Abstract
Pathogens and the diseases they cause have been among the most important selective forces experienced by humans during their evolutionary history. Although adaptive alleles generally arise by mutation, introgression can also be a valuable source of beneficial alleles. Archaic humans, who lived in Europe and Western Asia for more than 200,000 years, were probably well adapted to this environment and its local pathogens. It is therefore conceivable that modern humans entering Europe and Western Asia who admixed with them obtained a substantial immune advantage from the introgression of archaic alleles. Here we document a cluster of three Toll-like receptors (TLR6-TLR1-TLR10) in modern humans that carries three distinct archaic haplotypes, indicating repeated introgression from archaic humans. Two of these haplotypes are most similar to the Neandertal genome, and the third haplotype is most similar to the Denisovan genome. The Toll-like receptors are key components of innate immunity and provide an important first line of immune defense against bacteria, fungi, and parasites. The unusually high allele frequencies and unexpected levels of population differentiation indicate that there has been local positive selection on multiple haplotypes at this locus. We show that the introgressed alleles have clear functional effects in modern humans; archaic-like alleles underlie differences in the expression of the TLR genes and are associated with reduced microbial resistance and increased allergic disease in large cohorts. This provides strong evidence for recurrent adaptive introgression at the TLR6-TLR1-TLR10 locus, resulting in differences in disease phenotypes in modern humans.
Introduction
Modern humans dispersing out of Africa were confronted with new environmental challenges including novel foods, pathogens, and a different climate. They also encountered other human forms, and there is accumulating evidence that interbreeding with Neandertals and Denisovans contributed alleles to the modern human gene pool.1, 2, 3, 4 Two recent studies have provided genome-wide maps of archaic haplotypes in present-day people that are likely to have been introduced by introgression from Neandertals.5, 6 The discovery of introgression from now-extinct human forms into modern humans entering Eurasia raises the possibility that the arriving modern humans might have benefitted from the introduction of alleles that existed in archaic humans who were well adapted to the environment.1, 2, 3, 7 A few cases of such adaptive introgression have been described, usually involving genes that influence systems interacting directly with the environment. For example, the introgression of a Denisovan haplotype in EPAS1 (MIM: 603349) has recently been shown to confer altitude adaptation in Tibetans,8 and single-locus studies have identified adaptive introgression in genes involved in immunity and metabolism including genes of the major histocompatibility locus (MHC), SLC16A11 (MIM: 615765), OAS1 (MIM: 164350), and STAT2 (MIM: 600556).9, 10, 11, 12
Three of the ten human toll-like receptors (TLR10 [MIM: 606270], TLR1 [MIM: 601194], and TLR6 [MIM: 605403]) encoded in a cluster on chromosome 4 are among the top 1% of genes with the highest Neandertal introgression (Table S1). This TLR cluster is particularly interesting because of its high introgression probability reflecting the appreciable length and frequency of the Neandertal-like haplotype, and because of the critical role that these three TLRs play innate immunity. The innate immune system provides a first line of defense against pathogens and is involved in the early detection of micro-organisms as well as in the activation of the adaptive immune response.13, 14 In humans, TLR1, TLR6, and TLR10 occur on the cell surface and are known to detect bacterial, fungal, and parasite components including flagellin and glycolipids. They are essential for eliciting the inflammatory and anti-microbial responses as well as for activating an adaptive immune response.13 In agreement with their crucial role, TLRs have been shown to be highly conserved, although cell-surface TLRs are less strictly constrained than intracellular TLRs.15 In fact, the region encompassing the TLR6-TLR1-TLR10 genes has been shown to have some signatures of recent positive selection in certain non-African populations,15, 16 suggesting that they might be involved in local immune-system adaptation. However, the presence of introgressed archaic haplotypes confounds the signatures of recent positive selection17 and can lead to erroneous inference of adaptive evolution. We therefore explored in detail both the evidence for introgression and the evidence for natural selection at the TLR6-TLR1-TLR10 cluster.
Material and Methods
Identifying the Introgressed Region in Present Day Human Genomes
To identify potentially introgressed archaic-like haplotypes in present-day human genomes, we used the genome-wide Neandertal introgression maps from Sankararaman et al.5 and Vernot et al.6 The introgression map presented by Sankararaman et al. provides the probability that SNPs at polymorphic positions in modern humans arose on the Neandertal lineage and introgressed into modern humans. Vernot et al. used the S∗ statistic18 to detect putatively introgressed regions in modern humans and compared the candidate regions to the reference Neandertal genome to define introgressed Neandertal blocks in modern humans.
To delineate the introgressed region of interest, we used the per-SNP introgression probabilities from Sankararaman et al.5 for all Asian and all European individuals. In the region encompassing the three TLR genes and an additional region 50 kb up- and downstream (chromosome 4: 38,723,860–38,908,438; Figure 1), we computed the difference between the Neandertal probabilities for pairs of neighboring SNPs relative to the distance between them. We then defined the range of our preliminary introgressed region based on the SNPs with the largest difference in Neandertal probabilities in the introgression maps for both Asians and Europeans. The largest increase was observed at position chr4: 38,757,064 in both the European and Asian maps. The largest decrease in probabilities differed between the European and Asian maps. We used the Asian map (SNP at position chr4: 38,907,701) because it yielded the longer potentially introgressed region. The maximum difference in the European map is at chr4: 38,821,916.
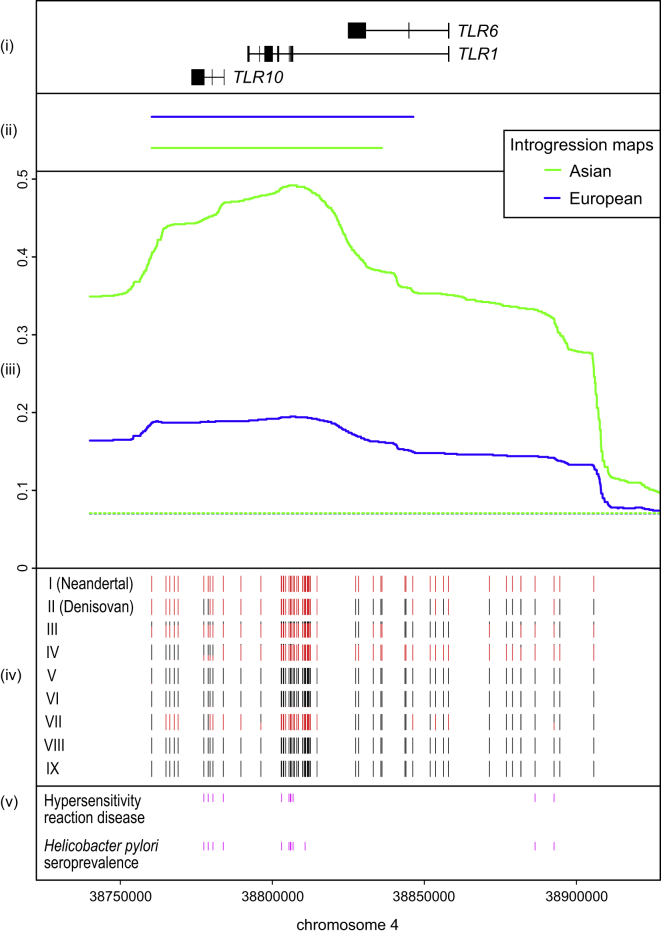
Figure 1.
