Abstract
Canonical Wnt/β-catenin signaling plays a major role in various biological contexts, such as embryonic development, cell proliferation, and cancer progression. Previously, a connection between p38 mitogen-activated protein kinase (MAPK) signaling and Wnt-mediated activation of β-catenin was implied but poorly understood. In the present study, we investigated potential cross talk between p38 MAPK and Wnt/β-catenin signaling. Here we show that a loss of p38 MAPK α/β function reduces β-catenin nuclear accumulation in Wnt3a-stimulated primary vascular smooth muscle cells (VSMCs). Conversely, active p38 MAPK signaling increases β-catenin nuclear localization and target gene activity in multiple cell types. Furthermore, the effect of p38 MAPK α/β on β-catenin activity is mediated through phosphorylation of a key p38 MAPK target, myocyte enhancer factor 2 (MEF2). Here we report a p38 MAPK-mediated, phosphorylation-dependent interaction between MEF2 and β-catenin in multiple cell types and primary VSMCs that results in (i) increased β-catenin nuclear retention, which is reversed by small interfering RNA (siRNA)-mediated MEF2 gene silencing; (ii) increased activation of MEF2 and Wnt/β-catenin target genes; and (iii) increased Wnt-stimulated cell proliferation. These observations provide mechanistic insight into a fundamental level of cross talk between p38 MAPK/MEF2 signaling and canonical Wnt signaling.
INTRODUCTION
Characterization of the canonical Wnt signaling pathway over the last 2 decades has revealed a fundamental role in many physiological and pathophysiological processes. Molecular defects in Wnt genes or their associated downstream effectors, most notably β-catenin, often have profound consequences linked with a myriad of developmental disorders and human diseases, including those involving hippocampal development, epithelial tube formation, and cancer (1–5).
The canonical Wnt pathway involves a family of 19 Wnt ligands, which are cysteine-rich glycoproteins that bind to the Frizzled receptor proteins, of which there are 10 family members. The ligand-receptor interaction comprises part of a larger signaling complex containing other receptor-related proteins, such as the low-density lipoprotein receptor-related protein 5 (LRP5) and LRP6 single-pass transmembrane proteins. β-Catenin, a bifunctional protein that serves as a component of the cell adhesion machinery in combination with E-cadherin and α-catenin, also performs an essential nodal function in the canonical Wnt pathway downstream of the receptor complex. In brief, without active Wnt signaling, β-catenin is phosphorylated by glycogen synthase kinase 3 (GSK3) and casein kinase I (CKI) in an adenomatous polyposis coli (APC)/axin “destruction complex,” which facilitates interaction with β-transducin repeat-containing E3 ubiquitin protein ligase (β-TrCP) and subsequent ubiquitin-mediated proteasomal degradation (6–8). Conversely, pathway activation by the Wnt-Frizzled interaction dismantles the destruction complex, leading to enhanced levels of cellular β-catenin and subsequent accumulation in both the cytoplasm and, particularly, the nuclear compartment. In combination with transcription factors, such as lymphoid enhancer-binding factor (LEF)/T-cell factor (TCF), and several other nuclear protein interactions, β-catenin acts as a powerful regulator of Wnt target genes, such as the cyclin D1 (9), c-Myc (10), axin2 (11), and c-Jun (12) genes, in a wide range of tissues (13–15).
Nuclear accumulation of β-catenin is a central tenet of the canonical Wnt pathway; however, the nuclear β-catenin level has largely been assumed to result from destruction complex disassembly and cytoplasmic accumulation. Consideration of β-catenin nuclear localization as a potential regulatory step in canonical Wnt signaling, and also how β-catenin is retained in the nucleus, has been unclear (16, 17). For a pathway that fulfills such a prominent role in many cellular processes, it seems unlikely that the facile cytoplasmic accumulation of β-catenin due to suppressed degradation is sufficient for precise regulation of the nuclear levels, especially in view of the fact that this step is heavily regulated for many transcriptional regulators (18, 19). Indeed, some studies have suggested that additional control of β-catenin localization occurs in a nuclear localization signal (NLS)- and importin-independent manner and by association with various proteins; however, the precise mechanism is still unknown (16, 20, 21).
Here we report a nexus of control of β-catenin nuclear localization by cross talk with the p38 mitogen-activated protein kinase (MAPK)/myocyte enhancer factor 2 (MEF2) signaling pathway that is dependent on a direct protein-protein interaction with MEF2 and on intact p38 MAPK activity in primary vascular smooth muscle and several cell lineages. These observations define a novel mechanism of β-catenin regulation with important implications for canonical Wnt signaling pathway modulation.
MATERIALS AND METHODS
Cell culture.
A10, COS7, HEK 293T, and C3H/10T1/2 cells were obtained from the American Type Culture Collection (ATCC). Cells were maintained in Dulbecco's modified Eagle medium (DMEM) with high glucose (HyClone), supplemented with 10% fetal bovine serum (FBS; HyClone) and 1% penicillin-streptomycin (Invitrogen), according to ATCC recommendations. Primary vascular smooth muscle cells (VSMCs) were isolated and cultured from mouse aortae as previously described (22). All mouse experiments were approved by the York University Animal Care Committee in accordance with Canadian Council of Animal Care regulations. Cells were grown in serum-free DMEM containing no FBS for the time specified for each experiment. Cells were maintained in an incubator at 95% humidity, 5% CO2, and 37°C. Drug and protein treatments were completed for the indicated times at the indicated concentrations.
Transfections.
For reporter gene assays and protein overexpression, cells were transfected by use of TurboFect (Fisher Scientific) according to the recommended instructions. Small interfering RNA (siRNA) was transfected according to TurboFect recommendations. Cells were transfected for 3 h, followed by medium replacement, and were harvested or had the serum withdrawn 24 to 48 h later, unless otherwise indicated. Myc(N)-β-catenin was N-terminally truncated by 89 amino acids (aa) prior to tagging to prevent GSK3β phosphorylation and degradation, and the resulting construct was named β-catenin (act) (activated).
Gene silencing.
A β-catenin short hairpin RNA (shRNA) (target sequence AACATGCAGTTGTCAATTTGA) or its scrambled control (target sequence GCAATTCTGAAGTCTGATATA) was inserted into the pSilencer 3.0 H1 vector per the manufacturer's instructions. siRNAs (Sigma-Aldrich) for MEF2A (siRNA 1, SASI_Mm01_00120788; and siRNA 2, SASI_Mm01_00120789), MEF2C (SASI_Mm01_00092891), and β-catenin (SASI_Rn01_00099923) and a universal scrambled RNA were used at a concentration of 25 nM. Double-siRNA experiments used a 25 nM concentration of each siRNA (total concentration, 50 nM), and equivalent controls used 50 nM scrambled RNA.
Antibodies and reagents.
Rabbit polyclonal MEF2A and MEF2C antibodies have been described previously (23, 24). Actin, c-Jun, and dsRed were purchased from Santa Cruz. β-Catenin antibody was purchased from Cell Signaling Technologies. MEF2D antibody was purchased from BD Biosciences. c-Myc antibody was purchased from the developmental studies hybridoma bank. Flag, glutathione S-transferase (GST), and actin (α-smooth muscle) were purchased from Sigma-Aldrich.
Purified recombinant mouse Wnt3a protein was purchased from Abcam. A p38 MAPK inhibitor (SB203580) and its inactive analog (SB202474) were purchased from Calbiochem. Purified recombinant human platelet-derived growth factor BB (PDGF) was purchased from Sigma-Aldrich.
In vitro kinase assay and mass spectrometric (MS) analysis of protein phosphorylation.
Three micrograms of either purified recombinant GST, GST-MEF2C, or 6×His–β-catenin was mixed with 0.5 μg purified recombinant GST-p38 MAPK (Millipore) and with [γ-32P]ATP and incubated at 30°C for 30 min. Samples were denatured in SDS loading buffer at 95°C for 5 min, and proteins were separated by 10% SDS-PAGE and exposed on X-ray film (GE Healthcare) for 16 h at −80°C to detect 32P incorporation.
In order to observe protein phosphorylation by liquid chromatography-MS (LC-MS), in vitro p38 MAPK treatment was repeated with ATP in place of [γ-32P]ATP. The protein mixture was separated by SDS-PAGE. Coomassie blue-stained gel bands whose molecular weights corresponded to those of ATF2 and β-catenin were cut away and then destained with acetonitrile (Fisher Scientific). The gel bands were treated with 9 mM dithiothreitol in 100 mM ammonium bicarbonate (Sigma-Aldrich) at 60°C for 15 min and then alkylated with iodoacetamide (Sigma-Aldrich) in the dark for 30 min at room temperature. Each band was treated with 1 μg of porcine trypsin (Promega) at 37°C for 16 h.
Tryptic peptides were resolved by nanoflow high-pressure liquid chromatography (nanoflow-HPLC) over 60-min acetonitrile gradients on a NanoLC-Ultra 2D-Nanoflex cHiPLC system (AB Sciex). A 0.5-mm-long, 200-μm-diameter trap and a 150-mm-long, 75-μm-diameter analytical column were used. The trap and column were both packed with 3-μm-diameter C18 particles with 120-Å pores. Mobile phase A was 0.1% formic acid in water (vol/vol), and mobile phase B was 0.1% formic acid in acetonitrile (vol/vol) (Honeywell). The pump was operated at 300 nl/min with the following mobile phase B contents: 2% (vol/vol) at 0 min, 35% at 35 min, 80% from 35.5 to 38.5 min, and 2% from 39 to 60 min. Peptides were detected on an Orbitrap Elite mass spectrometer (Thermo Scientific).
The mass spectrometer was operated in data-dependent acquisition mode, with survey scans having a resolution fixed at 60,000. The data acquisition method incorporated an inclusion list whose m/z values corresponded to fully tryptic β-catenin peptide and phosphopeptide ions containing putative p38 MAPK phosphosites based on the kinase consensus sequence. Predicted phosphosites S191, S246, and S605 were represented in the sequences SPQMVSAIVR, MLGSPVDSVLFYAITTLHNLLLHQEGAK, and GLNTIPLFVQLLYSPIENIQR, respectively.
Protein extraction and Western blot analysis.
Cells were collected using NP-40 lysis buffer (0.5% [vol/vol] Nonidet P-40, 50 mM Tris-HCl [pH 8], 150 mM NaCl, 10 mM sodium pyrophosphate, 1 mM EDTA [pH 8], and 0.1 M NaF) containing 10 μg/ml (each) leupeptin and aprotinin, 5 μg/ml pepstatin A, 0.2 mM phenylmethylsulfonyl fluoride, and 0.5 mM sodium orthovanadate. Protein extracts were denatured in SDS loading buffer at 95°C for 5 min and then run in a 10% SDS-PAGE gel, transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore), and blocked in 5% skim milk for 1 h prior to antibody incubation.
Coimmunoprecipitation and calf intestinal alkaline phosphatase (CIP) treatment.
For coimmunoprecipitation, cells were harvested and proteins extracted as described above. Immunoprecipitation was performed using an ImmunoCruz Optima kit (Santa Cruz Biotechnology) according to the manufacturer's instructions. Eluates were analyzed by Western blotting as described above.
For CIP treatment, 10 U of CIP was added to 1 mg of cell lysate and incubated for 30 min at 37°C, using an ImmunoCruz Optima kit.
GST pulldown assay.
