Summary
Gram-negative bacteria survive harmful environmental stressors by modifying their outer membrane. Much of this protection is afforded upon remodeling of the lipid A region of the major surface molecule lipopolysaccharide (LPS). For example, the addition of cationic substituents, such as 4-amino-4-deoxy-L-arabinose (L-Ara4N) and phosphoehthanolamine (pEtN) at the lipid A phosphate groups is often induced in response to specific environmental flux stabilizing the outer membrane. The work herein represents the first report of pEtN addition to P. aeruginosa lipid A. We have identified the key pEtN transferase which we named EptAPa and characterized its strict activity on only one position of lipid A, contrasting from previously studied EptA enzymes. We further show that transcription of eptAPa is regulated by zinc via the ColRS two-component system instead of the PmrAB system responsible for eptA regulation in E. coli and S. enterica. Further, although L-Ara4N is readily added to the same position of lipid A as pEtN under certain environmental conditions, ColR specifically induces pEtN addition to lipid A in lieu of L-Ara4N when Zn2+ is present. The unique, specific regulation of eptAPa transcription and enzymatic activity described in this work demonstrates the tight yet inducible control over LPS modification in P. aeruginosa.
Keywords: lipid A, EptA, phosphoethanolamine, ColRS, Pseudomonas, outer membrane, lipopolysaccharide, LPS
Introduction
Pseudomonas aeruginosa ubiquitously inhabits soil and water sources and is known for its intrinsic tolerance to potentially toxic contaminants such as heavy metals (Caille et al., 2007). It is also a formidable opportunistic pathogen frequently acquired in healthcare facilities due to its persistence in sinks, showers and many non-aquatic abiotic surfaces (Lyczak et al., 2000; Kerr and Snelling, 2009; Chai et al., 2014). Once inside a human host P. aeruginosa thrives in a variety of tissue types resulting in acute skin, eye, and burn wound infections (Lyczak et al., 2000). Perhaps most notably, however, are chronic P. aeruginosa infections that persist within the lungs of cystic fibrosis patients for years and are recalcitrant to most antimicrobial treatment (Kerr and Snelling, 2009; Moskowitz and Ernst, 2010).
The outer membrane of Gram-negative bacteria like P. aeruginosa acts as a protective barrier to prevent binding and uptake of toxic molecules. The major component of the outer leaflet, lipopolysaccharide, interfaces with the environment and is often remodeled to protect the cell from environmental stressors. Lipopolysaccharide is composed of three distinct domains: a lipid A anchor, a core sugar region, and an outer polysaccharide known as O-antigen (Whitfield and Trent, 2014). The lipid A domain is a potent immunostimulant, hence its other name, “endotoxin” (Needham et al., 2013). Furthermore, the negatively charged lipid A molecule is a prime target of cationic antimicrobial peptides that destabilize the outer membrane resulting in cell lysis (Vaara and Vaara, 1981; Needham and Trent, 2013).
Modification of the canonical, hexa-acylated, bis-phosphorylated lipid A molecule produced in Gram-negatives (Fig 1A; black) alter its chemical properties to bolster membrane integrity. A repertoire of modification enzymes is responsible for the dynamic structure of P. aeruginosa lipid A. Previous work from our laboratory recently revealed that P. aeruginosa has an LpxT lipid A kinase that adds an additional phosphate group to the 1- or the 4' position under standard laboratory growth conditions (Fig. 1A; brown)(Nowicki et al., 2014). Hydroxylation of the secondary acyl chains can also occur by one of two LpxO enzymes, although the purpose for this modification remains to be elucidated (Fig. 1B; orange) (King et al., 2009). In addition to these modifications, the toxicity of P. aeruginosa lipid A can be affected by altering the acylation pattern due to activity of the PagL deacylase or the PagP palmitoyltransferase (Fig. 1B; pink and green) (Ernst, 1999; Ernst et al., 2003; Ernst et al., 2006; Thaipisuttikul et al., 2014). Aside from influencing endotoxicity, lipid A modifications can contribute to antimicrobial peptide resistance (Needham and Trent, 2013). Addition of 4-amino-4-deoxy-L-arabinose (L-Ara4N) to either phosphate group of P. aeruginosa lipid A by the enzyme ArnT is one such strategy (Fig. 1B; blue)(Bhat et al., 1990; Fernández et al., 2013). Palmitoylation has also been shown to increase antimicrobial resistance (Thaipisuttikul et al., 2014).
Fig 1.
Lipid A structure of P. aeruginosa with and without inducible modifications. A) Canonical, hexa-acylated, bis-phosphorylated lipid A structure is shown in black. Phosphorylation of the lipid A phosphate groups by LpxT, which occurs in standard growth media, is indicated in brown. B) Inducible modifications to P. aeruginosa lipid A are indicated in color, including addition of a palmitate chain by PagP (green), removal of the 3-hydroxydecanoate acyl chain by PagL (pink), hydroxylation of the C12 secondary acyl chain(s) by LpxO (orange), addition of L-Ara4N at the lipid A phosphate groups by ArnT (blue), and pEtN addition by EptA (red).
Transcription of lipid A modification enzymes is often induced through two-component system signaling (Needham and Trent, 2013). Signal transduction occurs when a bacterial sensor kinase autophosphorylates in response to an external stimulus and transfers this phosphate group to a response regulator protein. The resulting conformation in the response regulator promotes DNA binding at target promoters to alter gene expression (Rodrigue et al., 2000). The two well-conserved two-component systems PhoPQ and PmrAB play a major role in modulating lipid A modification enzyme expression in P. aeruginosa. PhoPQ activates pagP and arnT transcription in response to limiting Mg2+ (McPhee et al., 2006), while PmrAB induces arnT transcription upon sensing either limiting Mg2+ or subinhibitory cationic antimicrobial peptide concentrations (McPhee et al., 2003; McPhee et al., 2006).
Aside from PhoPQ and PmrAB, three additional two-component systems in P. aeruginosa are involved in lipid A modification and resistance to the cationic antimicrobial peptide polymyxin. These include ParRS and CprRS, which are both activated by various antimicrobial peptides (Fernandez et al., 2010; Fernandez et al., 2012), and ColRS, which remains largely unstudied in P. aeruginosa but plays a role in heavy metal tolerance and overall membrane stability in P. putida and P. fluorescens (de Weert et al., 2006; Hu and Zhao, 2007; Ainsaar et al., 2014). A recent study investigated the role of CprRS and ColRS in P. aeruginosa polymyxin resistance. This report revealed that these two systems contribute to the elevated polymyxin resistance observed in P. aeruginosa phoQ mutants, and also suggested that ColRS and CprRS regulate additional unknown factors required for this resistance (Miller et al., 2011; Gutu et al., 2013). Given the complexities of two-component systems in P. aeruginosa, there are still significant questions regarding LPS modifications and their impact on antimicrobial resistance.
