Abstract
Tetrahymena thermophila, a member of the Ciliates, represents a class of organisms distantly related from commonly used model organisms in cell biology, and thus offers an opportunity to explore potentially novel mechanisms and their evolution. Ciliates, like all eukaryotes, possess a complex network of organelles that facilitate both macromolecular uptake and secretion. The underlying endocytic and exocytic pathways are key mediators of a cell’s interaction with its environment, and may therefore show niche-specific adaptations. Our laboratory has taken a variety of approaches to identify key molecular determinants for membrane trafficking pathways in Tetrahymena. Studies of Rab GTPases, dynamins, and sortilin-family receptors substantiate the widespread conservation of some features but also uncover surprising roles for lineage-restricted innovation.
A LIMITATION OF MODEL ORGANISM-BASED CELL BIOLOGY
Much of the rich detail we possess about cell biological structures and pathways comes from studies in a handful of well-domesticated model organisms (Boehm and Bonifacino, 2002; Kourtis and Tavernarakis, 2008; Westermann, 2010; Kimble, 2011), which are exploited for their inherent biological characteristics and using the extensive tools that have been developed by large communities of researchers (Cherry et al., 2012; Lamesch et al., 2012; McQuilton et al., 2012; Bult et al., 2013; Thompson et al., 2013). An important generalization that has emerged from many studies is that a multitude of cell biological mechanisms and structures are conserved between model organisms (Ferro-Novick and Jahn, ’94; Finger, ’98; Ashburner et al., 2000; Gönczy, 2008; Simons and Mlodzik, 2008). One might conclude, but we would argue incorrectly, that structures or mechanisms that are found only in a subset of organisms (i.e., “lineage-restricted”) are of secondary interest for understanding biology. In addition, there are strong reasons to question the criteria that are commonly used by many cell biologists to define a feature as “universal.”
Historically, the most heavily employed model organisms have been animal or fungal (Davis, 2004). As a result, many models in cell biology are heavily biased, if not exclusively based, on data from organisms that belong to a single eukaryotic lineage (Hedges, 2002). This is because animals and fungi both belong to the Opisthokont clade (Baldauf and Palmer, ’93; Wainright et al., ’93), just one of what are currently considered to be five major surviving eukaryotic lineages (Parfrey et al., 2006). Plants belong to a 2nd clade (Archeaplastidia) (Turner et al., ’99; Reyes-Prieto et al., 2007); an implication is that a comparison of cell biological data between Arabidopsis and yeast, or between Arabidopsis and humans, would be a better strategy for distinguishing between universal versus lineage-specific cellular features, than a comparison between yeast and humans. The remaining three clades (Excavata (Simpson and Patterson, ’99), Ameobozoa (Cavalier-Smith et al., 2004), and SAR (Burki et al., 2007)); consist largely of unicellular organisms, both parasitic and free-living, which are very poorly represented among the pantheon of model organisms (Baldauf, 2008). An exception are some parasites (e.g., Toxoplasma, Trypanosomes) that have won attention because of their importance as pathogens (Kissinger et al., 2003; Aslett et al., 2010; Hunter and Sibley, 2012; Stephens et al., 2012). Addressing this asymmetry (an issue highlighted eloquently by Dacks and Field) in the sampling of cellular biodiversity by cell biologists could produce a more accurate understanding both of conserved features, and the variation that exists within them, and of features that are limited to specific lineages. A broader, taxonomy-informed sampling would therefore provide a more accurate picture of the relative contributions made by inheritance versus innovation in the emergence of modern cells. In its most ambitious form, such an initiative would entail developing a broad array of new model organisms, an exciting but challenging goal.
CILIATES AS MODEL ORGANISMS
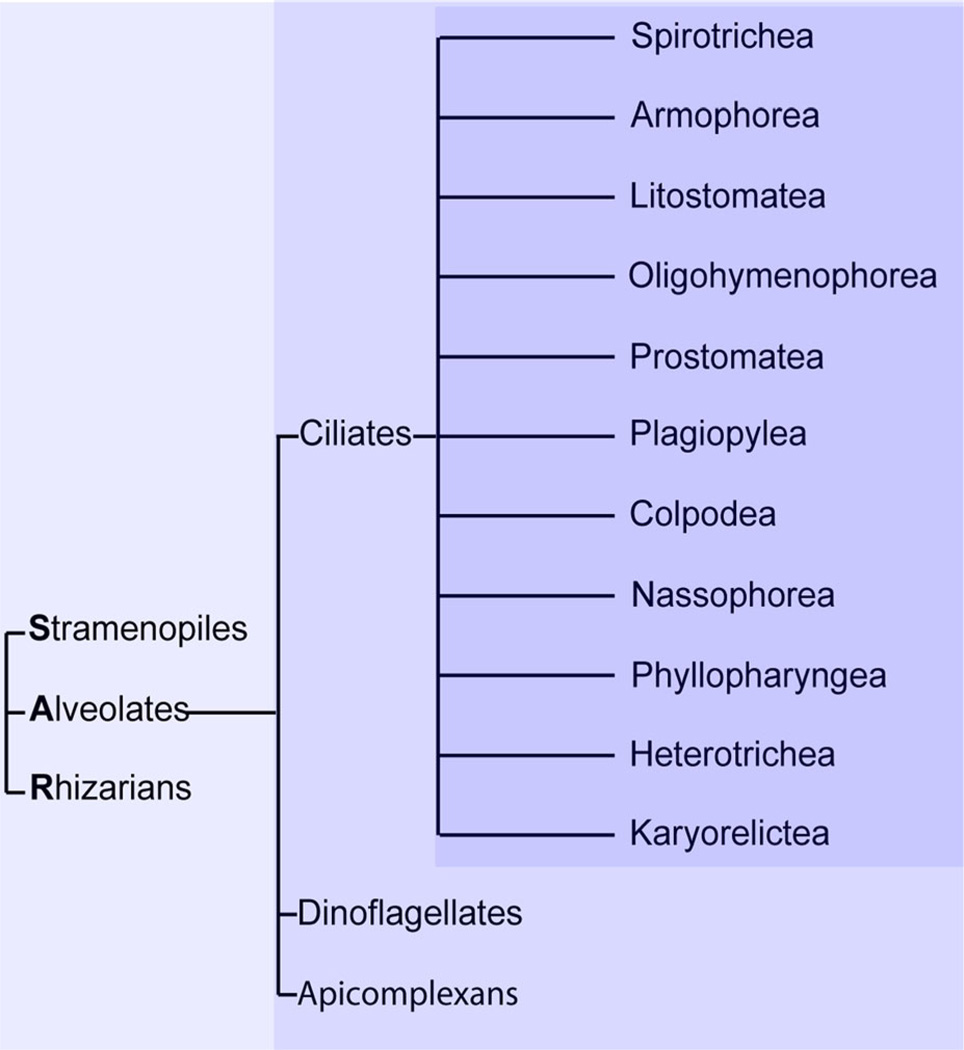
An intermediate approach to the same aims, which does not require developing new model organisms, is to exploit existing model organisms falling within underexplored, that is, non-Opisthokont, clades. Ciliates represent an attractive group of organisms for expanding cell biological studies to divergent lineages. Ciliates, which belong to the Stramenopiles–Alveolates–Rhizaria (SAR) lineage, are further classified together with Dinoflagellates and Apicomplexa as Alveolates, which refers to a sub-plasma membrane compartment called alveoli (Stelly et al., ’91; Plattner et al., 2012; Kono et al., 2013). Unlike the parasitic Apicomplexa (Sibley, 2004), many Ciliates are free-swimming protozoa, which as a group show remarkable morphological diversity (Lynn, 2001). Ciliates fall into 11 branches (Fig. 1) (Lynn, 2003). Importantly, defined laboratory strains and a wide range of experimental tools have already been established for two Ciliates in the Oligohymenophorea branch, Tetrahymena thermophila and Paramecium tetraurelia, and these can be used for rigorously pursing cell biological questions (Turkewitz et al., 2002; Collins and Gorovsky, 2005; Beisson et al., 2010). Excitingly, tool kits are also being developed for Ciliates belonging to other branches, such as Oxytricha trifallax (Nowacki et al., 2008; Fang et al., 2012; Swart et al., 2013) and Stentor coeruleus.
Figure 1.
Ciliates comprise 11 sub-branches, and fall within the Stramenopile/Alveolate/Rhizaria (SAR) lineage. These relationships are illustrated as a cladogram, showing classification but not evolutionary distances. Ciliates, along with Dinoflagellates and Apicomplexans, are Alveolate members of the SAR group (Stramenopiles–Alveolates–Rhizaria), one of the five major eukaryotic lineages (Lynn, 2003; Parfrey et al., 2006; Baldauf, 2008). The Ciliates fall into 11 classes (Lynn, 2003).
As with researchers using other model organisms, the aim for those using Ciliates has traditionally been to exploit features that offer unique experimental advantages (Gibbons and Rowe, ’65), and the most spectacular successes have come from nuclear dimorphism. Ciliates, though unicellular, maintain two nuclei: a diploid germline micronucleus (MIC) together with its derived, and dramatically modified, polyploid macronucleus (MAC) (Prescott, ’94). The mechanisms involved in generating the MAC in T. thermophila were exploited to yield Nobel Prize-winning discoveries, namely telomere structure and telomerase (Blackburn, 2010) as well as catalytic RNA (Cech, 2004), which are appreciated as fundamental cellular features. An interesting variation on this theme is represented by ongoing work that exploits the fact that the MIC must import a different cohort of cytoplasmic proteins compared to the MAC, since the MIC is silent while the MAC is actively transcribed. This line of work aims to define how nuclear pores (the conduits for macromolecules in and out of the nucleus) have been remodeled in Ciliates, to refine their selectivity (Malone et al., 2008; Iwamoto et al., 2009). Thus nuclear dimorphism can be harnessed to identify deeply conserved features but also to understand how conserved features can be refined through lineage-specific innovations.
Our laboratory is interested in the endomembrane system, which consists of a network of membranous compartments, including the plasma membrane, interconnected via vesicle-mediated traffic of lipids and proteins (Bonifacino and Glick, 2004), and which is a hallmark of eukaryotic cells. Like any complex cellular feature, the endomembrane system in any given cell must consist of a combination of broadly conserved structures and mechanisms together with lineage-restricted elements that arose more recently (Dacks and Field, 2007). For many pathways, mechanistic studies in Opisthokonts have led to identification of key determinants (McMahon and Mills, 2004; Robinson, 2004; Behnia and Munro, 2005; Stenmark, 2009). Due to the availability of sequenced genomes from many species, it has recently become possible to pursue rigorous phylogenetic analysis of such determinants to ask whether orthologous genes are present in divergent lineages, which is expected for conserved features deriving from an ancient eukaryotic ancestor (Pereira-Leal and Seabra, 2001; Jekely, 2003; Koumandou et al., 2007, 2011). The identification and analysis of lineage-restricted features would help to fill in our picture of cellular organization (Pereira-Leal and Teichmann, 2005; Kienle et al., 2009). One reason is that lineage-restricted elements may reveal adaptive pressures, particularly because both exocytosis (secretion) and endocytosis (uptake), which depend on the endomembrane system, are important in the interaction of cells with their environments.
ENDOCYTOSIS AND EXOCYTOSIS IN TETRAHYMENA
Morphological studies indicate that Tetrahymena maintain a large set of distinct membrane trafficking pathways (Frankel, 2000). These include at least four pathways of endocytosis by which plasma membrane and/or extracellular material is internalized into the cytoplasm via vesicles. First, clathrin-coated vesicles form at parasomal sacs, which are small invaginations adjacent to cilia (Allen, ’67; Nilsson and van Deurs, ’83; Elde et al., 2005). Because the plasma membrane overlays the alveoli that are organized as an extensive sheet of abutting cisternae (Fig. 2A, bottom), the parasomal sacs offer one of the few known places in these cells where the plasma membrane can make direct contact with the cytoplasm (Satir and Wissig, ’82). A second dramatic form of endocytosis is phagocytosis of bacteria-sized particles, which occurs at the base of a specialized anterior oral apparatus lined with beating cilia that sweep in food particles such as bacteria (Fig. 2A (top), B) (Nilsson, ’79; Frankel, 2000). Endocytosis at other distinct sites is involved in membrane recovery following exocytosis (Plattner et al., ’93) of secretory granules called mucocysts, and following fusion (Hausmann and Allen, 76) of old food vacuoles (“defecation”) at a defined posterior structure called the cytoproct (also see Fig. 2B) (Allen and Wolf, 79).
