Zentella et al. show that DELLAs are modified by the O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) SECRET AGENT (SEC) in Arabidopsis. O-GlcNAcylation of the DELLA protein REPRESSOR OF ga1-3 (RGA) inhibits RGA binding to four of its interactors—PHYTOCHROME-INTERACTING FACTOR3 (PIF3), PIF4, JASMONATE-ZIM DOMAIN1, and BRASSINAZOLE-RESISTANT1 (BZR1)—that are key regulators in light, jasmonate, and brassinosteroid signaling pathways, respectively.
Keywords: O-GlcNAcylation of DELLA, DELLA-interacting proteins, hormone signaling, gibberellin signaling, O-GlcNAc transferase, SECRET AGENT
Abstract
The DELLA family of transcription regulators functions as master growth repressors in plants by inhibiting phytohormone gibberellin (GA) signaling in response to developmental and environmental cues. DELLAs also play a central role in mediating cross-talk between GA and other signaling pathways via antagonistic direct interactions with key transcription factors. However, how these crucial protein–protein interactions can be dynamically regulated during plant development remains unclear. Here, we show that DELLAs are modified by the O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) SECRET AGENT (SEC) in Arabidopsis. O-GlcNAcylation of the DELLA protein REPRESSOR OF ga1-3 (RGA) inhibits RGA binding to four of its interactors—PHYTOCHROME-INTERACTING FACTOR3 (PIF3), PIF4, JASMONATE-ZIM DOMAIN1, and BRASSINAZOLE-RESISTANT1 (BZR1)—that are key regulators in light, jasmonate, and brassinosteroid signaling pathways, respectively. Consistent with this, the sec-null mutant displayed reduced responses to GA and brassinosteroid and showed decreased expression of several common target genes of DELLAs, BZR1, and PIFs. Our results reveal a direct role of OGT in repressing DELLA activity and indicate that O-GlcNAcylation of DELLAs provides a fine-tuning mechanism in coordinating multiple signaling activities during plant development.
To adapt to its growing environment, plant development requires constant coordination among complex hormonal signaling networks in response to external cues (Jaillais and Chory 2010). DELLAs have emerged as central regulators that integrate internal and external signals to enhance plant survival in different environments (Sun 2011; Xu et al. 2014; Daviere and Achard 2016). However, it remains unclear how DELLA activity can be dynamically modulated to allow flexible plant growth. DELLAs were first identified as key negative regulators of the phytohormone gibberellin (GA) signaling pathway (Peng et al. 1997; Silverstone et al. 1998). GA promotes vegetative and reproductive growth in plants via regulation of cell division, expansion, and differentiation (Hauvermale et al. 2012). The GA signal is perceived by its nuclear receptor, GA-INSENSITIVE DWARF1 (GID1) (Ueguchi-Tanaka et al. 2005). Acting downstream from GID1, DELLAs repress GA signaling and restrict plant growth by causing transcriptional reprogramming (Sun 2011; Xu et al. 2014). The Arabidopsis genome encodes five DELLAs (REPRESSOR OF ga1-3 [RGA], GA-INSENSITIVE [GAI], RGA-LIKE1 [RGL1], RGL2, and RGL3), with RGA and GAI playing the major role in repressing GA-induced vegetative growth (Dill and Sun 2001; King et al. 2001). Lacking a canonical DNA-binding domain, DELLAs regulate gene expression by interacting with other transcription factors (Zentella et al. 2007; Sun 2011; Xu et al. 2014). GA promotes GID1–DELLA interaction, resulting in the rapid degradation of DELLAs via the ubiquitin–proteasome pathway mediated by the ubiquitin E3 ligase SCFSLY1/GID2 (McGinnis et al. 2003; Ueguchi-Tanaka et al. 2005; Griffiths et al. 2006; Murase et al. 2008). DELLAs contain a conserved N-terminal DELLA domain essential for its interaction with GID1, followed by a more diverse region rich in Ser and Thr residues (PolyS/T) and a conserved C-terminal GRAS domain that confers the transcription regulator function (Silverstone et al. 1998; Griffiths et al. 2006).
Recent studies revealed that DELLAs mediate cross-talk between GA and multiple signaling pathways by antagonizing or enhancing functions of many key regulators via direct protein–protein interactions (Sun 2011; Xu et al. 2014; Daviere and Achard 2016). Known DELLA-interacting proteins (DIPs) include several distinct families of transcription factors/regulators. The following are several examples of DELLA-inhibited DIPs. The DELLA-interacting basic helix–loop–helix (bHLH) transcription factors PIFs (phytochrome-interacting factors) are key regulators in light signaling controlling hypocotyl growth (de Lucas et al. 2008; Feng et al. 2008). DELLAs also antagonize the function of a GRAS protein, SCARECROW-LIKE 3 (SCL3), which promotes GA-induced root and hypocotyl elongation and radial patterning (Zhang et al. 2011). DELLAs promote JA-mediated root growth inhibition and plant defense by binding to the JA signaling repressors JAZs (Hou et al. 2010; Yang et al. 2012). In addition, DELLAs inhibit ethylene-induced apical hook formation by interacting with ETHYLENE INSENSITIVE3 (EIN3), an ethylene signaling activator (An et al. 2012). DELLAs also inhibit brassinosteroid (BR)-induced hypocotyl elongation by binding to BRASSINAZOLE-RESISTANT1 (BZR1), a BR signaling activator (Bai et al. 2012). In the case of PIFs, EIN3, and BZR1, DELLAs block DNA binding of these transcription factors to their target gene promoters (de Lucas et al. 2008; Feng et al. 2008; An et al. 2012; Bai et al. 2012). In contrast, DELLAs inhibit the function of JAZs by preventing JAZ interaction with MYC2 (Hou et al. 2010). These findings indicate that protein–protein interaction is a central regulatory mechanism in DELLA-modulated plant development.
We reasoned that, in response to endogenous and external cues, post-translational modifications of DELLA might modulate DELLA activity during plant development more dynamically than proteolysis. In fact, a recent study showed that salt stress conditions induce small ubiquitin-like modifier (SUMO) conjugation (SUMOylation) of DELLA, which sequesters GID1 independent of GA and leads to accumulation of non-SUMOylated DELLA to restrict plant growth (Conti et al. 2014). In addition, DELLAs contain predicted sites for both phosphorylation and O-GlcNAcylation (Supplemental Fig. 1). Some evidence suggests that phosphorylation of DELLA increases its stability. For example, a casein kinase I, EARLIER FLOWERING1 (EL1) in rice, was shown to repress GA signaling by enhancing DELLA stability, although the molecular mechanism is unclear (Dai and Xue 2010). The first clue for the possible involvement of O-GlcNAcylation in regulating GA signaling came from the identification of a putative O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT) in Arabidopsis, SPINDLY (SPY), as a GA signaling repressor; the loss-of-function spy alleles partially rescue the dwarf phenotype of the GA-deficient ga1 mutant (Jacobsen and Olszewski 1993; Jacobsen et al. 1996; Silverstone et al. 1997, 2007). OGTs catalyze the transfer of an O-GlcNAc monosaccharide from UDP-GlcNAc to the hydroxyl oxygen on Ser and Thr residues of the target proteins (Hanover et al. 2010; Hart et al. 2011). Consequently, O-GlcNAcylation may interfere or compete with phosphorylation, providing another layer of regulation of the target proteins. Based on sequence comparison, both SPY and its paralog, SECRET AGENT (SEC), in Arabidopsis were predicted to encode OGTs (Jacobsen et al. 1996; Hartweck et al. 2002). However, only SEC, but not SPY, has been demonstrated to display OGT enzyme activity in Escherichia coli assays (Hartweck et al. 2002). Interestingly, phylogenetic analysis of the OGT catalytic domain sequences indicates that metazoans contain a single OGT that is SEC-like, whereas vascular plants and mosses have two putative OGTs (a SEC-like and a SPY-like) (Olszewski et al. 2010). SPY and SEC in Arabidopsis appear to have related functions essential for embryogenesis because the double sec spy mutant is embryo-lethal (Hartweck et al. 2002), which is similar to the knockout OGT mutants in mice and Drosophila (Shafi et al. 2000; Gambetta et al. 2009). However, previous studies suggested that SEC might not function in GA signaling because the sec single mutants display subtle phenotypes (Hartweck et al. 2002, 2006). In contrast, considering the strong GA-related phenotypes of spy and predicted O-GlcNAc sites in DELLA, it has been speculated that SPY may O-GlcNAcylate DELLA to enhance DELLA activity.
In this study, we directly tackled the questions of whether DELLAs are O-GlcNAcylated and whether SPY and/or SEC are involved in this process. We found that the Arabidopsis DELLA protein RGA is O-GlcNAcylated in planta by mass spectrometry (MS) analysis and metabolic labeling assay using an azidosugar precursor. Surprisingly, the levels of O-GlcNAcylated RGA are not reduced by the loss-of-function spy mutation, whereas RGA O-GlcNAcylation is abolished in a null sec mutant. Using a transient coexpression system in tobacco, we further confirmed that RGA is O-GlcNAcylated by SEC but not by SPY. Our genetic studies reveal that SEC and SPY play distinct roles in regulating GA signaling; SEC is a positive regulator, whereas SPY is a negative regulator of GA responses. In addition, in vitro pull-down assays indicate that RGA O-GlcNAcylation by SEC inhibits binding of RGA to four DELLA interactors (BZR1, JAZ1, PIF3, and PIF4). This is the first direct detection of O-GlcNAcylation of an endogenous protein target by a specific OGT in plants. Our study also uncovered a direct role of OGT (SEC) and O-GlcNAcylation in fine-tuning plant development by modulating DELLA interactions with key regulators in multiple signaling pathways. In addition, considering the tight correlation between nutrient status and the levels of OGT's donor substrate UDP-GlcNAc (Hanover et al. 2010; Hart et al. 2011), O-GlcNAcylation of DELLAs might help to coordinate the nutrient status of the plant with its growth and development.
