ABSTRACT
Tetherin (BST2, CD317, or HM1.24) is a host cellular restriction factor that prevents the release of enveloped viruses by mechanically linking virions to the plasma membrane. The precise arrangement of tetherin molecules at the plasma membrane site of HIV-1 assembly, budding, and restriction is not well understood. To gain insight into the biophysical mechanism underlying tetherin-mediated restriction of HIV-1, we utilized cryo-electron tomography (cryo-ET) to directly visualize HIV-1 virus-like particles (VLPs) and virions tethered to human cells in three dimensions (3D). Rod-like densities that we refer to as tethers were seen connecting HIV-1 virions to each other and to the plasma membrane. Native immunogold labeling showed tetherin molecules located on HIV-1 VLPs and virions in positions similar to those of the densities observed by cryo-ET. The location of the tethers with respect to the ordered immature Gag lattice or mature conical core was random. However, tethers were not uniformly distributed on the viral membrane but rather formed clusters at sites of contact with the cell or other virions. Chains of tethered HIV-1 virions often were arranged in a linear fashion, primarily as single chains and, to a lesser degree, as branched chains. Distance measurements support the extended tetherin model, in which the coiled-coil ectodomains are oriented perpendicular with respect to the viral and plasma membranes.
IMPORTANCE Tetherin is a cellular factor that restricts HIV-1 release by directly cross-linking the virus to the host cell plasma membrane. We used cryo-electron tomography to visualize HIV-1 tethered to human cells in 3D. We determined that tetherin-restricted HIV-1 virions were physically connected to each other or to the plasma membrane by filamentous tethers that resembled rods ∼15 nm in length, which is consistent with the extended tetherin model. In addition, we found the position of the tethers to be arbitrary relative to the ordered immature Gag lattice or the mature conical cores. However, when present as multiple copies, the tethers clustered at the interface between virions. Tethered HIV-1 virions were arranged in a linear fashion, with the majority as single chains. This study advances our understanding of tetherin-mediated HIV-1 restriction by defining the spatial arrangement and orientation of tetherin molecules at sites of HIV-1 restriction.
INTRODUCTION
Tetherin (BST2, CD317, or HM1.24) is an interferon-inducible host cellular restriction factor that blocks virus release by mechanically linking virions to the host cell's plasma membrane (1, 2). Tetherin acts on a wide range of enveloped viruses, including members of the Retroviridae, Filoviridae, Arenaviridae, Herpesviridae, Paramyxoviridae, and Togaviridae families (3–14). Viruses have in turn evolved a series of distinct mechanisms to counteract tetherin (1–4, 8, 13, 15–17). HIV-1 is able to circumvent tetherin-mediated entrapment through the disruption of tetherin trafficking by the viral protein Vpu. Vpu antagonism of tetherin has been implicated in viral expansion and dissemination in vivo (18–21).
The ability of tetherin to engage diverse families of enveloped viruses and the evolution of distinct mechanisms to counteract tetherin suggests that tetherin-mediated restriction does not require specific interactions between tetherin and viral proteins (22). Rather, it is the incorporation of tetherin into the viral and cellular membranes that is important. This is best exemplified by the ability of an artificial tetherin dimer (composed of a transferrin receptor transmembrane domain, a coiled-coil dimer of dystrophia myotonic protein kinase, and the C-terminal glycosylphosphatidylinositol [GPI] modification signal from the urokinase plasminogen activator) to restrict HIV particle release (23). Although it is clear that tetherin acts as a linker between viral and cellular membranes, the arrangement of tetherin on chains of virions is not entirely understood at the structural level.
Tetherin is a type II integral membrane protein with a molecular mass between 27 and 36 kDa. It contains a short N-terminal cytoplasmic tail (CT), a single-pass transmembrane domain (TM), a coiled-coil ectodomain, and a GPI anchor (24, 25) (as illustrated in Fig. S1 in the supplemental material). The restriction of virus release requires the presence of at least one of three conserved cysteine amino acids (C53, C63, and C91) and membrane insertion of the TM and the GPI anchor (23, 26). The structure of the mammalian tetherin ectodomain dimer has been solved by X-ray crystallography under oxidized and reduced buffer conditions. Each monomer consists of a continuous alpha helix arranged in parallel orientation, forming a coiled-coil dimer 14.5 to 16.5 nm in length (27–30). Based on X-ray structures of the mammalian ectodomains, researchers have proposed two main models to predict how tetherin cross-links HIV-1 to the plasma membrane (22, 31), with evidence to support both (see Fig. S1 in the supplemental material). In the extended or axial model, the TM and GPI are in trans-configuration, with the coiled-coil ectodomain oriented perpendicular with respect to viral and plasma membranes (29, 32). For the parallel or equatorial model, the TM and GPI are in cis configuration with the coiled-coil ectodomain situated in parallel register between the viral membrane and the plasma membrane. Based on high-resolution structures of the human tetherin ectodomain, one would predict the distance separating the viral membrane from the host cell's plasma membrane to be 16.5 nm for the extended model and 2.5 nm for the parallel model (30). Studies have demonstrated biochemically that the extended tetherin homodimer physically restricts HIV-1 release (23, 33, 34). However, structural information on tetherin-restricted HIV-1 confirming the extended tetherin model is lacking.
Ultrastructural analysis of HIV-1 tethered to human cells has been limited chiefly to examinations of chemically fixed and heavy-metal-stained samples by either thin-section transmission electron microscopy (TEM) (1, 9, 23, 32, 35–38) or scanning electron microscopy (SEM) (2, 23, 39). In previous studies, clusters of HIV-1 virions retained on the surface of infected or transfected cells expressing tetherin were consistently observed. In a few selected studies, researchers occasionally observed electron-dense material resembling stalks between virions (1, 9, 38). However, the molecular composition of these densities and their dimensions and orientation were not clearly defined. Immunoelectron microscopy studies have shown tetherin staining on viral membranes and on the host cell plasma membrane near sites of HIV-1 budding (23, 32, 37, 38). Electron-dense fibrous material linking HIV-1 to the host cell membrane (37) and to virus-containing compartments in macrophages was observed (36). However, the length of these tethers (between 50 and 100 nm), which was inferred from TEM two-dimensional (2D) projection images and not 3D volumes, exceeded the length predicted by either tetherin model by an order of 3- to 6-fold.
Despite progress made over the past few years, several fundamental questions regarding the ultrastructure of tethered HIV-1 remain unanswered, mainly: (i) how are tetherin molecules and domains oriented, (ii) what is the distance separating tethered virions from the cell and between tethered virions, and (iii) how are tethered HIV-1 virions arranged in 3D at the plasma membrane. In this study, we directly visualized HIV-1 virions tethered to human cells by cryo-electron tomography (cryo-ET). HIV-1 virions were linked by tethers in a linear, unbranched fashion and less often as branched chains. Rod-like densities or tethers connecting virions to each other and to the plasma membrane were seen; measurements of these tethers support the extended tetherin model. These findings provide strong support for tetherin's role as a proteinaceous link between virions and the plasma membrane and provide additional insight into the formation of tetherin-linked chains.
MATERIALS AND METHODS
Cell culture and plasmids.
HT1080 and HeLa cell lines were obtained from the American Type Culture Collection (ATCC) and maintained in high-glucose pyruvate-Dulbecco modified Eagle medium (DMEM) (GIBCO, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS), 5 mM l-glutamine, and 5 mM penicillin-streptomycin. Subconfluent cultures (40 to 70%) were grown directly on gold Quantifoil R2/1 or R2/2 TEM grids (Quantifoil Micro Tools GmbH, Jena, Germany) and on glass-bottom MatTek dishes (MatTek Corporation, Ashland, MA). HT1080 cells were grown on MatTek dishes and on TEM grids coated with collagen (0.2 mg/ml in 0.2% acetic acid). HeLa cells were grown on MatTek dishes coated with poly-d-lysine or on TEM grids that were incubated overnight in DMEM supplemented with 10% FBS, 5 mM l-glutamine, and 5 mM penicillin-streptomycin. TEM grids were inspected under a light microscope to ensure the integrity of the holey carbon film, which subsequently was reinforced by evaporating a 4- to 5-nm layer of carbon on top of it using a Denton Benchtop Turbo apparatus (Denton Vacuum, Moorestown, NJ). Prior to collagen coating or overnight incubation in media, the TEM grids were sterilized by plasma cleaning followed by immersion in 70% ethanol.
HIV-1 VLPs were generated by cotransfecting cells with a 3:1 ratio of pVRC-3900 (a plasmid encoding codon-optimized HIV-1 PR55Gag) and GagOpt-mCherry. GagOpt-mCherry was constructed by swapping the mCherry gene into Gag-yellow fluorescent protein (YFP) using BamHI and NotI sites (40). The plasmid pEGFP-tetherin (encoding an N-terminal enhanced green fluorescent protein [EGFP]-linked tetherin) was made by cloning tetherin in-frame from pFLAG-Tetherin into SalI and BamHI sites of pEGFP-C1 (Clontech Laboratories Inc.). The plasmids pNLenv1ΔU (encoding an HIV-1 env- and vpu-deficient provirus) and pEGFP-C3A-tetherin were kindly provided by Klaus Strebel (NIAID, Bethesda, MD) (41, 42). Cells were allowed to grow on TEM grids for 24 h prior to transfection using jetPRIME (Polyplus-Transfection Inc., New York, NY) according to the manufacturer's instructions. HT1080 cells were transfected overnight with an 8:1 ratio of pNLenv1ΔU to pEGFP-tetherin or pEGFP-C3A-tetherin. HeLa cells were transfected overnight with 500 ng of pNLenv1ΔU.
Viral release assay.
HT1080 cells were transfected with pEGFP-tetherin or pEGFP-C3A-tetherin titration (10, 25, 75, and 125 ng) and 1 μg pNLenv1ΔU. Transfected HT1080 cell supernatants were harvested, clarified by low-speed centrifugation, and subsequently concentrated by ultracentrifugation through a 20% sucrose cushion (100,000 × g for 2 h at 4°C). Viral pellets were lysed in 1× radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors (RIPA-PI). HT1080 cells were washed with phosphate-buffered saline (PBS) prior to detachment using prewarmed EDTA (0.2 g/liter EDTA-Na4 in PBS; Invitrogen). Cells then were pelleted by low-speed centrifugation, washed with PBS, and lysed with 1× RIPA-PI for 30 min at 4°C. Lysates were clarified by centrifugation at 15,000 × g for 30 min at 4°C. Analysis of cell lysates and concentrated supernatants was performed by Western blotting using anti-p24 hybridoma 183-H12-5C (obtained from Bruce Chesboro and Hardy Chen through the National Institutes of Health [NIH] AIDS Research and Reference Reagent Program) supernatants (1:1,000) and rabbit anti-tetherin antisera (1:2,000) (37).