The Introgressed Region Encompassing the Genes TLR10, TLR1, and TLR6 on Chromosome 4
The genetic length of the region (chr4: 38,760,338–38,905,731; hg19) based on three recombination maps ranges from 0.04 cM to 0.1 cM (HapMap, 0.1 cM; deCode, 0.04 cM; and Hinch African American map, 0.08 cM).
(i) The gene structures for TLR10, TLR1, and TLR6 are displayed.
(ii) Predicted Neandertal haplotypes from the Neandertal ancestry map by Vernot et al.6 for Asians (green) and Europeans (blue).
(iii) Neandertal introgression posterior probabilities across polymorphic positions for Asians (blue solid lines) and Europeans (green solid lines) from the Neandertal ancestry map by Sankararaman et al.5 The chromosomal average introgression posterior probabilities are given as green and blue dashed lines.
(iv) Sharing of the Neandertal allele across archaic-like SNPs within seven core haplotypes: the Neandertal and Denisovan sequences and the seven modern human core haplotypes. The frequencies of Neandertal and Yoruba alleles for the archaic-like SNPs within each core haplotype are colored in red and black, respectively.
(v) Significantly associated SNPs with Helicobacter seroprevalence or allergic disease overlapping archaic-like SNPs are displayed.19, 20
Within this region we defined potentially archaic-like SNPs as those where the Neandertal or Denisovan genomes differ from the 109 Yoruba individuals in the 1000 Genomes dataset.21 We refined the putatively introgressed region by selecting the archaic-like SNPs that are within the preliminary region and closest to the borders of this region. The final putatively introgressed region covers 143 kb of chromosome 4 (chr4: 38,760,338–38,905,731; Figure 1) and contains 61 archaic-like SNPs. This region overlaps two haplotypes identified by Vernot et al.6—one identified in Europeans and the other in Asians.
To further explore the extent of the introgressed haplotype, we computed linkage disequilibrium (LD) between the 61 archaic-like SNPs that define the introgressed region and the three nearest neighboring archaic-like SNPs up- and downstream of the defined 143 kb region. We included in the analysis all 1000 Genomes individuals22 that carried at least one archaic-like allele for at least one of the 61 archaic-like SNPs within the TLR region (Figure S1).
Introgression Scores for Each Gene
We calculated introgression summary scores for each of the 18,017 CCDS genes by averaging the posterior probabilities provided by Sankararaman et al.5 for all sites within each CCDS gene in each 1000 Genomes individual, and then calculating an average for all Europeans (EUR), all Asians (ASN), and the combined set of these two populations (EUR-ASN). Genes were ranked according to their averaged score (Table S1).
Haplotype Network Analysis
To explore the relationships between haplotypes in this locus, we constructed a haplotype network based on the haplotypes of all 1000 Genomes individuals (phase III21) and the Altai Neandertal and Denisovan genome sequences (Figure S2)23. We used all 1,997 SNPs that were observed in more than one chromosome in the 1,000 Genomes dataset21 and where the Neandertal and the Denisovan were homozygous. We excluded 11 SNPs where either the Neandertal or the Denisovan were heterozygous. We merged the resulting 2,609 unique haplotypes into core haplotypes that differed by <150 nucleotides (i.e., ∼1/1,000 base pairs in the region was allowed to differ between two chromosomes). This resulted in a set of seven modern human core haplotypes (Figures 1, 2, and S2, Table S2) and two archaic haplotypes (that are simply the Neandertal and Denisovan genome sequences). We generated a consensus sequence for each modern human core haplotype by computing the majority allele at each position.
Figure 2.
Divergence of the Identified Core Haplotypes at TLR10, TLR1, and TLR6
(A) Neighbor-joining tree on the inferred sequences of seven modern human core haplotypes (III–IX), and the Neandertal (I), Denisovan (II), orangutan, and chimpanzee genome sequences. Bootstrap values (1,000 replicates) for the topology are provided in blue squares at each node. The pie charts show the frequency in the four continental population groups of each modern human core haplotype. Next to each pie chart, the frequency of haplotypes among 1000 Genomes individuals assigned to the corresponding core haplotype is displayed. For core haplotype-defining positions in each branch, see Table S2.
(B) Pair-wise average nucleotide distance between haplotypes in core haplotypes.
Neighbor-Joining Tree
We computed a neighbor-joining tree for the entire 143 kb region based on the nucleotide distance between the sequences of the seven modern human core haplotypes together with the Neandertal, Denisovan, chimpanzee, and orangutan sequences using the R package ape.24 We obtained the orthologous sequences for chimpanzee and orangutan by using the liftover software.25 Branch support was obtained from 1,000 bootstraps (Figure 2A).
Diversity in Populations and Core Haplotypes
We computed the mean pair-wise difference between all haplotype pairs within each core haplotype and for all populations and combinations of populations (Figure S3, Table S3).
Recombination Rates
We obtained the average recombination rate from three maps (sex-averaged: deCode 1.5 cM/Mb, Marshfield 2.4 cM/Mb, Genethon 2.1 cM/Mb26, 27, 28) for the TLR region from UCSC.25 There is considerable variation in estimated recombination rates across the region. Two of the maps28, 29 show a 5- to 12-fold higher recombination rate than the chromosome average in the region overlapping TLR10 (Figures S5A and S5B). A third map30 based on African genomes did not show as extreme an increase in recombination rate (Figure S5C), which might be partially due to the absence of the archaic-like core haplotypes in individuals of African origin.
The Probability of a 143 kb Introgressed Segment due to Incomplete Lineage Sorting
Recombination reduces the length of introgressed haplotypes over time, and introgression events are quite recent when compared with the divergence between modern humans and Neandertals. Introgressed segments are therefore on average longer than those that arise due to incomplete lineage sorting (ILS). We calculated the probability that a haplotype of 143 kb in length is not broken by recombination since the common ancestor of modern humans and Neandertals and/or Denisovans (i.e., is due to ILS), according to the approach described in Huerta-Sánchez et al.8 In brief, we model the expected length of segments of ILS given the local recombination rate, time since the divergence from the common ancestor, and generation time, and compare this to the length of the observed introgressed haplotype.
The variable parameters used are provided in Table S4A. For all calculations we used a generation time (g) of 25 years and the lowest average recombination rate estimate for the TLR region from the available maps (r = 1.5 × 10−8) because this yields longer haplotypes and is therefore most conservative.
Estimates of branch lengths since divergence times (in years) are taken from Prüfer et al.3 and are already corrected for fossil age. To ensure that our estimates are as conservative as possible, we always use the lowest possible branch lengths (in years) from the estimated ranges given in Prüfer et al.3 because these allow the least time for recombination.
We computed the expected length L for two recent estimates of the human mutation rate; μ = 1 × 10−9 and μ = 0.5 × 10−9 per base pair per year: L = 1/(r × (t1+t2)/g) and the probability of a length of at least 143 kb as 1 − GammaCDF(143000, shape = 2, rate = 1/L) given these different parameters; with GammaCDF being the gamma distribution function. The expected lengths of haplotypes resulting from ILS and the probabilities of observing a haplotype of 143 kb are shown in Table S4B.
FST Analysis
In order to quantify allele frequency differences between populations, we computed pairwise FST for all genome-wide SNPs (1) between populations from different continents (Africa, Asia, Europe) and (2) from populations within a continent (Asia and Europe).
FST values were computed using vcftools based on the Weir and Cockerham calculation.31 For (1) we included 100 random unrelated individuals (1000 Genomes phase III) representing a sample from each continent. For (2) we used 50 unrelated individuals per population. We compared the FST values for two different sets of SNPs: first, the 61 archaic-like SNPs that were present in any of the three archaic-like core haplotypes (III, IV, VII) in the region (Table S5) and second, the four tag SNPs presented in Barreiro et al.15 and Laayouni et al.16 (Table S6).