Results
RGA is O-GlcNAcylated in Arabidopsis
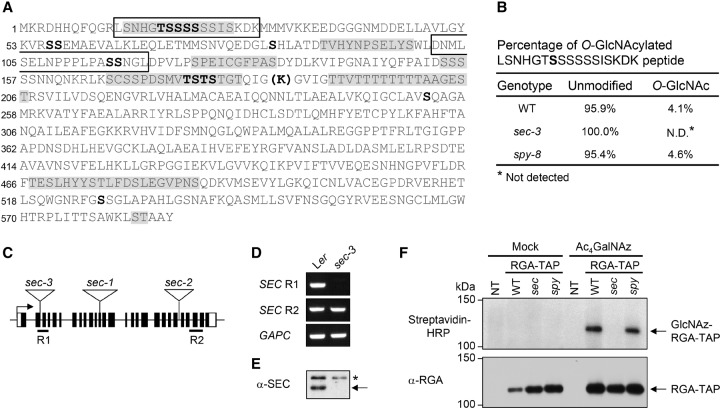
To test whether the Arabidopsis DELLA protein RGA is O-GlcNAcylated by SPY, we first determined whether RGA is O-GlcNAcylated in planta using collisionally activated dissociation (CAD)-MS and electron transfer dissociation (ETD)-MS analyses (Udeshi et al. 2008). For these analyses, we used the 35S:RGA-TAP transgenic Arabidopsis in the rga-24 ga1-3 mutant background; RGA-TAP is functional in rescuing the null rga phenotype (Zentella et al. 2007). RGA-TAP was tandem affinity-purified from Arabidopsis and digested with the endoproteinase trypsin or AspN followed by CAD-MS and ETD-MS analyses. We identified two O-GlcNAcylated peptides containing three GlcNAcylated Ser residues; one of these peptides (from trypsin digestion) is near the N terminus, and the other (from AspN digestion) is in the PolyS/T region (Fig. 1A; Supplemental Fig. 2; Supplemental Table 1).
Figure 1.
RGA is O-GlcNAc-modified by SEC in Arabidopsis. (A) Summary of O-GlcNAc modifications in RGA as determined by ETD-MS analyses. The two boxes highlight the peptides for which O-GlcNAcylation was identified using RGA-TAP isolated from 35S:RGA-TAP transgenic Arabidopsis in rga-24 ga1-3: mono- or di-O-GlcNAcylated (first box near the N terminus) or mono-O-GlcNAcylated (second box). Additional O-GlcNAc modifications were identified using Flag-RGA and Flag-RGAGKG purified from tobacco in which Flag-RGA/RGAGKG and SEC were coexpressed transiently. Bold type indicates modifications confirmed by accurate tandem MS (MS/MS). Sequences shaded in gray indicate that there is at least one modification in that peptide, but the specific amino acid could not be determined (see Supplemental Table 1 for detailed information; see Supplemental Fig. 6 for representative MS spectra). Within the poly-T stretch, there are up to six O-GlcNAc modifications. The bold K in parentheses indicates the position where an extra Lys residue was inserted to create an additional trypsin cleavage site. (B) MS analysis shows similar O-GlcNAc levels in the RGA peptide LSNHGTg[S]SSSSISKDK in wild type and spy-8, but no such modification was detected in sec-3. RGA-TAP was purified from 35S:RGA-TAP transgenic Arabidopsis lines in rga-24 ga1-3 (wild-type [WT] for both SPY and SEC) or spy-8 rga-24 ga1-3, or sec-3 rga-24 ga1-3 backgrounds (see Supplemental Fig. 2 for the MS spectra). (Unmodified) Not O-GlcNAcylated. (C) Scheme of the SEC locus showing the insertion position of the Ds element in sec-3 and T-DNA in sec-1 and sec-2. The black boxes represent exons, and the white boxes indicate 5′ and 3′ untranslated regions. R1 and R2 represent the positions of the RT–PCR products used to detect the SEC transcript. (D) RT–PCR analyses show the absence of wild-type SEC transcript (R1) in sec-3 but the presence of truncated mRNA downstream from the Ds insertion (R2). GAPC (At3g04120), a housekeeping gene, served as a control (Dill et al. 2004). (E) Immunoblot showing the absence of the SEC protein in sec-3. The blot contains total proteins extracted from seedlings and probed with an anti-SEC antiserum. The asterisk marks the nonspecific band. (F) The Ac4GalNAz feeding and click reaction assays show that RGA-TAP in the wild-type and spy-8 backgrounds is O-GlcNAcylated but not in the sec-3 mutant. The blots contain approximately equal amounts of RGA-TAP proteins that were purified from Ac4GalNAz- or mock-treated RGA-TAP transgenic lines in wild-type, sec-3, or spy-8 backgrounds and labeled with biotin-alkyne by the click reaction. The nontransgenic (NT) rga-24 ga1-3 plant was included as a negative control. (Top blot) Streptavidin-horseradish peroxidase (HRP) was used to detect biotinylated O-GlcNAz-labeled RGA-TAP. (Bottom blot) Anti-RGA antibodies were used to detect the amount of RGA-TAP in each lane.
RGA is O-GlcNAcylated by SEC but not by SPY
To determine whether SPY O-GlcNAcylates RGA, ETD-MS analysis was performed using RGA-TAP purified from the spy-8 rga-24 ga1-3 mutant background. However, the levels of O-GlcNAc modification in RGA were not affected by spy-8 (Fig. 1B; Supplemental Fig. 2), suggesting that SEC may modify RGA. To test this possibility, we obtained the sec-3 mutant (in the Landsberg erecta [Ler] ecotype background), which contains a Ds transposon insertion at the junction between exon 2 and intron 2 of SEC (Fig. 1C). This sec-3 allele is a null mutant, as it does not produce any wild-type SEC mRNA or protein (Fig. 1D,E). 35S:RGA-TAP was introduced into sec-3 by genetic crossing, and RGA-TAP was purified for ETD-MS analysis. Importantly, no O-GlcNAcylated RGA peptides were detected (Fig. 1B; Supplemental Fig. 2), suggesting that SEC O-GlcNAcylates RGA.
To verify the MS results, we labeled GlcNAcylated proteins in transgenic RGA-TAP Arabidopsis seedlings (in wild-type [for both SPY and SEC], sec-3, and spy-8 backgrounds) by incubating with peracetylated azidoacetylgalactosamine (Ac4GalNAz), a synthetic azide functionalized sugar that is metabolized by endogenous sugar salvage enzymes to create “UDP-GlcNAz,” an azide-bearing nucleotide sugar cofactor accepted by OGT (Vocadlo et al. 2003; Boyce et al. 2011). Boyce et al. (2011) showed that Ac4GalNAz treatment of mammalian cells allows efficient labeling of OGT target proteins with O-GlcNAz modifications, which can be further tagged with biotin through a “click chemistry” reaction between azide and alkyne-biotin. Figure 1F shows that RGA-TAP purified from Ac4GalNAz-treated wild-type and spy-8 seedlings were labeled by alkyne-biotin, as detected by immunoblot analysis using horseradish peroxidase (HRP)-conjugated streptavidin, whereas RGA-TAP from Ac4GalNAz-treated sec-3 seedlings was not labeled. These results confirm that RGA is O-GlcNAcylated in Arabidopsis by SEC and further suggest that SPY is not necessary for RGA O-GlcNAcylation.
