Abstract
Microorganisms shape, and are shaped by, their environment. In host-microbe associations, this environment is defined by tissue chemistry, which reflects local and organism-wide physiology, as well as inflammatory status. We review how, in the squid-vibrio mutualism, both partners shape tissue chemistry, revealing common themes governing tissue homeostasis in animal-microbe associations.
Keywords: symbiosis, nutrition, innate immunity, MAMPs, stress, development, bioluminescence
1. Introduction
Animals and plants have evolved in a sea of microbes [43]. Some of these microbes form beneficial and co-evolved alliances with specific host tissues, from which both partners may derive benefits such as nutrient acquisition or defense [43]. In recent years, it has become clear that the composition of an organism’s microbiota is structured by the host’s tissue environment [23]. Host-tissue chemistry reflects both underlying physiological function and inflammatory status (for an in-depth review of tissue homeostasis, refer to [9]). The balance of these two factors varies, even within tissues of the same organ system. For example, the intestinal epithelium of animals is much more tolerant of inflammation-inducing microbial products in the colon (a major site of immune maturation), than in the ileum (the primary site of nutrient absorption) [49]. Thus, tissue chemistry and physiology can present significant selective pressures on the evolution of mutualism. In turn, the interaction of the normal microbiota with the innate immune system is a key mechanism that shapes the tissue environment, and mediates the activation of adaptive immunity [32]. From this perspective, the relationship between the innate immune response and the microbiota is at the core of the acquisition, development, and maintenance of microbial symbionts.
Invertebrate animal models have long been critical to the study of innate immunity/microbe interactions, beginning in the 1800’s, when Metchnikoff first observed phagocytosis in starfish larvae. Since this time, these invertebrate models have provided a window into the mechanisms by which the innate immune system maintains homeostasis, and have already revealed cornerstones of the chemical dialogue that underlies stability: for example, toll-like receptors, a major class of sensors of bacterial products, were first discovered during studies of the fruit fly [26]. It is likely that the strategies of innate immune recognition used by other branches of invertebrate phylogeny, such as annelids, nematodes, and mollusks, have much to reveal about the conserved strategies of mutualism [53, 21]. In this review, we focus on the molluscan branch of metazoan phylogeny and, specifically, on what the squid Euprymna scolopes has revealed about how its innate immune system and its co-evolved microbial symbiont, Vibrio fischeri, orchestrate the initiation, development, and maintenance of mutualism. A discussion of the mechanisms underlying these processes is not within the scope of the present review; for that, we recommend recent articles on the immune systems of squids and other invertebrates [53, 21, 62]. We focus instead on how the squid-vibrio mutualism has contributed to our understanding of the chemical ‘language’ that forms the negotiation between a microbial symbiont and its host’s innate immune response.
1.1 The squid-vibrio model
E. scolopes forms a specific association with the Gram-negative, bioluminescent, marine gamma-proteobacterium V. fischeri. The mutualism takes place in the epithelium-lined crypts of a specialized anatomical structure of the squid called the light organ [73] (Fig. 1A). The functional basis of this mutualism is the bioluminescence provided by V. fischeri, which is used for host behaviors. The light organ shares much of the anatomical and physiological features of an eye: a lens and reflector manipulate the intensity and direction of bioluminescence (Fig. 1B), and the supporting epithelial tissue is photoreceptive [44] and expresses genes involved in eye development [58]. Unlike the eye, which collects and perceives light from the environment, the light organ manipulates the bioluminescence of a dense (108 CFU/mature light organ) population of V. fischeri [63]. Only V. fischeri can colonize this tissue habitat, and a persistent, monospecific population of the symbiont is maintained in the light organ throughout the squid’s ~9-month life. The squid’s immune response is orchestrated primarily by both a population of circulating macrophage-like immune cells, called hemocytes, and tissue-specific responses to microbe-associated molecular patterns (MAMPs) [45, 62]. In the following sections, we review our current understanding of the chemical selection for V. fischeri that occurs during the initiation, development and maintenance of mutualism.
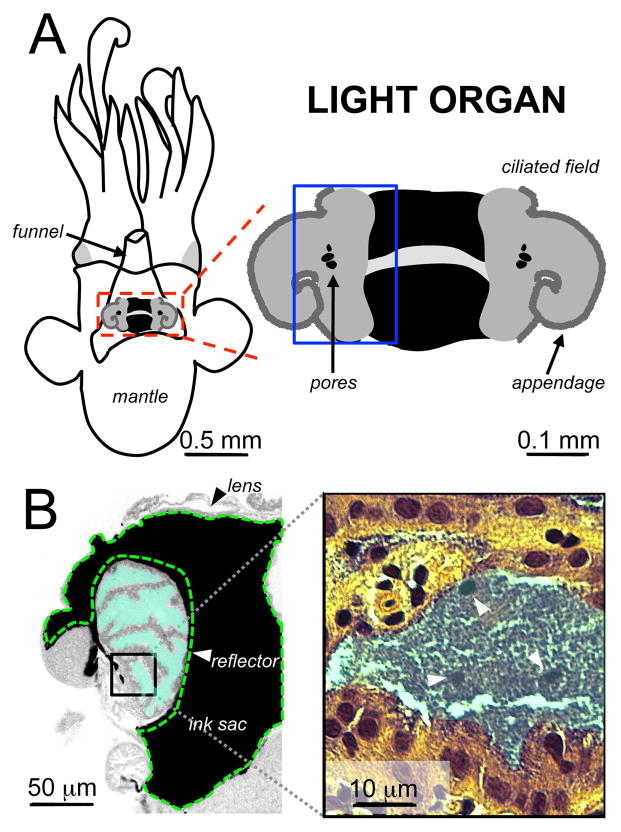
Figure 1. The anatomy of the juvenile squid-vibrio mutualism.
A) The squid’s light organ is the anatomical structure that maintains the bioluminescent symbiont V. fischeri and modulates light output. The organ is located underneath the mantle of the squid, just atop the funnel: a structure used to move water into and out of the mantle cavity. The immature light organ (boxed in red, and enlarged at right) has bilateral ciliated fields and appendages. At the base of the appendages are pores, leading into the crypts of the light organ. B) A cross-section of the light organ, boxed in blue in (A), shows that the symbiont (teal) is maintained in extracellular crypts lined with a polarized epithelium that is photoreceptive. Structures surrounding the crypts, such as the reflector (indicated in green dashed lines), ink sac, and lens manipulate the light produced by the symbiont for host behaviors.