For GST pulldown assays, GST-MEF2C and 6×His–β-catenin fusion proteins were obtained using standard protocols. Briefly, a cloned pGex2T (MEF2C) or pET28a (β-catenin) vector was transformed into Escherichia coli BL21, and protein production was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 6 h. GST-MEF2C- and 6×His–β-catenin-expressing cells were sonicated, and the proteins were purified with glutathione-agarose (Sigma-Aldrich) and nickel-agarose (Qiagen) beads, respectively. Protein concentrations were estimated by SDS-PAGE and Coomassie blue staining, using bovine serum albumin (BSA) for comparative estimation. Five micrograms of GST–full-length MEF2C (or the molar equivalent of the smaller GST, GST-MEF2C N terminus, or GST-MEF2C C terminus), 5 μg 6×His–β-catenin, and 25 μl glutathione-agarose beads (50% slurry) were incubated in 600 μl NETN buffer (100 mM NaCl, 20 mM Tris-HCl [pH 8], 0.5 mM EDTA, 0.5% [vol/vol] NP-40) overnight at 4°C. Beads were washed three times with phosphate-buffered saline (PBS) and eluates analyzed by Western blotting.
Reporter gene assays.
Transcriptional assays were performed using luciferase reporter plasmids. Cells were harvested using 20 mM Tris-HCl (pH 7.4) and 0.1% Triton X-100. Values obtained using a Lumat LB (Berthold) luminometer were normalized to renilla luciferase expression in the same cells, and values were expressed as fold activation by arbitrarily setting the control condition to 1. Bars represent the means (n = 3), and error bars represent the standard errors of the means (n = 3). Independent sample t tests were performed on experiments with two conditions, while one-way analysis of variance (ANOVA) followed by Tukey's highly significant difference (HSD) post hoc analysis was performed on experiments with more than two conditions (SPSS software). P values are shown with respect to controls, unless otherwise indicated.
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde in PBS for 10 min and then permeabilized with 0.3% Triton X-100 in PBS for 5 min at room temperature. Cells were blocked using 10% goat serum in PBS for 30 min at 37°C and then incubated overnight with β-catenin antibody diluted (1:500) in 1.5% goat serum in PBS at 4°C. Cells were washed three times with PBS (10 min each) and then incubated with the specified tetramethyl rhodamine isothiocyanate (TRITC) or fluorescein isothiocyanate (FITC) antibody (Sigma-Aldrich) diluted (1:500) in 1.5% goat serum for 2 h at room temperature, with 4′,6-diamidino-2-phenylindole (DAPI) nuclear staining (1:10,000; Sigma-Aldrich) in the last 15 min. Cells were washed three times with PBS (10 min each) and then left in PBS at room temperature for confocal microscopy imaging. Images were captured using either a Carl Zeiss Axio Observer.Z1 (Photometrics Evolve 512 EMCCD camera) or LSM 700 AxioObserver microscope. Cells were imaged under 40× (EC Plan-Neofluar; 1.30 numerical aperture [NA] in oil), 63× (Plan-Apochromat; 1.40 NA in oil), and 100× (Alpha Plan-Apochromat; 1.46 NA in oil) objectives, using ZEN image acquisition software. Images were taken as a z-stack and projected orthogonally by maximum intensity. Relative β-catenin nuclear/cytosolic fluorescence ratios were quantified using ImageJ with the Fiji image processing package. The nucleus was traced using DAPI or Hoechst 33342 for reference, and the following procedure and calculations were performed. The fluorescence intensities of 3 areas in the field with no cells were averaged and multiplied by the nuclear area to calculate background fluorescence. The integrated density of the nucleus was subtracted from this value and divided by the nuclear area to calculate the average nuclear intensity. Three areas of cytosol that were evenly spaced around the nucleus were measured and the background subtracted as described above, and the resulting value was divided by the area to calculate the average cytosolic intensity. The average nuclear intensity was divided by the average cytosolic intensity to calculate the nuclear/cytosolic ratio for the cell.
In vitro binding assay.
Two micrograms of purified recombinant GST-MEF2C (Abnova) was incubated with 0.4 μg of purified recombinant GST-p38 MAPK (Millipore) in kinase buffer (NEB) with 200 μM ATP (NEB), as specified, for 3 h at 30°C. Five micrograms of purified recombinant 6×His–β-catenin was added, along with 30 μl glutathione-agarose beads (50% slurry) and 600 μl NETN buffer (100 mM NaCl, 20 mM Tris-HCl [pH 8], 0.5 mM EDTA, 0.5% NP-40), and incubated for 1 h at room temperature. Beads were washed three times with Tris-buffered saline (TBS), heated with SDS loading buffer for 5 min at 95°C, run in a 10% SDS-PAGE gel, and subjected to Western blotting.
MTT cell proliferation assay.
A Vybrant MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] cell proliferation assay kit was purchased from Life Technologies and used according to the manufacturer's instructions. Cells were transfected for 3 h and recovered for 24 h prior to the MTT assay procedure. Bars represent the means (n = 3), and error bars represent the standard errors of the means (n = 3). One-way ANOVA followed by Tukey's HSD post hoc analysis was performed for statistical analysis (SPSS software). P values are shown with respect to controls, unless otherwise indicated.
RESULTS
p38 MAPK modulates Wnt-dependent β-catenin activity and nuclear localization.
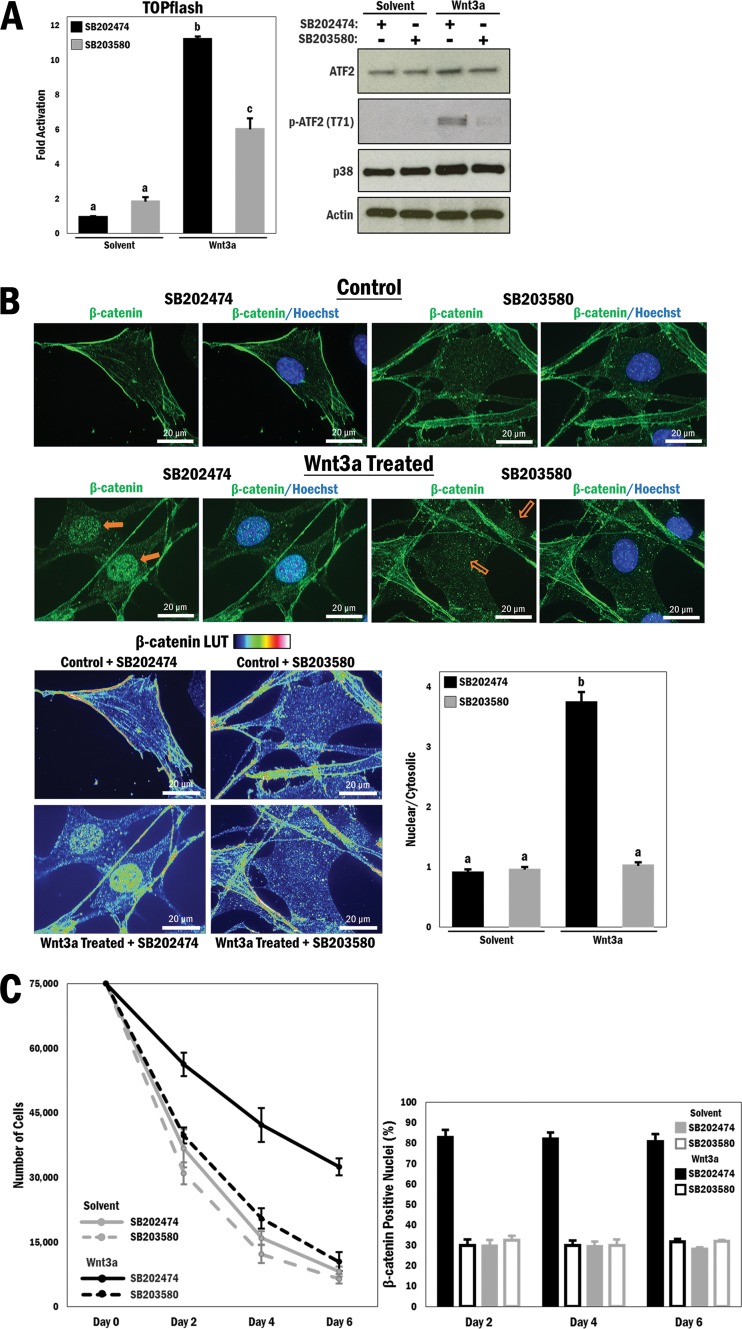
In order to determine whether p38 MAPK does play a role in regulating β-catenin, we utilized a well-characterized synthetic reporter gene named TOPflash, which responds to Wnt-dependent β-catenin activation and contains 7 TCF/LEF consensus binding sites driving luciferase expression (25). Initially, the A10 vascular smooth muscle cell line was transfected with TOPflash and then cultured under low-serum conditions, followed by treatment with a p38 MAPK inhibitor (SB203580; 10 μM) or its inactive analog (SB202474; 10 μM). Under these baseline conditions, canonical Wnt signaling was activated by treatment with purified Wnt3a protein (200 ng/ml). Wnt3a stimulation with the inactive inhibitor increased TOPflash activity 11-fold (P ≤ 0.0001), but this was significantly reduced in cells treated with Wnt3a and the active p38 MAPK inhibitor (P ≤ 0.0001) (Fig. 1A, left panel). Primary vascular smooth muscle cells (VSMCs) isolated from mouse aortae were similarly treated with the p38 MAPK inhibitor following serum withdrawal and were stimulated with the Wnt3a protein. Cell lysates were subjected to Western blot analysis to show p38 MAPK activity by analyzing a known substrate, activating transcription factor 2 (ATF2), which is phosphorylated at Thr69/71 by p38 MAPK (26) (Fig. 1A, right panel). Wnt3a stimulation has indeed previously been shown to lead to p38 MAPK activation and thus to promote ATF2 phosphorylation (27). Moreover, we reported a derepression of p38 MAPK by pharmacological GSK3 inhibition (28). Thus, there seem to be a number of possible mechanisms by which the activity of p38 MAPK may be modulated by Wnt signaling.
FIG 1.
