Abstract
Background
Although genetic studies have reported a number of loci associated with cutaneous melanoma (CM) risk, a comprehensive synopsis of genetic association studies published in the field and systematic meta-analysis for all eligible polymorphisms have not been reported.
Methods
We systematically annotated data from all genetic association studies published in the CM field (n = 145), including data from genome-wide association studies (GWAS), and performed random-effects meta-analyses across all eligible polymorphisms on the basis of four or more independent case–control datasets in the main analyses. Supplementary analyses of three available datasets derived from GWAS and GWAS-replication studies were also done. Nominally statistically significant associations between polymorphisms and CM were graded for the strength of epidemiological evidence on the basis of the Human Genome Epidemiology Network Venice criteria. All statistical tests were two-sided.
Results
Forty-two polymorphisms across 18 independent loci evaluated in four or more datasets including candidate gene studies and available GWAS data were subjected to meta-analysis. Eight loci were identified in the main meta-analyses as being associated with a risk of CM (P < .05) of which four loci showed a genome-wide statistically significant association (P < 1 × 10−7), including 16q24.3 (MC1R), 20q11.22 (MYH7B/PIGU/ASIP), 11q14.3 (TYR), and 5p13.2 (SLC45A2). Grading of the cumulative evidence by the Venice criteria suggested strong epidemiological credibility for all four loci with genome-wide statistical significance and one additional gene at 9p23 (TYRP1). In the supplementary meta-analyses, a locus at 9p21.3 (CDKN2A/MTAP) reached genome-wide statistical significance with CM and had strong epidemiological credibility.
Conclusions
To the best of our knowledge, this is the first comprehensive field synopsis and systematic meta-analysis to identify genes associated with an increased susceptibility to CM.
CONTEXT AND CAVEATS
Prior knowledge
Previous studies, including genome-wide association studies, have identified genetic variants associated with cutaneous melanoma (CM).
Study design
Data were collected from all genetic association studies in the CM field. Meta-analyses were performed for all polymorphisms identified in four or more independent case–control datasets. Supplementary meta-analyses were done for polymorphisms for which three datasets from GWAS were available. The epidemiological credibility of statistically significant associations between loci and CM was measured using the Human Genome Epidemiology Network Venice criteria.
Contribution
This is the first field synopsis and meta-analysis of genetic variants associated with CM. The main meta-analyses identified eight loci statistically significantly associated with CM, five of which had genome-wide statistical significance and strong epidemiological credibility. Supplementary analyses identified one additional loci with genome-wide statistical significance and strong epidemiological credibility for an association with CM. The accumulated evidence was reported on the publicly accessible regularly updated MelGene website (www.MelGene.org).
Implications
This study comprehensively reports on the genome-wide statistical significance and epidemiogical credibility of CM susceptibility genes. MelGene will provide a potential forum to report genes associated with CM as they are identified in future genetic studies.
Limitations
Some studies may have been excluded from the analyses and random errors may be found in some of the study entries. Because the meta-analyses were based on summary-level data, some potential confounders and other types of bias could not be accounted for, and both gene–gene and gene–environment interactions could not be assessed. Also, as is standard of systematic meta-analyses, publication bias could not be ruled out.
From the Editors
The incidence of cutaneous melanoma (CM) has steadily increased among populations of European descent during the past five decades. Metastatic melanoma is the leading cause of skin cancer–related mortality and is characterized by a poor prognosis and limited treatment options (1,2). As early detection of CM provides the best opportunity for cure, identification of high-risk individuals is an important preventative strategy for reducing mortality. Susceptibility to CM is likely determined by an interaction of environmental risk factors including excessive exposure to ultraviolet radiation (3) and genetically controlled phenotypic traits such as nevus propensity, red or blonde hair, light-colored eyes, fair skin, and limited tanning ability (4,5). The heritability, that is, the contribution of genetic factors to CM risk, has been estimated to be 18%–55% (6,7). Approximately, 5%–10% of patients exhibit an autosomal-dominant hereditary form of the disease, which can be caused by rare highly penetrant gene mutations, such as in the cyclin-dependent kinase inhibitor 2A gene (CDKN2A) on chromosome 9p21, the cyclin-dependent kinase 4 gene (CDK4) on chromosome 12q14, and other potentially novel loci (8,9). However, for the majority of CM patients, the genetic architecture is more complex, involving numerous genetic risk factors of relatively high frequencies but low penetrance (10,11). Among the low-penetrance genes, the most consistent association with CM has been reported for the gene encoding melanocortin-1 receptor (MC1R) on chromosome 16q24.3 (12,13). Other putative risk alleles involved in various cellular pathways such as pigmentation, DNA repair, oxidation stress, apoptosis, cell growth, and melanocyte differentiation and migration have also been implicated in melanoma susceptibility (14).
Within the last 15 years, approximately 150 genetic association studies including two GWAS (15,16) have been published that claim or refute associations between CM risk and putative melanoma genes. Although highly valuable for the understanding of the underlying pathogenesis and for the assessment of risk prediction, the increasing amount of information has become difficult to evaluate and interpret. To assess the evidence and strength of genetic associations with CM, we have collected and comprehensively cataloged all genetic association studies published in the field and conducted systematic meta-analyses for all eligible polymorphisms. Systematic field synopses and meta-analyses have previously been performed for other diseases using similar study designs (17–22). Furthermore, we have graded the epidemiological validity of nominally statistically significant meta-analysis results by applying the Venice criteria proposed by the Human Genome Epidemiology Network. Detailed summaries of all association studies and meta-analysis results are available on a regularly updated publicly available online database, MelGene (23), which currently highlights the most promising genetic loci associated with CM risk.
Methods
Collection and Management of Eligible Studies
Search Strategy.
