Abstract
Bioorthogonal reaction development has classically focused on bond-forming ligation reactions. In this report, we seek to expand the functional repertoire of such chemistries by introducing a new bond-cleaving reaction between N-oxide and boron reagents. The reaction features a large dynamic range of reactivity, showcasing second-order rate constants as high as 2.3 × 103 M−1s−1 using diboron reaction partners. Diboron displays minimal cell toxicity at millimolar concentrations, penetrates cell membranes, and effectively reduces N-oxides inside mammalian cells. This new bioorthogonal tool comprising miniscule components is well-suited for activating molecules within cells under chemical control. Additionally, we demonstrate that the metabolic diversity of nature enables the use of naturally occurring functionalities that display inherent biocompatibility alongside abiotic components for organism-specific applications.
Keywords: N-oxide, diboron, biorthogonal chemistry, uncaging, small molecule activator
Graphical Abstract
A new bioorthogonal bond-cleaving reaction between N-oxide and boron reagents is introduced. The reaction features second-order rate constants as high as 2.3 × 103 M−1s−1 using diboron reaction partners. Diboron displays minimal cell toxicity at millimolar concentrations, penetrates cell membranes, and reduces N-oxides inside mammalian cells. This new bioorthogonal tool comprising miniscule components is well-suited for activating molecules within cells under chemical control.
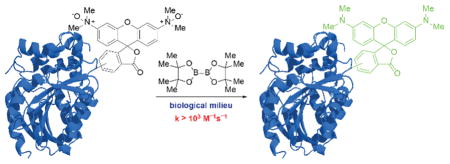
The field of bioorthogonal chemistry strives to meet the demands for tools that can facilitate the molecular analysis of biological processes. Its foundational applications were in chemical targeting of biomolecules with probes and affinity reagents, both in cultured cells and live organisms, and in building hybrid biomolecules for therapeutic applications.[1] In the context of these pursuits, chemists have compiled a reaction compendium that includes the Staudinger ligation,[2] copper-mediated[3] and metal-free[4] azide-alkyne cycloadditions, and tetrazine ligations.[5]
An emerging application of bioorthogonal chemistry is to modulate the activity of a target molecule under the control of a small molecule switch.[6] Small molecule-based chemical uncaging strategies are a compelling complement to photochemical activation methods[7] in applications demanding tissue penetrance, subcellular targeting at the multi-cell level, or cellular pretargeting. To this end, reactions that impart new functionality on their targets beyond the attachment of probes are particularly useful but remain relatively rare in the compendium—recent advances in transition metal[6a, 8] and non-metal-mediated[6b, 9] uncaging strategies notwithstanding.
The tetrazine ligation has recently been adapted by Chen and co-workers for bioorthogonal uncaging of modified lysine residues on target proteins within cells[6b] and by Robillard and co-workers for activation of prodrugs in vitro;[9d] notably, however, the reaction’s performance in the context of ligation reactions remains unmatched. In many ways, current bioorthogonal methods reflect an evolutionary history of selection for bond-forming reactions. Perhaps smaller, faster, and more tolerant bond-cleaving reactions could be constructed anew from a bioorthogonal set outside the ligation-centric compendium. Such reactions would fulfill the demand for novel triggers, which in combination with a generic immolative linker,[9a–c, 9e] would comprise a general scheme for the uncaging of biological effectors. We thus turned to new sources for bioorthogonal reaction development.
Organism-specific variations in biological reactivity space allow the potential discovery of bioorthogonal reaction partners not just from outside but also within biology. Natural organisms possess considerable metabolic diversity, such that functional groups endogenous to one species (e.g., terminal alkynes[10]) can be orthogonal in another. Trimethylamine N-oxide (TMAO, 1) provides another case in point. Elasmobranchs,[11] deep-sea teleosts,[12] and other osmoconforming marine invertebrates acquire high concentrations of urea to maintain high internal osmolalities. Chaotropic effects of urea are countered by proportionally high concentrations of the potent kosmotrope TMAO.[13] In deep-sea fish where the kosmotropic effects of TMAO are crucial for countering high hydrostatic pressures, its concentration can reach up to 400 mM.[12] Even at extreme levels, N-oxides are inert to biomolecules. Critically, for most organisms, including humans, TMAO is not a component of osmoregulation. Very low concentrations of this analyte (~1 μM) is present in serum as a metabolic byproduct of gut microbiota-derived trimethylamine.[14] By refining our notion of bioorthogonality and looking to nature for inspiration, we have identified N-oxides as potential reaction partners with inherent biocompatibility.