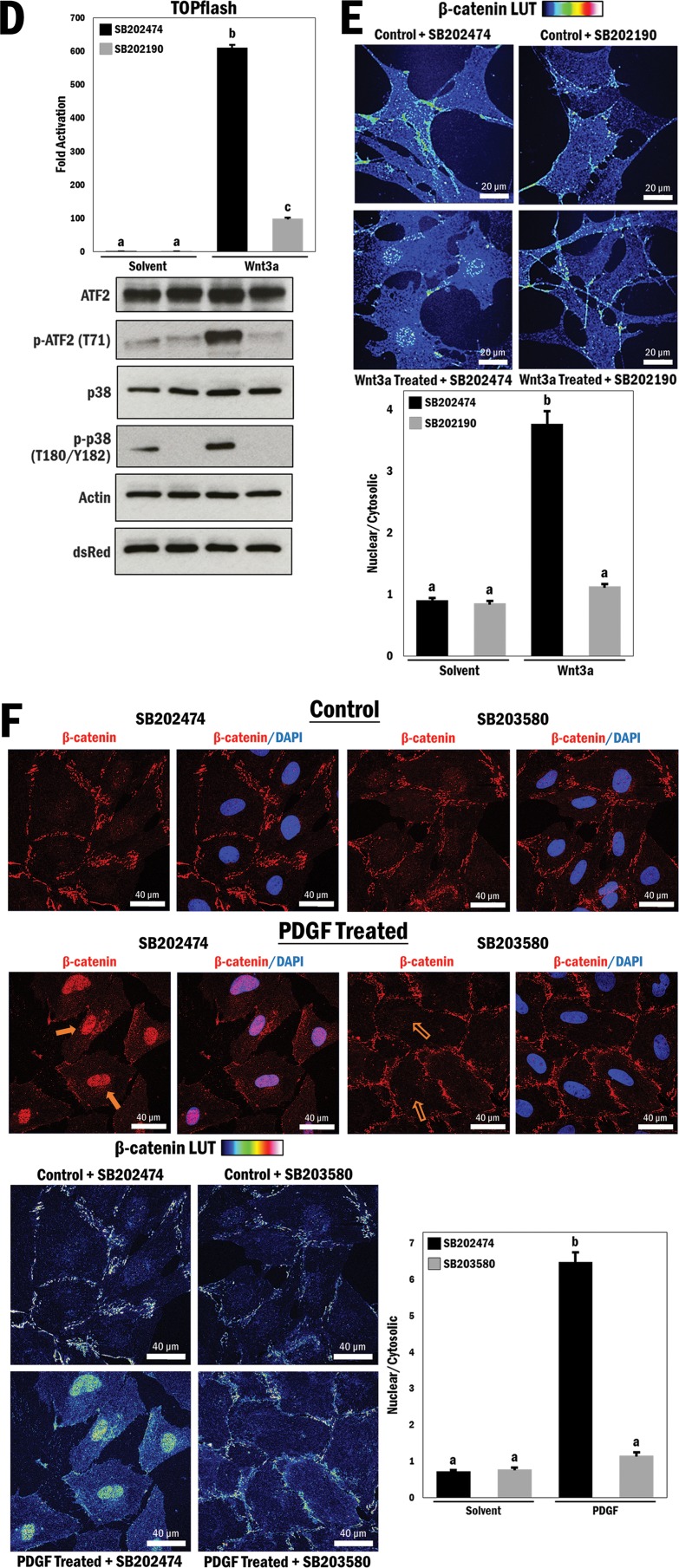
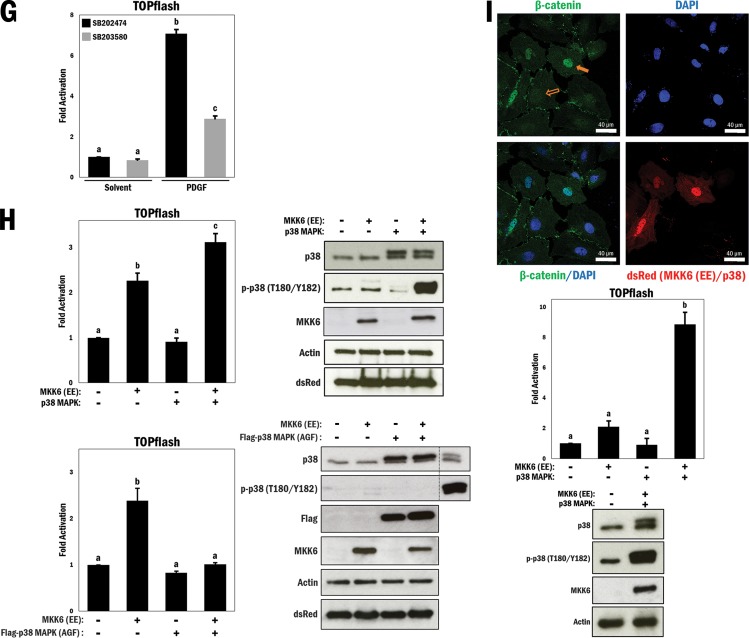
p38 MAPK regulates Wnt-dependent β-catenin transactivation properties and nuclear localization in primary vascular smooth muscle cells. (A) (Left) A10 cells were transfected with the TOPflash reporter gene for 3 h, followed by treatment with different combinations of Wnt3a (200 ng/ml), solvent (PBS; control), SB203580 (p38 MAPK inhibitor; 10 μM), and SB202474 (control; 10 μM) for 16 h in serum-free medium, as indicated. b and c, P ≤ 0.0001. (Right) Primary VSMCs underwent serum withdrawal (12 h) and were pretreated with SB203580 or SB202474 (10 μM) for 45 min and then stimulated with Wnt3a (200 ng/ml) or solvent (PBS) for 4 h. Cell lysates were subjected to Western blot analysis as indicated. (B) (Top) Primary VSMCs underwent serum withdrawal for 12 h and were pretreated with SB203580 or SB202474 (10 μM) for 45 min and then stimulated with Wnt3a (200 ng/ml) or solvent (PBS) for 4 h. Cells were fixed and stained with Hoechst 33342 (blue; nucleus) and FITC (green; β-catenin). Filled arrows indicate the presence of nuclear β-catenin, and empty arrows indicate a lack thereof. (Bottom left) A rainbow (blue to red) lookup table (LUT) was used to indicate relative β-catenin intensities. (Bottom right) Relative nuclear/cytosolic fluorescence ratios were determined (n = 33, 32, 31, and 32, from left to right). b, P ≤ 0.0001. (C) (Left) A10 cells were seeded at 75,000 cells, underwent serum withdrawal (12 h), and then were pretreated with SB203580 or SB202474 (10 μM) for 45 min and stimulated with Wnt3a (200 ng/ml) or solvent (PBS). This was repeated every 2 days for a total of 6 days, and cells were counted at each interval. (Right) A10 cells were cultured and treated as described for the left panel, fixed and stained at each interval for the nucleus (Hoechst 33342; blue) and β-catenin (FITC; green), and counted to determine the number of cells positive for nuclear β-catenin for each condition and time interval (n = 158, 210, 145, 146, 159, 201, 149, 160, 175, 167, 123, and 145, from left to right). (D and E) Primary VSMCs were prepared as described for panels A and B, respectively, but were treated with an alternative p38 MAPK inhibitor (SB202190; 10 μM) or its control (SB202474; 10 μM) (n = 26, 39, 34, and 35, from left to right). b, P ≤ 0.0001. (F) A10 cells were prepared as described for panel B but were stimulated with either PDGF (20 ng/ml) or solvent (0.1% BSA in 4 mM HCl) for 4 h prior to fixation and staining with DAPI and TRITC (n = 35, 28, 36, and 24, from left to right). b, P ≤ 0.0001. (G) A10 cells were transfected with the TOPflash reporter gene for 3 h, followed by treatment with different combinations of PDGF (20 ng/ml), solvent (0.1% BSA in 4 mM HCl), SB203580 (10 μM), and SB202474 (10 μM) for 16 h in serum-free medium, as indicated. b and c, P ≤ 0.0001. (H) (Top left) TOPflash reporter gene activity was measured in primary VSMCs transfected with different combinations of active MKK6 (EE) and p38 MAPK, as indicated. b and c, P ≤ 0.01. (Top right) Corresponding Western blots to show relative overexpression. (Bottom left) TOPflash reporter gene activity was measured in primary VSMCs transfected with different combinations of active MKK6 (EE) and mutant Flag-p38 MAPK (AGF), as indicated. b, P ≤ 0.01. (Bottom right) Corresponding Western blots to show relative overexpression. A10 cell lysates were used as controls for expression of p38 and p-p38. (I) (Top) A10 cells were cotransfected with MKK6 (EE) and p38 MAPK and marked for ectopic expression (dsRed; red). DAPI was used for nuclear staining, and FITC was used for β-catenin staining (green). (Middle) TOPflash reporter gene activity was measured in A10 cells transfected with different combinations of p38 MAPK and active MKK6 (EE), as indicated. b, P ≤ 0.0001. (Bottom) Western blot analysis to show activation of p38 MAPK.
Since nuclear accumulation is fundamental for β-catenin activity, we subsequently tested whether the effect of p38 MAPK inhibition on β-catenin activity was due to a change in its cellular localization. Exploratory experiments with A10 cells indicated that modulation of p38 MAPK activity affected β-catenin localization (see Fig. S1 in the supplemental material), and this was confirmed in primary VSMCs by confocal immunofluorescence microscopy under the conditions described above. Initially, β-catenin was localized to the nucleus due to Wnt3a stimulation, in contrast to the case in control cells that were maintained in serum-depleted medium to establish baseline levels of signaling (Fig. 1B, top panels). Strikingly, inhibition of p38 MAPK resulted in a large reduction in β-catenin nuclear accumulation. This dramatic change in β-catenin localization was illustrated by a color scheme (blue to red) dependent on fluorescence intensity (Fig. 1B, bottom left panel) and by quantification of relative nuclear/cytosolic fluorescence for all of the images taken, which indicated a nearly 4-fold reduction in nuclear β-catenin levels in Wnt3a-stimulated cells treated with the p38 MAPK inhibitor (P ≤ 0.0001) (Fig. 1B, bottom right panel). These results prompted us to hypothesize that p38 MAPK activity is required for Wnt3a-mediated β-catenin nuclear localization.
We next decided to analyze whether there was a biological effect of p38 MAPK inhibition on Wnt3a-stimulated cells. To this end, we performed a cell survival assay in which A10 cells were equally seeded at 75,000 cells and grown in serum-free medium overnight prior to being stimulated with purified Wnt3a and treated with either the p38 MAPK inhibitor or its inactive analog every 2 days over a period of 6 days; cells were counted at these intervals, and the results are displayed in Fig. 1C (left panel). Treatment with Wnt3a and the inactive p38 MAPK inhibitor resulted in a significantly higher cell count than that for control cells that underwent serum withdrawal over the course of the experiment. Interestingly, cells that were stimulated with Wnt3a and treated with the active p38 MAPK inhibitor had a much lower cell count, nearly as low as that for cells that underwent serum withdrawal. A parallel experiment was performed in which β-catenin localization was determined using immunofluorescence. The number of cells that were positive for nuclear β-catenin were counted for each condition throughout the course of 6 days, and the results showed that approximately 80% of cells that were stimulated with Wnt3a and treated with the inactive p38 MAPK inhibitor were positive for nuclear β-catenin, while only approximately 30% of Wnt3a-stimulated cells in the presence of the active p38 MAPK inhibitor were positive (Fig. 1C, right panel). These results demonstrate that the loss of nuclear β-catenin localization observed in p38 MAPK-inhibited cells had a profound effect on cell proliferation and therefore has important biological implications.
To further validate these data, we used a different p38 MAPK inhibitor (SB202190; 10 μM) with a different mechanism of inhibition (prevents p38 MAPK phosphorylation by upstream mitogen-activated protein kinase kinases [MKKs]) in order to determine if these data would be replicated. Primary smooth muscle cells were transfected with the TOPflash reporter gene and cultured under low-serum conditions prior to being treated with different combinations of Wnt3a and p38 MAPK inhibitor, similar to the experiment described for Fig. 1A. Wnt3a stimulation resulted in a large increase in reporter gene activity, and similarly to that by SB203580, inhibition of p38 MAPK by SB202190 resulted in a significant reduction in reporter gene activity (P ≤ 0.0001) (Fig. 1D, top panel). Western blot analysis of the cell lysates confirmed the activation of p38 MAPK by Wnt3a stimulation and its inhibition by SB202190 by analyzing the phosphorylation status of ATF2 (Fig. 1D, bottom panel). We further demonstrated that this effect is correlated with β-catenin localization, and we performed an immunofluorescence assay using this p38 MAPK inhibitor. Again, inhibition of p38 MAPK by SB202190 resulted in a loss of nuclear β-catenin localization in Wnt3a-stimulated cells, with a >3-fold decrease in the nuclear/cytosolic ratio of β-catenin with p38 MAPK inhibition (P ≤ 0.0001) (Fig. 1E; see Fig. S2 in the supplemental material).
To pursue this idea differently, we asked if platelet-derived growth factor (PDGF), which is a known activator of p38 MAPK signaling (29) and has been shown to promote nuclear β-catenin localization (30), might also be modulated by p38 MAPK inhibition. In this experiment, A10 cells were treated with purified PDGF (20 ng/ml). Compared to that in control cells, PDGF stimulation enhanced the nuclear accumulation of β-catenin >6-fold, and this effect was also counteracted by p38 MAPK inhibition (P ≤ 0.0001) (Fig. 1F). To determine whether this similar pattern in localization had an effect on β-catenin transactivation, we expressed the TOPflash reporter gene and stimulated cells with Wnt3a and a p38 MAPK inhibitor (SB203580). PDGF treatment resulted in a significant increase in reporter activity, although it was lower than that observed with Wnt3a treatment (P ≤ 0.0001) (Fig. 1G). Importantly, p38 MAPK inhibition resulted in a loss of reporter gene activity, similar to the effects seen with Wnt3a. Based on the data collected thus far, we concluded that a loss of p38 MAPK function has an effect on both β-catenin cellular localization and transactivation properties and that pharmacological inhibition of p38 MAPK can abrogate canonical Wnt-mediated β-catenin activity.
