Abstract
Background
Anti-cytokine autoantibodies (ACAAs) are pathogenic in a handful of rare immunodeficiencies. However, the prevalence and significance of other ACAAs across immunodeficiencies have not yet been described.
Objective
We sought to profile ACAAs in a diverse cohort of serum samples from patients with immunodeficiency and assess the sensitivity and specificity of protein microarrays for ACAA identification and discovery.
Methods
Highly multiplexed protein microarrays were designed and fabricated. Blinded serum samples from a cohort of 58 patients with immunodeficiency and healthy control subjects were used to probe microarrays. Unsupervised hierarchical clustering was used to identify clusters of reactivity, and after unblinding, significance analysis of microarrays was used to identify disease-specific autoantibodies. A bead-based assay was used to validate protein microarray results. Blocking activity of serum containing ACAAs was measured in vitro.
Results
Protein microarrays were highly sensitive and specific for the detection of ACAAs in patients with autoimmune polyendocrine syndrome type I and pulmonary alveolar proteinosis, detecting ACAA levels consistent with those in the published literature. Protein microarray results were validated by using an independent bead-based assay. To confirm the functional significance of these ACAAs, we tested and confirmed the blocking activity of select ACAAs in vitro.
Conclusion
Protein microarrays are a powerful tool for ACAA detection and discovery, and they hold promise as a diagnostic for the evaluation and monitoring of clinical immunodeficiency.
Keywords: Anti-cytokine autoantibodies, immunodeficiency, autoimmune polyendocrine syndrome type I, pulmonary alveolar proteinosis, thymoma, protein microarray
In patients with a handful of immunodeficiencies, recent work has shown that the presence of serum autoantibodies against endogenous cytokines accurately distinguishes discrete disease entities.1,2 Important examples include autoimmune polyendocrine syndrome type I (APS-1) associated with antibodies against type I interferons and TH17-related cytokines,3–5 pulmonary alveolar proteinosis (PAP) caused by antibodies against GM-CSF,6–9 and acquired susceptibility to mycobacterial infection associated with antibodies to IFN-γ.10–12 These anti-cytokine autoantibodies (ACAAs) are thought to be an integral component of disease pathogenesis because of 3 strong lines of evidence: epidemiologic specificity, in vitro blocking activity, and biologic plausibility. ACAAs against other cytokines have also been described in the sera of healthy subjects.13 These observations raise the possibility that ACAAs might be a ubiquitous component of immune homeostasis in health and disease.
Current tools to measure ACAAs include ELISA-based4,14 and bead-based5 assays. Additionally, new tools, such as a liquidphase luciferase assay, allow for detection of 10s of ACAAs simultaneously from a single sample.15,16 However, key questions in the field remain regarding the landscape of ACAAs in immune homeostasis and whether ACAAs contribute to the pathophysiology of other immunodeficiencies. New proteomic tools are needed to query for autoantibodies against hundreds of cytokines, chemokines, and growth factors in health and disease.17,18 Previously, our laboratory has developed protein microarrays for the detection of ACAAs in patients with the autoimmune disease systemic lupus erythematosus and identified enrichment of ACAAs against B cell–activating factor in patients.19 We also showed that arrays could detect ACAAs against IFN-α in patients with APS-1 and ACAAs against IFN-γ in patients with atypical mycobacterial infection.19 Here, using a cohort of patients referred to the immunodeficiency service at Cambridge University Hospital, we describe the further development and validation of a highly multiplexed tool for the detection and discovery of ACAAs using microliter quantities of serum or plasma.
METHODS
Protein microarrays
Protein microarrays were printed by using a Bio-Rad ChipWriter Compact robotic microarrayer and ChipWriter Pro software (Bio-Rad Laboratories, Hercules, Calif), as described previously.20 Briefly, 104 purified biomolecules were purchased from multiple vendors and printed in triplicate at dilutions of 200 µg/mL onto nitrocellulose-coated glass slides (Maine Manufacturing, Sanford, Me). A complete list of molecules and their vendors can be found in Fig E1 in this article’s Online Repository at www.jacionline.org.
Arrays were first blocked in 5% milk (Bio-Rad Laboratories) in PBS plus 0.1% Tween (PBST; Sigma-Aldrich, St Louis, Mo) for 1 hour and then washed 3 times in PBST. Arrays were probed with serum diluted 1:150 in 10% FCS in PBST. Arrays were subjected to three 5-minute washes in PBST. Serum reactivity was detected by using an Alexa Fluor 647–conjugated goat antihuman IgG secondary antibody (Jackson Laboratory, Bar Harbor, Me) diluted to 0.375 µg/mL in PBST for 45 minutes. Arrays were washed 3 times in PBST and dried under negative pressure.
Arrays were scanned with an Axon microarray scanner and processed with GenePix 6 software (Molecular Devices, Sunnyvale, Calif). Local background (defined as the mean of pixels surrounding each unique circular feature of interest) was subtracted from mean fluorescence intensity (MFI). Means were calculated across replicates. Those values that were negative because of background subtraction were made zero.
Statistics
Unsupervised hierarchical clustering was performed by using a Pearson correlation with average linkage clustering with the program Multiple Experiment Viewer.21 Significance analysis of microarrays (SAM) was performed as previously described, with significantly different reactivities defined by a false discovery rate of less than 0.1% after 10,000 permutations of the data.22 Pearson correlation coefficients, receiver operating characteristic curves, and linear regression were generated with Prism 6 software (GraphPad Software, San Diego, Calif). For blocking experiments, unpaired t tests of stimulation indices were calculated by using Prism 6 software (GraphPad Software).
Multiplex particle-based flow cytometry
Recombinant human cytokines (IL-1β, IL-6, IL-10, IL-12p40, IL-12p70, IL-17A, IL-17F, IL-18, IL-22, IL-23, IL-26 monomer, IL-26 dimer, IFN-α, IFN-β, IFN-γ, TNF-α, and GM-CSF; R&D Systems, Minneapolis, Minn) were covalently coupled to carboxylated beads (Bio-Plex; Bio-Rad Laboratories). Beads were first activated with 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride (Thermo Fisher Scientific, Waltham, Mass) in the presence of N-hydroxysuccinimide (Thermo Fisher Scientific), according to the manufacturer’s instructions, to form amine-reactive intermediates. The activated beads were incubated with the corresponding cytokines at a concentration of 20 µg/mL in the reaction mixture for 3 hours at 37°C on a rotator. Beads were washed and stored in blocking buffer (PBS, 1% BSA, and 0.05% NaN3). Cytokine-coupled beads were incubated with plasma or serum from patients for 1 hour in 96-well filter plates (Multi Screen HTS; Millipore, Temecula, Calif) at 37°C in the dark on a horizontal shaker. Fluids were aspirated with a vacuum manifold, and beads were washed 3 times with PBS/0.05% Tween 20. Beads were incubated for 30 minutes with a phycoerythrin-labeled anti-human IgG-Fc antibody (Leinco/Biotrend, St Louis, Mo), washed as described, and resuspended in 100 µL of PBS/Tween. Samples were then analyzed on a Bio-Plex platform by using Bio-Plex Manager 6.1 software (Bio-Rad Laboratories). Successful coupling of the cytokines to their respective bead sets was verified with specific mAbs.
