Abstract
Lupus is an autoimmune disease characterized by the development of antinuclear autoantibodies and immune complex-mediated tissue damage. T cells in lupus patients appear to undergo apoptosis at an increased rate, and this enhanced T cell apoptosis has been postulated to contribute to lupus pathogenesis by increasing autoantigen load. However, there is no direct evidence to support this hypothesis. In this study, we show that an Lck-cre transgene, which increases T cell apoptosis as a result of T cell specific expression of cre recombinase, accelerates the development of autoantibodies and nephritis in lupus-prone (NZB×NZW)F1 mice. Although the enhanced T cell apoptosis in Lck-cre transgenic mice resulted in an overall decrease in the relative abundance of splenic CD4+ and CD8+ T cells, the proportion of activated CD4+ T cells was increased and no significant change was observed in the relative abundance of suppressive T cells. We postulate that the Lck-cre transgene promoted lupus by enhancing T cells apoptosis, which, in conjunction with the impaired clearance of apoptotic cells in lupus-prone mice, increased the nuclear antigen load and accelerated the development of anti-nuclear autoantibodies. Furthermore, our results also underscore the importance of including cre-only controls in studies using the cre-lox system.
Keywords: T-cell apoptosis, autoantibodies, lupus, cre toxicity, (NZB × NZW)F1 mice
Introduction
Lupus is a chronic autoimmune disease associated with the enhanced survival and hyperactivation of autoreactive B cells and T cells. Perhaps paradoxically though, T cells from lupus patients also appear to undergo apoptosis at an increased rate compared to that in healthy controls.1–5 This increase in T cell apoptosis has been postulated to contribute to the pathogenesis of lupus by increasing the load of autoantigens. In human lupus patients, increased rates of T cell apoptosis has been correlated with higher disease activity.1 However, these studies are correlative and do not demonstrate a cause-effect relationship. Likewise, direct evidence to support the hypothesis that enhanced T cell apoptosis promotes lupus in mice is also lacking. In fact, whereas injection of apoptotic B cells into lupus prone (NZB × NZW)F1 mice has been shown to accelerate the development of lupus, injection of apoptotic T cells failed to elicit this response.6 Indeed, it has also been proposed that the increased rate of T cell apoptosis in lupus patients could be an epiphenomenon.6
Lupus is also associated with the inefficient clearance of nuclear antigens that can be released when cells undergo apoptosis. Both lupus patients and murine models for lupus exhibit impaired clearance of endogenous apoptotic bodies.7–9 Induction of macrophage apoptosis, which further attenuates macrophage-dependent clearance of apoptotic debris while simultaneously increasing nuclear antigen load, has been shown to exacerbate autoimmunity in lupus-prone mice.10 Additionally, mutations in genes such as Tim-4, Dnase1, Sap, C1q, and Mer, which encode proteins that play a role in the clearance of apoptotic cells, result in the development of lupus in mice, even when the mutations are harbored on a non-autoimmune genetic background.8,11,12 Altogether, these data suggest that inefficient clearance of apoptotic cells leads to the exposure of the immune system to intracellular antigens, including nuclear antigens, which drives the selection of autoreactive immune cells and promotes the development of anti-nuclear antibodies, including anti-chromatin IgG and anti-dsDNA IgG.
The use of the cre-lox system to generate tissue- or cell type- specific knockouts has become commonplace, and several recent publications investigating gene function in mouse models of lupus have employed this system.13–15 However, one caveat to this approach is that expression of cre-recombinase can result in significant DNA damage and chromosome rearrangements, leading to cell growth defects and increased apoptosis both in vitro and in vivo.16–22 This increase in cell death is thought to be due to single- and double-stranded breaks that result from cre-mediated cleavage at cryptic and pseudo loxP sites, which are estimated to occur at a frequency of about 1.2 sites per megabase (Mb) in the mouse genome.23–26 Although cre recombinase expression can cause DNA damage and apoptosis, most studies using the cre-lox system do not include cre only controls, which would allow one to determine if these (or other) cre-mediated side-effects had any impact on the phenotype being studied.22,27
In this study, we report that an Lck cre transgene accelerates the development of autoantibodies and lupus nephritis in (NZB × NZW)F1 mice. As has been reported previously, we found that in Lck-cre transgenic mice, the expression of cre recombinase driven from the T cell specific Lck promoter resulted in a significant increase in T cell apoptosis.28–30 In the context of the lupus prone (NZB × NZW)F1 genetic background, we hypothesize that this increase in apoptosis overwhelms the inefficient apoptotic cell clearance in these mice. The resulting accumulation of apoptotic debris would increase the exposure of immune cells to nuclear antigens, which would subsequently accelerate the development of autoimmunity. This interpretation of our results is fully consistent with the hypothesis that enhanced T cell apoptosis in lupus-prone individuals can promote the development of lupus. Furthermore, our results emphasize the critical importance of cre-only controls in experiments using the cre-lox system.
Materials and Methods
Generation of the NZB.Lck-cre and NZW.ERαfl/fl congenic lines
Congenic NZB.Lck-cre mice were generated by backcrossing FVB.Lck-cre mice,31 a kind gift from Dr. Rene Opavsky of the University of Nebraska Medical Center, onto the NZB background in conjunction with marker assisted selection as we have described previously.32 To eliminate FVB alleles that might impact the lupus phenotype of (NZB × NZW)F1 mice, we genotyped mice from each backcross generation using 120 polymorphic markers spanning the 19 murine autosomes (detailed in Table 1). PCR genotyping was performed as we have described previously32 using oligonucleotide primers specific for simple sequence length polymorphisms (SSLP) between the FVB and NZB strains. Primer sequences were obtained from the Mouse Genome Informatics database (www.informatics.jax.org). Mice carrying the Lck-cre transgene were identified via a PCR-based protocol obtained from The Jackson Laboratory (http://jaxmice.jax.org/strain/003802.html), and utilized two primer pairs; the first detected the Lck-cre transgene (F: 5’-CAGTCAGGAGCTTGAATCCCACGA-3’, R: 5’-TAATCGCCATCTTCCAGCAG-3’), and the second amplified a 324 bp region of the interleukin 2 gene as positive control (F: 5’-CTAGGCCACAGAATTGAAAGATCT-3’, R: 5’-GTAGGTGGAAATTCTAGCATCATCC-3’).
Table 1.
SSLP markers used in the genotyping of the NZB.Lck-cre straina
| Marker (location in cM, Mbp) | Marker (location in cM, Mbp) | Marker (location in cM, Mbp) |
|---|---|---|
| D1Mit316 (2.46, 10.4) | D6Mit133 (62.18, 127.3) | D13Mit266 (14.44, 36.9) |
| D1Mit123 (17.67, 39.1) | D6Mit14 (77.64, 145.7) | D13Mit13 (30.06, 56.5) |
| D1Mit214 (28.97, 58.2)b | D6Mit373 (77.70, 147.1) | D13Mit202 (47.43, 91.5) |
| D1Mit75a (29.16, 58.7) b | D13Mit106 (47.75, 92.7) | |
| D1Mit249 (30.56, 60.8) b | D7Mit267 (17.09, 29.5) | D13Mit74 (56.92, 105.9) |
| D1Mit303 (31.79, 62.9) b | D7Mit248 (27.8, 59.3) | |
| D1Mit132 (39.51, 77.1) | D7Mit93 (44.86, 79.0) | D14Mit126 (11.94, 21.9) |
| D1Mit440 (44.98, 88.8) | D7Mit31 (49.01, 87.5) | D14Mit55 (Syntenic)c |
| D1Mit495 (55.79, 127.6) | D7Mit282 (63.44, 118.2) | D14Mit5 (31.49, 59.7) |
| D1Mit47 (65.12, 152.9) | D7Mit109 (81.84, 136.5) | D14Mit68 (37.61, 72.5) |
| D1Mit111 (76.73, 169.0) | D7Mit223 (88.85, 144.6) | D14Mit228 (Syntenic)d |
| D1Mit155 (98.20, 194.4) | D14Mit170 (56.16, 106.3) | |
| D8Mit155 (2.14, 4.98) | ||
| D2Mit1 (2.23, 3.8) | D8Mit289 (16.47, 28.7) | D15Mit13 (1.84, 3.5) |
| D2Mit242 (31.72, 57.3) | D8Mit190 (22.16, 36.0) | D15Mit252 (8.54, 22.6) |
| D2Mit395 (59.97, 119.5) | D8Mit45 (42.16, 87.3) | D15Mit123 (29.31, 37.3) |
| D2Mit285 (75.41, 152.9) | D8Mit215 (62.63, 115.9) | D15Mit67 (32.17, 70.2) |
| D2Mit411 (80.04, 159.6) | D15Mit70 (38.02, 81.2) | |
| D2Mit145 (86.75, 166.4) | D9Mit90 (17.80, 32.5) | D15Mit161 (52.78, 97.0) |
| D9Mit129 (24.45, 43.9) | ||
| D3Mit178 (14.27, 30.4) | D9Mit336 (35.39, 65.6) | D16Mit131 (3.41, 7.3) |
| D3Mit67 (24.50, 53.2) | D9Mit269 (47.15, 87.9) | D16Mit169 (Syntenic)e |
| D3Mit311 (40.14, 93.0) | D9Mit347 (55.11, 103.3) | D16Mit139 (37.28, 65.7) |
| D3Mit106 (49.08, 112.0) | D9Mit19 (71.55, 120.4) | D16Mit52 (53.73, 92.7) |
| D3Mit319 (56.89, 128.3) | ||
| D3Mit320 (66.75, 143.6) | D10Mit80 (3.66, 11.6) | D17Mit101 (15.17, 29.3) |
| D3Mit147 (74.02, 148.7) | D10Mit213 (9.75, 20.4) | D17Mit180 (26.71, 51.4) |
| D10Mit3 (16.53, 29.2) | D17Mit53 (38.64, 67.2) | |
| D4Mit227 (4.43, 10.0) | D10Mit20 (34.83, 67.0) | D17Mit93 (45.20, 73.8) |
| D4Mit237a (22.38, ND) | D10Mit95 (47.06, 92.5) | D17Mit155 (55.02, 84.5) |
| D4Mit288 (31.66, 56.8) | D10Mit180 (66.65, 118.2) | |
| D4Mit9 (43.34, 95.1) | D18Mit68 (11.96, 21.4) | |
| D4Mit337 (61.59, 127.5) | D11Mit71 (4.70, 6.9) | D18Mit177 (21.39, 41.0) |
| D4Mit251 (69.05, 136.9) | D11Mit231 (21.92, 35.7) | D18Mit91 (29.67, 55.4) |
| D4Mit256 (86.16, 155.0) | D11Mit5 (40.59, 67.2) | D18Mit185 (42.78, 69.0) |
| D11Mit285 (54.64, 89.9) | D18Mit48 (51.89, 76.8) | |
| D5Mit145 (3.37, 7.3) | D11Mit336 (73.75, 110.6) | D18Mit144 (57.79, 85.5) |
| D5Mit353 (21.25, 40.4) | ||
| D5Mit201 (39.55, 75.1) | D12Mit182 (5.52, 10.9) | D19Mit68 (3.38, 3.64) |
| D5Mit24 (54.68, 112.3) | D12Mit60 (15.54, 34.8) | D19Mit96 (15.54, 21.8) |
| D5Mit370 (65.23, 126.8) | D12Mit54 (22.76, 53.9) | D19Mit46 (29.06, 32.9) |
| D5Mit143 (89.80, 151.0) | D12Mit91 (30.06, 71.7) | D19Mit88 (32.23, 37.3) |
| D12Mit158 (38.14, 82.6) | D19Mit90 (35.97, 42.2) | |
| D6Mit116 (11.50, 21.2) | D12Mit118 (44.93, 91.5) | D19Mit33 (51.76, 56.2) |
| D6Mit268 (15.12, 34.7) | D12Mit101 (51.55, 102.5) | |
| D6Mit123 (27.76, ND) | D12Nds2 (62.22, 113.9) | |
| D6Mit36 (48.93, 104.5) |
Abbreviations: cM, centiMorgan; Mb, Megabase pair; ND, Not determined
SSLP marker locations and oligo sequences were obtained from http://www.informatics.jax.org.
