Abstract
Platinum chemotherapeutic agents, such as cisplatin (cis-diamminedichloroplatinum(II)), can act as radiosensitizers when bound covalently to nuclear DNA in cancer cells. This radiosensitization is largely due to an increase in DNA damage induced by low-energy secondary electrons, produced in large quantities by high-energy radiation. We report the yields of single- and double-strand breaks (SSB and DSB) and interduplex cross-links (CL) induced by electrons of 1.6–19.6 eV (i.e., the yield functions) incident on 5 monolayer (ML) films of cisplatin–DNA complexes. These yield functions are compared with those previously recorded with 5 ML films of unmodified plasmid DNA. Binding of five cisplatin molecules to plasmid DNA (3197 base pairs) enhances SSB, DSB, and CL by factors varying, from 1.2 to 2.8, 1.4 to 3.5, and 1.2 to 2.7, respectively, depending on electron energy. All yield functions exhibit structures around 5 and 10 eV that can be attributed to enhancement of bond scission, via the initial formation of core-excited resonances associated with π → π* transitions of the bases. This increase in damage is interpreted as arising from a modification of the parameters of the corresponding transient anions already present in nonmodified DNA, particularly those influencing molecular dissociation. Two additional resonances, specific to cisplatin-modified DNA, are formed at 13.6 and 17.6 eV in the yield function of SSB. Furthermore, cisplatin binding causes the induction of DSB by electrons of 1.6–3.6 eV, i.e., in an energy region where a DSB cannot be produced by a single electron in pure DNA. Breaking two bonds with a subexcitation-energy electron is tentatively explained by a charge delocalization mechanism, where a single electron occupies simultaneously two σ* bonds linking the Pt atom to guanine bases on opposite strands.
1. INTRODUCTION
In the past decade, the processes induced by the abundant low-energy electrons (LEE) produced by high-energy ionizing radiation have attracted considerable attention1–3 Most biologically significant studies focused on the interaction of LEE with the genome, since the loss of its integrity can be detrimental to cells. In many of these investigations the yields of specific damages, including single- and double-strand breaks (SSB and DSB)1–8 and cross-links (CL),9 as well as the formation of neutral species7,10 and stable anions1,2 has been measured as a function of electron energy within the range 0–20 eV. Whereas the electron energy dependencies of DNA conformational changes (i.e., formation of SSB, DSB, and CL) has been observed in films of bacterial DNA,1,3 the yield functions for other specific products were mainly obtained in experiments with films of short synthetic DNA strands, e.g., self-assembled monolayer oligonucleotides,1–3,11 with lengths ranging from 3 to 40 bases. In most experiments DNA was bombarded with LEE in ultrahigh vacuum (UHV),1,12 but recent developments have enabled the investigation of LEE-induced DNA damage at standard atmospheric temperature and pressure.3
Well-defined maxima in the yield functions of SSB, DSB, and CL were interpreted as due to the formation of transient negative ions (TNI), which decayed into “destructive” channels such as dissociative electron attachment (DEA) and auto-ionization leaving a DNA site in an electronically excited dissociative state.1 Much of our present understanding of the processes involved in these condensed-phase studies has been possible by referring to the results obtained with gaseous basic subunits of DNA.12–18 Furthermore, theoretical calculations of LEE scattering from DNA and attachment to the molecule have established the occurrence of basic quantum phenomena such as electron transfer19,20 and diffraction.20 More recently, LEE have been found to play an important role in concomitant chemoradiation therapy (CRT).21 Their involvement in DNA radiosensitization by Pt drugs in CRT has been explained by Rezaee et al.22,23 and in a recent review article.24
CRT (i.e., the combination of chemotherapy and radiotherapy) has been an important modality for the treatment of solid tumors for decades.25 Cisplatin [Pt-(NH3)2Cl2], whose structure appears in Figure 1, was first introduced in chemotherapy in the 1970s. Since then, cisplatin and its analogues, oxaliplatin and carboplatin, have become the most frequently used agents in CRT.26 The choice of the Pt drug usually depends on the type of cancer being treated by CRT.26 For the present research we chose cisplatin because of its structural simplicity and the availability of basic information on its effect on DNA configuration.
Figure 1.
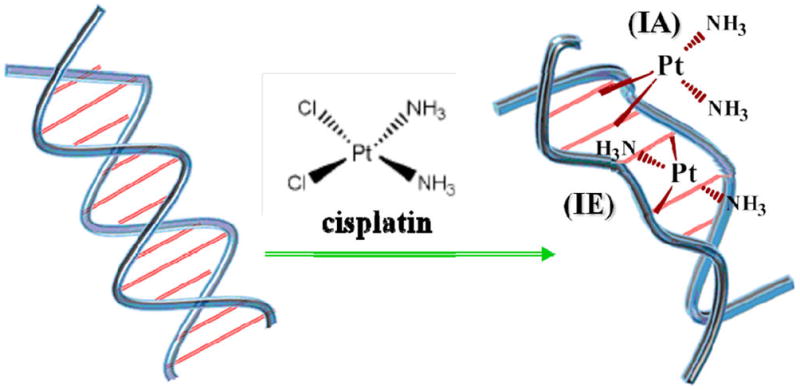
Schematic representation of the DNA double helix (left) and the change in its conformation caused by intrastrand (IA) or interstrand (IE) cross-linking by cisplatin (right). In the final product of the reaction of cisplatin with DNA, the chlorine–Pt bonds are replaced by guanine–Pt or adenine–Pt bonds.
The biochemical basis of cisplatin chemotherapy is well-known. The molecule preferentially reacts with the N7 of purine bases, leading to DNA binding with 60% intrastrand CL at GpG, 30% intrastrand CL at ApG, 10% CL at GpNpG, and <2% NpN interstrand CL, where N = A or G.27 At low Pt drug ratios (i.e., <1 Pt per 500 base pairs (bp)), such as those used in the present experiments and in clinical protocols, the percentage of interstrand CL is much larger, of the order of 30–40%.28 Formation of these CL leads to the distortion of the DNA conformation by unwinding, bending, and destabilizing the double helix, as shown schematically in Figure 1. These modifications subsequently inhibit DNA repair in addition to affecting transcription and replication.29 However, how the presence of cisplatin and other Pt drugs in cells increases radiation-induced lesions, including radiation damage to DNA, remains unclear. Since the general rationale of improving cancer treatment with CRT includes the possibility that a chemotherapeutic agent could act as radiosensitizer, in addition to being chemotherapeutic, the degree and nature of any synergy between radiation and the drug must be carefully investigated.30
Two nonexclusive mechanisms have been proposed for explaining the synergy between cisplatin and radiation in CRT:31 inhibition of repair of radiation-induced damage to DNA and an increase of cellular DNA damage, caused by additional immediate species created by the primary radiation, both occurring when cisplatin is covalently bound to DNA. Although originally difficult to trace, the latter hypothesis has been recently experimentally confirmed.21–23 Zheng et al. first reported that bombarding solid films of DNA–cisplatin complexes with electrons of 1, 10, 100, and 60 000 eV significantly promotes strand breaks by factors varying from 1.3 to 4.21 The enhancement in damage was much higher at low energies; e.g., at mole ratio of 2:1 SSB were enhanced by a factor of 3.6 at 10 eV compared to a factor of 2 at 60 keV. These findings led Zheng et al. to propose that the radiosensitization of DNA by high-energy radiation is essentially mediated by the action of secondary LEE. This hypothesis was confirmed later by Rezaee et al.,22,23 who measured enhancement of damage induced by 0.5, 10, and 10 000 eV electrons to DNA bound to cisplatin, oxaliplatin, and carboplatin. Most strikingly, a single 0.5 eV electron was able to induce a DSB in DNA–Pt complexes,23 whereas in pure DNA, electrons of at least 5 eV are needed to break two adjacent strands.5
Distortion of the DNA configuration and the changes in the electron affinity at the Pt binding site were proposed as modifications responsible for an increase in the cross section for temporary electron capture leading to TNI formation and an enhancement of the magnitude of potentially destructive decay channels, including DEA and autodetachment into dissociative electronic excited states.21–23 However, to confirm the occurrence of TNI in these DNA–Pt drug complexes and investigate their modification by Pt drug, the electron-energy dependence of the damage induced in these complexes must be measured. Such information is particularly useful to specify the type, energy, and magnitude of the TNI involved and their decay channels, so as to obtain more precise information on the mechanisms of radio-sensitization.