IFN-α neutralization assay
The ability of IFN-α to upregulate LPS-induced production of TNF-α by PBMCs was used to test for the neutralizing activity of sera containing hightiter blocking antibodies to IFN-α.
Anti–IFN-α–positive sera, negative control sera, or FBS were preincubated for 30 minutes in a 96-well plate (Corning, Corning, NY) at a dilution of 1:5 in RPMI 1640. IFN-α (2000 IU per well, INTRONA; Merck, Kenilworth, NJ) or LPS (1 µg/mL; BioLabs, ■■■) alone or both stimuli were added. Human PBMCs (separated from blood of a healthy control subject by using Ficoll-Hypaque gradient centrifugation) were then added at a concentration of 1 × 105 cells per well and incubated for 24 hours (37°C in a 5% CO2 atmosphere).
Supernatants were obtained and cytokines were measured on a Luminex analyzer (TNF-α, R&D Systems Fluorokine MAP and Bio-Plex, Bio-Rad Laboratories), according to the manufacturer’s recommendations.
GM-CSF neutralization assay
GM-CSF–mediated upregulation of LPS-induced production of IL-6 by U937 cells was used to test for the neutralizing activity of sera containing high-titer blocking antibodies to GM-CSF.
Anti–GM-CSF–positive sera, negative control sera, or FBS were preincubated for 30 minutes in a 96-well plate without stimulus or in the presence of GM-CSF (100 ng/mL; ImmunoTools, Friesoythe, Germany), IFN-γ (2 × 104 IU/mL; Immukin; Boehringer Ingelheim, Ingelheim am Rhein, Germany), or LPS (1 µg/mL, BioLabs) alone or in combination as indicated at a dilution of 1:5 in complete Dulbecco modified Eagle medium (Gibco, Carlsbad, Calif). U937 cells (ATCC CRL1593.2) were added at 1 × 105 cells per well and incubated for 24 hours (37°C in a 5% CO2 atmosphere).
Supernatants were obtained, and IL-6 levels were measured with a Luminex analyzer (R&D Systems Fluorokinemap and Bio-Plex, Bio-Rad Laboratories), according to the manufacturer’s recommendations.
IL-12 neutralization assay
The ability of IL-12 to synergistically upregulate the production of IFN-γ from PBMCs on costimulation with IL-18 was used to measure the neutralizing activity of sera containing high-titer blocking antibodies to IL-12.
Anti–IL-12–positive sera, negative control sera, or FBS were preincubated for 30 minutes in a 96-well plate without stimulus or in the presence of IL-12 (20 ng/mL, R&D Systems) or IL-18 (25 ng/mL, R&D Systems) or in combination, as indicated, at a dilution of 1:5 in RPMI 1640 in 96-well F plates (Corning). Human PBMCs (separated from blood of a healthy control subject by means of Ficoll-Hypaque gradient centrifugation) were then added at a concentration of 1 × 105 cells per well and incubated for 24 hours (37°C in a 5% CO2 atmosphere).
Supernatants were obtained, and IFN-γ concentrations were measured with a standard ELISA (IFN-γ; Pelikine; Sanquin, Amsterdam, The Netherlands), according to the manufacturer’s recommendations.
Accession numbers
Protein microarray data were deposited into the Gene Expression Omnibus and are freely available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE62599.
Human samples and study approval
Serum samples were collected from patients with immunodeficiency and control subjects after obtaining verbal consent and anonymized for subsequent investigation at the Immunodeficiency Service at Addenbrooke’s Hospital, Cambridge University Hospitals, Cambridge, United Kingdom.
RESULTS
Protein microarrays identify distinct clusters of anticytokine autoantibody reactivity in a blinded cohort of patients with immunodeficiency disorders
Protein microarrays were fabricated by using a robotic microarray printer and printed on nitrocellulose-coated glass microscope slides. One hundred four purified biomolecules, including known autoantigens, as well as cytokines, chemokines, growth factors, and receptors, were printed on the array, forming discrete spot features (Fig 1, A, and see Fig E1, A). All targets were printed in triplicate.
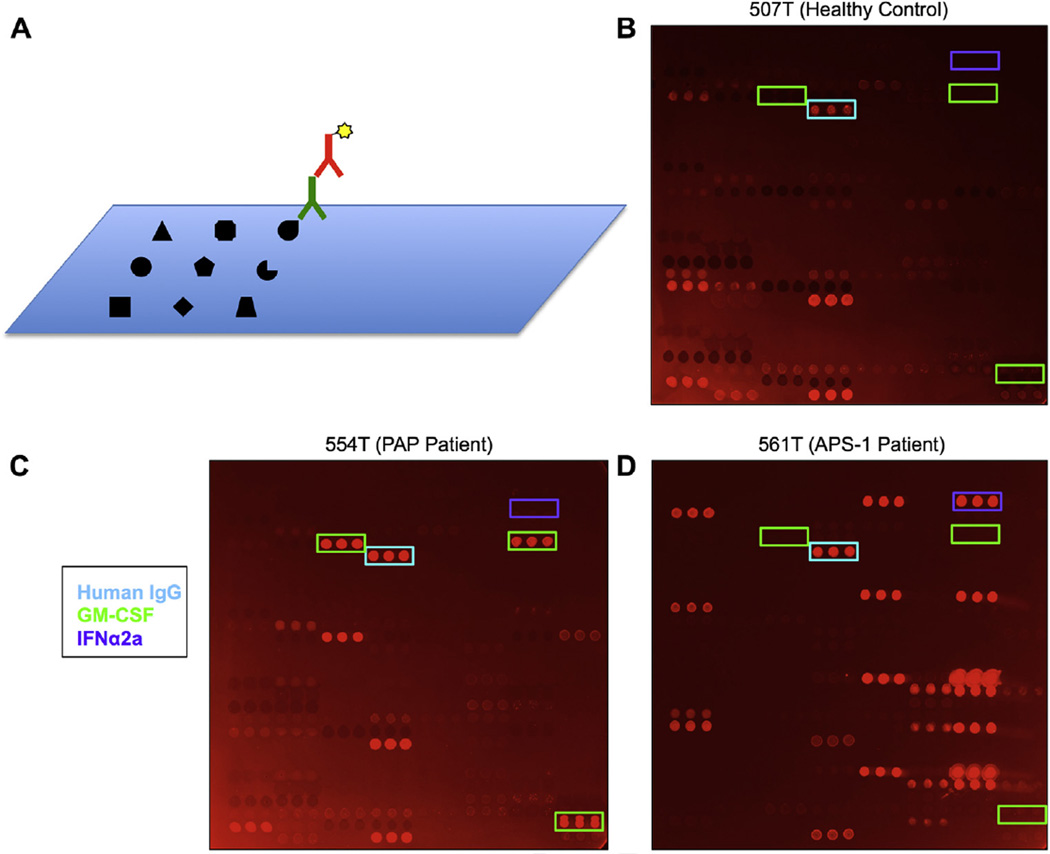
FIG 1.