We searched the PubMed database (24) by using the search terms “melanoma AND associat* AND gene*” for studies published on or before March 31, 2009, after which daily PubMed searches were performed (Figure 1). In addition, we searched the references of included publications and the table of contents in relevant journals on genetics, dermatology, and oncology. We also searched PubMed using the keywords “melanoma AND [name of every gene identified in included publications].” Furthermore, we screened the Human Genome and Epidemiology Network Navigator (25) for additional publications as well as the Melanoma Molecular Maps Project (26), a large online database, which reports published melanoma genetic association studies.
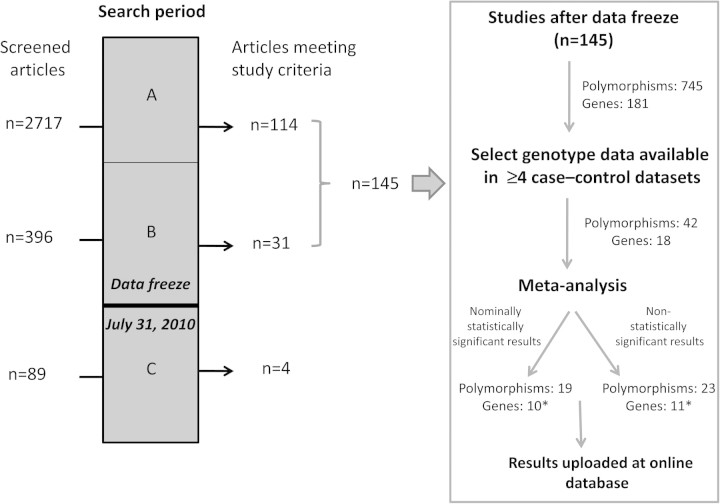
Figure 1.
Overview of the literature search and selection strategy of published genetic association studies of cutaneous melanoma. A) An initial PubMed search for “melanoma AND associat* AND gene*” was performed to identify articles published on or before March 31, 2009. Screening of those as well as of cross-references in relevant articles and additional search of the Human Genome and Epidemiology Network Navigator (25) and the Melanoma Molecular Maps Project (26) databases identified publications that fulfilled our inclusion criteria. B) Following the initial screening, daily PubMed searches for “melanoma AND associat* AND gene*” were performed to identify additional articles published before July 31, 2010 (the data freeze). C) The publication search continued after the date of the data freeze and yielded four additional articles until October 14, 2010. Data from articles of this period have not been included in the meta-analyses of the current article but are included in the online database. Asterisks indicate that the number of nominally statistically significant and non-statistically significant genes does not sum to the total number of genes because genes encoding cyclin-dependent kinase 2A, the melanocortin-1 receptor, and the vitamin D receptor contained polymorphisms showing both statistically significant (P < .05) and non-statistically significant genetic variants identified by meta-analysis.
Inclusion and Exclusion Criteria.
Studies included in MelGene had to assess the association between a polymorphism and CM. We included all relevant case–control studies that have been published in a peer-reviewed journal. We did not include those without healthy control subjects (ie, studies that examined genetic associations with phenotypic variables among individuals with CM alone or among different types of cancer). Studies with a family-based approach were listed in the qualitative gene summary overview on MelGene but were not subjected to meta-analysis. We excluded highly penetrant mutations only present in patients but not found in control subjects. We included studies on polymorphisms with three or more alleles (such as the HLA and MICA genes) in the qualitative gene summaries, but considered them for meta-analysis only if genotype frequencies of one allele compared with all other alleles were consistently reported. Abstracts from conference proceedings or scientific meetings were excluded. The criteria to establish a diagnosis had to be a histologically proven CM, either invasive or in situ. We excluded studies of patients with non-CM (including uveal melanoma) or with metastatic disease of an unknown primary cancer. Whenever an article studied more than one phenotype, only CM-specific data were included. Although publication in the English language was part of the criteria for this study, no articles published in a language other than English were identified.
Genotype and Allele Distributions.
We used the National Center for Biotechnology Information Single Nucleotide Polymorphism Database identifiers when provided (rs numbers). If an rs number was not specified in the respective publications, we generally used the most common definition provided in the primary publications. Genotype distributions were extracted from eligible publications for each polymorphism and listed on MelGene. Whenever allele frequencies, but not genotype frequencies, were reported in the original articles, we calculated the genotype frequencies on the basis of the reported allele frequencies and sample sizes, assuming that no deviations from Hardy–Weinberg equilibrium occurred, unless reported otherwise. We contacted the authors of publications with missing genotype data by email and if no response was received, the respective studies were labeled as no data provided on MelGene, unless there was information on odds ratios (ORs) and/or corresponding 95% confidence intervals (CIs) calculated on the basis of allelic contrasts. Approximately, 3% of all genotypes remained unavailable. For studies of overlapping populations, we included only one study in the respective meta-analyses, and whenever possible, the study with the largest sample size was included.
GWAS and GWAS-Replication Studies.
Because of the absence of publicly available CM-GWAS datasets, we extracted the allele frequencies or per-allele odds ratios (15,16) from the original GWAS publications and included them in the main meta-analyses when applicable. We also included data from GWAS-replication studies, that is, studies assessing the association of melanoma with selected variants derived from GWAS on CM-related traits including hair, eye, and skin pigmentation; basal cell carcinoma; and melanocytic nevi (27–31). To capture all the important information from the limited number of GWAS and GWAS-replication studies, we also performed supplementary meta-analyses on polymorphisms for which only three datasets from CM-GWAS and/or GWAS-replication datasets were available (27–29).
Statistical Analyses
Meta-analyses.
