Abstract
Hepatocellular carcinoma (HCC) is the most common primary tumor of liver and the fifth most common cancer in the world. Lung is the most frequent site for extra hepatic metastasis from hepatocellular carcinoma, while the cause and mechanism of it is still poor understood. Here, we identify that the expression of miR-195 is markedly impaired in the lung metastasis cell lines of HCC. The result of Real-time PCR reveals the expression of miR-195 is significantly downregulated in 92 HCC tissues. Low expression of miR-195 is associated with tumor size, portal vein thrombosis, TNM stage and patients survival. Luciferase reporter and ELISA assay prove that hematogenous metastasis related genes including FGF2 and VEGFA are the target genes of miR-195. Overexpression of miR-195 in HCC cell line BEL-7402 markedly inhibits the capability of migration and invasion. Taken together, our results suggest that miR-195, a tumor suppressor miRNA, contributes to the lung metastasis of HCC by negatively regulating FGF2 and VEGFA, providing key implications of miR-195 for the therapeutic intervention of HCC.
Keywords: Hepatocellular carcinoma, miR-195, metastasis, VEGFA, FGF2
Introduction
Metastasis, the most deadly progress of cancer, is a process that cells break off from primary tumor and travel to other organ or part of body. Although treatments including surgery, radiosurgery, chemotherapy, biological therapy, hormone therapy are used to treat metastasis cancer, they are rarely able to cure metastasis cancer [1]. HCC, as one of the most frequent human malignancies, has several characters such as difficulty of early detection, chemoresistance, radioresistance and high activity of angiogenesis, which account for the rapid recurrence and poor survival [2]. A large number of investigation on HCC have identified a variety of molecules, such as p53, MMPs, VEGF, Ras, Wnt-1, which are associated with the metastasis of HCC [3-7]. However, protein and mechanism associated with lung metastasis of HCC are still blurring.
microRNAs (miRNAs) are a class of small noncoding RNA that regulate the expression of many target gene via mRNA degradation or translation inhibition [8]. miRNAs play important role in processes such as differentiation, proliferation and apoptosis [9]. Emerging evidence confirm that the key enzymes involved in the miRNA biosynthesis pathway and some specific miRNAs are widely deregulated in carcinogenesis and cancer progress. miRNAs such as miR-10a, miR-144 and miR-200 which are downregulated or deletion in cancers [10-12]. In contrast, oncogenic miRNAs including miR-93, miR-183 and miR-224 are often overexpressed in many tumors [13-15]. Recently, there is a growing research interest on the role of miR-195 in tumorigenesis and cancer therapy. Some researchers prove that overexpression of miR-195 can markedly inhibit lung cancer proliferation, migration and invasion by targeting myb [16]. In colorectal, miR-195 represses cell proliferation, colony formation and invasion through targeting CARMA3 [17]. In the recent studies, several target genes of miR-195 associated with HCC have been identified: Wnt3a [18], Pcmt1 [19], VAV2, CDC42 and VEGF [20].
Angiogenesis is a complex process essential for the growth, invasion, and metastasis of various malignant cancers [21]. Tumor angiogenesis gives cancer cells more oxygen, nutrients and survival factors and provide a route for tumor cell egress [22]. So controlling tumor-associated angiogenesis is a promising tactic in limiting cancer progression. FGF2 and VEGFA, as the most important angiogenic factors, are significantly elevated in HCC tissues [23,24]. Besides, high expression of them is correlating with tumor invasiveness and prognosis of patients [25,26]. Antiangiogenesis drugs such as Bevacizumab and Gefitinib which have been widely used to treat cancer, are able to significantly inhibit tumor growth and metastasis [27,28]. However, treatment with antiangiogenesis drugs for a period of time often results in drug resistance and cause treatment failure. Therefore, understanding the mechanism of angiogenesis and exploring novel compounds to overcome this resistance is urgently required [29,30].
In this study, we identify miR-195 is important miRNA related with lung metastases HCC by analyzing miRNA microarray of lung metastases cell lines. We also detected the expression of miR-195 in 92 HCC tissues by Real-time PCR and identify the target genes by luciferase activity assay. In summary, our findings enrich the understanding of HCC metastasis and provide a novel direction for the HCC therapy.
Materials and methods
Tissue samples and cell lines
92 pairs of primary HCC tissues and matched normal liver tissues were obtained from patients in the First Affiliated Hospital of Zhengzhou University from 2008-2010. The clinicopathologic data of all patients including gender, age, hepatitis history, tumor size, lymph node stage, portal vein thrombosis, AFP level and TNM stage. All samples in this study were collected and snap-frozen immediately in liquid nitrogen and store at -80°C. Informed consent was obtained from all patients and protocols used for the tissue were approved by the Ethical Committee of the First Affiliated Hospital of Zhengzhou University. Human hepatocellular carcinoma cell lines BEL-7402 was purchased from the Cell Bank of Chinese Academy of Sciences. Cells were cultured in RPMI-1640 medium (Invitrogen, USA) with 10% fetal bovine serum (FBS) (Hyclone, USA) at 37°C in 5% CO2 incubator. To generate miR-195 overexpressing cell lines, cells were infected with lentivirus-coated pre-miR-195 or negative control (Genechem, China) following the manufacturer’s instructions. After 3 days, positive cells were select with puromycin antibiotic (Sigma, USA).