In the present work, we measure the energy dependence of the loss of the initial supercoiled form of 3197 bp plasmid DNA and the formation of SSB, DSB, and CL induced by the impact of 1.6–19.6 eV electrons on films of cisplatin–DNA complexes. The results are compared with those recently obtained from unmodified plasmid DNA with the same apparatus and under identical conditions.9 The respective yields are analyzed by agarose gel electrophoresis1,3 as a function of electron fluence. The fluence–response curves for the various yields are recorded at intervals of 1–2 eV to obtain the yield functions. The two significant signatures of electron resonances around 5 and 10 eV present in the in the yield functions of unmodified DNA remain in yield functions of the complex, which also exhibit new resonances at 14 and 18 eV. Furthermore, the magnitudes of the yields recorded from thin films of the cisPt–DNA complexes are increased by factors of 1.2–3.7 compared to those of unmodified DNA.
The comparison between the results obtained from films of DNA, with and without bound cisPt, allows the nature and energy of the major TNI in cisplatin–DNA complexes and their contribution in enhancing LEE damage to be established. Our results suggest that modifications of the configuration of DNA induced by the binding of cisPt, and the presence of new Pt bonds in the molecule, change the resonance parameters of existing TNI and produce new TNI associated with the binding site. These changes also enhance existing molecular dissociation pathways. The ensemble of the results indicate that radio-sensitization of DNA by cisplatin can be, at least partially, interpreted by an increase in the number and magnitude of LEE-induced reactions.
2. EXPERIMENTS
The present experimental procedures and the LEE irradiator have been described in detail previously.1,9 This section summarizes the preparation of plasmid DNA and the parameters of LEE irradiation. It provides more details on the platination reactions, measurement of Pt binding to DNA, preparation of cisplatin–DNA films, and quantification of different configurations of the DNA and Pt–DNA in the films.
Preparation of Plasmid DNA
Plasmid DNA (pGEM-3Zf(−), 3197 bp) was extracted from E. coli DH5 and purified with a HiSpeed plasmid Maxi kit (QIAGEN). The purified plasmid DNA was eluted in buffer TE (10 mM Tris and 1 mM EDTA); it consisted of 97% supercoiled, 1% CL, and 2% concatemeric forms. The concentration of DNA was determined by ultraviolet (UV) absorption at 260 nm with a BioTek Epoch spectrophotometer, according to 1 OD = 50 ng/μL. The quantity of impurities including protein and salts in the plasmid was estimated by measuring the ratio of UV absorption from 260 to 280 nm. The ratio was 1.88, which corresponds to a purity greater than 85%.32
Platination of Plasmid DNA
Cisplatin [cis-diamminedichloroplatinum(II)] was purchased from Sigma-Aldrich with a stated purity of 99.9% and used without further purification. Cisplatin dissolved in distilled deionized water (ddH2O) was added to the DNA solution at a ratio of 16 molecules per DNA and incubated in the dark at room temperature for 2 h. The unbound Pt compounds, Tris molecules from the DNA buffer TE and complexes of Tris linked to cisplatin, were separated with a Sephadex-G50 column. Sephadex G-50 is a suitable medium for separation of molecules having a molecular weight larger than 3 × 104 g mol−1 from molecules with a molecular weight smaller than 1500 g mol−1.33 Similarly, the TE buffer was separated from DNA using the G-50 medium for the irradiation experiments in order to obtain DNA with only its structural salt. Compared to previous preparations,21,34,35 the lower temperature of reaction and generally improved experimental conditions36 maximize the amount of DNA complexes retained in the initial supercoiled conformation (i.e., the cisplatin–DNA complexes contained 95% supercoiled DNA).
Analysis of Platinum–DNA Binding
Whereas in previous work the concentration of platinum bound to DNA was estimated from the added amount of platinum in the solution,21 in the present experiment, the average number of cisplatin molecules per plasmid was accurately measured by inductively coupled plasma mass spectroscopy (ICP-MS, Agilent 7500CE).37 A cisplatin-concentration calibration curve was recorded from the amount of Pt from cisplatin dissolved in ddH2O at known concentrations. The efficiency of cisplatin binding to DNA was found to be 33%. Cisplatin was dissolved in ddH2O at a concentration to a ratio of cisplatin to DNA in the complexes of 5:1.
Preparation of Cisplatin–DNA Films
The Pt–DNA samples were deposited on substrates consisting of 450 ± 50 nm films of Ta evaporated onto a 0.4 mm thick silicon wafer. The surface of Ta was cleaned in pure ethanol and ddH2O and dried with a flow of dry nitrogen. 10 μL of cisplatin–DNA solution consisting of 320 ng of DNA was deposited onto the cleaned Ta surface. After freezing at −65 °C for 7 min in a glovebox containing an atmosphere of pure dry nitrogen, the samples were lyophilized (freeze-dried) under a pressure of 7 mTorr by a hydrocarbon-free turbomolecular pump for 2 h. Assuming that the molecules were uniformly distributed on the surface with a radius of 2 ± 0.5 mm and the density of DNA was 1.7 g cm−3,38 the average thickness of the film was approximately 15 nm (5 monolayers (ML)). Such a thickness has been widely used in DNA-LEE experiments.1,3 Based on the average film thickness of 15 nm, which is of the order of the penetration depth (5–20 nm) of 4–15 eV electrons in liquid water or amorphous ice,39 most electrons impinging on the film should be transmitted to the metal substrate. However, due to local variations in thickness, some electrons are expected to be retained a sufficiently long time for the film to remain charged during the experiment. As recently shown, film charging does not affect the yields obtained from the slope at zero fluence of exposure–response curves.40
LEE Irradiation
The samples were transferred to the LEE irradiation chamber,9 which was subsequently evacuated for ~24 h to reach a base pressure of 2 × 10−8 Torr at ambient temperature. By changing the potential between sample target (ground) and the center of the filament of the LEE gun, the electron energy was varied from 2 to 20 ± 0.3 eV. The energy scale was calibrated by taking, as the zero electron energy reference, the onset of electron transmission through the films. At this onset, the potential between the point of emission of the filament and the substrate was 0.4 V, indicating the potential change due to the different work functions between the connections in our apparatus and the cisplatin–DNA vacuum interface. Hence, the absolute electron energy was obtained by subtracting 0.4 V from each voltage measured between the filament and the metal substrate.
At 11 different energies, the films were bombarded with an incident electron beam of 6 nA and exposures of 5–60 s. Six measurements were performed for each point of a fluence–response curve. At each electron energy, 66 samples were irradiated for a total of 726 irradiations and analyses. In addition, 12 control samples were inserted into the irradiator chamber, but not irradiated, to serve as the zero fluence data points in exposure curves.
Quantification of the DNA and Pt–DNA Films
The Pt–DNA films were recovered from the Ta substrates with 10 μL of ddH2O. Quantification of the different structural forms of DNA in the samples was performed by agarose gel electrophoresis as previously described.1,9 The forms analyzed were the supercoiled (SC), concatemeric, nicked circular (C), linear (L), and interduplex CL configurations, which correspond to the initial configuration of the plasmid, two interlaced circular DNAs, SSB, DSB, and cross-linking between the SC and C configurations, respectively.9 The samples and the agarose gels were stained with SYBR Green I at concentrations of 100× and 10000×, respectively. The samples were run on 1% agarose gel in 1X TAE buffer at 100 V for 7 min following by 75 V for 68 min (5 V cm−1). The gels were then scanned by a STORM 860 laser scanner (Molecular Dynamics) adjusted for the blue fluorescent mode at an excitation wavelength of 450 nm and PMT voltage of 800 in the normal sensitivity mode. The binding efficiencies of SYBR Green I for the same amount (80 ng) of supercoiled and linear DNA was measured, and the correction factor was determined to be 1.4. This factor arises from the weaker binding of SYBR Green I to supercoiled DNA than to the nicked circular and linear forms.
