Abstract
Orchidaceae are one of the largest families of flowering plants, with over 27,000 species described and all orchids are listed in CITES. Moreover, the seedlings of orchid species from the same genus are similar. The objective of DNA barcoding is rapid, accurate, and automated species identification, which may be used to identify illegally traded endangered species from vegetative specimens of Paphiopedilum (Venus slipper), a flagship group for plant conservation with high ornamental and commercial values. Here, we selected eight chloroplast barcodes and nrITS to evaluate their suitability in Venus slippers. The results indicate that all tested barcodes had no barcoding gap and the core plant barcodes showed low resolution for the identification of Venus slippers (18.86%). Of the single-locus barcodes, nrITS is the most efficient for the species identification of the genus (52.27%), whereas matK + atpF-atpH is the most efficient multi-locus combination (28.97%). Therefore, we recommend the combination of matK + atpF-atpH + ITS as a barcode for Venus slippers. Furthermore, there is an upper limit of resolution of the candidate barcodes, and only half of the taxa with multiple samples were identified successfully. The low efficiency of these candidate barcodes in Venus slippers may be caused by relatively recent speciation, the upper limit of the barcodes, and/or the sampling density. Although the discriminatory power is relatively low, DNA barcoding may be a promising tool to identify species involved in illegal trade, which has broad applications and is valuable for orchid conservation.
Introduction
DNA barcoding uses short DNA sequences to identify species [1,2]. Barcoding is a practical, simple, and quick method compared to traditional methods, but there are pros and cons for DNA barcoding [3–10]. Because of its potential application in several areas of biology, such as species identification, biodiversity assessment, plant conservation, trade control to biomedicine, forensics, and many other applications, DNA barcoding has undergone significant development and growth and hundreds of articles have been published. Therefore, many biologists and other end users have positive attitudes towards DNA barcoding.
Because of the frequent structural variation, low mutation rate, and horizontal gene transfer of plant mitochondrial genome [11,12], greater attention was paid to the chloroplast DNA barcodes in plants. A series of chloroplast fragments have been recommended as barcodes, such as the coding regions, accD, matK, ndhJ, rbcL, rpoC1, rpoB, and ycf5, and noncoding regions, atpF-atpH, psbK-psbI, trnH-psbA, and the trnL intron. Because plant chloroplast genes have a lower mutation rate than animal mitochondrial genes, a multi-locus approach is generally adopted for plant barcodes [2,13–19]. For example, Kress et al. [2] proposed that the commonly used ITS spacer and the highly variable trnH-psbA region be used in combination to identify flowering plants. Chase et al. [15] outlined two three-region options, rpoC1 + rpoB + matK and rpoC1 + matK + trnH-psbA. Finally, the CBOL Plant Working Group [18] recommended the combination of rbcL and matK as a core plant barcode.
Although there is considerable debate regarding DNA barcoding, the technique remains under active development and many plant groups have been tested, such as Aspalathus (Fabaceae) [20], Crocus and Sisyrinchium (Iridaceae) [21,22], Carex (Cyperaceae) [23], Tolpis (Asteraceae) [24], Picea (Pinaceae) [25], Alnus (Betulaceae) [26], Lemnaceae [27], Panax (Araliaceae) [28], Ligustrum (Oleaceae) [29], ferns [30], Prunus (Rosaceae) [31], Gaultheria (Ericaceae) [32], Juglandaceae [33], Pedicularis (Orobanchaceae) [34], Bromeliaceae [35], Parnassia (Parnassiaceae) [36], Lysimachia (Myrsinaceae) [37], Gossypium (Malvaceae) [38], Thymus (Lamiaceae) [39], Populus (Salicaceae) [40], Podocarpaceae [41], and Angelica (Umbelliferae) [42]. The resolution varied greatly among different plant lineages; for example, the resolution of single- and multi-locus was only 60% in Carex [23]. However, in other groups, the selected loci showed high resolution and the combination of ITS + trnH-psbA can discriminate 90.0% species of Parnassia [36], whereas ITS2 can resolve 98.93% of cotton species [38].
Orchidaceae are one of the largest families of flowering plants and all orchids are listed in CITES. However, to date the barcoding of orchids is rather limited in number and scope [43–51]. Lahaye et al. [44] proposed matK as barcode for the identification of the flowering plants based on data from >1,000 species of Mesoamerican and South Africa orchids. In addition, the species in some genera have been sparsely sampled, for example, Yao et al. [47] studied 17 species of Dendrobium, while Parveen et al. [48] sampled only eight species of Paphiopedilum. Because these studies were based on relatively sparse sampling, the question remains: when more samples are added to these large, diverse genera, will the resolution remain high? Orchid DNA barcoding is far from resolved and more samples and genera should be tested and Paphiopedilum provides an opportunity to explore these questions.
Paphiopedilum Pfitzer (Venus slipper) is the largest genus of slipper orchids, with 96 accepted species (data collected from KBG, 01/2014) and is an ideal group to evaluate the suitability of candidate barcodes for the conservation of plants. Almost all species of the genus have showy flowers and long flowering periods, often up to several months and have been cultivated widely since the 19th Century [52,53]. However, the ornamental and commercial value of the genus has caused over-collection and illegal poaching and trade [54,55]. For example, Paphiopedilum lawrenceanum has 120 years of cultivation history, but there are no wild populations because of over-collection [56]. Paphiopedilum vietnamense was only discovered in 1997 and is critically endangered in nature [57,58] and all of the species distributed in Vietnam are disappearing rapidly [54]. In addition, the customs and quarantine inspectors often cannot differentiate between rare and common species when not in flower [52]. The young seedlings of Paphiopedilum are very similar and are difficult to differentiate. Thus, morphological assessments are time-consuming, expensive, and require skilled labor [59]. Therefore, DNA barcoding might be used to solve these problems.
In this study, our objectives are as follows: 1) test the performance of the core plant barcode in Venus slippers; 2) evaluate the discriminatory power of nine single-loci (accD, matK, rbcL, rpoC2, ycf1, atpF-atpH, atpI-atpH, ITS) and multi-locus combinations with dense taxon sampling and test whether an upper limit exists in the barcodes; and 3) discuss the factors that affect barcoding success.