To determine whether population differentiation deviates from neutral expectations, the FST value of each SNP was compared to the empirical distributions of FST values of (1) all genome-wide SNPs, (2) all genome-wide archaic-like SNPs (SNPs present in one or more non-Africans that match the Neandertal or Denisovan genome sequences), and (3) all genome-wide archaic-like SNPs exclusively found in Asian individuals.
We repeated the FST analyses using all 155 SNPs differentiating the core haplotypes in the region and confirmed the results using solely the archaic-like SNPs.
To correct for multiple testing, we computed the false discovery rate for all comparisons using the Benjamini-Hochberg procedure.
SNPs with Evidence for Positive Selection Presented Previously
Because previous studies on this region did not take into account the (then unknown) archaic origin of some haplotypes, we re-evaluated the signatures of positive selection for haplotype-defining SNPs reported by Barreiro et al.15 and Laayouni et al.16 and their relationship to the core haplotypes we identified. Frequency information for SNPs presented in Barreiro et al. and Laayouni et al. were computed using all individuals of the 1000 Genomes dataset (phase III) and are shown in Tables S6B–S6D.
Expression Analysis in Multiple Tissues from the GTEx Dataset
To determine whether the archaic haplotypes have any effect on gene regulation, we analyzed expression data from 52 tissues provided by the GTEx.32 Between 4 and 210 individuals were available for each tissue. We identified nine shared archaic-like SNPs (chr4: 38,777,471; 38,778,903; 38,783,848; 38,802,913; 38,803,063; 38,805,983; 38,806,019; 38,806,096; 38,806,827) from the Illumina OMNI 5M SNP Array32 used to genotype each individual, and we used these to tag the archaic-like core haplotypes. All nine SNPs are in perfect linkage with one another.
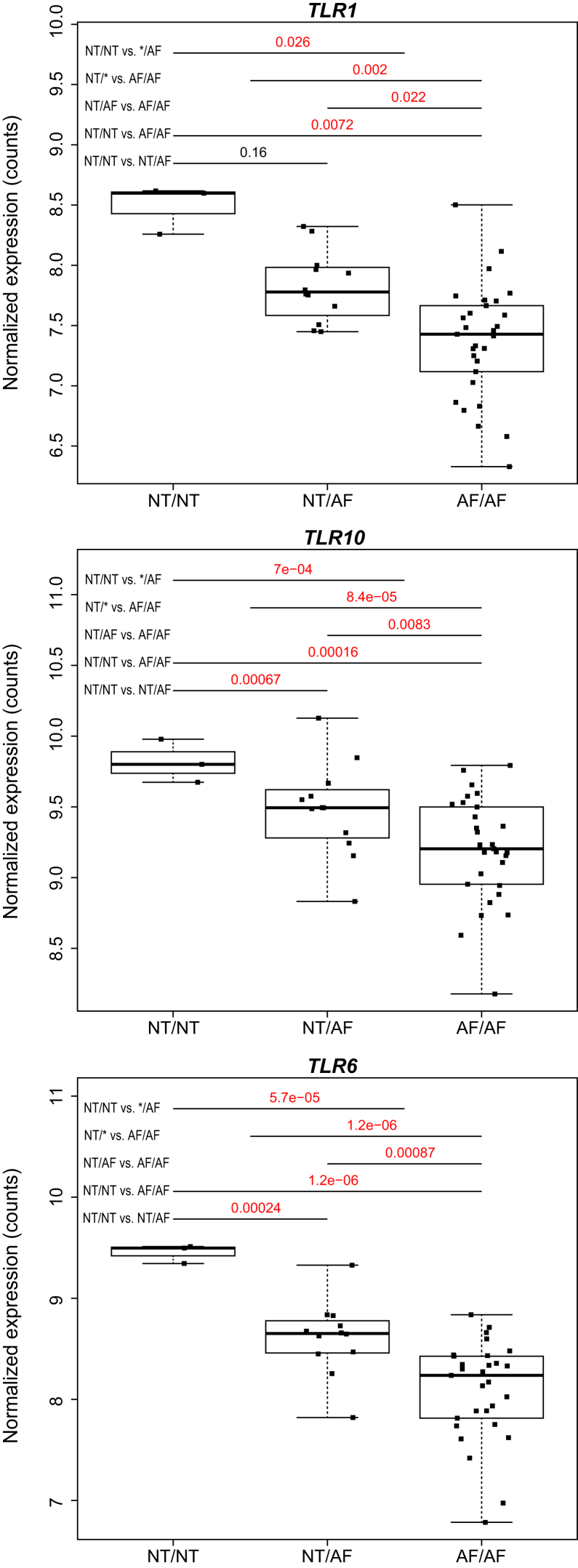
In each tissue, we used the DESeq2 package to test for allele-specific differential expression of TLR1, TLR6, and TLR10 between those individuals with and without archaic-like core haplotypes.33 The expression (in normalized counts) of each of the three TLR genes and the corresponding differential expression p values are shown in Figure 3 (additional information in Figures S4 and S7 and Table S7).
Figure 3.
Genotype-Dependent Expression of the TLR Genes
The normalized expression in EBV-transformed lymphocytes (individuals in black cubes and entire distribution shown as boxplot; y axis) across GTEx individuals with different genotypes at shared archaic-like SNP positions (x axis: NT, Neandertal allele; AF, African allele) for each TLR gene. Differential expression p values between genotypes are displayed in the upper part of each plot. Median and quartiles are shown and whiskers show the range of expression values.
Expression Analysis using a Lymphoblastoid Cell Line Dataset
We also analyzed gene expression using data from lymphoblastoid cell lines from 421 of the Yoruban and European individuals from the 1000 Genomes cohort.34
We tested for allele-specific expression differential expression using the 32 archaic-like alleles that are shared by the archaic-like core haplotypes (III, IV, VII) and not seen in the other modern-human core haplotypes. The archaic-like SNPs private to core haplotypes IV and VII were not polymorphic in the genomes of any of the individuals, probably because of their lower frequency and the absence of Asian individuals in this dataset. Of the 32 archaic-like SNPs that are shared by the archaic-like core haplotypes (III, IV, VII) and not seen in the other modern-human core haplotypes, 22 were variable in the 1000 Genomes individuals for whom expression data was available. We extracted high-quality mapped reads (MQ > 30) from the Geuvadis RNA sequencing project. For each individual we evaluated expression of TLR10, TLR1, and TLR6 by assigning to each gene those reads with overlapping map coordinates. We normalized the number of reads per individual with the DESeq package35 and computed differential expression between the individuals carrying the archaic-like allele and all other individuals via a Mann-Whitney U test. The FDR values are shown in Table S7.
Transcription Factor Analysis
We computed the number of transcription factor binding sites (obtained from the ENCODE project36) in 150-kb sliding windows with a step size of 10 kb across chromosome 4. We identified those SNPs that are shared by the archaic-like core haplotypes (III, IV, VII) but different from the other modern human core haplotypes and that overlap ENCODE-defined transcription factor binding sites (Table S8). A total of 49 binding sites for 28 different TFs overlap the archaic-specific SNPs.