The different DNA bands on the gel were analyzed by the ImageQuant 5.0 (Molecular Dynamics) software. Images of the bands and their intensity profiles are shown in the Supporting Information. Instead of using the automated peak-analysis program, each band in a given scan of the gel was deconvoluted manually and separated from the residual baseline, to obtain the most accurate percentage value possible for each configuration. Also, the binding of cisplatin to DNA caused the linear DNA (DSB) band to migrate close to that of the concatemeric configuration, which could result in an overevaluation of the yields of the L form. A better separation of the DSB from the concatemeric band was observed in the electrophoresis with 1.3% agarose gel (see Supporting Information). Thus, in the present study, the integration of DSB peak was corrected from further analysis at 1.3% agarose gel. With the improved preparation of cisplatin–DNA complexes and deconvolution procedure of the DSB peak, smaller yields of L-DNA were measured than previously,21,35 in agreement with the recent results of Rezaee et al.,22 who also deconvoluted the L band from the concatemeric band.
3. RESULTS
Exposure–response curves were recorded at 11 energies in the range 2–20 eV, for the formation of CL, SSB, DSB, and loss of supercoiled (LS) in cisplatin–DNA films. As an example of these curves, the ones recorded at an incident electron energy of 9.6 eV are shown in Figure 2. They are similar to those previously recorded from nonmodified DNA films.9 The yields of CL, SSB, and LS fit an exponential function, whereas those of DSB can be represented with a linear curve. Since the probability of breaking two bonds on opposite DNA strands with two or more electron collisions is vanishingly small under the present experimental conditions, this linearity indicates that the DSB are induced by a single electron.23 The effective yields per electron per molecule for each product are obtained by dividing the slope at zero fluence by the amount of supercoiled DNA in the control sample. The yields recorded within the 1.6–19.6 eV range are listed in Table 1.
Figure 2.
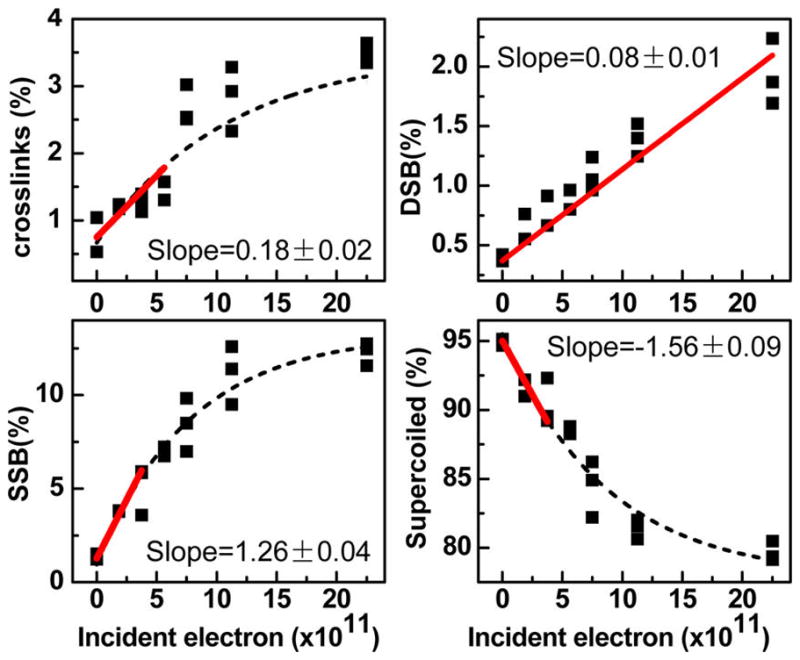
Exposure–response curves for the formation of cross-links (CL), DSB, SSB, and loss of supercoiled (LS) DNA induced by 9.6 eV electron impact on 5 monolayer (ML) films of cisplatin–DNA complexes. The slope for DSB is taken from a linear fit, whereas the other slopes are those at zero fluence, obtained from the dashed exponential fit.
Table 1.
Effective Yields (in 10−15 electron−1 molecule−1) of Cross-Links, SSB, DSB, and Loss of Initial Supercoiled DNA Induced by 1.6–19.6 eV electrons in 5 ML Thick Cisplatin–DNA Filmsa
| energy (eV) | configuration
|
|||
|---|---|---|---|---|
| cross-links | linear (DSB) | circular (SSB) | supercoiled | |
| 1.6 | 6.4 ± 1.1 | 3.6 ± 0.5 | 55.6 ± 5.4 | −64.9 ± 10.7 |
| 3.6 | 11.8 ± 2.1 | 4.1 ± 0.7 | 86.0 ± 12.2 | −106.4 ± 12.9 |
| 4.6 | 14.0 ± 1.1 | 5.1 ± 1.0 | 100.0 ± 16.5 | −123.6 ± 19.3 |
| 5.6 | 15.0 ± 2.1 | 5.9 ± 0.6 | 68.8 ± 9.7 | −88.2 ± 9.7 |
| 7.6 | 11.3 ± 0.6 | 5.9 ± 0.6 | 90.3 ± 4.3 | −136.5 ± 18.3 |
| 9.6 | 19.3 ± 2.1 | 8.6 ± 1.1 | 135.5 ± 8.6 | −167.7 ± 9.8 |
| 11.6 | 16.9 ± 1.5 | 6.1 ± 0.9 | 82.8 ± 7.5 | −93.5 ± 8.6 |
| 13.6 | 17.2 ± 2.3 | 6.7 ± 0.6 | 91.4 ± 8.6 | −105.4 ± 8.6 |
| 15.6 | 10.4 ± 1.2 | 6.3 ± 0.5 | 63.4 ± 11.8 | −76.3 ± 12.9 |
| 17.6 | 12.0 ± 1.2 | 7.9 ± 0.6 | 102.1 ± 9.7 | −136.6 ± 11.8 |
| 19.6 | 16.1 ± 2.1 | 9.6 ± 1.0 | 87.1 ± 7.5 | −108.6 ± 9.7 |
The error bars are the standard deviations for five identical experiments.
Figure 3 displays the yields of CL, DSB, SSB, and LS as a function of incident electron energy obtained from films of cisplatin–DNA complexes and unmodified DNA; the latter were previously recorded under conditions identical to those of the present experiment.9 In the range 1.6–19.6 eV, the presence of five cisplatin molecules covalently bound to each plasmid increases considerably the yields of CL, SSB, DSB, and LS, but the signatures of energy dependence for each form are different (Figure 3). The yield functions of SSB and LS exhibit similar line shapes, as expected, since most of the damage to supercoiled DNA results in SSB. In these functions, peaks are superimposed upon a large background signal, which increases when cisplatin is bound to DNA. The most prominent peak in all eight yield functions lies at 9.6 eV. Another noticeable peak is found at 4.6 eV in the SSB and LS yield functions and around 6 eV in those of CL and DSB. Interestingly, the 4.6 and 9.6 eV peaks clearly appear in both the cisplatin–DNA and pure DNA data. Conversely, the 5.6 eV peak in the CL curve, which is absent or not resolved in the pure DNA yield function, becomes clearly visible when cisplatin is bound to the plasmid. As seen in Figure 3, the SSB and LS yield functions of cisplatin–DNA contains two additional well-revolved peaks at 13.6 and 17.6 eV, indicating for the first time that cisplatin produces additional LEE–DNA interactions. Compared to SSB, the formation of CL and DSB are minor products (Table 1).
Figure 3.
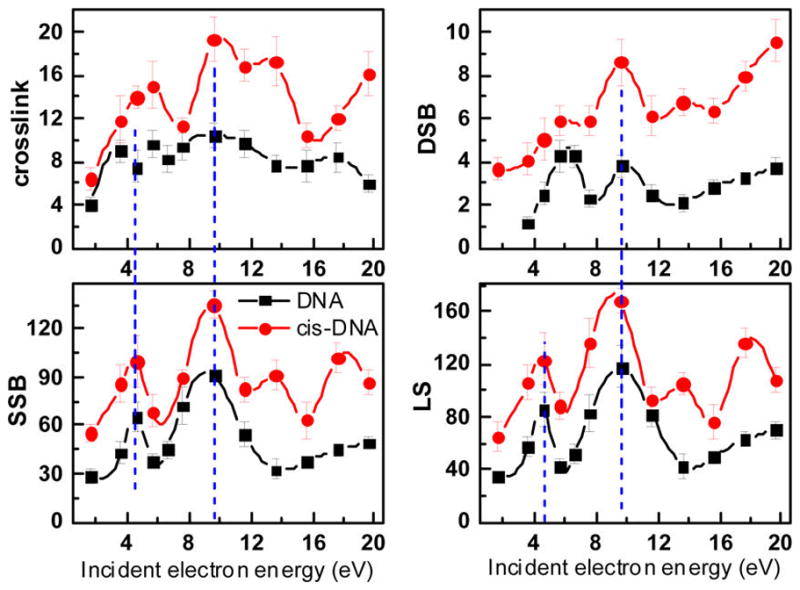
Effective yields (× 10−15/electron·plasmid) for the formation of CL, DSB, SSB, and LS obtained from DNA (■)10 and cisplatin–DNA (●) films of 5 ML thickness irradiated with electrons of 1.6–19.6 eV. The incident current was 6 nA in all experiments.
