Abstract
HIV-1 infection results in a chronic illness because long-term highly active antiretroviral therapy can lower viral titers to an undetectable level. However, discontinuation of therapy rapidly increases virus burden. Moreover, patients under highly active antiretroviral therapy frequently develop various metabolic disorders, neurocognitive abnormalities, and cardiovascular diseases. We have previously shown that exosomes containing trans-activating response (TAR) element RNA enhance susceptibility of undifferentiated naive cells to HIV-1 infection. This study indicates that exosomes from HIV-1-infected primary cells are highly abundant with TAR RNA as detected by RT-real time PCR. Interestingly, up to a million copies of TAR RNA/μl were also detected in the serum from HIV-1-infected humanized mice suggesting that TAR RNA may be stable in vivo. Incubation of exosomes from HIV-1-infected cells with primary macrophages resulted in a dramatic increase of proinflammatory cytokines, IL-6 and TNF-β, indicating that exosomes containing TAR RNA could play a direct role in control of cytokine gene expression. The intact TAR molecule was able to bind to PKR and TLR3 effectively, whereas the 5′ and 3′ stems (TAR microRNAs) bound best to TLR7 and -8 and none to PKR. Binding of TAR to PKR did not result in its phosphorylation, and therefore, TAR may be a dominant negative decoy molecule in cells. The TLR binding through either TAR RNA or TAR microRNA potentially can activate the NF-κB pathway and regulate cytokine expression. Collectively, these results imply that exosomes containing TAR RNA could directly affect the proinflammatory cytokine gene expression and may explain a possible mechanism of inflammation observed in HIV-1-infected patients under cART.
Keywords: cytokinesis, exosome (vesicle), human immunodeficiency virus (HIV), macrophage, protein kinase RNA-activated (PKR), toll-like receptor (TLR), TAR RNA
Introduction
Since its discovery in 1981, human immunodeficiency virus type-1 (HIV-1) has caused substantial deaths in both developed and emerging countries (1). As of 2011, an estimated 25 million people have died of acquired immunodeficiency syndrome (AIDS) caused by HIV-1 infection; 34 million are currently living with HIV-1 infection worldwide, and there is not yet an effective vaccine (1). Moreover, 2.5 million new infections worldwide and ∼50,000 in the United States alone have been reported each year (2). Long-term highly active antiretroviral therapy can halt the virus replication in blood to an undetectable level. However, discontinuation of therapy rapidly increases the virus burden (3, 4). For reasons not well understood, patients under highly active antiretroviral therapy frequently develop various metabolic disorders and also HIV-associated neurocognitive disorders (5–9).
HIV-1 virions contain a 9.7-kb dimeric positive-sense single-stranded RNA genome, comprising a total of the following nine genes: structural (env, gag, and pol), regulatory (tat and rev), and accessory (nef, vif, vpr, and vpu) (10). In addition, the HIV-1 genome also encodes several small, regulatory non-coding RNAs (ncRNAs)4 and probably microRNAs (miRNAs) that can regulate host gene expression (11–17). Tat is an essential regulatory protein that directs efficient elongation of the HIV-1 genome. It binds to an RNA stem-loop structure, the trans-activating response element (TAR) at the 5′ ends of HIV-1 transcripts, and recruits a positive transcription elongation factor b (P-TEFb) to increase the production of full-length viral RNA (18, 19). The minimum TAR-binding sequence of Tat has been mapped to a basic domain of 10 amino acids, comprising mostly Arg and Lys residues. The regulatory activity requires the 47 N-terminal residues, which interact with components of the transcription complex to function together as a transcriptional activation domain (18–20).
Exosomes are membrane-bound vesicles produced by many types of cells (21, 22). Exosomes contain classical membrane marker proteins such as tetraspanins, adhesion proteins, and metalloproteinases that distinguish them from other vesicular structures such as apoptotic vesicles (23). Exosomes are considered to play an important role in intercellular communication either by target cell uptake or by inducing cell signaling via membrane receptors (24, 25). In addition to membrane proteins, exosomes also carry mRNA, as well as ncRNA (i.e. miRNA), and are thought to affect gene regulation in the target cells (24, 26). Viruses such as Epstein-Barr virus and Kaposi sarcoma-associated herpesvirus encode miRNAs that are exported out of the infected cell via exosomes (27–30), and exosomally transported miRNAs are functional in recipient cells and alter their cellular fate (27, 30, 31). We have the first set of evidence to show that HIV-1-infected cells produce exosomes that alter naive target cells to make the latter more susceptible to HIV-1 infection. Numerous reports have demonstrated unique compositions of exosomes, including viral proteins and miRNAs (14, 22, 30–32). HIV-1-derived ncRNAs are considered as potential candidate regulators of expression for many cellular genes (15, 16, 33, 34). One example is the HIV-1 TAR element (stem-loop structure, 57 bases) produced in appreciable quantities in vitro and in vivo (35). Although the presence of HIV-1 viral miRNA in cells is controversial (36, 37), our data suggest that part of TAR RNA is processed into the typical double strand pre-miRNA structure as well as processed miRNA, which can be successfully isolated from infected cells (15). In 2008, the Provost and co-workers (17) also acquired evidence of the TAR miRNA in HIV-1-infected cells. Later, Jeang and co-workers found TAR-specific sequences and 125 other HIV-1 ncRNAs in the total RNA pools from HIV-1-infected cells and reported that the TAR RNA was the most abundant ncRNA (12). Recently, Schopman et al. (11) detected numerous small RNAs that correspond to the HIV-1 RNA genome. Finally, multiple experimental data indicate that the exosomes play key roles in the miRNA transfer to recipient cells (26, 38, 39).
Our recent finding that patient samples contain viral and host miRNAs in circulation has increased our interest in exosomes functioning as potential modulators of viral spread. This phenomenon could have important implications in explaining the systemic manifestation of AIDS and the large scale destruction of multiple cells in the body. For example, HIV-1 could exert effects on the central nervous system (CNS) without crossing the blood-brain barrier through several mechanisms (40, 41). This study looks into the various components of HIV-1-derived exosomes and how they may be putative factors for increased virulence. Thus, the study has the potential to greatly contribute to our understanding of HIV-1 pathogenesis in cells, including macrophages and those of the CNS. In this study, we have demonstrated that an abundance of extracellular TAR RNA is present in exosomes both in the infected primary cell culture supernatants and in the blood during an in vivo infection. Furthermore, incubation with TAR RNA-containing vesicles resulted in a significant secretion of proinflammatory cytokines suggesting a possible mechanism of inflammation and neuropathogenesis in HIV-1 infection. The putative mechanism by which TAR RNA is likely involved in activation of the recipient cells will be discussed.
Experimental Procedures
Cells and Viruses
The parental uninfected Jurkat, CEM, and U937 cells were obtained from ATCC (Manassas, VA). HIV-1-infected J1.1, ACH2, and U1 cells were from the AIDS Reagent Program (National Institutes of Health). The cells were cultured in RPMI 1640 medium containing 10% filtered fetal bovine serum (FBS), 1% l-glutamine, and 1% streptomycin/penicillin (Quality Biological, Gaithersburg, MD). The peripheral blood mononuclear cells (PBMCs) and purified macrophages were either purchased from Lonza or obtained as a buffy coat from the National Institutes of Health and grown in RPMI 1640 medium. PBMCs were isolated from peripheral blood from healthy anonymous donors using Ficoll gradient centrifugation and then expanded in medium containing 1 μg/ml PHA-L and 30 IU/ml recombinant human IL-2. After 2 days of cultivation the cells were washed and then cultured in the medium containing 30 IU/ml rhIL-2 without PHA-L. All cells were incubated at 37 °C in the presence of 5% CO2. The HEK293-derived HEK-Blue hTLR3 cells containing a secreted embryonic alkaline phosphatase (SEAP) reporter gene, used for measuring TLR3 activation, were obtained from InvivoGen (San Diego) and cultured in HEK-Blue Detection medium following the manufacturer's protocol.
The stocks of T cell-tropic NL4-3 and dual-tropic 89.6 HIV-1 were used for infection of activated PBLs or macrophages, respectively (800 ng of p24 for 40 × 106 cells/ml). PBLs were separated from human PBMCs using incubation with PHA (1 μg/ml) for 24 h. After 48 h of incubation with 25 units/ml IL-2, the PBLs were infected with HIV-1 and then cultured for 2 weeks with IL-7 to transfer T cells to the quiescent phase. The human monocytes obtained from PBMC samples were cultured for 1 week with M-CSF (10 ng/ml) for differentiation to monocyte-derived macrophages (MDM). The MDM were infected with HIV-1 dual-tropic strain 89.6 and then cultured for 2 weeks.
Human Cytokine Assay
CEM cells were maintained in complete RPMI 1640 medium (consisting of 10% FBS, 1% l-glutamine, and 1% streptomycin/penicillin) at 37 °C and 5% CO2. Three ml of cell culture (containing 5 × 105 cells per ml of media) were placed in each of 8 wells in a 12-well plate. The samples were then treated with either DOTAP TAR wild type (WT) RNA, DOTAP TAR-D mutant RNA, or exosomes from CEM or ACH2 cells with concentrations of 1, 5, and 10 μg/ml, respectively, for 24 h. Similarly, primary human macrophages were differentiated by incubating with 10 ng/ml M-CSF and phorbol 12-myristate 13-acetate for 1 week before incubating 106 cells with 104 copies of either exosomal RNA, TAR (WT), or mutated TAR D RNA packaged into DOTAP. After 48 h, supernatants were analyzed for the presence of 23 cytokine proteins using human cytokine antibody array. Cytoplasmic and nuclear extractions were obtained using low salt buffer and detergent for cytoplasmic extracts followed by high salt for nuclear extracts. Antibodies used for Western blots were obtained from Abcam (p-eIF2α, ab4837 (1:2500)) and Santa Cruz Biotechnology (PKR, sc707 (1:200), p-PKR, sc16565 (1:200), and eIF2α, sc133132 (1:200).
Preparation of Whole Cell Extract and Size-exclusion Chromatography
The pellets of uninfected Jurkat and HIV-1-infected J1.1 cells were washed with PBS without Ca2+ and Mg2+, resuspended in lysis buffer containing 50 mm Tris-HCl (pH 7.5), 120 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 50 mm NaF, 0.2 mm Na3VO4, 1 mm DTT, and one Complete protease inhibitor mixture tablet/50 ml (Roche Applied Science, Mannheim, Germany), and incubated on ice for 20 min with gentle vortexing. Lysates were centrifuged at 4 °C at 10,000 rpm for 10 min, and protein concentrations in the supernatants were determined using the Bradford protein assay (Bio-Rad). For each cell line, 2.5 mg of protein was equilibrated in chromatography running buffer (0.2 m Tris-HCl (pH 7.5), 0.5 m NaCl, and 5% glycerol) and run on a Superose 6 10/300 size-exclusion chromatography column (GE Healthcare, Uppsala, Sweden) using the ÄKTA purifier system (GE Healthcare). After sample injection, running buffer was set at a flow rate of 0.3 ml/min, and 0.5-ml fractions of the flow-through were collected at 4 °C for a total of 70 fractions. Fractions (10–55) from the infected J1.1 cell lysate were analyzed for HIV-1 using qRT-PCR, and the high molecular weight fraction number 15 from both uninfected and infected cell lysates was tested for the presence of CD63 (an exosome marker) by Western blot following concentration and capture by a mixture of NT080 and NT082 Nanotrap particles.