Activation of p38 MAPK results in increased β-catenin nuclear localization and Wnt-responsive gene activity.
In view of the loss-of-function data described above, we carried out p38 MAPK gain-of-function experiments to test if nuclear β-catenin levels and transactivation properties would be enhanced. First, we ectopically expressed p38 MAPK and a constitutively active form of its upstream kinase, dual-specificity MKK6 [MKK6 (EE)], in primary VSMCs and looked at TOPflash reporter gene activity. Overexpression of MKK6 alone increased reporter gene activity (P ≤ 0.01), while coexpression of MKK6 and p38 MAPK increased reporter gene activity significantly more than expression of MKK6 alone (P ≤ 0.01) (Fig. 1H, top left panel), despite the fact that these cells were cultured under high-serum conditions and would have high baseline p38 MAPK activity. Activation of p38 MAPK was confirmed by Western blotting (Fig. 1H, top right panel). To test whether the increase in reporter gene activity was specific to p38 MAPK and not a different target of MKK6, these cells were again transfected with MKK6, but with an inactive form of p38 MAPK in which the MKK6 phospho-acceptor sites are mutated (T180A/Y182F [p38 MAPK AGF]). Overexpression of the p38 MAPK AGF mutant did not have an effect on TOPflash reporter activity, leading us to conclude that the effects seen with ectopic MKK6 expression were through p38 MAPK (P ≤ 0.01) (Fig. 1H, bottom left panel). Expression patterns were confirmed by Western blotting, and levels of phospho-p38 were unchanged compared to those for a positive A10 lysate control, indicating that the p38 MAPK mutant was unaffected by MKK6 expression (Fig. 1H, bottom right panel).
We next assessed A10 cells that were transfected with MKK6 and p38 MAPK, marked by cotransfection with dsRed, and stained for endogenous β-catenin (FITC; green) and nuclei (DAPI; blue). MKK6/p38 MAPK-positive cells showed more nuclear β-catenin localization than that of untransfected cells in the same field (Fig. 1I, top panel). In addition, cells cotransfected with both MKK6 and p38 MAPK with TOPflash showed a synergistic increase in reporter gene activity (P ≤ 0.0001) (Fig. 1I, middle panel). This indicates that activation of p38 MAPK potently enhances β-catenin transactivation properties, an effect that was not observed using the FOPflash control reporter gene, which contains 6 mutated TCF/LEF sites (see Fig. S3 in the supplemental material). p38 MAPK activation was confirmed by Western blotting (Fig. 1I, bottom panel).
To determine if the effects of p38 MAPK signaling on β-catenin localization and activity are limited to smooth muscle cells, we repeated the gain-of-function experiments with both 10T1/2 (mouse embryonic fibroblast) and HEK 293T (human embryonic kidney) cells. Our observations confirmed by multiple techniques that the effects on β-catenin localization and activity by p38 MAPK hyperactivation were common to these other cell types as well (P ≤ 0.05 and P ≤ 0.01) (see Fig. S4 and S5 in the supplemental material, respectively). Taken together, these data support the conclusion that p38 MAPK plays a potent role in modulating Wnt-dependent β-catenin localization and transactivation properties in primary and several other cell lineages.
β-Catenin is not a direct p38 MAPK substrate.
We initially supposed that p38 MAPK might directly phosphorylate β-catenin and that this phosphorylation is required for β-catenin accumulation in the nucleus. To assess this idea, we performed an in vitro kinase assay using purified β-catenin, p38 MAPK, and [γ-32P]ATP. Compared to a known p38 MAPK substrate used as a positive control (MEF2C), weak β-catenin phosphorylation was apparent, but we suspected this to be a background signal (see Fig. S6 in the supplemental material). To further explore this, an in silico analysis of the β-catenin primary amino acid sequence revealed three possible p38 MAPK phosphorylation sites: S191, S246, and S605. These residues were individually mutated to alanine and the resulting mutants used in TOPflash reporter assays with ectopically expressed MKK6 [MKK6 (EE)] and p38 MAPK. There was no change in the p38 MAPK responsiveness of β-catenin with the engineered mutations (see Fig. S7), indicating that p38 MAPK may not directly phosphorylate β-catenin. Lastly, our group has extensive experience in detecting phosphorylation events by mass spectrometry (31–33), and despite robust detection of ATF2 phosphorylation in our parallel assays, there was no evidence of corresponding p38 MAPK-mediated β-catenin phosphopeptides by highly sensitive LC-MS/MS (see Fig. S8). Therefore, we concluded that β-catenin is not a direct substrate of p38 MAPK.
MEF2 is a direct β-catenin interaction partner.
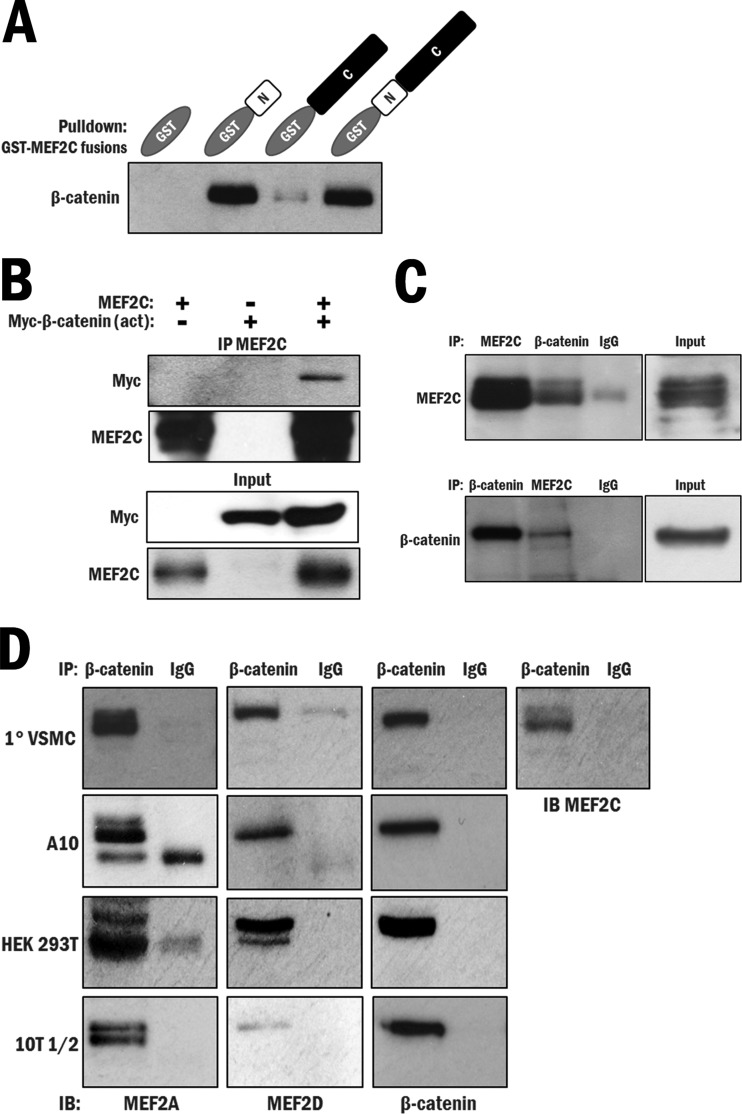
Previous observations from our group led us to perform the next set of experiments. The first was that MEF2 was reported by us and others to be potently regulated by p38 MAPK (34–37). The second was that we observed a high-confidence interaction between the highly conserved N terminus of MEF2 and β-catenin in a yeast two-hybrid screen for which we previously reported a novel interaction between MEF2 and big mitogen-activated protein kinase 1 (BMK1) (38), histone deacetylase 4 (HDAC4) (33), and protein phosphatase 1α (PP1α) (39). Also, another group independently documented an interaction between MEF2 and β-catenin in a hepatocellular carcinoma cell line (40). We next sought to further validate the interaction between β-catenin and MEF2 biochemically. To determine if the β-catenin–MEF2 interaction was direct, bacterially produced GST-MEF2C and 6×His–β-catenin fusion proteins were used in pulldown assays (see Fig. S9 in the supplemental material). This analysis indicated that β-catenin directly interacts with GST–full-length MEF2C, and also with GST-MEF2C N terminus (aa 1 to 86), with a high affinity. A weak, probably insignificant interaction with GST-MEF2C C terminus (aa 87 to 465) was observed, and no interaction was observed with a GST-only control (Fig. 2A; repeated in Fig. S10). To determine if the interaction occurs in a cellular context, GSK3 degradation-resistant, “activated” β-catenin [β-catenin (act)] and MEF2C were expressed in CV-1 origin SV40-7 (COS7) cells, and their interaction was confirmed by coimmunoprecipitation assays (Fig. 2B). Subsequently, we tested for an endogenous interaction between MEF2 and β-catenin in whole-cell lysates of the A10 cell line, in which we previously detected expression of both proteins. Western analysis of the MEF2C immunoprecipitate for β-catenin and the reverse, in which β-catenin was immunoprecipitated and probed for MEF2C, resulted in detection of the respective interacting partners, indicating a robust interaction between MEF2C and β-catenin (Fig. 2C).
FIG 2.
MEF2 directly interacts with β-catenin in vitro and in multiple tissue types. (A) An in vitro GST pulldown assay was performed using purified 6×His–β-catenin and either GST (control) or an N-terminal (aa 1 to 86), C-terminal (aa 87 to 465), or full-length (aa 1 to 465) GST-MEF2C fusion protein. (B) A coimmunoprecipitation (IP) assay was performed in COS7 cells, using MEF2C antibody to precipitate ectopically expressed MEF2C and determine whether an interaction with ectopically expressed, degradation-resistant Myc–β-catenin (act) occurs. (C) A coimmunoprecipitation assay was performed in A10 cells, using both MEF2C and β-catenin antibodies for immunoprecipitation to test for an interaction between the endogenous proteins. (D) Coimmunoprecipitation assays were performed using β-catenin antibody to test an interaction between endogenous β-catenin and either MEF2A, -C, or -D in primary VSMCs, A10 cells, HEK 293T cells, and 10T1/2 cells. IB, immunoblot.
We were further interested in determining if this interaction occurs with other MEF2 family proteins, since the region shown to interact with β-catenin is the highly conserved N-terminal MADS-MEF2 domain that is common to all MEF2 proteins, and if this interaction occurs in other cell types. Readily detectable endogenous interactions between β-catenin and MEF2A, as well as between β-catenin and MEF2D, were observed in primary vascular smooth muscle whole-cell lysates (Fig. 2D, top panel). This result was repeated with the A10, HEK 293T, and 10T1/2 cell lines (Fig. 2D, second, third, and fourth panels, respectively). Overall, these data establish that β-catenin interacts with MEF2A, -C, and -D, most likely through the highly conserved N-terminal region, and that the interaction occurs in several cell lines of different lineages and in primary vascular smooth muscle cells. We therefore hypothesized that MEF2 potentially enhances β-catenin accumulation in the nucleus by direct physical interaction.
MEF2–β-catenin interaction enhances β-catenin accumulation in the nucleus.
