Abstract
Periodontitis is an infectious inflammatory disease that destroys the tooth-supporting tissues. It is caused by the formation of subgingival biofilms on the surface of the tooth. Characteristic bacteria associated with subgingival biofilms are the Gram-negative anaerobes Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola, collectively known as the “red complex” species. Inter-epithelial junctions ensure the barrier integrity of the gingival epithelium. This may however be disrupted by the biofilm challenge. The aim of this in vitro study was to investigate the effect of subgingival biofilms on the expression of inter-epithelial junctions by gingival epithelia, and evaluate the relative role of the red complex. Multi-layered human gingival epithelial cultures were challenged with a 10-species in vitro subgingival biofilm model, or its variant without the red complex, for 3 h and 24 h. A low-density array microfluidic card platform was then used for analyzing the expression of 62 genes encoding for tight junctions, gap junctions, adherens junctions, and desmosomes. Although there was a limited effect of the biofilms on the expression of tight, adherens and gap junctions, the expression of a number of desmosomal components was affected. In particular, Desmoglein-1 displayed a limited and transient up-regulation in response to the biofilm. In contrast, Desmocollin-2, Desmoplakin and Plakoglobin were down-regulated equally by both biofilm variants, after 24 h. In conclusion, this subgingival biofilm model may down-regulate selected desmosomal junctions in the gingival epithelium, irrespective of the presence of the “red complex.” In turn, this could compromise the structural integrity of the gingival tissue, favoring bacterial invasion and chronic infection.
Keywords: desmosomes, epithelial junctions, gingival epithelium, immune response, oral biofilms, periodontal diseases, subgingival, virulence
Introduction
Periodontal diseases are caused by microbial biofilms that colonize the tooth surfaces and instigate an inflammatory response by the juxtaposed gingival tissue. The microbial species constituting these biofilms are part of the endogenous oral microbiota. Shifts in the tissue micro-environmental conditions may favor the uncontrolled growth of certain species, which now act as pathobionts by establishing a dysbiotic interaction with the host.1-4 The initial host response is a biological mechanism aimed at preventing bacterial colonization and establishment.5 Yet, in the case of dysbiosis,1,6 an excessive inflammatory response may cause tissue destruction, which manifests as periodontitis.7 The development of a “subgingival” biofilm is a primary etiological agent of a dysbiotic host response. In classical studies, the increase in numbers and proportions of the tree “red complex” species (Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia) in subgingival biofilms has been highly associated with the presence of periodontitis.8
The epithelium of the gingival sulcus or periodontal pocket is a first line of defense against the developing biofilm, by constituting a physical barrier, secreting chemo-attractants for neutrophils and permitting their trafficking to the site where the biofilm is established.9-11 The integrity of all epithelial tissues is ensured by several cell-to-cell molecular adhesion and sealing complexes, including tight junction, adherens junctions, gap junctions and desmosomes.12-14 The expression of gap and tight junctions has been well documented in the gingiva.15-17 Therefore, unimpaired expression of these molecular complexes in gingival epithelial tissues is crucial for maintaining their integrity. Once tissue integrity is disturbed by biofilm-derived noxious stimuli, the associated bacteria may be permitted to invade into the deeper periodontal tissue, triggering an inflammatory response and establishing chronic infection.
Therefore, this study aimed to investigate the effects of an in vitro 10-species subgingival biofilm model (designated as “BF”) on the gene expression of all known tight junctions, desmosomes, gap junctions and adherens junctions, in a multi-layered gingival epithelial cell culture,18-20 by using a low-density array microfluidic card platform.14,21 A further aim was to evaluate the involvement of the 3 “red-complex” species in the observed effects, by excluding them from the composition of the biofilm (designated as “BF-RC”). This in vitro experimental system resembles rather closely the in vivo interface between the gingival epithelium and the microbial biofilm.