In Gram-negatives such as E. coli, Helicobacter pylori, Campylobacter jejuni, and Neisseria gonorrhoeae addition of phosphoethanolamine (pEtN) groups to lipid A by the enzyme EptA also promote polymyxin resistance and virulence (Tran et al., 2006; Cullen and Trent, 2010; Herrera et al., 2010; Cullen et al., 2011; Cullen et al., 2013; Hobbs et al., 2013). Although P. aeruginosa has pEtN transferase orthologs, pEtN addition has not been observed in P. aeruginosa lipid A prepared from cells grown under conditions that induce this modification in other organisms or from P. aeruginosa clinical isolates. We investigated whether or not P. aeruginosa lipid A could be modified with pEtN, and if so, under what conditions. Here we report that P. aeruginosa gene PA14_39020 is a functional pEtN transferase which we have named EptAPa that strictly modifes the 4' phosphate group of lipid A. We also demonstrate that zinc acts as a signal to induce eptAPa transcription via the ColRS two-component system (Fig. 7). While transcription of eptAPa is upregulated in response to Zn2+, arnT transcription is downregulated, suggesting that mechanisms are in place to mediate strict control over specific lipid A modifications. The existence of eptAPa reveals the potential for greater diversity in Pseudomonas lipid A structure and the versatility of the outer membrane.
Fig 7.
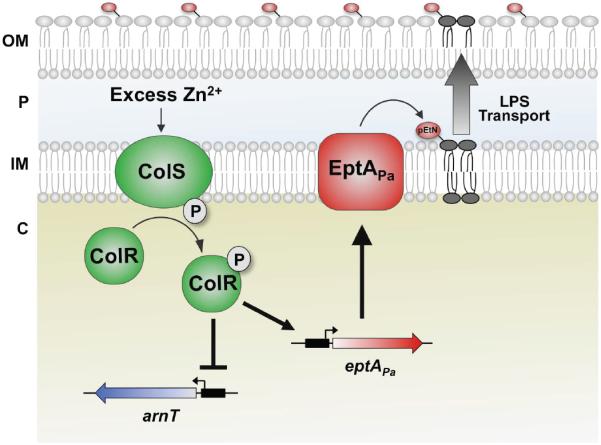
Proposed model of pEtN addition to P. aeruginosa lipid A. Upon sensing excess Zn2+, the ColS sensor kinase (green) autophosphorylates and transfers a phosphate group to the response regulator ColR (green). ColR then acts as a transcription factor, inducing transcription of eptAPa (red) while inhibiting that of arnT (blue). EptAPa protein is synthesized and transfers pEtN to the 4′-phosphate group of lipid A in the inner membrane. Lipid A is then transported to the bacterial cell surface. Following transport to the outer membrane, the 3-hydroxydecanoate acyl chain is removed by PagL (indicated in the model). In some instances, PagP can modify the lipid A (not shown). Cellular components are labelled as follows: OM, outer membrane; P, periplasm; IM, inner membrane; C, cytoplasm).
Results
P. aeruginosa has a functional EptA enzyme
In silico analysis identified three P. aeruginosa eptA orthologs with significant identity to the S. enterica eptA (pmrC) ortholog (Lee et al., 2004). These include PA14_58610 (24% identity, E-value 3e-28), PA14_21210 (43%, E-value 1e-149) and PA14_39020 (43%, E-value 6e-149). Since pEtN-modified P. aeruginosa lipid A has not been previously reported, we tested whether these orthologs could function as a lipid A pEtN transferase by expressing each gene in trans in the E. coli eptA mutant (W3110ΔeptAEc). 32P-labeled lipid A was prepared and separated by TLC from these and relevant control strains including wild-type strain W3110 and W3110ΔeptAEc+empty vector.
While no pEtN is detected in lipid A prepared from W3110 or W3110ΔeptAEc + empty vector (Fig. S1A, lanes 1 and 2), expression of both eptAEc and PA14_39020 resulted in pEtN-modified lipid A (Fig. S1A, lanes 3 and 6). MALDI-TOF mass spectrometry (MS) analysis also confirmed that while W3110ΔeptAEc expressing empty vector had no pEtN (Fig. S1B), lipid A prepared from W3110ΔeptAEc+PA14_39020 (peptAPa) was modified with pEtN, as evidenced by the ion of m/z 1920.4 (Fig. S1C; predicted [M-H]- at m/z 1920.2).
Since EptAPa can add pEtN to E. coli lipid A, we next determined whether EptAPa modified P. aeruginosa lipid A. 32P-labeled lipid A from wild-type strain PA14, PA14+empty vector or PA14+eptAPa was analyzed. TLC separation of lipid A clearly demonstrated an altered profile of PA14 expressing peptAPa (Fig. 2A, lane 3) relative to PA14 or PA14+empty vector (Fig. 2A, lanes 1 and 2). Penta-acylated, palmitoylated and L-Ara4N-modified lipid A species were observed by MALDI-TOF MS analysis of lipid A isolated from PA14+empty vector (Fig. 2B). Whereas expression of eptAPa in PA14 resulted in abundant ions of m/z 1489.0 and 1727.3 which correspond to the pEtN-modified lipid A species (Fig. 2C; predicted [M-H]- at m/z 1489.9 and 1728.1, respectively). These results reveal that P. aeruginosa has a functional lipid A pEtN-transferase enzyme capable of modifying both E. coli and P. aeruginosa LPS.
Fig 2.
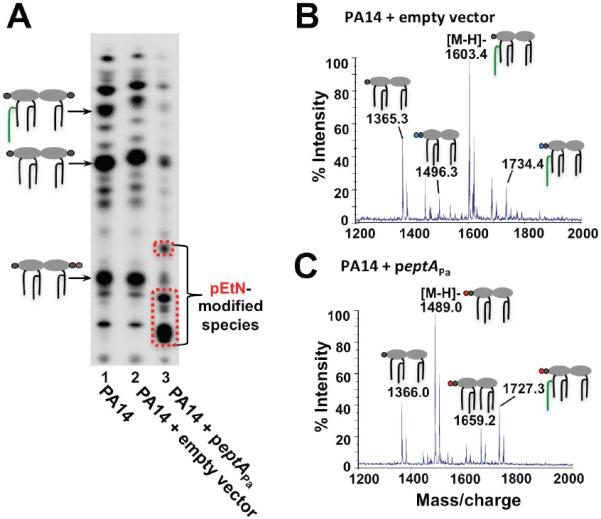
Heterologous expression of a P. aeruginosa eptA ortholog results in pEtN addition to the lipid A. A) Cells were grown in MOPS minimal medium. Major 32P-labeled lipid A species are indicated with a cartoon corresponding to the lipid A structure; colors of modification groups are the same as those used in Fig. 1. Expression of PA14_39020 (eptAPa) in P. aeruginosa results in modified lipid A species. B) MALDI-TOF MS analysis of PA14 + empty vector grown in MOPS minimal medium reveals no pEtN addition to the molecule, while C) analysis of PA14 + peptAPa shows pEtN modification of the lipid A. The fractions most representative of pEtN modification are shown.
EptAPa adds pEtN strictly to the lipid A 4' phosphate group
Previous work from our laboratory has demonstrated that in P. aeruginosa, S. enterica and E. coli the position of lipid A modification can be important due to potential competition with other modification groups. For example in S. enterica and E. coli EptA preferentially adds pEtN to the lipid A 1-phosphate group, which is the sole site of phosphorylation by the kinase LpxT (Herrera et al., 2010). Environmental conditions that activate eptA transcription simultaneously inhibit LpxT activity to prevent competition (Herrera et al., 2010; Kato et al., 2012). However, since both LpxT and ArnT enzymes in P. aeruginosa can act on either lipid A phosphate group (Bhat et al., 1990; Nowicki et al., 2014) we questioned whether EptAPa also has dual positional activity. To determine this we removed the 1- or the 4'-phosphate group of lipid A by heterologous expression of Fransicella novicida LpxE or LpxF phosphatases (Wang et al., 2004; Wang et al., 2006), respectively, and tested the ability of EptAPa to modify lipid A. This experiment was done in E. coli strain BN2 (Needham et al., 2013) since its lipid A is penta-acylated, and LpxF can only act on penta-acylated lipid A (Wang et al., 2006). BN2 also lacks some lipid A modification machinery including LpxT to facilitate easier analysis of the lipid A profiles.
