Summary
Influenza is a contagious respiratory acute viral disease characterized by a short incubation period, high fever and respiratory and systemic symptoms.
The burden of influenza is very heavy. Indeed, the World Health Organization (WHO) estimates that annual epidemics affect 5-15% of the world's population, causing up to 4-5 million severe cases and from 250,000 to 500,000 deaths.
In order to design anti-influenza molecules and compounds, it is important to understand the complex replication cycle of the influenza virus. Replication is achieved through various stages. First, the virus must engage the sialic acid receptors present on the free surface of the cells of the respiratory tract. The virus can then enter the cells by different routes (clathrin-mediated endocytosis or CME, caveolae-dependent endocytosis or CDE, clathrin-caveolae-independent endocytosis, or macropinocytosis). CME is the most usual pathway; the virus is internalized into an endosomal compartment, from which it must emerge in order to release its nucleic acid into the cytosol. The ribonucleoprotein must then reach the nucleus in order to begin the process of translation of its genes and to transcribe and replicate its nucleic acid. Subsequently, the RNA segments, surrounded by the nucleoproteins, must migrate to the cell membrane in order to enable viral assembly. Finally, the virus must be freed to invade other cells of the respiratory tract. All this is achieved through a synchronized action of molecules that perform multiple enzymatic and catalytic reactions, currently known only in part, and for which many inhibitory or competitive molecules have been studied. Some of these studies have led to the development of drugs that have been approved, such as Amantadine, Rimantadine, Oseltamivir, Zanamivir, Peramivir, Laninamivir, Ribavirin and Arbidol. This review focuses on the influenza lifecycle and on the currently available drugs, while potential antiviral compounds for the prevention and treatment of influenza are considered in the subsequent review.
Key words: Influenza, Influenza life cycle, Antiviral, Licensed drugs, Amantadine, Rimantadine, Oseltamivir, Zanamivir, Peramivir, Laninamivir, Ribavirin, Arbidol
Introduction
Influenza is a contagious acute respiratory viral disease characterized by a short incubation period, high fever, respiratory (e.g. runny and stuffy nose) and systemic symptoms (e.g. muscle or body aches) [1]. Most of the people affected by influenza recover in a few days or, at most, in 2 weeks. However, some patients may develop complications that may be very serious. The most common complications are bronchitis, pneumonia, ear infections, etc. People with underlying diseases, such as asthma, subjects with Chronic Obstructive Pulmonary Disease (COPD) or individuals with heart disease are at high risk of complications [2]. Related complications, such as myositis, acute encephalopathy or Reye's syndrome, are rare [3]. Reye's syndrome is classically characterized by rashes, vomiting and liver damage. It can typically occur during viral illness in children who have been taking aspirin for a long period [4].
Pneumonia can be caused by bacterial superinfection also called secondary pneumonia or viral pneumonitis [5, 6]. This is characterized by the complex interactions of host-co-infecting pathogens [7], and, in particularly frail and debilitated subjects, can result in the impairment of physical capabilities, dysregulation of immune responses and a delayed return to homeostasis [5, 6].
The burden of influenza is very heavy. Indeed, the World Health Organization (WHO) estimates that annual epidemics affect 5-15% of the world's population, causing up to 4-5 million severe cases and from 250,000 to 500,000 deaths [7]. The European Centre for Disease Prevention and Control (ECDC) estimates that approximately 10% of Europeans are infected each year [8]. Furthermore, the US government estimates that 5-20% of US residents catch influenza each year [9].
Influenza viruses belong to the Ortomyxoviridae family [10], with the other two genera being Isavirus and Thogotovirus, and have the ability to change their surface antigens relatively frequently [11]. When a major variation occurs, if the virus adapts to humans during zoonotic spill-over, widespread diffusion of the virus is possible, resulting in a pandemic [12]. The most severe influenza pandemic was that of 1918, which caused 500 million cases and from 50 to 100 million deaths [13, 14]. During that devastating pandemic, the treatment of patients suffering from influenza was empirical and symptomatic, and was intended primarily to relieve fever and pain (e.g. aspirin administration), while epinephrine was used to treat forms of secondary pneumonia [15]. Only in the 1960s did the first antiviral drug against influenza, namely Amantadine, become available in the US [16, 17], while in 1993 another drug, Rimantadine, was authorized [18]. Later, in 1999, the anti-neuraminidase (NA) medications Zanamivir and Oseltamivir were both licensed in the US [19].
Influenza viruses belong to the Ortomyxoviridae family [10], with the other two genera being Isavirus and Thogotovirus, and have the ability to change their surface antigens relatively frequently [11]. When a major variation occurs, if the virus adapts to humans during zoonotic spill-over, widespread diffusion of the virus is possible, resulting in a pandemic [12]. The most severe influenza pandemic was that of 1918, which caused 500 million cases and from 50 to 100 million deaths [13, 14]. During that devastating pandemic, the treatment of patients suffering from influenza was empirical and symptomatic, and was intended primarily to relieve fever and pain (e.g. aspirin administration), while epinephrine was used to treat forms of secondary pneumonia [15]. Only in the 1960s did the first antiviral drug against influenza, namely Amantadine, become available in the US [16, 17], while in 1993 another drug, Rimantadine, was authorized [18]. Later, in 1999, the anti-neuraminidase (NA) medications Zanamivir and Oseltamivir were both licensed in the US [19].
Since 1999, much knowledge concerning viral replication has been acquired, and new experimental hypotheses have been advanced for the development of new flu drugs and new protocols for both prevention and treatment. Anti-influenza drugs are an important complement to vaccination, which is the most efficacious weapon against the disease. In this review, it therefore seemed useful to deal with the issue of new/potential antiviral medications against influenza infections, especially in the light of the most recent scientific advances.
Biology of influenza viruses
Regarding the antigenic characteristics of the core proteins (nucleoproteins [NP] and Matrix proteins [M proteins]), three influenza virus types have been identified: A, B and C. Given the relevance of Influenza A Virus (IAV) to human pathology, we will provide a brief overview of its biology and life-cycle and underline the main differences among the three virus types in terms of structural and molecular biology.
The IAV particle varies in the range of 80-120 nm and is pleomorphic, being usually spherical, though cordlike forms with a diameter of 40-100 nm and a length in the range of 300 nm-20 μm can occur [11, 20, 21]. Transition from spherical to tubular form is not well understood: what is known so far is that M1 [22, 23], M2 [23, 24] and NP [25] could play a role in determining and modulating this process. Besides genetic traits, also the phenotype of the host cell, in terms of shape and polarization, seems to influence the viral form [21, 26]. Influenza B has a similar shape, being structurally indistinguishable from IAV [11], while Influenza C virus is usually filamentous and 500 nm long [11]. The IAV viral particle is an envelope made up of lipid rafts and spikes of two main types of glycoproteins: hemagglutinin (HA) accounts for about 80% (about 500 molecules) and NA for about 17% (about 100 molecules); M2 is the least abundant protein, with only 16-20 copies per virion [11, 27].
The particle of influenza B virus contains four proteins, namely HA, NA, NB, and BM2 [11, 28], while the particle of influenza C virus is made up of a major surface glycoprotein (HEF, hemagglutinin-esterase-fusion protein) and a minor surface glycoprotein (CM2). These surface glycoproteins form ordered hexagonal arrays [11, 29-32].
Underneath the viral membrane, M1 tightly binds the vRNPs, which consist of RNA strands (usually single strands but in certain cases double strands) [11] wrapped around NP and NEP, with a terminal polymerase ternary complex (PA, PB1, PB2). The genome is small (about 13-16 kilobases) and contains seven or eight pieces of segmented negative-sense RNA (eight segments for IAV and influenza B, seven for influenza C), each piece of RNA containing either one or two genes of 890 to 2,341 nucleotides each [11]. The genome codes for at least 11 proteins: hemagglutinin (HA) of about 76-77 kDa, NA of about 60 kDa, NP of about 60 kDa, M1 of about 28 kDa, M2 of about 15 kDa, non-structural protein type 1 (NS1) of about 26 kDa, non-structural protein type 2 (NS2) (also known as NEP: nuclear export protein) of about 11 kDa, PA of about 85 kDa, PB1 (polymerase basic type 1) of about 88 kDa, PB1-F2 of about 80 kDa [33-35] and PB2 (polymerase basic type 2) of about 91 kDa [36].
In particular, segments 1-3 code for the ternary polymerase complex (PB1, PB2 and PA, respectively), segment 4 for HA, segment 5 for NA, segment 6 for NP, segment 7 for the matrix proteins, and segment 8 for the non-structural proteins [11]. On the basis of the gene structures, segments can be divided into three classes: intronless, intron-containing and unspliced, and introncontaining and spliced. On the basis of the kinetics of the gene expression, they can be classified into "early" (segments 1-3, 5 and the unspliced segment 8 transcript) and "late" (segments 4, 6, 7 and spliced segment 8) classes. Usually, structural and functional/kinetic classifications do not correspond [37, 38].
Moreover, the presence of overlapping genes and different splicing mechanisms may give rise to further accessory proteins, such as PB1-N40 [39], PA-X [39-40], PA-N155 [41], PA-N182 [42], M42 [43], and NS3 [43], which have been discovered and characterized only recently. Another accessory protein, NEG8 ORF, has been predicted [44].
The viral proteome thus reveals unexpected dynamism and complexity [43-44]; Matsuoka and coll. have designed a comprehensive map of influenza interactions, termed FluMap, which contains 960 factors and 456 reactions as of April 2012 [47].
Replication cycle of influenza viruses
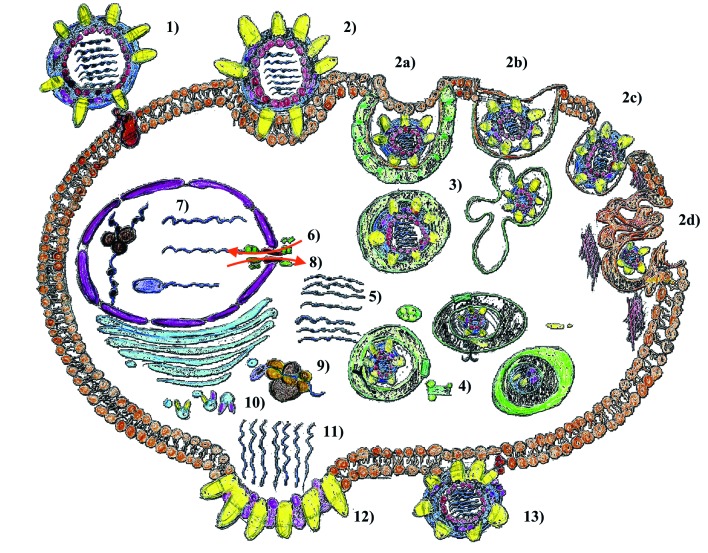
The replication cycle of the virus (Fig. 1) is a complex, highly dynamic, biological process which consists of the following steps: 1) attachment of the virion to target cells and receptor binding (virus adsorption); 2) internalization into cellular regions by means of clathrin-mediated endocytosis (CME), caveolae-dependent endocytosis (CDE), clathrin-caveolae-independent endocytosis, and macropinocytosis; 3) endosomal trafficking via endosomes / caveosome / macropinosome / lysosomes to the perinuclear compartment; 4) pH-dependent fusion of viral and endosomal / organellar membranes; 5) uncoating; 6) nuclear importation; 7) transcription and replication; 8) nuclear exportation; 9) protein synthesis; 10) post-translational processing and trafficking; 11) viral progeny assembly and packaging; 12) budding; and 13) release (modified from [48]).
Fig. 1.
Schematic representation of the replication cycle of the influenza: 1) attachment of the virion to target cells and receptor binding (virus adsorption); 2) internalization into cellular regions by means of clathrin-mediated endocytosis (CME), caveolae-dependent endocytosis (CDE), clathrin-caveolae-independent endocytosis, and macropinocytosis; 3) endosomal trafficking via endosomes / caveosome / macropinosome / lysosomes to the perinuclear compartment; 4) pH-dependent fusion of viral and endosomal / organellar membranes; 5) uncoating; 6) nuclear importation; 7) transcription and replication; 8) nuclear exportation; 9) protein synthesis; 10) post-translational processing and trafficking; 11) viral progeny assembly and packaging; 12) budding; and 13) release. For further details, the reader is referred to the text.
The cells infected by the influenza virus are: alveolar and bronchial epithelial tissue (BET) cells, alveolar macrophages (AM), lung epithelial tissue (LET) cells and, in particular, type II pneumocytes, plasmacytoid dendritic cells (pDCs) and natural killer cells (NKs) [49, 50].
Influenza virus is able to activate Endoplasmic Reticulum (ER) stress, caspase pathway [51] or to finely tune host secreted molecules, such as lung mucins [52], in order to avoid being trapped and subsequently eliminated. Moreover, it recruits host factors and misuses them [53]. Although the mechanisms of influenza virus replication are not fully understood, scientific projects for new drugs against influenza cannot ignore the biological cycle of this virus.
INFLUENZA VIRUS ENTRY
The first important event during infection in humans is the attachment of influenza virions to the apical cell surface (event known also as virus adsorption). Indeed, the entry of the Influenza virus into target cells is an essential process whereby viral genomes are delivered from extracellular virions to sites of transcription/replication in the cell nucleus [54]. During this phase, thanks to the surface glycoprotein HA, the virus interacts with (-2,3)- or (-2,6)-linked sialic acid receptors [55]. The physicochemical conformation of these receptors is not identical in different species of animals – humans, seals, birds, pigs, horses, etc., which are the natural reservoir of the virus. The vast majority of human receptors are located in the upper respiratory tract, but man also possesses receptors typical of birds, which are located deep in the respiratory tract [56].