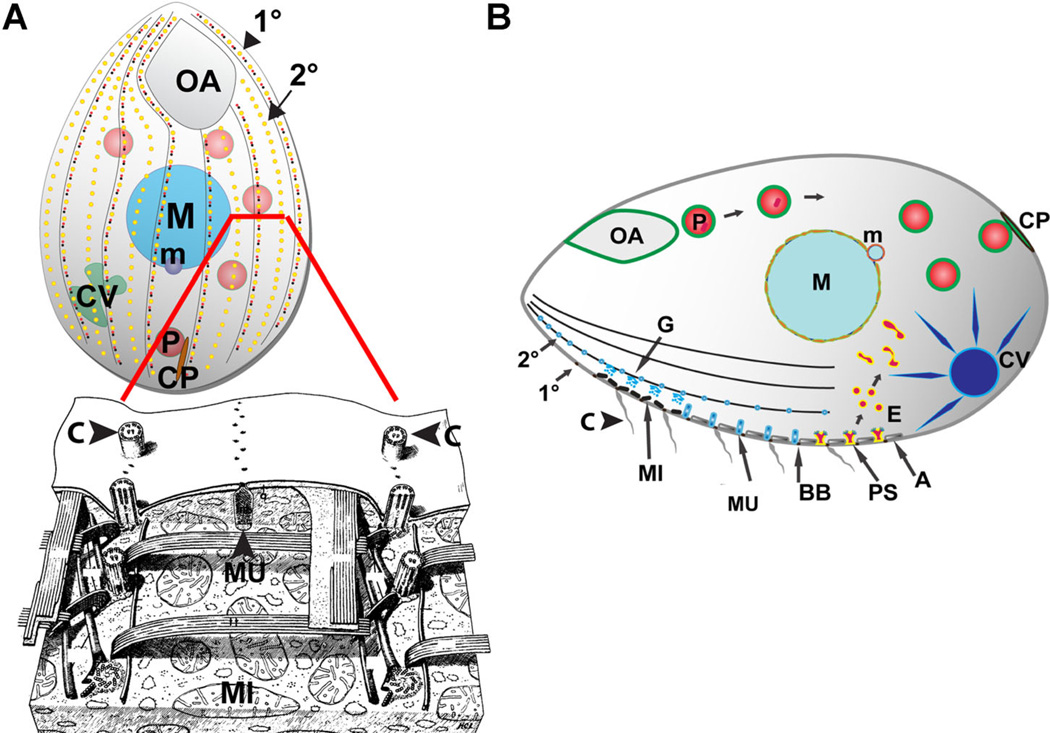
Figure 2.
Morphology and organization of Tetrahymena thermophila. Figure and legend adapted from Figure 1A and B of Bright et al. (2010) and Figure 22 of Allen (‘67). (A) Top: (Bright et al., 2010). Cartoon of a Tetrahymena cell (length ~50 µM). All surface features of Tetrahymena, with the exception of the oral apparatus (OA) (the site of phagocytic ingestion) and cytoproct (CP) (the site of cellular defecation) occur at repeated positions and therefore form linear arrays, organized by cytoskeletal “ribs” that run the length of the cell, known as 1° and 2° meridians. (A) Bottom: (Allen, ’67). A reconstruction to portray structures positioned just below the cell surface, as seen from outside of a cell in which the plasma membrane and alveoli have been stripped away from the front half. The reconstruction more clearly illustrates the positioning of cilia (C) in longitudinal linear 1° meridians, which can be seen running from top to bottom on either side of the image. Although not prominent in the reconstruction, alveolae are found just beneath the plasma membrane. Alveolae are a monolayer of cisternae underlying the plasma membrane, and function at least in part as a compartment for storage of mobilizable calcium. The reconstruction illustrates a mucocyst (MU) docked at a junction between alveolae. The mucocyst sits on a 2° meridian, positioned between cilia-bearing 1° meridians. (B) Prominent structures involved in membrane trafficking in T. thermophila. Phagocytosis begins at the oral apparatus (OA), resulting in formation of phagosomes (P) that, after undergoing multiple fusion and fission events, eventually egest undigested material via exocytic fusion at the cytoproct (CP). Clathrin-mediated endocytosis occurs at parasomal sacs (PS), giving rise to endocytic vesicles (E). These endocytic vesicles coalesce in tubulovesicular endosomes in the cell posterior. Outbound membrane trafficking, including proteins to be secreted, involves the endoplasmic reticulum (not shown) and Golgi(G), which are present as single cisterna or short stacks near the cell periphery, close to mitochondria (MI). Some of the secretory cargo is packaged into mucocysts (MU) which dock and subsequently undergo exocytosis at sites on 1° and 2° meridians. Another prominent organelle is the water-pumping contractile vacuole (CV). Other structures shown: alveolae (A). macronucleus (polyploid somatic nucleus, M). Micronucleus (diploid germline nucleus, m). Ciliary basal bodies (BB) and the cilia (C) that grow from them. Rows of cilia cover the entire cell surface, but only a subset are shown here for clarity. An excellent review of these structures is provided by Frankel (2000).
Exocytosis, or the fusion of cytoplasmic vesicles with the plasma membrane, also occurs via several known routes. The best studied are the aforementioned mucocysts. Hundreds of mucocysts dock in linear arrays at the plasma membrane where they can undergo release triggered by an extracellular stimulus (Tiedtke, ’76; Satir, ’77; Chilcoat et al., ’96). In addition to the regulated secretion of mucocysts, a pathway of constitutive release has also been inferred, though not identified at the morphological level, based on the rapid release of newly synthesized secretory proteins from non-stimulated cells (Bowman and Turkewitz, 2001; Madinger et al., 2010). Tetrahymena are additionally equipped to secrete hydrolytic enzymes, which appears to involve secretory lysosomes (Kiy et al., ’93). Other pathways of membrane traffic can also be inferred from morphological data, though few details are known. A dramatic example is the remodeling of the oral apparatus, which is necessary to allow the exchange of meiotic nuclear products in conjugating cells (Wolfe, ’82). By comparing these pathways with known pathways in other organisms, one may be able to infer or demonstrate potentially novel features that arose within Ciliates and/or the SAR lineage.
WHAT RAB GTPases TELL US ABOUT MEMBRANE TRAFFIC IN TETRAHYMENA
In broad terms, functionally analogous features of the endomembrane system in different lineages may be related to one another in several different ways, and we consider three classes. First, analogous features may be truly homologous, that is, both maintained in a relatively unchanged state during inheritance from a common ancestor. Second, such homologous features may be descended from a common ancestral feature, but have undergone significant modifications in one or both lineages being compared. Third, features that serve the same function in two modern lineages could be evolutionarily unrelated, arising from different origins and by different mechanisms, but have undergone functional convergence. Both the 2nd and 3rd classes involve innovations relative to the ancestral state, which may be at the level of novel genes or higher-order assemblies. It is reasonable to expect, given the current sampling bias, that many features currently considered to be conserved throughout eukaryotes will show greater variation when examined in divergent lineages. This in turn can provide insights into fundamental aspects of cellular evolution, such as physical and genetic constraints on cellular features. In the discussion below, we outline studies from our laboratory in Tetrahymena that address both conservation and innovation, and which provide illustrations of how the latter have shaped membrane traffic in these cells.
For studying the evolution of the endomembrane network, large gene families that serve as compartmental determinants have provided the most informative tools (Dacks et al., 2009). Rab GTPases, which represent the most numerous group of small GTPases in eukaryotes, are encoded by gene families in which each member is associated with one or a small number of endomembrane compartments (Stenmark, 2009) The association of Rabs with their target membranes, and with effectors, is controlled by GTP binding and hydrolysis. A Rab confers compartmental identity by recruiting effectors that in turn control the trafficking of proteins and lipids (i.e., vesicular traffic) to and from that compartment (Segev, 2001; Grosshans et al., 2006) In many cases, the number of Rabs expressed in an organism correlates with the complexity of the organism’s endomembrane network (Pereira-Leal and Seabra, 2001). Budding yeast, renowned for their relative simplicity, express 12 Rabs, whereas humans express about 63 (Stenmark and Olkkonen, 2001). Importantly, structures that are homologous in yeast and humans are associated with orthologous Rabs, consistent with the idea that the common ancestor of fungi and animals already possessed both those structures and the associated Rabs, and both structures and Rabs have been inherited in a conservative fashion (Pereira-Leal and Seabra, 2001). This corresponds to classes 1 or 2, in the paragraph above. Given this context, expanding the detailed study of Rabs to divergent lineages could yield important insights into both the organization and evolution of the endomembrane network (Brighouse et al., 2010).
Global analysis of the Rabs in T. thermophila, by our laboratory as well as by Saito-Nakano et al., revealed that this single-celled organism expresses roughly the same number of Rabs as humans (Bright et al., 2010; Saito-Nakano et al., 2010). In addition, Tetrahymena express a set of >20 Rab-like proteins (J. Briguglio, manuscript in preparation), which differ from authentic Rabs in lacking a predicted addition site for a C-terminal prenyl moiety that is important for targeting to membranes. The number of Rabs is extravagant relative to many single-celled organisms (Stenmark and Olkkonen, 2001), but at least some other protozoa have Rab families of comparable size. Trichomonas vaginalis and Entamoeba histolytica, for example, encode Rab families containing 65 (Lal et al., 2005) and 95 (Saito-Nakano et al., 2005) members, respectively; these numbers should be taken as estimates since annotation of Rabs is not straightforward (http://www.rabdb.org/about/). Do these organisms maintain endomembrane networks of comparable of greater complexity than in human cells? One layer of complexity in higher organisms is tissue-specific expression (Ayala et al., ’89; Gurkan et al., 2005; Zhang et al., 2007). Similarly, single-celled eukaryotes could show stage-specific expression (e.g., parasites could express different Rabs in different hosts, while free living cells could adjust for the vagaries of their environment) (Quevillon et al., 2003; Lal et al., 2005). Tetrahymena, in particular, adapt quickly and dramatically to different environmental conditions, for example changing both shape and swimming behavior when starved for food (Nelsen and Debault, 78). Further changes occur if cells of a complementary mating type are present, eventually leading to mating that involves extensive membrane reorganization (Wolfe and Grimes, 79). Importantly, a database of T. thermophila gene expression (http://tfgd.ihb.ac.cn/) allows one to interrogate the levels for any transcript for a variety of different conditions (Miao et al., 2009; Xiong et al., 2013). Taking advantage of this resource, we could conclude that nearly all Tetrahymena Rabs were expressed, many among the ~20% most highly expressed genes in this organism (Bright et al., 2010). The concurrent expression of ~58 Rabs within a single cell, was consistent with pathways of membrane traffic comparable in complexity to those in animal cells.
For practical reason, we focused on Rabs transcribed in vegetative cells and expressed these as GFP fusions, to assign them to different endomembrane compartments and pathways. Because few molecular markers are established in these cells, the assignments were based on previous ultra-structural studies and further validation with functional probes (e.g., the styryl dye FM4-64 as an endocytic tracer, and India Ink and fluorescent bacteria as ingestible markers for phagosomes). The same Rabs were analyzed by phylogenetic criteria to ask, as described above, whether structures analogous with those in fungi and animals were associated with the orthologous Rabs, consistent with those structures being related by common descent (Bright et al., 2010). Experimental data, initially from animals and fungi, had identified a “core” group of highly conserved Rabs corresponding to eight different pathways of membrane traffic, and inferred to be present in an ancestral eukaryotic cell (Pereira-Leal and Seabra, 2001). Those findings were subsequently extended by broad sequence-based phylogenetic surveys, but rarely including characterization of the proteins (Elias et al., 2012). From the sequence-based plus localization survey of Tetrahymena Rabs, several different pictures emerged.