Protein microarrays for ACAA detection. A, Schematic representation of cytokine microarrays. Cytokines, chemokines, growth factors, and traditional autoantigens are printed onto a nitrocellulose-coated microscope slide. Arrays are probed with serum (green) and detected by using fluorophore-conjugated secondary antibody (red). B–D, Representative images of protein microarrays probed with serum from a healthy control subject (Fig 1, B), a patient with PAP (Fig 1, C), and a patient with APS-1 (Fig 1, D). A control IgG feature printed on the array is boxed in cyan, GM-CSF features are boxed in green, and IFN-α2a features are boxed in purple.
To determine whether protein microarrays could accurately detect known or novel autoantibodies in serum, we probed arrays with serum samples from an immunodeficiency cohort collected at Cambridge University Hospital. This cohort consists of patients with immunodeficiency disorders of diverse cause referred from throughout Europe and includes patients with the diseases PAP and APS-1 and susceptibility to mycobacterial infection, each of which are conditions with described ACAAs (Table I and see Fig E2 in this article’s Online Repository at www.jacionline.org). Sera were anonymized and provided with barcoded identification, and protein microarrays were then probed and analyzed. Only after data analysis was complete was unblinding to clinical presentation performed.
TABLE I.
Clinical characteristics of the cohort
| Disease | No. of patients (n = 58) |
|---|---|
| APS-1 | 5 |
| APS-1 phenotype without known AIRE mutation | 2 |
| Atypical mycobacterial infection | 6 |
| Candidiasis | 6 |
| Thymoma | 3 |
| PAP | 11 |
| Suspected PAP | 4 |
| Other (see Fig E2 for a complete listing of other immunodeficiencies) | 15 |
| Healthy control | 6 |
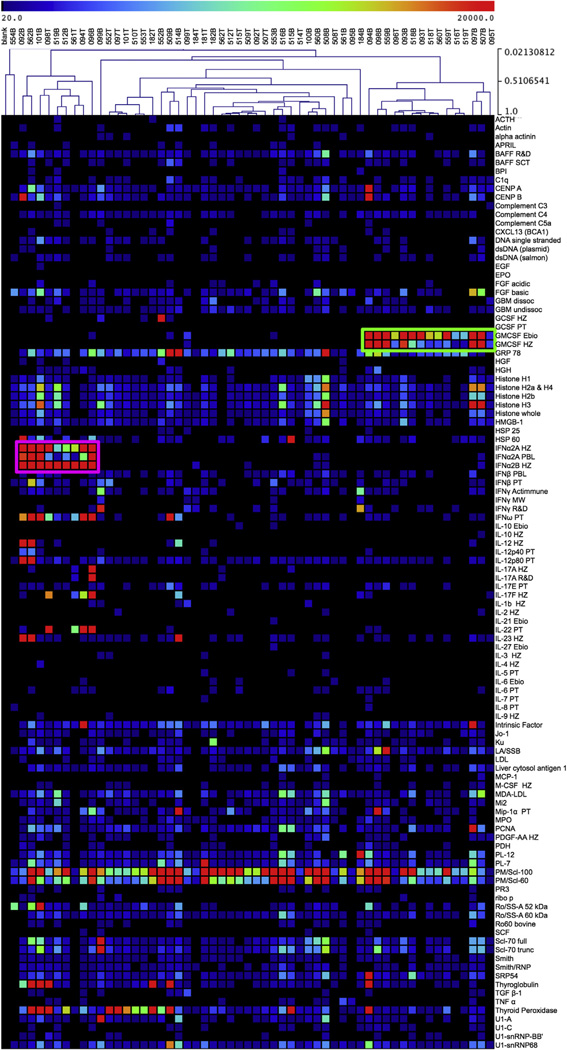
Protein microarrays showed a diversity of reactivity to various autoantigens, cytokines, and chemokines across samples (Figs 1, B–D, and 2). After unsupervised hierarchical clustering, distinct patterns of reactivity were clearly observed in at least 2 groups by means of clustering linkage. The first group demonstrated a marked reactivity to GM-CSF purchased from multiple vendors (Fig 2, green box), which led us to hypothesize that these patients had PAP.
FIG 2.
Unsupervised hierarchical clustering demonstrates 2 distinct groups of cytokine reactivity. Fifty-eight serum samples from patients with immunodeficiency and healthy control subjects were probed on protein microarrays. Samples were hierarchically clustered in an unsupervised fashion by using a Pearson correlation and plotted with samples along the x-axis, and autoantigens, cytokines, and chemokines were plotted along the y-axis. Clusters of high reactivity to GM-CSF (green box) and type I interferons (purple box) were identified.
The second group identified by means of unsupervised hierarchical clustering demonstrated marked reactivity to type I interferons, such as IFN-α2A, IFN-α2B, and IFN-ω, which were also purchased from multiple commercial vendors (Fig 2, purple box). This led us to hypothesize that these samples were derived from patients with APS-1 or thymoma. Intriguingly, we observed 2 distinct subgroups within this reactivity group. The first subgroup demonstrated reactivity to the TH1-associated cytokines IL-12 and IL-23, whereas the second subgroup demonstrated reactivity to the TH17-associated cytokines IL-17A, IL-17F, and IL-22.
In addition, we observed reactivity to a number of traditional autoantigens. Autoantigen reactivity was broadly distributed throughout the cohort, although we observed nonstatistically significant clustering of reactivities to features, such as histone components; nuclear autoantigens, such as the U1 small nuclear ribonucleoprotein complex (U1-snRNP); and endocrine autoantigens, such as thyroglobulin, thyroid peroxidase, and intrinsic factor. Reactivity to endocrine autoantigens was enriched in samples with reactivity to type I interferons. The finding of endocrine-associated autoantibodies supported the designation of these samples as derived from patients with APS-1.
Protein microarray ACAA profiling accurately assigns clinical disease
Once data analysis was complete, we were unblinded to the clinical history of individual subjects within the cohort. Of 16 patients with PAP, the arrays correctly identified all 16 as those samples with the highest reactivity against GM-CSF (Fig 2, boxed in green). Protein microarrays also identified 9 samples with high type I interferon reactivity (boxed in purple). We observed that all 5 patients with APS-1 in the cohort (101B, 096B, 094T, 561T, and 098T) fell within this assignment. The other 4 patients with high type I interferon reactivity all had chronic candidiasis, a key clinical feature of APS-1, which in patients with APS-1 has been proposed to be caused by neutralizing ACAAs to the TH17 cytokines IL-17A, IL-17F, and IL-22.3,5 Two of these 4 patients (092B and 512B) had coincident thymoma, for which antibodies against type I interferons and TH1 and TH17 cytokines have been described.16,23 The other 2 patients were clinically suspected of having APS-1; however, a mutation in autoimmune regulator (AIRE) was not identified by means of gene sequencing, and imaging results for thymoma were negative. Given the specificity of type I interferon ACAAs for APS-1 and the clinical similarity of these patients with those with APS-1, we speculate that these patients might harbor an undescribed mutation in AIRE or a mutation in a gene that might phenocopy APS-1.