Capture of CD63-positive Vesicles with Nanotrap Particles
A 15-μl slurry (30%) mixture of NT080 and NT082 Nanotrap particles (equal parts) was incubated with either 500 μl of cell culture supernatants or the same volume of size-exclusion chromatography fraction number 15 from either Jurkat or J1.1 cell lysates or corresponding cell culture supernatants for 1 h at 4 °C. The Nanotrap particle-bound materials were then washed with 500 μl of cold PBS or chromatography running buffer and then resuspended with 20 μl of Laemmli buffer (for SDS-PAGE) or 50 μl of PBS and 150 μl of TRIzol (Invitrogen) for RNA isolation.
Staining and Western Blot
Whole cell extracts or Nanotrap particle-bound materials were resuspended in Laemmli buffer (15 μl), heated at 95 °C for 5 min, and spun (15,000 rpm) for 5 min. The entire or partial volumes were loaded onto a 4–20% Tris-glycine SDS gel, run at 200 V, and transferred onto PVDF membranes. Gels were Coomassie-stained with 40% methanol, 7% glacial acetic acid, and Coomassie Brilliant Blue (Bio-Rad, R-250). Membranes were blocked with D-PBS containing 0.1% Tween 20 and 3% BSA and incubated overnight at 4 °C with the appropriate primary antibody (α-Nef, α-p24, α-gp41 (AIDS Reagent Program, National Institutes of Health) or anti-CD63 (Abcam)). Membranes were then incubated with the appropriate HRP-conjugated secondary antibodies and developed the next day using enhanced chemiluminescence.
Kinase Assay
Immunoprecipitation and in vitro kinase assays were carried out using either whole cell extract or column fraction from infected or transfected cells. Briefly, for immunoprecipitation, low and molecular weight complex fractions were immunoprecipitated at 4 °C overnight with IKK-β antibody. The next day, complexes were precipitated with A/G beads (Calbiochem) for 4 h at 4 °C. Immunoprecipitated samples were washed three times with appropriate TNE buffer (10 mm Tris, 100 mm NaCl, 1 mm EDTA) and kinase buffer. The reaction mixtures (30 μl) contained the following final concentrations: 40 mm β-glycerophosphate (pH 7.4), 7.5 mm MgCl2, 7.5 mm EGTA, 5% glycerol, [γ-32P]ATP (0.4 mm, 1 μCi), 50 mm NaF, 1 mm orthovanadate, and 0.1% (v/v) β-mercaptoethanol. Phosphorylation reactions were performed with immunoprecipitated material and γ-32P-labeled GST-IκBα (0.5 μg) or histone H1 (0.5 μg) as a substrate in threonine tyrosine kinase buffer containing 50 mm HEPES (pH 7.9), 10 mm MgCl2, 6 mm EGTA, and 2.5 mm dithiothreitol. Reactions were incubated at 30 °C for 1 h, stopped by the addition of 1 volume of Laemmli sample buffer containing 5% β-mercaptoethanol, and run on 4–20% SDS-polyacrylamide gel. Gels were subjected to autoradiography and quantification using PhosphorImager software (Amersham Biosciences).
Detection and Quantitation of HIV-1 RNA in Nanotrap Particle-bound Exosomes by Quantitative RT-PCR
We have previously shown that exosomes prepared using Optiprep density gradient centrifugation of HIV-1-infected J1.1 cell supernatant contain TAR RNA (42). To determine whether the specific Nanotrap particles could capture exosomes that are also derived from the J1.1 cell supernatants, the exosome preparations were similarly incubated with the 30% slurry of five different Nanotrap particles for 1 h at 4 °C on a shaker. After washing, total RNA was extracted via the addition of 3 volumes of TRIzol reagent (Invitrogen) to the pelleted particle-bound exosome materials resuspended in PBS. Total RNA was then isolated according to the manufacturer's protocol. RNA was precipitated with isopropyl alcohol and 1 μl of glycogen for molecular biology (Roche Applied Science, Mannheim, Germany). The RNA pellets were reconstituted in 1× TE buffer. A total of 0.5 μg of RNA from the RNA fraction was treated with 0.25 mg/ml DNase I RNase-free (Roche Applied Science, Mannheim, Germany) for 60 min in the presence of 5 mm MgCl2, followed by heat inactivation at 65 °C for 15 min. A 200–250-ng aliquot of total RNA was used to generate cDNA with the GoScript Reverse Transcription System (Promega, Madison, WI) using TAR-specific reverse primer TAR Reverse (+42 to +62): 5′-AGC AGT GGG TTC CCT AGT TAG-3′ or oligo(dT) reverse primer. Quantitative real time PCR analysis was performed with 2 μl of undiluted RT reaction mixtures. The iQ SYBR Green Supermix (Bio-Rad) was used with the primers specific for 1) HIV-1 TAR (42): TARfll-F, 5′-GGTCTCTCTGGTTAGACC-3′, and TARfll-R as above amplified 60 nucleotides; TAR sequence or 2) HIV-1 env gene, Gag1483-F, 5′-AAGGGGAAGTGACATAGCAG-3′, and Gag1625-R, 5′-GCTGGTAGGGCTATACATTCTTAC-3′ amplified 143 nucleotide fragments of the HIV-1 gag gene. Serial dilutions of DNA from 8E5 cells (CEM cell line containing a single copy of HIV-1 LAV provirus per cell) were used as the quantitative standards. Real time PCRs were carried out at least in triplicate as described above. Primers for cytokine expression were as follows: IL-6-F, 5′-GGT ACA TCC TCG ACG GCA CT3′; IL-6-R, 5′ GTG CCT CTT TGC TTT CAC3′; TNF-B-F, 5′-CCC ATG GCA TCC TGA AAC-3′, and TNF-B-R, 5′-GGA GGC CTG GAA TCC AAT-′.
TLR3 Reporter Assay
The HEK293-based reporter cell line, HEK-Blue hTLR3 (InvivoGen), was used to detect activation of TLR3 either by HIV-1 or exosomes containing TAR RNA from HIV-1-infected J1.1 cells. HEK-Blue hTLR3 cells (5 × 104 cells/well) containing a SEAP reporter gene were cultured at 37 °C in 5% CO2 in 96-well plates in SEAP detection medium in the presence of HIV-1 89.6 (1, 10, and 100 ng of p24/well), exosomes from either HIV-1-infected J1.1 cells or uninfected Jurkat cells (0.1, 1, and 10 μg/well, each), or poly(I-C) (10, 50, and 250 ng/ml). At 18 h of post-culture, cells were analyzed for the level of absorbance at 600 nm using the GloMax Multi Detection System (Promega). Readings were normalized to PBS-negative controls.
Animal Studies
All mice used in this study were maintained within the National Center for Biodefense and Infectious Disease's breeding colony (George Mason University, Manassas, VA). All experiments were carried out in bio-safety level 3 (BSL-3) facilities and in accordance with the Guide for the Care and Use of Laboratory Animals (Committee on Care and Use of Laboratory Animals of The Institute of Laboratory Animal Resources, National Research Council, National Institutes of Health Publication 86-23, revised 1996). NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ mice were obtained from The Jackson Laboratory (007799, Bar Harbor, ME) and were humanized as described previously (43). Briefly, three groups of three neonatal animals were sub-lethally irradiated. The next day, mice were intraperitoneally injected (100 μl) with ∼1 × 105 human cord blood-derived CD34+ hematopoietic stem cells (2C-101B, Lonza, Walkersville, MD). At 3 months post-engraftment, animals were subcutaneously infected with the dual-tropic HIV-1 89.6 (100 μl, 5 ng of p24/μl).
ELISA
Concentrations of the chemokines in culture supernatants of cells treated with or without DOTAP packaged WT TAR, TAR D mutant, or exosomal RNA were measured at 48 h post-treatment by a cytokine array ELISA (R&D Systems, Minneapolis, MN) according to the manufacturer's protocol.
Statistical Analysis
Quantitative data were analyzed by two-way analysis of variance (OriginPro version 8.0) and Student's t test (Microsoft Excel). Standard deviation was calculated in all quantitative experiments for at least three independent preparations. The difference was considered to be statistically significant at p ≤ 0.05. The results are shown as the mean ± S.D. for each group.
Results
TAR RNA Is Detected in Culture Supernatants of HIV-1-infected Primary Human Cells and in Blood from HIV-1-infected Humanized Mice
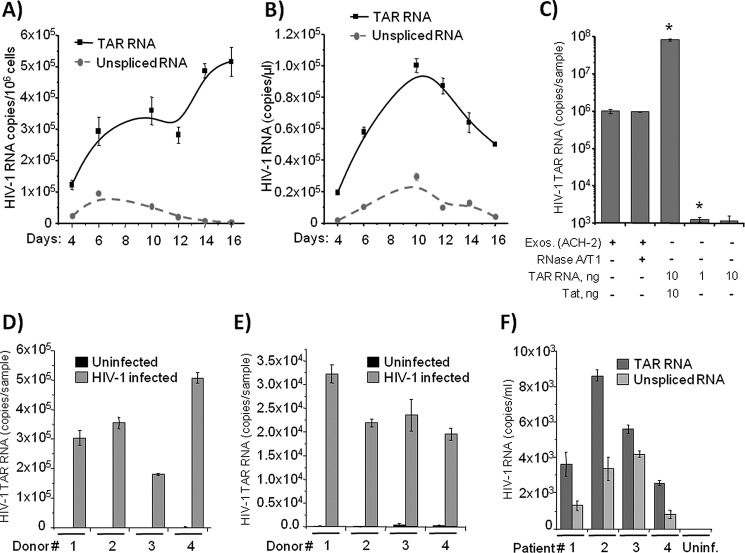
Earlier studies have demonstrated that HIV-1 produces a large pool of its own non-coding RNAs with the TAR region representing the predominant species (44). In fact, only a small fraction of TAR RNA is utilized by the virus to regulate its own and the host gene transcription. Therefore, we speculated that the vast excess of TAR RNA exerts its regulatory influence on bystander cells via the extracellular milieu. To test this hypothesis, we first analyzed the ratio of coding HIV-1 RNA and TAR within infected primary human T cells and in extracellular media. The purified human T cells were infected with HIV-1 NL4-3 and cultured for 16 days. The cells and culture supernatants were harvested every 2–3 days and subjected to quantitative RT-PCR analysis. As shown in Fig. 1A, up to 5 × 105 copies of unprocessed stem loop TAR RNA and ∼5-fold lower number of unspliced gag RNA were detected in a million infected T cells. The levels of intracellular TAR RNA increased as a function of time, whereas levels of the coding HIV-1 RNA gradually decreased over time, suggesting that TAR RNA accumulation may be an indicator of infection toward latency. Quantitation of extracellularly released TAR RNA revealed up to 90,000 RNA copies in a microliter of culture supernatant from infected cells (Fig. 1B). Again, a much lower number of unspliced RNA copies was present in the supernatants.
FIGURE 1.