2. Initiation: the winnowing process
Symbiosis is initiated in a multi-step process, beginning when the newly hatched squid harvests V. fischeri cells from the bacterioplankton. Ecological studies have demonstrated that V. fischeri is enriched in the squid’s habitat, but still represents only a minor constituent of the total population [40]. Therefore, the challenge for a juvenile squid is two-fold: it must first sample sufficient seawater to encounter V. fischeri, and then select cells of this specific symbiont from among the many kinds of other microbes that have been collected, in a process named ‘winnowing’ [56]. Winnowing begins outside of the light organ, in bilateral ciliated fields that are unique to the uncolonized, or aposymbiotic, state. This complex, step-wise progression occurs over a distance of only a hundred microns, highlighting the importance of understanding the nanoscale structure of host-tissue microenvironments. In this section, we review what is known of the chemical exchange that takes place between the partners during winnowing.
2.1 Making first contact
Upon hatching, ventilation of the mantle cavity brings bacterioplankton-rich seawater in contact with the surface of the light organ. Peptidoglycan (PGN) fragments of cell wall released by these bacteria induce a non-specific shedding of sialylated host mucus [52] (Fig. 2A). The acidic glycosylation of the mucins produces a matrix with a pH of ~6.3 [37], in which the bacterioplankton are entrapped. Host antimicrobial peptides/proteins such as galaxin and peptidoglycan recognition-protein 2 (PGRP2), establish an early negative selection against Gram-positive microbes in the mucus [30, 74]. Combined with other antimicrobial constituents secreted into the mucus, such as the oxygen carrier and phenoloxidase, hemocyanin [38] and, as yet, uncharacterized factors, the chemistry that defines this first site of host contact results in the exclusion of many seawater bacteria.
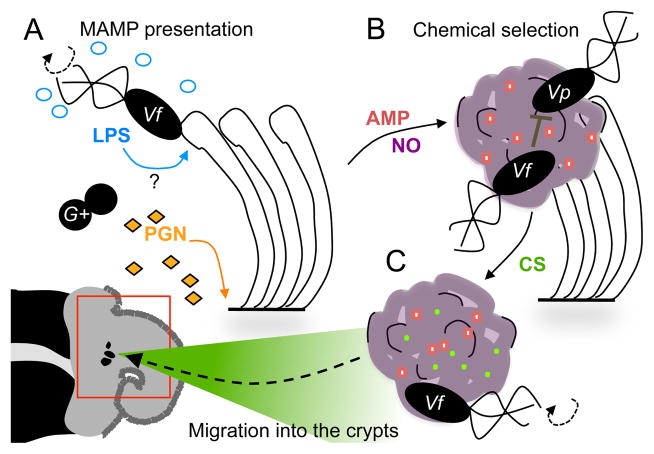
Figure 2. The winnowing of the symbiont.
The events described in this figure take place in the external ciliated epithelium of the light organ (lower left, indicated by red box). A) The first contact of host and symbiont is accompanied by the presentation of microbial products. Along the surface of the light organ’s ciliated epithelium, peptidoglycan (PGN), shed by seawater bacteria, induces mucus production (purple). V. fischeri bind to the cilia. The binding event may also promote presentation to host tissues of lipopolysaccharide (LPS), which are released by V. fischeri during rotation of its sheathed flagella. B) The mucus is a site of chemical selection. In addition to sialylated mucins (pH 6.3), antimicrobial peptides (AMP) and nitric oxide (NO) shape the chemistry of the mucus matrix. The mucus is an environment that not only excludes Gram-positive microbes (G+), but also allows V. fischeri to out-compete any co-aggregating Gram-negative ones such as Vibrio parahaemolyticus. C) The mucus chemistry prepares V. fischeri for colonization. Contact with V. fischeri induces the expression of host enzymes that result in chitin sugars (CS) to accumulate in the mucus matrix. The exposure to CS, as well as to NO, primes V. fischeri for migration into, and survival within, the light organ crypts. Specifically, after CS priming, V. fischeri chemotaxes towards a gradient of the sugar that emanates from the pores (indicated at center of red box), which serve as the entrance to the crypts.
2.2 Establishing dominance in the mucus
Aided by the PGN-triggered mucus, V. fischeri cells that are drawn into the mantle cavity during ventilation adhere directly to the cilia on the organ’s surface [1] (Fig. 2A). As a result of this adhesion, and perhaps the release of LPS during the rotation of the symbiont’s sheathed flagellum [7], the transcription of additional antimicrobial factors such as lysozyme, chitinase, legumin, ferritin, and peptidase is induced in light-organ tissues [37]. A subset of these antimicrobials accumulates in the mucus, and may represent another, symbiont-induced, stage of winnowing (Fig. 2B). Those V. fischeri cells that encode the gene rscS, a regulator of capsule formation that is present only in certain squid-specific strains [83, 42], will adhere to each other, forming aggregates [75]. The mucus may also be a site of competition between the small number of microbial species that can withstand its particular chemistry: some other Gram-negative bacteria, like V. parahaemolyticus, also form aggregates, but only in the absence of V. fischeri [55], suggesting a positive role of the symbiont in driving specificity. Future characterization of both mucus chemistry and the signaling between host and symbiont will reveal the mechanisms by which this environment is defined as the initial site of selection.
2.3 Preparing for colonization
While in the aggregate, exposure to host-derived nitric oxide (NO), and chitin-derived sugars (CS) prepares, or ‘primes’, V. fischeri cells for the next steps of colonization [37, 78] (Fig. 2C). NO is present in the mucus at levels that are below the lethal threshold for V. fischeri [15], yet sufficient to induce a response that coordinates resistance to subsequent exposure to NO and iron-linked oxidative stress [79, 19, 69, 78]. CS are derived from the host glycan chitin: a polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) residues. Hydrolysis of chitin by host enzymes creates a chemotactic gradient that emanates from pores at the base of the ciliated fields. This gradient primes the symbiont’s chemotactic migration from the ciliated fields to these pores, and then into the light organ [41, 37]. Thus, the mucus is both a site of selection and a platform that coordinates the transition of symbionts from the seawater habitat to the host’s tissues.
2.4 How does the squid-vibrio association contribute to our understanding of the initial selection for cooperation?
The complexity of light-organ colonization highlights a theme among co-evolved animal- microbe associations: the host tailors its tissue chemistry to achieve specificity [12]. Other examples of this strategy include (i) secretion of antimicrobial peptides on the epithelial surface of hydra, which facilitates the colonization by a specific and robust microbial community from the surrounding water [22], and (ii) the pregnancy-driven changes in the tissue chemistry of the mammalian gut and vaginal canal, which shift the composition of the maternal microbiota responsible for neonatal colonization [23]. Moreover, the presentation of MAMPs by the symbiont can induce changes in mucus chemistry, creating a site of selection and priming for colonization. Similar studies of the vaginal mucosa of mammals [23], or the egg capsule of invertebrate larvae, may reveal that both symbiont-induced changes in mucus chemistry and priming of the symbiont for colonization are common strategies by which animal hosts select their initial microbial colonizers.