Immunofluorescence microscopy.
HT1080 cells were seeded on collagen-coated MatTek dishes at a density of 1.2 × 105 cells per dish. HT1080 cells were transfected with an 8:1 ratio of pNLenv1ΔU to either pEGFP-tetherin or pEGFP-C3A-tetherin. HeLa cells were transfected overnight with 500 ng pNLenv1ΔU. Cells were washed with PBS and subsequently fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) for 10 min at room temperature. Following fixation, cells were extensively washed with PBS and permeabilized for 10 min with 0.2% Triton X-100. Cells were blocked for 30 min with Dako protein block (Dako, Carpinteria, CA) followed by primary and secondary antibody staining at the appropriate concentrations in Dako antibody diluent. Cells were stained with primary antibody for 1.5 h, followed by secondary antibody for 1 h at room temperature. Finally, 4′,6-diamidino-2-phenylindole (DAPI) was used to stain chromatin of labeled cell samples. Immunofluorescence microscopy (IFM) was performed using the DeltaVision imaging station (Applied Precision/GE Healthcare, Marlborough, MA).
Correlative light electron microscopy (CLEM).
Transfected HT1080 cells grown on gold London Finder Quantifoil R2/2 TEM grids maintained in uncoated MatTek dishes were imaged in real time with a DeltaVision imaging station equipped with a 20× objective lens (Olympus). Images were analyzed with Volocity 6.3 (PerkinElmer, Waltham, MA) software. After acquiring transmitted and fluorescence images (15 to 30 min), the TEM grids were cryo-immobilized. The letters and numbers of the London Finder grid were used to track areas of the grid imaged by light microscopy and then by cryo-EM/ET. Fluorescence light micrographs (mCherry-Gag) were superimposed on selected regions of the cryo-EM montages (100× nominal magnification) after scaling and rotating images in Adobe Photoshop CS6.
Immuno-TEM.
Live cells grown on gold TEM grids were immunolabeled with rabbit anti-tetherin antiserum (37) and 6-nm colloidal gold conjugated to goat anti-human IgG or protein G (Aurion) as described by previously (43).
Cryo-ET.
Whole cells were cryo-immobilized by rapid immersion in liquid ethane using a Cryoplunge 3 system (Gatan, Pleasanton, CA). TEM grids were gently dipped in warm PBS, and a 4-μl aliquot of bovine serum albumin (BSA) conjugated to 10- or 20-nm gold nanoparticles (Sigma-Aldrich, St. Louis, MO) was applied to the surface of the TEM grid for 30 to 45 s prior to cryoplunging. Cryogrids were stored short term in liquid nitrogen and transferred into a 914 high-tilt liquid nitrogen cryotransfer tomography holder (Gatan) maintained at −172 to −192°C. Cryogrids were imaged with a JEOL JEM-2200FS 200-kV field emission TEM (JEOL, Ltd., Japan) equipped with an in-column Omega energy filter with a slit width of 20 eV. Montages (100×, 4,000×, and 10,000× nominal magnification) and tilt series were collected using SerialEM (44). Single-axis tilt series were acquired at 2° tilt increments over an angular range of −62° to 62°. Tomograms were collected with a defocus range of 4- to 8-μm underfocus and under low-dose conditions so that the total electron dose was limited to 110 to 150 e−/Å2. Tilt series images of HT1080 cells were recorded with a Gatan US 4000 4096 by 4096 charge-coupled-device (CCD) camera (Gatan, Inc., Pleasanton, CA) at a nominal magnification of 30,000×, for a pixel size of 0.737 or 0.764 nm. Tilt series images of HeLa cells were recorded with a Direct Electron DE-20 5120 by 3840 camera (Direct Electron, LP, San Diego, CA) at 12 frames per s and at a nominal magnification of 10,000×, for a pixel size of 0.614 nm. Images collected with the DED-20 camera were motion corrected using open-source python scripts (DE_combine_references.py and DE_process_frames.py) provided by the manufacturer.
Tomographic reconstructions (binned by 2) were generated with IMOD (45) using the r-weighted back-projection algorithm (46) and denoised by nonlinear anisotropic diffusion (47). Tomographic data collected with the DE-20 camera was contrast transfer function (CTF) corrected by phase inversion (48) using IMOD. Tomograms were segmented manually with the 3D software platform Amira (FEI Visualization Science Group) (49). Distance measurements, including the length of segmented tethers and the space between tethered membranes (viral or cellular), were made in 3D with the Amira software package.
Flow cytometry.
A total of 2.0 × 105 HT1080 and HeLa cells per well were propagated overnight in six-well cell culture dishes. On the following day, HT1080 cells were transfected with 125 ng pEGFP-tetherin and HeLa cells with 100 ng pEGFP-C1, and they were incubated overnight prior to analysis. Cell monolayers were washed with prewarmed PBS and detached using EDTA. Cells then were pelleted and washed repeatedly with ice-cold PBS. Cells were resuspended in PBS containing 2% bovine serum albumin (BSA) and allowed to incubate on ice for 10 min prior to the addition of primary antibody (rabbit α-tetherin) for 1 h at 4°C. Cells then were pelleted by low-speed centrifugation and washed twice with PBS containing 2% BSA, followed by the addition of allophycocyanin (APC)-conjugated anti-rabbit F(ab′)2 (Jackson IR, West Grove, PA) for 30 min at 4°C. Cells were assayed for tetherin cell surface expression by flow cytometry using a FACSCanto II (BD Biosciences). Subsequent data analyses were performed using FlowJo 10.1 (Tree Star).
RESULTS
CLEM and cryo-ET of HIV-1 VLPs tethered to human cells.
For this study, we set out to characterize the ultrastructure of HIV-1 virions tethered to human cells by cryo-ET. HT1080 cells were used as a model system to study the structural basis of tetherin-mediated restriction, as these cells contain long extensions, such as lamella, lamellipodia, and microspikes, that are sufficiently thin for cryo-ET (50). Furthermore, this cell line can produce HIV-1 and does not constitutively express tetherin. Several groups have utilized this cell line to study the molecular basis of tetherin-mediated restriction of HIV (1, 23, 51, 52). Viral release assays confirmed transfection with pEGFP-tetherin inhibited the release of HIV-1 in a concentration-dependent manner (see Fig. S2A in the supplemental material). Transfection with pEGFP-C3A-tetherin, a previously characterized tetherin mutant that does not form the disulfide bonds between monomers that contribute to dimer stability (26, 53), failed to prevent HIV-1 release (see Fig. S2A). The examination of transfected HT1080 cells by IFM revealed puncta of EGFP-tetherin, EGFP-C3A-tetherin, and the HIV-1 Gag capsid protein p24 on the plasma membrane, including thin cellular protrusions (see Fig. S2B). HeLa cells transfected with pNLenv1ΔU exhibited the enrichment of HIV-1 p24 and tetherin in plasma membrane puncta consistent with the presence of tethered virion clustering (see Fig. S2B). Relative cellular tetherin expression levels of pEGFP-tetherin-transfected HT1080 cells were 2.4 times greater than endogenous tetherin expression in HeLa cells (see Fig. S2C). HT1080 cells transfected with pEGFP-tetherin exhibited a 2.6-fold increase in tetherin cell surface levels compared to endogenous tetherin expression in HeLa cells (see Fig. S2C).
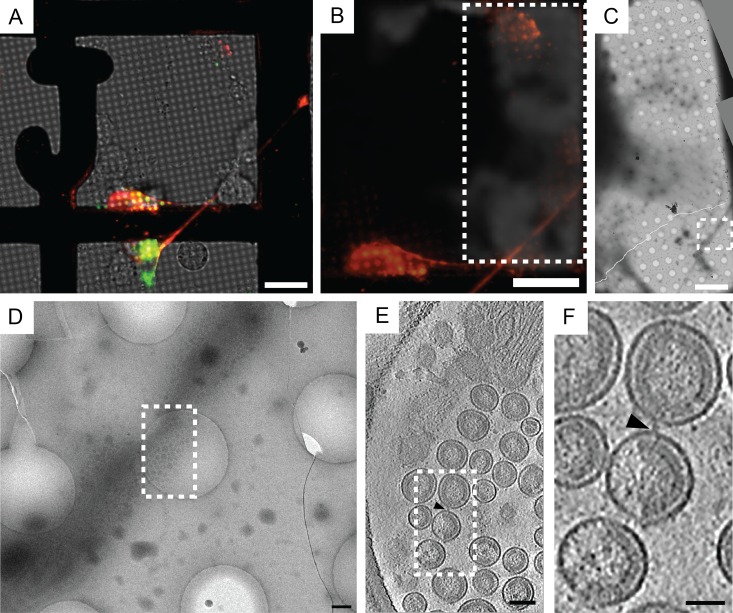
Fluorescently labeled HIV-1 VLPs tethered to human cells were imaged by CLEM. Briefly, cells were grown on collagen-coated gold London Finder TEM grids, imaged in real time, and then cryo-immobilized by rapid immersion in liquid ethane. Cotransfected cells bore puncta of mCherry-Gag and EGFP-tetherin along thin cellular extensions (Fig. 1A and B). Areas of the TEM grid imaged by live-cell microscopy then were imaged by cryo-EM, making use of the finder grid letters to collect montages for identifying large clusters of HIV-1 VLPs attached to the cells (Fig. 1C and D). Clusters of VLPs were imaged by cryo-ET, thus providing nanometer-scale resolution in 3D (Fig. 1E and F; also see Movie S1 in the supplemental material). Spherical VLPs ranging from 81 to 173 nm in diameter contained ordered immature Gag lattice, with the gross morphology being similar to that of previous reports (54). As reported in previous cryo-ET studies (55, 56), we observed sheets of Gag polyprotein below the plasma membrane in F-actin-rich cell extensions, indicating that assembly, budding, and retention to the plasma membrane occurred along the thin cellular extensions (Fig. 2C and D; also see Movie S1). Upon close inspection of tomographic slices, rod-like densities, which are referred to as tethers, were seen connecting HIV-1 VLPs to each other and to the cell plasma membrane. Tethers connecting HIV-1 VLPs to each other and to the plasma membrane were observed consistently in cryospecimens prepared and imaged on separate dates (Fig. 2A to H).
