Abstract
Tetrahymena thermophila is a ciliate with hundreds of cilia primarily used for cellular motility. These cells propel themselves by generating hydrodynamic forces through coordinated ciliary beating. The coordination of cilia is ensured by the polarized organization of basal bodies (BBs), which exhibit remarkable structural and molecular conservation with BBs in other eukaryotes. During each cell cycle, massive BB assembly occurs and guarantees that future Tetrahymena cells gain a full complement of BBs and their associated cilia. BB duplication occurs next to existing BBs, and the predictable patterning of new BBs is facilitated by asymmetric BB accessory structures that are integrated with a membrane-associated cytoskeletal network. The large number of BBs combined with robust molecular genetics merits Tetrahymena as a unique model system to elucidate the fundamental events of BB assembly and organization.
Keywords: Tetrahymena, Ciliate, Basal body, Centriole, Microtubule
Introduction: the organism
Tetrahymena thermophila is a free-swimming ciliate that utilizes hundreds of motile cilia for hydrodynamic force-generation. Tetrahymena belong to the superphylum Alveolata which also contains the parasitic Apicomplexans and the aquatic Dinoflagellates and together compose one of the largest groups of the kingdom Protozoa [1]. Tetrahymena are relatively large ovoid (20 μm wide and 35 μm long) single cells that contain 18–21 longitudinal rows of regularly spaced cilia (~30 per row; Fig. 1). Each cilium is nucleated and stabilized by a conventional basal body (BB). In addition, a single ciliated feeding structure, called an oral apparatus, contains 150 BBs segregated into four membranelles (tetra–“four’’ hymena–“membrane’’) and defines the organism’s anterior–posterior polarity. These cells divide every 3 h in a process that requires massive BB duplication to ensure that each daughter cell inherits an equal complement of cilia. Tetrahymena genetics allow for the generation of genomic knock-outs, knock-ins, and inducible promoter systems. Additionally, a sequenced and annotated genome was recently published [2]. With sophisticated molecular genetics, defined axes of organismal polarity and a tightly controlled linear arrangement of duplicating BBs, Tetrahymena is an outstanding cellular model for investigating the basic mechanisms of polarized BB assembly, stability, and organization.
Fig. 1.
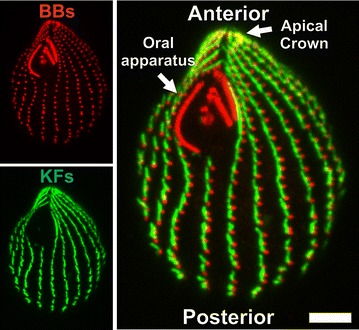
Polarized organization of Tetrahymena BBs. BBs are labeled in red (α-centrin, [27]) and kinetodesmal fibers are labeled in green (α-KF, [44]). The merged image highlights the organized ciliary array, the oral apparatus, and the apical crown which demarcates anterior–posterior polarity. Scale bar 5 μm
Basic Tetrahymena basal body structure
Tetrahymena BBs are structurally similar to BBs in other eukaryotes. Mature Tetrahymena BBs are 500–600 nm in length and 180–220 nm in diameter [3]. The length of the BB comprises the typical triplet microtubule blades that are arranged into a cylinder with ninefold radial symmetry (Fig. 2a). The proximal end of the BB possesses three structures that establish and maintain the cylindrical organization. First, the A- and C-tubules of adjacent triplet microtubules are connected by an A–C linkage (Fig. 2a). Second, the proximal 60–90 nm of the BB contains a cartwheel structure composed of a central hub and nine spokes that connect to the A-tubule of each triplet microtubule blade (Fig. 2b). Importantly, the cartwheel is retained through the BB lifecycle, perhaps to ensure BB stability as these BBs must resist mechanical forces from beating cilia. Third, an electron-dense “collar” asymmetrically wraps around one side of the triplet microtubules (Fig. 2a). Above the cartwheel, the BB lumen encloses an electron-dense structure whose function remains poorly understood (Fig. 2b; [3]). The distal end of the BB is capped by the terminal plate (the Tetrahymena transition zone), which consists of two electron-dense opaque sheets that cross the lumen of the BB (Fig. 2b; [3]). While the core structure of the BB is largely conserved across phylogeny, ciliates, including Tetrahymena, utilize a unique assemblage of accessory structures that position and anchor BBs at the cell cortex.
Fig. 2.