Figure 4 exhibits from top to bottom the respective enhancement factors (EF) of CL, DSB, SSB, and LS as a function of electron energy. The EF is the ratio between the yields obtained from cisplatin–DNA complexes to those obtained from nonmodified DNA. The EF denotes the increased sensitivity of DNA to LEE, induced by cisplatin inter- and intrastrand cross-linking. Within statistical variations, the EF for SSB and LS display two maxima around 13.6 and 17.6 eV. In the case of CL, only one peak can be discerned at 13.6 eV. A peak at 13.6 eV may also be present in the EF of DSB, which are the largest among all lesions, especially at low energies.
Figure 4.
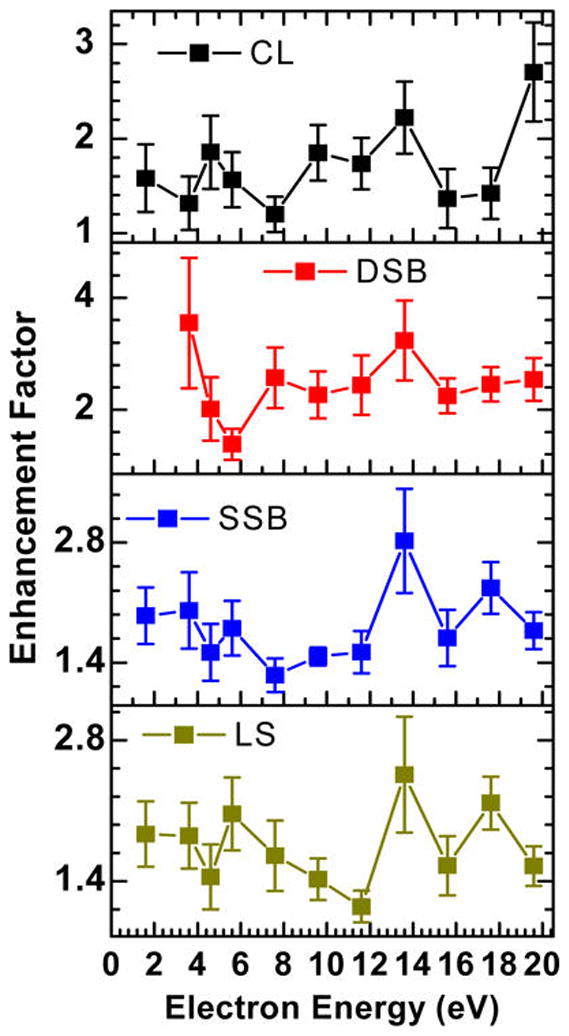
Enhancement factors (EF) of DNA damages including CL, DSB, SSB, and LS for cisplatin–DNA complexes as a function of incident electron energy. The deviations are the quadrature statistical errors.
4. DISCUSSION
A. TNI Manifold of DNA
Numerous experiments with plasmid (i.e., bacterial) DNA have established that there exist at least four transient anions in the 0–15 eV range that decay into destructive channels causing strand breaks in the molecule.1,3,5–7,9,24 Two of these are shape resonances located at 0.8 and 2.2 eV in the yield function of SSB, with cross sections varying from 10−13 to 10−15 cm2 within the 0.5–4.5 eV range.5 The energies of these resonances correlate with those determined from gas-phase electron scattering from the bases.41 From such comparisons and theoretical calculations, it was deduced that the incoming electron is first captured in an unfilled π* orbital of the bases and then transferred to an unfilled σ* orbital of the C–O bond within the backbone of DNA, causing rupture of the sugar–phosphate chain. No DSB is formed below 5 eV in normal plasmid DNA.5
The other two resonances have been located at 4.6 and 9.6 eV in the SSB yield function and at 6.1 and 9.6 eV in the yield function of DSB. They were characterized9 as core-excited resonances arising from capture of the incident electron by the electron affinity of electronic excited states of the bases,42,43 particularly those associated with n → π* and π → π* transitions. In this case, a SSB is also formed by transfer of the additional electron to the phosphate group, but following electronic excitation of the base. Occupation of a dissociative σ* previously unfilled orbital of the C–O bond ruptures the sugar–phosphate backbone, as in the case of the 0.8 and 2.2 eV shape resonances. The mechanism for breaking two bonds on adjacent chains of DNA (i.e., a DSB) with a single LEE has not been precisely determined yet.
In unmodified DNA, these core excited resonances appear as two strong maxima in the yield functions of SSB, DSB, and LS drawn from solid squares in Figure 3. The yield function for CL is more uniform but consistent with the possible presence of broad maxima around 10 and 18 eV. These CL arise from binding of the SC to the C configuration from radical formation at various positions within unmodified or circular DNA.9 Their mechanism of formation could therefore be different (e.g., the TNI involved) from that leading to rupture of the DNA backbone and thus produce a different yield function as seen in Figure 3.
Details of the SSB yields below 2 eV have not been recorded and do not appear in Figure 3. It should be noted, however, that in this energy range the yields of SSB are substantial; they are considered to arise from the mechanism described in the beginning of this subsection, which involves the 0.8 and 2.2 eV shape resonances. In Figure 3, this “background” yield appears to be progressively replaced by the signal from overlapping of the 4.6 and 9.6 eV resonance peaks, although the possibility of direct electronic excitation leading to bond dissociation cannot be completely rejected.
B. Increasing SSB and CL Yields with Cisplatin
According to the curves drawn from the filled circles in Figure 3, the presence of cisplatin bound to the DNA increases the yields of all configurations throughout the 1.6–19.6 eV range. This enhancement of LEE-induced DNA damage is energy dependent as evidenced by the ratios of specific damages recorded with and without cisplatin bound to DNA (Figure 4). The formation of SSB via electron transfer from a shape resonance of the bases in the 1.6–3.6 eV region is enhanced on average by a factor of about 2, whereas the EF corresponding to the formation of such breaks via the 4.6 and 9.6 eV core-excited resonances (Figure 3) followed by a similar electron transfer has an EF close to 1.6. The two strong peaks with EF reaching 2.8 and 2.4 reflect the appearance of new resonances in the SSB yield function at 13.6 and 17.6 eV seen in Figure 3, respectively, and clearly show the significant implication of TNI in the enhancement of DNA damage by cisplatin.
Whereas in unmodified DNA the 4.6 and 9.6 eV resonances are difficult to perceive in the CL yield function (Figure 3), they are clearly visible with the binding of cisplatin. It indicates that these TNI play a major role in increasing the radicals responsible for CL formation in cisplatin–DNA complexes. The formation of radicals from the radiolysis of single-stranded cisplatin–DNA adducts has been investigated by Behmand et al.44 The oligonucleotide TTTTTGTGTTT (T = thymine, G = guanine) with and without a cisplatin cross-link between the two guanines was reacted with hydrated electrons in a water solution irradiated with γ-rays. The two Pt–G bonds were found to be ruptured simultaneously by hydrated electrons giving, for example, about 35% detached cisPt at 1 kGy for a solution containing 0.03 μM oligonucleotide.44 Very little single breakage of a Pt–G bond breaking was observed. Furthermore, the hydrated electrons induced damage to thymines and guanines in the oligonucleotide.45 Cisplatin detachment generated both unmodified guanine and damaged guanine, in equimolar amounts. At 1 kGy, a net average of 2.5 thymines and 1 guanine were damaged for each platinum lost from the oligonucleotide. It was suggested that the hydrated electron could be initially captured by a thymine base and transferred by base to base electron hopping to the guanine site,46 where the cisplatin moiety detaches from the oligonucleotide via DEA. It is also possible for the hydrated electron to interact directly with the platinum–guanine adduct and induce detachment of the cisplatin moiety, again via DEA.44 The TNI responsible for these damages is probably akin to the one found by Kopyra et al.47 near 0 eV in gaseous cisplatin. Such a resonance in solution would have much lower energy owing to polarization of the molecular orbitals of electrons and vibrational and phonon modes of surrounding water molecules (i.e., polaron formation); binding of cisplatin to DNA could also lower the resonance energy. Thus, according to the results of Behmand et al.,44,45 the formation of TNI at the site of cisplatin could considerably increase the formation of radicals owing to detachment of the Pt drug and damage to the bases. Assuming that the core-excited resonances at 4.6, 9.6, and 13.6 eV have a behavior similar to the lowest energy TNI in cisplatin–DNA complexes, these higher energy anion states could also strongly amplify the formation of radicals from DNA, in the presence of cisplatin. As shown previously,48 radical formation within DNA could result in a SSB and its reaction with a nearby intact DNA could lead to a C–SC cross-links. Such CL could also result from a radical created on the SC configuration, which would react with circular DNA. The enhancement of interduplex cross-linking at the resonance energies would be reflected by peaks in the CL yields and EF functions as seen in Figure 3 and the upper curve of Figure 4, respectively.