With the ontology enrichment software FUNC,37 we tested whether transcription factors with binding sites overlapping archaic-like SNPs show evidence for functional enrichment in the gene ontology compared to 124 other transcription factors tested by the ENCODE consortium.36, 37, 38
Overlap with GWAS Phenotype Association Studies
We identified 79 SNPs that show significant association with any reported phenotype within the 143 kb TLR region using GWASdb39 (Table S9). We selected the two phenotypes (Helicobacter pylori seroprevalence and susceptiblity to allergic disease) for which the associated SNPs show overlap with multiple archaic-like SNPs. A principal-component analysis (PCA) was used to cluster the seven modern human core haplotypes based on allele sharing with these two GWAS phenotypes (Figure 4).
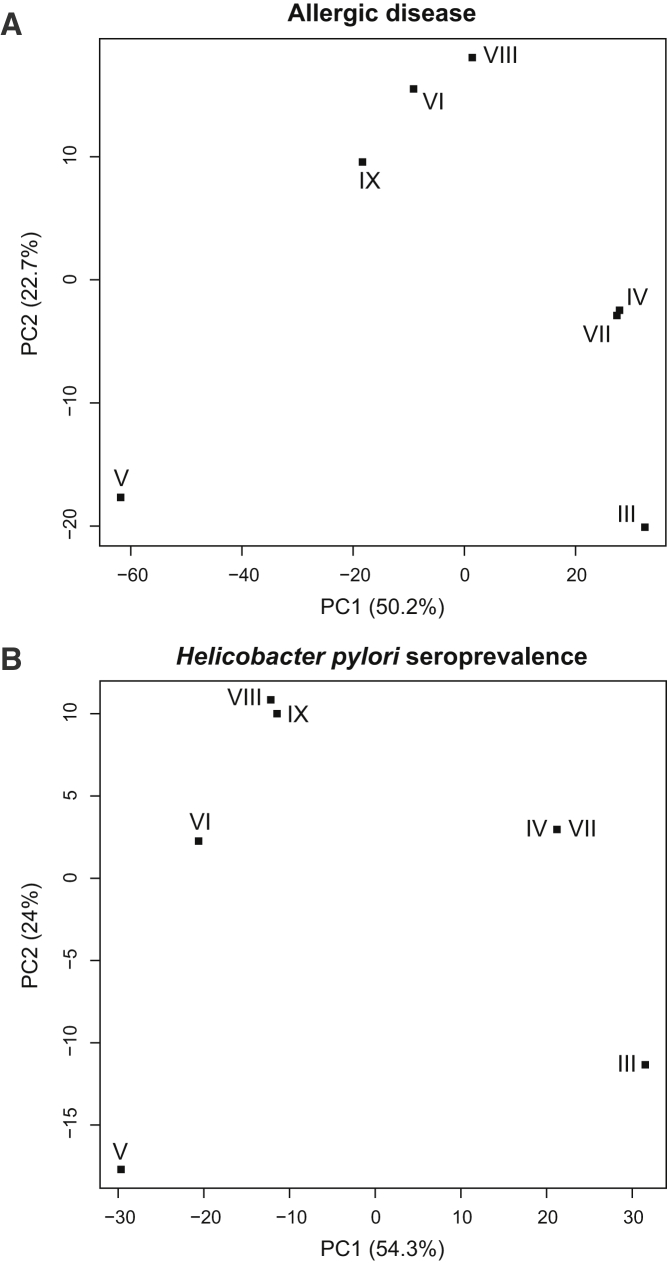
Figure 4.
Clustering of the Seven Core Haplotypes Based on Allele Sharing with GWAS SNPs
Principal-component analysis based on the sequence distance between modern human core haplotypes III–IX for the GWAS SNPs reported to be significantly associated with allergic disease (A)20 and Helicobacter pylori seroprevalence (B).19
Results
Identifying Introgressed Haplotypes
We identified an extended region encompassing the genes TLR10, TLR1, and TLR6 on chromosome 4 in both genome-wide maps of Neandertal introgression5, 6 (Figure 1, Table S1, Material and Methods). Because the two introgression maps did not agree completely on the length of the introgressed region, we used consecutive SNPs with the largest differences in introgression probabilities in the Neandertal introgression map5 to mark the start and end of a putatively introgressed region that is 143 kb in length (hg19; chr4: 38,760,338–38,905,731; Material and Methods). We also observe a significant drop in LD (D′, see Figure S1) between SNPs within the inferred introgressed region and SNPs outside the region, suggesting that the introgressed haplotype common in present-day populations does not extend beyond the defined 143 kb region.
To determine the haplotype structure of this TLR region, we used the inferred 1000 Genomes phase III haplotypes for 2,535 individuals from 26 populations.21 By clustering haplotypes that differ by fewer than 1/1,000 bases, we identified seven distinct core haplotypes in modern humans (Material and Methods, Figure S2, Table S2).
We computed pairwise distances between these seven modern human core haplotypes and the genome sequences of the Neandertal and Denisovan2, 3 and found that three of the modern human core haplotypes (III, IV, and VII) are found almost exclusively in individuals outside Africa (969/971 carriers are non-African) and are more similar to the archaic genome sequences than to any modern human core haplotype (Figures 2A and 2B). In fact, their distances to the archaic genome sequences are smaller than the distances between any pair of modern human core haplotypes. This suggests a recent common ancestor for the archaic haplotypes and modern human haplotypes III, IV, and VII that is younger than the common ancestor of the other pairs of modern human core haplotypes, as expected from archaic introgression.
Strikingly, this analysis also indicates that there are three distinct archaic-like haplotypes at this locus. Of the three putatively introgressed core haplotypes, III and IV are most similar to the Altai Neandertal genome (haplotype I, Figure 2). All 49 Neandertal-like alleles that define core haplotype III are shared with core haplotype IV, which has 12 additional archaic-like alleles. Core haplotype VII is most similar to the Denisovan sequence (haplotype II, Figures 2 and S2). We find 137 individuals homozygous for core haplotype III and two individuals homozygous for core haplotype IV (III: Table S11; IV: HG02388, NA18625), which indicates that these haplotypes were not introduced by errors in phasing. We did not identify any individual that was homozygous for core haplotype VII, probably because of its low frequency.
Evidence for Introgression
An alternative explanation for the persistence of three archaic-like haplotypes is incomplete lineage sorting (ILS). Both introgression and ILS can result in the presence of ancestral alleles in present-day human genomes and in a genomic region where the haplotype phylogeny does not match the expected phylogeny.40 The region described here was identified in two different genome-wide maps of Neandertal introgression via methods that are not sensitive to normal segments of ILS.5, 6 Nevertheless, we explored a number of key signatures that allow us to distinguish ILS from introgression.