Phylogenetic analysis from our laboratory indicated that, a minority of the Tetrahymena Rabs (~27%) could be robustly identified as orthologs of Rabs in other organisms (Bright et al., 2010). This conclusion (but see below for other predictions) suggests that only a minority of the Tetrahymena Rabs and/or membrane trafficking pathways correspond to Class 1 described above. The conserved Tetrahymena Rabs belong to six of the “core” groups considered to be conserved throughout eukaryotes, including ER-Golgi trafficking, endocytosis, endocytic recycling, late endocytosis, and retrograde Golgi trafficking. The remaining core groups, corresponding to Golgi-related and regulated exocytic pathways, appear to have been lost in Tetrahymena. This is unexpected because, as previously mentioned, Tetrahymena possess a prominent pathway of regulated exocytosis from mucocysts, so the absence of any conserved regulated exocytic Rabs suggests a lineage-specific origin for these organelles (Class 3).
Localizing the conserved Rabs (as GFP fusions) confirmed some expectations but also provided surprises. For example, all four of the retrograde Golgi Rabs displayed almost identical localizations, corresponding with the known distribution of the Golgi in this organism, so that the expansion of this Rab subfamily may reflect a dosage requirement. There may also be a lineage-restricted contribution, since one highly divergent Rab also showed Golgi localization. Other pathways may have an even more substantial lineage-restricted contribution. For example, nine Rabs were assigned to endocytic pathways based on their colocalization with FM4-64. Of these nine only three had clear orthologs in other organisms, suggesting that the remaining Rabs arose within the Tetrahymena lineage. These Rabs displayed similar but not equivalent localizations (e.g., variable overlap with FM4-64), so are likely to be specialized for different endocytic subcompartments. Thus, both the Golgi and endosomes in Tetrahymena appear to have derived from some combination of Classes 1 and 2 histories. Intriguingly, four other Tetrahymena Rabs, which did not show any detectable overlap with FM4-64, nonetheless showed robust orthology to conserved endocytic Rabs in other organisms; three of these localized to phagocytic structures (Bright et al., 2010). This case, in which sequence-conserved Rabs have apparently acquired functions different from their “core” group, serves as cautionary reminder of the potential pitfalls of inferring function based solely on sequence when interrogating divergent lineages.
One important consideration, as mentioned previously, is that sequence analysis itself can be ambiguous. The assignment of the Tetrahymena Rabs by Saito-Nakano et al. (2010) utilized reciprocal Blast searches, rather than the parsimony scores and tree-building approaches used by our group. Saito-Nakano et al. identified orthologs for some Tetrahymena Rabs that, in contrast, appeared lineage-restricted based on our analysis. These differences underscore the point that it can be difficult to derive robust phylogenies in divergent lineages, a problem that may be mitigated with broader sequencing outside of the Opisthokonts. If Saito-Nakano et al.’s conclusions are correct, the endocytic pathway in Tetrahymena is more substantially, though still incompletely, derived from conserved Rabs (Class 1) than inferred by our laboratory’s work. While both surveys indicate that endocytic pathways in Tetrahymena likely contain innovations that arose within this lineage, the nature of those innovations remains unclear. Ultimately, these surveys highlight the benefit of predictions based on multiple analytic approaches, but underline the need for experimental validation.
IDENTIFYING CONCRETE EXAMPLES OF INNOVATION
Whereas a broad survey of Rabs suggested that a number of pathways in Tetrahymena include non-conserved features, a concrete example of molecular innovation, in the form of a novel dynamin described below, was identified in studies of clathrin-mediated endocyosis (CME) in this organism (Elde et al., 2005). CME is widespread in eukaryotes and has historically been among the most intensely studied pathways of membrane traffic (Rappoport, 2008; Traub, 2009). In addition to the clathrin coat, CME involves a host of other proteins, including a complex adapter that links the cargo to be internalized with membrane invagination machinery. This adapter function is provided by the AP-2 heterotetramer, one of a family of AP complexes whose members function in a variety of compartments (Fig. 3A, bottom) (Boehm and Bonifacino, 2002; Robinson, 2004; McMahon and Boucrot, 2011). In Tetrahymena, CME occurs at parasomal sacs, located anterior to each ciliary basal body (Fig. 3A, top) (Nilsson and van Deurs, ’83; Elde et al., 2005). Since basal bodies are regularly spaced along cytoskeletal ribs called meridians (Satir and Wissig, ’82), the Tetrahymena cortex contains a globally ordered array of endocytic portals (Fig. 3B), which facilitates their analysis by microscopy.
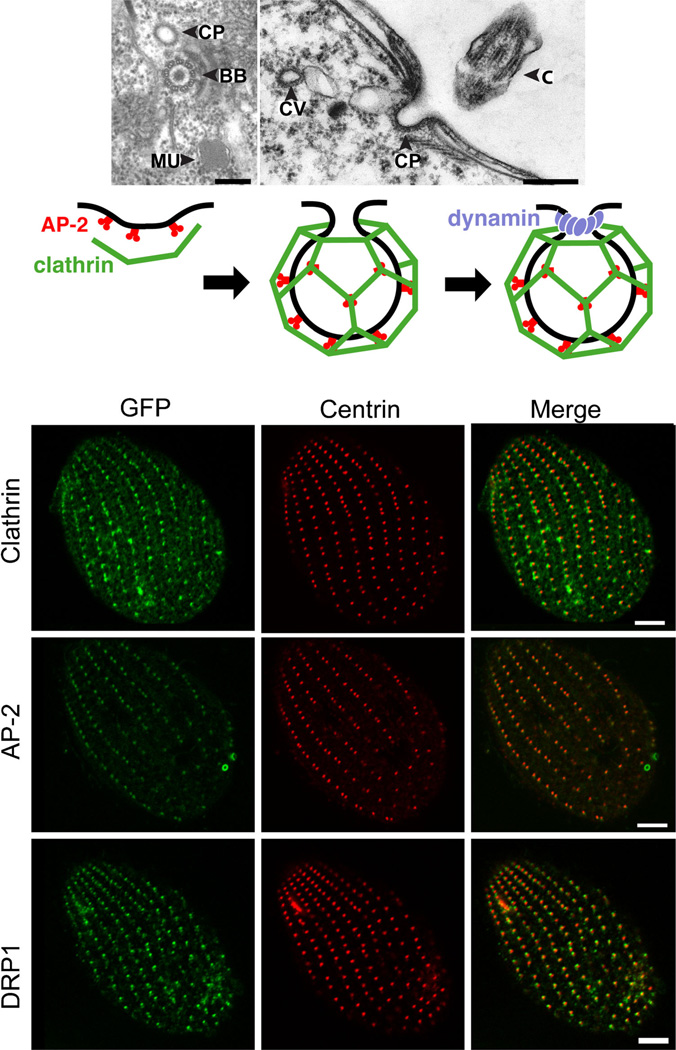
Figure 3.
Proteins involved in clathrin-mediated endocytosis in Tetrahymena localize to parasomal sacs. Figure and legend adapted from Figures 1B, 2A, 3D, 4C, and 5B of Elde et al. (2005). (A) Upper panel: In Tetrahymena, coated pits (CP) are found at parasomal sacs near the base of cilia, as shown in tangential (left) and cross (right) sections. BB, ciliary basal body; C, cilium; CV, coated vesicle; MU, mucocyst. Bars = 200 nm. (Lower panel) Schematic diagram of clathrin-mediated vesicle formation in animals. AP-2 (red) serves as an adapter. It can interact with receptors destined for internalization while also recruiting clathrin (green) to the plasma membrane. Clathrin assembly at those sites enables membrane invagination. Dynamin (blue) assembles at the neck of a nascent vesicle to promote membrane fission. (B) Left column: Cells expressing GFP-fusions of clathrin (top), AP-2 (middle), and Drp1p (bottom). These fusion proteins localize to linear arrays corresponding to parasomal sacs. The distribution of parasomal sacs was inferred by immunofluorescence using an antibody against centrin, which is used here as a marker for basal bodies (middle column). Centrin localizes to the basal bodies that lie immediately posterior to parasomal sacs. Bar = 5 µm. The merge between columns 1 and 2 is shown in the 3rd column.
T thermophila has a single clathrin (heavy chain) gene as well as unique genes encoding each of the AP-2 subunits (Elde et al., 2005). Exploiting the known distribution of parasomal sacs, both clathrin and AP-2, as GFP fusions, were shown to localize to these sites (Fig. 3B, top and middle); in contrast, GFP-tagged AP-1 and −4 complexes localized to other cellular loci. A functional role for clathrin at parasomal sacs could be demonstrated by conditionally expressing a truncated clathrin construct, the “hub” domain, which acts as a dominant-negative inhibitor in other organisms (Liu et al., ’98). In Tetrahymena, expression of the hub domain inhibited endocytic uptake, which was measured by uptake of the styryl dye, FM1-43 (Elde et al., 2005). Therefore, the phylogenetic and functional relationships between T. thermophila clathrin and AP-2, and the homologous proteins in other organisms, reinforced the conclusion that CME is a deeply conserved pathway in eukaryotes.
Another central player in CME is actin. Actin contributes to several stages in metazoan endocytosis (Merrifield et al., 2002; Robertson et al., 2009) and is also involved in fungi (Kaksonen et al., 2003; Toshima et al., 2006). Surprisingly, standard approaches to inhibiting actin function had no effect on FM1-43 uptake at parasomal sacs in T thermophila (Elde et al., 2005), though they dramatically inhibited actin-dependent phagocytosis at the oral apparatus (Elde et al., 2005; Williams et al., 2006). These results suggested that actin plays little if any role in CME in Tetrahymena, though it is also possible that phagocytosis and CME involve different actin isoforms, and that those involved in CME are resistant to standard pharmacologic inhibition. Consistent with this possibility, the related Ciliate P. tetraurelia expresses nine actin isoforms associated with distinct localizations and functions (Sehring et al., 2010). Moreover, actin sequences are altogether relatively divergent in Ciliates (Cupples and Pearlman, ’86; Drouin et al., ’95), so the loss of broadly conserved drug binding sites is plausible, especially since Tetrahymena actins appear to have lost binding sites for phalloidin (Hirono et al., ’89). In addition, some expression profiling data in Tetrahymena suggest that proteins involved in actin dynamics are co-regulated with proteins involved in endocytosis; for example, the gene most tightly coregulated with the µ subunit of AP-2 is a homolog of fibrin, an actin cross-linker (Nusblat et al., 2012). In summary, the current functional data suggest that Tetrahymena CME is relatively actin-independent, which would be highly novel and imply that other mechanisms have arisen to compensate for the loss of actin function in this pathway but more experiments are needed to test alternative explanations. Interestingly there is also evidence of actin-independent endocytosis during specific life stages in trypanosomes (García-Salcedo et al., 2004), which may involve mechanisms that arose separately from those in ciliates.
Remarkably CME in T. thermophila requires a protein that is related, but in an unexpected fashion, to a key protein in animal cell CME (Elde et al., 2005). The large GTPase called classical dynamin, as first shown in fruit flies (van der Bliek and Meyerowitz, ’91), polymerizes around the neck of endocytic invaginations to form a collar that is required for releasing the invagination in the form of a vesicle, that is, vesicle fission (Hinshaw and Schmid, ’95; Merrifield et al., 2005) (Fig. 3A, bottom). Consequently, blocking dynamin function strongly inhibits CME (van der Bliek and Meyerowitz, ’91; McCluskey et al., 2013). Classical dynamin also recruits effectors including actin-binding proteins, which may contribute to its late endocytic role (Mettlen et al., 2009). Notwithstanding this critical role, it is not clear whether the role of dynamin in CME is ancestral, since there is only limited evidence that dynamin homologs participate in CME in most non-Opisthokont lineages (Elde et al., 2005). One strong possibility is that classical dynamin was recruited to CME relatively recently, in evolutionary terms, from an ancestral protein that served a different function, which may have been mitochondrial scission (Liu et al., 2012).