TAR RNA is abundant in the exosomes from HIV-1-infected primary cells. A, purified T cells from healthy donors were stimulated with PHA/IL-2 and then infected with HIV-1 NL4-3. Cells were lysed on 4, 6, 10, 12, 14, and 16 days post-infection; total RNA was isolated and subjected to RT reaction and then to SYBR Green real time PCR with the primers specific for HIV-1 TAR and gag to quantitate numbers of copies of TAR and unspliced HIV-1 RNA. Error bars indicate ± S.D. of three independent measurements. B, cell supernatants from the HIV-1-infected T cells used in A were harvested at 4, 6, 10, 12, 14, and 16 days post-infection and then incubated with a mixture of Nanotrap particles NT080 and NT082 (to trap exosomes) for 1 h at 4 °C. The exosomes bound by the Nanotrap particles were lysed; total RNA was isolated and subjected to qRT-PCR using specific primers as in A. Error bars indicate ±S.D. of three independent measurements. C, trapping of exosomes and RNP complex by nanoparticles. One milliliter of infected ACH2 supernatants was incubated with/without RNase A/T1 mix (RNase A 800 ng/ml and RNase T1 2500 units/ml; Pharmingen) for 1 h at 37 °C. Samples were then treated with 25 μl of 30% slurry of NT080 + 82 overnight at 4 °C. Ten nanograms of purified Tat (76, 77) and purified TAR were incubated with beads overnight (total of 100 μl) at 4 °C. Also, purified TAR RNA (1 and 10 ng) was incubated with beads overnight at 4 °C. All samples were washed the next day with PBS (two times), and RNA was isolated for TAR RT-qPCR. Error bars indicate ±S.D. of three independent measurements. Asterisks indicate p ≤ 0.01 between indicated samples and exosomal samples. D, PBLs were separated from human PBMCs from four donors using incubation with PHA (1 μg/ml) for 24 h. After 48 h of incubation with 25 units/ml IL-2, the PBLs were infected with HIV-1 strain 89.6 and then cultured for 1 week with ART mixture (lamivudine/emitricitabine, indinavir, and tenofovir; 10 μm of each). Then the cells were cultivated with IL-7 (1 ng/ml) to transfer T cells to the quiescent phase. Culture supernatants were then harvested, and exosomes were separated with NT080/NT082 Nanotrap particle mixture as described in B. Total RNA was extracted, and TAR and unspliced viral RNA were quantitated as described in A. Error bars indicate ±S.D. of three independent measurements. E, human monocytes obtained from four donors were cultured for 1 week with 10 ng/ml M-CSF for differentiation of MDM. The MDM were infected with HIV-1 dual-tropic strain 89.6 and then cultured for 2 weeks (for the 1st week with ART mixture as described in D). The exosomes were then captured from culture media as described in D; RNA was extracted and HIV-1 TAR and unspliced RNA were quantitated using qRT-PCR. Error bars indicate ±S.D. of three independent measurements. F, serum specimens from four HIV-1-infected patients who had undetectable plasma HIV-1 RNA on cART were obtained from The Washington D. C. Metropolitan Site of Women's Interagency HIV Study. The exosomes were captured from 1.0 ml of serum as described in C; RNA was extracted and HIV-1 TAR, and unspliced RNA were quantitated using RT-qPCR. Error bars indicate ±S.D. of three independent measurements.
Along these lines, we recently asked whether TAR could potentially be free outside of the cells and not associated with exosomes. We previously had shown that TAR is exclusively present in exosomes using either unpurified supernatants from infected cells or purified exosomes (42). Here, we further asked whether the NT080 + 82 used in our assay could potentially bind to the free RNA or RNP complex. Therefore, we designed an experiment where supernatants from infected cells (±RNase A/T1) were incubated with supernatants prior to binding nanoparticles. The beads were incubated overnight and washed the next day, and RNA was isolated for RT-qPCR. We also utilized purified the Tat-TAR complex as a positive control, as we have previously shown that free Tat is able to bind to NT080 + 82 (45). Tat is the most specific TAR-binding protein. Results in Fig. 1C show that TAR RNA was not present outside of the exosomes, as RNase A/T1 mix did not reduce the presence of TAR from the media. We also found that free TAR is not able to bind to NT080 + 82 at similar concentrations; this further implies that the nanoparticles used here are able to bind to either exosomes or RNP complex (i.e. Tat-TAR). Therefore, if TAR RNP complexes exist in vivo, they would be trapped by these nanoparticles through Tat but not TAR RNA.
To further test the presence of TAR RNA in the extracellular environment of naturally HIV-1 target cells, we utilized an 89.6 dual-tropic HIV-1 virus. We separated both PBL and MDM from four PBMC samples of four healthy donors and then infected the cells. After infection, PBLs were cultured for 2 weeks with 1 ng/ml IL-7 to transfer T cells to the quiescent phase to reduce the level of production of HIV-1 virions. We observed from 2 × 105 to 5 × 105 copies of TAR RNA in the 250-μl sample of culture medium of infected PBLs from all tested donors (Fig. 1D).
To test whether the HIV-1-infected macrophages also secrete exosome-packaged TAR RNA as compared with T cells, we cultured MDM for 2 weeks in the media containing 10 ng/ml of M-CSF. Quantitative analysis of TAR RNA in the fraction of Nanotrap-captured exosomes revealed from 2.0 × 104 to 3.5 × 104 RNA copies per sample (Fig. 1E). These data indicate that both HIV-1-infected T lymphocytes and macrophages constitutively produce exosomes that contain TAR RNA.
Our earlier analysis employing primers that would amplify other HIV-1 mRNAs, including vif, vpr, tat, rev, vpu, and nef, also revealed that in chronically HIV-1-infected cell lines TAR RNA was in vast excess over all viral mRNAs (<5% of TAR), and the results were not due to virus contamination (42). Our data are also consistent with a recent publication describing the presence of TAR miRNA in HIV-1-infected cells (37). Although this study reports that the percentage of the desired 22 ± 2-nucleotide size viral miRNA is low, the smaller sized viral miRNAs were more commonly found in the HIV-1-infected long term cultures (see Figs. 2 and 3 and also supplemental Fig. 3 in Ref. 37). We asked next whether TAR RNA can be detected in the exosomes from the serum of aviremic HIV-1-infected patients. To perform this test, we isolated exosomes from 1.0-ml serum specimens from four patients with undetectable viral load, who were treated with combinational antiretroviral therapy (cART) and were virally suppressed for at least 2 years. Samples were obtained from the Metropolitan Consortium of the Women's Interagency HIV Study Collaborative Study Group, Washington, D. C. Quantitative RT-PCR analysis of the RNA from concentrated exosomes revealed from 3000 to 8000 copies of TAR RNA (Fig. 1F). Interestingly, despite the undetectable viral load in all tested patients, we found 0.5–4.0 thousand copies of unspliced HIV-1 RNA in exosomes, implying that the viral genomic RNA can be secreted from the cells via exosomes, although virion levels are virtually undetectable.
FIGURE 2.
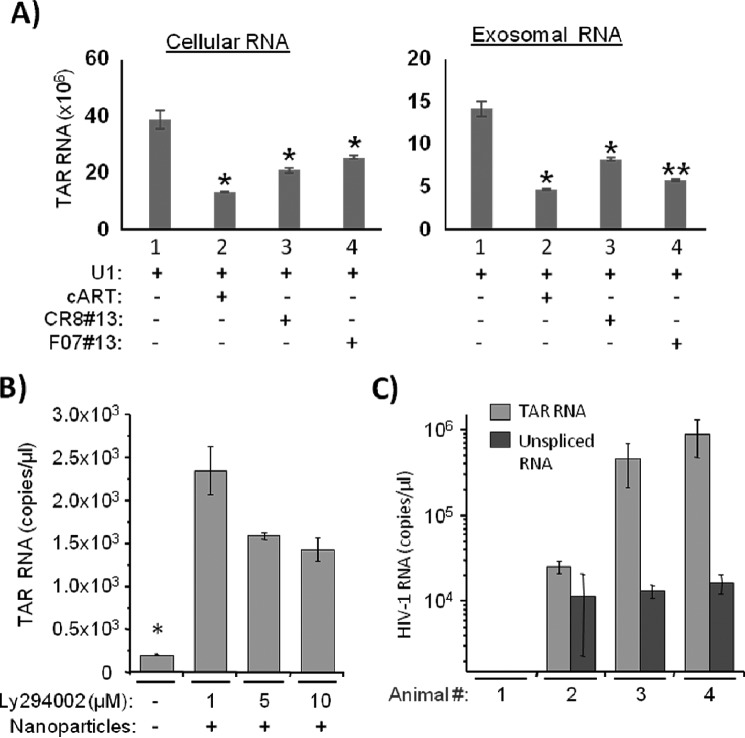
Exosome packaging of TAR RNA is dependent on its expression level in HIV-1-infected cells. A, promonocytic chronically HIV-1-infected U1 cells were treated with cART mixture (described in Fig. 1) for 5 days and subsequently treated with two different transcription inhibitors, F07 number 13 and CR8 number 13 (1 μm each), for 24 h. Exosomes were then isolated using NT080 and NT082 nanoparticles overnight at 4 °C and then tested for quantity of TAR RNA by qRT-PCR. Total RNA was also isolated from treated cells for TAR RNA analysis. Error bars indicate ±S.D. of three independent measurements. Asterisk indicates p ≤ 0.05. B, chronically HIV-1-infected T cells J1.1 were treated with three different concentrations (1, 5, and 10 μm) of PI3K inhibitor LY294002, and the supernatants (500 μl) were tested for TAR RNA by qRT-PCR following pulldown with a mixture of Nanotrap particles NT080 and NT082 as described in Fig. 1A. Error bars indicate ±S.D. of three independent measurements. Asterisk indicates p ≤ 0.05. C, three groups (2–4) of three humanized mice were subcutaneously infected with the dual tropic HIV-1 89.6 (100 μl). Total RNA was isolated from the blood plasma of infected mice (25–50 μl) after 14 days of infection. The exosomes were captured using Nanotrap particle mixture NT080/NT082. Copy numbers of both TAR RNA and unspliced HIV-1 RNA were measured using qRT-PCR as described in Fig. 1A. Results are presented as mean ± S.D. of at least three independent measurements.
FIGURE 3.
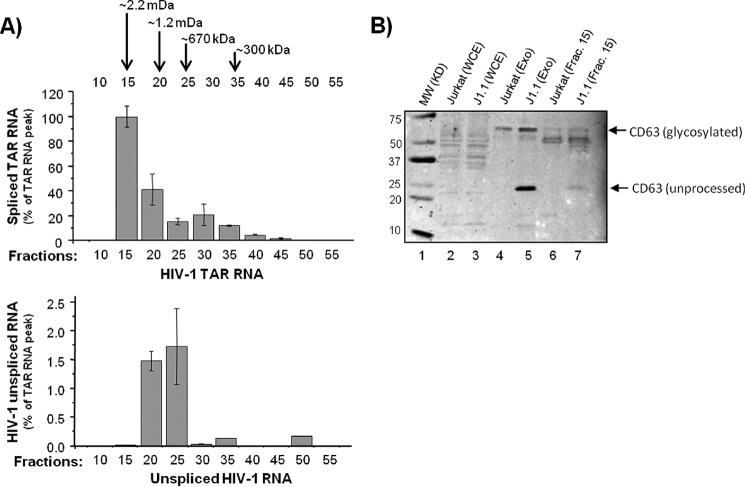
TAR RNA is a part of the endogenous fractions of pre-released exosomes. A, whole cell extracts from uninfected Jurkat and HIV-1-infected J1.1 cells were prepared and then separated at 2.5 mg of total protein run on a Superose 6 size-exclusion chromatography column in the presence of 500 mm salt buffer. No detergent was used during fractionation. A total of 55 fractions (500 μl each) from each cell extract was collected. Every 5th fraction (from fraction 10 to 55) from the J1.1 extract was subjected to isolation of total RNA followed RT reaction and real time PCR of cDNA with the primers specific for either TAR RNA (upper panel) or for unspliced HIV-1 RNA (gag-specific primers; lower panel). B, 500 μl of the chromatography fraction 15 from Jurkat (lane 6) and J1.1 (lane 7) cell extracts were incubated for 30 min with a mixture of Nanotrap particles NT080 and NT082. Exosomes were separated by centrifugation and then tested for the presence of CD63 (an exosome marker) by Western blot. Exosomes were similarly isolated from the Jurkat (lane 4) and J1.1 (lane 5) supernatants using the Nanotrap particles and tested for the CD63 marker protein by Western blot. The whole cell extracts from Jurkat (lane 2) and J1.1 (lane 3) were also separated onto a 4–20% Tris-glycine SDS-polyacrylamide gel and immunoblotted with anti-CD63 antibody.