Whereas the enhancement of SSB above 13 eV appears to arise essentially from new TNI created by the presence of cisplatin, below that energy the EF seem to result from modification of the parameters of TNI already present in unmodified DNA. It is well established that the resonance parameters of transient anions are influenced by their environment.1,49,50 Alternation of the surroundings of a site of formation of a TNI can modify the electron capture probability, the transitory anion’s lifetime, its energy, and the number of decay channels; furthermore, in a large molecule such as DNA, the probability of electron transfer from one site to another can also change. When a cisplatin molecule binds to DNA, two major modifications occur: the helix unwinds and bends51,52 as shown schematically in Figure 1, and the presence of a highly polarizable Pt atom in close proximity to the strands creates an attractive potential for electrons. The latter can certainly lower the energy and the number of decay channels of TNI formed close to cisplatin, but a priori, both modifications can affect all parameters mentioned previously. Unwinding and bending of the helix also modifies the strength of the bonds within DNA, which may also influence the yields of various damages.
It is therefore difficult to predict the magnitude of the change in the yields of SSB without quantum mechanical calculations taking these many parameters into consideration. Such calculations are presently limited to small segments of single-stranded DNA consisting, at the most, of three basic units (i.e., a base, sugar, or phosphate group).20 Furthermore, in these theoretical models double-stranded DNA is not considered, and the electron is captured only by the ground state of the segment.20 The latter excludes any estimate of the yields of SSB arising from core-excited resonances.
However, with a simplified potential energy curve diagram, such as that shown in Figure 5, it can be shown that the introduction of an attractive potential by cisplatin between the strands leads to an enhancement of C–O bond cleavage at the 3′- and 5′-positions of the DNA backbone. The upper curve II represents the potential energy of the TNI in unmodified DNA, at the phosphate site, along the dissociative σ* orbital to which an electron is transferred from a base.19,24 Since this transfer is much faster than vibrational relaxation of the molecule, the Born–Oppenheimer approximation is valid and the electron first occupies a segment of the potential energy curve lying in the Franck–Condon region. As time progresses, C–O separation increases causing rupture of the bond, unless the electron autodetaches leaving the CO moiety in the ground state, which is not dissociative (full curve I in Figure 5). However, when the C–O separation reaches the value Rc, the electron becomes bound to the oxygen atom; it can no longer detach, and dissociation necessarily occurs. If the energy of the σ* bond is lowered by the attractive potential created by Pt, its potential energy curve along the C–O coordinate is lowered in energy (curve III in Figure 5). Curve crossing between the ground state of the C–O moiety and the TNI state is also reduced to the value Rc′, as well as the time available for autoionization of the σ* electron. The probability of autodetachment depends exponentially on Rc,53,54 so that the probability of anion survival without electron autodetachment is considerably increased, as well as that of DEA at the 3′- and 5′-positions of the backbone. Since in the 0.5–15 eV range the yields of SSB arise via rupture of the C–O σ* bond in the DNA chain, this mechanism may lie at the basis of the observed enhancement of SSB in cisplatin–DNA complexes below 13 eV and could very well play a role in increasing direct DEA to the bases or other subunits, in the presence of cisplatin, so as to produce additional radicals causing cross-linking.
Figure 5.
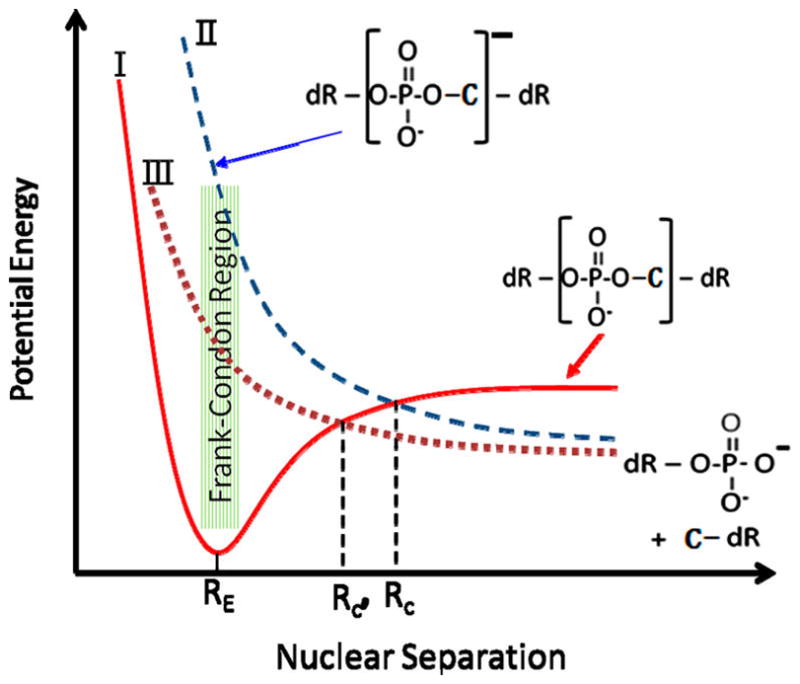
Schematic diagram of hypothetical potential energy curves for (I) the ground-state of CO in the backbone of DNA, (II) same ground state with an electron added in the lowest usually-unfilled σ* orbital in the absence of cisplatin, and (III) same as (II) in the presence of cisplatin. RE is the equilibrium internuclear C–O distance in the ground state. Rc and Rc′ are the crossing points of II and III with the potential energy curve of the ground state I, in unmodified and modified DNA, respectively. The binding of cisplatin lowers potential energy curve II and shifts the crossing point toward RE (Rc′ < Rc) resulting in enhanced DEA and cleavage of the C–O bond.
C. Modification of the DSB Yield Function by Cisplatin: Breaking Two Strands with a Single Electron
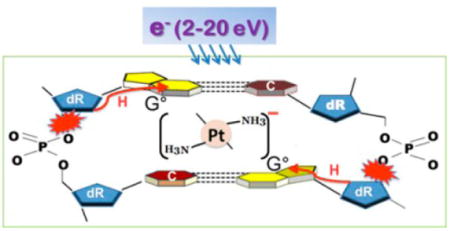
The 6.1 eV core-excited resonance in the upper right segment of Figure 3 does not appreciably enhance DSB formation (i.e., EF = 1.4 in Figure 4), whereas for the next one at 9.6 eV the EF lies around 2.4. The possible appearance of the new resonance at 13.6 eV, due to the binding of cisplatin to DNA, increases the EF for DSB to 3.2, which is the second highest value recorded in the present experiments. The EF for DSB increases considerably at low energies, since a single electron with an energy below 5 eV cannot create two adjacent strand breaks in unmodified DNA,5 whereas in modified DNA, an electron with such energies can induce a DSB with magnitudes similar to those found in the 5–15 eV range.23 We note that owing to the energy spread of the electron beam the EF at 3.6 eV remains finite. DSB formation below 5 eV has been attributed to the formation of a shape resonance at the site of cisplatin, when the latter links two adjacent strands as shown in Figure 1 and in more detail in Figure 6.
Figure 6.