FIG 1.
Correlative fluorescence light microscopy and cryo-ET of HIV-1 VLPs tethered to a human cell. (A) Transfected HT1080 cells grown on a gold London Finder TEM grid were imaged in real time by fluorescence microscopy, after which the TEM grid was cryo-immobilized and imaged by cryo-EM. The image is composed of separate layers to reflect EGFP-tetherin (green), mCherry-Gag (red), and transmitted light. (B) Image of the mCherry-Gag (red) channel was superimposed over a low-magnification cryo-EM image of the TEM grid. The dashed white box indicates the area of the TEM grid imaged by cryo-EM, which is equivalent to the area shown in panel C. (C) Cryo-EM montage of the cells and HIV-1 VLPs attached to a thin cellular extension. The dashed white box corresponds to panel D. (D) Magnified view of the dashed white box from panel C (enlarged by a factor of 10). The dashed white box over the HIV-1 VLPs attached to the thin cellular extension in panel D indicates the position where the tilt series was collected. (E) Tomographic slice (7.64 nm) through HIV-1 VLPs tethered to the surface of the cellular extension. The dashed white box corresponds to the enlarged image shown in panel F, which was magnified by a factor of 3.5. The black triangle points to a tether attached to 2 VLPs. Scale bars are 25 μm (A and B), 10 μm (C), 500 nm (D), 100 nm (E), and 50 nm (F).
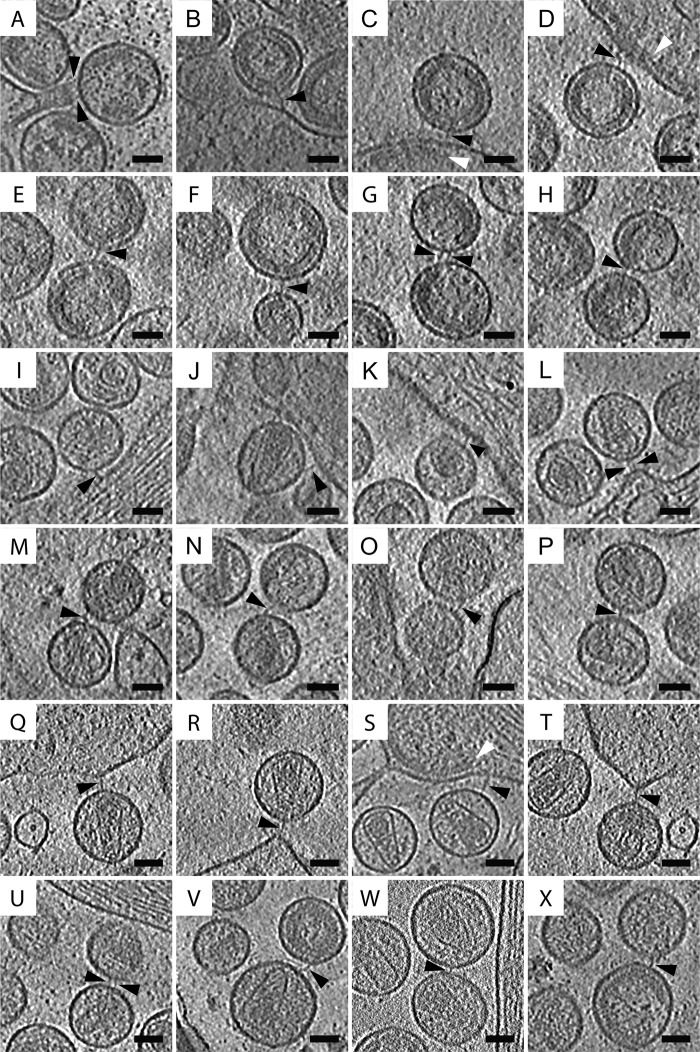
FIG 2.
Filamentous tethers attached to HIV-1 VLPs and HIV-1 virions resolved by cryo-ET. (A to P) HT1080 cells transfected with pEGFP-tetherin and cDNA encoding HIV-1 Gag or pNLenv1ΔU were imaged by cryo-ET. Tomographic slices (7.37 nm or 7.64 nm) of tethers (as indicated by the black triangles) connecting HIV-1 VLPs to the plasma membrane (A to D), HIV-1 VLPs to HIV-1 VLPs (E to H), HIV-1 virions to the plasma membrane (I to L), and HIV-1 virions to HIV-1 virions (M to P). (Q to X) Endogenous tethers were observed in HeLa cells transfected with pNLenv1ΔU. Shown are tomographic slices (7.37 nm) of endogenous tethers connecting HIV-1 virions to the plasma membrane (Q to T) and HIV-1 virions to HIV-1 virions (U to X). Sheets of Gag below the plasma membrane are indicated by white triangles. Scale bars, 50 nm.
Cryo-ET of HIV-1 tethered to human cells.
HT1080 cells cotransfected with pEGFP-tetherin and pNLenv1ΔU (deficient in env and vpu) were examined by cryo-ET (Fig. 3A to D; also see Movie S2 in the supplemental material). As with VLPs, HIV-1 virions attached to thin cellular extensions were observed. Regions of Gag polyprotein situated below the plasma membrane, which have been reported in previous cryo-ET studies (55, 56), were visualized at the point of tethering (Fig. 3C, white triangle). This suggests that assembly, budding, and tethering of HIV-1 to the plasma membrane occur at the same location. Spherical virions with diameters ranging from 64 to 150 nm were observed, with gross morphology similar to that seen in previous reports (57, 58). Although most particles visualized were mature, immature virions and some with the irregular cores of maturation intermediates also were apparent (56, 59–63).
FIG 3.
Tetherin homodimers are required for formation of filamentous tethers. (A to D) HT1080 cells cotransfected with pNLenv1ΔU and pEGFP-tetherin possess HIV-1 virions connected to the cell by filamentous tethers. (A and B) Cryo-EM montage of HIV-1 virions attached to a long cellular extension emanating from the cell (as indicated by the term “Cell” and by the white dashed line). The dashed black boxes in panels A and B indicate the positions at which the tilt series were collected. (C) Tomographic slice (7.64 nm) of HIV-1 virions tethered to a thin cellular extension. HIV-1 budding from the cell was observed, as indicated by a white triangle pointing to the Gag polyprotein situated underneath the plasma membrane. (D) Enlargement of the tomographic slice, corresponding to the black dashed box in panel C. Tethers are seen connecting HIV-1 virions to each other and to the plasma membrane (as indicated by the black triangle). (E to G) Released HIV-1 virions produced from HT1080 cells cotransfected with pEGFP-C3A-tetherin and pNLenv1ΔU were imaged by cryo-ET. (E and F) Cryo-EM montage of an HT1080 cell, as indicated by the term “Cell” and by the dashed white line drawn around the periphery of the cell, and released HIV-1. Dashed black boxes are positioned over the areas where the tilt series were collected. (G) Tomographic slice (7. 64 nm) of released HIV-1 virions. Filamentous rods were rarely seen between these HIV-1 virions. Immature and mature HIV-1 virions as well as maturation intermediates were observed. Scale bars are 2 μm (A and E), 1 μm (B), 500 nm (F), 100 nm (C and G), and 50 nm (D).
Tethers connecting HIV-1 virions to each other and to the plasma membrane were seen in cryosamples (Fig. 2I to P and 3; also see Movie S2 in the supplemental material). These tethers were similar to those identified on HIV-1 VLPs. Cells transfected with pEGFP-C3A-tetherin and pNLenv1ΔU did not have large clusters of HIV-1 (Fig. 3E to G). Released virions also were imaged by cryo-ET, revealing densities on the viral membranes but generally lacking visible tethers between viral particles (Fig. 3E to G; also see Movie S3). In contrast, the majority of virions produced by cells transfected with HIV-1 pEGFP-tetherin and pNLenv1ΔU were connected, by a tether, to either a viral or cellular membrane.
To visualize endogenous tethers we used HeLa cells, which constitutively express tetherin (1). HeLa cells were transfected with pNLenv1ΔU and imaged by cryo-ET. HIV-1 virions connected to one another and to the plasma membrane by filamentous tethers were observed (Fig. 2Q to X and 4). The endogenous tethers observed in tomograms of HeLa cells resembled the tethers seen in tomograms of HT1080 cells transfected with pEGFP-tetherin. The length of the endogenous tethers was identical to that of recombinant tethers (Table 1). Extended layers of the Gag polyprotein were situated underneath the plasma membrane, and budding HIV-1 particles were observed near sites of HIV-1 restriction (Fig. 2S and 4B). As best illustrated in segmented tomograms, HIV-1 virions connected to the cell and to other virions by endogenous tethers were arranged in a linear fashion, which resemble beads on a string (see Fig. 6C).
FIG 4.