RNA isolation and real-time PCR
microRNAs were extracted from tissues or cells using Trizol Agent (Invitrogen, USA) and mirVana microRNA isolation system (Ambion, USA), according to the manufacturer’s instructions. The expression of miRNAs were determined by the TaqMan stem-loop RT-PCR method with mirVana miRNA Detection Kit and specific primers (Applied Biosystems, USA). The expression of miR-195 was calculated using comparative CT method and U6 small nuclear RNA as internal control. The changes ratio were calculated using the 2-ΔΔCT method [31].
Luciferase reporter assay
The wild type 3’-untranslated region (WT-UTR) and binding site mutation type 3’UTR (MT-UTR) of FGF2 and VEGFA were constructed in the pMIR-reporter vector (Ambion, USA). Cells (104/well) were seeded in 24-well plates, and co-transfected with 50 nM miR-195 mimics or miRNA control, 100 ng 3’UTR or empty vector and 10 ng pGL3-TK control vector (Promega, USA). 24 hours later, cells were lysed and the luciferase activity was measured with Dual Luciferase Reporter Assay kit (Promega, USA) following the manufacturer’s guideline.
Enzyme-linked immunosorbent assay (ELISA)
1×105 miR-195 overexpressing stable cell lines or control cells were seeded in 6-well plate. After 36 hours, cell supernatant were collected and centrifuged at 1000 rpm for 10 minutes at 4°C to discard cell debris. The expression of FGF2 and VEGFA in 200 μl cell supernatant were detected with anti-human FGF2 ELISA kit and anti-human VEGFA ELISA kit (USCN, China). All the experiments were repeated three times in triplicates.
Transwell cell migration and invasion assay
For cell migration assay, miR-195 or control stable cell lines (1×105) were resuspended in 100 μl serum-free medium and added to the upper chamber of 24-well transwell insert (Corning Costar, USA). The lower chamber was filled with complete medium containing 10% FBS as chemoattractant. For invasion assay, cells were placed in the matrigel (BD, USA) coated (1:10 dilution) upper chamber. Cells were incubated for 24 h at 37°C, then cells were fixed with in ice-cold methanol for 15 min, and stained with 0.5% crystal violet solution for 15 min. Cells on the upper surface of membrane were removed with a cotton swap. Cells were captured by a microscope. Finally, the crystal violet was eluted with 33% acetic acid, and the OD of the stained cells was measured at 570 nm using a microplate reader.
microRNA expression analysis
microarray data of parental HCC PLC, MHCC97H cell lines and lung metastases PLC, MHCC97H cell lines (GSE43445) was analyzed by GEO2R. The differential expression of microRNAs were selected based on a P<0.05.
Statistical analysis
The statistical analysis was performed using the SPSS 15.0 software. All data was represented with mean ± SD from at least three independent experiments. One way ANOVA was used to determine the association between the expression of miR-195 and the clinicopathological characters. Overall survival curve calculated according to the Kaplan-Meier method using log rank test. Difference between groups was determined by pair-sample t test. Significance was defined as *P<0.05.
Results
Expression profile of microRNA microarray
microRNA microarray data of HCC metastasis was searched in the NCBI GEO database. Fortunately, one data GSE43445 which contained two HCC cell lines and two lung metastases cell lines derived the former was found. The downregulated and upregulated miRNAs were observed after analyzing with GEO2R software. As shown in Table 1, the 20 downregulated microRNAs and whole 4 upregulated were listed. From these results, we found miR-195 cluster which contained miR-15, miR-16 and miR-195 was significantly decreased in the lung metastases cell lines. So in this study, we focused our attention on miR-195.
Table 1.
miRNAs differentially expressed in lung metastasis hepatocarcinoma cell lines and primary cell lines
| miRNAs downregualted in metastasis cells (Top 20) | p value | Log FC |
|
| ||
| hsa-miR-518d | 0.0000182 | -8.555 |
| hsa-miR-19a | 0.0001138 | -4.6105 |
| hsa-miR-584 | 0.0002695 | -8.6355 |
| hsa-miR-630 | 0.0007755 | -8.691 |
| hsa-miR-21 | 0.0029415 | -4.31 |
| hsa-miR-29b | 0.0033382 | -3.0145 |
| hsa-miR-195 | 0.004605 | -3.1025 |
| hsa-let-7g | 0.0055515 | -2.775 |
| hsa-let-7f | 0.0065571 | -2.7305 |
| hsa-miR-18b | 0.006994 | -3.042 |
| hsa-miR-7 | 0.0105305 | -3.5765 |
| hsa-miR-15a | 0.011107 | -3.448 |
| hsa-miR-20a | 0.0134737 | -3.0815 |
| hsa-miR-26b | 0.0156391 | -3.6585 |
| hsa-miR-374 | 0.0161879 | -3.022 |
| hsa-miR-18a | 0.0178521 | -3.2355 |
| hsa-miR-19b | 0.0255466 | -3.776 |
| hsa-miR-106b | 0.0281044 | -2.8175 |
| hsa-miR-27a | 0.0361368 | -2.9455 |
| hsa-miR-16 | 0.0370817 | -2.6205 |
|
| ||
| miRNAs upregualted in metastasis cells | p value | Log FC |
|
| ||
| hsa-miR-124a | 0.0007219 | 9.3475 |
| hsa-miR-184 | 0.026537 | 0.5265 |
| hsa-miR-516-5p | 0.038165 | 0.685 |
| hsa-miR-632 | 0.0382034 | 0.9665 |
Expression of miR-195 in HCC tissues
Expression of miR-195 in 92 pairs of HCC tissues was analyzed by real-time PCR. The relative expression of miR-195 was calculated using 2-ΔΔCT method after normalizing with U6 small nuclear RNA. As shown in Figure 1A, miR-195 expression was significantly downregulated in tumor tissues comparing with the adjacent normal tissues (P<0.01).
