Abstract
Gelsolin (GSN), which is a Ca2+-dependent actin filament severing and capping protein, plays a critical role in the cancer progress and has the potential for providing a novel thread for cancer therapy. In current study, we demonstrate the roles of GSN on anti-apoptosis of hepatocarcinoma cells by transcriptome RNA-seq method. Then flow cytometry (FCM), in-cell immunoblotting and transmission electron microscopy (TEM) were used to examine the GSN regulatory cell apoptosis. The results revealed GSN significantly suppresses apoptosis-associated functional categories through down-regulating apoptosis-associated genes in 5 apoptosis terms and 6 relevant KEGG pathways. FCM showed a significant lower apoptotic rate in GSN-SMMC7721 (P<0.05). In-cell immunoblotting detected discrepant expression of the apoptosis factors among GSN expressed/shRNA transfectants (P<0.05). TEM observed the discernible apoptosis morphology. Above results suggest a negative relationship between GSN expression and hepatocarcinoma cell apoptosis. GSN overexpression suppresses apoptosis while down-regulated GSN promotes apoptosis. The possible mechanism could be associated with the regulation of GSN on the apoptosis-associated pathways and the apoptosis factors caspase 3 and bcl-2.
Keywords: Hepatocarcinoma, transcriptomics, RNA-seq, gelsolin, anti-apoptosis
Introduction
Hepatocellular carcinoma (HCC) is the most common form of primary cancer of liver. In terms of numbers, HCC is the sixth most malignant cancer worldwide (estimated 711,000 cases annually); however, due to its poor prognosis (survival rates 3-5% in cancer registries), HCC is the third most common cause of cancer death [1,2]. In our previous study, we revealed that GSN is a potential HCC serum biomarker through iTRAQ-MALDI-MS/MS technology [3]. Moreover, other reports also suggested GSN plays important roles in the biological processes of cancerization [4,5]. As for the correlations between GSN and HCC, however, were still unclear.
As a Ca2+-regulated actin filament severing and capping, gelsolin (GSN) is a widespread, polyfunctional regulator of cell structure and metabolism [6]. Research data showed that GSN was ubiquitously expressed in various kinds of cells [7-11], in spite of the variations of expression levels during cell differentiation [12,13] and carcinogenesis [5,14-24]. A recent understanding of the functions and regulatory mechanisms of GSN might lead to new considerations of this protein as a potential biomarker and/or therapeutic target of tumor. High levels of GSN expression were thought to be an independent marker for tumor recurrence and progression [25]. Actually, increased GSN expression was found to correlate with invasion in HCC [26] and small cell lung cancer [27], and higher tumor grade in renal cell carcinoma [28]. In human hepatocarcinoma HepG2 cells, overexpression of GSN inhibited the nuclear localization of p53, repressed apoptosis and negatively regulated transcriptional activity of a reporter construct in the cytoplasm [29]. Intriguingly, in urothelial and oral carcinomas, GSN exhibits a biphasic expression profile, being down-regulated in premalignant lesions but increased in higher grade lesions. 24Furthermore, the co-expression of GSN with erb-B2 and epidermal growth factor receptor (EGFR) is a predictor of poor prognosis in breast cancer [30]. It is likely that the role of GSN differs during the course of tumor progression, and in more advanced disease GSN may cooperate with other oncogenic factors to accelerate disease progression.
Recent development of the next-generation sequencing technologies makes RNA-seq become a powerful approach in defining the molecular mechanism of specific diseases with the advantage of analyzing abnormal transcriptome at genome-wide level [31,32]. Previous applications of RNA-seq with greater efficiency and higher resolution [33] than other expression profiling technologies included yeast [34], viruses [35], tissues [36] and cell lines [37]. For cancer expression profiling, the reprogramming of the transcriptome leads to aberrant cellular behavior and thus directly contributes to cancer progression [38]. Monitoring the cancer transcriptome not only enables us to fill in the gap between gene regulation and cancer cell behavior, but also allows us to identify the underlying mechanism [39,40].
Despite the prevalence of using RNA-seq to study various cancer pathogenetic mechanisms [41-43], the deep transcriptome descriptions of HCC and the roles of GSN in HCC are scarcely mentioned. We therefore applied RNA-seq technology to analyze the transcriptomics and to determine the roles of GSN in hepatocellular carcinoma. Through bioinformatics analysis, we revealed the role of GSN on suppressing HCC apoptosis and explored the possible regulatory mechanism.
Materials and methods
Cell culture and recombinant plasmid DNA
HCC cell line SMMC7721 cells were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. Stable SMMC7721 cell lines in which GSN were either up-regulated by plasmids containing full length of GSN cDNA or down-regulated by the short hairpin RNA recombinant plasmids, and their control transfectants induced by empty vector plasmids were all established by our institute. RT-PCR and in-cell immunoblotting were used to determine the GSN expression levels and were carried out in triplicate.
Transcriptome sequencing
Stable GSN expressed cells (GSN-SMMC7721) and parent SMMC7721 were used to explore the cell transcription. RNA was extracted from about 1×107 cells respectively, according to the manufacturer’s instruction, and quantified by NanoDrop 2000 (Thermo-Fisher Scientific). The whole transcriptome RNA-seq was constructed using Ion Total RNA-Seq Kit v2, Ion PI™ Chip kit v2, Ion PI™ Template OT2 200 Kit v2, and Ion PI™ Sequencing 200 Kit v2 based on the Life Technologies Corporation’s guide. We performed two runs of sample repetition. In brief, mRNA was purified using oligo-dT beads from 100 μg of total RNAs for each sample and fragmented into small fragments. The cleaved RNA fragments were reverse-transcribed into first strand cDNA, followed by second-strand cDNA synthesis. After end-repair procedure, a single ‘A’ base was added to cDNA fragments at 3’end. The cDNAs were then ligated to adapters, enriched by PCR to generate the final cDNA library. After amplifying sequencing template, RNA-seq was performed using Ion proton system (Life Technologies Corporation) with the standard protocol.
FCM analysis
GSN overexpressed (GSN-SMMC7721), shRNA down-regulated (shGSN-SMMC7721), and their empty vector transfected cells (NC1-SMMC7721 and NC2-SMMC7721) were used to examine the GSN-related apoptosis. After trypsinized, approximately 1×106 cells were collected by centrifugation at 1200 g for 5 min, and then washed in PBS, followed by resuspension in apoptosis detection buffer for about 0.5 h in dark. Cells were stained by propidium iodide (PI) and Annexin-V, and analyzed by FCM using Cell quest software (BD Biosciences). The experiment was carried out in triplicate.