Results
The effect of BF or BF-RC on the gene expression of tight-, gap- and adherens-junctions by multi-layered gingival epithelial cultures was investigated. Prior to that, it was confirmed that there was no significant quantitative difference in the individual bacterial composition between the 2 biofilm variants, with the obvious exception of the 3 “red complex” species, which had been omitted from the inoculum of BF-RC.19 In addition, it was confirmed that neither BF, nor BF-RC elicited any strong cytotoxic effects on these gingival epithelial cultures.19
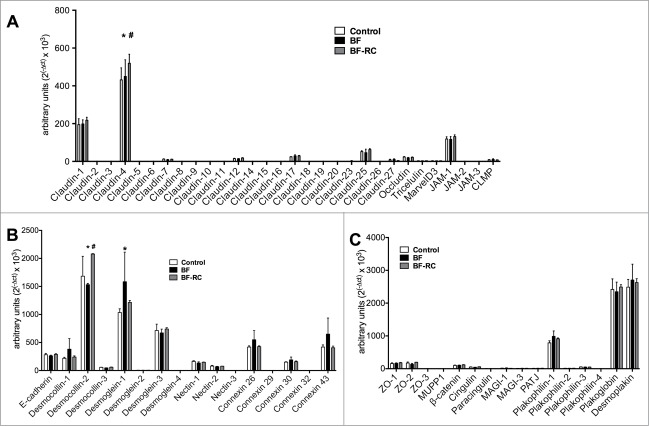
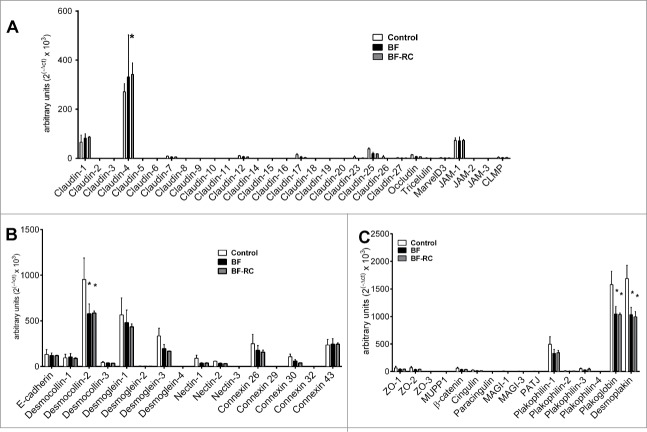
Among the 30 tight junction genes studied, the most highly expressed by the gingival epithelial cells were, sequentially, Claudin-4, Claudin-1, JAM-1, Claudin-25, Claudin-17, Occludin and Claudin-12 (Figs. 1A and 2A). On the contrary, Claudin-8, Claudin-18, Claudin-19, Claudin-20 and JAM-2 were not expressed in this epithelial culture under any of the experimental conditions. The effect of the biofilm challenge was further considered on the regulation of the expressed genes, after 3 h and 24 h. It was found that BF did not affect the expression of any of the studied tight junction genes. Absence of the “red complex” from the biofilm (BF-RC) resulted in significantly higher Claudin-4 expression compared to the control or the BF at 3 h (Fig. 1A), whereas at 24 h its expression was significantly higher only compared to the control group (Fig. 2A). Although these upregulations proved to be significant, they were rather low numerically, ranging at increases of 14%–36% over the control.
Figure 1.
Junctions gene expression profile in multi-layered gingival epithelial cells cultures, assayed by Taqman low-density array microfluidic card. The cell cultures were challenged for 3 h with BF or BF-RC, and thereafter the gene expression of transmembrane tight junction proteins (A), desmosomes, adherens junctions, and gap junction proteins (B), as well as junctional adaptor proteins (C) were assayed. Bars represent mean values ± SEM from 3 independent cell cultures in each group. Two-way ANOVA was used to calculate the differences between groups. Asterisks (*) represent statistically significant difference compared to the control group, whereas hash tags (#) represent statistically significant difference compared BF (P < 0.05).
Figure 2.
Junctions gene expression profile in multi-layered gingival epithelial cells cultures, assayed by Taqman low-density array microfluidic card. The cell cultures were challenged for 24 h with BF or BF-RC, and thereafter the gene expression of transmembrane tight junction proteins (A), desmosomes, adherens junctions, and gap junction proteins (B), as well as junctional adaptor proteins (C) were assayed. Bars represent mean values ±SEM from 3 independent cell cultures in each group. Two-way ANOVA was used to calculate the differences between groups. Asterisks (*) represent statistically significant difference compared to the control group, whereas hash tags (#) represent statistically significant difference compared BF (P < 0.05).
