ABSTRACT
The oncogenic herpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) is known to encode four viral interferon regulatory factors (vIRF1 to -4) to subvert the host antiviral immune response, but their detailed DNA-binding profiles as transcription factors in the host remain uncharacterized. Here, we first performed genome-wide vIRF2-binding site mapping in the human genome using chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq). vIRF2 was capable of binding to the promoter regions of 100 putative target genes. Importantly, we confirmed that vIRF2 can specifically interact with the promoters of the genes encoding PIK3C3, HMGCR, and HMGCL, which are associated with autophagosome formation or tumor progression and metastasis, and regulate their transcription in vivo. The crystal structure of the vIRF2 DNA-binding domain (DBD) (referred to here as vIRF2DBD) showed variable loop conformations and positive-charge distributions different from those of vIRF1 and cellular IRFs that are associated with DNA-binding specificities. Structure-based mutagenesis revealed that Arg82 and Arg85 are required for the in vitro DNA-binding activity of vIRF2DBD and can abolish the transcription regulation function of vIRF2 on the promoter reporter activity of PIK3C3, HMGCR, and HMGCL. Collectively, our study provided unique insights into the DNA-binding potency of vIRF2 and suggested that vIRF2 could act as a transcription factor of its target genes in the host antiviral immune response.
IMPORTANCE The oncogenic herpesvirus KSHV is the etiological agent of Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. KSHV has developed a unique mechanism to subvert the host antiviral immune responses by encoding four homologues of cellular interferon regulatory factors (vIRF1 to -4). However, none of their DNA-binding profiles in the human genome have been characterized until now, and the structural basis for their diverse DNA-binding properties remain poorly understood. In this study, we performed the first genome-wide vIRF2-binding site mapping in the human genome and found vIRF2 can bind to the promoter regions of 100 target cellular genes. X-ray structure analysis and functional studies provided unique insights into its DNA-binding potency and regulation of target gene expression. Our study suggested that vIRF2 could act as a transcription factor of its target genes and contribute to KSHV infection and pathogenesis through versatile functions.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV8), is the etiological agent of Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and multicentric Castleman's disease (MCD) (1–3). Moreover, KS is the first tumor to be associated with HIV infection and the most common AIDS-associated malignancy in sub-Saharan Africa and the second most common cancer in HIV-infected patients (2–5). KSHV has developed a unique mechanism to subvert host antiviral immune responses by encoding four homologues (viral interferon regulatory factors [vIRF1 to -4]) of cellular interferon regulatory factors (c-IRFs) with a cluster of KSHV open reading frames (ORFs), K9, K11/11.1, K10.5/10.6, and K10, respectively (6–8). vIRFs have been demonstrated to be effective inhibitors of interferon signaling and regulators of cellular oncogenic pathways, which may contribute to KSHV infection, as well as pathogenesis (4, 9, 10).
vIRF2 was previously demonstrated to be a constitutively expressed viral protein, consisting of 163 amino acids, encoded by a single ORF, K11.1 (5). Recently, the full-length vIRF2 protein, consisting of 680 amino acids, which was encoded by a 2.2-kb spliced transcript representing two exons of ORFs K11.1 and K11, has been confirmed (11, 12). vIRF2 (here representing the full length of vIRF2 unless otherwise indicated) is presented in both the cytoplasm and nucleus of infected cells during KSHV lytic replication, and its expression can be induced by interferon (IFN) treatment (3, 13). vIRF2 shares about 13% sequence homology with human IRFs, mostly located in the DNA-binding region, and also shows some similarity to the IRF interaction domain (IAD) found in IRF8/interferon consensus sequence binding protein (ICSBP) (14, 15).
Similarly to vIRF1, vIRF2 can function as a modulator of the innate immune response by negatively regulating the transactivation of the interferon-stimulated response element (ISRE) promoter by IRF-1, as well as activation of the IFN reporter promoter by either IRF-3 or IRF-1, but not by IRF-7 (12). Moreover, vIRF2 targets components of interferon-stimulated gene factor 3 (ISGF3) complexes, STAT1, and IRF-9, which results in the inhibition of ISG expression (16). Thus, vIRF2 appears to be able to inhibit both early and later steps of the antiviral IFN signaling pathway (9). Similar to other vIRFs, vIRF2 does not contain a unique tryptophan pentad repeat in the DNA-binding domain, which is important in binding the promoters of IFNs and IFN-inducible genes for all cellular IRFs (17, 18). Previous studies showed that vIRFs were unable to bind to the consensus DNA sequence of human IRFs (6). The inhibitory role of vIRF2 in regulating IFN signaling seemed to rely on its interaction with other regulatory factors, but not direct transcriptional regulation. For example, vIRF2 was shown to recruit caspase 3 to IRF-3 and thus to accelerate the caspase-dependent process of IRF-3 turnover, leading to an inefficient antiviral response (19).
However, some evidence has suggested that the short-form vIRF2 (K11.1) was capable of directly binding to a consensus NF-κB binding site, but not to an ISRE (8). vIRF1 has been shown to bind some DNA oligonucleotides in vitro and can interact with the K3-viral dihydrofolate reductase-viral interleukin 6 promoter region in the KSHV genome (20). The recent structure of the vIRF1 DNA-binding domain (DBD) (referred to here as vIRF1DBD) in complex with a double-stranded DNA from the KSHV genome suggested that vIRF1 might function as a transcription factor on operator regions within the viral genome (20, 21). Interestingly, in a thermal-stability shift assay (TSSA), the vIRF1 DBD can bind DNA, whereas the full-length vIRF1 cannot (21). Although the DNA-binding character of vIRF1 was preliminarily defined in the KSHV genome, there is still a question whether vIRFs can function as bona fide transcription factors in the host. Due to the important function of vIRF2 in the KSHV life cycle, we are curious to know whether the full length of vIRF2 can bind to DNA and whether vIRF2 can act as a transcription factor in humans. In this study, we combined human genome-wide vIRF2-binding site mapping by chromatin immunoprecipitation coupled to high-throughput sequencing (ChIP-seq) and structural analysis of vIRF2DBD to address the potential function of vIRF2 as a DNA-binding transcription factor.
MATERIALS AND METHODS
ChIP-seq and ChIP-PCR.
