Abstract
Electronic health records (EHR) provide a comprehensive resource for discovery, allowing unprecedented exploration of the impact of genetic architecture on health and disease. The data of EHRs also allow for exploration of the complex interactions between health measures across health and disease. The discoveries arising from EHR based research provide important information for the identification of genetic variation for clinical decision-making. Due to the breadth of information collected within the EHR, a challenge for discovery using EHR based data is the development of high-throughput tools that expose important areas of further research, from genetic variants to phenotypes. Phenome-Wide Association studies (PheWAS) provide a way to explore the association between genetic variants and comprehensive phenotypic measurements, generating new hypotheses and also exposing the complex relationships between genetic architecture and outcomes, including pleiotropy. EHR based PheWAS have mainly evaluated associations with case/control status from International Classification of Disease, Ninth Edition (ICD-9) codes. While these studies have highlighted discovery through PheWAS, the rich resource of clinical lab measures collected within the EHR can be better utilized for high-throughput PheWAS analyses and discovery. To better use these resources and enrich PheWAS association results we have developed a sound methodology for extracting a wide range of clinical lab measures from EHR data. We have extracted a first set of 21 clinical lab measures from the de-identified EHR of participants of the Geisinger MyCode™ biorepository, and calculated the median of these lab measures for 12,039 subjects. Next we evaluated the association between these 21 clinical lab median values and 635,525 genetic variants, performing a genome-wide association study (GWAS) for each of 21 clinical lab measures. We then calculated the association between SNPs from these GWAS passing our Bonferroni defined p-value cutoff and 165 ICD-9 codes. Through the GWAS we found a series of results replicating known associations, and also some potentially novel associations with less studied clinical lab measures. We found the majority of the PheWAS ICD-9 diagnoses highly related to the clinical lab measures associated with same SNPs. Moving forward, we will be evaluating further phenotypes and expanding the methodology for successful extraction of clinical lab measurements for research and PheWAS use. These developments are important for expanding the PheWAS approach for improved EHR based discovery.
1. Introduction
Precision medicine aims to find clinical treatments based on the phenotypic and genetic makeup of each individual. Electronic health records (EHR) are a powerful resource for the investigation of common and rare disease, with the potential for discovery that will lead to meaningful and data-driven individualized patient care. Accessing de-identified EHR data linked to DNA biorepositories has already proved useful for a wide range of genetic association discovery efforts, such as through the Electronic Medical Records and Genomics (eMERGE) network1.
In PheWAS, the association between thousands of phenotypes and any number of single nucleotide polymorphisms (SNPs) are evaluated in a high-throughput manner to identify new hypotheses, biologically relevant associations, and the identification of potential pleiotropy, highlighting important connections between networks of phenotypes and genetic architecture2–4. To date, de-identified EHR data coupled with genetic data have been used for multiple PheWAS, primarily through using International Classification of Disease, Ninth Edition (ICD-9) based case/control status for identifying significant associations between medical record diagnoses and genetic data5–8.
There are other data within the EHR that can also be used for high-throughput PheWAS research, with one of the most readily available additional sources of data being clinical lab measures. Clinical lab measures are an important part of clinical decision-making, providing clues and measures of a variety of conditions as well as important reflections of health. Many of these lab measures are found in multiple diagnoses, for example, blood cell count information is important for a variety of clinical conditions and diagnoses. To date, high-throughput use of clinical lab measures from the EHR have been underutilized for multiple reasons. These include the variability and error in the units recorded that can occur across measurements, error that can occur in the collected laboratory result, change in laboratory assays, sensitivity of different assays, lack of documentation for fasting, and changes in biological function due to treatment of injury or disease (e.g. medication use). Even with these challenges there is an opportunity for further discovery by using more of the comprehensive clinical lab data available within the EHR for both high-quality phenotype algorithm development as well as expanding EHR based PheWAS beyond the use of ICD-9 based case/control status. The clinical lab measures of the EHR can more closely reflect the impact of genetic variation on phenotype, and some phenotypes observed from the clinical lab data collected in EHR are not reflected at all in case/control diagnoses or common to multiple case/control diagnoses. Using a wide range of clinical lab measures within the PheWAS framework also creates a series of results to compare and contrast with the findings of ICD-9 based PheWAS, providing a complementary set of information pertinent to health and disease and genetic association studies, enriching the interpretation and exploration of ICD-9 based PheWAS. There is also the potential for improved power for association analyses, as case numbers for ICD-9 based PheWAS can be very low depending on the ICD-9 based diagnosis compared to larger sample sizes for quantitative clinical lab measures.