Underlined markers denote those used for linkage analysis of the Lck-cre transgene
Position is undetermined, however additional positional data in the MGI database places this marker on chromosome 14 between 16.4 – 18.6 cM
Position is undetermined, however additional positional data in the MGI database places this marker on chromosome 14 at approximately 46 cM
Position is undetermined, however additional positional data in the MMDBJ database places this marker on chromosome 16 between 32.5 – 36.5 cM
Marker assisted selection was also used to generate the NZW.ERαfl/fl strain, which is homozygous for an estrogen receptor alpha (ERα) allele in which exon 3 is flanked by loxP sites. This strain was obtained by backcrossing B6.ERαfl/fl mice, obtained from Ken Korach, onto the NZW background. 33 To accomplish this, genotypes were determined using 105 PCR-based SSLP markers that are polymorphic between the B6 and NZW strains (detailed in Table 2). The presence of the ERα floxed allele was assessed with PCR using REDTaq ReadyMix PCR Reaction Mix (Sigma-Aldrich, St. Louis, MO) and a primer set that detected the insertion of the loxP sites (F: 5’- GACTCGCTACTGTGCCGTGTGC-3’; R: 5’-CTTCCCTGGCATTACCACTTCTCCT-3’). 34 At the N6 backcross generation, the genetic backgrounds for both NZB.Lck-cre and NZW.ERαfl mice were assessed at the DartMouse™ Speed Congenic Core Facility at Dartmouth Medical School. DartMouse uses the Illumina, Inc. (San Diego, CA) GoldenGate Genotyping Assay to interrogate 1449 SNPs spread throughout the genome. The raw SNP data were analyzed using DartMouse’s SNaP-Map™ and Map-Synth™ software, to determine the SNP genotype, and thus strain of origin of SNP alleles, in each mouse.
Table 2.
SSLP markers used in the genotyping of the NZW.ERαfl straina
| Marker (location in cM, Mbp) | Marker (location in cM, Mbp) | Marker (location in cM, Mbp) |
|---|---|---|
| D1Mit316 (2.46, 10.4) | D7Mit267 (17.09, 29.5) | D12Mit60 (15.54, 34.8) |
| D1Mit171 (15.40, 36.7) | D7Mit83 (32.84, 51.8) | D12Mit285 (23.46, 54.6) |
| D1Mit215 (39.91, 78.2) | D7Mit350 (47.43, 83.6) | D12Mit158 (38.14, 82.6) |
| D1Mit440 (44.98, 88.8) | D7Mit323 (54.45, 100.9) | D12Mit118 (44.93, 91.5) |
| D1Mit1001 (56.44, 129.0) | D7Mit109 (81.84, 136.5) | D12Nds2 (62.22, 113.9) |
| D1Mit159 (69.03, 159.7) | ||
| D1Mit426 (84.32, ND) | D8Mit155 (2.14, 5.0) | D13Mit275 (14.50, 37.3) |
| D1Mit155 (98.20, 194.4) | D8Mit190 (22.16, 36.0) | D13Mit13 (30.06, 56.5) |
| D8Mit178 (34.43, 71.0) | D13Mit78 (67.21, 118.8) | |
| D2Mit66 (49.45, 84.8) | D8Mit211 (52.00, 102.7) | |
| D2Mit14a (49.68, 85.9) | D8Mit88 (61.66, 114.8) | D14Mit98 (7.08, 16.5) |
| D2Mit480 (56.65, 109.9) | D8Mit49 (72.38, 124.1) | D14Mit60 (24.60, 47.1) |
| D2Mit395 (59.97, 119.5) | D14Mit28 (34.66, 67.7) | |
| D2Mit148 (100.49, 178.8) | D9Mit90 (17.80, 32.5) | D14Mit228 (Syntenic)d |
| D9Mit129 (24.45, 43.9) | D14Mit170 (56.16, 106.3) | |
| D3Mit203 (10.82, 26.9) | D9Mit191 (25.52, 46.7) | |
| D3Mit167 (15.64, 32.3) | D9Mit96 (27.75, 50.8) | D15Mit13 (1.84, 3.5) |
| D3Mit51 (Syntenic)b | D9Mit142 (30.13, 56.3) | D15Mit115 (22.92, 56.3) |
| D3Mit106 (49.08, 112.0) | D9Mit48 (31.63, 57.9) | D15Mit159 (41.96, 87.5) |
| D3Mit320 (66.75, 143.6) | D9Mit208 (33.65, 62.3) | D15Mit161 (52.78, 97.0) |
| D3Mit89 (80.79, 156.8) | D9Mit269 (47.15, 87.9) | |
| D9Mit355 (51.41, 98.8) | D16Mit131 (3.41, 7.3) | |
| D4Mit227 (4.43, 10.0) | D9Mit19 (71.55, 120.4) | D16Mit60 (23.27, 32.7) |
| D4Mit193 (13.99, 32.2) | D16Mit139 (37.28, 65.7) | |
| D4Mit17 (33.96,63.4) | D10Mit166 (2.06, 3.9) | D16Mit189 (46.83, 82.5) |
| D4Mit9 (43.34, 95.1) | D10Mit298 (2.78, 8.8) | D16Mit52 (53.73, 92.7) |
| D4Mit308 (57.66, 124.2) | D10Mit152 (3.77, 12.2) | |
| D4Mit42 (82.64, 151.6) | D10Mit248 (5.21, 14.2) | D17Mit81 (15.80, 30.9) |
| D10Mit106 (11.67, 24.4) | D17Mit51 (19.74, ND) | |
| D5Mit348 (11.97, 24.9) | D10Mit36 (24.87, 48.7) | D17Mit10 (Syntenic)e |
| D5Mit391 (24.24, 44.7) | D10Mit20 (34.83, 67.0) | D17Mit217 (35.26, 65.9) |
| D5Mit309 (42.22, 79.5) | D10Mit230 (45.28, ND) | D17Mit93 (45.20, 73.8) |
| D5Mit314 (53.25, 109.7) | D10Mit95 (47.06, 92.5) | |
| D10Mit233 (61.58, 114.4) | D18Mit177 (21.39, 41.0) | |
| D6Mit138 (1.81, 4.5) | D18Mit186 (45.63, 72.0) | |
| D6Mit268 (15.12, 34.7) | D11Mit231 (21.92,35.7) | D18Mit144 (57.79, 85.5) |
| D6Mit123 (27.76, ND) | D11Mit4 (41.87, 68.6) | |
| D6Mit284 (41.11, 92.6) | D11Mit285 (54.64, 89.9) | D19Mit96 (15.54, 21.8) |
| D6Mit328 (52.62, 112.8) | D11Mit48 (82.96, 118.1) | D19Mit90 (35.97, 42.2) |
| D6Mit198 (Syntenic)c | D19Mit103 (48.46, 53.8) | |
| D6Mit14 (77.64, 145.7) | ||
| D6Mit374 (64.60, 134.1) |
Abbreviations: cM, centiMorgan; Mb, Megabase pair; ND, Not determined
SSLP marker locations and oligo sequences were obtained from the MGI database at http://www.informatics.jax.org.
Position is undetermined, however additional positional data in the MGI database places this marker on chromosome 3 between 25.1 – 26.2 cM
Position is undetermined, however additional positional data in the MMDBJ database places this marker on chromosome 6 between 53.75 – 67.0 cM
Position is undetermined, however additional positional data in the MGI database places this marker on chromosome 14 at approximately 46 cM
Position is undetermined, however additional positional data in the MGI database places this marker on chromosome 17 at approximately 24.5 cM
The Institutional Animal Care and Use Committee of the University of Nebraska Medical Center approved all procedures involving mice. Mice were housed under controlled temperature, humidity, and lighting conditions in an animal facility accredited by the American Association for Accreditation of Laboratory Animals Care operating in accordance with the standards highlighted in Guide for the Care and Use of Laboratory Animals). Mice were provided Harlan irradiated rodent diet #7904 (Harlan Teklad, WI, USA) and allowed to feed ad libitum.
Generation and analysis of experimental mice
At the N7 backcross generation, NZB.Lck-cre mice were crossed with NZB.ERα+/− mice, which are NZB congenic mice heterozygous for a targeted deletion of exon 2 of ERα.32,35 The genotype for ERα was determined via PCR using two primer sets. One set amplified a region of the neomycin cassette used to disrupt exon 2 deletion (F: 5′-TGAATGAACTGCAGGACGAG-3′; R: 5′-AATATCACGGGTAGCCAACG-3′) and the other amplified a region of ERα exon 5 (F: 5′-CTACGGCCAGTCGGGCAT-3′; R: 5′ AGACCTGTAGAAGGCGGGAG-3′) as a positive control. 35 The resulting female NZB.Lck-cre;ERα+/− offspring were crossed with NZW.ERαfl/fl male mice. The genotypes with respect to the Lck-cre transgene, ERα knockout allele (exon 2 deletion), and the ERα floxed allele (floxed exon 3) were determined using the aforementioned PCR assays.
Beginning at six weeks of age, mice were monitored fortnightly for the development of albuminuria using Albustix (Bayer Corp., In, USA). Incidence of albuminuria was defined as two consecutive readings of 2+ (>100 mg/dL). Beginning at two months of age, serum was isolated from peripheral blood collected monthly via saphenous vein. Mice were euthanized by CO2 asphyxiation when they appeared moribund, or had reached one year of age.
Histological Analysis
Upon sacrifice, kidneys were collected and fixed overnight in 10% neutral buffered formalin. Kidneys were then processed, paraffin-embedded and sectioned. Sections were stained with periodic acid and Schiff’s reagent (Sigma Aldrich) and mounted with Permount (Thermo Fisher). Stained sections were analyzed for evidence of glomerulonephritis via light microscopy as described previously.32
Analysis of the efficiency of cre-mediated deletion of the ERαfl allele
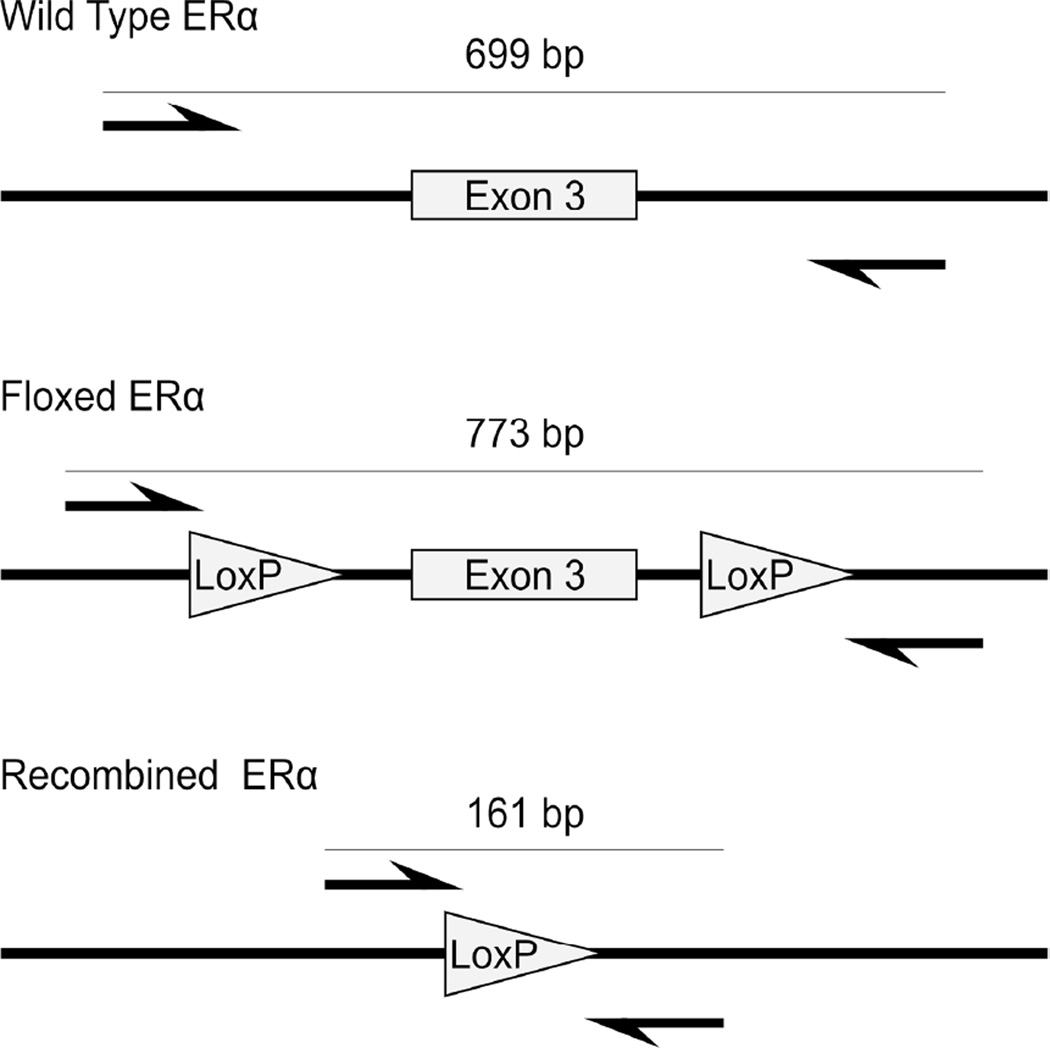
To directly determine the efficiency of cre-mediated deletion of the ERα fl allele in splenic T cells, we designed a quantitative PCR assay. As described previously, the floxed allele of ERα consisted of an ERα allele in which exon 3 is flanked by loxP sites. Upon cre-mediated recombination, sequences between the loxP sites, including exon 3, are physically excised, resulting in the ERαEx3− allele. We designed qPCR primers flanking the loxP sequences (ERαDelF & ERαDelR) which amplified a 161 bp product from the ERαEx3− allele only (Figure 1).