HA is a homotrimeric integral type 1 membrane cylinder- like glycoprotein, approximately 13.5 nanometres long. HA monomers are synthesized as precursors (HA0) containing a hydrophobic signal sequence. After being translated, they are glycosylated and cleaved into two smaller subunits: namely, HA1 of 50 kDa and HA2 of 27 kDa, which are linked by a disulfide bridge. Each subunit is characterized by a long, central, α-helix connected to the membrane by HA2 and surmounted by HA1, a spherical head containing the sialic acid binding sites (receptor binding sites or RBSs, also known as receptor binding pocket or RBP) [57]. The apolar domain of HA2, near the cleavage site, is known as the "fusion peptide" or HAfp23 [58, 59], since it is characterized by a domain of highly conserved N-terminal 23 residues. HAfp23 has a helical-hairpin structure consisting of two tightly-packed helices, which are fundamental to inducing the negative curvature ("fusogenic conformation" of HA) [60].
There are at least eighteen HA subtypes [61]. These are further subdivided into two groups: group 1 comprises H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18 (from waterfowl, the last two having been recently isolated from bats in Guatemala and Peru) [61, 62]; group 2 comprises H3, H4, H7, H10, H14, and H15 [62]. However, a recent experiment has proved that the virus can also enter into cells whose surface has been completely depleted of sialic acid-based glycoproteins and glycolipids [63]. This seems to suggest alternative entry routes. Indeed, H17 and H18 do not bind to sialic acid and their receptor is still unknown [64].
Entry is essentially through receptor-mediated endocytosis [65], though an alternative uptake pathway, namely macropinocytosis, has quite recently been discovered [66-68]. Receptor-mediated endocytosis can be CME or mediated by lipid rafts: CDE or non-clathrin non-caveolae endocytosis [69-71].
CME is the most common pathway through which the virion is internalized. The clathrin triskelion is made up of three heavy chains, which constitute the backbone of the polyhedral structure, and of three light chains, which finely tune the assembly / disassembly of the triskelion [72]. Adaptor proteins, such as AP-2 [73], epsin- 1 [74], epsin-15, recognize specific internalization signals located on cargo receptors, and take part in the formation of clathrin-coated pits (CCPs).
Caveolae are small flask-like infoldings of the membrane, with high levels of cholesterol and glycosphingolipids and with caveolin as the integral membrane scaffolding structure. The remaining entry routes have been investigated less. In clathrin caveolae-independent endocytosis, the Ras-phosphoinositide 3-kinase (PI3K) signalling pathway may play a major role [75].
The PI3K inactive complex (PI3K p110-p85 heterodimer) moves to the plasma membrane, where the SH2 (Src Homology 2) domains of p85 engage the phosphotyrosine residues present in receptor-associated proteins, causing a molecular rearrangement of the complex, in such a way that p110 is now enzymatically active and can recruit / produce intracellular second messenger, such as PtdIns(3,4,5)P3 (phosphatidylinositol-(3,4,5)- trisphosphate), from PtdIns(4,5)P2 (phosphatidylinositol-( 4,5)-bisphosphate). Subsequently, several effector proteins with pleckstrin homology (PH) domains mediate an array of different signalling cascades: namely, activation of Akt, mTOR (mammalian Target Of Rapamycin), PKC (Protein Kinase C), PTEN (Phosphatase and tensin homolog) pathways [76].
Macropinocytosis can be activated by cell growth factors such as the Epidermal Growth Factor (EGF) or Nerve Growth Factor (NGF) or can be induced by phorbol esters. Macropinocytosis is Rac1-, PAK1- (p21-activated kinase type 1), cholesterol- and actin-dependent, since it extensively reorganizes the plasma membrane and implies energy mechanisms, such as Na+/H+ exchanger (NHE) activity [77, 78]. Also other pathogens such as Ebolavirus [79, 80], arenavirus [81], adenovirus [82, 83], HPV (Human Papillomavirus) [84], HIV (Human Immunodeficiency Virus) [85], Escherichia coli [86], Trypanosoma cruzi [87], vaccinia virus [88], African Swine Fever virus [89], RSV (Respiratory Syncytial Virus) [90] and HCV (Hepatitis C Virus) [91], among the others, use macropinocytosis as additional entry pathways.
These different routes (CME, CDE, non-clathrin noncaveolae endocytosis and macropinocytosis) result in different internalization yields and may be related to the different degrees of virulence and pathogenicity of influenza strains [92].
ENDOSOMAL TRAFFICKING
After attaching to the respiratory cell surface, the influenza virus, as already mentioned, can penetrate into the cell through more than one mechanism, and the final result is that the virions are internalized in an endosomal compartment [93]. According to the previous routes, virions can be packaged into CCPs and clathrin-coated vesicles (CCVs) or into caveosomes / macropinosomes. Subsequently, they are further processed into early endosomes (EEs) and into late endosomes (LEs) [94].
Endosomal trafficking usually follows influenza virus uptake. Virions are taken up into Rab-5/Vps4 and Rab- 7/LAMP (lysosomal-associated membrane protein) positive endosomes. Rab-5/Vps4 and Rab-7 are Rab guanosine triphosphatases (GTPases) that are responsible for the processes of trafficking and fusion for EEs and LEs, respectively [95-97].
LAMPs are a series of proteins – 3 isoforms are currently known, namely, LAMP1 (known also as CD107a), LAMP2, and LAMP3 (DC-LAMP, TSC403 or CD208) – that are involved in endosomal maturation [98]. Moreover, the expression of LAMP3 has been found to be predictive of the host's response to influenza [99].
Cullin-3 (Cul3) belongs to the Cullin-RING-ligase family (CRL). It functions as a scaffolding protein in the Bric-a-brac, Tramtrack, Broad-complex (BTB)-Cul3- Rbx1 ubiquitin E3 ligase complex and is involved in LE maturation and cargo trafficking [100, 101].
Endosomal trafficking is a complex multifaceted process that can be divided into three stages: stage I, which is actin-dependent, stage II, which is dynein- and microtubule- dependent, and stage III, which is microtubuledependent [102].
INFLUENZA ENVELOPE FUSION
In order for the virus to emerge from the endosome and for ribonucleoprotein (RNP) to be released into the cytosol, the envelope must fuse with the endosome membrane. This part of the life-cycle of the virus requires the acidity of the lumen of the endosome to decrease to a pH value of about 5 [103, 104]. In this process, a crucial role is played by M2, which not only acts on ion channels, thereby allowing acidification of the interior of the virus [105], but also alters its conformation, resulting in changes of the curvature of the viral envelope [106, 107]. This leads to two important events, namely: the dissociation of the M1 protein from RNP and a dramatic change in the conformation of HA [108], which can expose its fusion peptide. This peptide may allow fusion of the viral envelope and the endosome membrane and the release of RNP into the cytoplasm. The cleavage of HA occurs through a proteasic enzymatic action [109]. Proteases that can cause the cleavage of HA are widely distributed throughout the human body, and the precise role that each plays in cutting the HA is not exactly known. Nevertheless, proteases are known to belong to two main classes, namely trypsin-like enzymes or furinlike serine enzymes [109]. These enzymes are produced by the cells, particularly those of the respiratory system, in the presence of inflammation (proinflammatory cytokines/ chemokines, neutrophils, etc.) [110].
The release of RNP into the cytosol is also enabled by the host's aggresome processing machinery (made up of dynein, dynactin and myosin II) [111]. Further molecules and pathways could take part in this process.
M2 is more than just a simple ion channel. Indeed, it plays a multifaceted role in the entire viral life-cycle, as the mechanism of proton permeation, conductance and acidification is crucial to each different step and activity of the virus. After endocytosis, the low endosomal pH (in the range 5-6) activates the M2 channel prior to HA-mediated fusion, and favours dissociation of the M1 protein from the vRNPs, thereby facilitating the entry of RNPs into the nucleus. M2 is fundamental to genome unpacking and the release of the viral RNA during viral uncoating [112]. Moreover, M2 enables the viral RNA to package and facilitates virion scission and budding during viral maturation and assembly, replication and infection: during transport to the cell surface, in the trans-Golgi network (TGN) membrane, M2 acts as a proton-leak channel and prevents the activation of de novo synthesized HA by regulating the pH of the Golgi apparatus of the host cell and equilibrating its pH with the pH of the viral interior. Indeed, if it were not elevated in this way, the low pH would cause a conformational rearrangement of HA, whose intracellular cleavage would prematurely inactivate the new influenza viral progeny [113, 114]. M2, together with other proteins such as NS1 and HA, could play an additional role of fine-tuning the apoptosis of infected cells, thus favouring viral replication [115].
NS1 is involved in several biological tasks, such as mRNA splicing and translation, cell survival, and immune defence. In particular, as far as the type I interferon (IFN-I)-mediated response is concerned, it interacts with PACT/PRKRA (Protein activator of the interferon- induced protein kinase / Protein kinase, interferoninducible double stranded RNA dependent activator), which is an important cofactor for the IFN-I response elicited by the viral RNA-sensor RIG-I (Retinoic acid- Inducible Gene I). Therefore, it blocks PACT/RIG-Imediated activation of IFN-I [116, 117]. Moreover, it binds latent protein kinase PKR (Protein Kinase R, also known as Protein kinase RNA-activated or interferoninduced, double-stranded RNA-activated protein kinase, or eukaryotic translation initiation factor 2-alpha kinase 2 – EIF2AK2), whose activation would inhibit viral protein translation and synthesis [118], and also TRIM25 (tripartite motif-containing protein 2) [119, 120]. Recently, it has been shown to interact with an array of host proteins, such as interleukin-6 receptor (IL-6R), MHC class I HLA-B, cathepsin B, ubiquitin, and adenosine deaminase acting on RNA (ADAR1) [121].
With regard to M2 ion channel activity, the heart of this mechanism is the HxxxW motif of the inner transmembrane (TM) residues [122-125]. In this HxxxW motif, Histidine 37 putatively acts as the pH sensor and, when the pH is low, the protonation of the imidazolic ring destabilizes TM packing because of electrostatic repulsion. Tryptophan 41, which acts as a primary gate, rotates, becomes unlocked from Aspartic acid 44 ("the channel lock") and, being now parallel to the axis of the pore, makes the protons flow. By contrast, Valine 27 acts as a secondary gate (the so-called "Valine 27 valve"); its importance has been confirmed only recently by the multi-scale simulation carried out by Liang and coll. [126]. On the basis of the exact role of the Histidine 37 tetrad, two models have been proposed: the shutter model, in which the biprotonated charge of Histidine 37 does not change during the proton flux (proton diffusion is coupled with water wire via the Grotthuss mechanism), and the shuttle model, in which the protonation status of Histidine 37 is subject to changes during excess proton transfer [127]. However, the exact mechanism of highly selective transport of protons is not known.
Acidification is enhanced by the viral activation of the PI3K cascade [76].
NUCLEAR IMPORTATION
The next event in replication is the importation of RNP into the nucleus. The trafficking of RNP into the cytoplasm is achieved extremely rapidly, by means of a mechanism of diffusion, without the intervention of microtubules, intermediate filaments or actin filaments, through the nuclear pore complex (NPC). Some important components of the NPC are the nucleoporins Nup37, Nup43, Nup45 [128], Nup50 [129], Nup54 [130], Nup58 [128], Nup62 [131], Nup75, Nup88, Nup93, Nup98, Nup107, Nup133 [128], Nup153 [132-135], Nup160 [128], Nup214 [130], NuTF2 (Nuclear Transport Factor 2) and Nup358/RanBP2 [128].
Nup37, Nup88, Nup96, Nup107, Nup133 and Nup160 belong to the so-called Nup107 subcomplex [136], while NupL1 (Nup45/Nup58), Nup54, and Nup62 belong to the Nup62 subcomplex [137]. Other components are Magoh, ALADIN, Tpr (Translocated promoter region), EJC (exon junction complex), NLP1/CG1 (Nucleoporin- Like Protein type 1), Seh1, Rae1/Gle2 and POM121 (nuclear envelope pore membrane protein type 121) [128].
When RNPs reach the nuclear membrane, nuclear importation is mediated by the binding of the nucleoprotein with the alpha importins, which then bind importin-β [129]. Simultaneously, the importins must interact with PA, PB1 and PB2 [130], and this affects the interaction of the RNP with the same importins [131]. Specifically, subunits PB1 and PA are imported by Ran- BP5 or karyopherin beta3 (also known as importin beta3, or importin 5), whilst subunit PB2 is imported by importin alpha-3 or importin alpha-7. NPs are imported by importin alpha-1 [132-141]. Other molecules that take part in nuclear importation are Hsp70 and Hsp90 [142-144]. In the nucleus, the importins detach from RNP. Although it is not clear, nor is the mechanism known at the molecular level, dissociation of the RNP should occur after separation from the importins. Thus, after the spreading of RNP in the nucleoplasm, transcription and replication can initiate.
TRANSCRIPTION AND REPLICATION
The transcription and replication of viral nucleic acid are not fully understood. However, the phenomena involve coordinated and differentiated RNA, NP and RdRp activities. The package that consists of the genomic segments (sRNA), the unit of trimeric polymerase and the nucleoprotein is the elementary replication unit of the influenza virus (vRNP). Therefore, in the nucleoplasm, vRNP needs to trigger the first round of its replication cycle, i.e. copying its genomic information onto mRNAs. Subsequently, mRNA exportation occurs in the cytoplasm.
The influenza polymerase is a heterotrimeric ~250 kD complex. It plays central roles in the viral life-cycle and is directly responsible for RNA synthesis for both viral replication and transcription. Moreover, it recruits host factors such as DnaJA1/Hsp40 [145].
The PA subunit interacts with host factors such as the mini-chromosome maintenance complex (MCM, a putative DNA helicase) and hCLE/CGI-99 [146, 147]. The PB2 subunit binds to the host RNA cap (7-methylguanosine triphosphate (m(7)GTP)) and supports the endonuclease activity of PA in order to "snatch" the cap from host pre-mRNAs [148, 149]. Moreover, PB2 interacts with the acetyl-CoA found in eukaryotic histone acetyltransferases (HATs) [150].
Viral mRNA synthesis is initiated by a cis-acting viral RNA polymerase, which is part of the vRNP structure and is bound to the vRNA promoter. However, mRNAs are not able to translate the genetic message efficiently; indeed, they need to be capped. Specifically, the virus must use the pre-mRNA of the cell and, for this purpose, a coordinated process mediated by PB2, PA and the cellular Polymerase II (Pol II) is necessary. Very briefly, PB2 binds mRNAs with cellular Pol II and PA, which, by means of an endonuclease mechanism, generates capped-mRNAs, which are translatable at the ribosomal level.