Materials and Methods
Plant sampling
We used the data in Guo et al. [60] for our analysis with two unknown samples excluded. A total of 107 samples representing 77 Paphiopedilum species were used to test the species resolution, 22 of which were represented by two or more individuals and varieties were treated as samples within the same species. These data were supplemented with additional data from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) (S1 Table) to test the upper limit of the barcodes. In total, 359 ITS sequences, 116 matK sequences, 60 ycf1 sequences, and 44 rbcL sequences were downloaded from GenBank.
Data analysis
The sequences were aligned with BioEdit [61] and refined manually. First, we analyzed the data from Guo et al. [60]. We evaluated the resolution of eight single-locus DNA regions (accD, matK, rbcL, rpoC2, ycf1, atpF-atpH, atpI-atpH), six selected two-locus combinations (rbcL + accD, rbcL + matK, ycf1 + rpoC2, ycf1 + atpF-atpH, rpoC2 + atpF-atpH, matK + atpF-atpH), two three-locus combinations (rbcL + matK + atpF-atpH, trnS-trnfM + atpI-atpH + atpF-atpH), and the combined eight cpDNA regions. Then, we evaluated the resolution of ITS and three cpDNA sequence regions (matK, rbcL, ycf1) with the data downloaded from GenBank. The analysis was performed with the SpeciesIdentifier 1.7.7 program from the TaxonDNA software package [62]. The inter- and intra-specific genetic divergences were calculated following Meyer and Paulay [63] and were used to determine whether a barcoding gap exists. The best match/best close match was used to assess the correct identification of the species [62]. To assess the haplotype accumulation in different datasets, we calculated the accumulation curves for haplotypes in the cpDNA and ITS of Paphiopedilum with the SPIDER package in R [64]. Neighbor-joining analysis of the eight combined cpDNAs was performed in MEGA6 [65], with the Kimura-2-parameter distance option and 1000 replicates.
Results
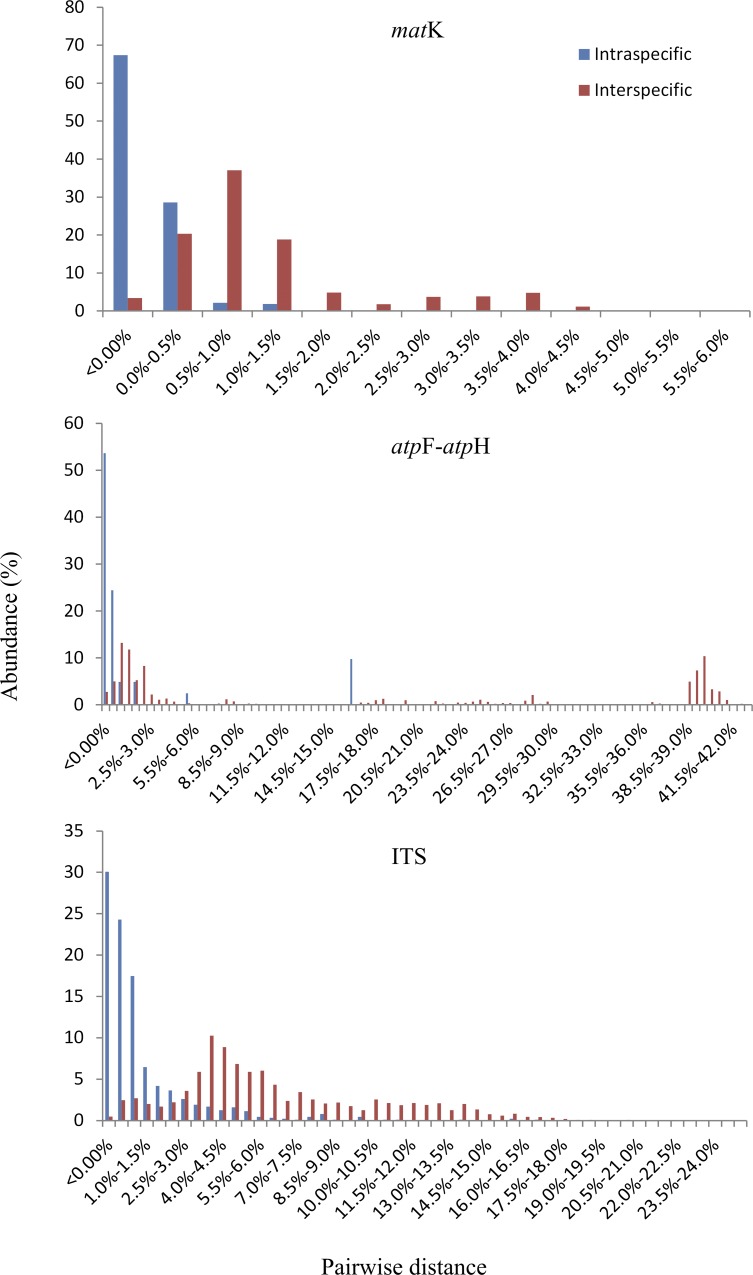
The number of sequences analyzed and the sequence lengths are listed in Table 1. The attendant datasets included approximately 70–90% of the accepted species of Venus slipper. The species were best represented by the ITS dataset (72/85), followed by matK (55/84), and ycf1 (52/79), but other datasets have lower intra-species sampling. The intra- and interspecific distance ranges overlapped and all tested barcodes had no barcoding gap (Fig 1). The summary of the single- and multi-locus barcode resolution is listed in Table 2. The ITS has the highest discriminatory power of the single-locus barcodes (52.27%) and approximately half the attendant sequences were identified successfully. In the single-locus analysis of the five coding cpDNA regions, rpoC2 has the highest resolution (25.74%), followed by ycf1, matK, and accD (22.42%, 15.88%, and 14.01%, respectively), whereas rbcL has the lowest discrimination rate (3.77%). Of the three intergenic regions, atpF-atpH has the highest resolution (22.42%), followed by atpI-atpH, and trnS-trnfM (19.62% and 13.33%, respectively). Of the multi-locus combinations, except the two two-locus combinations, those with rbcL have relatively lower resolutions (14.14% and 18.86%) and the discriminatory power of the other combinations is similar, ranging from 25.74% to 29.52%. The resolution did not increase significantly with the addition of sequence length.