T. thermophila expresses eight dynamin homologs (DRP1-8), reflecting a significant expansion in the gene family within this lineage (Elde et al., 2005). Two of the dynamins localized, as GFP fusions, to parasomal sacs, and of these DRP1 was studied in detail (Fig. 3B, bottom panel). Immuno-EM analysis of Drp1p showed that it localized to the sides of clathrin-coated areas within parasomal sacs, consistent with a role in CME. Gene knockout resulted in cell death, but such cells could be rescued by introducing an exogenous copy of DRP1 under the transcriptional control of a cadmium-inducible promoter. Using these cells, it was possible to show that reducing the expression of DRP1 led to a decrease in CME, measured as above by FM1-43 uptake (Elde et al., 2005). These results indicate that, in contrast to the absence of endocytic dynamins in many non-Opisthokonts, a protein in this family is required for CME in Tetrahymena. This could potentially suggest an ancient role in endocytosis for dynamins, which were subsequently lost in many lineages.
Phylogenetic analysis of the Tetrahymena DRPs revealed a different and more interesting story. A minority of the Tetrahymena DRPs were closely related to dynamins in many other species but most, including DRP1, were found on branches in which the only other members were Tetrahymena or other Ciliate genes (Fig. 4). In other words, the endocytic dynamins of Tetrahymena belong to lineage-restricted branches that arose via expansions (i.e., gene duplication and retention) that occurred after Ciliates had branched from other eukaryotes (Elde et al., 2005). Duplicated gene copies can, by acquiring mutations, also acquire functions that are different from the ancestral gene (Kellis et al., 2004; Taylor and Raes, 2004). The hypothesis suggested by the phylogeny was that a duplicated dynamin in Tetrahymena was recruited to sites of CME, and over time acquired an essential role in that pathway. The same events may have occurred in the animal lineage.
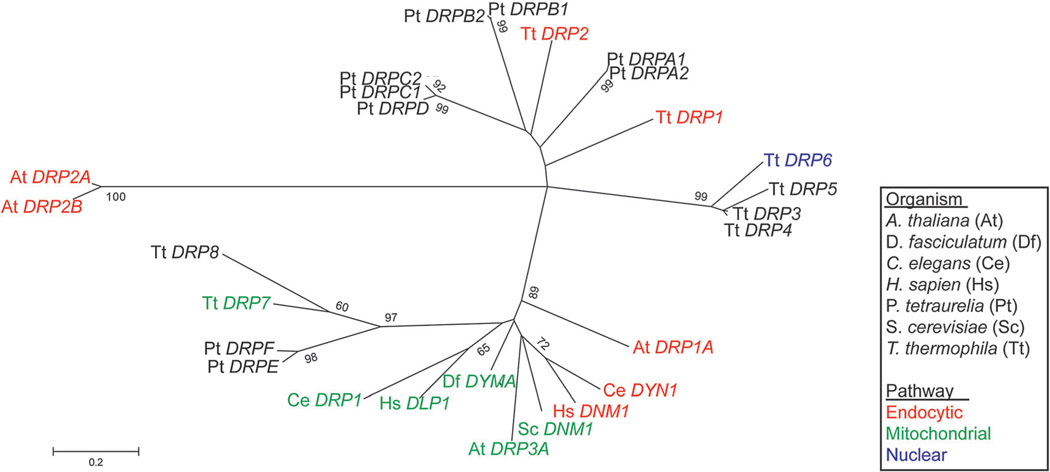
Figure 4.
Endocytic dynamins appear on multiple unrelated branches. An updated maximum likelihood tree of eukaryotic dynamin-related proteins (DRPs) from Elde et al. (2005). Whereas mitochondria-associated dynamins appear to have a single origin, endocytic dynamins are found on multiple, distantly related branches. The tree was constructed from an alignment of the dynamin N domain sequences from which gaps were deleted. The bootstrap consensus tree was inferred from 1,000 replicates; only values >55% are shown.
A testable inference of this hypothesis of parallel recruitment was that mechanisms responsible for targeting dynamin to clathrin-coated pits are likely to differ between animals and Ciliates. To investigate this issue, the targeting motif in Drp1p was identified by expressing chimeras between Drp1p and a 2nd T. thermophila dynamin that localizes to the nuclear envelope, called Drp6p. These experiments revealed that a 28-residue motif in Drp1p, when swapped with the corresponding region of Drp6p, was sufficient to target the otherwise Drp6p protein to parasomal sacs (Elde et al., 2005). Classical dynamin in animals is targeted to coated pits via a pleckstrin homology (PH) domain that binds to specific phosphatidyl inositol head groups (Vallis et al., ’99). The 28 amino acid motif in Drp1p is not predicted to form a PH domain; consistent with this, purified Drp1p showed no binding to phosphatidyl inositides on blots (unpublished) (note that the phylogenetic analysis discussed above was based solely on the most highly conserved domains of dynamins, so was not influenced by divergent targeting domains). Taken together, the results suggest that dynamins were recruited at least twice, in independent lineages, to play essential roles in endocytosis. CME in Tetrahymena therefore appears to be based upon a core of deeply conserved proteins but with at least one innovation that is surprisingly similar to one that developed in animals. That is not to say that the precise roles played by dynamins during CME in the two lineages are identical, and indeed Drp1p lacks the PRD domain present in classical dynamin that is involved in regulating the actin cytoskeleton (Elde et al., 2005). It is tempting to speculate that Drp1p provides some function in Tetrahymena CME that is provided by actin in other lineages.
INDEPENDENT ORIGINS OF REGULATED PEPTIDE SECRETION?
A striking potential case of cellular innovation has emerged from detailed studies of mucocysts in Tetrahymena. Elaborate secretory organelles are found in many Ciliates (Rosati and Modeo, 2003) and have been extensively studied in the form of Tetrahymena mucocysts (Turkewitz et al., 2000; Turkewitz, 2004) and the corresponding organelles in Paramecium, known as trichocysts (Vayssié et al., 2000). The historical use of two different names stemmed from the very distinct morphologies of mucocysts and trichocysts (Hausmann, 78), but more recent molecular work clearly indicates that they are homologous organelles (Adoutte et al., ’84; Turkewitz et al., ’91; Shih and Nelson, ’92; Madeddu et al., ’95; Chilcoat et al., ’96; Gautier et al., ’96; Cowan et al., 2005). Mucocysts and trichocysts belong to a very broad class of secretory organelles more generally termed extrusomes. Extrusomes embody a mode of secretion in which specialized vesicles first accumulate in the cytoplasm. In response to extracellular signals the vesicles can extrude their contents via membrane fusion, called regulated exocytosis (Kelly, ’85). For the Ciliates, the ability to quickly release large amounts of stored extrusome contents has enabled sophisticated strategems (Wessenberg and Antipa, 70; Harumoto, ’94; Sugibayashi and Harumoto, 2000) for predation or defense (Rosati and Modeo, 2003). The term extrusome has also been used to describe secretory organelles in a variety of other eukaryotes (Hausmann, 78), but whether these are evolutionary related to those in Ciliates has not been investigated. From our perspective, a particularly interesting class of secretory vesicles with extrusome-like features are the dense core granules in animals, since there has been extensive molecular and mechanistic analysis of these organelles (Tooze et al., 2001; Crivellato et al., 2006; Hou et al., 2009).
A defining feature of dense core secretory granules in animals, which is shared by Ciliate mucocysts and trichocysts, is an electron-dense protein core (Fig. 5A) (Tokuyasu and Scherbaum, ’65; Bannister, 72; Michael et al., ’87). In both animals and Ciliates, the cores are formed by protein assembly that is controlled by proteolytic processing (Adoutte et al., ’84; Turkewitz et al., ’91; Steiner et al., ’96) Moreover, the core proteins in Ciliates and animals are similar in their acidity (Chilcoat et al., ’96), their capacity for low-affinity calcium binding (Turkewitz et al., ’91; Verbsky and Turkewitz, ’98) and their tendency to aggregate (Gorr et al., ’89; Chanat and Huttner, ’91) within the secretory pathway (Rambourg et al., ’88; Rahaman et al., 2009) The last of these properties is likely to be a key mechanism involved in protein sorting to dense core granules and mucocysts/trichocysts, respectively (Arvan and Castle, ’98; Jain et al., 2000; Bowman et al., 2009). Notwithstanding these and other similarities, there is no evidence that dense core granules and mucocysts/trichocysts arose from the same ancestral organelle (Elde et al., 2007). The genes encoding the principal core proteins in mucocysts, called the GRLs (Chilcoat et al., ’96; Cowan et al., 2005), have no identifiable homologs in animals. The same is true for a 2nd major family of proteins in mucocysts, called GRTs (Fig. 5B) (Bowman et al., 2005), to which we return below. It is also clear that Ciliates have no homologs to the best-studied enzymes responsible for proteolytic processing of granule core proteins in animals, the prohormone convertases, or to key animal proteins involved in regulated exocytosis itself, like the calcium sensor synaptotagmin (unpublished) (Elde et al., 2007). The likelihood is that similar functions are being performed by unrelated genes in Ciliates, as already suggested by the absence of a Ciliate Rab in the regulated exocytosis subgroup (Bright et al., 2010). Consistent with this idea, forward genetic analysis in Paramecium identified a set of Ciliate-specific genes that are required for regulated exocytosis (Cohen and Beisson, ’80; Bonnemain et al., ’92; Klauke et al., ’98; Froissard et al., 2004). Taken together, the evidence suggest that the regulated secretory organelles of Ciliates and animals had largely independent evolutionary histories, but converged on similar features (Class 3) (Elde et al., 2007). But to understand the origins of secretory organelles in Ciliates, one needs to understand the mechanisms underlying their formation, rather than relying on a catalog of the contents.
Figure 5.
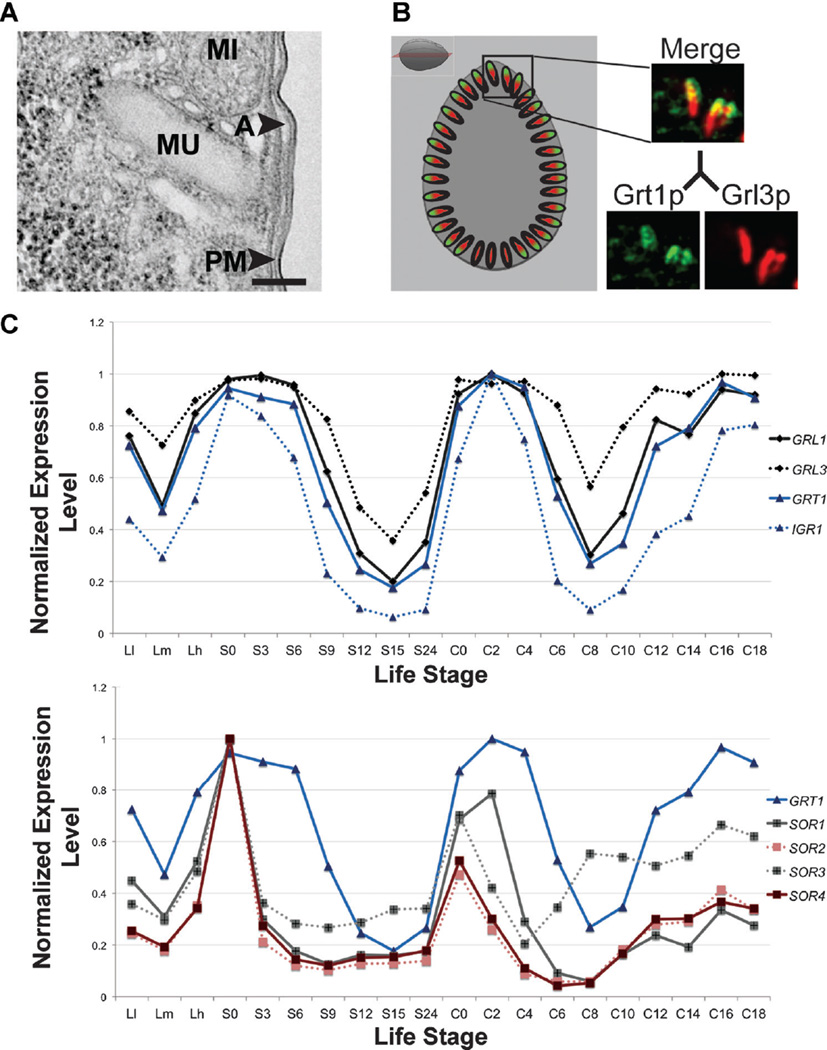
Expression profiling identifies potential mucocyst biogenesis genes. Figure and legend adapted from Figures 2A, 3A, and 5C of Briguglio et al. (2013). (A) Electron micrograph of a mucocyst (MU) docked at the plasma membrane illustrating the characteristic elongated shape and electron dense core. Other structures shown include plasma membrane (PM), Alveolus (A), and mitochondrion (MI). Bar = 200 nM. (B) Simultaneous visualization of mucocysts, by indirect immunofluorescence (IF), using monoclonal antibodies against a GRL family protein (Grl3p) and a GRT family protein (Grt1p). In a cross section through the cell (see cartoon to right for reference), Grl3p (red) is found throughout the mucocyst while Grt1p is concentrated at the tip which docks at the plasma membrane. (C) The expression profiles derived from the Tetrahymena Functional Genomics Database illustrate the three peak expression profile that is characteristic of mucocyst genes. The expression profiles of the four Tetrahymena sortilins (SOR1-4, bottom) are similar to those of genes (GRL1, GRL3, GRT1, and IGR1, top) encoding mucocyst cargo proteins. Each expression profile is normalized to that gene’s maximum expression level. Points on the x-axis correspond to successive time-points and represent growing, starved, and mating cultures, including three different culture densities (low (Ll), medium (Lm), and high (Lh)), 7 samples taken during 24 hr of starvation, and 10 samples subsequently taken during 18 hr following conjugation (Xiong et al., 2013).