Figure 1.
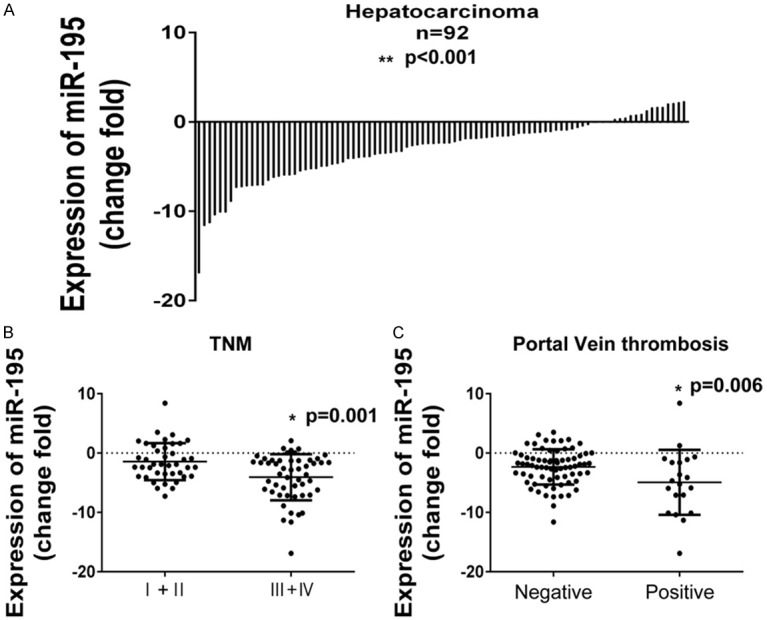
The expression of miR-195 in HCC tissue. A. Levels of miR-195 in 92 primary HCC tissues compared with the adjacent normal tissues. B. The expression of miR-195 in different TNM stage (I+II VS III+IV). C. The different expression of miR-195 in tissues with and without portal vein thrombosis.
Further analysis was performed in order to elucidate the relationship between miR-195 expression levels and clinicopathologic characters. Results revealed that expression of miR-195 was negatively correlated with age, young patients had a lower level of miR-195 (-4.07±4.35) comparing with the old (-2.22±3.26). Besides, the expression of miR-195 was also negatively with tumor size (P=0.048), portal vein thrombosis (P=0.006) and TNM stage (P=0.001). In contrast, there was no correlation between miR-195 expression and gender, hepatitis history, lymph node metastases and AFP level (Table 2). Figure 1B and 1C showed the expression of miR-195 in differential status of portal vein thrombosis and TNM stage. Moreover, the 5-year overall survival of patients with a low level of miR-195 was markedly decreased, indicating miR-195 was a prognostic factor of miR-195 (Figure 2).
Table 2.
Association between miR-195 expression and clinical characteristics of hepatocellular carcinoma
| Clinical Characteristics | NO | Change Fold (Mean ± SD) | P value# |
|---|---|---|---|
| Gender | |||
| Male | 69 | -2.66±3.66 | 0.281 |
| Female | 23 | -3.64±4.14 | |
| Age | |||
| ≥50 | 58 | -2.22±3.26 | 0.022* |
| <50 | 34 | -4.07±4.35 | |
| Hepatitis History | |||
| Yes | 27 | -3.12±4.30 | 0.722 |
| No | 65 | -2.81±3.58 | |
| Tumor Size | |||
| ≥5 cm | 45 | -3.70±3.63 | 0.048* |
| <5 cm | 47 | -2.14±3.81 | |
| LN metastasis | |||
| Yes | 15 | -3.27±3.95 | 0.686 |
| No | 77 | -2.83±3.77 | |
| Portal Vein Thrombosis | |||
| Yes | 20 | -4.93±5.47 | 0.006** |
| No | 72 | -2.34±2.98 | |
| AFP | |||
| High | 46 | -3.52±3.74 | 0.119 |
| Low | 46 | -2.29±3.77 | |
| TNM | |||
| I, II | 41 | -1.45±3.12 | 0.001** |
| III, IV | 51 | -4.07±3.89 |
One Way ANOVA was used;
P<0.05;
P<0.01.
Figure 2.
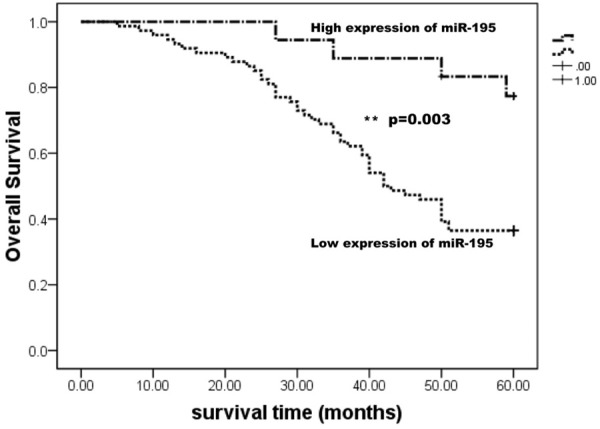
Kaplan-Meier survival curves of patients with HCC based on miR-195 expression status. Patients with low expression of miR-195 have a poor prognosis compared with high expression group.