First, given the time since divergence between modern and archaic humans, ILS is expected to be restricted to short genomic regions due to the long-term effects of recombination. We calculated the probability that a haplotype of 143 kb in length is not broken by recombination since the common ancestor of modern humans and Neandertals and/or Denisovans according to the approach used by Huerta-Sánchez et al.8 Using conservative estimates for the age of divergence of the Neandertal and Denisovan (Table S4A) we show that a 143-kb archaic-like haplotype is highly unlikely under ILS (p value < 5.6 × 10−14; Table S4B, Material and Methods). An unusually low recombination rate in the region is also not a plausible explanation because the recombination rate across the region has been estimated to between 1.5 and 2.4 cM/Mb by different methods26, 27, 28—a rate that is higher than the genome-wide average of 1.3 cM/Mb.41
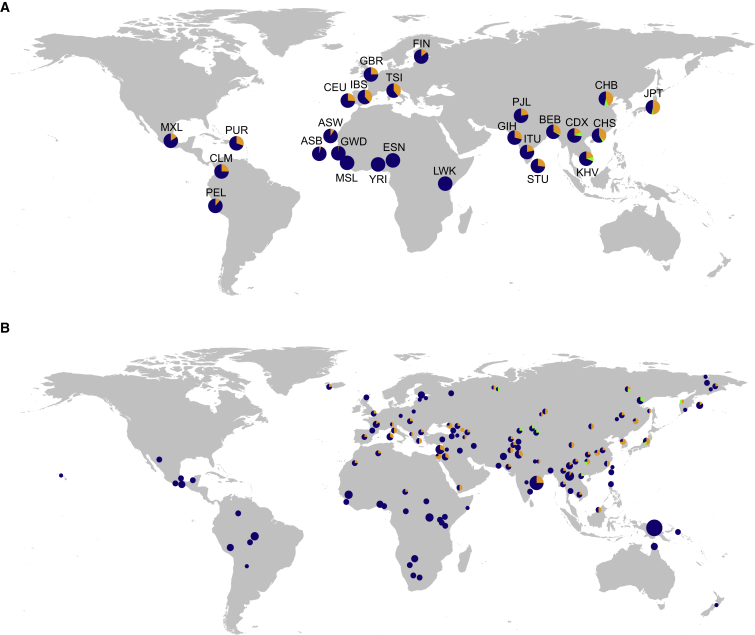
Further, evidence in favor of introgression is provided by the geographic distribution of archaic-like core haplotypes III, IV, and VII. We expect that alleles present due to ILS should be shared across human populations, whereas Neandertal and Denisovan introgressed alleles should be largely absent in Africa. In the 1000 Genomes Phase III data, all three archaic-like haplotypes are largely restricted to populations outside Africa (Figures 2A and 5A; Table S10). Core haplotype III is present in all non-African populations, and we also observe it in two chromosomes from two Northwest Gambian individuals (excluding the potentially admixed African Americans). Northwest African populations have been shown to have experienced recent gene flow from non-Africans42 that might have carried archaic introgressed alleles into these populations.43 This suggests that the presence of this haplotype in these Gambian individuals might be explained by recent back-to-Africa migration. Core haplotype IV is restricted to specific Asian groups, and core haplotype VII is present in only two South Asian individuals (Figures 2A and 5A; Table S10). Haplotypes III and IV are also present among 271 geographically diverse individuals from the Simons Genome Diversity Panel with similar geographic distribution (Figure S5B, Table S12). The geographic distribution of these archaic-like haplotypes is consistent with recent studies that suggest that there might have been at least two pulses of Neandertal introgression: one into the ancestors of all non-Africans, and one into the ancestors of present-day Asians.44, 45 It is also compatible with a low level of Denisovan ancestry in mainland Asia.46 We note that two early modern humans (dated to ∼7,000–8,000 years before present) from Europe (Stuttgart and Loschbour)47 also carry TLR haplotypes similar to Neandertal core haplotype III and an early modern human from Asia (Ust’-Ishim, ∼45,000 years)48 carries a haplotype that is most similar to core haplotype V—the major non-introgressed haplotype in modern humans.
Figure 5.
Geographic Distribution of the Neandertal-like TLR Haplotypes
World map showing the frequencies of Neandertal-like core haplotypes in the 1000 Genomes dataset (A) and the Simons Genome Diversity Panel (B). In (B), the size of each pie is proportional to the number of individuals within a population. Core haplotypes (III, orange; IV, green; non-archaic core haplotypes V, VI, VIII, IX, blue) are colored.
Finally, an expected signature of introgression (but not ILS) is low diversity within the introgressed haplotypes, because they have a recent origin—the introgression event. To assess core haplotype diversity, we calculated mean pairwise differences between all haplotypes within each of the seven core haplotypes (Material and Methods; Figure S3, Table S3). Consistent with an introgressed origin, the three archaic-like core haplotypes (III, IV, and VII) show lower diversity in non-Africans than the most common modern human core haplotype (V). We also note that core haplotype V has high diversity in Africa. This makes it unlikely that core haplotypes III, IV, and VIII were due to ILS and specifically lost in Africans, because this would have reduced the overall diversity at this locus.
A number of independent lines of evidence therefore support the introgression of at least three distinct archaic-like haplotypes at the TLR6-TLR1-TLR10 locus in modern non-African populations.
Evidence for Positive Selection in Modern Humans
The most frequent non-introgressed haplotype (V) is at high frequency in all 1000 Genomes populations (39%–88%), whereas the other non-introgressed haplotypes (VI, VIII, and IX) are present at less than 1% frequency and are predominantly found in African individuals (Figures 2 and 5, Table S10). Among the introgressed haplotypes, Neandertal-like core haplotype III is at intermediate frequency in all non-African populations (11%–51%) and Neandertal-like core haplotype IV is present at a frequency of 2%–10% in Asians. The Denisovan-like core haplotype VII is present in only two Southern Asian individuals (HG03750 and HG0411). The proportion of Neandertal-derived ancestry in non-Africans has been estimated to be between 1.5% and 2.1%, whereas Denisovan ancestry in mainland Asia and Native Americans has been estimated at well below 0.5%.3 The presence of three distinct archaic haplotypes, with two of them at frequencies substantially greater than 2%, is therefore surprising and suggests that the archaic-like alleles might have been advantageous in modern humans.
Signatures of recent population-specific positive selection on the TLR6-TLR1-TLR10 cluster were reported by Barreiro et al.15 and Laayouni et al.16 based on the distribution of allele frequencies (Tajima’s D and Fay and Wu’s H statistics),49, 50 population differentiation (FST),51 allele-specific nucleotide diversity (DIND test),15 and long-range LD (LRH test).52 However, the presence of introgressed haplotypes was not known at that time. Introgression confounds many of these signatures of recent positive selection because introgressed haplotypes show particular patterns of population differentiation and because even under neutrality, introgressed haplotypes affect the patterns of allele-specific diversity and allele-specific long-range linkage disequilibrium. It is therefore necessary to re-evaluate the signatures of selection at this locus while accounting for introgression. To determine whether the frequencies of Neandertal-like core haplotypes III and IV (up 10% and 51% in certain Eurasian populations; Table S10) are higher than expected from drift alone, we measured allele frequency differentiation between pairs of populations using FST,53 for all 61 archaic-like SNPs. To assess significance, we compared these to the genome-wide empirical distribution of FST values for putatively introgressed archaic-like SNPs (Material and Methods, Table S5). The differentiation between African and non-African populations for the SNPs shared by haplotypes III and IV is unusually high when compared to the empirical FST distribution of archaic-like SNPs (p value = 0.01 Africa-Asia and p value = 0.04–0.05 Africa-Europe, Table S5A). Results are similar for core haplotype IV (Table S5A). However, because divergence between African and non-African genomes was used as a factor in identifying the region as having high proportion of Neandertal introgression,5 this high population differentiation is not necessarily unexpected. However, we also observe moderate population differentiation between Asians and Europeans for both core haplotype III (p value = 0.08–0.23) and core haplotype IV (p value = 0.11; Table S5A), which is not explained by the ascertainment bias.