In-cell immunoblotting
GSN-SMMC7721, shGSN-SMMC7721, NC1-SMMC7721 and NC2-SMMC7721 were seeded on 96-well plate until they reached logarithmic phase, then the cells were immediately fixed, permeabilized and blocked at room temperature. After incubated with primary antibodies overnight at 4°C, the cells were sequentially incubated with corresponding second infrared labeled antibodies for 2 h in dark place. Odyssey Infrared Imaging System (LI-COR Biosciences GmbH) was used to obtain the image and analyze the target protein expression, which was calculated as the ratio of the intensity of target protein to that of GAPDH. The experiments were carried out in triplicate.
TEM observations
GSN down-regulated cells induced by shRNA transfection were used to observe the GSN-affected apoptosis morphology. Hitachi H7650 was employed and operated at 200 KV, and was attached to a high-angle annular dark-field detector for determining the apoptosis morphology of cells. The observed cells were dropped on a copper mesh coated with an amorphous carbon film for TEM observations. The experiment was carried out in triplicate.
Statistical analysis
Results are expressed as means ± standard error. Statistical analyses were performed using SPSS statistics software (SPSS). P-value <0.05 is considered statistically significant.
Results
Transcriptome sequencing gained superior sequences
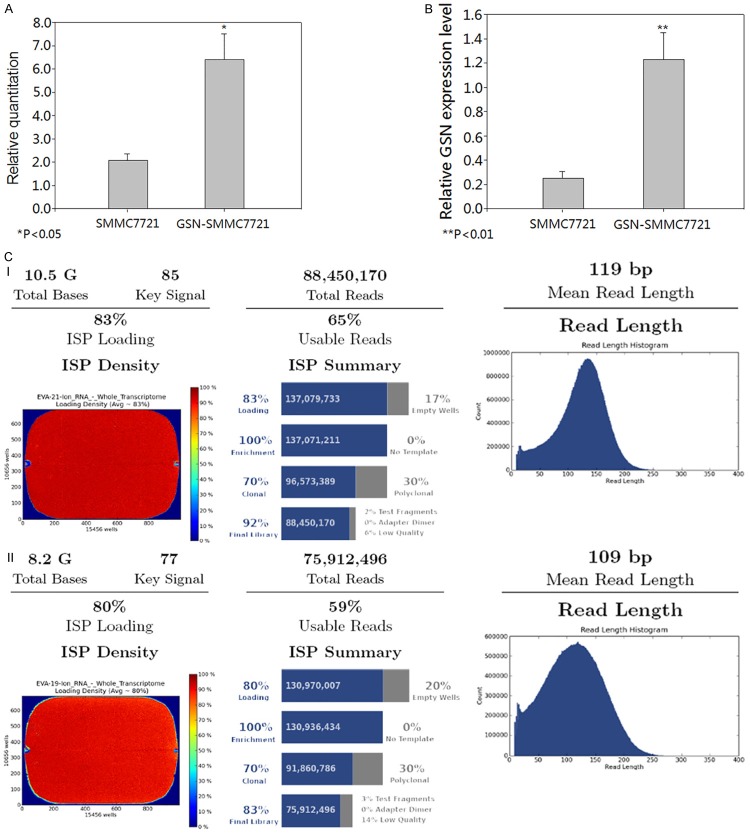
GSN expression recombinant plasmid was transfected into hepatocarcinoma SMMC7721 cell line and then treated with puromycin for the selection of stable cell lines. We performed real-time PCR and in-cell immunoblotting to confirm the overexpression of GSN. The experiments were carried out in triplicate. The target GSN expression was calculated as the ratio of the intensity of target to that of GAPDH. As expected, GSN level was increased in the GSN-SMMC7721 cell line (Figure 1A, 1B). Then, RNA-seq was carried out to analyze the transcriptional differences. We performed two runs of sample repetition. On completion, 75.9 million 109 bp long sequencing reads and 88.4 million 119 bp long sequencing reads were generated in two runs of repetition, and that corresponded to an 8.2G and 10.5G raw sequence data respectively (Figure 1CI, 1CII). Samples in both above repetitive runs of transcriptome RNA-seq gained >50 M superior sequences that could be utilized in downstream analysis.
Figure 1.
Stable transfected cell validation and RNA-seq. A. Real-time PCR for stable transfected cell validation. B. In-cell immunoblotting for stable transfected cell validation. The red and green channel respectively stands for GSN and GAPDH expression. C. Description for two repetitive runs (I & II) of RNA-seq. Both above repetitive runs of transcriptome RNA-seq gained >50 M superior sequences.
Enrichment analysis confirmed 341 mutual DEGs
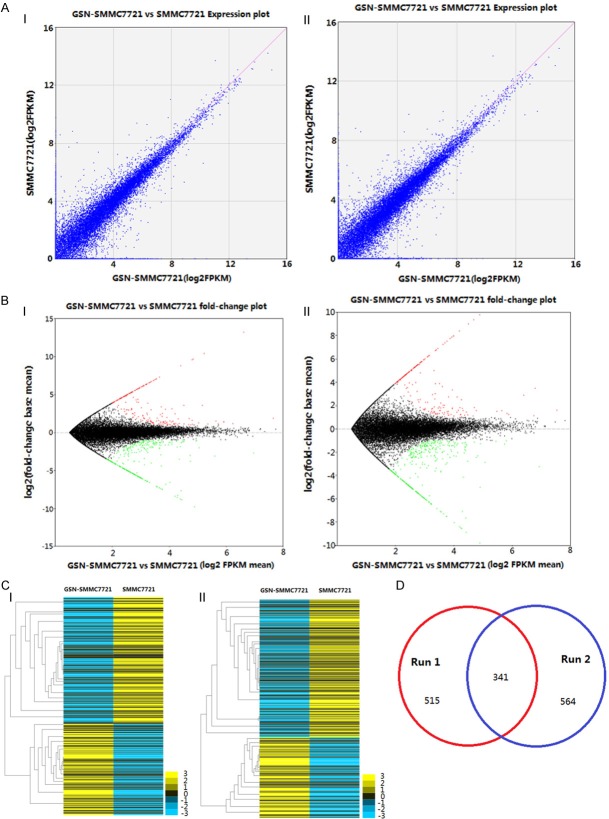
To estimate gene expression abundance, we conducted an enrichment analysis to estimate the gene expression and identify significantly dysregulated genes. The results showed that approximate 50000 genes were detected in each run and the DEGs were 515 and 546 respectively. The normalized gene expression was measured by Fragments Per Kilobase of exon per Million fragments mapped (FPKM) (Figure 2A, 2B). In addition, the clustering analysis indicated that the transcriptome of GSN overexpressed SMMC7721 cells was distinct from the parental control (Figure 2C). The overlapping of the mutual DEGs between two runs reached up to 341 and was shown as a Venn diagram in Figure 2D.