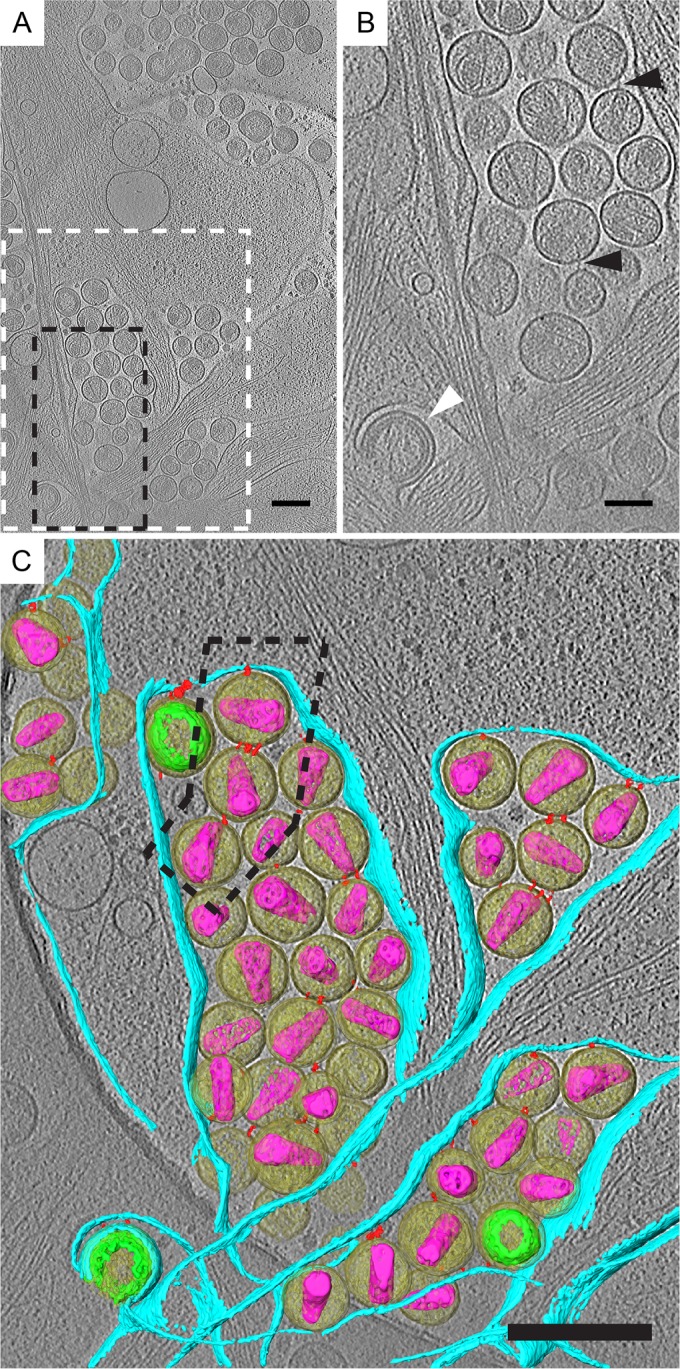
Direct visualization of endogenous tethers on HIV-1 attached to HeLa cells by cryo-ET. HeLa cells transfected with pNLenv1ΔU were imaged by cryo-ET. (A) Tomographic slice (7.37 nm) of HIV-1 virions attached to plasma membrane. Immature and mature virions, maturation intermediates, and HIV budding from the plasma membrane were observed. (B) Enlarged tomographic slice. The dashed black box in panel A corresponds to panel B. Filamentous tethers attached to HIV-1 virions are indicated by black triangles. HIV-1 budding from the plasma membrane is indicated by the white triangle. (C) Segmented tomogram corresponding to the dashed white box in panel A. Elements shown include HIV-1 virions (yellow), immature Gag polyprotein (green), mature cores (purple), tethers (red), and the plasma membrane (cyan). Tethered HIV-1 elements are arranged as single chains, as indicated by the dashed black box around 3 HIV-1 virions connected to the cell. Scale bars are 200 (A and C) and 100 nm (B).
TABLE 1.
Distance between tethered membranes
| Target of measurementa | Distanceb (nm) |
|||
|---|---|---|---|---|
| Mean | SD | Range | n | |
| HT1080 cells | ||||
| Segmented tethers | 15.3 | 5.93 | 4.56–36.6 | 219 |
| Segmented membranes | 17.9 | 6.71 | 6.66–36.6 | 219 |
| HIV-cell | 19.9 | 6.88 | 6.66–35.0 | 62 |
| HIV-HIV | 17.1 | 6.50 | 6.82–36.6 | 157 |
| Mature-cell | 19.7 | 7.09 | 6.66–35.0 | 49 |
| Immature-cell | 18.2 | 4.19 | 10.3–24.2 | 13 |
| Mature-mature | 17.8 | 7.24 | 6.82–36.6 | 90 |
| Mature-immature | 16.0 | 5.83 | 7.79–30.5 | 52 |
| Immature-immature | 17.9 | 6.31 | 7.78–29.0 | 15 |
| HeLa cells | ||||
| Segmented tethers | 15.6 | 4.78 | 5.98–23.4 | 29 |
| Segmented membranes | 18.6 | 6.77 | 9.64–30.5 | 29 |
For the length of segmented tethers, only the segmented tethers were measured.
Distance between tethered virions. Measurements were made along the segmented tethers between the viral or cellular membranes.
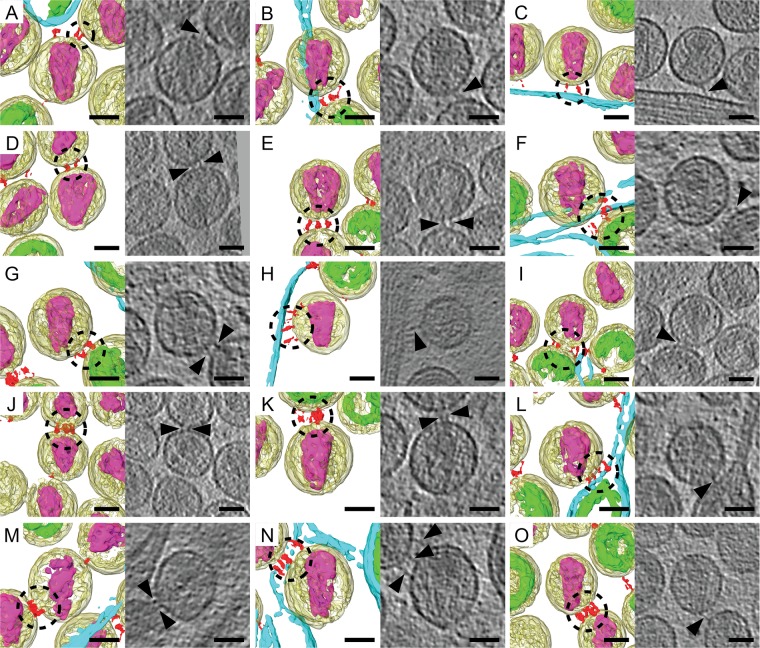
FIG 6.
Spatial arrangement of multiple tethers on mature HIV-1 virions. Shown are example images of manually segmented tomographic reconstructions and corresponding tomographic slices (1.53 nm) of mature HIV-1 virions that possess multiple tethers. The tomograms were rotated in 3D so that the mature conical cores all were oriented in the same direction with the broad end of the core at 12:00. The following elements were segmented: HIV-1 virions (yellow), immature Gag polyprotein (green), mature cores (purple), tethers (red), and the plasma membrane (cyan). When present as multiple copies on the same mature virion, the tethers form clusters of 2 tethers (A to D), 3 tethers (E to H), 4 tethers (I to N), and 5 tethers (O), as indicated by the dashed black circles. The black triangles point to tethers seen in the tomographic slices. Scale bars are 50 nm.
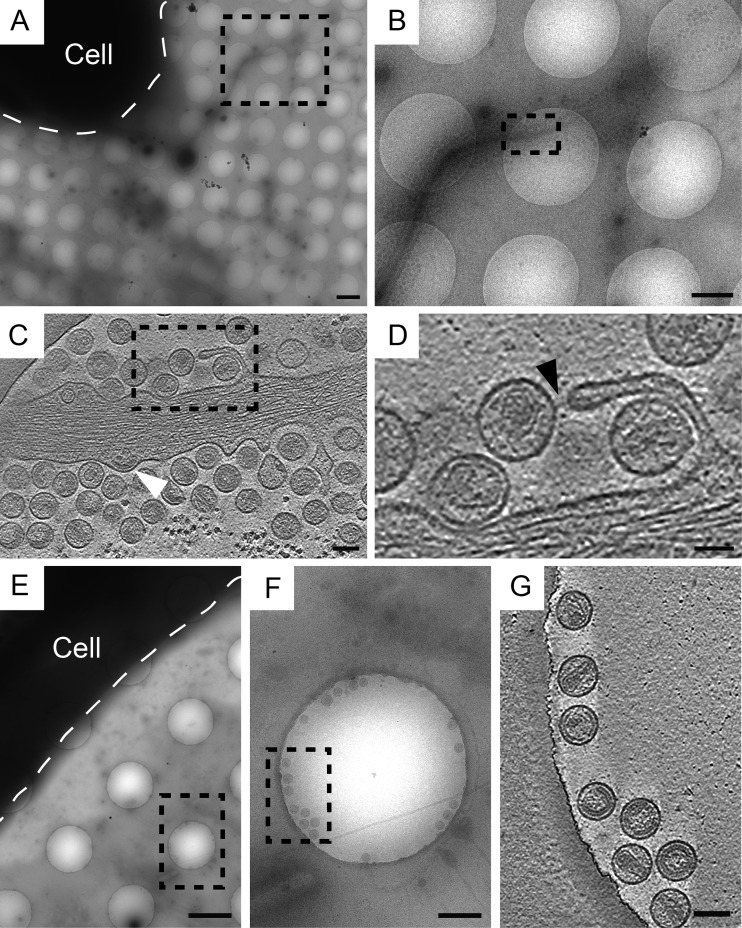
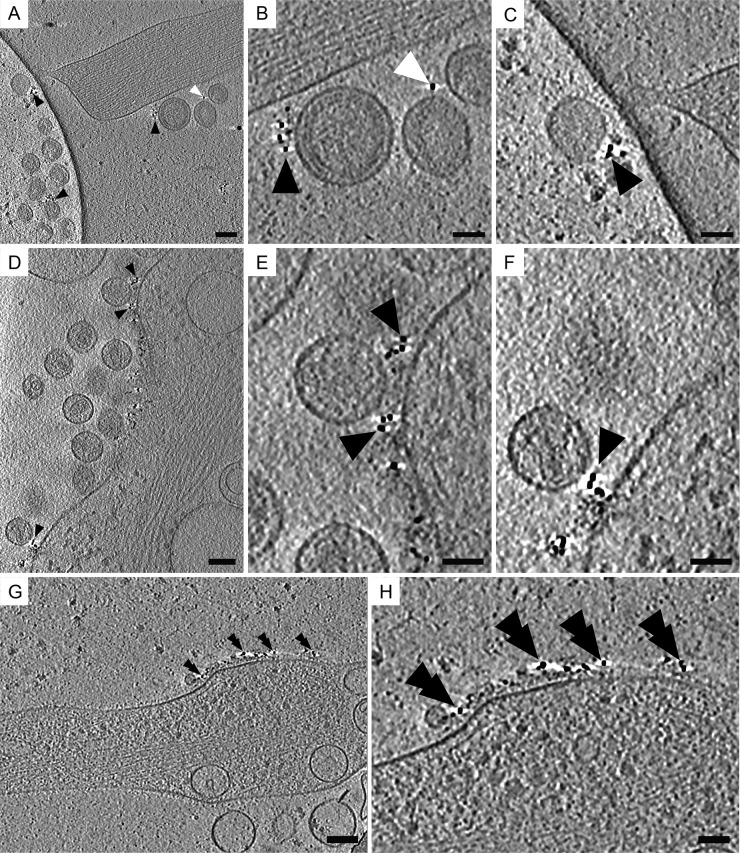
We utilized native immunogold labeling to confirm the localization of tetherin molecules on transfected cells and on virions (43). This method involves labeling live cells with antibodies prior to cryo-immobilization or chemical fixation, which preserved the integrity of the epitope of interest and the arrangement of macromolecules on the surface of cells and viruses (64). For these experiments we utilized a previously characterized antitetherin antiserum generated against tetherin's extracellular coiled-coil ectodomain (corresponding to amino acids 43 to 179) (37). Cryo-ET of native immunogold-labeled specimens showed tetherin located on HIV-1 VLPs, HIV-1 virions, and the plasma membrane (Fig. 5). Tetherin staining was not uniformly distributed over the cell but was very punctate, as seen by fluorescence microscopy (see Fig. S2B in the supplemental material). Patches of immunogold were visualized on thin cellular extensions devoid of HIV-1 virions (Fig. 5G and H). Bundles of F-actin were oriented parallel to the plasma membrane near patches of immunolabeled tetherin (Fig. 5). Immunogold labeling also was seen on vesicles tethered to HIV-1 VLPs and virions (data not shown). These results confirm the presence of tetherin on HIV-1 VLPs, virions, and transfected cells. Previous immuno-TEM studies of chemically fixed, embedded, or cryosectioned material have reported similar distributions of tetherin on transfected or HIV-1-infected cells (23, 32, 37, 38).