Potential target genes of miR-195
To identify effectors of miR-195, two databases were used (TargetScan, MIRDB). Overlap analysis between miR-195 target genes and angiogenesis genes was performed, as shown in Figure 3A, FGF2 and VEGFA were identified. Figure 3B displayed the interaction model between miR-195 and FGF2 or VEGFA 3’UTR. These results indicating that miR-195 may participate in lung metastases of HCC by regulating FGF2 and VEGFA.
Figure 3.
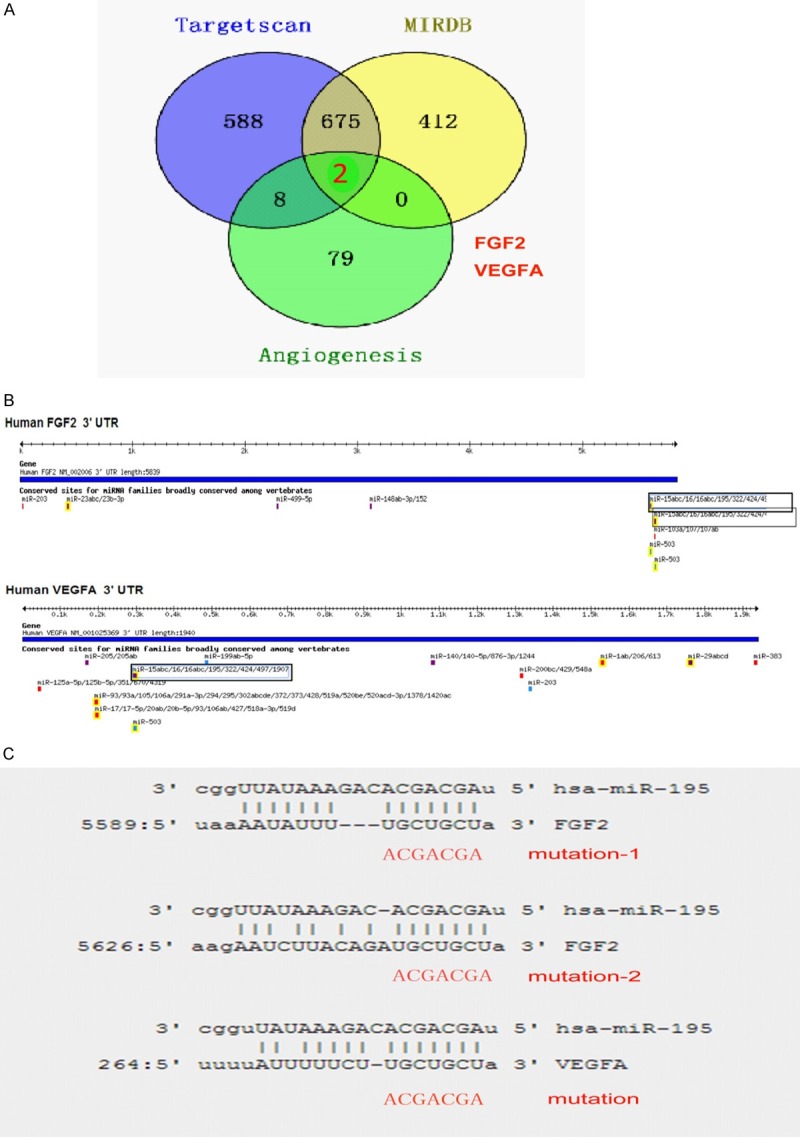
Potential targets genes of miR-195. A. Genes overlap analysis between miR-195 target genes predicted by two prediction algorithms and angiogenesis related genes reported by Qiagen Company (Angiogenesis PCR Array). B. Putative miR-195 binding sites in the 3’UTR of FGF2 and VEGFA. C. The conserved miR-195 binding sequence and the mutated sequence in the 3’UTR of FGF2 and VEGFA.
FGF2 and VEGFA are the target genes of miR-195
In order to confirm whether FGF2 and VEGFA were the target genes, luciferase reporter assay was performed. Figure 3C showed the putative binding sites of miR-195. Plasmids containing the wild type binding sites 3’UTR or mutated binding sites 3’UTR were clone into a luciferase reporter vector. HCC cell lines BEL-7402 were used for analyzing luciferase activity. Results revealed that miR-195 mimics significantly suppressed the luciferase activity of FGF2 WT-UTR. Luciferase activity of plasmids with one mutant binding site (FGF2 had two putative binding site, MT1 and MT2) only had partly recovery. While, overexpression of miR-195 had no effect on the luciferase activity of MT-all-UTR in which all the binding sites were mutated (Figure 4A). Similarly, miR-195 transfection also repressed the luciferase activity of VEGFA WT-UTR. When co-transfected with VEGFA MT-UTR, there was no reduction in luciferase activity was observed (Figure 4B).
Figure 4.
