Abstract
Interactions between nuclear and mitochondrial gene products are critical for eukaryotic cell function. Nuclear genes encoding mitochondrial-targeted proteins (N-mt genes) experience elevated rates of evolution, which has often been interpreted as evidence of nuclear compensation in response to elevated mitochondrial mutation rates. However, N-mt genes may be under relaxed functional constraints, which could also explain observed increases in their evolutionary rate. To disentangle these hypotheses, we examined patterns of sequence and structural evolution in nuclear and mitochondrial-encoded oxidative phosphorylation proteins from species in the angiosperm genus Silene with vastly different mitochondrial mutation rates. We found correlated increases in N-mt gene evolution in species with fast-evolving mitochondrial DNA. Structural modeling revealed an overrepresentation of N-mt substitutions at positions that directly contact mutated residues in mitochondrial-encoded proteins, despite overall patterns of conservative structural evolution. These findings support the hypothesis that selection for compensatory changes in response to mitochondrial mutations contributes to the elevated rate of evolution in N-mt genes. We discuss these results in light of theories implicating mitochondrial mutation rates and mitonuclear co-evolution as drivers of speciation and suggest comparative and experimental approaches that could take advantage of heterogeneity in rates of mtDNA evolution across eukaryotes to evaluate such theories.
Keywords: mitochondrial mutations, electron transport, mitonuclear interactions, evolutionary genomics, cytonuclear
Introduction
In eukaryotes, gene products from the mitochondrial and nuclear genomes must interact intimately with each other to meet the energetic demands of the cell. When such mitonuclear interactions are disrupted, organismal health and fitness can be drastically compromised (Wallace 1992, 2005; Dowling 2014), and mitochondrial function also plays a general role in aging (Linnane et al. 1989; Larsson 2010; Lane 2011). However, the combination of high mutation rates in many eukaryotic lineages (Denver et al. 2000; Lynch et al. 2008) and inefficient selection (resulting from uniparental inheritance and the associated lack of sexual recombination) predisposes mitochondrial DNA (mtDNA) to accumulation of deleterious mutations (Muller 1932; Lynch and Blanchard 1998; Neiman and Taylor 2009). Consequently, rates of evolution are often much higher in mtDNA than in the nucleus (Brown et al. 1979; Lynch 1997; Lynch et al. 2006; Lynch et al. 2008). Nuclear gene products that are targeted to the mitochondria (N-mt genes) are thought to undergo compensatory evolution in order to offset the deleterious metabolic effects of mitochondrial mutations. In support of this hypothesis, N-mt genes often experience higher substitution rates or exhibit a history of positive selection when compared with genes that are not involved in direct interactions with the mitochondria (Levin et al. 2014). This has been observed for protein-coding genes that form the chimeric oxidative phosphorylation (OXPHOS) complexes responsible for electron transport (Schmidt et al. 2001; Rand et al. 2004; Willett and Burton 2004; Mishmar et al. 2006; Shen et al. 2010; Osada and Akashi 2012) and ribosomal proteins that must interact with mtDNA-encoded rRNAs (Barreto and Burton 2013; Sloan et al. 2014). Moreover, there is evidence of co-evolution between nuclear-encoded tRNA synthetases and mtDNA-encoded tRNAs (Hoekstra et al. 2013; Meiklejohn et al. 2013). It has also been proposed that the recruitment of novel nuclear-encoded OXPHOS subunits in eukaryotes may have been a form of structural compensation for the unstable mitochondrial-encoded subunits (van der Sluis et al. 2015).
However, the extent to which elevated rates of nucleotide substitution in N-mt genes are a direct consequence of compensation for high mitochondrial mutation rates remains unclear. Alternative hypotheses to explain the rapid sequence evolution in N-mt genes include that these genes have lower expression levels (Nabholz et al. 2013; Adrion et al. 2015) and may be under fewer functional constraints than their mitochondrial counterparts (Barreto and Burton 2013; Zhang and Broughton 2013; Sloan et al. 2014; Adrion et al. 2015; Pett and Lavrov 2015). An ideal comparative strategy for disentangling these competing hypotheses would be to compare closely related species with fast and slow rates of mtDNA evolution. If mitochondrial mutation rates and selection for compensatory responses drive evolutionary rates in N-mt genes, then lineages with fast mtDNA evolution should show faster N-mt gene evolution compared to lineages with slowly evolving mtDNA. If differences in functional constraints are solely responsible, then N-mt genes should not differ in evolutionary rates between lineages with slow- vs. fast-evolving mtDNA. Unfortunately, the potential for mitonuclear compensation has overwhelmingly been studied in bilaterian animal lineages, which consistently have high rates of mtDNA evolution. Land plants represent attractive and underutilized alternatives for investigating the evolution of mitonuclear interactions (Moison et al. 2010; Greiner and Bock 2013; Sloan 2015) because their rates of mitochondrial sequence evolution tend to be much slower than in the nucleus (Wolfe et al. 1987; Drouin et al. 2008; Smith and Keeling 2015). Plants in the genus Silene (Caryophyllaceae) offer an exceptional model to test such comparative predictions, as most species have slow mtDNA substitution rates characteristic of other angiosperms, but closely related species within the genus have experienced recent, rapid accelerations in mtDNA evolution, which are believed to reflect increases in the underlying mtDNA mutation rate (Mower et al. 2007; Sloan et al. 2009; Rautenberg et al. 2012; Sloan et al. 2012a). These latter species have mtDNA substitution rates on par with those observed in mammals.
An additional way to evaluate the hypothesis of nuclear compensation, while disentangling the effects of relaxed selection, is to incorporate structural information on mitonuclear protein complexes. In mammals with rapid mtDNA evolution, N-mt genes are thought to compensate for mitochondrial changes by undergoing amino acid changes at residues that contact altered mitochondrial residues (Osada and Akashi 2012), and estimates of positive selection differ between mitochondrial-nuclear interacting residues and mitochondrial-mitochondrial interacting residues (Schmidt et al. 2001; Aledo et al. 2014). Although extreme mitochondrial rate accelerations have been found in multiple independent angiosperm lineages (Cho et al. 2004; Parkinson et al. 2005; Mower et al. 2007; Skippington et al. 2015), how these changes influence nuclear evolution through structural compensation has yet to be investigated.
Here, we use transcriptomic data and modeled 3D structures of OXPHOS proteins in Silene species with highly divergent rates of mitochondrial evolution to examine the hypothesis that compensatory evolution in the nucleus is driven by elevated mitochondrial mutation rates. First, we consider general molecular evolution in OXPHOS complexes to quantify the selection pressures acting on protein sequences and structures in species with elevated mtDNA substitution rates. Second, we use the ratio of nonsynonymous to synonymous substitutions (dN/dS) to determine if nuclear-encoded OXPHOS proteins have higher rates of amino acid substitution in species with fast-evolving mtDNA than in slower species. Finally, we test for compensatory evolution by determining if nuclear substitutions occur in protein structural regions that physically interact with changing mitochondrial residues.
Materials and Methods
OXPHOS protein sequence analysis and structural modeling
Transcriptome assemblies have already been described (Sloan et al. 2014), and Illumina reads have been deposited into the NCBI Sequence Read Archive for S. conica (SRX353031) and S. noctiflora (SRX353048), which have experienced recent accelerations in mtDNA evolution, and for S. latifolia (SRX353047) and S. vulgaris (SRX353050), which have slower rates of mtDNA evolution. Complete mitochondrial genomes from each of these species have also been sequenced and annotated (Sloan et al. 2010; Sloan et al. 2012a). Transcriptome and mitogenome assemblies were searched for genes encoding OXPHOS complex subunits using NCBI tBLASTn v2.2.29 and Arabidopsis thaliana protein sequences from the Arabidopsis Mitochondrial Protein Database as queries (Heazlewood et al. 2004). For the N-mt genes, N-terminal targeting peptides were predicted using the TargetP 1.1 server (Emanuelsson et al. 2007) and removed prior to alignment. For the mtDNA-encoded proteins, RNA editing sites were predicted using PREP-Mt (Mower 2005) with a cutoff value of 0.2, and edited sequences were used in alignments.