We have recently obtained key insights into mechanisms of mucocyst biogenesis in Tetrahymena by exploiting expression profiling via the aforementioned TFGD (Xiong et al., 2013). We initially observed that all known mucocyst genes in Tetrahymena are coregulated, sharing a three-peak expression profile in growing, starved, and conjugating cultures (Fig. 5C, top). By using this profile as a screen, we then found a large number of putative mucocyst-associated genes, some of which had Opisthokont homologs. The latter included genes encoding VPS10 domain-containing proteins (Briguglio et al., 2013) (Fig. 5C, bottom), called sortilins in animals (Petersen et al., ’97), which were first identified in yeast as type 1 transmembrane receptors. More specifically, S. cerevisiae Vps10p in yeast is involved in the sorting of hydrolytic enzymes into vesicles for transport from the TGN to the vacuole, which is the lysosome equivalent in fungi (Marcusson et al., ’94; Cereghino et al., ’95; Deloche et al., 2001). Sortilins in animals, which expanded into a gene family (Hermey, 2009), can serve a similar role in sorting to lysosome-related organelles (Herda et al., 2012). Sortilin-family proteins may also play additional roles in mammalian secretory granule formation, discussed briefly at the end.
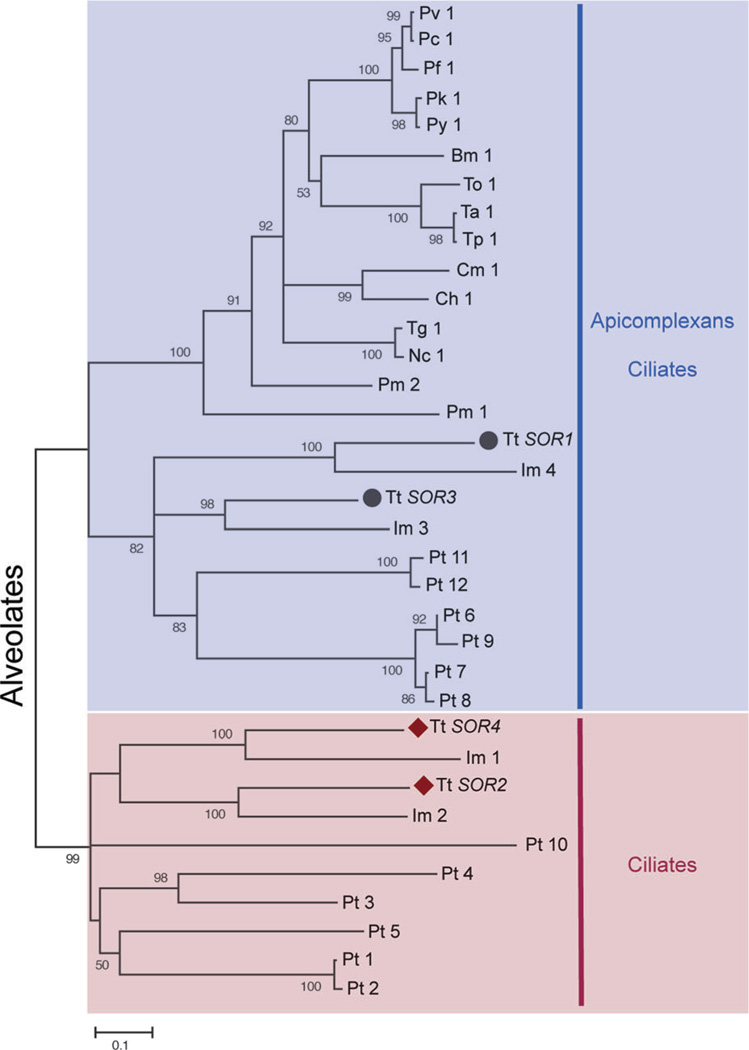
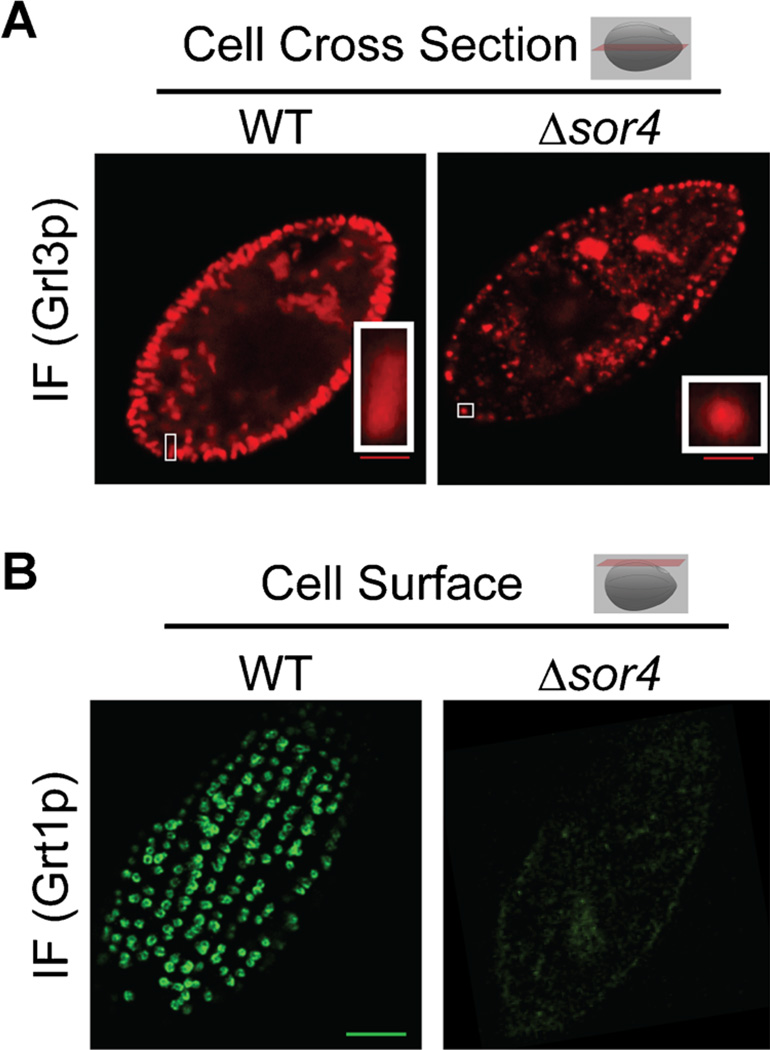
Four sortilins are encoded in the Tetrahymena genome (Briguglio et al., 2013). Phylogenetic analysis divided them into two clades; SOR1 and 3 belong to a clade including non-Ciliate sequences, while SOR2 and 4 belong to a Ciliate-restricted clade (Fig. 6). To test for potential roles in mucocyst formation, each gene was individually disrupted. The disruption of either lineage-restricted sortilin (2 or 4) resulted in clear mucocyst phenotypes; most dramatically, SOR4 knockout cells (Δsor4) failed to extrude any mucocyst content when stimulated. The Δsor4 cells still accumulated mucocysts (Fig. 7A), but these were deficient in at least two classes of cargo proteins. The first was the aforementioned GRT family (Fig. 7B), and the 2nd was an aspartyl cathepsin (Briguglio et al., 2013) shown to play a key role in proteolytic processing during mucocyst formation (S. Kumar, ms. in preparation). These results suggested that, in Tetrahymena, the absence of a sortilin receptor resulted in a failure to sort normal mucocyst contents to the proper compartment. Indeed, Sor4p could be co-immunoprecipitated with Grt proteins, consistent with the idea that it acts as a sorting receptor for these mucocyst components. Interestingly, the sorting of the other major class of mucocyst proteins, the Grls, did not depend on SOR4 function (Briguglio et al., 2013).
Figure 6.
The sortilin family of VPS10 domain-containing receptors has expanded in Tetrahymena. Figure 2B and legend directly from Briguglio et al. (2013). The maximum likelihood tree illustrates a phylogeny of VPS10 domain-containing receptors (sortilins) in Alveolates, the taxonomic group consisting of Ciliates, Apicomplexans, and Dinoflagellates. Two of the T. thermophila sortilins, marked by black circles, cluster with the sortilins from other Alveolates. In contrast, T. thermophila SOR2 and SOR4, marked by maroon diamonds, belong to an expansion of sortilins restricted to Ciliates. Babesia microti (Bm), Cryptosporidium hominis (Ch), Cryptosporidium muris (Cm), Ichthyophthirius multifiliis (Im), Neospora caninum (Nc), Paramecium tetraurelia (Pt), Perkinsus marinus (Pm), Plasmodium berghei (Pb), Plasmodium cynomolgi (Pc), Plasmodium falciparum (Pf), Plasmodium knowlesi (Pk), Plasmodium vivax(Pv), Plasmodium yoelii yoelii(Py), Tetrahymena thermophila (Tt), Theileria annulata (Ta), Theileria orientalis (To), Theileria parva (Tp), and Toxoplasma gondii (Tg).
Figure 7.
Disruption of SOR4 results in the loss of specific mucocyst cargo proteins and aberrant mucocyst morphology. Figure and legend adapted from Figures 3B and 5B of Briguglio et al. (2013). (A) Indirect immunofluorescent visualization of the core protein Grl3p in a cross section of Δsor4 cells reveals that these mutant cells (right) still produce mucocysts, which can be seen at the periphery of the cell, but the mucocysts exhibit an abnormally round morphology compared to the wildtype (left, also compare insets). Bar = 5 µm (B) Indirect immunofluorescent visualization of the tip protein Grt1p in WT cells (left) shows that Grt1p accumulates in the array of docked mucocysts. Here, an optical section at the cell surface is shown, illustrated by the red plane in the cartoon. In contrast, there is only background staining of Grt1p in Δsor4 cells (right), consistent with a failure to sort Grt1p to mucocysts in the absence of Sor4p. Bar = 5 µm
Our recent data, combined with insights from studies in Paramecium (de Loubresse, ’93; Vayssié et al., 2001) and another Ciliate, Pseudomicrothorax dubius (Peck et al., 2012), suggest that the two classes of mucocyst proteins are transported by different mechanisms. The first depends on the aggregation of core components, and is responsible for delivery of Grl proteins, while the second depends on classical receptor-mediated sorting of Grt proteins. Surprisingly, the activity of Sor4p as a mucocyst sorting receptor suggests that the formation of mucocysts and lysosomes relies on shared cellular machinery. Lending further support to this idea, we found that the Tetrahymena AP-3 adaptor is highly coregulated with known mucocyst-associated genes, including SOR4. The AP-3 adaptor, related to but different from the AP-2 adaptor discussed above for CME, is required for sortilin-mediated sorting to lysosome-related organelles (LROs) in Opisthokonts (Raposo et al., 2007). Does this mean that mucocysts represent a class of LROs? Consistent with this idea, another sortilin is essential for LRO formation in the Apicomplexan Toxoplasma gondii, which like Ciliates is an Alveolate (Sloves et al., 2012). However, given the evidence that Grl protein sorting to Tetrahymena mucocysts is Sor4p-independent (Briguglio et al., 2013), it tempting to speculate that mucocyst formation evolved from the coalescence of an LRO-related pathway with a second route of membrane trafficking. In future work, one way to potentially address this question will be to ask whether Grl and Grt proteins are being delivered to mucocysts in separate vesicles, reflecting separate origins. This possibility is directly raised by electron microscopy studies on trichocyst formation in Pseudomicrothorax (Peck et al., 2012).