3. Development: transitioning in the environment
Colonization, defined as the proliferation of symbiont cells in the light-organ crypts, sets in motion an irreversible program of host development and morphogenesis. The signal for this transition is presented at dawn, following the first night of colonization [35, 16]. Whereas the transcriptional response to first contact correlates with physiological changes that support symbiont acquisition, a study profiling host transcription after colonization (>18 h post inoculation) suggests both bioluminescence-dependent and -independent morphological responses [10]. For example, MAMPs presented in the crypts by V. fischeri (e.g., PGN and lipopolysaccharide) induce an irreversible morphogenesis of the light organ [35] that marks the beginning of the symbiont’s accommodation to the crypt environment. Symbiont bioluminescence not only serves as the functional basis of the mutualism [76, 72], but also is required for full morphogenesis [36]. In the following section, we review the chemistry and physiology that characterize both symbiont-induced host-tissue morphogenesis, and the selection for symbiont bioluminescence.
3.1 Creating barriers: symbionts induce irreversible morphogenesis
Symbionts that successfully migrate into the light organ, and present MAMPs there, attenuate the production of NO in the crypts, and induce mucus shedding [15, 52] (Fig. 3A). The attenuation of NO by both PGN and lipopolysaccharide (LPS) signaling is accompanied by the trafficking of hemocytes into the ciliated-fields [2] (Fig. 3B). MAMP signaling from the crypts orchestrates the subsequent apoptosis and regression of these structures [34, 45] (Fig. 3B). Although the crypts remain open to the environment via the pores, the loss of the ciliated fields creates a significant physical restriction to subsequent colonization [73] (Fig. 3A). Moreover, the symbiont’s presence induces global changes in the protein composition of host symbiotic tissues [17], and cues the secretion of the enzymes galaxin [30], PGRP2 [74], and alkaline phosphatase [61] into the crypts (Fig. 3B). Thus, MAMP signaling by V. fischeri re-shapes light-organ anatomy to effectively ‘shut the door’ behind those few cells winnowed from the environment, as well as shift the chemical composition of the crypts. It remains to be seen whether this symbiont-induced chemical shift contributes to the specificity and stability of colonization by V. fischeri.
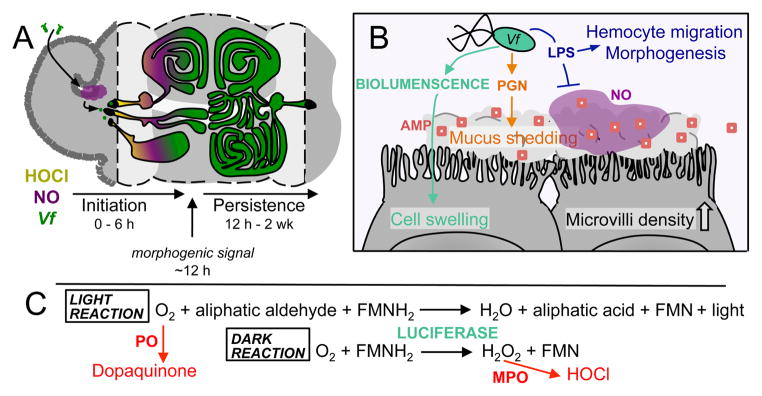
Figure 3. Accommodations of host and symbiont in the light-organ crypts.
A) V. fischeri (Vf, green), attenuates host production of hypochlorous acid (HOCl, yellow), and nitric oxide (NO, purple), upon transit into the crypts. The process of initial colonization occurs within the first 6 h after contact between host and symbiont. Around 12 h post-colonization, the presentation of MAMPs (PGN & LPS) by the symbiont has delivered an irreversible morphogenic signal to light-organ tissues. During subsequent development of the persistent association, the ciliated fields of the light organ regress, and the crypt structure becomes more complex. By 2 weeks the ducts leading to the crypts have condensed from three to one, and continued colonization by the symbiont has induced a physiological state in host tissues that prevents secondary colonization. B) Symbiont-derived chemical cues orchestrate physiological and morphological changes in the light organ. Bioluminescence (teal) induces swelling of the crypt epithelium. Peptidoglycan (PGN) induces release of mucus, and the secretion of antimicrobial factors (AMP, red squares) into the crypts. Lipopolysaccharide (LPS), attenuates the local production of NO in the crypts, and induces the migration of hemocytes into the ciliated appendages, which is followed by apoptosis and regression of these structures. C) V. fischeri luciferase is a mixed-function oxidase that produces light upon the oxidation of FMNH2 and an aliphatic aldehyde by oxygen. Oxygen is also a substrate of the dopaquinone-producing host enzyme phenoloxidase (PO). In the absence of sufficient aliphatic aldehyde, the oxidation of FMNH2 and reduction of oxygen produce toxic hydrogen peroxide (H2O2), but no light. Hydrogen peroxide is a substrate of the toxic hypochlorous acid-producing host enzyme myeloperoxidase (MPO). Greater production of light by the symbiont may reduce the host’s production of antimicrobial compounds (red pathways), facilitating the persistence of the symbionts in the crypts.
3.2 Establishing a colonization: bioluminescence allows persistence in the crypts
The few cells that survive winnowing populate the crypt by doubling approximately 15 times within the first 12 h of colonization (i.e., ~1 generation per hour [63]). The resulting increase in symbiont population density induces bioluminescence: an enzymatic process that consumes molecular oxygen and chemical reducing equivalents [18]. The expression of the bioluminescence-producing enzyme, luciferase, is transcriptionally regulated by the population-density dependent accumulation of quorum-signaling pheromones in V. fischeri ([6]; for a more in-depth review of the transcriptional regulation of bioluminescence, consult reference [48]). This regulation is subject to additional layers of transcriptional and post-translational control that are linked to the metabolic state of the symbiont [48, 18]. In addition, the metabolic demands of bioluminescence must be accommodated to those of growth to maintain both a robust colonization and an appropriate level of light production throughout the first days of symbiosis [72]. Symbionts that grow but do not produce sufficient bioluminescence cannot maintain a stable colonization of the light organ [72]. Thus, the balance between growth and bioluminescence in the crypts is key to maintaining the stability of the association.