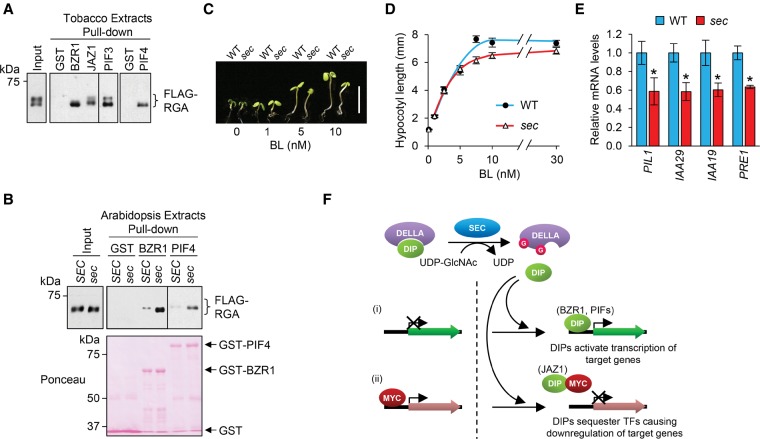
To investigate further whether RGA could serve as a substrate of SPY, we tested whether SPY and SEC bind to RGA by coimmunoprecipitation (co-IP) assays using a transient coexpression system in leaves of Nicotiana benthamiana. By agroinfiltration, we transiently expressed HA-RGA alone or coexpressed it with Myc-tagged SEC, SPY, or GUS (a negative control) (Zhang et al. 2011). Co-IP was then performed using anti-Myc agarose beads. Figure 2A shows that HA-RGA was pulled down by anti-Myc beads when it was coexpressed with Myc-SPY or Myc-SEC but not when it was expressed alone or coexpressed with Myc-GUS. These co-IP results indicate that both SPY and SEC interact with RGA. Interestingly, when coexpressed with SEC but not with SPY, the RGA electrophoretic mobility was shifted to a slightly higher molecular mass (by ∼2–3 kDa, equivalent to ∼10–15 GlcNAc residues) compared with the RGA alone sample (Fig. 2A,B). A GalT on-blot assay (Hartweck et al. 2002) was then performed to verify whether this RGA mobility shift in the RGA + SEC sample was caused by high levels of O-GlcNAcylation and whether the lack of RGA mobility shift in the RGA + SPY sample was due to lower levels or a lack of O-GlcNAcylation (Fig. 2C). For the GalT assay, Flag-RGA was expressed alone or coexpressed with Myc-tagged SEC or SPY in tobacco, and Flag-RGA was purified using anti-Flag beads. Flag-RGA (instead of HA-RGA) was used here because we were able to purify this fusion protein more efficiently using anti-Flag beads. The GalT assay labels GlcNAc-terminal glycans with [3H]-galactose. To distinguish between N- and O-linked glycans, alkaline β elimination treatment was then used to remove only O-linked modifications; N-linked GlcNAc refers to attachment of GlcNAc to the nitrogen of the Asn side chain. Figure 2C shows that only the shifted RGA band (in the RGA + SEC sample) was labeled by GalT, whereas the nonshifted RGA (in the RGA alone or RGA + SPY samples) was not labeled. β Elimination removed the labeled RGA-TAP signal in the RGA + SEC sample. Therefore, although RGA interacts with both SPY and SEC, RGA is only O-GlcNAcylated by SEC. Gel mobility assays in the tobacco transient expression system further showed that SEC also O-GlcNAcylates the other four AtDELLAs (GAI, RGL1, RGL2, and RGL3) although to a lesser extent (Supplemental Fig. 3A). Similar to RGA, coexpression of SPY did not cause any mobility shift in these AtDELLAs (Supplemental Fig. 3B). To rule out the possibility that other plant factors are required for SEC modification of RGA, we coexpressed SEC and RGA in E. coli, and a Gal-T assay showed that RGA was O-GlcNAcylated in the presence of SEC (Supplemental Fig. 3C).
Figure 2.
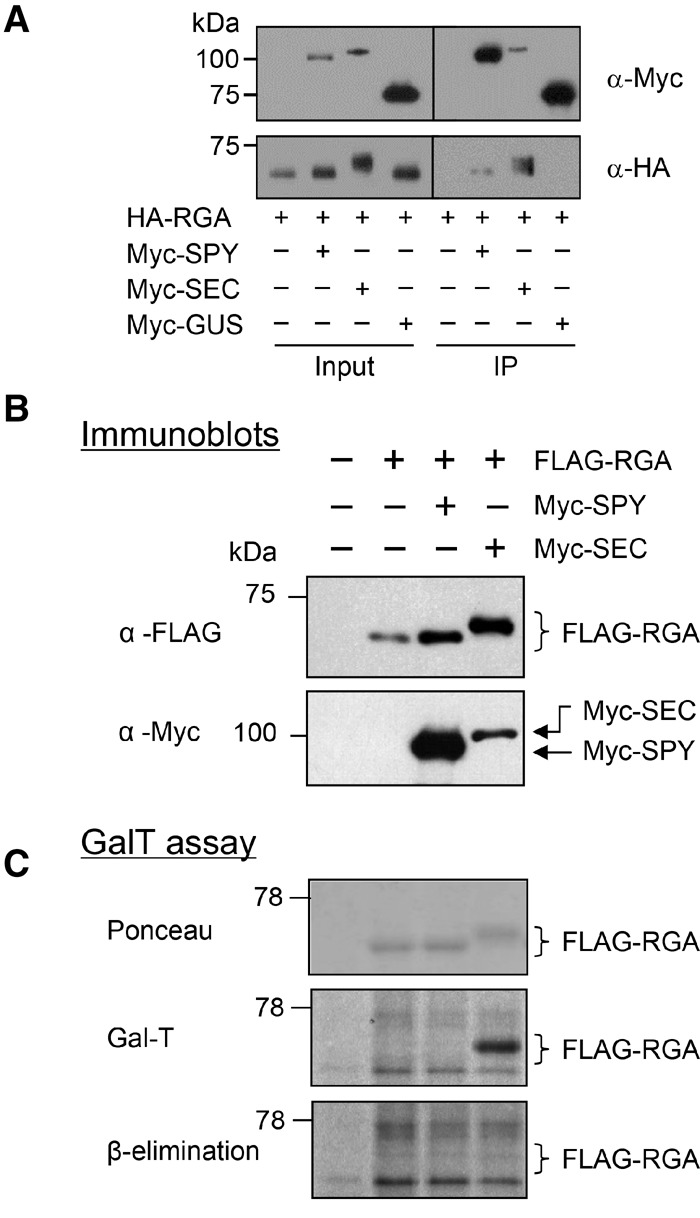
Both SEC and SPY bind to RGA, but only SEC O-GlcNAcylates RGA in the tobacco transient expression system. (A) Co-IP of RGA with SEC and SPY in planta. HA-RGA was transiently expressed alone or coexpressed with Myc-SPY, Myc-SEC, or Myc-GUS-NLS in N. benthamiana. Total protein extracts were immunoprecipitated with anti-Myc agarose beads; input and immunoprecipitation (IP) samples were analyzed by immunoblotting using anti-HA and anti-Myc antibodies separately. (B,C) RGA is O-GlcNAcylated by SEC but not by SPY in the tobacco transient expression system. Flag-RGA was expressed alone or coexpressed with Myc-SPY or Myc-SEC in N. benthamiana (as labeled above the blot in B). (B) Immunoblot analysis using total protein extracts and anti-Flag or anti-Myc antibodies; coexpression of RGA and SEC causes a detectable mobility shift of RGA. (C) A GalT assay followed by β elimination confirmed that the mobility-shifted RGA in the RGA + SEC sample is caused by O-GlcNAcylation. The blot contains equal amounts of affinity-purified Flag-RGA proteins, as indicated by the Ponceau-S-stained blot image. GalT labeled both N- and O-linked GlcNAcylated proteins with [3H]-galactose. The Flag-RGA signal (in the RGA + SEC sample) disappeared after β elimination. The remaining signals on the blot are N-GlcNAcylated proteins. Brackets indicate the position of unmodified and modified RGA. In A–C, 6% SDS-PAGE long (12-cm) gels were used to show differential electrophoretic mobility of GlcNAcylated RGA versus unmodified RGA proteins.
To determine whether spy and sec affect overall protein O-GlcNAcylation levels in Arabidopsis, immunoblot analysis was performed using an anti-O-GlcNAc antibody (CTD110.6) (Comer et al. 2001) and total protein extracts from wild type, spy-8, sec-3, and SEC-OE (a SEC overexpression line that is described in detail below). sec-3 and SEC-OE displayed reduced and elevated levels of protein O-GlcNAcylation, respectively (Supplemental Fig. 3E). In contrast, spy-8 showed patterns similar to wild type. These results suggest that SEC plays a major role in protein O-GlcNAcylation, whereas SPY may modify target proteins at lower levels or does not have OGT activity. Although CTD110.6 was reported to have a high specificity to O-GlcNAc (Comer et al. 2001), it can still cross-react with N-GlcNAc (Isono 2011). Therefore, the signals detected by CTD110.6 in the sec-3 sample could be N-GlcNAcylated proteins.
To identify the interaction domains in RGA and SEC, we performed yeast two-hybrid (Y2H) assays using full-length and various truncated RGA and SEC sequences. Full-length RGA, RGA-CT1, RGA-NT3, and RGA-NT4 interacted strongly with SEC (Fig. 3A,B; Supplemental Fig. 4A), suggesting that amino acids 107–285 in RGA (containing the PolyS/T, LHR1, and NLS [nuclear localization signal]) are important for SEC binding. RGA-NT4 showed slightly stronger interaction with SEC than full-length RGA, suggesting that the C-terminal region of RGA may interfere with SEC binding in this yeast system. SEC contains an N-terminal protein interaction domain with tetratricopeptide repeats (TPRs) and a C-terminal OGT catalytic domain (Olszewski et al. 2010). We found that the TPR domain of SEC is essential for RGA binding, whereas the OGT domain did not interact with RGA (Fig. 3C; Supplemental Fig. 4B).
Figure 3.