Table 1. Sequence information of the genes used in the study.
| Data sets | N of sequences/ N of species | Species represented by multiple individuals | Sequence length (bp) | Alignment length (bp) |
|---|---|---|---|---|
| accD | 107/77 | 22 | 669–699 | 723 |
| matK | 107/77 | 22 | 600–609 | 619 |
| matK_1 | 223/84 | 55 | 591–609 | 609 |
| rbcL | 106/76 | 22 | 485 | 485 |
| rbcL_1 | 147/77 | 27 | 485 | 485 |
| ycf1 | 107/77 | 22 | 1525–1777 | 2041 |
| ycf1_1 | 167/79 | 52 | 1525–1777 | 2047 |
| rpoC2 | 101/73 | 21 | 2721–2736 | 2781 |
| trnS-trnfM | 105/75 | 22 | 701–785 | 903 |
| atpI-atpH | 107/77 | 22 | 467–624 | 914 |
| atpF-atpH | 107/77 | 22 | 180–423 | 577 |
| ITS | 352/85 | 72 | 588–689 | 739 |
Fig 1. Distribution of the relative abundance of intra- and interspecific K2P for the candidate barcode marker.
Table 2. Identification success of analyzed barcodes using SpeciesIdentifier 1.7.7 program under ‘best match’ and ‘best close match’ methods (Meier et al. 2006).
| Barcode | No. Sequences | Best match (%) | Best close match (%) | Threshold (%) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Correct | Ambiguous | Incorrect | Correct | Ambiguous | Incorrect | No match | |||
| rbcL (A) | 106 | 4 (3.77) | 96 (90.56) | 6 (5.66) | 4 (3.77) | 96 (90.56) | 6 (5.66) | 0 (0.00) | 0.41 |
| rbcL_1 | 150 | 10 (6.66) | 132 (88.00) | 8 (5.33) | 10 (6.66) | 132 (88.00) | 8 (5.33) | 10 (6.66) | 0.4 |
| accD (B) | 107 (107) | 15 (14.01) | 81 (75.70) | 11 (10.28) | 15 (14.95) | 80 (74.76) | 10 (9.34) | 2 (1.86) | 0.41 |
| matK (C) | 107 (107) | 17 (15.88) | 74 (69.15) | 16 (14.95) | 17 (15.88) | 72 (67.28) | 13 (12.14) | 5 (4.67) | 0.65 |
| matK_1 | 223 | 73 (32.73) | 134 (60.08) | 16 (7.17) | 73 (32.73) | 133 (59.64) | 14 (6.27) | 3 (1.34) | 0.49 |
| ycf1 (D) | 107 (107) | 24 (22.42) | 50 (46.72) | 33 (30.84) | 24 (22.42) | 44 (41.12) | 20 (18.69) | 19 (17.75) | 0.14 |
| ycf1_1 | 167 | 52 (31.13) | 64 (38.32) | 51 (30.53) | 52 (31.13) | 64 (38.32) | 50 (29.94) | 1 (0.59) | 5.59 |
| rpoC2 (E) | 101 (107) | 26 (25.74) | 42 (41.58) | 33(32.67) | 26 (25.74) | 42 (41.58) | 23 (22.77) | 10 (9.90) | 0.17 |
| trnS-trnfM (F) | 105 (107) | 14 (13.33) | 85 (80.95) | 6 (5.71) | 12 (11.42) | 71 (67.61) | 6 (5.71) | 16 (15.23) | 0.21 |