The high energy cost of bioluminescence predicts that a subset of symbiont cells might ‘cheat’ their neighbors by shifting metabolic resources toward growth at the expense of bioluminescence. However, this outcome is prevented by a remarkable mechanism of host surveillance capable of detecting the bioluminescent status of each cell within the symbiont population [33]. There are essentially two mechanisms by which a mutation could lead to the loss of bioluminescence: an altered production (luxIR) or functionality (luxAB) of luciferase, or the loss of its substrate, tetradecanal, by mutation of luxC,D or E (Fig. 3C). The loss of luciferase enzyme function would also alter the balance of oxygen consumption by the symbiont. In the presence of free oxygen, the host enzyme phenoloxidase/hemocyanin catalyzes the production of the bactericidal melanin precursor dopaquinone from tyrosine derivatives [38]. Thus, a symbiont unable to consume oxygen at a typical rate might be detected and killed by an ambient increase in phenoloxidase activity [38] (Fig. 3C). Alternately, tetradecanal limitation leads to an incomplete reduction of oxygen by luciferase to hydrogen peroxide, rather than to water and a photon of light [27]. Not only is hydrogen peroxide toxic, but it also is a substrate for the host enzyme halide peroxidase, a dianisidine peroxidase that catalyzes the formation of bacteriocidal hypochlorous acid (HOCl) from hydrogen peroxide [71]. Halide peroxidase is abundant in the crypt, and coats the outside of the symbionts [80]. Thus, it is possible that aldehyde-underproducers would be detected and killed by increased halide peroxidase substrate availability (Fig. 3C). In support of this latter prediction, the hydrogen-peroxide consuming enzyme catalase is required for V. fischeri to maintain a stable colonization of the light organ [77]. In addition, squid-specific strains of V. fischeri only produce sufficient aldehyde to support bioluminescence in symbiosis [5], and non-squid specific strains of V. fischeri that produce abundant aldehyde in culture can be evolved by passage through the squid reduce their aldehyde level to one resembling that of symbiotic isolates [67]. We predict that further investigation will reveal how phenoloxidase, halide peroxidase, or other host enzymes detect within the population dark cells that arise from either class of mutations.
3.3 Constructing the environment: symbionts create a functional tissue physiology
The bioluminescence and growth of the symbiont population also induces reversible physiological/morphological changes in the crypt epithelium, such as cell swelling [76], and an increase in the density of microvilli at epithelial surfaces in contact with symbionts [39]. The function of these physiological changes is not fully understood, but they may be evidence of the host’s provision of nutrients to the symbiont. Among the carbon sources provided in the crypts are (i) amino acids [25], (ii) glycerophospholipids [81], and (iii) chitin-derived sugars (CS) [68]. The crypt matrix contains millimolar levels of several amino acids and peptides, which are used by the symbiont to support growth. Because certain amino-acid auxotrophs of V. fischeri fail to colonize the crypts to normal levels [25], it appears that these nutrients are not provided in a growth-balanced proportion. While the significance of the composition of amino acids provided to the symbionts is not known, perhaps it is designed to support specific metabolic functions of the symbionts.
Symbionts also incorporate host lipids into their membranes [81], leading to an unusual enrichment of long-chain unsaturated aliphatic acids. A similar incorporation of eukaryotic lipids into Vibrio cholerae [24, 59], and Enterococcus faecalis [64] provides protection from host-associated stressors, suggesting that this modification might also confer stress resistance to symbiotic V. fischeri. Finally, as the light organ matures, chitin-associated sugars (CS) become available [68]. Host-derived CS have profound consequences on the metabolism and physiology of the crypt, which will be discussed in Section 4, below. However, in the immature crypts, the stability of colonization is positively correlated with the strength of the transcription regulator NagC, whose repression is relieved by symbiont CS catabolism [47]. Thus, the host selects for symbionts that do not respond to CS during the initial phase of symbiosis, but subsequently require CS catabolism in the mature association. Efforts are currently underway to understand how CS catabolism contributes to the stability of light-organ colonization, and to model the metabolism of symbiotic V. fischeri at both stages of light-organ development. These efforts will help reveal how host-derived nutrition supports the association’s homeostasis during the development of the symbiosis.
3.4 Making an indelible first impression: the development of a host refractory state
A key transition point in development is reached ~2 weeks after the host-symbiont interaction is initiated: if a non-bioluminescent strain of V. fischeri has been present in the crypts, it will have been eliminated by the host [33]. Conversely, the persistent colonization of the light organ with normal, bioluminescent symbionts leaves an ‘imprint’; experimental curing after at least 5 days of colonization reveals that bioluminescent V. fischeri cells induce a physiological state in the light organ that is refractory to immediate re-colonization [33]. In addition, while adult hemocytes within the crypt can contain engulfed V. fischeri cells (S. Nyholm, personal communication), the continual exposure to V. fischeri apparently ‘educates’ the circulating population of these immune cells. Interestingly, symbiosis-dependent differences are apparent in the proteome of hemocytes derived from adult (>4-week old) animals [66]. In addition, protracted interaction with V. fischeri expressing the major outer membrane porin OmpU decreases the binding to, and phagocytosis of the symbiont [57], suggesting that symbiont physiology may also play a role. Notably, the V. cholerae ompU homolog is induced in a low-pH environment, leading to an increased acidic-bile resistance in this enteric pathogen [60]. Further study of the mechanisms of hemocyte education, as well as the physiological characteristics of bacteria recognized by educated hemocytes, may reveal just how the symbiont modulates the activity of the host’s circulating immune cells.
3.5 How does the squid-vibrio partnership contribute to our understanding of the development of mutualism?
The squid-vibrio association was one of the first examples of MAMP-directed tissue morphogenesis [35], and subsequent studies in this system have reinforced the theme that microbial metabolites can direct normal host-tissue maturation. Work with mammals provides many examples of how initial, even transient, dysbiosis of the microbiota during development can have lasting detrimental consequences on host health, due to mis-regulation of pattern-recognition and microbe-dependent tissue development. For example, antibiotic exposure at birth affects ileal immunity in mice, leading to inflammation, metabolic imbalances and increased adiposity [13]. In this context, the resilience of the symbiont to the constraints of episodic changes in the host environment is a key determinant of stability. The squid-vibrio association has revealed that the metabolic and physiological accommodations of the symbiont make it resilient to the host-tissue environment and, perhaps, tailor the immune defenses. This discovery is echoed by the resistance of the intestinal symbiont Bacteroides thetaiotaomicron to host antimicrobial peptides, which contributes to the resilience of this abundant member of the gut microbiota in the intestinal lumen [14]. Squid-derived nutrition also structures the development of symbiont metabolism, much like the changing profile of mammalian mucin glycans selects for microbes that are able to metabolize plant starches, even before these starches have entered the infant diet. The resilience of V. fischeri is linked to its metabolic repertoire: the host’s selection initially favors those symbionts that repress CS-responsive genes during the early stages of symbiosis; then, it depends on the ability of the symbiont to express these genes in the mature association. Perhaps genetic manipulation of primary colonizers of the gut will reveal that an expanded metabolic repertoire promotes symbiont resilience over the course of host development.