The ChIP protocol was adapted from Rockland Immunochemicals Inc. with some modifications. Briefly, cells were cross-linked in the medium with 1% formaldehyde for 30 min at room temperature and quenched with 0.125 M glycine. After cross-linking, the cells were washed with phosphate-buffered saline (PBS) twice and resuspended in 1 ml of buffer A (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.2% Triton X-100, 1 mM dithiothreitol [DTT], 0.5 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) for 10 min at 4°C. The extracted nuclei were pelleted by centrifugation at 2,000 rpm for 5 min. The nuclei were lysed in SDS lysis buffer (50 mM HEPES, 1 mM EDTA, 1% SDS, 1 mM PMSF) for 10 min on ice. The lysates were subjected to sonication to obtain ∼200-bp fragments of DNA and to centrifugation at 13,000 × g at 4°C for 10 min to obtain the supernatants. The supernatants were diluted 1:10 with RIPA buffer. The samples were precleared with pretreated protein A or G beads for 2 h at 4°C and then incubated with anti-Flag antibody overnight at 4°C. After extensive washing with RIPA buffer, wash buffer (20 mM Tris-HCl, pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 1 mM PMSF), and TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) (each 4 times), the beads were resuspended in TE buffer. The resuspended beads were subjected to RNase A and proteinase K digestion, and then the cross-linking was reversed at 65°C for 8 to 10 h. DNA was recycled with a DNA purification kit (Tiangen). The enriched DNA fragments were subjected to quantitative PCR (qPCR) or library construction. The library construction and sequencing procedures were performed by the Omics Core, Chinese Academy of Sciences, following Illumina's construction.
Bioinformatics analysis of ChIP-seq data.
The ChIP-seq data were aligned with the human genome (hg19) using Bowtie2 (22). The output files were subjected to peak calling with MACS (model-based analysis of ChIP-seq), as described previously (23). All the parameters were default. The P value cutoff for the peak detection was 10−5. The input group was used as a control. The results were visualized with IGV software (24). The peak information was annotated with PeakAnalyzer (25). The distribution of peaks in relation to genes was calculated by PAVIS (26). De novo motif results were generated by HOMER, and all the parameters were default (27).
Quantitative real-time PCR.
Cells were collected and lysed in TRIzol buffer (Life technology), and RNA was isolated according to the manufacturer's instructions. Reverse transcription was performed with a cDNA reverse transcription kit (Toyobo). Real-time (RT)-PCR was performed with a SYBR green master mix kit (Toyobo). Relative mRNA levels were normalized to the actin gene and calculated by the ΔΔCT method. The primers for quantitative RT (qRT)-PCR and ChIP-PCR are listed in Table S3 in the supplemental material.
Cloning, protein expression, and purification.
pCDH-SF-vIRF2 was constructed by inserting full-length vIRF2 into the pCDH-SF vector. Mutants of pCDH-SF-vIRF2 (R82A, R85A, and R82A/R85A) were constructed by site-directed mutagenesis based on the KOD-plus kit (Toyobo) protocol. pET28-vIRF2DBD was constructed by subcloning the gene fragment encoding the vIRF2 DBD (residues 7 to 115) from pCDH-SF-vIRF2 into the expression vector pET28(+) with a cleavable N-terminal 6×His tag. 3× Motif1-Luc and 3× Motif2-Luc were constructed by inserting annealed DNA fragments containing 3 tandem copies of a representative vIRF2-binding motif 1 or 2 sequence into the pGL6-TA vector (Beyotime, China). PIK3C3p-Luc, HMGCRp-Luc, and HMGCLp-Luc were constructed by inserting the indicated promoter region (see Table S3 in the supplemental material) of each gene into the pGL3-enhancer vector (Promega). All the primers used in vector construction are listed in Table S3 in the supplemental material.
pET28-vIRF2DBD was transformed into an Escherichia coli BL21(DE3) star expression strain (Invitrogen). For protein expression, bacterial cells were grown to mid-log phase in LB medium at 37°C in the presence of 50 mg/ml kanamycin. Induction of the culture was then carried out by adding 0.2 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) at 20°C. The cells were pelleted after 20 h by centrifugation at 8,000 rpm for 10 min at 4°C. The cell pellet was then resuspended in buffer containing 20 mM Tris (pH 8.0), 500 mM NaCl, 2 mM β-mercaptoethanol, and 1 mM PMSF and lysed with a high-pressure homogenizer at 4°C. The cell debris and membranes were pelleted by centrifugation at 16,000 rpm for 60 min at 4°C. The soluble N-terminally hexahistidine-tagged vIRF2DBD was purified by affinity chromatography with nickel-nitrilotriacetic acid resin (Bio-Rad). The protein was further purified by gel filtration chromatography (Superdex 75; GE Healthcare) equilibrated in buffer containing 20 mM Tris (pH 8.0), 500 mM NaCl, and 2 mM DTT using an Äkta Purifier System (Amersham). The highly purified protein fractions were pooled and concentrated to 4 mg/ml and quantified with the Bio-Rad protein assay kit for crystallization trials by ultrafiltration in an Amicon cell (Millipore, CA, USA).
The selenomethionine (Se-Met) protein was produced in minimal medium supplemented with 100 mg/liter (each) lysine, phenylalanine, and threonine and 50 mg/liter (each) isoleucine, leucine, valine, and selenomethionine. The production and purification of Se-Met vIRF2DBD were the same as those described above.
Crystallization, data collection, structure determination, and refinement.
Crystallization screens were performed with Hampton Research and Qiagen kits using the sitting-drop vapor diffusion method at room temperature and 4°C. The Se-Met vIRF2DBD crystal was obtained in a mixture (solution) of 30% (vol/vol) polyethylene glycol (PEG) 400, 0.1 M 2-(cyclohexylamino)ethanesulfonic acid (CHES), pH 9.5, at room temperature. The crystal quality was optimized by adjusting the concentrations of the precipitant and buffer. The best crystal was obtained in a solution of 25% (vol/vol) PEG 400, 0.1 M CHES, pH 9.5, after 4 days at room temperature.
All of the data were collected on beamline 3W1A at the Beijing Synchrotron Radiation Facility (BSRF) with a mounted MAR-165 charge-coupled device (CCD) detector. Before data collection, the crystals were soaked in the reservoir solution supplemented with 20% (vol/vol) glycerol for a few seconds and then flash-frozen in liquid nitrogen. All the data were processed with HKL2000 (28). The Se-Met crystal structure of the vIRF2DBD was determined by the single-wavelength anomalous-dispersion (SAD) method. The selenium atoms were located with the program Shelxd (29) and then used to calculate the initial phases in Shelxe. The phases from Shelxe were improved in Resolve (30) and then used in Buccaneer for model building. Coot (31) and Phenix.refine (32) were used for manual building and refinement, respectively. The refinement statistics and model parameters are provided below (see Table 2). The program PyMOL (http://www.pymol.sourceforge.net/) was used to prepare the structural figures. Superpositions of the structures were performed in Coot using secondary-structure matching (33).
TABLE 2.