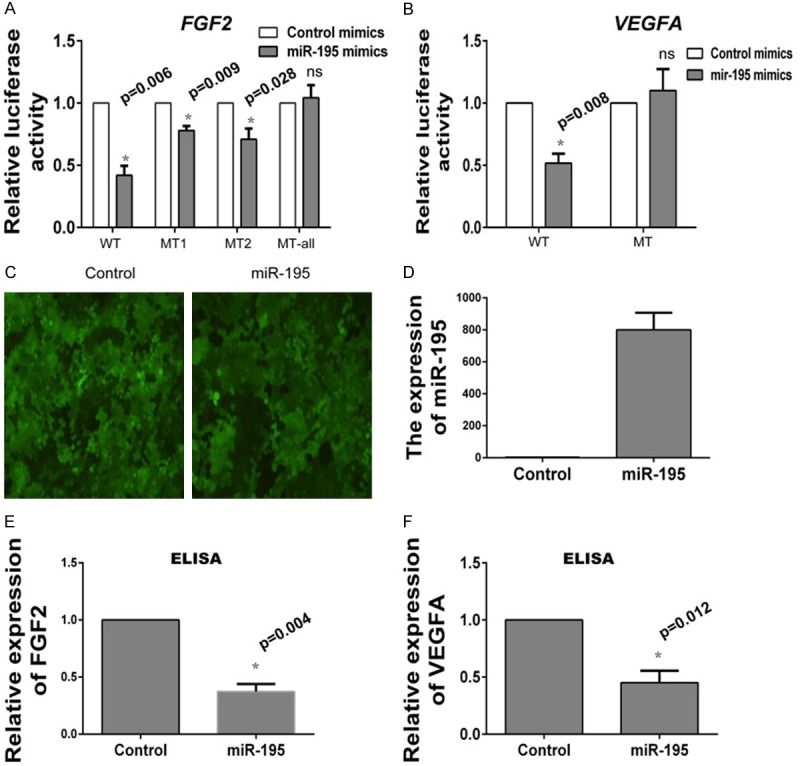
FGF2 and VEGFA are the target of miR-195. A. miR-195 targets the wild type but not the mutant 3’UTR of FGF2. miR-195 mimics or control mimics are co-transfected with reporter vectors containing WT or MT FGF2 3’UTR in BEL-7402 cell lines. B. miR-195 targets the wild type but not the mutant 3’UTR of VEGFA. miR-195 mimics or control mimics are co-transfected with reporter vectors containing WT or MT VEGFA 3’UTR in BEL-7402 cell lines. C. The infection efficiency in BEL-7402 cell lines is examined by fluorescence microscope. D. Overexpression of miR-195 in stable cell lines is demonstrated by Real-time PCR. E. The protein expression of FGF2 in cell supernatant is detected by ELISA assay. F. The protein expression of VEGFA in cell supernatant is detected by ELISA assay.
Next we gained miR-195 overexpressing BEL-7402 cell lines by infecting cells with lentivirus coated pre-miR-195. Cells infection efficiency was detected by a fluorescence microscope (Figure 4C), and the overexpression of miR-195 was examined by Real-time PCR. Figure 4D confirmed miR-195 expression level was significantly elevated in BEL-7402 miR-195 overexpressing cell lines. Then, the protein expression of FGF2 and VEGFA in cell supernant was examined using ELISA assay. The results showed that miR-195 overexpression markedly inhibited endogenous expression of FGF2 (Figure 4E) and VEGFA protein (Figure 4F).
miR-195 inhibit the migration and invasion of BEL-7402 cell lines
To investigate the function of miR-195 in HCC, transwell assay was performed. Migration assay showed that overexpression of miR-195 in BEL-7402 significantly inhibited cell migration (Figure 5A). Similarly, the invasion ability of BEL-7402 cell decreased after overexpressing miR-195 (Figure 5B).
Figure 5.
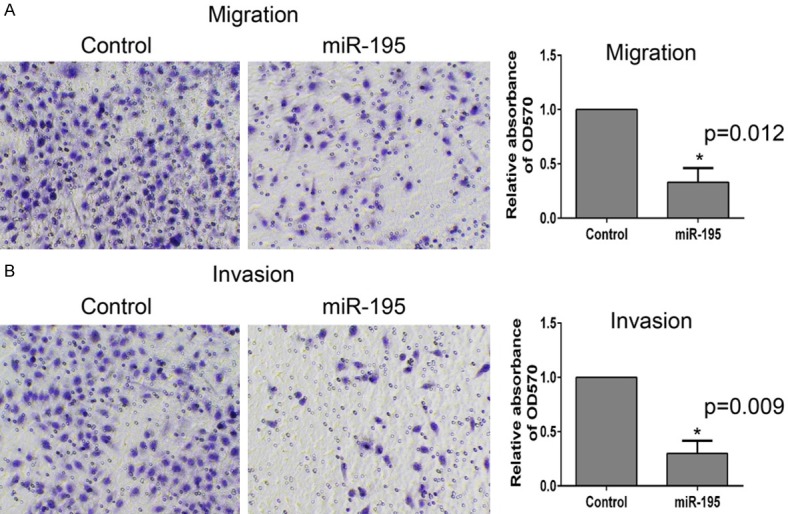
miR-195 inhibit the migration and invasion of BEL-7402. A. In vitro migration assay is performed (described in Materials and methods) to detect the function of miR-195. B. In vitro invasion assay is performed to detect the function of miR-195.