FIG 5.
Cryo-ET of native immunolabeled tetherin associated with HIV-1 VLPs and HIV-1 virions. Protein G conjugated to 6-nm colloidal gold distributed along the plasma membrane, on tethered HIV-1 VLPs, and on HIV-1 virions. (A) Tomographic slices (7.64 nm) of immunolabeled HT1080 cells transfected with plasmids encoding mCherry Gag (1:3 ratio of mCherry Gag and codon-optimized Gag) and pEGFP-tetherin. (B) Enlarged image of the dashed white box in panel A, which was magnified by a factor of 3. Shown is immunogold staining of tethered HIV-1 VLPs attached to the cell, as indicated by black triangles, and between other HIV-1 VLPs, as indicated by white triangles. (C) Enlarged image of the dashed black box in panel A, which was magnified by a factor of 3. (D) Tomographic slice (7.37 nm) of HIV-1 virions attached to the plasma membrane of an HT1080 cell (transfected with pNLenv1ΔU and pEGFP-tetherin). Black triangles point to immunogold staining. (E) Enlarged image of the dashed white box in panel D (magnified by a factor of 3). (F) Enlarged image of the dashed black box in panel D, which was magnified by a factor of 3. (G) Tomographic slice (7.64 nm) of a thin cellular extension devoid of HIV-1 virions but containing tetherin, as indicated by black double triangles. (H) Enlargement of area in panel G, which was magnified by a factor of 2. Scale bars are 100 nm (A, D, and G) and 50 nm (B, C, E, F, and H).
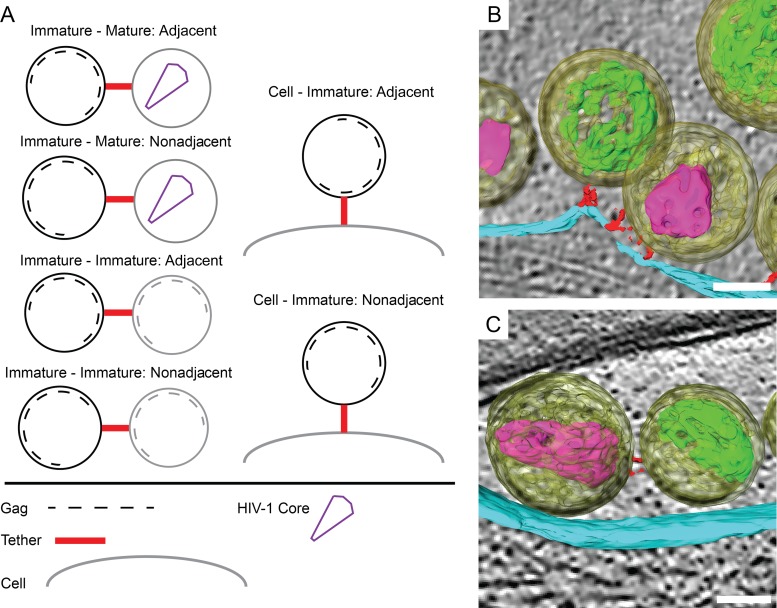
Spatial distribution of tethers on mature and immature HIV-1 virions.
To gain insight into the biophysical basis of tetherin-mediated restriction of HIV, we characterized the spatial distribution of tethers on HIV-1. Tomograms of HIV-1 tethered to thin cellular extensions were manually segmented with the Amira software package (49). In the analysis, mature virions (106 virions) possessed 1 to 8 tethers, with an average of 2 tethers per virion. The position of the tether relative to the conical core's 2-fold axis of symmetry varied from virion to virion and from tether to tether (Fig. 6). When present as multiple copies, tethers were not uniformly distributed over the viral membrane but rather formed clusters at sites of contact between other HIV-1 virions or the cell plasma membrane (Fig. 6).
The distribution of tethers attached to 43 immature HIV-1 virions was characterized in segmented tomograms. Tethers were seen making contact with the viral membrane adjacent to the Gag polyprotein or in areas that were devoid of Gag (Fig. 7). The number of tethers adjacent to Gag (54%) was close to that of the tethers not adjacent to Gag (46%) (Table 2). The results of this analysis indicate EGFP tethers are present on the immature viral membrane without respect to the Gag polyprotein or to areas of the viral membrane lacking an underlying Gag polyprotein.
FIG 7.
Spatial relationship between tethers and the immature HIV-1 Gag lattice. (A) Tethers connected to immature HIV-1 virions were classified according to the arrangement of tethers relative to the Gag lattice. Tethers attached to an area of the viral membrane directly adjacent to the Gag lattice or nonadjacent to the Gag lattice were observed. (B) Example of an adjacent tether connecting an immature virion (yellow) to the plasma membrane (cyan). The tethers (red) are proximal to the Gag polyprotein (green). (C) Example of a nonadjacent tether adjoining an immature virion to a mature virion (mature core [purple]). The tether is making contact with the viral membrane in an area lacking an ordered immature Gag polyprotein. Scale bars are 50 nm.
TABLE 2.
Arrangement of tethers relative to Gag
| Class | Virions | Tethers | Adjacent | Nonadjacent |
|---|---|---|---|---|
| Cell-immature | 11 | 19 | 8 | 11 |
| Immature-immature | 11 | 28 | 16 | 12 |
| Immature-mature | 26 | 59 | 32 | 26 |
| Total | 48 | 106 | 57 | 49 |
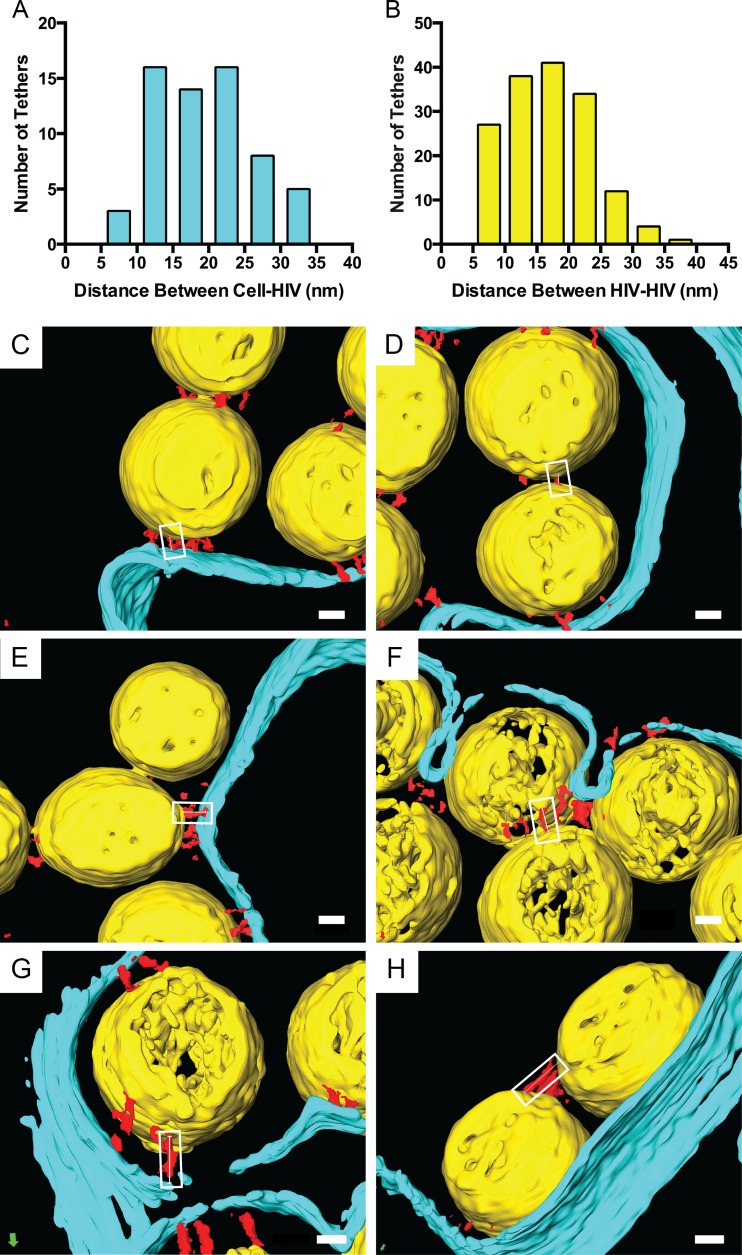
Distance measurements of tethers cross-linking HIV-1 virions to each other and to the cell.
To determine whether tetherin restricts HIV-1 release by adopting the extended conformation, the length of segmented tethers (recombinant tethers [15.3 ± 5.93 nm] or endogenous tethers [15.6 ± 4.78 nm]) and distance between tethered virions was measured (Table 1 and Fig. 8). The average distance between HIV-1 virions tethered to an HT1080 cell was 19.9 ± 6.82 nm, which was slightly greater than the distance between two tethered virions, 17.1 ± 6.50 nm. The average length of the segmented tethers and distance between membranes is close to the length of the mammalian ectodomain homodimer (27–30), which is predicted for the extended tetherin model (see Fig. S1 in the supplemental material). The histogram analysis showed a range of distances between HIV-1 tethered to cells of 7 to 35 nm and a range of 6 to 37 nm for virus-to-virus tethers (Fig. 8A and B). We did not observe tethers greater than 37 nm in length, as seen in selected images of chemically fixed samples imaged by conventional TEM (32, 36, 37). The distance between tethered virions was the same for mature (17.8 ± 7.24 nm) and immature virions (17.9 ± 6.31 nm), which suggests that HIV-1 maturation does not influence the length of the tether.