According to a model proposed by Fodor [151], a trans-acting polymerase activity is necessary for vRNA replication. Because of the negative polarity of vRNA, positive RNA (cRNA) must be synthesized from the mRNA templates. The replication of vRNA could then be performed by a trans-acting RNA polymerase, which may be distinct from the polymerase in the packages of RNPs. Various hypotheses have been formulated regarding the proteins, for instance NPs, that trigger trans-acting polymerase [152, 153]. Although this assumption is contradicted by other studies, which have shown viral replication in the absence of NP [154], the presence of newly synthesised NPs appears to be important in stabilizing the length of the segments of viral RNA. Furthermore, other studies have suggested that NEP could regulate viral RNA synthesis [155]. The importation of newly synthesised nucleoproteins and polymerases is also very important in order to assemble nucleoprotein with the negative-sense viral RNA. This event is not completely understood and requires coordinated interaction between Nuclear Localisation Signals (NLSs), PA, PB1, PB2, NP and importins [156-158]. In addition, as recent research has demonstrated [159, 160], NP plays a major role in the assembly of vRNP by interacting with polymerase subunits and is involved in the nuclear and cytoplasmatic transportation of vRNP. Specifically, NPs are oligomerized in ring structures, which interact with viral RNA segments and also with polymerase trimers. This latter interaction allows the viral RNA to twist into a double helix with the polymerase complex at one termination, and with a loop of RNA at another termination [161-164].
To progress, the infection requires a coordinated twoway flow between proteins synthesized in the cytoplasm (for instance M1 protein and NP) and the viral RNA that has been replicated in the nucleus. In this way, the RNP, which is full of vRNA, NP, M1, PA, PB1, PB2, and the non-structural proteins can reconstitute in the nucleus.
The transcription factory is made up of components such as SFPQ/PSF (splicing factor proline-glutamine rich [164], though its entire molecular anatomy is not known. As already mentioned, splicing mechanisms are crucial to increasing the dynamism and complexity of the influenza proteome. Specifically, the RED-SMU1 spliceosomal complex interacts with PB1 and PB2 and is responsible for the splicing of NS1. Other components of the spliceosomal complex are: EJC, SR (serine/ arginine-rich proteins, such as SRp40, SRp55 and pre-mRNA Splicing Factor type 2 / Alternative Splicing Factor – SF2/ASF), NS1-BP and hnRNP K (heterogeneous nuclear ribonucleoproteins K) [165-168].
Transcription and replication are enhanced by activation of the PI3K cascade via NS1 [76].
NUCLEAR EXPORTATION
Subsequently, through more than one mechanism, such as that of small proteins capable of passing through the nuclear pores easily, NS1 and NEP/NS2, the RNP complexes can be exported into the cytoplasm.
The export machinery is a factory made up of several components: E1B/AP5, Rae1/mRNP41 [132], NXF1-TAP (Nuclear RNA export factor type 1) [133], CBC (nuclear cap-binding complex, in particular the 80 kDa subunit), REF/Aly, TREX, P15-INXT, YB-1 (Y-box-binding protein type 1), CRM1/XPO1 (exportin type 1) [169], Rcc1 (Regulator of chromosome condensation type 1), CHD3 (chromodomain helicase DNA-binding protein type 3), RMB15B [170], RanBP3 [171], DDX19B, an ATP-dependent RNA helicase [172], and Hsc70 (heat shock cognate protein 70) [173] among others.
Briefly, two main export routes can be described: the NXF1-dependent and the CRM1-dependent pathways. RNPs are exported via CRM1, whilst HA, NA are transported by NXF1.
CRM1 pathway includes: Nup88, Nup214, Rcc1, Ran- BP3, CHD3, Hsc70, NS2, among others [174, 175].
POST-TRANSLATIONAL PROCESSING AND TRAFFICKING
After being translated, proteins are transported to the Golgi network, where they are modified. Modifications, such as glycosylation of HA and NA, palmitoylation/Sacylation of HA and M2, phosphorylation of NS1, and SUMOylation of M1, NS1, NP, PB1 and NEP by SUMO- 1/2/3 (Small Ubiquitin-like Modifier type 1/2/3) are essential steps in the production of functionally active viral proteins [176]. These tasks are performed by kinases, such as Cyclin-Dependent Kinases (CDKs) and extracellular signal-regulated kinases (ERKs) [177].
Proteins are retrogradely transported by COPI and Rab8 from the trans-Golgi apparatus to the cis-Golgi and ER [178].
The quality of protein folding and the correctness of posttranslational processing are checked by proteins such as malectin, calnexin, calreticulin and ERp57 [179-183].
APOPTOSIS PATHWAY
The exportation of RNPs is favoured as the infection progresses, which results in activation of the mechanism of spontaneous cell death (apoptosis) through the activation of caspase 3 [184]. By altering the nuclear membrane, the activation of caspase 3 increases perviousness through the nuclear pore [185, 186]. Indeed, caspase signalling pathways play an important role in the activation, replication, propagation and pathogenicity of the influenza virus, and are therefore related to the severity of influenza symptoms and its clinical burden [187]. The virus finely tunes and modulates the host cellular proteins involved in the processes of regulation and control of the induction of apoptosis [188].
The units of RNP released from the nucleus are concentrated in the perinuclear cytoplasm [189], particularly in the region of the centre for organizing microtubules (MTOC) [190] and, subsequently, in the area of recycling endosomes (REs) [191]. Interaction with the REs allows the RNPs to interact better with the network of microtubules, and thus to orient themselves and to travel towards the cell membrane [192]. The exportation of RNPs is a complicated mechanism that requires wellsynchronized timing, and results in an accumulation of RNPs on the apical surface of the cell in the late stages of viral multiplication.
In the late stages of viral replication, the accumulation of HA molecules on the cell membrane, probably by activating the mitogen-activated protein kinase (MAPK) [193], increases the exportation of RNPs, which, through a still unknown mechanism, is oriented towards the apical surface of the cell.
Subsequently, as each unit of RNP contains only one of the eight segments of the viral genome, it is particularly important that the different segments be assembled properly. The viral RNAs themselves mediate this process through a "hierarchical assembly" signalling mechanism [194].
BUDDING AND RELEASE
As the infection progresses, the apical membrane becomes rich in viral proteins, which together initiate the budding of the virus around the complex of RNPs, at the regions of the membrane where the extruded HA, the NA, and M1 and M2 proteins are concentrated [195]. Viral proteins are then delivered to the plasma membrane and assembled. Here, Rab11a [196] and HRB (HIV Rev-binding protein) [197] play an important role. HA is able to initiate the process of budding, but not to complete it. This requires the mediation of NA, M1 and M2 proteins [195]. It is also important to consider that, during the formation of the positive curvature of the cell membrane, the suitably assembled units of RNPs move toward the distal part of the viral bud, so that they can be properly wrapped by the viral envelope. As the budding process progresses, a stalk is generated which holds the virion to the cell. The viral envelope must then detach itself from the cell membrane. The M2 protein appears to be crucial to this process. Indeed, it is capable of generating a positive curvature of the membrane), which is necessary in order to enable the spherical virions to split off [195]. However, the virus is not yet free; it is bound to the cell by the binding of HA molecules with the units of sialic acid of the membrane surface. NA molecules must therefore detach the sialic acid from the cell surface in order to accomplish viral budding.
The enzymatic mechanism of influenza virus sialidase has been studied by Taylor, who showed that the enzyme catalysis process is particularly complex and consists of four steps. During the first step the α-sialoside is distorted from a chair conformation to a pseudoboat conformation when the sialoside binds to the sialidase. The second step leads to sialosyl cation, an oxocarbocation intermediate. The third step is the formation of Neu5Ac, as α-anomer. The fourth step involves its mutarotation and the subsequent release of the thermodynamicallystable β-Neu5Ac [198]. Finally, these steps lead to sialic acid hydrolysis.
Currently, 11 isoforms of NA are known; NA10 and NA11 have recently been isolated from bats and are not able to cleave sialic acid. Their precise role and mechanism are still unknown [54].
NA is further classified into two groups: group 1 (N1, N4, N5 and N8) and group 2 (N2, N3, N6, N7 and N9), based upon primary sequence [199]. Group 1 NAs contain a 150-cavity (formed by amino acids 147–151 of the 150-loop), an exposed pocket near the active catalytic site, whereas group 2 NAs lack this cavity [200].
Budding occurs via a VPS4 and VPS28 independent pathway [201, 202].
Currently available drugs
M2 INHIBITORS
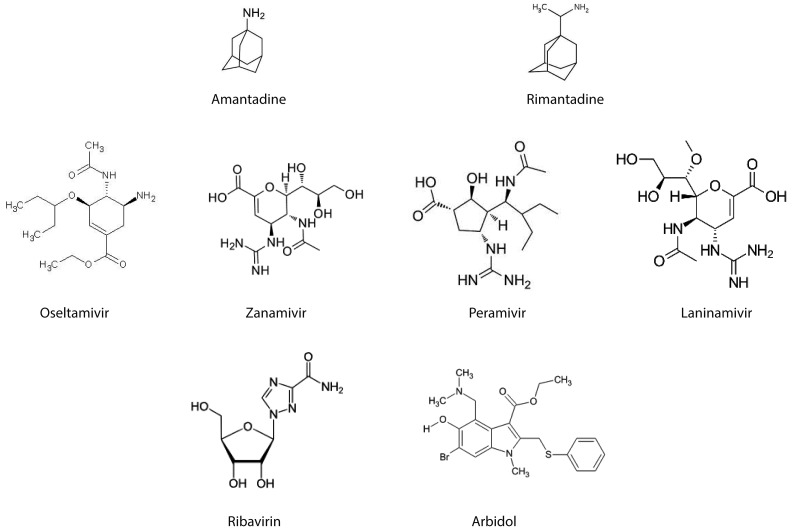
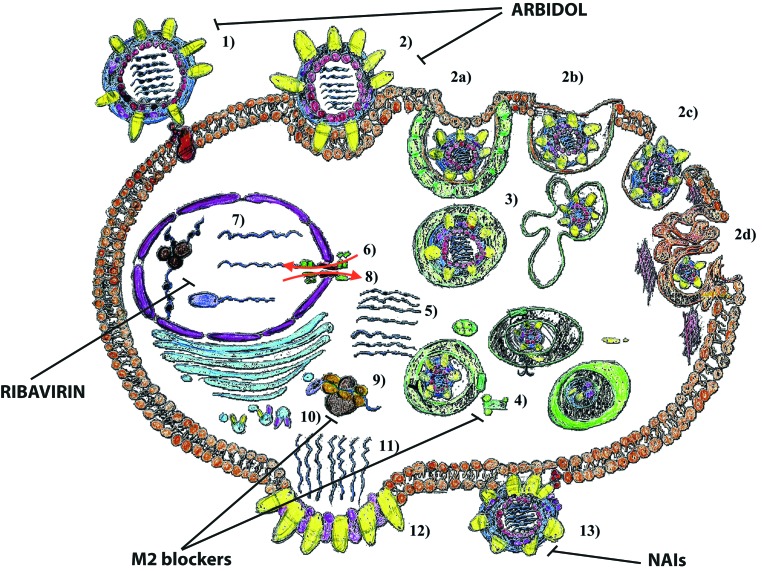
Amantadine and Rimantadine (Figs. 2, 3) were the first generation of influenza antiviral agents [203]. Amantadine (1-aminoadamantane) is a derivative of the hydrocarbon tricyclo[3.3.1.1.3,7]decane. Amantadine can be administered either as a hydrochloride derivative (Symmetrel), as 100 mg tablets or syrup, or as its effective derivative Rimantadine (α-methyl-1- adamantanemethylamine hydrochloride, Flumadine). At high concentrations, Amantadine and Rimantadine nonspecifically raise the pH within cellular endosomes, thus inhibiting or retarding the acid-induced conformational change in viral HA. At low concentrations, Amantadine and Rimantadine specifically inhibit the ion-channel activity of the M2 protein [204].
Fig. 2.
Chemical structure of the available licensed anti-influenza drugs: M2 blockers (Amantadine, Rimantadine), Neuraminidase inhibitors (NAIs; Oseltamivir, Zanamivir, Peramivir, Laninamivir), Ribavirin and Arbidol.
Fig. 3.
Schematic representation of the sites of action of anti-influenza licensed drugs. The steps of the replication cycle of the influenza virus are the following: 1) attachment of the virion to target cells and receptor binding (virus adsorption); 2) internalization into cellular regions by means of clathrin-mediated endocytosis (CME), caveolae-dependent endocytosis (CDE), clathrin-caveolae-independent endocytosis, and macropinocytosis; 3) endosomal trafficking via endosomes / caveosome / macropinosome / lysosomes to the perinuclear compartment; 4) pH-dependent fusion of viral and endosomal / organellar membranes; 5) uncoating; 6) nuclear importation; 7) transcription and replication; 8) nuclear exportation; 9) protein synthesis; 10) post-translational processing and trafficking; 11) viral progeny assembly and packaging; 12) budding; and 13) release. Abbreviations: NAIs (neuraminidase inhibitors).
Crossing the brain-blood barrier and being present in the cerebrospinal fluid (CSF) with a concentration around 75% of serum level, Amantadine can also be used to treat Parkinson's disease [205], depression and obsessivecompulsive disorder (OCD) [206], Huntington's disease [207], attention deficit hyperactivity disorder (ADHD) and other neuropsychiatric diseases [208], cocaine abuse and dependence [209], HCV [210], Creutzfeldt- Jakob's disease [211], Borna's disease [212], herpes and post herpes zoster neuralgia (PHN) [213].
This variety of uses seems to suggest that Amantadine, besides blocking the M2 channel, acts on an array of receptors, from the dopaminergic receptors to noradrenergic, serotoninergic, cholinergic, and N-Methyl-Daspartate (NMDA) receptors [214, 215].
After being rapidly adsorbed, with an excellent bioavailability profile (usually in the range 86-94%) [216], the drug reaches peak plasma levels within 4 hours [216]. The plasma elimination half-life is about 11–15 h in patients with normal renal function. It has a plasma protein binding of about 67% [216].