Figure 2.
DEGs and cluster analysis for two runs of RNA-seq. A. The scatter plot for global expression in two runs. The normalized gene expression was measured by FPKM. B. log2 ratioplot for all the genes in two runs. The red and green dots indicated that up- and down-regulated DEGs were significant at P values less than 0.05. C. Hierarchical clustering of DEGs in two runs. D. Venn diagram to illustrate the overlapped DEGs between two runs.
DEGs involved in 116 functional categories.
To better understand the function of DEGs, we conducted an enrichment analysis of Gene Ontology for the dysregulated genes. To identify the functional categories, we first performed GO categories using online tools from WEGO. Three hundred forty one DEGs involved in 63 GO classifications including cellular component, molecular function, and biological process were enriched (Figure 3A). A further analysis of functional annotation was achieved by studying the enrichment of DEGs in Cog categories, which revealed 20 function classifications of consensus sequences (Figure 3B). For GO categories provided informative DEGs in each gene ontology term and liable to cancer-specific analysis, we counted the significantly up- and down-regulated DEGs in terms from the component ontology with corrected p-value better than 1. In total, all DEG were categorized into approximately 116 functional categories, containing 40 up-regulated GO categories and 28 down-regulated GO categories (Figure 4).
Figure 3.
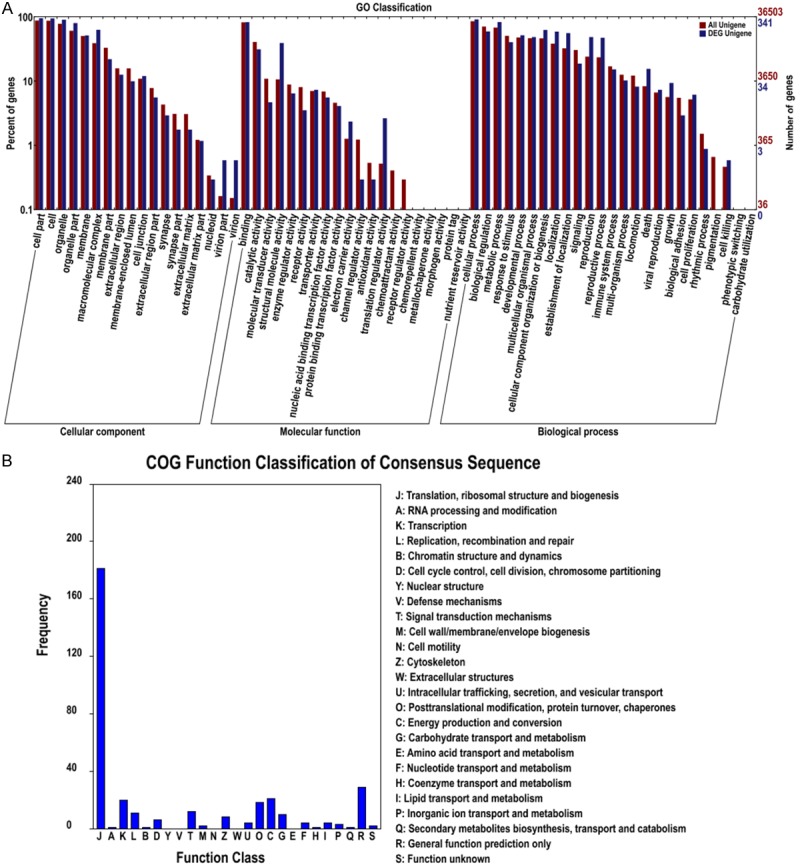
Functional enrichment analyses of differentially expressed genes. A. GO annotation map for DEGs. GO classifications including cellular component, molecular function, and biological process. B. COG classification map for DEGs.
Figure 4.
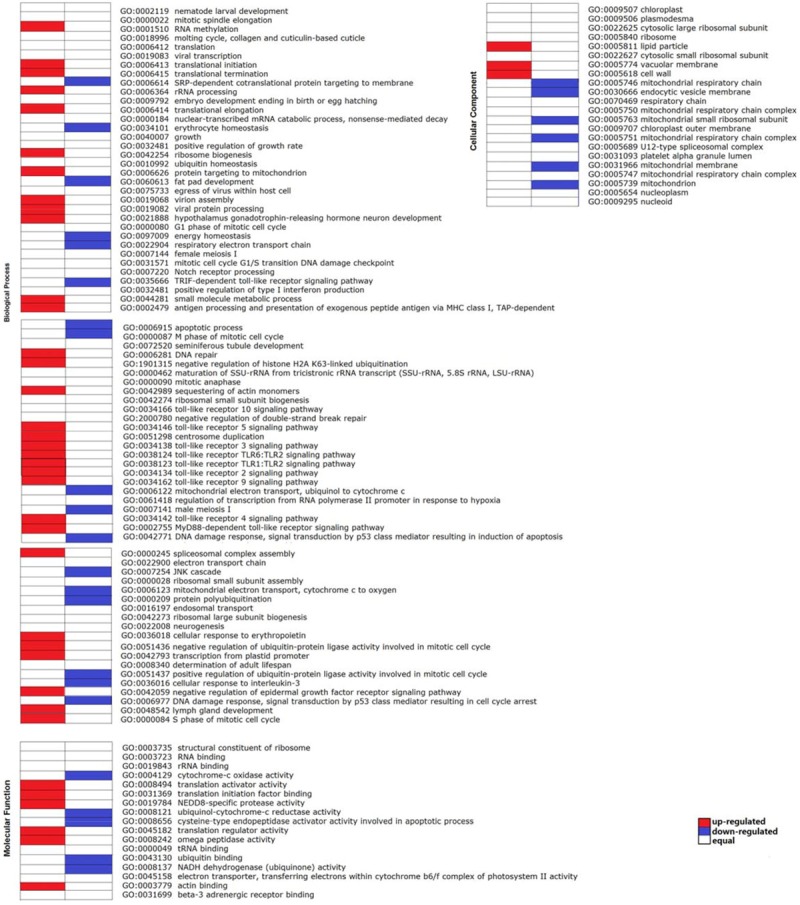
Analysis of significantly up- and down-regulated DEGs in GO categories. Terms from the component ontology with corrected p-value better than 1. The red or blue rectangles indicated over 60% up- or down-regulated DEGs were in the corresponding terms.