Figure 1.
Schematics of the genomic region surrounding exon 3 of ERα are shown for the wild type allele, floxed ERα allele, and the floxed ERα allele which has undergone cre-mediated recombination. The arrows indicate the location of annealing of the ERαDelF and ERαDelR primers.
Quantitative PCR was performed on DNA isolated from splenic CD4+ T cells. To collect CD4+ T cells, spleens were harvested from 14 week (100 days) old and 38 week (270 days) old mice, macerated in MACS buffer (1x PBS with 0.5% BSA and 2mM EDTA), and passed through a 70 µm nylon mesh to create a single-cell suspension of splenocytes. Cells were centrifuged and erythrocytes were lysed using ACK lysis buffer (Thermo Fisher, Waltham MA). CD4+ T cells were isolated with the murine CD4+ T cell Isolation Kit II (Miltenyi, San Diego, CA), and AutoMACS cell separator (Miltenyi), according to the manufacturer’s protocol. Genomic DNA from isolated CD4+ T cells was obtained using Qiagen’s DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) and quantitated using a NanoDrop 1000 Spectrophotometer. qPCR reactions containing 5ng of DNA were performed using Power SYBR Green PCR Master Mix (Thermo Fisher), were run on an Applied Biosystems 7500 real time PCR system, and analyzed with SDS software v1.4. To quantitate the efficiency of cre-mediated deletion, two sets of primers and independent standard curves were designed: The ERαDel primer pair amplified the genomic region that had undergone cre-mediated deletion of ERα exon 3 (F: 5’-TGGAATGAGACTTGTCTATCTTCG-3’, R: 5’-AACCAAGGAGAACAGAGAGACT-3’). A second primer pair, which served as a positive control, amplified a portion of exon 5 of ERα (F: 5’-GGAAGGCCGAAATGAAATGGG-3’, R: 5’-CCAACAAGGCACTGACCATC-3’). To quantify amplification resulting from each primer pair, a standard curve was generated using a serial dilution of a DNA sample from a mouse containing constitutive deletion of the floxed exon 3 (ERαex3−/ex3−). For each T cell DNA sample, the abundance of the allele in which exon3 was successfully deleted was determined using the standard curve, and this amount was then normalized to the abundance of ERα exon 5 , to correct for any slight differences in the quantity of input DNA.
Serological Analysis
Autoantibody levels and total serum IgG and IgM levels were determined by ELISA using serum isolated from peripheral blood. All samples were assayed in duplicate. Total serum IgG and IgM concentrations were determined as we have described previously using isotype-specific alkaline phosphatase antibodies (Southern Biotech, Birmingham, Alabama, USA. ELISAs to quantify anti-dsDNA IgG, anti-dsDNA IgM, and anti-chromatin IgG were conducted as described previously using isotype-specific horseradish peroxidase-conjugated or alkaline phosphatase-conjugated antibodies (Alpha Diagnostics Corp., San Antonio, TX, USA). 36, 37 Quantitative measurements of antibody concentrations for experimental samples were obtained using a standard curve generated from serial dilution of a positive control that consisted of serum pooled from female (NZB × NZW)F1 mice with heavy albuminuria. Autoantibody data is expressed as arbitrary Units/mL. All optical density measurements were obtained using a BioRad 680 microplate reader and Microplate Manager Software version 5.2.1. Experimental sera were diluted serially 1:10,000 to 1:50,000-fold for determination of total serum IgG and IgM levels and 1:50 to 1:1000 for autoantibody levels.
Flow Cytometric Analysis of T cell Compartments
To analyze splenic and thymic T cell populations, females and males were sacrificed at 3 months and 5 months, respectively. Spleens and thymuses were collected and macerated in MACS buffer and passed through a 70µm filter to create a single-cell suspension. RBCs were lysed with ACK Lysis buffer, and total cell counts were obtained using the Countess Automated Cell Counter. The antibodies used for flow cytometry were: CD3e-Pacific Blue (500A2), CD4-PE (RM4–5), CD8-APC (53–6.7), CD69-FITC (H1.2F3), CD134-Biotin (OX-86), CD62L–APC (MEL-14), and CD25-APC.Cy7 (PC61). All antibodies were obtained from BD Biosciences (San Jose, CA). Biotinylated antibodies were detected using FITC-conjugated streptavidin (BD Biosciences). Thymic and splenic single- and double-positive T cell populations and rates of apoptosis were evaluated by labeling cells with CD4, CD8, Annexin-V, and PI. Splenic T-regulatory (Treg) cells were evaluating by examining the CD3+CD4+CD8−CD25hiCD62Llo cell population. Splenic activated T cells were evaluated by examining CD4+CD69+, CD4+CD62Llo, and CD4+CD134+ cell populations. Labeled cells were assayed on an LSR II bench-top flow cytometer, and data was analyzed using FACSDiva (8.0) and plotted using FlowJo X (10.0.7r2) software.
Analysis of Previously Published Papers Using Lck-Cre Constructs
PubMed (http://www.ncbi.nlm.nih.gov/pubmed) was queried using the search term ‘Lck cre’ within the 5 year span of 2008 to 2013. Among the 41 results, 39 studies were deemed relevant because they used any one of the existing Lck-cre transgenic strains to generate a T cell specific gene deletion. This subset of published studies was examined in detail to determine if Lck-cre only controls were reported as part of the experimental design.
Statistical Analysis
All statistics were calculated using SPSS software (IBM, Armonk, NY; Version 23). Initially, statistical analysis of data from male and female mice was performed separately because of the intrinsic gender differences that exist in the (NZB × NZW)F1 phenotype. However, similar results were obtained from virtually all analyses of female and male mice. Therefore, unless specified otherwise, the two genders were combined and analyzed together to simplify the analysis and reporting of the results. Linkage of the Lck-cre transgene to markers on chromosome 1 was evaluated using the Chi-squared test. The impact of the Lck-cre transgene on time-to-event data including lifespan and incidence of albuminuria was examined using Kaplan-Meier survival analysis and the log-rank test. Median values are reported for all time-to-event data. Incidence of premature mortality and development of albuminuria was conducted using Fisher’s exact test. For all antibody concentrations, the efficiency of cre deletion, and flow cytometric analyses, mean values along with the standard error of the mean, are reported, and statistical significance was calculated using Student’s t-test. Two-sided p-values less than or equal to 0.05 were considered to be evidence of statistical significance and are provided in the text.
Results
Production of NZB.Lck-cre and NZW.ERαfl/fl congenic strains using marker assisted selection
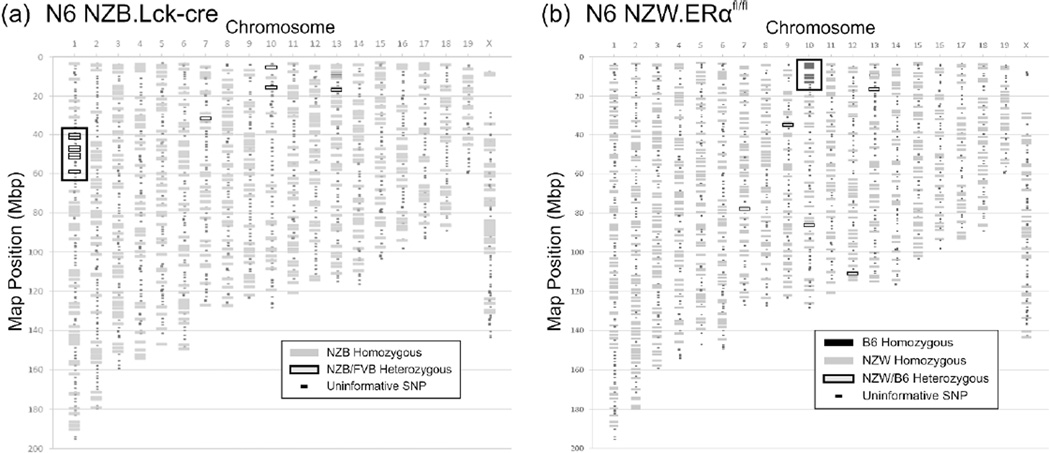
Congenic NZB.Lck-cre and NZW.ERαfl/fl strains were produced using a marker-assisted selection strategy which initially employed SSLP markers. At the N6 backcross generation, the uniformity of the genetic background of each strain was confirmed by genotyping with a higher density array of SNP markers. At the N6 generation, the NZB.Lck-cre mice were identical to the NZB strain, with the exception of a genomic region on chromosome 1 between 40 and 63 Mb. All NZB.Lck-cre transgenic mice were heterozygous for FVB alleles in this interval, suggesting that this region likely contained the Lck-cre transgene (Figure 2(a)). This interpretation was confirmed by further genotyping of a large panel of transgenic and non-transgenic NZB.Lck-cre congenic mice with additional SSLP markers on chromosome 1 (D1Mit214, 58.2 Mb; D1Mit75a, 58.7 Mb, D1Mit249, 60.8 Mb; D1Mit303, 62.9 Mb). We observed complete concordance between genotype in this interval on chromosome 1 and the presence or absence of the Lck-cre transgene (p<7.7×10−29). Importantly, this segment of chromosome 1 does not overlap with any known lupus-susceptibility loci 38–40 , and thus the impact of the residual FVB alleles linked to the Lck-cre transgene on the lupus phenotype is likely to be negligible. By the N6 generation, the genetic background of the NZW.ERαfl/fl strain was fully congenic, except for a 20 Mb segment derived from the B6 strain on chromosome 10 flanking the floxed ERα allele (Figure 2(b)). This region does not contain any known lupus susceptibility alleles.41,42
Figure 2.
Marker-assisted selection produced congenic NZB Lck-cre and NZW.ERαfl/fl mice at the N6 backcross generation. Both NZB.Lck-cre (a) and NZW.ERαfl/fl (b) mice were genotyped via SNPs using the DartMouse Speed Congenic Core Facility. The genomic regions containing the Lck-cre transgene in NZB mice and the floxed ERα allele in NZW mice are indicated by boxes. In a subset of the N6 mice genotyped from each congenic strain, several individual SNPs (indicated as grey boxes with black borders) on different chromosomes were called as heterozygous (NZB/FVB or NZW/B6, respectively). However, in the N5 congenic parent and the N4 congenic grandparent of these N6 congenic mice, these same SNPs were genotyped as homozygous (NZB/NZB or NZW/NZW, respectively). Thus, we conclude that the designations of these N6 congenic mice as heterozygous at these few SNPs represent miscalls.