We describe here our preliminary algorithmic development for high-throughput extraction of clinical lab measures from de-identified data linked to genetic data from the Geisinger Health System (GHS) MyCode™ Biorepository. For these analyses we used our approach to extract 21 clinical lab measurements with some of the largest sample sizes within the EHR from, or derived from, blood: alanine amino transferase (ALT), albumin aspartate aminotransferase (AST), carbon dioxide (CO2), cholesterol, creatinine, free T3, free T4, glucose, hemoglobin A1C (Hb-A1C), high density lipoprotein (HDL), insulin-like growth factor (IgF-1), low density lipoprotein (LDL), platelets, urine protein, red blood cell counts (RBC), thyroglobulin antibody (TgAb), thyroid peroxidase antibody (TPO), thyroid stimulating hormone (TSH), triglycerides (TG), white blood cell counts (WBC). We also extracted body mass index measurements (BMI). We calculated the median value for each of 12,039 individuals and performed comprehensive genome-wide association analyses (GWAS) with these measurements in European-Americans within the MyCode Biorepository, and then explored associations with highly-significant SNPs from the GWAS with an ICD-9 diagnosis code based PheWAS. These preliminary analyses show the success of our approach, and the ultimate success possible in high-throughput extraction of a wide range of clinical lab measurements from the EHR.
2. Methods
2.1 Study Participants
In this study we used de-identified genetic and phenotypic data from MyCode™ biorepository of Geisinger Heath System (GHS). MyCode is a biorepository that stores blood samples and Electronic Health Record (EHR) data from consented individuals for research to improve patient healthcare. GHS is located in central Pennsylvania, which is a primarily European American (EA) population with 95.7% of individuals in the study of European decent. Thus we only focused on individuals from EA ancestry for these analyses.
2.2 Genotypic data and Quality Control
GHS MyCode subjects were genotyped using the Illumina HumanOmniExpressExome Bead Chips, with coverage of a total of 964,193 SNPs. We performed Genotype Quality Control (QC) procedures to account of genotyping error prior to association testing using the R programming statistical package9 and PLINK software10. We filtered out the missing data using 99% genotype and sample call rates and minor allele frequency (MAF) threshold of 1%. Also, relatedness between the individuals was calculated by Identity by Descent (IBD) and related samples were dropped using kinship coefficient of 0.125. After these QC steps and MAF filter, the genotypic data consisted of 635,525 SNPs and 12,278 samples. While individuals within GHS are primarily from EA populations, we calculated principle components to further correct for global ancestry in our associations using EIGENSOFT11.
2.3 Clinical Lab Extraction
We extracted a total of 21 clinical lab measurements from, or derived from, blood of participants in the study. We selected an initial set of lab measurements to extract by choosing the measures with the large sample sizes that we have commonly used for other phenotype algorithm development using data from the EHR. All the summary information on the clinical lab measurements is provided in Table 1. We also extracted data to calculate body mass index (BMI). BMI is known to have confounding effects on various metabolic traits and many of the clinical lab measurements could be affected by BMI of study participants. We extracted height and weight to calculate BMI. We calculated the median value for each of these clinical lab measures over the course of all visits of each individual.
Table 1.
Summary of clinical lab measures, and any transformation of the variable before analysis
| Phenotype | Median | Mean | Min/Max | SD | % Male | % Female |
|---|---|---|---|---|---|---|
| Alanine Amino Transferase (ALT) (Log Transform) | 3.09 | 3.10 | 1.79/4.48 | 0.43 | 41.47 | 58.53 |
| Albumin | 4.30 | 4.26 | 3.45/5.30 | 0.26 | 41.19 | 58.81 |
| Aspartate Aminotransferase (AST) (Log Transform) | 3.13 | 3.16 | 2.35/4.04 | 0.26 | 41.23 | 58.77 |
| Carbon Dioxide (CO2) | 27.00 | 26.97 | 21.29/34 | 1.98 | 42.06 | 57.94 |
| Cholesterol | 183.00 | 184.55 | 110/320 | 33.56 | 43.13 | 56.87 |
| Creatinine | 57.03 | 57.45 | 21.07/105.76 | 13.58 | 63.36 | 36.64 |
| Free T3 | 2.90 | 2.94 | 1.64/5.19 | 0.53 | 24.48 | 75.52 |
| Free T4 (Log Transform) | 0.19 | 0.22 | 0.009/0.63 | 0.14 | 29.95 | 70.05 |
| Glucose (Log Transform) | 4.60 | 4.64 | 4.14/5.12 | 0.18 | 41.83 | 58.17 |
| Hemoglobin A1C (Hb-A1C) | 6.20 | 6.50 | 4.09/10.80 | 1.17 | 43.01 | 56.99 |
| High Density Lipoprotein (HDL) (Log Transform) | 3.87 | 3.88 | 3.13/4.70 | 0.27 | 43.31 | 56.69 |
| Insulin-like Growth Factor (IgF-1) (Log Transform) | 4.96 | 4.90 | 3.08/6.08 | 0.62 | 41.67 | 58.33 |
| Low Density Lipoprotein (LDL) | 102.00 | 104.08 | 48/237.5 | 28.44 | 43.17 | 56.83 |
| Platelets | 238.50 | 243.80 | 112/526 | 60.16 | 40.67 | 59.33 |
| Urine Protein (Log Transform) | 2.40 | 2.60 | 0.09/6.53 | 1.24 | 44.26 | 55.74 |
| Red Blood Cell Count (RBC) | 4.48 | 4.48 | 3.27/6.09 | 0.43 | 40.75 | 59.25 |
| Thyroglobulin antibody (TgAb) | 20.00 | 26.83 | 1.70/305 | 31.95 | 21.12 | 78.88 |
| Thyroid Peroxidase Antibody (TPO) (Log Transform) | 2.30 | 3.27 | 0.09/7.94 | 1.74 | 18.38 | 81.62 |