The drug, after being poorly metabolized and being widely distributed, is almost completely excreted via glomerular filtration and tubular secretion: this implies a dose adjustment when administered to patients with renal failure or to the elderly, such as reducing the daily dose of 100 mg, instead of 200 mg.
Acting on muscarinic receptors, some patients may experience anti-muscarinic adverse effects such as orthostatic hypotension, gastrointestinal discomfort (nausea, vomiting, anorexia), congestive heart failure [216]. Moreover, because Amantadine has some Central Nervous System (CNS) stimulatory properties, adults may complain of confusion, disorientation, jitteriness, anxiety, mood disorders, slurred speech, insomnia, ataxia, tremors, and, rarely, nightmares, oculogyric episodes. These symptoms are usually more common (up to 15- 30%) when the drug is used for different weeks for prophylactic purpose. When instead used for treatment (less than a week), it is better tolerated. In rare cases, seizures, hallucinosis/hallucinations, coma, acute psychosis and cardiac arrhythmia may occur, usually in patients with underlying psychiatric comorbidities [216, 217]. Adult Respiratory Distress Syndrome (ARDS) has been rarely and anecdotally reported [218].
Crossing the placenta and being present in breast milk, it is teratogenic at least in animals, even though safety has not been established in pregnant women: for precaution sake, it belongs to class C. Pregnancy is recognized as one of the risk factors for catching influenza and untoward outcomes (higher morbidity, hospitalization and mortality rates), but cannot benefit from adamantanes.
Also in children and in the elders, the effectiveness in preventing, treating and shortening the duration of influenza A appears to be limited, according to a recent systematic review [219].
Rimantadine can be administered as 100 mg tablets. It reaches peak plasma concentration after 3-6 hours. The plasma half-life is long (24-36 hours). Rimantadine is more metabolized than Amantadine: only 25% is secreted unchanged [216]. It has a plasma protein binding of about 40%.
Rimantadine is characterized by lower rates of ADRs [220]: compared to Amantadine, it is better tolerated by children and the elderly.
Unfortunately, the use of M2 inhibitors has been limited by the emergence of drug-resistant strains of influenza viruses [221], such as the mutations of pore-facing residues (V27A, A30T, S31N, G43E), mutations of close interhelical residues located at the N-terminal half of the channel (L26F), and mutations of far interhelical residues far located at the C-terminal half of the channel (L38F) [221-222].
NA INHIBITORS
In recent times, the most widely used antivirals against influenza have been the inhibitors of Neuroaminidase: namely Oseltamivir and Zanamivir (Figs. 2, 3). From a chemical point of view, NA inhibitors can be classified into: sialic acid derivatives (or 5,6-dihydro-4H-pyran derivatives), benzoic acid derivatives, cyclohexene derivatives, cyclopentane derivatives, pyrrolidine derivatives and natural products. The first NA inhibitors were representatives of the first chemical class, being unsaturated syalic acid analogs, such as DANA and FANA, which were initially described by Meindl and Tuppy in 1969 [223].
NA is a homotetrameric enzyme of about 220-240 kDa that is essential to the reproduction of the influenza virus. Indeed, it exerts at least three crucial actions. First, neuraminidase frees the virus from the respiratory mucus and allows it to reach the cells of the respiratory mucosa more easily. The coordinated action of HA, NA, M1 and M2 is required during the phase of viral budding. Finally, NA is required in order to release the virus from the cell surface by cutting the molecules of sialic acid that still anchor the virus to the cell surface by means of HA after completion of the replication cycle. This action also facilitates separation of the self-aggregated virions of the viral progeny. Burnet and coll. [224] first had the idea that an inhibitor of NA could be an effective antiviral agent, but only when the crystal structure of NA and its complex with neuroaminic acid were defined by Coleman in 1993 was it possible for von Itzstein [225] to synthesize a neuroaminic acid derivative with an enhanced affinity for influenza NA. This compound is Zanamivir (4-guanidino-Neu5Ac2en, or 4-GU-DANA). Its mechanism of action, which is identical to that of Oseltamivir, is characterized by the fact that the molecule mimics sialic acid; thus, it enters into competition with the acid and reversibly binds the molecules of viral NA. Zanamivir was designed to concentrate locally in the respiratory tract, while Oseltamivir (GS4104) was designed to have a high bioavalability (80%) concentration after oral administration. Indeed, Oseltamivir is very well absorbed from the gastrointestinal tract and is rapidly metabolised to active Oseltamivir carboxylate (GS4071) ([3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)- 1-cyclohexene-1-carboxylate phosphate) in the liver by hepatic carboxyl-esterase 1. The active metabolite is distributed throughout the body, including the upper and lower respiratory tract, middle ear, tracheal lining, nasal mucosa and lungs [226]. Plasma half-life varies from 1-2 to 3-4 hours, being 6-10 hours for the carboxylate form [226]. Oseltalmivir is extensively metabolized (more than 95%).
Oseltamivir and Zanamivir result comparable in terms of clinical profile and superior to the adamantanes [227, 228]. Regarding the clinical effectiveness of the two drugs, a recent meta-analysis by Jefferson et al. [229] revealed that both drugs had a modest therapeutic effect in healthy adults and that prophylactic treatment with either Zanamivir or Oseltamivir was effective in preventing the disease.
Zanamivir is supplied in "Rotadisks" with four blisters containing 5 mg of powder each. Five Rotadisks are packaged with a Diskhaler inhalation device. Oseltamivir is available as 75 mg capsules or as an oral suspension containing 12 mg/mL (Oseltamivir phosphate) [217].
ADRs are usually nausea, vomiting. Recently, CNS toxicity in infants younger than one year of age has been reported [217].
While adjustments are necessary in subjects suffering from renal failure, no adjustments are needed in patients with hepatic failure or mild obesity. Oseltamivir can be administered to pregnant women [230].
While the majority of currently circulating influenza viral strains are resistant to amantadanes, the resistance of both H1N1 and H3N2 strains and of type B viruses to Oseltamivir and Zanamivir is very low. For this reason, the US CDC and the WHO recommend their use for the treatment and prevention of seasonal and pandemic influenza [231, 232].
Other licensed drugs
OTHER NAIS
Two analogues of Zanamivir and Oseltamivir, which are currently licensed in Japan and other Asian countries, are Peramivir and Laninamivir octanoate hydrate (CS- 8958) (Figs. 2, 3).
The former is only administered intravenously because of its very poor bioavailability [233]. It has been licensed in Japan and in South Korea, but was only temporarily approved for emergency use in the USA during the H1N1 pandemic [234].
The latter drug is very promising because of its long-acting inhibitory effect [235]. Inavir was launched in Japan in October 2010 as a 20-mg dry powder inhaler. It is a prodrug that is converted in the airway to laninamivir (R-125489), the active NAI and is retained at high concentrations for at least 10 days after a single inhalation of 40 mg. Only 15% of the drug is orally bioavailable [223]. Commonly reported ADRs were psychiatric, gastrointestinal and CNS disorders [223]. A single inhalation dose makes Inavir a quite convenient drug, even though children and young adolescents could not inhale it properly [232].
RIBAVIRIN
Considering the compounds targeted against the transcription and replication of vRNA, one of the first developed drug is Ribavirin (Figs. 2, 3). Ribavirin, also known as the trade name of Virazole, is the guanosine nucleoside analog: 1β-D-ribofuranosyl-1,2,4,-triazole- 3-carboxamide. Its mechanism of action is not completely known. However, Inosine 5'-monoposphate dehydrogenase (IMPDH) appears to be the principal target of the molecule [234]. This inhibition diminishes the intracellular concentration of GTP (Guanosine-5'- triphosphate), and this would stop viral protein synthesis and limit the vRNA replication. Crotty et al also demonstrated that Ribavirin is a vRNA lethal mutagen, resembling guanosine or adenosine and causing mutations in RNA replication [235]. However, the need of high doses of the drug to have good clinical results have limited the use of Ribavirin as anti-influenza drug, and a recent revision of the literature by Chan-Tack et al. suggests that there are not conclusive results about the beneficial use of Virazole for treatment of influenza [236].
Ribavirin can be administered orally, by aerosolization, rarely by intravenous route [237]. ADR is dependent on the administration route, being extravascular haemolytic anemia if the drug is delivered intravenously, a bronchospasm if aerosolized [216].
ARBIDOL
There are several potentially effective drugs, which act as HA inhibitors. However, only one medication, the small indole derivative Arbidol (ARB) or Umifenovir (Figs. 2, 3), or ethyl-6-bromo-4-[(dimethylamino)methyl]-5-hydroxy- 1-methyl-2[(phenylthio)methyl]-indole-3-carboxylatehydrochloride monohydrate, has been licensed [232-248].
ARB was created by the Center for Drug Chemistry in Moscow [246], has been licensed in Russia 20 years ago and since 2006 has been used in China for the prophylaxis and treatment of pneumonia caused by influenza viruses A and B [234]. ARB probably exerts a multiple antiviral action: a direct virucidal effect, a block of the virus at the level of cell-entry (attachment and internalization), and impairment of viral replication, because of its ability to bind with proteins and lipids [232-248]. Several studies have demonstrated that ARB is also effective against other enveloped and non-enveloped viruses, such as Hepatitis B Virus (HBV), HCV, RSV, some Picornavirus (such as rhinovirus 14), Poliovirus 1, Coxsackievirus B5), parainfluenza type 3 (PIV3), as well as the avian coronavirus, infectious bronchitis virus, Chikungunya virus, Reovirus, Hantaan virus, Vesicular stomatitis virus (VSV) and Marek disease virus, an avian oncogenic herpesvirus [234].
It is metabolized in the liver and redistributed in the body tissues.
The principal biotransformation pathways include sulfoxidation, dimethylamine N-demethylation, glucuronidation, and sulfate conjugation. The major metabolite is sulfinylarbidol, followed by unmetabolized arbidol, Ndemethylsulfinylarbidol, and sulfonylarbidol. CYP3A4 is the major isoform involved in ARB metabolism, whereas the other P450s and flavin-containing monooxygenases (FMOs) play minor roles. Plasma half-life is long (up to 25 hours) [243].
Conclusions
In recent decades, few antiviral drugs against influenza virus infections have been available. This has limited their use in human and animal outbreaks. Indeed, antiviral drugs used during seasonal and pandemic outbreaks have usually been administered as mono-therapy and, sometimes, in an uncontrolled manner in farm animals. This has led to the emergence of viral strains that are resistant, especially to the compounds of the amantadane family. For this reason, it is particularly important to develop new antiviral drugs against influenza viruses. Indeed, although vaccination is currently the most effective means of mitigating the effects of influenza epidemics and can delay the spread of new pandemic viruses, as maintained by the Advisory Committee on Immunization Practice (ACIP), antiviral drugs can be very useful in allowing manufacturers to prepare large quantities of pandemic vaccines. In addition, antiviral drugs are particularly valuable in complicated cases of influenza, particularly in hospitalized patients and in individuals at risk, such as the elderly or patients with chronic respiratory diseases. In such cases, it would be particularly desirable to have more antivirals and to administer them in an appropriate manner [249, 250].
Acknowledgements
The authors thank Dr. Bernard Patrick for revising the manuscript.