Random-effects summary odds ratios and 95% confidence intervals (32) were calculated on the basis of study-specific unadjusted odds ratios and 95% confidence intervals using allelic contrasts for all variants with case–control genotype data available from at least four independent datasets in the main meta-analyses, and for variants with only three case–control datasets derived from CM-GWAS and GWAS-replication studies in the supplementary meta-analyses. The main meta-analyses were first performed on all datasets regardless of patient ancestry. Summary odds ratios and 95% confidence intervals were also calculated after stratification for different ancestries if three or more such datasets existed and were applicable only to datasets of European ancestry. In addition, for this study, dominant and recessive models were assessed by meta-analysis on all eligible polymorphisms following the same inclusion and exclusion criteria. Genome-wide statistical significance (P < 1 × 10−7) was determined after including all eligible datasets without further adjustment for multiple comparisons. All statistical tests were two-sided. Meta-analysis results are displayed on MelGene for each eligible polymorphism as forest plots and as cumulative meta-analyses summarizing the evolvement of the summary effect estimate over time.
Sensitivity Analyses and Between-Study Heterogeneity.
The sensitivity analyses of the main meta-analyses results entailed calculating summary odds ratios and 95% confidence intervals for all studies excluding the initial report and after excluding studies violating Hardy–Weinberg equilibrium in control subjects according to a χ2 test implemented in the Hardy–Weinberg R package (P ≤ .05). When cell counts were below five, Fisher exact test was used. Between-study heterogeneity was assessed by calculating the I 2 heterogeneity metric. I 2 is estimated by the Q statistic (I 2 = [(Q − df)/Q] × 100) and describes the percentage of variability in point estimates that is because of heterogeneity rather than sampling error (33,34). Contrary to the Q statistic, I 2 does not depend on the number of studies and can be compared across meta-analysis results calculated from different sample sizes. Generally, values greater than 50% represent high heterogeneity (33,34). When there are few studies, both Q and I 2 carry considerable uncertainty and should be interpreted cautiously (35).
Estimating the Credibility of Statistically Significant Associations.
Each nominally statistically significant result of the main meta-analyses was graded on the basis of the Human Genome Epidemiology Network Venice criteria for the assessment of cumulative evidence of genetic associations (36,37). These criteria take into account the amount of evidence (sample size measured as the number of minor alleles), consistency of replication (heterogeneity across studies measured as I 2), and protection from bias [the bias reason, in particular including sensitivity analysis as outlined above, assessments of the strength of the association, small-study bias (38), and evidence for an excess of statistically significant results (39)]. On the basis of the analysis, the overall epidemiological credibility was graded as strong (grade A), moderate (grade B), or weak (grade C). For more details on these criteria, see the Supplementary Methods (available online).
The MelGene Database
Demographic details of the studies included in our meta-analyses, genotype data, forest plots, and cumulative meta-analyses results can be found on the publicly available continuously updated MelGene website (http://www.melgene.org) (23).
Results
Literature Searches
After an initial screening of 3113 articles, 145 individual publications reporting on 745 genetic variants in 181 different genes were included on MelGene at the time of the data freeze on July 31, 2010 (Figure 1), including two CM-GWAS (15,16). Both the number of polymorphisms evaluated across all publications and the combined sample size per publication have steadily increased in the past 20 years, with a median of 12 polymorphisms (interquartile range [IQR] = 1–37) and 985 combined patients and control subjects (IQR = 330–1781) between 1992 and 2002, and a median of 82 polymorphisms (IQR = 56–195) and 12613 combined patients and control subjects (IQR = 10 272–20 501) between 2002 and 2010. The most extensively studied genes include MC1R, the vitamin D receptor (VDR), and the agouti signaling protein (ASIP) (18%, 7%, and 6% of the publications listed on MelGene, respectively) (Supplementary Figure 1, available online).
Results of the Main Meta-analyses
Of the 745 polymorphisms included on MelGene, 42 variants in 18 loci fulfilled our criteria for meta-analysis with data available from at least four independent case–control datasets. The meta-analyses were conducted on the basis of a median of six independent case–control datasets (IQR = 5–9). For the number of combined patients and control subjects in specific datasets, see Supplementary Table 1 (available online). Of the variants analyzed, 19 (45%) of 42 in eight loci had nominally statistically significant associations with CM (P < .05) after inclusion of all ethnicities (Table 1 and Supplementary Table 1, available online). These loci comprise the chromosomal region 5p15.33 containing the genes encoding telomerase reverse transcriptase (TERT) and cleft lip and palate-associated transmembrane protein 1-like protein (CLPTM1L); 5p13.2 containing the gene that encodes solute carrier family 45 member 2 (SLC45A2); 9p23 containing the gene for tyrosinase-related protein 1 (TYRP1); 9p21.3 containing CDKN2A; 11q14.3 containing the gene encoding tyrosinase (TYR); 12q13.11 containing VDR; 16q24.3 containing MC1R; and 20q11.22 containing the gene ASIP, and genes encoding the myosin heavy chain 7B (MYH7B) and the phosphatidylinositol glycan anchor biosynthesis class U protein (PIGU). Stratification for European ancestry when applicable did not change the overall results (the ancestry-specific meta-analysis results are displayed on the respective forest plots in Supplementary Figure 2, available online). The MelGene website continues to be updated with newly published studies (23).
Table 1.