4. Persistence: daily and circadian rhythms
Once established in a morphologically mature light organ, the symbiont population is maintained in a dynamic, daily cycle for the life of the host [4]. This period of ‘persistence’ constitutes 90% of the lifetime of the squid [33]. Remarkably, the host maintains a monospecific colonization of V. fischeri throughout this period, and does so while preventing the loss of bioluminescence over approximately 3000 microbial generations [82]. From the first day, the light organ is subject to a diel biological rhythm, in which photoreceptive tissues perceive the dawn light cue, and cause expulsion of the crypt contents, including the majority of the symbiont population [4] (Fig. 4). The remaining symbionts rapidly repopulate the crypts, and the majority of the resulting culture is expelled anew the following morning [63]. After a bioluminescent population has been established, it will be maintained throughout subsequent host maturation: a process that occurs over several weeks, and during which time the anatomical features of the light organ develop and the crypts increase in morphological complexity [46].
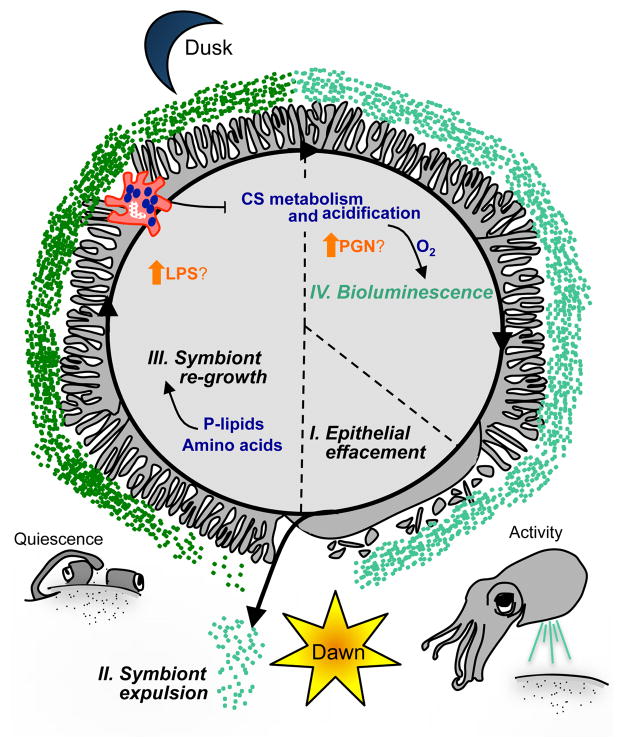
Figure 4. Biological rhythms in the host and symbiont.
The daily cycle of symbiosis is characterized by 4 stages (I – IV). The physiological and behavioral attributes of this rhythm are a combination of symbiont-independent (grey/black), and symbiont-dependent (colored) events, depicted as the temporal change in the interface between the crypt epithelium and the symbionts. (I) Just before dawn, the microvilli at the apical surfaces of the epithelial cells efface. (II) With the dawn light cue, the contents of the light-organ crypts are expelled. If an animal is colonized, this event clears the crypts of ~95% of the symbiont population. (III) The host microvilli reappear and the remaining symbionts re-populate during the day: a period of behavioral quiescence for the squid. Host-derived nutrients, including amino acids and phospholipids (P-lipids) support this bacterial proliferation. Just before dusk, several events take place: lipopolysaccharide (LPS) levels in the crypt are predicted to increase due to a decrease in LPS-degrading, host alkaline phosphatase production; in addition, chitin sugar (CS)-bearing hemocytes migrate into symbiont-colonized light organ tissues. (IV) At dusk, the symbiont population ferments CS, producing acid, and releasing oxygen from the host carrier-protein, hemocyanin. The oxygen released from hemocyanin fuels bioluminescence, while the acidification of the crypts may lead to an increase in PGN levels due to its ability to decrease peptidoglycan recognition-protein 2 activity. The host is active in the water column during this night phase, and uses symbiont bioluminescence in its behaviors.
The establishment and maintenance of the mature symbiotic state is driven both by the host’s daily response to environmental cues, as well as by circadian rhythms (Fig. 4). In animals like squid, the circadian rhythm is orchestrated by the central nervous system, which coordinates the peripheral oscillatory circuits, or clocks, of individual organ systems [3]. The circadian clock is entrained by both transcriptional and redox-responsive post-translational regulation [31, 20]. In this section, we review the development of daily and entrained rhythms in the squid-vibrio symbiosis, and the consequences of these rhythms on tissue physiology and biochemistry.
4.1 Initial rhythms
Beginning with the onset of the association, the daily expulsion and subsequent re-growth of the symbiont population establish a biological rhythm [4]. The expulsion process occurs even in the absence of the symbiont, but is triggered only by a light stimulus to the squid’s eyes [54]. These factors indicate that expulsion is a rhythmic, but not a light-entrained, or circadian, function. In contrast to expulsion, other symbiosis-dependent rhythms in the initial light-organ tissues show signs of circadian underpinnings. For example, the microvilli at the apical surfaces of the crypt epithelium are shed just before dawn [81] (Fig. 4). Cytoskeletal remodeling genes are also subject to a daily transcriptional rhythm, reflecting in part the daily effacement and renewal of the microvilli [81]. These initial light-dependent rhythms define the physiology upon which the symbiosis is established and, thus, the baseline from which the symbiont population can shift tissue homeostasis.
The symbiont adds to the existing rhythms of the light organ by bioluminescence-dependent and -independent processes. Symbiont bioluminescence entrains a tissue-specific circadian oscillation of the clock gene escry1 [29]. The regulatory targets of the bioluminescence-entrained light-organ clock have yet to be identified, but may include genes transcribed in response to bacterial bioluminescence, such as the eye specification genes eya, six, pac6 and dac [58], and genes encoding a predicted vasoconstrictor, octopressin, a matrix metalloprotease, and hemocyanin [10]. The two MAMPs, LPS and PGN, orchestrate a rhythm of alkaline phosphatase enzymatic activity [61], and may be required for the symbiont-dependent daily rhythm of immune-cell migration [68]. Further study of the light organ’s rhythmic biology may well reveal general physiological parameters that define tissue homeostasis.