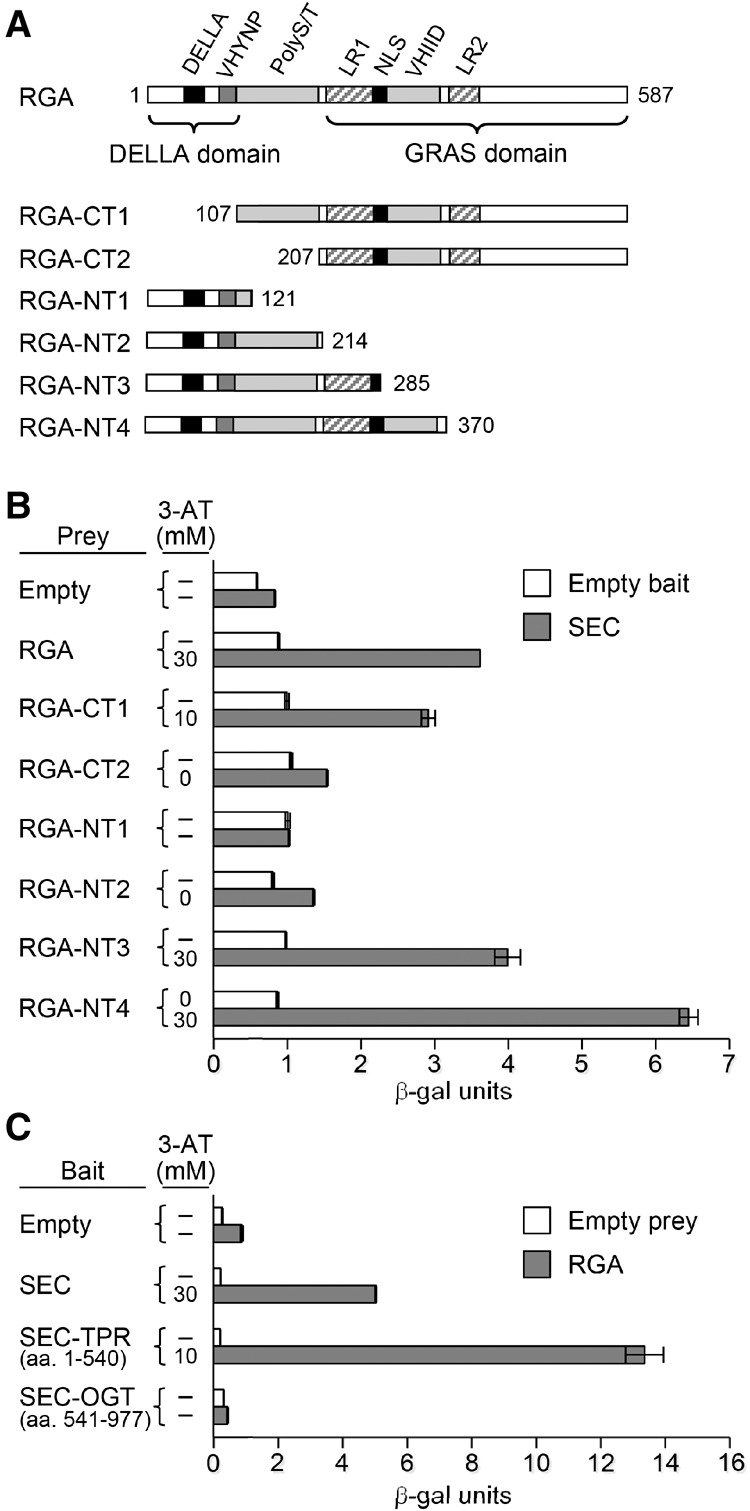
Mapping interaction domains in RGA and SEC by Y2H assays. (A) Protein structure diagram showing full-length RGA and the C terminus (CT) and N terminus (NT) of RGA constructs. Different motifs in RGA are labeled as DELLA, VHYNP, PolyS/T, LR (Leu heptad repeats), NLS, and VHIID. The numbers indicate which amino acid each RGA construct begins or ends with. (B) Y2H assays using the full-length SEC as the bait and various RGA constructs as prey. (C) Y2H assays using full-length RGA and SEC-TPR/SEC-OGT domains. In B and C, the degree of interaction was indicated by their ability to grow on His− plates with 0–40 mM 3-AT and by quantitative β-galactosidase assay. The dash means no growth on His− plates with 0 mM 3-AT. See Supplemental Figure 4 for the growth assays in different 3-AT concentrations.
sec-3 resembles reduced GA response mutants
To elucidate the role of SEC in regulating the function of DELLAs in GA signaling and plant development, we characterized phenotypes of the newly identified sec-3-null mutant. We found that the sec-3 mutant is recessive and has a 17% shorter hypocotyl than wild-type Ler (Fig. 4A,B). It also exhibited a clear mutant phenotype after flowering, most notably reduced final height (∼60% of wild type) due to decreased internode length (Fig. 4C–F). The reduced hypocotyl length and inflorescence stem length phenotypes of sec-3 were rescued by a SEC genomic clone, indicating that the dwarf phenotype is caused by the sec-3 mutation (Fig. 4A–D,G). In addition, sec-3 displayed slightly reduced silique length and fertility (i.e., average number of seeds per silique) (Supplemental Fig. 5D,E), phenotypes consistent with sec-3 being a reduced GA response mutant. We also found that SEC-OE lines that contain CaMV 35S promoter:SEC cDNA were ∼35% taller than wild-type Ler (Fig. 4H–J). We further performed a quantitative hypocotyl elongation assay to examine the GA responsiveness of sec-3 in the GA-deficient ga1-3 background and showed that sec-3 ga1-3 displayed a reduced GA response compared with ga1-3 (Fig. 5A,B). These results indicate that SEC is a positive regulator of GA signaling.
Figure 4.
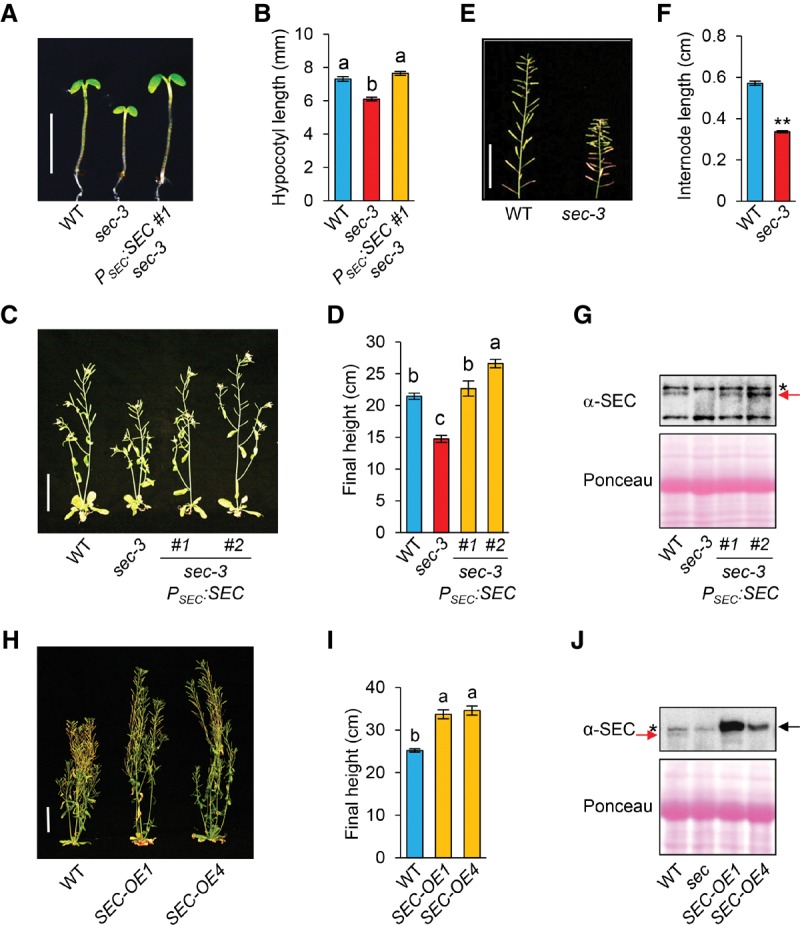
The sec-3 mutant displayed a semidwarf phenotype that was rescued by PSEC:SEC. (A,B) The shorter hypocotyl phenotype of sec-3 was rescued by the PSEC:SEC genomic sequence. Seedlings were grown under short-day conditions, and hypocotyl lengths were measured at day 5. n ≥ 13. Bar in A, 5 mm. (B) The average hypocotyl lengths. (C,D) The semidwarf phenotype of sec-3 was rescued by PSEC:SEC. (C) Eight-week-old plants are shown, and two representative transgenic lines are included. Bar, 5 cm. (D) The average final heights of sec-3 PSEC:SEC lines were determined using T2 BASTA-resistant plants. n ≥ 9. (E,F) sec-3 displayed reduced internode length in comparison with wild type (Ler). Bar in E, 5 cm. In F, average internode length was determined by measuring the length of the primary stem from siliques 1–20 and then dividing by 19. n ≥ 28. (G) Immunoblot analysis using anti-SEC antiserum confirmed expression of SEC (marked by a red arrow) in sec-3 PSEC:SEC lines. (*) Nonspecific background bands. Ponceau-S staining indicates equal loading. (H,I) Overexpression of SEC increased plant height. In H, 68-day-old plants of wild type and two independent SEC-OE carrying 35S:Myc-SEC are shown. Bar in H, 5 cm. (I) Average final plant heights. n ≥ 19. (J) The immunoblot shows elevated amounts of Myc-SEC protein (black arrow) in the SEC-OE lines compared with the levels of endogenous SEC (red arrow) in wild type. (*) Nonspecific background bands. In B, D, F, and I, data are means ± SE. Different letters or two asterisks above the bars indicate significant differences (P < 0.01).
Figure 5.