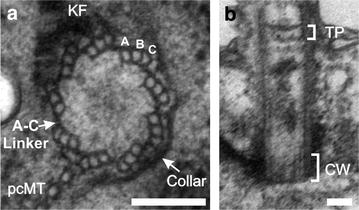
Tetrahymena BB structure. a Cross-sectional view of a proximal section of a Tetrahymena BB. Collar electron-dense collar; pcMT post-ciliary microtubules; KF kinetodesmal fiber; b longitudinal view of a BB; TP terminal plate; CW Cartwheel. Scale bars 100 nm
Additional BB structures or accessory structures
Tetrahymena BBs are endowed with accessory structures that coordinate BB positioning with cellular polarity and stabilize them against cilia-generated forces (Fig. 3). The location and composition of these structures depend on the BB population in the Tetrahymena cell. At the cell’s anterior pole, a ring of two closely positioned BBs, called dikinetids, begin each ciliary row and are associated with filaments of unknown composition called the apical filament ring [4]; together these structures are called the apical crown (Fig. 1). Within the oral apparatus, a dense microtubule meshwork organizes approximately 150 BBs into its four membranelles (Fig. 1; [5]). The majority of Tetrahymena BBs, however, are the cortical basal bodies that are required for cellular locomotion. Cortical BBs possess three major accessory structures: the post-ciliary microtubules, the transverse microtubules, and the kinetodesmal fiber (Fig. 3; [3]). Post-ciliary microtubules nucleate from the BB posterior face and radially project toward the posterior BB situated in the same ciliary row. Transverse microtubules originate from the BB anterior face and project upward and leftward (from the cell’s perspective) toward the cell cortex, where they overlap with the post-ciliary microtubules of anterior BB in the adjacent ciliary row. The kinetodesmal fiber is a striated structure that extends from the BB’s anterior face to the plasma membrane adjacent to the distal end of the anteriorly positioned BB within the same ciliary row. The kinetodesmal fiber also associates with the anterior BB’s post-ciliary microtubules [3]. By providing points of contact with the subcortical cytoskeletal network and neighboring BBs, accessory structures help establish and maintain the cellular organization and stability of BBs [3]. Moreover, these structures guide the placement of newly assembled BBs, suggesting that cortical BB accessory structures play an important role in cortical BB duplication [3, 6–8].
Fig. 3.
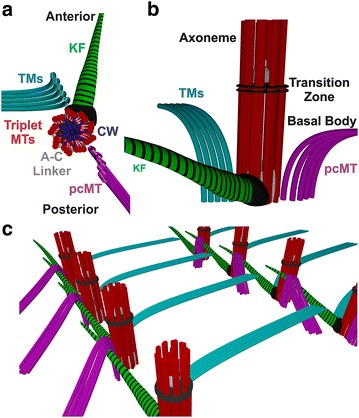
Schematic representation of Tetrahymena BBs and associated accessory structures. a 3D schematic of an individual cortical BB viewed from the interior of the cell. b An individual cortical BB viewed slightly offset from the anterior direction. c Image shows a portion of two ciliary rows highlighting the positioning of the three major accessory structures relative to neighboring BBs. pcMTs post-ciliary microtubules; KF kinetodesmal fiber; TMs transverse microtubules; CW cartwheel
Basal body origins
Tetrahymena cortical BBs arise next to existing BBs in what is called centriolar BB assembly. During assembly, a daughter BB forms orthogonally to a defined triplet microtubule at the anterior face of the proximal end of an existing mother BB [3]. New assembly commences with the formation of the cartwheel and a ring of short microtubules (called a pro-BB) that is separated from the mother BB by an amorphous electron-dense cloud [3]. As the pro-BB separates from the mother BB, the triplet microtubules elongate and tilt toward the apical surface to dock the BB distal end with Tetrahymena’s subcortical cytoskeletal network [3]. The pro-BB is positioned by the asymmetric localization of accessory structures on the mother BB, including the kinetodesmal fiber, which ensures that the new BB is appropriately spaced and positioned within the ciliary row [3]. Although cortical BBs assemble via the centriolar pathway, the origin of oral apparatus BBs is unclear and may arise from de novo assembly. Importantly, oral apparatus BB orientation, which is random early in development, coincides with BB linkage to an underlying microtubule network, representing a likely parallel to the process of BB orientation in vertebrate multi-ciliated cells [5, 9–14].