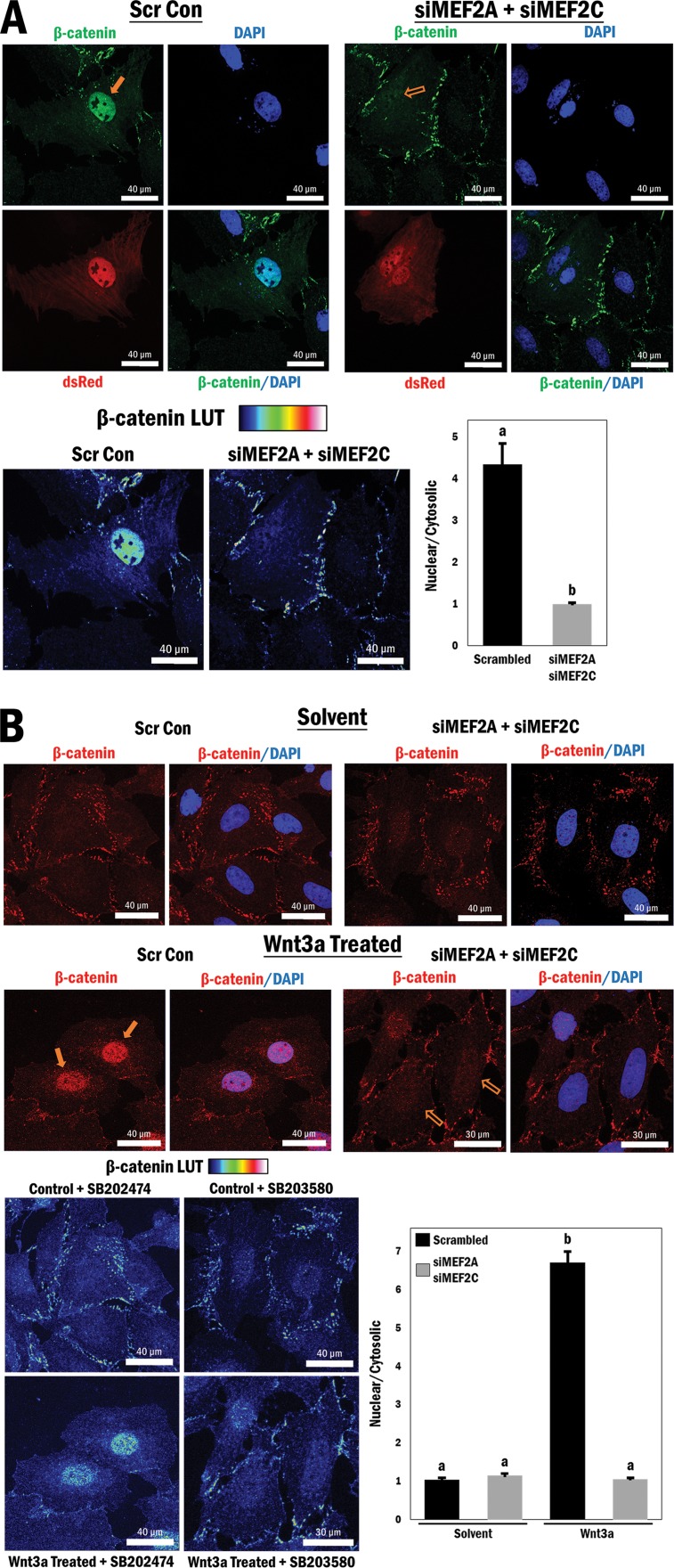
As alluded to above, previous studies have shown that phosphorylation of MEF2 by p38 MAPK results in potent activation of MEF2, and we have now shown that activation of p38 MAPK results in enhanced nuclear localization of β-catenin, even though it is not a direct substrate of p38 MAPK. Therefore, given the observation that β-catenin interacts with MEF2, we postulated that there might be a p38 MAPK-mediated, phospho-dependent interaction between β-catenin and MEF2 that enhances the nuclear accumulation of β-catenin. To investigate this idea, A10 cells were grown under high-serum conditions and transfected with siRNAs directed against MEF2A and MEF2C, which reduced the levels of these proteins (see Fig. S11 in the supplemental material), followed by immunofluorescence microscopy to detect β-catenin localization. Cells transfected with the siRNAs were marked (dsRed), and endogenous β-catenin localization was analyzed by antibody detection (FITC; green). Combined reduction of MEF2A and MEF2C expression resulted in a substantial and significant loss of nuclear β-catenin localization compared to that in cells transfected with the scrambled siRNA control (P ≤ 0.0001) (Fig. 3A).
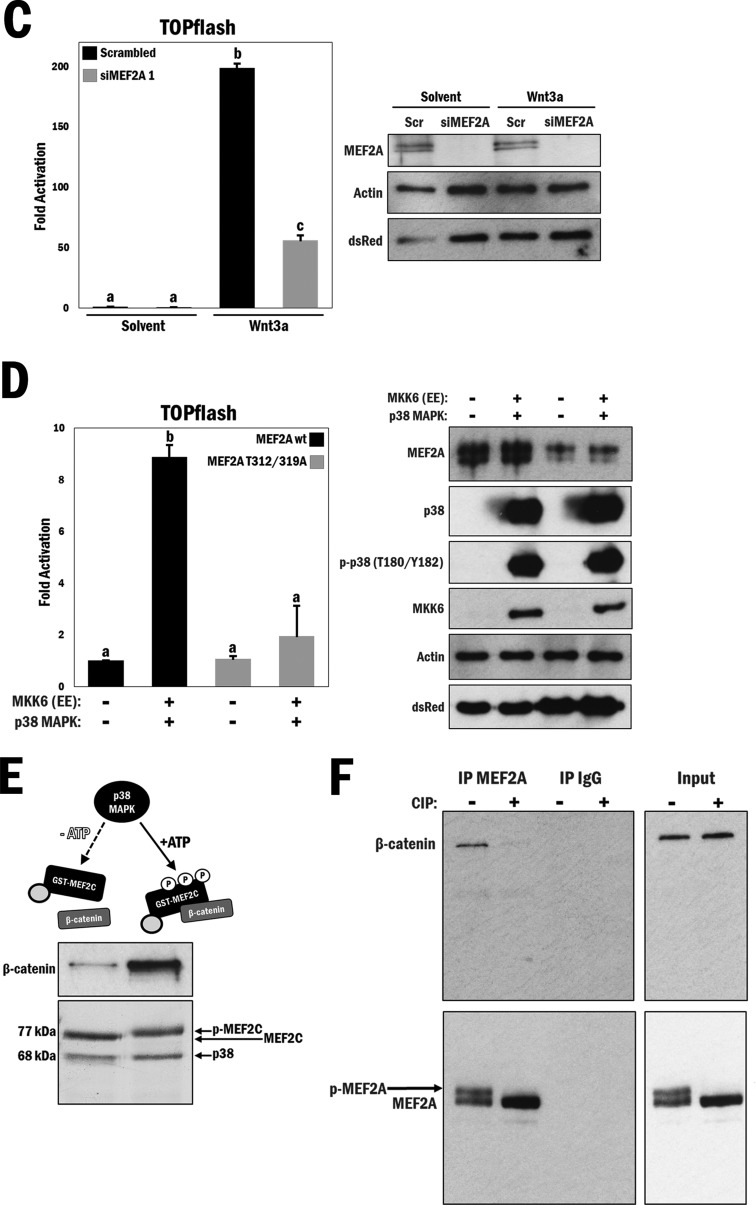
FIG 3.
MEF2 protein expression is required for β-catenin nuclear retention and enhanced by p38 MAPK-mediated phosphorylation of MEF2. (A) (Top) Cytomegalovirus (CMV)-driven dsRed was cotransfected with either scrambled siRNA (Scr Con) or siMEF2A/C RNA, followed by immunofluorescence staining of β-catenin (FITC) and the nucleus (DAPI), in A10 cells. (Bottom left) A rainbow LUT was used to indicate relative β-catenin intensities. (Bottom right) Relative nuclear/cytosolic fluorescence ratios were determined (n = 11 and 13, from left to right). b, P ≤ 0.0001. (B) (Top) Scrambled or siMEF2A/C RNA-transfected A10 cells underwent serum withdrawal for 12 h and were treated with either Wnt3a (200 ng/ml) or solvent (PBS; control) for 4 h, followed by immunofluorescence staining of β-catenin (TRITC) and the nucleus (DAPI). (Bottom left) A rainbow LUT was used to indicate relative β-catenin intensities. (Bottom right) Relative nuclear/cytosolic fluorescence ratios were determined (n = 28, 27, 27, and 28, from left to right). b, P ≤ 0.0001. (C) (Left) A10 cells were transfected with the TOPflash reporter and scrambled or siMEF2A RNA for 3 h and then cultured under serum conditions for 24 h, followed by treatment with either Wnt3a (200 ng/ml) or solvent (PBS) for 16 h in serum-free medium. b and c, P ≤ 0.0001. (Right) Corresponding Western blot analysis of cell lysates to confirm siRNA-mediated MEF2A depletion. (D) (Left) COS7 cells were transfected with the TOPflash reporter gene and different combinations of wild-type MEF2A, mutated MEF2A (T312/319A), MKK6 (EE), and p38 MAPK, as indicated. b, P ≤ 0.0001. (Right) Corresponding Western blot analysis. (E) Purified GST-MEF2C and GST-p38 MAPK were incubated at 30°C for 3 h, with or without ATP, as indicated in the figure, followed by addition of purified 6×His–β-catenin and GST-agarose beads for 1 h at room temperature. (Top) Immunoblot indicating relative amounts of β-catenin bound to MEF2C following precipitation. (Bottom) Representation of total MEF2C phosphorylation following 3 h of incubation with p38 MAPK and ATP. (F) A10 cell lysates (1 mg) were incubated with or without 10 U of CIP for 30 min at 37°C during immunoprecipitation using MEF2A antibody. Beads were then washed and analyzed for the corresponding β-catenin protein by Western blotting. Lysates without antibody-conjugated beads were similarly incubated as input samples.
Next, we tested if the loss of MEF2A and MEF2C might modulate Wnt-dependent β-catenin accumulation in the nucleus. To investigate this, siMEF2A- and siMEF2C-transfected cells were stimulated with the Wnt3a protein or solvent (control) prior to analysis of β-catenin subcellular localization. As expected, β-catenin nuclear accumulation was >6-fold higher in cells transfected with the siRNA control and treated with Wnt3a. Importantly, β-catenin failed to accumulate in the nuclei of Wnt3a-stimulated cells in which the MEF2 proteins were depleted (P ≤ 0.0001) (Fig. 3B). These data provide unequivocal evidence that, in this cellular context, MEF2 is absolutely required for canonical Wnt-mediated β-catenin nuclear accumulation.
As we observed in the earlier experiments illustrated in Fig. 1, loss of nuclear β-catenin localization resulted in a corresponding loss of β-catenin reporter gene activity, and we were interested to know if this was also the case with MEF2 gene silencing. Previous studies with C2C12 myoblasts showed that shRNA-mediated silencing of MEF2A results in a significant reduction in TOPflash activity under high-serum conditions (41), so we decided to assess the effects of MEF2 depletion in Wnt3a-stimulated A10 cells. We observed that Wnt3a treatment greatly enhanced TOPflash activity and that depletion of MEF2A by siRNA significantly reduced this activity in Wnt3a-stimulated cells, thus underscoring the observation that the presence of MEF2 is essential for Wnt-mediated β-catenin transactivation properties (P ≤ 0.0001) (Fig. 3C, left panel). Confirmation of MEF2A silencing by Western analysis is shown in Fig. 3C, right panel, and this experiment was repeated using a different siRNA targeting MEF2A, with similar results (see Fig. S12 in the supplemental material).
The affinity of the MEF2–β-catenin interaction and β-catenin transactivation are potently regulated by p38 MAPK phosphorylation of MEF2.
Based on the indirect effect of activated p38 MAPK on β-catenin localization, coupled with the interaction of β-catenin with MEF2, a direct p38 MAPK substrate, we hypothesized that the MEF2–β-catenin interaction might be phospho-MEF2 dependent and was responsible for the enhancement of Wnt reporter activity by p38 MAPK. This would establish a direct connection between β-catenin, p38 MAPK, and MEF2. To test this idea, we employed a MEF2A mutation that is incapable of being phosphorylated by p38 MAPK (T312A/T319A) in a TOPflash assay using COS7 cells, which do not contain any appreciable endogenous MEF2 proteins or activity. Wild-type MEF2A was coexpressed with MKK6 (EE) and p38 MAPK, which increased TOPflash activity 9-fold. However, when the MEF2A mutant was coexpressed in a similar manner, there was no apparent change in TOPflash activity, thereby indicating that the mechanism of activation of the Wnt reporter gene is through phosphorylation of MEF2 by p38 MAPK (P ≤ 0.0001) (Fig. 3D, left panel). Western blot analysis shows the expression patterns of the COS7 cell lysates (Fig. 3D, right panel).