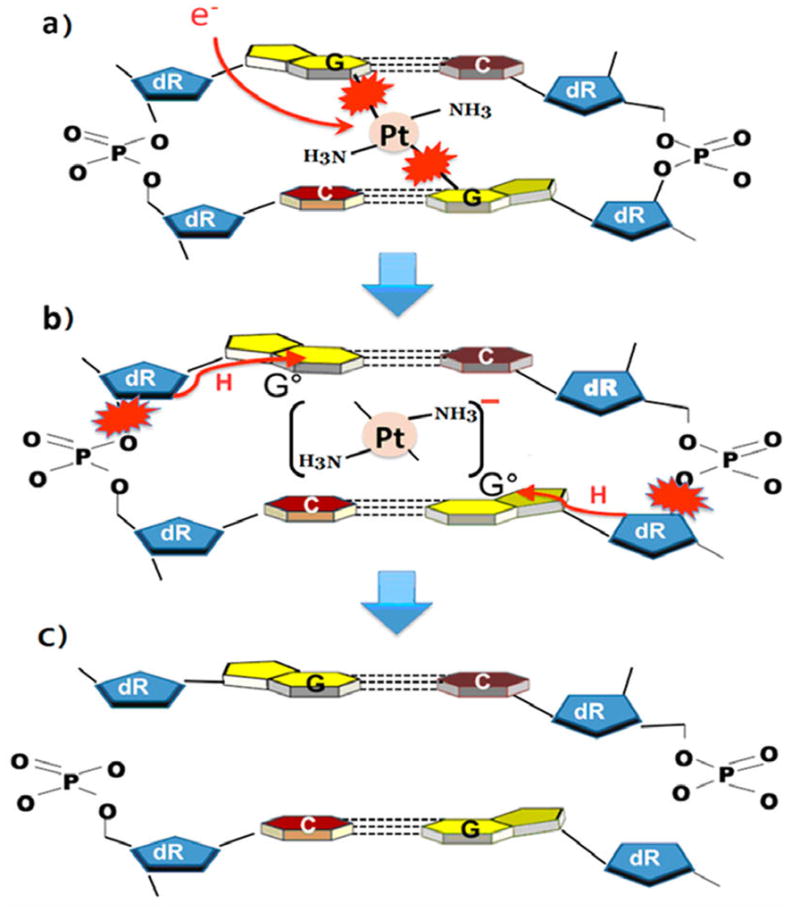
Possible mechanism for the formation of a DSB by a single electron, when cisplatin links two guanine (G) bases on opposite strands. (a) Electron capture into two dissociative orbitals between Pt and two G’s. (b) The transient anion thus formed dissociates, leaving the electron on the (NH3)2Pt moiety and causing simultaneous cleavage of the two Pt–G bonds. The resulting two guanine radicals (G•) abstract hydrogen from the backbones, causing cleavage of phosphodiester bonds on opposite strands. (c) Resulting DSB.
The breaking of two Pt bonds simultaneously has been related to the charge delocalization process found in small molecules by Polanyi and co-workers,55,56 in which the wave functions of a near 0 eV electron lie simultaneously on multiple orbitals, causing concerted bond breaking. In fact, according to the experiments of Kopyra et al.,47 near 0 eV electron attachment into σ* orbitals of gaseous cisplatin [i.e., (NH3)2PtCl2] can break simultaneously two Pt–Cl bonds with considerable probability, yielding Cl2−. Thus, the reaction requires no additional energy than that of the 2.4 eV electron affinity of Cl2. When cisplatin reacts with DNA to form an interstrand CL, Pt is linked to the N7 of guanines releasing two Cl atoms, as shown in Figure 6a. Owing to the symmetry of the structure, we expect the captured electron wave function to extend into the dissociative σ* orbitals with equal magnitude in modified DNA, as in the case of gaseous cisplatin. In other words, the bond starts to elongate with equal momentum toward the dissociation limit of both Pt–G bonds. However, if the electron would end up on one bond, the symmetry would be broken and it would be unlikely to induce the rupture of the two bonds. On the other hand, if the electron resides on the Pt–(NH3)2 moiety, which has an electron affinity larger than 3 eV,47 sufficient energy should become available to rupture the two Pt–G bonds. From entropy arguments in the case of cisPt,47 localization of the excess charge is more favorable on [Pt(NH3)2] than on Cl2. We therefore expect that in our case the additional electron has a higher probability to remain on the Pt(NH3)2 than to transfer to the base. Considering an electron affinity of 3 eV for [Pt(NH3)2] and the minimum energy of 1.6 eV of electrons in our experiments, we should have at least 4.6 eV available for the reaction leading to rupture of the two Pt–N bonds in cisPt–DNA, which is about twice the energy required to break two bonds in isolated cisPt. From these considerations, it seems plausible that an electron of 1.6–3.6 eV breaks simultaneously the two Pt–N bonds in the cisPt–DNA complex. In this case, the cleavage of Pt–G bonds produces guanine radicals (Figure 6b) via
In general, hydrogen abstraction from an adjacent deoxyribose by a guanine radical is considered to be an important initial reaction leading to SSB or DSB in DNA (Figure 6b).57,58 The helix of DNA being greatly distorted with the covalent binding of cisplatin,51,52 we speculate that such binding may shorten the distance of C5′–H or even C3′–H to the guanine radical and make H abstraction more feasible than in regular DNA, thus increasing the yields of SSB or DSB. Since at low Pt ratios, in supercoiled DNA, Pt drugs form a considerable quantity of interstrand CL between guanines in opposite strands,28 the simultaneous formation of two guanine radicals by a single LEE could induce a DSB by a single-hit process with considerable efficiency (Figure 6c). This dissociation process has been postulated to occur at energies much lower than that of the first electronically excited of DNA (i.e., in the energy region of the σ* shape resonance).23
Above the energy required for electronic excitation, the EF of DSB are still larger than those of the three other conformations with characteristic signature around 8 and 14 eV (Figure 4). This enhancement in DSB may be the result of additional resonances owing to the binding of cisplatin to DNA, similar to the low-energy TNI state invoked above or the result of modifications of the resonance parameters of existing TNI states. At this point, any discussion becomes highly speculative, since we do not know exactly how an electron of 5 eV or more can cause, within 10 bp, double cleavages on adjacent phosphate–sugar backbone. Zheng et al. proposed that interstrand electron transfer within DNA could possibly explain how a single electron hit could result in a DSB.59 They suggested that a TNI formed on the phosphate group decays by autoionization, leaving the CO unit in the backbone in an electronic excited state that dissociates breaking the CO bond. Simultaneously, the departing electron transfers to the opposite strand and forms a transient anion of the phosphate group causing a CO bond on the other chain to rupture by DEA. Such breaks could easily occur within a distance of 10 bp, causing DSB. Within this hypothesis, enhancement of DSB could have many causes related to the structural distortion of DNA (Figure 1), which could favor not only the capture of electrons by bases and intrastrand electron transfer to the backbone but also interstrand electron transfer.
5. SUMMARY
Thin films of cisplatin–DNA complexes were bombarded with 1.6–19.6 eV electrons in order to investigate the role of TNI and the secondary LEEs, generated in copious amounts by high energy radiation, in the radiosensitization of DNA by Pt chemotherapeutic agents. The resulting SSB, DSB, interduplex CL, and loss of the initial supercoiled cisplatin–DNA (3197 bp) configuration were analyzed by agarose gel electrophoresis. The effective yield of damage to cisplatin–DNA complexes at 11 different electron energies were obtained from the corresponding exposure–response curves.
The covalent binding of five cisplatin molecules per DNA considerably enhances all types of damages investigated. All yield functions exhibit strong peaks due to the decay of core-excited resonances into destructive channels. Binding of the drug increases the damage induced by pre-existing resonances by factors of 1.4–2.3, depending on resulting configuration and electron energy. The enhancement of damage via pre-existing dissociative anion states most likely results from modifications of the parameters of these transient states. The creation of new resonances by cisplatin gives enhancement factors varying from 2.2 to 3.2. The ensemble of the results indicates that radiosensitization of DNA by cisplatin can at least be partially interpreted by the preferential enhancement of dissociative channels of TNI and the creation of new dissociative TNI.