| atpI-atpH (G) | 107 (107) | 21 (19.62) | 61 (57.00) | 25 (23.36) | 21 (19.62) | 61 (57.00) | 24 (22.42) | 1 (0.93) | 14.65 |
| atpF-atpH (H) | 107 (107) | 24 (22.42) | 62 (57.94) | 21 (19.62) | 24 (22.42) | 62 (57.94) | 21 (19.62) | 0 (0.00) | 16.45 |
| ITS | 352 | 184 (52.27) | 113 (32.1) | 55 (15.62) | 183 (51.98) | 112 (31.81) | 54 (15.34) | 3 (0.85) | 4.86 |
| AB | 106 (107) | 15 (14.14) | 76 (71.69) | 15 (14.14) | 13 (12.26) | 59 (55.66) | 10 (9.43) | 24 (22.64) | 0 |
| AC | 106 (107) | 20 (18.86) | 62 (58.48) | 24 (22.64) | 20 (18.86) | 61 (57.54) | 24 (22.64) | 1 (0.94) | 0.54 |
| DE | 101 (107) | 28 (27.72) | 35 (34.65) | 38 (37.62) | 27 (26.73) | 34 (33.66) | 28 (27.72) | 12 (11.88) | 0.21 |
| DH | 107 (107) | 29 (27.1) | 35 (32.71) | 43 (40.18) | 29 (27.1) | 35 (32.71) | 42 (39.25) | 1 (0.93) | 3.74 |
| EH | 101 (107) | 29 (28.71) | 44 (43.56) | 28 (27.72) | 29 (28.71) | 44 (43.56) | 28 (27.72) | 0 (0.00) | 2.84 |
| CH | 107 (107) | 31 (28.97) | 45 (42.05) | 31 (28.97) | 31 (28.97) | 44 (42.05) | 32 (28.97) | 0 (0.00) | 8.26 |
| ACH | 106 (107) | 30 (28.3) | 43 (40.56) | 33 (31.13) | 30 (28.3) | 43 (40.56) | 33 (31.13) | 0 (0.00) | 5.86 |
| ABCDE | 101 (107) | 26 (25.74) | 32 (31.68) | 43(42.57) | 26 (25.74) | 29 (28.71) | 31 (30.69) | 15 (14.85) | 0.2 |
| FGH | 105 (107) | 31 (29.52) | 41 (39.04) | 33 (31.42) | 31 (29.52) | 41 (39.04) | 32 (30.47) | 1 (0.95) | 4.13 |
| ABCDEFGH | 100 (107) | 29 (28.99) | 26 (26.00) | 45 (45.00) | 29 (28.99) | 26 (26.00) | 41 (41.00) | 4(4.00) | 1.21 |
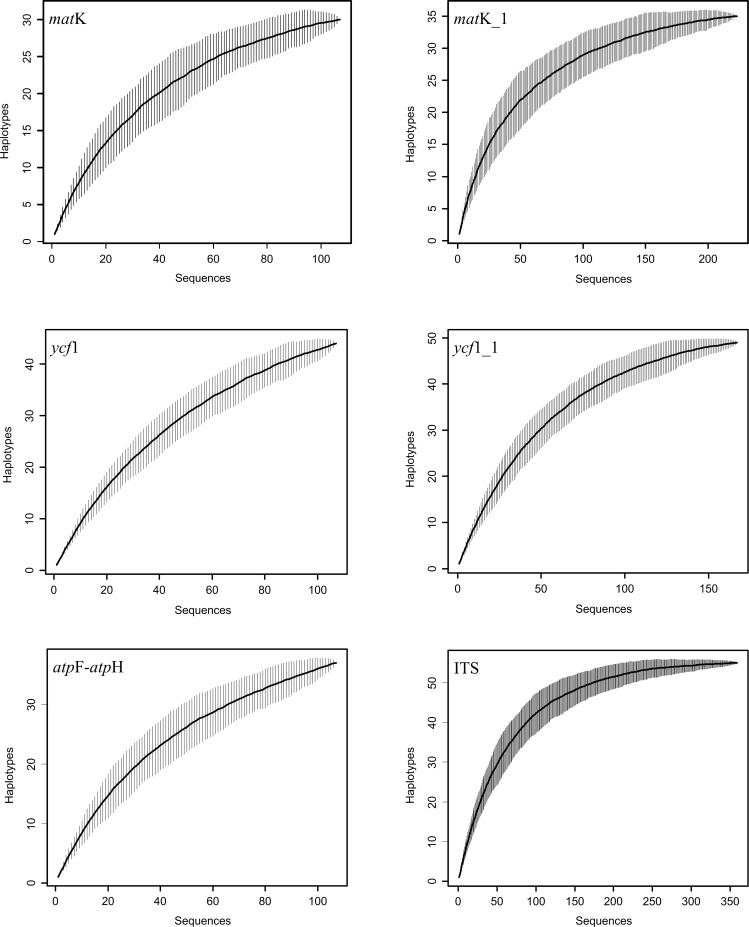
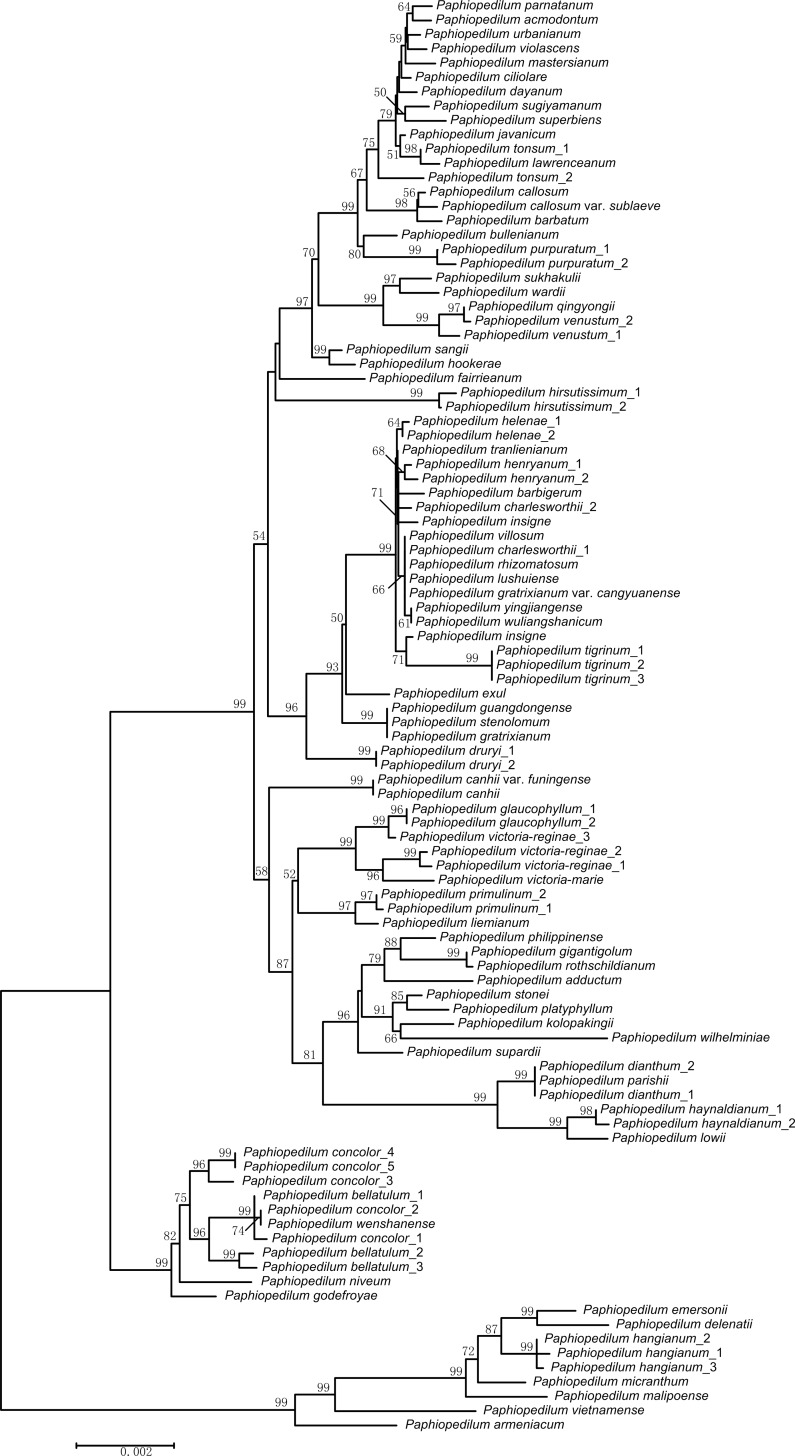
To eliminate the error induced by the sampling, we calculated the resolution of the taxa with the sequences downloaded from GenBank and the single-locus resolution increased significantly (Table 2), such that the resolution of matK increased from 15.88% to 32.73% and the resolution of ycf1 increased from 22.42% to 31.13%. In addition, the accumulation curves for the haplotypes in the cpDNA and ITS indicated saturation of the candidate markers with the addition of the sequences from GenBank, which indicates the upper limit of the attendant barcodes (Fig 2). The tree topology of the NJ tree was congruent with that reported in previous studies [60,66]. However, several species represented by two or more individuals did not form monophyletic groups (Fig 3).