4.2 The mature daily rhythm
The initial biological rhythms of the light organ take on an additional metabolic and physiological complexity during post-embryonic development [4, 68]. In parallel with the development of the symbiotic rhythm, the host’s behavior shifts: the squid begins to bury in the daytime, and emerge only at night to hunt in the open water [70]. It is not known to what extent this behavioral transition influences the physiology of the light organ, or the animal as a whole; however, it is likely that the daily burying behavior is accompanied by a period of metabolic quiescence, as the animal reduces its respiratory ventilation while buried. Along with the establishment of this behavioral rhythm, light-organ structures such as the lens, ink sac, reflector and crypt epithelium undergo a period of morphogenesis [46]. Behavioral and morphological development is complete at ~ 4 weeks, marking the onset of the mature symbiotic state [33, 11].
Coincident with light-organ maturation, the symbiont’s metabolism of carbon develops a daily rhythm. Reflecting this rhythm, transcripts related to host-associated chitin synthesis and breakdown, as well to symbiont CS catabolism, increase in abundance in light-organ tissues at night [81]. CS are provided to the symbiont through the migratory rhythm of hemocytes [68], which carry granules of chitin, a polymeric ‘storage’ form of CS, and enzymes required to hydrolyze the chitin into soluble fragments [28]. The provision of CS is positively correlated with the nocturnal migration of hemocytes: a behavior that begins in the first days of symbiosis [68]. However, in the immature crypts, the symbiont neither senses nor catabolizes CS, suggesting that crypt maturation, or perhaps education of the hemocytes, is required for the nocturnal delivery of CS to the symbiont.
4.3 Biochemical cycling in the mature symbiosis
The consequence of the symbiont’s metabolic rhythm is a nocturnal acidification of the crypts of adult animals (Fig. 4). Fermentation of CS at night is sufficient to acidify the crypt (nocturnal pH ~5.7) [38, 68]. In vitro work has demonstrated that this acidification decreases the activities of two secreted immunity proteins, alkaline phosphatase and PGRP2 [61, 74]. Typically, alkaline phosphatase dephosphorylates LPS, while PGRP2 degrades PGN, decreasing the ability of these MAMPs to signal animal cells; thus, acidification may lead to an increase in MAMP stability and, thus, signaling in the crypt. In support of this hypothesis, proteomic studies of the crypt contents also indicate an abundance of oxidative stress-associated host and symbiont enzymes [65], possibly a marker of tissue inflammation. Alternately, oxidative stress in the crypt may reflect the partitioning of oxygen between symbiont luciferase and reactive-oxygen producing host enzymes [71, 38]. Supporting the latter hypothesis, the affinity of the host’s oxygen carrier-protein, hemocyanin, is negatively correlated with acidification [38], potentiating the delivery of oxygen to the symbiont at night (Fig. 4). Further characterization of the source of reactive-oxygen species in the crypts promises to inform our understanding of the mechanisms by which the host selects for nocturnal bioluminescence, and also identify additional rhythmic attributes of host-tissue chemistry.
4.4 What does the squid-vibrio symbiosis reveal about biological rhythms and mutualism?
Biological rhythms are a ubiquitous characteristic of animals. Underlying these rhythms are the light-entrained oscillation of circadian clocks, as well as multiple light-cued behaviors. Increasingly, light-independent physiological inputs, such as the cellular redox state [20], and microbiota-dependent immune rhythms [50], have been tied to the regulation of the circadian clock. It remains to be seen how many of the mechanisms emerging to describe the microbe-dependent maintenance of tissue homeostasis, such as the modulation of gut-associated macrophage function by n-butyrate [8], or the modulation of dendritic-cell antigen presentation by the skin microbiota [51], are coordinated by diel or circadian biological rhythms.
5. Concluding remarks
When interpreted as a negotiation between host and symbiont, the squid-vibrio mutualism reveals a chemical language that is selective, dynamic, and rhythmic. From the host’s perspective, the squid has demonstrated chemical strategies to select for the acquisition and maintenance of a beneficial microbe that are conserved throughout the metazoans, such as the immune-dependent structuring of the environment by antimicrobial peptides, NO, and host-associated glycans. Future studies of immune-system signaling in the squid could reveal novel strategies evolved by animals to maintain tissue homeostasis and prevent inflammation via induction of these conserved chemical responses. From the perspective of the microbe, V. fischeri provides an example of how regulation of metabolic functions, in addition to MAMP signaling, tailors tissue chemistry and immune function to support a highly specific and stable colonization. For example, the regulation of CS catabolism is central to the acquisition, development and maintenance of bioluminescent V. fischeri in the squid light organ. As studies of gut tract and other microbiota shift from the descriptive to the mechanistic, application of a broad perspective, encompassing studies of invertebrate model systems such as the squid, will help decipher the reciprocal and interconnected nature of symbiont stability and host tissue homeostasis.