Cancer-specific functional filter highlighted a GSN effect on cell apoptosis
Interestingly, although we identified approximate numbers of up- and down-regulated terms in the functional enrichment analysis, we observed clues of significant GO categories for GSN overexpressed SMMC7721 cells, suggesting that the increased GSN might functionally important for cancer progression. For example, the DEGs expression in the GO terms for “apoptotic process” is mainly significantly suppressed, the same trend can also be found in the gene catalog of “DNA damage response, signal transduction by p53 class mediator resulting in induction of apoptosis”, “JNK cascade”, “cellular response to interleukin-3”, and “cysteine-type endopeptidase activator activity involved in apoptotic process”, which indicates that GSN might affect apoptosis of cancer cells.
Then we focused on the apoptosis-related DEGs annotation and particular pathway so as to seek for the evidences of GSN’s regulatory effect in HCC apoptosis. The results showed that in all the 13 mutual apoptosis-related DEGs of two runs, 9 pro-apoptotic genes including PSME2, PTK2B, FOS, JUN, ITGB1, MAP2K7, MAP3K4, MAP3K12 and Rac1 were down-regulated, and RRM2B, as a anti-apoptotic gene was up-regulated (Table 1). Above 10 apoptosis-associated DEGs participated in the process of anti-apoptosis, and simultaneously plays important roles in 6 apoptosis-related pathways (antigen processing and presentation, natural killer cell mediated cytotoxicity, p53 signaling pathway, pathways in cancer, Jak-STAT signaling pathway and MAPK signaling pathway) (Table 2).
Table 1.
List of mutual apoptosis-related genes in DEGs annotation
| Gene ID | GeneSymbol | Regulated | Q value | Function |
|---|---|---|---|---|
| ENSG00000100600 | LGMN | -1.57053543 | 0 | negative regulation of neuron apoptotic process |
| ENSG00000225131 | PSME2 | -1.148863386 | 0.0080288 | regulation of apoptotic process |
| ENSG00000120899 | PTK2B | -1.821029859 | 0 | positive regulation of apoptotic process; |
| negative regulation of apoptotic process; | ||||
| positive regulation of JNK cascade | ||||
| ENSG00000048392 | RRM2B | 2.415037499 | 0.009938502 | negative regulation of apoptotic process |
| ENSG00000170345 | FOS | -4.185111405 | 0 | stress-activated MAPK cascade |
| ENSG00000177606 | JUN | -1.925999419 | 0.006229 | negative regulation of neuron apoptotic process; |
| positive regulation of neuron apoptotic process; | ||||
| stress-activated MAPK cascade | ||||
| ENSG00000269378 | ITGB1 | -1.123382416 | 0.000443 | positive regulation of apoptotic process |
| ENSG00000113594 | LIFR | -7.813781191 | 0 | negative regulation of muscle cell apoptotic process |
| ENSG00000076984 | MAP2K7 | -1.015941544 | 0 | MAP kinase kinase activity; |
| positive regulation of neuron apoptotic process | ||||
| ENSG00000085511 | MAP3K4 | -1.807354922 | 0.002673 | activation of MAPKK activity; |
| activation of MAPK activity; | ||||
| MAP kinase activity | ||||
| ENSG00000120129 | DUSP1 | -1.187202993 | 0 | inactivation of MAPK activity; |
| MAP kinase tyrosine/serine/threonine phosphatase activity; | ||||
| stress-activated MAPK cascade | ||||
| ENSG00000139625 | LGMN | -1.57053543 | 0 | MAPK cascade; MAP kinase activity; |
| activation of JNKK activity; | ||||
| positive regulation of JNK cascade; | ||||
| positive regulation of neuron apoptotic process | ||||
| ENSG00000249936 | PSME2 | -1.148863386 | 0.0080288 | positive regulation of JNK cascade; |
| negative regulation of apoptotic process; | ||||
| engulfment of apoptotic cell; | ||||
| apoptotic signalling pathway |
Table 2.
Apoptosis-related KEGG pathway
| Pathway | P-value | Pathway ID |
|---|---|---|
| Antigen processing and presentation | 4.9392e-01 | ko04612 |
| Natural killer cell mediated cytotoxicity | 7.8074e-01 | ko04650 |
| P53 signaling pathway | 9.4388e-01 | ko04115 |
| pathways in cancer | 9.7949e-01 | ko05200 |
| JAK-STAT signaling pathway | 9.8563e-01 | ko04630 |
| MAPK-JNK signaling pathway | 5.5878e-01 | ko04010 |
We further paid attention to mutual association between the above apoptosis pathways. The results suggested that among above 6 apoptosis-related pathways, except antigen processing and presentation pathway, Natural killer cell mediated cytotoxicity, JAK-STAT signaling pathway, P53 signaling pathway and pathways in cancer all interacts with MAPK-JNK signaling pathway. We also noticed that the regulation of MAPK-JNK pathway for apoptosis is carried out through P53 signaling pathway, which finally affects apoptosis through terminal apoptosis-associated factors such as caspase 3, bcl-2 and cytochrome C.
GSN inhibited hepatocarcinoma cell apoptosis
We then generated the stable GSN overexpression cell line (GSN-SMMC7721) and stable GSN knock down cell line(shGSN-SMMC7721), as well as their corresponding control cell lines (NC1- and NC2-SMMC7721) to performed apoptosis studies in vitro so as to evaluate the roles of GSN in hepatocarcinoma cells. FCM, in-cell immunoblotting and TEM were performed, and were carried out in triplicate.
The results of FCM showed the early, late and total apoptosis rates were 0.72±0.11, 3.48±0.25, and 4.20±0.18 respectively in GSN-SMMC7721. The early, late and total apoptosis rates were 9.97±1.01, 5.44±0.98, 15.41±1.26 respectively in shGSN-SMMC7721. The late and total apoptosis rates in GSN overexpressed SMMC7721 cells were lower than those of the NC1-SMMC7721 (P<0.05). And the early and total apoptosis rates in shGSN-SMMC7721 cells were higher than those of the NC2-SMMC7721 (P<0.05) (Figure 5). In-cell immunoblotting showed the expression of caspase 3 was down-regulated 0.625± 0.133 (P<0.05), and the expression of bcl-2 was up-regulated 4.138±0.857 (P<0.05) in GSN-SMMC7721 while comparing to NC1-SMMC7721. The expression of caspase 3 was up-regulated 1.908±0.257 (P<0.05), and the expression of bcl-2 was down-regulated 0.411±0.122 (P<0.05) in shGSN-SMMC7721 while comparing to NC2-SMMC7721. But the expression of cytochrome C was invariable while GSN was overexpressed/inhibited (Figure 6). Finally, TEM confirmed the discernible apoptosis morphology in the GSN shRNA transfectant, including apoptotic body, autophagy, early apoptosis, late apoptosis and oncosis (Figure 7).