Genetic variants associated with cutaneous melanoma after meta-analyses of at least four independent datasets (main meta-analyses) d
| Chromosome | Gene (nearest gene)† | Polymorphism | Location, bp‡ | Allele contrast | Ethnicity | No. of datasets | No. of subjects§ | MAF | OR (95% CI) | P | N minor | I 2 | Bias reason | Venice overall grade‖ |
| 5p15.33 | CLPTM1L | rs401681 | 1375087 | T vs C | All | 7 | 47 195 | 0.450 | 1.15 (1.08 to 1.22) | 9.6 × 10−6 | 42 771 | 27 | Low OR | C |
| 5p13.2 | SLC45A2 | rs16891982 | 33987450 | C vs G | All | 9 | 14 880 | 0.068 | 0.40 (0.33 to 0.47) | 4 × 10−27 | 1644 | 29 | NA | A |
| 9p23 | (TYRP1) | rs1408799 | 12662097 | T vs C | All | 4 | 43 166 | 0.258 | 0.86 (0.80 to 0.93) | 9.1 × 10−5 | 22 196 | 0 | NA | A |
| 9p21.3 | CDKN2A | rs3088440 | 21958159 | T vs C | All | 4 | 3867 | 0.077 | 1.25 (1.03 to 1.51) | .026 | 704 | 24 | F, HWE | C¶ |
| 11q14.3 | TYR | rs1126809 | 88657609 | A vs G | All | 6 | 46 939 | 0.295 | 1.22 (1.14 to 1.31) | 2.7 × 10−8 | 27 940 | 29 | NA | A¶ |
| 12q13.11 | VDR | rs1544410 | 46526102 | A vs G | All | 6 | 7440 | 0.400 | 0.89 (0.82 to 0.97) | .007 | 5857 | 24 | Low OR | C |
| 16q24.3 | MC1R | rs1805007 (R151C) | 88513618 | T vs C | All | 21 | 27 747 | 0.078 | 1.83 (1.56 to 2.15) | 2.1 × 10−13 | 4764 | 69 | NA | A¶ |
| 20q11.22 | MYH7B | rs1885120 | 33040650 | C vs G | All | 4 | 6787 | 0.073 | 1.59 (1.41 to 1.79) | 7.4 × 10−15 | 1306 | 0 | NA | A |
CI = confidence interval, F = statistical significance (P < .05) lost when first study was excluded, HWE = statistical significance (P < .05) lost when studies violating Hardy–Weinberg equilibrium in control subjects were excluded, I 2 = estimate of percentage of between-study heterogeneity that is beyond chance, Low OR = odds ratio less than 1.15, MAF = minor allele frequency in control subjects, NA = not applicable, N minor = number of minor alleles in patients and control subjects combined across all included datasets, OR = odds ratio. Allelic ORs, CIs and P values (two-sided) were calculated using the DerSimonian–Laird random-effects model (32).
“Gene” denotes the gene in the respective locus into which the listed SNP maps, whereas “nearest gene” refers to the most proximal gene in the respective locus if the SNP itself does not map into a gene region. It should be noted that these genes are not necessarily the genes that are functionally affected by the genetic association finding in this locus.
The location is on the basis of the Human Genome Build hg19 (40).
This number includes both patients and control subjects.
Each statistically significant meta-analysis result was graded according to the Human Genome Epidemiology Network Venice criteria. Venice grading: A = grade A (strong epidemiological credibility), B = grade B (modest epidemiological credibility), and C = grade C (weak epidemiological credibility).
If multiple polymorphisms showed a statistically significant association in the same locus, only the variant with the best Venice grading is listed here. When the Venice grading yielded equivalent scores the variant with the smallest P is listed.
Genetic Variants Nominally Statistically Significantly Associated With CM in the Main Meta-analyses
The median allelic summary odds ratio was 1.37 (range = 1.05–3.04) for all statistically significant polymorphisms (P < .05) on the basis of a median of seven independent datasets (IQR = 4–19; Supplementary Table 1, available online). To account for the impact of loci with multiple associated single-nucleotide polymorphisms (SNPs), the summary odds ratio was also calculated for the best result per gene (defined according to the SNP with the best Venice score and then according to P), yielding a median summary odds ratio of 1.25 (range = 1.12–2.50). Four loci achieved genome-wide statistical significance (P < 1 × 10−7), namely MC1R (rs1805007, rs1805008, and rs1805009), MYH7B/PIGU (rs1885120 and rs910873), TYR (rs1126809 and rs1393350), and SLC45A2 (rs16891982; Supplementary Table 1, available online). Dominant and recessive meta-analysis models were also calculated, but neither provided evidence for the association of additional loci with CM nor did the statistical significance of the associations change substantially (data not shown). Twenty-three SNPs in 11 genes did not show statistical significance for association with CM (P ≥ .05) upon meta-analysis, neither on combination of all ancestries nor after stratification for European ancestry (Supplementary Table 1, available online). The sample size was statistically significantly different by Wilcoxon rank sum test when compared with previously reported statistically significant findings (P < .001). For the sample sizes of the individual non-statistically significant polymorphisms, see Supplementary Table 1 (available online). Of note, none of these non-statistically significant polymorphisms have been reported as associated with CM in a GWAS (15,16), although coverage of some loci with the current genotyping platforms may not be optimal.
Supplementary Meta-analyses on Data From GWAS and GWAS-Replication Studies
To assess the evidence for the association of prominent GWAS-identified loci with CM for which less than four independent datasets were available, we performed supplementary meta-analyses on SNPs with only three available datasets (Table 2 and Supplementary Table 2, available online). The results of these analyses confirmed the genome-wide statistically significant association of the chromosome 16q24.3 locus (where MC1R lies), with additional markers identified in this region (rs258322 P = 4 × 10−18 and rs4785763 P = 5 × 10−20). Furthermore, evidence for a genome-wide statistical association with CM was confirmed for the locus on chromosome 20q11.23 containing the genes ASIP, MYH7B, and PIGU (rs1204552, P = 7.1 × 10−9), and for the locus on chromosome 9p21.3 containing the genes MTAP (encoding methylthioadenosine phosphorylase) and CDKN2A (rs2218220, rs1335510, rs10757257, and rs7023329; P = 3.0 × 10−11 to 5.9 × 10−9). On chromosome 20q11.23, rs1204552 is in strong linkage disequilibrium with rs1885120 of the ASIP/MYH7B/PIGU locus (r 2 = 0.68 on the basis of HapMap CEU) that showed genome-wide statistical significance in the main meta-analyses (Table 1), and thus likely represents the same underlying association signal. Regarding the CDKN2A/MTAP locus on chromosome 9p21.3, MTAP and CDKN2A are located in close vicinity to each other—both genes span a chromosomal interval of approximately 175 kb, but common variants in MTAP that show genome-wide statistically significant association in the supplementary meta-analyses are in no or only very weak linkage disequilibrium with common variants in CDKN2A [according to the HapMap CEU + TSI sample, release II + III, August 2010 (41)]. Influence on the statistical significance of the association of MTAP with CM by rare functional variants in CDKN2A occurring on different genetic backgrounds cannot be excluded. In this context, it is noteworthy that the sample of Bishop et al. (15) was enriched for familial early-onset patients (42).