Inefficient deletion of the ERαfl allele in Lck-cre transgenic mice
At the N7-N8 backcross generations, female NZB.Lck-cre;ERα+/− mice were crossed with male NZW.ERαfl/fl mice to generate (NZB × NZW)F1.Lck-cre;ERαfl/− mice that would be used to determine the impact of ERα-deletion in the T cell compartment on the lupus phenotype. The (NZB × NZW)F1.Lck-cre;ERαfl/+, (NZB × NZW)F1.ERαfl/−, and (NZB × NZW)F1.ERαfl/+ mice also produced from this cross would serve as controls. After generating these mice, we sought to determine the efficiency with which the floxed ERα allele was successfully deleted in T cells in Lck-cre transgenic (NZB × NZW)F1 mice. Using reporter gene-based analysis on different genetic backgrounds, the Lck-cre transgene has been previously reported to be expressed in approximately 90% of double positive (DP) thymocytes. 43 Similarly, Southern blot analysis has revealed that the Lck-cre transgene results in the deletion of floxed alleles in thymocytes with approximately 87%-99% efficiency.44
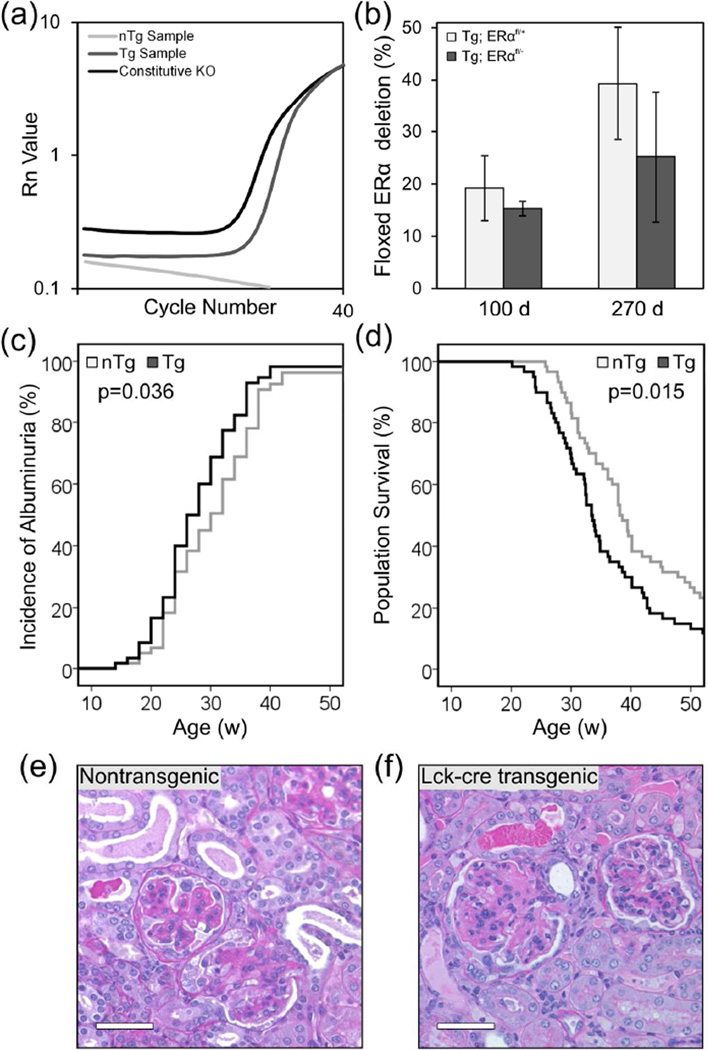
To determine what fraction of splenic CD4+ T cells carried the deleted ERαfl allele in (NZB × NZW)F1 Lck-cre transgenic mice, we developed a PCR assay using primers that amplified the ERα allele only if exon 3 has been successfully deleted (Figure 1). These primers amplified a 161 bp fragment of DNA from both Lck-cre transgenic ERαfl/+ and ERαfl/− samples, but failed to generate the significantly larger amplicons in non-transgenic samples that could be theoretically generated from either the unrecombined ERαfl allele (773 bp) or the non-floxed ERα allele (699 bp; Figure 3(a)). Thus, this assay specifically quantified the ERαfl allele that had successfully undergone cre-mediated deletion.
Figure 3.
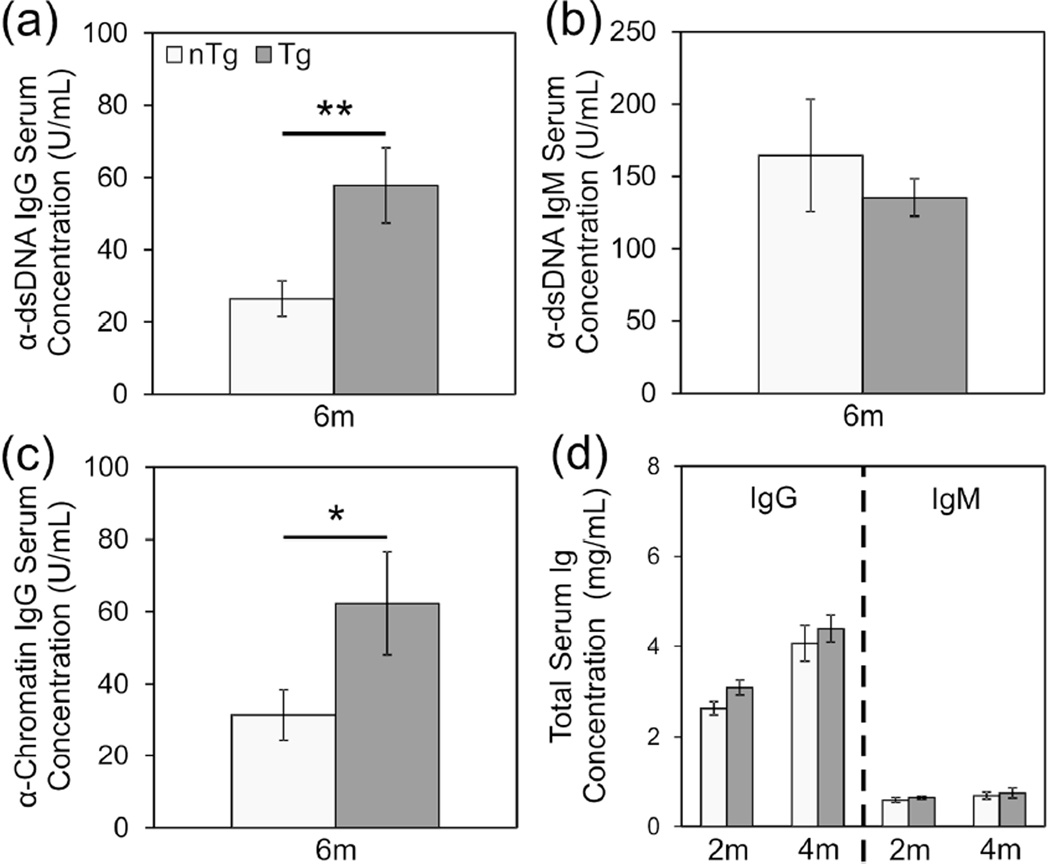
The Lck-cre transgene itself accelerated the development of lupus nephritis in (NZB × NZW)F1 mice. (a) qPCR amplification curves are shown for a representative DNA sample from a non-transgenic (nTg) mouse carrying the floxed allele of exon 3 of ERα, an Lck-cre transgenic (Tg) mouse carrying the floxed allele of exon 3 of ERα, and a mouse homozygous for a constitutive knockout of exon 3 of ERα. The calculated efficiency of cre-mediated deletion of the floxed allele (b) in Lck-cre transgenic ERαfl/+and ERαfl/− mice is shown (N=4–6 mice per genotype per time point). Additionally the incidence of albuminuria (c) and survival (d) of Lck-cre transgenic and non-transgenic (NZB × NZW)F1 mice is shown, along with representative histological images (e, f) of kidneys collected at the time of sacrifice (N=30 mice per genotype per gender). Scale bars represent 50 µm.
In contrast to what has been reported previously for the Lck-cre transgene, 28, 44 at both 14 weeks and 38 weeks days of age, the efficiency of cre-mediated deletion of ERα was found to be quite low. The ERαfl allele had been successfully deleted in only ~16% of splenic CD4+ T cells in 14 week old Lck-cre;ERαfl/− mice (Figure 3(b)). At this same time point, in Lck-cre;ERαfl/+ mice, the ERαfl allele had been deleted in ~22% of splenic CD4+ T cells. The efficiency of cre-mediated deletion was not different between the two groups (p=0.491), indicating that the apparently very low efficiency of the ERαfl allele in Lck-cre;ERαfl/− mice was not due to the resulting ERα−/− T cells being at a competitive disadvantage. Similar results were observed when mice were analyzed at 38 weeks of age. At this time point, the ERαfl allele had been deleted in ~24% and ~40% of splenic CD4+ T cells in 38 week old Lck-cre;ERαfl/− and Lck-cre;ERαfl/+ mice, respectively. As the mice aged, there was an increase in the proportion of splenic CD4+ T cells that had deleted the floxed allele, but this difference did not achieve significance (p>0.1). Importantly, at both 14 and 38 week time-points, on average, less than 25% of splenic CD4+ T cells in Lck-cre;ERαfl/− mice had deleted the ERαfl allele and were rendered ERα deficient. Because the majority of splenic CD4+ T cells in Lck-cre;ERαfl/− mice retained the functional ERαfl allele, the T cell compartment in these mice was essentially equivalent to that in the Lck-cre;ERαfl/+ mice. Thus, the Lck-cre;ERαfl/− mice could not be used to analyze the impact of T cell-specific deletion of ERα on the development of lupus in (NZB × NZW)F1 mice.
The Lck-cre transgene accelerates the development of lupus nephritis in (NZB × NZW)F1 mice
Although the Lck-cre/lox system could not be used to determine the impact of T cell-specific deletion of ERα on the lupus phenotype, during the course of evaluating the mice originally generated for that purpose, we observed that the Lck-cre transgene itself modulated the lupus phenotype in (NZB × NZW)F1 mice. Over the experimental time course, 92% of non-transgenic (NZB × NZW)F1 mice (ERαfl/− and ERαfl/+) developed albuminuria at a median age of 30 wks (Figure 3(c)). There was no difference in latency to onset of albuminuria between non-transgenic ERαfl/− and ERαfl/+ mice (p=0.57), so these two groups were pooled and are referred to hereafter as “non-transgenic mice”. Likewise, there was no difference in latency to onset of albuminuria between Lck-cre.ERαfl/− and Lck-cre.ERαfl/+ mice (p=0.84), so these two groups were also pooled and are referred to hereafter as “Lck-cre transgenic mice”. Over the experimental time course, 97% of Lck-cre transgenic (NZB × NZW)F1 mice developed albuminuria (p=0.28) at a median age of 26 weeks (p=0.036). Because all of the mice in both the Lck-cre transgenic and non-transgenic groups carried a floxed allele of ERα, the difference in median latency to onset of albuminuria between the Lck-cre transgenic and non-transgenic groups could not be attributed to the presence of the floxed ERα allele. Rather, these results suggested that the Lck-cre transgene accelerated the development of albuminuria.