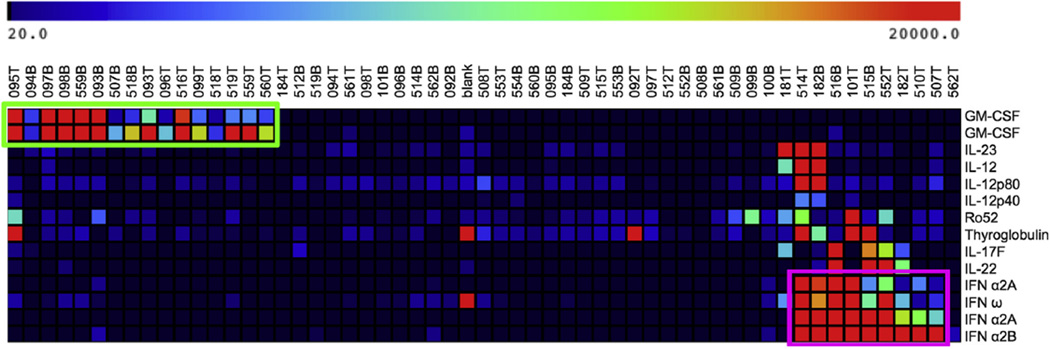
To identify additional autoantibodies that might be associated with either APS-1 or PAP, we performed 3-class SAM, a permutation-based algorithm designed for analysis of large data sets.22 We assigned groupings based on the presence of ACAAs against IFN-α, GM-CSF, or neither. With a false discovery rate of less than 0.1%, SAM identified the ACAAs discussed previously and additionally identified autoantibodies against the autoantigens Ro52 and thyroglobulin as enriched in patients with APS-1 (Fig 3). Reactivity to thyroglobulin, a thyroid autoantigen, is consistent with the systemic endocrine autoimmunity described in patients with APS-1. The presence of Ro52 autoantibodies, which are found prototypically in patients with systemic lupus erythematosus and Sjögren syndrome, suggests a possible shared autoimmune pathophysiology with APS-1 and identifies Ro52 as a candidate biomarker worthy of further study in patients with APS-1.24,25
FIG 3.
SAM identifies reactivities associated with ACAAs against type I interferons and GM-CSF. Three-class SAM was applied based on cytokine reactivity to type I interferons, GM-CSF, or neither. SAM identified 14 statistically significant differences. These targets include cytokines purchased from multiple vendors. GMCSF reactivity and type I interferon–associated reactivity are boxed in green and purple, respectively, for emphasis.
Validation of protein microarray profiling results on a second platform
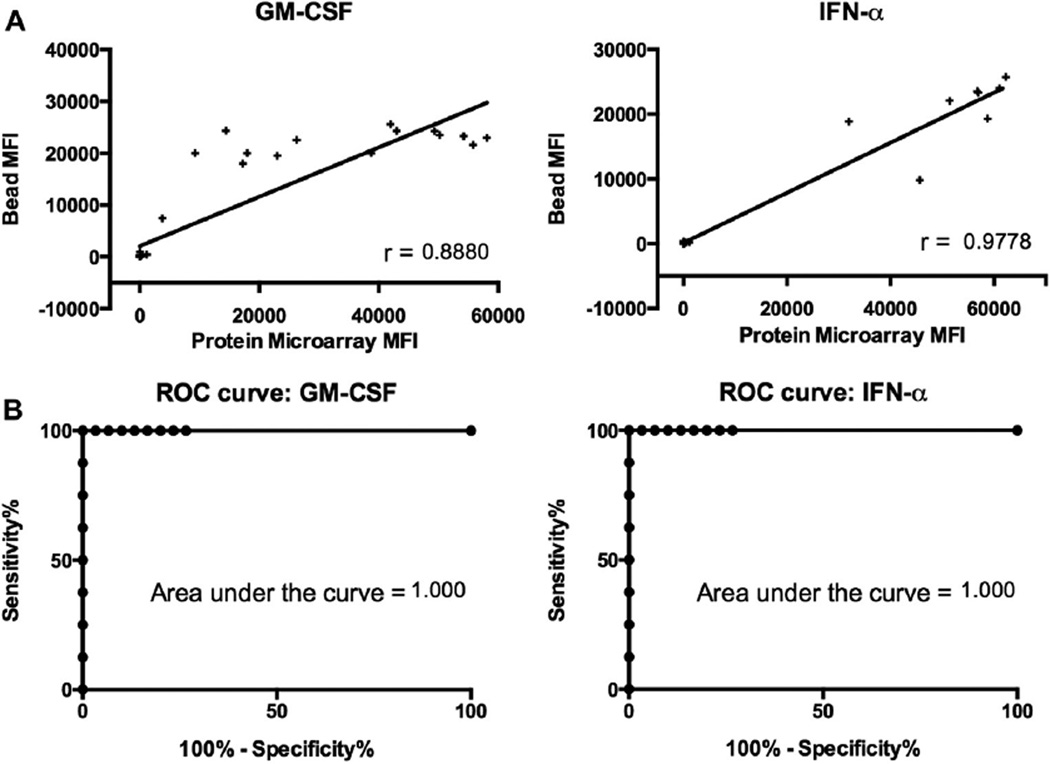
To validate our results, we compared protein microarray reactivities with results from a bead-based assay performed independently in the laboratory of one of the authors (R.D.). We compared ACAAs against GM-CSF and IFN-α between the 2 platforms. Pearson correlation coefficients were r values of 0.888 and 0.9778, respectively (Fig 4). Both correlations had P values of less than .005.
FIG 4.
Comparison of protein microarray performance with an established bead-based assay. A, Corresponding MFI values from protein microarrays and a bead-based assay were plotted for the targets GM-CSF and IFN-α, and Pearson correlation coefficients were determined. All P values were less than .005. B, Receiver operating characteristic (ROC) curves were generated to assess the performance of protein microarrays, with a bead-based assay used as the gold standard. P values were less than .05 for all areas under the curve.
To more precisely determine whether protein microarrays could sensitively and specifically identify samples with ACAAs, we built receiver operating characteristic curves for each of the ACAAs listed above using the bead-based assay as the gold standard. Receiver operating characteristic curves had areas under the curve of 1.000 for both GM-CSF and IFN-α.19 This high level of concordance between protein microarrays and an independently performed bead-based assay demonstrate that protein microarrays can detect ACAAs with a sensitivity and specificity comparable with those of current gold standard assays.
Sera containing select ACAAs block cytokine signaling in vitro
To test the functional activity of identified ACAAs, we performed in vitro blocking assays using sera from patients with ACAAs and healthy control subjects. We tested the ability of sera with ACAAs against GM-CSF, IFN-α, and IL-12 to block downstream cytokine production from heterologous PBMCs or U937 cells, as described below.