Figure 5.
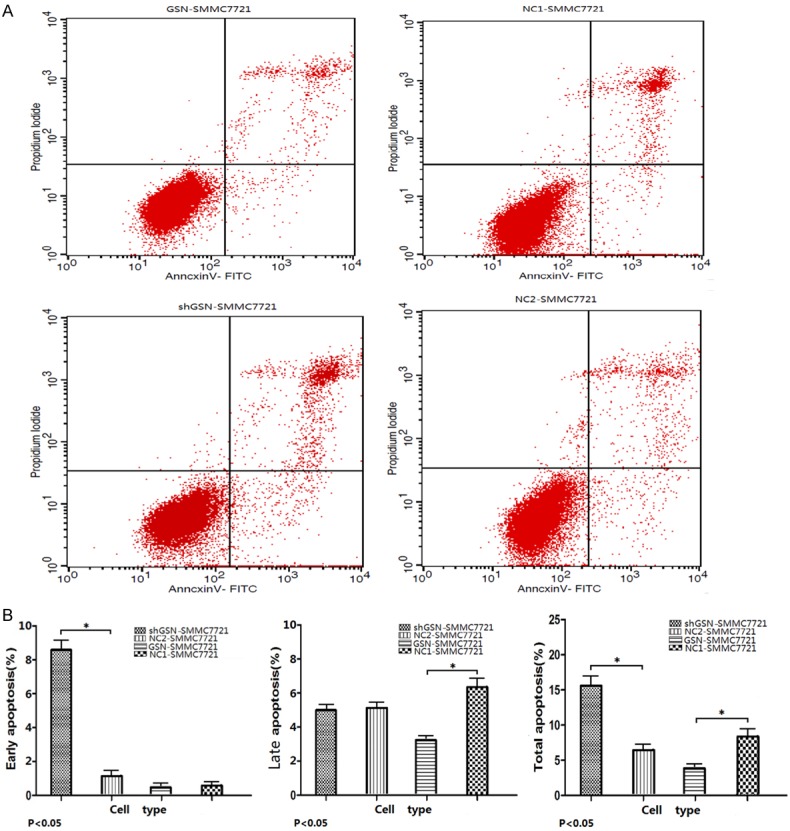
Apoptosis detection through flow cytometry. (A) GSN-SMMC7721, shGSN-SMMC7721, NC1-SMMC7721 and NC2-SMMC7721 were collected, suspended, and then stained by PI and Annexin-V, and finally analyzed by FCM. The experiments were carried out in triplicate. (B) Experiments were performed as in (A), the results displayed a remarkable lower apoptotic rate (including total and late apoptotic rate) in GSN up-regulated cells, and a higher apoptotic rate (including total and early apoptotic rate) in GSN down-regulated cells compared with the control cells (P<0.05).
Figure 6.
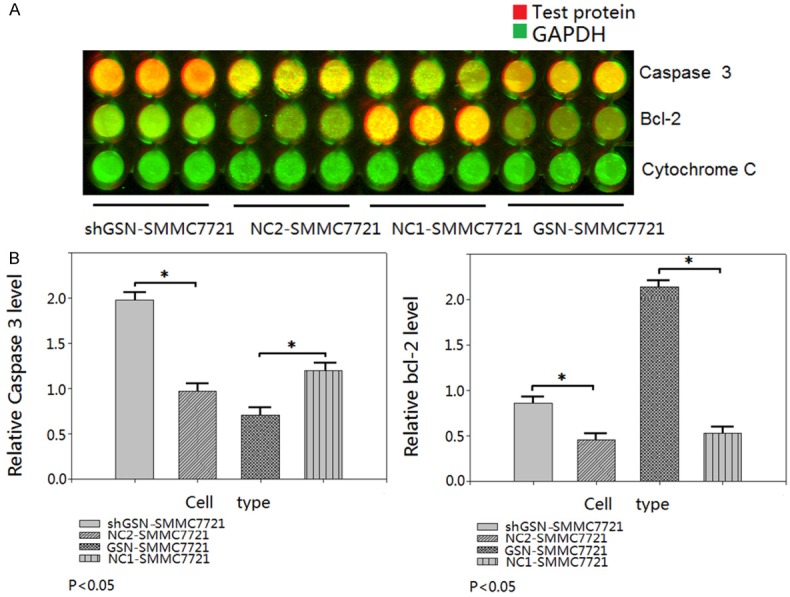
Expression of apoptosis factors caspase 3, bcl-2, and cytochrome C. (A) In-cell immunoblotting detected the expression level of caspase 3, bcl-2, and cytochrome C in GSN-SMMC7721, shGSN-SMMC7721, NC1-SMMC7721 and NC2-SMMC7721. The experiments were carried out in triplicate. (B) Experiments were performed as in (A), the results showed the expression of caspase 3 was down-regulated 0.625±0.133 (P<0.05), and the expression of bcl-2 was up-regulated 4.138±0.857 (P<0.05) in GSN-SMMC7721 while comparing to NC1-SMMC7721. The expression of caspase 3 was up-regulated 1.908±0.257 (P<0.05), and the expression of bcl-2 was down-regulated 0.411±0.122 (P<0.05) in shGSN-SMMC7721 while comparing to NC2-SMMC7721. But the expression of cytochrome C was invariable while GSN was overexpressed/inhibited.
Figure 7.
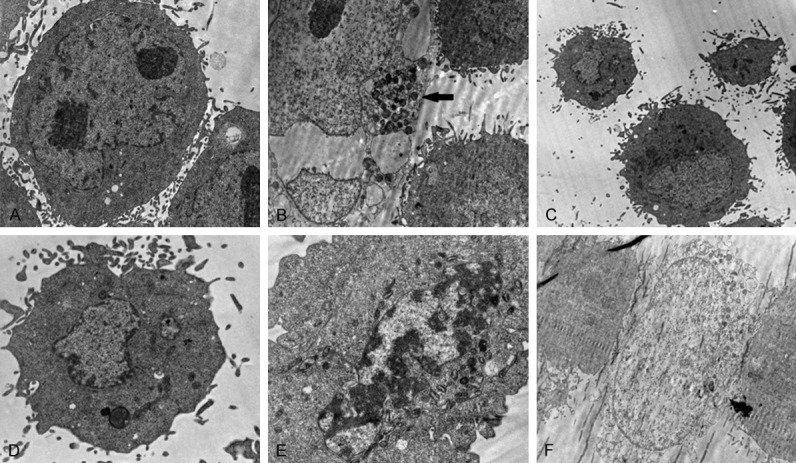
Observation of apoptosis morphology. A. Normal form. B. Apoptotic body. C. Autophagy. D. Early apoptosis. E. Late apoptosis. F. Oncosis. (Magnification ×10000).