Data collection and refinement statistics
| Statistica | Value for vIRF2DBDb |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9792 |
| Space group | P3221 |
| Unit cell parameters (Å) | a = b = 48.467; c = 195.088 |
| Resolution (Å) | 2.50 (2.54–2.50) |
| No. of unique reflections | 9,881 (490) |
| Completeness (%) | 100 (100) |
| Redundancy | 15.3 (10.9) |
| Mean I/σ(I) | 29.6 (4.5) |
| No. of molecules in asymmetric unit | 2 |
| Rmerge (%) | 13.4 (46.8) |
| Refinement | |
| Resolution range (Å) | 41.0–2.50 |
| Rwork/Rfree (%) | 21.2/24.8 |
| No. of residues/protein atoms | 201/1,699 |
| No. of water atoms | 85 |
| Average B factor | |
| Main chain (A/B) | 19.7/24.1 |
| Side chain (A/B) | 22.4/26.4 |
| Waters | 21.7 |
| Ramachandran plot (%) | |
| Most favored | 98.5 |
| Allowed | 1.50 |
| RMSD | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.257 |
I, average intensity of whole reflections; σ(I), deviation of I.
The values in parentheses are for the highest-resolution shell.
Gel EMSA.
vIRF2-binding motif 1 and motif 2 revealed by ChIP-seq were selected to verified their interaction with vIRF2DBD by electrophoretic mobility shift assays (EMSA). DNA probes were synthesized with the sequences of 3 tandem copies of vIRF2-binding motif 1 or 2 or 1 copy of LANA-binding motif 1 as a negative control. The probe sequences are listed in Table S3 in the supplemental material. Double-stranded oligonucleotides were prepared by mixing equimolar ratios of complementary oligonucleotides. They were annealed at 65°C for 10 min in a buffer (prepared with diethyl pyrocarbonate [DEPC]-sterilized water) containing 150 mM NaCl, 50 mM Tris (pH 8.0), 2 mM MgCl2, and 1 mM DTT and then cooled at room temperature for another 10 min obtain a final concentration of 1 mM.
vIRF2DBD-DNA complexes were prepared by adding the protein at 0, 4, 8, 16, and 24 μM final concentrations to the solution containing the DNA fragment samples (2 μM final concentration) and incubated for 30 min at room temperature. For each sample, free DNA and complexes were separated on a 10% acrylamide native gel run for 1 h at 200 V and 5 W at 4°C and visualized by Golden View (Biotium) staining.
Luciferase reporter assay.
293T cells (2 × 105) were seeded into 12-well plates the day before transfection. The cells were cotransfected with 1 μg luciferase reporter, 20 ng pRL-TK, and 2 μg pCDH-SF-GFP, pCDH-SF-vIRF2, pCDH-SF-vIRF2-R82A, pCDH-SF-vIRF2-R85A, or pCDH-SF-vIRF2-R82A/R85A plasmid using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Forty-eight hours posttransfection, cells were harvested and lysed in 100 μl cell lysis buffer (Promega) for luciferase reporter assays using the Promega dual-luciferase reporter assay system. All data sets were repeated multiple times, and the data shown are means of three independent experiments.
Accession numbers.
The atomic coordinates and structure factor files of vIRF2DBD have been deposited in the Protein Data Bank (PDB) with the entry code 4P55. The ChIP-seq data for vIRF2 are available under accession numbers SRX951274 and SRX951275 at the NCBI SRA database.
RESULTS
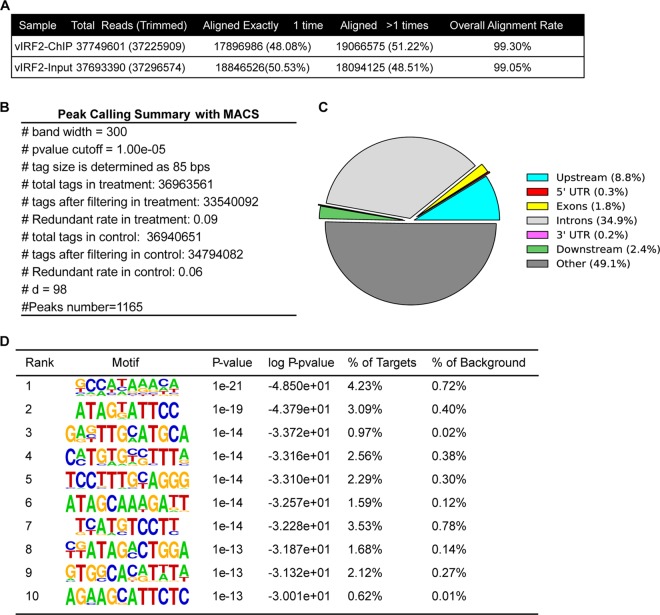
Human genome-wide mapping of KSHV vIRF2-binding sites.
To identify all vIRF2-binding sites in the human genome using an unbiased approach, ChIP-seq was performed in 293T cells expressing Flag-tagged vIRF2. pCDH-SF-vIRF2 was transfected into 293T cells, and after 24 h, the cells were harvested for ChIP using anti-Flag antibody. The DNA fragments enriched by ChIP or input control were then subjected to high-throughput sequencing according to Illumina's instructions. The ChIP-seq data were aligned to the human genome (hg19), and only uniquely mapped nonredundant reads were used in subsequent analyses (Fig. 1A). Peaks significantly enriched over the input control were determined using MACS, as described previously (23). In total, 1,165 peaks were identified (Fig. 1B), which suggested that full-length vIRF2 was indeed capable of binding DNA extensively. Even though most of the peaks were distributed in intergenic and intron regions (the ratios were 49.1% and 34.9%, respectively), there were 101 peaks (8.8%) located within the promoter regions of 100 annotated genes (Fig. 1C; see Table S1 in the supplemental material). The top 10 vIRF2 target promoters are listed in Table 1. Furthermore, we wondered whether there were consensus motifs within the 1,165 ChIP-seq peaks of vIRF2. The sequences of these peaks were extracted and subjected to de novo motif analysis with HOMER (22). The top 10 potential vIRF2-binding motifs are listed in Fig. 1D and Table S2 in the supplemental material. These potential vIRF2-binding motifs showed significant homology with those in several DNA-binding transcriptional factors (Fig. 1D; see Table S2 in the supplemental material), such as the transcriptional repressor CDX2 and the transcriptional enhancer factor TEAD4. More importantly, the motif 10# AGA(C)AGC(G)ATTCTC (no. 10 in Fig 1D) shared significant homology with several cellular-IRF secondary binding motifs (including IRF3 to IRF6) (see Table S2 in the supplemental material), suggesting a DNA-binding pattern conserved between vIRF2 and cellular IRFs.
FIG 1.
Genome-wide mapping of KSHV vIRF2-binding sites in 293T cells using ChIP-seq. (A) Summary of vIRF2 ChIP-seq experiments. (B) Peak-calling summary with MACS. (C) Genome distribution of KSHV vIRF2-binding sites. (D) Summary of the top 10 potential vIRF2-binding motifs generated by HOMER.
TABLE 1.