Perhaps most surprisingly, the frequency of the shared SNPs common to Neandertal-like core haplotypes III and IV also varies significantly between populations even within continents. In Europe, a North-South gradient is apparent with significantly high population differentiation between southern Europeans (Toscani and Iberians, with frequencies of 39.3% and 38.3%, respectively) and all other European groups (Finnish, British, and CEPH, with frequencies between 14.8% and 26.4%) (p value < 0.05; Tables S5B and S5C). The frequency of the introgressed haplotypes is higher in the Southern European populations (Figure 5). In Asia, the most Eastern populations (Japanese and Han Chinese, frequency 53.4% and 53.6%) show high differentiation from other Asian populations (frequency 21.7%–41.9%; p value < 0.05; Tables S5D and S5E). In addition, the 12 SNPs defining Asian-specific core haplotype IV show high population differentiation when Dai and Vietnamese (with frequencies 9.4% and 9.9%) are compared to other Asian populations (frequencies between 0% and 4.7%), when using putatively introgressed SNPs present exclusively in Asia as the background (p value < 0.05, Tables S5D and S5E). Overall, the frequency of the three introgressed haplotypes combined ranges between 14.8% and 39.3% in Europeans and between 21.7% and 53.5% in Asian populations, although the contribution of each haplotype (III or IV) varies substantially across Asian groups (Figure 5, Table S10).
Barreiro et al.15 previously reported signatures of positive selection tagged by two SNPs: rs4129009 and rs5743618. rs4129009 is a non-synonymous SNP in TLR10 that showed signatures of positive selection in East Asians. In our data, the putatively selected allele (C) is predominantly found on Neandertal-like core haplotype III (707 out of 741 C carrier haplotypes belong to core haplotype III; Table S6), and the observed signatures of selection in Barreiro et al.15 are therefore most likely explained by positive selection on the archaic core haplotype, as described above. rs5743618 is a non-synonymous SNP whose derived allele (C) reduces TRL1 signaling, resulting in a 60% reduction in the activity of NF-κB, and that in Barreiro et al.15 showed signatures of positive selection in Europeans. In our data, despite significant population differentiation between African and non-African populations (p value < 0.01), the proposed advantageous allele (C) of rs5743618 is present in all human populations and almost exclusively found on non-introgressed core haplotype V (994 out of 1,007 haplotypes with a C, 11 on core haplotype III, and 2 on core haplotype VI). The signatures of natural selection reported for this SNP are thus not due simply to the introgressed haplotypes. In fact, the unusually large population differentiation remains when we investigate the frequency of rs5743618 within only the non-introgressed core haplotype V (p value < 0.01; Table S6). This indicates that the signatures of selection for this SNP are independent of the changes in allele frequency of the introgressed archaic-like core haplotypes (Material and Methods). Additionally, Laayouni et al. reported parallel signatures of very recent adaptive evolution of the TLR6-TLR1-TLR10 cluster in Rroma and Romanian populations, possibly due to adaptation to Yesinia pestis during the plague.16 The SNP with the strongest signatures of positive selection (rs4833103) is intergenic, but three non-synonymous SNPs at the locus (rs4833095, rs11096957, and rs5743810) were shown to influence cytokine production induced by Y. pestis. The derived alleles at these four SNPs are at their highest frequency on non-introgressed core haplotype V (percentage of individuals carrying the derived allele on haplotype V: rs4833103 [A] = 99%; rs5743810 [A] = 72%; rs11096957 [T] = 93%; rs4833095 [T] = 95%) and show high population differentiation between European and non-European populations (Table S6). These alleles are either absent (rs4833103) or at low frequency (rs4833095 and rs11096957) on the archaic-like core haplotypes (III, IV, and VII), indicating that they are not of archaic origin. We note that the signals of selection reported by Barreiro et al.15 and Laayouni et al.16 might not be completely independent. Both groups identified SNP rs4833095, and both report SNPs with similar geographic frequency patterns. The absence of these apparently selected alleles on introgressed haplotypes and the recent age of the inferred selection on some of them16 provides evidence that there has been selection on both archaic- and non-archaic-like haplotypes.
Our results show that the TLR locus shows signatures of recent positive selection even after taking the introgression into account. Strikingly, they also provide evidence for multiple independent rounds of positive selection on both introgressed and non-introgressed haplotypes at the TLR6-TLR1-TLR10 cluster and possibly at quite different times. This region has been proposed to be a hotspot of positive selection in other great apes54, 55 and the presence of several positively selected alleles in modern human populations is therefore perhaps not surprising. However, the fact that several of these alleles have been acquired through adaptive introgression is quite interesting.
Functional Consequences of the Introgressed Alleles
There are 42 SNPs that distinguish all archaic-like core haplotypes from the other modern human core haplotypes. Of these shared archaic-like SNPs, 47% are located upstream of genes, 46% in introns, 2% in 5′ UTRs, 2% downstream of genes, 2% in non-coding exons, and 1% in 3′ UTRs (Material and Methods, Table S2). None of the archaic-like SNPs modifies the amino acid sequence of any TLR gene, but the 143 kb encompassing TLR6-TLR1-TLR10 is rich in regulatory elements, falling in the top 1% of similarly sized windows for transcription factor binding site density on chromosome 436 (Material and Methods, Table S8). In fact, the archaic-like SNPs overlap 49 well-characterized transcription factor binding sites, for 28 different transcription factors, suggesting a putative regulatory impact of the introgressed alleles.
To determine whether the regulatory effect is tissue specific, we obtained expression data for 52 tissues from between 4 and 210 individuals from the GTEx project.32 Using the individual genotype data, we identified four shared archaic-like SNPs that we used to tag the archaic-like core haplotypes. Individuals carrying the archaic-like haplotypes show a significantly higher expression of all three TLR genes in EBV-transformed lymphocytes (p < 0.05, Figure 3), whereas other tissues show differential expression of single TLR genes (Figure S4). Because most tissues do not show an expression change between individuals that is dependent on the archaic-like alleles, it seems likely that the regulatory effect is tissue specific. Using the genome sequences and expression data from lymphoblastoid cell lines from 421 individuals of European and African origin,34 we confirmed that all three TLR genes show significantly higher expression in individuals carrying archaic-like alleles in core haplotype III than in individuals carrying the non-introgressed modern human alleles (Figure S7, Table S7). This is consistent with the observation made in Mayerle et al.19 where increased expression of TLR1 was reported for individuals carrying the alleles that we show here are introgressed.
We further assessed the functional relevance of the archaic-like core haplotypes by using data from genome-wide association studies. A total of 79 SNPs, of which 13 are archaic-like and present on all three archaic haplotypes, are significantly associated with GWAS phenotypes within the 143 kb TLR region.39 Of the 37 SNPs significantly associated with Helicobacter pylori seroprevalence, 13 are archaic-like;19 of the 58 SNPs associated with susceptibility to allergic disease, 12 are archaic-like20 (Figure 1, Table S9). Clustering of the seven core haplotypes based on allele sharing with the GWAS SNPs significantly associated with these two phenotypes revealed high sequence differentiation between all three archaic-like core haplotypes and core haplotype V for both phenotypes (Figure 4, Material and Methods). The largest sequence differentiation is between the archaic-like core haplotype III and all non-archaic-like core haplotypes, which differ in 23 of the 37 SNPs underlying Helicobacter pylori seroprevalence and in 31 of the 58 SNPs associated with allergic disease (Table S9). Interestingly, archaic-like alleles are consistently associated with reduced Helicobacter pylori seroprevalence19 and with increased susceptibility to allergic disease.20
Discussion
Neandertals lived in Europe and Western Asia for more than 200,000 years and were probably well adapted to the environment and local pathogens. It is therefore conceivable that admixture with Neandertals contributed alleles that conferred a substantial immune advantage on modern humans expanding into Europe and Western Asia. The presence of not one but three distinct introgressed haplotypes, whose sequences are closer to the genomes of two different archaic humans, suggests that maintaining these introgressed alleles is likely to have been advantageous. In fact, the high frequencies that these alleles have reached in several human groups are inconsistent with neutral processes since the admixture between Neandertals and modern humans, around 50,000 years ago.56, 57 At least two of these introgressed haplotypes appear to have been advantageous in certain modern human populations.
