Abstract
An individual’s sex affects gene expression and many inflammatory diseases present in a sex-biased manner. Glucocorticoid receptors (GRs) are regulators of inflammatory genes, but their role in sex-specific responses is unclear. Our goal was to evaluate whether GR differentially regulates inflammatory gene expression in male and female mouse liver. Twenty-five percent of the 251 genes assayed by nanostring analysis were influenced by sex. Of these baseline sexually dimorphic inflammatory genes, 82% was expressed higher in female liver. Pathway analyses defined pattern-recognition receptors as the most sexually dimorphic pathway. We next exposed male and female mice to the proinflammatory stimulus LPS. Female mice had 177 genes regulated by treatment with LPS, whereas males had 149, with only 66% of LPS-regulated genes common between the sexes. To determine the contribution of GR to sexually dimorphic inflammatory genes we performed nanostring analysis on liver-specific GR knockout (LGRKO) mice in the presence or absence of LPS. Comparing LGRKO to GRflox/flox revealed that 36 genes required GR for sexually dimorphic expression, whereas 24 genes became sexually dimorphic in LGRKO. Fifteen percent of LPS-regulated genes in GRflox/flox were not regulated in male and female LGRKO mice treated with LPS. Thus, GR action is influenced by sex to regulate inflammatory gene expression.—Quinn, M. A., Cidlowski, J. A. Endogenous hepatic glucocorticoid receptor signaling coordinates sex-biased inflammatory gene expression.
Keywords: sexual dimorphism, liver, nanostring, inflammation, corticosteroid
It is widely appreciated that there are differences between males and females in regard to gene expression, and the regulation of these sex-specific differences results in functional differences in various physiologic processes, including immune function (1–3) and metabolism (4–6). Sex hormones acting through the estrogen receptor (ER), progesterone receptor (PR), and androgen receptor (AR) are thought to be key regulators of sexually dimorphic gene expression; however, non–sex hormones, such as growth hormone, also exert effects in a sex-biased manner (7, 8). The molecular pathways governing sex-biased gene expression are critical to understand, because these properties are thought to contribute to sexually dimorphic diseases. For example, women are more prone to autoimmune diseases, such as lupus (9), rheumatoid arthritis (10), autoimmune hepatitis, and primary biliary cirrhosis (11), whereas men are more prone to chronic kidney disease (12), cardiovascular disease (13), and hepatocellular carcinoma (14). Inflammation is a cornerstone of many of these diseases, suggesting that inflammatory gene expression is regulated uniquely between males and females.
Glucocorticoids are steroid hormones secreted in response to physiologic stresses such as infection. Once secreted, glucocorticoids exert potent anti-inflammatory and immunomodulatory effects via the glucocorticoid receptor (GR), a member of the nuclear receptor family of genes (15, 16). Glucocorticoids are the most widely prescribed drug for the treatment of inflammatory diseases because of their potent anti-inflammatory properties (17–21). Glucocorticoids, such as cortisol in humans and corticosterone in rodents, are endogenous anti-inflammatory modulators, and little consideration has been given to their ability to function uniquely in males and females. Synthetic glucocorticoids have been shown to act differentially between male and female in the rat liver (22) and brain (23, 24); however, the contribution of endogenous glucocorticoids and GRs in promoting sex differences in inflammatory gene expression is largely unknown.
In this study, we identified considerable sexual dimorphism in inflammatory gene expression in the mouse liver. Using nanostring technology, we focused our studies on a preset group of 250 classic inflammatory genes and found that female mice have a higher expression of inflammatory genes than do males. In basal conditions, the TLR pathway is one of the most highly sexually dimorphic for expression. Female mice mount a stronger functional hepatic inflammatory response than do males in response to TLR activation by LPS. Genetic ablation of hepatic GR reduces the number of inflammatory genes that exhibit sexually dimorphic expression under basal conditions, although some sexually dimorphic responses to LPS were still observed in liver-specific GR (LGRKO) knockout mice. That the deletion of the GR also results in sex-specific alterations in gene expression in response to LPS is a provocative finding. Our data indicate that the GR regulates sexually distinct inflammatory gene networks in homeostatic conditions and during an inflammatory challenge.
MATERIALS AND METHODS
Animal studies
Male and female C57BL/6J mice were purchased from the Jackson Laboratory (Bar Harbor, ME, USA). LGRKO mice were generated by crossing GRflox/flox (25) with mice expressing carbapenem-resistant Enterobacteriaceae (CRE) recombinase under the liver-specific promoter albumin. Male and female GRflox/flox CRE– mice were used as controls, and GRflox/flox CRE+ animals were used for the experimental groups. All animals maintained in a 12:12 h light/dark cycle and had ad libitum access to standard mouse chow and drinking water. To induce endotoxemia, LPS from Escherichia coli 0111:B4 was purchased from Sigma-Aldrich (St. Louis, MO, USA) and dissolved in pyrogen/pathogen-free PBS. Male and female wild-type (WT) and LGRKO mice were injected with 5 mg/kg, i.p. LPS for 4, 8, or 24 h, after which the animals were killed and the tissues were collected. The animals were 10–12 wk of age at the time of the experiments. All animal experiments were treated in accordance with the Guidelines of the Institutional Animal Care and Use Committee at the National Institute of Environmental Health Sciences.
RNA measurements
Quantitative RT-PCR
RNA was extracted from livers of mice following various experimental paradigms with the RNeasy RNA isolation kit (Qiagen, Valencia, CA, USA), according to the manufacturer’s protocol. Total RNA (100 ng) was reverse transcribed and amplified with the iScript One-Step RT-PCR kit for probes (Bio-Rad, Hercules, CA, USA). Real-time quantitative (q)PCR was performed with the Bio-Rad CFX96 sequence-detection system, with predesigned primer/probe sets from Applied Biosystem/Thermo Fisher Scientific, according to the manufacturer’s protocol. Relative fluorescent signal from each transcript was normalized to the housekeeping gene peptidylprolyl isomerase B (Ppib), which was invariant in our experimental conditions (the ΔCt method). Data are expressed as mRNA expression relative to PPIB mRNA expression (means ± sem).
Nanostring analysis
RNA was extracted as described earlier, and 50 ng was used to determine the absolute levels of gene expression. Hybridization and nCounter were performed according to the manufacturer’s protocol (Nanostring Technologies, Seattle, WA, USA). In brief, reactions were hybridized for 20 h at 65°, after which the products were used to run on the nCounter preparation station for removal of excess probes. Data were collected with the nCounter digital analyzer by counting individual barcodes. Data generated from the nCounter digital analyzer were examined with the nCounter digital analyzer software system v2.1.1 (Nanostring Technologies). Data were normalized to the geometric means of spiked-in positive controls (controls for assay efficiency) and spiked-in negative controls (normalized for background). The data were further normalized to the housekeeping genes Gapdh, Hprt, and Tubb5 and are reported as normalized RNA counts (means ± sem). Nanostring RNA counts were analyzed with the Partek Genomic Suite (Partek, Inc., St. Louis, MO, USA), to identify significantly regulated probe. We classified genes as sex-biased if their expression was significantly different between males and females (P < 0.05). Significantly regulated probes were sorted by using a Venn diagram generator (http://www.pangloss.com/seidel/Protocols/venn.cgi), and pathway analysis was performed with Ingenuity Pathway Analysis (IPA) software (Qiagen). Heat maps of nanostring data were generated with the Partek Genomic Suite.
Immunohistochemistry
To determine the subcellular localization of GRs after LPS treatment, we performed immunohistochemistry in male and female mice treated with PBS vehicle or LPS for 4 h (26). Slides were incubated with anti-rabbit polyclonal GR antibody overnight with an antibody derived in our lab (25, 27), visualized via biotinylated secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA), and developed with HRP-conjugated 3,3′-diaminobenzidine (DAB; Vector Laboratories, Burlingame, CA, USA). DAB signal was visualized with an inverted light microscope (Zeiss, Oberkochen, Germany).
