Abstract
This study was designed to identify significant differences in gene expression profiles of human papillomavirus (HPV)‐positive and HPV‐negative oropharyngeal squamous cell carcinomas (OPSCC) and to better understand the functional and biological effects of HPV infection in the premalignant pathway. Twenty‐four consecutive patients with locally advanced primary OPSCC were included in a prospective clinical trial. Fresh tissue samples (tumor vs. matched normal epithelium) were subjected to whole transcriptome analysis and the results validated on the same cohort with RT–quantitative real‐time PCR. In a separate retrospective cohort of 27 OPSCC patients, laser capture microdissection of formalin‐fixed, paraffin‐embedded tissue allowed RNA extraction from adjacent regions of normal epithelium, carcinoma in situ (premalignant) and invasive SCC tissue. The majority of patients showed evidence of high‐risk HPV16 positivity (80.4%). Predictable fold changes of RNA expression in HPV‐associated disease included multiple transcripts within the p53 oncogenic pathway (e.g. CDKN2A/CCND1). Other candidate transcripts found to have altered levels of expression in this study have not previously been established (SFRP1, CRCT1, DLG2, SYCP2, and CRNN). Of these, SYCP2 showed the most consistent fold change from baseline in premalignant tissue; aberrant expression of this protein may contribute to genetic instability during HPV‐associated cancer development. If further corroborated, this data may contribute to the development of a non‐invasive screening tool. This study is registered with the UK Clinical Research Network (ref.: 11945).
Keywords: Diagnosis by tumor markers and biomarkers, human papillomavirus, mRNA expression analysis, oropharyngeal carcinoma, Rb/p16‐related genes
Human papillomavirus (HPV) is strongly associated with the development of oropharyngeal carcinoma. Although over 150 genotypes have been described, HPV16 is considered responsible for ~95% of viral associated cancers at this site.1
Epidemiological evidence from the USA would suggest that HPV‐associated oropharyngeal squamous cell carcinomas (OPSCC) is rising at an ever‐increasing rate.2 If this published trend continues, the annual number of viral‐associated OPSCC cases will surpass cervical cancers by the year 2020. Global cancer statistics reflect this situation with a rise in incidence predominantly affecting younger adult males from developed nations.3
The roles of the two HPV16 oncoproteins E6 and E7 have been studied extensively and include, among others, inhibition of p53 and pRb (retinoblastoma) tumor suppressor proteins.4 This situation is quite different to HPV‐negative (HPV−) oropharyngeal SCC, where an irreversible p53 mutation will normally be present and may contribute to the poorer clinical outcomes observed in this patient cohort.5, 6, 7 This information has provided the basis for several ongoing clinical trials that investigate de‐escalation treatment protocols in HPV‐positive (HPV+) disease.8, 9
Oropharyngeal squamous cell carcinogenesis involves progressive transformation of normal epithelium into premalignant tissue (dysplasia/carcinoma in situ) and, ultimately, invasive cancer.10, 11, 12, 13 Although the presence of HPV subtypes within invasive oropharyngeal SCC has been evaluated in large epidemiological studies,1, 14 there is limited data on this subject in regions of confirmed dysplasia/carcinoma in situ. Prior studies have reported markedly variable HPV prevalence rates due to limitations of size and variable assay techniques.12, 15 Jayaprakash et al. recently published a meta‐analysis of 22 relevant articles, suggesting HPV16 to be present in ~25% of all dysplastic lesions within the oropharyngeal subsite. The same author concluded this to be a conservative estimate due to the inclusion of oral cavity SCC lesions in some of the studies (traditionally a subsite with low HPV16 prevalence).13
The majority (~75%) of patients with HPV+ OPSCC present at an advanced stage (III/IV) due to cystic nodal disease.7, 16 In view of this, investigation of premalignant molecular pathways represents an important research priority, with the ultimate aim to produce a non‐invasive screening tool.17
Materials and Methods
Study population
This project received formal approval from the National Research Ethics Service Committee of East of England (12/EE/44). After informed consent, 24 consecutive patients with OPSCC donated multiple fresh biopsy samples (from regions of macroscopically normal and invasive tumor material) at Cambridge University Hospitals National Health Service Foundation Trust between June 2011 and July 2013. A further 27 OPSCC patients were included from a retrospective cohort and assessed for evidence of carcinoma in situ (dysplastic) change surrounding invasive carcinoma (Fig. S1). Disease stage was classified using the TNM classification of malignant tumors.18 Data from this study were deposited in the National Institutes of Health Gene Expression Omnibus database under accession code GSE56142. The trial protocol can be downloaded from the UK Clinical Research Network (http://public.ukcrn.org.uk).
In all prospective fresh biopsy samples, tumor and the adjacent normal tissue were processed for DNA and RNA extraction. A consultant histopathologist with expertise in head and neck pathology reviewed each sample to ensure representative sampling (minimum of 75% cancer cells for malignant tissue). Whole transcriptome analysis used the Illumina Genome Analyzer IIx machine (HumanHT‐12 version 4 BeadChip; Illumina, San Diego, CA, USA) and the results were validated with RT–quantitative real‐time PCR (RT‐qPCR) (ViiA 7; Applied Biosystems, Hampton, NH, USA).
The retrospective formalin‐fixed paraffin‐embedded (FFPE) tissue cohort was subjected to laser capture microdissection (LCM). This allowed precise RNA extraction from areas of invasive cancer, carcinoma in situ, and normal epithelial tissue to facilitate RT‐qPCR analysis.
Human papillomavirus stratification
Human papillomavirus stratification methods included: consensus PGMY PCR, type‐specific HPV16 DNA PCR, and E6/E7 mRNA PCR, DNA sequencing, p16INK4A immunohistochemistry (IHC), and HPV DNA in situ hybridization.