Fig 2. Accumulation curves for haplotypes in cpDNA and ITS in Paphiopedilum.
Fig 3. Neighbor-joining tree of Paphiopedilum based on the combination of the eight cpDNAs.
Discussion
The efficiency of the chloroplast markers in Paphiopedilum
Compared to the study of Parveen et al. [48], the identification rate decreased with denser species sampling (Table 2). The single-locus resolution ranged from 3.77% (rbcL) to 50.69% (ITS) (ITS > rpoC2 > atpF-atpH > ycf1 > atpI-atpH > matK > accD > trnS-trnfM > rbcL), but the single-locus can assign these species to Venus slipper. ITS is the most efficient single-locus barcode, identifying half the attendant sequences correctly and could be used easily as a potential barcode for Venus slipper. For the five coding cpDNA regions, the efficiency of rbcL is too low, whereas ycf1 and rpoC2 are too long to be used as barcodes (Table 1). The resolution of matK is slightly higher than accD and matK is one of the most widely used phylogenetic markers with high variation. Therefore, we suggest matK as one of the coding cpDNA regions for the identification of the Venus slipper, which is consistent with the results of Lahaye et al. [44] and Parveen et al. [48]. For the three intergenic regions, atpF-atpH has the highest resolution and shortest length compared to the other two regions (Tables 1, 2), and should be selected as a potential barcode. Moreover, the resolution of the combination of matK and atpF-atpH is comparable with the other combinations.
For the other multi-locus combinations, the efficiency is similar, except for the two two-locus combination with relatively lower resolution (Table 2). The core plant barcode showed low efficiency in the Venus slipper (18.86%), which is much lower than the 72% obtained by the CBOL Plant Working Group [18] and this is not suitable to barcode the genus. In addition, the lengths of matK, atpF-atpH, and ITS (Table 1) are also suitable as potential barcodes, which could be sequenced with one primer. Therefore, we recommend the combination of matK + atpF-atpH + ITS as a barcode for Venus slipper during the preliminary stage.
Factors that affect species discrimination
Fazekas et al. [67] demonstrated that the resolution of the plant dataset is ~70%. The resolution of the present study is relatively low compared to other orchid barcoding studies [44,46–48,50,51] and also non-orchid plant groups [21,25,35,37,38,40,41,67,68]. According to the evolution of the Venus slipper and the sampling strategy of this study, the factors that affect the species discrimination may include the recent diversification of many species, the upper limit of the barcodes, and/or the sampling density.
The common ancestor of the Venus slipper dates to the early Miocene [69] and many species are recently diverged [60]. Recently diverged species are difficult to identify [70]. For example, the successful identification of Inga species is 69% and 32% in Araucaria [68]. Most species of Inga originated from recent radiations [68]. In Picea, the recently diversified species distributed in the Himalayan–Hengduan Mountains and northeastern Asia are also a challenge for barcoding [25]. In young species, gene flow may blur the delimitation of closely related species. Guo et al. [60] determined that reticulate evolution plays an important role in the speciation of Paphiopedilum and the rampant non-monophyly of the tested species [43,60] (Fig 3) indicates that the Venus slippers are a conundrum for DNA barcoding.
The upper limit of the chloroplast genes also constrains the success rate of species identification [67]. In our study, the combination of the eight cpDNAs together did not significantly improve the resolution of this genus (Table 2), which indicates that the addition of other cpDNAs may lead to correct identification, but would not improve efficiency. In addition, the accumulation curves for the haplotypes in matK, ycf1, and ITS show saturation, which suggests that the barcode efficiency reached the upper limit with increased sampling. There is no barcoding gap in the candidate barcodes of the genus (Fig 1). The barcoding gap does not exist in some other tested plant groups [22,25,44,71–73] and it also affects the upper limit of resolution in the Venus slipper and other untested plant groups. In Bromeliaceae, the two-locus (matK + rbcL) species discrimination is 43.48% and the addition of a third locus (trnH-psbA) did not show a significant improvement [35].
The sampling density may also affect the efficiency. Our study covers 70–90% of the accepted species of Venus slipper. Parveen et al. [48] only sampled eight species of Paphiopedilum, which represent no more than 8% of the accepted species and those eight species are strongly diverged; therefore, matK may identify the eight species correctly. In our study, the resolution of matK is 32.73% and after the saturation of the haplotype, with additional sampling of this genus, the efficiency may decrease. With more multiple representation species included, the resolution may be much higher before the accumulation curve of the single-locus barcode reaches saturation, similar to the single-locus resolution of matK, rbcL, and ycf1 increasing with the addition of sequences from GenBank (Table 2). Other studies showed high resolution with relatively small sampling. For example, Yao et al. [47] collected 17 species of Dendrobium and Hologlossum is a relatively small genus [46]. The rate of successful identification is low in species rich clades and several species-rich genera, such as Pouteria, Inga, Eschweilera, and Ocotea, showed little or no variation in cpDNA [74]. Furthermore, several studies with dense sampling showed low resolution [22,72,75]. For Sisyrinchium, the study sampled 185 accessions from 98 putative species and ITS only identified 30.61–38.78% of the species included [22], whereas Sun et al. [72] collected 148 accessions from 38 species and determined that matK could discriminate only 23.26% of Dioscorea taxa.