Overall, above results suggest a negative relationship between GSN expression and hepatocarcinoma cell apoptosis. Overexpressed GSN inhibits apoptosis and the knockdown of GSN promotes apoptosis.
Discussion
Transcriptome bridges the gap between the genetic codes and the functional molecule pathways. By comparing transcriptomics between normal and cancerous cells, it is able to determine what genes are turned on or off in disparate biological process and gain a deeper understanding of what constitutes a specific cell type and how changes in transcriptional activity may reflect or contribute to disease progression. Comparative transcriptomics has been applied to many human pathological studies such as neurodegenerative disease [44], retina defection [45], prostate cancer [46], and colorectal cancer [47]. For hepatocarcinoma transcriptome, Huang et al performed transcriptome analyses for 10 matched pairs of cancer and non-cancerous tissues from HCC patients on Solexa/Illumina GAII platform [48]. About 21.6 million sequencing reads and 10.6 million aligned reads were obtained and 1,378 significantly DEGs were annotated. Additionally, comprehensive functional analysis indicated that cell growth-related, metabolism-related and immune-related pathways are most significantly enriched by DEGs, pointing to a complex mechanism for HCC carcinogenesis. In another study, David et al detected five hundred DEGs from HCC cells, and the further gene ontology analysis indicated that the over-expressed genes were associated with inflammation, drug resistance and lipid metabolism [49].
Our study was mainly focusing on GSN regulatory HCC transcription through next-generation RNA-seq technology. The results revealed a regulator role of GSN in hepatoma cell apoptosis. We noticed 5 functional categories with 13 relevant DEGs, which relate to cell apoptosis, are mainly restrained and might play important roles in 6 apoptosis-related pathways. This observation provide hint for the potential mechanism of how GSN regulate apoptosis. Interestingly, among all of the 13 mutual apoptosis-related DEGs, 7 DEGs including PTK2B, FOS, JUN, RAC1, MAP2K7, MAP3K4, MAP3K12 enriched in MAPK-JNK signaling pathway are down-regulated.
Meanwhile, except antigen processing and presentation pathway, all other 4 apoptosis-related pathways such as Natural killer cell mediated cytotoxicity, JAK-STAT signaling pathway, P53 signaling pathway and pathways in cancer all interacts with MAPK-JNK signaling pathway. Both observations strongly suggest that GSN might suppress apoptosis through the cross-linking of above pathway, while MAPK-JNK pathway might played central roles in the GSN-regulated apoptosis.
We also noticed that the regulation of MAPK-JNK pathway for apoptosis is carried out through P53 signaling pathway, which finally affects apoptosis through terminal apoptosis-associated factors such as caspase 3, bcl-2 and cytochrome C. We detected the expression of above apoptosis factors through in-cell immunoblotting. The results suggest the discrepant expression of caspase 3 and bcl-2 in GSN up-/down-regulated hepatocarcinoma cells. But the expression of cytochrome C was not changed in the test. Previous reports elucidated the role of GSN as an anti-apoptotic protein in mitochondrial-dependent cell death due to the inositol lipids PI (4, 5) P2 and PI (3, 4) P2 prevent caspase 3 cleavage of GSN in vitro. Through the formation of a stable PI (4, 5) P2-GSN-Caspase 3 complex, PI (4, 5) P2-GSN strongly inhibited caspase-3 activity [50]. Except caspase 3, GSN affects actin-dependent VDAC, which is thought to be a target of bcl-2 family members [51]. Though the release of cytochrome C is considered as an important factor for mitochondria initiated apoptosis, and GSN is thought to be an apoptosis inhibitor by blocking mitochondrial membrane potential loss and cytochrome c release, we did not detect the expression change of cytochrome C. We guessed the result might be accord with the theory that the release of cytochrome C could not be relative to its expression level.
The structural changes of cytoskeletal proteins (such as GSN) in apoptosis circumstance leads to the transmogrification of cells, for instance, chromatin condensation, nuclear fragmentation, cell membrane blebbing, and apoptotic body formation. Early studies showed that there is a tight connection between skelemin change and the apoptosis morphology [52-55]. In our study, we observed the GSN-derived morphology of apoptosis through TEM. According to the previous reports, dissociation of GSN products a NH2-extreme of 352aa, which induces cells turned round, fallen off, and splintered in a Ca2+ independent mode. The observation of TEM suggests that as the regulator and effector of apoptosis, GSN might regulate cell apoptosis through acting on the cytoskeleton and actin activity.
In conclusion, GSN overexpression suppresses apoptosis while down-regulated GSN promotes apoptosis in human HCC cells. The regulation of GSN in the apoptosis of hepatoma cell mainly involves several important pathway such as natural killer cell mediated cytotoxicity, p53 signaling pathway, pathways in cancer, Jak-STAT signaling pathway and MAPK signaling pathway. Caspase 3 and bcl-2 are two pivotal apoptosis factors regulated by GSN. These findings suggest a role for GSN in HCC and a potential of using GSN for the treatment of this malignancy.
Acknowledgements
This research was supported by the National Natural Science Foundation of China, (No. 81260445, 30960332). The Fund of Key Laboratory of High-Incidence-Tumor Prevention & Treatment (No. GK2013-13-A-01 -02, GK2014-ZZ04). The Natural Science Foundation of Guangxi (No. 2013GXNSFBA019183).
Disclosure of conflict of interest
None.