Discussion
During the last past decades, a large number of studies have demonstrated that dysfunction of miRNAs can regulate tumor promotion and progression by leading to altered expression of tumor suppressors or oncogenes. Moreover, abnormal expression of miRNAs can serve as a prognostic indicator of patients’ survival [32]. Recently, researchers prove deregulation of miRNAs is associated with HCC metastasis by regulating epithelial to mesenchymal transition (EMT) [33,34]. In this study, we identify many deregulated miRNAs in HCC lung metastases cells lines. miRNAs such as miR-518d, miR-21, miR-let-7, miR-15, miR-16 and miR-195 are downregulated in lung metastases cell lines. Other miRNAs including miR-124a, miR-184, miR-516 and miR-623 are upregulated. Considering miR-15, miR-16 and miR-195 are belong to a microRNA cluster and miR-195 is conserved across mammalian [35], in this study, we focus on the potential cancer suppressor miR-195.
Previous studies have shown miR-195 acts as a tumor suppressor in several cancers including non-small cell lung cancer (NSCLC) [36], colorectal cancer [37], breast cancer [38], gastric cancer [39] and HCC [19]. Overexpression of miR-195 in cancer cells can inhibit tumor proliferation and metastasis by targeting genes such as CCND1 [40], PIK3CA [16], CDC42 [41] and MYC [42]. However, miR-195 is increased in melanoma [43] and leukemia [44]. In this paper, we examine miR-195 expression in 92 pairs HCC tissues. Results show that miR-195 is significantly downregulated in tumor tissues compared with the adjacent normal tissues. We also find aberrantly deregulated expression of miR-195 is associated with age, tumor size, TNM stage and survival rate. Transwell assay demonstrates that overexpressing miR-195 in HCC cell lines significantly repressed cancer migration and invasion. Moreover, our data reveals tissues with portal vein thrombosis have an obviously lower miR-195 expression level. These findings are consistent with the microarray results, indicating miR-195 is a crucial regulator of HCC lung metastasis.
Angiogenesis plays a privotal role in the occurrence, development and metastasis of HCC [45]. Repressing angiogenesis with specific factors or drugs can markedly inhibit tumor metastasis [46]. VEGFA is a crucial factor that specifically acts on endothelial cells and stimulate endothelial cell mitogenesis and cell migration. High expression of VEGFA is associated with the development and metastasis of cancers [47]. Silence of VEGFA expression in breast cancer markedly reduced cancer angiogenesis and metastasis [48]. FGF2 is a more potent angiogenic factor than VEGF and PDGF by acting as a promotion of endothelial cell proliferation and the physical organization of endothelial cells into tube-lime structures. Recent studies show that miR-503 inhibits tumor angiogenesis and growth by targeting FGF2 and VEGFA [49]. In this study, we analyze the overlap genes between miR-195 target genes and angiogenesis related gens, and identify two important genes: FGF2 and VEGFA. These hypothesis are further confirmed by luciferase reporter assay and ELISA assay.
In summary, our data suggest that miR-195 is a vital regulator in the lung metastasis of HCC. This function may be mediated by targeting angiogenesis related genes FGF2 and VEGFA. miR-195 has significant therapeutic potential as a molecular medicine for the treatment of HCC by regulating cancer angiogenesis.
Disclosure of conflict of interest
None.
References
- 1.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Yang SF, Chang CW, Wei RJ, Shiue YL, Wang SN, Yeh YT. Involvement of DNA damage response pathways in hepatocellular carcinoma. Biomed Res Int. 2014;2014:153867. doi: 10.1155/2014/153867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang YH, Dong YY, Wang WM, Xie XY, Wang ZM, Chen RX, Chen J, Gao DM, Cui JF, Ren ZG. Vascular endothelial cells facilitated HCC invasion and metastasis through the Akt and NF-kappaB pathways induced by paracrine cytokines. J Exp Clin Cancer Res. 2013;32:51. doi: 10.1186/1756-9966-32-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang N, Zhu M, Tsao SW, Man K, Zhang Z, Feng Y. Up-regulation of TIMP-1 by genipin inhibits MMP-2 activities and suppresses the metastatic potential of human hepatocellular carcinoma. PLoS One. 2012;7:e46318. doi: 10.1371/journal.pone.0046318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahsnig C, Mikula M, Petz M, Zulehner G, Schneller D, van Zijl F, Huber H, Csiszar A, Beug H, Mikulits W. ILEI requires oncogenic Ras for the epithelial to mesenchymal transition of hepatocytes and liver carcinoma progression. Oncogene. 2009;28:638–650. doi: 10.1038/onc.2008.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinway SN, Zanudo JG, Ding W, Rountree CB, Feith DJ, Loughran TP Jr, Albert R. Network modeling of TGFbeta signaling in hepatocellular carcinoma epithelial-to-mesenchymal transition reveals joint sonic hedgehog and Wnt pathway activation. Cancer Res. 2014;74:5963–5977. doi: 10.1158/0008-5472.CAN-14-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fan HX, Tang H. Complex interactions between microRNAs and hepatitis B/C viruses. World J Gastroenterol. 