Western blot analysis
To assess GR, p65, and TLR4 levels total protein lysates were prepared from male and female mice with radioimmunoprecipitation assay buffer (RIPA; Thermo Fisher Scientific, Grand Island, NY, USA) in the presence of protease inhibitors (Roche, Indianapolis, IN, USA). Total protein (40 μg) was mixed with 2× Laemmli buffer (Bio-Rad) containing 2-ME (Sigma-Aldrich), resolved on a 4–20% Tris-glycine gel (Bio-Rad), and transferred to a 0.2 μm nitrocellulose membrane (Bio-Rad). After protein transfer, membranes were blocked with Licor blocking buffer (Licor Biosystems, Lincoln, NE, USA) and incubated overnight in anti-rabbit monoclonal GR antibody (Abcam, Cambridge, MA, USA), anti-rabbit polyclonal p65 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-rat monoclonal TLR4 antibody (Santa Cruz Biotechnology), or anti-mouse monoclonal β-actin (EMD-Millipore, Bedford, MA, USA). All primary antibodies were used at a dilution of 1:1,000, except β-actin, which was used at 1:10,000. Protein was detected via fluorescent secondary antibody detection (1:10,000; Licor) and imaged on the Odyssey system (Licor). TLR4 and p65 were normalized to β-actin and expressed as a percentage of the male level (means ± sem).
IL-1β protein measurement
IL-1β was assayed via a commercially available ELISA (eBioscience, San Diego, CA, USA) according to the manufacturer’s protocol. Briefly, 100 mg of liver tissue was homogenized in RIPA buffer. Lysates were centrifuged at 14,000 rpm for 15 min at 4°C. IL-1β was detected in the lysates.
Corticosterone measurement
To determine the levels of circulating glucocorticoids after administration of LPS, we collected serum after from male and female WT mice. Corticosterone was determined via a commercially available ELISA kit (Arbor Assays, Ann Arbor, MI, USA), according to the manufacturer’s protocol.
Statistical analysis
To detect statistical significance between groups, Student’s t test was used (Prism 6; Graphpad, La Jolla, CA, USA). A normality test was performed on the t test for distribution and if the data failed the normality test, a Mann-Whitney post hoc test was performed. A 2-way ANOVA was performed when comparing 3 or more groups followed by a Sidak post hoc test to correct for multiple comparisons. Statistical significance was defined as P < 0.05.
RESULTS
Livers from male and female mice display sexually dimorphic inflammatory gene expression
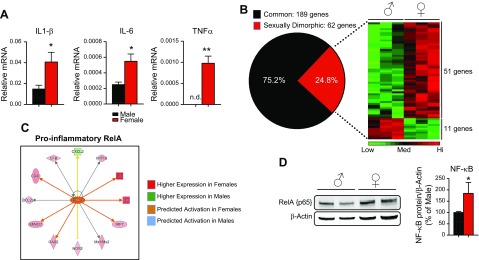
Given that a variety of sexually dimorphic diseases have an inflammatory component, we hypothesized that male and female mice express genes involved in the inflammatory pathway in a sex-specific fashion. To test this idea, we initially sought to determine whether there is sexual dimorphism in the expression of 3 classic proinflammatory genes in the mouse liver: IL-1β, IL-6, and TNFα. Quantitative PCR analysis showed that female mice had significantly higher expression of all 3 proinflammatory genes in the liver when compared to that in male liver (Fig. 1A). Based on these findings, we wanted to know whether this pattern is consistent across the large spectrum of inflammatory genes reported in the literature. To accomplish this goal, we used nanostring technology to show the global sex differences in inflammatory gene expression. Nanostring analysis can be considered a hybrid between high-throughput techniques, such as microarray or RNA sequencing, but with the accuracy of conventional qPCR. Nanostring analysis allows for the direct measurement of mRNA concentrations without the synthesis of cDNA or amplification of transcripts, thus avoiding the experimental biases introduced during these steps. This technology is highly sensitive, enabling the detection of a single RNA transcript that has high reproducibility with qPCR (28–31). Of the 251 genes present on the nanostring mouse inflammatory code set, we found 189 (75.2%) to be commonly expressed in male and female mouse liver and 62 (24.8%) to be sexually dimorphic in their expression (Fig. 1B). A heat map depiction of RNA counts in liver lysates revealed that female mice expressed 51 genes more highly than did males, whereas 11 genes were more highly expressed in males (P < 0.05). Approximately 25% of the genes in the inflammatory panel (250 genes) in the liver were influenced by the sex of the mouse, and most of these sexually dimorphic genes (82%) were enriched in the female vs. the male liver (Fig. 1B).
Figure 1.
Hepatic inflammatory genes are expressed in a sexually dimorphic manner. A) qPCR for IL-1β, IL-6, and TNFα from livers of male and female C57BL/6J mice. Data are gene vs. PPIB mRNA levels (means ± sem, n = 3 animals). B) Common and sexually dimorphic genes from nanostring analysis. Heat map of sexually dimorphic genes showing 51 genes have higher expression in female liver and 11 genes have higher expression in males (n = 3 animals). C) Network map of RelA target genes from nanostring data. Males did not show activation of this pathway, hence no blue indicators. D) Western blot analysis of RelA in male and female liver homogenates. RelA protein was normalized to β-actin and expressed as a percentage of that in male liver (n = 4–5 animals). n.d., nondetectable. *P < 0.05, **P < 0.01.
To define the biologic pathways that display sexual dimorphism, we performed IPA (Qiagen) on this dataset. The results revealed that the top 5 canonical pathways displaying sexual dimorphism are the role of pattern recognition receptors in recognition of bacteria and viruses; the role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis; the complement system; TREM1 (triggering receptor expressed on myeloid cells-1) signaling; and NF-κB signaling (Table 1). Because NF-κB (p65/RelA) is one of the most powerful regulators of the inflammatory response, we further examined this pathway in IPA. The female liver had higher expression of various RelA target genes, including cluster of differentiation 40 (Cd40), interferon regulatory factor-7 (Irf7) and nitric oxide synthase-2, inducible (Nos2), than did male liver, whereas the only RelA target gene showing higher male expression was chemokine (C-X-C motif) ligand-2 (Cxcl2) (Fig. 1C). This literature-based prediction of gene expression profile strongly suggests that RelA is more highly activated in female liver than in male liver. To understand why the RelA pathway has higher basal activity, we assayed its expression at the protein level. We observed that females had a 40–50% higher expression of RelA at the protein level than did males (Fig. 1D). That IPA identified inflammatory pathways as being the top sexually dimorphic pathways is not surprising, given that we performed our analysis on a focused set of inflammatory genes rather than using a genome-wide approach. Taken together, these results indicate that significant components of the inflammatory pathway in the liver are influenced by sex. Furthermore, the female liver is predicted to have a higher inflammatory milieu than male liver based on our in silico analysis of gene expression data sets. Of note, most of the genes assayed under homeostatic conditions were expressed at low levels, likely reflecting the absence of inflammation in male and female livers in the resting state. We speculate that the physiologic role of sexually dimorphic expression of hepatic inflammatory genes during the resting state is to prime the female liver to respond to inflammatory challenges (more robustly than in the male liver), rather than to promote inflammation in the resting female liver.
TABLE 1.
IPA of top hepatic sexually dimorphic pathways in mice
| Canonical pathway | P | Overlap |
|---|---|---|
| Role of pattern recognition receptors in recognition of bacteria and viruses | 3.67E−26 | 15.4 (18/117) |
| Role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis | 6.06E−22 | 6.9 (20/288) |
| Complement system | 8.99E−14 | 24.2 (8/33) |
| TREM1 signaling | 8.14E−13 | 13.2 (9/68) |
| NF-κB signaling | 3.90E−12 | 6.7 (11/64) |
Data are expressed as percentage of total group (number of genes affected/total number of genes in the group).