References
- 1.Huang G, Lau WY, Wang ZG, Pan ZY, Yuan SX, Shen F, Zhou WP, Wu MC. Antiviral therapy improves postoperative survival in patients with hepatocellular carcinoma: a randomized controlled trial. Ann Surg. 2015;261:56–66. doi: 10.1097/SLA.0000000000000858. [DOI] [PubMed] [Google Scholar]
- 2.Ochiai T, Ogino S, Ishimoto T, Toma A, Yamamoto Y, Morimura R, Ikoma H, Otsuji E. Prognostic impact of hepatectomy for patients with non-hepatitis b, non-hepatitis c hepatocellular carcinoma. Anticancer Res. 2014;34:4399–410. [PubMed] [Google Scholar]
- 3.He X, Wang Y, Zhang W, Li H, Luo R, Zhou Y, Liao CL, Huang H, Lv X, Xie Z, He M. Screening differential expression of serum proteins in AFP-negative HBV-related hepatocellular carcinoma using iTRAQ-MALDI-MS/MS. Neoplasma. 2014;61:17–26. [PubMed] [Google Scholar]
- 4.Mullauer L, Fujita H, Ishizaki A, Kuzumaki N. Tumor-suppressive function of mutated gelsolin in ras-transformed cells. Oncogene. 1993;8:2531–6. [PubMed] [Google Scholar]
- 5.Winston JS, Asch HL, Zhang PJ, Edge SB, Hyland A, Asch BB. Down regulation of gelsolin correlates with the progression to breast carcinoma. Breast Cancer Res Treat. 2001;65:11–21. doi: 10.1023/a:1006446108411. [DOI] [PubMed] [Google Scholar]
- 6.Li GH, Arora PD, Chen Y, McCulloch CA, Liu P. Multifunctional Roles of Gelsolin in Health and Diseases. Med Res Rev. 2012;32:999–1025. doi: 10.1002/med.20231. [DOI] [PubMed] [Google Scholar]
- 7.Chaponnier C, Kocher O, Gabbiani G. Modulation of gelsolin content in rat aortic smooth muscle cells during development, experimental intimal thickening and culture. An immunohistochemical and biochemical study. Eur J Biochem. 1990;190:559–65. doi: 10.1111/j.1432-1033.1990.tb15610.x. [DOI] [PubMed] [Google Scholar]
- 8.Scholz A, Hinssen H. Biphasic pattern of gelsolin expression and variations in gelsolin-actin interactions during myogenesis. Exp Cell Res. 1995;219:384–91. doi: 10.1006/excr.1995.1243. [DOI] [PubMed] [Google Scholar]
- 9.Kwiatkowski DJ. Predominant induction of gelsolin and actin-binding protein during myeloid differentiation. J Biol Chem. 1988;263:13857–62. [PubMed] [Google Scholar]
- 10.Tanaka M, Mullauer L, Ogiso Y, Fujita H, Moriya S, Furuuchi K, Harabayashi T, Shinohara N, Koyanagi T, Kuzumaki N. Gelsolin: A candidate for suppressor of human bladder cancer. Cancer Res. 1995;55:3228–32. [PubMed] [Google Scholar]
- 11.Chaponnier C, Gabbiani G. Gelsolin modulation in epithelial and stromal cells of mammary carcinoma. Am J Pathol. 1989;134:597–603. [PMC free article] [PubMed] [Google Scholar]
- 12.Paunio T, Kangas H, Kiuru S, Palo J, Peltonen L, Syvanen AC. Tissue distribution and levels of gelsolin mRNA in normal individuals and patients with gelsolin-related amyloidosis. FEBS Lett. 1997;406:49–55. doi: 10.1016/s0014-5793(97)00237-8. [DOI] [PubMed] [Google Scholar]
- 13.Ahn JS, Jang IS, Kim DI, Cho KA, Park YH, Kim K, Kwak CS, Chul Park S. Aging-associated increase of gelsolin for apoptosis resistance. Biochem Biophys Res Commun. 2003;312:1335–41. doi: 10.1016/j.bbrc.2003.11.061. [DOI] [PubMed] [Google Scholar]
- 14.Mullauer L, Fujita H, Ishizaki A, Kuzumaki N. Tumor-suppressive function of mutated gelsolin in ras-transformed cells. Oncogene. 1993;8:2531–6. [PubMed] [Google Scholar]
- 15.Gay F, Estornes Y, Saurin JC, Joly-Pharaboz MO, Friederich E, Scoazec JY, Abello J. In colon carcinogenesis, the cytoskeletal protein gelsolin is down-regulated during the transition from adenoma to carcinoma. Hum Pathol. 2008;39:1420–30. doi: 10.1016/j.humpath.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Choi YK, Kwon HJ, Yang HK, Choi JH, Kim DY. Downregulation of gelsolin and retinoic acid receptor beta expression in gastric cancer tissues through histone deacetylase 1. J Gastroenterol Hepatol. 2004;19:218–24. doi: 10.1111/j.1440-1746.2004.03336.x. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka M, Mullauer L, Ogiso Y, Fujita H, Moriya S, Furuuchi K, Harabayashi T, Shinohara N, Koyanagi T, Kuzumaki N. Gelsolin: A candidate for suppressor of human bladder cancer. Cancer Res. 1995;55:3228–32. [PubMed] [Google Scholar]
- 18.Dosaka-Akita H, Hommura F, Fujita H, Kinoshita I, Nishi M, Morikawa T, Katoh H, Kawakami Y, Kuzumaki N. Frequent loss of gelsolin expression in non-small cell lung cancers of heavy smokers. Cancer Res. 1998;58:322–7. [PubMed] [Google Scholar]
- 19.Lee HK, Driscoll D, Asch H, Asch B, Zhang PJ. Downregulated gelsolin expression in hyperplastic and neoplastic lesions of the prostate. Prostate. 1999;40:14–9. doi: 10.1002/(sici)1097-0045(19990615)40:1<14::aid-pros2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 20.Visapaa H, Bui M, Huang Y, Seligson D, Tsai H, Pantuck A, Figlin R, Rao JY, Belldegrun A, Horvath S, Palotie A. Correlation of Ki-67 and gelsolin expression to clinical outcome in renal clear cell carcinoma. Urology. 2003;61:845–50. doi: 10.1016/s0090-4295(02)02404-4. [DOI] [PubMed] [Google Scholar]
- 21.Ni XG, Zhou L, Wang GQ, Liu SM, Bai XF, Liu F, Peppelenbosch MP, Zhao P. The ubiquitinproteasome pathway mediates gelsolin protein downregulation in pancreatic cancer. Mol Med. 2008;14:582–9. doi: 10.2119/2008-00020.Ni. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noske A, Denkert C, Schober H, Sers C, Zhumabayeva B, Weichert W, Dietel M, Wiechen K. Loss of gelsolin expression in human ovarian carcinomas. Eur J Cancer. 2005;41:461–9. doi: 10.1016/j.ejca.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 23.Kim CS, Furuya F, Ying H, Kato Y, Hanover JA, Cheng SY. Gelsolin: A novel thyroid hormone receptor-beta interacting protein that modulates tumor progression in a mouse model of follicular thyroid cancer. Endocrinology. 2007;148:1306–12. doi: 10.1210/en.2006-0923. [DOI] [PubMed] [Google Scholar]
- 24.Shieh DB, Chen IW, Wei TY, Shao CY, Chang HJ, Chung CH, Wong TY, Jin YT. Tissue expression of gelsolin in oral carcinogenesis progression and its clinicopathological implications. Oral Oncol. 2006;42:599–606. doi: 10.1016/j.oraloncology.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 25.Rao J, Seligson D, Visapaa H, Horvath S, Eeva M, Michel K, Pantuck A, Belldegrun A, Palotie A. Tissue microarray analysis of cytoskeletal actin-associated biomarkers gelsolin and E-cadherin in urothelial carcinoma. Cancer. 2002;95:1247–57. doi: 10.1002/cncr.10823. [DOI] [PubMed] [Google Scholar]