Conclusions
The potential application of DNA barcoding promotes the development and growth of the method. In this study, we selected eight chloroplast barcodes and ITS to evaluate their suitability in Venus slippers with dense sampling. We found that ITS is the most efficient single-locus barcode, which can identify half the Venus slippers correctly, whereas the combination of matK + atpF-atpH is the most efficient multi-locus barcode. Therefore, we recommend the combination of matK + atpF-atpH + ITS as the barcode for Venus slipper. However, there is an upper limit of the barcodes tested; therefore, adding more fragments apparently cannot solve the problem. Because of recent diversification and a complex evolutionary history in the genus, low-copy nuclear genes may be used in the DNA barcoding of this genus for more precise identification.
This study sheds light on the barcoding of orchids in a more efficient manner, which can improve orchid conservation. In the future, additional horticultural forms may be cultivated, which will lessen the over-collection from the natural environment. However, based on the assessment of the markers commonly used for the standardized application of this technique, much work remains to be done.
Supporting Information
(DOC)
Data Availability
DNA sequences: All sequences are extracted from GenBank (the sequences No. are listed in S1 Table). Alignment of the sequences are available from Dryad (DOI: 10.5061/dryad.rc2mp).
Funding Statement
This research was supported by the National Natural Science Foundation of China (Grant No. 30730010, 31300179), the Chinese Academy of Sciences (the 100-Talent Project), and the General Financial Grant from the China Postdoctoral Science Foundation (Grant No. 2014M550712). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Philos Trans R Soc Lond B Biol Sci 270: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH (2005) Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci USA 102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipscomb D, Platnick N, Wheeler Q (2003) The intellectual content of taxonomy: a comment on DNA taxonomy. Trends Ecol Evol 18: 65–66. [Google Scholar]
- 4.Wheeler QD (2004) Taxonomic triage and the poverty of phylogeny. Philos Trans R Soc Lond B Biol Sci 359: 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Will KW, Rubinoff D (2004) Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics 20: 47–55. [DOI] [PubMed] [Google Scholar]
- 6.Ebach MC, Holdrege C (2005) DNA barcoding is no substitute for taxonomy. Nature 434: 697–697. [DOI] [PubMed] [Google Scholar]
- 7.Hebert PDN, Gregory TR (2005) The promise of DNA barcoding for taxonomy. Syst Biol 54: 852–859. [DOI] [PubMed] [Google Scholar]
- 8.Will KW, Mishler BD, Wheeler QD (2005) The perils of DNA barcoding and the need for integrative taxonomy. Syst Biol 54: 844–851. [DOI] [PubMed] [Google Scholar]
- 9.Ravikanth G, Srirama R, Ganeshaiah KN, Shaanker RU (2011) In pursuit of a universal barcode of plants: Peril of followers? Curr Sci 101: 269–271. [Google Scholar]
- 10.Collins RA, Cruickshank RH (2013) The seven deadly sins of DNA barcoding. Mol Ecol Resour 13: 969–975. 10.1111/1755-0998.12046 [DOI] [PubMed] [Google Scholar]
- 11.Palmer JD (1992) Mitochondrial DNA in plant systematics: applications and limitations In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular Systematics of Plants. New York: Chapman & Hall; pp. 36–49. [Google Scholar]
- 12.Adams KL, Palmer JD (2003) Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395. [DOI] [PubMed] [Google Scholar]
- 13.Chase MW, Salamin N, Wilkinson M, Dunwell JM, Kesanakurthi RP, Haidar N, et al. (2005) Land plants and DNA barcodes: short-term and long-term goals. Philos Trans R Soc Lond B Biol Sci 360: 1889–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newmaster SG, Fazekas AJ, Ragupathy S (2006) DNA barcoding in land plants: evaluation of rbcL in a multigene tiered approach. Can J Bot 84: 335–341. [Google Scholar]
- 15.Chase MW, Cowan RS, Hollingsworth PM, van den Berg C, Madrinan S, Petersen G, et al. (2007) A proposal for a standardised protocol to barcode all land plants. Taxon 56: 295–299. [Google Scholar]
- 16.Kress WJ, Erickson DL (2007) A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE 2: e508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fazekas AJ, Burgess KS, Kesanakurti PR, Graham SW, Newmaster SG, Husband BC, et al. (2008) Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well. PLoS ONE 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CBOL Plant Working Group (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106: 12794–12797. 10.1073/pnas.0905845106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ford CS, Ayres KL, Toomey N, Haider N, Stahl JV, Kelly LJ, et al. (2009) Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc 159: 1–11. [Google Scholar]
- 20.Edwards D, Horn A, Taylor D, Savolainen V, Hawkins JA (2008) DNA barcoding of a large genus, Aspalathus L. (Fabaceae). Taxon 57: 1317–1327. [Google Scholar]
- 21.Seberg O, Petersen G (2009) How many loci does it take to DNA barcode a crocus? PLoS ONE 4: e4598 10.1371/journal.pone.0004598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alves TLDS, Chauveau O, Eggers L, de Souza-Chies TT (2014) Species discrimination in Sisyrinchium (Iridaceae): assessment of DNA barcodes in a taxonomically challenging genus. Mol Ecol Resour 14: 324–335. 10.1111/1755-0998.12182 [DOI] [PubMed] [Google Scholar]