Table 2.
Genetic variants showing genome-wide statistically significant association on the basis of meta-analyses of three independent datasets from genome-wide association studies and replication studies on related traits (supplementary meta-analyses)*
| Chromosome | Gene (nearest gene) | Polymorphism | Location, bp† | Allele contrast | Ethnicity | No. of datasets | No. of subjects‡ | MAF | OR (95% CI) | P | N minor‡ | I 2 | Bias reason | Venice overall grade§ |
| 9p21.3 | MTAP | rs2218220 | 21756089 | T vs C | All | 3 | 13 005 | 0.487 | 0.84 (0.80 to 0.89) | 5.5 × 10−11 | 12 930 | 0 | NA | A |
| 9p21.3 | MTAP | rs1335510 | 21757803 | G vs T | All | 3 | 10 616 | 0.417 | 0.83 (0.78 to 0.89) | 5.9 × 10−9 | 8550 | 0 | NA | A |
| 9p21.3 | MTAP | rs10757257 | 21806564 | A vs G | All | 3 | 10 581 | 0.415 | 0.81 (0.76 to 0.86) | 3.0 × 10−11 | 8442 | 0 | NA | A |
| 9p21.3 | MTAP | rs7023329 | 21816528 | G vs A | All | 3 | 12 481 | 0.497 | 0.84 (0.80 to 0.89) | 3.1 × 10−10 | 12 194 | 0 | NA | A |
| 20q11.23 | LOC647979 | rs1204552 | 34638903 | A vs T | All | 3 | 4063 | 0.070 | 1.59 (1.36 to 1.87) | 7.1 × 10−9 | 716 | 3 | NA | B |
| 16q24.3 | CDK10 | rs258322 | 89755903 | A vs G | All | 3 | 8992 | 0.095 | 1.66 (1.48 to 1.86) | 4 × 10−18 | 2084 | 28 | NA | A |
| 16q24.3 | AFG3L1 | rs4785763 | 90066936 | A vs C | All | 3 | 9158 | 0.328 | 1.36 (1.27 to 1.45) | 5 × 10−20 | 6533 | 0 | NA | A |
CI = confidence interval, I 2 = estimate of percentage of between-study heterogeneity that is beyond chance, MAF = minor allele frequency in control subjects, NA = not applicable, N minor = number of minor alleles in patients and control subjects combined across all included datasets, OR = odds ratio. Allelic ORs, CIs, and P values (two-sided) were calculated using the DerSimonian–Laird random-effects model (32). “Gene” denotes the gene in the respective locus into which the listed SNP maps, whereas “nearest gene” refers to the most proximal gene in the respective locus if the SNP itself does not map into a gene region. It should be noted that these genes are not necessarily the genes that are functionally affected by the genetic association finding in this locus.
Location is on the basis of the Human Genome Build hg19 (40).
For inclusion of initial genome-wide association study data and replication data of related traits which only provided allelic odds ratios and 95% confidence intervals, we estimated genotype summary counts on the basis of the odds ratios and 95% confidence intervals for the purpose of the Venice grading, thus approximating numbers of the real genotype counts. This number includes both patients and control subjects.
Each statistically significant meta-analysis result was graded according to the Human Genome Epidemiology Network Venice criteria. Venice grading: A = grade A (strong epidemiological credibility), B = grade B (modest epidemiological credibility), and C = grade C (weak epidemiological credibility).
Grading of Associations
Table 1 lists the variant with the best graded result per genetic locus when applying the Venice interim criteria (36,37) to all meta-analysis results with at least nominal statistical significance for association with CM. For a list of all Venice-graded meta-analysis results and variants, see Supplementary Table 1 (available online). Five genetic loci were found to have strong epidemiological credibility (grade A) for at least one SNP (SLC45A2, TYRP1, TYR, MC1R, and MYH7B), whereas three loci (CDKN2A, VDR, and CLPTM1L) were found to have only weak credibility (grade C) because of criterion 3 (protection from bias). The main reasons for low grades in the last criterion (protection from bias) were the presence of a summary odds ratio less than 1.15 that can easily be dissipated even by relatively small biases in a meta-analysis of published data (eg, CLPTM1L and VDR), or loss of statistical significance after excluding the initial study and exclusion of Hardy–Weinberg equilibrium–violating datasets (eg, CDKN2A). In our supplementary meta-analyses of data from GWAS and GWAS-replication studies, four variants (rs10757257, rs2218220, rs1335510, and rs7023329) in the CDKN2A/MTAP locus showed a genome-wide statistically significant association with CM and furthermore, were assigned an overall strong epidemiological credibility with a grade of A (Table 2); thus, assuming one single underlying association signal in this region, the CDKN2A/MTAP locus has strong cumulative evidence for association with CM as well.