Previous studies highlighted the contribution of archaic human alleles to the immune systems of modern humans.9, 10, 11, 12 We show here that the TLR genes, which are critical components of the innate immune response, have also acquired advantageous alleles by admixture with archaic humans. These archaic alleles lead to significantly increased expression of TLR6, TLR1, and TLR10 in white blood cells, and in present-day people are associated with reduced Helicobacter pylori seroprevalence and increased susceptibility to allergies.19, 20 Taken together, this suggests that the introgressed alleles might enhance innate immune surveillance and reactivity against certain pathogens, but that this might also have increased hypersensitivity to non-pathogenic allergens, resulting in allergic diseases in present-day people. Given the evidence for selection on multiple TLR haplotypes in humans, including recent selection on non-introgressed haplotypes that are associated with changes in response to Y. pestis and Y. pseudotuberculosis,16 and also selection in other primates, it is possible that exposure to different pathogens might have favored different haplotypes at different times, resulting in higher overall diversity. Introgressed haplotypes are a particularly desirable source of diversity because they are highly divergent, viable, and perhaps even adaptive, in closely related populations.58 Immune-related loci, which are also enriched among targets of balancing selection,59 probably benefit substantially from the introduction of such genetic diversity.7, 59 The variation introduced by introgression from archaic humans might have been an important factor in the adaptation of modern humans to novel pathogens in challenging new environments.
Both adaptive introgression and local positive selection on modern human haplotypes have contributed to the evolution of the TLR6-TLR1-TLR10 locus in some human populations, affecting both gene expression and protein function. We note, however, that because pathogens evolve quickly, it is difficult to pinpoint the precise selective force or forces driving these changes. Further genome-wide screens linking Neandertal haplotypes to modern human phenotypes will provide more insight into how admixture with archaic humans has influenced modern human biology.
Acknowledgments
We thank Cesare de Filippo, Felix Key, Svante Pääbo, Kay Prüfer, João Teixeira, and two anonymous reviewers for helpful comments and discussions. We thank David Reich and Nick Patterson for access to the Simons Genome Diversity Panel data and Marike Schreiber for assistance with the figure preparation. Funding was provided by the Max Planck Society and the Deutsche Forschungsgemeinschaft SFB1052 “Obesity mechanisms” (project A02) (to J.K.).
Published: January 7, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Data include 7 figures and 12 tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.015.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
Ensembl Genome Browser, http://www.ensembl.org/index.html
Geuvadis Project, http://www.geuvadis.org/
GTEx Portal, http://www.gtexportal.org/home/
GWASdb, http://jjwanglab.org/gwasdb
OMIM, http://www.omim.org/
Supplemental Data
For each GTEx tissue, the normalized expression (y axis) across individuals with different genotypes at shared archaic-like SNP positions (x axis: NT = Neandertal allele, AF = African allele) for each TLR gene are displayed. Individuals are represented by black cubes and the entire distribution is shown as boxplot. Differential expression p values between genotypes are displayed in the upper part of each plot.
Haplogram for the 671 alleles for which the nine core haplotypes differ in their major allele. These are identified from 2,535 individuals in the 1000 Genomes dataset. For each variable position, the functional impact as reported by the Variant Effect Predictor (see McLaren et al.6) (ENSEMBL v.73) is provided. For missense mutations, SIFT and PolyPhen (see Adzhubei et al.7 and Kumar et al.8) predicted effects are given. Alleles private to one modern human core haplotype are displayed in column 16.
(A) The table displays the p values of FSTs for SNPs which differ between the most frequent core haplotype (V) and the archaic-like core haplotypes (III, IV, VII) within the FST distribution of (1) genome-wide SNPs, (2) genome-wide archaic SNPs, and (3) genome-wide archaic SNPs that are specific to Asians.
(B and C) The tables display the p values of FST values for SNPs which differ between the most frequent core haplotype (V) and the archaic-like core haplotypes (III, IV, VII) within the FST distribution of all genome-wide archaic-like SNPs (B), and the distribution of all genome-wide SNPs (C), for the comparisons between European populations.
(D and E) The tables display the p values of FST values for SNPs which differ between the most frequent core haplotype (V) and the archaic-like core haplotypes (III, IV, VII) within the FST distribution of genome-wide archaic-like SNPs (D), and the distribution of all genome-wide SNPs (E), for the comparisons between Asian populations.
p values with FDR < 0.05 are highlighted in yellow.
References
- 1.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich D., Green R.E., Kircher M., Krause J., Patterson N., Durand E.Y., Viola B., Briggs A.W., Stenzel U., Johnson P.L. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 2010;468:1053–1060. doi: 10.1038/nature09710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vernot B., Akey J.M. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 7.Ségurel L., Quintana-Murci L. Preserving immune diversity through ancient inheritance and admixture. Curr. Opin. Immunol. 2014;30:79–84. doi: 10.1016/j.coi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Huerta-Sánchez E., Jin X., Asan, Bianba Z., Peter B.M., Vinckenbosch N., Liang Y., Yi X., He M., Somel M. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–197. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abi-Rached L., Jobin M.J., Kulkarni S., McWhinnie A., Dalva K., Gragert L., Babrzadeh F., Gharizadeh B., Luo M., Plummer F.A. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams A.L., Jacobs S.B., Moreno-Macías H., Huerta-Chagoya A., Churchhouse C., Márquez-Luna C., García-Ortíz H., Gómez-Vázquez M.J., Burtt N.P., Aguilar-Salinas C.A., SIGMA Type 2 Diabetes Consortium Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mendez F.L., Watkins J.C., Hammer M.F. A haplotype at STAT2 introgressed from neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am. J. Hum. Genet. 2012;91:265–274. doi: 10.1016/j.ajhg.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez F.L., Watkins J.C., Hammer M.F. Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol. Biol. Evol. 2013;30:798–801. doi: 10.1093/molbev/mst004. [DOI] [PubMed] [Google Scholar]
- 13.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Quintana-Murci L., Clark A.G. Population genetic tools for dissecting innate immunity in humans. Nat. Rev. Immunol. 2013;13:280–293. doi: 10.1038/nri3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barreiro L.B., Ben-Ali M., Quach H., Laval G., Patin E., Pickrell J.K., Bouchier C., Tichit M., Neyrolles O., Gicquel B. Evolutionary dynamics of human Toll-like receptors and their different contributions to host defense. PLoS Genet. 2009;5:e1000562. doi: 10.1371/journal.pgen.1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laayouni H., Oosting M., Luisi P., Ioana M., Alonso S., Ricaño-Ponce I., Trynka G., Zhernakova A., Plantinga T.S., Cheng S.C. Convergent evolution in European and Rroma populations reveals pressure exerted by plague on Toll-like receptors. Proc. Natl. Acad. Sci. USA. 2014;111:2668–2673. doi: 10.1073/pnas.1317723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plagnol V., Wall J.D. Possible ancestral structure in human populations. PLoS Genet. 2006;2:e105. doi: 10.1371/journal.pgen.0020105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mayerle J., den Hoed C.M., Schurmann C., Stolk L., Homuth G., Peters M.J., Capelle L.G., Zimmermann K., Rivadeneira F., Gruska S. Identification of genetic loci associated with Helicobacter pylori serologic status. JAMA. 2013;309:1912–1920. doi: 10.1001/jama.2013.4350. [DOI] [PubMed] [Google Scholar]