- 26.Kimura K, Ojima H, Kubota D, Sakumoto M, Nakamura Y, Tomonaga T, Kosuge T, Kondo T. Proteomic identification of the macrophagecapping protein as a protein contributing to the malignant features of hepatocellular carcinoma. J Proteomics. 2013;78:362–73. doi: 10.1016/j.jprot.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 27.Shieh DB, Godleski J, Herndon JE 2nd, Azuma T, Mercer H, Sugarbaker DJ, Kwiatkowski DJ. Cell motility as a prognostic factor in stage I non small cell lung carcinoma: The role of gelsolin expression. Cancer. 1999;85:47–57. doi: 10.1002/(sici)1097-0142(19990101)85:1<47::aid-cncr7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 28.Visapää H, Bui M, Huang Y, Seligson D, Tsai H, Pantuck A, Figlin R, Rao JY, Belldegrun A, Horvath S, Palotie A. Correlation of Ki-67 and gelsolin expression to clinical outcome in renal clear cell carcinoma. Urology. 2003;61:845–50. doi: 10.1016/s0090-4295(02)02404-4. [DOI] [PubMed] [Google Scholar]
- 29.An JH, Kim JW, Jang SM, Kim CH, Kang EJ, Choi KH. Gelsolin negatively regulates the activity of tumor suppressor p53 through their physical interaction in hepatocarcinoma HepG2 cells. Biochem Biophys Res Commun. 2011;412:44–9. doi: 10.1016/j.bbrc.2011.07.034. [DOI] [PubMed] [Google Scholar]
- 30.Thor AD, Edgerton SM, Liu SQ, Moore DH, Kwiatkowski DJ. Gelsolin as a negative prognostic factor and effector of motility in erbB-2-positiveepidermal growth factor receptorpositive breast cancers. Clin Cancer Res. 2001;7:2415–24. [PubMed] [Google Scholar]
- 31.Mardis ER. The impact of next-generation sequencing technology ongenetics. Trends Genet. 2008;24:133–41. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzker ML. Sequencing technologies-the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 34.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walter NM, Joan AS. Genome-wide analyses of Epstein-Barr virus reveal conserved RNA structures and a novel stable intronic sequence RNA. BMC Genomics. 2013;14:543–558. doi: 10.1186/1471-2164-14-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 37.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 38.Wong KM, Hudson TJ, McPherson JD. Unraveling the genetics ofcancer: genome sequencing and beyond. Annu Rev Genomics Hum Genet. 2011;12:407–30. doi: 10.1146/annurev-genom-082509-141532. [DOI] [PubMed] [Google Scholar]
- 39.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY, Cao X, Barrette T, Tewari AK, Chee MS, Chinnaiyan AM, Rickman DS, Demichelis F, Gerstein MB, Rubin MA. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–7. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah SP, Köbel M, Senz J, Morin RD, Clarke BA, Wiegand KC, Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E, Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y, Zeng T, Glover JN, Vanderhyden B, Zhao C, Parkinson CA, Jimenez-Linan M, Bowtell DD, Mes-Masson AM, Brenton JD, Aparicio SA, Boyd N, Hirst M, Gilks CB, Marra M, Huntsman DG. Mutation of FOXL2 in Granulosa-Cell Tumors of the Ovary. N Engl J Med. 2009;360:2719–29. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 44.Courtney E, Kornfeld S, Janitz K, Janitz M. Transcriptome profiling in neurodegenerative disease. J Neurosci Methods. 2010;193:189–202. doi: 10.1016/j.jneumeth.2010.08.018. [DOI] [PubMed] [Google Scholar]
- 45.Farkas MH, Grant GR, Pierce EA. Transcriptome analyses to investigate the pathogenesis of RNA splicing factor retinitis pigmentosa. Adv Exp Med Biol. 2012;723:519–25. doi: 10.1007/978-1-4614-0631-0_65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren S, Peng Z, Mao JH, Yu Y, Yin C, Gao X, Cui Z, Zhang J, Yi K, Xu W. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long non coding RNAs and aberrant alternative splicings. Cell Research. 2012;22:806–21. doi: 10.1038/cr.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacteriumnucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Q, Lin B, Liu H, Ma X, Mo F, Yu W, Li L, Li H, Tian T, Wu D, Shen F, Xing J, Chen ZN. RNASeq analyses generate comprehensive transcriptomic landscape and reveal complex transcript patterns in hepatocellular carcinoma. Plos One. 2011;6:e26168. doi: 10.1371/journal.pone.0026168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho DW, Yang ZF, Yi K, Lam CT, Ng MN, Yu WC, Lau J, Wan T, Wang X, Yan Z, Liu H, Zhang Y, Fan ST. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PLos One. 2012;7:e37159. doi: 10.1371/journal.pone.0037159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azuma T, Koths K, Flanagan L, Kwiatkowski D. Gelsolin in complex with phosphatidylinositol4,5-bisphosphate inhibits Caspase-3 and -9 to retard apoptotic progression. J Biol Chem. 2000;275:3761–66. doi: 10.1074/jbc.275.6.3761. [DOI] [PubMed] [Google Scholar]
- 51.Kusano H, Shimizu S, Koya RC, Fujita H, Kamada S, Kuzumaki N, Tsujimoto Y. Human gelsolin prevents apoptosis by inhibiting apoptotic mitochondrial changes via closing VDAC. Oncogene. 2000;19:4807–14. doi: 10.1038/sj.onc.1203868. [DOI] [PubMed] [Google Scholar]
- 52.Jordan M, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 53.Holy TE, Ieibler S. Dynamic instability of microtubules as an efficient way to search in space. Proc Natl Acad Sci U S A. 1994;91:5682–5. doi: 10.1073/pnas.91.12.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen ZY, Xu LY, Li EM, Li JT, Chen MH, Shen J, Zeng Y. Ezrin, acin and cytoskeleton in apoptosis of esophageal epithelial cells induced by arsenic trioxide. Int J Mol Med. 2003;12:341–7. [PubMed] [Google Scholar]
- 55.Parlato S, Giammarioli AM, Logozzi M, Lozupone F, Matarrese P, Luciani F, Falchi M, Malorni W, Fais S. CD95 (APO-1/Fas) linkage to the actin cytoskeleton through ezrin in human T lymphocytes: a novel regulatory mechanism of the CD95 apoptotic pathway. EMBO J. 2000;19:5123–34. doi: 10.1093/emboj/19.19.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]