The gene expression of desmosomes, adherens junctions, and gap junction proteins was further determined (Figs. 1B and 2B). Only Connexin 32 (GJB1) was not expressed, whereas Desmoglein-2, Desmoglein-4, and Nectin-3 were expressed at low levels. The most highly expressed ones were Desmocollin-2, Desmoglein-1, Desmoglein-3, Connexin 26 and Connexin 43. After 3 h of challenge, Desmocollin-2 expression was significantly up-regulated in response to BF-RC, compared to BF or to the control, by approximately 20% (Fig. 1B). However, after 24 h, this was significantly downregulated by both biofilm variants, by approximately 40%, compared to the unchallenged control (Fig. 2B). Desmoglein-1 was significantly upregulated at 3 h by approximately 44% only in response to BF, but its expression resumed control levels after 24 h (Fig. 2B).
All studied junctional adaptor proteins were expressed by the gingival epithelial cultures. Most highly expressed were Desmoplakin, Plakoglobin, and Plakophilin-1. After 3 h of biofilm challenge, the gene expression of none of these proteins was regulated (Fig. 1C). However, after 24 h, the expression of Desmoplakin and Plakoglobin were significantly down-regulated in response to both biofilms by approximately 40% and 34%, respectively, whereas there were no significant differences between the 2 biofilm groups (Fig. 2C).
Discussion
The present study investigated the effect of in vitro multi-species subgingival biofilms on intra-epithelial junctions expression in multi-layered human gingival epithelial cultures, and evaluated the relative effects of the “red complex” species. While the development of this experimental model is highly relevant for studying the initial tissue responses associated with the pathogenesis of periodontal diseases,22 its potential limitation is that the biofilm comprises of relatively few cultivable species. This may under-represent the full diversity of the cultivable and uncultivable in vivo oral microbiome, given that a single periodontal pocket may foster more than a hundred different species,23 and that a dysbiotic environment induces multiple changes in the behavior of the constituent species.1,3,24
The rationale for this study is that intra-epithelial junctions are crucial for the integrity of the gingival tissue and consequently for the homeostasis and healthy status of the periodontium. Therefore, disruption of their expression may be detrimental for tissue integrity and bacterial invasion. In support of this, recent observations in the present experimental model showed that increased colonization (and potential invasion) of the superficial multi-layered gingival epithelium is associated with disruption signs of the epithelial cell borders, and nuclear degradation.18 Moreover, a recent proteomic analysis of the secreted proteins in this experimental model showed that several of the downregulated biological processes and networks are associated with disruption of epithelial tissue integrity and impaired tissue turnover.20
Among the 30 tight junction genes studied here, only Claudin-4 was affected by the biofilm lacking the 3 “red complex” species (BF-RC). Yet, the magnitude of this regulation was rather limited, and may thus not confer any biological relevance. Although Claudin-4 is expressed in healthy and diseased gingival epithelial tissue,25-27 there is as yet no evidence of its regulation by periodontal pathogens. In another experimental model using the same low-density microfluidic card assay, Claudin-4 expression was lower in air-liquid interface nasal epithelial cell cultures from chronic rhinosinusitis patients, than healthy individuals.21
None of the adherens or gap junction proteins' gene expression was regulated by the subgingival biofilm challenge in the present experimental system. However, the gene expression of 2 demosomal proteins, namely Desmocollin-2 and Desmoglein-1 were affected. In particular, Desmocollin-2 expression displayed a short-lived and weak up-regulation in response to BF-RC only, but after 24 h this was down-regulated by both biofilm variants, irrespective of the presence of the “red complex.” This reduced expression may denote compromised gingival tissue coherence and integrity. To our knowledge, there is at present no further information on the expression of Desmocollin-2 in the healthy or diseased periodontal tissues. Desmoglein-1 expression also displayed a short-lived but significant induction in response to BF at 3 h, which resumed control levels after 24 h. Desmoglein-1 is expressed by the healthy gingival epithelium,28 whereas its expression is downregulated in the periodontitis affected gingival tissue.29
Among the junctional adaptor proteins studied, the gene expression of only Desmoplakin and Plakoglobin, 2 desmosomal-associated proteins, were regulated. Desmoplakin expression has been demonstrated in the gingival epithelium,30-32 whereas Plakoglobin is known to structurally associate with Desmoglein-1.33 To date, there has been no study on the effects of the biofilm on the expression of these 2 proteins in gingival epithelium. The present study demonstrated that after 24 h of challenge with either biofilm, the expressions of both Desmoplakin and Plakoglobin were significantly reduced. Once again, this down-regulatory trend may denote an active loss of tissue integrity. Since, the regulatory effect of the 2 biofilms variants was of similar magnitude, the “red complex” may not hold a crucial role in this event.