Comparison With Previously Published Meta-analyses
Six meta-analyses in the CM field had been published as of July 31, 2010, including analysis of a total of 35 SNPs across nine different genetic loci (43–48). In comparison, 83 SNPs across 18 genetic loci were meta-analyzed for our study when combining main and supplementary meta-analyses. The median overall sample size for all SNPs was approximately 8400 combined patients and control subjects in the previous meta-analyses, whereas our study had a median of approximately 11 500 combined patients and control subjects for the same SNPs. For a detailed comparison with previous meta-analyses, see Supplementary Table 3 (49–52) and Supplementary Figure 3 (available online).
Discussion
We have conducted, to the best of our knowledge, the first comprehensive and systematic field synopsis of genetic association studies in CM. We more than doubled the number of meta-analyses on genetic variants thus far published in the field of CM. Our synopsis provides an integrated perspective of the accumulated evidence of genetic associations in CM, the results of which are accessible online at the dedicated, regularly updated searchable MelGene website. We identified eight independent genetic loci nominally statistically significantly associated with CM, of which six (MC1R, TYR, TYRP1, SLC45A2, ASIP/PIGU/MYH7B, and CDKN2A/MTAP) were found to have strong epidemiological credibility.
Whereas most studies included in the synopsis used a candidate gene approach, the findings of two recent GWAS have also been included, to the extent of the available data (15,16). Of the six loci with strong credibility, four (MC1R, TYR, TYRP1, and SLC24A5) were previously proposed by candidate gene studies, and two (20q11.22, ASIP/PIGU/MYH7B and 9p21.3, MTAP) were first evaluated by GWAS, although adjacent candidate genes had previously been considered (ASIP and CDKN2A, respectively). Our findings suggest a complementary role for candidate studies and GWAS for CM (53). In studies of other cancer phenotypes, few candidate variants have been validated by GWAS, whereas none of the newly discovered variants in the respective cancer phenotypes had previously been considered in the candidate era (54). It is possible that with larger sample sizes and more comprehensive platforms, some additional previously proposed candidate genes for CM may reach genome-wide statistical significance. Despite the indisputable successes of recent GWAS (15,16) and GWAS-related studies (27–31), the available platforms currently do not cover all human genetic variation. Therefore, it is likely that there are some CM susceptibility genes that are missed by current approaches but may be captured by more comprehensive coverage through extensive imputation using the 1000 genomes resource (55,56) and next-generation resequencing projects. In this context, MelGene provides a dynamic resource to evaluate the constantly evolving evidence for those genetic associations by use of a comprehensive and up-to-date meta-analysis approach.
The loci showing the most compelling risk effect estimates in the MelGene meta-analyses contain genes that seemingly play an important role in modulating pigmentary traits of the skin, hair, and/or eyes. However, these CM association findings have to be interpreted cautiously because many of these candidates were tested for association with melanoma because of their association to pigmentary phenotypes. MC1R on chromosome 16q24.3 encodes a cell-surface G-protein-coupled receptor that binds to alpha-melanocyte-stimulating hormone, leading to the production of the photoprotective brown/black eumelanin (57). MC1R loss-of-function polymorphisms have been reported to cause a shift of melanogenesis from eumelanin to the red and yellow pheomelanin, resulting in lighter skin, freckles, and red hair color (58,59). TYR on chromosome 11q14.3 encodes tyrosinase, which controls the rate-limiting step of melanin biosynthesis. Rs1126809 (R402Q), a common TYR SNP that shows the strongest association with CM in our meta-analyses, has been reported to reduce the catalytic activity of tyrosinase (60) and has been associated with blue vs green eyes, and skin sensitivity to the sun (61). The association signal of rs1393350, yielding the second strongest association in the MelGene analyses for this locus was reported as being independently associated with CM in a recent GWAS (15). Similar to TYR, TYRP1 on chromosome 9p23 encodes an enzyme of the pigmentary system required for the eumelanization of melanosomes (62). Germline mutations of TYRP1 are the cause of oculocutaneous albinism type 3, and coding variants associated with CM have been associated with eye and hair color (61). SLC45A2 on chromosome 5p13.2 encodes a melanocyte differentiation antigen that is overexpressed in melanoma cell lines (63). For the nonsynonymous SNP rs16891982 (F374L) that is associated with CM with genome-wide statistical significance in this study, the ancestral Leu allele has been found more frequently in Southern Europe and is associated with dark skin, eye, and hair color in people of European ancestry (64–66). Finally, the two statistically significant variants at chromosome 20q11.22 fall within the PIGU gene (also known as CDC91L1), which is involved in cell cycle control and within the MYH7B gene, respectively. These statistically significant variants at chromosome 20q11.22 may not be disease-modifying variants, but may be positional markers that highlight an unidentified melanoma risk locus adjacent to ASIP (16). Although the identification of associated SNPs in the vicinity of CDKN2A (MTAP, 9p21.3) (15) possibly reflect the effects of highly penetrant CDKN2A mutations via linkage disequilibrium not tagged by currently known common variants, the concurrence of low to moderate independent risk effects in the MTAP locus itself may directly influence melanocytic proliferation (42). This hypothesis is also supported by an association signal with nevi count in MTAP (29), suggesting a shared susceptibility of nevi and melanoma. Thus, there may be at least two genetically distinct effects influencing melanoma susceptibility, that is, those mainly influencing pigmentation and those associated with nevus development (67).