To test potential cellular mechanisms involved in the high level of TAR containing exosomes produced from HIV-1-infected cells, we analyzed effects of the inhibition of transcription of TAR RNA and down-regulation of endosomal pathway on the release of TAR-comprising exosomes from both myeloid cells and T lymphocytes. We used U1 cells treated with cART for 5 days and subsequently treated with two different transcription inhibitors. We have previously shown that a mimetic of Tat (F07 number 13) and an ATP analog blocking CDK9 (CR8 number 13) can inhibit viral transcription (46). Both F07 number 13 and CR8 number 13 was used to treat U1 cells, and exosomes were isolated after 24 and 72 h using nanoparticles. We also isolated RNA from both treated cells (donor) along with the exosomal RNAs. Results in Fig. 2A show a 2–3-fold decrease of TAR RNA levels from intracellular as well as exosomal RNA.
Because phosphatidylinositol 3-kinase (PI3K) has been shown to be involved in Nef-mediated release of multivesicular bodies (MVB) from cells (47), we then tested whether the inhibition of PI3K could reverse the HIV-mediated enhanced release of exosomes from the infected cells. We treated HIV-1-infected J1.1 cells with different concentrations of PI3K inhibitor LY294002, and we tested the release of exosomal TAR RNA in the supernatant by RT-qPCR. Interestingly, we observed that the release of exosomal TAR RNA was inhibited by LY294002 in a dose-dependent manner (Fig. 2B), and this is consistent with the published data that this drug has been shown to decrease the release of MVB containing immature MHC invariant chain (47), suggesting that HIV-1 Nef plays an important role in the release of exosomes from infected cells.
Finally, we asked whether TAR could be detected in an in vivo infection using a humanized mouse model. We infected three groups of humanized NSG mice with HIV-1 89.6 and tested the presence of TAR RNA and gag-specific unspliced RNA in blood 14 days post-infection. As shown in Fig. 2C, up to a million copies of TAR RNA were present in a microliter of infected mouse blood, although the gag-specific unspliced RNA was from 10- to 100-fold lower than TAR, indicating that TAR RNA was also produced in vivo, and potentially incorporated within exosomes, following HIV-1 infection. Collectively, these data demonstrate that HIV-1-derived TAR RNA can be detected as an extracellular species in a time-dependent manner in cell culture supernatants, as well as animals infected with HIV-1, and that may be carried within exosomes released from the infected cells.
TAR-RNA Present in the Infected Cells Can Be Separated from Viral Genomic RNA Using Size-exclusion Chromatography
Data presented above support the detection of an abundance of HIV-1 TAR RNA released from infected cells both in vitro and in vivo. We further attempted to verify whether TAR RNA was part of the endogenous exosomal fractions (pre-release), and we determined any possible modifications associated with these exosomes. To characterize exosomal and viral fractions, we utilized chromatography fractionation of intracellular complexes from infected cell extracts. We have previously successfully used this method to identify novel P-TEFb complexes from HIV-1-infected cells (48). The total cell extracts from chronically HIV-1-infected J1.1 cells were separated on a size-exclusion column (over 55 fractions) in the presence of 500 mm salt buffer, and each 5th fraction (from 10 to 55) was tested for the presence of HIV-1 RNA copies by qRT-PCR using primers specific for either TAR RNA or unspliced HIV-1 RNA. As shown in Fig. 3A (upper panel), the peak TAR RNA was detected in fraction 15 (large molecular weight complexes), and the peak unspliced viral RNA was detected in fraction 25 (lower panel). This is consistent with our previous report where p24 protein (or potentially viral particles) was also detected in fractions 25–30 (48). We then asked whether the exosomes purified from the extracellular environment had any similarities to intracellular exosomes by staining for CD63 expression. We have previously shown that exosomes from HIV-1-infected cells contain increased levels of CD63 (42). The results in Fig. 3B indicate that the level of both glycosylated and the unprocessed forms of CD63 proteins were highly enriched in exosomes isolated from HIV-1-infected supernatant (lane 5) compared with uninfected exosomes (lane 4). Similarly, more visible CD63 protein bands, especially the unprocessed form, were also observed from chromatography fraction 15 from the infected cell extract (Fig. 3B, lane 7) compared with the control extract (lane 6) following concentration by Nanotrap particles. Nanotrap particles are routinely used by us to either concentrate exosomes or viral particles from complex environments (45). As expected, the major CD63 bands were not highly visible in either infected or uninfected whole cell extract without the use of Nanotrap particles (Fig. 3B, lanes 2 and 3). Collectively, these results indicate that TAR RNA can be efficiently separated from genomic RNA or virus using a combination of chromatography and concentration with nanoparticles and that the unmodified CD63 levels are higher in the infected cells. This may further assist in better purification of exosomes from HIV-1-infected cells.
Effect of TAR RNA on Cytokine Production
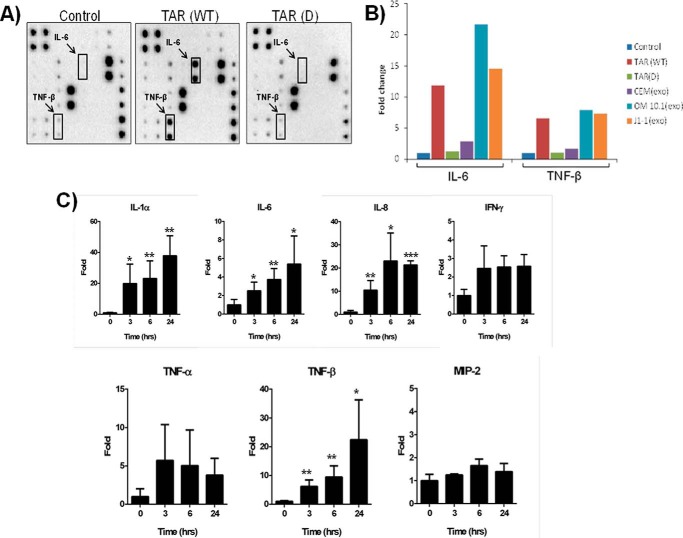
We have previously shown that TAR is present in exosomes where it contributes to survival of recipient cells (42). Here we asked whether TAR RNA alone was capable of activating cytokines in the recipient cells. To avoid any possible other cellular or viral miRNA that could activate cytokines, we packaged either wild type T7-TAR RNA (52 bases) or a mutated TAR D, which is missing most of the stem-loop structure, into DOTAP liposomal transfection reagent (Roche Applied Science). Primary macrophages were then treated with either DOTAP alone (control) or 104 copies of WT TAR or TAR mutant D. Supernatants were recovered after 48 h and analyzed for the presence of cytokine proteins. Although a number of cytokines, including IL-1β, INF-α, INF-γ, and MCP-1 were all increased more than 1–3-fold, two cytokines showed dramatic changes in expression pattern. Results in Fig. 4A show that two cytokines, IL-6 and TNF-β, were highly up-regulated in the treated cells. IL-6 is a pleiotropic cytokine that belongs to the four helical cytokine family, and macrophages from HIV-infected patients are capable of producing IL-6 (49, 50). IL-6 has been shown to stimulate T cell proliferation in vitro (51). TNF-β (LTA) is a member of the tumor necrosis factor family, shares its receptor with both TNF receptor-1 and -2, and induces its own expression in BV-2 microglial cells and macrophages to coordinate lymphocyte migration (52–55).
FIGURE 4.
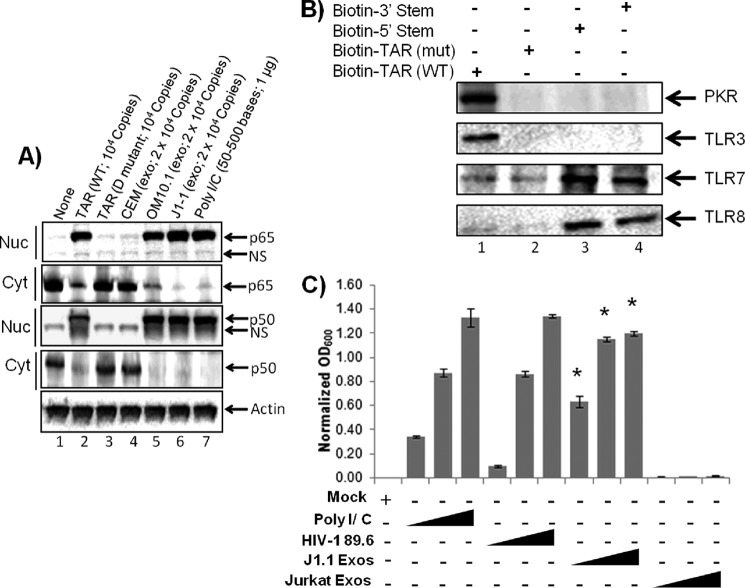
Exposure to TAR RNA alters cytokine profiles in primary monocyte-derived macrophages. A, primary human MDMs were differentiated by incubating in 10 ng/ml M-CSF and phorbol 12-myristate 13-acetate for 1 week before incubating 106 cells with 104 copies of either TAR WT or mutated TAR D RNA packaged into DOTAP liposomal transfection reagent or with DOTAP only (control). After 48 h, supernatants were analyzed for the presence of 23 cytokine proteins using human cytokine antibody array I. Cytokines that are highly up-regulated after treatment with TAR are indicated by enclosed rectangles. B, exosomes were first isolated from CEM, OM10.1, and J1.1 cell supernatants using Nanotrap particles as described in Fig. 1B; total RNA was purified and then packaged into DOTAP reagent as in A. The DOTAP (liposomes) containing extracellular RNA, as well as TAR (WT) and mutated TAR (D) RNA, were incubated with MDM for 48 h, and then supernatants were analyzed for 23 cytokines (data not shown) as in A. Only IL-6 and TNF data are shown as bar graphs. MDMs were incubated with DOTAP alone as a control. C, primary macrophages were treated with either DOTAP alone (control) or DOTAP/TAR wild type (104 copies/10 μl/experimental), and supernatants were assayed for the presence of cytokines after 0, 3, 6, and 24 h. Results are presented as mean ± S.D. of at least three independent measurements.
We next tested whether RNA from purified exosomes from CEM (uninfected), promyeloid OM10.1 (infected; contain TAR), or J1.1 (infected; contain TAR) cells could activate these two cytokines. Total RNA was purified from these exosomes, precipitated, and re-packaged with DOTAP reagent and then incubated with primary macrophages for 48 h. Data in Fig. 4B indicate that these two cytokines were also up-regulated with exosomal RNA obtained from HIV-1-infected OM10.1 and J1.1 cells. The level of induction was slightly different where TAR (WT) could activate up to 10-fold for the IL-6 and 6.5-fold for TNF-β. RNA from exosomes obtained from OM10.1 induced 22-fold for IL-6 and 7.5-fold for TNF-β. Similar trends of up-regulation of these cytokines were also observed for RNA obtained from J1.1 exosomes. Finally, we performed a similar experiment with exosomes containing TAR and scored for dynamic changes in cytokine secretion over time. Primary macrophages were treated with either DOTAP alone or DOTAP/TAR wild type, and supernatants were assayed for the presence of cytokines after 0, 3, 6, and 24 h. Data in Fig. 4C indicate that the number of cytokines, including IL-1β, IL-6, IL-8, TNF-α, TNF-β, and IFN, were up-regulated following TAR treatment.