| Thyroid Stimulating Hormone (TSH) (Log Transform) | 0.75 | 0.77 | 0.009/2.04 | 0.41 | 37.43 | 62.57 |
| Triglycerides (TG) (Log Transform) | 4.88 | 4.88 | 3.46/6.25 | 0.47 | 43.32 | 56.68 |
| White Blood Cell Count (WBC) (Log Transform) | 7.42 | 7.68 | 3.15/16.84 | 2.00 | 40.80 | 59.20 |
We extracted these clinical lab measurements as follows: First, we extracted each clinical laboratory measures from the de-identified EHR and log base ten transformed the results. We then standardized units within each clinical lab measure. Different Geisinger Health laboratories and Point of Care devices can have differing units of measure within the same Logical Observation Identifiers Names and Codes (LOINC) code and thus standardization and transformation of individual values needed to be performed so that all units were consistent across each clinical lab measure. We excluded measurements where the unit of measure reported on the result was different than the suggested unit of measure from LOINC when conversion was not possible. We then excluded results that were identified as implausible through a process of comparing individual level and population level medians greater than a deviation threshold determined by each LOINC code, +/− 3 standard deviations from the median. After the process of excluding these results, we transformed the results back to their original values. For TG, HDL, LDL, glucose levels and cholesterol, we omitted all observations that were not known to be fasting, i.e., observations with non-fasting or unknown fasting state. When calculating medians and standard deviation we accounted for the number of results over a patient’s lifetime to adjust where exclusions are applied. Using this approach, as proof-of-principle, we extracted 21 different clinical lab tests from the entire GHS cohort, ~1.25 million people.
After these clinical lab measurements were extracted and prepared for further analyses, we calculated the lifetime median value from each individual for each of the lab measures for those individuals we had genetic data for, for association testing. We then created histograms and calculated the population median and max values for each clinical lab measure. This identified any measurements with non-normal distributions, and identified some of the most extreme outliers. We only removed outliers for white blood cell counts, values > 20K cells per/uL. We used naturallog transformation to improve the normality of the distributions for glucose and platelet measures. Our summary information of the median and mean of each measure, and whether or not each lab measure was transformed before association testing, is listed in Table 1.
2.4 ICD-9 Based Case-Control Status
We used International Classification of Diseases, Ninth Revision (ICD-9) codes as the phenotypic data to define case-control status for PheWAS. Patients in MyCode were diagnosed with 7,039 different ICD-9 codes, these codes have been used at least once during clinic visits at GHS facilities. We defined cases as individuals with three or more visits of a specific ICD-9 code at the 5-digit code level (e.g. 250.12), no visits of an ICD-9 code were defined as a control. If an individual had one to three visits of an ICD-9 code, they were not included as a case or control (i.e. excluded from analysis). A total of 200 or more case subjects per ICD-9 code were required for inclusion in our association study. Using our criteria for inclusion/exclusion for cases and controls there were a total of 165 ICD-9 codes used for the case/control association testing.
2.5 Genetic Associations
To evaluate the association between SNPs and the 21 clinical lab measures, we used linear regression with an additive encoding for the SNPs. We used the software Platform for the Analysis, Translation and Organization of large-scale data (PLATO), freely available here: http://ritchielab.psu.edu/software/plato-download. We have implemented PLATO in DNANexus (https://www.dnanexus.com/) to use cloud-computing resources for analyses. Covariates in the models included for each association were gender, age, age2 (age-squared), body mass index (BMI), and the first 4 principal components. We made Manhattan plots of the results of these associations, as well as a table of the results passing our Bonferroni correction based p-value available in supplementary materials (http://ritchielab.psu.edu/publications/supplementarydata/psb-2016/clinical-measure-phewas). After filtering the clinical lab measure association results by our Bonferroni threshold (described further in methods), and performing a PheWAS using ICD-9 based case/control status for these SNPs, we compared and contrasted the results of the clinical lab measurements with results of case/control ICD-9 based PheWAS. Further, we annotated the results of the clinical lab measure association testing using Biofilter12, to add information about any genes that the SNPs from the p-value filtered results were in or near, as well as to annotate the SNPs with any known results from the NHGRI GWAS catalog. The NHGRI GWAS catalog contains results from published GWAS in the literature reaching genome-wide significance13. To have the magnitude and direction of effect values more comparable across the association results, we divided each beta by the standard deviation of the respective clinical lab measure for the association.