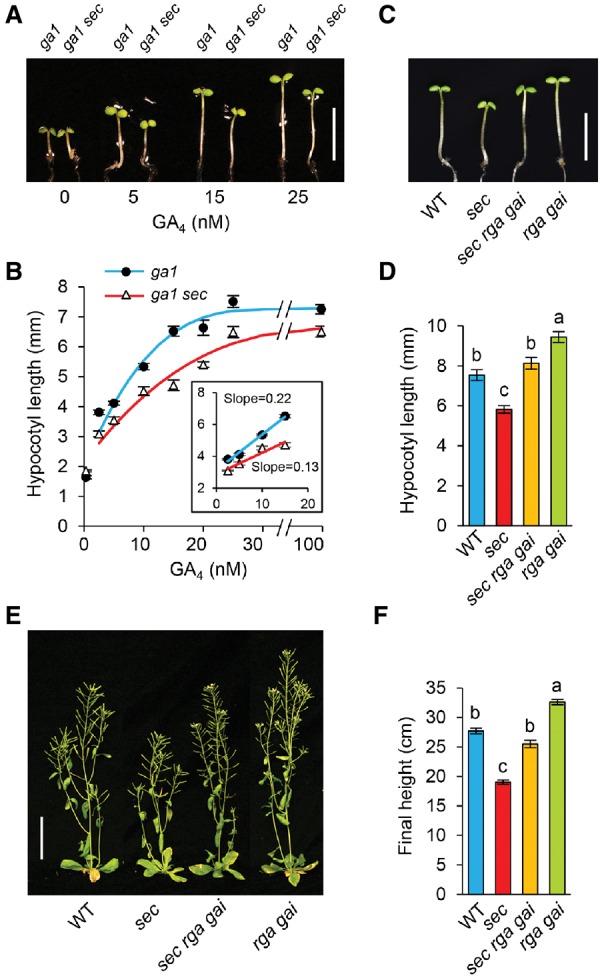
sec-3 displayed reduced GA responses, and its dwarf phenotypes were partially rescued by the double-null della mutation rga-24 gai-t6. (A,B) In the GA-deficient ga1-3 background, sec-3 caused a reduced GA response in hypocotyl growth. Hypocotyl lengths were measured at day 5. n ≥ 14. Bar in A, 5 mm. (B) GA response curves for ga1 and ga1 sec-3. (C–F) Double-null della mutant rga-24 gai-t6 partially rescued the semidwarf phenotype of sec-3. In C and D, hypocotyl lengths were measured at day 5. n ≥ 10. Bar in C, 5 mm. In E, 6-wk-old plants are shown. Bar, 5 cm. (F) Average final plant heights. n ≥ 16. In D and F, data are means ± SE. Different letters above the bars indicate significant differences (P < 0.01).
Our findings are surprising because previous studies reported that two null alleles, sec-1 and sec-2 (in the Wassilewskija and Columbia [Col-0] ecotypes, respectively) (Fig. 1C), showed subtle phenotypes and suggested that SEC does not function in GA signaling (Hartweck et al. 2002). The unique phenotype of the sec-3 allele (in the Ler ecotype) is likely caused by genetic background differences. Ler contains a mutation in the ERECTA (ER) locus, and the er mutation is known to enhance the dwarf phenotypes of the GA biosynthesis and response mutants (Fridborg et al. 2001). To determine whether er enhances the dwarf phenotype of sec, we generated sec-3 ER by crossing sec-3 (in Ler) with La-0. At the same time, the sec-2 er double-homozygous mutant was generated by crossing sec-2 (in Col-0) with Col-er. However, in both the er and ER backgrounds, sec-3 caused a more dramatic dwarfing phenotype than sec-2 (Supplemental Fig. 5A). These results indicate that other modifiers in Ler versus Col-0 backgrounds contribute to the phenotypic differences between sec-2 and sec-3.
Taken together, the O-GlcNAcylation of DELLAs by SEC and the semidwarf phenotype of sec-3 with reduced GA responses suggest that SEC activates GA signaling by inhibiting DELLA function via O-GlcNAcylation. Our results also indicate that SEC and SPY have distinct functions in GA signaling, with SEC being a positive regulator and SPY being a negative regulator.
To verify the hypothesis that SEC inhibits the function of DELLAs, epistasis analyses were performed to determine whether the sec-3 dwarf phenotype could be rescued by rga-24 gai-t6 double-null mutations (because RGA and GAI are the major AtDELLAs for inhibiting stem growth). We found that the sec-3 rga gai triple mutant is taller than sec-3 (40% increased in hypocotyl length and 34% increased in the final plant height) (Fig. 5C–F). These results are consistent with the notion that SEC promotes GA signaling by inhibiting DELLA function via O-GlcNAcylation. The incomplete rescue of sec-3 by GA treatment (Fig. 5B) or by rga gai (Fig. 5C–F) could be due to the pleiotropic effects of sec on other pathways independent of DELLAs and/or GA signaling.
Identification of O-GlcNAc sites in RGA by gel mobility shift assays and MS analyses
The differential gel mobility is a convenient indicator of the O-GlcNAcylation status of transiently expressed RGA and other AtDELLAs from tobacco (Fig. 2B; Supplemental Fig. 3A). Similarly, the endogenous RGA protein from the sec-3 mutant also displayed a slightly faster gel mobility than that of wild type, and PSEC:SEC restored the RGA mobility in sec-3 to that in wild type (Fig. 6A). According to the O-GlcNAc site prediction program YinOYang 1.2 (http://www.cbs.dtu.dk/services/YinOYang) (Gupta and Brunak 2002), 21 out of the 28 predicted O-GlcNAc sites in RGA are located in its N terminus, and the PolyS/T region alone contains 16 predicted O-GlcNAc sites (Supplemental Fig. 1). To test whether the PolyS/T region contains most of the O-GlcNAc sites, we examined two truncated rga proteins (rga-CT1 and rga-CT2) in transgenic Arabidopsis that carries PRGA:(rga-CT1)-Myc or PRGA:(rga-CT2)-Myc. rga-CT1 contains the PolyS/T and the GRAS domain (amino acid residues 107–587), whereas rga-CT2 contains only the GRAS domain (amino acid residues 207–587) (Fig. 3A). As predicted, rga-CT1 in the sec-3 background displayed a slightly faster mobility than in the wild-type SEC background, whereas rga-CT2 mobility was not affected by sec-3 (Fig. 6B). Moreover, β-hexosaminidase treatment, which cleaves GlcNAc residues, caused rga-CT1 in the wild-type SEC background to migrate faster (Fig. 6C), confirming that the slower mobility of rga-CT1 in the wild-type background was caused by GlcNAcylation. These results support that the PolyS/T region in RGA contains most of the native O-GlcNAc sites.
Figure 6.
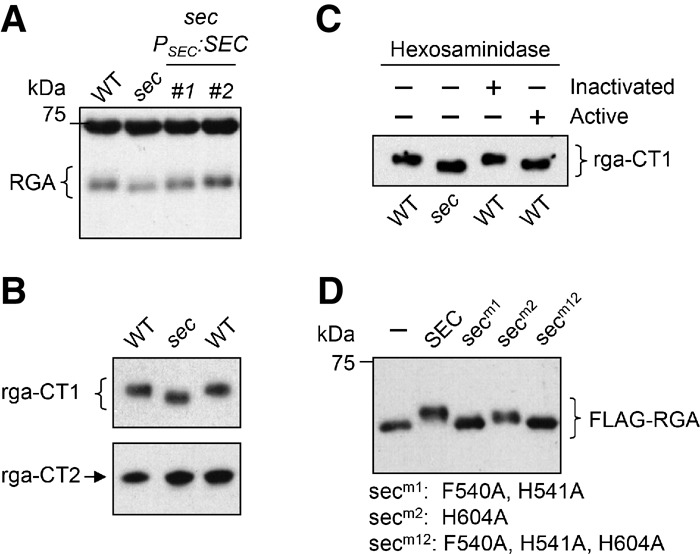
O-GlcNAc modifications map mostly to the DELLA domain and the PolyS/T region of RGA by the gel mobility assays. (A,B) The endogenous RGA (A) and rga-CT1 (B) from the sec-3 mutant background displayed faster mobility than from the wild-type SEC background, whereas rga-CT2 mobility (B) was not altered by sec-3. The rga-CT1 and rga-CT2 proteins were expressed in transgenic Arabidopsis that contained PRGA:(rga-CT1)-Myc or PRGA:(rga-CT2)-Myc. Immunoblots were probed with anti-RGA antibodies (for RGA in A) or an anti-Myc antibody (for Myc-tagged rga-CT1 and rga-CT2 in B). (C) Hexosaminidase treatment confirmed that the mobility shift for rga-CT1 in the wild-type SEC background was caused by GlcNAcylation. The immunoblot contains affinity-purified rga-CT1 from wild type or sec-3 that was untreated or treated with active or inactivated hexosaminidase. (D) Site-directed sec mutations abolished (secm1 and secm12) or reduced (secm2) the OGT activity by gel mobility shift assays when coexpressed with Flag-RGA in tobacco. The blot was probed with an anti-Flag antibody. In A, C, and D, 6% SDS-PAGE (12-cm) gels were used to show differential electrophoretic mobility of GlcNAcylated versus unmodified RGA/rga proteins. In B, 7% SDS-PAGE (12-cm) gels were used.