Effect of TAR on Nuclear Accumulation of NF-κB
We next asked whether components of NF-κB members were altered and/or moved to the nucleus after TAR treatment. This could potentially explain the increase in cytokine levels of treated cells. As above, we analyzed the effect of either purified TAR or TAR-containing RNA pool from exosomes on macrophages and obtained both cytoplasmic and nuclear extracts after 24 h of treatment. Results in Fig. 5A indicate that WT TAR, but not the mutant TAR, was able to initiate translocation of both p65 and p50 into the nucleus of recipient macrophages. A similar experiment performed with total RNA from exosomes that contained TAR (OM10.1 and J1.1 cells) revealed the presence of increased p65 and p50 in the nucleus of macrophages. As expected, exosomal RNA from uninfected CEM cells did not result in dramatic translocation of NF-κB subunits p65 or p50 (Fig. 5A).
FIGURE 5.
Effects of exosomal TAR RNA on NF-κB and TLR pathways. A, exosomes were prepared from CEM (uninfected), OM10.1 (HIV-1-infected), and J1.1 (HIV-1-infected) cell culture supernatants using Nanotrap particles. The total exosomal RNAs were purified and then mixed with DOTAP transfection reagent before incubating with MDM (7-day cultures). DOTAP liposome-packaged TAR WT or mutant TAR D RNAs were also incubated with macrophages; poly(I-C) was used as positive control. Nuclear and cytoplasmic extracts were prepared 24 h post-treatment, run on a 4–20% SDS-polyacrylamide gel, and then Western blotted for NF-κB components p65 and p50 both from cytoplasm and nuclear extracts. Actin antibody served as a control. B, biotin-labeled WT TAR, TAR D mutant, and TAR miRNAs (5′ stem or 3′ stem) were incubated with total MDM (5 × 105) extracts. Three hundred micrograms of extracts were incubated with labeled RNA (1 μg) overnight at 4 °C, and the next day 30% slurry of streptavidin-Sepharose beads were added. Samples were washed once with TNE 50 + 0.1% Nonidet P-40, then once with RIPA buffer, and then one final time with TNE50 + 0.1% Nonidet P-40 buffer. Bound samples were separated on 4–20% SDS-polyacrylamide gel and then Western-blotted for PKR, TLR3, TLR7, and TLR8. C, HEK-Blue hTLR3 cells containing a TLR3 activation-inducible SEAP reporter gene were incubated in a SEAP detection medium with HIV-1 89.6 virus (1, 10, and 100 ng of p24/well), exosomes from HIV-1-infected J1.1 cells (0.1, 1, and 10 μg/well), or exosomes from uninfected Jurkat cells (0.1, 1, and 10 μg/well). Similarly, cells were incubated with poly(I-C) (10, 50, and 250 ng/ml) as a positive control. After 18 h of incubation, absorbance (600 nm) of each sample was measured and normalized to PBS controls. Error bars indicate ±S.D. of three independent measurements. Asterisk indicates p ≤ 0.01.
The TLRs that recognize viral pathogen-associated molecular patterns are TLR3 (recognizes double-stranded RNA (dsRNA)), TLR7/8 (recognizes single-stranded RNA), and TLR9 (recognizes viral DNA) (56). TLR3 can also be activated by a viral mimic, a synthetic dsRNA called polyriboinosinic-ribocytidylic acid (poly(I-C)). A number of cells express all known TLRs and respond robustly to both poly(I-C) (TLR3 ligand) and LPS (TLR4 ligand) (57). It has previously been shown that miRNAs in addition to their role as gene expression regulators can also interact with TLRs (58). Here, we asked whether TAR RNA or TAR miRNA (processed 5′ stem or 3′ stem miRNAs) could also bind to TLRs and modulate gene expression of cytokines. We used PKR binding as a positive control, because it has been shown that TAR binds tightly to PKR in vitro (59, 60). Results in Fig. 5B indicate that biotin-labeled WT TAR, but not mutant TAR D, was capable of binding to TLR3 when incubated with primary human macrophage extracts. Also, the TAR miRNAs (5′ and 3′ TAR stems) were capable of binding to TLR7 and -8. It is important to note that we have previously shown a 10:1 ratio of intact TAR to TAR miRNA in exosomes (42). Furthermore, these findings suggest that TAR miRNA may be capable of binding to TLRs and contribute to activation of NF-κB and cytokine production in recipient cells. This is also consistent with a previous report where cellular miRNA is able to directly bind to TLR through access in endosomes (61).
We next asked whether binding to TLR could potentially activate gene transcription. Here a TLR3 reporter cell line, HEK-Blue hTLR3, was incubated with either exosomes from infected cells, uninfected cells, or virus (positive control). As shown in Fig. 5C, the exosomes from infected cells dose-dependently enhanced TLR3 activation in the target cells suggesting that exosomes containing TAR may contribute to TLR activation, as well as downstream cytokine production. Conversely, exosomes from uninfected cells did not readily activate TLR3 at any of the three concentrations tested. Moreover, SEAP activation by exosomes from the HIV-1-infected T cell J1.1 plateaued at the highest two concentrations. This observation was repeated in additional experiments (data not shown) and may indicate a saturation of the exosome receptors on the recipient cells. Lower level of TLR3 activation was observed with HIV-1, and as expected a higher level of activation was observed with poly(I-C).
Presence of a Small IKK-β Complex in TAR-containing Cells
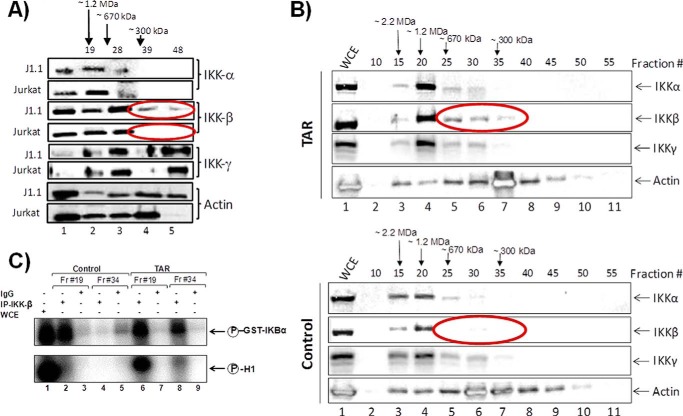
The IKK complex is the upstream component in the NFκB cascade that leads to the phosphorylation of IκBα and p65. The IKK complex that functions as the IκBα and p65 kinase is typically a heterotrimer that consists of IKK-α, IKK-β, and IKK-γ (NEMO) proteins. IKK-α and IKK-β possess kinase activity, whereas IKK-γ (NEMO) is required to stabilize the heterotrimeric complex. Results in Fig. 5A indicated that TAR wild type, and not the mutant TAR, was able to translocate both p65 and p50 into the nucleus of recipient cells. Here, we first asked whether HIV-infected cells could contain altered IKK complexes when using chromatography. We have previously observed a smaller IKK-β that is kinase activity from Rift Valley fever virus- and Venezuelan equine encephalitis virus-infected cells (62). Using Superose 6-fractionated uninfected Jurkat and infected J1 cells followed by Western blots, we observed the presence of a smaller IKK-β from HIV-1-infected cells (Fig. 6A), which may explain the translocation of both p50 and p65 into the nucleus of cells. We next asked whether TAR alone could be responsible for the presence of the smaller IKK-β form. We therefore transfected cells with either LTR-Luc plasmid (expressing TAR) or CMV-Luc (control). Extracts were isolated 48 h later and passed through chromatography. Data in Fig. 6B indicate the presence of the similar smaller form of the IKK-β in these transfected cells. We have observed similar data from latent PBMC samples under cART in collaboration with Dr. Fabio Romerio at Institute of Human Virology/Baltimore, MD (data not shown). Finally, we asked whether the IKK-β was functional in these fractions in an in vitro kinase assay using GST-IKBα as substrate. We also included histone H1 (a general signature of chromatin decompaction) in the reaction mix for any potential changes in substrate specificity. Results in Fig. 6C indicate that IKK-β was fully functional from Jurkat control transfected cells (large complex; fraction 19); however, the smaller complex (fraction 34) showed no specific phosphorylation (lanes 4 and 5). In contrast, IKK-β from LTR-transfected cells showed phosphorylation from both large and smaller complexes on IKBα substrate (fractions 19 and 34). Interestingly, histone H1 was phosphorylated only from cells containing TAR, indicating that there might be altered substrate specificity with the smaller IKK-β complex from infected cells. Collectively, these results suggest that TAR may be capable of binding to TLRs and contribute to activation of NFκB (through a novel smaller IKK-β complex) and cytokine production in cells.
FIGURE 6.
Effect of TAR on NFκB pathway. A, IKK-β complex components are altered in HIV-1-infected cells. Extracts from uninfected Jurkat and infected J1.1 cells were fractionated in a Superose 6 size-exclusion column (AKTA). A total of 70 fractions were obtained, and every fifth fraction was analyzed for IKK-α, -β, and -γ complexes and for β-actin. B, Jurkat cells were transfected with either an HIV-Luc (TAR+; 20 μg) or CMV-Luc (TAR−; 20 μg), and cell lysates were prepared for chromatography and Western blots. C, fractions 19 (large complex) and 34 (small complex) were used (500 μl) for immunoprecipitation with α-IKK-β antibody (10 μg), bound to A + G beads, washed, and used for kinase reaction, which included GST-IKBα (0.5 μg) or histone H1 (0.5 μg) as substrates and [γ-32P]ATP. Samples were run on SDS-polyacrylamide gel, dried, and exposed to a cassette.
Effect of Exosomal TAR on PKR Pathway
PKR is regulated by a variety of cellular and viral RNAs that contain duplex regions. The most well characterized activators of PKR are simple dsRNAs. Dimerization of PKR molecules following the binding with dsRNA plays a crucial role in the mechanism of activation of PKR (60–64). In the context of regular duplex RNA, a minimum of 30–33 bp of dsRNA is required to activate PKR autophosphorylation, and the maximal level of activation increases with duplex length (60, 63, 65). Here, we asked whether exosomal TAR was able to activate PKR in recipient cells.
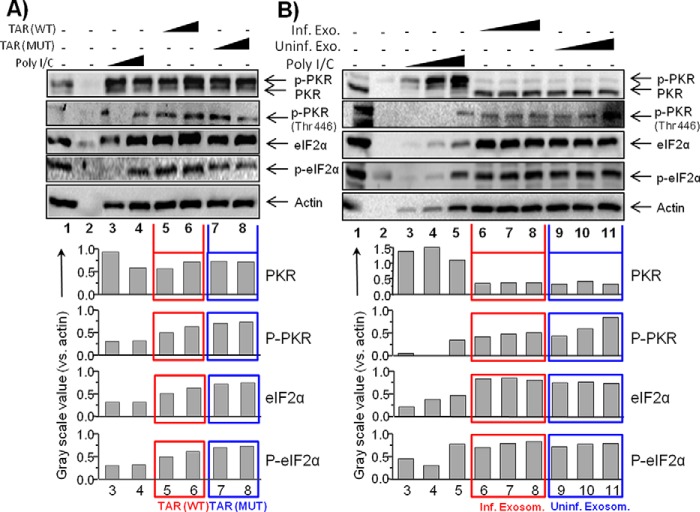
We packaged TAR with DOTAP and treated cells with increasing concentrations of either WT TAR or TAR mutant D. Following treatment, extracts were isolated for Western blots against PKR, phospho-PKR, eukaryotic translational initiation factor 2α (eIF2α), and phospho-elF2α. The purpose of these experiments was to ask whether TAR, which contains only 23-bp at its stem, could potentially activate PKR phosphorylation, phosphorylate eIF2α and inhibit translation. Results in Fig. 7A indicate that increasing concentrations of WT TAR, but not mutant TAR D, slightly increased PKR phosphorylation. We then asked whether exosomes from either infected or uninfected cells could potentially activate PKR phosphorylation in recipient cells. We treated cells with various concentrations of exosomes (from infected or uninfected cells) and Western-blotted for PKR and other factors. Results in Fig. 7B show that PKR was not activated after the treatment with TAR-containing exosomes, as compared with control uninfected exosomes. Collectively, these data imply that TAR binding to PKR may mainly act as a dominant negative molecule in cells.