2.6 Multiple Hypothesis Testing
In terms of independent SNPs used for multiple testing adjustment, independence of SNPs varies across different populations and appropriate measures are necessary14. In this study, we used a linkage-disequilibrium (LD) pruning approach to identify the number of independent SNPs used in our association testing. We used PLINK to prune the 635,525 SNPs based on pairwise linkage disequilibrium (LD) at r2 = 0.3 and that resulted in total of 174,401 SNPs. An r2 of 0.3 is estimated to be a reasonable threshold for finding independent SNPs based on pairwise LD14. For our clinical lab measures PheWAS, our Bonferoni threshold was α value divided by the number of independent tests: 0.05/(174,401 × 21) = 1.37 × 10−8. We used the same approach for multiple hypothesis correction for the ICD-9 based PheWAS, where we calculated LD between the 286 SNPs resulting in a total of 61 independent SNPs and thus a Bonferoni threshold: 0.05 / (61 × 165) = 4.9 × 10−6.
3. Results
3.1 Clinical Lab Measure GWAS
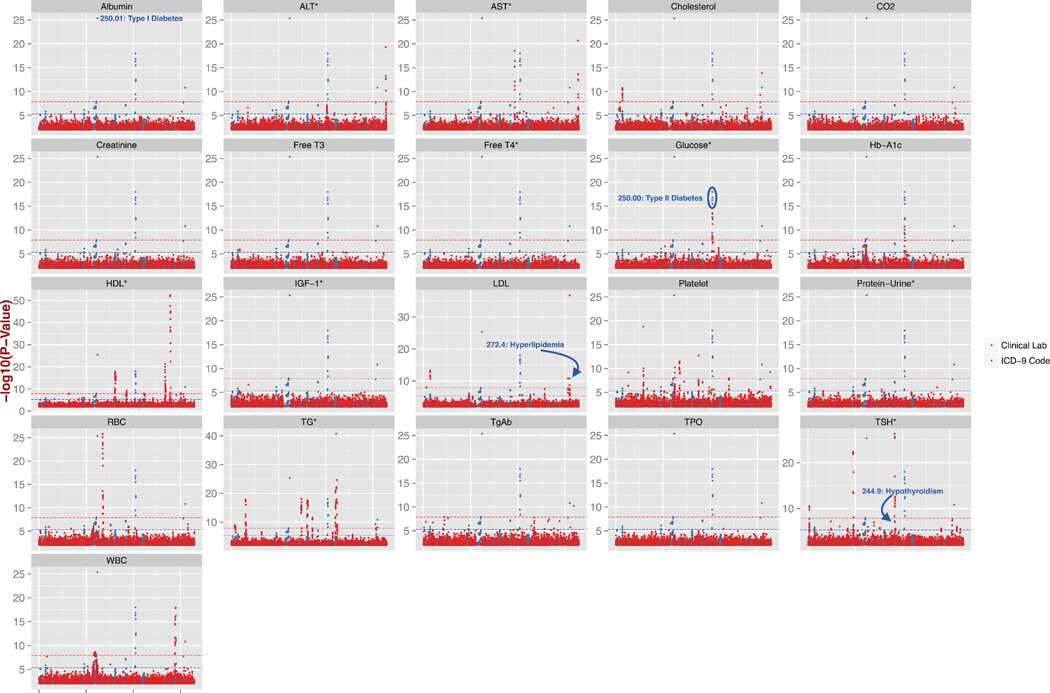
In this study, we first calculated the association between 635,525 SNPs and 21 clinical lab measurements using linear regression. Figure 1 shows the Manhattan plots from each of the clinical lab measure genome-wide association studies (GWAS), points indicated in red, for p-values < 0.01. We provide higher-resolution copies of the figures of this paper, as well as Quantile-Quantile plots for the associations in supplementary materials. A total of 286 SNPs were found significantly associated with our Bonferroni defined p-value threshold of 1.37 × 10−8, with a total of 344 SNP-clinical lab measure associations. A total of 163 out of 347 associations were found to be associated with same or similar previously reported phenotypic traits in the literature. Several associations were also for SNPs in high LD with SNPs for previously reported associations with the same or similar previously reported phenotypic trait. We observed that almost half of associations related to a previously reported association were with triglyceride and HDL-cholesterol levels, where we found 109 SNPs associated with triglycerides and 66 SNPs associated with HDL. For example, the association between SNP rs247616 downstream of CETP and HDL was the most significant association of all the GWAS we performed at p = 5.25 × 10−53, β = −0.22, this association has been previously reported in the literature15. CETP is protein-coding gene involved in cholesterol ester transfer from HDL to other lipoprotein18. The most significant association for TG was with SNP rs964184 downstream from ZPR1 with p = 1.9 × 10−41, β = −0.27, that has also been previously reported in the literature16,17.
Figure 1.
Manhattan plots of all 21 GWAS for clinical lab measures and the results of the following ICD-9 based PheWAS. For each of 21 clinical lab measures, the results of associations are marked as −log(10) of the p-value in red, with the abbreviation of each clinical lab measure indicated above each plot, abbreviations explained in Table 1. Plotted in blue are −log10 (p-value) from the associations of distinct ICD-9 code based case/control diagnoses. All results are from p-values < 0.01. The red dashed line in each Manhattan plot is at the Bonferroni corrected p-value of 1.37 × 10−8 for the clinical lab GWAS, and the blue dashed line is the Bonferroni corrected p-value 4.9 × 10−6 for the ICD-9 diagnoses based PheWAS.