The Lck-cre transgenic (NZB × NZW)F1 mice also displayed accelerated mortality compared to non-transgenic littermates. In both the Lck-cre transgenic and non-transgenic groups, ERα genotype (ERαfl/+ versus ERαfl/−) had no impact on median survival (data not shown; p>0.23). This finding, together with the low efficiency of cre mediated deletion, validated the grouping of mice based upon transgene status alone for all further analysis. The median lifespan of non-transgenic (NZB × NZW)F1 mice was 38.1 weeks, and by one year of age, 77% of non-transgenic mice had died (Figure 3(d)). By contrast, the Lck-cre-transgenic (NZB × NZW)F1 mice had a median survival of 33.4 weeks (p=0.015) and an 88% mortality rate by one year of age (p=0.15). The mean time between onset of albuminuria and morbidity was ~7–8 weeks in both non-transgenic and transgenic mice, indicating that once albuminuria developed, nephritis continued to progress at an identical rate in both groups. Altogether, these data indicate that the Lck-cre transgene hastens mortality by accelerating the disease onset rather than by modulating disease progression.
Histological analysis of sections of kidneys collected from mice at the time of sacrifice indicated that all (NZB × NZW)F1 mice, irrespective of Lck-cre transgene or ERα genotype, developed severe lupus nephritis. Kidneys of all mice showed evidence of glomerular matrix deposition, glomerular hypercellularity, wire-loop lesions, tubular dilation and the invasion of lymphocytes into the renal cortex (Figure 3(e, f)). The severity of nephritis at the time of sacrifice did not differ between transgenic and non-transgenic mice (p=0.88). Although both Lck-cre transgenic and non-transgenic mice ultimately developed severe nephritis, the accelerated mortality of Lck-cre-transgenic (NZB × NZW)F1 mice indicated that these mice developed severe nephritis at a significantly younger age than their non-transgenic littermates (p=0.015). Furthermore, these observations demonstrate that the early mortality in the Lck-cre transgenic (NZB × NZW)F1 mice is due to accelerated development of lupus and is not the result of some other transgene-associated pathology.
Lck-cre transgene accelerates the development of autoantibodies
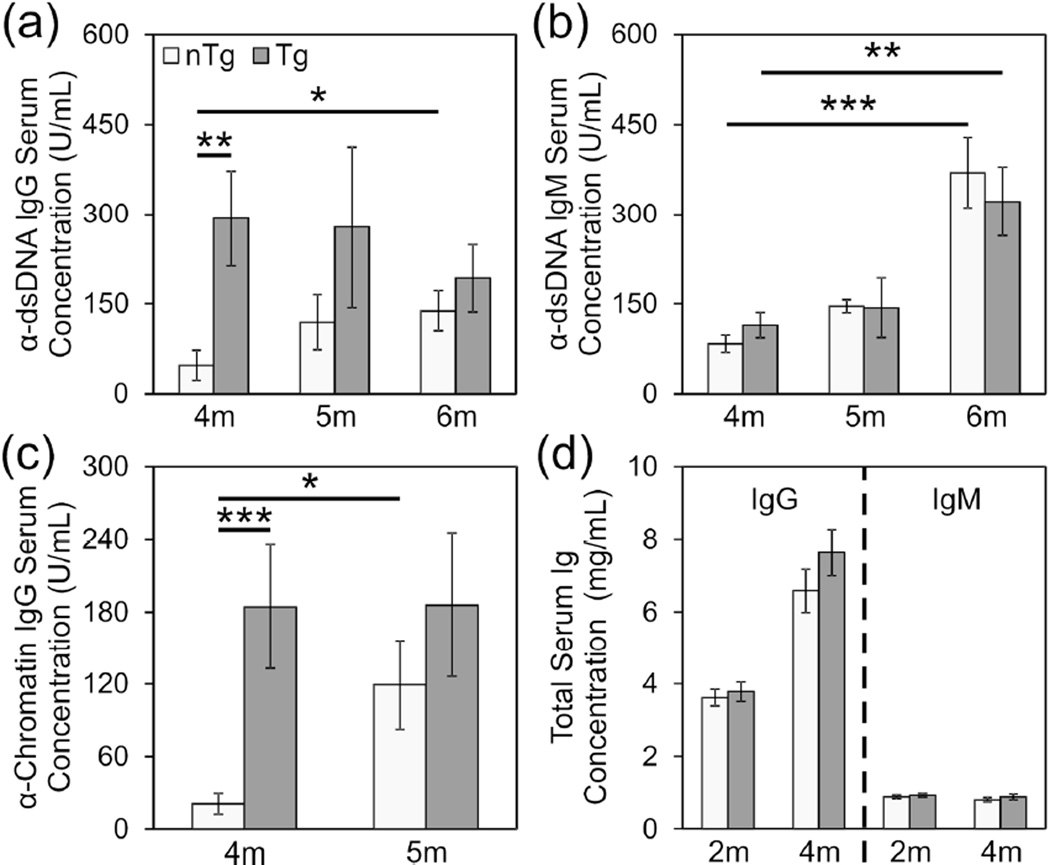
Because Lck-cre-transgenic (NZB × NZW)F1 mice displayed accelerated development of lupus nephritis, we evaluated the impact of the transgene on the time course of development of pathogenic anti-dsDNA IgG autoantibodies. Due to significant gender differences in absolute serum autoantibody concentrations as well as the time course of appearance of these autoantibodies, males and females were evaluated separately for this analysis. However, similar results were obtained from the independent analysis of each gender. Consistent with the observation that Lck-cre-transgenic female mice displayed accelerated albuminuria and mortality compared to non-transgenic females, transgenic females also developed high levels of pathogenic anti-ds-DNA IgG antibodies at a younger age than non-transgenic females; At 4 months of age, Lck-cre-transgenic (NZB × NZW)F1 females had significantly higher anti-dsDNA IgG concentrations than non-transgenic females (Figure 4(a); p=0.0054). After this time point, anti-dsDNA IgG levels in non-transgenic females increased significantly over time (p=0.049) whereas that in the Lck-cre transgenic females remained relatively constant (p=0.76). Consequently, at 5 and 6 months of age, no significant difference in anti-dsDNA IgG concentrations between Lck-cre transgenic and non-transgenic mice was detectable (p>0.24). The level of non-pathogenic anti-dsDNA IgM also increased in both the Lck-cre transgenic and non-transgenic mice over time (Figure 4(b); p=0.0068 and p=0.000031, respectively) but no difference in anti-dsDNA IgM concentrations was observed between groups at any time point (p>0.56).
Figure 4.
Lck-cre transgenic (NZB × NZW)F1 females develop high levels of autoantibodies earlier than non-transgenic females. Production of anti-dsDNA IgG (a), anti-dsDNA IgM (b) and anti-chromatin IgG (c) at 4, 5, and 6 months in female (NZB × NZW)F1 experimental mice were measured by ELISA. Total IgG and IgM (d) production at 2 months and 4 months is also shown. N=30 mice per genotype (*, p<0.05; **, p<0.01; ***, p<0.005).
Because anti-dsDNA IgG autoantibodies are thought to arise via epitope spreading in B cells that are initially chromatin reactive, we examined the impact of the Lck-cre transgene on the development of anti-chromatin IgG. As we had observed with anti-dsDNA IgG levels in young mice, Lck-cre transgenic females displayed significantly higher concentrations of anti-chromatin IgG than non-transgenic females at 4 months of age (Figure 4(c); p=0.0032). Here again, at 5 months of age, the anti-chromatin IgG concentrations in Lck-cre transgenic females remained relatively unchanged (p=0.62) whereas anti-chromatin IgG levels had increased significantly in non-transgenic female mice (p=0.014). Thus, at this time point, there was no significant difference in serum anti-chromatin IgG between Lck-cre transgenic and non-transgenic females (p=0.33). Total serum IgG and IgM did not differ between the Lck-cre transgenic and non-transgenic (NZB × NZW)F1 females (Figure 4(d); p>0.23). This observation indicated that the accelerated development of high levels of autoantibodies in the Lck-cre transgenic mice was not a reflection of a global increase in total serum IgG. Rather, the impact of the Lck-cre transgene was specific to anti-nuclear IgG autoantibodies.
Production of anti-dsDNA IgG and anti-chromatin IgG in (NZB × NZW)F1 males occurs later than that in females. Nevertheless, similar to what we observed in female mice, Lck-cre transgenic males developed significantly greater levels of pathogenic anti-dsDNA IgG than non-transgenic males at an earlier age; At six months of age, anti- dsDNA IgG levels were significantly greater in Lck-cre transgenic males compared to non-transgenic males (Figure 5(a); p=0.008). No difference was observed in the production of non-pathogenic anti-dsDNA IgM at this same time point (Figure 5(b); p=0.48). Likewise, we also observed that at 6 months of age, Lck-cre transgenic males displayed significantly higher anti-chromatin IgG levels than non-transgenic males (Figure 5(c), p=0.027). As was observed in female mice, there was no difference in the concentration of total serum immunoglobulin between transgenic and non-transgenic mice (Figure 5(d); p>0.10).
Figure 5.
Lck-cre transgenic (NZB × NZW)F1 males develop high levels of autoantibodies earlier than non-transgenic males. Production of anti-dsDNA IgG (a), anti-dsDNA IgM (b) and anti-chromatin IgG (c) at 6 months in male (NZB × NZW)F1 experimental mice were measured. Total IgG and IgM (d) production at 2 months and 4 months is also shown. N=30 mice per genotype (*, p<0.05; **, p<0.01).
The Lck-cre transgene alters developing thymic T cell populations and their survival
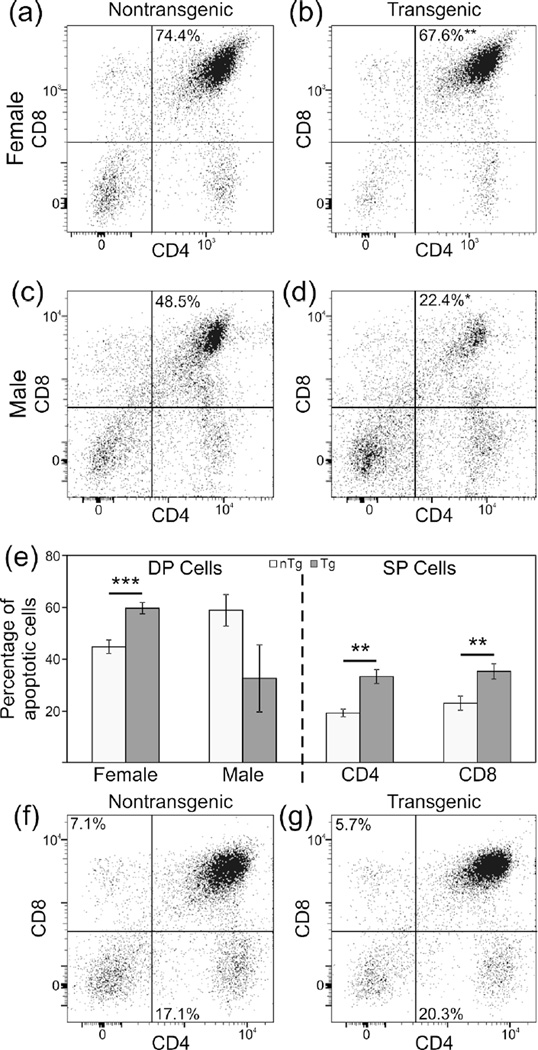
The Lck-cre transgene used in this study has previously been reported to induce T cell apoptosis, likely due to DNA damage resulting from cre recombinase mediated cleavage at lox-like sequences in the genome.25,28 We postulated that the accelerated development of autoantibodies and lupus nephritis in Lck-cre transgenic mice might be driven by increased exposure of transgenic mice to nuclear antigens as a result of increased T cell apoptosis. To explore this possibility, we first examined the impact of the Lck-cre transgene on thymic T cell development and survival in the context of the lupus-prone (NZB × NZW)F1 background. For this analysis, we examined thymocytes from female mice sacrificed at 3 months of age and male mice sacrificed at 5 months. These ages were selected because they are one month prior to the ages at which transgenic (NZB × NZW)F1 mice displayed higher autoantibody levels than their non-transgenic counterparts. Furthermore, at these ages, the mice did not display albuminuria or other evidence of lupus. Due to a gender difference in the proportion of DP T cells, males and females were analyzed separately. However, the nature of impact of the Lck-cre transgene on DP cells was identical in males and females.