Expression of eptAPa resulted in a pEtN-modified lipid A species that migrated below the unmodified bis-phosphorylated species (Fig. 3A, lane 2). Expression of either lpxE or lpxF caused a marked increase of mono-phosphorylated lipid A, which migrates near the top of the TLC plate (Fig. 3A, lanes 3 and 5). Simultaneous expression of eptAPa and lpxE resulted in the appearance of a species that migrated a distance between that of unmodified bis-phosphorylated lipid A and pEtN-modified lipid A (Fig. 3A, lane 4). MALDI-TOF MS confirmed this species to be pEtN-modified mono-phosphorylated lipid A (Fig. 3B). This same pEtN-modified species was not detected when LpxF was expressed with eptAPa (Fig 3A, lane 6). The lack of pEtN addition to 4'-dephosphorylated lipid A was further confirmed by MALDI-TOF MS as simultaneous expression of EptAPa and LpxF resulted in only a single ion of m/z 1506.1, corresponding to mono-phosphorylated lipid A (Fig. 3C; predicted [M-H]- at m/z 1507.1).
Fig 3.

EptAPa adds pEtN exclusively to the lipid A 4' phosphate group. A) Cells were grown in LB broth. Major 32P-labeled lipid A species are indicated with a cartoon corresponding to the lipid A structure; colors of modification groups are the same as those used in Fig. 1. Heterologous expression of eptAPa in BN2 results in a pEtN-modified species, while expression of either the lpxEFn or lpxFFn phosphatase results in an increased of monophosphorylated species. Co-expression of lpxEFn and eptAPa results in pEtN addition to the 1-dephosphorylated lipid A molecule, while no pEtN addition of 4' –dephosphorylated species is detected. B) MALDI-TOF analysis of lipid A isolated from BN2 coexpressing lpxEFn and eptAPa corroborates the presence of a monophosphorylated, pEtN-modified species. C) MALDI-TOF analysis of lipid A isolated from BN2 coexpressing lpxFFn and eptAPa reveals that when the 4' phosphate group is removed, pEtN addition does not occur.
EptAPa-dependent addition of pEtN to the 4' phosphate group of BN2 and PA14 lipid A was corroborated by ultraviolet photodissociation (UVPD) tandem MS (Figs. S2, S3 and S4). For all UVPD mass spectra, cleavage sites (7) and (8) provided evidence of the presence of a pEtN group. In addition, the glycosidic and cross-ring cleavages at cleavage sites (10) and (11) in the fragmentation map shown in Figure S2, sites (9)-(12) in Figure S3, and sites (9)-(12) and (17)-(21) in Figure S4 further support the location of the pEtN modification at the 4' phosphate group of each lipid A species. Taken together, these results demonstrate that EptAPa functions strictly at the 4' phosphate group, unlike any previously characterized EptA enzyme.
Extracellular zinc induces pEtN addition to lipid A
In S. enterica, modification of lipid A with pEtN is induced via the PmrAB two-component system in response to mildly acidic pH (Perez and Groisman, 2007), and indirectly via the PhoPQ system when Mg2+ is limiting or cationic antimicrobial peptides are present (Kox et al., 2000; Bader et al., 2005). Since none of these signals induced pEtN addition to P. aeruginosa lipid A (data not shown), we next tested transition metals including Fe3+ and Zn2+, which activate PmrAB in E. coli (Hagiwara et al., 2004; Lee et al., 2005). We also tested Ga3+ due to its chemical similarity to Fe3+, and Cd2+, which is closely related to Zn2+ (Laddaga and Silver, 1985; Kaneko et al., 2007). Transition metals Co2+, Cu2+, Mn2+, and Ni2+ were also tested for their ability to induce pEtN modification of lipid A since they are associated with biological catalysts and are commonly found in the environment (Andreini et al., 2008; Mathiyazhagan and Natarajan, 2011).
Lipid A was isolated from 32P-labeled P. aeruginosa grown in LB alone or supplemented with metal. Addition of extracellular Zn2+ but no other metal tested resulted in modified lipid A (Fig. 4A, lane 3). Zn2+-dependent modification was abolished when eptAPa was deleted from the genome and restored upon complementation with the native eptAPa promoter (Figs. 4A, lanes 4 and 5), suggesting that the changes observed were due to EptAPa activity. To determine whether Zn2+ induced transcription of eptAPa, cDNA was prepared from cells grown in the presence or absence of Zn2+. As shown by both quantitative and semi-quantitative reverse-transcriptase (RT) PCR, eptAPa gene expression is induced when Zn2+ is added to the growth media (Figs. 4B and S5). These results indicate that transcription of eptAPa is dependent on extracellular Zn2+
Fig 4.

Zn2+ induces transcription of eptAPa. A) Cells were grown in LB broth. Major 32P-labeled lipid A species are indicated with a cartoon corresponding to the lipid A structure; colors of modification groups are the same as those used in Fig. 1. Both heterologous expression of eptAPa as well as addition of 2mM ZnSO4 to the media results in pEtN addition to lipid A. This modification is not detectable in the eptAPa mutant, but restored upon complementation with peptAnprom. B) Relative gene expression of eptAPa and arnT in response to Zn2+. Transcription of eptAPa is induced by 2mM ZnSO4 approximately 21-fold. Zn2+ downregulates arnT transcription by >4-fold. Ratios were standardized relative to expression of the housekeeping control gene, clpX. C), D) and E). MALDI-TOF MS analysis of lipid A prepared from cells grown in LB broth. C) Analysis of PA14 + 2mM ZnSO4 reveals pEtN addition to lipid A, while D) ΔeptAPa + 2mM ZnSO4 shows no pEtN modification, but instead L-Ara4N addition. E) Complementation of ΔeptAPa with peptAnprom restores pEtN addition to the lipid A in response to Zn2+. The fractions most representative of pEtN modification are shown.
To confirm whether Zn2+-dependent induction of EptAPa synthesis resulted in pEtN addition to P. aeruginosa lipid A, MALDI-TOF MS analysis of lipid A isolated from cells grown with or without Zn2+ was performed. Wild-type P. aeruginosa lipid A was modified with pEtN in the presence of Zn2+ (Fig. 4C), while lipid A prepared from the PA14ΔeptAPa mutant showed no pEtN modification when Zn2+ was added to the media (Fig. 4D). Complementation of PA14ΔeptAPa using the native eptAPa promoter restored Zn2+-dependent pEtN addition to the lipid A (Fig. 4E). MS analysis revealed that in addition to pEtN addition, L-Ara4N-modified lipid A was present in PA14 grown with Zn2+ (Fig. 4C). We were therefore curious as to how Zn2+ might influence L-Ara4N addition to lipid A. Since Zn2+ affected the transcription of eptAPa, we assessed whether Zn2+ altered arnT gene expression by performing quantitative RT-PCR. While eptAPa transcription increased by 21-fold in the presence of 2mM Zn2+, arnT transcription was downregulated >4-fold (Fig. 4B). This result indicates that pEtN modification is selected for in the presence of Zn2+ while arnT expression, and thus L-Ara4N modification of lipid A, is downregulated.