Basal body life cycle and other functions
Tetrahymena undergo a closed mitosis where BBs do not function as centrioles in organizing a centrosome but rather remain docked at the cell cortex to organize cilia for the entire cell cycle. During mitosis, the two nuclei of Tetrahymena utilize distinct mechanisms to organize the microtubules of the mitotic micronucleus and amitotic macronucleus [15–19]. The micronuclear spindle microtubules are organized by a laminar structure analogous to the yeast spindle pole body while the macronuclear microtubules are nucleated from the nuclear envelope by a mysterious mechanism [20]. Importantly, because Tetrahymena BBs are solely used for locomotion and not mitosis, BB defects can be studied without perturbations that result in checkpoint arrest phenotypes. Existing mother BBs serve as sites of new BB assembly that occurs continuously throughout the cell cycle and increase in frequency before cell division [21–24]. The production of new BBs and their remarkably consistent integration into the polarized cell must be coupled with the dynamic and spatially controlled incorporation of proteins required for BB assembly.
Basal body components
Tetrahymena BBs are molecularly conserved with the BBs and centrioles of other eukaryotes. Forward and reverse genetic approaches have been used in Tetrahymena to discover and elucidate the molecular mechanisms of important BB components [25–28]. Furthermore, purified BBs from Tetrahymena were used in combination with proteomics and immuno-electron microscopy to identify and localize many BB components to their ultrastructural BB domains [29]. These studies highlight Tetrahymena as a powerful model system to study the molecules and mechanisms of basal body assembly and function.
The triplet microtubules are composed of canonical α and β tubulin, while γ tubulin and ε tubulin are required for BB assembly and maintenance [30–32]. In addition, the Tetrahymena genome possesses δ tubulin along with the ciliate specific η and κ tubulins, although the functions of these isoforms remain unclear [2]. Also present are the conserved UNIMOD proteins (SAS-6, CEP135/Bld10, and SAS-4/CPAP) in addition to other conserved proteins like POC1 and members of the centrin family [27–29, 33]. Overall, the molecular conservation of BB components combined with adaptable genetics has led to a number of novel BB findings.
Notable basal body findings
Tetrahymena has played a foundational role in our understanding of BB assembly, stability, and organization. Early studies capitalized on the polarized morphology of Tetrahymena BBs to study the propagation and maintenance of pre-existing BB order in the cell, which extended the pioneering studies of Paramecium ‘structural inheritance’ by Beisson and Sonneborn into other organisms [34, 35]. By mechanically inverting ciliary rows, Joseph Frankel and colleagues demonstrated that the Tetrahymena cortical architecture contains the epigenetic cues for placing new BBs within the polarized cell [35]. More recently, molecular–genetic and cytological studies identified a novel role for γ tubulin in regulating BB assembly [32]. Microtubule post-translational modifications are important for MT control and Tetrahymena was fundamental in the discovery and characterization of the MEC-17/α-TAT1 tubulin acetyl-transferase and the Tubulin Tyrosine Ligase-Like (TTLL) modifying enzymes that glutamylate and glycylate tubulin [36–40]. Tetrahymena has also played a large role in discovering a novel class of BB stability components and understanding their functions [27, 31, 41, 42]. Study of BB stability in Tetrahymena is advantageous because the cilia-generated forces experienced at the BB can be modulated experimentally [41]. Tetrahymena’s polarized cytology and ease of genetic manipulation have dramatically furthered our understanding of BB and tubulin biology.
Conclusions: strengths and future of basal body research in Tetrahymena
Coupled with new high-resolution microscopy technologies, an expanding arsenal of molecular genetic tools make Tetrahymena an immensely powerful system for the next wave of BB research. The combined use of established forward genetics with Next-Generation sequencing enables the discovery of new molecules and mutants for further dissection of BB assembly and organization. BB protein localization and turnover dynamics are accessible to study in Tetrahymena using live cell imaging of fluorescently tagged proteins [29, 43]. Moreover, high-resolution light microscopy and cryo-electron tomography with the numerous and easily purified BBs of Tetrahymena will link the molecular and structural studies amenable to this system. The future is bright for BB research using this evolutionarily divergent model organism to understand the most highly conserved and divergent features of BB biology.