A novel GWAS association found in this study was for DPP4 SNP rs2302872 associated with thyroid globulin antibody (TgAb). TgAb is a diagnostic measure used for thyroid related autoimmune disorders like Hashimoto’s disease and a measure to evaluate the treatment effectiveness of thyroid cancer. The SNP rs2302872 was associated with TgAb at p = 1.48 × 10−8, β = −1.28. DPP4 has known expression in cancerous thyroid tissue in comparison to no expression in a healthy thyroid tissue19. TgAb as a tumor marker for thyroid cancer has been controversial where many studies suggest no association as a tumor marker20,21 and others suggest TgAb levels can be used to identify increased risk of thyroid cancer22. In our case we see a potentially protective effect of this SNP in the direction of the association. In another thyroid measure, thyroid stimulating hormone (TSH), we report 4 novel loci on chromosome 5 mapped to PDE8B. There are other variants in PDE8B with known association with TSH23, but we found 4 polymorphisms (rs1351283, rs13158164, rs6885813, rs9686502) not in LD with the previously known variants with p-values of p = 1.16 × 10−18, β = −0.13; p = 2.60 × 10−14, β = −0.16; p = 4.74 × 10−14, β = −0.13; p=9.10 × 10−9, β = −0.08. We also found 7 novel SNPs associated with aspartate amino transferase (AST) levels, where all 7 variants mapped to MRC1, where the most significant association was for SNP rs35038329 with p = 2.87 × 10−19, β = 0.12. AST levels are used for the diagnosis of various liver diseases like hepatitis and cirrhosis. In a recent study variants in MRC1 were reported to be associated with treatment outcomes for hepatitis C24.
3.2 Targeted ICD-9 PheWAS
We selected the SNPs from the top associations of the clinical lab measure PheWAS (p-value < 1.37 × 10−8), resulting in 286 SNPs. We then performed an ICD-9 code based PheWAS with these SNPs, performing comprehensive associations testing between these SNPs and the ICD-9 based case/control status using logistic regression. Figure 1 shows in blue the ICD-9 based diagnosis associations with p-values less than 0.01. We found 39 associations passing our Bonferoni p-value threshold adjusted for the smaller number of SNPs for the ICD-9 base analyses, these additional association results are reported in detail within the supplementary materials. The most significant association was between SNP rs9273363 and the ICD-9 diagnosis 250.01 “Diabetes mellitus, Type I” with p = 4.39 × 10−26, β = −0.8. This SNP is located in the HLA region that is known to have high susceptibility for type 1 diabetes, and is one of the most high-risk polymorphisms in HLA region for Type I diabetes25. There were other significant associations with ICD-9 diagnoses such as SNPs associated with the diagnosis of 250.00 “Type II Diabetes” (original associated clinical lab measures of Glucose and Hb-A1C), 272.4 “Hyperlipidemia” (original associated clinical lab measure: Cholesterol, LDL, TG), and 244.9 “Hypothyroidism” (clinical lab measure: TSH).
3.3 Comparing Clinical Lab Measure GWAS and ICD-9 Based PheWAS
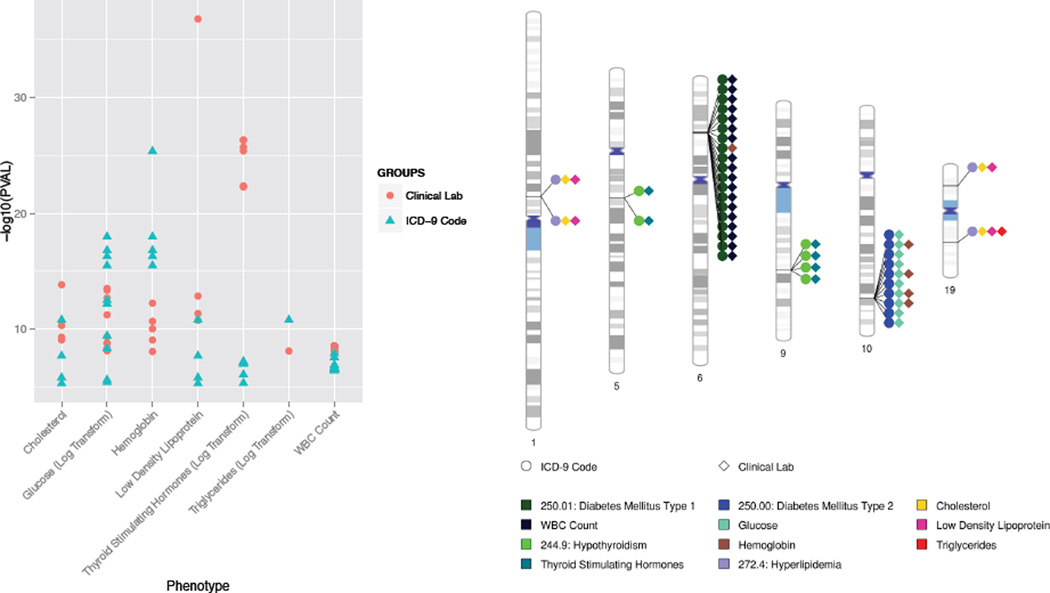
PheWAS frequently identifies cross-phenotype associations, where one SNP is associated with more than one phenotype. These cross-phenotype associations highlight potential relationships between the phenotypes, and can also identify pleiotropy. Thus, one of the potential benefits of using clinical lab measures in addition to ICD-9 codes is the addition of a complementary set of phenotypic information for exploring the multiple cross-phenotype associations that arise in PheWAS. In this study we started with SNPs highly associated with clinical lab measures, thus have compared and contrasted what the ICD-9 diagnoses were also associated with these clinical lab measures. Table 2 shows what the clinical lab measures were, and for SNPs associated with those specific clinical lab measures what the ICD-9 codes diagnoses were also associated those SNPs.