At this stage, a comparison with in vivo studies is worth considering. For instance, an immunohistochemical study using biopsies from clinically healthy gingiva and advanced periodontitis lesions demonstrated reduced E-cadherin, involucrin, Connexin 26 and Connexin 43 staining in the epithelial lining of the periodontal pocket, associated with alterations of filamentous actin expression.34 Hence, that study concluded that the profound perturbation of the lining epithelium in periodontitis compromises its ability to function as an effective barrier against microbial invasion. Although a different set of junctions was affected in the present in vitro epithelial tissue-biofilm interaction model, the findings point to a similar direction, namely the down-regulation of junctions necessary for tissue integrity. While in vivo studies provide direct insights into changes within the periodontitis-affected tissues, in vitro models such as the one employed here, can give answers to mechanistic questions, due to their highly controlled and reproducible nature. As such, we were able to show that the “red complex” species had minimal interference in junctions gene expression.
Finally, it should be acknowledged that this study screened for broad transcriptional changes in epithelial junctions expression, rather than their regulation on the protein level. The findings may allude to proteins that could be investigated in more detail. Collectively it is shown that the present subgingival biofilm model used as a polymicrobial challenge did not cause major alterations in the gene expression of tight, gap or adherens junctions over an experimental period of 24 h. Nevertheless, it down-regulated the expression of 3 desmosome-associated proteins, and this was not commensurate with the presence of the 3 “red complex” species. Hence, subgingival biofilms may down-regulate the transcription of selected desmosomal junctions in the gingival epithelium, an effect that may compromise structural tissue integrity and enable bacterial invasion, should this also prove to translate on the protein level in vivo.
Materials and methods
In vitro biofilm model
The 10-species in vitro “subgingival” biofilm model used in this study was grown as previously described.19,35,36 It consisted of the individual species Campylobacter rectus (OMZ 697), Fusobacterium nucleatum (OMZ 598), P. gingivalis ATCC 33277T (OMZ 925), Prevotella intermedia ATCC 25611T (OMZ 278), T. forsythia OMZ1047, T. denticola ATCC 35405T (OMZ 661), Veillonella dispar ATCC 17748T (OMZ 493), Actinomyces oris (OMZ 745), Streptococcus anginosus (OMZ 871), and Streptococcus oralis SK 248 (OMZ 607). This biofilm variant is referred to as “BF,” while its 7-species variant lacking P. gingivalis, T. forsythia and T. denticola (i.e. the “red complex”) is referred to as “BF-RC” in the manuscript text. These biofilms were grown in 24-well cell culture plates on sintered hydroxyapatite discs, in order to mimic the natural tooth-biofilm interface. The hydroxyapatite discs were pre-conditioned for 4 h with 800 μl of pasteurized human saliva diluted 1:1 in sterile saline, in order to establish a pellicle on their surface. Biofilm formation was initiated by inoculating on the pellicle-covered hydroxyapatite 1.6 ml of growth medium consisting of 60% saliva, 10% heat-inactivated human serum, 30% modified fluid universal medium (mFUM) 36,37 with 0.3% glucose, and 200 µl of a bacterial cell suspension containing equal volumes and densities from each strain. The volumes were not adjusted according to the size of each strain in the suspension. After 16.5 h of anaerobic incubation at 37°C, the medium was replenished, and 50 μl of T. denticola liquid culture were also added (OD550 = 1.0). Biofilms were grown anaerobically for further 48 h and during this period, the discs were “dip-washed” in saline 3 times daily for 1 min, and the medium was replenished once daily. After a total 64.5 h of incubation, the biofilm-grown hydroxyapatite discs were carefully placed onto the multi-layered gingival epithelial cell cultures (described below), mediated by a plastic ring to ensure a distance of 1 mm, and co-cultured for 3 h or 24 h. These time-points represent an earlier and a later host response to the biofilm. At each one of these 2 time-points, the discs were removed from the cultures and subsequently processed for analysis of bacterial composition by quantitative real-time Polymerase Chain Reaction (qPCR), as previously described.19,36 Three independent biofilms were performed per each experimental group. Pellicle pre-coated hydroxyapatite discs were used as controls. This pellicle derived from the same saliva batch and was processed according to the same protocol as the biofilm grown-discs, but omitting the bacterial suspensions. Three independent cell cultures were performed in each experimental group.