Discerning the evolutionary links between the mechanisms involved in formation of secretory granules versus mucocysts will ultimately require that we develop a more complete mechanistic picture of both, since many steps are still poorly understood. This point is underscored by recent experimental data. Receptors in the sortilin family have been implicated by genetic studies, in both mice and humans, as contributing to physiological responses that rely on peptides released from secretory granules (Clee et al., 2006; Goodarzi et al., 2007). In this context, sortilins may be acting as signaling receptors, but recent results suggest that these receptors in some cases could be directly involved in granule formation and not serving a retrieval function. That is, sortilin-family receptors may be playing a wider role in animal secretory granule formation than envisioned in current models. Similarly, AP-3 appears to be involved in secretory granule formation in Drosophila and mammalian cells, but the mechanisms involved are not known (Grabner et al., 2006; Asensio et al., 2010). These data, informed by our recent results in Tetrahymena, may indicate surprising mechanistic similarities between the formation of lysosomes and secretory organelles with dense cores, in multiple lineages. Since dense core granules are currently featured as a prime example of non-lysosomal post-TGN trafficking in mammalian cells, validating this new view would significantly change our conception of trafficking in the endomembrane network.
CONCLUSIONS
The endomembrane network is a hallmark of eukaryotic cells. Signal advances over the past 50 years have generated a profound albeit still incomplete understanding of mechanisms involved in establishing and maintaining the compartments within this network, including the details of both endocytic and exocytic pathways, with the vast majority of this work done in a phylogenetically narrow range of model organisms. Our lab and others have investigated a variety of membrane trafficking pathways in distantly related organisms, the Ciliates. Taken together, the results in Ciliates lead to some inferences that would be unsurprising for biologists in many subfields, but that are frequently overlooked by mainstream cell biologists. While the endomembrane network is clearly based on many elements that were already possessed by an ancient eukaryotic ancestor, the extent of conservation of those elements in any specific modern lineage cannot be unambiguously discerned by phylogenetic analysis nor by functional or morphological characterization, but rather requires a combination of approaches. This is because conserved proteins can evolve new functions, and because functionally analogous features can arise via multiple mechanisms. Developing a more nuanced view of the variation in both origins and mechanisms underlying cellular features will create new opportunities for discerning general principles, both physical and genetic, that underlie cellular organization.
Acknowledgments
We thank our current colleagues Cassandra Kontur and Santosh Kumar, as well as previous laboratory members whose work spawned the substance of this review. That work was supported by grants from the National Institutes of Health (General Medicine), the National Science Foundation, and the Chicago Biomedical Consortium. We also thank colleagues working with other Ciliates, particularly the Paramecium groups of Helmut Plattner, Jean Cohen, and Linda Sperling, whose work has been key in understanding pathways discussed here. APT is a member of the scientific advisory board of Tetragenetics, Inc.
The research described here on Tetrahymena sortilins was supported by a Catalyst Grant from the Chicago Biomedical Consortium and by NSF MCB-1051985 to A.P.T. J.S.B. was supported by a National Institutes of Health Training Grant T32 GM007191 and by the NSF through a Graduate Research Fellowship. This manuscript is dedicated to Joseph Frankel, a beacon of Tetrahymena cell biology, on the occasion of his retirement from the University of Iowa.
LITERATURE CITED
- Adoutte A, De Loubresse NG, Beisson J. Proteolytic cleavage and maturation of the crystalline secretion products of Paramecium. J Mol Biol. 1984;180:1065–1081. doi: 10.1016/0022-2836(84)90271-7. [DOI] [PubMed] [Google Scholar]
- Allen RD. Fine structure, reconstruction and possible functions of components of the cortex of Tetrahymena pyriformis . J Protozool. 1967;14:553–565. doi: 10.1111/j.1550-7408.1967.tb02042.x. [DOI] [PubMed] [Google Scholar]
- Allen RD, Wolf RW. Membrane recycling at the cytoproct of Tetrahymena. J Cell Sci. 1979;35:217–227. doi: 10.1242/jcs.35.1.217. [DOI] [PubMed] [Google Scholar]
- Arvan P, Castle D. Sorting and storage during secretory granule biogenesis: looking backward and looking forward. Biochem J. 1998;332:593–610. doi: 10.1042/bj3320593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensio CS, Sirkis DW, Edwards RH. RNAi screen identifies a role for adaptor protein AP-3 in sorting to the regulated secretory pathway. J Cell Biol. 2010;191:1173–1187. doi: 10.1083/jcb.201006131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool forthe unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M, Aurrecoechea C, Berriman M, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala J, Olofsson B, Tavitian A, Prochiantz A. Developmental and regional regulation of rab3: a new brain specific “ras-like” gene. J Neurosci Res. 1989;22:241–246. doi: 10.1002/jnr.490220303. [DOI] [PubMed] [Google Scholar]
- Baldauf SL. An overview of the phylogeny and diversity of eukaryotes. J Syst Evol. 2008;46:263–273. [Google Scholar]
- Baldauf SL, Palmer JD. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister LH. The structure of trichocysts in Paramecium caudatum . J Cell Sci. 1972;11:899–929. doi: 10.1242/jcs.11.3.899. [DOI] [PubMed] [Google Scholar]
- Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nat Cell Biol. 2005;438:597–604. doi: 10.1038/nature04397. [DOI] [PubMed] [Google Scholar]
- Beisson J, Bétermier M, Bré M-H, et al. Paramecium tetraurelia: the renaissance of an early unicellular model. Cold Spring Harb Protoc 2010. 2010 doi: 10.1101/pdb.emo140. http://www.ncbi.nlm.nih.gov/pubmed/?term=Paramecium+tetraurelia%3A+the+renaissance+of+an+early+unicellular+model. [DOI] [PubMed]
- Blackburn EH. Telomeres and telomerase: the means to the end (Nobel lecture) Angew Chem Int Ed Engl. 2010;49:7405–7421. doi: 10.1002/anie.201002387. [DOI] [PubMed] [Google Scholar]
- Boehm M, Bonifacino JS. Genetic analyses of adaptin function from yeast to mammals. Gene. 2002;286:175–186. doi: 10.1016/s0378-1119(02)00422-5. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonnemain H, Gulik-Krzywicki T, Grandchamp C, Cohen J. Interactions between genes involved in exocytotic membrane fusion in paramecium. Genetics. 1992;130:461–470. doi: 10.1093/genetics/130.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Turkewitz AP. Analysis of a mutant exhibiting conditional sorting to dense core secretory granules in Tetrahymena thermophila . Genetics. 2001;159:1605–1616. doi: 10.1093/genetics/159.4.1605. http://link.springer.com/chapter/10.1007/978-0-387-93877-6_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Elde NC, Morgan G, Winey M, Turkewitz AP. Core formation and the acquisition of fusion competence are linked during secretory granule maturation in Tetrahymena. Traffic. 2005;6:303–323. doi: 10.1111/j.1600-0854.2005.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Cowan AT, Turkewitz AP. Biogenesis of dense-core secretory granules. Traffic Inside Cells. 2009:183–209. [Google Scholar]
- Brighouse A, Dacks JB, Field MC. Rab protein evolution and the history of the eukaryotic endomembrane system. Cell Mol Life Sci. 2010;67:3449–3465. doi: 10.1007/s00018-010-0436-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright LJ, Kambesis N, Nelson SB, Jeong B, Turkewitz AP. Comprehensive analysis reveals dynamic and evolutionary plasticity of Rab GTPases and membrane traffic in Tetrahymena thermophila . PLoS Genet. 2010;6:e1001155. doi: 10.1371/journal.pgen.1001155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briguglio JS, Kumar S, Turkewitz AP. Lysosomal sorting receptors are essential for secretory granule biogenesis in Tetrahymena. J Cell Biol. 2013;203:537–550. doi: 10.1083/jcb.201305086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, Eppig JT, Blake JA, et al. The mouse genome database: genotypes, phenotypes, and models of human disease. Nucleic Acids Res. 2013;41:D885–D891. doi: 10.1093/nar/gks1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, et al. Phylogenomics reshuffles the eukaryotic supergroups. PLoS ONE. 2007;2:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao E, Oates B. Molecular phylogeny of Amoebozoa and the evolutionary significance of the unikont Phalansterium. Eur J Protistol. 2004;40:21–48. http://www.sciencedirect.com/science/article/pii/S0932473904000045. [Google Scholar]
- Cech TR. Self-splicing and enzymatic activity of an intervening sequence RNA from Tetrahymena. Biosci Rep. 2004;24:362–385. doi: 10.1007/s10540-005-2738-3. [DOI] [PubMed] [Google Scholar]
- Cereghino JL, Marcusson EG, Emr SD. The cytoplasmic tail domain of the vacuolar protein sorting receptor Vps10p and a subset of VPS gene products regulate receptor stability, function, and localization. Mol Biol Cell. 1995;6:1089–1102. doi: 10.1091/mbc.6.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanat E, Huttner WB. Milieu-induced, selective aggregation of regulated secretory proteins in the trans-Golgi network. J Cell Biol. 1991;115:1505–1519. doi: 10.1083/jcb.115.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JM, Hong EL, Amundsen C, et al. Saccharomyces genome database: the genomics resource of budding yeast. Nucleic Acids Res. 2012;40:D700–D705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcoat ND, Melia SM, Haddad A, Turkewitz AP. Granule lattice protein 1 (Grl1p), an acidic, calcium-binding protein in Tetrahymena thermophila dense-core secretory granules, influences granule size, shape, content organization, and release but not protein sorting or condensation. J Cell Biol. 1996;135:1775–1787. doi: 10.1083/jcb.135.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clee SM, Yandell BS, Schueler KM, et al. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet. 2006;38:688–693. doi: 10.1038/ng1796. [DOI] [PubMed] [Google Scholar]
- Cohen J, Beisson J. Genetic analysis of the relationships between the cell surface and the nuclei in Paramecium tetraurelia . Genetics. 1980;95:797–818. doi: 10.1093/genetics/95.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K, Gorovsky MA. Tetrahymena thermophila. Curr Biol. 2005;15:R317–R318. doi: 10.1016/j.cub.2005.04.039. [DOI] [PubMed] [Google Scholar]
- Cowan AT, Bowman GR, Edwards KF, Emerson JJ, Turkewitz AP. Genetic, genomic, and functional analysis of the granule lattice proteins in Tetrahymena secretory granules. Mol Biol Cell. 2005;16:4046–4060. doi: 10.1091/mbc.E05-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crivellato E, Nico B, Bertelli E, Nussdorfer GG, Ribatti D. Dense-core granules in neuroendocrine cells and neurons release their secretory constituents by piecemeal degranulation (review) Int J Mol Med. 2006;18:1037–1046. [PubMed] [Google Scholar]
- Cupples CG, Pearlman RE. Isolation and characterization of the actin gene from Tetrahymena thermophila . Proc Natl Acad Sci USA. 1986;83:5160–5164. doi: 10.1073/pnas.83.14.5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacks JB, Field MC. Evolution of the eukaryotic membrane-trafficking system: origin, tempo and mode. Proc Natl Acad Sci USA. 2007;120:2977–2985. doi: 10.1242/jcs.013250. http://www.ncbi.nlm.nih.gov/pubmed/?term=dacks+origin+tempo+mode. [DOI] [PubMed] [Google Scholar]