The ColRS two-component system induces pEtN addition to lipid A
Since inducible lipid A modification genes like eptA are commonly regulated by two-component systems, our next goal was to determine the system responsible for eptAPa transcriptional activation in response to Zn2+. The ColRS system has recently been shown to respond to transition metals including Zn2+ (Ainsaar et al., 2014). Our first approach was therefore to investigate the presence of potential ColR binding sites in the eptAPa promoter. A consensus ColR binding site has been determined for promoters of genes within the ColR regulon in P. putida (Kivistik et al., 2009). Using the Virtual Footprint online analysis tool (Münch et al., 2005), we found three potential ColR binding sites within the eptAPa promoter region with close agreement to this consensus sequence (Fig 5A), suggesting that ColR binds to the eptAPa promoter. It was then tested whether overexpression of colR could induce eptAPa transcription by semi-quantitative RT-PCR of cDNA. Response regulators pmrA and phoP, which directly or indirectly regulate eptA transcription in S. enterica, were also tested. Only overexpression of colR resulted in detectable transcription of eptAPa (Fig 5B). Lipid A was modified with pEtN upon overexpression of colR, as demonstrated by both TLC separation of 32Plabeled lipid A (Fig 5C, lane 4) and MALDI-TOF MS analysis of PA14 + pcolR (Fig 5D).
Fig 5.

The two-component system response regulator ColR activates eptAPa transcription. A) Putative eptAPa promoter ColR binding sites are in bold and boxed; nucleotides that deviate from the conserved recognition sequence in P. putida ((T/C)(T/C)NA(C/G)NN(T/C)TTTTT(C/G)AC) are indicated in red. The number of base pairs between ColR sites or upstream of the start codon is indicated. B) Semi-quantitative RT-PCR of cDNA prepared from cells grown in MOPS minimal medium. While eptAPa is not transcribed in PA14, expression of colR in trans results in eptAPa transcription. C) Lipid A was isolated from 32P-labeled cells grown in MOPS minimal medium and separated by TLC. Only expression of the colR response regulator, and not pmrA or phoP, results in pEtN modification of lipid A. D) MALDI-TOF MS analysis of PA14 + pcolR grown in MOPS minimal medium reveals pEtN-modified lipid A. The fraction most representative of pEtN modification is shown.
To determine whether ColR induction of eptAPa transcription is dependent on Zn2+, a PA14 colR deletion mutant was generated and assessed for transcription of eptAPa in the presence or absence of Zn2+ by both quantitative and semi-quantitative RT-PCR analysis. Although a Zn2+ concentration of 2mM had been used in the initial Zn2+ assay experiments, the PA14ΔcolR mutant was sensitive to 2mM Zn2+. Instead, 1mM Zn2+ was used, which was sufficient to visualize pEtN modification in PA14 (Fig. 6A, lane 2). Minimal eptAPa transcription was detected in response to Zn2+ upon deletion of colR; complementation of this mutant restored Zn2+-dependent eptAPa transcription by >4-fold (Fig. 6B, Fig. S5). While 1mM Zn2+ induced pEtN modification of PA14 lipid A (Fig. 6A, lane 2), lipid A from the PA14ΔcolR was not modified with pEtN in response to Zn2+, as determined by both TLC separation of 32P-labeled lipid A and MALDI-TOF MS analysis (Fig. 6A, lane 3 and Fig 6C). pEtN addition was restored upon complementation of the colR mutant with pcolRnprom (Fig. 6A, lane 4 and Fig 6D). A PA14 colS mutant and complemented mutant were also tested for pEtN addition to lipid A in response to Zn2+ by TLC separation of 32P-labeled lipid A. As for the colR mutant, lipid A modification with pEtN was not detected in the colS mutant grown in the presence of 1mM Zn2+, but was restored upon complementation of colS with pcolSnprom (Fig. S6, lanes 3 and 4). These results demonstrate that the ColRS system induces pEtN addition to lipid A upon sensing Zn2+.
Fig 6.
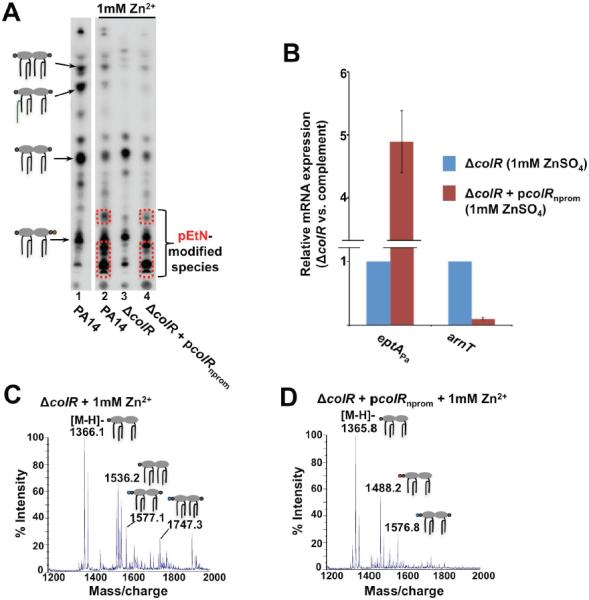
Deletion of colR results in loss of Zn2+-induced pEtN modification of P. aeruginosa lipid A. A) Lipid A was isolated from 32P-labeled cells grown in LB broth and separated by TLC. While pEtN modification of lipid A is detectable for PA14 + 1mM ZnSO4, no such modification occurs in PA14ΔcolR in response to Zn2+. Modification is restored in the complemented mutant. B) Relative gene expression of eptAPa and arnT in response to Zn2+ in the ΔcolR mutant or complemented mutant. Transcription of eptAPa in the presence of 1mM ZnSO4 is induced >4-fold in a ColR-dependent manner. An approximately 10-fold decrease in arnT transcription in the presence of 1mM ZnSO4 is also dependent on ColR. Ratios were standardized relative to expression of the housekeeping control gene, clpX. C) and D). MALDI-TOF MS analysis of lipid A prepared from cells grown in LB broth. C) No pEtN modification is detected in the PA14ΔcolR mutant grown in LB + 1mM ZnSO4. D) Complementation of PA14ΔcolR with pcolRnprom restores the Zn2+-dependent pEtN addition to the lipid A. The fractions most representative of pEtN modification are shown.
We also investigated whether the downregulation of arnT transcription in the presence of Zn2+ was dependent on the ColR response regulator. Gene expression of arnT was analyzed by quantitative RT-PCR in the PA14 colR mutant and complemented strains in the presence of 1mM Zn2+. Transcription of arnT was reduced by approximately 10-fold upon complementation of colR (Fig. 6B). This result indicates that ColR activates transcription of eptAPa in response to Zn2+ while downregulating arnT transcription.