We subsequently assessed the affinities of the interactions of p38 MAPK-phosphorylated and unphosphorylated forms of MEF2C with purified β-catenin. These data clearly show that phosphorylation of purified MEF2C by p38 MAPK greatly enhances the MEF2C–β-catenin interaction in vitro (Fig. 3E).
To probe the phospho-dependency of the MEF2–β-catenin interaction further, we incubated A10 whole-cell lysates with CIP, which catalyzes the removal of phosphate groups on peptides. Cell lysates were incubated with CIP at 37°C (10 U enzyme in 1 mg lysate for 30 min) during immunoprecipitation using the MEF2A antibody. Compared with control cells that were incubated at 37°C without the addition of CIP, there was a marked reduction of corresponding β-catenin protein (Fig. 3F, top panels). Western blot analysis of MEF2A immunoprecipitates and input protein confirmed the loss of an upper band for MEF2A with CIP treatment, indicating successful MEF2A dephosphorylation, which correlated with the reduction in the interaction with β-catenin (Fig. 3F, bottom panels). Based on these experiments, we concluded that p38 MAPK-mediated phosphorylation of MEF2 promotes the association of MEF2 and β-catenin.
MEF2 and β-catenin associate and cooperate in the activation of the c-jun promoter.
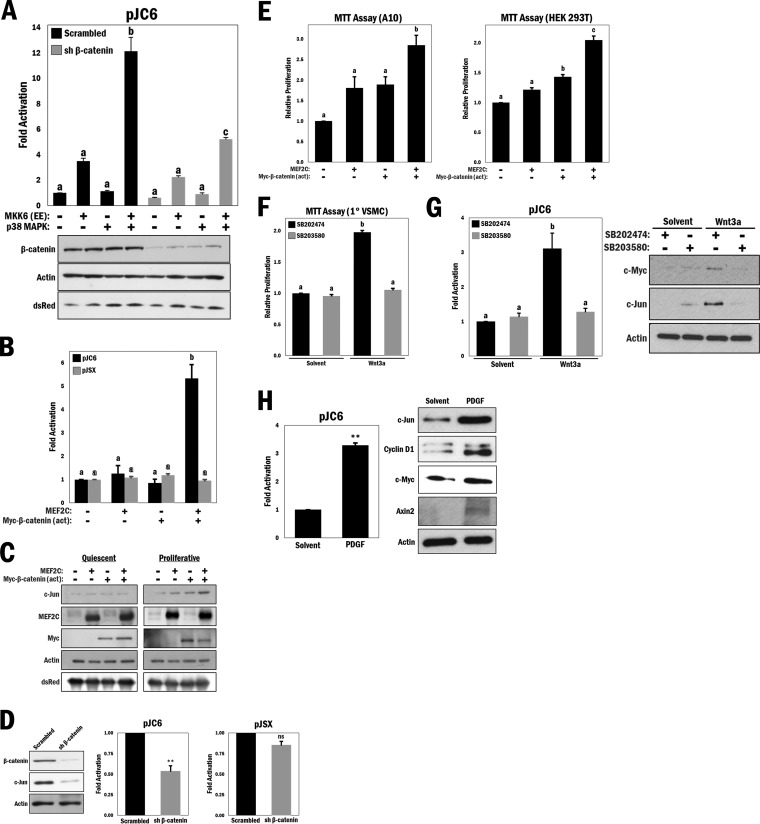
The effects observed with the TOPflash reporter gene illustrate that p38 MAPK/MEF2 signaling can cooperate with β-catenin in transcriptional activation. We used a known MEF2 target gene, c-jun, to determine the cooperation between MEF2 and β-catenin. To investigate potential selectivity with MEF2, we used the c-jun promoter region, which has been shown by us and others to be potently regulated by a single MEF2 consensus site [(C/T)TA(A/T)4TA(G/C)] (42, 43). In order to test the effects of β-catenin cooperativity on MEF2-dependent c-jun expression, we ectopically expressed the pCJ6 reporter gene (a c-jun promoter-based reporter gene), MKK6 (EE), and p38 MAPK in A10 cells. As expected, a pronounced increase in c-jun promoter activity was observed upon MKK6/p38 MAPK activation (P ≤ 0.0001) (Fig. 4A). Strikingly, shRNA-mediated silencing of β-catenin expression resulted in a significant decrease in c-jun promoter activation (P ≤ 0.0001), leading us to conclude that β-catenin can synergistically contribute to the activation of c-jun through MEF2. c-Jun fulfills a prominent cellular role as part of the activator protein 1 (AP-1) complex, which is known to promote cell proliferation by its induction of cyclin D1 (44) and downregulation of p53 (45). This experiment was repeated using an siRNA targeting β-catenin at a different sequence, with similar results (see Fig. S13 in the supplemental material).
FIG 4.
MEF2 and β-catenin cooperate to promote cell proliferation in a p38 MAPK-dependent manner, and p38 MAPK regulates Wnt target gene protein levels. (A) (Top) c-Jun reporter gene (pJC6) activity was measured in A10 cells with various combinations of ectopically expressed MKK6 (EE) and p38 MAPK and either scrambled shRNA (control) or shRNA β-catenin, as indicated in the figure. b and c, P ≤ 0.0001. (Bottom) Corresponding Western blot analysis of cell lysates. (B) A10 cells were transfected with different combinations of MEF2C and Myc–β-catenin (act) and cotransfected with either pJC6 or pJSX (analogous promoter region containing mutations in the MEF2 binding site), as indicated. b, P ≤ 0.001. (C) Western blot analysis of either quiescent (serum withdrawal for 12 h) or proliferative (maintained with 10% FBS) A10 cells that were transfected with different combinations of MEF2C and Myc–β-catenin (act), as indicated. (D) Western blot analysis of A10 cells transfected with scrambled shRNA (control) or shRNA β-catenin, with comparative pJC6 and pJSX reporter activities. **, P ≤ 0.01; ns, not significant. (E) MTT cell proliferation assay in A10 (b, P ≤ 0.01) and HEK 293T (b and c, P ≤ 0.01) cells transfected with different combinations of MEF2C and Myc–β-catenin (act), as indicated. (F) MTT cell proliferation assay in primary VSMCs that underwent serum withdrawal (12 h) and were pretreated with SB203580 or SB202474 (10 μM) for 45 min and then stimulated with Wnt3a (200 ng/ml) or solvent (PBS) for 4 h. b, P ≤ 0.0001. (G) (Left) A10 cells were transfected with the pJC6 reporter for 3 h, followed by treatment with either Wnt3a (200 ng/ml), solvent (PBS), SB203580 (10 μM), or SB202474 (10 μM) for 16 h in serum-free medium, as indicated. b, P ≤ 0.001. (Right) Primary VSMCs were treated similarly to the manner described for panel F, and lysates were subjected to Western blot analysis as indicated. (H) (Left) pJC6 reporter assay in A10 cells transfected for 3 h and then treated with PDGF (20 ng/ml) or solvent (0.1% BSA in 4 mM HCl) for 16 h in serum-free medium. **, P ≤ 0.01. (Right) Western blot analysis of primary VSMCs that underwent serum withdrawal for 12 h and were treated with PDGF (20 ng/ml) or solvent (0.1% BSA in 4 mM HCl) for 4 h.
In view of this, we further investigated the cellular implications of the cooperativity between MEF2 and β-catenin on c-jun expression. Ectopically expressed combinations of MEF2C and degradation-resistant β-catenin (act) with the pCJ6 reporter revealed that coexpression of MEF2C and β-catenin (act) resulted in a significant and synergistic increase in c-jun promoter activity (P ≤ 0.001) (Fig. 4B, black bars) in A10 cells, an effect not observed when the MEF2 site was mutated (pJSX reporter gene) (Fig. 4B, gray bars). We further analyzed c-Jun protein levels, since c-Jun expression has been shown to be regulated by canonical Wnt signaling through a TCF/LEF binding motif on its endogenous promoter, which is truncated in the pJC6 reporter (12). Based on reports in the literature, induction of quiescence results in a reduction of c-Jun (46, 47), and this was observed even with coexpression of both MEF2C and β-catenin (act) (Fig. 4C, left panel). However, under serum-stimulated conditions, which we would predict to allow cooperativity, coexpression of MEF2C and β-catenin (act) resulted in increased c-Jun protein levels (Fig. 4C, right panel). Conversely, in a loss-of-function approach, shRNA-mediated knockdown of β-catenin resulted in decreased c-Jun protein levels (Fig. 4D, left panel). Under the same conditions, c-jun promoter activity was significantly decreased following depletion of β-catenin, but that of the pJSX reporter was not (P ≤ 0.01) (Fig. 4D, middle and right panels). The experiments for Fig. 4D were repeated using an siRNA targeting β-catenin to confirm that the effect was target specific (see Fig. S14 in the supplemental material). To assess the functional implications of this cooperativity, we performed MTT cell proliferation assays. In both A10 and HEK 293T cells, coexpression of MEF2C and β-catenin (act) resulted in a significant increase in cell proliferation compared to that with either MEF2C or β-catenin (act) transfected alone (P ≤ 0.01 and P ≤ 0.0001, respectively) (Fig. 4E).
To establish a role for p38 MAPK regulation of Wnt-mediated cell proliferation, we performed another MTT assay with primary VSMCs that were cultured in serum-free medium and then stimulated with Wnt3a in the presence of an active or inactive p38 MAPK inhibitor. The results showed a significant increase in cell proliferation following Wnt3a stimulation that was completely abolished by p38 MAPK inhibition (P ≤ 0.0001) (Fig. 4F). Additionally, Wnt3a stimulation increased the promoter activity of c-jun (P ≤ 0.001) (Fig. 4G, left panel), as well as the protein levels of c-Jun and another Wnt target, c-Myc, which were all reduced in the presence of the p38 MAPK inhibitor (Fig. 4G, right panel).
Since our data regarding PDGF stimulation showed increased nuclear β-catenin localization and Wnt reporter gene activity, we determined the effects on other Wnt targets. c-Jun reporter gene activity was increased with PDGF stimulation (P ≤ 0.01) (Fig. 4H, left panel), as were c-Jun protein levels. Furthermore, this treatment also caused increases in the protein levels of other Wnt targets, i.e., cyclin D1 (9), c-Myc (10), and axin2 (11) (Fig. 4H, right panel).
Based on the collective data, we conclude that p38 MAPK is a key regulator of canonical Wnt signaling by promoting a phospho-dependent interaction between MEF2 and β-catenin to enhance cooperative transcriptional activity and cell proliferation.