According to the present results, cisplatin enhances preferentially the formation of cluster damage to DNA (i.e., CL and DSB) induced by the direct effect of ionizing radiation. Interduplex CL and DSB belong to the most critical radiobiological lesions, since they are difficult to repair by the cell. Unrepaired and misrepaired lesions can have severe biological consequences, such as loss of genetic information, mutation, promotion of genomic instability, and cell death.60 In the case of malignant cells, such lesions play an important role in cancer treatment by chemoradiation therapy. As recently shown in animal studies with cisplatin and a liposomal formulation of the drug,30 a fundamental understanding the biological action of LEEs with22,23 and without1 chemical modification of DNA can lead to the development of more efficient clinical protocols and radiosensitizing chemotherapeutic agents.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the Canadian Institutes of Health Research (MOP81356), the China Award Program of Minjiang Scholar Professorship, National Basic Research Program of China (973 Program: 2013CB632405), the Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT0818), and the NNSF of China (21077023). Thanks are extended to Professor Darel Hunting and Dr. Andrew Bass for corrections and helpful comments.
Footnotes
Notes
The authors declare no competing financial interest.
Analysis of 1% and 1.3% agarose gel eletrophoresis of cisplatin–DNA complexes used in this study. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Sanche L. Low-Energy Electron Interaction with DNA: Bond Dissociation and Formation of Transient Anions, Radicals and Radical Anions, Radicals in Nucleic Acids. In: Greenberg M, editor. Radical and Radical Ion Reactivity in Nucleic Acid Chemistry. Wiley; Hoboken, NJ: 2009. pp. 239–294. [Google Scholar]
- 2.Arumainayagam CR, Lee HL, Nelson RB, Haines DR, Gunawardane RP. Low-Energy Electron-induced Reactions in Condensed Matter. Surf Sci Rep. 2010;65:1–44. [Google Scholar]
- 3.Alizadeh E, Sanche L. Precursors of Solvated Electrons in Radiobiological Physics and Chemistry. Chem Rev. 2012;112:5578–5602. doi: 10.1021/cr300063r. [DOI] [PubMed] [Google Scholar]
- 4.Boudaıffa B, Cloutier P, Hunting D, Huels MA, Sanche L. Resonant Formation of DNA Strand Breaks by Low-Energy (3 to 20 eV) Electrons. Science. 2000;287:1658–1660. doi: 10.1126/science.287.5458.1658. [DOI] [PubMed] [Google Scholar]
- 5.Martin F, Burrow PD, Cai Z, Cloutier P, Hunting D, Sanche L. DNA Strand Breaks Induced by 0–4 eV Electrons: The Role of Shape Resonances. Phys Rev Lett. 2004;93:068101. doi: 10.1103/PhysRevLett.93.068101. [DOI] [PubMed] [Google Scholar]
- 6.Boulanouar O, Fromm M, Bass AD, Cloutier P, Sanche L. Absolute Cross Section for Loss of Supercoiled Topology Induced by 10 eV Electrons in Highly Uniform/DNA/1,3-diaminopropane Films Deposited on Highly Ordered Pyrolitic Graphite. J Chem Phys. 2013;139:055104. doi: 10.1063/1.4817323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Aleksandrov A, Orlando TM. Probing Low-Energy Electron Induced DNA Damage Using Single Photon Ionization Mass Spectrometry. Int J Mass Spectrom. 2008;277:314–320. [Google Scholar]
- 8.Kumar SV, Pota T, Peri D, Dongre AD, Rao B. Low Energy Electron Induced Damage to Plasmid DNA pQE30. J Chem Phys. 2012;137:045101. doi: 10.1063/1.4737182. [DOI] [PubMed] [Google Scholar]
- 9.Luo X, Zheng Y, Sanche L. DNA Strand Breaks and Crosslinks Induced by Transient Anions in the Range 2–20 eV. J Chem Phys. 2014;140:155101. doi: 10.1063/1.4870519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abdoul-Carime H, Sanche L. Fragmentation of Short Single DNA Strands by 1–30 eV Electrons: Dependence on Base Identity and Sequence. Int J Radiat Biol. 2002;78:89–99. doi: 10.1080/09553000110090016. [DOI] [PubMed] [Google Scholar]
- 11.Solomun T, Seitz H, Sturm H. DNA Damage by Low-Energy Electron Impact: Dependence on Guanine Content. J Phys Chem B. 2009;113:11557–11559. doi: 10.1021/jp905263x. [DOI] [PubMed] [Google Scholar]
- 12.Baccarelli I, Bald I, Gianturco FA, Illenberger E, Kopyra J. Electron-Induced Damage of DNA and Its Components: Experiments and Theoretical Models. Phys Rep. 2011;508:1–44. [Google Scholar]
- 13.Kopyra J. Low Energy Electron Attachment to the Nucleotide Deoxycytidine Monophosphate: Direct Evidence for the Molecular Mechanisms of Electron-Induced DNA Strand Breaks. Phys Chem Chem Phys. 2012;14:8287–8289. doi: 10.1039/c2cp40847c. [DOI] [PubMed] [Google Scholar]
- 14.Baccarelli I, Gianturco FA, Grandi A, Sanna N, Lucchese RR, Bald I, Kopyra J, Illenberger E. Selective Bond Breaking in Beta-D-ribose by Gas-phase Electron Attachment Around 8 eV. J Am Chem Soc. 2007;129:6269–77. doi: 10.1021/ja070542h. [DOI] [PubMed] [Google Scholar]
- 15.Ptasińska S, Denifl S, Grill V, Märk TD, Scheier P, Gohlke S, Huels MA, Illenberger E. Bond-Selective H− Ion Abstraction from Thymine. Angew Chem, Int Ed. 2005;44:1647–1650. doi: 10.1002/anie.200461739. [DOI] [PubMed] [Google Scholar]
- 16.Abdoul-Carime H, Langer J, Huels MA, Illenberger E. Decomposition of Purine Nucleobases by Very Low Energy Electrons. Eur Phys J D. 2005;35:399–404. [Google Scholar]
- 17.Huber D, Beikircher M, Denifl S, Zappa F, Matejcik S, Bacher A, Grill V, Märk TD, Scheier P. High Resolution Dissociative Electron Attachment to Gas Phase Adenine. J Chem Phys. 2006;125:084304. doi: 10.1063/1.2336775. [DOI] [PubMed] [Google Scholar]
- 18.Ptasińska S, Denifl S, Grill V, Märk TD, Illenberger E, Scheier P. Bond- and Site-Selective Loss of H− from Pyrimidine Bases. Phys Rev Lett. 2005;95:093201. doi: 10.1103/PhysRevLett.95.093201. [DOI] [PubMed] [Google Scholar]
- 19.Berdys J, Anusiewicz I, Skurski P, Simons J. Damage to Model DNA Fragments from Very Low-Energy (<1 eV) Electrons. J Am Chem Soc. 2004;126:6441–6447. doi: 10.1021/ja049876m. [DOI] [PubMed] [Google Scholar]
- 20.Caron L, Sanche L. In: Low-Energy Electron Scattering from Molecules, Biomolecules and Surfaces. Čársky P, Čurik R, editors. CRC Press; Boca Raton, FL: 2012. [Google Scholar]
- 21.Zheng Y, Hunting DJ, Ayotte P, Sanche L. Role of Secondary Electrons in the Concomitant Chemoradiation Therapy of Cancer. Phys Rev Lett. 2008;100:198101. doi: 10.1103/PhysRevLett.100.198101. [DOI] [PubMed] [Google Scholar]
- 22.Rezaee M, Hunting D, Sanche L. New Insights into the Mechanism Underlying the Synergistic Action of Ionizing Radiation with Platinum Chemotherapeutic Drugs: the Role of Low-Energy Electrons. Int J Radiat Oncol Biol Phys. 2013;87:847–853. doi: 10.1016/j.ijrobp.2013.06.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rezaee M, Alizadeh E, Cloutier P, Hunting D, Sanche L. A Single Subexcitation-Energy Electron Can Induce a Double-Strand Break in DNA Modified by Platinum Chemotherapeutic Drugs. ChemMedChem. 2014;9:1145–1149. doi: 10.1002/cmdc.201300462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng Y, Sanche L. Low Energy Electrons in Nanoscale Radiation Physics: Relationship to Radiosensitization and Chemoradiation Therapy. Rev Nanosci Nanotechnol. 2013;2:1–28. [Google Scholar]