Top 10 vIRF2 target gene promoters
| Chromosome | Peak start | Peak end | Distance | Closest TSS_ID | Symbol | Fold enrichment |
|---|---|---|---|---|---|---|
| chr18 | 39535630 | 39535949 | 406 | ENST00000585528 | PIK3C3 | 9.04 |
| chr9 | 45727093 | 45727502 | 190 | ENST00000377531 | FAM27A | 8.66 |
| chr12 | 3046048 | 3046414 | −613 | ENST00000541678 | TULP3 | 8.21 |
| chr4 | 154681548 | 154681850 | −312 | ENST00000347063 | RNF175 | 8.21 |
| chr1 | 26604759 | 26605136 | −719 | ENST00000270792 | SH3BGRL3 | 8.21 |
| chr18 | 3409366 | 3409684 | −2,81 | ENST00000552383 | TGIF1 | 8.21 |
| chr20 | 206728 | 207044 | −1,13 | ENST00000246105 | DEFB129 | 8.14 |
| chr2 | 97651402 | 97651709 | 745 | ENST00000327896 | FAM178B | 8.12 |
| chr5 | 139778872 | 139779135 | −2,95 | ENST00000360839 | ANKHD1 | 7.56 |
| chrX | 48910547 | 48910871 | −392 | ENST00000597275 | CCDC120 | 7.56 |
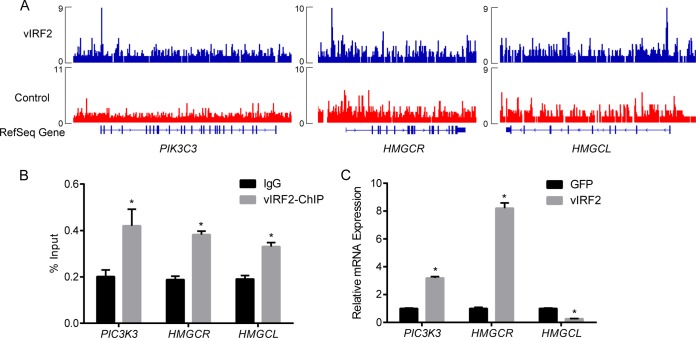
Binding of vIRF2 to the target gene promoter region as shown by ChIP-qPCR and qRT-PCR.
Since vIRF2 was shown by ChIP-seq to bind to the promoter regions of 100 genes (see Table S1 in the supplemental material), we speculated that vIRF2 is involved in the transcriptional regulation of these genes. For cross validation of the ChIP-seq result, we verified a number of vIRF2 target promoter sequences by ChIP-qPCR (see Fig. S1 in the supplemental material). As examples, a schematic of the vIRF2-bound sites within the promoter regions of PIK3C3, HMGCR (encoding HMG-coenzyme A [CoA] reductase), and HMGCL (encoding HMG-CoA lyase) are shown (Fig. 2A). We showed that DNA fragments representing the promoter regions of PIK3C3, HMGCR, and HMGCL were significantly enriched using anti-Flag antibody, but not in the control IgG (Fig. 2B; see Fig. S1 in the supplemental material), in the ChIP-qPCR assay, which confirmed the binding of vIRF2 to their promoter regions.
FIG 2.
vIRF2 regulates transcription of genes bound by itself in the promoter region. (A) Schematic of the vIRF2-bound sites within the promoter regions of PIK3C3, HMGCR, and HMGCL. The vIRF2 read depth is plotted as reads per million. (B) Binding of vIRF2 to the promoter regions of PIK3C3, HMGCR, and HMGCL was validated by ChIP-qPCR. The data are shown as means plus standard errors of the mean (SEM); n = 3. *, P < 0.05. (C) mRNA levels of PIK3C3 and HMGCR were significantly upregulated, while that of HMGCL was significantly downregulated, by vIRF2 expression measured by qRT-PCR. GFP, green fluorescent protein. The data are shown as means plus SEM; n = 3. *, P < 0.05.
We then asked whether vIRF2 was involved in the transcription regulation of the genes. pCDH-SF-vIRF2 or control pCDH-SF-GFP was transfected into 293T cells, and the cells were harvested for qRT-PCR 48 h postinfection. We showed that the mRNA levels of PIK3C3 and HMGCR were significantly upregulated, while HMGCL was significantly downregulated, by vIRF2 expression (Fig. 2C). Based on these data, we speculated that vIRF2 can downregulate or upregulate the transcription of its target genes through cooperation with different cotranscription factors in vivo.
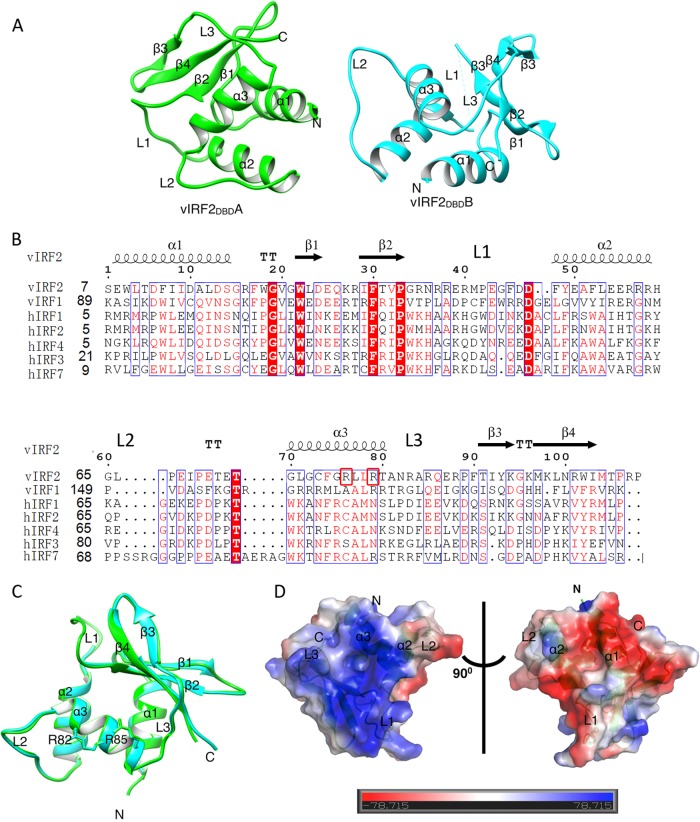
Overall structure of vIRF2DBD.