Glossary
Abbreviations
- A
Alanine;
- ACIP
Advisory Committee on Immunization Practice;
- ADAR1
adenosine deaminase acting on RNA type 1;
- ADHD
Attention Deficit and Hyperactivity Disorder;
- ADR
Adverse Drug Reactions;
- AM
alveolar macrophage;
- AP-2
Adaptor Protein 2;
- ARB
Arbidol;
- ARDS
Adult Respiratory Distress Syndrome;
- ASF
Alternative Splicing Factor;
- BET
Bronchial Epithelial Tissue;
- BM2
Type B Influenza Virus Matrix Protein 2;
- BTB
Bric-a-brac, Tramtrack, Broad-complex;
- CBC
cap-binding complex;
- CCP
Clathrin-Coated Pit;
- CCV
Clathrin-Coated Vesicle;
- CD
Cluster of Differentiation;
- CDC
Center for Disease Control and Prevention;
- CDE
Caveole-Dependent Endocytosis;
- CDK
Cyclin-Dependent Kinase;
- CHD3
chromodomain-helicase-DNA-binding protein type 3;
- CME
Clathrin-Mediated Endocytosis;
- CNS
Central Nervous System;
- COPD
Chronic Obstructive Pulmonary Disease;
- CRL
Cullin-RING-Ligases;
- CRM1
Chromosome Region Maintenance type 1;
- cRNA
complementary RNA (positive RNA necessary as template for viral RNA replication, or template RNA);
- CSF
Cerebrospinal Fluid;
- DANA
2,3-didehydro-2-deoxy-N-acetylneuraminic acid;
- E
Glutamic acid;
- ECDC
European Centre for Disease Prevention and Control;
- EE
Early Endosome;
- EGF
Epidermal Growth Factor;
- EJC
Exon Junction Complex;
- ER
Endoplasmic Reticulum;
- ERK
extracellular signal-regulated kinase;
- F
phenylalanine;
- FANA
2-deoxy-2,3-dehydro-Ntrifluoroacetylneuraminic acid;
- FMO
flavin-containing monooxygenase;
- G
Glycine;
- GTP
Guanosine-5'-triphosphate;
- GTPase
guanosine triphosphatase;
- HA
Hemagglutinin;
- HAfp23
Hemagglutinin fusion peptide 23;
- HAT
histone acetyltransferase;
- HBV
Hepatitis B Virus;
- HCV
Hepatitis C Virus;
- HEF
Hemagglutinin-Esterase-Fusion protein;
- HIV
Human Immunodeficiency Virus;
- hnRNP K
heterogeneous nuclear ribonucleoproteins K;
- HPV
Human Papillomavirus;
- HRB
HIV Rev-binding protein;
- Hsc
Heat shock cognate protein;
- HSP
Heat Shock Protein;
- IAV
Influenza A Virus;
- IFN-I
type I interferon;
- IL
interleukin;
- IL6R
interleukin 6 receptor;
- IMPDH
Inosine 5'-monoposphate dehydrogenase;
- kD
kilodalton;
- L
Leucine;
- LAMP
lysosomal-associated membrane protein;
- LE
Late Endosome;
- LET
Lung Epithelial Tissue;
- M protein
Matrix protein;
- M1
Matrix protein 1;
- M2
Matrix Protein 2;
- m(7)GTP
RNA cap 7-methylguanosine triphosphate;
- MAPK
Mitogen-Activated Protein Kinase;
- MCM
mini-chromosome maintenance complex;
- MHC
Major Histocompatibility Complex;
- mRNA
messenger RNA;
- mRNAs
messenger RNA segment;
- mRNP
messenger ribonucleoprotein;
- MTOC
MicroTubules-Organizing Centre;
- mTOR
mammalian Target Of Rapamycin;
- N
Asparagine;
- NA
Neuroaminidase;
- NAIs
Neuroaminidase inhibitors;
- NEP
Nuclear Export Protein;
- NES
Nuclear Export Signal;
- NGF
Nerve Growth Factor;
- NHE
Na+/H+ exchanger;
- NK
Natural Killer cell;
- NLP1
Nucleoporin-Like Protein type 1;
- NLSs
Nuclear Localization Signals;
- NMDA
N-Methyl-D-aspartate;
- NP
Nucleoprotein;
- NPC
Nuclear Pore Complex;
- NS1
Non-Structural protein type 1;
- NS1-BP
NS1 Binding Protein;
- NS2
Non-Structural protein type 2;
- NS3
Non-Structural protein type 3;
- Nup
Nucleoporin;
- NuTF2
Nuclear Transport Factor 2;
- NXF1
Nuclear RNA export factor type 1;
- OCD
Obsessive- Compulsive Disorder;
- ORF
Open Reading Frame;
- PA
Acidic Polymerase;
- PACT
Protein ACTivator of the interferon-induced protein kinase;
- PAK
p21-activated Kinase;
- PB1
Basic Polymerase 1;
- PB2
Basic Polymerase 2;
- pDC
plasmacytoid dendritic cell;
- PH
pleckstrin homology;
- PHN
Post- Herpetic Neuralgia;
- PI3K
phosphoinositide 3-kinase;
- PIV
Para-Infliuenza Virus;
- PKC
Protein Kinase C;
- PKR
Protein Kinase R (also known as Protein kinase RNA-activated, or interferon-induced, double-stranded RNA-activated protein kinase, or eukaryotic translation initiation factor 2-alpha kinase 2 – EIF2AK2);
- Pol II
Polymerase II;
- POM121
nuclear envelope pore membrane protein type 121;
- PRKRA
Protein kinase, interferon-inducible double stranded RNA dependent activator;
- PtdIns(3,4,5)P3
phosphatidylinositol-(3,4,5)-trisphosphate;
- PtdIns(4,5)P2
phosphatidylinositol-(4,5)- bisphosphate;
- PTEN
Phosphatase and tensin homolog;
- qPCR
quantitative Polymerase Chain Reaction;
- RanBP
Ran Binding Protein;
- RBP
Receptor Binding Pocket;
- RBS
Receptor Binding Site;
- Rcc1
Regulator of chromosome condensation type 1;
- RdRp
RNA-dependent RNA polymerase complex;
- REs
Recycling Endosomes.;
- RIG-I
Retinoic acid-Inducible Gene 1;
- RNA
Ribonucleic Acid;
- RNP
Ribonucleoprotein;
- RSV
Respiratory Syncytial Virus;
- S
Serine;
- SF
pre-mRNA Splicing Factor;
- SFPQ/PSF
splicing factor proline-glutamine rich;
- SFRS
Serine/arginine-rich splicing factor;
- SH2
Src Homology 2;
- siRNA
short interfering RNA;
- SR
Serine/arginine-Rich protein;
- sRNA
genomic segments of RNA;
- SUMO
Small Ubiquitin-like Modifier;
- T
Threonine;
- TGN
Trans-Golgi Network;
- TM
transmembrane;
- Tpr
Translocated promoter region;
- TRIM
tripartite motif-containing protein;
- US
United States of America;
- V
Valine;
- Vps
Vacuolar protein sorting;
- vRNP
viral Ribonucleoprotein;
- VSV
Vesicular stomatitis virus;
- WHO
World Health Organization;
- XPO1
exportin 1;
- YB-1
Y-box-binding protein type 1.
References
- 1.Schutten M, Baalen C, Zoeteweij P, et al. The influenza virus: disease, diagnostics, and treatment. MLO Med Lab Obs. 2013;45:38–40. [PubMed] [Google Scholar]
- 2. CDC , author. Seasonal Influenza: Flu Basics. Document available at: http://www.cdc.gov/flu/about/disease/index.htm. Accessed on 01 August 2014.
- 3.Toovey S. Influenza-associated central nervous system dysfunction: a literature review. Travel Med Infect Dis. 2008;6:114–124. doi: 10.1016/j.tmaid.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 4.Davis LE, Koster F, Cawthon A. Neurologic aspects of influenza viruses. Handb Clin Neurol. 2014;123:619–645. doi: 10.1016/B978-0-444-53488-0.00030-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Short KR, Habets MN, Hermans PW, et al. Interactions between streptococcus pneumoniae and influenza virus: a mutually beneficial relationship? Future Microbiol. 2012;7:609–624. doi: 10.2217/fmb.12.29. [DOI] [PubMed] [Google Scholar]
- 6.McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 7.Cauldwell AV, Long JS, Moncorgé O, et al. Viral determinants of influenza a virus host range. J Gen Virol. 2014;95:1193–1210. doi: 10.1099/vir.0.062836-0. [DOI] [PubMed] [Google Scholar]
- 8. WHO , author. Influenza (seasonal). Fact sheet N°211 March 2014. Available at: http://www.who.int/mediacentre/factsheets/fs211/en/. Accessed on 1st August 2014.
- 9. ECDC , author. Seasonal Influenza. Document available at: http://www.ecdc.europa.eu/en/healthtopics/seasonal_influenza/Pages/index.aspx. Accessed on 10th June 2014.
- 10. FLU.gov , author. Seasonal Flu. Document available at: http://www.flu.gov/about_the_flu/seasonal/. Accessed on 1st August 2014.
- 11.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J, Deng YM. Influenza virus antigenic variation, host antibody production and new approach to control epidemics. Virol J. 2009;6:30–30. doi: 10.1186/1743-422X-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Labella AM, Merel SE. Influenza. Med Clin North Am. 2013;97:621–645. doi: 10.1016/j.mcna.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Taubenberger JK. The origin and virulence of the 1918 "spanish" influenza virus. Proc Am Philos Soc. 2006;150:86–112. [PMC free article] [PubMed] [Google Scholar]
- 15.Patterson KD, Pyle GF. The geography and mortality of the 1918 influenza pandemic. Bull Hist Med. 1991;65:4–21. [PubMed] [Google Scholar]
- 16. Stanford University , author. The medical and scientific conceptions of influenza. Document available at: http://www.stanford.edu/group/virus/uda/fluscimed.html. Accessed on 14th June 2014.
- 17.Davies WL, Grunert RR, Haff RF, et al. Antiviral activity of 1-adamantanamine (amantadine) Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 18.Dolin R, Reichman RC, Madore HP, et al. A controlled trial of amantadine and rimantadine in the prophylaxis of influenza A infection. N Engl J Med. 1982;307:580–584. doi: 10.1056/NEJM198209023071002. [DOI] [PubMed] [Google Scholar]
- 19. Center for disease Control and Prevention (CDC) , author. Advisory Committee on Immunization Practice (ACIP). Prevention and control of influenza. MMWR Recommendations and Reports. 2000;49:1–38. [PubMed] [Google Scholar]
- 20.Noda T, Sagara H, Yen A, et al. Architecture of ribonucleoprotein complexes in influenza a virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 21.Noda T. Native morphology of influenza virions. Front Microbiol. 2012;2:269–269. doi: 10.3389/fmicb.2011.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elleman CJ, Barclay WS. The M1 matrix protein controls the filamentous phenotype of influenza A virus. Virology. 2004;321:144–153. doi: 10.1016/j.virol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 23.Roberts PC, Lamb RA, Compans RW. The M1 and M2 proteins of influenza a virus are important determinants in filamentous particle formation. Virology. 1998;240:127–137. doi: 10.1006/viro.1997.8916. [DOI] [PubMed] [Google Scholar]
- 24.Rossman JS, Jing X, Leser GP, et al. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J Virol. 2010;84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bialas KM, Bussey KA, Stone RL, et al. Specific nucleoprotein residues affect influenza virus morphology. J Virol. 2014;88:2227–2234. doi: 10.1128/JVI.03354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roberts PC, Compans RW. Host cell dependence of viral morphology. Proc Natl Acad Sci U S A. 1998;95:5746–5751. doi: 10.1073/pnas.95.10.5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samji T. Influenza A: understanding the viral life cycle. Yale J Biol Med. 2009;82:153–159. [PMC free article] [PubMed] [Google Scholar]
- 28.Cross TA. Flu BM2 structure and function. Nat Struct Mol Biol. 2009;16:1207–1209. doi: 10.1038/nsmb1209-1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pleschka S, Klenk HD, Herrler G. The catalytic triad of the influenza C virus glycoprotein HEF esterase: characterization by site-directed mutagenesis and functional analysis. J Gen Virol. 1995;76:2529–2537. doi: 10.1099/0022-1317-76-10-2529. [DOI] [PubMed] [Google Scholar]
- 30.Kollerova E, Betáková T. Influenza viruses and their ion channels. Acta Virol. 2006;50:7–16. [PubMed] [Google Scholar]
- 31.Fischer WB, Sansom MS. Viral ion channels: structure and function. Biochim Biophys Acta. 2002;1561:27–45. doi: 10.1016/s0304-4157(01)00009-0. [DOI] [PubMed] [Google Scholar]
- 32.Lamb RA, Choppin PW. The gene structure and replication of influenza virus. Annu Rev Biochem. 1983;52:467–506. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- 33.Košík I, Hollý J, Russ G. PB1-F2 expedition from the whole protein through the domain to aa residue function. Acta Virol. 2013;57:138–148. doi: 10.4149/av_2013_02_138. [DOI] [PubMed] [Google Scholar]
- 34.Krumbholz A, Philipps A, Oehring H, et al. Current knowledge on PB1-F2 of influenza A viruses. Med Microbiol Immunol. 2011;200:69–75. doi: 10.1007/s00430-010-0176-8. [DOI] [PubMed] [Google Scholar]
- 35.Košík I, Krejnusová I, Práznovská M, et al. The multifaceted effect of PB1-F2 specific antibodies on influenza a virus infection. Virology. 2013;447:1–8. doi: 10.1016/j.virol.2013.08.022. [DOI] [PubMed] [Google Scholar]
- 36.Lamb RA, Krug RM. Orthomyxoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fundamental virology. Philadelphia: Williams & Williams; 2001. pp. 725–769. [Google Scholar]
- 37.Skehel JJ. Polypeptide synthesis in influenza virus-infected cells. Virology. 1972;49:23–36. doi: 10.1016/s0042-6822(72)80004-7. [DOI] [PubMed] [Google Scholar]
- 38.Skehel JJ. Early polypeptide synthesis in influenza virus-infected cells. Virology. 1973;56:394–399. doi: 10.1016/0042-6822(73)90320-6. [DOI] [PubMed] [Google Scholar]
- 39.Hayden FG, Aoki FY. Amantadine, rimatadine, and related agents. In: Barriere SL, editor. Antimicrobial therapy and vaccines. Baltimore: Williams & Williams; 1999. pp. 1344–1365. [Google Scholar]
- 40.Wise HM, Barbezange C, Jagger BW, et al. Overlapping signals for translational regulation and packaging of influenza a virus segment 2. Nucleic Acids Res. 2011;39:7775–7790. doi: 10.1093/nar/gkr487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wise HM, Hutchinson EC, Jagger BW, et al. Identification of a novel splice variant form of the influenza a virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012;8:e1002998–e1002998. doi: 10.1371/journal.ppat.1002998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yewdell JW, Ince WL. Virology. Frameshifting to PA-X influenza. Science. 2012;337:164–165. doi: 10.1126/science.1225539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muramoto Y, Noda T, Kawakami E, et al. Identification of novel influenza a virus proteins translated from PA mRNA. J Virol. 2013;87:2455–2462. doi: 10.1128/JVI.02656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vasin AV, Temkina OA, Egorov VV, et al. Molecular mechanisms enhancing the proteome of influenza A viruses: an overview of recently discovered proteins. Virus Res. 2014;185:53–63. doi: 10.1016/j.virusres.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 45.Sabath N, Morris JS, Graur D. Is there a twelfth protein-coding gene in the genome of influenza A? A selection-based approach to the detection of overlapping genes in closely related sequences. J Mol Evol. 2011;73:305–315. doi: 10.1007/s00239-011-9477-9. [DOI] [PubMed] [Google Scholar]
- 46.Kummer S, Flöttmann M, Schwanhäusser B, et al. Alteration of protein levels during influenza virus H1N1 infection in host cells: a proteomic survey of host and virus reveals differential dynamics. PLoS One. 2014;9:e94257–e94257. doi: 10.1371/journal.pone.0094257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsuoka Y, Matsumae H, Katoh M, et al. A comprehensive map of the influenza a virus replication cycle. BMC Syst Biol. 2013;7:97–97. doi: 10.1186/1752-0509-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edinger TO, Pohl MO, Stertz S. Entry of influenza A virus: host factors and antiviral targets. J Gen Virol. 2014;95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 49.Mao H, Tu W, Qin G, et al. Influenza virus directly infects human natural killer cells and induces cell apoptosis. J Virol. 2009;83:9215–9222. doi: 10.1128/JVI.00805-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun X, Whittaker GR. Entry of influenza virus. Adv Exp Med Biol. 2013;790:72–82. doi: 10.1007/978-1-4614-7651-1_4. [DOI] [PubMed] [Google Scholar]
- 51.Roberson EC, Tully JE, Guala AS, et al. Influenza induces endoplasmic reticulum stress, caspase-12-dependent apoptosis, and c-Jun N-terminal kinase-mediated transforming growth factor-β release in lung epithelial cells. Am J Respir Cell Mol Biol. 2012;46:573–581. doi: 10.1165/rcmb.2010-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbier D, Garcia-Verdugo I, Pothlichet J, et al. Influenza A induces the major secreted airway mucin MUC5AC in a protease- EGFR-extracellular regulated kinase-Sp1-dependent pathway. Am J Respir Cell Mol Biol. 2012;47:149–157. doi: 10.1165/rcmb.2011-0405OC. [DOI] [PubMed] [Google Scholar]
- 53.Ehrhardt C, Marjuki H, Wolff T, et al. Bivalent role of the phosphatidylinositol- 3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol. 2006;8:1336–1348. doi: 10.1111/j.1462-5822.2006.00713.x. [DOI] [PubMed] [Google Scholar]