- 20.Hinds D.A., McMahon G., Kiefer A.K., Do C.B., Eriksson N., Evans D.M., St Pourcain B., Ring S.M., Mountain J.L., Francke U. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet. 2013;45:907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- 24.Paradis E., Claude J., Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 25.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Broman K.W., Murray J.C., Sheffield V.C., White R.L., Weber J.L. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am. J. Hum. Genet. 1998;63:861–869. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dib C., Fauré S., Fizames C., Samson D., Drouot N., Vignal A., Millasseau P., Marc S., Hazan J., Seboun E. A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature. 1996;380:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 28.Kong A., Gudbjartsson D.F., Sainz J., Jonsdottir G.M., Gudjonsson S.A., Richardsson B., Sigurdardottir S., Barnard J., Hallbeck B., Masson G. A high-resolution recombination map of the human genome. Nat. Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- 29.Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F., Peltonen L., International HapMap 3 Consortium Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinch A.G., Tandon A., Patterson N., Song Y., Rohland N., Palmer C.D., Chen G.K., Wang K., Buxbaum S.G., Akylbekova E.L. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prüfer K., Muetzel B., Do H.H., Weiss G., Khaitovich P., Rahm E., Pääbo S., Lachmann M., Enard W. FUNC: a package for detecting significant associations between gene sets and ontological annotations. BMC Bioinformatics. 2007;8:41. doi: 10.1186/1471-2105-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li M.J., Wang P., Liu X., Lim E.L., Wang Z., Yeager M., Wong M.P., Sham P.C., Chanock S.J., Wang J. GWASdb: a database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2012;40:D1047–D1054. doi: 10.1093/nar/gkr1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogers J., Gibbs R.A. Comparative primate genomics: emerging patterns of genome content and dynamics. Nat. Rev. Genet. 2014;15:347–359. doi: 10.1038/nrg3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu A., Zhao C., Fan Y., Jang W., Mungall A.J., Deloukas P., Olsen A., Doggett N.A., Ghebranious N., Broman K.W., Weber J.L. Comparison of human genetic and sequence-based physical maps. Nature. 2001;409:951–953. doi: 10.1038/35057185. [DOI] [PubMed] [Google Scholar]
- 42.Henn B.M., Botigué L.R., Gravel S., Wang W., Brisbin A., Byrnes J.K., Fadhlaoui-Zid K., Zalloua P.A., Moreno-Estrada A., Bertranpetit J. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sánchez-Quinto F., Botigué L.R., Civit S., Arenas C., Avila-Arcos M.C., Bustamante C.D., Comas D., Lalueza-Fox C. North African populations carry the signature of admixture with Neandertals. PLoS ONE. 2012;7:e47765. doi: 10.1371/journal.pone.0047765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim B.Y., Lohmueller K.E. Selection and reduced population size cannot explain higher amounts of Neandertal ancestry in East Asian than in European human populations. Am. J. Hum. Genet. 2015;96:454–461. doi: 10.1016/j.ajhg.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vernot B., Akey J.M. Complex history of admixture between modern humans and Neandertals. Am. J. Hum. Genet. 2015;96:448–453. doi: 10.1016/j.ajhg.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qin P., Stoneking M. Denisovan ancestry in East Eurasian and Native American populations. Mol. Biol. Evol. 2015;32:2665–2674. doi: 10.1093/molbev/msv141. [DOI] [PubMed] [Google Scholar]
- 47.Lazaridis I., Patterson N., Mittnik A., Renaud G., Mallick S., Kirsanow K., Sudmant P.H., Schraiber J.G., Castellano S., Lipson M. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature. 2014;513:409–413. doi: 10.1038/nature13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fu Q., Li H., Moorjani P., Jay F., Slepchenko S.M., Bondarev A.A., Johnson P.L., Aximu-Petri A., Prüfer K., de Filippo C. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fay J.C., Wu C.I. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–1413. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weir B.S., Cockerham C.C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 52.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 53.Wright S. Genetical structure of populations. Nature. 1950;166:247–249. doi: 10.1038/166247a0. [DOI] [PubMed] [Google Scholar]
- 54.Enard D., Depaulis F., Roest Crollius H. Human and non-human primate genomes share hotspots of positive selection. PLoS Genet. 2010;6:e1000840. doi: 10.1371/journal.pgen.1000840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quach H., Wilson D., Laval G., Patin E., Manry J., Guibert J., Barreiro L.B., Nerrienet E., Verschoor E., Gessain A. Different selective pressures shape the evolution of Toll-like receptors in human and African great ape populations. Hum. Mol. Genet. 2013;22:4829–4840. doi: 10.1093/hmg/ddt335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sankararaman S., Patterson N., Li H., Pääbo S., Reich D. The date of interbreeding between Neandertals and modern humans. PLoS Genet. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fu Q., Mittnik A., Johnson P.L., Bos K., Lari M., Bollongino R., Sun C., Giemsch L., Schmitz R., Burger J. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr. Biol. 2013;23:553–559. doi: 10.1016/j.cub.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hedrick P.W. Adaptive introgression in animals: examples and comparison to new mutation and standing variation as sources of adaptive variation. Mol. Ecol. 2013;22:4606–4618. doi: 10.1111/mec.12415. [DOI] [PubMed] [Google Scholar]
- 59.Key F.M., Teixeira J.C., de Filippo C., Andrés A.M. Advantageous diversity maintained by balancing selection in humans. Curr. Opin. Genet. Dev. 2014;29:45–51. doi: 10.1016/j.gde.2014.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For each GTEx tissue, the normalized expression (y axis) across individuals with different genotypes at shared archaic-like SNP positions (x axis: NT = Neandertal allele, AF = African allele) for each TLR gene are displayed. Individuals are represented by black cubes and the entire distribution is shown as boxplot. Differential expression p values between genotypes are displayed in the upper part of each plot.
Haplogram for the 671 alleles for which the nine core haplotypes differ in their major allele. These are identified from 2,535 individuals in the 1000 Genomes dataset. For each variable position, the functional impact as reported by the Variant Effect Predictor (see McLaren et al.6) (ENSEMBL v.73) is provided. For missense mutations, SIFT and PolyPhen (see Adzhubei et al.7 and Kumar et al.8) predicted effects are given. Alleles private to one modern human core haplotype are displayed in column 16.
(A) The table displays the p values of FSTs for SNPs which differ between the most frequent core haplotype (V) and the archaic-like core haplotypes (III, IV, VII) within the FST distribution of (1) genome-wide SNPs, (2) genome-wide archaic SNPs, and (3) genome-wide archaic SNPs that are specific to Asians.
(B and C) The tables display the p values of FST values for SNPs which differ between the most frequent core haplotype (V) and the archaic-like core haplotypes (III, IV, VII) within the FST distribution of all genome-wide archaic-like SNPs (B), and the distribution of all genome-wide SNPs (C), for the comparisons between European populations.
(D and E) The tables display the p values of FST values for SNPs which differ between the most frequent core haplotype (V) and the archaic-like core haplotypes (III, IV, VII) within the FST distribution of genome-wide archaic-like SNPs (D), and the distribution of all genome-wide SNPs (E), for the comparisons between Asian populations.
p values with FDR < 0.05 are highlighted in yellow.