The Lck-cre transgene has been shown previously to result in a slight reduction in the number of DP thymic T cells.28,45,46 Consistent with these reports, we observed that the proportion of DP thymic T cells in female Lck-cre transgenic (NZB × NZW)F1 mice was significantly less than that in non-transgenic littermates; DP T cells represented 67.6% of thymocytes in Lck-cre transgenic mice and 74.4% in non-transgenic mice (Figure 6(a, b); p=0.0053). This trend was also observed in male (NZB × NZW)F1 mice, where the proportion of DP T cells in the thymus was also reduced in Lck-cre transgenic mice compared to non-transgenic mice (22.3% versus 48.5% Figure 6(c, d); p=0.021). Additionally, the proportion of DP thymocytes undergoing apoptosis in Lck-cre transgenic females (59.7%) was significantly different than that observed in non-transgenic females (44.8%) (Figure 6(e); p=0.0025). Surprisingly, the proportion of thymic DP cells undergoing apoptosis in Lck-cre transgenic males was actually lower than that in non-transgenic mice (32.7% versus 58.9%), although this difference fell short of significance (Figure 6(e); p=0.065).
Figure 6.
Relative abundance of some T cell subsets are reduced whereas rates of apoptosis are increased in the thymus glands of Lck-cre transgenic (NZB × NZW)F1 mice. Proportions of DP thymocytes in non-transgenic and Lck-cre transgenic females (a and b, respectively) and males (c and d, respectively) are displayed. Rates of apoptosis for thymic DP and SP populations are shown (e). Population proportions for thymic CD4 and CD8 SP populations are displayed for nontransgenic (f) and Lck-cre transgenic mice (g). N=6–12 mice per genotype (*, p<0.05; **, p<0.01; ***, p<0.005).
Because DP thymocytes are the immediate precursors of CD4+ and CD8+ single positive cells, we also analyzed the impact of the Lck-cre transgene on these thymocyte populations. Between Lck-cre transgenic and non-transgenic groups, there was no significant difference in the relative abundance of either CD4+ thymocytes (20.3% versus 17.3%) or CD8+ thymocytes (5.7% versus 5.7%; Figure 6(f, g); p>0.2). However, there was a significant difference between Lck-cre transgenic and non-transgenic mice with respect to the proportion of apoptotic CD4+ thymocytes (33.3% versus 19.4%) and apoptotic CD8+ thymocytes (35.4% versus 23.1%; Figure 6(e); p<0.01).
The Lck-cre transgene alters splenic T cell subsets, apoptosis and activation
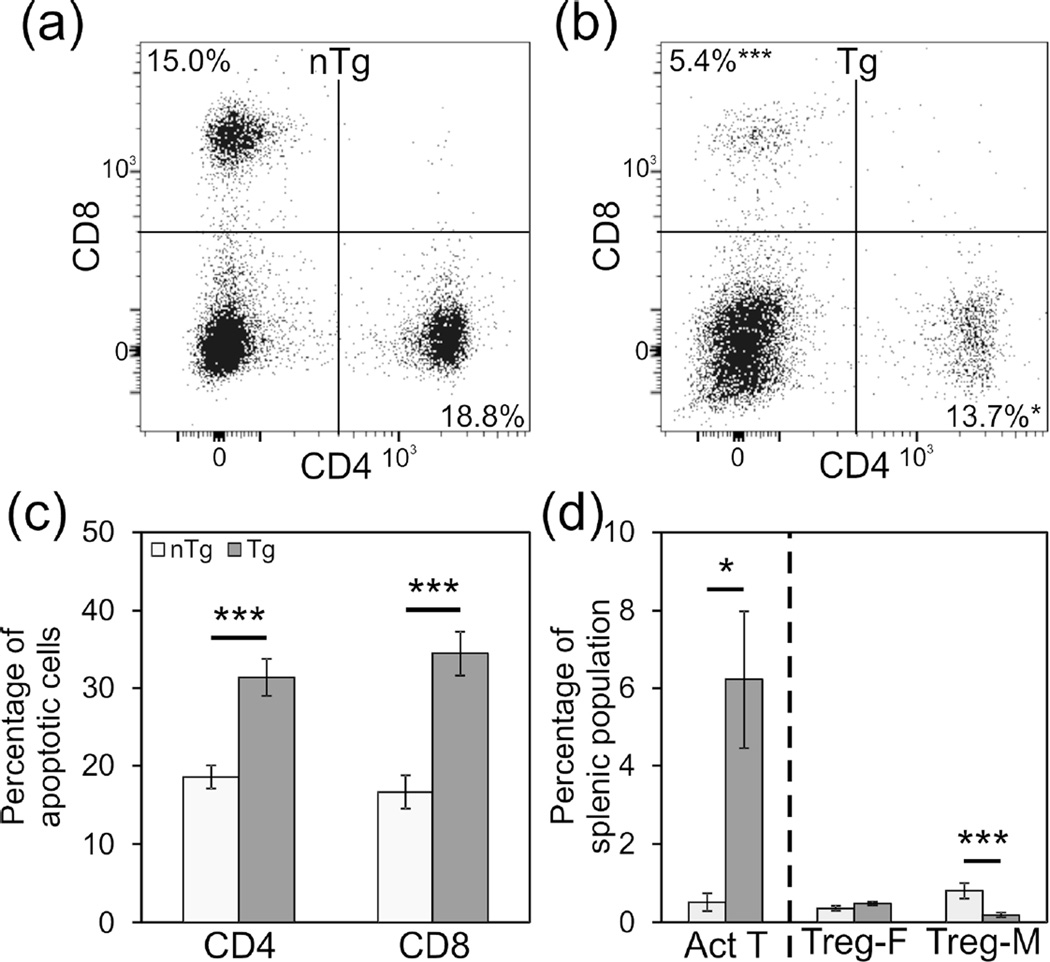
The impact of the Lck-cre transgene on mature T cells in the spleen of (NZB × NZW)F1 mice was also examined. Here again, mice were evaluated prior to both the development of high levels of autoantibodies and the onset of autoimmune disease—at 3 months of age in females and at 5 months of age in males. In non-transgenic mice, CD4+ T cells and CD8+ T cells represented 18.8% and 15.0% of splenocytes, respectively (Figure 7(a)). By contrast, in Lck-cre transgenic mice the relative abundance of each of these subsets was significantly lower than that in non-transgenic mice; 13.7% of splenocytes were CD4+ T cells and 5.4% were CD8+ T cells (Figure 7(b); p<0.05). In addition to impacting the abundance of CD4+ and CD8+ splenic T cells, the Lck-cre transgene also appeared to promote apoptosis in both subsets. In non-transgenic mice, 18.5% of CD4+ T cells and 16.6% of CD8+ T cells appeared to be apoptotic (Figure 7(c)). By contrast, in Lck-cre transgenic mice, approximately 31.38% of CD4+ T cells and 34.4% of CD8+ T cells were apoptotic (Figure 7(c)). In both subsets of splenic T cells, the proportion of apoptotic cells in the transgenic mice differed significantly from that in the transgenic mice (p<0.005).
Figure 7.
Relative abundance of some T cell subsets are altered and rates of apoptosis are increased in the spleens of Lck-cre transgenic (NZB × NZW)F1 mice. Representative plots displaying splenic CD4+ and CD8+ populations for non-transgenic (a) and Lck-cre transgenic (b) mice are shown. Rates of apoptosis for single positive T cells, measured by Annexin V and PI staining, are also displayed (c). The relative abundance of activated T cells is displayed as a percentage of total CD4+ cells, and for Tregs is displayed as the percentage of total cells (d). N=6–12 mice per genotype (*, p<0.05; ***, p<0.005).
We also evaluated the effect of the Lck-cre transgene on activated T cells, Tregs, and iNKT cells, three specific splenic T cell subsets whose relative abundance is known to impact lupus pathogenesis. Once again, for these analyses, T cells were evaluated from 3 month old female mice and from 5 month old male mice. Consistent with the accelerated development of autoantibodies and lupus nephritis in the Lck-cre transgenic (NZB × NZW)F1 mice, the spleens of young transgenic mice contained significantly more activated T cells; In young non-transgenic mice, 0.52% of CD4+ splenocytes were also CD69+ whereas in aged matched Lck-cre transgenic mice, 6.23% of CD4+ splenocytes were also CD69+ (Figure 7(d); p=0.033). This increase in activated splenic CD4+ T helper cells in the Lck-cre transgenic mice is particularly striking given the fact that the transgene decreased the relative proportion of splenic CD4+ T cells overall. In non-transgenic males, the relative abundance of splenic T regulatory cells was 0.81%, which was significantly greater than the 0.19% seen in Lck-cre transgenic males (Figure 7(d); p=0.002). However, there was no significant difference in the relative abundance of splenic T regulatory cells in Lck-cre transgenic females compared to non-transgenic females (0.36% vs. 0.49%; Figure 7(d); p=0.134). Thus, the Lck-cre transgene was not associated with a consistent alteration in T regulatory cell populations. Finally, there was no significant difference in the relative abundance of splenic iNKT cells in non-transgenic males as compared to Lck-cre transgenic males (1.0% vs 2.0%) or in non-transgenic females as compared to Lck-cre transgenic females (1.1% vs 1.6%; data not shown; p>0.17).
Discussion
Here we report that an Lck-cre transgene, which results in a high level of expression of cre recombinase in the T lineage, accelerates the development of autoimmunity in lupus-prone (NZB × NZW)F1 mice. Lck-cre transgenic (NZB × NZW)F1 mice developed high levels of anti-chromatin IgG and anti-dsDNA IgG autoantibodies at a significantly younger age than their non-transgenic counterparts. Consistent with the observed premature development of high levels of pathogenic anti-dsDNA IgG, Lck-cre transgenic (NZB × NZW)F1 mice also developed albuminuria at a younger age and had a shorter median survival time than non-transgenic mice. At the time of sacrifice, the kidneys of all mice, irrespective of transgene genotype, showed clear evidence of severe nephritis. Altogether, these observations demonstrate that the Lck-cre transgene accelerated the development of lupus in (NZB × NZW)F1 mice and that the early mortality in these mice was not the result of some other transgene-associated pathology.
Our studies suggest that the Lck-cre transgene hastened the development of fatal lupus nephritis in (NZB × NZW)F1 mice by accelerating the emergence of pathogenic autoantibodies. The onset of autoimmunity, as indicated by the development of high levels of anti-chromatin and anti-dsDNA IgG autoantibodies, occurred about 1 month earlier in Lck-cre transgenic females compared to non-transgenic females (4 months vs 5 months). However, once autoimmunity develops, the rate of disease progression in Lck-cre transgenic and non-transgenic females appears to be very similar. In both Lck-cre transgenic and non-transgenic females, median time to development of albuminuria is ~2.5 months after high levels of anti-dsDNA IgG autoantibodies appear (6.5 months (26wks) of age in Lck-cre transgenic females and 7.5 months (30 weeks) of age in non-transgenic females). Mortality due to severe nephritis develops in Lck-cre transgenic and non-transgenic females ~7–8 weeks after the onset of albuminuria; Median survival is 8.3 months (33 weeks) in Lck-cre transgenic females and 9.5 months (38 wks) non-transgenic females. These data suggest that the Lck-cre transgene promotes lupus by accelerating the initial development of autoantibodies in (NZB × NZW)F1 mice, and not by altering the relative timing of subsequent events during lupus pathogenesis.