- 23.Starr JR, Naczi RFC, Chouinard BN (2009) Plant DNA barcodes and species resolution in sedges (Carex, Cyperaceae). Mol Ecol Resour 9: 151–163. 10.1111/j.1755-0998.2009.02640.x [DOI] [PubMed] [Google Scholar]
- 24.Mort ME, Crawford DJ, Archibald JK, O'Leary TR, Santos-Guerra A (2010) Plant DNA barcoding: a test using Macaronesian taxa of Tolpis (Asteraceae). Taxon 59: 581–587. [Google Scholar]
- 25.Ran J-H, Wang P-P, Zhao H-J, Wang X-Q (2010) A test of seven candidate barcode regions from the plastome in Picea (Pinaceae). J Integr Plant Biol 52: 1109–1126. 10.1111/j.1744-7909.2010.00995.x [DOI] [PubMed] [Google Scholar]
- 26.Ren B-Q, Xiang X-G, Chen Z-D (2010) Species identification of Alnus (Betulaceae) using nrDNA and cpDNA genetic markers. Mol Ecol Resour 10: 594–605. 10.1111/j.1755-0998.2009.02815.x [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Wu Y, Yan Y, Ermakova M, Kerstetter R, Messing J (2010) DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol 10: 205 10.1186/1471-2229-10-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuo Y, Chen Z, Kondo K, Funamoto T, Wen J, Zhou S (2010) DNA Barcoding of Panax Species. Planta Med 77: 182–187. 10.1055/s-0030-1250166 [DOI] [PubMed] [Google Scholar]
- 29.Gu J, Su J-X, Lin R-Z, Li R-Q, Xiao P-G (2011) Testing four proposed barcoding markers for the identification of species within Ligustrum L. (Oleaceae). J Syst Evol 49: 213–224. [Google Scholar]
- 30.Li F-W, Kuo L-Y, Rothfels CJ, Ebihara A, Chiou W-L, Windham MD, et al. (2011) rbcL and matK earn two thumbs up as the core DNA barcode for ferns. PLoS ONE 6: e26597 10.1371/journal.pone.0026597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quan X, Zhou S-L (2011) Molecular identification of species in Prunus sect. Persica (Rosaceae), with emphasis on evaluation of candidate barcodes for plants. J Syst Evol 49: 138–145. [Google Scholar]
- 32.Ren H, Lu L, Wang H, Li D-Z (2011) DNA barcoding of Gaultheria L. in China (Ericaceae: Vaccinioideae). J Syst Evol 49: 411–424. [Google Scholar]
- 33.Xiang X-G, Zhang J-B, Lu A-M, Li R-Q (2011) Molecular identification of species in Juglandaceae: a tiered method. J Syst Evol 49: 252–260. [Google Scholar]
- 34.Yu W-B, Huang P-H, Ree RH, Liu M-L, Li D-Z, Wang H (2011) DNA barcoding of Pedicularis Linn.(Orobanchaceae): evaluating four universal barcode loci in a large and hemiparasitic genus. J Syst Evol 49: 425–437. [Google Scholar]
- 35.Maia VH, Mata CSd, Franco LO, Cardoso MA, Cardoso SRS, Hemerly AS, et al. (2012) DNA Barcoding Bromeliaceae: achievements and Pitfalls. PLoS ONE 7: e29877 10.1371/journal.pone.0029877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J-B, Wang Y-P, Möller M, Gao L-M, Wu D (2012) Applying plant DNA barcodes to identify species of Parnassia (Parnassiaceae). Mol Ecol Resour 12: 267–275. 10.1111/j.1755-0998.2011.03095.x [DOI] [PubMed] [Google Scholar]
- 37.Zhang C-Y, Wang F-Y, Yan H-F, Hao G, Hu C-M, Ge X-J (2012) Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae). Mol Ecol Resour 12: 98–108. 10.1111/j.1755-0998.2011.03076.x [DOI] [PubMed] [Google Scholar]
- 38.Ashfaq M, Asif M, Anjum ZI, Zafar Y (2013) Evaluating the capacity of plant DNA barcodes to discriminate species of cotton (Gossypium: Malvaceae). Mol Ecol Resour 13: 573–582. 10.1111/1755-0998.12089 [DOI] [PubMed] [Google Scholar]
- 39.Federici S, Galimberti A, Bartolucci F, Bruni I, De mattia F, Cortis P, et al. (2013) DNA barcoding to analyse taxonomically complex groups in plants: the case of Thymus (Lamiaceae). Bot J Linn Soc 171: 687–699. [Google Scholar]
- 40.Feng J, Jiang D, Shang H, Dong M, Wang G, He X, et al. (2013) Barcoding poplars (Populus L.) from western China. PloS ONE 8: e71710 10.1371/journal.pone.0071710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Little DP, Knopf P, Schulz C (2013) DNA barcode identification of Podocarpaceae—The second largest conifer family. PLoS ONE 8: e81008 10.1371/journal.pone.0081008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yuan Q-J, Zhang B, Jiang D, Zhang W-J, Lin T-Y, Wang N-H, et al. (2015) Identification of species and materia medica within Angelica L. (Umbelliferae) based on phylogeny inferred from DNA barcodes. Mol Ecol Resour 15: 358–371. 10.1111/1755-0998.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrison CL, Hovatter K, Eackles M, Spidle AP, King TL (2005) Molecular identification of Cypripedioid orchids in international trade. Selbyana 26: 196–216. [Google Scholar]
- 44.Lahaye R, Van der Bank M, Bogarin D, Warner J, Pupulin F, Gigot G, et al. (2008) DNA barcoding the floras of biodiversity hotspots. Proc Natl Acad Sci USA 105: 2923–2928. 10.1073/pnas.0709936105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Farrington L, MacGillivray P, Faast R, Austin A (2009) Investigating DNA barcoding options for the identification of Caladenia (Orchidaceae) species. Aust J Bot 57: 276–286. [Google Scholar]
- 46.Xiang X-G, Hu H, Wang W, Jin X-H (2011) DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNA barcode candidates. Mol Ecol Resour 11: 1012–1021. 10.1111/j.1755-0998.2011.03044.x [DOI] [PubMed] [Google Scholar]
- 47.Yao H, Song J-Y, Ma X-Y, Liu C, Li Y, Xu H-X, et al. (2009) Identification of Dendrobium Species by a Candidate DNA Barcode Sequence: the Chloroplast psbA-trnH Intergenic Region. Planta Med 75: 667–669. 10.1055/s-0029-1185385 [DOI] [PubMed] [Google Scholar]
- 48.Parveen I, Singh HK, Raghuvanshi S, Pradhan UC, Babbar SB (2012) DNA barcoding of endangered Indian Paphiopedilum species. Mol Ecol Resour 12 82–90. 10.1111/j.1755-0998.2011.03071.x [DOI] [PubMed] [Google Scholar]