FIGURE 7.
Effect of exosomal TAR RNA on PKR. A, uninfected CEM T cells (7.5 × 105) were treated with DOTAP TAR or DOTAP TAR mutant at varying concentrations (5 × 104/1 μg and 10 × 104/10 μg) for 24 h. Total cell extracts were isolated and used for Western blot using antibodies against PKR, p-PKR, eIF2α, p-eIF2α, and actin as a reference protein. Grayscale values of each Western blot band were quantified using ImageJ software, and results are presented as a ratio of analyzed protein to actin in the same sample. B, similar to A, CEM cells were treated with the exosomes (1, 5, or 10 μg) from either CEM (uninfected) or ACH2 (HIV-1-infected) cells for 24 h. Total cell extracts were isolated for Western blot with PKR, p-PKR, eIF2α, p-eIF2α, and actin antibodies. The grayscale values of each Western blot band were measured as described in A.
Inhibition of Exosome Formation in Infected Cells Affects HIV-1 Replication in Bystander Cells in Vitro and in Vivo
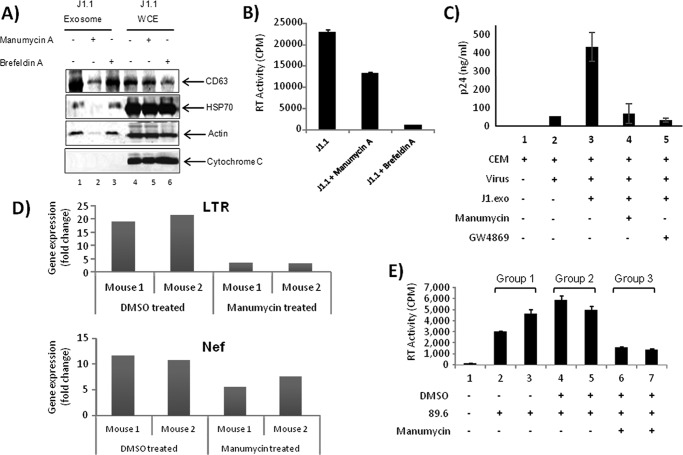
Next, we tested whether inhibition of exosome production from infected cells could affect viral replication in recipient cells to further validate the bystander effect of exosomes. We used two drugs, namely manumycin A and brefeldin A, that have been demonstrated to down-regulate release of proteins such as TNFR1, CD63, and CD81 in exosomes (66, 67). Although both drugs may have inhibitory effects on exosome release, brefeldin A is also known to specifically inhibit extracellular release of viruses (68, 69). Infected cells were treated with either inhibitor, and exosomes were purified. Results in Fig. 8A show that manumycin A, but not brefeldin A treatment, inhibited exosome production in HIV-1-infected cells. This is apparent by the loss of exosome markers, including HSP70 and actin in manumycin A-treated cells (Fig. 8A, lane 2). To test whether these drugs inhibit HIV-1 production in cells, we analyzed RT activity of culture supernatants from the treated cells and found that brefeldin A greatly inhibited virus production in infected cells compared with manumycin A treatment (Fig. 8B). This is in contrast to the effect of the inhibitors on exosome production (compare Fig. 8, A and B) suggesting that there are distinct pathways by which the host cell releases virus versus exosomes. We also asked whether prior exposure of naive cells to exosomes from manumycin A-treated cells could make the naive cells less susceptible to infection. We treated recipient CEM cells with exosomes isolated from the culture medium of manumycin A-treated J1.1 cells and then infected with HIV-1 89.6 strain. As shown in Fig. 8C, p24 ELISA was not detected in CEM cells and was markedly reduced in the cells treated with manumycin as compared with the cells pretreated with exosomes from untreated J1.1 without manumycin.
FIGURE 8.
Inhibition of exosome release from HIV-1-infected cells make naive recipient cells less susceptible to HIV-1 infection. A, chronically HIV-1-infected J1.1 cells were treated with 1 μm MVB pathway inhibitors manumycin A or brefeldin A and tested for exosome production by Western blot using antibodies specific for exosomal proteins, CD63 and HSP70. Antibody for actin and cytochrome c were used as control. B, supernatants from inhibitor-treated or untreated J1.1 cells used in A were utilized in RT assay for quantification of virus. Results are presented as mean ± S.D. of at least three independent measurements. C, naive CEM cells were pre-treated with exosomes (10 μg) from either manumycin-treated, GW4869-treated (100 nm), or untreated infected J1.1 cells prior to addition of HIV-1 89.6 and scored for p24 activity. D, humanized NSG mice were divided into two groups and treated with either DMSO or manumycin A (5 mg/kg) and then infected with 100 μl (5 ng/μl of p24) of HIV-1 89.6. Animals were then euthanized, and blood, brain, and other tissues were collected. Brains were harvested, and white matter (midbrain) from each animal (1 mm) were diced and digested with trypsin. Half of the samples were processed for qRT-PCR using LTR-specific (upper panel) and Nef-specific (lower panel) primers. E, other half-samples of midbrain were co-cultured with CEM cells (5 × 105) for 9 days and processed for the quantitation of virus using RT assay from the supernatants. Results are presented as mean ± S.D. of three independent measurements.
We next asked whether manumycin A treatment could potentially contribute to lower viral spread in vivo. The NSG humanized mice (43, 70) were infected with HIV-1 89.6 either with or without manumycin A treatment, and we focused on detecting infected human monocytes/macrophages. Thus, the brain tissues of the animals were tested. We expect that the human cells in the brain most likely originated from the periphery, because transplantation of CD34 human cells occurred post-brain and blood-brain barrier development. For the exosome experiment, we first injected animals with human stem cells and allowed the cells to differentiate for 3 months. Animals were divided into three experimental groups as follows: animals received no treatment; DMSO; or manumycin (day 0). On day 1, animals were infected with 89.6 virus, and further treatment was continued every 3 days for a period of 2 weeks. Samples were processed for the presence of viral RNA in various tissues (data not shown) and blood samples. Results in Fig. 8D indicate that the levels of both LTR and Nef RNA were down-regulated in manumycin-treated animals. Finally, we obtained brain tissue from another set of animals (white matter, 1 mm) and trypsin-digested and co-cultured the remaining cells with CEM uninfected cells to rescue the functional virus. Results in Fig. 8E indicate that manumycin-treated animals showed considerably less virus in co-culture experiments. Collectively, these results imply that exosomes may increase the susceptibility of recipient uninfected cells to infection.
Discussion
Exosomally transported miRNAs are known to affect the functional fate of recipient cells (31). Earlier, we have shown that the HIV-1 TAR RNA is incorporated into exosomes from different HIV-1-infected cells and patient sera along with the host miRNA machinery proteins Dicer and Drosha (42). We also found that the exosomes from infected cells delivered TAR RNA to the naive undifferentiated target cells, down-regulated apoptosis by lowering Bim and CDK9 proteins in recipient cells, and increased their susceptibility to HIV-1 infection (42). Our new data indicate that the TAR element in exosomes entering recipient uninfected cells can alter the PKR (as a decoy) and increase cytokine expression through the NF-κB pathway. This phenomenon may partly explain the observed inflammatory signals that are present in patients who are undergoing cART treatment.
Compared with the uninfected cells, exosomes derived from HIV-1-infected cells are known to contain many unique host molecules (42). Although some viral proteins and RNAs have also been reported to incorporate into exosomes, it is not clear whether any viral proteins or RNAs directly contribute to the mechanism of exosomal effects on HIV-mediated diseases. We characterized the proteome of exosomes from HIV-1-infected and -uninfected cells via liquid chromatography tandem mass spectrometry (LC-MS/MS). On average, we identified 225–230 proteins incorporated into exosomes from both HIV-1-infected and -uninfected cell types, and 120 proteins were found to be common between both cell types (supplemental Tables 1 and 2). Functional classification of common proteins revealed an abundance of cellular metabolism-related proteins (enzymes), followed by signaling, translation, and nucleic acid-binding components and also other proteins such as cytoskeleton, ion transport, small molecule binding, membrane proteins, and nuclear cytoplasmic transport and migration proteins. Interestingly, after subtracting the common proteins, 105 proteins were found to be unique in exosomes from infected cells. Close examination of the unique protein products from infected exosomes shows the presence of various “helper proteins,” including RAB8B, NDRG1, and PARP1, that are known to aid in different pathological consequences (supplemental Table 3). For example, the RAB8B protein may aid in recipient cell uptake of exosomal content as this protein is known to function in intracellular vesicle transport by aiding in the docking and/or fusion of vesicles with their target membranes (71). The NDRG1 protein is a signaling protein that shuttles between the cytoplasm and the nucleus necessary for axonal survival (72). It has been shown to act as a novel Rab4a effector protein that alters the kinetics of transferrin recycling in cells (73). Finally, the PARP1 protein when unloaded into recipient cells may potentiate the apoptotic pathway after being activated in the recipient nucleus. It is well established that PARP1 activation is required for translocation of apoptosis-inducing factor from the mitochondria to the nucleus and that apoptosis-inducing factor is necessary for PARP1-dependent cell death (74). Also, recently, we obtained some preliminary data indicating that exosomes from infected cells of non-T-cell origin (microglia) contain most of these cellular proteins, as well as viral proteins including Tat, Vpr, and Nef.5 Of note, we have also observed the presence of Tat and Vpr protein in the exosomes from HIV-1-infected promyeloid OM10.1 cells but not from infected T cell lines. Our future efforts will focus on the comparison between T cell-derived and myeloid exosomes with regards to the protein contents (cellular and viral) and their functions in recipient cells.
The main focus of this study was to elucidate the role of HIV-1 TAR-RNA incorporated into the exosomes. Several sets of data indicated that an abundance of extracellular RNA is present in the exosomes under both in vitro and in vivo conditions, and more importantly, they may directly affect the functionality of the uninfected cells. For example, we have observed that TAR-RNA in exosomes from HIV-1-infected cells markedly enhanced proinflammatory cytokines such as IL-6 and TNF-β in both human primary macrophages and mouse neuronal cells. These results strongly suggest that a similar phenomenon could also occur in vivo where exosomes containing TAR RNA cross the blood-brain barrier and stimulate microglia and other cells to secrete proinflammatory cytokines. Although the mechanism of exosomal TAR effects on HIV-mediated CNS disease is not clear, some of our results indicate that TAR may likely bind to PKR and/or potentially to TLRs to stimulate cytokine secretion through activating NF-κB components.