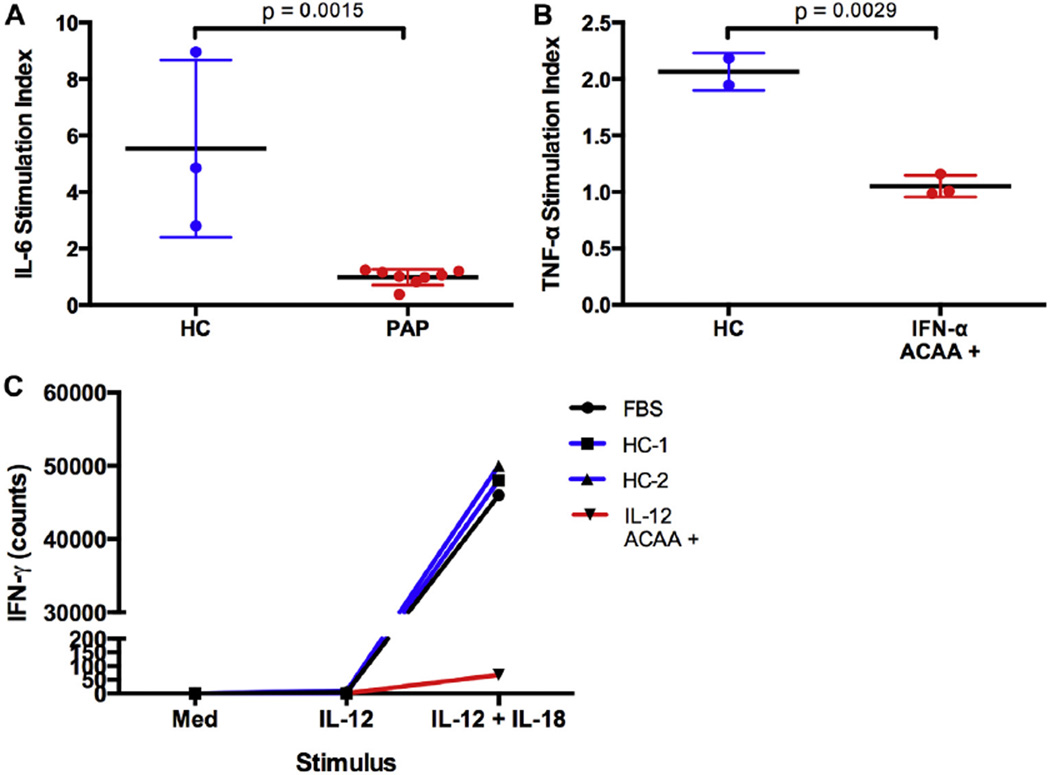
To test GM-CSF blocking activity, we incubated U937 cells with sera from either patients with PAP or healthy control subjects. Cultures were stimulated with LPS alone or LPS and GM-CSF. IL-6 release was measured from the supernatants by using ELISA as a readout for signaling activity. After stimulation with LPS alone, low IL-6 levels were detected in the supernatants of all samples. However, when LPS and GM-CSF were added together, we observed a synergistic upregulation of IL-6 release in healthy control but not PAP cultures (Fig 5, A). To quantify this effect, we plotted the ratio of IL-6 released after stimulation with LPS and GM-CSF and divided by IL-6 released after LPS stimulation alone. The difference between healthy control and PAP indices was statistically significant (P = .0015). These results demonstrate that sera containing ACAAs against GMCSF but not sera from healthy control subjects have the ability to block GM-CSF signaling in vitro.
FIG 5.
Sera from patients with ACAAs block cytokine signaling in vitro. A, U937 cells were incubated in the presence of sera from healthy control subjects or patients with PAP and stimulated with LPS alone or LPS and GM-CSF. IL-6 concentrations were measured by the using Luminex analyzer. An IL-6 stimulation index was calculated as described in the text. B, Heterologous PBMCs were incubated in the presence of LPS alone or LPS and IFN-α. TNF-α concentration was measured by using a Luminex analyzer. A TNF-α stimulation index was calculated as described in the text. C, Heterologous PBMCs were incubated in the presence of medium, IL-12, or IL-12 and IL-18. IFN-γ levels in the supernatant were measured by using ELISA and plotted. Given the limited sample size, statistical analysis of a stimulation index was not performed.
To test the blocking activity of IFN-α ACAA-containing sera, we measured supernatant levels of TNF-α released from heterologous PBMCs. It is known that LPS induces production of TNF-α from PBMCs.26 We found that stimulation by the combination of LPS and IFN-α resulted in a synergistic increase in TNF-α production above that induced by LPS alone. In the presence of sera containing IFN-αACAAs but not the presence of sera from healthy control subjects, this synergistic increase in TNF-α levels was blocked (Fig 5, B). To quantify this effect, we plotted the ratio of TNF-α released after stimulation with LPS and IFN-α and divided by TNF-α released after LPS stimulation alone (P =.0029). These data show that sera containing IFN-α ACAAs but not those from healthy control subjects have the ability to block IFN-α signaling in vitro.
Finally, we tested the ability of IL-12 ACAA–containing sera to block the activity of IL-12. To do so, we used the ability of IL-12 to synergistically upregulate the production of IFN-γ from heterologous PBMCs when costimulated with IL-18.27 This effect was blocked by the single IL-12 ACAA–containing serum sample available for testing (Fig 5, E). Given the small sample size, statistical analysis was not performed. To test whether blocking activity was restricted to the serum or whether an additional cell-intrinsic defect was present, we performed a mixing experiment. Washed PBMCs from an IL-12 ACAA–positive patient regained their ability to upregulate IFN-γ secretion in the presence of serum from a healthy control subject, and washed PBMCs from a healthy control subject lost their ability to upregulate IFN-γ in the presence of patient serum (data not shown). These experiments demonstrate that in the single patient available for testing, IL-12 ACAA–containing serum can block IL-12 signaling in vitro and that this IL-12 signaling defect is intrinsic to the serum component in the conditions tested.
Protein microarrays detect novel ACAAs
Protein microarrays also identified individual samples with interesting or novel ACAA reactivities by using a threshold of reactivity of 2 SDs greater than the mean of all features. For example, sample 552B, a patient with multiple sclerosis and borderline neutropenia receiving IFN-β treatment, demonstrated high reactivity to granulocyte colony-stimulating factor, a critical cytokine in granulopoiesis.28 ACAAs to granulocyte colony-stimulating factor have been described in a cohort of patients with Felty syndrome, which is characterized by the triad of rheumatoid arthritis, neutropenia, and splenomegaly,29 but have not been described in patients with multiple sclerosis. Two other patients with PAP demonstrated high reactivity to basic fibroblast growth factor, an important growth factor involved in angiogenesis, wound healing, and tissue repair.30 To our knowledge, basic fibroblast growth factor autoantibodies have not been previously described, although we identified sporadic reactivity against this cytokine in cohorts of previously screened patients with lupus.19
Additionally, we identified ACAA reactivities not previously described in the literature. Serum samples 092B and 562B demonstrated prominent reactivity to IL-23. Sample 092B was derived from a patient with thymoma and chronic candidiasis, and sample 562B was derived from a patient with a congenital immunodeficiency of unknown cause with normal AIRE sequencing and chest imaging results. To confirm these results, we tested 39 samples for IL-23 ACAAs using our bead-based assay. Samples 092B and 562B were the only 2 samples with MFIs 2 SDs greater than the mean. Comparing the bead-based MFIs of these 2 samples against all others yielded a P value of less than .0001, confirming our array-based results (see Fig E3 in this article’s Online Repository at www.jacionline.org).