2014;20:13477–13492. doi: 10.3748/wjg.v20.i37.13477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 10.Stadthagen G, Tehler D, Hoyland-Kroghsbo NM, Wen J, Krogh A, Jensen KT, Santoni-Rugiu E, Engelholm LH, Lund AH. Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice. PLoS Genet. 2013;9:e1003913. doi: 10.1371/journal.pgen.1003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun L, Bian G, Meng Z, Dang G, Shi D, Mi S. MiR-144 Inhibits Uveal Melanoma Cell Proliferation and Invasion by Regulating c-Met Expression. PLoS One. 2015;10:e0124428. doi: 10.1371/journal.pone.0124428. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Wong CM, Wei L, Au SL, Fan DN, Zhou Y, Tsang FH, Law CT, Lee JM, He X, Shi J, Wong CC, Ng IO. MiR-200b/200c/429 subfamily negatively regulates Rho/ROCK signaling pathway to suppress hepatocellular carcinoma metastasis. Oncotarget. 2015;6:13658–13670. doi: 10.18632/oncotarget.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta K, Hoshino H, Wang J, Ono S, Iida Y, Hata K, Huang SK, Colquhoun S, Hoon DS. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;6:3211–3224. doi: 10.18632/oncotarget.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung WK, He M, Chan AW, Law PT, Wong N. Wnt/beta-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015;362:97–105. doi: 10.1016/j.canlet.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Cui R, Meng W, Sun HL, Kim T, Ye Z, Fassan M, Jeon YJ, Li B, Vicentini C, Peng Y, Lee TJ, Luo Z, Liu L, Xu D, Tili E, Jin V, Middleton J, Chakravarti A, Lautenschlaeger T, Croce CM. MicroRNA-224 promotes tumor progression in nonsmall cell lung cancer. Proc Natl Acad Sci U S A. 2015;112:E4288–4297. doi: 10.1073/pnas.1502068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yongchun Z, Linwei T, Xicai W, Lianhua Y, Guangqiang Z, Ming Y, Guanjian L, Yujie L, Yunchao H. MicroRNA-195 inhibits non-small cell lung cancer cell proliferation, migration and invasion by targeting MYB. Cancer Lett. 2014;347:65–74. doi: 10.1016/j.canlet.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Qian L, Li X, Yan J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony-formation and invasion through targeting CARMA3. Mol Med Rep. 2014;10:473–478. doi: 10.3892/mmr.2014.2178. [DOI] [PubMed] [Google Scholar]
- 18.Yang Y, Li M, Chang S, Wang L, Song T, Gao L, Hu L, Li Z, Liu L, Yao J, Huang C. MicroRNA-195 acts as a tumor suppressor by directly targeting Wnt3a in HepG2 hepatocellular carcinoma cells. Mol Med Rep. 2014;10:2643–2648. doi: 10.3892/mmr.2014.2526. [DOI] [PubMed] [Google Scholar]
- 19.Amer M, Elhefnawi M, El-Ahwany E, Awad AF, Gawad NA, Zada S, Tawab FM. HsamiR-195 targets PCMT1 in hepatocellular carcinoma that increases tumor life span. Tumour Biol. 2014;35:11301–11309. doi: 10.1007/s13277-014-2445-4. [DOI] [PubMed] [Google Scholar]
- 20.Wang R, Zhao N, Li S, Fang JH, Chen MX, Yang J, Jia WH, Yuan Y, Zhuang SM. MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology. 2013;58:642–653. doi: 10.1002/hep.26373. [DOI] [PubMed] [Google Scholar]
- 21.Tsirakis G, Pappa CA, Kanellou P, Stratinaki MA, Xekalou A, Psarakis FE, Sakellaris G, Alegakis A, Stathopoulos EN, Alexandrakis MG. Role of platelet-derived growth factor-AB in tumour growth and angiogenesis in relation with other angiogenic cytokines in multiple myeloma. Hematol Oncol. 2012;30:131–136. doi: 10.1002/hon.1014. [DOI] [PubMed] [Google Scholar]
- 22.Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 23.Murakami K, Kasajima A, Kawagishi N, Sekiguchi S, Fujishima F, Watanabe M, Sato Y, Ohuchi N, Sasano H. The prognostic significance of vasohibin 1-associated angiogenesis in patients with hepatocellular carcinoma. Hum Pathol. 2014;45:589–597. doi: 10.1016/j.humpath.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Wang ZL, Liang P, Dong BW, Yu XL, Yu de J. Prognostic factors and recurrence of small hepatocellular carcinoma after hepatic resection or microwave ablation: a retrospective study. J Gastrointest Surg. 2008;12:327–337. doi: 10.1007/s11605-007-0310-0. [DOI] [PubMed] [Google Scholar]
- 25.Pang R, Poon RT. Angiogenesis and antiangiogenic therapy in hepatocellular carcinoma. Cancer Lett. 2006;242:151–167. doi: 10.1016/j.canlet.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Yang J, Yuan D, Wang J, Zhao J, Wang L. Effects of basic fibroblast growth factor on angiogenin expression and cell proliferation in H7402 human hepatoma cells. J Genet Genomics. 2009;36:399–407. doi: 10.1016/S1673-8527(08)60129-0. [DOI] [PubMed] [Google Scholar]
- 27.Jary M, Borg C, Bouche O, Kim S, Andre T, Bennouna J. [Anti-angiogenic treatments in metastatic colorectal cancer: Does a continuous angiogenic blockade make sense?] . Bull Cancer. 2015;102:758–71. doi: 10.1016/j.bulcan.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Preusser M, Marosi C. Antiangiogenic treatment of meningiomas. Curr Treat Options Neurol. 2015;17:359. doi: 10.1007/s11940-015-0359-0. [DOI] [PubMed] [Google Scholar]