- 54.Air GM. Influenza virus-glycan interactions. Curr Opin Virol. 2014;7:128–133. doi: 10.1016/j.coviro.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shinya K, Ebina M, Yamada S, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 56.Russell RJ, Kerry PS, Stevens DJ, et al. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc Natl Acad Sci U S A. 2008;105:17736–17741. doi: 10.1073/pnas.0807142105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sączyńska V. Influenza virus hemagglutinin as a vaccine antigen produced in bacteria. Acta Biochim Pol. 2014;61:561–572. [PubMed] [Google Scholar]
- 58.Smrt ST, Draney AW, Lorieau JL. The influenza hemagglutinin fusion domain is an amphipathic helical-hairpin that functions by inducing membrane curvature. J Biol Chem. 2014 Nov 14;pii:jbc.M114.611657–jbc.M114.611657. doi: 10.1074/jbc.M114.611657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burke DF, Smith DJ. A Recommended numbering scheme for influenza A HA subtypes. PLoS One. 2014;9:e112302–e112302. doi: 10.1371/journal.pone.0112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza a virus from bats. Proc Natl Acad Sci U S A. 2012;109:4269–4274. doi: 10.1073/pnas.1116200109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9:e1003657–e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vanderlinden E, Naesens L. Emerging antiviral strategies to interfere with influenza virus entry. Med Res Rev. 2014;34:301–339. doi: 10.1002/med.21289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stray SJ, Cummings RD, Air GM. Influenza virus infection of desialylated cells. Glycobiology. 2000;10:649–658. doi: 10.1093/glycob/10.7.649. [DOI] [PubMed] [Google Scholar]
- 64.Lakadamyali M, Rust MJ, Zhuang X. Endocytosis of influenza viruses. Microbes Infect. 2004;6:929–936. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vries E, Tscherne DM, Wienholts MJ, et al. Dissection of the influenza a virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7:e1001329–e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rossman JS, Leser GP, Lamb RA. Filamentous influenza virus enters cells via macropinocytosis. J Virol. 2012;86:10950–10960. doi: 10.1128/JVI.05992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Whittaker GR. Influenza entry pathways in polarized MDCK cells. Biochem Biophys Res Commun. 2014;450:234–239. doi: 10.1016/j.bbrc.2014.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nunes-Correia I, Eulálio A, Nir S, et al. Caveolae as an additional route for influenza virus endocytosis in MDCK cells. Cell Mol Biol Lett. 2004;9:47–60. [PubMed] [Google Scholar]
- 69.Sieczkarski SB, Whittaker GR. Influenza virus can enter and infect cells in the absence of clathrin-mediated endocytosis. J Virol. 2002;76:10455–10464. doi: 10.1128/JVI.76.20.10455-10464.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young A. Structural insights into the clathrin coat. Semin Cell Dev Biol. 2007;18:448–458. doi: 10.1016/j.semcdb.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Wilbur JD, Hwang PK, Brodsky FM. New faces of the familiar clathrin lattice. Traffic. 2005;6:346–350. doi: 10.1111/j.1600-0854.2005.00277.x. [DOI] [PubMed] [Google Scholar]
- 72.Heilker R, Manning-Krieg U, Zuber JF, et al. In vitro binding of clathrin adaptors to sorting signals correlates with endocytosis and basolateral sorting. EMBO J. 1996;15:2893–2899. [PMC free article] [PubMed] [Google Scholar]
- 73.Fire E, Brown CM, Roth MG, et al. Partitioning of proteins into plasma membrane microdomains. Clustering of mutant influenza virus hemagglutinins into coated pits depends on the strength of the internalization signal. J Biol Chem. 1997;272:29538–29545. doi: 10.1074/jbc.272.47.29538. [DOI] [PubMed] [Google Scholar]
- 74.Chen C, Zhuang X. Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc Natl Acad Sci U S A. 2008;105:11790–11795. doi: 10.1073/pnas.0803711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fujioka Y, Tsuda M, Hattori T, et al. The Ras-PI3K signaling pathway is involved in clathrin-independent endocytosis and the internalization of influenza viruses. PLoS One. 2011;6:e16324–e16324. doi: 10.1371/journal.pone.0016324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ayllon J, García-Sastre A, Hale BG. Influenza a viruses and PI3K: are there time, place and manner restrictions? Virulence. 2012;3:411–414. doi: 10.4161/viru.20932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Marchant DJ, Bilawchuk L, Nish G, Hegele RG. Virus-induced signaling influences endosome trafficking, virus entry, and replication. Methods Enzymol. 2014;534:65–76. doi: 10.1016/B978-0-12-397926-1.00004-4. [DOI] [PubMed] [Google Scholar]
- 78.Pascua PN, Lee JH, Song MS, et al. Role of the p21-activated kinases (PAKs) in influenza a virus replication. Biochem Biophys Res Commun. 2011;414:569–574. doi: 10.1016/j.bbrc.2011.09.119. [DOI] [PubMed] [Google Scholar]
- 79.Nanbo A, Imai M, Watanabe S, et al. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoproteindependent manner. PLoS Pathog. 2010;6:e1001121–e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saeed MF, Kolokoltsov AA, Albrecht T, et al. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110–e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Iwasaki M, Ngo N, Torre JC. Sodium hydrogen exchangers contribute to arenavirus cell entry. J Virol. 2014;88:643–654. doi: 10.1128/JVI.02110-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Amstutz B, Gastaldelli M, Kälin S, et al. Subversion of CtBP1- controlled macropinocytosis by human adenovirus serotype 3. EMBO J. 2008;27:956–969. doi: 10.1038/emboj.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kälin S, Amstutz B, Gastaldelli M, et al. Macropinocytotic uptake and infection of human epithelial cells with species B2 adenovirus type 35. J Virol. 2010;84:5336–5350. doi: 10.1128/JVI.02494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schelhaas M, Shah B, Holzer M, et al. Entry of human papillomavirus type 16 by actin-dependent, clathrin- and lipid raftindependent endocytosis. PLoS Pathog. 2012;8:e1002657–e1002657. doi: 10.1371/journal.ppat.1002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carter GC, Bernstone L, Baskaran D, et al. HIV-1 infects macrophages by exploiting an endocytic route dependent on dynamin, Rac1 and Pak1. Virology. 2011 Jan 20;409(2):234–250. doi: 10.1016/j.virol.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 86.Nagasawa S, Ogura K, Tsutsuki H, et al. Uptake of shigatoxigenic escherichia coli subAB by HeLa cells requires an actin – and lipid raft-dependent pathway. Cell Microbiol. 2014;16:1582–1601. doi: 10.1111/cmi.12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barrias ES, Reignault LC, Souza W, et al. Trypanosoma cruzi uses macropinocytosis as an additional entry pathway into mammalian host cell. Microbes Infect. 2012;14:1340–1351. doi: 10.1016/j.micinf.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 88.Mercer J, Helenius A. Vaccinia virus uses macropinocytosis and apoptotic mimicry to enter host cells. Science. 2008;320:531–535. doi: 10.1126/science.1155164. [DOI] [PubMed] [Google Scholar]
- 89.Sánchez EG, Quintas A, Pérez-Núñez D, et al. African swine fever virus uses macropinocytosis to enter host cells. PLoS Pathog. 2012;8:e1002754–e1002754. doi: 10.1371/journal.ppat.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krzyzaniak MA, Zumstein MT, Gerez JA, et al. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 2013;9:e1003309–e1003309. doi: 10.1371/journal.ppat.1003309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Matsuda M, Suzuki R, Kataoka C, et al. Alternative endocytosis pathway for productive entry of hepatitis C virus. J Gen Virol. 2014;95:2658–2667. doi: 10.1099/vir.0.068528-0. [DOI] [PubMed] [Google Scholar]
- 92.Conto F, Covan S, Arcangeletti MC, et al. Differential infectious entry of human influenza A/NWS/33 virus (H1N1) in mammalian kidney cells. Virus Res. 2011;155:221–230. doi: 10.1016/j.virusres.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 93.Liu SL, Wu QM, Zhang LJ, et al. Three-dimensional tracking of rab5- and rab7 – associated infection process of influenza virus. Small. 2014 Jun 27; doi: 10.1002/smll.201400944. doi: 10.1002/smll.201400944. [DOI] [PubMed] [Google Scholar]
- 94.Huotari J, Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sieczkarski SB, Whittaker GR. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic. 2003;4:333–343. doi: 10.1034/j.1600-0854.2003.00090.x. [DOI] [PubMed] [Google Scholar]
- 96.Costello DA, Whittaker GR, Daniel S. Variation of pH sensitivity, acid stability, and fusogenicity of three influenza H3 subtypes. J Virol. 2014 Oct 15;pii:JVI.01927-14–JVI.01927-14. doi: 10.1128/JVI.01927-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khor R, McElroy LJ, Whittaker GR. The ubiquitin-vacuolar protein sorting system is selectively required during entry of influenza virus into host cells. Traffic. 2003;4:857–868. doi: 10.1046/j.1398-9219.2003.0140.x. [DOI] [PubMed] [Google Scholar]
- 98.Zhou Z, Xue Q, Wan Y, et al. Lysosome-associated membrane glycoprotein 3 is involved in influenza a virus replication in human lung epithelial (A549) cells. Virol J. 2011;8:384–384. doi: 10.1186/1743-422X-8-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Davenport EE, Antrobus RD, Lillie PJ, et al. Transcriptomic profiling facilitates classification of response to influenza challenge. J Mol Med (Berl) 2014 Oct 28; doi: 10.1007/s00109-014-1212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hubner M, Peter M. Cullin-3 and the endocytic system: new functions of ubiquitination for endosome maturation. Cell Logist. 2012;2:166–168. doi: 10.4161/cl.20372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Huotari J, Meyer-Schaller N, Hubner M, et al. Cullin-3 regulates late endosome maturation. Proc Natl Acad Sci U S A. 2012;109:823–828. doi: 10.1073/pnas.1118744109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li S, Sieben C, Ludwig K, et al. pH-Controlled two-step uncoating of influenza virus. Biophys J. 2014;106:1447–1456. doi: 10.1016/j.bpj.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pinto LH, Lamb RA. The M2 Proton Channels of Influenza A and B Viruses. J Biol Chem. 2006;281:8997–8997. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 104.Martyna A, Rossman Alterations of membrane curvature during influenza virus budding. J Biochem Soc Trans. 2014;42:1425–1428. doi: 10.1042/BST20140136. [DOI] [PubMed] [Google Scholar]
- 105.Fuhrmans M, Marrink SJ. Molecular view of the role of fusion peptides in promoting positive membrane curvature. J Am Chem Soc. 2012;134:1543–1552. doi: 10.1021/ja207290b. [DOI] [PubMed] [Google Scholar]
- 106.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 107.Bentz J, Mittal A. Architecture of the influenza hemagglutinin membrane fusion site. Biochim Biophys Acta. 2003;1614:24–35. doi: 10.1016/s0005-2736(03)00160-3. [DOI] [PubMed] [Google Scholar]
- 108.Kido H, Okumura Y, Takahashi E, et al. Role of host cellular proteases in the pathogenesis of influenza and influenza- induced multiple organ failure. Biochim Biophys Acta. 2012;1824:186–194. doi: 10.1016/j.bbapap.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 109.Zhirnov PO, Matrosovich TY, Matrosovich MN, et al. Aprotinin, a protease inhibitor, suppresses proteolytic activation of pandemic H1N1v influenza virus. Antivir Chem Chemoth. 2011;21:169–174. doi: 10.3851/IMP1715. [DOI] [PubMed] [Google Scholar]
- 110.Seki M, Kohno S, Newstead MW, et al. Critical role of IL-1 receptor-associated kinase-M in regulating chemokine-dependent deleterious inflammation in murine influenza pneumonia. J Immunol. 2010;184:1410–1418. doi: 10.4049/jimmunol.0901709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Banerjee I, Miyake Y, Nobs SP, et al. Influenza a virus uses the aggresome processing machinery for host cell entry. Science. 2014;346:473–477. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 112.Sakaguchi T, Leser GP, Lamb RA. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J Cell Biol. 1996;133:733–747. doi: 10.1083/jcb.133.4.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ciampor F, Cmarko D, Cmarková J, et al. Influenza virus M2 protein and haemagglutinin conformation changes during intracellular transport. Acta Virol. 1995;39:171–181. [PubMed] [Google Scholar]
- 114.Zhirnov OP, Klenk HD. Influenza a virus proteins NS1 and hemagglutinin along with M2 are involved in stimulation of autophagy in infected cells. J Virol. 2013;87:13107–13114. doi: 10.1128/JVI.02148-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gu RX, Liu LA, Wei DQ. Structural and energetic analysis of drug inhibition of the influenza A M2 proton channel. Trends Pharmacol Sci. 2013;34:571–580. doi: 10.1016/j.tips.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 116.Tawaratsumida K, Phan V, Hrincius ER, et al. Quantitative proteomic analysis of the influenza a virus nonstructural proteins NS1 and NS2 during natural cell infection identifies PACT as an NS1 target protein and antiviral host factor. J Virol. 2014;88:9038–9048. doi: 10.1128/JVI.00830-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Li S, Min JY, Krug RM, et al. Binding of the influenza a virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 118.Khaperskyy DA, Emara MM, Johnston BP, et al. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog. 2014;10:e1004217–e1004217. doi: 10.1371/journal.ppat.1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ludwig S, Wolff T. Influenza a virus TRIMs the type I interferon response. Cell Host Microbe. 2009;5:420–421. doi: 10.1016/j.chom.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 120.Gack MU, Albrecht RA, Urano T, et al. Influenza a virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ngamurulert S, Limjindaporn T, Auewaraku P. Identification of cellular partners of Influenza a virus (H5N1) nonstructural protein NS1 by yeast two-hybrid system. Acta Virol. 2009;53:153–159. doi: 10.4149/av_2009_03_153. [DOI] [PubMed] [Google Scholar]
- 122.Kolocouris A, Spearpoint P, Martin SR, et al. Comparisons of the influenza virus A M2 channel binding affinities, antiinfluenza virus potencies and NMDA antagonistic activities of 2-alkyl-2-aminoadamantanes and analogues. Bioorg Med Chem Lett. 2008;18:6156–6160. doi: 10.1016/j.bmcl.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 123.Kozakov D, Chuang GY, Beglov D, et al. Where does amantadine bind to the influenza virus M2 proton channel? Trends Biochem Sci. 2010;35:471–475. doi: 10.1016/j.tibs.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Laohpongspaisan C, Rungrotmongkol T, Loisruangsin A, et al. How amantadine and rimantadine inhibit proton transport in the M2 protein channel. J Mol Graph Model. 2008;27:342–348. doi: 10.1016/j.jmgm.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 125.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Liang R, Li H, Swanson JM, et al. Multiscale simulation reveals a multifaceted mechanism of proton permeation through the influenza A M2 proton channel. Proc Natl Acad Sci U S A. 2014;111:9396–1401. doi: 10.1073/pnas.1401997111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Babcock HP, Chen C, Zhuang X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J. 2004;87:2749–2758. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Influenza life cycle pathway map. FluMap interactive view. Flu- Map: a comprehensive influenza life cycle map. Available at: http://www.influenza-x.org/flumap/Pathway.html. Accessed on 1st August 2014.