DISCUSSION
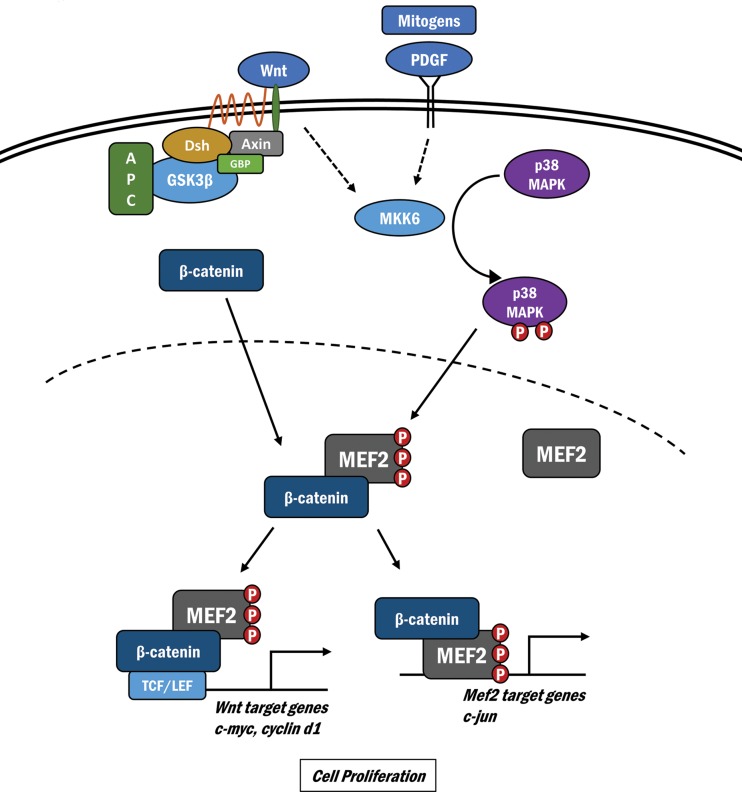
A prolific literature concerning the genetic, developmental, and biochemical bases of canonical Wnt signaling has positioned the β-catenin transcriptional regulatory protein as an indispensable determinant of embryonic development, stem cell biology, and also postnatal physiology and pathophysiology in a variety of organisms ranging from Caenorhabditis elegans to humans (6, 8, 48). Here we report a novel, critical step in the nuclear function of β-catenin-mediated transcriptional control that results from a protein-protein interaction between β-catenin and MEF2 and is potently enhanced by p38 MAPK signaling. This phospho-dependent interaction consolidates β-catenin accumulation in the nucleus, leading to a substantial activation of Wnt-dependent target genes (Fig. 5).
FIG 5.
Schematic illustrating the cross talk between Wnt/β-catenin and p38 MAPK/MEF2 signaling. Wnt-mediated inactivation of the GSK3–β-catenin destruction complex results in suppressed degradation of β-catenin. Activation of p38 MAPK signaling results in phosphorylation and activation of MEF2 in the nucleus, which enhance its physical interaction with nuclear β-catenin. This results in nuclear β-catenin retention, synergistic activation of the Wnt target genes c-myc and c-jun, and increased cell proliferation.
There is ample evidence of kinase-mediated phosphorylation modulating the affinity of protein-protein interactions in the canonical Wnt pathway, resulting in important outcomes for Wnt-dependent target gene activation. Interestingly, while it is widely acknowledged that CKI/GSK3 phosphorylation of β-catenin leads to the conformational formation of a binding site for β-TrCP, the E3 ligase that mediates ubiquitination and subsequent degradation of β-catenin, it is perhaps not as well recognized that these kinases also phosphorylate axin and APC, leading to their tighter association with β-catenin (49, 50). Thus, phospho-dependent protein interactions are a signature of β-catenin function, and the association with MEF2 that we report here falls into this category. Moreover, it may be more than an interesting coincidence that two serine/threonine phosphatases, PP1 and PP2A, which target the β-catenin destruction complex (51, 52) to reduce β-catenin degradation, have also been characterized by us and others as MEF2 phosphatases (39, 53). Whether MEF2 can recruit PP1 or PP2A to the β-catenin complex remains to be investigated.
While the prevalent dogma in the literature is that β-catenin stabilization results in enhanced nuclear levels, how β-catenin is retained in the nucleus is unclear (16, 17). One study reported that Wnt-induced β-catenin stabilization is insufficient for its nuclear accumulation and further proposed that Wnt activation of the Rac1 GTPase is also necessary to promote nuclear localization (54). Previous studies indicated that β-catenin translocates into the nucleus in an NLS- and importin-independent manner, possibly by direct interaction with nuclear pore proteins. Conversely, β-catenin nuclear extrusion has been linked to its association with APC (16), Axin (21), and Ran binding protein 3 (RanBP3) (20). It is of interest to consider the possibility that p38 MAPK/MEF2 signaling could alter the balance of the cell-adhesive and transcriptional properties of β-catenin. In our studies, the dramatically enhanced levels of nuclear β-catenin with p38 MAPK activation appeared to concomitantly reduce β-catenin amounts within the plasma membrane-associated adhesion complex. As is the case with many proteins that partition between the cytoplasm and the nucleus, β-catenin localization is dynamic and is probably the product of several processes involving both shuttling and nuclear retention. Here we present evidence suggesting that a p38 MAPK-dependent interaction with the MEF2 transcription factor enhances β-catenin nuclear retention in a number of cell lineages and in primary VSMCs.
An implication of the above discussion is that the p38 MAPK/MEF2 signaling pathway dynamically intersects with the Wnt pathway in MEF2-dependent target tissues. To date, MEF2 has been found to play important roles in cardiac (55, 56), skeletal (57–59), and smooth (60, 61) muscle cells, neurons (62), B cells (63, 64), and osteoblasts (65). Many of these tissues are also profoundly influenced by Wnt signaling. For example, in C2C12 myoblasts, Wnt4, generally recognized as a noncanonical Wnt, has been shown to increase myogenesis by activation of canonical Wnt/β-catenin signaling and upregulation of the muscle regulatory factors (MRFs) Myf5, myogenin, and muscle regulatory factor 4 (MRF4) (66). MEF2 is concomitantly required in these tissues during skeletal myogenesis by its direct interaction with the aforementioned MRF myogenin and MyoD to cooperatively activate the myogenic program (58). Moreover, the mef2c gene is controlled by these MRFs through an E box in its regulatory region, whose expression is essential for the maintenance of the myogenic program (67). In view of the profound role that MEF2 plays in both embryonic skeletal muscle and adult muscle stem (satellite) cells, it is tempting to speculate that p38 MAPK/MEF2 may intersect with Wnt signaling in these contexts. It will be interesting to observe if, in MEF2-dependent tissues, Wnt pathway modulation is a component of MEF2 function or whether both pathways are in fact mutually codependent.
A litany of highly insightful studies since the early 1980s have implicated β-catenin and its associated regulators as drivers of various human cancers (68). Moreover, the identification of several β-catenin-interacting transcriptional regulators has provided a mechanism by which proliferative growth-promoting genes are regulated (69, 70). Aberrations in components of this pathway of growth control have been linked directly to colorectal cancers, Wilm's tumors in pediatric renal cancer, metastasis in some lung cancers, and a variety of others (68). In addition, Wnt pathway hyperactivity has been reported for certain breast cancer types without any apparent mutations in components of the pathway (71, 72). These observations raise the question of whether “drugging” the canonical Wnt pathway may be efficacious in certain cancers (68). Moreover, our observations suggest that pharmacological targeting of the p38 MAPK pathway might serve as an unconventional way to blunt Wnt/β-catenin signaling in certain cancer cells. Of course, the efficacy of targeting a kinase that plays a role in other cellular processes is unclear at this point, but the existence of a number of well-characterized cell-permeating small-molecule inhibitors of p38 MAPK should make this a tractable question to address in the near future.
Here we report a previously uncharacterized aspect of canonical Wnt/β-catenin signaling through cross talk with the p38 MAPK/MEF2 signaling pathway, which may have implications in all muscle types, neurons, B cells, and osteogenic cells, as well as in Wnt-dependent cancer cells. These observations raise the possibility of therapeutic intervention to target canonical Wnt signaling in a variety of pathologies.
Supplementary Material
ACKNOWLEDGMENTS
J.C.M. is supported by the McLaughlin Research Chair.
We thank Tetsuaki Miyake for useful discussions pertaining to this research.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00832-15.
REFERENCES
- 1.Kadzik RS, Cohen ED, Morley MP, Stewart KM, Lu MM, Morrisey EE. 2014. Wnt ligand/Frizzled 2 receptor signaling regulates tube shape and branch-point formation in the lung through control of epithelial cell shape. Proc Natl Acad Sci U S A 111:12444–12449. doi: 10.1073/pnas.1406639111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JLR, Grosschedl R. 2000. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development 127:469–482. [DOI] [PubMed] [Google Scholar]
- 3.Ikeya M, Lee SMK, Johnson JE, McMahon AP, Takada S. 1997. Wnt signalling required for expansion of neural crest and CNS progenitors. Nature 389:966–970. doi: 10.1038/40146. [DOI] [PubMed] [Google Scholar]
- 4.Roose J, Huls G, van Beest M, Moerer P, van der Horn K, Goldschmeding R, Logtenberg T, Clevers H. 1999. Synergy between tumor suppressor APC and the β-Catenin-Tcf4 target Tcf1. Science 285:1923–1926. doi: 10.1126/science.285.5435.1923. [DOI] [PubMed] [Google Scholar]
- 5.Chan EF, Gat U, McNiff JM, Fuchs E. 1999. A common human skin tumour is caused by activating mutations in β-catenin. Nat Genet 21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 6.Wodarz A, Nusse R. 1998. Mechanisms of Wnt signaling in development. Annu Rev Cell Dev Biol 14:59–88. doi: 10.1146/annurev.cellbio.14.1.59. [DOI] [PubMed] [Google Scholar]
- 7.Logan CY, Nusse R. 2004. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 8.Moon RT, Kohn AD, De Ferrari GV, Kaykas A. 2004. Wnt and β-catenin signalling: diseases and therapies. Nat Rev Genet 5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- 9.Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. 1999. The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc Natl Acad Sci U S A 96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He T, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. 1998. Identification of c-MYC as a target of the APC pathway. Science 281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 11.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, Bodmer WF, Moyer MP, Riecken EO, Buhr HJ, Hanski C. 1999. Target genes of β-catenin–T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci U S A 96:1603–1608. doi: 10.1073/pnas.96.4.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishitani T, Ninomiya-Tsuji J, Nagai SI, Nishita M, Meneghini M, Barker N, Waterman M, Bowerman B, Clevers H, Shibuya H, Matsumoto K. 1999. The TAK1-NLK-MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature 399:798–802. doi: 10.1038/21674. [DOI] [PubMed] [Google Scholar]
- 14.Tago KI, Nakamura T, Nishita M, Hyodo J, Nagai SI, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, Akiyama T. 2000. Inhibition of Wnt signaling by ICAT, a novel β-catenin-interacting protein. Genes Dev 14:1741–1749. [PMC free article] [PubMed] [Google Scholar]
- 15.Takemaru KI, Yamaguchi S, Lee YS, Zhang Y, Carthew RW, Moon RT. 2003. Chibby, a nuclear β-catenin-associated antagonist of the Wnt/Wingless pathway. Nature 422:905–909. doi: 10.1038/nature01570. [DOI] [PubMed] [Google Scholar]
- 16.Henderson BR, Fagotto F. 2002. The ins and outs of APC and β-catenin nuclear transport. EMBO Rep 3:834–839. doi: 10.1093/embo-reports/kvf181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stadeli R, Hoffmans R, Basler K. 2006. Transcription under the control of nuclear Arm/β-catenin. Curr Biol 16:R378–R385. doi: 10.1016/j.cub.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Hill CS. 2009. Nucleocytoplasmic shuttling of Smad proteins. Cell Res 19:36–46. doi: 10.1038/cr.2008.325. [DOI] [PubMed] [Google Scholar]
- 19.Shrum CK, Defrancisco D, Meffert MK. 2009. Stimulated nuclear translocation of NF-κB and shuttling differentially depend on dynein and the dynactin complex. Proc Natl Acad Sci U S A 106:2647–2652. doi: 10.1073/pnas.0806677106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hendriksen J, Fagotto F, van der Velde H, van Schie M, Noordermeer J, Fornerod M. 2005. RanBP3 enhances nuclear export of active β-catenin independently of CRM1. J Cell Biol 171:785–797. doi: 10.1083/jcb.200502141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong F, Varmus H. 2004. Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of β-catenin. Proc Natl Acad Sci U S A 101:2882–2887. doi: 10.1073/pnas.0307344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray JL, Leach R, Herbert JM, Benson M. 2001. Isolation of vascular smooth muscle cells from a single murine aorta. Methods Cell Sci 23:185–188. doi: 10.1023/A:1016357510143. [DOI] [PubMed] [Google Scholar]