Discussion
Changes in the environment require bacterial outer membrane remodeling, including LPS structural changes, to promote membrane stability (Whitfield and Trent, 2014). L-Ara4N addition to lipid A phosphate groups contributes to cationic antimicrobial peptide resistance in P. aeruginosa, E. coli and S. enterica (Lee et al., 2004; Herrera et al., 2010; Fernández et al., 2013). The addition of the amine-containing residue pEtN can also result in increased peptide resistance (Tran et al., 2006; Herrera et al., 2010), and in some organisms is a crucial factor for host infection (Cullen et al., 2013; Hobbs et al., 2013). Lipid A modifications in P. aeruginosa have been well-studied, yet despite the existence of eptA orthologs, pEtN addition has never been observed. Due to the importance of pEtN lipid A modification in other organisms, we investigated the functionality and regulation of P. aeruginosa eptA orthologs. In this report, we identify and characterize a functional P. aeruginosa lipid A pEtN transferase and determine that Zn2+ induces transcription of eptAPa via the Pseudomonas-specific ColRS system (Fig. 7).
Overexpression of three P. aeruginosa eptA orthologs in E. coli revealed that PA14_39020 (eptAPa) was able to modify lipid A with pEtN (Fig. S1A and C). It is likely that the other two orthologs add pEtN to other targets in the cell. Based on its homology to S. enterica CptA, PA14_58610 may be the enzyme responsible for adding pEtN to the core of P. aeruginosa LPS (Kooistra et al., 2003; Tamayo et al., 2005). While a very minor amount of lipid A modification is detected by TLC separation of lipid A upon expression of PA14_21210 in PA14, lipid A is probably not the primary target of this enzyme. It is possible that this enzyme modifies an as yet unidentified target, as pEtN transferase enzymes in other organisms have been shown to modify structural proteins of the flagellum and pilus (Hegge et al., 2004; Cullen and Trent, 2010), and in doing so has some very minor, non-specific activity toward lipid A. This activity toward lipid A, however, is so minor that is cannot be detected by mass spectrometry analysis (data not shown).
We characterized the site-specificity of pEtN addition to lipid A due to the potential for competition with other modification groups. Whereas pEtN addition occurs specifically or preferentially at the 1-phosphate group of lipid A in H. pylori (Tran et al., 2004) and S. enterica (Herrera et al., 2010), respectively, analysis of EptAPa activity in E. coli revealed that this enzyme acts solely at the 4' position (Figs. 3, S2). EptAPa activity thus differs from ArnT and LpxT enzymes in P. aeruginosa that can modify either lipid A phosphate group (Bhat et al., 1990; Nowicki et al., 2014).
Investigation of conditions that induce pEtN modification revealed that excess Zn2+ acts as the activating signal for eptAPa transcription. Pseudomonas species are readily found in the soil and aqueous environments which can be contaminated with metals due to waste runoff from mines, smelting, and other industrial facilities (Teitzel and Parsek, 2003; Raja et al., 2006; Mathiyazhagan and Natarajan, 2011). In such environments, Pseudomonas can be exposed to high levels of metal pollutants and has thus evolved the ability to alter gene expression to promote metal tolerance (Perron et al., 2004; Ha et al., 2004; Hu and Zhao, 2007; Caille et al., 2007). Excess Zn2+ may also be relevant in healthcare settings as concentrations up to 1mM can leach out from latex catheters and gloves (Perron et al., 2004; Ballesta et al., 2006). Deletion of eptAPa, however, does not result in increased sensitivity to Zn2+ or to Cd2+, Ga3+, Fe3+, Co2+, Cu2+, Mn2+ or Ni2+. Under laboratory settings Zn2+-induced pEtN addition to lipid A has no effect on polymyxin resistance, biocide tolerance, or biofilm formation (data not shown). As the ColRS system was previously implicated to play a role in polymyxin resistance, unidentified genes other than eptAPa within the ColRS regulon are likely involved in this resistance. The fact that P. aeruginosa has evolved regulatory mechanisms to control pEtN addition to lipid A, however, suggests the importance of this modification for conditions we have not yet identified.
Extracellular metals are sensed by one of three two-component systems in Pseudomonas species: CzcRS (Perron et al., 2004), CopRS (Caille et al., 2007) and ColRS (Hu and Zhao, 2007; Ainsaar et al., 2014). Both CzcRS (activated by Zn2+) and CopRS (activated by Cu2+) induce expression of the heavy metal efflux pump CzcCBA while downregulating the OprD porin, leading to decreased carbapenem and imipenem uptake (Perron et al., 2004; Caille et al., 2007). In P. putida, the ColRS system senses Zn2+, Fe3+, Mn2+ and Cd2+, and mutants of colR and colS display lower tolerance to these metals (Ainsaar et al., 2014). We have now determined Zn2+ to be an activating signal for ColRS in P. aeruginosa. Additionally, the PA14 colR and colS mutants are more sensitive to Zn2+ than wild-type, suggesting a role for the ColRS system in Zn2+ tolerance in P. aeruginosa. While the ColRS system is important for metal tolerance in P. putida and deletion of multiple genes in the ColRS regulon results in increased metal sensitivity, no individual ColRS-regulated gene has a major contribution to metal tolerance (Ainsaar et al., 2014). It is likely that multiple genes in the P. aeruginosa ColRS regulon are involved in Zn2+ tolerance, which could explain why the eptAPa isogenic mutant does not have any growth defect in media with Zn2+.
Our demonstration of pEtN-modified lipid A via the ColRS system in response to Zn2+ (Fig. 7) reveals that lipid A remodeling in P. aeruginosa is more complex than previously realized. Extracellular Zn2+ specifically induces transcription of eptAPa and not arnT through the ColR response regulator, demonstrating coordinated control over lipid A modifications. In addition to selectively inducing expression of eptAPa Zn2+ downregulates arnT transcription by over 4-fold (Fig. 4B). This is interesting given that ArnT-mediated L-Ara4N modification typically plays a more significant role in virulence and antimicrobial peptide resistance than eptA in organisms possessing both modification enzymes (Tamayo et al., 2005; Herrera et al., 2010). While eptAPa does not seem to be involved in metal or polymyxin resistance, there is likely an evolutionary reason for this targeted induction of eptAPa transcription. Our findings demonstrate the tight control of P. aeruginosa lipid A modification systems, and suggest the need for further studies to better elucidate the mechanisms involved in outer membrane remodeling and its contribution to bacterial persistence and versatility.
Experimental Procedures
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table S1. E. coli strains were cultured in LB broth or agar (Difco) at 37°C. P. aeruginosa strains were grown on LB agar plates, and initial liquid cultures were grown overnight in LB broth at 37°C. The next day, P. aeruginosa cultures were diluted to an OD600 of ~0.05 in either LB broth or in morpholinepropanesulfonic acid (MOPS)-buffered minimal medium (50mM MOPS, 93mM NH4Cl, 43mM NaCl, 2mM KH2PO4) supplemented with 3.5μM FeSO4•7H2O, 20mM sodium succinate, and 1mM MgSO4. Chloramphenicol was used at a concentration of 30μg/mL for E. coli. Ampicillin or carbenicillin was used at a concentration of 100μg/mL or 300μg/mL for E. coli or P. aeruginosa, respectively. For growth of P. aeruginosa in medium with added metals, LB was used to prevent metals from crashing out of solution. For the initial screen of lipid A modifications, metal salts were added in the following concentrations: 2mM ZnSO4, 0.2mM CdSO4, 0.1mM Ga(III)NO3, 0.2mM FeSO4, 0.1mM CoCl2, 2mM CuSO4, 2mM MnSO4, and 2mM NiSO4. The highest concentration of metal that did not significantly reduce growth (defined as a >50% reduction) in liquid medium was used, with the exception of Fe3+, for which a lower, more physiologically relevant concentration was used based on concentrations known to induce lipid A modification in other organisms (Herrera et al., 2010).