As shown earlier, our MS analyses using RGA-TAP purified from Arabidopsis identified only two O-GlcNAcylated peptides that are outside of the PolyS/T region in RGA (Fig. 1A; Supplemental Table 1). Additional O-GlcNAcylated peptides should be present in the Arabidopsis RGA because we showed that its PolyS/T region contains most of the O-GlcNAc sites in the native protein, and the differential gel mobility of RGA isolated from wild type and sec-3 cannot be explained by the addition of two O-GlcNAc residues in wild type (Fig. 6). The reason why we failed to identify additional O-GlcNAc sites in RGA isolated from Arabidopsis is likely because of the low abundance of this modification in general and because most of the PolyS/T region lacks trypsin or AspN cleavage sites. To identify additional potential GlcNAc sites in RGA, we performed ETD-MS analysis using Flag-RGA that was expressed alone or coexpressed with SEC in tobacco leaves. To detect the O-GlcNAcylated peptide that spans the poly-T track within the PolyS/T region, we introduced an extra trypsin cleavage site by inserting a K residue between G185 and G186 in RGA (named RGAGKG) (Fig. 1A). Gel mobility shift assay showed that coexpression of SEC caused a similar mobility shift in Flag-RGAGKG in comparison with Flag-RGA (Supplemental Fig. 3D). In total, 30 O-GlcNAc sites (located in 17 peptides) were identified in Flag-RGA and Flag-RGAGKG isolated from tobacco. Fifteen of them were mapped to specific Ser or Thr residues (Fig. 1A; Supplemental Fig. 6; Supplemental Table 1). The majority of the O-GlcNAc sites are located in the DELLA domain and the PolyS/T region (Fig. 1A; Supplemental Fig. 1C). In the absence of SEC, up to six O-GlcNAc sites in RGA were detected (Supplemental Table 1), much less abundant than in the SEC coexpression sample, indicating that RGA was modified to a lesser extent by endogenous OGTs in tobacco. Although transient expression of SEC in this tobacco system may cause hyper-GlcNAcylation of RGA, this system allowed us to quickly identify most (if not all) potential GlcNAc sites in RGA. Most of the identified GlcNAc sites in RGA match the prediction by the YinOYang program (Supplemental Fig. 1). More importantly, both O-GlcNAcylated peptides identified in the Arabidopsis sample were also modified in the tobacco transient expression system, suggesting that the modification sites found using the tobacco system are potential O-GlcNAc sites in Arabidopsis.
OGT activity in SEC is essential for RGA O-GlcNAcylation and SEC-dependent stem growth
Recent determination of the ternary complex structure of the human OGT with UDP and a peptide substrate revealed that H498–H499 and H558 play a key role in maintaining its catalytic activity (Lazarus et al. 2011). The H498A or H558A mutation dramatically reduces the human OGT activity to <10%, and the H499A mutation decreases the OGT activity to 30%–60% of wild type. In silico rendering of the three-dimensional (3D) structure of SEC using the Protein Homology/AnalogY recognition engine version 2.0 (Phyre2; http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index) (Kelley and Sternberg 2009) revealed that the active site of SEC superimposed well with that of the human OGT (Supplemental Fig. 7A), and two of the three key residues mentioned above are conserved in SEC (H541 and H604) (Supplemental Fig. 7B). Just like the human OGT, the F540A/H541A mutations in SEC (secm1) abolished its OGT activity in the RGA mobility shift assay (Fig. 6D). The H604A mutation in secm2 also decreased the OGT activity, indicated by the reduced mobility shift of RGA (Fig. 6D). secm1+m2 (secm12) did not cause any mobility shift of RGA, similar to secm1. Y2H assays confirmed that secm12 still interacts with RGA, like wild-type SEC (Supplemental Fig. 8A), indicating that these sec mutations block O-GlcNAcylation without affecting RGA binding.
To test the effects of mutant sec proteins in planta, we generated transgenic Arabidopsis lines (in the sec-3 mutant background) that carry 35S:SEC, 35S:secm1, 35S:secm2, or 35S:secm12. Phenotype analysis was performed using homozygous transgenic lines that expressed similar levels of sec proteins (Supplemental Fig. 8B–D). Unlike 35S:SEC that rescued stem growth, 35S:secm1 and 35S:secm12 failed to rescue the dwarf phenotype of sec-3, whereas 35S:secm2 slightly increased the plant height of sec-3 (11%) (Supplemental Fig. 8C,D). These results indicate that the OGT activity of SEC is required for SEC's function in promoting stem elongation in Arabidopsis.
O-GlcNAcylation of RGA inhibits RGA binding to its interactors
In animal systems, O-GlcNAcylation can affect protein localization, stability, protein–protein interaction, or protein activity (Bond and Hanover 2013). The sec-3 allele does not increase RGA mRNA or protein accumulation in comparison with wild type (Supplemental Fig. 9A,B), suggesting that SEC does not inhibit RGA gene expression or destabilize RGA protein. We also monitored the effects of sec-3 and SEC-OE on RGA nuclear localization by introducing PRGA:GFP-RGA (Silverstone et al. 2001) into these mutant backgrounds. Supplemental Figure 9C shows that sec-3 or SEC-OE does not affect RGA localization to the nucleus.
DELLA proteins have been shown recently to antagonize several key regulators in multiple pathways via direct protein–protein interactions (Sun 2011; Xu et al. 2014). To test whether O-GlcNAcylation of RGA by SEC reduces the binding affinity of RGA to its interacting proteins, we performed in vitro pull-down assays. In these assays, O-GlcNAcylated and “unmodified” Flag-RGA proteins were transiently expressed separately in the tobacco system in the presence or absence of SEC, respectively. Four DIPs—BZR1, JAZ1, PIF3, and PIF4—were expressed individually in E. coli as glutathione S-transferase (GST) fusion proteins and purified using glutathione sepharose. These purified GST fusion proteins were incubated with a mixture of tobacco protein extracts containing equal amounts of O-GlcNAcylated and unmodified RGA. Similar pull-down assays were also performed using GST as a negative control. Figure 7A shows that GST-tagged BZR1, JAZ1, PIF3, and PIF4 pulled down more unmodified RGA (top band) than O-GlcNAcylated RGA (bottom band).
Figure 7.
Preferential binding of DELLA interactors to unmodified RGA versus O-GlcNAcylated RGA. (A,B) In vitro pull-down assays using tobacco (A) or Arabidopsis (B) protein extracts. Recombinant GST, GST-BZR1, GST-JAZ1, GST-PIF3, and GST-PIF4 bound to glutathione sepharose beads were used separately to pull down Flag-RGA from a mixture of tobacco protein extracts containing equal amounts of O-GlcNAcylated and unmodified Flag-RGA (A) or protein extracts from PRGA:Flag-RGA transgenic Arabidopsis in a SEC sly1-10 rga-24 or sec-3 sly1-10 rga-24 background (B). The Arabidopsis input samples were pulled down separately because the Flag-RGA mobility difference between SEC and sec backgrounds is not as dramatic as in the tobacco sample. Immunoblots contained input tobacco (A) or Arabidopsis (B) extracts and pulled down samples. In A, 6% SDS-PAGE (12-cm) gels were used to separate the O-GlcNAcylated and unmodified Flag-RGA proteins. In B, 7% SDS-PAGE 10-cm gels were used. Flag-RGA was detected using an anti-Flag antibody. In , the Ponceau-stained blot image below the immunoblot shows equal amounts of GST and GST fusion proteins between the SEC and sec samples. (C,D) sec-3 displayed a reduced BR response in hypocotyl growth. Seedlings were grown in medium containing 2 µM propiconazole (a BR biosynthesis inhibitor) and varying concentrations of brassinolide (BL). Hypocotyl lengths were measured at day 5. n ≥ 25. Bar, 5 mm. Average hypocotyl lengths are means ± SE. (E) Quantitative RT–PCR (qRT–PCR) analysis showing that transcript levels of PHYTOCHROME-INTERACTING FACTOR3-LIKE1 (PIL1) and INDOLE 3-ACETIC ACID-INDUCIBLE29 (IAA29) (PIF target genes) as well as IAA19 and PACLOBUTRAZOL RESISTANCE1 (PRE1) (common target genes of BZR1, PIF, and DELLA) were reduced in sec-3 compared with those in wild type. Average values of three stably expressed genes (GAPC, PP2A, and ROC1) were used to normalize samples (Dill et al. 2004; Czechowski et al. 2005; Hu et al. 2008). Data are means ± SE of three biological replicas and two technical repeats. (*) P < 0.05. (F) Model for the role of DELLA O-GlcNAcylation by SEC. O-GlcNAcylation of DELLA by SEC reduces binding affinity of DELLA to DIPs. (i) Without DELLA binding, DIPs (e.g., BZR1 and PIFs) activate transcription of their target genes. (ii) Alternatively, DIPs (e.g., JAZ1) interact with other transcription factors (TFs; such as MYC2) to down-regulate target gene expression.