IL-23 is composed of p19 and p40 subunits, the latter of which is shared with IL-12. Indeed, on both our protein microarrays and bead-based assay, serum samples 092B and 562B were reactive against IL-12 in addition to IL-23. Given the known association of IL-12 ACAAs in patients with thymoma and a recent case report in a patient with Burkholderia species infection, we hypothesized that these IL-23ACAAs bind the shared p40 subunit.16,23,31 Using both protein microarrays and our bead-based assay, we successfully detected ACAAs in samples 562B and 092B against the shared IL-12p40 subunit (Fig 2). These results suggest that IL-12 ACAAs might cross-react against the related cytokine IL-23 through recognition of the shared p40 subunit.
Novel reactivity was also observed against macrophage inflammatory protein 1α (MIP-1α). Serum samples 514B and 098B both demonstrated reactivity to MIP-1α, which was defined as greater than 2 SDs above the mean of all feature reactivities. After unblinding, we found that sample 514B was derived from a healthy control subject; however, this sample also demonstrated autoantibodies against the autoantigens PM/Scl-60, PM/Scl-100, and GRP-78, suggesting possible undiagnosed or not yet clinically apparent autoimmunity. This patient also harbored moderate ACAAs against IL-23 and IL-12, although at lower levels than the patients discussed above. The other MIP-1α–reactive sample (098B) was obtained from a patient with PAP and contained autoantibodies against GM-CSF and the autoantigens PM/Scl-60 and PM/Scl-100. These data demonstrate that protein microarrays can detect novel ACAAs, although further studies will be required to determine their clinical significance.
DISCUSSION
The burgeoning study of ACAAs has already provided key contributions to our understanding of human immunity and immunodeficiency. Studies of diseases, such as PAP, APS-1, and acquired susceptibility to nontuberculous mycobacterial infection, have demonstrated the fundamental roles in immunity of their ACAA targets: GM-CSF; IL-17A, IL17-F, and IL-22; and IFN-γ, respectively. These insights arose from careful clinical observation of “experiments of nature” combined with a rich knowledge of cytokine biology.32
A complementary strategy for the discovery of novel ACAAs is an unbiased multiplexed approach. Using protein microarray technology, we have developed a tool that allows for the screening of an order of magnitude more ACAAs than current technologies permit, probing for hundreds of ACAAs simultaneously by using only a few microliters of serum. Protein microarrays are a powerful tool for the discovery of novel ACAAs in patients with immunodeficiency and autoimmunity and can be applied to any disease context in which serum is available.17 Additionally, ACAAs have been described in healthy subjects, and protein microarrays provide the opportunity to study how ACAAs can regulate immune homeostasis by using a proteomic approach.
Here we have shown that protein microarrays can accurately detect ACAAs in patients with PAP and APS-1 from a cohort of patients with complex immunodeficiency. We included cytokine antigens on the array from multiple vendors to ensure the accuracy of our platform. Although MFI values from different vendors can vary to some degree because of discrepancies, such as differences in expression systems, there remains a high degree of correlation between different vendors with accurate classification of disease throughout. We then validated our platform by correlating our results to an established bead-based assay.
Because of their high-throughput nature and ease of use, protein microarrays also hold potential as a diagnostic tool in the clinic, allowing screening for multiple ACCAs simultaneously with a single platform. Given the high coincidence of autoimmunity and immunodeficiency demonstrated in this cohort, protein microarray diagnostics could simultaneously measure ACAA and traditional autoantibody levels, alerting clinicians to organ-specific autoimmunity and monitoring ACAA-disease progression and response.33,34
The results described here provide the broadest view to date of the landscape of ACAAs in patients with immunodeficiency, and they identify targets worthy of further investigation. Our cohort consists of a diverse group of patients with immunodeficiency. One of the most striking observations is the robust nature of the reactivity signal in patients with APS-1 and PAP, which is in agreement with previously published work.4,6 In almost all samples from patients with APS-1 and PAP, the MFI of type I interferon and GM-CSF ACAAs, respectively, approached the upper limits of detection on our instrument. Although MFI is not designed here as a precise quantitative measurement, the MFIs of these reactivities were also markedly higher than those of other well-studied clinical autoantibodies, such as the antithyroglobulin and anti-thyroid peroxidase autoantibodies found in patients with autoimmune thyroiditis.
A second observation is that unlike APS-1 and PAP, we do not find groups of high-titer ACAAs to be of a uniform pattern across all immunodeficiency states. Although this cohort lacks statistical power to rule out ACAAs as enriched in many of the individual immunodeficiencies tested, the extensive list of cytokines tested here across many different immunodeficiencies suggests that the ACAAs tested might not be a general phenomenon of immunodeficiency. Although our arrays screened for ACAAs against 50 cytokines, chemokines, and other secreted proteins, it is possible that some ACAA targets were not included in our arrays. Furthermore, it is also possible that with increased sample size of individual immunodeficiencies, other unique ACAA patterns might emerge. We are currently developing expanded protein microarrays to test both possibilities.
A third conclusion from our results is the description of previously undescribed high-titer ACAAs. We found ACAAs against IL-23 in 2 patients with type I interferon reactivity and have shown that sera from these patients also react against the shared p40 subunit of both IL-12 and IL-23. We have not excluded the possibility that these ACAAs additionally bind IL-23p19, although a previous study of patients with thymoma assayed for but did not identify IL-23p19 ACAAs.16 We also observed high levels of traditional autoantibodies in IL-12/IL-23–reactive samples. Our observations hold potential clinical relevance because an mAb directed against IL-12p40 is approved for the treatment of psoriasis and has demonstrated efficacy in the treatment of Crohn disease.35,36
We also identified previously undescribed ACAAs against MIP-1α in the sera of 2 subjects, although a clinical presentation could not be correlated to these ACAAs. The finding of ACAAs against MIP-1α in serum from a healthy control subject parallels the finding of sporadic classical autoantibodies in healthy subjects.37 Whether such ACAAs have physiologic or diagnostic importance cannot be determined from our experiments.
Lastly, we tested the functional activity of ACAAs by assaying the capacity of serum from patients to block cytokine signaling. We designed our assays for IFN-α, GM-CSF, and IL-12 blocking activity to measure the ability of serum to block downstream cytokine secretion from heterologous cells. Previous studies have used downstream signal transducer and activator of transcription phosphorylation as a readout for blocking activity.5,16,38 The consistency of our results with published findings that used other assays is reassuring.
Pressing questions still remain about the cause and function of ACAAs.3 First, what gives rise to the autoantibody patterns found in patients with diseases such as APS-1 and PAP? Possible causes of ACAAs include autoimmunization events or specific defects in tolerance. Second, what is the function of ACAAs? In patients with PAP, passive transfer experiments in nonhuman primates, as well as the clinical phenotype of both human subjects and mice with defects in the GM-CSF receptor, offer convincing evidence in support of the pathophysiology of GM-CSF ACAAs.7,39–42 Additionally, in the case of PAP and susceptibility to atypical mycobacterial infection associated with ACAAs against IFN-γ, pilot studies have shown efficacy for B-cell depletion therapy with rituximab.43–45 Such studies provide supporting evidence toward a pathophysiologic role for these ACAAs; however, a functional role for many high-titer ACAAs in health and disease is still lacking.