DNA and RNA preparation
Before preparing P. aeruginosa genomic DNA from an overnight culture in LB broth, two washes with 0.1M NaCl were performed. Genomic DNA was prepared using the Easy-DNA Kit (Invitrogen). Total RNA was extracted from cells grown to an OD600 of ~0.6 using the RNeasy Mini Kit (Qiagen), according to the manufacturer's instructions. To eliminate residual DNA contamination, total RNA was treated with DNase from the RNase-Free DNase Set (Qiagen). cDNA synthesis was performed using SuperScript III Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions.
Recombinant DNA methods
Plasmid DNA was isolated with the QIAprep Spin Miniprep Kit (Qiagen). Chromosomal DNA for insertion into plasmid constructs was amplified using either the DNA polymerase PfuTurboR (Stratagene) or Takara Ex Taq (Takara). PCR products were separated on an agarose gel and purified using the QIAquick Gel Extraction Kit (Qiagen). All primers were purchased from Integrated DNA Technologies (Table S2). Restriction endonucleases, T4 DNA ligase, and Antarctic Phosphatase used in this study for generation of plasmid constructs were purchased from New England Biolabs and used according to the manufacturer's instructions.
Generation of chromosomal gene deletion mutants
In-frame, markerless gene deletions were generated in P. aeruginosa by homologous recombination using the suicide plasmid pEX18Gm. ~1Kb DNA fragments flanking the target gene up or downstream were amplified using primers listed in Table S2. An assembly PCR was then carried out to stitch together these flanking regions. Assembly PCR fragments were digested with restriction endonucleases EcoRI and HindIII or BamHI, and ligated into pEX18Gm. The suicide plasmid constructs, pEX18-eptAdel, pEX18-colRdel or pEX18-colSdel were introduced into P. aeruginosa via conjugation with E. coli strain SM10. Deletion mutants were then screened for as described previously (Hoang et al., 1998). Deletions were confirmed by PCR.
Plasmid constructs
To construct pPA14_39020 (pACeptAEc), pPA14_58610 and pPA14_21210, each gene was amplified along with the native RBS and cloned into the medium copy vector pACYC184 using EcoRV and SalI restriction endonucleases. For generation of peptAPa, pcolR, ppmrA and pphoP, each gene and its native RBS were amplified and digested with EcoRI and HindIII to clone into pEX1.8. All constructs were confirmed by sequencing. The eptAPa and colR genes were amplified along with their native promoters and cloned into the medium copy vector pEX1.8 (Pearson et al., 1997) by digestion with SalI or SalI and HindIII, respectively, generating peptAnprom and p colRnprom. For generation of pcolSnprom, the colRS promoter was first amplified and cloned into pEX1.8 by digestion with BamHI and EcoRI. The colS coding sequence was then amplified and cloned into pEX1.8 (containing the colRS promoter) with EcoRI and HindIII.
Isolation and analysis of labeled lipid A
Overnight cultures were diluted to an OD600 of ~0.05 in 5mL of fresh medium (as indicated within each figure legend) and labeled with 2.5μCi/mL 32Pi (Perkin-Elmer). Cells were harvested at an OD600 of ~1.0, and lipid A was isolated by mild acid hydrolysis as described previously (Zhou et al., 1999; Tran et al., 2004). 32Pi-labeled lipid A species were spotted on a TLC plate at ~5,000 cpm per lane (10,000 cpm for E. coli), and run in a solvent prepared in a 50:50:16:5 (v/v) ratio of chloroform, pyridine, 88% formic acid, and water, respectively. TLC plates were dried, set on a phosphor screen overnight and imaged using a phosphor-imager (BioRad PMI).
Large scale lipid A isolation and MALDI-TOF mass spectrometry
For large scale lipid A analysis, 250mL cultures were grown at 37°C to an OD600 of ~1.0 in the medium indicated. Lipid A was prepared by mild acid hydrolysis, washed and resuspended in chloroform/methanol/water (2:3:1, v/v), as described previously (Hankins et al., 2013). The sample was run through a DEAE cellulose column, washed with chloroform/methanol/water (2:3:1, v/v), and eluted in individual fractions with chloroform/methanol/increasing concentrations of ammonium acetate, as described previously (Odegaard et al., 1997; Hankins et al., 2013). Typically, hydrophilic or monophosphorylated species fractionate at lower ammonium acetate concentrations (flow-through, wash, 60 or 120mM elution fractions), while more hydrophobic, unmodified or phosphate-modified species fractionate at the highest concentration, 480mM ammonium acetate. MALDI-TOF mass spectrometry was performed as described using a MALDI-TOF/TOF mass spectrometer (ABI 4700 Proteomics Analyser) (Hankins et al., 2013).
ESI and UVPD mass spectrometry
Lipid A was isolated and prepared as described above for MALDI-TOF analysis. All mass spectrometry experiments were executed on a Thermo Scientific Orbitrap Elite mass spectrometer (Bremen, Germany) modified to perform ultraviolet photodissociation (UVPD). The mass spectrometer was equipped with a 193-nm Coherent ExciStar XS excimer laser (Santa Clara, CA) and operated in the negative ion mode using a previously described set-up (Shaw et al., 2013). Briefly, solutions containing 1–5 μM lipid A in 50:50 methanol/ chloroform were directly infused using an electrospray ionization (ESI) source at a flow rate of 3 μl/min. The ESI voltage was set to 4 kV. UVPD mass spectra were collected using 10 laser pulses per spectrum (at 4–5 mJ/pulse) and were interpreted as described previously (Madsen et al., 2011).
Quantitative PCR methods
Primers for semi-quantitative and quantitative PCR (qPCR) were designed using the Primer-BLAST tool (NCBI) and are listed in Table S2. Semi-quantitative PCR was performed by amplifying cDNA obtained from samples cultured in the conditions or with the ppmrA, pphoP and pcolR expression constructs as indicated, using primers specific for eptAPa or clpX as a reference gene (Palmer et al., 2005). qPCR was performed in a OneStep thermocycler (Applied Biosystems) using Power SYBR Green PCR Master Mix (Applied Biosystems), according to the manufacturer's instructions, as described previously (Pfaffl, 2001; Nowicki et al., 2014).
Supplementary Material
Acknowledgements
Funding from NIH (grants AI064184, AI076322 to M.S.T. & GM103655 to J.S.B.), the Welch Foundation (F-1155 to J.S.B.), the Army Research Office (grant W911NF-12-1-0390 to M.S.T.) and the Cystic Fibrosis Foundation (grant Trent13G0 to M.S.T.) is gratefully acknowledged.