- Dacks JB, Peden AA, Field MC. Evolution of specificity in the eukaryotic endomembrane system. Int J Biochem Cell Biol. 2009;41:330–340. doi: 10.1016/j.biocel.2008.08.041. [DOI] [PubMed] [Google Scholar]
- Davis RH. Timeline: the age of model organisms. Nat Rev Genet. 2004;5:69–76. doi: 10.1038/nrg1250. [DOI] [PubMed] [Google Scholar]
- de Loubresse NG. Early steps of the secretory pathway in Paramecium: ultrastructural, immunocytochemical, and genetic analysis of trichocyst biogenesis. Adv Cell Mol Biol Membr. 1993;2A:27–59. [Google Scholar]
- Deloche O, Yeung BG, Payne GS, Schekman R. Vps10p transport from the trans-Golgi network to the endosome is mediated by clathrin-coated vesicles. Mol Biol Cell. 2001;12:475–485. doi: 10.1091/mbc.12.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin G, Moniz de Sá M, Zuker M. The Giardia lamblia actin gene and the phylogeny of eukaryotes. J Mol Evol. 1995;41:841–849. doi: 10.1007/BF00173163. [DOI] [PubMed] [Google Scholar]
- Elde NC, Morgan G, Winey M, Sperling L, Turkewitz AP. Elucidation of clathrin-mediated endocytosis in tetrahymena reveals an evolutionarily convergent recruitment of dynamin. PLoS Genet. 2005;1:e52. doi: 10.1371/journal.pgen.0010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Long M, Turkewitz AP. A role for convergent evolution in the secretory life of cells. Trends Cell Biol. 2007;17:157–164. doi: 10.1016/j.tcb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Elias M, Brighouse A, Gabernet-Castello C, Field MC, Dacks JB. Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J Cell Sci. 2012;125:2500–2508. doi: 10.1242/jcs.101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S, Jahn R. Vesicle fusion from yeast to man. Nature. 1994;370:191–193. doi: 10.1038/370191a0. [DOI] [PubMed] [Google Scholar]
- Finger FP. Spatial regulation of exocytosis: lessons from yeast. J Cell Biol. 1998;142:609–612. doi: 10.1083/jcb.142.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. Cell biology of Tetrahymena thermophila . Methods Cell Biol. 2000;62:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- Froissard M, Keller A-M, Dedieu J-C, Cohen J. Novel secretory vesicle proteins essential for membrane fusion display extracellular-matrix domains. Traffic. 2004;5:493–502. doi: 10.1111/j.1600-0854.2004.00194.x. [DOI] [PubMed] [Google Scholar]
- García-Salcedo JA, Pérez-Morga D, Gijón P, et al. A differential role for actin during the life cycle of Trypanosoma brucei . EMBO J. 2004;23:780–789. doi: 10.1038/sj.emboj.7600094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier MC, Sperling L, Madeddu L. Cloning and sequence analysis of genes coding for paramecium secretory granule (trichocyst) proteins. A unique protein fold for a family of polypeptides with different primary structures. J Biol Chem. 1996;271:10247–10255. doi: 10.1074/jbc.271.17.10247. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ. Dynein: a protein with adenosine triphosphatase activity from cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- Gönczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- Goodarzi MO, Lehman DM, Taylor KD, et al. SORCS1: a novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes. 2007;56:1922–1929. doi: 10.2337/db06-1677. [DOI] [PubMed] [Google Scholar]
- Gorr SU, Shioi J, Cohn DV. Interaction of calcium with porcine adrenal chromogranin A (secretory protein-I) and chromogranin B (secretogranin I) Am J Physiol Endocrinol Metab. 1989;257:E247–E254. doi: 10.1152/ajpendo.1989.257.2.E247. [DOI] [PubMed] [Google Scholar]
- Grabner CP, Price SD, Lysakowski A, Cahill AL, Fox AP. Regulation of large dense-core vesicle volume and neurotransmitter content mediated by adaptor protein 3. Proc Natl Acad Sci USA. 2006;103:10035–10040. doi: 10.1073/pnas.0509844103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci USA. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkan C, Lapp H, Alory C, et al. Large-scale profiling of Rab GTPase trafficking networks: the membrome. Mol Biol Cell. 2005;16:3847–3864. doi: 10.1091/mbc.E05-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harumoto T. The role of trichocyst discharge and backward swimming in escaping behavior of paramecium from Dileptus margaritifer . J Eukaryot Microbiol. 1994;41:560–564. [Google Scholar]
- Hausmann K. Extrusive organelles in protists. Int Rev Cytol. 1978;52:197–276. doi: 10.1016/s0074-7696(08)60757-3. [DOI] [PubMed] [Google Scholar]
- Hausmann K, Allen RD. Membrane behavior of exocytic vesicles. II. Fate of the trichocyst membranes in Paramecium after induced trichocyst discharge. J Cell Biol. 1976;69:313–326. doi: 10.1083/jcb.69.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Herda S, Raczkowski F, Mittrücker H-W, et al. The sorting receptor Sortilin exhibits a dual function in exocytic trafficking of interferon-γ and granzyme A in T cells. Immunity. 2012;37:854–866. doi: 10.1016/j.immuni.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Hermey G. The Vps10p-domain receptor family. Cell Mol LifeSci. 2009;66:2677–2689. doi: 10.1007/s00018-009-0043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Hirono M, Kumagai Y, Numata O, Watanabe Y. Purification of Tetrahymena actin reveals some unusual properties. Proc Natl Acad Sci USA. 1989;86:75–79. doi: 10.1073/pnas.86.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JC, Min L, Pessin JE. Insulin granule biogenesis, trafficking and exocytosis. Vitam Horm. 2009;80:473–506. doi: 10.1016/S0083-6729(08)00616-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, Sibley LD. Modulation of innate immunity by Toxoplasma gondii virulence effectors. Nat Rev Microbiol. 2012;10:766–778. doi: 10.1038/nrmicro2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Mori C, Kojidani T, et al. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr Biol. 2009;19:843–847. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- Jain RK, Joyce PB, Gorr SU. Aggregation chaperones enhance aggregation and storage of secretory proteins in endocrine cells. J Biol Chem. 2000;275:27032–27036. doi: 10.1074/jbc.M000095200. [DOI] [PubMed] [Google Scholar]
- Jekely GSR. Small GTPases and the evolution of the eukaryotic cell. BioEssays. 2003;25:1129–1138. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae . Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kelly R. Pathways of protein secretion in eukaryotes. Science. 1985;230:25–32. doi: 10.1126/science.2994224. [DOI] [PubMed] [Google Scholar]
- Kienle N, Kloepper TH, Fasshauer D. Differences in the SNARE evolution of fungi and metazoa. Biochem Soc Trans. 2009;37:787–791. doi: 10.1042/BST0370787. [DOI] [PubMed] [Google Scholar]
- Kimble J. Molecular regulation of the mitosis/meiosis decision in multicellular organisms. Cold Spring Harb Perspect Biol. 2011;3:a002683. doi: 10.1101/cshperspect.a002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissinger JC, Gajria B, Li L, Paulsen IT, Roos DS. ToxoDB: accessing the Toxoplasma gondii genome. Nucleic Acids Res. 2003;31:234–236. doi: 10.1093/nar/gkg072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiy T, Vosskühler C, Rasmussen L, Tiedtke A. Three pools of lysosomal enzymes in Tetrahymena thermophila . Exp Cell Res. 1993;205:286–292. doi: 10.1006/excr.1993.1088. http://www.ncbi.nlm.nih.gov/pubmed/?term=three+pools+of+lysosomal+enzymes+in+tetrahymena. [DOI] [PubMed] [Google Scholar]
- Klauke N, Kissmehl R, Plattner H, Haga N, Watanabe T. An exocytotic mutant of Paramecium caudatum: membrane fusion without secretory contents release. Cell Calcium V. 1998;23:349-L–360-L. doi: 10.1016/s0143-4160(98)90030-6. [DOI] [PubMed] [Google Scholar]
- Kono M, Prusty D, Parkinson J, Gilberger TW. The apicomplexan inner membrane complex. Front Biosci. 2013;18:982–992. doi: 10.2741/4157. [DOI] [PubMed] [Google Scholar]
- Koumandou VL, Dacks JB, Coulson RMR, Field MC. Control systems for membrane fusion in the ancestral eukaryote; evolution of tethering complexes and SM proteins. BMC Evol Biol. 2007;7:29. doi: 10.1186/1471-2148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumandou VL, Klute MJ, Herman EK, et al. Evolutionary reconstruction of the retromer complex and its function in Trypanosoma brucei . J Cell Sci. 2011;124:1496–1509. doi: 10.1242/jcs.081596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N. Autophagy and cell death in model organisms. Cell Death Differ. 2008;16:21–30. doi: 10.1038/cdd.2008.120. [DOI] [PubMed] [Google Scholar]
- Lal K, Field MC, Carlton JM, Warwicker J, Hirt RP. Identification of a very large Rab GTPase family in the parasitic protozoan Trichomonas vaginalis . Mol Biochem Parasitol. 2005;143:226–235. doi: 10.1016/j.molbiopara.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, et al. The Arabidopsis information resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012;40:D1202–D1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SH, Marks MS, Brodsky FM. A dominant-negative clathrin mutant differentially affects trafficking of molecules with distinct sorting motifs in the class II major histocompatibility complex (MHC) pathway. J Cell Biol. 1998;140:1023–1037. doi: 10.1083/jcb.140.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-W, Su AI, Schmid SL. The evolution of dynamin to regulate clathrin-mediated endocytosis. BioEssays. 2012;34:643–647. doi: 10.1002/bies.201200033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DH. Ciliophora. Chichester, UK: John Wiley & Sons, Ltd; 2001. [Google Scholar]
- Lynn DH. Morphology or molecules: how do we identify the major lineages of ciliates (Phylum Ciliophora) Eur J Protistol. 2003;39:356–364. [Google Scholar]
- Madeddu L, Gautier MC, Vayssié L, Houari A, Sperling L. A large multigene family codes for the polypeptides of the crystalline trichocyst matrix in Paramecium. Mol Biol Cell. 1995;6:649–659. doi: 10.1091/mbc.6.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madinger CL, Collins K, Fields LG, Taron CH, Benner JS. Constitutive secretion in Tetrahymena thermophila . Eukaryot Cell. 2010;9:674–681. doi: 10.1128/EC.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Falkowska KA, Li AY, et al. Nucleus-specific importin alpha proteins and nucleoporins regulate protein import and nuclear division in the binucleate Tetrahymena thermophila . Eukaryot Cell. 2008;7:1487–1499. doi: 10.1128/EC.00193-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- McCluskey A, Daniel JA, Hadzic G, et al. Building a better dynasore: the dyngo compounds potently inhibit dynamin and endocytosis. Traffic. 2013;4:1272–1289. doi: 10.1111/tra.12119. http://www.ncbi.nlm.nih.gov/pubmed/24025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Mills IG. COPand clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- McQuilton P, St Pierre SE, Thurmond J FlyBase Consortium. FlyBase 101—the basics of navigating FlyBase. Nucleic Acids Res. 2012;40:D706–D714. doi: 10.1093/nar/gkr1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrincoated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, et al. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael J, Carroll R, Swift HH, Steiner DF. Studies on the molecular organization of rat insulin secretory granules. J Biol Chem. 1987;262:16531–16535. [PubMed] [Google Scholar]
- Nelsen EM, Debault LE. Transformation in Tetrahymena pyriformis: description of an inducible phenotype. J Protozool. 1978;25:113–119. doi: 10.1111/j.1550-7408.1978.tb03880.x. [DOI] [PubMed] [Google Scholar]
- Nilsson JR. Phagotrophy in Tetrahymena . In: Hutner SH, editor. Biochemistry and Physiology of Protozoa. Vol. 2. New York: Academic Press; 1979. pp. 339–379. [Google Scholar]