The Lck-Cre transgene was associated with an increase in apoptosis in T cells in (NZB × NZW)F1 mice, and this increased apoptosis is likely the result of a significant increase in DNA damage that is caused by cre expression.16–18 This cre toxicity is the result of cre-mediated DNA cleavage at pseudo-loxP sites, which are found relatively abundantly throughout the murine genome.24 The increased T cell apoptosis in Lck-cre (NZB × NZW)F1 mice lead to a decrease in the relative abundance of certain T cell subsets. Increased apoptosis was observed in CD4+ and CD8+ T cells in both the thymus and the spleen of Lck-cre transgenic (NZB × NZW)F1 mice. Importantly, these increases in T cell apoptosis in Lck-cre transgenic (NZB × NZW)F1 mice were detectable in females as early as 3 months of age, which is well before the development of high levels of pathogenic autoantibodies and onset of albuminuria. The relative timing of these events in Lck-cre transgenic (NZB × NZW)F1 mice indicates that the enhanced T cell apoptosis could not simply be a consequence of the development of autoimmunity in these mice. Rather, our observations raise the possibility that the increase in T cell apoptosis in young Lck-cre transgenic (NZB × NZW)F1 mice could be the cause of the accelerated autoimmunity in these mice.
Enhanced apoptosis in CD4+ and CD8+ T cells in the thymus of Lck-cre transgenic (NZB × NZW)F1 mice did not seem to result in any change in the relative abundance of these subsets within the thymus. By contrast, in the spleens of Lck-cre transgenic (NZB × NZW)F1 mice, we observed a significant increase in apoptosis in CD4+ and CD8+ T cells as well as a significant decrease in the relative proportion of these T cell subsets. Despite the overall decrease in splenic CD4+ T cells in Lck-cre transgenic (NZB × NZW)F1 mice, the relative proportion of activated CD4+ helper T cells in these mice was significantly greater than that in age matched non-transgenic(NZB × NZW)F1 mice. This early increase in abundance of activated CD4+T helper cells is consistent with the accelerated development of autoantibodies and lupus in Lck-cre transgenic (NZB × NZW)F1 mice. Importantly, no consistent reductions in the relative proportion of T regulatory or iNKT cells were seen in Lck-cre transgenic (NZB × NZW)F1 mice. These data indicate that the accelerated autoimmunity in Lck-cre transgenic (NZB × NZW)F1 mice was not due to a decrease in the relative abundance of suppressive T cells. However, it is possible that the Lck-cre transgene might impair the function of these cells without altering their relative abundance.
We postulate that the accelerated development of autoimmunity in Lck-cre transgenic (NZB × NZW)F1 mice is due to enhanced nuclear antigen load as a result of cre-induced T cell apoptosis. Lupus-prone (NZB × NZW)F1 mice display impaired clearance of apoptotic debris. In the context of this genetic background, enhanced apoptosis in young (NZB × NZW)F1 mice would likely lead to the accumulation of apoptotic debris and an increase in the nuclear antigen load, which would, in turn, promote the earlier development of anti-chromatin and anti-dsDNA IgG autoantibodies and thereby accelerate the development of lupus. This interpretation is consistent not only with the fact that Lck-cre transgenic (NZB × NZW)F1 mice developed autoantibodies at a younger age than their non-transgenic counterparts, but also with the observation that the enhanced T cell apoptosis in Lck-cre transgenic mice was detectable at least one month before high levels of anti-chromatin and anti-dsDNA IgG autoantibodies in the Lck-cre transgenic mice. Furthermore, the observation that total serum IgG levels in the Lck-cre transgenic mice do not differ from that in non-transgenic mice suggests that the early development of antinuclear IgG in Lck-cre transgenic mice is the result of a specific, antigen driven process and is not due to a generic abnormality in class switching or production of IgG in these mice.
It was previously reported that apoptotic B cells but not T cells could accelerate lupus in (NZB × NZW)F1 mice. 6 However, in this study, apoptotic cells from a single mature B cell line and a single T cell leukemia line were administered via i.p. injection. Thus, it is not clear to what extent this previous study reflects the potential of endogenous T cells that undergo apoptosis in vivo to accelerate lupus. It should also be noted that in this previous study, apoptotic cells were injected either in a single bolus or over a two week interval. This experimental design might reflect physiological scenarios in which there is a short term, acute induction of apoptosis, but it might not accurately reflect the impact of a chronic increase in apoptotic cell burden. In contrast to this previously published work, our study reflects a scenario in which there is chronic exposure of the immune system to T cells that are undergoing apoptosis in situ during lupus pathogenesis. Our results support the hypothesis that enhanced T cell apoptosis can accelerate the development of autoimmunity in lupus prone (NZB × NZW)F1 mice.
In the (NZB × NZW)F1 mice, we found that the efficiency of cre-mediated deletion of ERα was very low. In fact, the efficiency of deletion was significantly lower than what has been reported for this Lck-cre strain in previous studies. 28, 44,47 One possible interpretation of these results might be that ERα deficient T cells are at a significant disadvantage and that these cells are outcompeted over time by cells in which the floxed allele of ERα was not deleted. However, this interpretation is not consistent with our observation that the efficiency of ERα deletion is also very low in ERαfl/+ mice, which would retain a wild type ERα allele even after deletion of the floxed allele. Based upon this observation, we posit that the apparent low efficiency of cre-mediated deletion in Lck-cre transgenic (NZB × NZW)F1 mice is a reflection of the fact that most of the T cells that express cre in these mice undergo apoptosis and are thus eliminated from the population. Thus, we hypothesize that the majority of cells that deleted the floxed allele of ERα underwent apoptosis as a result of cre toxicity. Although Lck-cre expression in non-autoimmune backgrounds also causes T cell apoptosis, many cre-expressing cells in such mice survive, allowing for a high efficiency of cre-mediated deletion in mature T cells in these strains. 28, 47 By contrast, our results suggest that in lupus-prone (NZB × NZW)F1 mice, there is strong selective pressure against the cre expressing T cells, ultimately resulting in a very low efficiency of cre-mediated deletion. One explanation for this apparent decreased fitness of cre-expressing cells in lupus prone mice might be a reduced ability to repair cre-mediated DNA damage; indeed, lymphocytes and lymphoblastoid cell lines from lupus patients have been shown to have a reduced DNA repair capacity.48
One potential caveat to the interpretation of the results presented herein stems from the fact that in our study we utilized a single Lck-cre transgenic line. Thus we cannot truly separate the impact of the Lck-cre transgene itself from any potential impact of the transgene insertion site. Consequently, we cannot formally exclude the possibility that the disruption of endogenous sequences as a result of the transgene insertion is actually what is responsible for the acceleration of autoimmunity in the (NZB × NZW)F1 mice. Although insertional mutagenesis due to transgene integration can occur, this phenomenon seems unlikely to explain the results observed in this study because the region of the genome containing the transgene integration site does not contain any known lupus susceptibility alleles 38, 40. Furthermore, we also note that the Lck-cre transgenic mice in this study are hemizygous for the Lck-cre transgene. Thus, in order for any gene disruption caused by transgene insertion to impact autoimmunity in these mice, this gene disruption would be required to act in a dominant manner. Based upon these considerations, together with the fact that the Lck-cre transgene used in this study is known to induce T cell apoptosis, and T cell apoptosis has been associated with lupus pathogenesis, we posit that the simplest interpretation our results is that the Lck-cre transgene itself, rather than the integration site, accelerates the development of autoimmunity in lupus prone (NZB × NZW)F1 mice. Because both Lck-cre transgenic and non-transgenic mice are heterozygous for the floxed allele of ERα, none of the observed differences between Lck-cre transgenic and non-transgenic mice can be attributed to the ERαfl allele. Nevertheless, it is also worth noting that the median ages for the onset of albuminuria and mortality in non-transgenic (NZB × NZW)F1.ERαfl/− and (NZB × NZW)F1.ERαfl/+ mice in this study are similar to what we have reported previously regarding these parameters in ERα+/− and ERα+/+ (NZB × NZW)F1 mice 32. These observations indicate that the floxed allele of ERα had no impact on the lupus phenotype.
Our results also clearly demonstrate the necessity of incorporating “cre-only” controls in studies using the cre-lox system. This need is further underscored by a recent publication that demonstrates that induced expression of cre-recombinase alone in lymphoma-bearing p53−/− mice caused apoptosis of lymphoma cells and resulted in complete tumor regression.49 Despite the fact that it is well established that cre recombinase can have off-target effects, resulting in DNA damage, chromosomal aberrations, and apoptosis in the tissue in which cre is expressed,19–22 many studies fail to employ “cre-only” controls that would allow one to control for any impact of the cre driver itself on the phenotype being examined. In fact, our examination of the 35 studies published between 2008 and 2013 in which an Lck-cre transgene was used, only 4 studies (11.4%) included cre-only controls.50–53 Of the 31 studies that did not include a cre-only control, only 1 study explicitly discussed the possibility that the observed phenotype could be due to the off-target effects of cre recombinase.54 This possibility appears to have been dismissed although no data was provided to support this conclusion. The Lck-cre strain we used has not been shown to cause autoimmunity in other studies. However, this is likely due to the fact that other studies using the Lck-cre strain were performed on non-autoimmune strain backgrounds. Because these non-autoimmune strains do not have the intrinsic defect in the clearance of apoptotic debris that is seen in lupus prone (NZB × NZW)F1 mice, their phagocytic system must satisfactorily compensate for the increase load of apoptotic materials. Thus, our results emphasize not only the need for cre-only controls, but the need for such controls in the context of the same genetic background on which the studies are being performed.
Acknowledgements
We gratefully acknowledge Victoria Smith and Dr. Philip Hexley of the University of Nebraska Medical Center Flow Cytometry Research Facility, who greatly contributed to the acquisition and analysis of flow cytometric data. We also thank Robert Grove and Kim Bynoté for conducting additional genotyping during backcrossing. Finally, we thank Chandan BK for helping to collect total IgG concentration data.
Funding & conflicting interest statement
This work was supported by NIH R01 AI075167 (KAG) and a pre-doctoral research assistantship awarded by the University of Nebraska Medical Center (RKN).
Abbreviations
- ERα
estrogen receptor alpha
- SSLP
simple sequence length polymorphism
- Mb
Megabase
- DP
double positive
- iNKT
invariant natural killer T cell
Footnotes
The authors declare that they have no competing interests.