References
- Ainsaar K, Mumm K, Ilves H, Hõrak R. The ColRS signal transduction system responds to the excess of external zinc, iron, manganese, and cadmium. BMC Microbiol. 2014;14:162. doi: 10.1186/1471-2180-14-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. JBIC J Biol Inorg Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, et al. Recognition of Antimicrobial Peptides by a Bacterial Sensor Kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Ballesta S, Conejo MC, García I, Rodríguez-Martínez JM, Velasco C, Pascual A. Survival and resistance to imipenem of Pseudomonas aeruginosa on latex gloves. J Antimicrob Chemother. 2006;57:1010–1012. doi: 10.1093/jac/dkl072. [DOI] [PubMed] [Google Scholar]
- Bhat R, Marx A, Galanos C, Conrad RS. Structural studies of lipid A from Pseudomonas aeruginosa PAO1: occurrence of 4-amino-4-deoxyarabinose. J Bacteriol. 1990;172:6631–6636. doi: 10.1128/jb.172.12.6631-6636.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caille O, Rossier C, Perron K. A copper-activated two-component system interacts with zinc and imipenem resistance in Pseudomonas aeruginosa. J Bacteriol. 2007;189:4561–4568. doi: 10.1128/JB.00095-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai H, Allen WE, Hicks RP. Spectroscopic investigations of the binding mechanisms between antimicrobial peptides and membrane models of Pseudomonas aeruginosa and Klebsiella pneumoniae. Bioorg Med Chem. 2014;22:4210–4222. doi: 10.1016/j.bmc.2014.05.040. [DOI] [PubMed] [Google Scholar]
- Cullen TW, Giles DK, Wolf LN, Ecobichon C, Boneca IG, Trent MS. Helicobacter pylori versus the Host: Remodeling of the Bacterial Outer Membrane Is Required for Survival in the Gastric Mucosa. PLoS Pathog. 2011;7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, O'Brien JP, Hendrixson DR, Giles DK, Hobb RI, Thompson SA, et al. EptC of Campylobacter jejuni Mediates Phenotypes Involved in Host Interactions and Virulence. Infect Immun. 2013;81:430–440. doi: 10.1128/IAI.01046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen TW, Trent MS. A link between the assembly of flagella and lipooligosaccharide of the Gram-negative bacterium Campylobacter jejuni. Proc Natl Acad Sci. 2010;107:5160–5165. doi: 10.1073/pnas.0913451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK. Specific Lipopolysaccharide Found in Cystic Fibrosis Airway Pseudomonas aeruginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- Ernst RK, Adams KN, Moskowitz SM, Kraig GM, Kawasaki K, Stead CM, et al. The Pseudomonas aeruginosa Lipid A Deacylase: Selection for Expression and Loss within the Cystic Fibrosis Airway. J Bacteriol. 2006;188:191–201. doi: 10.1128/JB.188.1.191-201.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Hajjar AM, Tsai JH, Moskowitz SM, Wilson CB, Miller SI. Pseudomonas aeruginosa lipid A diversity and its recognition by Toll-like receptor 4. J Endotoxin Res. 2003;9:395–400. doi: 10.1179/096805103225002764. [DOI] [PubMed] [Google Scholar]
- Fernández L, Alvarez-Ortega C, Wiegand I, Olivares J, Kocíncová D, Lam JS, et al. Characterization of the polymyxin B resistome of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:110–119. doi: 10.1128/AAC.01583-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, Hancock REW. Adaptive Resistance to the “Last Hope” Antibiotics Polymyxin B and Colistin in Pseudomonas aeruginosa Is Mediated by the Novel Two-Component Regulatory System ParR-ParS. Antimicrob Agents Chemother. 2010;54:3372–3382. doi: 10.1128/AAC.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez L, Jenssen H, Bains M, Wiegand I, Gooderham WJ, Hancock REW. The Two-Component System CprRS Senses Cationic Peptides and Triggers Adaptive Resistance in Pseudomonas aeruginosa Independently of ParRS. Antimicrob Agents Chemother. 2012;56:6212–6222. doi: 10.1128/AAC.01530-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, et al. Polymyxin Resistance of Pseudomonas aeruginosa phoQ Mutants Is Dependent on Additional Two-Component Regulatory Systems. Antimicrob Agents Chemother. 2013;57:2204–2215. doi: 10.1128/AAC.02353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha U-H, Kim J, Badrane H, Jia J, Baker HV, Wu D, Jin S. An in vivo inducible gene of Pseudomonas aeruginosa encodes an anti-ExsA to suppress the type III secretion system: PtrA inhibits type III secretion system. Mol Microbiol. 2004;54:307–320. doi: 10.1111/j.1365-2958.2004.04282.x. [DOI] [PubMed] [Google Scholar]
- Hagiwara D, Yamashino T, Mizuno T. A Genome-wide view of the Escherichia coli BasS-BasR two-component system implicated in iron-responses. Biosci Biotechnol Biochem. 2004;68:1758–1767. doi: 10.1271/bbb.68.1758. [DOI] [PubMed] [Google Scholar]
- Hankins JV, Madsen JA, Needham BD, Brodbelt JS, Trent MS. The Outer Membrane of Gram-Negative Bacteria: Lipid A Isolation and Characterization. In: Delcour AH, editor. Bacterial Cell Surfaces. Humana Press; [Accessed May 15, 2014]. 2013. pp. 239–258. http://link.springer.com/protocol/10.1007/978-1-62703-245-2_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegge FT, Hitchen PG, Aas FE, Kristiansen H, Løvold C, Egge-Jacobsen W, et al. Unique modifications with phosphocholine and phosphoethanolamine define alternate antigenic forms of Neisseria gonorrhoeae type IV pili. Proc Natl Acad Sci U S A. 2004;101:10798–10803. doi: 10.1073/pnas.0402397101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CM, Hankins JV, Trent MS. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol Microbiol. 2010;76:1444–1460. doi: 10.1111/j.1365-2958.2010.07150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ, Schweizer HP. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- Hobbs MM, Anderson JE, Balthazar JT, Kandler JL, Carlson RW, Ganguly J, et al. Lipid A's Structure Mediates Neisseria gonorrhoeae Fitness during Experimental Infection of Mice and Men. mBio. 2013;4 doi: 10.1128/mBio.00892-13. e00892-13-e00892-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N, Zhao B. Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol Lett. 2007;267:17–22. doi: 10.1111/j.1574-6968.2006.00505.x. [DOI] [PubMed] [Google Scholar]
- Kaneko Y, Thoendel M, Olakanmi O, Britigan BE, Singh PK. The transition metal gallium disrupts Pseudomonas aeruginosa iron metabolism and has antimicrobial and antibiofilm activity. J Clin Invest. 2007;117:877–888. doi: 10.1172/JCI30783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Chen HD, Latifi T, Groisman EA. Reciprocal Control between a Bacterium's Regulatory System and the Modification Status of Its Lipopolysaccharide. Mol Cell. 2012;47:897–908. doi: 10.1016/j.molcel.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr KG, Snelling AM. Pseudomonas aeruginosa: a formidable and ever-present adversary. J Hosp Infect. 2009;73:338–344. doi: 10.1016/j.jhin.2009.04.020. [DOI] [PubMed] [Google Scholar]
- King JD, Kocíncová D, Westman EL, Lam JS. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun. 2009;15:261–312. doi: 10.1177/1753425909106436. [DOI] [PubMed] [Google Scholar]
- Kivistik PA, Kivi R, Kivisaar M, Horak R. Identification of ColR binding consensus and prediction of regulon of ColRS two-component system. BMC Mol Biol. 2009;10:46. doi: 10.1186/1471-2199-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra O, Bedoux G, Brecker L, Lindner B, Carballo PS, Haras D, Zähringer U. Structure of a highly phosphorylated lipopolysaccharide core in the ΔalgC mutants derived from Pseudomonas aeruginosa wild-type strains PAO1 (serogroup O5) and PAC1R (serogroup O3) Carbohydr Res. 2003;338:2667–2677. doi: 10.1016/j.carres.2003.07.004. [DOI] [PubMed] [Google Scholar]