In this study, we also asked whether TAR RNA levels increased over time in primary cell infections. Interestingly, we found that TAR, at both intracellular and extracellular levels, increases over time. This is consistent with our previous observations that TAR may directly contribute to HIV-1 latency through transcriptional gene silencing (15, 16). We and multiple other colleagues have shown that HIV-1-infected cells contain 1–5 copies of viral genome, yet the number of TAR RNA molecules range from 103 to 105 depending on the infection. Therefore, a fundamental question for us has been on what happens to the majority of TAR RNA that may not be used for establishing latency in cells through transcriptional gene silencing. Our results show that TAR is present in high copy numbers in recipient cells. The binding can activate a new form of IKK-β, which may contribute to the release of free p50 and p65 NF-κB and ultimately contribute to cytokine activation. Therefore, TAR in exosomes may modulate activation and cytokine production in uninfected cells (Fig. 4). Our data strongly suggest that exosome-packaged full-size TAR RNA (∼90%), as well as TAR miRNA (∼10%), may bind to TLRs in recipient cells and activate the NF-κB pathway. A similar analogous finding has been shown for TNF-α release and TLR8 signaling (75). The final net effect would be to keep the uninfected recipient cells in an active state to potentially allow the incoming virus to enter and replicate efficiently in these cells or simply contribute to a cytokine “storm” phenotype.
Overall, we have demonstrated in this study that an abundance of HIV-1 TAR-RNA (potentially with other viral proteins) carried by the exosomes could play an important role in pathogenesis in HIV-1-infected individuals. Our future studies will focus more on better characterization of both 5′ and 3′ ends of the TAR and TAR miRNAs (which may contribute differentially to RIG-I and TLR pathways), the potential presence of TAR/Gag RNA in exosomes, and the mechanisms of exosome effects using animal experiments and decipher the kinetics of repeated exposure of cells to exosomes both in vitro and in vivo contributing to potential immune exhaustion and/or senescence over time.
Author Contributions
G. C. S., M. S., A. S., R. B., and S. P. carried out most of experiments, contributed to cell culture work, Western blots, RNA isolation, RT-qPCR, and data analysis. M. C. C., R. M. H., and M. A. Z. contributed to performing the cytokine experiments. B. L. contributed to use of nanoparticles. Z. A. K. and N. E. H. contributed data for Figs. 1 and 2. M. Y. contributed samples from Georgetown University. S. I. and F. K. contributed to the overall direction and coordination of the study as well as design of experiments, quantitative analyses, and data interpretation.
Supplementary Material
Acknowledgments
We thank the members of the Kashanchi laboratory, including Kinza Noor and Yao Akpamagbo, for experiments and assistance with the manuscript. We thank Dr. Aarthi Narayanan for MS experiments. We thank Dr. Jay Rappaport (Temple University) for the suggestion of experiments related to PKR and the RNA sizes. We also grateful to the National Institutes of Health AIDS Research and Reference Reagent Program for the contribution of important reagents. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: U1, J1.1 and ACH-2 cells from Dr. Thomas Folks; HIV-1 89.6 dual-tropic viral strain from Dr. Ronald Collman; and anti-HIV-1 p24 mouse monoclonal antibody from Dr. Michael Malim. The antiretrovirals lamivudine/emtricitabine, tenofovir, indinavir, raltegravir, and maraviroc were also from AIDS Research and Reference Reagent Program. Four clinical samples utilized in this study were provided by the Washington, D. C., Metropolitan Site of the Women's Interagency HIV Study Collaborative Study Group (Principal Investigator, Mary Young). The Women's Interagency HIV Study is supported by National Institutes of Health Grant U01-AI-34994, from NIAID.
This work was supported by Department of Energy Grant DE-SC0001599, National Institutes of Health Grants AI078859, AI074410, and AI043894 (to F. K.) and Grant NS086453 (to G. C. S.), and in part by NIAID, NCI, and the National Institute on Drug Abuse. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. S1 and Tables S1–S3.
F. Kashanchi, S. Punya, and S. Iordanskiy, unpublished data.
- ncRNA
- non-coding RNA
- TAR
- trans-activating response
- qRT
- quantitative RT
- PBL
- peripheral blood lymphocyte
- miRNA
- microRNA
- MDM
- monocyte-derived macrophage
- cART
- combinational antiretroviral therapy
- PBMC
- peripheral blood mononuclear cell
- MVB
- multivesicular body
- SEAP
- secreted embryonic alkaline phosphatase
- RNP
- ribonucleoprotein
- TLR
- toll-like receptor
- DOTAP
- N-[1-(2,3-dioleoyloxy)]-N,N,N-trimethylammonium propane methylsulfate.
References
- 1.UNAIDS (2012) UNAIDS Report on the Global AIDS Epidemic 2012. http://www.unaids.org/sites/default/files/media_asset/20121120_UNAIDS_Global_Report_2012_with_annexes_en_1.pdf
- 2.Prejean J., Song R., Hernandez A., Ziebell R., Green T., Walker F., Lin L. S., An Q., Mermin J., Lansky A., Hall H. I., and HIV Incidence Surveillance Group (2011) Estimated HIV incidence in the United States, 2006–2009. PLoS ONE 6, e17502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laird G. M., Eisele E. E., Rabi S. A., Lai J., Chioma S., Blankson J. N., Siliciano J. D., and Siliciano R. F. (2013) Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 9, e1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Sluis R. M., Jeeninga R. E., and Berkhout B. (2013) Establishment and molecular mechanisms of HIV-1 latency in T cells. Curr. Opin. Virol. 3, 700–706 [DOI] [PubMed] [Google Scholar]
- 5.Kiage J. N., Heimburger D. C., Nyirenda C. K., Wellons M. F., Bagchi S., Chi B. H., Koethe J. R., Arnett D. K., and Kabagambe E. K. (2013) Cardiometabolic risk factors among HIV patients on antiretroviral therapy. Lipids Health Dis. 12, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feleke Y., Fekade D., and Mezegebu Y. (2012) Prevalence of highly active antiretroviral therapy associated metabolic abnormalities and lipodystrophy in HIV infected patients. Ethiop. Med. J. 50, 221–230 [PubMed] [Google Scholar]
- 7.Deminice R., Vassimon H. S., Machado A. A., de Paula F. J., Monteiro J. P., and Jordao A. A. (2013) Plasma homocysteine levels in HIV-infected men with and without lipodystrophy. Nutrition 29, 1326–1330 [DOI] [PubMed] [Google Scholar]
- 8.Heaton R. K., Franklin D. R., Ellis R. J., McCutchan J. A., Letendre S. L., Leblanc S., Corkran S. H., Duarte N. A., Clifford D. B., Woods S. P., Collier A. C., Marra C. M., Morgello S., Mindt M. R., Taylor M. J., et al. (2011) HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: differences in rates, nature, and predictors. J. Neurovirol. 17, 3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McArthur J. C., Brew B. J., and Nath A. (2005) Neurological complications of HIV infection. Lancet Neurol. 4, 543–555 [DOI] [PubMed] [Google Scholar]
- 10.Coffin J. M., Cann A. J., Chen I., Luciw P., Hirsch M. S., and Curran J. W. (2013) in Fields Virology (Fields B. N., Knipe D. M., Howley P. M., eds) 6th Ed., pp. 1767–1997, Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 11.Schopman N. C., Willemsen M., Liu Y. P., Bradley T., van Kampen A., Baas F., Berkhout B., and Haasnoot J. (2012) Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res. 40, 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung M. L., Bennasser Y., Watashi K., Le S. Y., Houzet L., and Jeang K. T. (2009) Pyrosequencing of small non-coding RNAs in HIV-1-infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acids Res. 37, 6575–6586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan A., Kehn-Hall K., Bailey C., and Kashanchi F. (2011) Analysis of the roles of HIV-derived microRNAs. Expert Opin. Biol. Ther. 11, 17–29 [DOI] [PubMed] [Google Scholar]
- 14.Bennasser Y., Le S. Y., Yeung M. L., and Jeang K. T. (2004) HIV-1 encoded candidate micro-RNAs and their cellular targets. Retrovirology 1, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klase Z., Kale P., Winograd R., Gupta M. V., Heydarian M., Berro R., McCaffrey T., and Kashanchi F. (2007) HIV-1 TAR element is processed by Dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol. Biol. 8, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klase Z., Winograd R., Davis J., Carpio L., Hildreth R., Heydarian M., Fu S., McCaffrey T., Meiri E., Ayash-Rashkovsky M., Gilad S., Bentwich Z., and Kashanchi F. (2009) HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouellet D. L., Plante I., Landry P., Barat C., Janelle M. E., Flamand L., Tremblay M. J., and Provost P. (2008) Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res. 36, 2353–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei P., Garber M. E., Fang S. M., Fischer W. H., and Jones K. A. (1998) A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92, 451–462 [DOI] [PubMed] [Google Scholar]
- 19.Laspia M. F., Rice A. P., and Mathews M. B. (1989) HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 59, 283–292 [DOI] [PubMed] [Google Scholar]
- 20.Luo Y., Madore S. J., Parslow T. G., Cullen B. R., and Peterlin B. M. (1993) Functional analysis of interactions between Tat and the trans-activation response element of human immunodeficiency virus type 1 in cells. J. Virol. 67, 5617–5622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Février B., and Raposo G. (2004) Exosomes: endosomal-derived vesicles shipping extracellular messages. Curr. Opin. Cell Biol. 16, 415–421 [DOI] [PubMed] [Google Scholar]
- 22.Record M., Subra C., Silvente-Poirot S., and Poirot M. (2011) Exosomes as intercellular signalosomes and pharmacological effectors. Biochem. Pharmacol. 81, 1171–1182 [DOI] [PubMed] [Google Scholar]
- 23.Théry C., Zitvogel L., and Amigorena S. (2002) Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2, 569–579 [DOI] [PubMed] [Google Scholar]
- 24.Lotvall J., and Valadi H. (2007) Cell to cell signalling via exosomes through esRNA. Cell Adh. Migr. 1, 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camussi G., Deregibus M. C., Bruno S., Cantaluppi V., and Biancone L. (2010) Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 78, 838–848 [DOI] [PubMed] [Google Scholar]
- 26.Skog J., Würdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T. Jr., Carter B. S., Krichevsky A. M., and Breakefield X. O. (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pegtel D. M., van de Garde M. D., and Middeldorp J. M. (2011) Viral miRNAs exploiting the endosomal-exosomal pathway for intercellular cross-talk and immune evasion. Biochim. Biophys. Acta 1809, 715–721 [DOI] [PubMed] [Google Scholar]
- 28.Barth S., Meister G., and Grässer F. A. (2011) EBV-encoded miRNAs. Biochim. Biophys. Acta 1809, 631–640 [DOI] [PubMed] [Google Scholar]