- 129.Pumroy RA, Nardozzi JD, Hart DJ, et al. Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin α5. J Biol Chem. 2012;287:2022–2031. doi: 10.1074/jbc.M111.315838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tafforeau L, Chantier T, Pradezynski F, et al. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. J Virol. 2011;85:13010–13018. doi: 10.1128/JVI.02651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Munier S, Rolland T, Diot C, et al. Exploration of binary virushost interactions using an infectious protein complementation assay. Mol Cell Proteomics. 2013;12:2845–2855. doi: 10.1074/mcp.M113.028688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Satterly N, Tsai PL, Deursen J, et al. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc Natl Acad Sci U S A. 2007;104:1853–1858. doi: 10.1073/pnas.0610977104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Read EK, Digard P. Individual influenza a virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J Gen Virol. 2010;91:1290–1301. doi: 10.1099/vir.0.018564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen J, Huang S, Chen Z. Human cellular protein nucleoporin hNup98 interacts with influenza a virus NS2/nuclear export protein and overexpression of its GLFG repeat domain can inhibit virus propagation. J Gen Virol. 2010;91:2474–2484. doi: 10.1099/vir.0.022681-0. [DOI] [PubMed] [Google Scholar]
- 136.Beck M, Glavy JS. Toward understanding the structure of the vertebrate nuclear pore complex. Nucleus. 2014;5:119–123. doi: 10.4161/nucl.28739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shi F, Xie Y, Shi L, et al. Viral RNA polymerase: a promising antiviral target for influenza A virus. Curr Med Chem. 2013;20:3923–3934. doi: 10.2174/09298673113209990208. [DOI] [PubMed] [Google Scholar]
- 138.Hudjetz B, Gabriel G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 2012;8:e1002488–e1002488. doi: 10.1371/journal.ppat.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chou YY, Heaton NS, Gao Q, et al. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 2013;9:e1003358–e1003358. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Resa-Infante P, Gabriel G. The nuclear import machinery is a determinant of influenza virus host adaptation. ResaBioessays. 2013;35:23–27. doi: 10.1002/bies.201200138. [DOI] [PubMed] [Google Scholar]
- 141.Gabriel G, Herwig A, Klenk HD. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza a virus. PLoS Pathog. 2008;4:e11–e11. doi: 10.1371/journal.ppat.0040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Manzoor R, Kuroda K, Yoshida R, et al. Heat shock protein 70 modulates influenza a virus polymerase activity. J Biol Chem. 2014;289:7599–7614. doi: 10.1074/jbc.M113.507798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang C, Yang Y, Zhou X, et al. The NS1 protein of influenza a virus interacts with heat shock protein Hsp90 in human alveolar basal epithelial cells: implication for virus-induced apoptosis. Virol J. 2011;8:181–181. doi: 10.1186/1743-422X-8-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Naito T, Momose F, Kawaguchi A, et al. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cao M, Wei C, Zhao L, et al. DnaJA1/Hsp40 is co-opted by influenza a virus to enhance its viral RNA polymerase activity. J Virol. 2014 Sep 24;pii:JVI.02475-14–JVI.02475-14. doi: 10.1128/JVI.02475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pérez-González A, Rodriguez A, Huarte M, et al. hCLE/CGI- 99, a human protein that interacts with the influenza virus polymerase, is a mRNA transcription modulator. J Mol Biol. 2006;362:887–900. doi: 10.1016/j.jmb.2006.07.085. [DOI] [PubMed] [Google Scholar]
- 147.Kawaguchi A, Nagata K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 2007;26:4566–4575. doi: 10.1038/sj.emboj.7601881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bier K, York A, Fodor E. Cellular cap-binding proteins associate with influenza virus mRNAs. J Gen Virol. 2011;92:1627–1634. doi: 10.1099/vir.0.029231-0. [DOI] [PubMed] [Google Scholar]
- 149.Reich S, Guilligay D, Pflug A, et al. Structural insight into capsnatching and RNA synthesis by influenza polymerase. Nature. 2014 Nov 19; doi: 10.1038/nature14009. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 150.Hatakeyama D, Shoji M, Yamayoshi S, et al. A novel functional site in the PB2 subunit of influenza a virus essential for acetyl- CoA interaction, RNA polymerase activity, and viral replication. J Biol Chem. 2014;289:24980–24994. doi: 10.1074/jbc.M114.559708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hutchinson EC, Fodor E. Transport of the Influenza Virus Genome from Nucleus to Nucleus. Viruses. 2013;5:2424–2446. doi: 10.3390/v5102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.York A, Fodor E. Biogenesis, assembly, and export of viral messenger ribonucleoproteins in the influenza a virus infected cell. RNA Biol. 2013;10:1274–1282. doi: 10.4161/rna.25356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fodor E. The RNA polymerase of influenza A virus: mechanisms of viral transcription and replication. Acta Virol. 2013;57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 154.Momose F, Basler CF, O'Neill RE, et al. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol. 2001;75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Naito T, Momose F, Kawaguchi A, et al. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol. 2007;81:1339–1349. doi: 10.1128/JVI.01917-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vreede FT, Brownlee GG. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J Virol. 2007;81:2196–2204. doi: 10.1128/JVI.02187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Perez JT, Varble A, Sachidanandam R, et al. Influenza a virusgenerated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A. 2010;107:11525–11530. doi: 10.1073/pnas.1001984107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hutchinson EC, Fodor E. Nuclear import of the influenza a virus transcriptional machinery. Vaccine. 2012;30:7353–7358. doi: 10.1016/j.vaccine.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 159.Loucaides EM, Kirchbach JC, Foeglein A, et al. Nuclear dynamics of influenza a virus ribonucleoproteins revealed by live-cell imaging studies. Virology. 2009;394:154–163. doi: 10.1016/j.virol.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Resa-Infante P, Jorba N, Zamarreno N, et al. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One. 2008;3:e3904–e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Portela A, Digard P. The influenza virus nucleoprotein: a multifunctional RNA-binding protein pivotal to virus replication. J Gen Virol. 2002;83:723–734. doi: 10.1099/0022-1317-83-4-723. [DOI] [PubMed] [Google Scholar]
- 162.Amorim MJ, Read EK, Dalton RM, et al. Nuclear export of influenza a virus mRNAs requires ongoing RNA polymerase II activity. Traffic. 2007;8:1–11. doi: 10.1111/j.1600-0854.2006.00507.x. [DOI] [PubMed] [Google Scholar]
- 163.Moeller A, Kirchdoerfer RN, Potter CS, et al. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zheng W, Tao YJ. Structure and assembly of the influenza a virus ribonucleoprotein complex. FEBS Lett. 2013;587:1206–1214. doi: 10.1016/j.febslet.2013.02.048. [DOI] [PubMed] [Google Scholar]
- 165.Su CY, Cheng TJ, Lin MI, et al. High-throughput identification of compounds targeting in2l6enza RNA-dependent RNA polymerase activity. Proc Natl Acad Sci U S A. 2010;107:19151–19156. doi: 10.1073/pnas.1013592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Landeras-Bueno S, Jorba N, Pérez-Cidoncha M, et al. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011;7:e1002397–e1002397. doi: 10.1371/journal.ppat.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Tsai PL, Chiou NT, Kuss S, et al. Cellular RNA binding proteins NS1-BP and hnRNP K regulate influenza a virus RNA splicing. PLoS Pathog. 2013;9:e1003460–e1003460. doi: 10.1371/journal.ppat.1003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dubois J, Terrier O, Rosa-Calatrava M. Influenza viruses and mRNA splicing: doing more with less. MBio. 2014;5:e00070–e00114. doi: 10.1128/mBio.00070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Perwitasari O, Johnson S, Yan X, et al. Verdinexor, a novel selective inhibitor of nuclear export, reduces influenza a virus replication in vitro and in vivo. J Virol. 2014;88:10228–10243. doi: 10.1128/JVI.01774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Simon JP, Ivanov IE, Shopsin B, et al. The in vitro generation of post-Golgi vesicles carrying viral envelope glycoproteins requires an ARF-like GTP-binding protein and a protein kinase C associated with the Golgi apparatus. J Biol Chem. 1996;271:16952–16961. doi: 10.1074/jbc.271.28.16952. [DOI] [PubMed] [Google Scholar]
- 171.Predicala R, Zhou Y. The role of Ran-binding protein 3 during influenza a virus replication. J Gen Virol. 2013;94:977–984. doi: 10.1099/vir.0.049395-0. [DOI] [PubMed] [Google Scholar]
- 172.Akarsu H, Burmeister WP, Petosa C, et al. Crystal structure of the M1 protein-binding domain of the influenza a virus nuclear export protein (NEP/NS2) EMBO J. 2003;22:4646–4655. doi: 10.1093/emboj/cdg449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Watanabe K, Takizawa N, Noda S, et al. Hsc70 regulates the nuclear export but not the import of influenza viral RNP: a possible target for the development of anti-influenza virus drugs. Drug Discov Ther. 2008;2:77–84. [PubMed] [Google Scholar]
- 174.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 175.Ma K, Roy AM, Whittaker GR. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology. 2001;282:215–220. doi: 10.1006/viro.2001.0833. [DOI] [PubMed] [Google Scholar]
- 176.Santos A, Pal S, Chacón J, et al. SUMOylation affects the interferon blocking activity of the influenza A nonstructural protein NS1 without affecting its stability or cellular localization. J Virol. 2013;87:5602–5620. doi: 10.1128/JVI.02063-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wurzer WJ, Planz O, Ehrhardt C, et al. Caspase 3 activation is essential for efficient influenza virus propagation. EMBO J. 2003;22:2717–2728. doi: 10.1093/emboj/cdg279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Marjuki H, Alam MI, Ehrhardt C, et al. Membrane accumulation of influenza a virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J Biol Chem. 2006;281:16707–16715. doi: 10.1074/jbc.M510233200. [DOI] [PubMed] [Google Scholar]
- 179.Galli C, Bernasconi R, Soldà T, et al. Malectin participates in a backup glycoprotein quality control pathway in the mammalian ER. PLoS One. 2011;6:e16304–e16304. doi: 10.1371/journal.pone.0016304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Wang N, Glidden EJ, Murphy SR, et al. The cotranslational maturation program for the type II membrane glycoprotein influenza neuraminidase. J Biol Chem. 2008;283:33826–33837. doi: 10.1074/jbc.M806897200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pearse BR, Gabriel L, Wang N, et al. A cell-based reglucosylation assay demonstrates the role of GT1 in the quality control of a maturing glycoprotein. J Cell Biol. 2008;181:309–320. doi: 10.1083/jcb.200712068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hebert DN, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 183.Daniels R, Kurowski B, Johnson AE, et al. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 184.Pleschka S, Wolff T, Ehrhardt C, et al. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol. 2001;3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 185.Iwai A, Shiozaki T, Miyazaki T. Relevance of signaling molecules for apoptosis induction on influenza a virus replication. Biochem Biophys Res Commun. 2013;441:531–537. doi: 10.1016/j.bbrc.2013.10.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Eisfeld AJ, Kawakami E, Watanabe T, et al. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol. 2011;85:6117–6126. doi: 10.1128/JVI.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kawaguchi A, Matsumoto K, Nagata K. YB-1 functions as a porter to lead influenza virus ribonucleoprotein complexes to microtubules. J Virol. 2012;86:11086–11095. doi: 10.1128/JVI.00453-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Momose F, Sekimoto T, Ohkura T, et al. Apical transport of influenza a virus ribonucleoprotein requires Rab11-positive recycling endosome. PLoS One. 2011;6:e21123–e21123. doi: 10.1371/journal.pone.0021123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jo S, Kawaguchi A, Takizawa N, et al. Involvement of vesicular trafficking system in membrane targeting of the progeny influenza virus genome. Microbes Infect. 2010;12:1079–1084. doi: 10.1016/j.micinf.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 190.Marjuki H, Alam MI, Ehrhardt C, et al. Membrane accumulation of influenza a virus hemagglutinin triggers nuclear export of the viral genome via protein kinase Calpha-mediated activation of ERK signaling. J Biol Chem. 2006;281:16707–16715. doi: 10.1074/jbc.M510233200. [DOI] [PubMed] [Google Scholar]