LPS challenge leads to sexually dimorphic hepatic responses in mice
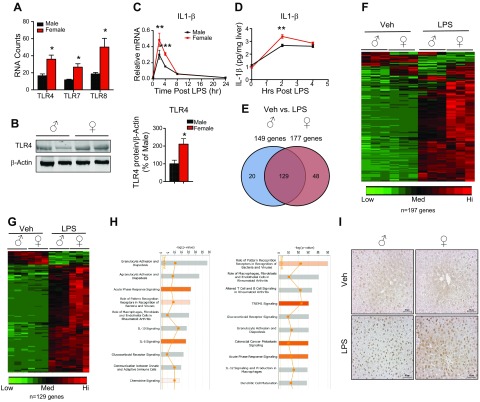
Recognizing that female mice had a higher hepatic inflammatory milieu than male mice in basal conditions prompted us to ask whether this phenomenon would lead to functional differences in inflammatory responses between the sexes. To investigate this, we focused on the “recognition of bacteria and viruses by pattern-recognition receptors,” pathway, as it was the sexually dimorphic pathway ranked number 1 by IPA of the nanostring inflammatory gene codeset. Pattern recognition receptors are a family of proteins including TLRs, RIG (retinoic acid-inducible gene)-like receptors, NOD (nucleotide-binding oligomerization domain)-like receptors and C-type lectin receptors, which bind to pathogen-associated molecular patterns (PAMPs), eliciting an inflammatory response to eliminate foreign pathogens [reviewed in Takeuchi and Akira (32)]. Of the 9 TLR family members assayed, we observed that TLR4, -7, and -8 are sexually dimorphic in expression, with all 3 being more highly expressed in female mice (Fig. 2A). We further confirmed the higher expression of TLR4 in the female via qPCR (Supplemental Fig. 1A). We did not observe sexually dimorphic expression of the other 6 TLR family members or of the TLR adaptor molecular MyD88 (Supplemental Fig. 1F, G). Furthermore, we confirmed sexually dimorphic expression of TLR4 at the protein level via Western blot analysis and found that female mice expressed higher levels of TLR4 protein in liver lysates than did males (Fig. 2B).
Figure 2.
Male and female mount unique hepatic inflammatory responses after LPS challenge. A) RNA counts from nanostring analysis of TLR4, -7, and -8 expression in male and female liver. Data are RNA counts (means ± sem, n = 3 animals). B) Western blot analysis of TLR4 in male and female liver homogenates. TLR4 protein was normalized to β-actin and expressed as a percentage of that in male liver (n = 4 animals). C) Quantitative PCR for IL-1β in male and female mouse liver after 2, 4, 8, and 24 h LPS (5 mg/kg) treatment. Data are expressed as relative mRNA to PPIB (means ± sem, n = 3–5 animals). D) IL-1β ELISA in liver homogenates from male and female LPS-treated mice. Data are expressed as IL-1β pg/mg of liver tissue (means ± sem, n = 3–4 animals). E) Venn diagram of genes significantly altered by LPS in male and female mice. F) Heat map of nanostring counts from male and female mice treated with LPS for 4 h. Red indicates high-expressing genes whereas green indicates low-expressing genes. G) Heat map of nanostring counts from the 129 commonly regulated genes in male and female in response to LPS. H) Pathway analysis of the 10 canonical pathways most regulated by LPS in male and female mouse liver. I) Immunohistochemical analysis of GR in paraffin-embedded liver sections from male and female mice treated with LPS. Brown immunoreactivity represents GR. Scale bars, 50 μm. *P < 0.05, **P < 0.01, ***P < 0.001 vs. male.
To explore the functional consequences of higher TLR4 expression on gene expression, we injected male and female mice with a nonlethal dose of the TLR4 ligand LPS for 2, 4, 8, and 24 h and recorded the IL-1β mRNA and protein detected in liver homogenates via qPCR and ELISA, respectively. We observed higher expression of IL-1β at the transcript level in the female liver, but we did not observe a sexually dimorphic expression of IL-1β at the protein level under basal conditions (Fig. 2C). We suggest that IL-1β is sexually dimorphic in expression at the transcriptional level in the liver; however, the dimorphism does not result in increased protein expression caused by translational repression (33). In fact, regulation of expression at the translational level has been thought to be a major mode of regulating inflammatory gene and protein expression in the innate immune system [(34), reviewed in Carpenter et al. (35)]. Nevertheless, both male and female mice robustly induced IL-1β message and protein after LPS treatment (Fig. 2C, D). Both IL-1β message and protein were more highly induced in the female liver at 2 h after LPS injection, suggesting a higher sensitivity to LPS-induced inflammation in female mice. The more robust acute response of IL-1β protein in female mice challenged with LPS at 2 h was probably caused by the higher basal levels of IL-1β mRNA. These findings suggest a priming mechanism in females in the homeostatic condition that responds more rapidly to an inflammatory stimulus. Moreover, the higher LPS induction of IL-1β mRNA indicates that, in fact, the IL-1β loci are more transcriptionally active in females during both basal and inflammatory conditions.
We next explored the extent of the sex-biased gene expression elicited by LPS by performing nanostring analysis of male and female mice treated with vehicle or LPS for 4 h. Our initial observation was that females had more significantly altered genes than did males after LPS treatment (177 genes in females; 149 in males) (Fig. 2E). To determine whether LPS elicits a sexually distinct response, we performed a 2-way comparison of genes significantly altered by LPS in male and female mice. LPS altered 129 genes that were present in common in both sexes. The males were sensitized to regulate a cohort of 20 additional genes, whereas 48 LPS-regulated genes were found in the females. The male-sensitive genes included chemokines such as Ccl21 and the endoplasmic reticulum stress sensor DNA damage-inducible transcript (Ddit)-3 (Supplemental Fig. 2B), whereas the female-sensitive genes included a variety of proinflammatory mediators such as IL-18 receptor accessory protein (IL-18RAP) and NADPH oxidase 1 (Nox1) (Supplemental Fig. 2C). The overall directionality of gene regulation in response to LPS was determined by generating heat maps of nanostring RNA counts, which revealed that both male and female mice induced the majority (∼90%) of the inflammatory genes after 4 h of LPS administration and repressed only a small fraction (Fig. 2F); however, expression of the most commonly regulated genes was induced to a greater extent in the female mice (Fig. 2G; Supplemental Fig. 2A). These findings suggest that higher TLR4 expression in female mice provides increased sensitivity to LPS, leading to a more robust acute response to an LPS challenge.
To determine whether sex-biased genes regulated by LPS administration reside in functional networks, we performed IPA on the significantly regulated genes by LPS administration in male and female mice. Examination of the top 10 canonical pathways altered by LPS in male and female mice indicated that the sex-specific responses to LPS could be categorized into functional pathways. For example, males show selective regulation of granulocyte and agranulycte adhesion and diapedesis, whereas females specifically had altered TREM1 and GR signaling (Fig. 2H). It was not surprising to see that the GR pathway was activated after LPS injection, as several rodent models of endotoxemia have shown acute activation of the hypothalamic–pituitary–adrenal axis, leading to an increase in circulating corticosteroids (36–40). However, we found it intriguing that the GR pathway was ranked 8 on our LPS-regulated pathways in male mice, whereas it was ranked 5 in the females. Given that IPA identified the GR pathway to be differentially regulated between male and female mice after LPS administration, we wanted to know whether this effect is caused by differential activation of GR. To achieve this goal, we performed immunohistochemistry for GR in male and female mice treated with either vehicle or LPS for 4 h. We observed faint nuclear and cytoplasmic staining of GRs in both male and female vehicle-treated mice (Fig. 2I). Upon LPS administration, however, we observed an increased nuclear signal of GR in livers of both males and females. Of note, the staining intensity of GR was higher in males treated with LPS than in LPS-treated females. In contrast to males, in which almost all GR signal was in the nucleus after LPS injection, we observed the presence of cytoplasmic staining of GR in females treated with LPS. These data indicate that GR is more strongly activated in males than in females after an inflammatory stimulus. To determine whether the higher degree of nuclear GR staining induced by LPS was caused by differences in ligand availability, we measured circulating corticosterone in male and female mice after LPS injection. Although the amount of circulating corticosterone was robustly up-regulated in both males and females after LPS injection, we did not observe any sex differences in corticosterone secretion in response to LPS (Supplemental Fig. 2D). These findings indicate that the female mice exhibited a more robust acute hepatic inflammatory profile after LPS treatment than did the males and that perhaps the differential regulation of inflammatory genes is mediated by GR.
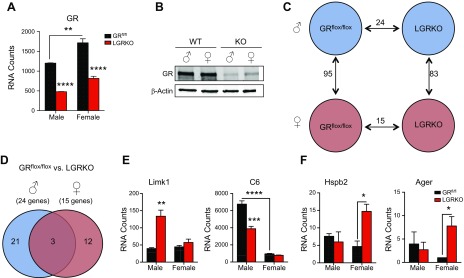
Hepatic GR deficiency alters basal inflammatory gene expression in a sex-specific manner
Given the observation of a higher level of nuclear GR in males in response to LPS, we wanted to determine whether GR has a role in the phenomenon of sexually dimorphic hepatic inflammatory gene expression. We generated LGRKO mice by crossing GRflox/flox mice (25) with CRE+ mice, driven by the liver-specific albumin promoter (41). Both male and female LGRKO mice had an ∼60% reduction in GR mRNA via nanostring analysis (Fig. 3A), an ∼80% reduction according to real-time qPCR (Supplemental Fig.1B), and an ∼80% decrease in GR protein (Fig. 3B). Furthermore, when liver expression of AR and ER was assayed in LGRKO mice, we observed no significant changes in sex hormone receptor expression in response to hepatic GR ablation (Supplemental Fig. 3A). Consistent with our previous findings, a large degree of sexual dimorphism in basal expression of inflammatory genes between GRflox/flox male and GRflox/flox female control mice (95 genes) was observed (Fig. 3C). We found a 62% overlap of sexually dimorphic inflammatory genes in WT and GRflox/flox mice (Supplemental Fig. 3B). Moreover, the role of pattern recognition receptors in the recognition of bacteria and viruses’ pathway remained the sexually dimorphic pathway ranked 1 in GRflox/flox mice and in our LGRKO mice (Supplemental Fig. 3B, C). Twenty-four genes were differentially expressed in male GRflox/flox vs. male LGRKO mice, and female mice had 15 genes altered by hepatic GR deletion. To assess whether GR ablation in the liver leads to sex-specific changes in gene expression we performed a 2-way comparison of significantly altered genes in male and female LGRKO mice (Fig. 3D). Most of the genes dysregulated by hepatic GR deletion were sex-specific (21 genes in males and 12 genes in females) with only 3 genes being commonly dysregulated. For example, LIM domain kinase-1 (Limk1) was specifically derepressed in male LGRKO mice, whereas Limk1 expression in females was unaffected by GR ablation (Fig. 3E). GR deletion lowered the expression of the highly sexually dimorphic gene C6 in male mice only, rendering its expression in male LGRKO mice similar to that of control female mice. In contrast, all genes altered in female LGRKO mice were derepressed. The absence of hepatic GR signaling in females led to induction of heat shock 27 kDa protein (Hspb2) and advanced glycosylation end product–specific receptor (Ager) uniquely in females, with no changes observed in male LGRKO mice (Fig. 3F). These data indicate that GR targets sex-specific inflammatory genes in the livers of mice.
Figure 3.
Hepatic GR regulates sexually distinct inflammatory gene expression. A) RNA counts of GR in male and female GRflox/flox and LGRKO mice. Data are RNA counts (means ± sem, n = 3 animals). B) Western blot analysis of GR protein expression in male and female GRflox/flox and LGRKO mice. C) Nanostring analysis of male and female GRflox/flox and LGRKO mice showing significantly altered genes in each comparison. Differences in gene expression that reached P < 0.05 were significant. D) Venn diagram of nanostring results overlaying male GRflox/flox to male LGRKO and female GRflox/flox to female LGRKO mice. E) RNA counts of male-specific GR genes dysregulated by GR ablation. Data are represented as RNA counts (means ± sem, n = 3 animals). F) RNA counts of female-specific GR genes dysregulated by GR ablation. Data are represented as RNA counts (means ± sem, n = 3 animals). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
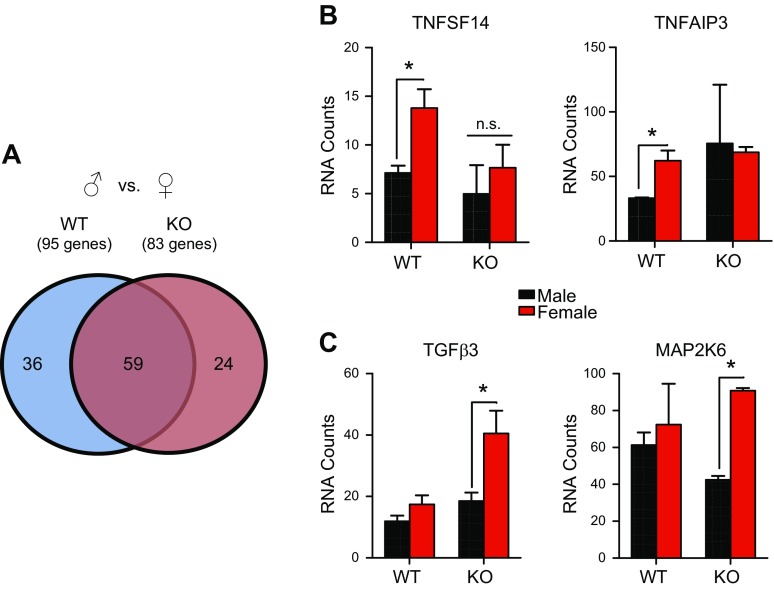
Based on these findings, we hypothesized that hepatic GR actively contributes to the phenomenon of sex-biased inflammatory gene expression. Deletion of GR reduced the number of inflammatory genes that displayed sexual dimorphism in the liver of mice (83 genes in LGRKO vs. 95 genes in GRflox/flox) (Fig. 4A). To characterize GR’s contribution to basal expression of sexually dimorphic genes, we compared the sexually dimorphic genes in control GRflox/flox mice with those exhibiting sexually dimorphism in LGRKO mice. We discovered unique sexually dimorphic genes in each genotype, as well as a common core set of sexually dimorphic genes (n = 29 genes). These genes were characterized into 3 functional categories. The first consisted of 59 genes that were sexually dimorphic independent of GR (overlapped area of Venn diagram). The second category comprised sexually dimorphic genes unique to GRflox/flox mice (left side of Venn diagram). This category represents genes that depend on the presence of GR to be expressed in a sexually dimorphic manner. For example, TNF superfamily, member 13 (Tnfsf14) and TNFα-induced protein-3 (Tnfaip3) mRNA transcripts were more highly expressed in the female liver, but GR ablation led to comparable expression of these genes in male and female livers (Fig. 4B). The last category consisted of 24 genes (right side of Venn diagram) that did not display sexually dimorphic expression in GRflox/flox mice but exhibited sex bias only in the absence of GR. This category represents de novo sexually dimorphic genes, such as TGFβ3 and MAPK kinase-6 (Map2k6), which become sexually dimorphic in expression only in the absence of GR signaling (Fig. 4C). TGFβ3 and Map2k6, along with most of the other genes in this category, were derepressed specifically in female LGRKO mice but not in male LGRKOs. These data indicate a suppressive function of these genes in female mice. Together, these findings revealed that hepatic GR regulates basal inflammatory gene expression in a sex-biased manner, to produce sexually distinct inflammatory milieus in male and female mouse livers. Furthermore, the role of liver-expressed GR in sexually dimorphic inflammatory gene expression is complex, with GR promoting sex differences of certain genes while inhibiting sexually dimorphic expression of other genes.
Figure 4.
A) Venn diagram of male GRflox/flox mice vs. female GRflox/flox and male LGRKO vs. female LGRKO. B) Genes that lose sexually dimorphic expression in the absence of GR. C) Genes that become sexually dimorphic in expression in LGRKO (n = 3 animals). n.s., not significant. *P < 0.05.
Hepatic GR signaling promotes sex-specific inflammatory responses to LPS
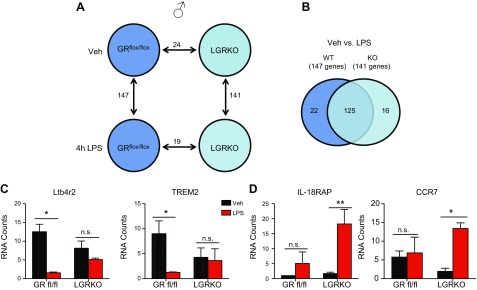
We next sought to find whether GR deletion leads to functional sex-specific alterations in gene expression after LPS challenge. LPS injection of 3-mo-old male control GRflox/flox mice for 4 h altered 147 genes (Fig. 5A), similar to the data observed in WT C57BL/6J mice (Fig. 2E). In male LGRKO mice, LPS treatment altered a comparable number of genes (n = 141) as was altered in the GRflox/flox mice (n = 147). A comparison of LPS-treated male GRflox/flox mice and LPS-treated LGRKO mice revealed 19 genes to be differentially expressed.
Figure 5.
GR regulates male-specific inflammatory programs during LPS challenge. A) Nanostring analysis of male GRflox/flox and LGRKO treated with either vehicle or LPS (5 mg/kg) for 4 h showing significantly regulated genes between various comparisons. B) Venn diagram of male GRflox/flox and LGRKO treated with vehicle or LPS. C) Male-specific glucocorticoid target genes regulated in GRflox/flox but not LGRKO in response to LPS challenge. D) Male-specific genes that become induced in response to LPS treatment in male LGRKO mice (n = 3 animals). n.s., not significant. *P < 0.05, **P < 0.01.
We determined whether GR deletion from the male liver alters the gene set regulated by LPS by performing 2-way comparison analysis of significantly altered genes after LPS treatment in male GRflox/flox mice and male LGRKO mice. The results revealed 125 genes commonly regulated in both male GRflox/flox and male LGRKO mice treated with LPS (Fig. 5B), whereas 22 genes required the presence of GR to be responsive to LPS. For example, leukotriene B4 receptor -2 (Ltb4r2) and Trem2 were down-regulated after LPS treatment of GRflox/flox mice but were unaltered in LGRKO mice treated with LPS (Fig. 5C). The requirement for GR for LPS-induced repression of Ltb4r2 was confirmed via real-time qPCR (Supplemental Fig.1C). These data suggest that GR negatively regulates these genes in vivo after LPS challenge. We also found genes that were not altered by LPS treatment in GRflox/flox animals that were significantly regulated in LPS-treated LGRKO mice (Fig. 5B). The LPS-regulated genes in the male LGRKO mice were proinflammatory mediators, such as IL-18RAP and chemokine (C-C motif) receptor-7 (Ccr7) (Fig. 5D) (Ccr7 was independently validated via quantitative real-time PCR; Supplemental Fig.1D), suggesting that GR, either directly or indirectly, blunted the induction of these genes during an inflammatory challenge in male mice.
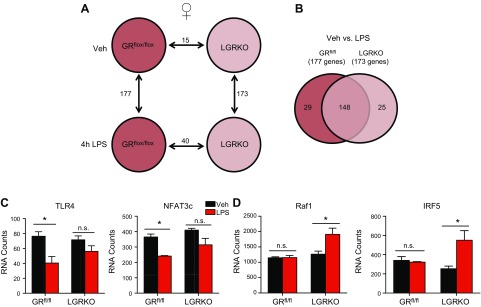
We characterized the effects of GR ablation on inflammatory gene expression regulated by LPS in female mice. Female GRflox/flox mice showed regulation of more genes after LPS treatment than did male GRflox/flox mice (177 vs. 147, respectively) (Figs. 5A and 6A), similar to WT C57BL/6J (Fig. 2E). Females showed regulation of a comparable number of genes in both the presence and absence of GR after LPS treatment (173 genes in GRflox/flox; 177 genes in LGRKO) (Fig. 6A). Of the LPS-regulated genes, 40 were differentially expressed in female GRflox/flox and LGRKO mice treated with LPS, twice as many as were observed in the males in the same comparison. Overlaying the list of LPS-regulated genes in female GRflox/flox mice with that of LPS-regulated genes in female LGRKO mice revealed that most of the genes were regulated in both control and LGRKO mice (148 genes; Fig. 6B, overlapping area of Venn diagram). Similar to our observations in males, a cohort of genes such as TLR4 and nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent-3 (Nfatc3) (Fig. 6C) required the presence of GR to be regulated in response to LPS in female mice (29 genes; Fig. 6B, left side of Venn diagram). Last, a cohort of genes that were typically not regulated by LPS in female mice, such as Raf1 and Irf5 (Fig. 6D), exhibited altered expression after LPS injection in the absence of GR (25 genes; Fig. 6B, right side of Venn diagram). The acquired induction of Irf5 after LPS in LGRKO was independently confirmed via real-time qPCR (Supplemental Fig. 1E). The genes requiring the presence of GR to be regulated during LPS challenge and the genes regulated by LPS in the absence of GR differed between the males and females (Figs. 5 and 6; Supplemental Fig. 4). The collective data indicate that GR has a pleiotropic effect on the regulation of sex-specific expression of inflammatory genes, with this pathway promoting sexually dimorphic expression of inflammatory gene expression on particular loci, while hindering the presence of sex-biased expression at other gene loci.
Figure 6.
GR regulates female-specific inflammatory programs during LPS challenge. A) Nanostring analysis of female GRflox/flox and LGRKO treated with either vehicle or LPS for 4 h, showing significantly regulated genes between various comparisons. B) Venn diagram of female GRflox/flox and LGRKO treated with vehicle or LPS. C) Female-specific glucocorticoid target genes regulated in GRflox/flox but not LGRKO in response to LPS challenge. D) Female-specific genes activated in response to LPS treatment LGRKO mice (n = 3 animals). n.s., not significant. *P < 0.05.
DISCUSSION
The liver is one of the most highly sexually dimorphic organs in regard to gene expression, but the molecular determinants of this phenomenon have yet to be fully elucidated. Several master regulators have been identified as contributors to sex-biased gene expression. The signal transducer and activator of transcription-5 (STAT5)/growth hormone axis is well appreciated for mediating sex-biased effects on gene expression because of its sexually dimorphic secretion pattern (42–45). Hepatic nuclear factor-4α (HNF4α), a liver-enriched transcription factor, is another master regulator of hepatic gene sexual dimorphism. Genetic deletion of this transcription factor in mice caused ∼50% of sexually dimorphic genes to be dysregulated in male mice, whereas only ∼10% of sex-biased genes were altered in female HNF4α-knockout mice (46). In our study, endogenous GR signaling also promoted sexually distinct inflammatory gene expression profiles.
Inflammation is a core component of cancer and autoimmunity, both sexually dimorphic diseases of the liver. This observation indicates that regulation of inflammatory gene expression is inherently different in males and females. Consistent with this notion, we found a high degree of sex-biased regulation of inflammatory gene expression in the livers of male and female mice. Nanostring technology showed that the female mouse liver exhibited a higher proinflammatory profile of gene expression than the male mouse liver in normal homeostatic conditions (Fig. 1). Uncovering the mechanisms responsible for this inflammatory phenotype in females is critical in understanding the susceptibility to autoimmune liver diseases in female. Because glucocorticoids are one of the primary endogenous regulators of inflammation, we hypothesized that this pathway targets inflammatory gene expression differentially in males and females and found sex-specific alterations in inflammatory gene expression in the absence of glucocorticoid signaling (Figs. 3, 4). The notion of differential glucocorticoid sensitivity in females could account for the higher expression and predicted activation of RelA/NF-κB that we observed in our in silico analysis, given that NF-κB is a well-known molecular target for glucocorticoid-mediated antagonism (47–49).
TLR4 has recently been implicated in several autoimmune diseases, such as rheumatoid arthritis (50), psoriasis (51), and Sjögren’s syndrome (52–54). Of the sexually dimorphic pathways we identified, the hepatic TLR4 system was highly sexually dimorphic in expression. The expression of several TLRs is sexually dimorphic in the mouse liver, consistent with previous findings in peritoneal macrophages (55, 56). Similar to macrophages, we found higher expression of TLR4 in female vs. male liver lysates; however, unlike in the macrophages we did not observe sex-biased expression of TLR2 or -3 but instead found higher expression of TLR7 and -8 in the female liver. Consistent with previous studies (55), we found that our female mice regulated more genes than males challenged with LPS (Fig. 2). Moreover, TLR4 was found to be a female, not a male, glucocorticoid target gene in the liver of mice challenged with LPS (Fig. 5). Of note, however, LPS has been shown to induce the expression of TNFα more potently in the lungs of male C57bl/6 mice (57), indicating that the sex-specific effects of LPS are tissue specific.
The mechanisms that drive sex-specific effects of glucocorticoids are still unclear, but we propose several candidate hypotheses. First is sex-specific activation of the GR. We observed a stronger nuclear translocation of the GR in males after administration of LPS compared to LPS-challenged female mice (Fig. 2). Although the difference was not caused by differential synthesis of glucocorticoids, given that we observed similar levels of circulating levels of corticosterone after LPS challenge, we cannot rule out differences in hepatic ligand availability. In fact, the enzyme responsible for converting inactive glucocorticoids to the active hormone 11β-HSD1 has been shown to be sexually dimorphic in expression in the rat liver, with males expressing higher levels (58). This sexually dimorphic expression of 11β-HSD1 is caused by the negative regulation of expression by the estrogen in females (59) and the effects of growth hormone (58).
Another candidate mechanism is that sex hormones inherent to males and females may converge on the GR pathway leading to sex-specific effects of glucocorticoids. Although little is known of the relationship between glucocorticoids and testicular hormones, there are several reports of ovarian hormone and GR crosstalk (22, 23, 60, 61). In the rat brain, glucocorticoids have been shown to bind to the GR at different affinities between males and females (22, 23, 60, 61). Differential binding of glucocorticoids is altered by ovariectomy, leading to an increase in binding sites in the hypothalamus compared with those in intact females (23). These studies may offer an explanation as to why we observed a stronger glucocorticoid translocation in males after LPS administration, compared with LPS-treated females, when equal levels of ligand were observed. Furthermore, in our past study, ovariectomy increased the efficacy of dexamethasone to improve survival in a rat model of LPS-induced endotoxemia (22). The ovary produces a myriad of hormones, predominantly estrogen and progesterone. Estradiol antagonizes glucocorticoid action in the mouse. Estradiol, via ERα blunts GR-mediated induction of glucocorticoid-induced leucine zipper (GILZ), a classic GR target gene that mediates many anti-inflammatory effects of glucocorticoids (60). Estradiol antagonism of glucocorticoid-mediated gene regulation has been further shown to occur, not only at the GILZ loci, but also on the global scale. Microarray analyses of human uterine epithelial cells indicate a plethora of genes involved in the inflammatory response are target genes of both dexamethasone and estradiol, and coadministration of these 2 hormones leads to dysregulated expression of these genes (61). Moreover, the presence of estradiol expands the GR transcriptome from ∼1800 to ∼5300 genes in human uterine epithelial cells. Whether estradiol treatment leads to alterations in the GR cistrome or GR uses the ER as a coactivator/corepressor remains to be elucidated, but this is an active area of research.
An alternative mechanism that drives sexually dimorphic actions of glucocorticoids may occur via crosstalk with sex-specific transcription factors and coregulators. B-cell CLL/lymphoma-6 (Bcl6), for example, is a male-specific corepressor (62). Bcl6 has been shown to promote sex-specific gene expression by targeting STAT5 genes, thereby giving rise to male-biased STAT5-mediated gene regulation (7). It is known that GR activation leads to induction of Bcl6 (63–65), suggesting that glucocorticoids have the potential to alter this male-specific pathway.
Our study indicates that GR contributes to sexually dimorphic gene expression in the mouse liver. The overwhelming number of females diagnosed with autoimmune hepatitis over males (7:1) makes it crucial to understand factors that may limit the efficacy of synthetic glucocorticoids in females. Understanding the molecular mechanisms responsible for this phenomenon could prove beneficial to the development of sex-specific corticosteroid therapies for the treatment of inflammatory diseases.
Supplementary Material
Acknowledgments
The authors thank Dr. D. W. Cain for technical assistance and critical reading of the manuscript and the U.S. National Institutes of Health (NIH) National Institute of Environmental Health Sciences (NIEHS) Genomics Core for help with the nanostring analysis. This research was supported by the Intramural Research Program of the NIH NIEHS. The authors declare no conflicts of interest.
Glossary
- AR
androgen receptor
- Bcl
B-cell CLL/lymphoma
- CRE
carbapenem-resistant Enterobacteriaceae
- CCR
chemokine (C-C motif) receptor
- DAB
3,3′-diaminobenzidine
- ER
estrogen receptor
- GILZ
glucocorticoid-induced leucine zipper
- GR
glucocorticoid receptor
- HNF
hepatic nuclear factor
- IL-18RAP
IL-18 receptor accessory protein
- LGRKO
liver-specific GR knockout (mice)
- LIMK
LIM domain kinase
- LTB4R
leukotriene B4 receptor
- MAPA2K
mitogen-activated protein kinase kinase
- NFATC
nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent
- PPIB
peptidylprolyl isomerase B
- qPCR
quantitative PCR
- SOCS
suppressor of cytokine signaling
- STAT
signal transducer and activator of transcription
- TREM1
triggering receptor expressed on myeloid cells-1
- WT
wild-type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1.Yamamoto Y., Saito H., Setogawa T., Tomioka H. (1991) Sex differences in host resistance to Mycobacterium marinum infection in mice. Infect. Immun. 59, 4089–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamason R., Zhao P., Rawat R., Davis A., Hall J. C., Chae J. J., Agarwal R., Cohen P., Rosen A., Hoffman E. P., Nagaraju K. (2006) Sexual dimorphism in immune response genes as a function of puberty. BMC Immunol. 7, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghazeeri G., Abdullah L., Abbas O. (2011) Immunological differences in women compared with men: overview and contributing factors. Am. J. Reprod. Immunol. 66, 163–169 [DOI] [PubMed] [Google Scholar]
- 4.Mittelstrass K., Ried J. S., Yu Z., Krumsiek J., Gieger C., Prehn C., Roemisch-Margl W., Polonikov A., Peters A., Theis F. J., Meitinger T., Kronenberg F., Weidinger S., Wichmann H. E., Suhre K., Wang-Sattler R., Adamski J., Illig T. (2011) Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 7, e1002215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittendorfer B. (2005) Sexual dimorphism in human lipid metabolism. J. Nutr. 135, 681–686 [DOI] [PubMed] [Google Scholar]
- 6.Shen W., Punyanitya M., Silva A. M., Chen J., Gallagher D., Sardinha L. B., Allison D. B., Heymsfield S. B. (2009) Sexual dimorphism of adipose tissue distribution across the lifespan: a cross-sectional whole-body magnetic resonance imaging study. Nutr. Metab. (Lond.) 6, 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Laz E. V., Waxman D. J. (2012) Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol. 32, 880–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro B. H., Agrawal A. K., Pampori N. A. (1995) Gender differences in drug metabolism regulated by growth hormone. Int. J. Biochem. Cell Biol. 27, 9–20 [DOI] [PubMed] [Google Scholar]
- 9.Lahita R. G. (1999) The role of sex hormones in systemic lupus erythematosus. Curr. Opin. Rheumatol. 11, 352–356 [DOI] [PubMed] [Google Scholar]
- 10.Kovacs W. J., Olsen N. J. (2011) Sexual dimorphism of RA manifestations: genes, hormones and behavior. Nat. Rev. Rheumatol. 7, 307–310 [DOI] [PubMed] [Google Scholar]
- 11.McFarlane I. G., Heneghan M. A. (2004) Autoimmunity and the female liver. Hepatol. Res. 28, 171–176 [DOI] [PubMed] [Google Scholar]
- 12.Ishikawa I., Maeda K., Nakai S., Kawaguchi Y. (2000) Gender difference in the mean age at the induction of hemodialysis in patients with autosomal dominant polycystic kidney disease. Am. J. Kidney Dis. 35, 1072–1075 [DOI] [PubMed] [Google Scholar]
- 13.Bubb K. J., Khambata R. S., Ahluwalia A. (2012) Sexual dimorphism in rodent models of hypertension and atherosclerosis. Br. J. Pharmacol. 167, 298–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El-Serag H. B., Rudolph K. L. (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132, 2557–2576 [DOI] [PubMed] [Google Scholar]
- 15.Oakley R. H., Cidlowski J. A. (2011) Cellular processing of the glucocorticoid receptor gene and protein: new mechanisms for generating tissue-specific actions of glucocorticoids. J. Biol. Chem. 286, 3177–3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oakley R. H., Cidlowski J. A. (2013) The biology of the glucocorticoid receptor: new signaling mechanisms in health and disease. J. Allergy Clin. Immunol. 132, 1033–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quax R. A., Manenschijn L., Koper J. W., Hazes J. M., Lamberts S. W., van Rossum E. F., Feelders R. A. (2013) Glucocorticoid sensitivity in health and disease. Nat. Rev. Endocrinol. 9, 670–686 [DOI] [PubMed] [Google Scholar]
- 18.Clark A. R., Belvisi M. G. (2012) Maps and legends: the quest for dissociated ligands of the glucocorticoid receptor. Pharmacol. Ther. 134, 54–67 [DOI] [PubMed] [Google Scholar]
- 19.Beck I. M., Vanden Berghe W., Vermeulen L., Yamamoto K. R., Haegeman G., De Bosscher K. (2009) Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr. Rev. 30, 830–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyata M., Lee J. Y., Susuki-Miyata S., Wang W. Y., Xu H., Kai H., Kobayashi K. S., Flavell R. A., Li J. D. (2015) Glucocorticoids suppress inflammation via the upregulation of negative regulator IRAK-M. Nat. Commun. 6, 6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhen T., Cidlowski J. A. (2005) Antiinflammatory action of glucocorticoids: new mechanisms for old drugs. N. Engl. J. Med. 353, 1711–1723 [DOI] [PubMed] [Google Scholar]
- 22.Duma D., Collins J. B., Chou J. W., Cidlowski J. A. (2010) Sexually dimorphic actions of glucocorticoids provide a link to inflammatory diseases with gender differences in prevalence. Sci. Signal. 3, ra74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner B. B., Weaver D. A. (1985) Sexual dimorphism of glucocorticoid binding in rat brain. Brain Res. 343, 16–23 [DOI] [PubMed] [Google Scholar]
- 24.Elaković I., Djordjevic A., Adzic M., Djordjevic J., Radojčić M., Matić G. (2011) Gender-specific response of brain corticosteroid receptors to stress and fluoxetine. Brain Res. 1384, 61–68 [DOI] [PubMed] [Google Scholar]
- 25.Oakley R. H., Ren R., Cruz-Topete D., Bird G. S., Myers P. H., Boyle M. C., Schneider M. D., Willis M. S., Cidlowski J. A. (2013) Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proc. Natl. Acad. Sci. USA 110, 17035–17040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinn M., Ueno Y., Pae H. Y., Huang L., Frampton G., Galindo C., Francis H., Horvat D., McMillin M., Demorrow S. (2012) Suppression of the HPA axis during extrahepatic biliary obstruction induces cholangiocyte proliferation in the rat. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G182–G193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cidlowski J. A., Bellingham D. L., Powell-Oliver F. E., Lubahn D. B., Sar M. (1990) Novel antipeptide antibodies to the human glucocorticoid receptor: recognition of multiple receptor forms in vitro and distinct localization of cytoplasmic and nuclear receptors. Mol. Endocrinol. 4, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 28.Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T., James J. J., Maysuria M., Mitton J. D., Oliveri P., Osborn J. L., Peng T., Ratcliffe A. L., Webster P. J., Davidson E. H., Hood L., Dimitrov K. (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317–325 [DOI] [PubMed] [Google Scholar]
- 29.Reis P. P., Waldron L., Goswami R. S., Xu W., Xuan Y., Perez-Ordonez B., Gullane P., Irish J., Jurisica I., Kamel-Reid S. (2011) mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 11, 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malkov V. A., Serikawa K. A., Balantac N., Watters J., Geiss G., Mashadi-Hossein A., Fare T. (2009) Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res. Notes 2, 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan M., Vaes E., Mombaerts P. (2011) Regulation of the probability of mouse odorant receptor gene choice. Cell 147, 907–921 [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 [DOI] [PubMed] [Google Scholar]
- 33.Radwan M., Stiefvater R., Grunert T., Sharif O., Miller I., Marchetti-Deschmann M., Allmaier G., Gemeiner M., Knapp S., Kovarik P., Müller M., Strobl B. (2010) Tyrosine kinase 2 controls IL-1ß production at the translational level. J. Immunol. 185, 3544–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schott J., Reitter S., Philipp J., Haneke K., Schäfer H., Stoecklin G. (2014) Translational regulation of specific mRNAs controls feedback inhibition and survival during macrophage activation. PLoS Genet. 10, e1004368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carpenter S., Ricci E. P., Mercier B. C., Moore M. J., Fitzgerald K. A. (2014) Post-transcriptional regulation of gene expression in innate immunity. Nat. Rev. Immunol. 14, 361–376 [DOI] [PubMed] [Google Scholar]
- 36.Hoekstra M., Frodermann V., van den Aardweg T., van der Sluis R. J., Kuiper J. (2013) Leukocytosis and enhanced susceptibility to endotoxemia but not atherosclerosis in adrenalectomized APOE knockout mice. PLoS One 8, e80441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Q. H., Hruby V. J., Tatro J. B. (1998) Systemic alpha-MSH suppresses LPS fever via central melanocortin receptors independently of its suppression of corticosterone and IL-6 release. Am. J. Physiol. 275, R524–R530 [DOI] [PubMed] [Google Scholar]
- 38.Kinoshita P. F., Yshii L. M., Vasconcelos A. R., Orellana A. M., Lima Lde. S., Davel A. P., Rossoni L. V., Kawamoto E. M., Scavone C. (2014) Signaling function of Na,K-ATPase induced by ouabain against LPS as an inflammation model in hippocampus. J. Neuroinflammation 11, 218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Silverman M. N., Mukhopadhyay P., Belyavskaya E., Tonelli L. H., Revenis B. D., Doran J. H., Ballard B. E., Tam J., Pacher P., Sternberg E. M. (2013) Glucocorticoid receptor dimerization is required for proper recovery of LPS-induced inflammation, sickness behavior and metabolism in mice. Mol. Psychiatry 18, 1006–1017 [DOI] [PubMed] [Google Scholar]
- 40.Hansen M. K., Nguyen K. T., Fleshner M., Goehler L. E., Gaykema R. P., Maier S. F., Watkins L. R. (2000) Effects of vagotomy on serum endotoxin, cytokines, and corticosterone after intraperitoneal lipopolysaccharide. Am. J. Physiol. Regul. Integr. Comp. Physiol. 278, R331–R336 [DOI] [PubMed] [Google Scholar]
- 41.Postic C., Shiota M., Niswender K. D., Jetton T. L., Chen Y., Moates J. M., Shelton K. D., Lindner J., Cherrington A. D., Magnuson M. A. (1999) Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 [DOI] [PubMed] [Google Scholar]
- 42.Jansson J. O., Edén S., Isaksson O. (1985) Sexual dimorphism in the control of growth hormone secretion. Endocr. Rev. 6, 128–150 [DOI] [PubMed] [Google Scholar]
- 43.Udy G. B., Towers R. P., Snell R. G., Wilkins R. J., Park S. H., Ram P. A., Waxman D. J., Davey H. W. (1997) Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl. Acad. Sci. USA 94, 7239–7244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laz E. V., Wiwi C. A., Waxman D. J. (2004) Sexual dimorphism of rat liver nuclear proteins: regulatory role of growth hormone. Mol. Cell. Proteomics 3, 1170–1180 [DOI] [PubMed] [Google Scholar]
- 45.Waxman D. J. (2000) Growth hormone pulse-activated STAT5 signalling: a unique regulatory mechanism governing sexual dimorphism of liver gene expression. Novartis Found. Symp. 227, 61–74, discussion 75–81 [DOI] [PubMed] [Google Scholar]
- 46.Holloway M. G., Miles G. D., Dombkowski A. A., Waxman D. J. (2008) Liver-specific hepatocyte nuclear factor-4alpha deficiency: greater impact on gene expression in male than in female mouse liver. Mol. Endocrinol. 22, 1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caldenhoven E., Liden J., Wissink S., Van de Stolpe A., Raaijmakers J., Koenderman L., Okret S., Gustafsson J. A., Van der Saag P. T. (1995) Negative cross-talk between RelA and the glucocorticoid receptor: a possible mechanism for the antiinflammatory action of glucocorticoids. Mol. Endocrinol. 9, 401–412 [DOI] [PubMed] [Google Scholar]
- 48.Ray A., Prefontaine K. E. (1994) Physical association and functional antagonism between the p65 subunit of transcription factor NF-kappa B and the glucocorticoid receptor. Proc. Natl. Acad. Sci. USA 91, 752–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scheinman R. I., Gualberto A., Jewell C. M., Cidlowski J. A., Baldwin A. S. Jr (1995) Characterization of mechanisms involved in transrepression of NF-kappa B by activated glucocorticoid receptors. Mol. Cell. Biol. 15, 943–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abdollahi-Roodsaz S., Joosten L. A., Koenders M. I., Devesa I., Roelofs M. F., Radstake T. R., Heuvelmans-Jacobs M., Akira S., Nicklin M. J., Ribeiro-Dias F., van den Berg W. B. (2008) Stimulation of TLR2 and TLR4 differentially skews the balance of T cells in a mouse model of arthritis. J. Clin. Invest. 118, 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Rodriguez S., Arias-Santiago S., Perandrés-López R., Castellote L., Zumaquero E., Navarro P., Buendía-Eisman A., Ruiz J. C., Orgaz-Molina J., Sancho J., Zubiaur M. (2013) Increased gene expression of Toll-like receptor 4 on peripheral blood mononuclear cells in patients with psoriasis. J. Eur. Acad. Dermatol. Venereol. 27, 242–250 [DOI] [PubMed] [Google Scholar]
- 52.Kawakami A., Nakashima K., Tamai M., Nakamura H., Iwanaga N., Fujikawa K., Aramaki T., Arima K., Iwamoto N., Ichinose K., Kamachi M., Ida H., Origuchi T., Eguchi K. (2007) Toll-like receptor in salivary glands from patients with Sjögren’s syndrome: functional analysis by human salivary gland cell line. J. Rheumatol. 34, 1019–1026 [PubMed] [Google Scholar]
- 53.Kwok S. K., Cho M. L., Her Y. M., Oh H. J., Park M. K., Lee S. Y., Woo Y. J., Ju J. H., Park K. S., Kim H. Y., Park S. H. (2012) TLR2 ligation induces the production of IL-23/IL-17 via IL-6, STAT3 and NF-kB pathway in patients with primary Sjogren’s syndrome. Arthritis Res. Ther. 14, R64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spachidou M. P., Bourazopoulou E., Maratheftis C. I., Kapsogeorgou E. K., Moutsopoulos H. M., Tzioufas A. G., Manoussakis M. N. (2007) Expression of functional Toll-like receptors by salivary gland epithelial cells: increased mRNA expression in cells derived from patients with primary Sjögren’s syndrome. Clin. Exp. Immunol. 147, 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scotland R. S., Stables M. J., Madalli S., Watson P., Gilroy D. W. (2011) Sex differences in resident immune cell phenotype underlie more efficient acute inflammatory responses in female mice. Blood 118, 5918–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calippe B., Douin-Echinard V., Delpy L., Laffargue M., Lélu K., Krust A., Pipy B., Bayard F., Arnal J. F., Guéry J. C., Gourdy P. (2010) 17Beta-estradiol promotes TLR4-triggered proinflammatory mediator production through direct estrogen receptor alpha signaling in macrophages in vivo. J. Immunol. 185, 1169–1176 [DOI] [PubMed] [Google Scholar]
- 57.Card J. W., Carey M. A., Bradbury J. A., DeGraff L. M., Morgan D. L., Moorman M. P., Flake G. P., Zeldin D. C. (2006) Gender differences in murine airway responsiveness and lipopolysaccharide-induced inflammation. J. Immunol. 177, 621–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Low S. C., Chapman K. E., Edwards C. R., Wells T., Robinson I. C., Seckl J. R. (1994) Sexual dimorphism of hepatic 11 beta-hydroxysteroid dehydrogenase in the rat: the role of growth hormone patterns. J. Endocrinol. 143, 541–548 [DOI] [PubMed] [Google Scholar]
- 59.Low S. C., Assaad S. N., Rajan V., Chapman K. E., Edwards C. R., Seckl J. R. (1993) Regulation of 11 beta-hydroxysteroid dehydrogenase by sex steroids in vivo: further evidence for the existence of a second dehydrogenase in rat kidney. J. Endocrinol. 139, 27–35 [DOI] [PubMed] [Google Scholar]
- 60.Whirledge S., Cidlowski J. A. (2013) Estradiol antagonism of glucocorticoid-induced GILZ expression in human uterine epithelial cells and murine uterus. Endocrinology 154, 499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whirledge S., Xu X., Cidlowski J. A. (2013) Global gene expression analysis in human uterine epithelial cells defines new targets of glucocorticoid and estradiol antagonism. Biol. Reprod. 89, 66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer R. D., Laz E. V., Su T., Waxman D. J. (2009) Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol. 23, 1914–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddy T. E., Pauli F., Sprouse R. O., Neff N. F., Newberry K. M., Garabedian M. J., Myers R. M. (2009) Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 19, 2163–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sevilla L. M., Bayo P., Latorre V., Sanchis A., Pérez P. (2010) Glucocorticoid receptor regulates overlapping and differential gene subsets in developing and adult skin. Mol. Endocrinol. 24, 2166–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stojadinovic O., Lee B., Vouthounis C., Vukelic S., Pastar I., Blumenberg M., Brem H., Tomic-Canic M. (2007) Novel genomic effects of glucocorticoids in epidermal keratinocytes: inhibition of apoptosis, interferon-gamma pathway, and wound healing along with promotion of terminal differentiation. J. Biol. Chem. 282, 4021–4034 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.