- 49.Tsai C-C, Chiang Y-C, Lin Y-S, Liu W-L, Chou C-H (2012) Plastid trnL intron polymorphisms among Phalaenopsis species used for identifying the plastid genome type of Phalaenopsis hybrids. Sci Hort 142: 84–91. [Google Scholar]
- 50.Kim HM, Oh S-H, Bhandari GS, Kim C-S, Park C-W (2014) DNA barcoding of Orchidaceae in Korea. Mol Ecol Resour 14: 499–507. 10.1111/1755-0998.12207 [DOI] [PubMed] [Google Scholar]
- 51.Xu S, Li D, Li J, Xiang X, Jin W, Huang W, et al. (2015) Evaluation of the DNA Barcodes in Dendrobium (Orchidaceae) from mainland Asia. PLoS ONE 10: e0115168 10.1371/journal.pone.0115168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koopowitz H, Comstock J, Woodin C (2008) Tropical Slipper Orchids: Paphiopedilum and Phragmipedium Species and Hybrids. Portland, Oregon: Timber Press, Inc. [Google Scholar]
- 53.Liu ZJ, Chen SC, Chen LJ, Lei SP (2009) The Genus Paphiopedilum in China. Beijing: Science Press. [Google Scholar]
- 54.Averyanov L, Cribb P, Loc PK, Hiep NT (2003) Slipper Orchids of Vietnam. Royal Botanic Gardens, Kew: Compass Press Limited. [Google Scholar]
- 55.Dixon KW, Kell SP, Barrett RL, Cribb PJ (2003) Orchid Conservation. Kota Kinabalu, Sabah: Natural History Publications; (Borneo). [Google Scholar]
- 56.Cribb PJ (1998) The Genus Paphiopedilum. Kota Kinabalu and Kew: Natural History Publications. [Google Scholar]
- 57.Cribb P (2005) 512. Paphiopedilum Vietnamense. Curtis's Bot Magazine 22: 12–18. [Google Scholar]
- 58.Roberts DL, Dixon KW (2008) Orchids. Curr Biol 18: R325–R329. 10.1016/j.cub.2008.02.026 [DOI] [PubMed] [Google Scholar]
- 59.Pečnikar ŽF, Buzan EV (2014) 20 years since the introduction of DNA barcoding: from theory to application. J Appl Genet 55: 43–52. 10.1007/s13353-013-0180-y [DOI] [PubMed] [Google Scholar]
- 60.Guo Y-Y, Luo Y-B, Liu Z-J, Wang X-Q (2015) Reticulate evolution and sea-level fluctuations together drove species diversification of slipper orchids (Paphiopedilum) in Southeast Asia. Mol Ecol 24: 2838–2855. 10.1111/mec.13189 [DOI] [PubMed] [Google Scholar]
- 61.Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 62.Meier R, Shiyang K, Vaidya G, Ng P (2006) DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol 55: 715–728. [DOI] [PubMed] [Google Scholar]
- 63.Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. Plos Biol 3: 2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown SDJ, Collins RA, Boyer S, Lefort M- C, Malumbres-Olarte J, Vink CJ, et al. (2012) Spider: an R package for the analysis of species identity and evolution, with particular reference to DNA barcoding. Mol Ecol Resour 12: 562–565. 10.1111/j.1755-0998.2011.03108.x [DOI] [PubMed] [Google Scholar]
- 65.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chochai A, Leitch IJ, Ingrouille MJ, Fay MF (2012) Molecular phylogenetics of Paphiopedilum (Cypripedioideae; Orchidaceae) based on nuclear ribosomal ITS and plastid sequences. Bot J Linn Soc 170: 176–196. [Google Scholar]
- 67.Fazekas AJ, Kesanakurti PR, Burgess KS, Percy DM, Graham SW, Barrett SCH, et al. (2009) Are plant species inherently harder to discriminate than animal species using DNA barcoding markers? Mol Ecol Resour 9: 130–139. 10.1111/j.1755-0998.2009.02652.x [DOI] [PubMed] [Google Scholar]
- 68.Hollingsworth ML, Clark AA, Forrest LL, Richardson J, Pennington RT, Long DG, et al. (2009) Selecting barcoding loci for plants: evaluation of seven candidate loci with species-level sampling in three divergent groups of land plants. Mol Ecol Resour 9: 439–457. 10.1111/j.1755-0998.2008.02439.x [DOI] [PubMed] [Google Scholar]
- 69.Guo YY, Luo YB, Liu ZJ, Wang XQ (2012) Evolution and biogeography of the slipper orchids: Eocene vicariance of the conduplicate genera in the Old and New World tropics. PLoS ONE 7: e38788 10.1371/journal.pone.0038788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Velzen R, Weitschek E, Felici G, Bakker FT (2012) DNA barcoding of recently diverged species: relative performance of matching methods. PLoS ONE 7: e30490 10.1371/journal.pone.0030490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arca M, Hinsinger DD, Cruaud C, Tillier A, Bousquet J, Frascaria-Lacoste N (2012) Deciduous trees and the application of universal DNA barcodes: a case study on the circumpolar Fraxinus. PLoS ONE 7: e34089 10.1371/journal.pone.0034089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun X-Q, Zhu Y-J, Guo J-L, Peng B, Bai M-M, Hang Y.-Y (2012) DNA Barcoding the Dioscorea in China, a vital group in the evolution of Monocotyledon: use of matK gene for species discrimination. PLoS ONE 7: e32057 10.1371/journal.pone.0032057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Simeone MC, Piredda R, Papini A, Vessella F, Schirone B (2013) Application of plastid and nuclear markers to DNA barcoding of Euro-Mediterranean oaks (Quercus, Fagaceae): problems, prospects and phylogenetic implications. Bot J Linn Soc 172: 478–499. [Google Scholar]
- 74.Gonzalez MA, Baraloto C, Engel J, Mori SA, Pétronelli P, Hector A (2009) Identification of Amazonian trees with DNA barcodes. PLoS ONE 4: e7483 10.1371/journal.pone.0007483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, Burgess KS, et al. (2014) Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep? Mol Ecol 23: 4737–4756. 10.1111/mec.12837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
DNA sequences: All sequences are extracted from GenBank (the sequences No. are listed in S1 Table). Alignment of the sequences are available from Dryad (DOI: 10.5061/dryad.rc2mp).