References
- 1.Dhir V, Singh AP, Aggarwal A, Naik S, Misra R. Increased T-lymphocyte apoptosis in lupus correlates with disease activity and may be responsible for reduced T-cell frequency: a cross-sectional and longitudinal study. Lupus. 2009;18:785–791. doi: 10.1177/0961203309103152. [DOI] [PubMed] [Google Scholar]
- 2.Funauchi M, Sugiyama M, SukYoo B, Ikoma S, Ohno M, Kinoshita K, et al. A possible role of apoptosis for regulating autoreactive responses in systemic lupus erythematosus. Lupus. 2001;10:284–288. doi: 10.1191/096120301680416977. [DOI] [PubMed] [Google Scholar]
- 3.Grondal G, Traustadottir KH, Kristjansdottir H, Lundberg I, Klareskog L, Erlendsson K, et al. Increased T-lymphocyte apoptosis/necrosis and IL-10 producing cells in patients and their spouses in Icelandic systemic lupus erythematosus multicase families. Lupus. 2002;11:435–442. doi: 10.1191/0961203302lu223oa. [DOI] [PubMed] [Google Scholar]
- 4.Perniok A, Wedekind F, Herrmann M, Specker C, Schneider M. High levels of circulating early apoptic peripheral blood mononuclear cells in systemic lupus erythematosus. Lupus. 1998;7:113–118. doi: 10.1191/096120398678919804. [DOI] [PubMed] [Google Scholar]
- 5.Wang H, Xu J, Ji X, Yang X, Sun K, Liu X, et al. The abnormal apoptosis of T cell subsets and possible involvement of IL-10 in systemic lupus erythematosus. Cell Immunol. 2005;235:117–121. doi: 10.1016/j.cellimm.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 6.Trebeden-Negre H, Weill B, Fournier C, Batteux F. B cell apoptosis accelerates the onset of murine lupus. Eur J Immunol. 2003;33:1603–1612. doi: 10.1002/eji.200323665. [DOI] [PubMed] [Google Scholar]
- 7.Licht R, Dieker JWC, Jacobs CWM, Tax WJM, Berden JHM. Decreased phagocytosis of apoptotic cells in diseased SLE mice. J Autoimmun. 2004;22:139–145. doi: 10.1016/j.jaut.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann M, Voll RE, Zoller OM, Hagenhofer M, Ponner BB, Kalden JR. Impaired phagocytosis of apoptotic cell material by monocyte-derived macrophages from patients with systemic lupus erythematosus. Arthritis Rheum. 1998;41:1241–1250. doi: 10.1002/1529-0131(199807)41:7<1241::AID-ART15>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 9.Potter PK, Cortes-Hernandez J, Quartier P, Botto M, Walport MJ. Lupus-prone mice have an abnormal response to thioglycolate and an impaired clearance of apoptotic cells. J Immunol. 2003;170:3223–3232. doi: 10.4049/jimmunol.170.6.3223. [DOI] [PubMed] [Google Scholar]
- 10.Denny MF, Chandaroy P, Killen PD, Caricchio R, Lewis EE, Richardson BC, et al. Accelerated macrophage apoptosis induces autoantibody formation and organ damage in systemic lupus erythematosus. J Immunol. 2006;176:2095–2104. doi: 10.4049/jimmunol.176.4.2095. [DOI] [PubMed] [Google Scholar]
- 11.Dieker JW, van der Vlag J, Berden JH. Deranged removal of apoptotic cells: its role in the genesis of lupus. Nephrol Dial Transplant. 2004;19:282–285. doi: 10.1093/ndt/gfg485. [DOI] [PubMed] [Google Scholar]
- 12.Rodriguez-Manzanet R, Sanjuan MA, Wu HY, Quintana FJ, Xiao S, Anderson AC, et al. T and B cell hyperactivity and autoimmunity associated with niche-specific defects in apoptotic body clearance in TIM-4-deficient mice. Proc Natl Acad Sci U S A. 2010;107:8706–8711. doi: 10.1073/pnas.0910359107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao X, Yang R, Yan M, Li Y, Du Y, Raman I, et al. Inducible expression of kallikrein in renal tubular cells protects mice against spontaneous lupus nephritis. Arthritis Rheum. 2013;65:780–791. doi: 10.1002/art.37798. [DOI] [PubMed] [Google Scholar]
- 14.Hwang SH, Lee H, Yamamoto M, Jones LA, Dayalan J, Hopkins R, et al. B cell TLR7 expression drives anti-RNA autoantibody production and exacerbates disease in systemic lupus erythematosus-prone mice. J Immunol. 2012;189:5786–5796. doi: 10.4049/jimmunol.1202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teichmann LL, Ols ML, Kashgarian M, Reizis B, Kaplan DH, Shlomchik MJ. Dendritic cells in lupus are not required for activation of T and B cells but promote their expansion, resulting in tissue damage. Immunity. 2010;33:967–978. doi: 10.1016/j.immuni.2010.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loonstra A, Vooijs M, Beverloo HB, Allak BA, van Drunen E, Kanaar R, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci U S A. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silver DP, Livingston DM. Self-excising retroviral vectors encoding the Cre recombinase overcome Cre-mediated cellular toxicity. Mol Cell. 2001;8:233–243. doi: 10.1016/s1097-2765(01)00295-7. [DOI] [PubMed] [Google Scholar]
- 18.Janbandhu VC, Moik D, Fassler R. Cre recombinase induces DNA damage and tetraploidy in the absence of loxP sites. Cell Cycle. 2014;13:462–470. doi: 10.4161/cc.27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci U S A. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higashi AY, Ikawa T, Muramatsu M, Economides AN, Niwa A, Okuda T, et al. Direct hematological toxicity and illegitimate chromosomal recombination caused by the systemic activation of CreERT2. J Immunol. 2009;182:5633–5640. doi: 10.4049/jimmunol.0802413. [DOI] [PubMed] [Google Scholar]
- 21.Jeannotte L, Aubin J, Bourque S, Lemieux M, Montaron S, Provencher St-Pierre A. Unsuspected effects of a lung-specific Cre deleter mouse line. Genesis. 2011;49:152–159. doi: 10.1002/dvg.20720. [DOI] [PubMed] [Google Scholar]
- 22.Naiche LA, Papaioannou VE. Cre activity causes widespread apoptosis and lethal anemia during embryonic development. Genesis. 2007;45:768–775. doi: 10.1002/dvg.20353. [DOI] [PubMed] [Google Scholar]
- 23.Zheng B, Sage M, Sheppeard EA, Jurecic V, Bradley A. Engineering mouse chromosomes with Cre-loxP: range, efficiency, and somatic applications. Mol Cell Biol. 2000;20:648–655. doi: 10.1128/mcb.20.2.648-655.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Semprini S, Troup TJ, Kotelevtseva N, King K, Davis JR, Mullins LJ, et al. Cryptic loxP sites in mammalian genomes: genome-wide distribution and relevance for the efficiency of BAC/PAC recombineering techniques. Nucleic Acids Res. 2007;35:1402–1410. doi: 10.1093/nar/gkl1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 26.Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 27.Lee JY, Ristow M, Lin X, White MF, Magnuson MA, Hennighausen L. RIP-Cre revisited, evidence for impairments of pancreatic beta-cell function. J Biol Chem. 2006;281:2649–2653. doi: 10.1074/jbc.M512373200. [DOI] [PubMed] [Google Scholar]
- 28.Guo F, Cancelas JA, Hildeman D, Williams DA, Zheng Y. Rac GTPase isoforms Rac1 and Rac2 play a redundant and crucial role in T-cell development. Blood. 2008;112:1767–1775. doi: 10.1182/blood-2008-01-132068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gubbels Bupp MR, Edwards B, Guo C, Wei D, Chen G, Wong B, et al. T cells require Foxo1 to populate the peripheral lymphoid organs. Eur J Immunol. 2009;39:2991–2999. doi: 10.1002/eji.200939427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephensen CB, Borowsky AD, Lloyd KC. Disruption of Rxra gene in thymocytes and T lymphocytes modestly alters lymphocyte frequencies, proliferation, survival and T helper type 1/type 2 balance. Immunology. 2007;121:484–498. doi: 10.1111/j.1365-2567.2007.02595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci U S A. 1995;92:12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bynoté KK, Hackenberg JM, Korach KS, Lubahn DB, Lane PH, Gould KA. Estrogen receptor-alpha deficiency attenuates autoimmune disease in (NZB x NZW)F1 mice. Genes Immun. 2008;9:137–152. doi: 10.1038/sj.gene.6364458. [DOI] [PubMed] [Google Scholar]
- 33.Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development. 2000;127:4277–4291. doi: 10.1242/dev.127.19.4277. [DOI] [PubMed] [Google Scholar]
- 34.Hewitt SC, O'Brien JE, Jameson JL, Kissling GE, Korach KS. Selective disruption of ER{alpha} DNA-binding activity alters uterine responsiveness to estradiol. Mol Endocrinol. 2009;23:2111–2116. doi: 10.1210/me.2009-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lubahn DB, Moyer JS, Golding TS, Couse JF, Korach KS, Smithies O. Alteration of reproductive function but not prenatal sexual development after insertional disruption of the mouse estrogen receptor gene. Proc Natl Acad Sci U S A. 1993;90:11162–11166. doi: 10.1073/pnas.90.23.11162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohan C, Adams S, Stanik V, Datta SK. Nucleosome: a major immunogen for pathogenic autoantibody-inducing T cells of lupus. J Exp Med. 1993;177:1367–1381. doi: 10.1084/jem.177.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burlingame RW, Rubin RL. Subnucleosome structures as substrates in enzyme-linked immunosorbent assays. J Immunol Methods. 1990;134:187–199. doi: 10.1016/0022-1759(90)90380-e. [DOI] [PubMed] [Google Scholar]
- 38.Drake CG, Babcock SK, Palmer E, Kotzin BL. Genetic analysis of the NZB contribution to lupus-like autoimmune disease in (NZB x NZW)F1 mice. Proc Natl Acad Sci U S A. 1994;91:4062–4066. doi: 10.1073/pnas.91.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drake CG, Rozzo SJ, Hirschfeld HF, Smarnworawong NP, Palmer E, Kotzin BL. Analysis of the New Zealand Black contribution to lupus-like renal disease Multiple genes that operate in a threshold manner. J Immunol. 1995;154:2441–2447. [PubMed] [Google Scholar]
- 40.Rozzo SJ, Vyse TJ, Drake CG, Kotzin BL. Effect of genetic background on the contribution of New Zealand black loci to autoimmune lupus nephritis. Proc Natl Acad Sci U S A. 1996;93:15164–15168. doi: 10.1073/pnas.93.26.15164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vyse TJ, Morel L, Tanner FJ, Wakeland EK, Kotzin BL. Backcross analysis of genes linked to autoantibody production in New Zealand White mice. J Immunol. 1996;157:2719–2727. [PubMed] [Google Scholar]
- 42.Kotzin BL, Palmer E. The contribution of NZW genes to lupus-like disease in (NZB x NZW)F1 mice. J Exp Med. 1987;165:1237–1251. doi: 10.1084/jem.165.5.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J, Petrie HT. Activation kinetics and off-target effects of thymus-initiated cre transgenes. PLoS One. 2012;7:e46590. doi: 10.1371/journal.pone.0046590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orban PC, Chui D, Marth JD. Tissue- and site-specific DNA recombination in transgenic mice. Proc Natl Acad Sci U S A. 1992;89:6861–6865. doi: 10.1073/pnas.89.15.6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y, Li H, Liu H, Zhang M, Hua Z, Ji H, et al. LKB1 regulates TCR-mediated PLCgamma1 activation and thymocyte positive selection. EMBO J. 2011;30:2083–2093. doi: 10.1038/emboj.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gupta A, Hunt CR, Pandita RK, Pae J, Komal K, Singh M, et al. T-cell-specific deletion of Mof blocks their differentiation and results in genomic instability in mice. Mutagenesis. 2013;28:263–270. doi: 10.1093/mutage/ges080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reiss C, Haneke T, Volker HU, Spahn M, Rosenwald A, Edelmann W, et al. Conditional inactivation of MLH1 in thymic and naive T-cells in mice leads to a limited incidence of lymphoblastic T-cell lymphomas. Leuk Lymphoma. 2010;51:1875–1886. doi: 10.3109/10428194.2010.510360. [DOI] [PubMed] [Google Scholar]
- 48.Bassi C, Xavier D, Palomino G, Nicolucci P, Soares C, Sakamoto-Hojo E, et al. Efficiency of the DNA repair and polymorphisms of the XRCC1, XRCC3 and XRCC4 DNA repair genes in systemic lupus erythematosus. Lupus. 2008;17:988–995. doi: 10.1177/0961203308093461. [DOI] [PubMed] [Google Scholar]
- 49.Li Y, Choi PS, Casey SC, Felsher DW. Activation of cre recombinase alone can induce complete tumor regression. PLoS One. 2014;9:e107589. doi: 10.1371/journal.pone.0107589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung SH, Seki K, Choi BI, Kimura KB, Ito A, Fujikado N, et al. CXC chemokine receptor 4 expressed in T cells plays an important role in the development of collagen-induced arthritis. Arthritis Res Ther. 2010;12:R188. doi: 10.1186/ar3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gaidarenko O, Xu Y. Transcription activity is required for p53-dependent tumor suppression. Oncogene. 2009;28:4397–4401. doi: 10.1038/onc.2009.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, Johnson SM, Fedoriw Y, Rogers AB, Yuan H, Krishnamurthy J, et al. Expression of p16(INK4a) prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong WF, Nakazato M, Watanabe T, Kohu K, Ogata T, Yoshida N, et al. Over-expression of Runx1 transcription factor impairs the development of thymocytes from the double-negative to double-positive stages. Immunology. 2010;130:243–253. doi: 10.1111/j.1365-2567.2009.03230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang HC, Han L, Jabeen R, Carotta S, Nutt SL, Kaplan MH. PU.1 regulates TCR expression by modulating GATA-3 activity. J Immunol. 2009;183:4887–4894. doi: 10.4049/jimmunol.0900363. [DOI] [PMC free article] [PubMed] [Google Scholar]