- 23.Ornatsky OI, Andreucci JJ, McDermott JC. 1997. A dominant-negative form of transcription factor MEF2 inhibits myogenesis. J Biol Chem 272:33271–33278. doi: 10.1074/jbc.272.52.33271. [DOI] [PubMed] [Google Scholar]
- 24.Ornatsky OI, McDermott JC. 1996. MEF2 protein expression, DNA binding specificity and complex composition, and transcriptional activity in muscle and nonmuscle cells. J Biol Chem 271:24927–24933. doi: 10.1074/jbc.271.40.24927. [DOI] [PubMed] [Google Scholar]
- 25.Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. 2003. Zebrafish Prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol 13:680–685. doi: 10.1016/S0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 26.Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. 1995. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem 270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- 27.Bikkavilli RK, Feigin ME, Malbon CC. 2008. p38 mitogen-activated protein kinase regulates canonical Wnt-beta-catenin signaling by inactivation of GSK3beta. J Cell Sci 121:3598–3607. doi: 10.1242/jcs.032854. [DOI] [PubMed] [Google Scholar]
- 28.Dionyssiou MG, Nowacki NB, Hashemi S, Zhao J, Kerr A, Tsushima RG, McDermott JC. 2013. Cross-talk between glycogen synthase kinase 3β (GSK3β) and p38MAPK regulate myocyte enhancer factor 2 (MEF2) activity in skeletal and cardiac muscle. J Mol Cell Cardiol 54:35–44. doi: 10.1016/j.yjmcc.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Matsumoto T, Yokote K, Tamura K, Takemoto M, Ueno H, Saito Y, Mori S. 1999. Platelet-derived growth factor activates p38 mitogen-activated protein kinase through a Ras-dependent pathway that is important for actin reorganization and cell migration. J Biol Chem 274:13954–13960. doi: 10.1074/jbc.274.20.13954. [DOI] [PubMed] [Google Scholar]
- 30.Uglow EB, Slater S, Sala-Newby GB, Aguilera-Garcia CM, Angelini GD, Newby AC, George SJ. 2003. Dismantling of cadherin-mediated cell-cell contacts modulates smooth muscle cell proliferation. Circ Res 92:1314–1321. doi: 10.1161/01.RES.0000079027.44309.53. [DOI] [PubMed] [Google Scholar]
- 31.Alli NS, Yang EC, Miyake T, Aziz A, Collins-Hooper H, Patel K, McDermott JC. 2013. Signal-dependent fra-2 regulation in skeletal muscle reserve and satellite cells. Cell Death Dis 4:e692. doi: 10.1038/cddis.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox DM, Du M, Marback M, Yang EC, Chan J, Siu KW, McDermott JC. 2003. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J Biol Chem 278:15297–15303. doi: 10.1074/jbc.M211312200. [DOI] [PubMed] [Google Scholar]
- 33.Du M, Perry RL, Nowacki NB, Gordon JW, Salma J, Zhao J, Aziz A, Chan J, Siu KW, McDermott JC. 2008. Protein kinase A represses skeletal myogenesis by targeting myocyte enhancer factor 2D. Mol Cell Biol 28:2952–2970. doi: 10.1128/MCB.00248-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. 1997. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- 35.Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, Olson EN, Ulevitch RJ, Han J. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol Biol Cell 19:21–30. doi: 10.1128/MCB.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ornatsky OI, Cox DM, Tangirala P, Andreucci JJ, Quinn ZA, Wrana JL, Prywes R, Yu YT, McDermott JC. 1999. Post-translational control of the MEF2A transcriptional regulatory protein. Nucleic Acids Res 27:2646–2654. doi: 10.1093/nar/27.13.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Angelis L, Zhao J, Andreucci JJ, Olson EN, Cossu G, McDermott JC. 2005. Regulation of vertebrate myotome development by the p38 MAP kinase-MEF2 signaling pathway. Dev Biol 283:171–179. doi: 10.1016/j.ydbio.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 38.Yang CC, Ornatsky OI, McDermott JC, Cruz TF, Prody CA. 1998. Interaction of myocyte enhancer factor 2 (MEF2) with a mitogen-activated protein kinase, ERK5/BMK1. Nucleic Acids Res 26:4771–4777. doi: 10.1093/nar/26.20.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perry RL, Yang C, Soora N, Salma J, Marback M, Naghibi L, Ilyas H, Chan J, Gordon JW, McDermott JC. 2009. Direct interaction between myocyte enhancer factor 2 (MEF2) and protein phosphatase 1α represses MEF2-dependent gene expression. Mol Cell Biol 29:3355–3366. doi: 10.1128/MCB.00227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai XL, Zhang Q, Ye LY, Liang F, Sun X, Chen Y, Hu QD, Fu QH, Su W, Chen Z, Zhuang ZP, Liang TB. 2015. Myocyte enhancer factor 2C regulation of hepatocellular carcinoma via vascular endothelial growth factor and Wnt/β-catenin signaling. Oncogene 34:4089–4097. doi: 10.1038/onc.2014.337. [DOI] [PubMed] [Google Scholar]
- 41.Snyder CM, Rice AL, Estrella NL, Held A, Kandarian SC, Naya FJ. 2013. MEF2A regulates the Gtl2-Dio3 microRNA mega-cluster to modulate WNT signaling in skeletal muscle regeneration. Development 140:31–42. doi: 10.1242/dev.081851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon JW, Pagiatakis C, Salma J, Du M, Andreucci JJ, Zhao J, Hou G, Perry RL, Dan Q, Courtman D, Bendeck MP, McDermott JC. 2009. Protein kinase A-regulated assembly of a MEF2-HDAC4 repressor complex controls c-Jun expression in vascular smooth muscle cells. J Biol Chem 284:19027–19042. doi: 10.1074/jbc.M109.000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han TH, Prywes R. 1995. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol Cell Biol 15:2907–2915. doi: 10.1128/MCB.15.6.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wisdom R, Johnson RS, Moore C. 1999. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J 18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. 1999. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev 13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tullai JW, Tacheva S, Owens LJ, Graham JR, Cooper GM. 2011. AP-1 is a component of the transcriptional network regulated by GSK-3 in quiescent cells. PLoS One 6:e20150. doi: 10.1371/journal.pone.0020150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lallemand D, Spyrou G, Yaniv M, Pfarr CM. 1997. Variations in Jun and Fos protein expression and AP-1 activity in cycling, resting and stimulated fibroblasts. Oncogene 14:819–830. doi: 10.1038/sj.onc.1200901. [DOI] [PubMed] [Google Scholar]
- 48.Lien WH, Fuchs E. 2014. Wnt some lose some: transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes Dev 28:1517–1532. doi: 10.1101/gad.244772.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, He X. 2008. Wnt/β-catenin signaling: new (and old) players and new insights. Curr Opin Cell Biol 20:119–125. doi: 10.1016/j.ceb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimelman D, Xu W. 2006. β-Catenin destruction complex: insights and questions from a structural perspective. Oncogene 25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 51.Luo W, Peterson A, Garcia BA, Coombs G, Kofahl B, Heinrich R, Shabanowitz J, Hunt DF, Yost HJ, Virshup DM. 2007. Protein phosphatase 1 regulates assembly and function of the β-catenin degradation complex. EMBO J 26:1511–1521. doi: 10.1038/sj.emboj.7601607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su Y, Fu C, Ishikawa S, Stella A, Kojima M, Shitoh K, Schreiber EM, Day BW, Liu B. 2008. APC is essential for targeting phosphorylated β-catenin to the SCFβ-TrCP ubiquitin ligase. Mol Cell 32:652–661. doi: 10.1016/j.molcel.2008.10.023. [DOI] [PubMed] [Google Scholar]
- 53.Li J, Chen J, Ricupero CL, Hart RP, Schwartz MS, Kusnecov A, Herrup K. 2012. Nuclear accumulation of HDAC4 in ATM deficiency promotes neurodegeneration in ataxia telangiectasia. Nat Med 18:783–791. doi: 10.1038/nm.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X, Tu X, Joeng KS, Hilton MJ, Williams DA, Long F. 2008. Rac1 activation controls nuclear localization of β-catenin during canonical Wnt signaling. Cell 133:340–353. doi: 10.1016/j.cell.2008.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backs J, Worst BC, Lehmann LH, Patrick DM, Jebessa Z, Kreusser MM, Sun Q, Chen L, Heft C, Katus HA, Olson EN. 2011. Selective repression of MEF2 activity by PKA-dependent proteolysis of HDAC4. J Cell Biol 195:403–415. doi: 10.1083/jcb.201105063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Phan D, van Rooij E, Wang DZ, McAnally J, Qi X, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. 2008. The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J Clin Invest 118:124–132. doi: 10.1172/JCI33255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu N, Nelson BR, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. 2014. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci U S A 111:4109–4114. doi: 10.1073/pnas.1401732111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Molkentin JD, Black BL, Martin JF, Olson EN. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 59.Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. 1995. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- 60.Pagiatakis C, Gordon JW, Ehyai S, McDermott JC. 2012. A novel RhoA/ROCK-CPI-17-MEF2C signaling pathway regulates vascular smooth muscle cell gene expression. J Biol Chem 287:8361–8370. doi: 10.1074/jbc.M111.286203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin Q, Lu J, Yanagisawa H, Webb R, Lyons GE, Richardson JA, Olson EN. 1998. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 125:4565–4574. [DOI] [PubMed] [Google Scholar]
- 62.Salma J, McDermott JC. 2012. Suppression of a MEF2-KLF6 survival pathway by PKA signaling promotes apoptosis in embryonic hippocampal neurons. J Neurosci 32:2790–2803. doi: 10.1523/JNEUROSCI.3609-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khiem D, Cyster JG, Schwarz JJ, Black BL. 2008. A p38 MAPK-MEF2C pathway regulates B-cell proliferation. Proc Natl Acad Sci U S A 105:17067–17072. doi: 10.1073/pnas.0804868105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilker PR, Kohyama M, Sandau MM, Albring JC, Nakagawa O, Schwarz JJ, Murphy KM. 2008. Transcription factor Mef2c is required for B cell proliferation and survival after antigen receptor stimulation. Nat Immunol 9:603–612. doi: 10.1038/ni.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stephens AS, Stephens SR, Hobbs C, Hutmacher DW, Bacic-Welsh D, Woodruff MA, Morrison NA. 2011. Myocyte enhancer factor 2C, an osteoblast transcription factor identified by dimethyl sulfoxide (DMSO)-enhanced mineralization. J Biol Chem 286:30071–30086. doi: 10.1074/jbc.M111.253518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernardi H, Gay S, Fedon Y, Vernus B, Bonnieu A, Bacou F. 2011. Wnt4 activates the canonical β-catenin pathway and regulates negatively myostatin: functional implication in myogenesis. Am J Physiol Cell Physiol 300:C1122–C1138. doi: 10.1152/ajpcell.00214.2010. [DOI] [PubMed] [Google Scholar]
- 67.Wang DZ, Valdez MR, McAnally J, Richardson J, Olson EN. 2001. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development 128:4623–4633. [DOI] [PubMed] [Google Scholar]
- 68.Polakis P. 2012. Drugging Wnt signalling in cancer. EMBO J 31:2737–2746. doi: 10.1038/emboj.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Behrens J, von Kries JP, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. 1996. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 70.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. 1996. XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell 86:391–399. doi: 10.1016/S0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 71.Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC. 2000. β-Catenin, a novel prognostic marker for breast cancer: its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A 97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung GG, Zerkowski MP, Ocal IT, Dolled-Filhart M, Kang JY, Psyrri A, Camp RL, Rimm DL. 2004. β-Catenin and p53 analyses of a breast carcinoma tissue microarray. Cancer 100:2084–2092. doi: 10.1002/cncr.20232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.