To rule out the potential artifact due to hyper-O-GlcNAcylation of RGA by overexpression of SEC in the tobacco system, we generated PRGA:Flag-RGA transgenic Arabidopsis lines with or without the sec-3 mutation. These transgenic lines also contain the F-box sly1 mutation that promotes accumulation of RGA (McGinnis et al. 2003). Protein extracts of these Flag-RGA Arabidopsis lines were used to repeat the in vitro pull-down assays. Figure 7B shows that Flag-RGA (not GlcNAcylated) from sec-3 displayed stronger binding to GST-BZR1 and GST-PIF4 than Flag-RGA (GlcNAcylated) from wild type. These results support the idea that O-GlcNAcylation of RGA by SEC interferes with RGA interactions with DIPs (e.g., BZR1, JAZ1, and PIFs), which releases DIPs to activate or repress transcription of their target genes (Fig. 7F). Based on this model, the sec-3 mutant should display reduced BR- and PIF-dependent responses. Indeed, sec-3 showed a reduced BR response in hypocotyl elongation (Fig. 7C,D). Additionally, sec-3 had a shorter hypocotyl length than wild type (Fig. 4A,B), which is regulated by PIF3, PIF4, and PIF5 (Nozue et al. 2007; Soy et al. 2012). We also examined the effect of sec-3 on transcript levels of several target genes of BZR1, PIFs, and DELLAs. PHYTOCHROME-INTERACTING FACTOR3-LIKE1 (PIL1) and INDOLE 3-ACETIC ACID-INDUCIBLE29 (IAA29) are well-known target genes of PIF3, PIF4, and PIF5; transcription of PIL1 and IAA29 is induced by PIFs (Hornitschek et al. 2009, 2012; Soy et al. 2012). As predicted by our model, PIL1 and IAA29 mRNA levels were ∼1.7-fold lower in the sec-3 mutant than in wild type (Fig. 7E). In addition, transcript levels of IAA19 and PACLOBUTRAZOL RESISTANCE1 (PRE1), two common target genes of BZR1, PIF, and DELLAs (Bai et al. 2012; Gallego-Bartolome et al. 2012; Oh et al. 2014), were decreased ∼1.6-fold in sec-3 compared with those in wild type (Fig. 7E). Consistent with these results, we found that overexpression of SEC (in the SEC-OE line) caused a 1.9-fold to 1.5-fold increase in PIL1, IAA29, and IAA19 mRNA levels than in wild type (Supplemental Fig. 9D). These results support our model depicted in Figure 7F. This model also predicted that sec-3 should display an enhanced JA response because DELLAs interfere with the JA signaling repressor JAZ1. However, sec-3 did not show increased JA response in root growth (Supplemental Fig. 10). This could be because of pleiotropic effects of sec on other cellular pathways. Taken together, these physiological and gene expression studies provide evidence for the role of SEC in fine-tuning activities of multiple signaling pathways.
Discussion
Previous studies of the animal OGTs show that many OGT targets function in transcription regulation, including RNA polymerase II and numerous transcription factors (Slawson and Hart 2011). In contrast, the function and targets of OGT in plants are largely unknown. A recent study suggests that O-GlcNAcylation of an RNA-binding protein (TaGRP2) plays a role in floral induction in response to vernalization in wheat (Xiao et al. 2014). However, this model will need to be verified by direct detection of O-GlcNAcylated TaGRP2 by MS analysis and identification and characterization of the OGT in wheat. Our study reveals that the Arabidopsis OGT, SEC, promotes hypocotyl and internode elongation by reducing the activity of the master growth repressors DELLAs via O-GlcNAcylation. DELLAs have been shown to regulate plant development by antagonizing the functions of key transcription factors/regulators in multiple signaling pathways via direct protein–protein interactions (Xu et al. 2014; Daviere and Achard 2016). Our in vitro pull-down assays showed that O-GlcNAc modification of the Arabidopsis DELLA protein RGA by SEC inhibits interactions between RGA and four DIPs (BZR1, JAZ1, PIF3, and PIF4). Consistent with this finding, the sec mutant displayed subtle defects in GA-, BR-, and PIF-dependent hypocotyl elongation and showed reduced expression of several target genes of BZR1, PIFs, and DELLAs. These data indicate that O-GlcNAcylation of DELLAs by SEC is a common mechanism to inhibit the function of DELLAs in order to integrate activities in multiple cellular pathways (Fig. 7F). Two other post-translational modifications, phosphorylation and SUMOylation, have been reported to regulate DELLA accumulation (Dai and Xue 2010; Conti et al. 2014). It will be important to determine whether these protein modifications affect one another in regulating DELLA function.
Based on our model, the sec mutant should also display an enhanced JA response. Although this was not observed in our root length assay, this finding is not too surprising because SEC is likely to regulate many other cellular pathways, which could result in pleiotropic effects of sec. Hart et al. (2011) suggested that O-GlcNAcylation serves as a “rheostat” that fine-tunes activities in multiple cellular pathways, unlike the on or off effects of phosphorylation on specific target proteins. Therefore, in the sec mutant background, it may be difficult to tease out its specific effect on a particular pathway. This may explain the relatively mild defects of sec in the GA and BR responses. Nevertheless, the overall stem growth defect of the sec mutant is dramatic (40% reduction from wild type), indicating the importance of SEC in modulating plant growth. Our biochemical, physiological, and gene expression studies further provide direct evidence for a clear role of SEC in modulating multiple signaling pathways. This study is the first step in elucidating the function of SEC as an integrator of multiple plant signaling pathways. Future work will elaborate on other protein targets of SEC and additional pathways that it regulates.
It is intriguing that SEC and SPY in Arabidopsis appear to play distinct roles in GA signaling, although the embryo-lethal phenotype of the sec spy double mutant indicates that these two genes have a closely related function during embryogenesis (Hartweck et al. 2002). Similar to SEC, SPY also interacts with DELLA directly (Fig. 2A). However, unlike SEC, we failed to detect any O-GlcNAcylation of DELLA by SPY in the tobacco transient expression system (Fig. 2B,C; Supplemental Fig. 3B). Moreover, the amount of O-GlcNAcylated DELLA was unaltered in the Arabidopsis spy mutant background compared with wild type (Fig. 1B,F). Furthermore, we did not detect changes in overall protein O-GlcNAcylation in the spy mutant (Supplemental Fig. 3E). Although we cannot rule out the possibility that SPY may O-GlcNAcylate DELLAs at a much lower level than SEC, these results suggest that SPY is not required for O-GlcNAcylation of DELLAs. It is possible that binding of DELLAs to SPY may target SPY to specific chromatin locations in the nucleus to modify other proteins. A similar mechanism has been reported in mammalian systems. For example, the corepressor mSin3A, a component of a histone deacetylase complex, targets OGT to promoters to modify and inactivate RNA polymerase II and transcription factors (Yang et al. 2002). Alternatively, SPY could be a novel glycosyl transferase that modifies DELLA proteins differently from SEC. Although SPY was predicted to be an OGT, its putative catalytic domain shares lower amino acid sequence identity (24%) to the human OGT than SEC does (41%). In addition, SPY lacks a conserved His (H541 in SEC and H499 in human OGT) that is important for the OGT activity (Supplemental Fig. 7). Further biochemical studies are needed to elucidate the enzymatic function of SPY and its role in GA signaling and plant development.
Protein O-GlcNAcylation by OGT in animals is a dynamic post-translational modification, and the reverse process is catalyzed by the O-GlcNAcase (OGA) (Hart et al. 2011). In plants, it is unclear whether protein O-GlcNAcylation is a reversible process because no OGA orthologs have been identified. It is possible that the sequences of the plant OGA genes are too divergent from the animal OGAs and therefore cannot be identified by sequence comparison analysis. Alternatively, OGA is absent in plant genomes, and O-GlcNAc residues are removed by hexosaminidases or O-GlcNAcylation is an irreversible process in plants. In animals, protein O-GlcNAcylation by OGT is affected by the level of its donor substrate, UDP-GlcNAc, which is derived from glucose via the hexosamine biosynthetic pathway (Hanover et al. 2010; Hart et al. 2011). The synthesis of UDP-GlcNAc requires additional key metabolites, including glutamine, acetyl CoA, ATP, and UDP. Protein O-GlcNAcylation in animals plays a critical role in coordinating the metabolic and nutrient status of the cell with gene expression and cell cycle control (Slawson and Hart 2011; Bond and Hanover 2013). Reduced nutrients are often correlated with an overall decreased O-GlcNAcylation of proteins (Zhang et al. 2003; Bond and Hanover 2013). Our study on O-GlcNAcylation of DELLAs by SEC reveals how multiple signaling pathways and plant growth are regulated by this type of post-translational modification. DELLAs play a key role in mediating interactions between GA and other hormone signaling pathways in response to external biotic and abiotic signals. Identification of DELLAs as OGT targets provides a gateway to investigate the function of protein O-GlcNAcylation in modulating plant development and responses to environmental stresses. Unlike the drastic effect of protein degradation induced by GA-GID1, O-GlcNAc modification of DELLAs reduces the activities of DELLAs in order to fine-tune multiple signaling activities according to the metabolic status of the cell. Future studies are needed to determine whether the plant OGT (SEC) coordinates the nutrient status of the plant with DELLA activity to modulate plant development.
Materials and methods
Plant materials, plasmid construction, plant transformation, and statistical analyses
All mutant and transgenic lines in this study were derived from Arabidopsis thaliana ecotype Ler, with the exception of sec-2 and er-105 (Col-er) that were derived from the Col-0 ecotype (Hartweck et al. 2002; Torii et al. 1996). The line La-0 is Landsberg wild type without the er mutation. Mutants spy-8, rga-24 gai-t6, and sly1-10 and transgenic lines carrying PRGA:GFP-RGA and 35S:RGA-TAP have been described previously (Silverstone et al. 1997, 2001; Dill and Sun 2001; McGinnis et al. 2003; Zentella et al. 2007). The sec-3 mutant was obtained from Cold Spring Harbor Laboratory (http://www.genetrap.cshl.edu) (Sundaresan et al. 1995). The homozygous sec-3 mutant was identified using the kanamycin-resistant marker in the Ds insertion, confirmed by PCR (Supplemental Table 2), and backcrossed twice to Ler. Procedures for generating DNA constructs, plant transformation, and plant growth conditions are described in Supplemental Material. Primers used in this study are listed in Supplemental Table 2. All statistical analyses were performed with the statistical package JMP Pro 10.0.2 (SAS Institute, Inc.) using Student's t-tests.