- 29.Yang J, Qin G, Luo M, Chen J, Zhang Q, Li L, Pan L, Qin S. Reciprocal positive regulation between Cx26 and PI3K/Akt pathway confers acquired gefitinib resistance in NSCLC cells via GJIC-independent induction of EMT. Cell Death Dis. 2015;6:e1829. doi: 10.1038/cddis.2015.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao W, Liu Y, Zhang R, Zhang B, Wang T, Zhu X, Mei L, Chen H, Zhang H, Ming P, Huang L. Homoharringtonine induces apoptosis and inhibits STAT3 via IL-6/JAK1/STAT3 signal pathway in Gefitinib-resistant lung cancer cells. Sci Rep. 2015;5:8477. doi: 10.1038/srep08477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Vosa U, Vooder T, Kolde R, Fischer K, Valk K, Tonisson N, Roosipuu R, Vilo J, Metspalu A, Annilo T. Identification of miR-374a as a prognostic marker for survival in patients with early-stage nonsmall cell lung cancer. Genes Chromosomes Cancer. 2011;50:812–822. doi: 10.1002/gcc.20902. [DOI] [PubMed] [Google Scholar]
- 33.Huang JY, Zhang K, Chen DQ, Chen J, Feng B, Song H, Chen Y, Zhu Z, Lu L, De W, Wang R, Chen LB. MicroRNA-451: epithelial-mesenchymal transition inhibitor and prognostic biomarker of hepatocelluar carcinoma. Oncotarget. 2015;6:18613–30. doi: 10.18632/oncotarget.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yau WL, Lam CS, Ng L, Chow AK, Chan ST, Chan JY, Wo JY, Ng KT, Man K, Poon RT, Pang RW. Over-expression of miR-106b promotes cell migration and metastasis in hepatocellular carcinoma by activating epithelialmesenchymal transition process. PLoS One. 2013;8:e57882. doi: 10.1371/journal.pone.0057882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finnerty JR, Wang WX, Hebert SS, Wilfred BR, Mao G, Nelson PT. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010;402:491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Qu J, Xu F, Guo Y, Wang Y, Yu H, Qian B. MiR-195 suppresses non-small cell lung cancer by targeting CHEK1. Oncotarget. 2015;6:9445–56. doi: 10.18632/oncotarget.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tan YG, Zhang YF, Guo CJ, Yang M, Chen MY. Screening of differentially expressed microRNA in ulcerative colitis related colorectal cancer. Asian Pac J Trop Med. 2013;6:972–976. doi: 10.1016/S1995-7645(13)60174-1. [DOI] [PubMed] [Google Scholar]
- 38.Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, Chang G, Li X, Li Q, Wang S, Wang W. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014;9:e96228. doi: 10.1371/journal.pone.0096228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–657. doi: 10.1111/j.1440-1746.2008.05666.x. [DOI] [PubMed] [Google Scholar]
- 40.Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50:113–121. doi: 10.1002/hep.22919. [DOI] [PubMed] [Google Scholar]
- 41.Wang YS, Wang HY, Liao YC, Tsai PC, Chen KC, Cheng HY, Lin RT, Juo SH. MicroRNA-195 regulates vascular smooth muscle cell phenotype and prevents neointimal formation. Cardiovasc Res. 2012;95:517–526. doi: 10.1093/cvr/cvs223. [DOI] [PubMed] [Google Scholar]
- 42.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 43.Bhattacharya A, Schmitz U, Wolkenhauer O, Schonherr M, Raatz Y, Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene. 2013;32:3175–3183. doi: 10.1038/onc.2012.324. [DOI] [PubMed] [Google Scholar]
- 44.Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva-Jr WA, Falcao RP, Zago MA. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz J Med Biol Res. 2007;40:1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 45.Ye G, Qin Y, Lu X, Xu X, Xu S, Wu C, Wang X, Wang S, Pan D. The association of reninangiotensin system genes with the progression of hepatocellular carcinoma. Biochem Biophys Res Commun. 2015;459:18–23. doi: 10.1016/j.bbrc.2015.02.030. [DOI] [PubMed] [Google Scholar]
- 46.Arakawa Y, Shimada M, Utsunomiya T, Imura S, Morine Y, Ikemoto T. Effects of pegylated interferon alpha2b on metastasis of hepatocellular carcinoma. J Surg Res. 2012;172:95–101. doi: 10.1016/j.jss.2010.07.023. [DOI] [PubMed] [Google Scholar]
- 47.Park ST, Kim BR, Park SH, Lee JH, Lee EJ, Lee SH, Rho SB. Suppression of VEGF expression through interruption of the HIF1alpha and Akt signaling cascade modulates the antiangiogenic activity of DAPK in ovarian carcinoma cells. Oncol Rep. 2014;31:1021–1029. doi: 10.3892/or.2013.2928. [DOI] [PubMed] [Google Scholar]
- 48.Zhang J, Wang P, Zhu J, Wang W, Yin J, Zhang C, Chen Z, Sun L, Wan Y, Wang X, Chen G, Liu Y. SPARC expression is negatively correlated with clinicopathological factors of gastric cancer and inhibits malignancy of gastric cancer cells. Oncol Rep. 2014;31:2312–2320. doi: 10.3892/or.2014.3118. [DOI] [PubMed] [Google Scholar]
- 49.Zhou B, Ma R, Si W, Li S, Xu Y, Tu X, Wang Q. MicroRNA-503 targets FGF2 and VEGFA and inhibits tumor angiogenesis and growth. Cancer Lett. 2013;333:159–169. doi: 10.1016/j.canlet.2013.01.028. [DOI] [PubMed] [Google Scholar]