Prospective cohort
Oropharyngeal fresh tissue samples from normal and invasive malignant regions (maximum 25 mg) were DNA extracted using a protocol published from this unit.19 As previously described, L1 DNA PCR analysis of tumor DNA (50–100 ng) involved the PGMY09/11 primer set with all negative samples subjected to further amplification using GP5+/GP6+ primers.20, 21 DNA bands identified after agarose gel electrophoresis were excised, purified using QiaQuick Gel Extraction columns (Qiagen, Venlo, Netherlands, UK) and sequenced directly (Source Bioscience, Cambridge, UK). The E6/E7 DNA and cDNA PCR analysis involved primers specific for HPV16 E6/E7.22 For all fresh tissue biopsies, parallel FFPE samples enabled p16INK4a IHC (see below).
Retrospective cohort
The p16INK4a IHC was carried out on FFPE tissue using a mouse mAb (BD Biosciences, Franklin Lakes, NJ, USA).23 DNA in situ hybridization consisted of a probe directed against high‐risk HPV subtypes 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, and 66 (INFORM HPV III; Ventana, Tucson, AZ, USA).24 Genomic DNA was extracted from 9 × 3.5‐μm FFPE sections using a QIAamp tissue kit in accordance with manufacturer's guidelines (Qiagen). DNA was eluted in autoclaved and nuclease‐free H2O and stored at −20°C. Concentration and purity of DNA was assessed by spectrophotometry. Samples had an absorbance ratio (260/280 nm) in the range of 1.8–2.0, and were diluted with H2O to a concentration of 1–25 ng/μL prior to PCR. The L1 and E6/E7 DNA PCR was carried out as for fresh tissue samples (above).
Clinical and histopathological data for all prospective and retrospective subjects are shown in Figures 1 and S1, respectively.
Figure 1.
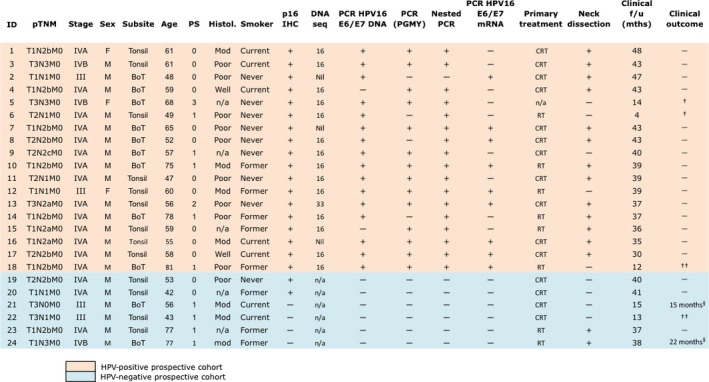
Clinical and histopathological data for the prospective cohort. Human papillomavirus (HPV)‐positive status was defined as evidence of HPV16 L1/E6/E7 DNA or HPV16 E6/E7 mRNA +/− p16INK 4A expression (>70% tumor cell staining). †Non‐malignant cause of death. ††Malignant cause of death. §Locoregional recurrence. BoT, base of tongue; CRT, chemoradiotherapy; F, female; f/u, follow‐up; Histol., histology; IHC, immunohistochemistry; M, male; Mod, moderately differentiated squamous cell carcinoma (SCC); mths, months; n/a, not applicable; Poor, poorly differentiated SCC; PS, Eastern Co‐Operative Group(48) physiological performance status; RT, radiotherapy; Well, well differentiated SCC.
RNA sequencing
The 24 OPSCC patients provided multiple fresh biopsy samples at the time of diagnostic or therapeutic surgery. All biopsies were selected on the basis of their RNA integrity number after histopathological review.
Messenger RNA‐seq cDNA libraries were prepared from ~400 ng total RNA. In brief, mRNA was isolated using polydT oligonucleotides connected to magnetic beads, fragmented using elevated temperature and divalent cations, and converted to cDNA using reverse transcriptase. DNA polymerase I and random primers were then used to convert single‐stranded cDNA into double‐stranded cDNA. This was blunt end repaired with Klenow DNA polymerase and T4 before adenylation of the 3′‐end of the fragment. A final purification step used gel electrophoresis, with fragments cut out in the range 200–300 bp. These fragments were amplified by PCR and sequenced using the Illumina Cluster Station and Genome Analyzer (Illumina). Paired‐end sequence analysis (51 cycles per end) was carried out with primers specific to the ends of the bridge‐amplified cDNA fragments to obtain 51 nucleotides of sequence from each end of all cDNA fragments.25
Each array on the HumanHT‐12 version 4 Expression BeadChip (GPL10558) targets more than 31 000 annotated genes with more than 47 323 oligonucleotide probes derived from the NCBI Reference Sequence Release 38 and other sources. Raw reads from normal epithelium (control) and tumor samples were processed using the GeneSifter Analysis Edition (Geospiza, Seattle, WA, USA) pipeline. Expression values for annotated genes were calculated from the aligned data by adding the number of reads linked to all exons and splicing events for a given gene and dividing that parameter value normalized by the total number of mapped reads in a sample. Two‐way anova identified target sequences with significant differential expression between normal and tumor tissue and further stratified by HPV status.26
Reverse transcription–qPCR
Validation of expression data by RT‐qPCR analysis used the SYBR green method with an Applied Biosystems ViiA 7 Fast Real‐time PCR system. Primers (Eurofins MWG Operon, Ebersberg, Germany) were optimized with β‐actin as a control gene and then with the transcript region of interest. When the optimal primer concentration produced a linear response to input cDNA concentration (range, <1–150 ng), samples were analyzed in triplicate for each tested transcript.
Statistical analysis
Statistical calculations were carried out using spss version 21 (SPSS, Chicago, IL, USA). To identify 80% of clinically relevant genes from the Illumina analysis, we based our power calculation on data supplied from Laborde et al.25 A minimal sample size of 10 subjects in each group was required if the false‐discovery rate (FDR) was set at 0.5% and the desired mean log2 fold change >1 (×10 change from baseline).27 Pearson's regression coefficient was used to investigate any correlation between the Illumina and RT‐qPCR analysis.28 Reverse transcription−qPCR data were analyzed by the 2−ΔΔCT technique, as described previously.29 In summary, the average Ct was derived for the three replicate analyses of the reference gene (β‐actin), and this was subtracted from the average Ct value from the three replicate analyses for the genes of interest. Expression differences between the HPV+ and HPV− tumors were compared using these normalized ΔCt values and the observed differences subjected to a Student's t‐test. Rates of disease‐free survival (DFS) were estimated by means of the Kaplan–Meier method and were compared by the log–rank test. A multivariate model was developed using Cox regression to investigate the effect of clinical factors on disease outcome (HPV16, SYCP2, p16INK4a, SFRP1, T stage, N stage, sex, physiological performance status, oropharyngeal subsite, histology grade, smoking, concurrent chemotherapy, and age).
Results
Human papillomavirus stratification
In total, 18/24 of the fresh biopsy samples (prospective cohort) and 23/27 of the FFPE samples (retrospective cohort) were classified as HPV+, defined by evidence of HPV16 L1/E6/E7 DNA, HPV16 E6/E7 mRNA, or HPV DNA in situ hybridization episomal/integrative staining pattern. Immunohistochemical analysis for expression of p16INK4a was shown for all HPV+ OPSCC samples but also present for 3 out of 10 OPSCC samples categorised as HPV− (Figs. 1,S1).
Gene expression differences with HPV status in OPSCC
Among the 47 323 oligonucleotide probes on the DNA microarray, 223 differentially expressed genes were statistically significant in classifying HPV+ versus HPV− OPSCC (P < 0.01; FDR, 0.5%; Figs. 2a,b,S2). As expected, one of the most significantly expressed genes in HPV+ tumor tissue was CDKN2A, which encodes for p16INK4A. This cellular protein may be upregulated as a result of oncogenic HPV E7 inhibiting activity of pRb.30 Other genes noted to have significant differential expression in the HPV+ group and potential relevance in malignant disease are highlighted in Figure S3.
Figure 2.
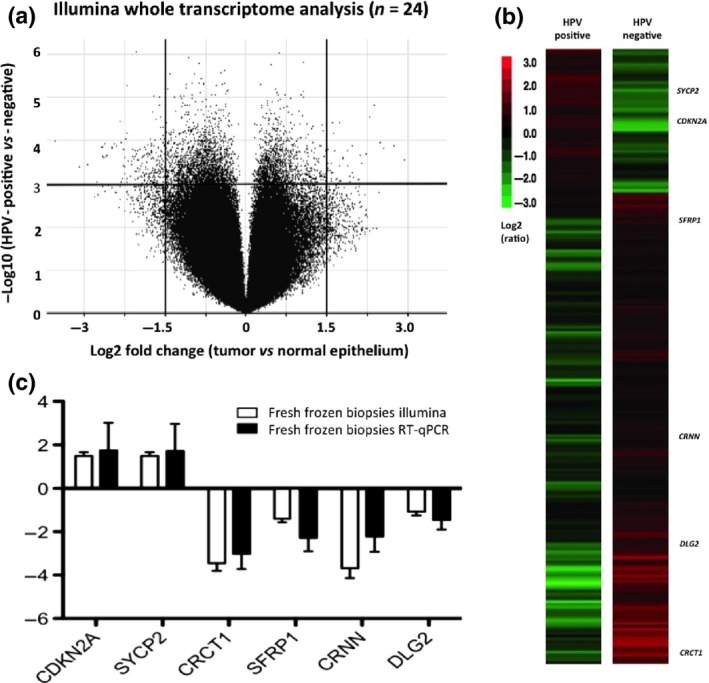
(a) Two‐way anova identified target sequences with significant differential expression between normal and tumor tissue and further stratified by human papillomavirus (HPV) status. In the volcano plot, the x‐axis is the log2 fold change value (tumor vs. normal epithelium) and the y‐axis is the −log10 odds value (HPV‐positive vs. HPV‐negative cohort). The two vertical lines represent 1.5 log2 (×15) fold change as the threshold cut‐off, both downregulated (left side) and upregulated (right side). The horizontal line represents a log odds value, P < 0.01 (false‐discovery rate of 0.5%), as the threshold cut‐off. (b) Hierarchical clustered heatmap of 223 genes displaying significant differential expression. As expected, CDKN2A ranked highly in the full transcriptome analysis. Five other genes that may have clinical relevance in HPV‐positive malignant disease are SYCP2,SFRP1, CRCT1, CRNN, and DLG2. (c) Validation graph for expression analysis (Illumina vs. RT–real‐time quantitative PCR [RT‐qPCR]; Pearson correlation coefficient, r = 0.905; P = 0.013; Kolmogorov–Smirnov test of normality, P > 0.10).
Reverse transcription–qPCR
A subset of differentially expressed genes from the Illumina platform analysis was confirmed by RT‐qPCR (selected on the basis of clinical relevance in malignant disease). The six target transcripts were: CDKN2A, SYCP2, SFRP1, DLG2, CRNN, and CRCT1. A high level of agreement existed between the Illumina and RT‐qPCR analysis (Pearson's correlation coefficient, r = 0.905; P‐value = 0.013; Kolmogorov–Smirnov test of normality, P > 0.10) (Fig. 2c).
In the prospective fresh tissue cohort (Fig. S4), HPV+ OPSCC expression levels of CDKN2A and SYCP2 were increased with an average log2 fold change of 1.7 (P < 0.01; 95% confidence interval [CI], 1.0–3.2) and 1.8 (P < 0.01; 95% CI, 1.35–3.35), respectively. Significant decreased expression was found for CRCT1 (log2, −3.0; P < 0.001; 95% CI, 2.2–4.4], SFRP1 (log2, −2.3; P < 0.01; 95% CI, 1.7–2.8), CRNN [log2, −2.2; P < 0.01; 95% CI, 1.7–2.9), and DLG2 (log2, −1.5; P < 0.01; 95% CI, 1.4–2.2].
Laser capture microdissection from the retrospective FFPE cohort allowed analysis of log2 fold change in adjacent regions of normal epithelium, invasive malignancy, and carcinoma in situ (Figs. 3a,S5,S6). The results largely reflect the analysis from fresh frozen samples for the HPV+ cohort; SYCP2 (log2 fold change, 3.1; P < 0.01; 95% CI, 1.8–4.4) and SFRP1 (log2 fold change, −0.97; 95% CI, −0.57–1.61) showed the largest differential expression from normal epithelium to premalignant (carcinoma in situ) tissue. In the same cohort, 70% of patients displayed significantly elevated expression of CDKN2A in the premalignant region (log2 fold change, >1.5). Figure 3(b) shows comparative data for both fresh and FFPE tissue designated within the HPV+ category.
Figure 3.
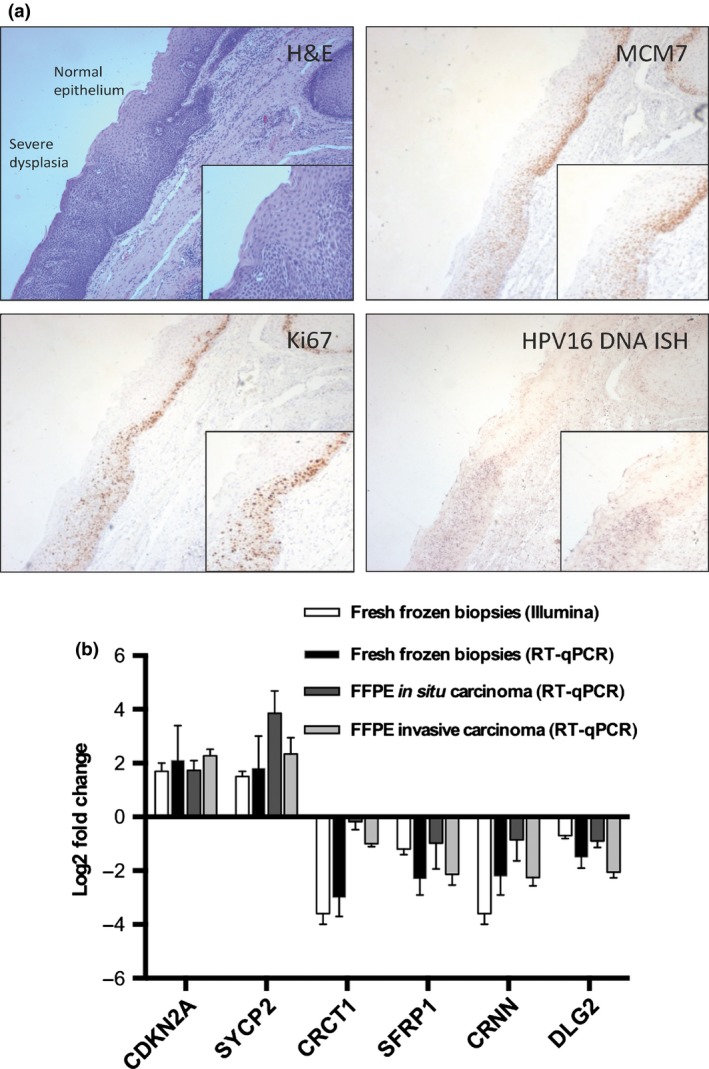
(a) H&E staining showing the junction between normal epithelium and severe dysplasia in human papillomavirus (HPV)‐positive tonsil squa‐mous cell carcinoma (top left). Although some flattening of cells at the apical surface may exist, the majority of the abnormal epithelium has a poorly differentiated dense basaloid appearance with nuclear pleomorphism. MCM7 (top right) and Ki67 (bottom left) staining are present in more than two‐thirds of the abnormal epithelium. In situ hybridization reveals an elevated level of HPV 16 DNA (bottom right) in the dysplastic epithelium when compared to the normal region (magnification, ×100; inset, ×150; Infinity capture software). ISH, in situ hybridization. (b) Reverse transcription–real‐time quantitative PCR (RT‐qPCR) analysis for all HPV‐positive sample types showing significant differential expression of SYCP2 (log2 fold change, 3.1 [95% confidence interval, 1.8–4.4]; P < 0.01) in premalig‐nant (in situ) tissue.
Clinical outcomes
The average DFS for all patients was 43.7 months (+/− SE, 2.5). For the HPV16+ cohort, the DFS period increased to 47.3 months (+/− SE, 2.3). Significant expression of SYCP2+ (log2 fold change, >1.5) and SFRP1+ (log2 fold change, <1.5) cohorts showed an average DFS period of 49.6 months (+/− SE, 2.2) and 40.1 months (+/− SE, 3.8), respectively. A multivariate proportional hazards model using Cox regression analysis revealed HPV16, SYCP2, smoking, and physiological performance status to have significant influence on DFS (Figs. 4,S7).
Figure 4.
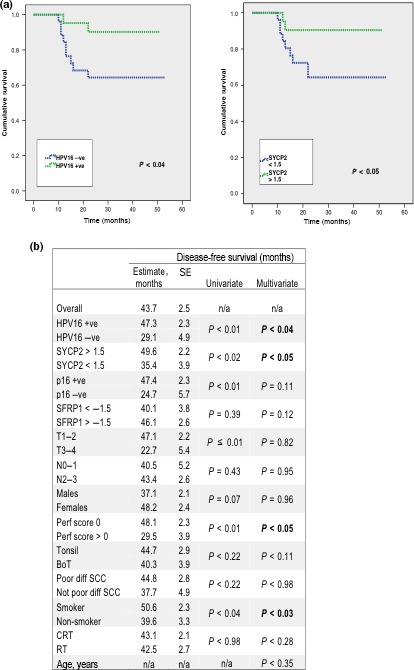
(a) Disease‐free survival calculations from Kaplan–Meier survival analysis of patients with human papillomavirus (HPV)‐positive oropharyngeal carcinoma. (b) A multivariate model was developed using Cox regression to investigate the effect of clinical parameters on disease‐free survival. Strati‐fication by HPV16, SYCP2, physiological perfor‐mance status, and smoking were all retained in the final model [bold text p < 0.05]. +ve, positive; −ve, negative; BoT, base of tongue; CRT, chemoradiotherapy; n/a, not applicable; SCC, squamous cell carcinoma; (SCC); mths, months; Perf, physiological performance status; Poor diff, poorly differentiated SCC; RT, radiotherapy.
To evaluate the prognostic accuracy of SYCP2 expression in regions of HPV+ in situ carcinoma, we constructed a receiver operating characteristic (ROC) curve (Figs. S8,S9). The area under the ROC curve was found to be 0.86 (+/− SE, 0.08; 95% CI, 0.71–0.99), indicating a good discriminating power when compared to control subjects. Sensitivity and specificity estimates over a range of cut‐off points suggest optimal results were obtained for the log2 fold range 1.5–3.0 (sensitivity, 70%; specificity, 95%). Similar testing of SFRP1 expression revealed the area under the ROC curve to be 0.64 (+/− SE, 0.11; 95% CI, 0.42–0.86), indicating a poor discriminating power when compared to controls.
Discussion
In this prospective observational study, we describe the use of mRNA massive parallel sequencing technology to investigate HPV+/− OPSCC tumors versus matched normal tissue. The data obtained were then validated by RT‐qPCR and the results used on retrospective FFPE tissue to investigate premalignant change surrounding areas of invasive carcinoma.
To our knowledge, comparatively few studies have investigated expression profiles in HPV‐associated OPSCC,29, 31, 32, 33 and even fewer have focused on precancer pathways.34 In particular, further characterization of a pre‐malignant state in the development of HPV‐associated OPSCC would be of clinical value as it infers the potential for a screening test (similar to the cervical carcinoma model). In HNSCC surgical excisions, dysplastic epithelium is often found adjacent to the cancer,10 but the exact nature of the disease in these regions is poorly defined.12, 13
Within our retrospective LCM cohort, 70% of patients showed significantly increased expression of CDKN2A (a proxy marker for HPV infection) in regions of carcinoma in situ relative to normal epithelium. This is higher than the HPV16 estimate (~25%) provided by Jayaprakash et al.13 and may be consistent with the hypothesis that HPV plays a significant role in the early phase of oropharyngeal carcinogenesis. Of course, high‐risk HPV infection can be a transient phenomenon and detection alone may not be sufficient to provide a causal association. Many studies have previously shown the presence of HPV subtypes even in normal oral cavity tissue.35, 36, 37 Gillison et al. (2012) carried out the largest epidemiological study on this topic (~6000 subjects), and estimated the prevalence for high‐risk HPV subtypes to be 6.9%, of which ~1% can be attributed to HPV16.38
It should still be noted that high‐risk HPV subtypes in normal oral cavity samples have significantly lower prevalence than the estimates reported by Jayaprakesh et al. (and largely confirmed in this present study). Available published reports also suggest a higher prevalence of HPV16 within areas of in situ malignant change when stratified by the male sex (×2 compared to females) and between areas of transformation from normal through to dysplastic epithelium (×3).13
In our prospective cohort of 24 OPSCC patients, RNA massive parallel sequencing data provided a statistically significant association for 223 target transcripts stratified by tumor and HPV status. Gene ontology data revealed the majority of transcripts to have limited clinical relevance but a focused analysis of oncological pathways produced a number of possible transcript candidates e.g. SYCP2, SFRP1, DLG2, CRNN, and CRCT1. Of these, SYCP2 showed the most significant change from baseline in premalignant retrospective FFPE tissue, superseding the performance of CDKN2A (encoding for p16INK2A).
The elevated expression of SYCP2 in HPV‐associated tumor tissue has previously been noted in three expression analysis studies.29, 32, 39 SYCP2 is a testis‐specific human gene and aberrant expression in HPV+ cancers may contribute to the genomic instability induced by high‐risk HPVs and subsequent oncogenic change.31 A hypothetical model applied to HPV+ oropharyngeal carcinoma is provided in Figure 5. The Wisconsin Alumni Group Foundation has included SYCP2 as one of three target biomarkers in the development for OPSCC.40 To date, no other study has revealed its elevated expression in premalignant tissue. The use of SYCP2 as a prognostic indicator is also of interest, given our DFS data, however, further research will be required to discern if this is truly independent of HPV16 expression.
Figure 5.
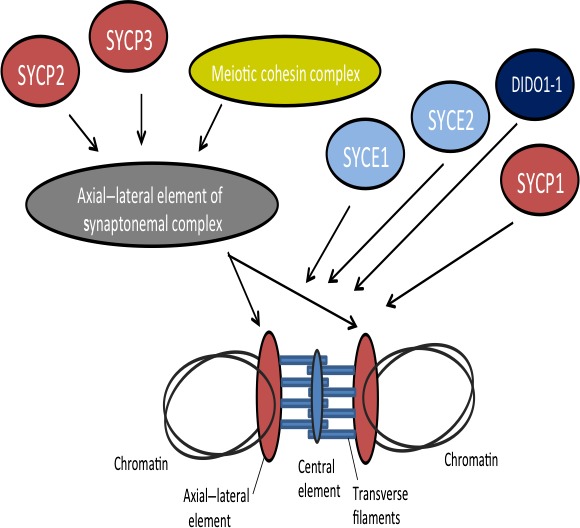
Hypothetical model for SYCP2 with regard to human papillomavirus‐positive oropharyngeal squamous cell carcinomas. SYCP2 is a protein that is involved in linkage chromosomes through the synaptonemal complex, binding DNA at the scaffold attachment regions and driving the prophase of meiosis. Alterations in the gene expression of SYCP2 have been associated with impaired meiosis.
The p16INK4a protein is an inhibitor of cyclin‐dependent kinase and has increased expression with elevated levels of HPV E7, however, many units have reported a concern regarding false positive results.41 Within our HPV16 negative cohort (10/51), three patients had elevated expression of p16INK4a, which may concur with this analysis. At present, stratification of OPSCC tumors by p16INK4a alone is still the preferred approach by the majority of oncology centers. In an era of OPSCC de‐escalation treatments, which are based on a viral etiology, this may incur a risk of undertreating a small proportion of patients falsely considered as HPV+.(48) The use of stepwise algorithms, which combine different HPV assays, may compensate for the limitations of individual tests and should now be considered in clinical settings.42
In this study, positive smoking status proved to have a significant adverse effect on DFS, regardless of HPV category (multivariable analysis, P < 0.03). The negative impact of smoking in HPV+ OPSCC has previously been highlighted by several randomized trials with post‐hoc analysis of HPV status.6, 43, 44 All studies indicated that the degree of tobacco exposure at diagnosis and during treatment directly correlates with the risk of disease progression and death from malignancy. This may indicate that smoking confers additional tumor mutations in the HPV+ cohort, leading to more aggressive disease and inferior responses to available curative‐intent therapies.
However, important questions remain about how to quantify smoking risk to enable comparison between studies. All the major de‐escalation trials have largely adopted the arbitrary cut‐off point proposed by O'Sullivan et al.45 in which ‘smokers’ are defined as having >10 pack year history. Perhaps more reliable information can be obtained from Laborde et al.,25 who recently published transcription profile data in OPSCC patients stratified by both HPV and smoking status. This indicated that two genes involved in the p53 DNA damage repair pathway, ATR and CHEK2, display patterns of increased expression associated with HPV− OPSCC smokers only.
This study is limited by its exclusive focus on whole transcriptome analysis; we recognize the need to integrate DNA sequence analysis in future projects.11 DNA analysis will indicate changes that have occurred to the DNA sequence, whereas mRNA sequence analysis clarifies the effect of those changes. This critically important process therefore identifies which mutations and rearrangements could be the best diagnostic and prognostic indicators. Of course, many current studies display the importance of DNA sequencing in establishing mutations associated with cancer development. The final choice of six target transcripts in this study may also be open to debate as it is primarily based on gene ontology data supplied through the KEGG46 pathway network.
In conclusion, developments in whole‐genome sequencing and mRNA analysis are rapidly creating an opportunity to provide personalized information on genetic and functional aspects of malignant tumors. With regard to HPV+ OPSCC, the investigation of differentially expressed genes in normal, premalignant, and malignant tissue may reveal unique pathways that can explain their different natural history and biological properties. The data from this study reveal SYCP2 as a potentially significant biomarker; if corroborated on a larger scale this may contribute to the development of a non‐invasive screening tool, e.g. mouthwash or brush biopsy. Current epidemiological data would suggest it is not sufficient to simply screen for OPSCC by the presence of HPV16 alone (due to a ~1% carriage rate in the general adult population). Clearly, further well‐designed prospective studies are required to confirm this data and also to establish if other biomarkers may have future significance.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Fig. S1. Clinical and histopathological data for retrospective cohort of oropharyngeal squamous cell carcinoma patients. Human papillomavirus (HPV)‐positive status was defined as evidence of HPV16 L1/E6/E7 DNA or episomal/integrative pattern on HPV DNA in situ hybridization +/− p16INK4A expression (>70% tumor cell staining). †Non‐malignant cause of death. ††Malignant cause of death. #Locoregional recurrence. BoT, base of tongue; CRT, chemoradiotherapy; F, female; f/u, follow‐up; IHC, immunohistochemistry; ISH, in situ hybridization; M, male; Mod, moderately differentiated SCC; mths, months; n/a, not applicable; pal., palate; Poor, poorly differentiated SCC; RT, radiotherapy; SCC, squamous cell carcinoma; Well, well differentiated SCC.
Fig. S2. Full heatmap of 223 transcripts that are differentially expressed between human papillomavirus (HPV)‐positive and HPV‐negative oropharyngeal squamous cell carcinoma tumors (P < 0.01; false‐discovery rate 0.5%).
Fig. S3. Gene ontology data for the 6/223 transcripts with possible oncological relevance.
Fig. S4. Reverse transcription–real‐time quantitative PCR data for prospective human papillomavirus (HPV)16‐positive and HPV16‐negative cohorts (fresh tissue). When the optimal primer concentration produced a linear response to input cDNA, samples were analyzed in triplicate for each tested transcript (CDKN2A, CRCT1, SYCP2, SFRP1, CRNN, and DLG2). β‐Actin (ACTB) was used as the housekeeping gene.
Fig. S5. Reverse transcription–real‐time quantitative PCR data for retrospective cohort (laser capture microdissection invasive vs. normal tissue). Formalin‐fixed paraffin‐embedded tissue samples were subjected to laser capture microdissection to enable RNA extraction from representative regions of invasive carcinoma and adjacent normal tissue.
Fig. S6. Reverse transcription–real‐time quantitative PCR data for retrospective cohort (laser capture microdissection carcinoma in situ vs. normal tissue). Formalin‐fixed paraffin‐embedded tissue samples were subjected to laser capture microdissection to enable RNA extraction from representative regions of in situ carcinoma (premalignant change) and adjacent normal tissue.
Fig. S7. Disease‐free survival – multivariable and univariable statistical analysis. A proportional hazards model using Cox regression analysis revealed human papillomavirus (HPV)16, SYCP2, smoking, and physiological performance status to have significant influence on disease‐free survival.
Fig. S8. Receiver operating characteristic (ROC) curve to evaluate prognostic accuracy of SYCP2/SFRP1 biomarker expression in regions of human papillomavirus‐positive (HPV+) in situ carcinoma. All control subjects were patients undergoing tonsillectomy for benign pathology (see Fig. S9). A further analysis incorporated data from the HPV− oropharyngeal squamous cell carcinomas cohort.
Fig. S9. Reverse transcription–real‐time quantitative PCR data for the prospective control cohort. Fresh tonsil biopsy samples were subjected to RNA extraction (epithelium and adjacent stroma region).
Acknowledgments
We thank the patients from Cambridge University Hospital National Health Service Foundation Trust for volunteering clinical samples. This study was funded by Cancer Research UK (A14962).
Cancer Sci 106 (2015) 1568–1575
Funding Information
Cancer Research UK (A14962).
References
- 1. Mehanna H, Beech T, Nicholson T et al The prevalence of human papillomavirus in oropharyngeal and nonoropharyngeal head and neck cancer: a systematic review and meta‐analysis of trends by time and region. Head Neck 2012; 35: 747–55. [DOI] [PubMed] [Google Scholar]
- 2. Chaturvedi AK, Engels EA, Pfeiffer RM et al Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol 2011; 29: 4294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaturvedi AK, Anderson WF, Lortet‐Tieulent J et al Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol 2013; 31: 4550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hoffmann M, Ihloff AS, Gorogh T, Weise JB, Fazel A, Krams M. p16(INK4a) overexpression predicts translational active human papillomavirus infection in tonsillar cancer. Int J Cancer 2010; 127: 1595–602. [DOI] [PubMed] [Google Scholar]
- 5. Oliver M, Eeles R, Holstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat 2002; 19: 607–14. [DOI] [PubMed] [Google Scholar]
- 6. Gillison ML, Zhang Q, Jordan R et al Tobacco smoking and increased risk of death and progression for patients with p16‐positive and p16‐negative oropharyngeal cancer. J Clin Oncol 2012; 10: 2102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Licitra L, Perrone F, Bossi P et al High‐risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol 2006; 24: 5630–6. [DOI] [PubMed] [Google Scholar]
- 8. Masterson L, Moualed D, Liu ZW et al De‐escalation treatment protocols for human papillomavirus‐associated oropharyngeal squamous cell carcinoma: a systematic review and meta‐analysis of current clinical trials. Eur J Cancer 2014; 50: 2636–48. [DOI] [PubMed] [Google Scholar]
- 9. Masterson L, Moualed D, Masood A et al De‐escalation treatment protocols for human papillomavirus‐associated oropharyngeal squamous cell carcinoma. Cochrane Database Syst Rev 2014; 2: CD010271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Zeeburg HJ, Graveland AP, Brink A et al Generation of precursor cell lines from preneoplastic fields surrounding head and neck cancers. Head Neck 2013; 35: 568–74. [DOI] [PubMed] [Google Scholar]
- 11. Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer 2011; 11: 9–22. [DOI] [PubMed] [Google Scholar]
- 12. Miller CS, Johnstone BM. Human papillomavirus as a risk factor for oral squamous cell carcinoma: a meta‐analysis, 1982–1997. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2001; 91: 622–35. [DOI] [PubMed] [Google Scholar]
- 13. Jayaprakash V, Reid M, Hatton E et al Human papillomavirus types 16 and 18 in epithelial dysplasia of oral cavity and oropharynx: a meta‐analysis, 1985–2010. Oral Oncol 2011; 47: 1048–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Näsman A, Nordfors C, Holzhauser S et al Incidence of human papillomavirus positive tonsillar and base of tongue carcinoma: a stabilisation of an epidemic of viral induced carcinoma? Eur J Cancer 2015; 51(1): 55–61. [DOI] [PubMed] [Google Scholar]
- 15. Chaudhary AK, Singh M, Sundaram S, Mehrotra R. Role of human papillomavirus and its detection in potentially malignant and malignant head and neck lesions: updated review. Head Neck Oncol 2009; 25(1): 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ang K, Harris J, Wheeler R et al Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010; 363: 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Psyrri A, Sasaki C, Vassilakopoulou M, Dimitriadis G, Rampias T. Future directions in research, treatment and prevention of HPV‐related squamous cell carcinoma of the head and neck. Head Neck Pathol 2012; 6(Suppl 1): S121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sobin LH, Gospodarowicz MK, Wittekind C. TNM Classification of Malignant Tumors, 7th edn Oxford: Wiley‐Blackwell, 2009. [Google Scholar]
- 19. Winder DM, Ball SL, Vaughan K et al Sensitive HPV detection in oropharyngeal cancers. BMC Cancer 2009; 9: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gravitt PE, Peyton CL, Alessi TQ et al Improved amplification of genital human papillomaviruses. J Clin Microbiol 2000; 38: 357–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J Gen Virol 1995; 76: 1057–62. [DOI] [PubMed] [Google Scholar]
- 22. Sotlar K, Stubner A, Diemer D et al Detection of high‐risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT‐polymerase chain reaction. J Med Virol 2004; 74: 107–16. [DOI] [PubMed] [Google Scholar]
- 23. Masterson L, Winder D, Marker A et al Investigating the role of human papillomavirus in squamous cell carcinoma of the temporal bone. Head Neck Oncol 2013; 5(2): 22. [Google Scholar]
- 24. Grogan T, Nitta H, Pestic‐Dragovich L, Pang L, Ji J. Interpretation Guide for Ventana INFORM® HPVzr Probes In Situ Hybridization (ISH) Staining of Cervical Tissue. Tucson, AZ, USA: Ventana Medical Systems, Incorporated, 2006. [Google Scholar]
- 25. Laborde RR, Wang VW, Smith TM et al Transcriptional profiling by sequencing of oropharyngeal cancer. Mayo Clin Proc 2012; 87: 226–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 2001; 98: 5116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li CI, Su PF, Guo Y, Shyr Y. Sample size calculation for differential expression analysis of RNA‐seq data under Poisson distribution. Int J Comput Biol Drug Des 2013; 6: 358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sabo E, Meitner PA, Tavares R et al Expression analysis of Barrett's esophagus‐associated high‐grade dysplasia in laser capture microdissected archival tissue. Clin Cancer Res 2008; 14: 6440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Slebos RJ, Yi Y, Ely K et al Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res 2006; 12(3): 701–9. [DOI] [PubMed] [Google Scholar]
- 30. Syrjanen S. HPV infections and tonsillar carcinoma. J Clin Pathol 2004; 57: 449–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pannone G, Santoro A, Papagerakis S, Lo Muzio L, De Rosa G, Bufo P. The role of human papillomavirus in the pathogenesis of head & neck squamous cell carcinoma: an overview. Infect Agent Cancer 2011; 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pyeon D, Newton MA, Lambert PF et al Fundamental differences in cell cycle deregulation in human papillomavirus‐positive and human papillomavirus‐negative head/neck and cervical cancers. Cancer Res 2007; 67: 4605–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walter V, Yin X, Wilkerson MD et al Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE 2013; 8(2): e56823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mooren J, Gültekin S, Straetmans J et al P16INK4A immunostaining is a strong indicator for high‐risk‐HPV‐associated oropharyngeal carcinomas and dysplasias, but is unreliable to predict low‐risk‐HPV‐infection in head and neck papillomas and laryngeal dysplasias. Int J Cancer 2014; 134: 2108–17. [DOI] [PubMed] [Google Scholar]
- 35. Beachler DC, Viscidi R, Sugar EA et al A longitudinal study of human papillomavirus 16 L1, e6, and e7 seropositivity and oral human papillomavirus 16 infection. Sex Transm Dis 2015; 42: 93–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kreimer AR, Bhatia RK, Messeguer AL, González P, Herrero R, Giuliano AR. Oral human papillomavirus in healthy individuals: a systematic review of the literature. Sex Transm Dis 2010; 37: 386–91. [DOI] [PubMed] [Google Scholar]
- 37. Kreimer AR, Pierce Campbell CM, Lin HYFW et al Incidence and clearance of oral human papillomavirus infection in men: the HIM cohort study. Lancet 2013; 382: 877–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gillison ML, Broutian T, Pickard RK et al Prevalence of oral HPV infection in the United States, 2009–2010. JAMA 2012; 307: 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. Identification of differentially expressed genes in HPV‐positive and HPV‐negative oropharyngeal squamous cell carcinomas. Eur J Cancer 2007; 43: 415–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lambert P, Newton M, Ahiquist P. HPV‐Positive Biomarkers for Cervical and Head and Neck Cancers. 2015. [Cited 01 April 2015.] Available from URL: http://www.warf.org/technologies/summary/P07312US.cmsx.
- 41. Schache AG, Liloglou T, Risk JM et al Evaluation of human papilloma virus diagnostic testing in oropharyngeal squamous cell carcinoma: sensitivity, specificity, and prognostic discrimination. Clin Cancer Res 2011; 17: 6262–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Robinson M, Schache A, Sloan P, Thavaraj S. HPV specific testing: a requirement for oropharyngeal squamous cell carcinoma patients. Head Neck Pathol 2012; 6(Suppl 1): S83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Posner MR, Lorch JH, Goloubeva O et al Survival and human papillomavirus in oropharynx cancer in TAX 324: a subset analysis from an international phase III trial. Ann Oncol 2011; 22: 1071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rischin D, Young RJ. Prognostic significance of p16INK4A and human papillomavirus in patients with oropharyngeal cancer treated on TROG 02.02 phase III trial. J Clin Oncol 2010; 28: 4142–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. O'Sullivan B, Huang SH, Siu LL et al Deintensification candidate subgroups in human papillomavirus‐related oropharyngeal cancer according to minimal risk of distant metastasis. J Clin Oncol, 2013; 31: 543–50. [DOI] [PubMed] [Google Scholar]
- 46. Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014; 42: D199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oken MM, Creech RH, Tormey DC et al Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol 1982; 5: 649–55. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Clinical and histopathological data for retrospective cohort of oropharyngeal squamous cell carcinoma patients. Human papillomavirus (HPV)‐positive status was defined as evidence of HPV16 L1/E6/E7 DNA or episomal/integrative pattern on HPV DNA in situ hybridization +/− p16INK4A expression (>70% tumor cell staining). †Non‐malignant cause of death. ††Malignant cause of death. #Locoregional recurrence. BoT, base of tongue; CRT, chemoradiotherapy; F, female; f/u, follow‐up; IHC, immunohistochemistry; ISH, in situ hybridization; M, male; Mod, moderately differentiated SCC; mths, months; n/a, not applicable; pal., palate; Poor, poorly differentiated SCC; RT, radiotherapy; SCC, squamous cell carcinoma; Well, well differentiated SCC.
Fig. S2. Full heatmap of 223 transcripts that are differentially expressed between human papillomavirus (HPV)‐positive and HPV‐negative oropharyngeal squamous cell carcinoma tumors (P < 0.01; false‐discovery rate 0.5%).
Fig. S3. Gene ontology data for the 6/223 transcripts with possible oncological relevance.
Fig. S4. Reverse transcription–real‐time quantitative PCR data for prospective human papillomavirus (HPV)16‐positive and HPV16‐negative cohorts (fresh tissue). When the optimal primer concentration produced a linear response to input cDNA, samples were analyzed in triplicate for each tested transcript (CDKN2A, CRCT1, SYCP2, SFRP1, CRNN, and DLG2). β‐Actin (ACTB) was used as the housekeeping gene.
Fig. S5. Reverse transcription–real‐time quantitative PCR data for retrospective cohort (laser capture microdissection invasive vs. normal tissue). Formalin‐fixed paraffin‐embedded tissue samples were subjected to laser capture microdissection to enable RNA extraction from representative regions of invasive carcinoma and adjacent normal tissue.
Fig. S6. Reverse transcription–real‐time quantitative PCR data for retrospective cohort (laser capture microdissection carcinoma in situ vs. normal tissue). Formalin‐fixed paraffin‐embedded tissue samples were subjected to laser capture microdissection to enable RNA extraction from representative regions of in situ carcinoma (premalignant change) and adjacent normal tissue.
Fig. S7. Disease‐free survival – multivariable and univariable statistical analysis. A proportional hazards model using Cox regression analysis revealed human papillomavirus (HPV)16, SYCP2, smoking, and physiological performance status to have significant influence on disease‐free survival.
Fig. S8. Receiver operating characteristic (ROC) curve to evaluate prognostic accuracy of SYCP2/SFRP1 biomarker expression in regions of human papillomavirus‐positive (HPV+) in situ carcinoma. All control subjects were patients undergoing tonsillectomy for benign pathology (see Fig. S9). A further analysis incorporated data from the HPV− oropharyngeal squamous cell carcinomas cohort.
Fig. S9. Reverse transcription–real‐time quantitative PCR data for the prospective control cohort. Fresh tonsil biopsy samples were subjected to RNA extraction (epithelium and adjacent stroma region).