Table 2.
Phenotypes for SNPs significantly associated with clinical lab measures also significantly associated with highly related ICD-9 diagnoses
| Clinical Lab Measure | ICD-9 Diagnoses |
|---|---|
| Cholesterol | 272.4: Hyperlipidemia |
| Glucose | 250.00: Type II Diabetes Mellitus |
| Hemoglobin A1C | 250.01: Type I Diabetes Mellitus; 250.00: Type II Diabetes Mellitus |
| Low Density Lipoprotein (LDL) | 272.4: Hyperlipidemia |
| Thyroid Stimulating Hormones | 244.9: Hypothyroidism |
| Triglycerides | 272.4: Hyperlipidemia |
| White Blood Cell Count | 250.01: Type I diabetes mellitus |
First we identified results between the two clinical lab measures and ICD-9 diagnoses were highly related between the two sets of associations. For instance, for lipid related phenotypes and diagnoses we found the SNP rs445925 in APOC1 associated with LDL levels (p = 1.75 × 10−37, β = 0.30) also associated with the ICD-9 diagnosis 272.4 “Hyperlipidemia” (p = 1.5 × 10−11, β = 0.30). We also found the SNP rs602633 associated with LDL levels (p= 8.03 × 10−10, β = 0.12) and the ICD-9 diagnosis 272.4 (p = 4.55 × 10−6, β = 0.18). A SNP in LD with rs599839 (r2=1) has known associations with LDL concentrations26 and coronary artery disease27.
For thyroid diagnoses and TSH levels we also found related phenotypic associations for specific SNPs. For example, a cluster of variants in PTSC2 were associated with thyroid stimulating hormone levels (TSH) and also significantly associated with the ICD-9 diagnosis 244.9 “Hypothyroidism”. Another PTSC2 SNP rs10759944 was associated with TSH levels (p = 4.12 × 10−26, β = 0.16) and the ICD-9 diagnosis 244.9 “Hypothyroidism” (p = 9.48 × 10−8, β = 0.21). The SNP is in LD with rs965513 (r2 =0.9), a SNP with a known association with thyroid cancer28,29.
For Type 2 diabetes we had another significant finding with the known Type II diabetes risk gene TCF7L2, with variants in TCF7L2 associated with the ICD-9 diagnosis 250.00 “Type II diabetes” and clinical lab measures related to Type II diabetes including glucose levels and Hb-A1C levels.
We did find potentially novel pleiotropic associations with a cluster of SNPs in LD on chromosome 6. These SNPs were associated with WBC as well as Hb-A1C, and also associated with the ICD-9 based diagnosis of 250.01 “Type I diabetes”. While diabetes has an impact on Hb-A1C levels, the associations with WBC are more novel. White blood cell counts have been found to impact insulin sensitivity and diabetes development30.
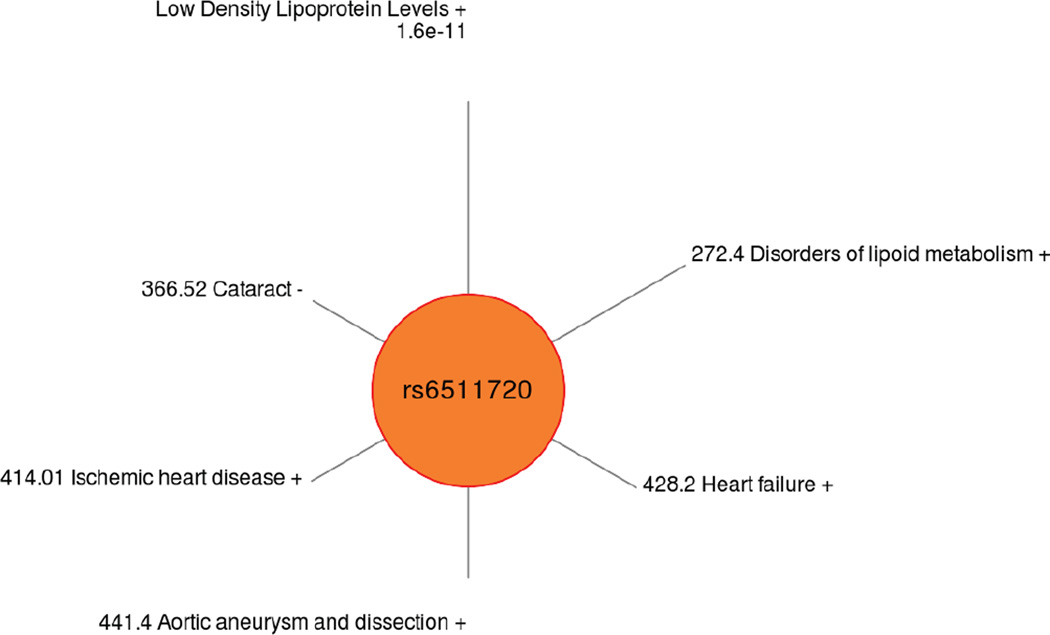
Using a less stringent cutoff for p-value for ICD-9 codes, which are often under powered associations due to the number of cases, Figure 3 shows an example of the ICD-9 PheWAS results for LDLR SNP rs6511720. The associated clinical lab measure was LDL levels, a previously reported association in the literature, and Figure 3 shows the spectrum of PheWAS results associated with this SNP with p < 0.01, a series of comorbidities related to cholesterol levels.