FIG 8.
Distances between tethered HIV-1 virions and the cell. (A and B) Histograms of segmented 3D reconstructions of HIV-1 virions tethered to the cell or other virions. Measurements were made along tethers (red) connecting HIV-1 virions (yellow) to each other or to the plasma membrane (cyan), as indicated by the white boxes and thin white lines. Example images of the following types of HIV-1 tethers are shown: 7.5 to 10 nm (C and D), 17.5 to 20 nm (E and F), and 27.5 to 30 nm (G and H). Values for different classes (means [SD]) were the following: HIV-cell, 19.9 nm (6.88 nm), range of 6.66 to 35.0 nm, n = 62; HIV-HIV, 17.1 nm (6.50 nm), range of 6.82 to 36.6 nm, n = 157. Scale bars are 20 nm.
Spatial arrangement of HIV-1 virions tethered to the cell and to other virions.
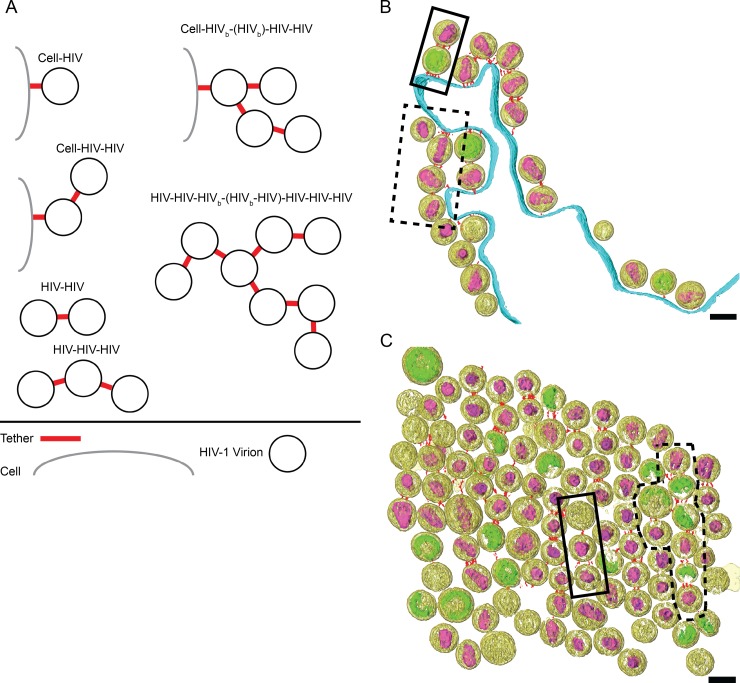
Clusters of HIV-1 VLPs and virions attached to the cell have been imaged by conventional TEM (1, 9, 23, 32, 36–38) and SEM (2, 23, 39). However, it is not clear how HIV-1 virions are attached to each other or how the virions are arranged. We sought to better understand how tethered virions were organized in relation to the cell and to other virions. For segmented tomograms, groups of tethered virions were classified according to the number of virions tethered together and the number of connections between virions (Fig. 9 and Table 3; also see Table S3 in the supplemental material). Tethered HIV-1 virions possessed between 1 and 3 nodes, or connections, consisting of at least one tether. Based on our analysis, we concluded that tethered HIV-1 virions most frequently are arranged in a linear fashion. Tethered HIV-1 virions were observed primarily as single chains (77%) and less often as branched chains (23%). This suggests that the formation of branch points is relatively rare and implies that particles bud repeatedly from a tetherin-enriched plasma membrane site in a generally linear fashion.
FIG 9.
Tethered HIV-1 virions are arranged as either single chains or branched chains. (A) HIV-1 virions connected to the cell plasma membrane or to other virions were classified according to the number of connections between particles. Examples of segmented tomograms of tethered HIV-1 virions are arranged as either single chains as indicated by solid black boxes or branched chains as indicated by black dashed boxes. (B) HIV-1 virions tethered to the cell plasma membrane; cell-HIV-HIV and cell-HIVb-(HIVb)-HIV-HIV are shown. (C) Cluster of HIV-1 virions tethered to each other; HIV-HIV-HIV and HIV-HIV-HIVb-(HIVb-HIV)-HIV-HIV-HIV groups are shown. Shown are the segmented volumes of HIV-1 virions (yellow), immature Gag lattice (green), mature conical cores (purple), tethers (red), and plasma membrane (cyan). For visualization purposes the bottom portion of the tomogram shown in panel C was cropped. Scale bars are 100 nm.
TABLE 3.
Arrangement of tethered HIV-1
| Class | n | % |
|---|---|---|
| Single chain | 20 | 77 |
| Cell-HIV | 8 | 30 |
| HIV-HIV | 12 | 46 |
| Branched chain | 6 | 23 |
| Cell-HIV | 3 | 12 |
| HIV-HIV | 3 | 12 |
DISCUSSION
The discovery of tetherin as the Vpu-responsive host restriction factor that retains particles on the plasma membrane of cells was made in 2008 by the Bieniasz group (1). Since that time, a number of important aspects of how tetherin functions have been elucidated. Tetherin functions as a proteinaceous link between the plasma membrane and virions, as revealed by biochemical and electron microscopic analysis (23, 37). The ectodomain of tetherin forms a coiled-coil dimer, and dimerization is essential to tetherin's function in restriction (26–29). Tetherin does not interact with viral proteins in attaching particles to cells and to each other but rather inserts into the lipid envelope of virions. This property somehow is facilitated by the ability of tetherin to compartmentalize to very punctate microdomains where HIV virions bud (51, 65). Intricate biochemical and genetic studies have established that the most likely orientation of the tetherin dimer is oriented in an axial or extended configuration in which either the transmembrane domains or GPI anchors insert into the viral membrane, with some preference for the GPI ends of the dimers to be within the viral membrane versus the cell membrane (23, 32–34). Biophysical studies indicate the purified coiled-coil tetherin ectodomain dimer exists as an L-shaped rod of 15 to 18 nm in length and exhibits flexibility in solution (27). In this study, we provide ultrastructural confirmation of this size prediction as well as additional insights into tetherin's mechanism of restriction of HIV-1 using cryo-ET.
Previous EM studies of HIV-1 tethered to human cells have relied mainly on imaging chemically fixed, embedded, sectioned, and stained tissue (1, 9, 32, 37, 38, 66). Our group has used immuno-EM techniques to definitively show that tetherin is present between the plasma membrane and tethered particles, as well as within the virus-containing compartment (VCC) of human macrophages (36, 37). We observed some electron-dense structures that could be consistent with tetherin in these studies, including some very extended filamentous connections that would be inconsistent with the length of tetherin dimers. Although useful, standard TEM techniques could not provide high-resolution or 3D information of the tethers restricting HIV-1 release, and we considered that the very long apparent tethers could have been a shrinkage/processing artifact. Cryo-ET offers several advantages over conventional imaging methods, mainly the acquisition of high-resolution 3D spatial information of unstained frozen-hydrated biological specimens (67, 68). Cryo-immobilization by rapid immersion into liquid ethane preserves the ultrastructure of thin eukaryotic cellular appendages and isolated viruses (69–72). This was best illustrated by the application of whole-cell cryo-ET to study HIV-1 assembly events as well as maturation (55, 56, 59). In this study, the use of whole-cell cryo-ET permitted the direct visualization of tethers composed of tetherin connecting HIV-1 VLPs and virions to each other and to the cell, which would not have been resolvable by light microscopy or conventional EM.
We observed that HIV-1 VLPs and HIV-1 virions were connected to each other and to the plasma membrane by rod-like bodies, referred to as tethers. The tethers were present only on cells transfected with tetherin. As expected, these tethers were rarely seen on released HIV-1 virions produced from cells transfected with C3A-tetherin, a tetherin mutant that does not restrict HIV-1 release. Immunogold staining was performed in parallel experiments, providing corroborative evidence that the filamentous structures imaged by cryo-ET indeed contain tetherin molecules. This represents the first definitive and high-resolution visualization of the proteinaceous tethers between cells and virions and provided us, for the first time, with 3D data suitable for further analysis.
We observed a range of 1 to 8 filamentous tethers bound to each HIV-1 virion, with an average of 2 tethers per particle. The number of filamentous tethers visualized by cryo-ET is on the same order of magnitude as the number (on average, 4 to 7) of tetherin homodimers on HIV assembly sites as estimated by superresolution light microscopy (33) but is one order of magnitude less than that estimated by quantitative Western blot analysis (34). One possibility is that the filamentous tethers observed by cryo-ET consist of multiple closely associated tetherin homodimers. Biophysical and structural studies have shown that the isolated tetherin ectodomain can oligomerize in solution and can form a tetramer consisting of two antiparallel homodimers (28, 30). However, distance measurements of cryo-ET data suggest that the extended tetherin homodimer (and not the tetramer) is responsible for restricting HIV release. In addition, tethers were not evenly distributed over the viral membrane but formed discrete clusters at contact points between membranes. This agrees with observations made by superresolution light microscopy that 80% of the tetherin-positive HIV budding sites on transfected HeLa cells contained a single cluster of mEosFP-tetherin (33).
The arrangement of tethers, relative to the position of the Gag polyprotein in immature HIV-1, was characterized in segmented tomographic reconstructions. When interpreting the results of this analysis, one can consider the possibility that the EGFP tag could disrupt the Gag lattice or putative protein-protein interaction between tetherin and the Gag protein. In addition, Venkatesh and Bieniasz have shown that the GPI anchor of tetherin is inserted preferably (by a factor of 3 to 5) into the viral membrane (34). However, cryo-ET results from the HeLa cells that express endogenous levels of tetherin indicate that the EGFP tag does not disrupt putative protein-protein interactions or ordering of the Gag lattice. We also reasoned that if the tetherin CT tail was interacting strongly with the Gag polyprotein or being excluded by steric hindrance, one would expect that the ratio of adjacent to nonadjacent tethers for HIV-1 tethered to the cell would be different from that of HIV-1 tethered to other virions. This was not the case, as the ratio of adjacent to nonadjacent tethers was similar for the different classes of immature HIV-1, namely, 21:29 for immature HIV-1 tethered to the plasma membrane, 57:43 for immature HIV-1 tethered to immature HIV, and 27:23 for immature HIV-1 tethered to mature HIV-1.