Acknowledgments
The authors thank Margaret McFall-Ngai for helpful discussions. J.A.S. received support from the National Institutes of Health, National Research Service Award AI55397. The work was also funded by NIH grants RR12294/OD11024 and GM099507 to EGR and M. McFall-Ngai, and AI050661 to M. McFall-Ngai and EGR
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Altura MA, Heath-Heckman EA, Gillette A, Kremer N, Krachler AM, Brennan C, et al. The first engagement of partners in the Euprymna scolopes–Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol. 2013;15:2937–50. doi: 10.1111/1462-2920.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ. Attenuation of host NO production by MAMPs potentiates development of the host in the squid–vibrio symbiosis. Cell Microbiol. 2011;13:527–37. doi: 10.1111/j.1462-5822.2010.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bass J, Takahashi JS. Circadian integration of metabolism and energetics. Science. 2010;330:1349–54. doi: 10.1126/science.1195027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boettcher K, Ruby E, McFall-Ngai M. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol. 1996;179:65–73. [Google Scholar]
- 5.Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172:3701–6. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boettcher KJ, Ruby EG. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J Bacteriol. 1995;177:1053–8. doi: 10.1128/jb.177.4.1053-1058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan CA, Hunt JR, Kremer N, Krasity BC, Apicella MA, McFall-Ngai MJ, et al. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. eLife. 2014;3:e01579. doi: 10.7554/eLife.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111:2247–52. doi: 10.1073/pnas.1322269111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chovatiya R, Medzhitov R. Stress, inflammation, and defense of homeostasis. Mol Cell. 2014;54:281–8. doi: 10.1016/j.molcel.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, Snir E, et al. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A. 2008;105:11323–8. doi: 10.1073/pnas.0802369105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Claes MF, Dunlap PV. Aposymbiotic culture of the sepiolid squid Euprymna scolopes: role of the symbiotic bacterium Vibrio fischeri in host animal growth, development, and light organ morphogenesis. J Exp Zool. 2000;286:280–96. [PubMed] [Google Scholar]
- 12.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–62. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–21. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cullen T, Schofield W, Barry N, Putnam E, Rundell E, Trent M, et al. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science. 2015;347:170–5. doi: 10.1126/science.1260580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol. 2004;6:1139–51. doi: 10.1111/j.1462-5822.2004.00429.x. [DOI] [PubMed] [Google Scholar]
- 16.Doino JA, McFall-Ngai M. A transient exposure to symbiosis-competent bacteria induces light organ morphogenesis in the host squid. Biol Bull. 1995:347–55. doi: 10.2307/1542152. [DOI] [PubMed] [Google Scholar]
- 17.Doino Lemus J, McFall-Ngai MJ. Alterations in the proteome of the Euprymna scolopes light organ in response to symbiotic Vibrio fischeri. Appl Environ Microbiol. 2000;66:4091–7. doi: 10.1128/aem.66.9.4091-4097.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn AK. Vibrio fischeri metabolism: Symbiosis and beyond. Adv Microb Physiol. 2012;61:37. doi: 10.1016/B978-0-12-394423-8.00002-0. [DOI] [PubMed] [Google Scholar]
- 19.Dunn AK, Karr EA, Wang Y, Batton AR, Ruby EG, Stabb EV. The alternative oxidase (AOX) gene in Vibrio fischeri is controlled by NsrR and upregulated in response to nitric oxide. Mol Microbiol. 2010;77:44–55. doi: 10.1111/j.1365-2958.2010.07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–64. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erkosar B, Storelli G, Defaye A, Leulier F. Host-intestinal microbiota mutualism: ‘learning on the fly’. Cell Host Microbe. 2013;13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Fraune S, Bosch TC. Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc Natl Acad Sci U S A. 2007;104:13146–51. doi: 10.1073/pnas.0703375104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert SF. A holobiont birth narrative: the epigenetic transmission of the human microbiome. Front Genet. 2014;5:282. doi: 10.3389/fgene.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles DK, Hankins JV, Guan Z, Trent MS. Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol Microbiol. 2011;79:716–28. doi: 10.1111/j.1365-2958.2010.07476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graf J, Ruby EG. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A. 1998;95:1818–22. doi: 10.1073/pnas.95.4.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal- ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–79. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 27.Hastings J, Balny C. The oxygenated bacterial luciferase-flavin intermediate. Reaction products via the light and dark pathways. J Biol Chem. 1975;250:7288–93. [PubMed] [Google Scholar]
- 28.Heath-Heckman EA, McFall-Ngai MJ. The occurrence of chitin in the hemocytes of invertebrates. Zoology. 2011;114:191–8. doi: 10.1016/j.zool.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. mBio. 2013;4:e00167–13. doi: 10.1128/mBio.00167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heath-Heckman EA, Gillette AA, Augustin R, Gillette MX, Goldman WE, McFall-Ngai MJ. Shaping the microenvironment: evidence for the influence of a host galaxin on symbiont acquisition and maintenance in the squid-vibrio symbiosis. Environ Microbiol. 2014;16:3669–82. doi: 10.1111/1462-2920.12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoyle NP, O’Neill JS. Oxidation-reduction cycles of peroxiredoxin proteins and non-transcriptional aspects of timekeeping. Biochemistry. 2015;54:184–93. doi: 10.1021/bi5008386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janeway CA. Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor Laboratory Press; 1989. Approaching the asymptote? Evolution and revolution in immunology; pp. 1–13. [DOI] [PubMed] [Google Scholar]
- 33.Koch EJ, Miyashiro TI, McFall-Ngai MJ, Ruby EG. Features governing symbiont persistence in the squid-vibrio association. Mol Ecol. 2013;23:1624–34. doi: 10.1111/mec.12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koropatnick T, Goodson MS, Heath-Heckman EA, McFall-Ngai M. Identifying the cellular mechanisms of symbiont-induced epithelial morphogenesis in the squid-vibrio association. Biol Bull. 2014;226:56–68. doi: 10.1086/BBLv226n1p56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–8. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- 36.Koropatnick TA, Kimbell JR, McFall-Ngai MJ. Responses of host hemocytes during the initiation of the squid-vibrio symbiosis. Biol Bull. 2007;212:29–39. doi: 10.2307/25066578. [DOI] [PubMed] [Google Scholar]