Our study does have some limitations. First, despite the use of different strategies to identify eligible studies for MelGene, some studies may have been erroneously excluded. Also, although manual data extraction from the included studies followed an established protocol, it is possible that some study entries contain random errors. In addition, our analyses focused on allelic contrasts. However, we also performed an analysis of genotypic models that yielded similar results. Another limitation of our study is that lack of access to individual data precludes more refined analyses and adjustment for potential confounders and other types of bias, for example, pigmentation status, age, and sex which may potentially lead to false-positive or false-negative meta-analysis results, or different magnitudes of effect. Furthermore, lack of individual data does not allow the assessment of potential gene–gene and gene–environment interactions. We assessed potential sources of bias by extensive sensitivity analyses on the data summary level, but our study is limited because potential undetectable bias can never be ruled out, particularly publication bias. Although we tested for small-study effects, publication bias cannot be entirely excluded in retrospective meta-analyses. Finally, several of the nominally statistically significant associations may still be false-positive results. However, this is unlikely for those associations that have strong credibility on the basis of the Venice criteria and those that have reached levels of genome-wide statistical significance.
In summary, we present a systematic and comprehensive assessment of the current evidence of genetic epidemiology research in CM. The putative risk factors that have emerged from the meta-analyses—accessible on MelGene—represent the most promising CM susceptibility genes described to date. The integration of results from future small- and large-scale genetic association studies in MelGene will further expand our knowledge of the underlying genetic mechanisms of CM.
Supplementary Material
Footnotes
F. Chatzinasiou and C. M. Lill contributed equally to this work.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20(suppl 6):vi1–vi7. doi: 10.1093/annonc/mdp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tucker MA, Goldstein AM. Melanoma etiology: where are we? Oncogene. 2003;22(20):3042–3052. doi: 10.1038/sj.onc.1206444. [DOI] [PubMed] [Google Scholar]
- 4.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer. 2005;41(14):2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 5.Gandini S, Sera F, Cattaruzza MS, et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer. 2005;41(1):28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Risch N. Genetic epidemiology of cancer: interpreting family and twin studies and their implications for molecular genetic approaches. Cancer Epidemiol Biomark Prev. 2001;10(7):733–741. [PubMed] [Google Scholar]
- 7.Shekar SN, Duffy DL, Youl P, et al. A population-based study of Australian twins with melanoma suggests a strong genetic contribution to liability. J Invest Dermatol. 2009;129(9):2211–2219. doi: 10.1038/jid.2009.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8(1):15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 9.Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet. 2007;44(2):99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meyle KD, Guldberg P. Genetic risk factors for melanoma. Hum Genet. 2009;126(4):499–510. doi: 10.1007/s00439-009-0715-9. [DOI] [PubMed] [Google Scholar]
- 11.Lin J, Hocker TL, Singh M, Tsao H. Genetics of melanoma predisposition. Br J Dermatol. 2008;159(2):286–291. doi: 10.1111/j.1365-2133.2008.08682.x. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy C, ter Huurne J, Berkhout M, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001;117(2):294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 13.Palmer JS, Duffy DL, Box NF, et al. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000;66(1):176–186. doi: 10.1086/302711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fargnoli MC, Argenziano G, Zalaudek I, Peris K. High- and low-penetrance cutaneous melanoma susceptibility genes. Expert Rev Anticancer Ther. 2006;6(5):657–670. doi: 10.1586/14737140.6.5.657. [DOI] [PubMed] [Google Scholar]
- 15.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41(8):920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown KM, Macgregor S, Montgomery GW, et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat Genet. 2008;40(7):838–840. doi: 10.1038/ng.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 18.Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40(7):827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- 19.Lill CM, Roehr JT, McQueen MB, et al. The PDGene Database. Alzheimer Research Forum. http://www.pdgene.org/. Accessed July 31, 2010. [Google Scholar]
- 20.Castaldi PJ, Cho MH, Cohn M, et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Hum Mol Genet. 2010;19(3):526–534. doi: 10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vineis P, Manuguerra M, Kavvoura FK, et al. A field synopsis on low-penetrance variants in DNA repairs genes and cancer susceptibility. J Natl Cancer Inst. 2009;101(1):24–36. doi: 10.1093/jnci/djn437. [DOI] [PubMed] [Google Scholar]
- 22.Dolan SM, Hollegaard MV, Merialdi M, et al. Synopsis of preterm birth genetic association studies: the preterm birth genetics knowledge base (PTBGene) Public Health Genomics. 2010;13(7–8):514–523. doi: 10.1159/000294202. [DOI] [PubMed] [Google Scholar]
- 23.Chatzinasiou F, Lill CM, Kypreou K, et al. The MelGene database. doi: 10.1038/jid.2014.491. http://www.melgene.org/ [DOI] [PubMed] [Google Scholar]
- 24.PubMed: U.S. National Library of Medicine National Institutes of Health. http://www.ncbi.nlm.nih.gov/pubmed. [Google Scholar]
- 25.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40(2):124–125. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 26.MMMP: Melanoma Molecular Map Project. doi: 10.1097/CMR.0b013e328300c50b. http://www.mmmp.org/MMMP. Accessed October 14, 2010. [DOI] [PubMed] [Google Scholar]
- 27.Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer. 2009;125(4):909–917. doi: 10.1002/ijc.24327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, Sulem P, Stacey SN, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet. 2008;40(7):886–891. doi: 10.1038/ng.161. [DOI] [PubMed] [Google Scholar]