Authors’ contributions
BAB, DFG, and CGP wrote and edited the manuscript. BAB and DFG generated the figures. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Michael McMurray, Jeff Moore, and Adam Soh for comments and edits. CGP is supported by NIH-NIGMS GM0099820, the Boettcher Webb-Waring Foundation and the Pew Biomedical Scholars Program.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- BB
basal body
Contributor Information
Brian A. Bayless, Email: brian.bayless@ucdenver.edu
Domenico F. Galati, Email: domenico.galati@ucdenver.edu
Chad G. Pearson, Email: chad.pearson@ucdenver.edu
References
- 1.Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57(4):953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, model eukaryote. PLoS Biol. 2006;4(9):e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen RD. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J. Cell Biol. 1969;40(3):716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jerka-Dziadosz M. Cytoskeleton-related structures in tetrahymena thermophila: microfilaments at the apical and division-furrow rings. J. Cell Sci. 1981;51:241–253. doi: 10.1242/jcs.51.1.241. [DOI] [PubMed] [Google Scholar]
- 5.Williams NE, Frankel J. Regulation of microtubules in Tetrahymena. I. Electron microscopy of oral replacement. J. Cell Biol. 1973;56(2):441–457. doi: 10.1083/jcb.56.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearson CG. Choosing sides–asymmetric centriole and basal body assembly. J. Cell Sci. 2014;127(Pt 13):2803–2810. doi: 10.1242/jcs.151761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerka-Dziadosz M, Koll F, Wloga D, Gogendeau D, Garreau de Loubresse N, Ruiz F, et al. A Centrin3-dependent, transient, appendage of the mother basal body guides the positioning of the daughter basal body in Paramecium. Protist. 2013;164(3):352–368. doi: 10.1016/j.protis.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Dippell RV. How ciliary Basal bodies develop. Sci. 1967;158(3800):527. doi: 10.1126/science.158.3800.527. [DOI] [PubMed] [Google Scholar]
- 9.Sorokin SP. Centriole formation and ciliogenesis. Aspen Emphysema Conf. 1968;11:213–216. [PubMed] [Google Scholar]
- 10.Dirksen ER. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J. Cell Biol. 1971;51(1):286–302. doi: 10.1083/jcb.51.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J. Cell Sci. 1968;3(2):207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 12.Kalnins VI, Porter KR. Centriole replication during ciliogenesis in the chick tracheal epithelium. Z Zellforsch Mikrosk Anat. 1969;100(1):1–30. doi: 10.1007/BF00343818. [DOI] [PubMed] [Google Scholar]
- 13.Steinman RM. An electron microscopic study of ciliogenesis in developing epidermis and trachea in the embryo of Xenopus laevis. Am J. l Anat. 1968;122(1):19–55. doi: 10.1002/aja.1001220103. [DOI] [PubMed] [Google Scholar]
- 14.Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J. Cell Biol. 2011;195(1):19–26. doi: 10.1083/jcb.201106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endo M, Sugai T. Amitotic division of the macronucleus in Tetrahymena thermophila: DNA distribution by genomic unit. Zool Sci. 2011;28(7):482–490. doi: 10.2108/zsj.28.482. [DOI] [PubMed] [Google Scholar]
- 16.Kushida Y, Takaine M, Nakano K, Sugai T, Numata O. Dynamic Change of Cellular Localization of Microtubule-Organizing Center During Conjugation of Ciliate Tetrahymena thermophila. Zool Sci. 2015;32(1):25–32. doi: 10.2108/zs140149. [DOI] [PubMed] [Google Scholar]
- 17.Davidson L, LaFountain JR., Jr Mitosis and early meiosis in Tetrahymena pyriformis and the evolution of mitosis in the phylum Ciliophora. Bio Syst. 1975;7(3–4):326–336. doi: 10.1016/0303-2647(75)90010-6. [DOI] [PubMed] [Google Scholar]
- 18.LaFountain JR, Jr, Davidson LA. An analysis of spindle ultrastructure during prometaphase and metaphase of micronuclear division in Tetrahymena. Chromosom. 1979;75(3):293–308. doi: 10.1007/BF00293473. [DOI] [PubMed] [Google Scholar]
- 19.LaFountain JR, Jr, Davidson LA. An analysis of spindle ultrastructure during anaphase of micronuclear division in Tetrahymena. Cell Motil. 1980;1(1):41–61. doi: 10.1002/cm.970010105. [DOI] [PubMed] [Google Scholar]
- 20.Wolfe J, Hunter B, Adair WS. A cytological study of micronuclear elongation during conjugation in Tetrahymena. Chromosom. 1976;55(4):289–308. doi: 10.1007/BF00292827. [DOI] [PubMed] [Google Scholar]
- 21.Perlman BS. Basal body addition in ciliary rows of Tetrahymena pyriformis. J. Exp Zool. 1973;184(3):365–368. doi: 10.1002/jez.1401840310. [DOI] [PubMed] [Google Scholar]