- 191.Hutchinson EC, Kirchbach JC, Gog JR, et al. Genome packaging ininfluenza a virus. J Gen Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 192.Rossman JS, Lamb RA. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Calder LJ, Wasilewski S, Berriman JA, et al. Structural organization of a filamentous influenza a virus. Proc Natl Acad Sci U S A. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Fournier E, Moules V, Essere B, et al. A supramolecular assembly formed by influenza a virus genomic RNA segments. Nucleic Acids Res. 2012;40:2197–2209. doi: 10.1093/nar/gkr985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Rossman JS, Jing X, Leser GP, et al. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Eisfeld AJ, Kawakami E, Watanabe T, et al. RAB11A is essential for transport of the influenza virus genome to the plasma membrane. J Virol. 2011;85:6117–6126. doi: 10.1128/JVI.00378-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Eisfeld AJ, Neumann G, Kawaoka Y. Human immunodeficiency virus rev-binding protein is essential for influenza a virus replication and promotes genome trafficking in late-stage infection. J Virol. 2011;85:9588–9598. doi: 10.1128/JVI.05064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Taylor G. Sialidases: structures, biological significance and therapeutic potential. Curr Opin Struct Biol. 1996;6:830–837. doi: 10.1016/s0959-440x(96)80014-5. [DOI] [PubMed] [Google Scholar]
- 199.Air GM, Laver WG. The neuraminidase of influenza virus. Proteins. 1989;6:341–356. doi: 10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 200.Greenway KT, LeGresley EB, Pinto BM. The influence of 150-cavity binders on the dynamics of influenza a neuraminidases as revealed by molecular dynamics simulations and combined clustering. PLoS One. 2013;8:e59873–e59873. doi: 10.1371/journal.pone.0059873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Watanabe R, Lamb RA. Influenza virus budding does not require a functional AAA+ ATPase, VPS4. Virus Res. 2010;153:58–63. doi: 10.1016/j.virusres.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bruce EA, Medcalf L, Crump CM, et al. Budding of filamentous and non-filamentous influenza a virus occurs via a VPS4 and VPS28-independent pathway. Virology. 2009;390:268–278. doi: 10.1016/j.virol.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 203.Hayden FG, Aoki FY. Amantadine, rimatadine, and related agents. In: Barriere SL, editor. Antimicrobial Therapy and Vaccines. Baltimore: Williams & Williams; 1999. pp. 1344–1365. [Google Scholar]
- 204.Wang C, Takeuchi K, Pinto LH, et al. Ion channel activity of influenza a virus M2 protein: characterization of the amantadine block. J Virol. 1993:675585–675594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Connolly BS, Lang AE. Pharmacological treatment of Parkinson disease: a review. JAMA. 2014;311:1670–1683. doi: 10.1001/jama.2014.3654. [DOI] [PubMed] [Google Scholar]
- 206.Stryjer R, Budnik D, Ebert T, et al. Amantadine augmentation therapy for obsessive compulsive patients resistant to SSRIs-an open-label study. Clin Neuropharmacol. 2014;37:79–81. doi: 10.1097/WNF.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 207.Videnovic A. Treatment of Huntington disease. Curr Treat Options Neurol. 2013;15:424–438. doi: 10.1007/s11940-013-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Hosenbocus S, Chahal R. Amantadine: a review of use in child and adolescent psychiatry. J Can Acad Child Adolesc Psychiatry. 2013;22:55–60. [PMC free article] [PubMed] [Google Scholar]
- 209.Soares BG, Lima MS, Reisser AA, et al. Dopamine agonists for cocaine dependence. Cochrane Database Syst Rev. 2001;(4):CD003352–CD003352. doi: 10.1002/14651858.CD003352. [DOI] [PubMed] [Google Scholar]
- 210.Smith JP, Riley TR, Bingaman S, et al. Amantadine therapy for chronic hepatitis C: a dose escalation study. Am J Gastroenterol. 2004;99:1099–1104. doi: 10.1111/j.1572-0241.2004.30798.x. [DOI] [PubMed] [Google Scholar]
- 211.Braham J. Amantadine in the treatment of Creutzfeldt-Jakob disease. Arch Neurol. 1984;41:585–586. [PubMed] [Google Scholar]
- 212.Ohlmeier MD, Zhang Y, Bode L, et al. Amantadine reduces mania in borna disease virus-infected non-psychotic bipolar patients. Pharmacopsychiatry. 2008;41:202–203. doi: 10.1055/s-2008-1078748. [DOI] [PubMed] [Google Scholar]
- 213.Colgan R, Michocki R, Greisman L, et al. Antiviral drugs in the immunocompetent host: part I. Treatment of hepatitis, cytomegalovirus, and herpes infections. Am Fam Physician. 2003;67:757–762. [PubMed] [Google Scholar]
- 214.Ruigrok RW, Hirst EM, Hay AJ. The specific inhibition of influenza a virus maturation by amantadine: an electron microscopic examination. J Gen Virol. 1991;72:191–194. doi: 10.1099/0022-1317-72-1-191. [DOI] [PubMed] [Google Scholar]
- 215.Gu RX, Liu LA, Wei DQ. Structural and energetic analysis of drug inhibition of the influenza A M2 proton channel. Trends Pharmacol Sci. 2013;34:571–580. doi: 10.1016/j.tips.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 216.Ison MG, Hayden FG. Therapeutic options for the management of influenza. Curr Opin Pharmacol. 2001;1:482–490. doi: 10.1016/s1471-4892(01)00084-4. [DOI] [PubMed] [Google Scholar]
- 217.Allen UD, Aoki FY, Stiver HG. The use of antiviral drugs for influenza: recommended guidelines for practitioners. Can J Infect Dis Med Microbiol. 2006;17:273–284. doi: 10.1155/2006/165940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Cattoni J, Parekh R. Acute respiratory distress syndrome: a rare presentation of amantadine toxicity. Am J Case Rep. 2014;15:1–3. doi: 10.12659/AJCR.889931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Alves Galvão MG, Rocha Crispino Santos MA, Alves da Cunha AJ. Amantadine and rimantadine for influenza A in children and the elderly. Cochrane Database Syst Rev. 2014;(11):CD002745–CD002745. doi: 10.1002/14651858.CD002745.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Keyser LA, Karl M, Nafziger AN, et al. Comparison of central nervous system adverse effects of amantadine and rimantadine used as sequential prophylaxis of influenza A in elderly nursing home patients. Arch Intern Med. 2000;160:1485–1488. doi: 10.1001/archinte.160.10.1485. [DOI] [PubMed] [Google Scholar]
- 221.Hsieh HP, Hsu JT. Strategies of development of antiviral agents directed against influenza virus replication. Curr Pharm Des. 2007;13:3531–3542. doi: 10.2174/138161207782794248. [DOI] [PubMed] [Google Scholar]
- 222.Sheu TG, Fry AM, Garten RJ, et al. Dual resistance to adamantanes and oseltamivir among seasonal influenza A(H1N1) viruses: 2008-2010. J Infect Dis. 2011;203:13–17. doi: 10.1093/infdis/jiq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ison MG. Clinical use of approved influenza antivirals: therapy and prophylaxis. Influenza Other Respir Viruses. 2013;7(Suppl 1):7–13. doi: 10.1111/irv.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Burnet FM, McCrea JF, Stone JD. Modification of Human Red Cells by Virus Action. I. The Receptor Gradient for Virus Action in Human Red Cells. Br J Exp Pathol. 1946;27:228–236. [PMC free article] [PubMed] [Google Scholar]
- 225.Itzstein M, Wu WY, Kok GB, et al. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 226.Tullu MS. Oseltamivir. J Postgrad Med. 2009;55:225–230. doi: 10.4103/0022-3859.57411. [DOI] [PubMed] [Google Scholar]
- 227.Burch J, Paulden M, Conti S, et al. Antiviral drugs for the treatment of influenza: a systematic review and economic evaluation. Health Technol Assess. 2009;13:1–265. doi: 10.3310/hta13580. [DOI] [PubMed] [Google Scholar]
- 228.Jackson RJ, Cooper KL, Tappenden P, et al. Oseltamivir, zanamivir and amantadine in the prevention of influenza: a systematic review. J Infect. 2011;62:14–25. doi: 10.1016/j.jinf.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 229.Jefferson T, Jones MA, Doshi P, et al. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database Syst Rev. 2014;(4):CD008965–CD008965. doi: 10.1002/14651858.CD008965.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Beigi RH, Venkataramanan R, Caritis SN. Oseltamivir for influenza in pregnancy. Semin Perinatol. 2014 Sep 30;pii:S0146- 0005(14)00103–S0146- 0005(14)00107. doi: 10.1053/j.semperi.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. CDC , author. Influenza antiviral medications: summary for clinicians (current for the 2013-14 influenza season) Document available at: http://www.cdc.gov/flu/professionals/antivirals/summaryclinicians.htm. accessed on 24th July 2014.
- 232. WHO , author. Global alert and Response (GAR) Antiviral drugs for pandemic (H1N1) 2009: definitions and use. Document available at: http://www.who.int/csr/disease/swineflu/frequently_asked_questions/antivirals/definitions_use/en/ accessed on 24th July 2014).
- 233.Barroso L, Treanor J, Gubareva L, et al. Efficacy and tolerability of the oral neuraminidase inhibitor peramivir in experimental human influenza: randomized, controlled trials for prophylaxis and treatment. Antivir Ther. 2005;10:901–910. [PubMed] [Google Scholar]
- 234.Birnkrant D, Cox E. The emergency use authorization of peramivir for treatment of 2009 h1n1 influenza. N Engl J Med. 2009;361:2204–2207. doi: 10.1056/NEJMp0910479. [DOI] [PubMed] [Google Scholar]
- 235.Koyama K, Ogura Y, Nakai D, et al. Identification of bioactivating enzymes involved in the hydrolysis of laninamivir octanoate, a long-acting neuraminidase inhibitor, in human pulmonary tissue. Drug Metab Dispos. 2014;42:1031–1038. doi: 10.1124/dmd.114.057620. [DOI] [PubMed] [Google Scholar]
- 236.Kashiwagi S, Yoshida S, Yamaguchi H, et al. Safety of the long-acting neuraminidase inhibitor laninamivir octanoate hydrate in post-marketing surveillance. Int J Antimicrob Agents. 2012;40:381–388. doi: 10.1016/j.ijantimicag.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 237.Katsumi Y, Otabe O, Matsui F, et al. Effect of a single inhalation of laninamivir octanoate in children with influenza. Pediatrics. 2012;129:e1431–e1436. doi: 10.1542/peds.2011-2054. [DOI] [PubMed] [Google Scholar]
- 238.Yoshiba S, Okabe H, Ishizuka H. Pharmacokinetics of laninamivir after a single administration of its prodrug, laninamivir octanoate, a long-acting neuraminidase inhibitor, using an easy-to-use inhaler in healthy volunteers. J Bioequiv Availab. 2011;3:001–004. [Google Scholar]
- 239.Streeter DG, Witkowski JT, Khare GP, et al. Mechanism of action of 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide (Virazole), a new broad-spectrum antiviral agent. Proc Natl Acad Sci U S A. 1973;70:1174–1178. doi: 10.1073/pnas.70.4.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Crotty S, Cameron C, Andino R. Ribavirin's antiviral mechanism of action: lethal mutagenesis? J Mol Med (Berl) 2002;80:86–95. doi: 10.1007/s00109-001-0308-0. [DOI] [PubMed] [Google Scholar]
- 241.Chan-Tack KM, Murray JS, Birnkrant DB. Use of ribavirin to treat influenza. N Engl J Med. 2009;361:1713–1714. doi: 10.1056/NEJMc0905290. [DOI] [PubMed] [Google Scholar]
- 242.Uchide N, Ohyama K, Toyoda H. Current and future anti-influenza virus drugs. The Open Antimicrobial Agents Journal. 2010;2:34–48. [Google Scholar]
- 243.Deng P, Zhong D, Yu K, et al. Pharmacokinetics, metabolism, and excretion of the antiviral drug arbidol in humans. Antimicrob Agents Chemother. 2013;57:1743–1755. doi: 10.1128/AAC.02282-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Shi L, Xiong H, He J, et al. Antiviral activity of arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo. Arch Virol. 2007;152:1447–1455. doi: 10.1007/s00705-007-0974-5. [DOI] [PubMed] [Google Scholar]
- 245.Mola A, Peduto A, Gatta A, et al. Structure-activity relationship study of arbidol derivatives as inhibitors of chikungunya virus replication. Bioorg Med Chem. 2014 Sep 16;pii:S0968-0896(14)00649-X–S0968-0896(14)00649-X. doi: 10.1016/j.bmc.2014.09.013. [DOI] [PubMed] [Google Scholar]
- 246.Brooks MJ, Sasadeusz JJ, Tannock GA. Antiviral chemotherapeutic agents against respiratory viruses: where are we now and what's in the pipeline? Curr Opin Pulm Med. 2004;10:197–203. doi: 10.1097/00063198-200405000-00009. [DOI] [PubMed] [Google Scholar]
- 247.Blaising J, Lévy PL, Polyak SJ, et al. Arbidol inhibits viral entry by interfering with clathrin-dependent trafficking. Antiviral Res. 2013;100:215–219. doi: 10.1016/j.antiviral.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 248.Wei F, Li JL, Ling JX, et al. Establishment of SYBR greenbased qPCR assay for rapid evaluation and quantification for anti-Hantaan virus compounds in vitro and in suckling mice. Virus Genes. 2013;46:54–62. doi: 10.1007/s11262-012-0834-6. [DOI] [PubMed] [Google Scholar]
- 249.Santesso N, Hsu J, Mustafa R, et al. Antivirals for influenza: a summary of a systematic review and meta-analysis of observational studies. Influenza Other Respir Viruses. 2013;7(Suppl 2):76–81. doi: 10.1111/irv.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.He G, Qiao J, Dong C, et al. Amantadine-resistance among H5N1 avian influenza viruses isolated in Northern China. Antiviral Res. 2008;77:72–76. doi: 10.1016/j.antiviral.2007.08.007. [DOI] [PubMed] [Google Scholar]