Quantitative RT–PCR (qRT–PCR) analyses, immunoblot analyses, co-IP, and Y2H assays
qRT–PCR and immunoblot analyses were performed as described in the Supplemental Material. Transient expression of HA-RGA with or without Myc-tagged SPY, SEC, or GUS-NLS using the 35S promoter in tobacco leaves was performed as described before (Zhang et al. 2011), and subsequent co-IP assays are described in the Supplemental Material. The Y2H assays were performed as described previously (Dill et al. 2004) with slight modifications (see the Supplemental Material).
Protein purifications for click reaction, GalT assays, and MS
RGA-TAP was tandem affinity-purified from the 35S:RGA-TAP rga-24 ga1-3 transgenic lines in the wild-type SEC and SPY backgrounds (labeled as “WT”) (Zentella et al. 2007) or the spy-8 and sec-3 backgrounds using His-Bind resin (EMD Millipore) followed by anti-cMyc-agarose beads (Sigma-Aldrich). His-Flag-RGA protein transiently expressed in tobacco was tandem affinity-purified using a His-Bind resin followed by anti-Flag-M2-agarose beads (Sigma-Aldrich). Detailed protein purification procedures and GalT assays are provided in the Supplemental Material.
GalNAz labeling and click reaction for detecting O-GlcNAcylated RGA
Eight-day-old 35S:RGA-TAP transgenic Arabidopsis seedlings in liquid culture (1/2 MS, 1% sucrose) were treated with 100 µM Ac4GalNAz (synthesized as described in Boyce et al. 2011) or mock-treated for 48 h. RGA-TAP was tandem affinity-purified, labeled with biotin-alkyne using Click-iT protein analysis detection kit (Invitrogen) following the manufacturer's protocol, and detected by protein blot analyses using HRP-streptavidin (Jackson ImmunoResearch Laboratories) and anti-RGA antibodies separately.
Identification of O-GlcNAc sites by liquid chromatography (LC)-ETD tandem MS (MS/MS) analyses
RGA-TAP and His-Flag-RGA proteins produced from Arabidopsis and tobacco, respectively, were affinity-purified and digested with Trypsin or Asp-N. The peptides were separated by high-pressure LC (HPLC) and analyzed online by MS/MS analysis using a modified ETD-enabled high-resolution LTQ-FT or LTQ-Orbitrap mass spectrometer (Thermo Scientific) (Udeshi et al. 2008; Berk et al. 2013). A detailed procedure of sample processing and MS analysis is described in the Supplemental Material.
N-acetylhexosaminidase assay and pull-down assays
RGA-CT1 from transgenic Arabidopsis bound to anti-cMyc agarose beads was incubated with active or heat-inactivated β-D-N-acetylhexosaminidase (V-Lab). Removal of β-linked GlcNAc was monitored by immunoblot analysis. In vitro pull-down assays were performed using recombinant GST fusion proteins and tobacco or Arabidopsis extracts containing equal amounts of unmodified and GlcNAcylated Flag-RGA. Detailed protocols are described in the Supplemental Material.
In silico predictions of O-GlcNAc sites and 3D protein structures
The YinOYang 1.2 program (Gupta and Brunak 2002) was used to identify putative O-GlcNAcylation sites in DELLAs. Renditions of the 3D structures of SEC and SPY were generated using the online tool Phyre2 (Kelley and Sternberg 2009). The template for SEC and SPY was the crystal structure of the human OGT (Protein Data Bank ID 3PE3).
Supplementary Material
Acknowledgments
We thank Philip Benfey, Zhen-Ming Pei, and Jim Siedow for helpful comments on the manuscript. We also thank Jeongmoo Park for providing pGST-PIF3, Emily Jie-Ning Yang for her assistance in Y2H assays, and Jonathan Dayan and the Duke University Light Microscopy Core Facility for their help with confocal microscopy. This work was supported by the National Science Foundation (IOS-0641548 and MCB-0923723 to T.-P.S., and MCB-0516690, MCB-0820666, and MCB-1158089 to N.E.O.), U.S. Department of Agriculture (2014-67013-21548 to T.-P.S.), and National Institutes of Health (R01 GM100051 to T.-P.S., and R01 GM037537 to D.F.H.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.270587.115.
References
- An F, Zhang X, Zhu Z, Ji Y, He W, Jiang Z, Li M, Guo H. 2012. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res 22: 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun T-P, Wang Z-Y. 2012. Brassinosteroid, gibberellin, and phytochrome signalling pathways impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk JM, Maitra S, Dawdy AW, Shabanowitz J, Hunt DF, Wilson KL. 2013. O-GlcNAc regulates emerin binding to BAF in a chromatin- and lamin B-enriched ‘niche’. J Biol Chem 288: 30192–30209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond MR, Hanover JA. 2013. O-GlcNAc cycling: a link between metabolism and chronic disease. Annu Rev Nutr 33: 205–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Carrico IS, Ganguli AS, Yu SH, Hangauer MJ, Hubbard SC, Kohler JJ, Bertozzi CR. 2011. Metabolic cross-talk allows labeling of O-linked β-N-acetylglucosamine-modified proteins via the N-acetylgalactosamine salvage pathway. Proc Natl Acad Sci 108: 3141–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. 2001. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal Biochem 293: 169–177. [DOI] [PubMed] [Google Scholar]
- Conti L, Nelis S, Zhang C, Woodcock A, Swarup R, Galbiati M, Tonelli C, Napier R, Hedden P, Bennett M, et al. 2014. Small ubiquitin-like modifier protein SUMO enables plants to control growth independently of the phytohormone gibberellin. Dev Cell 28: 102–110. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Xue HW. 2010. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J 29: 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daviere JM, Achard P. 2016. A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant 9: 10–20. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez-Falcon M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S. 2008. A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484. [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T-P. 2001. Synergistic de-repression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A, Thomas SG, Hu J, Steber CM, Sun T-P. 2004. The Arabidopsis F-box protein SLEEPY1 targets GA signaling repressors for GA-induced degradation. Plant Cell 16: 1392–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridborg I, Kuusk S, Moritz T, Sundberg E. 2001. The Arabidopsis protein SHI represses gibberellin responses in Arabidopsis and barley. Plant Physiol 127: 937–948. [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolome J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadi D, Blazquez MA. 2012. Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci 109: 13446–13451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J. 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325: 93–96. [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun T-P, et al. 2006. Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput 7: 310–322. [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC. 2010. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta 1800: 80–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. 2011. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem 80: 825–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. 2002. Two O-Linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics 161: 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Genger RK, Grey WM, Olszewski NE. 2006. SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J Exp Bot 57: 865–875. [DOI] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM. 2012. Gibberellin signaling: a theme and variations on DELLA repression. Plant Physiol 160: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. 2009. Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711. [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. 2010. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell 19: 884–894. [DOI] [PubMed] [Google Scholar]
- Hu J, Mitchum MG, Barnaby N, Ayele BT, Ogawa M, Nam E, Lai W-C, Hanada A, Alonso JM, Ecker JR, et al. 2008. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell 20: 320–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isono T. 2011. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One 6: e18959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. 1993. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci 93: 9292–9296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Chory J. 2010. Unraveling the paradoxes of plant hormone signaling integration. Nat Struct Mol Biol 17: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371. [DOI] [PubMed] [Google Scholar]
- King K, Moritz T, Harberd N. 2001. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics 159: 767–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. 2011. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469: 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis KM, Thomas SG, Soule JD, Strader LC, Zale JM, Sun T-P, Steber CM. 2003. The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15: 1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun T-P, Hakoshima T. 2008. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. 2007. Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361. [DOI] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. 2014. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. ELife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM. 2010. O-GlcNAc protein modification in plants: evolution and function. Biochim Biophys Acta 1800: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. 1997. The Arabidopsis GAI gene defines a signalling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. 2000. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci 97: 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Casamitjana Martínez E, Sun T-P. 1997. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-P. 1998. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun T-P. 2001. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng T-S, Swain S, Dill A, Jeong SY, Olszewski NE, Sun T-P. 2007. Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Hart GW. 2011. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer 11: 678–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soy J, Leivar P, Gonzalez-Schain N, Sentandreu M, Prat S, Quail PH, Monte E. 2012. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis. Plant J 71: 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. 2011. The molecular mechanism and evolution of the GA–GID1–DELLA signaling module in plants. Curr Biol 21: R338–R345. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. 1995. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev 9: 1797–1810. [DOI] [PubMed] [Google Scholar]
- Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. 1996. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8: 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi ND, Compton PD, Shabanowitz J, Hunt DF, Rose KL. 2008. Methods for analyzing peptides and proteins on a chromatographic timescale by electron-transfer dissociation mass spectrometry. Nat Protoc 3: 1709–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. 2005. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698. [DOI] [PubMed] [Google Scholar]
- Vocadlo DJ, Hang HC, Kim EJ, Hanover JA, Bertozzi CR. 2003. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc Natl Acad Sci 100: 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, et al. 2014. O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nature Commun 5: 4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X. 2014. Shedding light on integrative GA signaling. Curr Opin Plant Biol 21: 89–95. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang F, Kudlow JE. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110: 69–80. [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci 109: E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. 2007. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Su K, Yang X, Bowe DB, Paterson AJ, Kudlow JE. 2003. O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115: 715–725. [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Ogawa M, Fleet CM, Zentella R, Hu J, Heo J-O, Lim J, Kamiya Y, Yamaguchi S, Sun T-P. 2011. SCARECROW-LIKE 3 promotes gibberellin signaling by antagonizing DELLA in Arabidopsis. Proc Natl Acad Sci 108: 2160–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.