We investigated whether the proteins encoded by N-mt genes and mitochondrial genes are experiencing accelerated evolution in species with fast-evolving mtDNA. An amino acid alignment for each N-mt and mitochondrial gene was generated using MUSCLE v3.7 (Edgar 2004) and then converted back into nucleotides to produce in-frame alignments (supplement 1). Pairwise nonsynonymous (dN) and synonymous (dS) divergences per site were estimated for each gene as described previously using either fast (S. conica vs. S. noctiflora) or slow (S. latifolia vs. S. vulgaris) species pairs (Sloan et al. 2014). Briefly, codeml in PAML v4.8 (Yang 2007) was used with an F1x4 model to estimate dN and dS for concatenated genes from each OXPHOS complex (number of N-mt and mitochondrial genes analyzed in complex I: 29, 9; II: 7, 0; III: 10, 1; IV: 6, 3; V: 11, 5) and to implement a likelihood ratio test to compare dN/dS for concatenated gene sets between the fast and slow species pairs.
For structural comparisons (see below), the amino acid sequence for the common ancestor of the four Silene species was inferred using codeml in PAML. For each protein sequence, homologs in outgroups were identified by performing a tBLASTn search of the Agrostemma githago transcriptome (SRX352988) and mitochondrial genome (SRX1089472) and the Beta vulgaris genome (Dohm et al. 2013). Amino acid sequences from Arabidopsis thaliana and the identified templates were also used as outgroups. This analysis was performed with a constrained topology with the four Silene species coded as a polytomy. Because the amino acid sequences were identical for S. latifolia and S. vulgaris in nearly all proteins, the common ancestor sequence was usually identical to the S. latifolia/S. vulgaris sequence.
For both mtDNA-encoded and N-mt genes from the four Silene species, 3D structural protein models were predicted using MODELLER v9.14 following the “basic” protocol outlined by Eswar et al. (2006). Proteins were only modeled if the best template structures identified by MODELLER showed at least 40% amino acid sequence identity with the Silene sequences (retained models had 40–82% amino acid identity; supplement 1). Template structures were taken from Bos taurus: 1v54 served as the template for cyctochrome c oxidase proteins and the complexes thereof (Tsukihara et al. 2003), 1pp9 served as the template for cytochrome bc1 proteins and the complexes thereof (Huang et al. 2005), and 1h8e served as the template for F(1)-ATPase proteins and the complexes thereof (Menz et al. 2001). This screening process resulted in generation of structural models for five mitochondrial-encoded proteins (COX1, COX2, COX3, COB, and ATP1) and eight nuclear-encoded proteins (COX5B, COX6A, COX6B, CYC1, QCR7, UCR1, ATP2, and ATP3). Residues with no homologous position in the template sequence (i.e., those with gaps in the sequence alignment) were removed prior to modeling. For each protein sequence, 100 models were generated using a random seed.
Structural comparisons of thermodynamic stability
To test if species with fast rates of mtDNA sequence evolution also had fast rates of structural evolution, structures of Silene proteins were compared to the inferred ancestral state using the difference in thermodynamic stability (ΔΔG) as calculated by the EnergyDifference function of FoldX v3.0 (Guerois et al. 2002). FoldX uses a description of the structure based on all atoms to determine protein stability and can estimate the relative stability between structures. The repair function option was not used, as this did not alter results in earlier tests. All models were derived from the homologous template structures via the same protocol. ΔΔG values were estimated based on the average of 50 comparisons of replicate structural models. Differences among species were then statistically tested using an ANOVA as implemented with the aov function in R v2.15.0 (R Development Core Team 2012).
Quantifying conservative structural evolution in individual subunits
To test whether observed amino acid substitutions in the fast-evolving mitochondrial proteins subunits of S. conica and S. noctiflora have been relatively conservative with respect to protein structure and thermodynamic stability, an in silico alanine scan was performed on the mitochondrial structural models using the alascan function in FoldX. In this process, we calculated the ΔΔG associated with individually mutating each residue to alanine based on ten replicate structural models of each ancestral Silene mitochondrial protein. This in silico analysis was performed in order to determine which individual residues most contributed to protein stability and to provide results comparable to conventional alanine-scanning mutagenesis (Weiss et al. 2000). Sequences from S. conica and S. noctiflora were analyzed separately, and for each species, amino acid positions were classified as either “conserved” if they were identical to the inferred Silene ancestral sequence or “variable” if they were not. To test whether these types of sites differed significantly in ΔΔG, t-tests or ANOVAs (and Tukey post-hoc tests where appropriate) were implemented with the aov function in R.
We also tested whether mitochondrial proteins in fast-evolving species underwent more or less change in thermodynamic stability than would be expected by chance (i.e. in the absence of selection for protein stability) given the number of inferred residues that changed. For both S. conica and S. noctiflora, the underlying DNA sequence of the ancestral protein was randomly mutated until the actual observed number of amino acid substitutions (9–32) had taken place. In this way, 1000 randomly mutated sequences were generated for each mitochondrial protein using a custom Python script (supplement 2). Structural models were then generated for each of these sequences using MODELLER just as with the authentic sequences. The simulated evolved variants were compared to the ancestral structure using the EnergyDifference function of FoldX to estimate ΔΔG as described above. Observed ΔΔG values were compared to the null distribution generated from the 1000 randomly mutated sequences to test for significance.
Modeling thermodynamic stability of interacting mitochondrial and nuclear subunits
We defined interacting pairs of mitochondrial and nuclear subunits as those having contact residues in their template structure, which were identified using the Contact Map Analysis webserver and default settings (Sobolev et al. 2005). This server defines a contact for any two residues that have a minimum contact area of 0.2 Å2. Twelve mitonuclear pairs (COX1-COX5B, COX1-COX6A, COX1-COX6B, COX2-COX6B, COX3-COX5B, COX3-COX6A, COX3-COX6B, COB-CYC1, COB-QCR7, COB-UCR1, ATP1-ATP2, and ATP1-ATP3) were then modeled using the inferred ancestral Silene sequences against the original templates following the MODELLER protocol implemented in HOMCOS (Fukuhara and Kawabata 2008). Contact sites were identified for the resulting models of mitonuclear pairs using the Contact Map Analysis webserver.
To investigate how amino acid substitutions have affected thermodynamic stability of mitonuclear protein pairs, the BeAtMusic webserver was used to predict the change in binding affinity (ΔΔGB) between the mitochondrial and nuclear subunit upon mutation of a single residue (Dehouck et al. 2013). In silico mutagenesis of single residues was performed by mutating residues into the other 19 possible residues (including the actual observed substitution) and calculating the resulting ΔΔGB. A two-way ANOVA was implemented using the aov function in R to test for the effects of residue type and genome (mitochondrial vs. nuclear) on ΔΔGB. This analysis was performed for all residues and then repeated to consider only contact residues.
Investigating spatial and thermodynamic characteristics of substitutions
We first assessed whether contact residues in mitochondrial and nuclear subunits were more likely than noncontact residues to be associated with variable residues (for S. conica and S. noctiflora), and the relative proportion of variable, contact residues was evaluated using a Fisher’s exact test as implemented using the fisher.test function in R. Also, by restricting the analysis to exposed residues, we removed the confounding effect that surface residues may be more likely to tolerate mutations (Aledo et al. 2014). We classified residues as buried if surface accessible area in single-protein models (normalized to the area that residues would have in an extended, unfolded polypeptide chain) was less than 0.05, following the protocol of Aledo et al. (2012).
We also tested whether nuclear-encoded residues that contacted a variable mitochondrial-encoded residue were more likely to undergo a change themselves. Nuclear-encoded residues were categorized as variable vs. conserved, and as contacting vs. not contacting a variable mitochondrial residue. Fisher’s exact tests were then used to evaluate whether there was an excess of substitutions at nuclear residues that contacted a variable mitochondrial residue. These tests were repeated with a reduced dataset that only contained exposed residues and an even further reduced dataset that only contained contact residues.
Finally, targeted mutagenesis was performed in silico on ancestral reconstructions of mitonuclear protein pairs to determine if nuclear substitutions offset thermodynamic instability caused by mitochondrial substitutions. In these experiments, the BuildModel function of FoldX was used to introduce multiple mutations and the AnalyseComplex function was used to calculate the resulting ΔΔGB values. Mutations were first introduced at each of the eight pairs of contact residues that were variable in both mitochondrial and nuclear-encoded subunits for S. conica (S. noctiflora was excluded since no such pairs were found). Mutations were introduced solely for the mitochondrial residue, the nuclear residue, or for both residues simultaneously. Mutations were also introduced simultaneously at all variable contact residues, either for the mitochondrial and nuclear subunits separately or for both at once.
Results
Despite fast mtDNA evolution, mitochondrial protein sequences and structures evolve conservatively
For mitochondrial-encoded OXPHOS genes, pairwise dN and dS values were on average 27- and 44-fold higher, respectively, for the S. conica & S. noctiflora comparison relative to the S. latifolia & S. vulgaris comparison. Given the similar divergence times between these species pairs (Rautenberg et al. 2012), these dN and dS values confirm the dramatically faster rate of mtDNA evolution in S. conica and S. noctiflora (Table 1). There was no indication of relaxed or positive selection on protein sequence in the fast-evolving species (dN/dS ≪ 1), suggesting a primary role of purifying selection. In complexes III, IV, and V, no statistical difference was observed between mitochondrial dN/dS between the species pairs, while in complex I the slow species pair had a dN/dS value that was significantly higher (2.72-fold) than in the fast species pair (Table 1).
Table 1.
dN, dS, and dN/dS for mitochondrial genes in fast and slow Silene species pairs, with associated P-values from a likelihood ratio test comparing dN/dS between slow and fast species pairs.
| Complex | Slow (S. latifolia & S. vulgaris) | Fast (S. conica & S. noctiflora) | P | ||||
|---|---|---|---|---|---|---|---|
| dN | dS | dN/dS | dN | dS | dN/dS | ||
| I | 0.002 | 0.008 | 0.196 | 0.040 | 0.554 | 0.072 | 0.022 |
| III | 0.002 | 0.010 | 0.203 | 0.063 | 1.181 | 0.054 | 0.198 |
| IV | 0.004 | 0.033 | 0.108 | 0.047 | 0.903 | 0.052 | 0.093 |
| V | 0.002 | 0.028 | 0.053 | 0.083 | 0.806 | 0.103 | 0.270 |
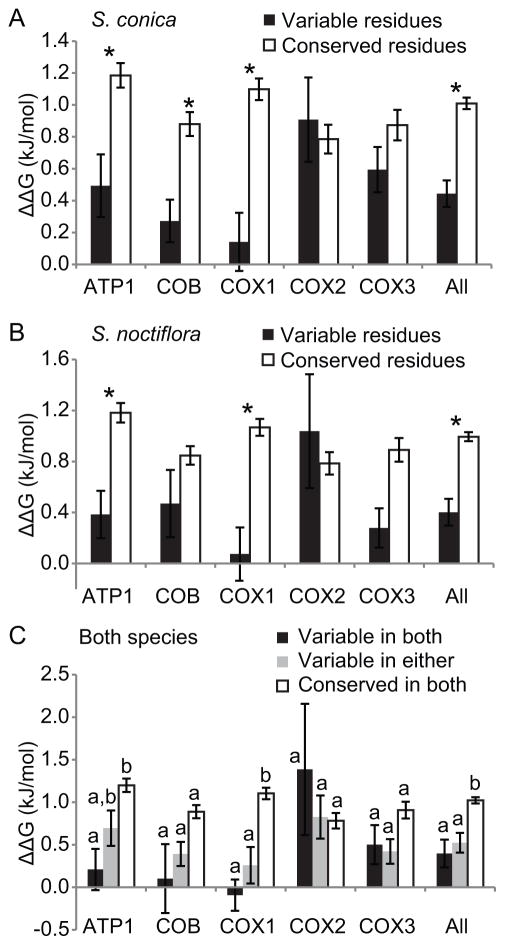
In S. conica (Fig. 1A) and S. noctiflora (Fig. 1B), in silico mutations to alanine resulted in a 2.28- and 2.47-fold smaller increase in energy for variable residues compared to conserved residues when all mitochondrial proteins were combined (P < 0.001 for each species, t-tests), and for several proteins when considered independently. Moreover, mutating residues that were variable in both S. conica and S. noctiflora tended to produce the smallest changes in thermodynamic stability compared with residues that only underwent a change in one species (Fig. 1C), although this trend was not significant (adjusted P = 0.708, ANOVA with Tukey). Supporting this finding, ΔΔG from the ancestral state was similar for the fast species compared with the slow species (supplement 3; P = 0.246, ANOVA). However, using raw ΔΔG values may be confounding because ΔΔG can increase (resulting in a less stable structure) or decrease (resulting in a more stable structure), and fast-evolving species might experience more change in either direction. Therefore, we also compared absolute ΔΔG values and found similar results (supplement 3; P = 0.863).
Figure 1.
Differences in thermodynamic stability (ΔΔG, ± SEM) resulting from an in silico alanine scan of modeled structures from mitochondrial-encoded proteins of (A) Silene conica, (B) S. noctiflora, or (C) both species. Asterisks indicate significant differences between conserved and variable residues when compared to the Silene ancestral sequence. Letters in (C) indicate significant groupings.
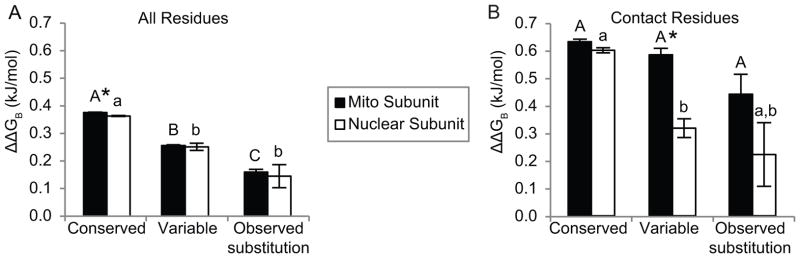
Comparisons of randomly mutated sequences also supported a role of conservative evolution in mitochondrial protein structure. ΔΔG values between protein structures that were modeled based on the inferred ancestral protein sequences and the extant protein sequences were more stable on average for the observed S. conica and S. noctiflora sequences than when changes were introduced randomly (Table 2, supplement 4). Echoing this theme, the results of the mutagenesis scan conducted on mitonuclear protein pairs also indicated conservative structural evolution (Fig. 2). Overall, mutating contact residues caused a 1.77-fold greater ΔΔGB than mutating noncontact residues (P < 0.001, t-test), validating the identification of contact residues and the methodology for measuring ΔΔGB. Mutating variable residues resulted in a smaller increase in ΔΔGB than conserved residues, for both mitochondrial (1.47-fold smaller) and nuclear (1.45-fold smaller) subunits (Fig. 2A). These calculations were averaged over all 19 possible amino acid substitutions that could occur at each site. Considering specific substitutions that were actually observed in either S. conica or S. noctiflora sequences resulted in even lower ΔΔGB values for both mitochondrial residues (2.43-fold) and nuclear residues (2.51-fold). This effect was significant for the mitochondrial residues (adjusted P < 0.001) but not for the nuclear residues (adjusted P = 0.326). When contact residues were considered separately, a similar trend towards conservative evolution was observed in the nuclear residues, where mutating variable residues resulted in a smaller ΔΔGB than mutating conserved residues (Fig. 2B, P < 0.001). However, when mitochondrial contact residues were considered, there was a weaker trend (resulting in a significant interaction between residue type and genome; P < 0.001): There were no statistical differences in ΔΔGB between variable and conserved sites, even when actual, observed substitutions were considered (adjusted P = 0.526). As a result, mutating mitochondrial residues resulted in ΔΔGB values that were 1.84-fold higher than those for nuclear residues when only variable, contact residues were considered (adjusted P < 0.001).
Table 2.
ΔΔG between Silene species and ancestral proteins, either as observed or the mean based on 1000 randomly mutated variants of the ancestral sequence, with associated P-values.
| Silene conica | Silene noctiflora | |||||
|---|---|---|---|---|---|---|
| observed ΔΔG | mean ΔΔG of randomly mutated variants | P | observed ΔΔG | mean ΔΔG of randomly mutated variants | P | |
|
| ||||||
| ATP1 | 19.135 | 72.363 | 0.022 | 12.710 | 58.795 | 0.030 |
| COB | −0.965 | 40.333 | 0.022 | −8.553 | 22.111 | 0.052 |
| COX1 | 17.870 | 62.431 | 0.058 | −10.721 | 26.783 | 0.044 |
| COX2 | −1.438 | 20.927 | 0.116 | −5.610 | 10.706 | 0.142 |
| COX3 | −2.652 | 31.716 | 0.016 | −11.060 | 22.930 | 0.004 |
Figure 2.
Resulting differences in binding affinity (ΔΔGB, ± SEM) between modeled mitonuclear pairs of OXPHOS proteins from Silene conica and S. noctiflora when residues from three different categories were mutated in silico: 1) conserved residues between the ancestral and extant Silene sequences; 2) variable residues between the ancestral and extant Silene sequences, and; 3) for the actual observed substitutions in the modern Silene species. (A) Considering all protein residues; (B) considering contact residues only. Asterisks indicate differences between mitochondrial and nuclear-encoded subunits. Letters indicate significant groupings between residue categories for either mitochondrial (uppercase) or nuclear (lowercase) subunits.
However, there were exceptions to these overall trends of conservative structural evolution when each protein was considered individually. For example, in COX2, which lacked many variable sites, there was no systematic difference in ΔΔG in either species when mutating variable vs. conserved residues to alanine (P = 0.55, 0.66). In addition, among the S. conica QCR7 residues that contact COB, in silico mutations resulted in a 2.00-fold greater increase in ΔΔGB for variable sites than for conserved sites, contrary to overall results (P < 0.001, supplement 5).
N-mt OXPHOS genes have elevated dN/dS in species with fast mtDNA evolution
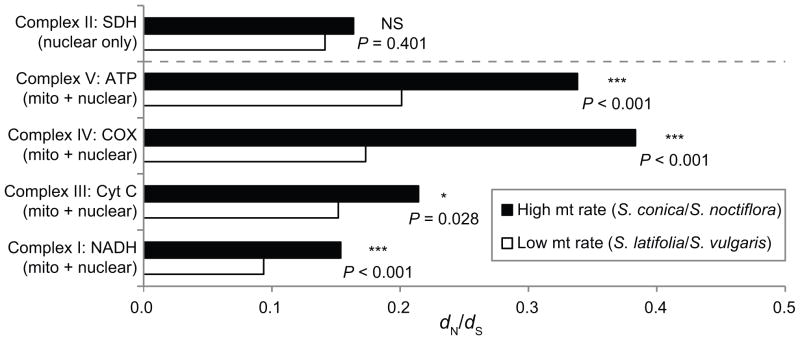
The dN/dS ratio for N-mt genes was elevated between 1.41- and 2.2-fold in chimeric OXPHOS complexes of the species pair with fast-evolving mtDNA (S. conica/S. noctiflora) compared to the species pair with more slowly-evolving mtDNA (S. latifolia/S. vulgaris; Fig. 3, supplement 6). This pattern was stronger in complexes IV and V than in complexes I and III. As expected, there was no significant difference between species pairs for complex II, which is composed entirely of nuclear-encoded proteins in Silene (Sloan et al. 2012a). To provide an additional example, we investigated the mitochondrial-targeted caseinolytic protease (CLP), which is composed entirely of four nuclear-encoded proteins (CLPP2 and three CLPX subunits; van Wijk 2015) and found no significant difference (P = 0.601) in pairwise dN/dS in S. conica & S. noctiflora (0.165) vs. S. latifolia & S. vulgaris (0.150; Supplement 6). The dN/dS ratio was less than one for all comparisons, suggesting a primary role of purifying selection.
Figure 3.
The dN/dS ratio for nuclear-encoded OXPHOS genes, calculated by comparing Silene species with slow (S. latifolia & S. vulgaris) or fast (S. conica & S. noctiflora) rates of mtDNA evolution. Asterisks indicate significance between the fast vs. slow comparison for each OXPHOS complex.
Nuclear compensatory evolution is common in species with fast mtDNA evolution but specific to certain protein-protein interactions
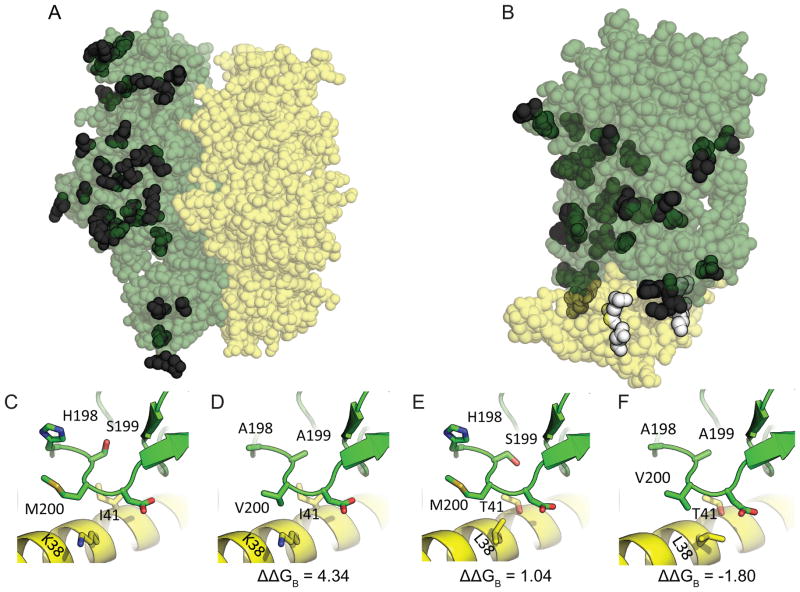
In some mitonuclear protein pairs, mitochondrial amino acid substitutions were restricted to non-contact residues (e.g., ATP1-ATP2, Fig. 4A), preventing an analysis of association with residues in the nuclear subunit. In contrast, other pairs possessed both mitochondrial and nuclear contact residues that underwent changes, implying the possibility of nuclear compensatory evolution (e.g., COB-QCR7, Fig. 4B). Overall when S. conica and S. noctiflora were considered together, substitutions preferentially occurred at contact sites in nuclear-encoded proteins (odds ratio, OR = 2.405, P = 0.011, Fisher’s exact test) but not in mitochondrial-encoded proteins (OR = 1.025, P = 0.923). The same result was obtained when S. conica was analyzed separately (OR = 2.823, P = 0.018 for nuclear proteins; OR = 0.956, P = 1.000 for mitochondrial encoded proteins), and there was a similar but non-significant trend when analyzing only S. noctiflora (OR = 1.929, P = 0.237 for nuclear proteins; OR = 1.139, P = 0.639 for mitochondrial proteins). Similar results were also found when the analysis was restricted to only consider exposed residues (Table 3). Also, nuclear residues were over six times more likely to undergo a substitution when contacting a mitochondrial residue that also underwent a change than when contacting a conserved mitochondrial residue (Table 3). However, we did not observe this pattern when analyzing S. noctiflora individually, as there were no substitutions at nuclear-encoded residues that contacted a changing mitochondrial-encoded residue, which likely reflected the very small number of changes at mitochondrial-encoded contact residues in this species. Finally, when mitonuclear pairs were considered individually, it was clear that specific interactions (e.g., COX1-COX5B and COB-QCR7) were driving these overall trends, while others (e.g., ATP1-ATP2 and ATP1-ATP3) showed little possibility of correlated nuclear substitutions because of the lack of substitutions at mitochondrial-encoded contact residues (supplement 7).
Figure 4.
Modeled structures for OXPHOS mitonuclear protein pairs based on ancestral Silene sequences for (A) ATP1-ATP2 and (B) COB-QCR7. (C) – (F) show residues in white indicated in (B), with (C) corresponding to the ancestral Silene sequences, (D) resulting from introducing the substitutions inferred to have taken place in the mitochondrial-encoded contact residues of S. conica, (E) resulting from introducing the substitutions inferred to have taken place in the nuclear-encoded contact residues of S. conica, and (F) resulting from introducing both mitochondrial and nuclear-encoded substitutions in the contact residues simultaneously. In (A) and (B) variable residues are opaque and conserved residues are transparent, while mitochondrial- and nuclear-encoded are green and yellow, respectively (or dark grey and light grey in the print version). In (C) – (F), changing variable contact residues are displayed as “sticks” while other residues are displayed as “ribbons”.
Table 3.
Odds ratios and P-values for statistical associations between categories of residues (Fisher’s exact tests).
| Do substitutions preferentially occur at contact residues?1 | Do nuclear substitutions preferentially contact variable mitochondrial residues?2 | ||
|---|---|---|---|
| Mito | Nuc | ||
| S. conica | 0.856, P = 0.626 | 2.551, P = 0.043 | 19.386, P < 0.001 |
| S. noctiflora | 0.998, P = 1.000 | 1.625, P = 0.401 | – |
| Both spp. | 0.910, P = 0.705 | 2.102, P = 0.025 | 6.105, P < 0.001 |
Analysis restricted to exposed residues.
Analysis restricted to contact residues.
Specific examples of nuclear substitutions that offset structural instability caused by mitochondrial substitutions were found through in silico targeted mutagenesis. For COX1-COX5B, mutating the mitochondrial-encoded COX1-I508F by itself resulted in a ΔΔGB of 7.08 kJ/mol compared to the ancestral state. However, when one of its nuclear-encoded contacts, COX5B-I29V, was simultaneously mutated, this destabilizing effect was ameliorated, resulting in a ΔΔGB of 2.05 kJ/mol. As another example, in COB-QCR7 mutating all mitochondrial-encoded COB contact residues that were inferred to have undergone a substitution (H198A, S199A, M200V, S363F; Fig. 5D) or all such nuclear-encoded QCR7 contact residues (I41T, K38L; Fig. 5E) caused elevated ΔΔGB (4.34 and 1.04 kJ/mol, respectively). However, when both sets of residues were mutated together, ΔΔGB was −1.80 kJ/mol (Fig. 5F), indicating favorable epistatic effects between mitochondrial and nuclear subunits. Unfortunately, it was not feasible to perform statistical analyses on the targeted mutagenesis experiments given small sample sizes, and other examples showing the opposite trends were also found (supplement 8).
Discussion
Purifying selection dominates OXPHOS sequence and structural evolution
Several results indicate conservative evolution of mitochondrial OXPHOS proteins. This finding is consistent with the general conservation of OXPHOS protein structure and function across the eukaryotic tree (Moyes and Hood 2003). Substitutions in the species with fast-evolving mtDNA appear to be generally restricted to residues with little effect on the thermodynamic stability of proteins. The low dN/dS values for both mitochondrial and N-mt genes (Table 1 and Fig. 3) suggest selection for structural or functional novelty is not driving mitochondrial OXPHOS evolution. Documented cases of adaptive evolution in these core proteins have been linked to major shifts in physiology, metabolism, or ecological niche (Grossman et al. 2004; Castoe et al. 2008). While these characteristics have not been examined in detail for Silene with divergent mtDNA substitution rates, given their recent divergence from species with slowly-evolving mtDNA only ~6 million years ago (Rautenberg et al. 2012), it is unlikely that a major shift has occurred in these core features.
Compensatory evolution in the nucleus vs. relaxed functional constraints and implications for mitonuclear ecology
Given the general pattern of conservative evolution in mitochondrial OXPHOS subunits, a role for nuclear compensatory evolution in Silene with fast-evolving mtDNA would indicate that even small changes in mitochondrial-encoded subunits could lead to altered selection on the nuclear genome. An alternate hypothesis explaining elevated substitution rates in N-mt genes is that relaxed functional constraints and lower gene expression levels may lead to accelerated changes in N-mt genes (Barreto and Burton 2013; Nabholz et al. 2013; Zhang and Broughton 2013; Sloan et al. 2014; Adrion et al. 2015; Pett and Lavrov 2015).
By using closely related species with radically divergent rates of mtDNA substitution, we were able to assess whether mitochondrial mutation rates affect accelerated evolution in N-mt genes, while controlling for differences in functional constraint across genes. The dN/dS ratio of N-mt genes was higher in species with faster mtDNA substitution rates (Fig. 3), suggesting that mitochondrial mutation rates are, at least in part, responsible for elevated amino acid substitution rates in N-mt genes. This finding has implications for theories detailing a role for mitonuclear interactions and nuclear compensatory evolution in broad evolutionary processes such as adaptation to local environments or metabolic needs (Castoe et al. 2008) and key transitions in life history traits such as reproductive mode and onset of aging (Havird et al. 2015; Hill 2015). In particular, mitonuclear mismatch has been proposed to play an important role in reproductive isolation between species (Gershoni et al. 2009; Burton and Barreto 2012), and this might extend to Silene species with elevated mtDNA mutation rates. A natural extension of our work will be to investigate mitonuclear interactions and OXPHOS activity in hybrids generated within Silene clades with rapidly evolving mtDNA and compare those to hybrids involving species with more typical slow substitution rates.
Previous analyses of the effects of mitonuclear interactions on plant reproduction have focused on cytoplasmic male sterility (CMS), which is typically caused by expression of chimeric open reading frames (ORFs) in the mitochondrial genome that disrupt pollen production, while nuclear-encoded proteins in the pentatricopeptide repeat (PPR) family that bind to mitochondrial mRNA transcripts act as restorers of male fertility (Schnable and Wise 1998; Bentolila et al. 2002; Fujii et al. 2011). However, examples have been identified of species that exhibit CMS but lack obvious candidate ORFs in their mitochondrial genomes, suggesting alternative mechanisms may be involved (Darracq et al. 2011; Sloan et al. 2012b). Our data provide a possible alternative mechanism: amino acid substitutions in mitochondrial-encoded OXPHOS subunits generate incompatibilities with interacting nuclear-encoded subunits, leading to CMS. Further characterization of mitonuclear interactions – particularly in species with rapidly evolving mitochondrial genomes – could assess this possibility.
Structural analysis of mitonuclear protein pairs also supported a role for nuclear compensation, as nuclear substitutions were preferentially found at residues that interact with mitochondrial-encoded residues. It is possible that this correlation is not indicative of nuclear compensation, but rather contact residues in mitonuclear proteins may be under relaxed functional constraints or could be changing for entirely different reasons. However, we did not find a correlation between interacting residues and substitutions in mitochondrial proteins, suggesting the underlying cause is only affecting nuclear residues, as would be expected for compensatory selection in response to elevated mitochondrial substitution rates.
Importantly, our support for nuclear compensation is not at odds with the functional constraint hypothesis (Barreto and Burton 2013; Nabholz et al. 2013; Zhang and Broughton 2013; Sloan et al. 2014; Adrion et al. 2015; Pett and Lavrov 2015). When dN/dS of nuclear-encoded ribosomal proteins were compared in fast and slow Silene (Sloan et al. 2014), the effect of where the protein was targeted to in the cell (mitochondrial vs. cytosol) was much larger (~10-fold higher for mitochondrial-targeted proteins) than the effect of whether the species had elevated mtDNA substitution rates (~1.5-fold higher in fast species). This latter effect size was on the same order that we observed for N-mt OXPHOS genes (overall 1.76-fold higher in faster Silene). This relatively small effect size of mtDNA evolutionary rate indicates that functional constraints or gene expression levels could still play a major role in the elevated substitution rates observed in N-mt genes. For example, Nabholz et al. (2013) found that variation in gene expression levels alone could explain differences between dN/dS in N-mt vs. mitochondrial OXPHOS genes, among multiple animal lineages, and among OXPHOS complexes within a lineage. Therefore, compensatory changes within the nucleus may be relatively rare, which is supported from previous studies usually identifying few such sites based on signatures of positive selection (Osada and Akashi 2012; Finch et al. 2014; Morales et al. 2015). Although these sites may be rare, they could still be functionally important, and identifying such sites could require a thorough, comparative experimental design.
In an effort to implement such a design, we tested whether nuclear substitutions in Silene might offset structural instability introduced by mitochondrial mutations. While specific examples were found that appeared to support this prediction, the limited sample of substitutions prevented us from conducting statistical analyses of the resulting changes in thermodynamic stability. However, extending such an analysis to a lineage with well-curated sequences from N-mt and mitochondrial OXPHOS genes from many species would allow for the evaluation of such a prediction. Mammals are obvious candidates, as resources are available from many species, mitochondrial mutation rates are high across the group (resulting in many substitutions), and solved crystal structures for OXPHOS complexes exist from mammals. One prediction would be that mutating nuclear residues simultaneously with mitochondrial residues should result in a lower ΔΔGB than mutating either set without the other.
Variation in OXPHOS evolution among and within lineages
Here, we used closely related species within Silene that experience vastly different rates of mtDNA evolution to evaluate the hypothesis that nuclear-encoded OXPHOS proteins must compensate for changes in mtDNA-encoded proteins. Although mtDNA substitution rates are often described as being slow in plants and fast in animals, this is a gross oversimplification. In addition to Silene, several other plant lineages contain species with highly variable rates of mtDNA evolution (Cho et al. 2004; Parkinson et al. 2005; Mower et al. 2007), and low mtDNA mutation rates are common in certain animals lineages (Hellberg 2006). Even among relatively recently evolved groups such as mammals, substantial heterogeneity exists in mtDNA substitution rates (Nabholz et al. 2008, 2009), with recent accelerations in primates and bats linked to metabolic adaptations for brain activity and flight (Goldberg et al. 2003; Grossman et al. 2004; Shen et al. 2010). Moreover, it is increasingly clear that variation in mutation rate is prevalent throughout some of the most deeply divergent lineages in the eukaryotic tree (Smith et al. 2012; Smith et al. 2014; Smith and Keeling 2015). Extending our comparative approach to other eukaryotes and continued characterization of mtDNA evolution in disparate lineages could further evaluate and refine the nuclear compensation hypothesis.
In addition to variation in mitochondrial evolutionary rates among lineages, significant variation exists within a genome between different genes. As an extreme example, genes in the mitochondrial genome of Ajuga were shown to experience a 340-fold range in synonymous substitution variation (Zhu et al. 2014), and similar variation exists among Silene mitochondrial genes (Sloan et al. 2009). Moreover, variation in evolutionary rate among genes within the plastid genome is also common (Jansen et al. 2007; Sloan et al. 2014), with recent evidence supporting correlations between evolutionary rates in nuclear and plastid subunits that form chimeric protein complexes (Zhang et al. 2015). In our study, the effect of mtDNA evolutionary rate on N-mt evolution was most pronounced in complexes IV and V, which also have the highest fraction of mitochondrial-encoded subunits (over 30% in both), suggesting that nuclear compensation may be most critical in complexes with the most significant mitochondrial contributions. Complex II serves as a negative control for such mitonuclear interactions (Burton et al. 2013), because it is composed entirely of nuclear-encoded subunits in many eukaryotic lineages including Silene, and we accordingly found no disparity among species pairs in dN/dS of complex II genes. In addition, we did not find any significant differences for the mitochondrial-targeted CLP, which is composed entirely nuclear-encoded subunits, and the opposite trend (lower dN/dS) has been observed for a sample of 140 nuclear-encoded ribosomal proteins targeted to the cytosol (not the mitochondria or plastids) in S. conica and S. noctiflora (Sloan et al. 2014). Therefore, it is unlikely that the observed differences in dN/dS for N-mt OXPHOS genes are the result of some general difference in species biology that is unrelated to mitochondrial function (e.g., effective population size).
Accordingly, we found substantial variation in signals for nuclear structural compensation among OXPHOS complexes. Although dN/dS analyses identified the strongest compensatory signals in complexes with the highest proportion of mtDNA-encoded proteins (complexes IV and V), structural results for nuclear compensation were mainly driven by interactions in complexes III and IV (specifically to mtDNA-encoded COX1 and COB subunits), while no support was found for structural compensation in complex V. This apparent discrepancy could be for several reasons. First, while five out of 16 ATP subunits are encoded by mtDNA, only one mitochondrial and two nuclear subunits had sufficient sequence identity for structural modeling, suggesting that many critical mitonuclear interacting residues were not evaluated in our structural analyses. Second, restricting our definition of nuclear compensation to residues that physically contact mitochondrial residues is a crude generalization that likely misses many compensatory changes (e.g. allosteric changes). Finally, we note that the greatest evidence for structural compensation comes from two mitochondrial subunits (cob and cox1) that, along with cox3, make up the core set of genes that are universally maintained in the mitochondria of all described eukaryotes with a functional electron transport chain (Gray et al. 2004; Barbrook et al. 2010; Allen 2015). Furthermore, co-introgression of N-mt genes along with introgressed mitochondrial genes was most pronounced for N-mt genes whose products directly interact with COX1 (Beck et al. 2015). This suggests that nuclear compensation may be most critical in response to mitochondrial changes that alter core properties of electron transport.
Supplementary Material
Acknowledgments
We thank members of the Sloan, Snow, and Muller labs at CSU for providing helpful comments on this study. We also thank Deb Triant and the lab of Doug Taylor for their efforts in generating the original Silene and Agrostemma transcriptome data. This work was funded by NSF grants MCB-1412260 and MCB-1022128, as well as a National Institutes of Health Postdoctoral Fellowship (F32GM116361) to JCH.
Footnotes
Data Archival Location
The sequence data from the Silene transcriptomes and mitogenomes are deposited in the NCBI’s Ssequence Read Archive (accession #: SRX1089472, SRX352988, SRX353050, SRX353047, SRX353048, SRX353031; and see Table S1 of Sloan et al. 2014). Raw data are provided in the supplementary material.
Contributor Information
Justin C. Havird, Email: justin.havird@colostate.edu.
Daniel B. Sloan, Email: dan.sloan@colostate.edu.
Literature cited
- Adrion JR, White PS, Montooth KL. The roles of compensatory evolution and constraint in aminoacyl tRNA synthetase evolution. Mol Biol Evol. 2015 doi: 10.1093/molbev/msv206. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aledo JC, Valverde H, Ruiz-Camacho M. Thermodynamic stability explains the differential evolutionary dynamics of cytochrome b and COX I in mammals. J Mol Evol. 2012;74:69–80. doi: 10.1007/s00239-012-9489-0. [DOI] [PubMed] [Google Scholar]
- Aledo JC, Valverde H, Ruiz-Camacho M, Morilla I, Lopez FD. Protein-protein interfaces from cytochrome c oxidase I evolve faster than nonbinding surfaces, yet negative selection is the driving force. Genome Biol Evol. 2014;6:3064–3076. doi: 10.1093/gbe/evu240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1500012112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbrook AC, Howe CJ, Kurniawan DP, Tarr SJ. Organization and expression of organellar genomes. Philos Trans R Soc Lond B Biol Sci. 2010;365:785–797. doi: 10.1098/rstb.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreto FS, Burton RS. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 2013;30:310–314. doi: 10.1093/molbev/mss228. [DOI] [PubMed] [Google Scholar]
- Beck EA, Thompson AC, Sharbrough J, Brud E, Llopart A. Gene flow between Drosophila yakuba and Drosophila santomea in subunit V of cytochrome c oxidase: A potential case of cytonuclear co-introgression. Evolution. 2015 doi: 10.1111/evo.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci U S A. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial-DNA. Proc Natl Acad Sci U S A. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RS, Barreto FS. A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol Ecol. 2012;21:4942–4957. doi: 10.1111/mec.12006. [DOI] [PubMed] [Google Scholar]
- Burton RS, Pereira RJ, Barreto FS. Cytonuclear genomic interactions and hybrid breakdown. Ann Rev Ecol Evol Syst. 2013;44:281–302. [Google Scholar]
- Castoe TA, Jiang ZJ, Gu W, Wang ZO, Pollock DD. Adaptive evolution and functional redesign of core metabolic proteins in snakes. PLoS One. 2008;3:e2201. doi: 10.1371/journal.pone.0002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Mower JP, Qiu YL, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci U S A. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darracq A, Varre JS, Marechal-Drouard L, Courseaux A, Castric V, Saumitou-Laprade P, Oztas S, Lenoble P, Vacherie B, Barbe V, Touzet P. Structural and content diversity of mitochondrial genome in beet: a comparative genomic analysis. Genome Biol Evol. 2011;3:723–736. doi: 10.1093/gbe/evr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y, Kwasigroch JM, Rooman M, Gilis D. BeAtMuSiC: Prediction of changes in protein-protein binding affinity on mutations. Nucleic Acids Res. 2013;41:W333–339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Vassilieva LL, Thomas WK. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science. 2000;289:2342–2344. doi: 10.1126/science.289.5488.2342. [DOI] [PubMed] [Google Scholar]
- Dohm JC, Minoche AE, Holtgrawe D, Capella-Gutierrez S, Zakrzewski F, Tafer H, Rupp O, Sorensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schulz B, Stadler PF, Schmidt T, Gabaldon T, Lehrach H, Weisshaar B, Himmelbauer H. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris) Nature. 2013;505:546–549. doi: 10.1038/nature12817. [DOI] [PubMed] [Google Scholar]
- Dowling DK. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim Biophys Acta. 2014;1840:1393–1403. doi: 10.1016/j.bbagen.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Drouin G, Daoud H, Xia J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol Phylogenet Evol. 2008;49:827–831. doi: 10.1016/j.ympev.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics. 2006;Chapter 5(Unit 5):6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch TM, Zhao N, Korkin D, Frederick KH, Eggert LS. Evidence of positive selection in mitochondrial complexes I and V of the African elephant. PLoS One. 2014;9:e92587. doi: 10.1371/journal.pone.0092587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Bond CS, Small ID. Selection patterns on restorer-like genes reveal a conflict between nuclear and mitochondrial genomes throughout angiosperm evolution. Proc Natl Acad Sci U S A. 2011;108:1723–1728. doi: 10.1073/pnas.1007667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara N, Kawabata T. HOMCOS: a server to predict interacting protein pairs and interacting sites by homology modeling of complex structures. Nucleic Acids Res. 2008;36:W185–189. doi: 10.1093/nar/gkn218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni M, Templeton AR, Mishmar D. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays. 2009;31:642–650. doi: 10.1002/bies.200800139. [DOI] [PubMed] [Google Scholar]
- Goldberg A, Wildman DE, Schmidt TR, Huttemann M, Goodman M, Weiss ML, Grossman LI. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci U S A. 2003;100:5873–5878. doi: 10.1073/pnas.0931463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- Greiner S, Bock R. Tuning a menage a trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. Bioessays. 2013;35:354–365. doi: 10.1002/bies.201200137. [DOI] [PubMed] [Google Scholar]
- Grossman LI, Wildman DE, Schmidt TR, Goodman M. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 2004;20:578–585. doi: 10.1016/j.tig.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- Havird J, Hall MD, Dowling D. Mitochondria, mutations and sex: a new hypothesis for the evolution of sex based on mitochondrial mutational erosion. Bioessays. doi: 10.1002/bies.201500057. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell. 2004;16:241–256. doi: 10.1105/tpc.016055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellberg ME. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol Biol. 2006;6:24. doi: 10.1186/1471-2148-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill GE. Mitonuclear ecology. Mol Biol Evol. 2015;32:1917–1927. doi: 10.1093/molbev/msv104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra LA, Siddiq MA, Montooth KL. Pleiotropic effects of a mitochondrial-nuclear incompatibility depend upon the accelerating effect of temperature in Drosophila. Genetics. 2013;195:1129–1139. doi: 10.1534/genetics.113.154914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang LS, Cobessi D, Tung EY, Berry EA. Binding of the respiratory chain inhibitor antimycin to the mitochondrial bc1 complex: a new crystal structure reveals an altered intramolecular hydrogen-bonding pattern. J Mol Biol. 2005;351:573–597. doi: 10.1016/j.jmb.2005.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Muller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal JR, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N. Mitonuclear match: Optimizing fitness and fertility over generations drives ageing within generations. Bioessays. 2011;33:860–869. doi: 10.1002/bies.201100051. [DOI] [PubMed] [Google Scholar]
- Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- Levin L, Blumberg A, Barshad G, Mishmar D. Mito-nuclear co-evolution: the positive and negative sides of functional ancient mutations. Front Genet. 2014;5:448. doi: 10.3389/fgene.2014.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnane AW, Marzuki S, Ozawa T, Tanaka M. Mitochondrial DNA mutations as an important contributor to ageing and degenerative diseases. Lancet. 1989;1:642–645. doi: 10.1016/s0140-6736(89)92145-4. [DOI] [PubMed] [Google Scholar]
- Lynch M. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 1997;14:914–925. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- Lynch M, Blanchard JL. Deleterious mutation accumulation in organelle genomes. Genetica. 1998;102–103:29–39. [PubMed] [Google Scholar]
- Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- Lynch M, Sung W, Morris K, Coffey N, Landry CR, Dopman EB, Dickinson WJ, Okamoto K, Kulkarni S, Hartl DL, Thomas WK. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiklejohn CD, Holmbeck MA, Siddiq MA, Abt DN, Rand DM, Montooth KL. An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 2013;9:e1003238. doi: 10.1371/journal.pgen.1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menz RI, Walker JE, Leslie AG. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene. 2006;378:11–18. doi: 10.1016/j.gene.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Moison M, Roux F, Quadrado M, Duval R, Ekovich M, Le DH, Verzaux M, Budar F. Cytoplasmic phylogeny and evidence of cyto-nuclear co-adaptation in Arabidopsis thaliana. Plant J. 2010;63:728–738. doi: 10.1111/j.1365-313X.2010.04275.x. [DOI] [PubMed] [Google Scholar]
- Morales HE, Pavlova A, Joseph L, Sunnucks P. Positive and purifying selection in mitochondrial genomes of a bird with mitonuclear discordance. Mol Ecol. 2015;24:2820–2837. doi: 10.1111/mec.13203. [DOI] [PubMed] [Google Scholar]
- Mower JP. PREP-Mt: predictive RNA editor for plant mitochondrial genes. BMC Bioinformatics. 2005;6:96. doi: 10.1186/1471-2105-6-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyes CD, Hood DA. Origins and consequences of mitochondrial variation in vertebrate muscle. Annu Rev Physiol. 2003;65:177–201. doi: 10.1146/annurev.physiol.65.092101.142705. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Some genetic aspects of sex. Am Nat. 1932;66:118–138. [Google Scholar]
- Nabholz B, Ellegren H, Wolf JB. High levels of gene expression explain the strong evolutionary constraint of mitochondrial protein-coding genes. Mol Biol Evol. 2013;30:272–284. doi: 10.1093/molbev/mss238. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. Strong variations of mitochondrial mutation rate across mammals--the longevity hypothesis. Mol Biol Evol. 2008;25:120–130. doi: 10.1093/molbev/msm248. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N. The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol. 2009;9:54. doi: 10.1186/1471-2148-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M, Taylor DR. The causes of mutation accumulation in mitochondrial genomes. Proc Biol Sci. 2009;276:1201–1209. doi: 10.1098/rspb.2008.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada N, Akashi H. Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol Biol Evol. 2012;29:337–346. doi: 10.1093/molbev/msr211. [DOI] [PubMed] [Google Scholar]
- Parkinson CL, Mower JP, Qiu YL, Shirk AJ, Song K, Young ND, DePamphilis CW, Palmer JD. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 2005;5:73. doi: 10.1186/1471-2148-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett W, Lavrov D. Mito-nuclear interactions in the evolution of animal mitochondrial tRNA metabolism. Genome Biol Evol. 2015 doi: 10.1093/gbe/evv124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–653. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Rautenberg A, Sloan DB, Alden V, Oxelman B. Phylogenetic relationships of Silene multinervia and Silene Section Conoimorpha (Caryophyllaceae) Syst Bot. 2012;37:226–237. [Google Scholar]
- Schmidt TR, Wu W, Goodman M, Grossman LI. Evolution of nuclear- and mitochondrial-encoded subunit interaction in cytochrome c oxidase. Mol Biol Evol. 2001;18:563–569. doi: 10.1093/oxfordjournals.molbev.a003836. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Wise RP. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 1998;3:175–180. [Google Scholar]
- Shen YY, Liang L, Zhu ZH, Zhou WP, Irwin DM, Zhang YP. Adaptive evolution of energy metabolism genes and the origin of flight in bats. Proc Natl Acad Sci U S A. 2010;107:8666–8671. doi: 10.1073/pnas.0912613107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1504491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB. Using plants to elucidate the mechanisms of cytonuclear co-evolution. New Phytol. 2015;205:1040–1046. doi: 10.1111/nph.12835. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Chuckalovcak JP, Wu M, McCauley DE, Palmer JD, Taylor DR. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PloS Biol. 2012a:10. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Alverson AJ, Storchova H, Palmer JD, Taylor DR. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 2010;10:274. doi: 10.1186/1471-2148-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Muller K, McCauley DE, Taylor DR, Storchova H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 2012b;196:1228–1239. doi: 10.1111/j.1469-8137.2012.04340.x. [DOI] [PubMed] [Google Scholar]
- Sloan DB, Oxelman B, Rautenberg A, Taylor DR. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol. 2009:9. doi: 10.1186/1471-2148-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan DB, Triant DA, Wu M, Taylor DR. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol Biol Evol. 2014;31:673–682. doi: 10.1093/molbev/mst259. [DOI] [PubMed] [Google Scholar]
- Smith DR, Hua JM, Lee RW, Keeling PJ. Relative rates of evolution among the three genetic compartments of the red alga Porphyra differ from those of green plants and do not correlate with genome architecture. Mol Phylogenet Evol. 2012;65:339–344. doi: 10.1016/j.ympev.2012.06.017. [DOI] [PubMed] [Google Scholar]
- Smith DR, Jackson CJ, Reyes-Prieto A. Nucleotide substitution analyses of the glaucophyte Cyanophora suggest an ancestrally lower mutation rate in plastid vs mitochondrial DNA for the Archaeplastida. Mol Phylogenet Evol. 2014;79:380–384. doi: 10.1016/j.ympev.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1422049112. [DOI] [PMC free article] [PubMed]
- Sobolev V, Eyal E, Gerzon S, Potapov V, Babor M, Prilusky J, Edelman M. SPACE: a suite of tools for protein structure prediction and analysis based on complementarity and environment. Nucleic Acids Res. 2005;33:W39–43. doi: 10.1093/nar/gki398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Shimokata K, Katayama Y, Shimada H, Muramoto K, Aoyama H, Mochizuki M, Shinzawa-Itoh K, Yamashita E, Yao M, Ishimura Y, Yoshikawa S. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci U S A. 2003;100:15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, Frauenfeld J, Berninghausen O, Neupert W, Herrmann JM, Beckmann R. Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol. 2015;7:1235–1251. doi: 10.1093/gbe/evv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk KJ. Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol. 2015;66:75–111. doi: 10.1146/annurev-arplant-043014-115547. [DOI] [PubMed] [Google Scholar]
- Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–1212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss GA, Watanabe CK, Zhong A, Goddard A, Sidhu SS. Rapid mapping of protein functional epitopes by combinatorial alanine scanning. Proc Natl Acad Sci U S A. 2000;97:8950–8954. doi: 10.1073/pnas.160252097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett CS, Burton RS. Evolution of interacting proteins in the mitochondrial electron transport system in a marine copepod. Mol Biol Evol. 2004;21:443–453. doi: 10.1093/molbev/msh031. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang F, Broughton RE. Mitochondrial-nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorylation genes? Genome Biol Evol. 2013;5:1781–1791. doi: 10.1093/gbe/evt129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Sabir J, Blazier JC, Jansen RK. Coordinated rates of evolution between interacting plastid and nuclear genes in Geraniaceae. Plant Cell. 2015;27:563–573. doi: 10.1105/tpc.114.134353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Guo W, Jain K, Mower JP. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 2014;31:1228–1236. doi: 10.1093/molbev/msu079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.