- 29.Falchi M, Bataille V, Hayward NK, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet. 2009;41(8):915–919. doi: 10.1038/ng.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stacey SN, Sulem P, Masson G, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41(8):909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rafnar T, Sulem P, Stacey SN, et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat Genet. 2009;41(2):221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 33.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 34.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335(7626):914–916. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37(1):120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 37.Khoury MJ, Bertram L, Boffetta P, et al. Genome-wide association studies, field synopses, and the development of the knowledge base on genetic variation and human diseases. Am J Epidemiol. 2009;170(3):269–279. doi: 10.1093/aje/kwp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harbord RM, Egger M, Sterne JA. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat Med. 2006;25(20):3443–3457. doi: 10.1002/sim.2380. [DOI] [PubMed] [Google Scholar]
- 39.Ioannidis JP, Trikalinos TA. An exploratory test for an excess of significant findings. Clin Trials. 2007;4(3):245–253. doi: 10.1177/1740774507079441. [DOI] [PubMed] [Google Scholar]
- 40.The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. URL: http://genome.ucsc.edu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 42.Yeh I, Bastian BC. Genome-wide associations studies for melanoma and nevi. Pigment Cell Melanoma Res. 2009;22(5):527–528. doi: 10.1111/j.1755-148X.2009.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerstenblith MR, Shi J, Landi MT. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res. 2010;23(5):587–606. doi: 10.1111/j.1755-148X.2010.00730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mocellin S, Nitti D. Vitamin D receptor polymorphisms and the risk of cutaneous melanoma: a systematic review and meta-analysis. Cancer. 2008;113(9):2398–2407. doi: 10.1002/cncr.23867. [DOI] [PubMed] [Google Scholar]
- 45.Randerson-Moor JA, Taylor JC, Elliott F, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. Eur J Cancer. 2009;45(18):3271–3281. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gandini S, Raimondi S, Gnagnarella P, Doré JF, Maisonneuve P, Testori A. Vitamin D and skin cancer: a meta-analysis. Eur J Cancer. 2009;45(4):634–641. doi: 10.1016/j.ejca.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 47.Raimondi S, Sera F, Gandini S, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008;122(12):2753–2760. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 48.Mocellin S, Verdi D, Nitti D. DNA repair gene polymorphisms and risk of cutaneous melanoma: a systematic review and meta-analysis. Carcinogenesis. 2009;30(10):1735–1743. doi: 10.1093/carcin/bgp207. [DOI] [PubMed] [Google Scholar]
- 49.Duffy DL, Zhao ZZ, Sturm RA, Hayward NK, Martin NG, Montgomery GW. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J Invest Dermatol. 2010;130(2):520–528. doi: 10.1038/jid.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24(9):1291–1306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 51.Landi MT, Kanetsky PA, Tsang S, et al. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst. 2005;97(13):998–1007. doi: 10.1093/jnci/dji176. [DOI] [PubMed] [Google Scholar]
- 52.Debniak T, Gapska P, Serrano-Fernandez P, et al. Modest association of malignant melanoma with the rs910873 and rs1885120 markers on chromosome 20: a population-based study. Melanoma Res. 2010;20(2):159–160. doi: 10.1097/CMR.0b013e32833716e6. [DOI] [PubMed] [Google Scholar]
- 53.Siontis KC, Patsopoulos NA, Ioannidis JP. Replication of past candidate loci for common diseases and phenotypes in 100 genome-wide association studies. Eur J Hum Genet. 2010;18(7):832–837. doi: 10.1038/ejhg.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ioannidis JP, Castaldi P, Evangelou E. A compendium of genome-wide associations for cancer: critical synopsis and reappraisal. J Natl Cancer Inst. 2010;102(12):846–858. doi: 10.1093/jnci/djq173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durbin RM, Abecasis GR, et al. 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ashley EA, Butte AJ, Wheeler MT, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375(9725):1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beaumont KA, Shekar SN, Newton RA, et al. Receptor function, dominant negative activity and phenotype correlations for MC1R variant alleles. Hum Mol Genet. 2007;16(18):2249–2260. doi: 10.1093/hmg/ddm177. [DOI] [PubMed] [Google Scholar]
- 58.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11(3):328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 59.Flanagan N, Healy E, Ray A, et al. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet. 2000;9(17):2531–2537. doi: 10.1093/hmg/9.17.2531. [DOI] [PubMed] [Google Scholar]
- 60.Tripathi RK, Strunk KM, Giebel LB, Weleber RG, Spritz RA. A polymorphism of the human tyrosinase gene is associated with temperature-sensitive enzymatic activity. Gene Expr. 1991;1(2):103–110. [PMC free article] [PubMed] [Google Scholar]
- 61.Sulem P, Gudbjartsson DF, Stacey SN, et al. Two newly identified genetic determinants of pigmentation in Europeans. Nat Genet. 2008;40(7):835–837. doi: 10.1038/ng.160. [DOI] [PubMed] [Google Scholar]
- 62.Sturm RA, Teasdale RD, Box NF. Human pigmentation genes: identification, structure and consequences of polymorphic variation. Gene. 2001;277(1–2):49–62. doi: 10.1016/s0378-1119(01)00694-1. [DOI] [PubMed] [Google Scholar]
- 63.Harada M, Li YF, El-Gamil M, Rosenberg SA, Robbins PF. Use of an in vitro immunoselected tumor line to identify shared melanoma antigens recognized by HLA-A*0201-restricted T cells. Cancer Res. 2001;61(3):1089–1094. [PubMed] [Google Scholar]
- 64.Graf J, Hodgson R, van Daal A. Single nucleotide polymorphisms in the MATP gene are associated with normal human pigmentation variation. Hum Mutat. 2005;25:278–284. doi: 10.1002/humu.20143. [DOI] [PubMed] [Google Scholar]
- 65.Fernandez LP, Milne RL, Pita G, et al. SLC45A2: a novel malignant melanoma-associated gene. Hum Mutat. 2008;29(9):1161–1167. doi: 10.1002/humu.20804. [DOI] [PubMed] [Google Scholar]
- 66.Guedj M, Bourillon A, Combadières C, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum Mutat. 2008;29(9):1154–1160. doi: 10.1002/humu.20823. [DOI] [PubMed] [Google Scholar]
- 67.Whiteman DC, Watt P, Purdie DM, Hughes MC, Hayward NK, Green AC. Melanocytic nevi, solar keratoses, and divergent pathways to cutaneous melanoma. J Natl Cancer Inst. 2003;95(11):806–812. doi: 10.1093/jnci/95.11.806. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.