- Kox LF, Wösten MM, Groisman EA. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 2000;19:1861–1872. doi: 10.1093/emboj/19.8.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laddaga RA, Silver S. Cadmium uptake in Escherichia coli K-12. J Bacteriol. 1985;162:1100–1105. doi: 10.1128/jb.162.3.1100-1105.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Hsu F-F, Turk J, Groisman EA. The PmrA-Regulated pmrC Gene Mediates Phosphoethanolamine Modification of Lipid A and Polymyxin Resistance in Salmonella enterica. J Bacteriol. 2004;186:4124–4133. doi: 10.1128/JB.186.13.4124-4133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LJ, Barrett JA, Poole RK. Genome-Wide Transcriptional Response of Chemostat-Cultured Escherichia coli to Zinc. J Bacteriol. 2005;187:1124–1134. doi: 10.1128/JB.187.3.1124-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect Inst Pasteur. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- Madsen JA, Cullen TW, Trent MS, Brodbelt JS. IR and UV photodissociation as analytical tools for characterizing lipid A structures. Anal Chem. 2011;83:5107–5113. doi: 10.1021/ac103271w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiyazhagan N, Natarajan D. [Accessed October 28, 2014];Bioremediation on Effluents from Magnesite and Bauxite Mines using Thiobacillus Spp and Pseudomonas Spp. J Bioremediation Biodegrad. 2011 02 http://www.omicsonline.org/2155-6199/2155-6199-2-115.digital/2155-6199-2-115.html. [Google Scholar]
- McPhee JB, Bains M, Winsor G, Lewenza S, Kwasnicka A, Brazas MD, et al. Contribution of the PhoP-PhoQ and PmrA-PmrB Two-Component Regulatory Systems to Mg2+-Induced Gene Regulation in Pseudomonas aeruginosa. J Bacteriol. 2006;188:3995–4006. doi: 10.1128/JB.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, Hancock REW. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa: PmrA-PmrB of Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, et al. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother. 2011;55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz SM, Ernst RK. The role of Pseudomonas lipopolysaccharide in cystic fibrosis airway infection. Subcell Biochem. 2010;53:241–253. doi: 10.1007/978-90-481-9078-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinforma Oxf Engl. 2005;21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc Natl Acad Sci. 2013;110:1464–1469. doi: 10.1073/pnas.1218080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham BD, Trent MS. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol. 2013;11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki EM, O'Brien JP, Brodbelt JS, Trent MS. Characterization of P seudomonas aeruginosa LpxT reveals dual positional lipid A kinase activity and coordinated control of outer membrane modification: Identification of P. aeruginosa LpxT. Mol Microbiol. 2014 doi: 10.1111/mmi.12796. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odegaard TJ, Kaltashov IA, Cotter RJ, Steeghs L, Ley P, van der, Khan S, et al. Shortened hydroxyacyl chains on lipid A of Escherichia coli cells expressing a foreign UDP-N-acetylglucosamine O-acyltransferase. J Biol Chem. 1997;272:19688–19696. doi: 10.1074/jbc.272.32.19688. [DOI] [PubMed] [Google Scholar]
- Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic Fibrosis Sputum Supports Growth and Cues Key Aspects of Pseudomonas aeruginosa Physiology. J Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JP, Pesci EC, Iglewski BH. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J Bacteriol. 1997;179:5756–5767. doi: 10.1128/jb.179.18.5756-5767.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JC, Groisman EA. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol. 2007;63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron K, Caille O, Rossier C, Delden C, Van, Dumas J-L, Köhler T. CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa. J Biol Chem. 2004;279:8761–8768. doi: 10.1074/jbc.M312080200. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja CE, Anbazhagan K, Selvam GS. Isolation and Characterization of A Metal-resistant Pseudomonas Aeruginosa Strain. World J Microbiol Biotechnol. 2006;22:577–585. [Google Scholar]
- Rodrigue A, Quentin Y, Lazdunski A, Méjean V, Foglino M. Cell signalling by oligosaccharides. Two-component systems in Pseudomonas aeruginosa: why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, et al. Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J Am Chem Soc. 2013;135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R, Choudhury B, Septer A, Merighi M, Carlson R, Gunn JS. Identification of cptA, a PmrA-regulated locus required for phosphoethanolamine modification of the Salmonella enterica serovar typhimurium lipopolysaccharide core. J Bacteriol. 2005;187:3391–3399. doi: 10.1128/JB.187.10.3391-3399.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitzel GM, Parsek MR. Heavy Metal Resistance of Biofilm and Planktonic Pseudomonas aeruginosa. Appl Environ Microbiol. 2003;69:2313–2320. doi: 10.1128/AEM.69.4.2313-2320.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaipisuttikul I, Hittle LE, Chandra R, Zangari D, Dixon CL, Garrett TA, et al. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol Microbiol. 2014;91:158–174. doi: 10.1111/mmi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Karbarz MJ, Wang X, Raetz CRH, McGrath SC, Cotter RJ, Trent MS. Periplasmic cleavage and modification of the 1-phosphate group of Helicobacter pylori lipid A. J Biol Chem. 2004;279:55780–55791. doi: 10.1074/jbc.M406480200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran AX, Whittimore JD, Wyrick PB, McGrath SC, Cotter RJ, Trent MS. The lipid A 1-phosphatase of Helicobacter pylori is required for resistance to the antimicrobial peptide polymyxin. J Bacteriol. 2006;188:4531–4541. doi: 10.1128/JB.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaara M, Vaara T. Outer Membrane Permeability Barrier Disruption by Polymyxin in Polymyxin-Susceptible and -Resistant Salmonella typhimurium. Antimicrob Agents Chemother. 1981;19:578–583. doi: 10.1128/aac.19.4.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Karbarz MJ, McGrath SC, Cotter RJ, Raetz CRH. MsbA Transporter-dependent Lipid A 1-Dephosphorylation on the Periplasmic Surface of the Inner Membrane: TOPOGRAPHY OF FRANCISELLA NOVICIDA LpxE EXPRESSED IN ESCHERICHIA COLI. J Biol Chem. 2004;279:49470–49478. doi: 10.1074/jbc.M409078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McGrath SC, Cotter RJ, Raetz CRH. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4'-phosphatase LpxF. J Biol Chem. 2006;281:9321–9330. doi: 10.1074/jbc.M600435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weert S, e, Dekkers LC, Bitter W, Tuinman S, Wijfjes AHM, Boxtel R, van, Lugtenberg BJJ. The two-component colR/S system of Pseudomonas fluorescens WCS365 plays a role in rhizosphere competence through maintaining the structure and function of the outer membrane. FEMS Microbiol Ecol. 2006;58:205–213. doi: 10.1111/j.1574-6941.2006.00158.x. [DOI] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and Export of Bacterial Lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Lin S, Cotter RJ, Raetz CR. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-L-arabinose, phosphoethanolamine and palmitate. J Biol Chem. 1999;274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