- 29.Gourzones C., Gelin A., Bombik I., Klibi J., Vérillaud B., Guigay J., Lang P., Témam S., Schneider V., Amiel C., Baconnais S., Jimenez A. S., and Busson P. (2010) Extracellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol. J. 7, 271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Würdinger T., and Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verweij F. J., van Eijndhoven M. A., Hopmans E. S., Vendrig T., Wurdinger T., Cahir-McFarland E., Kieff E., Geerts D., van der Kant R., Neefjes J., Middeldorp J. M., and Pegtel D. M. (2011) LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 30, 2115–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lenassi M., Cagney G., Liao M., Vaupotic T., Bartholomeeusen K., Cheng Y., Krogan N. J., Plemenitas A., and Peterlin B. M. (2010) HIV Nef is secreted in exosomes and triggers apoptosis in bystander CD4+ T cells. Traffic 11, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omoto S., Ito M., Tsutsumi Y., Ichikawa Y., Okuyama H., Brisibe E. A., Saksena N. K., and Fujii Y. R. (2004) HIV-1 nef suppression by virally encoded microRNA. Retrovirology 1, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennasser Y., Le S. Y., Benkirane M., and Jeang K. T. (2005) Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity 22, 607–619 [DOI] [PubMed] [Google Scholar]
- 35.Collin D., van Heijenoort C., Boiziau C., Toulmé J. J., and Guittet E. (2000) NMR characterization of a kissing complex formed between the TAR RNA element of HIV-1 and a DNA aptamer. Nucleic Acids Res. 28, 3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeffer S., Zavolan M., Grässer F. A., Chien M., Russo J. J., Ju J., John B., Enright A. J., Marks D., Sander C., and Tuschl T. (2004) Identification of virus-encoded microRNAs. Science 304, 734–736 [DOI] [PubMed] [Google Scholar]
- 37.Whisnant A. W., Bogerd H. P., Flores O., Ho P., Powers J. G., Sharova N., Stevenson M., Chen C. H., and Cullen B. R. (2013) In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. MBio 4, e000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zomer A., Vendrig T., Hopmans E. S., van Eijndhoven M., Middeldorp J. M., and Pegtel D. M. (2010) Exosomes: fit to deliver small RNA. Commun. Integr. Biol. 3, 447–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 40.Banks W. A., Kastin A. J., and Akerstrom V. (1997) HIV-1 protein gp120 crosses the blood-brain barrier: role of adsorptive endocytosis. Life Sci. 61, PL119–125 [DOI] [PubMed] [Google Scholar]
- 41.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., and Wood M. J. (2011) Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29, 341–345 [DOI] [PubMed] [Google Scholar]
- 42.Narayanan A., Iordanskiy S., Das R., Van Duyne R., Santos S., Jaworski E., Guendel I., Sampey G., Dalby E., Iglesias-Ussel M., Popratiloff A., Hakami R., Kehn-Hall K., Young M., Subra C., et al. (2013) Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J. Biol. Chem. 288, 20014–20033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van Duyne R., Cardenas J., Easley R., Wu W., Kehn-Hall K., Klase Z., Mendez S., Zeng C., Chen H., Saifuddin M., and Kashanchi F. (2008) Effect of transcription peptide inhibitors on HIV-1 replication. Virology 376, 308–322 [DOI] [PubMed] [Google Scholar]
- 44.Althaus C. F., Vongrad V., Niederöst B., Joos B., Di Giallonardo F., Rieder P., Pavlovic J., Trkola A., Günthard H. F., Metzner K. J., and Fischer M. (2012) Tailored enrichment strategy detects low abundant small noncoding RNAs in HIV-1-infected cells. Retrovirology 9, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaworski E., Saifuddin M., Sampey G., Shafagati N., Van Duyne R., Iordanskiy S., Kehn-Hall K., Liotta L., Petricoin E. 3rd, Young M., Lepene B., and Kashanchi F. (2014) The use of Nanotrap particles technology in capturing HIV-1 virions and viral proteins from infected cells. PLoS ONE 9, e96778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Duyne R., Guendel I., Jaworski E., Sampey G., Klase Z., Chen H., Zeng C., Kovalskyy D., El Kouni M. H., Lepene B., Patanarut A., Nekhai S., Price D. H., and Kashanchi F. (2013) Effect of mimetic CDK9 inhibitors on HIV-1-activated transcription. J. Mol. Biol. 425, 812–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stumptner-Cuvelette P., Jouve M., Helft J., Dugast M., Glouzman A. S., Jooss K., Raposo G., and Benaroch P. (2003) Human immunodeficiency virus-1 Nef expression induces intracellular accumulation of multivesicular bodies and major histocompatibility complex class II complexes: potential role of phosphatidylinositol 3-kinase. Mol. Biol. Cell 14, 4857–4870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narayanan A., Sampey G., Van Duyne R., Guendel I., Kehn-Hall K., Roman J., Currer R., Galons H., Oumata N., Joseph B., Meijer L., Caputi M., Nekhai S., and Kashanchi F. (2012) Use of ATP analogs to inhibit HIV-1 transcription. Virology 432, 219–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breen E. C., Rezai A. R., Nakajima K., Beall G. N., Mitsuyasu R. T., Hirano T., Kishimoto T., and Martinez-Maza O. (1990) Infection with HIV is associated with elevated IL-6 levels and production. J. Immunol. 144, 480–484 [PubMed] [Google Scholar]
- 50.Stone S. F., Price P., Keane N. M., Murray R. J., and French M. A. (2002) Levels of IL-6 and soluble IL-6 receptor are increased in HIV patients with a history of immune restoration disease after HAART. HIV Med. 3, 21–27 [DOI] [PubMed] [Google Scholar]
- 51.Lotz M., Jirik F., Kabouridis P., Tsoukas C., Hirano T., Kishimoto T., and Carson D. A. (1988) B cell stimulating factor 2/interleukin 6 is a costimulant for human thymocytes and T lymphocytes. J. Exp. Med. 167, 1253–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suen W. E., Bergman C. M., Hjelmström P., and Ruddle N. H. (1997) A critical role for lymphotoxin in experimental allergic encephalomyelitis. J. Exp. Med. 186, 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ngo V. N., Korner H., Gunn M. D., Schmidt K. N., Riminton D. S., Cooper M. D., Browning J. L., Sedgwick J. D., and Cyster J. G. (1999) Lymphotoxin α/β and tumor necrosis factor are required for stromal cell expression of homing chemokines in B and T cell areas of the spleen. J. Exp. Med. 189, 403–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matusevicius D., Navikas V., Söderström M., Xiao B. G., Haglund M., Fredrikson S., and Link H. (1996) Multiple sclerosis: the proinflammatory cytokines lymphotoxin-α and tumour necrosis factor-α are upregulated in cerebrospinal fluid mononuclear cells. J. Neuroimmunol. 66, 115–123 [DOI] [PubMed] [Google Scholar]
- 55.Navikas V., He B., Link J., Haglund M., Söderström M., Fredrikson S., Ljungdahl A., Höjeberg J., Qiao J., Olsson T., and Link H. (1996) Augmented expression of tumour necrosis factor-α and lymphotoxin in mononuclear cells in multiple sclerosis and optic neuritis. Brain 119, 213–223 [DOI] [PubMed] [Google Scholar]
- 56.Kawai T., and Akira S. (2007) Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 57.Suh H. S., Zhao M. L., Choi N., Belbin T. J., Brosnan C. F., and Lee S. C. (2009) TLR3 and TLR4 are innate antiviral immune receptors in human microglia: role of IRF3 in modulating antiviral and inflammatory response in the CNS. Virology 392, 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eiring A. M., Harb J. G., Neviani P., Garton C., Oaks J. J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C. J., Becker H., Chandler J. C., Andino R., Cortes J., Hokland P., et al. (2010) miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell 140, 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edery I., Petryshyn R., and Sonenberg N. (1989) Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell 56, 303–312 [DOI] [PubMed] [Google Scholar]
- 60.Carpick B. W., Graziano V., Schneider D., Maitra R. K., Lee X., and Williams B. R. (1997) Characterization of the solution complex between the interferon-induced, double-stranded RNA-activated protein kinase and HIV-I trans-activating region RNA. J. Biol. Chem. 272, 9510–9516 [DOI] [PubMed] [Google Scholar]
- 61.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G. J., Zanesi N., Crawford M., Ozer G. H., Wernicke D., Alder H., et al. (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–E2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amaya M., Voss K., Sampey G., Senina S., de la Fuente C., Mueller C., Calvert V., Kehn-Hall K., Carpenter C., Kashanchi F., Bailey C., Mogelsvang S., Petricoin E., and Narayanan A. (2014) The role of IKK-β in Venezuelan equine encephalitis virus infection. PLoS ONE 9, e86745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heinicke L. A., Wong C. J., Lary J., Nallagatla S. R., Diegelman-Parente A., Zheng X., Cole J. L., and Bevilacqua P. C. (2009) RNA dimerization promotes PKR dimerization and activation. J. Mol. Biol. 390, 319–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Husain B., Mukerji I., and Cole J. L. (2012) Analysis of high-affinity binding of protein kinase R to double-stranded RNA. Biochemistry 51, 8764–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Langland J. O., Cameron J. M., Heck M. C., Jancovich J. K., and Jacobs B. L. (2006) Inhibition of PKR by RNA and DNA viruses. Virus Res. 119, 100–110 [DOI] [PubMed] [Google Scholar]
- 66.Islam A., Shen X., Hiroi T., Moss J., Vaughan M., and Levine S. J. (2007) The brefeldin A-inhibited guanine nucleotide-exchange protein, BIG2, regulates the constitutive release of TNFR1 exosome-like vesicles. J. Biol. Chem. 282, 9591–9599 [DOI] [PubMed] [Google Scholar]
- 67.Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., Bernad A., and Sánchez-Madrid F. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pal R., Mumbauer S., Hoke G. M., Takatsuki A., and Sarngadharan M. G. (1991) Brefeldin A inhibits the processing and secretion of envelope glycoproteins of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 7, 707–712 [DOI] [PubMed] [Google Scholar]
- 69.Macovei A., Zitzmann N., Lazar C., Dwek R. A., and Branza-Nichita N. (2006) Brefeldin A inhibits pestivirus release from infected cells, without affecting its assembly and infectivity. Biochem. Biophys. Res. Commun. 346, 1083–1090 [DOI] [PubMed] [Google Scholar]
- 70.Van Duyne R., Pedati C., Guendel I., Carpio L., Kehn-Hall K., Saifuddin M., and Kashanchi F. (2009) The utilization of humanized mouse models for the study of human retroviral infections. Retrovirology 6, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen D., Guo J., and Gahl W. A. (1997) RAB GTPases expressed in human melanoma cells. Biochim. Biophys. Acta 1355, 1–6 [DOI] [PubMed] [Google Scholar]
- 72.Kalaydjieva L., Gresham D., Gooding R., Heather L., Baas F., de Jonge R., Blechschmidt K., Angelicheva D., Chandler D., Worsley P., Rosenthal A., King R. H., and Thomas P. K. (2000) N-myc downstream-regulated gene 1 is mutated in hereditary motor and sensory neuropathy-Lom. Am. J. Hum. Genet. 67, 47–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kachhap S. K., Faith D., Qian D. Z., Shabbeer S., Galloway N. L., Pili R., Denmeade S. R., DeMarzo A. M., and Carducci M. A. (2007) The N-Myc down regulated Gene1 (NDRG1) is a Rab4a effector involved in vesicular recycling of E-cadherin. PLoS ONE 2, e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu S. W., Wang H., Poitras M. F., Coombs C., Bowers W. J., Federoff H. J., Poirier G. G., Dawson T. M., and Dawson V. L. (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297, 259–263 [DOI] [PubMed] [Google Scholar]
- 75.Bernard M. A., Zhao H., Yue S. C., Anandaiah A., Koziel H., and Tachado S. D. (2014) Novel HIV-1 miRNAs stimulate TNF-α release in human macrophages via TLR8 signaling pathway. PLoS ONE 9, e106006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bohan C. A., Kashanchi F., Ensoli B., Buonaguro L., Boris-Lawrie K. A., and Brady J. N. (1992) Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 2, 391–407 [PMC free article] [PubMed] [Google Scholar]
- 77.Kashanchi F., Duvall J. F., and Brady J. N. (1992) Electroporation of viral transactivator proteins into lymphocyte suspension cells. Nucleic Acids Res. 20, 4673–4674 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.