Figure 3.
Spectrum of phenotypic associations for LDLR SNP rs6511720, for PheWAS p-values < 0.01. This SNP was originally associated in our study with the clinical lab measure of LDL.
4. Discussion
The goal of this study was preliminary work in the process of accessing clinical lab measurements in a high-throughput way and developing algorithms, methodologies, and ultimately an analysis pipeline to be able to use a wide range of clinical lab measure for PheWAS. We have shown here that we can successfully extract clinical lab measures for association research and use these measures for association testing. The process of extracting and preparing these clinical lab measurements has been informative. Some of the challenges with these measurements, surmounted through our research, may provide information to inform better practices for collecting these data within the clinic in terms of standardization, which could benefit patients and clinicians as well as researchers. Facing the challenges of using clinical lab measurements is also providing preliminary information for how to address the challenge of accessing medication information within the EHR for use in research, which has many of the same issues as clinical lab measurements but additional challenges for research use.
We did identify association results replicating previously published associations indicating our clinical lab measure extraction is functional, as well as a number of novel associations. We intend moving forward to do an expanded study including additional clinical lab measurements, including additional measures that have been previously studied to continue expanding our proof-of-principle results, as well as a wide array of additional measures little studied in genetic association testing. The development of algorithms for obtaining summary information about these measures, when moving to hundreds of measures, will be important for quick evaluation of these phenotypes, and sub-setting of data based on specific criteria. Of further importance will be better use of the longitudinal nature of these clinical lab measures, in health and disease, as we are currently using median values in our association testing.
A challenge within PheWAS is to understand if we find associations due to correlated phenotypes (such as glucose levels and diabetes), or if we find associations related to the impact of genetic variation on more than one phenotype (pleiotropy). A future direction is to expand our use of clinical lab measurements in addition to the use of ICD-9 codes in PheWAS to help provide more insight into the findings we have for both clinical lab measurements and ICD-9 code based case/control status to begin to understand more of the complex relationship between genetic architecture and the complex networks of signaling and phenotypic outcomes. Further, with the longitudinal nature of the EHR, we can leverage more of these data for longitudinal analyses. Clinical laboratory measures fluctuate for an individual in health and disease, and also with medication usage and age, and this complexity can be leveraged in future association testing for further discovery.
Clinical lab measurements provide an important area of exploration for PheWAS. The results of using more phenotypic measurements in a high-throughput way can enrich and expand our results of PheWAS based on ICD-9 code case/control status. Further, potential pleiotropy identified through cross-phenotype associations could show new important relationships between phenotypes through an expansion of phenotypic data available for PheWAS. Identifying a wide range of standardized and “cleaned” clinical lab measurements can also be used in the future to subset individuals based on clinical lab measure criteria before association testing. These approaches will also open the door to using more of the longitudinal nature of clinical lab measurements in future PheWAS analyses. These clinical lab measurements could also prove useful for continued development of high-quality phenotypic algorithms. The discoveries with these expanded PheWAS could prove important for discovery that leads to improvements in precision medicine as well as drug development.
Supplementary Material
Figure 2.
(a) Comparison of significant SNPs between clinical lab measures and ICD-9 code PheWAS. The x-axis has the clinical lab measures and y-axis shows its association p-value with the SNP, where red dots are the top SNPs from clinical lab PheWAS and blue triangle are the same SNPs associated with ICD-9 diagnoses. Table 2 lists what the ICD-9 diagnoses were for each of the clinical lab measures. (b) In this chromosomal ideogram, lines link SNP chromosomal locations to colored diamonds (representing clinical lab measures) or circles (representing ICD-9 diagnoses) showing the cross-phenotype associations for the SNPs identified first with associations with clinical lab measures.