- 22.Kaczanowski A. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J. Exp Zool. 1978;204(3):417–430. doi: 10.1002/jez.1402040313. [DOI] [PubMed] [Google Scholar]
- 23.Nanney DL. Patterns of basal body addition in ciliary rows in Tetrahymena. J. Cell Biol. 1975;65(3):503–512. doi: 10.1083/jcb.65.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Galati DF, Abuin DS, Tauber GA, Pham AT, Pearson CG. Automated image analysis reveals the dynamic 3-dimensional organization of multi-ciliary arrays. Biol Open. In Press. [DOI] [PMC free article] [PubMed]
- 25.Jerka-Dziadosz M, Jenkins LM, Nelsen EM, Williams NE, Jaeckel-Williams R, Frankel J. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev Biol. 1995;169(2):644–661. doi: 10.1006/dbio.1995.1176. [DOI] [PubMed] [Google Scholar]
- 26.Galati DF, Bonney S, Kronenberg Z, Clarissa C, Yandell M, Elde NC, et al. DisAp-dependent striated fiber elongation is required to organize ciliary arrays. J. Cell Biol. 2014;207(6):705–715. doi: 10.1083/jcb.201409123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, et al. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16(8):3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Culver BP, Meehl JB, Giddings TH, Jr, Winey M. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol biol cell. 2009;20(6):1865–1877. doi: 10.1091/mbc.E08-08-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, et al. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 2007;178(6):905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 2002;158(7):1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross I, Clarissa C, Giddings TH, Jr, Winey M. epsilon-tubulin is essential in Tetrahymena thermophila for the assembly and stability of basal bodies. J. Cell Sci. 2013;126(Pt 15):3441–3451. doi: 10.1242/jcs.128694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang Y, Tsao CC, Gorovsky MA. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J. Cell Biol. 2005;171(6):1035–1044. doi: 10.1083/jcb.200508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vonderfecht T, Cookson MW, Giddings TH, Jr, Clarissa C, Winey M. The two human centrin homologues have similar but distinct functions at Tetrahymena basal bodies. Mol Biol Cell. 2012;23(24):4766–4777. doi: 10.1091/mbc.E12-06-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beisson J, Sonneborn TM. Cytoplasmic Inheritance of the Organization of the Cell Cortex in Paramecium Aurelia. Proc Natl Acad Sci U.S.A. 1965;53:275–282. doi: 10.1073/pnas.53.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng SF, Frankel J. 180 degrees rotation of ciliary rows and its morphogenetic implications in Tetrahymena pyriformis. Proceedings of the Natl Acad Sci U.S.A. 1977;74(3):1115–1119. doi: 10.1073/pnas.74.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia L, Hai B, Gao Y, Burnette D, Thazhath R, Duan J, et al. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 2000;149(5):1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, et al. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Sci. 2005;308(5729):1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 38.Wloga D, Rogowski K, Sharma N, Van Dijk J, Janke C, Edde B, et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008;7(8):1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wloga D, Webster DM, Rogowski K, Bre MH, Levilliers N, Jerka-Dziadosz M, et al. TTLL3 Is a tubulin glycine ligase that regulates the assembly of cilia. Dev Cell. 2009;16(6):867–876. doi: 10.1016/j.devcel.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 40.Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, et al. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biology : CB. 2006;16(21):2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Bayless BA, Giddings TH, Jr, Winey M, Pearson CG. Bld10/Cep135 stabilizes basal bodies to resist cilia-generated forces. Mol Biol Cell. 2012;23(24):4820–4832. doi: 10.1091/mbc.E12-08-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson CG, Osborn DP, Giddings TH, Jr, Beales PL, Winey M. Basal body stability and ciliogenesis requires the conserved component Poc1. J. Cell Biol. 2009;187(6):905–920. doi: 10.1083/jcb.200908019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson CG, Giddings TH, Jr, Winey M. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol Biol Cell. 2009;20(3):904–914. doi: 10.1091/mbc.E08-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams NE, Honts JE, Dress VM, Nelsen EM, Frankel J. Monoclonal antibodies reveal complex structure in the membrane skeleton of Tetrahymena. J. Eukaryot Microbiol. 1995;42(4):422–427. doi: 10.1111/j.1550-7408.1995.tb01606.x. [DOI] [PubMed] [Google Scholar]