- 37.Kremer N, Philipp EE, Carpentier M-C, Brennan CA, Kraemer L, Altura MA, et al. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe. 2013;14:183–94. doi: 10.1016/j.chom.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kremer N, Schwartzman J, Augustin R, Zhou L, Ruby EG, Hourdez S, et al. The dual nature of haemocyanin in the establishment and persistence of the squid–vibrio symbiosis. Proc Biol Sci. 2014:281. doi: 10.1098/rspb.2014.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamarcq LH, McFall-Ngai MJ. Induction of a gradual, reversible morphogenesis of its host’s epithelial brush border by Vibrio fischeri. Infect Immun. 1998;66:777–85. doi: 10.1128/iai.66.2.777-785.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee KH, Ruby EG. Effect of the squid host on the abundance and distribution of symbiotic Vibrio fischeri in nature. Appl Environ Microbiol. 1994;60:1565–71. doi: 10.1128/aem.60.5.1565-1571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, DeLoney-Marino CR, McFall-Ngai MJ, et al. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol. 2012;78:4620–6. doi: 10.1128/AEM.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. A single regulatory gene is sufficient to alter bacterial host range. Nature. 2009;458:215–8. doi: 10.1038/nature07660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 2013;110:3229–36. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. The secret languages of coevolved symbioses: Insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol. 2012;24:3–8. doi: 10.1016/j.smim.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McFall-Ngai M, Nyholm SV, Castillo MG. Semin Immunol. Elsevier; 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes–Vibrio fischeri symbiosis; pp. 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McFall-Ngai MJ. The importance of microbes in animal development: Lessons from the squid-vibrio symbiosis. Annu Rev Microbiol. 2014:68. doi: 10.1146/annurev-micro-091313-103654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyashiro T, Klein W, Oehlert D, Cao X, Schwartzman J, Ruby EG. The N-acetyl-d-glucosamine repressor NagC of Vibrio fischeri facilitates colonization of Euprymna scolopes. Mol Microbiol. 2011;82:894–903. doi: 10.1111/j.1365-2958.2011.07858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miyashiro T, Ruby EG. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014 doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 50.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–27. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 51.Naik S, Bouladoux N, Linehan JL, Han S-J, Harrison OJ, Wilhelm C, et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–8. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol. 2002;68:5113–22. doi: 10.1128/AEM.68.10.5113-5122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nyholm SV, Graf J. Knowing your friends: invertebrate innate immunity fosters beneficial bacterial symbioses. Nat Rev Microbiol. 2012;10:815–27. doi: 10.1038/nrmicro2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nyholm SV, McFall-Ngai MJ. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull. 1998;195:89–97. doi: 10.2307/1542815. [DOI] [PubMed] [Google Scholar]
- 55.Nyholm SV, McFall-Ngai MJ. Dominance of Vibrio fischeri in secreted mucus outside the light organ of Euprymna scolopes: the first site of symbiont specificity. Appl Environ Microbiol. 2003;69:3932–7. doi: 10.1128/AEM.69.7.3932-3937.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–42. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 57.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol. 2009;11:483–93. doi: 10.1111/j.1462-2920.2008.01788.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peyer SM, Pankey MS, Oakley TH, McFall-Ngai MJ. Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mech Develop. 2014;131:111–26. doi: 10.1016/j.mod.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pride AC, Herrera CM, Guan Z, Giles DK, Trent MS. The outer surface lipoprotein VolA mediates utilization of exogenous lipids by Vibrio cholerae. mBio. 2013;4:e00305–13. doi: 10.1128/mBio.00305-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Provenzano D, Klose KE. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci U S A. 2000;97:10220–4. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rader BA, Kremer N, Apicella MA, Goldman WE, McFall-Ngai MJ. Modulation of symbiont lipid A signaling by host alkaline phosphatases in the squid-vibrio symbiosis. mBio. 2012;3:e00093–12. doi: 10.1128/mBio.00093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rader BA, Nyholm SV. Host/microbe interactions revealed through “omics” in the symbiosis between the Hawaiian bobtail squid Euprymna scolopes and the bioluminescent bacterium Vibrio fischeri. Biol Bull. 2012;223:103–11. doi: 10.1086/BBLv223n1p103. [DOI] [PubMed] [Google Scholar]
- 63.Ruby EG, Asato LM. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol. 1993;159:160–7. doi: 10.1007/BF00250277. [DOI] [PubMed] [Google Scholar]
- 64.Saito HE, Harp JR, Fozo EM. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol. 2014;80:6527–38. doi: 10.1128/AEM.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schleicher TR, Nyholm SV. Characterizing the host and symbiont proteomes in the association between the bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS One. 2011;6:e25649. doi: 10.1371/journal.pone.0025649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schleicher TR, VerBerkmoes NC, Shah M, Nyholm SV. Colonization state influences the hemocyte proteome in a beneficial squid–vibrio symbiosis. Mol Cell Proteomics. 2014;13:2673–86. doi: 10.1074/mcp.M113.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuster BM, Perry LA, Cooper VS, Whistler CA. Breaking the language barrier: experimental evolution of non-native Vibrio fischeri in squid tailors luminescence to the host. Symbiosis. 2010;51:85–96. [Google Scholar]
- 68.Schwartzman JA, Koch E, Heath-Heckman EA, Zhou L, Kremer N, McFall-Ngai MJ, et al. The chemistry of negotiation: Rhythmic, glycan-driven acidification in a symbiotic conversation. Proc Natl Acad Sci U S A. 2015;112:566–71. doi: 10.1073/pnas.1418580112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK. The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ Microbiol. 2011;13:2855–64. doi: 10.1111/j.1462-2920.2011.02558.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Singley C. Euprymna scolopes. Cephalopod life cycles: species accounts. 1983;1:69. [Google Scholar]
- 71.Small AL, McFall-Ngai MJ. Halide peroxidase in tissues that interact with bacteria in the host squid Euprymna scolopes. J Cell Biochem. 1999;72:445–57. [PubMed] [Google Scholar]
- 72.Studer SV, Schwartzman JA, Ho JS, Geske GD, Blackwell HE, Ruby EG. Non-native acylated homoserine lactones reveal that LuxIR quorum sensing promotes symbiont stability. Environ Microbiol. 2013;16:2623–3. doi: 10.1111/1462-2920.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sycuro LK, Ruby EG, McFall-Ngai M. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol. 2006;267:555–68. doi: 10.1002/jmor.10422. [DOI] [PubMed] [Google Scholar]
- 74.Troll JV, Bent EH, Pacquette N, Wier AM, Goldman WE, Silverman N, et al. Taming the symbiont for coexistence: a host PGRP neutralizes a bacterial symbiont toxin. Environ Microbiol. 2010;12:2190–203. doi: 10.1111/j.1462-2920.2009.02121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Visick KL. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol. 2009;74:782–9. doi: 10.1111/j.1365-2958.2009.06899.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol. 2000;182:4578–86. doi: 10.1128/jb.182.16.4578-4586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Visick KL, Ruby EG. The periplasmic, group III catalase of Vibrio fischeri is required for normal symbiotic competence and is induced both by oxidative stress and by approach to stationary phase. J Bacteriol. 1998;180:2087–92. doi: 10.1128/jb.180.8.2087-2092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Dufour YS, Carlson HK, Donohue TJ, Marletta MA, Ruby EG. H-NOX–mediated nitric oxide sensing modulates symbiotic colonization by Vibrio fischeri. Proc Natl Acad Sci U S A. 2010;107:8375–80. doi: 10.1073/pnas.1003571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid–vibrio symbiosis. Mol Microbiol. 2010;78:903–15. doi: 10.1111/j.1365-2958.2010.07376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weis VM, Small AL, McFall-Ngai MJ. A peroxidase related to the mammalian antimicrobial protein myeloperoxidase in the Euprymna-Vibrio mutualism. Proc Natl Acad Sci U S A. 1996;93:13683–8. doi: 10.1073/pnas.93.24.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiassé PR, Schaefer AL, Koroleva I, et al. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A. 2010;107:2259–64. doi: 10.1073/pnas.0909712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wollenberg MS, Ruby EG. Phylogeny and fitness of Vibrio fischeri from the light organs of Euprymna scolopes in two Oahu, Hawaii populations. ISME. 2012;6:352–62. doi: 10.1038/ismej.2011.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol. 2006;62:1586–600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]