- 25.Saif MW, Diato RB. Fluoropyrimidines as Radiation Sensitizers. In: Choy H, editor. Chemoradiation in Cancer Therapy. Humana Press; Totowa, NJ: 2003. pp. 23–45. [Google Scholar]
- 26.DeVita VT Jr, Lawrence TS, Rosenberg SA, editors. Cancer: Principles and Practice of Oncology. Lippincott Williams & Wilkins; Philadelphia: 2011. [Google Scholar]
- 27.Wang D, Lippard SJ. Cellular Processing of Platinum Anticancer Drugs. Nat Rev Drug Discovery. 2005;4:307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 28.Vrana O, Boudny V, Brabec V. Superhelical Torsion Controls DNA Interstrand Cross-linking by Antitumor Cis-diamminedichloroplatinum(II) Nucleic Acids Res. 1996;24:3918–3925. doi: 10.1093/nar/24.20.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung Y, Lippard SJ. Direct Cellular Responses to Platinum-induced DNA Damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 30.Tippayamontri T, Kotb R, Paquette B, Sanche L. Efficacy of Cisplatin and Lipoplatin in Combined Treatment with Radiation of a Colorectal Tumor in Nude Mouse. Anticancer Res. 2013;33:3005–3014. [PubMed] [Google Scholar]
- 31.Seiwert TY, Salama JK, Vokes EE. The Concurrent Chemoradiation Paradigm-General Principles. Nat Clin Pract Oncol. 2007;4:86–100. doi: 10.1038/ncponc0714. [DOI] [PubMed] [Google Scholar]
- 32.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons Inc; New York: 2003. [Google Scholar]
- 33.Gel Filtration - Principles and Methods Handbook. GE Healthcare; Pataskala, OH: 2007. [Google Scholar]
- 34.Malina J, Vrana O, Brabec V. Mechanistic Studies of the Modulation of Cleavage Activity of Topsoisomerase I by DNA Adducts of Mono- and Bi-functional PtII Complexes. Nucleic Acids Res. 2009;37:5432–5442. doi: 10.1093/nar/gkp580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usami N, Kobayashi K, Furusawa Y, Frohlich H, Lacombe S, Sech CL. Irradiation of DNA Loaded with Platinum Containing Molecules by Fast Atomic Ions C(6+) and Fe(26+) Int J Radiat Biol. 2007;83:569–576. doi: 10.1080/09553000701447130. [DOI] [PubMed] [Google Scholar]
- 36.Razaee M, Alizadeh E, Hunting D, Sanche L. DNA Platination Thin Films for Use in Chemoradiation Therapy Studies. Bioinorg Chem Appl. 2012;2012:923914. doi: 10.1155/2012/923914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouwers EEM, Tibben M, Rosing H, Schellens JHM, Beijnen JH. The Application of Inductively Coupled Plasma Mass Spectrometry in Clinical Pharmacological Oncology Research. Mass Spectrosc Rev. 2008;27:67–100. doi: 10.1002/mas.20159. [DOI] [PubMed] [Google Scholar]
- 38.Fashman GD, editor. Handbook of Biochemistry and Molecular Biology. CRC Press; Boca Raton, FL: 1995. [Google Scholar]
- 39.Meesungnoen J, Jay-Gerin JP, Filali-Mouhim A, Mankhetkorn S. Low-Energy Electron Penetration Range in Liquid Water. Radiat Res. 2002;158:657. doi: 10.1667/0033-7587(2002)158[0657:leepri]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Rezaee M, Cloutier P, Bass AD, Hunting DJ, Sanche L. Absolute Cross Section for Low-energy Damage to Condensed Macromolecules: a Case Study of DNA. Phys Rev E. 2012;86:031913. doi: 10.1103/PhysRevE.86.031913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aflatooni K, Gallup GA, Burrow PD. Electron Attachment Energies of the DNA Bases. J Phys Chem A. 1998;102:6205. [Google Scholar]
- 42.Lévesque PL, Michaud M, Cho W, Sanche L. Absolute Vibrational and Electronic Cross Sections for Low-Energy Electron (2–12 eV) Scattering from Condensed Pyrimidines. J Chem Phys. 2005;122:224704-1–7. doi: 10.1063/1.1854121. [DOI] [PubMed] [Google Scholar]
- 43.Bazin M, Michaud M, Sanche L. Absolute Cross Sections for Electronic Excitations of Cytosine by Low Energy Electron Impact. J Chem Phys. 2010;133:155104. doi: 10.1063/1.3505301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Behmand B, Cloutier P, Girouard S, Wagner RJ, Sanche L, Hunting DJ. Hydrated Electrons Reacting with High Specificity with Cisplatin Bound to Single-Stranded DNA. J Phys Chem B. 2013;117:15994–15999. doi: 10.1021/jp4089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Behmand B, Wagner RJ, Sanche L, Hunting DJ. Cisplatin Intrastrand Adducts Sensitize DNA to Base Damage by Hydrated Electrons. J Phys Chem B. 2014;118:4803–4808. doi: 10.1021/jp5014913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito T, Rokita SE. Criteria for Efficient Transport of Excess Electrons in DNA. Angew Chem. 2004;116:1875–1878. doi: 10.1002/anie.200353038. [DOI] [PubMed] [Google Scholar]
- 47.Kopyra J, Koening-Lehmann C, Bald I, Illenberger E. A Single Slow Electron Triggers the Loss of Both Chlorine Atoms from the Anticancer Drug Cisplatin: Implications for Chemoradiation Therapy. Angew Chem, Int Ed. 2009;48:7904–7907. doi: 10.1002/anie.200903874. [DOI] [PubMed] [Google Scholar]
- 48.Cai Z, Cloutier P, Sanche L, Hunting D. DNA Interduplex Crosslinks Induced by AlKα X rays under Vacuum. Radiat Res. 2005;164:173–179. doi: 10.1667/rr3408. [DOI] [PubMed] [Google Scholar]
- 49.Abdoul-Carime H, Sanche L. Sequence Specific Damage to Oligonucleotides Induced by 3–30 eV Electrons. Radiat Res. 2001;156:151–157. doi: 10.1667/0033-7587(2001)156[0151:ssdibt]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 50.Mozejko P, Bass AD, Parenteau L, Sanche L. Intrinsic and Extrinsic Factors in Anion Electron Stimulated Desorption: D− from Deuterated Hydrocarbons Condensed on Kr and Water Ice Films. J Chem Phys. 2004;121:10181–10189. doi: 10.1063/1.1807813. [DOI] [PubMed] [Google Scholar]
- 51.Todd RC, Lippard SJ. Inhibition of Transcription by Platinum Antitumor Compounds. Metallomics. 2009;4:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jamieson ER, Lippard SJ. Stucture, Recognition, and Processing of Cisplatin-DNA Adducts. Chem Rev. 1999;99:2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 53.O’Malley TF. Theory of Dissociative Attachment. Phys Rev. 1966;150(1):14–29. [Google Scholar]
- 54.Massey HSW. Negative Ions. Cambridge University Press; London: 1976. p. 741. [Google Scholar]
- 55.Ning Z, Polanyi JC. Charge Delocalization Induces Reaction in Molecular Chains at a Surface. Angew Chem, Int Ed. 2013;52:320–324. doi: 10.1002/anie.201207819. [DOI] [PubMed] [Google Scholar]
- 56.Huang K, Leung L, Lim T, Ning Z, Polanyi JC. Single-Electron Induces Double-reaction by Charge Delocalization. J Am Chem Soc. 2013;135:6220–6225. doi: 10.1021/ja400612c. [DOI] [PubMed] [Google Scholar]
- 57.Dizdaroglu M, Jaruga P. Mechanisms of Free Radical-induced Damage to DNA. Free Radical Res. 2012;46:382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 58.Kumar A, Sevilla MD. Proton-coupled Electron Transfer (PCET) in DNA on Formation of Radiation Produced Ion Radicals. Chem Rev. 2010;110:7002–7023. doi: 10.1021/cr100023g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng Y, Sanche L. Influence of Organic Ions on DNA Damage Induced by 1 eV to 60 keV Electrons. J Chem Phys. 2010;133:155102. doi: 10.1063/1.3505046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Driscoll M, Jeggo PA. The Role of Double-Strand Break Repair - Insights from Human Genetics. Nat Rev Gen. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