When present in linear chains, the tethers often were present on opposite poles of the viral envelope, suggesting that virion budding had occurred sequentially through a single tetherin-enriched microdomain. The orientation of the tethers relative to the mature conical core (narrow or broad end), on the other hand, was random. Tethers present on immature HIV-1 virions were situated on the viral membrane at areas both adjacent and nonadjacent to the underlying Gag polyprotein, again suggesting that the distribution of tethers on the viral envelope relative to Gag is random. Based on our results, it is likely that tetherin remains attached to the viral membrane without any direct interaction with the immature Gag lattice, remains so during the remodeling of the core that accompanies viral maturation, and has no influence on the resulting orientation of the conical core in the mature virion.
An important aspect of this study is the confirmation of the extended ectodomain dimer model obtained through biochemical and genetic studies (23, 33, 34). The average length of segmented tethers in our study was 15.3 nm, and the distance between tethered HIV-1 virions was slightly longer at 17.9 nm. We observed a range of distances between 7 and 37 nm separating tethered virions from each other and from the cell plasma membrane. One explanation for the presence of shorter tethers is the conformational flexibility of the tetherin molecule. Bending of the coiled-coil ectodomain is thought to enable the tetherin homodimer to remain attached to the HIV-1 virions during assembly and budding (29). However, flexibility of the tetherin coiled-coil ectodomain alone does not explain the presence of the occasional tethers observed that were greater than 18 nm in length. A possible explanation for the longer tethers is that they include a deformation or extension of the lipid bilayer of the virion or plasma membrane in addition to the proteinaceous tether. Altogether, however, these data strongly support the established model of an extended, parallel homodimer of tetherin as the basic unit forming the connections between viral and cellular membranes and between tethered virions in chains.
Another intriguing aspect of tetherin biology that is highlighted by the current study is the extended linear conformation of chains of virions created by tetherin. We were able to characterize these chains in more detail than was previously possible by generating 3D volumes of particle chains by cryo-ET. We found that approximately three-quarters of the chains were unbranched. The unbranched chain predominance and the polar appearance of tethers support a model of budding and restriction in which particles repeatedly bud from a common plasma membrane microdomain in the presence of a concentration of tetherin dimers, producing a beads-on-a-string, linear appearance. Occasionally a branch point in the chain is observed, which must be formed by two adjacent budding virions separately tethered to a particle that has completed the budding process, as depicted in Fig. 9A (also see Fig. S3 in the supplemental material). Based on the number of branched chains observed, we propose that the probability of tethers being incorporated into multiple assembly sites is a relatively uncommon event.
A somewhat unexpected finding was the appearance of multiple separate tethers in some images, such as those depicted in Fig. 6. Although most commonly there were only 1 to 2 tethers observed (Fig. 2), we documented in some cases 4 or more distinct tethers. This suggests that in most cases the tetherin dimers coalesce around 1 to 2 physical connections but can occasionally form multiple distinct proteinaceous links between membranes.
In summary, the current study defined the 3D ultrastructure of HIV-1 tethered to human cells through the use of cryo-ET, native immunolabeling, and CLEM. The ability of tetherin to surround and capture budding HIV-1 virions implies that tetherin dimers undergo dramatic conformational changes during assembly and budding prior to the formation of the final rod-like tether. Future cryo-ET studies may allow individual conformational intermediates of tetherin to be better defined to provide a more complete picture of the restriction of particle release at this unique late stage of the viral life cycle.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeannette Taylor of the Emory University Robert P. Apkarian Integrated Electron Microscopy Core for assistance and Fredrick Leon for help with image processing.
This work was supported in part by Emory University, Children's Healthcare of Atlanta, the Center for AIDS Research at Emory University (P30 AI050409), and the Georgia Research Alliance to E.R.W.; NSF grant 0923395 to E.R.W; the James B. Pendleton Charitable Trust to P.S. and E.R.W.; and public health service grants F32GM112517 to J.D.S and NIH AI058828 to P.S.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01880-15.
REFERENCES
- 1.Neil SJ, Zang T, Bieniasz PD. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 2.Van Damme N, Goff D, Katsura C, Jorgenson RL, Mitchell R, Johnson MC, Stephens EB, Guatelli J. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245–252. doi: 10.1016/j.chom.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondeau C, Pelchen-Matthews A, Mlcochova P, Marsh M, Milne RS, Towers GJ. 2013. Tetherin restricts herpes simplex virus 1 and is antagonized by glycoprotein M. J Virol 87:13124–13133. doi: 10.1128/JVI.02250-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia B, Serra-Moreno R, Neidermyer W, Rahmberg A, Mackey J, Fofana IB, Johnson WE, Westmoreland S, Evans DT. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog 5:e1000429. doi: 10.1371/journal.ppat.1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jouvenet N, Neil SJ, Zhadina M, Zang T, Kratovac Z, Lee Y, McNatt M, Hatziioannou T, Bieniasz PD. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J Virol 83:1837–1844. doi: 10.1128/JVI.02211-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaletsky RL, Francica JR, Agrawal-Gamse C, Bates P. 2009. Tetherin-mediated restriction of filovirus budding is antagonized by the Ebola glycoprotein. Proc Natl Acad Sci U S A 106:2886–2891. doi: 10.1073/pnas.0811014106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong WS, Irie T, Yoshida A, Kawabata R, Kadoi T, Sakaguchi T. 2012. Inhibition of virus-like particle release of Sendai virus and Nipah virus, but not that of mumps virus, by tetherin/CD317/BST-2. Hiroshima J Med Sci 61:59–67. [PubMed] [Google Scholar]
- 8.Mansouri M, Viswanathan K, Douglas JL, Hines J, Gustin J, Moses AV, Fruh K. 2009. Molecular mechanism of BST2/tetherin downregulation by K5/MIR2 of Kaposi's sarcoma-associated herpesvirus. J Virol 83:9672–9681. doi: 10.1128/JVI.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neil SJ, Sandrin V, Sundquist WI, Bieniasz PD. 2007. An interferon-alpha-induced tethering mechanism inhibits HIV-1 and Ebola virus particle release but is counteracted by the HIV-1 Vpu protein. Cell Host Microbe 2:193–203. doi: 10.1016/j.chom.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radoshitzky SR, Dong L, Chi X, Clester JC, Retterer C, Spurgers K, Kuhn JH, Sandwick S, Ruthel G, Kota K, Boltz D, Warren T, Kranzusch PJ, Whelan SP, Bavari S. 2010. Infectious Lassa virus, but not filoviruses, is restricted by BST-2/tetherin. J Virol 84:10569–10580. doi: 10.1128/JVI.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ooi YS, Dube M, Kielian M. 2015. BST2/tetherin inhibition of alphavirus exit. Viruses 7:2147–2167. doi: 10.3390/v7042147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones PH, Maric M, Madison MN, Maury W, Roller RJ, Okeoma CM. 2013. BST-2/tetherin-mediated restriction of chikungunya (CHIKV) VLP budding is counteracted by CHIKV non-structural protein 1 (nsP1). Virology 438:37–49. doi: 10.1016/j.virol.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Luo S, He S, Zhang M, Wang P, Li C, Huang W, Hu B, Griffin GE, Shattock RJ, Hu Q. 2015. Tetherin restricts HSV-2 release and is counteracted by multiple viral glycoproteins. Virology 475:96–109. doi: 10.1016/j.virol.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 14.Zenner HL, Mauricio R, Banting G, Crump CM. 2013. Herpes simplex virus 1 counteracts tetherin restriction via its virion host shutoff activity. J Virol 87:13115–13123. doi: 10.1128/JVI.02167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Tortorec A, Neil SJ. 2009. Antagonism to and intracellular sequestration of human tetherin by the human immunodeficiency virus type 2 envelope glycoprotein. J Virol 83:11966–11978. doi: 10.1128/JVI.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, Wilson SJ, Landford WC, Virgen B, Gregory D, Johnson MC, Munch J, Kirchhoff F, Bieniasz PD, Hatziioannou T. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67. doi: 10.1016/j.chom.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez LA, Yang SJ, Hauser H, Exline CM, Haworth KG, Oldenburg J, Cannon PM. 2010. Ebola virus glycoprotein counteracts BST-2/Tetherin restriction in a sequence-independent manner that does not require tetherin surface removal. J Virol 84:7243–7255. doi: 10.1128/JVI.02636-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dave VP, Hajjar F, Dieng MM, Haddad E, Cohen EA. 2013. Efficient BST2 antagonism by Vpu is critical for early HIV-1 dissemination in humanized mice. Retrovirology 10:128. doi: 10.1186/1742-4690-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sato K, Misawa N, Fukuhara M, Iwami S, An DS, Ito M, Koyanagi Y. 2012. Vpu augments the initial burst phase of HIV-1 propagation and downregulates BST2 and CD4 in humanized mice. J Virol 86:5000–5013. doi: 10.1128/JVI.07062-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homann S, Smith D, Little S, Richman D, Guatelli J. 2011. Upregulation of BST-2/Tetherin by HIV infection in vivo. J Virol 85:10659–10668. doi: 10.1128/JVI.05524-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liberatore RA, Bieniasz PD. 2011. Tetherin is a key effector of the antiretroviral activity of type I interferon in vitro and in vivo. Proc Natl Acad Sci U S A 108:18097–18101. doi: 10.1073/pnas.1113694108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neil SJ. 2013. The antiviral activities of tetherin. Curr Top Microbiol Immunol 371:67–104. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Caballero D, Zang T, Ebrahimi A, McNatt MW, Gregory DA, Johnson MC, Bieniasz PD. 2009. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139:499–511. doi: 10.1016/j.cell.2009.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishikawa J, Kaisho T, Tomizawa H, Lee BO, Kobune Y, Inazawa J, Oritani K, Itoh M, Ochi T, Ishihara K, Hirano T. 1995. Molecular cloning and chromosomal mapping of a bone marrow stromal cell surface gene, BST2, that may be involved in pre-B-cell growth. Genomics 26:527–534. doi: 10.1016/0888-7543(95)80171-H. [DOI] [PubMed] [Google Scholar]