References
- 1.Crawford DC, et al. eMERGEing progress in genomics-the first seven years. Front. Genet. 2014;5:184. doi: 10.3389/fgene.2014.00184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tyler AL, Crawford DC, Pendergrass SA. Detecting and Characterizing Pleiotropy: New Methods for Uncovering the Connection Between the Complexity of Genomic Architecture and Multiple phenotypes; Pac. Symp. Biocomput. Pac. Symp. Biocomput; 2014. pp. 183–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pendergrass SA, et al. Phenome-Wide Association Studies: Embracing Complexity for Discovery. Hum. Hered. doi: 10.1159/000381851. [DOI] [PubMed] [Google Scholar]
- 4.Hebbring SJ. The challenges, advantages and future of phenome-wide association studies. Immunology. 2014;141:157–165. doi: 10.1111/imm.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny JC, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinforma. Oxf. Engl. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hebbring SJ, et al. A PheWAS approach in studying HLA-DRB1*1501. Genes Immun. 2013;14:187–191. doi: 10.1038/gene.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Namjou B, et al. Phenome-wide association study (PheWAS) in EMR-linked pediatric cohorts, genetically links PLCL1 to speech language development and IL5-IL13 to Eosinophilic Esophagitis. Front. Genet. 2014;5:401. doi: 10.3389/fgene.2014.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny JC, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Development Core Team. R: A Language and Environment for Statistical Computing. 2008 at< http://www.R-project.org>. [Google Scholar]
- 10.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pendergrass SA, et al. Biofilter 2.0 – Using Biological Knowledge for Advanced Filtering, Annotation, and Model Development for Genomic Analysis. doi: 10.1186/1756-0381-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sobota RS, et al. Addressing Population-Specific Multiple Testing Burdens in Genetic Association Studies: Population-Specific Genome-Wide Thresholds. Ann. Hum. Genet. 2015;79:136–147. doi: 10.1111/ahg.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith EN, et al. Longitudinal Genome-Wide Association of Cardiovascular Disease Risk Factors in the Bogalusa Heart Study. PLoS Genet. 2010;6:e1001094. doi: 10.1371/journal.pgen.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko A, et al. Amerindian-specific regions under positive selection harbour new lipid variants in Latinos. Nat. Commun. 2014;5:3983. doi: 10.1038/ncomms4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano K, et al. Disease-associated marked hyperalphalipoproteinemia. Mol. Genet. Metab. Rep. 2014;1:264–268. doi: 10.1016/j.ymgmr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozóg J, et al. Expression of DPP4 gene in papillary thyroid carcinoma. Endokrynol. Pol. 2006;57(Suppl A):12–17. [PubMed] [Google Scholar]
- 20.Rago T, et al. Clinical features of thyroid autoimmunity are associated with thyroiditis on histology and are not predictive of malignancy in 570 patients with indeterminate nodules on cytology who had a thyroidectomy. Clin. Endocrinol. (Oxf.) 2007;67:363–369. doi: 10.1111/j.1365-2265.2007.02892.x. [DOI] [PubMed] [Google Scholar]
- 21.Holm L-E, Blomgren H, Löwhagen T. Cancer Risks in Patients with Chronic Lymphocytic Thyroiditis. N. Engl. J. Med. 1985;312:601–604. doi: 10.1056/NEJM198503073121001. [DOI] [PubMed] [Google Scholar]
- 22.Kim ES, et al. Thyroglobulin Antibody Is Associated with Increased Cancer Risk in Thyroid Nodules. Thyroid. 2010;20:885–891. doi: 10.1089/thy.2009.0384. [DOI] [PubMed] [Google Scholar]
- 23.Arnaud-Lopez L, et al. Phosphodiesterase 8B Gene Variants Are Associated with Serum TSH Levels and Thyroid Function. Am. J. Hum. Genet. 2008;82:1270–1280. doi: 10.1016/j.ajhg.2008.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng C-Y, et al. Association of MRC-1 and IL-28B with the treatment outcome of hepatitis C: a case control study. BMC Gastroenterol. 2014;14:113. doi: 10.1186/1471-230X-14-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nguyen C, Varney MD, Harrison LC, Morahan G. Definition of High-Risk Type 1 Diabetes HLA-DR and HLA-DQ Types Using Only Three Single Nucleotide Polymorphisms. Diabetes. 2013;62:2135–2140. doi: 10.2337/db12-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandhu MS, et al. LDL-cholesterol concentrations: a genome-wide association study. The Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deloukas P, et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2012;45:25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ai L, et al. Associations between rs965513/rs944289 and papillary thyroid carcinoma risk: a meta-analysis. Endocrine. 2014;47:428–434. doi: 10.1007/s12020-014-0256-4. [DOI] [PubMed] [Google Scholar]
- 29.He H, et al. Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. Proc. Natl. Acad. Sci. 2015;112:6128–6133. doi: 10.1073/pnas.1506255112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vozarova B, et al. High White Blood Cell Count Is Associated With a Worsening of Insulin Sensitivity and Predicts the Development of Type 2 Diabetes. Diabetes. 2002;51:455–461. doi: 10.2337/diabetes.51.2.455. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.