The N-terminal region (residues 7 to 114) of vIRF2 was indicated as a putative DNA-binding domain (here referred to as vIRF2DBD) through Phyre prediction (34) and sequence alignment with vIRF1 and cellular IRFs (Fig. 3A). To gain more insight into the vIRF2 DNA-binding property, we solved the crystal structure of vIRF2DBD (PDB ID 4P55) by the SAD method using Se-Met-labeled protein and refined it to a final Rwork/Rfree factor of 0.21/0.25 at 2.50-Å resolution, with the space group P3221 (Table 2). There were two molecules in an asymmetric unit, labeled vIRF2DBDA and vIRF2DBDB (Fig. 3B; see Fig. S2 in the supplemental material [shown in green and cyan, respectively]). Each monomer consisted of four antiparallel β-strands (β1↑-β2↓-β3↑-β4↓) and three α-helices (α1 to α3) connected by three long loops (L1 to L3) and three short loops (Fig. 3A and B). The structure of vIRF2DBD adopted a winged helix-turn-helix (HTH) conformation, a feature commonly found in vIRF1, cellular IRFs, and other transcriptional factors (21, 35, 36). The structures of the two monomers were very similar (Fig. 3B), and superimposition of the two molecules yielded a low root mean square deviation (RMSD) of 0.36 Å for 95 aligned C-α carbon atoms. A notable difference between the two monomers was the absence of electronic density of the residues Arg37-Gly45 in vIRF2DBDB, corresponding to the L1 region, which can probably be attributed to the relatively high flexibility of the region (see Fig. S1 in the supplemental material). Here, vIRF2DBDA was used for structural alignment and analysis (see below).
FIG 3.
Structural characteristics of vIRF2DBD. (A) Overall view of vIRF2DBD structure shown as a cartoon. There are two molecules (named vIRF2DBDA, in green, and vIRF2DBDB, in cyan) in the asymmetric unit. (B) Structure-based sequence alignment of vIRF2DBD with vIRF1 and human IRF DBDs, performed using clustal X (version 1.81) and ESPript 2.2 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The conserved residues are boxed in blue, and identical and low-conserved residues are shaded in red or appear as red letters, respectively. Arg82 and Arg85 are boxed in red. (C) Superimposition of the two molecules in the asymmetric unit, showing they are essentially the same and the conformations of the putative specificity-determining arginines (Arg82 and Arg85) in α3 of vIRF2 for DNA binding are identical. (D) Molecular surface representation of vIRF2DBD (blue, +7.8 KT; red, −7.8 KT, where KT is the Boltzmann energy at room temperature) colored according to their local electrostatic potentials.
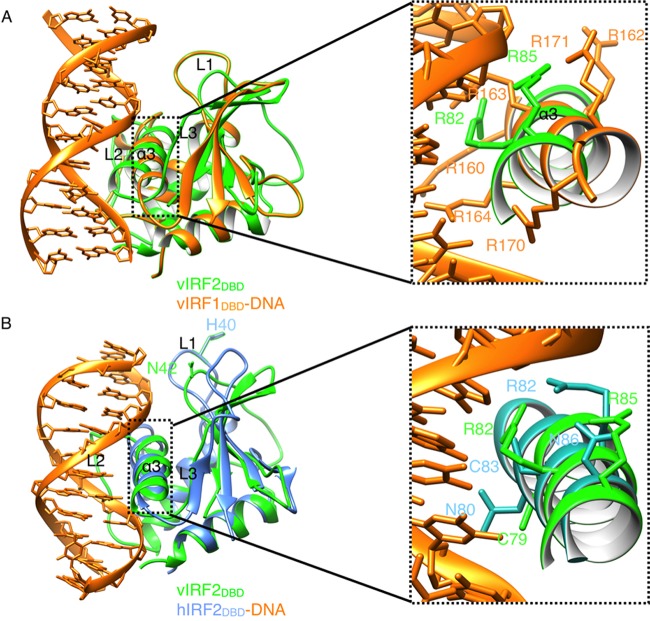
A DALI search (http://ekhidna.biocenter.helsinki.fi/dali_server) (37) for globally similar proteins revealed that vIRF2DBD had significant structural similarity to the DNA-binding domains of vIRF1 and human IRFs (RMSD, 2.0 to 3.2 Å). The top two closest homologues were the vIRF1DBD-DNA complex (PDB ID 4HLY), with a Z-score of 10.6 and an RMSD of 2.9 Å for 106 C-α atoms, and the human IRF2DBD-DNA complex (PDB ID 2IRF), with a Z-score of 10.2 and an RMSD of 2.4 Å for 109 C-α atoms. vIRF2DBD shared 23.1% and 18.2% amino acid sequence identity with vIRF1DBD and IRF2DBD, respectively. The structures of the vIRF1DBD-DNA and IRF2DBD-DNA complexes were compared with that of vIRF2DBD as described below.
Structural features of vIRF2DBDinvolved in DNA binding.
In vIRF2DBD, the positively charged protuberances were mainly formed by α2 and α3 and by L1 and L3 (Fig. 3C), similar to the distribution of the charged surface patches in vIRF1DBD and IRF2DBD. However, there are remarkable shifts and conformation changes in loops L1 to L3 of vIRF2 that may be involved in DNA binding, compared with those regions in the vIRF1DBD and IRF2DBD structures (Fig. 4A and B), which may reflect their distinct DNA-binding characteristics.
FIG 4.
Structural comparisons of vIRF2DBD with vIRF1-DNA and IRF2-DNA complexes. (A) Structural superposition of DBDs between vIRF2 (green) and the vIRF1-DNA complex (orange; PDB ID 4HLY). There were six arginine residues (orange sticks) in DNA recognition helix α3 in vIRF1 (Arg163 and Arg164 can interact with DNA through hydrogen bonds), while there were only two arginine residues (green sticks) potentially involved in DNA binding in α3 of vIRF2DBD. Arg147 and Asn149 in L2 of vIRF1DBD, which made main-chain interactions with the phosphate backbone, corresponded to residues Arg61 and Arg63 in α2 of vIRF2DBDA, respectively. (B) Structural superposition of DBDs between vIRF2 (green) and the IRF2-DNA complex (blue; PDB ID 2IRF). The DNA-binding residues Asn80, Arg82, Cys83, and Asn86 (blue sticks) in α3 of IRF2 correspond to residues Cys79, Gly81, Arg82, and Arg85 (green sticks) in vIRF2DBD.
Although no significant changes in the position and orientation of the canonical DNA recognition helix α3 in the major grooves of vIRF1DBD and IRF2DBD (RMSD, 0.57 and 0.43 Å for C-α carbon atoms, respectively), the DNA specificity-determining residues in this helix may be distinct. There were only 2 (Arg82 and Arg85 [discussed below]) and up to 6 arginine residues in α3 of vIRF2DBD and vIRF1DBD (Fig. 3B and 4A), respectively, which indicates their different cellular DNA-binding capacities and specificities. Superimposition of the putative DNA-binding residues Arg82 and Arg85 in the two monomers of vIRF2DBD showed they are highly conserved and have identical conformations (Fig. 3B). However, among the four molecules in the asymmetric unit of apo-form vIRF1DBD (PDB ID 4HLX), the DNA-binding residues Arg163 and Arg171 seemed to be highly flexible and different in conformation. Moreover, superimposition of vIRF2DBD with the vIRF1DBD-DNA complex revealed bad steric clashes with the DNA conformation (Fig. 4A), which may also be caused by their different DNA-binding specificities.