- 25.Kupzig S, Korolchuk V, Rollason R, Sugden A, Wilde A, Banting G. 2003. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4:694–709. doi: 10.1034/j.1600-0854.2003.00129.x. [DOI] [PubMed] [Google Scholar]
- 26.Andrew AJ, Miyagi E, Kao S, Strebel K. 2009. The formation of cysteine-linked dimers of BST-2/tetherin is important for inhibition of HIV-1 virus release but not for sensitivity to Vpu. Retrovirology 6:80. doi: 10.1186/1742-4690-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinz A, Miguet N, Natrajan G, Usami Y, Yamanaka H, Renesto P, Hartlieb B, McCarthy AA, Simorre JP, Gottlinger H, Weissenhorn W. 2010. Structural basis of HIV-1 tethering to membranes by the BST-2/tetherin ectodomain. Cell Host Microbe 7:314–323. doi: 10.1016/j.chom.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schubert HL, Zhai Q, Sandrin V, Eckert DM, Garcia-Maya M, Saul L, Sundquist WI, Steiner RA, Hill CP. 2010. Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations. Proc Natl Acad Sci U S A 107:17951–17956. doi: 10.1073/pnas.1008206107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swiecki M, Scheaffer SM, Allaire M, Fremont DH, Colonna M, Brett TJ. 2011. Structural and biophysical analysis of BST-2/tetherin ectodomains reveals an evolutionary conserved design to inhibit virus release. J Biol Chem 286:2987–2997. doi: 10.1074/jbc.M110.190538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Wang J, Jia X, McNatt MW, Zang T, Pan B, Meng W, Wang HW, Bieniasz PD, Xiong Y. 2010. Structural insight into the mechanisms of enveloped virus tethering by tetherin. Proc Natl Acad Sci U S A 107:18428–18432. doi: 10.1073/pnas.1011485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swiecki M, Omattage NS, Brett TJ. 2013. BST-2/tetherin: structural biology, viral antagonism, and immunobiology of a potent host antiviral factor. Mol Immunol 54:132–139. doi: 10.1016/j.molimm.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Fitzpatrick K, Skasko M, Deerinck TJ, Crum J, Ellisman MH, Guatelli J. 2010. Direct restriction of virus release and incorporation of the interferon-induced protein BST-2 into HIV-1 particles. PLoS Pathog 6:e1000701. doi: 10.1371/journal.ppat.1000701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann M, Rocha S, Mangeat B, Blanchet F, Uji IH, Hofkens J, Piguet V. 2011. Quantitative multicolor super-resolution microscopy reveals tetherin HIV-1 interaction. PLoS Pathog 7:e1002456. doi: 10.1371/journal.ppat.1002456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venkatesh S, Bieniasz PD. 2013. Mechanism of HIV-1 virion entrapment by tetherin. PLoS Pathog 9:e1003483. doi: 10.1371/journal.ppat.1003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee SK, Potempa M, Swanstrom R. 2012. The choreography of HIV-1 proteolytic processing and virion assembly. J Biol Chem 287:40867–40874. doi: 10.1074/jbc.R112.399444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu H, Wang JJ, Qi M, Yoon JJ, Chen X, Wen X, Hammonds J, Ding L, Spearman P. 2012. Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe 12:360–372. doi: 10.1016/j.chom.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammonds J, Wang JJ, Yi H, Spearman P. 2010. Immunoelectron microscopic evidence for Tetherin/BST2 as the physical bridge between HIV-1 virions and the plasma membrane. PLoS Pathog 6:e1000749. doi: 10.1371/journal.ppat.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Habermann A, Krijnse-Locker J, Oberwinkler H, Eckhardt M, Homann S, Andrew A, Strebel K, Krausslich HG. 2010. CD317/tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. J Virol 84:4646–4658. doi: 10.1128/JVI.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Casartelli N, Sourisseau M, Feldmann J, Guivel-Benhassine F, Mallet A, Marcelin AG, Guatelli J, Schwartz O. 2010. Tetherin restricts productive HIV-1 cell-to-cell transmission. PLoS Pathog 6:e1000955. doi: 10.1371/journal.ppat.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derdowski A, Ding L, Spearman P. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J Virol 78:1230–1242. doi: 10.1128/JVI.78.3.1230-1242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan MA, Aberham C, Kao S, Akari H, Gorelick R, Bour S, Strebel K. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J Virol 75:7252–7265. doi: 10.1128/JVI.75.16.7252-7265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schubert U, Bour S, Willey RL, Strebel K. 1999. Regulation of virus release by the macrophage-tropic human immunodeficiency virus type 1 AD8 isolate is redundant and can be controlled by either Vpu or Env. J Virol 73:887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi H, Strauss JD, Ke Z, Alonas E, Dillard RS, Hampton CM, Lamb KM, Hammonds JE, Santangelo PJ, Spearman PW, Wright ER. 2015. Native immunogold labeling of cell surface proteins and viral glycoproteins for cryo-electron microscopy and cryo-electron tomography applications. J Histochem Cytochem 63:780–792. doi: 10.1369/0022155415593323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 45.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 46.Gilbert PF. 1972. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. II. Direct methods. Proc R Soc Lond B Biol Sci 182:89–102. doi: 10.1098/rspb.1972.0068. [DOI] [PubMed] [Google Scholar]
- 47.Frangakis AS, Hegerl R. 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135:239–250. doi: 10.1006/jsbi.2001.4406. [DOI] [PubMed] [Google Scholar]
- 48.Xiong Q, Morphew MK, Schwartz CL, Hoenger AH, Mastronarde DN. 2009. CTF determination and correction for low dose tomographic tilt series. J Struct Biol 168:378–387. doi: 10.1016/j.jsb.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pruggnaller S, Mayr M, Frangakis AS. 2008. A visualization and segmentation toolbox for electron microscopy. J Struct Biol 164:161–165. doi: 10.1016/j.jsb.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 50.Pollanen J, Hedman K, Nielsen LS, Dano K, Vaheri A. 1988. Ultrastructural localization of plasma membrane-associated urokinase-type plasminogen activator at focal contacts. J Cell Biol 106:87–95. doi: 10.1083/jcb.106.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammonds J, Ding L, Chu H, Geller K, Robbins A, Wang JJ, Yi H, Spearman P. 2012. The tetherin/BST-2 coiled-coil ectodomain mediates plasma membrane microdomain localization and restriction of particle release. J Virol 86:2259–2272. doi: 10.1128/JVI.05906-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia X, Weber E, Tokarev A, Lewinski M, Rizk M, Suarez M, Guatelli J, Xiong Y. 2014. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. eLife 3:e02362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welbourn S, Kao S, Du Pont KE, Andrew AJ, Berndsen CE, Strebel K. 2015. Positioning of cysteine residues within the N-terminal portion of the BST-2/tetherin ectodomain is important for functional dimerization of BST-2. J Biol Chem 290:3740–3751. doi: 10.1074/jbc.M114.617639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fuller SD, Wilk T, Gowen BE, Krausslich HG, Vogt VM. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol 7:729–738. doi: 10.1016/S0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 55.Carlson LA, de Marco A, Oberwinkler H, Habermann A, Briggs JA, Krausslich HG, Grunewald K. 2010. Cryo electron tomography of native HIV-1 budding sites. PLoS Pathog 6:e1001173. doi: 10.1371/journal.ppat.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woodward CL, Cheng SN, Jensen GJ. 2014. Electron cryo-tomography studies of maturing HIV-1 particles reveal the assembly pathway of the viral core. J Virol 89:1267–1277. doi: 10.1128/JVI.02997-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Briggs JA, Grunewald K, Glass B, Forster F, Krausslich HG, Fuller SD. 2006. The mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15–20. doi: 10.1016/j.str.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 58.Benjamin J, Ganser-Pornillos BK, Tivol WF, Sundquist WI, Jensen GJ. 2005. Three-dimensional structure of HIV-1 virus-like particles by electron cryotomography. J Mol Biol 346:577–588. doi: 10.1016/j.jmb.2004.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Z, Dobro MJ, Woodward CL, Levandovsky A, Danielson CM, Sandrin V, Shi J, Aiken C, Zandi R, Hope TJ, Jensen GJ. 2013. Unclosed HIV-1 capsids suggest a curled sheet model of assembly. J Mol Biol 425:112–123. doi: 10.1016/j.jmb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keller PW, Adamson CS, Heymann JB, Freed EO, Steven AC. 2011. HIV-1 maturation inhibitor bevirimat stabilizes the immature Gag lattice. J Virol 85:1420–1428. doi: 10.1128/JVI.01926-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Marco A, Muller B, Glass B, Riches JD, Krausslich HG, Briggs JA. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog 6:e1001215. doi: 10.1371/journal.ppat.1001215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Krausslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A 106:11090–11095. doi: 10.1073/pnas.0903535106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J 26:2218–2226. doi: 10.1038/sj.emboj.7601664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse-Locker J, Griffiths G. 1996. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J 15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 65.Billcliff PG, Rollason R, Prior I, Owen DM, Gaus K, Banting G. 2013. CD317/tetherin is an organiser of membrane microdomains. J Cell Sci 126:1553–1564. doi: 10.1242/jcs.112953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hammonds J, Wang JJ, Spearman P. 2012. Restriction of retroviral replication by tetherin/BST-2. Mol Biol Int 2012:424768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lucic V, Rigort A, Baumeister W. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol 202:407–419. doi: 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diebolder CA, Koster AJ, Koning RI. 2012. Pushing the resolution limits in cryo electron tomography of biological structures. J Microsc 248:1–5. doi: 10.1111/j.1365-2818.2012.03627.x. [DOI] [PubMed] [Google Scholar]
- 69.Medalia O, Beck M, Ecke M, Weber I, Neujahr R, Baumeister W, Gerisch G. 2007. Organization of actin networks in intact filopodia. Curr Biol 17:79–84. doi: 10.1016/j.cub.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 70.Adrian M, Dubochet J, Lepault J, McDowall AW. 1984. Cryo-electron microscopy of viruses. Nature 308:32–36. doi: 10.1038/308032a0. [DOI] [PubMed] [Google Scholar]
- 71.Booy FP, Ruigrok RW, van Bruggen EF. 1985. Electron microscopy of influenza virus. A comparison of negatively stained and ice-embedded particles. J Mol Biol 184:667–676. [DOI] [PubMed] [Google Scholar]
- 72.Fernandez-Busnadiego R, Asano S, Oprisoreanu AM, Sakata E, Doengi M, Kochovski Z, Zurner M, Stein V, Schoch S, Baumeister W, Lucic V. 2013. Cryo-electron tomography reveals a critical role of RIM1alpha in synaptic vesicle tethering. J Cell Biol 201:725–740. doi: 10.1083/jcb.201206063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.