In summary, we have developed and validated a tool for the discovery and study of ACAAs in patients with immunodeficiency. We have demonstrated a high level of accuracy of the platform and used it to profile a cohort of patients with diverse immunodeficiency. In doing so, we have generated a broad view of the landscape of ACAAs and traditional autoantibodies in immunodeficiency to serve as a resource for future investigation and have identified novel ACAAs worthy of further study. Future efforts focused on screening cohorts of specific diseases for ACAAs with this technology are needed to better understand the complicated role of ACAAs in both health and disease.
Supplementary Material
Key messages.
We have profiled anti-cytokine, anti-chemokine, and traditional autoantibodies in patients with diverse immunodeficiencies at the deepest level to date.
Protein microarrays accurately detect ACAAs and will be an important tool for the discovery of novel ACAAs.
Sera from patients with ACAAs against IFN-α and GMCSF block signaling of their respective cytokines in vitro.
Acknowledgments
Research reported in this publication was supported by the National Institutes of Health (NIH) under award number T32GM007365. P.J.U. is the recipient of a Donald E. and Delia B. Baxter Foundation Career Development Award, a gift from the Floren Family Trust, and a gift from the Ben May Charitable Trust (Mobile, Alabama) and is supported by National Heart, Lung, and Blood Institute (NHLBI) Proteomics contract N01-HV-00242, HSN288201000034C, NIH grants (1 U19-AI1110491, 1 U19-AI090019, 1 UH2 AR067676, 1 UM2 AR067678, and 1 UM1A110498), the Alliance for Lupus Research (grant no. 21858), and FP grant no. 261. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 261382. G.B.-M. was supported in part by grants from UNAM-DGAPA-PAPIIT (codes IN217312-3 and IN220815). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
D. S. Kumararatne has received grants from the National Institute for Health Research Cambridge Biomedical Centre, has received payment for lectures from Baxter, and has received travel support from CSL Behring and Shire.
Abbreviations used
- ACAA
Anti-cytokine autoantibody
- AIRE
Autoimmune regulator
- APS-1
Autoimmune polyendocrine syndrome type I
- MFI
Mean fluorescence intensity
- MIP-1α
Macrophage inflammatory protein 1α
- PAP
Pulmonary alveolar proteinosis
- PBST
PBS plus Tween
- SAM
Significance analysis of microarrays
Footnotes
Disclosure of potential conflict of interest: The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Browne SK. Anticytokine Autoantibody–Associated Immunodeficiency. Annu Rev Immunol. 2014;32:635–657. doi: 10.1146/annurev-immunol-032713-120222. [DOI] [PubMed] [Google Scholar]
- 2.Browne SK, Holland SM. Anticytokine autoantibodies in infectious diseases: pathogenesis and mechanisms. Lancet Infect Dis. 2010;10:875–885. doi: 10.1016/S1473-3099(10)70196-1. [DOI] [PubMed] [Google Scholar]
- 3.Kisand K, Bøe Wolff AS, Podkrajšek KT, Tserel L, Link M, Kisand KV, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meager A, Visvalingam K, Peterson P, Möll K, Murumägi A, Krohn K, et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med. 2006;3:e289. doi: 10.1371/journal.pmed.0030289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puel A, Döffinger R, Natividad A, Chrabieh M, Barcenas-Morales G, Picard C, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–297. doi: 10.1084/jem.20091983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitamura T, Tanaka N, Watanabe J, Uchida Kanegasaki S, Yamada Y, et al. Idiopathic pulmonary alveolar proteinosis as an autoimmune disease with neutralizing antibody against granulocyte/macrophage colony-stimulating factor. J Exp Med. 1999;190:875–880. doi: 10.1084/jem.190.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakagami T, Uchida K, Suzuki T, Carey BC, Wood RE, Wert SE, et al. Human GM-CSF autoantibodies and reproduction of pulmonary alveolar proteinosis. N Engl J Med. 2009;361:2679–2681. doi: 10.1056/NEJMc0904077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell BC, Whitsett JA, Nakata K. Pulmonary alveolar proteinosis. N Engl J Med. 2003;349:2527–2539. doi: 10.1056/NEJMra023226. [DOI] [PubMed] [Google Scholar]
- 9.Uchida K, Beck DC, Yamamoto T, Berclaz PY, Abe S, Staudt MK, et al. GM-CSF autoantibodies and neutrophil dysfunction in pulmonary alveolar proteinosis. N Engl J Med. 2007;356:567–579. doi: 10.1056/NEJMoa062505. [DOI] [PubMed] [Google Scholar]
- 10.Browne SK, Burbelo PD, Chetchotisakd P, Suputtamongkol Y, Kiertiburanakul S, Shaw PA, et al. Adult-onset immunodeficiency in Thailand and Taiwan. N Engl J Med. 2012;367:725–734. doi: 10.1056/NEJMoa1111160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Döffinger R, Helbert MR, Barcenas-Morales G, Yang K, Dupuis S, Ceron-Gutierrez L, et al. Autoantibodies to interferon-γ in a patient with selective susceptibility to mycobacterial infection and organ-specific autoimmunity. Clin Infect Dis. 2004;38:e10–e14. doi: 10.1086/380453. [DOI] [PubMed] [Google Scholar]
- 12.Höflich C, Sabat R, Rosseau S, Temmesfeld B, Slevogt H, Döcke WD, et al. Naturally occurring anti-IFN-gamma autoantibody and severe infections with Mycobacterium cheloneae and Burkholderia cocovenenans. Blood. 2004;103:673–675. doi: 10.1182/blood-2003-04-1065. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe M, Uchida K, Nakagaki K, Kanazawa H, Trapnell BC, Hoshino Y, et al. Anti-cytokine autoantibodies are ubiquitous in healthy individuals. FEBS Lett. 2007;581:2017–2021. doi: 10.1016/j.febslet.2007.04.029. [DOI] [PubMed] [Google Scholar]
- 14.Meager A, Wadhwa M. Detection of anti-cytokine antibodies and their clinical relevance. Expert Rev Clin Immunol. 2014;10:1029–1047. doi: 10.1586/1744666X.2014.918848. [DOI] [PubMed] [Google Scholar]