Similarly, boron-containing compounds fit the description that they are sometimes, yet infrequently, found in biological systems. Indeed, natural product antibiotics such as boromycin incorporate boron into their structures. Additionally, boronic acids are widely appreciated for their relatively benign toxicology profile[15] and have been incorporated into chemical biology tools such as the fluorogenic tetraserine-binding[16] and peroxide-responsive probes.[17] Given the positive qualities of this functional group, we wished to marry N-oxides with boronic acids to generate a new bioorthogonal reaction.
The hydroxydeboration reaction between TMAO and alkyl boranes was first discovered by Köster in 1967.[18] Its remarkable functional group tolerance and quantitative nature continue to render it a mild, reliable alternative to hydrogen peroxide-mediated deborylative oxidations under challenging circumstances in total syntheses.[19] Kabalka’s subsequent recognition that the dihydrate of TMAO is just as effective as its anhydrous form[20] offered the first hint that the reaction would work in aqueous media, a necessary criterion for bioorthogonality.
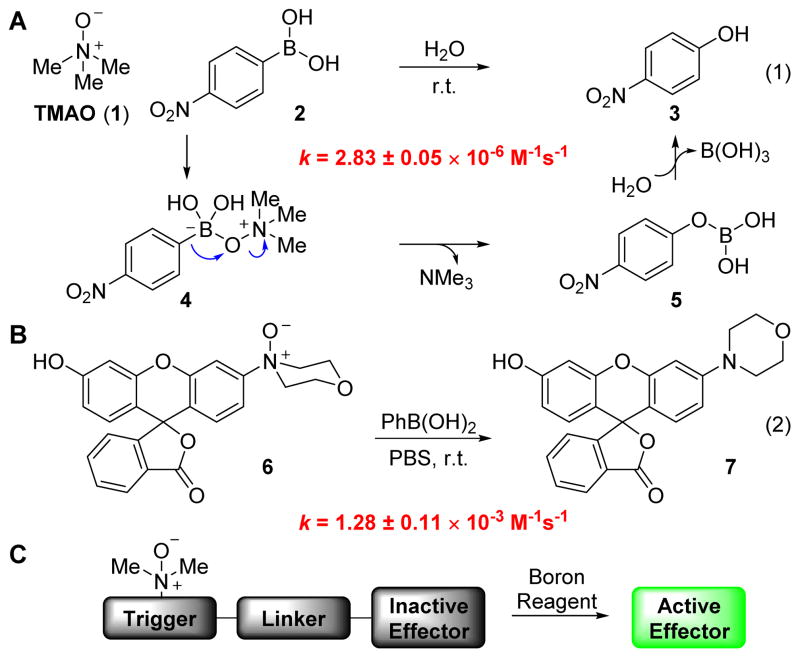
We first set out to determine the kinetic parameters for the reaction between TMAO (1) and p-nitrophenylboronic acid (2) in water. Reaction progress was monitored by UV/vis absorption of product p-nitrophenol (3) under pseudo-first order conditions. We determined a second-order rate constant of 2.83 ± 0.05 × 10−6 M−1s−1 for the reaction at room temperature, several orders of magnitude below current standards of bioorthogonal reactivity[1b–e] but nonetheless a starting point for kinetic optimization.
Conjecturing that the concomitant C–B bond migration and N–O bond cleavage events are rate limiting (Fig. 1A), we expected that the principal determinants of reaction rate would be the leaving group ability of the tertiary amine nucleofuge and the migratory aptitude of the boronic acid. Focusing first on the former, the reaction could indeed be accelerated by turning to N,N-dialkylaryl N-oxides, which produce superior leaving groups compared to trialkylamines.[21] Kinetic parameters for this reaction were measured by employing a fluorogenic N,N-dialkylaryl N-oxide 6,[22] obtained through mCPBA-mediated oxidation of the parent rhodol fluorophore.[23] The reaction of N-oxide 6 with phenylboronic acid in phosphate-buffered saline (PBS, pH 7.4) proceeded with a second-order rate constant of 1.28 ± 0.11 × 10−3 M−1s−1 (Fig. 1B). While gratified by the three orders of magnitude in rate acceleration relative to our baseline reaction using TMAO, we sought even faster rates for use in biological systems.
Figure 1.
Characterization of kinetic parameters. (A) The reaction between TMAO (1) and p-nitrophenylboronic acid (2) features biologically compatible reagents and byproducts. (B) Reaction of N,N-dialkylaniline-derived N-oxide 6 and phenylboronic acid is three orders of magnitude faster. (C) General scheme for the use of a triggering reaction for the uncaging of biological effectors.
We thus turned to the second factor: migratory aptitude of the boronic acid. We anticipated that significant gains could be achieved through direct weakening of the dissociating bond. Accordingly, we considered exchanging the migrating C–B bond for a B–B bond. The reduction of N-oxides by bis(pinacolato)diboron (B2pin2), first reported by Carter et al. in 2002,[24] exploits the cleavage of a weak B–B bond (68 kcal/mol) and formation of strong B–O bonds (125 kcal/mol) to provide an enthalpic driving force of ~180 kcal/mol. This powerful reaction, which superficially effects nothing more than deoxygenation, has seen scant use in the chemical literature.[25] We immediately sought to characterize the reaction kinetics and adapt it for biological systems.
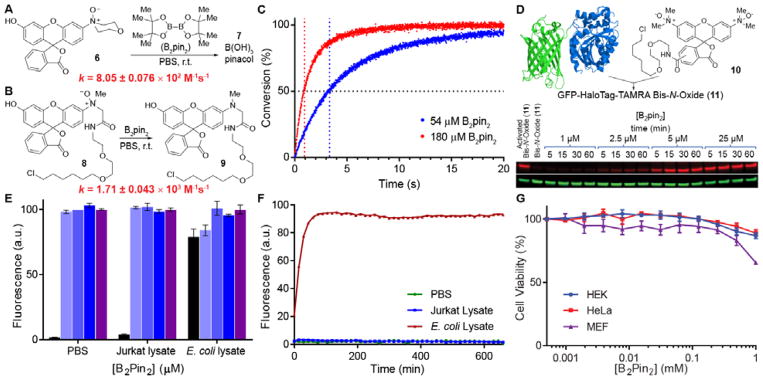
We first evaluated the kinetics of the reaction using N-oxide 6 (Fig. 2A). Impressively, fluorescence measurements obtained on a stopped-flow fluorometer under pseudo-first order conditions revealed a second-order rate constant of 8.05 ± 0.076 × 102 M−1s−1 in PBS (pH 7.4). We also synthesized HaloTag linker-bound profluorophore 8, designed for use in cell labeling studies (vide infra), and found the second-order rate constant for its reaction with B2pin2 to be even higher at 1.71 ± 0.043 × 103 M−1s−1, likely due to steric relaxation (Fig. 2B).
Figure 2.
Evaluating the bioorthogonality of diboron. (A) B2pin2 rapidly reduces N-oxide 6 in PBS (pH 7.4). The second-order rate constant was calculated from fluorescence measurements of fluorophore 7 under pseudo-first-order conditions with saturating concentrations of B2pin2. (B) Steric relaxation around the N-oxide enhances the reaction rate, and the second-order rate constant exceeds 103 M−1s−1. (C) N-oxide 9 conjugated to a 34 kDa HaloTag protein rapidly reacts with B2pin2 in PBS. Reaction conversion for the lowest and highest B2pin2 concentrations used in kinetics experiments are displayed. Data represent the mean of triplicate experiments. The vertical dotted lines trace out the half-lives of the reactions at the respective saturating concentrations of B2pin2. (D) 500 nM Bis-N-oxide profluorophore 10 was conjugated to a 64 kDa GFP-HaloTag fusion and treated with stoichiometric to slightly superstoichiometric quantities of B2pin2. The red and green channels represent TAMRA and GFP signals, respectively. (E) 1 μM N-oxide profluorophore 6 was reacted with 0 (■), 5 (
 ), 50 (
), 50 (
 ), or 500 (
), or 500 (
 ) μM B2pin2 in Jurkat and E. coli cell lysates consisting of 1 mg/mL protein and then normalized to the level of a positive control consisting of fluorophore 7 (
) μM B2pin2 in Jurkat and E. coli cell lysates consisting of 1 mg/mL protein and then normalized to the level of a positive control consisting of fluorophore 7 (
 ). Data represent the mean of triplicate experiments. Error bars represent a standard deviation. (F) Time-dependent fluorescence measurements reveal the stability of N-oxide profluorophore 6 to mammalian cell lysate. Data represent the mean of triplicate experiments. (G) Viability of three mammalian cell lines in a range of B2pin2 concentrations was evaluated after 24 h using an MTT assay. Data represent the mean of triplicate experiments. Error bars represent a standard deviation.
). Data represent the mean of triplicate experiments. Error bars represent a standard deviation. (F) Time-dependent fluorescence measurements reveal the stability of N-oxide profluorophore 6 to mammalian cell lysate. Data represent the mean of triplicate experiments. (G) Viability of three mammalian cell lines in a range of B2pin2 concentrations was evaluated after 24 h using an MTT assay. Data represent the mean of triplicate experiments. Error bars represent a standard deviation.
We then explored the reaction on a biomolecule. The 34-kDa HaloTag protein was ligated to compound 8 to produce HaloTag-8, which was purified by size exclusion chromatography and treated with B2pin2 under pseudo-first-order conditions. Analysis by stopped-flow fluorometry revealed a second-order rate constant of 2.30 ± 0.073 × 103 M−1s−1 (Fig. 2C). It should be noted, however, that pseudo-first-order kinetics performed under saturating conditions can obscure important information regarding the deactivation of the diboron reagent through sequestration or off-target reactivity. To address this issue, a 64-kDa GFP-HaloTag fusion protein was expressed, ligated to compound 10 to produce GFP-HaloTag-10. Conjugate 11 (500 nM) was then treated with stoichiometric to slightly superstoichiometric quantities of B2pin2 (1–25 μM) and analyzed by in-gel fluorescence imaging. Fig. 2D shows that 5–10 equiv. B2pin2 are necessary to fully reduce conjugated fluorophore 10 in < 15 min. Provided that 2 equiv. of reductant are theoretically required, this experiment validates the robustness of the reaction and suggests minimal off-target reactivity.
Having verified the compatibility of our N-oxide–diboron reaction with proteins, we explored the viability of the reaction in mammalian (Jurkat) cell lysate (Fig. 2E). Lysates were made to a final concentration of 1 mg/mL protein and variable concentrations of B2pin2 were reacted with 1 μM probe 6. Fluorescence intensities were then measured after 30 min. Consistent with prior kinetic data, adding just 5 equiv. of B2pin2 was sufficient in driving the reaction to completion in mammalian cell lysate within the allotted time.
An organism-centric approach to bioorthogonal reaction development, a central facet of our thesis, enables the implementation of reactions that would be overlooked under more stringent searches for abiotic reaction partners. As a point of emphasis, we also performed the preceding experiment in E. coli cell lysate. E. coli, as do several other facultative anaerobes, possesses an N-oxide reductase[26] and was predicted to be incompatible with our method. Indeed, the results bore this out: our negative control containing no B2pin2 displayed high levels of fluorescence (Fig. 2E). Time-dependent fluorescence measurements of N-oxide probe 6 in E. coli cell lysate confirmed the gradual reduction of N-oxides for which we measured a half-life of ~15 min (Fig. 2F). Gratifyingly, no degradation was observed in mammalian cell lysate over 11 h. Therefore, the N-oxide–diboron reaction is not suitable for all systems, but should still be powerful in many.
In order to ascertain the toxicity of diboron reagents to cells, we subjected three mammalian cell lines (HEK293T, HeLa and MEF) to the MTT cell viability assay (Fig. 2G). Each cell line was treated with 2-fold serial dilutions of B2pin2 starting at 1 mM in 0.5% DMSO/DMEM. The maximum concentration of B2pin2 was dictated by its solubility limitations in the medium. The MTT assays indicated that the human cell lines are relatively insensitive to diboron, at least up to the concentrations that were tested, and while MEFs showed some toxicity at higher diboron levels, the IC50 was >1 mM. No aberrations in cell morphology were observed for HEK293T cells upon treatment with 1 mM B2pin2 for 24 h (Fig. S5). Additionally, no gross changes in time to confluency were observed between cells treated with B2pin2 and DMSO vehicle.
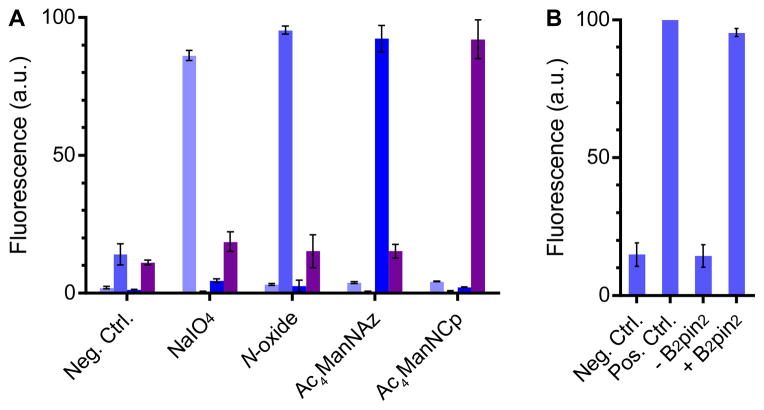
Next, we demonstrated the mutual orthogonality of our N-oxide–diboron reaction with a representative cohort of bioorthogonal reactions commonly in use today: the aminooxy-aldehyde condensation, the azide-cyclooctyne cycloaddition, and the tetrazine-cyclopropene ligation (Fig. 3). And to showcase the robustness of the new reaction, we executed this experiment amidst the complexity of a cellular system. Utilizing a combination of chemical, genetic, and metabolic engineering techniques, four populations of HEK293T cells were endowed with distinct cell surface modifications. Population 1 was modified by sodium periodate to display cell surface aldehydes derived from sialic acid moieties;[27] population 2 was modified with azides through the metabolic incorporation of Ac4ManNAz;[2] population 3 was modified with a cyclopropene through the metabolic incorporation of Ac4ManNCp;[28] and population 4 was modified by cell surface expression of an N-terminal HaloTag-EGFR fusion protein followed by the ligation of HaloTag-linked bis-N-oxide TAMRA profluorophore 10. Each cell population was labeled with a distinct combination of Hoechst 33342 and Syto 41 nuclear stains, combined, then treated with a reagent cocktail consisting of 10 mM aniline, 100 μM aminooxy–Alexa Fluor 488, 10 μM DIBAC-Cy5, 20 μM tetrazine-Cy7, and 100 μM B2pin2 in pH 6.7 PBS. B2pin2 was applied at a concentration of 100 μM to rigorously challenge the aminooxy condensation reaction, the reaction most likely to be perturbed by diboron reagents. The mélange of cells was then analyzed by flow cytometry.
Figure 3.
The N-oxide–diboron reaction is compatible with common bioorthogonal reactions. (A) HEK293T cell surfaces were modified with aldehydes, N-oxides, azides, or cyclopropenes, mixed, then treated with a cocktail of bioorthogonal reagents. Each population was analyzed by flow cytometry for reaction with aminooxy-Alexa Fluor 488 (
 ), B2pin2 (
), B2pin2 (
 ), DIBAC-Cy5 (
), DIBAC-Cy5 (
 ), and tetrazine-Cy7 (
), and tetrazine-Cy7 (
 ). The mean normalized fluorescence intensity of each fluorophore from three biological replicates is plotted. Error bars represent a standard deviation. (B) N-oxide-modified HEK293T cells were treated with aminooxy, DIBAC, and tetrazine reagents with or without diboron. N-oxides are not reactive toward hydroxylamines, cyclooctynes, or tetrazines.
). The mean normalized fluorescence intensity of each fluorophore from three biological replicates is plotted. Error bars represent a standard deviation. (B) N-oxide-modified HEK293T cells were treated with aminooxy, DIBAC, and tetrazine reagents with or without diboron. N-oxides are not reactive toward hydroxylamines, cyclooctynes, or tetrazines.
Impressively, each of the cell types was labeled highly selectively by its corresponding bioorthogonal partner, and the labeling efficiency was undiminished in the presence of the complete complement of reactive functional groups (Fig. 3). Notably, aminooxy functional groups are fully compatible with diboron, and the N-oxide functional group is neither reduced by nor reactive toward tetrazines, which are known to be susceptible to degradation through nucleophilic attack.
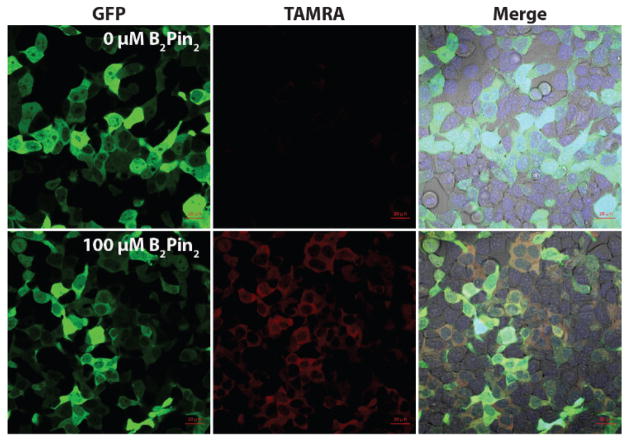
Finally, to demonstrate the utility of the N-oxide–diboron reaction in an intracellular uncaging application, we first had to determine the membrane permeability of each of the compounds and evaluate their bioorthogonality in live cells. We transiently transfected HEK293T cells with a cytosolic GFP-HaloTag fusion construct then treated the cells with 100 μM profluorophore 10. After three washes, the cells were treated with 1 mM, 100 μM, 10 μM, or 0 μM B2pin2 for 45 min (Figs. 4, S1). The TAMRA signal increased upon addition of B2pin2 and also co-localized with the GFP signal. These data indicate that both profluorophore 10 and diboron are cell permeable, the reaction can be performed intracellularly, and impressively, fluorophore activation can be observed at 10 μM diboron with full activation requiring at most 100 μM B2pin2, well below toxic levels.
Figure 4.
B2pin2 activates N-oxides on cytosolic proteins in mammalian cells. HEK293T cells transfected with GFP-HaloTag were incubated with 100 μM profluorophore 10, washed, treated with 0 or 100 μM B2pin2, then imaged by confocal microscopy after 45 min. The merge is a composite of GFP, TAMRA, and Hoechst 33342 fluorescence with a brightfield image.
Fast rates of ligand hydrolysis, even of bulky pinacol derivatives, enables the utility of diboron species even amidst diols present in the biological milieu. B2(OH)4 is as nontoxic and competent at reducing N-oxides (Figs. S2–S4) as B2pin2. In addition, tolerance of the reaction to sterically demanding ligand structures on the boron reagents indicates that even diboron species transiently ligated to saccharides or other cellular diols may still be competent at reducing N-oxides.
In conclusion, we sought new reactions to expand the functional repertoire of bioorthogonal chemistries beyond bond-forming ligation reactions. We initiated our search for new bioorthogonal reactivity by relaxing its definition. Rather than exploring chemical space completely outside the realm of biology, we focused our search for molecular components within organisms that are metabolically orthogonal to models of human disease. We could readily find novel reaction components with inherent biocompatibility as a result.
We have thus described a powerful new reaction between N-oxide and boron reagents. The reaction features fast reaction kinetics that rival the fastest bioorthogonal reactions currently in the literature while introducing functionality that has yet to gain significant attention. Due to the benignity of the reaction conditions toward live cells and its intracellular compatibility, this reaction holds much promise as a tool for small molecule activation of biomolecules in vivo.
We envision the role of this technology in spatially and temporally controlled chemical uncaging of important naturally-occurring trialkylamines such as epigenetically relevant dimethyllysine histone residues.[29] Additionally, this reaction acts as a powerful trigger, which, when interfaced to chemical or biological effectors through immolative linkers[9] containing an N-oxide, should enable the small molecule activation of enzymes and chemotherapeutics.
Supplementary Material
Acknowledgments
We acknowledge Dr. Paresh Agarwal, Dr. Daniel Liu, Peter Robinson, and Mason Appel for critical discussions, and Prof. Susan Marqusee for generous access to stopped flow instrumentation and HaloTag protein. J.K. acknowledges the Miller Institute for Basic Research in Science at UC Berkeley for a postdoctoral fellowship and research funding. This research was supported by the NIH (GM058867).
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- 1.a) Agarwal P, Bertozzi CR. Bioconjugate Chem. 2015;26:176–192. doi: 10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lang K, Chin JW. ACS Chem Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]; c) McKay Craig S, Finn MG. Chem Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Patterson DM, Nazarova LA, Prescher JA. ACS Chem Biol. 2014;9:592–605. doi: 10.1021/cb400828a. [DOI] [PubMed] [Google Scholar]; e) Sletten EM, Bertozzi CR. Angew Chem, Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Gong Y, Pan L. Tetrahedron Lett. 2015;56:2123–2132. [Google Scholar]
- 2.Saxon E, Bertozzi CR. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 3.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem, Int Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 4.Jewett JC, Bertozzi CR. Chem Soc Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a) Devaraj NK, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2008;19:2297–2299. doi: 10.1021/bc8004446. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Li J, Yu J, Zhao J, Wang J, Zheng S, Lin S, Chen L, Yang M, Jia S, Zhang X, Chen PR. Nat Chem. 2014;6:352–361. doi: 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]; b) Li J, Jia S, Chen PR. Nat Chem Biol. 2014;10:1003–1005. doi: 10.1038/nchembio.1656. [DOI] [PubMed] [Google Scholar]
- 7.a) Nguyen DP, Mahesh M, Elsässer SJ, Hancock SM, Uttamapinant C, Chin JW. J Am Chem Soc. 2014;136:2240–2243. doi: 10.1021/ja412191m. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gautier A, Deiters A, Chin JW. J Am Chem Soc. 2011;133:2124–2127. doi: 10.1021/ja1109979. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Lemke EA, Summerer D, Geierstanger BH, Brittain SM, Schultz PG. Nat Chem Biol. 2007;3:769–772. doi: 10.1038/nchembio.2007.44. [DOI] [PubMed] [Google Scholar]
- 8.Völker T, Meggers E. Curr Opin Chem Biol. 2015;25:48–54. doi: 10.1016/j.cbpa.2014.12.021. [DOI] [PubMed] [Google Scholar]
- 9.a) Azoulay M, Tuffin G, Sallem W, Florent JC. Bioorg Med Chem Lett. 2006;16:3147–3149. doi: 10.1016/j.bmcl.2006.03.073. [DOI] [PubMed] [Google Scholar]; b) Brakel Rv, Vulders RCM, Bokdam RJ, Grüll H, Robillard MS. Bioconjugate Chem. 2008;19:714–718. doi: 10.1021/bc700394s. [DOI] [PubMed] [Google Scholar]; c) Gorska K, Manicardi A, Barluenga S, Winssinger N. Chem Commun. 2011;47:4364–4366. doi: 10.1039/c1cc10222b. [DOI] [PubMed] [Google Scholar]; d) Versteegen RM, Rossin R, ten Hoeve W, Janssen HM, Robillard MS. Angew Chem, Int Ed. 2013;52:14112–14116. doi: 10.1002/anie.201305969. [DOI] [PubMed] [Google Scholar]; e) Matikonda SS, Orsi DL, Staudacher V, Jenkins IA, Fiedler F, Chen J, Gamble AB. Chem Sci. 2015;6:1212–1218. doi: 10.1039/c4sc02574a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Yamaguchi M, Park HJ, Ishizuka S, Omata K, Hirama M. J Med Chem. 1995;38:5015–5022. doi: 10.1021/jm00026a008. [DOI] [PubMed] [Google Scholar]; b) Minto RE, Blacklock BJ. Prog Lipid Res. 2008;47:233–306. doi: 10.1016/j.plipres.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhu X, Liu J, Zhang W. Nat Chem Biol. 2015;11:115–120. doi: 10.1038/nchembio.1718. [DOI] [PubMed] [Google Scholar]
- 11.Yancey P, Clark M, Hand S, Bowlus R, Somero G. Science. 1982;217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 12.Yancey PH, Gerringer ME, Drazen JC, Rowden AA, Jamieson A. Proc Natl Acad Sci. 2014;111:4461–4465. doi: 10.1073/pnas.1322003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yancey PH. Amer Zool. 2001;41:699–709. [Google Scholar]
- 14.a) Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, Hazen SL. Nat Med. 2013;19:576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, DuGar B, Feldstein AE, Britt EB, Fu X, Chung YM, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, Hazen SL. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trippier PC, McGuigan C. MedChemComm. 2010;1:183–198. [Google Scholar]
- 16.Halo TL, Appelbaum J, Hobert EM, Balkin DM, Schepartz A. J Am Chem Soc. 2009;131:438–439. doi: 10.1021/ja807872s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang MCY, Pralle A, Isacoff EY, Chang CJ. J Am Chem Soc. 2004;126:15392–15393. doi: 10.1021/ja0441716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Köster R, Morita Y. Liebigs Ann Chem. 1967;704:70–90. [Google Scholar]
- 19.a) Carruthers W, Cumming SA. J Chem Soc, Chem Commun. 1983:360–361. [Google Scholar]; b) Nicolaou KC, Tria GS, Edmonds DJ. Angew Chem Int Ed. 2008;47:1780–1783. doi: 10.1002/anie.200800066. [DOI] [PubMed] [Google Scholar]
- 20.Kabalka GW, Hedgecock HC. J Org Chem. 1975;40:1776–1779. [Google Scholar]
- 21.Zhu C, Wang R, Falck JR. Org Lett. 2012;14:3494–3497. doi: 10.1021/ol301463c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Niwa M, Hirayama T, Okuda K, Nagasawa H. Org Biomol Chem. 2014;12:6590–6597. doi: 10.1039/c4ob00935e. [DOI] [PubMed] [Google Scholar]; b) Hirayama T, Okuda K, Nagasawa H. Chemical Science. 2013;4:1250–1256. [Google Scholar]
- 23.Peng T, Yang D. Org Lett. 2010;12:496–499. doi: 10.1021/ol902706b. [DOI] [PubMed] [Google Scholar]
- 24.Carter CAG, John KD, Mann G, Martin RL, Cameron TM, Baker RT, Bishop KL, Broene RD, Westcott SA. In: Group 13 Chemistry: From Fundamentals to Applications. Shapiro PJ, Atwood DA, editors. American Chemical Society; Washington, D.C: 2002. pp. 70–87. [Google Scholar]
- 25.a) Kokatla HP, Thomson PF, Bae S, Doddi VR, Lakshman MK. J Org Chem. 2011;76:7842–7848. doi: 10.1021/jo201192c. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Gurram V, Akula HK, Garlapati R, Pottabathini N, Lakshman MK. Adv Synth Catal. 2015;357:451–462. doi: 10.1002/adsc.201400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.a) Sakaguchi M, Kawai A. Bull Jpn Soc Sci Fish. 1975;41:661–665. [Google Scholar]; b) Barrett EL, Kwan HS. Annu Rev Microbiol. 1985;39:131–149. doi: 10.1146/annurev.mi.39.100185.001023. [DOI] [PubMed] [Google Scholar]
- 27.Zeng Y, Ramya TNC, Dirksen A, Dawson PE, Paulson JC. Nat Methods. 2009;6:207–209. doi: 10.1038/nmeth.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Späte AK, Bußkamp H, Niederwieser A, Schart VF, Marx A, Wittmann V. Bioconjugate Chem. 2014;25:147–154. doi: 10.1021/bc4004487. [DOI] [PubMed] [Google Scholar]
- 29.a) Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) David Y, Vila-Perelló M, Verma S, Muir TW. Nat Chem. 2015;7:394–402. doi: 10.1038/nchem.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.