Another remarkable difference was the absence of a highly conserved histidine residue in L1 of vIRF2 (Fig. 4B), which has been shown to be an important determinant of differential cellular-IRF–DNA binding. This particular histidine made a water-mediated hydrogen bond to an adenine base in the minor groove in the cellular IRFs (36), but the corresponding residue was an asparagine in vIRF2DBD (Asn42) (Fig. 3A and 4B) and a proline in vIRF1DBD (Fig. 3A), which had no direct recognition in the minor DNA groove. In the IRF2DBD-DNA complex, the interaction with the major groove of the GAAA sequence was mediated by 4 residues (Asn80, Arg82, Cys83, and Asn86) in the recognition helix α3, and Arg82 and Asn86 form hydrogen bonds with the phosphate group to trigger DNA structure deformations by IRF2 binding. In vIRF2DBD, the corresponding residues were Cys79, Gly81, Arg82, and Arg85 in α3 (Fig. 3A and 4B). Therefore, by analyzing the conserved residues in this region for recognition of the DNA major groove, we suggest that there may be some similarities in the recognition sequences for IRF2 and vIRF2, with the positive Arg82 in both structures (Fig. 4B). However, their superimposition also revealed somewhat bad steric clashes, which indicated their different DNA-binding specificities (Fig. 4B).
The roles of Arg82 and Arg85 in DNA binding and transcription regulation of vIRF2.
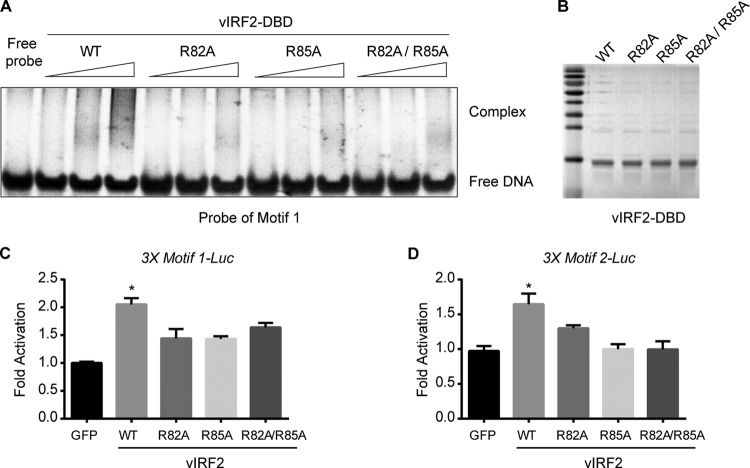
From the structure analysis of vIRF2DBD, we inferred that the vIRF1 DBD had potential DNA-binding properties and that Arg82, as well as Arg85, was critical for its DNA-binding ability. Gel EMSA was applied to determine the in vitro DNA-binding ability of vIRF2DBD to the top 2 ChIP-seq-based vIRF2-binding motifs, motif 1 and motif 2. DNA probes were synthesized with the sequences of 3 tandem copies of vIRF2-binding motif 1 or 2 or 1 copy of LANA-binding motif 1 as a negative control. We showed that vIRF2DBD induced strong gel shifts for probe of motif 1 and motif 2 but only a very weak shift, if any, for probe of LANA-binding motif (see Fig. S3A in the supplemental material). Moreover, the gel shifts of probes for motif 1 and motif 2 were strengthened in a vIRF2DBD dose-dependent manner (see Fig. S3A in the supplemental material). These data suggested that vIRF2DBD bound to the ChIP-seq-based vIRF2-binding motif efficiently in vitro. Then, we tested whether Arg82 and Arg85 were required for their DNA-binding abilities by a series of site-directed mutations. Our data showed that both a single mutant (R82A or R85A, respectively) and a double mutant induced much weaker gel shifts for probes of both motif 1 and motif 2 than wild-type (WT) vIRF2DBD (Fig. 5A and B; see Fig. S3B in the supplemental material), indicating these residues are critical for its in vitro DNA-binding ability. To further verify the binding of vIRF2 to its binding motifs, we constructed artificial 3× motif 1 and 3× motif 2 promoter luciferase reporters, and we also showed that vIRF2 can upregulate the artificial motif 1 and motif 2 promoter-driven luciferase reporter activities (Fig. 5C and D) by up to 2-fold, which suggested that vIRF2 might upregulate gene expression by binding to motif 1 or 2.
FIG 5.
Interaction of vIRF2DBD with consensus vIRF2-binding motifs in vitro. (A) Validation of direct vIRF2-DNA binding by EMSA using vIRF2DBD and the DNA probe representing vIRF2-binding motif 1. WT vIRF2DBD induced significant gel shift of probe 1 in a dose-dependent manner; however, the R82A, R85A, or R82A/R85A mutant induced much weaker gel shifts for probe 1 than wild-type vIRF2DBD. (B) Purified wild-type vIRF2 DBD and its mutants by Coomassie brilliant blue staining. (C and D) vIRF2 upregulated the artificial motif 1 and motif 2 promoter-driven luciferase reporter activity. 3× Motif1-Luc and 3× Motif2-Luc reporters were constructed by inserting annealed DNA fragments containing 3 tandem copies of representative vIRF2-binding motif 1 or 2 sequences into the pGL6-TA vector. WT vIRF2DBD induced up to 2-fold activation of the luciferase reporter compared to the control, while R82A, R85A, or R82A/R85A had a defect in upregulating reporter activity. The data are shown as means plus SEM; n = 3. *, P < 0.05.