Cell cultures
Stratified multi-layered gingival epithelial cell cultures in 24-well plate format (0.5 cm2 surface) were used (EpiGing, MatTek, Ashland, MA, USA) and maintained in culture in defined keratinocyte serum-free medium, supplemented with 0.05 mM calcium chloride and 200 mM L-glutamine (Gibco/Invitrogen, Lucerne, Switzerland). These cultures resemble morphologically the gingival epithelium, as they comprise of normal human gingival epithelial cells forming a highly differentiated multi-layered tissue with keratinized layers.
RNA extraction and cDNA synthesis
After completion of the experiments, the culture supernatants were removed and the multi-layered gingival epithelia were washed twice in phosphate buffer saline. Thereafter, they were lysed and total RNA was extracted by using the RNeasy Mini Kit (Qiagen). The concentration of the RNA was measured by a NanoDrop 1000 spectrophotometer (Thermoscientific). One μg of total RNA was then reverse transcribed into single-stranded cDNA by M-MLV Reverse Transcriptase, Oligo(dT)15 Primers, and PCR Nucleotide Mix (Promega), at 40°C for 60 min, and 70°C for 15 min. The resulting cDNA was stored at −20°C.
Gene expression analysis by TaqMan low-density array microfluidic cards
A total of 62 predesigned gene expression assays (Applied Biosystems) representing the junctional apparatus of epithelial cells were selected for the analyses performed in this study (Table 1).14,21 The probes were spanning over an exon-exon junction and amplified an amplicon length of maximal 200 nt. As housekeeping gene, GAPDH was used (Applied Biosystems assay ID: Hs99999905-m1). From the extracted total RNA, 400 ng were used per microfluidic card, and the reactions were run in a 7900HT Fast Real-Time PCR System (Applied Biosystems), using a TaqMan Universal PCR MasterMix (Applied Biosystems, 4304437). Arbitrary units representing gene expression were calculated with the following formula: arbitrary units = 2(−Δct) × 1000. Genes whose transcription was undetectable beyond 40 cycles under any of the experimental conditions were considered as non-expressed.
Table 1.
The 62 genes included in the microfluidic card mRNA expression array
| Gene symbol | Gene name | Assay ID | Expressed |
|---|---|---|---|
| OCLN | Occludin | Hs00170162_m1 | Yes |
| F11R | JAM-1 | Hs00170991_m1 | Yes |
| JAM2 | JAM-2 | Hs00221894_m1 | No |
| JAM3 | JAM-3 | Hs00230289_m1 | Yes |
| MARVELD2 | Tricellulin | Hs00376394_m1 | Yes |
| CLDN1 | Claudin-1 | Hs01076359_m1 | Yes |
| CLDN2 | Claudin-2 | Hs00252666_s1 | Yes |
| CLDN3 | Claudin-3 | Hs00265816_s1 | Yes |
| CLDN4 | Claudin-4 | Hs00976831_s1 | Yes |
| CLDN5 | Claudin-5 | Hs00533949_s1 | Yes |
| CLDN6 | Claudin-6 | Hs00607528_s1 | Yes |
| CLDN7 | Claudin-7 | Hs00600772_m1 | Yes |
| CLDN8 | Claudin-8 | Hs00273282_s1 | No |
| CLDN9 | Claudin-9 | Hs00253134_s1 | Yes |
| CLDN10 | Claudin-10 | Hs00199599_m1 | Yes |
| CLDN11 | Claudin-11 | Hs00194440_m1 | Yes |
| CLDN12 | Claudin-12 | Hs01082669_m1 | Yes |
| CLDN14 | Claudin-14 | Hs00273267_s1 | Yes |
| CLDN15 | Claudin-15 | Hs00204982_m1 | Yes |
| CLDN16 | Claudin-16 | Hs00198134_m1 | Yes |
| CLDN17 | Claudin-17 | Hs01043467_s1 | Yes |
| CLDN18 | Claudin-18 | Hs00212584_m1 | No |
| CLDN19 | Claudin-19 | Hs00381204_m1 | No |
| CLDN20 | Claudin-20 | Hs00378662_m1 | No |
| CLDN23 | Claudin-23 | Hs01013638_s1 | Yes |
| CLDND1 | Claudin-25 | Hs00219886_m1 | Yes |
| TMEM114 | Claudin-26 | Hs00418203_m1 | Yes |
| C1orf91 | Claudin-27 | Hs00963921_m1 | Yes |
| ASAM | CMLP | Hs00293345_m1 | Yes |
| GJA1 | Connexin-43 | Hs00748445_s1 | Yes |
| GJB1 | Connexin-32 | Hs00939759_s1 | No |
| GJB2 | Connexin-26 | