thus, this conditional knockout cell line is useful for studying the function of RCC1 at particular stages of the cell cycle.
FIGURE 1:
Establishment of the Aid-based RCC1 conditional knockout cell line. (A) Scheme of Aid-based RCC1 conditional knockout cell line generation. (B) Protein levels of Aid-RCC1 in Aid-based RCC1 conditional knockout cells after addition of IAA in the presence or absence of nocodazole. Wild-type DT40 (Cl18) cells or cells expressing RCC1 cDNA (RCC1tTA) under the control of a TET-responsive promoter were also examined. Whole-cell lysates were subjected to 5–20% SDS–PAGE, followed by Western blot analysis with affinity-purified polyclonal anti-RCC1 antibody. Anti-Ran was used as a negative control, and anti–histone H3 was used as the loading control. (C) Proliferation of Aid-based RCC1 conditional knockout cells after addition of TET and IAA. Live cells were counted after trypan blue staining. (D) Cell cycle distribution after depletion of RCC1. Aid-based RCC1 conditional knockout cells were maintained in the presence of TET at a final concentration of 2 μg/ml, and samples were collected at the indicated times after addition of 500 μM IAA. Incorporation of propidium iodide (x-axis, linear scale) and BrdU (y-axis, log scale) was analyzed by FACS. The boxes represent the populations of G1 phase cells, and the numbers indicate the percentages of G1 population.
The growth rates of this cell line in the presence and absence of TET were similar (Figure 1C), indicating that wild-type RCC1 was functionally replaced by Aid-RCC1. Therefore this conditional knockout cell line was cultured in the presence of TET (+TET, –IAA), and addition of IAA (+TET, +IAA) resulted in growth arrest and rapid cell death (Figure 1C). For further validation, we examined the cell cycle distribution and found that ∼50% of the cells were arrested at the G1 stage with the continuous presence of IAA (Figure 1D). Because loss of RCC1 presumably results in defective nuclear transport of many essential proteins, inhibition of various biological processes is possible at any stage of the cell cycle. However, because the Aid system showed remarkable G1 arrest, we focused on this phenotype.
RCC1 is not essential for mitotic progression but is required for nuclear reformation after mitosis
Microscopic examination of the morphology of RCC1-deficient cells frequently revealed cells with partially condensed, abnormally shaped nuclei (clover-shaped nuclei; Figure 2A). Corresponding to the G1 cell populations (Figure 1D), cells with clover-shaped nuclei were increased by the continuous presence of IAA (Figure 2B). Because these clover-shaped nuclei appeared to be condensed chromosomes, we hypothesized that this phenotype might result from abnormal mitosis. To rule out the possibility that this phenotype was caused by interphase defects, we synchronized the RCC1 conditional knockout cells in mitosis in the absence of IAA. Addition of IAA and subsequent release from mitosis converted most of the mitotic population to cells with clover-shaped nuclei (Figure 2C). Consistent with our hypothesis, this result indicates that cells with clover-shaped nuclei were formed during or after mitosis.
FIGURE 2:
Phenotype of RCC1-deficient cells. (A) Abnormal nuclear morphology (clover-shaped nuclei) of RCC1-deficient cells (right). Aid-based RCC1 conditional knockout cells were fixed before (left) and after (right) addition of 500 μM IAA and stained with DAPI. Scale bar, 10 μm. (B) Quantification of cell types after addition of IAA to Aid-based RCC1 conditional knockout cells. Samples were collected at the indicated times after addition of 500 μM IAA. (C) Quantification of cell types after release from mitotic arrest by nocodazole. Aid-based RCC1 conditional knockout cells were synchronized in mitosis by nocodazole treatment in the absence of IAA. Cells were washed for mitotic release in the presence or absence of IAA, and samples were collected at the indicated time-points (nocodazole release = time 0). (D) Live-cell imaging of Aid-based RCC1 conditional knockout cells expressing histone H2B-RFP (red) and GFP-tubulin (green) after IAA addition (bottom). Control cells are also shown (top). The numbers indicate time (minutes) from NEB. (E) Durations from NEB to the onset of anaphase for RCC1 conditional knockout cells, represented as box-and-whisker plots. (Control, n = 50, m = 25 min; +IAA, n = 22, m = 35 min). (F) Characterization of clover-shaped nuclei. RCC1 conditional knockout cells were stained with anti–phospho CDK1 substrates, DiOC6(3), and mAb414, respectively (left). Nuclei of GFP-lamin B2 (green)– and H2B-RFP (red)–expressing RCC1 conditional knockout cells were also observed microscopically (right).
To visualize directly the conversion of mitotic cells to cells with clover-shaped nuclei, we recorded the cell cycle progression of RCC1-deficient cells by live-cell imaging (Figure 2D and Supplemental Movie S1). Control cells took ∼30 min to progress from NEB to the onset of anaphase (Figure 2, D and E, and Supplemental Movie S2). In RCC1-deficient cells, after chromosomal condensation and alignment at the metaphase plate, chromosome segregation occurred normally. The time taken by RCC1-deficient cells to progress from NEB to the onset of anaphase was similar to that of the control cells (Figure 2, D and E, and Supplemental Movie S1). These observations suggest that RCC1 is not essential for mitotic progression. However, after mitotic exit, telophase/G1 cells began showing clover-shaped nuclei (from 54 to 72 min; Figure 2D, +IAA). We confirmed that these cells showed reduced CDK activity (Figure 2F). Considering these observations, we conclude that conversion to cells with clover-shaped nuclei occurs at telophase/G1 phase, just after the completion of mitosis.
Because normal nuclear reformation did not occur in RCC1-deficient cells (Figure 2D and Supplemental Movie S1), we examined their nuclear envelope. Presence of the nuclear membrane (lipid stained by 3,3’-dihexyloxacarbocyanine iodide [DiOC6(3)]), nuclear pore components (Nup recognized by mAb414), and lamin B2 was observed around the clover-shaped nuclei (Figure 2F).
The histone/DNA-binding region is essential for chromatin association of RCC1
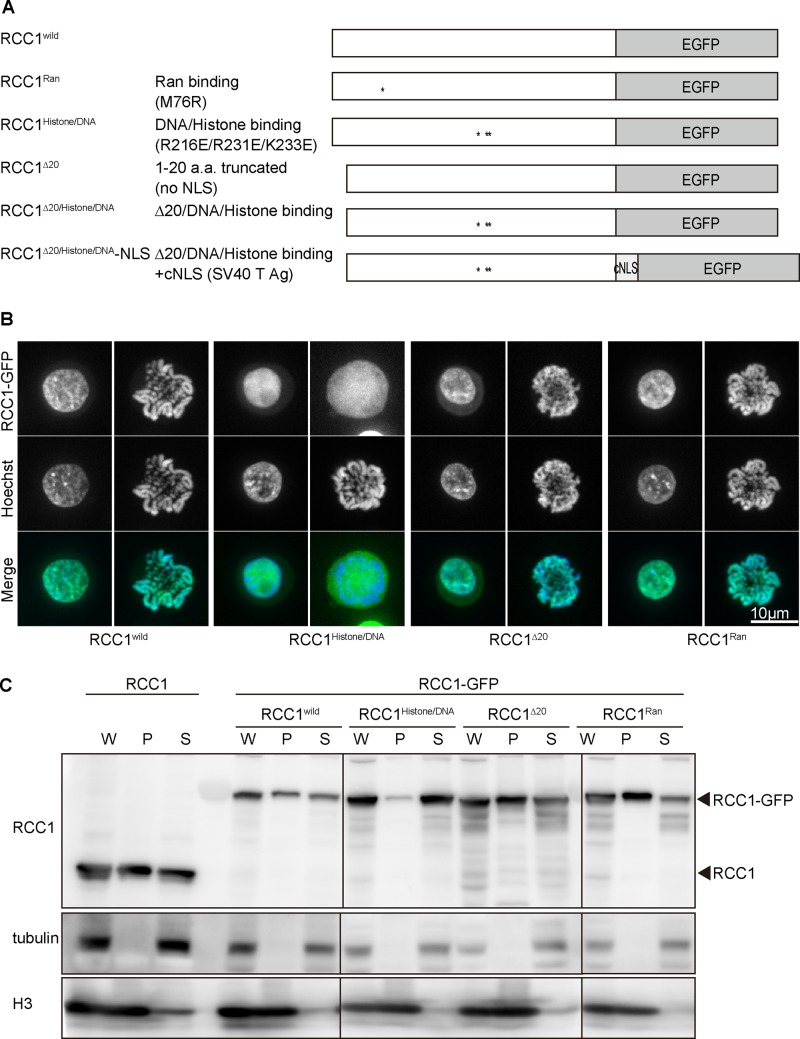
Next we examined the domains required for each RCC1 function. RCC1 contains multiple domains, including an NTD, a Ran-binding site, and a histone/DNA-binding site in the catalytic domain (Figure 3A). To evaluate the functional significance of these domains in vivo, we generated several mutants for each RCC1 domain (Figure 3A and Supplemental Figure S2), based on previous studies (Renault et al., 2001; Makde et al., 2010). RCC1∆20 (Δ1–20 amino acids) is an N-terminal–truncated mutant that lacks NLS. RCC1Ran (M76R) is expected to show defects in GEF activity because M76 is one of the critical Ran-binding sites (Renault et al., 2001), whereas RCC1histone/DNA (R216E/R231E/K223E) is expected to show defective histone/DNA binding (Makde et al., 2010).
FIGURE 3:
RCC1 histone/DNA-binding sites are required for chromatin association. (A) Diagrammatic representation of various RCC1 mutants. (B) Subcellular localization of mutant RCC1s in interphase and mitotic cells. RCC1-deficient cells expressing mutant RCC1-GFP (green) were stained with Hoechst (blue). (C) Biochemical fractionation of RCC1-deficient cells expressing mutant RCC1-GFP. Wild-type cells were used as controls. Whole-cell (W), chromatin (P), and soluble (S) fractions were analyzed by Western blot and probed with anti-RCC1 antibody. Tubulin was detected mainly in the soluble fraction, and histone H3 was detected mainly in the chromatin fraction.
To investigate the cellular localization of these mutants (Figure 3B and Supplemental Figure S3), we expressed green fluorescent protein (GFP)–tagged RCC1 mutants in RCC1-deficient cells. The wild-type RCC1 localized to the interphase nuclei and mitotic chromosomes; however, RCC1histone/DNA did not associate with mitotic chromosomes. In contrast, RCC1∆20 localized to interphase nuclei and mitotic chromosomes even though it lacks NLSs. As expected, the localization of RCC1Ran was comparable to that of wild-type RCC1 (Figure 3B and Supplemental Figure S3).
Although RCC1histone/DNA did not associate with mitotic chromosomes, it localized in interphase nuclei (Figure 3B and Supplemental Figure S3). To characterize the defects in the chromatin binding of RCC1histone/DNA in interphase nuclei, we separated the chromatin (P) and soluble (S) fractions by biochemical fractionation. Most RCC1histone/DNA was detected in the soluble fraction (Figure 3C), suggesting that RCC1histone/DNA does not associate with chromatin even in interphase nuclei. The distribution profile of other mutants was similar to that of wild-type RCC1 (Figure 3C). On the basis of these analyses, we concluded that chromatin binding of RCC1 is mediated by histone/DNA-binding sites in the catalytic domain, as predicted by its crystal structure (Makde et al., 2010).
Neither the chromatin-binding domain nor the NTD is essential for RCC1 function
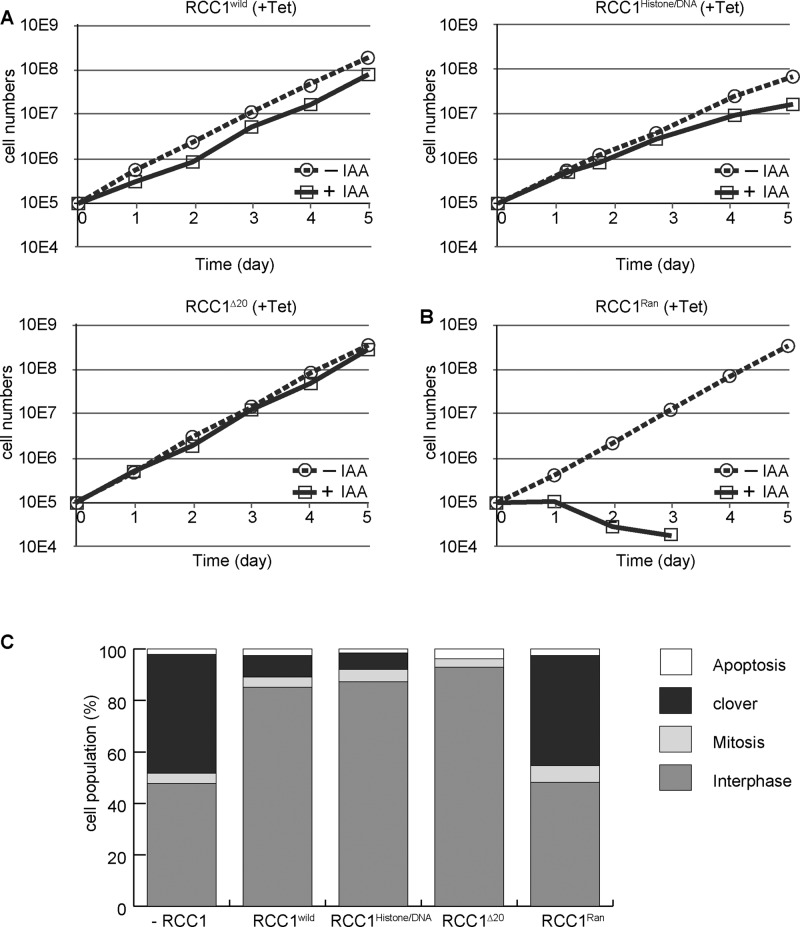
To assess the importance of each domain of RCC1, we examined the viability of each cell line in which wild-type RCC1 was replaced with mutant RCC1 (Figure 4A). We confirmed that the expression levels of RCC1 mutants were similar to that of endogenous RCC1 (Figure 3C). Introduction of wild-type RCC1 into RCC1-deficient cells restored cell growth (Figure 4A). Surprisingly, introduction of RCC1histone/DNA or RCC1∆20 could rescue the growth defect of RCC1-deficient cells (Figure 4A), indicating that either the chromatin-binding domain or the NTD is dispensable for RCC1 function. We also confirmed that the morphology of nuclei in RCC1histone/DNA- or RCC1∆20-expressing RCC1-deficient cells was normal (Figure 4C). In contrast, RCC1-deficient cells expressing RCC1Ran died after nuclear reformation defects (Figure 4B). We observed that 50% of RCC1Ran-expressing cells showed clover-shaped nuclei (Figure 4C). Because the RCC1Ran mutation causes defects in GEF activity, we conclude that GEF activity of RCC1 is essential for nuclear reformation; however, neither the chromatin-binding domain nor the NTD is required for RCC1 function.
FIGURE 4:
Neither the chromatin-binding domain nor the NTD is essential for RCC1 function. (A) Proliferation of RCC1-deficient cells expressing RCC1, RCC1histone/DNA, or RCC1∆20. (B) Proliferation of RCC1-deficient cells expressing RCC1Ran. (C) Quantification of cell types after expression of RCC1 mutants in RCC1-deficient cells. Samples were collected 5 h after addition of 500 μM IAA.
Nuclear localization of RCC1 is essential for its function
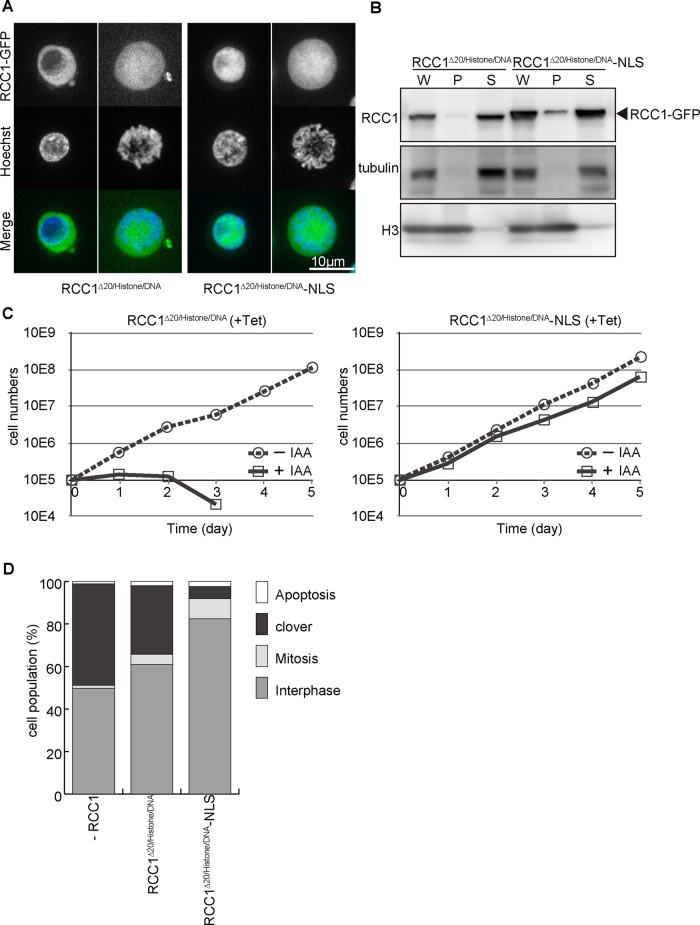
Because RCC1∆20 showed nuclear localization in the interphase without an NLS, we hypothesized that nuclear localization may be facilitated by chromatin association rather than the NLS. To test this hypothesis, we generated a double mutant for NTD and chromatin-binding domain (RCC1∆20/histone/DNA) and introduced this mutant into RCC1-deficient cells (Figure 5, A–D). Consistent with our hypothesis, RCC1∆20/histone/DNA did not localize in interphase nuclei and was not associated with mitotic chromosomes (Figure 5, A and B, and Supplemental Figure S4). In addition, RCC1∆20/histone/DNA did not rescue RCC1 deficiency (Figure 5, C and D). These analyses suggest that these two domains of RCC1 show functional overlap for its nuclear localization, which might be essential for its function.
FIGURE 5:
Nuclear localization of RCC1 is essential for its function. (A) Subcellular localization of RCC1∆20/histone/DNA or RCC1∆20/histone/DNA-NLS in interphase and mitotic cells. RCC1-deficient cells expressing each mutant RCC1-GFP (green) were stained with Hoechst (blue). (B) Biochemical fractionation of RCC1-deficient cells expressing RCC1∆20/histone/DNA or RCC1∆20/histone/DNA-NLS. Whole-cell (W), chromatin (P), and soluble (S) fractions were analyzed by Western blot with anti-RCC1 antibody. (C) Proliferation of RCC1-deficient cells expressing RCC1∆20/histone/DNA or RCC1∆20/histone/DNA-NLS. (D) Quantification of cell types after expression of RCC1∆20/histone/DNA or RCC1∆20/histone/DNA-NLS in RCC1-deficient cells. Samples were collected 5 h after addition of 500 μM IAA.
To test whether nuclear localization of RCC1 is crucial for its function, we added SV40-NLS to the C-terminal end of RCC1∆20/histone/DNA (RCC1∆20/histone/DNA-NLS) and expressed it in RCC1-deficient cells. Although RCC1∆20/histone/DNA-NLS did not show chromatin association (Figure 5, A and B, and Supplemental Figure S4), it was observed to localize in interphase nuclei (Figure 5A). Of importance, RCC1∆20/histone/DNA-NLS rescued RCC1 deficiency (Figure 5, C and D), suggesting that GEF activity is not affected in this double mutant. On the basis of these analyses, we conclude that nuclear localization of RCC1 is essential for its function.
Chromatin-binding activity during mitosis facilitates interphase nuclear localization of RCC1
Although our results suggest that nuclear localization of RCC1 is essential for its function, it is unclear how RCC1∆20 could localize in the nuclei in the absence of the NLS. Because the NTD and the chromatin-binding domain show functional overlap for nuclear localization of RCC1, it is possible that RCC1 translocates into the nucleus via its chromatin-binding activity. To visualize this process, we transiently expressed each RCC1 mutant and observed the nuclear import of newly synthesized RCC1-GFP by live-cell imaging.
RCC1histone/DNA translocated into the nuclei of cells within their first interphase, soon after its expression by using its NLS. After NEB, RCC1histone/DNA spread throughout the cell and dissociated from chromatin during mitosis; it then translocated back into the nuclei by using its NLS in the second interphase (Figure 6A, second panel). In contrast, RCC1∆20, which lacks an NLS, was not incorporated into the nuclei in the first interphase after its expression. However, it associated with chromatin during mitosis and remained in the nuclei at the second interphase, possibly due to its chromatin-binding activity (Figure 6A, third panel). The double mutant RCC1∆20/histone/DNA did not incorporate into the nuclei at any point (Figure 6A, fourth panel). On the basis of these results, we propose two pathways for nuclear localization of RCC1: the NLS-dependent and NLS-independent pathways (Figure 6B). The NLS-dependent pathway uses the N-terminal NLS, whereas the association of RCC1 with chromatin during mitosis mediates its nuclear localization at the next interphase in the NLS-independent pathway.
FIGURE 6:
Chromatin binding of RCC1 during mitosis facilitates its nuclear localization in interphase. (A) Temporal localization of each mutant RCC1-GFP after transient expression. Localization of newly synthesized RCC1-GFP was followed throughout the cell cycle by live-cell imaging. RCC1-GFP intensity within the cytosol and nucleus were quantified before (first interphase, blue) and after (second interphase, red) mitosis. N/C ratios are represented as box-and-whisker plots. (B) A model for nuclear localization of RCC1: NLS-dependent and -independent pathways. In the NLS-dependent pathway, N-terminal NLS is essential for the nuclear transport of RCC1, which is mediated by importin α3/β in a Ran-GTP–dependent manner. In the NLS-independent pathway, chromatin binding of RCC1 during mitosis plays an important role in its nuclear localization at the next interphase.
DISCUSSION
RCC1 has been identified as the gene product responsible for the phenotype of the tsBN2 cell line (Uchida et al., 1990). This cell line is a temperature-sensitive cell cycle–progression mutant produced by random mutagenesis (Nishimoto et al., 1978). The RCC1 protein disappears in these cells at the restrictive temperature for reasons yet unknown. At the same time, the possibility that heat stress could disrupt the Ran cycle in experiments using tsBN2 cells cannot be excluded completely (Furuta et al., 2004; Miyamoto et al., 2004). Although the tsBN2 cell has long been a major genetic tool, it was worth creating an alternative tool to analyze the function of RCC1 for the reasons mentioned. In this study, we established an efficient conditional knockout cell line in which auxin addition can rapidly eliminate RCC1. This is a useful tool to assess the function of RCC1 at specific stages of the cell cycle.
Different experimental approaches suggested that RCC1 contributes to multiple cellular functions through the Ran-GTP gradient around chromosomes, including mitotic spindle assembly (Kalab and Heald, 2008). However, our auxin-based RCC1 knockout DT40 cell line did not show significant defects in spindle shape or chromosome segregation. On the other hand, we found abnormally shaped nuclei after mitotic exit, leading us to conclude that RCC1 is not essential for mitotic progression in chicken DT40 cells but is required for nuclear reformation. It is possible that despite the rapid loss of RCC1, there was sufficient Ran-GTP for spindle formation, although not for nuclear reformation. Alternatively, DT40 cells may be less dependent on Ran-GTP for spindle assembly than some other experimental systems. In addition to the Ran-GTP gradient (Clarke and Zhang, 2008; Hasegawa et al., 2013), the chromosomal passenger complex (CPC; composed of inner centromere protein, Borealin/DasraA, Survivin, and Aurora B; Sampath et al., 2004) directly contributes to the spindle assembly (Zierhut and Funabiki, 2015). CPC, rather than the Ran-GTP gradient, may be the major contributor to spindle assembly in DT40 cells. In fact, we observed centromeric localization of Aurora B in RCC1-deficient cells (unpublished data).
A striking phenotype that we observed in DT40 cells involved defects in nuclear reformation. Although we did not observe significant changes in the nuclear envelope components that we tested (Figure 2F), it is possible that the integrity of the nuclear envelopes in RCC1 deficient cells was compromised. We could not validate the functionality of the nuclear envelope, as these cells did not show nuclear transport activity (unpublished data). In addition, given that RCC1-deficient cells exited mitosis at the normal time, it is also possible that the cells had acquired some defects during mitosis that were reflected in the terminal phenotype that we observed in the telophase/G1 phase.
To investigate each domain of RCC1, we characterized RCC1-deficicent cells expressing various RCC1 mutants. Our results are summarized in Supplemental Figure S5. Our analyses of RCC1 mutants revealed that the major chromatin-binding domain of RCC1 was not the NTD but the histone/DNA-binding site of the catalytic domain. Previous reports suggested that the NTD contributes to chromatin binding of RCC1 and that the NTD bound to chromatin DNA through α-methylation of the N-terminal serine 2 (Chen et al., 2007; Hitakomate et al., 2010). However, because chicken RCC1 does not have a consensus sequence for α-methylation (Supplemental Figure S2), the contribution of its NTD to chromatin binding might be less than that observed in other species.
We found that the histone/DNA-binding site was not essential for cellular proliferation. However, careful examination of growth curves indicated that RCC1-deficient cells expressing the histone/DNA mutant RCC1 showed slightly slower growth (Figure 4). Although the cell cycle distribution of these cells was similar to that of wild-type cells, it is possible that the histone/DNA-binding site contributes to additional RCC1 functions.
In summary, our data support a model in which parallel NLS-dependent and NLS-independent pathways promote the nuclear localization of RCC1 (Figure 6B). Wild-type cells possess both pathways, either of which is sufficient for survival. The NLS-dependent pathway is essential for nuclear import of newly synthesized RCC1 during interphase, and yet RanBP1 can inhibit cytosolic RCC1 and maintain the Ran-GTP gradient until NEB even if this pathway is compromised (Bischoff et al., 1995; Zhang et al., 2014). Once NEB occurs, RCC1 can bind to the chromatin through its chromatin-binding region and remain nuclear through this association indefinitely.
Conversely, the NLS-independent pathway is essential for chromatin binding during mitosis, which in turn promotes the correct localization of RCC1 in the subsequent interphase. In the absence of this pathway, we expect that cells will have difficulty in establishing the Ran-GTP gradient at the telophase/G1 transition. In this case, the small portion of active RCC1 (Ciciarello et al., 2010; Zhang et al., 2014) that is passively incorporated into nuclei at the end of mitosis might be enough to jump start the Ran-GTP gradient because most cytosolic RCC1 could be inhibited by RanBP1 after mitosis.
In summary, our genetic experiments indicate that these two pathways for RCC1 localization are redundant. In the event that one pathway is compromised, the other pathway still allows for cell growth and survival. This redundancy is consistent with the idea that because the nuclear localization of RCC1 is crucial for its function, both pathways may be active at every cell cycle stage to ensure proper nuclear localization of RCC1.
MATERIALS AND METHODS
Cell culture and cell line construction
DT40 cells were maintained in Dulbecco’s modified medium (Sigma-Aldrich, Tokyo, Japan) supplemented with 10% fetal bovine serum, 1% chicken serum, 100 μM β-mercaptoethanol, and penicillin–streptomycin (Gibco, Tokyo, Japan) in an atmosphere containing 5% CO2 at 38.5°C.
To disrupt the RCC1 gene, targeting vectors containing a selectable marker at the RCC1 gene locus were constructed (Supplemental Figure S1A, bold line). The 5′ genomic arm (chromosome 23, 2104841-2107474) was amplified by subjecting the genomic DNA to PCR using the primers GCTCTAGAGCATTTCACTGGAACAAACACG and TCCGGATCCCTGCTCCTTCCTCAGCCTCCTC. The 3′ genomic arm (chromosome 23, 2110389-2118112 for first knockout or 2113048-2118163 for second knockout) was cloned from the lambda Fix II DT40 genomic library by using RCC1 cDNA as a probe. For the TRE promoter RCC1 construct, the chicken RCC1 cDNA was amplified by PCR from chicken DT40 mRNA by using the primers TATAAGCTTATGTCTGGAAAGCGTGCTGCC and ATAGGATCCCAGCTCCGTGCCTTGTCCTTG and cloned into pUHD10-3.
Plasmid constructs were linearized and transfected with a Gene Pulser II electroporator (Bio-Rad, Tokyo, Japan) into DT40 cells. Histidinol (1 mg/ml), Zeocin (1 mg/ml), and puromycin (0.5 μg/ml) were used to select for stable transfectants. After selection, DNA from drug-resistant clones was extracted and analyzed by Southern blot after digestion with Bglll or EcoRV and then probed with L or R probe, respectively (Supplemental Figure S1B). To obtain Aid-based RCC1-conditional knockout cells, the construct (Nishimura et al., 2009) containing the Aid degron-tag at the N-terminal end of RCC1 was transfected, and stable transfectants were selected in 2 mg/ml G418.
RCC1 mutants were generated by site-directed mutagenesis and transfected into the Aid-based RCC1-conditional knockout cells. Stable transfectants were selected in 25 μg/ml blasticidin S.
Western blot analysis and antibodies
Whole-cell lysates were prepared and analyzed by 5–20% SDS–PAGE. Western blot analysis was performed by a standard protocol. Rabbit polyclonal anti-chicken RCC1 antibody (used at 1:10,000) was raised against hexahistidine-ggRCC1 and affinity purified. Anti-Ran antibody (used at 1:5000; BD Transduction Laboratories, Tokyo, Japan) and anti–α-tubulin antibody (DM1A, used at 1:1000; Sigma-Aldrich) were used. Anti–histone H3 antibody (used at 1:10.000) was a gift from H. Kimura (Tokyo Institute of Technology, Tokyo, Japan).
Fluorescence-activated cell sorting
Cells were treated with 20 μM bromodeoxyuridine (BrdU) for 20 min before fixation at the indicated time points. After fixation with ice-cold 70% ethanol, cells were stained with anti-BrdU (BD Biosciences, Tokyo, Japan) and propidium iodide and subjected to fluorescence-activated cell sorting (FACS) analysis.
Indirect immunofluorescence staining
Cells were cytospun onto slides and fixed in 3.7% paraformaldehyde in phosphate-buffered saline (PBS) at room temperature for 15 min. After permeabilization with 0.1% Triton X-100 in PBS at room temperature for 5 min, samples were blocked with 1% bovine serum albumin (BSA) in PBS. Cells were probed with primary antibodies against phospho-CDK substrate (9477; used at 1:1000; Cell Signaling Technology, Danvers, MA) or mAb414 (ab24609, used at 1:5000; Abcam, Cambridge, UK) diluted with 1% BSA in PBS at 37°C for 1 h. After washing, Cy3-conjugated secondary antibody diluted with 1% BSA in PBS was used, and DNA was stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Immunofluorescence images were collected with a Cool SNAP HQ camera (Roper Scientific, Tokyo, Japan) mounted on an Olympus IX71 inverted microscope with a 100× objective lens together with a filter wheel (Olympus, Tokyo, Japan). All subsequent analysis and processing of images were performed using MetaMorph software (Molecular Devices, Tokyo, Japan).
Live-cell imaging
For live-cell imaging, histone H2B–red fluorescent protein (RFP) and tubulin-GFP plasmids (Figure 2D and Supplemental Movies S1 and S2), histone H2B-RFP and GFP–lamin B2 plasmids (Figure 2F), or mutant RCC1-GFP plasmids (Figures 3B and 5A) were stably transfected into RCC1-deficient cells. Cells were stained with 1 μM DiOC6(3) (Figure 2F) or 100 ng/ml Hoechst 33342 (Figures 2F, 3B, and 5A) and observed using a confocal scanner box (CellVoyager CV1000; Yokogawa, Tokyo, Japan) with an oil immersion 100× objective lens in an atmosphere containing 5% CO2 at 37°C in the presence of 500 μM IAA. Time-lapse images were recorded at 3-min intervals with an exposure time of 0.1 s, and Z-sections (n = 30) were acquired at 0.3-μm steps for each time point (Figure 2D and Supplemental Movies S1 and S2).
To observe the temporal localization of newly synthesized RCC1-GFP, RCC1 mutants were transiently transfected using the Neon transfection system (Life Technologies, Carlsbad, CA). One hour after transfection, time-lapse images were recorded at 3-min intervals with an exposure time of 0.1 s. RCC1-GFP intensities within the cytosol and the nucleus (five points each) were quantified before (first interphase, blue) and after (second interphase, red) mitosis, and N/C ratios were represented as box-and-whisker plots (Figure 6A; n = 20).
Cell fractionation for biochemical analysis
RCC1-deficient cells expressing mutant RCC1-GFP were lysed in the lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 150 mM KCl, 2 mM MgCl2, 0.1% NP40; 1 mM dithiothreitol/protease inhibitor) on ice for 5 min. The soluble fractions (S) were taken from the supernatant, and pellet fractions (P) were washed twice with the lysis buffer. Whole-cell (W), chromatin (P), and soluble (S) fractions were analyzed by 5–20% SDS–PAGE. Western blot analysis was performed with indicated antibodies.
Quantification of cell types
RCC1-deficient cells expressing mutant RCC1s were cytospun onto slides. After fixation, the cells were stained with anti–histone H3pS10 (1:10,000 dilution; provided by H. Kimura) and DAPI. Histone H3P10–positive cells were counted as mitotic cells, and partially condensed, abnormally shaped nuclei were counted as clover-shaped nuclei. More than 500 cells were scored for each cell line.
Supplementary Material
Acknowledgments
We are very grateful to Mayumi Takahashi, Kaeko Nakaguchi, and Michiko Arii for technical assistance and Mary Dasso for useful suggestions and comments on the manuscript. This work was supported by Grants-in-Aid for Scientific Research (S) and for Scientific Research on Innovative Areas (Chromosome OS) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.F.
Abbreviations used:
- Aid
auxin-induced-degradation
- CDK
cyclin-dependent kinase
- CPC
chromosomal passenger complex
- DiOC6(3)
3,3’-dihexyloxacarbocyanine iodide
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- IAA
indole-3-acetic acid
- NEB
nuclear envelope breakdown
- NLS
nuclear localization signal
- NTD
N-terminal domain
- Nup
nuclear pore component
- RCC1
regulator of chromosome condensation 1
- TET
tetracycline
- TRE
tetracycline-responsive promoter.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E15-07-0497) on November 12, 2015.
REFERENCES
- Bischoff FR, Krebber H, Smirnova E, Dong W, Ponstingl H. Co-activation of RanGTPase and inhibition of GTP dissociation by Ran-GTP binding protein RanBP1. EMBO J. 1995;14:705–715. doi: 10.1002/j.1460-2075.1995.tb07049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Muratore TL, Schaner-Tooley CE, Shabanowitz J, Hunt DF, Macara IG. N-terminal alpha-methylation of RCC1 is necessary for stable chromatin association and normal mitosis. Nat Cell Biol. 2007;9:596–603. doi: 10.1038/ncb1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciciarello M, Roscioli E, Di Fiore B, Di Francesco L, Sobrero F, Bernard D, Mangiacasale R, Harel A, Schinina ME, Lavia P. Nuclear reformation after mitosis requires downregulation of the Ran GTPase effector RanBP1 in mammalian cells. Chromosoma. 2010;119:651–668. doi: 10.1007/s00412-010-0286-5. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Furuta M, Kose S, Koike M, Shimi T, Hiraoka Y, Yoneda Y, Haraguchi T, Imamoto N. Heat-shock induced nuclear retention and recycling inhibition of importin alpha. Genes Cells. 2004;9:429–441. doi: 10.1111/j.1356-9597.2004.00734.x. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Ryu SJ, Kalab P. Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J Cell Biol. 2013;200:151–161. doi: 10.1083/jcb.201206142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitakomate E, Hood FE, Sanderson HS, Clarke PR. The methylated N-terminal tail of RCC1 is required for stabilisation of its interaction with chromatin by Ran in live cells. BMC Cell Biol. 2010;11:43. doi: 10.1186/1471-2121-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Saiwaki T, Yamashita J, Yasuda Y, Kotera I, Shibata S, Shigeta M, Hiraoka Y, Haraguchi T, Yoneda Y. Cellular stresses induce the nuclear accumulation of importin alpha and cause a conventional nuclear import block. J Cell Biol. 2004;165:617–623. doi: 10.1083/jcb.200312008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore W, Zhang C, Clarke PR. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr Biol. 2002;12:1442–1447. doi: 10.1016/s0960-9822(02)01076-x. [DOI] [PubMed] [Google Scholar]
- Nemergut ME, Macara IG. Nuclear import of the Ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol. 2000;149:835–850. doi: 10.1083/jcb.149.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto T, Eilen E, Basilico C. Premature of chromosome condensation in a ts DNA- mutant of BHK cells. Cell. 1978;15:475–483. doi: 10.1016/0092-8674(78)90017-x. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods. 2009;6:917–922. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- Quensel C, Friedrich B, Sommer T, Hartmann E, Kohler M. In vivo analysis of importin alpha proteins reveals cellular proliferation inhibition and substrate specificity. Mol Cell Biol. 2004;24:10246–10255. doi: 10.1128/MCB.24.23.10246-10255.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Kuhlmann J, Henkel A, Wittinghofer A. Structural basis for guanine nucleotide exchange on Ran by the regulator of chromosome condensation (RCC1) Cell. 2001;105:245–255. doi: 10.1016/s0092-8674(01)00315-4. [DOI] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- Sampath SC, Ohi R, Leismann O, Salic A, Pozniakovski A, Funabiki H. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Seino H, Hisamoto N, Uzawa S, Sekiguchi T, Nishimoto T. DNA-binding domain of RCC1 protein is not essential for coupling mitosis with DNA replication. J Cell Sci. 1992;102:393–400. doi: 10.1242/jcs.102.3.393. [DOI] [PubMed] [Google Scholar]
- Talcott B, Moore MS. The nuclear import of RCC1 requires a specific nuclear localization sequence receptor, karyopherin alpha3/Qip. J Biol Chem. 2000;275:10099–10104. doi: 10.1074/jbc.275.14.10099. [DOI] [PubMed] [Google Scholar]
- Tooley CE, Petkowski JJ, Muratore-Schroeder TL, Balsbaugh JL, Shabanowitz J, Sabat M, Minor W, Hunt DF, Macara IG. NRMT is an alpha-N-methyltransferase that methylates RCC1 and retinoblastoma protein. Nature. 2010;466:1125–1128. doi: 10.1038/nature09343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Sekiguchi T, Nishitani H, Miyauchi K, Ohtsubo M, Nishimoto T. premature chromosome condensation is induced by a point mutation in the hamster RCC1 gene. Mol Cell Biol. 1990;10:577–584. doi: 10.1128/mcb.10.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MS, Arnaoutov A, Dasso M. RanBP1 governs spindle assembly by defining mitotic Ran-GTP production. Dev Cell. 2014;31:393–404. doi: 10.1016/j.devcel.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zierhut C, Funabiki H. Nucleosome functions in spindle assembly and nuclear envelope formation. BioEssays. 2015;37:1074–1085. doi: 10.1002/bies.201500045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.