- 15.Burbelo PD, Ching KH, Klimavicz CM, Iadarola MJ. Antibody profiling by luciferase immunoprecipitation systems (LIPS) J Vis Exp. 2009;(32) doi: 10.3791/1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burbelo PD, Browne SK, Sampaio EP, Giaccone G, Zaman R, Kristosturyan E, et al. Anti-cytokine autoantibodies are associated with opportunistic infection in patients with thymic neoplasia. Blood. 2010;116:4848–4858. doi: 10.1182/blood-2010-05-286161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenberg JM, Utz PJ. Protein microarrays: a new tool for the study of autoantibodies in immunodeficiency. Front Immunol. 2015;6:138. doi: 10.3389/fimmu.2015.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maecker HT, Lindstrom TM, Robinson WH, Utz PJ, Hale M, Boyd SD, et al. New tools for classification and monitoring of autoimmune diseases. Nat Rev Rheumatol. 2012;8:317–328. doi: 10.1038/nrrheum.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price JV, Haddon DJ, Kemmer D, Delepine G, Mandelbaum G, Jarrell JA, et al. Protein microarray analysis reveals BAFF-binding autoantibodies in systemic lupus erythematosus. J Clin Invest. 2013;123:5135–5145. doi: 10.1172/JCI70231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson WH, DiGennaro C, Hueber W, Haab BB, Kamachi M, Dean EJ, et al. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- 21.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, et al. TM4: a free, open-source system for microarray data management and analysis. Bio Techniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 22.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meager A, Wadhwa M, Dilger P, Bird C, Thorpe R, Newsom-Davis J, et al. Anti-cytokine autoantibodies in autoimmunity: preponderance of neutralizing autoantibodies against interferon-alpha, interferon-omega and interleukin-12 in patients with thymoma and/or myasthenia gravis. Clin Exp Immunol. 2003;132:128–136. doi: 10.1046/j.1365-2249.2003.02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghillani P, André C, Toly C, Rouquette AM, Bengoufa D, Nicaise P, et al. Clinical significance of anti-Ro52 (TRIM21) antibodies non-associated with anti-SSA 60kDa antibodies: results of a multicentric study. Autoimmun Rev. 2011;10:509–513. doi: 10.1016/j.autrev.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Husebye ES, Perheentupa J, Rautemaa R, Kämpe O. Clinical manifestations and management of patients with autoimmune polyendocrine syndrome type I. J Intern Med. 2009;265:514–529. doi: 10.1111/j.1365-2796.2009.02090.x. [DOI] [PubMed] [Google Scholar]
- 26.Duffy D, Rouilly V, Libri V, Hasan M, Beitz B, David M, et al. Functional analysis via standardized whole-blood stimulation systems defines the boundaries of a healthy immune response to complex stimuli. Immunity. 2014;40:436–450. doi: 10.1016/j.immuni.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Döffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27:277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 28.Lieschke GJ, Burgess AW. Granulocyte colony-stimulating factor and granulocyte-macrophage colony-stimulating factor (1) N Engl J Med. 1992;327:28–35. doi: 10.1056/NEJM199207023270106. [DOI] [PubMed] [Google Scholar]
- 29.Hellmich B, Csernok E, Schatz H, Gross WL, Schnabel A. Autoantibodies against granulocyte colony-stimulating factor in Felty’s syndrome and neutropenic systemic lupus erythematosus. Arthritis Rheum. 2002;46:2384–2391. doi: 10.1002/art.10497. [DOI] [PubMed] [Google Scholar]
- 30.Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16:107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Sim BT, Browne SK, Vigliani M, Zachary D, Rosen L, Holland SM, et al. Recurrent Burkholderia gladioli suppurative lymphadenitis associated with neutralizing anti-IL-12p70 autoantibodies. J Clin Immunol. 2013;33:1057–1061. doi: 10.1007/s10875-013-9908-z. [DOI] [PubMed] [Google Scholar]
- 32.Casanova JL, Abel L. The genetic theory of infectious diseases: a brief history and selected illustrations. Annu Rev Genomics Hum Genet. 2013;14:215–243. doi: 10.1146/annurev-genom-091212-153448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bussone G, Mouthon L. Autoimmune manifestations in primary immune deficiencies. Autoimmun Rev. 2009;8:332–336. doi: 10.1016/j.autrev.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 34.Todoric K, Koontz JB, Mattox D, Tarrant TK. Autoimmunity in immunodeficiency. Curr Allergy Asthma Rep. 2013;13:361–370. doi: 10.1007/s11882-013-0350-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths CE, Strober BE, van de Kerkhof P, Ho V, Fidelus-Gort R, Yeilding N, et al. Comparison of ustekinumab and etanercept for moderate-to-severe psoriasis. N Engl J Med. 2010;362:118–128. doi: 10.1056/NEJMoa0810652. [DOI] [PubMed] [Google Scholar]
- 36.Sandborn WJ, Gasink C, Gao LL, Blank MA, Johanns J, Guzzo C, et al. Ustekinumab induction and maintenance therapy in refractory Crohn’s disease. N Engl J Med. 2012;367:1519–1528. doi: 10.1056/NEJMoa1203572. [DOI] [PubMed] [Google Scholar]
- 37.Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, James JA, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349:1526–1533. doi: 10.1056/NEJMoa021933. [DOI] [PubMed] [Google Scholar]
- 38.Patel SY, Ding L, Brown MR, Lantz L, Gay T, Cohen S, et al. Anti-IFN-γ autoantibodies in disseminated nontuberculous mycobacterial infections. J Immunol. 2005;175:4769–4776. doi: 10.4049/jimmunol.175.7.4769. [DOI] [PubMed] [Google Scholar]
- 39.Dirksen U, Nishinakamura R, Groneck P, Hattenhorst U, Nogee L, Murray R, et al. Human pulmonary alveolar proteinosis associated with a defect in GM-CSF/IL-3/IL-5 receptor common beta chain expression. J Clin Invest. 1997;100:2211–2217. doi: 10.1172/JCI119758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dranoff G, Crawford AD, Sadelain M, Ream B, Rashid A, Bronson RT, et al. Involvement of granulocyte-macrophage colony-stimulating factor in pulmonary homeostasis. Science. 1994;264:713–716. doi: 10.1126/science.8171324. [DOI] [PubMed] [Google Scholar]
- 41.Stanley E, Lieschke GJ, Grail D, Metcalf D, Hodgson G, Gall JA, et al. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci U S A. 1994;91:5592–5596. doi: 10.1073/pnas.91.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishinakamura R, Nakayama N, Hirabayashi Y, Inoue T, Aud D, McNeil T, et al. Mice deficient for the IL-3/GM-CSF/IL-5 beta c receptor exhibit lung pathology and impaired immune response, while beta IL3 receptor-deficient mice are normal. Immunity. 1995;2:211–222. doi: 10.1016/1074-7613(95)90046-2. [DOI] [PubMed] [Google Scholar]
- 43.Browne SK, Zaman R, Sampaio EP, Jutivorakool K, Rosen LB, Ding L, et al. Anti-CD20 (rituximab) therapy for anti–IFN-γ autoantibody–associated nontuberculous mycobacterial infection. Blood. 2012;119:3933–3939. doi: 10.1182/blood-2011-12-395707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malur A, Kavuru MS, Marshall I, Barna BP, Huizar I, Karnekar R, et al. Rituximab therapy in pulmonary alveolar proteinosis improves alveolar macrophage lipid homeostasis. Respir Res. 2012;13:46. doi: 10.1186/1465-9921-13-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borie R, Debray MP, Laine C, Aubier M, Crestani B. Rituximab therapy in autoimmune pulmonary alveolar proteinosis. Eur Respir J. 2009;33:1503–1506. doi: 10.1183/09031936.00160908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.