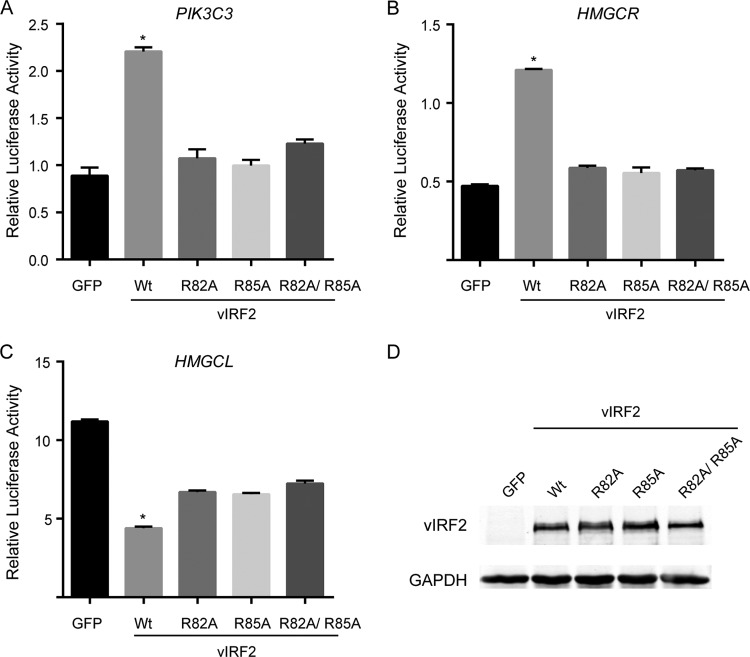
Since vIRF2 was involved in the transcription regulation of its target genes, we wondered whether its DNA-binding activity is required for its transcription regulation function. By constructing the promoter reporter for PIK3C3, HMGCR, and HMGCL, we showed that vIRF2 can significantly upregulate the reporter activity of PIK3C and HMGCR or downregulate the activity of HMGCL, which is similar to the result of qRT-PCR. These data suggest that vIRF2 may be directly involved in the transcriptional regulation of these genes. Moreover, all the mutations, i.e., R82A, R85A, and R82A/R85A, severely reduced the regulation function of vIRF2 (Fig. 6A to D). Similar to the reporter assay, we showed that mutation of either R82A, R85A, or R82A/R85A was not able to regulate mRNA levels of PIK3C3, HMGCR, and HMGCL (see Fig. S4 in the supplemental material). These data suggested that DNA-binding activity is required for the vIRF2 transcription regulation function. However, the promoter regions of PIK3C3, HMGCR, and HMGCL did not contain sequences similar to motif 1 or 2. Also, we were not able to find a new consensus vIRF2-binding motif within the promoter regions of PIK3C3, HMGCR, and HMGCL. Currently, it is unknown why vIRF2 upregulated the expression of PIK3C3 and HMGCR while it downregulated the expression of HMGCL. Since vIRF2 has only a DNA-binding domain and not a transcription activation domain, we thought that vIRF2 might regulate gene expression differentially by binding to certain sequences and recruit either a transcription coactivator or corepressor through currently unknown mechanisms.
FIG 6.
Roles of Arg82 and Arg85 involved in transcription regulation of vIRF2. (A to C) vIRF2 can significantly upregulate the reporter activity of PIK3C and HMGCR promoters or downregulate the activity of the HMGCL promoter, while all the mutations R82A, R85A, and R82A/R85A severely abolished the regulation function of vIRF2. The data are shown as means plus SEM; n = 3. *, P < 0.05. (D) Expression of the wild type and mutants of vIRF2 detected by Western blotting. The results showed Arg82 and Arg85 are required for transcription regulation of vIRF2. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
DISCUSSION
KSHV vIRF2 is a bona fide DNA-binding transcription factor.
vIRFs were believed to be unable to directly bind DNA as a result of the absence of the tryptophan residues that are essential for DNA binding (9). However, some evidence suggested that vIRFs might play unexpected roles in transcription regulation, and recent studies showed vIRF4 may compete with c-IRF4 for binding to the specific promoter region of the c-Myc gene (21, 38). A consensus sequence interacting with vIRF1 from a pool of random oligonucleotides in the KSHV genome was identified recently (21). However, it remained controversial whether vIRF1DBD interacted with DNA, whereas full-length vIRF1 did not. Similarly, the short-form vIRF2 was previously reported to interact with the NF-κB consensus binding site, but not the full-length protein (10). We should ask why only the DBD and not the full-length protein could interact with DNA and whether vIRFs could serve as bona fide DNA-binding transcription factors. One of the possible answers is that the natural confirmation of the vIRF DBD in the full-length protein setting was not allowed to interact with DNA. However, there was another possible answer, i.e., that the real vIRF binding sequence was not identified.
To investigate the function of vIRF2 as a transcription factor, we mapped the DNA-binding sites for vIRF2 in a genome-wide manner using ChIP-seq. This was the first overall DNA-binding profile for KSHV vIRFs in the human genome. We found that vIRF2 bound to the 100 promoter regions of ChIP-seq-based vIRF2 target genes. More importantly, we showed that vIRF2 either downregulated or upregulated the transcription of some of these target genes, suggesting a role of vIRF2 in transcription regulation. Mutation of Arg82 and Arg85 can abolish the regulatory function of vIRF2 on the promoter reporter activities of target genes. Our study provides essential evidence to show that vIRF2 could act as a transcription factor of its target genes through the DNA-binding activity.
vIRF2 may contribute to KSHV infection and pathogenesis through versatile functions.
The function of vIRF2 in subverting the antiviral innate immune response has been well characterized (12). It is possible that vIRF2 may have unexpected functions through transcription regulation of a panel of its target genes. The potential vIRF2-binding motifs from our ChIP-seq showed significant homology with those in several DNA-binding transcriptional factors involved in various functions, including several cellular-IRF secondary binding motifs. By ChIP-qPCR and qRT-PCR, we verified that vIRF2 can significantly upregulate the expression of PIK3C3 and HMGCR and downregulate HMGCL by targeting their promoter regions. PIK3C3 is a catalytic subunit of the phosphatidylinositol 3-kinase (PI3K) complex that mediates the formation of phosphatidylinositol 3-phosphate, which plays a key role in initiation and maturation of autophagosomes (39). HMGCR is the rate-limiting enzyme for cholesterol synthesis (40), while HMGCL is a mitochondrial enzyme that catalyzes the final step of leucine degradation and plays a key role in ketone body metabolism, which is critical for tumor progression and metastasis (41, 42). Therefore, vIRF2 may be involved in regulating cellular metabolism by regulating the expression of PIK3C3, HMGCR, and HMGCL after KSHV infection.
Variable loop conformations and arginine distribution in vIRFs and IRFs.
We have provided the high-resolution structure of vIRF2DBD to delineate the molecular basis for vIRF2 as a transcription factor. Structural comparisons of vIRF2DBD revealed that the conformations of L1 to L3 are significantly different from those in vIRF1 and IRFs (21, 36), which are highly disordered in solution and are involved in interactions with the DNA phosphate backbone. The varying intrinsic flexibility of loops and tails may serve as a mechanism to modulate the binding specificity of vIRF family members and to respond to a larger population of diverse promoter sites in IRFs. The structural superposition also showed the notable difference in number and distribution of DNA specificity-determining arginine residues in helix α3, which may cause their distinct cellular DNA-binding capacities and specificities. The in vitro and in vivo DNA-binding studies of vIRF2DBD gave strong support for its role in direct interaction with cellular DNA by its key residues, Arg82 and Arg85.
In summary, for the first time, we mapped the human genome-wide binding sites of vIRF2 by ChIP-seq and found the potential promoter regions of cellular genes that vIRF2 can bind. Structural analysis, combined with in vitro and in vivo functional studies, provided essential evidence that vIRF2 can act as a DNA-binding transcription factor of its target genes in various physiological processes during KSHV infection.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of beamline station 3W1A of the Beijing Synchrotron Radiation Facility (BSRF) for providing technical support and for many fruitful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01392-15.
REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi's sarcoma and its associated herpesvirus. Nat Rev Cancer 10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cousins E, Nicholas J. 2014. Molecular biology of human herpesvirus 8: novel functions and virus-host interactions implicated in viral pathogenesis and replication. Recent Results Cancer Res 193:227–268. doi: 10.1007/978-3-642-38965-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coscoy L. 2007. Immune evasion by Kaposi's sarcoma-associated herpesvirus. Nat Rev Immunol 7:391–401. doi: 10.1038/nri2076. [DOI] [PubMed] [Google Scholar]
- 5.Burysek L, Yeow WS, Pitha PM. 1999. Unique properties of a second human herpesvirus 8-encoded interferon regulatory factor (vIRF-2). J Hum Virol 2:19–32. [PubMed] [Google Scholar]
- 6.Jacobs SR, Damania B. 2011. The viral interferon regulatory factors of KSHV: immunosuppressors or oncogenes? Front Immunol 2:19. doi: 10.3389/fimmu.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs SR, Gregory SM, West JA, Wollish AC, Bennett CL, Blackbourn DJ, Heise MT, Damania B. 2013. The viral interferon regulatory factors of Kaposi's sarcoma-associated herpesvirus differ in their inhibition of interferon activation mediated by Toll-like receptor 3. J Virol 87:798–806. doi: 10.1128/JVI.01851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi YB, Sandford G, Nicholas J. 2012. Human herpesvirus 8 interferon regulatory factor-mediated BH3-only protein inhibition via Bid BH3-B mimicry. PLoS Pathog 8:e1002748. doi: 10.1371/journal.ppat.1002748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baresova P, Pitha PM, Lubyova B. 2013. Distinct roles of Kaposi's sarcoma-associated herpesvirus-encoded viral interferon regulatory factors in inflammatory response and cancer. J Virol 87:9398–9410. doi: 10.1128/JVI.03315-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baresova P, Pitha PM, Lubyova B. 2012. Kaposi sarcoma-associated herpesvirus vIRF-3 protein binds to F-box of Skp2 protein and acts as a regulator of c-Myc protein function and stability. J Biol Chem 287:16199–16208. doi: 10.1074/jbc.M111.335216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuld S, Cunningham C, Klucher K, Davison AJ, Blackbourn DJ. 2006. Inhibition of interferon signaling by the Kaposi's sarcoma-associated herpesvirus full-length viral interferon regulatory factor 2 protein. J Virol 80:3092–3097. doi: 10.1128/JVI.80.6.3092-3097.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang C, Lee J-S, Jung JU. 2008. Immune evasion in Kaposi's sarcoma associated oncogenesis. Semin Cancer Biol 18:423–436. doi: 10.1016/j.semcancer.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenner RG, Alba MM, Boshoff C, Kellam P. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J Virol 75:891–902. doi: 10.1128/JVI.75.2.891-902.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci U S A 93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimring JC, Goodbourn S, Offermann MK. 1998. Human herpesvirus 8 encodes an interferon regulatory factor (IRF) homolog that represses IRF-1-mediated transcription. J Virol 72:701–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutocheluh M, Hindle L, Areste C, Chanas SA, Butler LM, Lowry K, Shah K, Evans DJ, Blackbourn DJ. 2011. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor-2 inhibits type 1 interferon signalling by targeting interferon-stimulated gene factor-3. J Gen Virol 92:2394–2398. doi: 10.1099/vir.0.034322-0. [DOI] [PubMed] [Google Scholar]
- 17.Takaoka A, Tamura T, Taniguchi T. 2008. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci 99:467–478. doi: 10.1111/j.1349-7006.2007.00720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Royer WE Jr. 2010. Structural insights into interferon regulatory factor activation. Cell Signal 22:883–887. doi: 10.1016/j.cellsig.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Areste C, Mutocheluh M, Blackbourn DJ. 2009. Identification of caspase-mediated decay of interferon regulatory factor-3, exploited by a Kaposi sarcoma-associated herpesvirus immunoregulatory protein. J Biol Chem 284:23272–23285. doi: 10.1074/jbc.M109.033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park J, Lee MS, Yoo SM, Jeong KW, Lee D, Choe J, Seo T. 2007. Identification of the DNA sequence interacting with Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 1. J Virol 81:12680–12684. doi: 10.1128/JVI.00556-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hew K, Dahlroth SL, Venkatachalam R, Nasertorabi F, Lim BT, Cornvik T, Nordlund P. 2013. The crystal structure of the DNA-binding domain of vIRF-1 from the oncogenic KSHV reveals a conserved fold for DNA binding and reinforces its role as a transcription factor. Nucleic Acids Res 41:4295–4306. doi: 10.1093/nar/gkt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. 2010. PeakAnalyzer: genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang W, Loganantharaj R, Schroeder B, Fargo D, Li L. 2013. PAVIS: a tool for peak annotation and visualization. Bioinformatics 29:3097–3099. doi: 10.1093/bioinformatics/btt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Sheldrick GM. 2010. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr 66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC. 2000. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 35.Panne D, Maniatis T, Harrison SC. 2007. An atomic model of the interferon-beta enhanceosome. Cell 129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Ioannes P, Escalante CR, Aggarwal AK. 2011. Structures of apo IRF-3 and IRF-7 DNA binding domains: effect of loop L1 on DNA binding. Nucleic Acids Res 39:7300–7307. doi: 10.1093/nar/gkr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38(Suppl 2):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HR, Doğanay S, Chung B, Toth Z, Brulois K, Lee S, Kanketayeva Z, Feng P, Ha T, Jung JU. 2014. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 (vIRF4) targets expression of cellular IRF4 and the myc gene to facilitate lytic replication. J Virol 88:2183–2194. doi: 10.1128/JVI.02106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levine B, Mizushima N, Virgin HW. 2011. Autophagy in immunity and inflammation. Nature 469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marijanovic Z, Laubner D, Möller G, Gege C, Husen B, Adamski J, Breitling R. 2003. Closing the gap: identification of human 3-ketosteroid reductase, the last unknown enzyme of mammalian cholesterol biosynthesis. Mol Endocrinol 17:1715–1725. doi: 10.1210/me.2002-0436. [DOI] [PubMed] [Google Scholar]
- 41.Mack M, Schniegler-Mattox U, Peters V, Hoffmann GF, Liesert M, Buckel W, Zschocke J. 2006. Biochemical characterization of human 3-methylglutaconyl-CoA hydratase and its role in leucine metabolism. FEBS J 273:2012–2022. doi: 10.1111/j.1742-4658.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 42.Martinez-Outschoorn UE, Lin Z, Whitaker-Menezes D, Howell A, Lisanti MP, Sotgia F. 2012. Ketone bodies and two-compartment tumor metabolism: stromal ketone production fuels mitochondrial biogenesis in epithelial cancer cells. Cell Cycle 11:3956–3963. doi: 10.4161/cc.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.