- Nilsson JR, van Deurs B. Coated pits with pinocytosis in Tetrahymena. J Cell Sci. 1983;63:209–222. doi: 10.1242/jcs.63.1.209. [DOI] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, et al. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusblat AD, Bright LJ, Turkewitz AP. Conservation and innovation in Tetrahymena membrane traffic: proteins, lipids, and compartments. Methods Cell Biol. 2012;109:141–175. doi: 10.1016/B978-0-12-385967-9.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Barbero E, Lasser E, et al. Evaluating support for the current classification of eukaryotic diversity. PLoS Genet. 2006;2:e220. doi: 10.1371/journal.pgen.0020220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck RK, Swiderski B, Tourmel A-M. Secretory organelle biogenesis: trichocyst formation in a ciliated protozoan. Biology of the Cell. 2012;77:277–287. [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Pereira-Leal JB, Teichmann SA. Novel specificities emerge by stepwise duplication of functional modules. Genome Res. 2005;15:552–559. doi: 10.1101/gr.3102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CM, Nielsen MS, Nykjaer A, et al. Molecular identification of a novel candidate sorting receptor purified from human brain by receptor-associated protein affinity chromatography. J Biol Chem. 1997;272:3599–3605. doi: 10.1074/jbc.272.6.3599. [DOI] [PubMed] [Google Scholar]
- Plattner H, Knoll G, Pape R. Synchronization of different steps of the secretory cycle in Paramecium tetraurelia: trichocyst exocytosis, exocytosis-coupled endocytosis, and intracellular transport. Adv Cell Mol Biol Membr. 1993;2A:123–148. [Google Scholar]
- Plattner H, Sehring IM, Mohamed IK, et al. Calcium signaling in closely related protozoan groups (Alveolata): non-parasitic ciliates (Paramecium,Tetrahymena) vs. parasitic Apicomplexa (Plasmodium, Toxoplasma) Cell Calcium. 2012;51:351–382. doi: 10.1016/j.ceca.2012.01.006. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Spielmann T, Brahimi K, et al. The Plasmodium falciparum family of Rab GTPases. Gene. 2003;306:13–25. doi: 10.1016/s0378-1119(03)00381-0. [DOI] [PubMed] [Google Scholar]
- Rahaman A, Miao W, Turkewitz AP. Independent transport and sorting of functionally distinct protein families in Tetrahymena thermophila dense core secretory granules. Eukaryot Cell. 2009;8:1575–1583. doi: 10.1128/EC.00151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Hermo L. Formation of secretion granules in the Golgi apparatus of pancreatic acinar cells of the rat. Am J Anat. 1988;183:187–199. doi: 10.1002/aja.1001830302. [DOI] [PubMed] [Google Scholar]
- Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport JZ. Focusing on clathrin-mediated endocytosis. Biochem J. 2008;412:415. doi: 10.1042/BJ20080474. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Weber APM, Bhattacharya D. The origin and establishment of the plastid in algae and plants. Annu Rev Genet. 2007;41:147–168. doi: 10.1146/annurev.genet.41.110306.130134. [DOI] [PubMed] [Google Scholar]
- Robertson AS, Smythe E, Ayscough KR. Functions of actin in endocytosis. Cell Mol Life Sci. 2009;66:2049–2065. doi: 10.1007/s00018-009-0001-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Rosati G, Modeo L. Extrusomes in ciliates: diversification, distribution, and phylogenetic implications. J Eukaryot Microbiol. 2003;50:383–402. doi: 10.1111/j.1550-7408.2003.tb00260.x. [DOI] [PubMed] [Google Scholar]
- Saito-Nakano Y, Loftus BJ, Hall N, Nozaki T. The diversity of Rab GTPases in Entamoeba histolytica . Exp Parasitol. 2005;110:244–252. doi: 10.1016/j.exppara.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Saito-Nakano Y, Nakahara T, Nakano K, Nozaki T, Numata O. Marked amplification and diversification of products of ras genes from rat brain, Rab GTPases, in the ciliates Tetrahymena thermophila and Paramecium tetraurelia . J Eukaryot Microbiol. 2010;57:389–399. doi: 10.1111/j.1550-7408.2010.00503.x. [DOI] [PubMed] [Google Scholar]
- Satir B. Dibucaine-induced synchronous mucocyst secretion in Tetrahymena. Cell Biol Int Rep. 1977;1:69–73. doi: 10.1016/0309-1651(77)90012-1. [DOI] [PubMed] [Google Scholar]
- Satir BH, Wissig SL. Alveolar sacs of Tetrahymena: ultrastructural characteristics and similarities to subsurface cisterns of muscle and nerve. J Cell Sci. 1982;55:13–33. doi: 10.1242/jcs.55.1.13. [DOI] [PubMed] [Google Scholar]
- Segev N. Ypt/Rab GTPases: regulators of protein trafficking. Sci Signal. 2001;2001:re11–re11. doi: 10.1126/stke.2001.100.re11. [DOI] [PubMed] [Google Scholar]
- Sehring IM, Reiner C, Plattner H. The actin subfamily PtAct4, out of many subfamilies, is differentially localized for specific local functions in Paramecium tetraurelia cells. Eur J Cell Biol. 2010;89:509–524. doi: 10.1016/j.ejcb.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Shih SJ, Nelson DL. Proteolytic processing of secretory proteins in Paramecium: immunological and biochemical characterization of the precursors of trichocyst matrix proteins. J Cell Sci. 1992;103:349–361. doi: 10.1242/jcs.103.2.349. [DOI] [PubMed] [Google Scholar]
- Sibley LD. Intracellular parasite invasion strategies. Science. 2004;304:248–253. doi: 10.1126/science.1094717. [DOI] [PubMed] [Google Scholar]
- Simons M, Mlodzik M. Planar cell polarity signaling: from fly development to human disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AGB, Patterson DJ. The ultrastructure of Carpediemonas membranifera (Eukaryota) with reference to the “excavate hypothesis”. Eur J Protistol. 1999;35:353–370. [Google Scholar]
- Sloves P-J, Delhaye S, Mouveaux T, et al. Toxoplasma sortilin-like receptor regulates protein transport and is essential for apical secretory organelle biogenesis and host infection. Cell Host Microbe. 2012;11:515–527. doi: 10.1016/j.chom.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Steiner DF, Rouillé Y, Gong Q, et al. The role of prohormone convertases in insulin biosynthesis: evidence for inherited defects in their action in man and experimental animals. Diabetes Metab. 1996;22:94–104. [PubMed] [Google Scholar]
- Stelly N, Mauger JP, Claret M, Adoutte A. Cortical alveoli of Paramecium: a vast submembranous calcium storage compartment. J Cell Biol. 1991;113:103–112. doi: 10.1083/jcb.113.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stenmark H, Olkkonen VM. The Rab GTPase family. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-reviews3007. REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens NA, Kieft R, Macleod A, Hajduk SL. Trypanosome resistance to human innate immunity: targeting Achilles’ heel. Trends Parasitol. 2012;28:539–545. doi: 10.1016/j.pt.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugibayashi R, Harumoto T. Defensive function of trichocysts in Paramecium tetraurelia against heterotrich ciliate Climacostomum virens . Eur J Protistol. 2000;36:415–422. [Google Scholar]
- Swart EC, Bracht JR, Magrini V, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J. Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet. 2004;38:615–643. doi: 10.1146/annurev.genet.38.072902.092831. [DOI] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, et al. The million mutation project: a new approach to genetics in Caenorhabditis elegans . Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. http://www.ncbi.nlm.nih.gov/pubmed/23800452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedtke A. Capsule shedding in tetrahymena. Naturwissen-schaften. 1976;63:93. doi: 10.1007/BF00622415. [DOI] [PubMed] [Google Scholar]
- Tokuyasu K, Scherbaum OH. Ultrastructure of mucocysts and pellicle of Tetrahymena pyriformis . J Cell Biol. 1965;27:67–81. doi: 10.1083/jcb.27.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze SA, Martens GJ, Huttner WB. Secretory granule biogenesis: rafting to the SNARE. Trends Cell Biol. 2001;11:116–122. doi: 10.1016/s0962-8924(00)01907-3. [DOI] [PubMed] [Google Scholar]
- Toshima JY, Toshima J, Kaksonen M, et al. Spatial dynamics of receptor-mediated endocytic trafficking in budding yeast revealed by using fluorescent alpha-factor derivatives. Proc Natl Acad Sci USA. 2006;103:5793–5798. doi: 10.1073/pnas.0601042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- Turkewitz AP. Out with a bang! Tetrahymena as a model system to study secretory granule biogenesis. Traffic. 2004;5:63–68. doi: 10.1046/j.1600-0854.2003.00155.x. [DOI] [PubMed] [Google Scholar]
- Turkewitz AP, Madeddu L, Kelly RB. Maturation of dense core granules in wild type and mutant Tetrahymena thermophila . EMBO J. 1991;10:1979–1987. doi: 10.1002/j.1460-2075.1991.tb07727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkewitz AP, Chilcoat ND, Haddad A, Verbsky JW. Regulated protein secretion in Tetrahymena thermophila. Methods Cell Biol. 2000;62:347–362. doi: 10.1016/s0091-679x(08)61542-3. [DOI] [PubMed] [Google Scholar]
- Turkewitz AP, Orias E, Kapler G. Functional genomics: the coming of age for Tetrahymena thermophila . Trends Genet. 2002;18:35–40. doi: 10.1016/s0168-9525(01)02560-4. [DOI] [PubMed] [Google Scholar]
- Turner S, Pryer KM, Miao VP, Palmer JD. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J Eukaryot Microbiol. 1999;46:327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x. [DOI] [PubMed] [Google Scholar]
- Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- van der Bliek AM, Meyerowitz EM. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- Vayssié L, Skouri F, Sperling L, Cohen J. Molecular genetics of regulated secretion in paramecium. Biochimie. 2000;82:269–288. doi: 10.1016/s0300-9084(00)00201-7. [DOI] [PubMed] [Google Scholar]
- Vayssié L, Garreau de Loubresse N, Sperling L. Growth and form of secretory granules involves stepwise assembly but not differential sorting of a family of secretory proteins in Paramecium. J Cell Sci. 2001;114:875–886. doi: 10.1242/jcs.114.5.875. [DOI] [PubMed] [Google Scholar]
- Verbsky JW, Turkewitz AP. Proteolytic processing and Ca2+-binding activity of dense-core vesicle polypeptides in Tetrahymena. Mol Biol Cell. 1998;9:497–511. doi: 10.1091/mbc.9.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainright PO, Hinkle G, Sogin ML, Stickel SK. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260:340–342. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- Wessenberg H, Antipa G. Capture and Ingestion of Paramecium by Didinium nasutum . J Protozool. 1970;17:250–270. [Google Scholar]
- Westermann B. Mitochondrial dynamics in model organisms: what yeasts, worms and flies have taught us about fusion and fission of mitochondria. Semin Cell Dev Biol. 2010;21:542–549. doi: 10.1016/j.semcdb.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Williams NE, Tsao C-C, Bowen J, et al. The actin gene ACT1 is required for phagocytosis, motility, and cell separation of Tetrahymena thermophila . Eukaryot Cell. 2006;5:555–567. doi: 10.1128/EC.5.3.555-567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe J. The conjugation junction ofTetrahymena: its structure and development. J Morphol. 1982;172:159–178. doi: 10.1002/jmor.1051720204. [DOI] [PubMed] [Google Scholar]
- Wolfe J, Grimes GW. Tip transformation in Tetrahymena: a morphogenetic response to interactions between mating types*. J Protozool. 1979;26:82–89. [Google Scholar]
- Xiong J, Lu Y, Feng J, et al. Tetrahymena functional genomics database (TetraFGD): an integrated resource for Tetrahymena functional genomics. Database (Oxford) 2013;2013 doi: 10.1093/database/bat008. bat008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, et al. Thirty-one flavors of Drosophila rab proteins. Genetics. 2007;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]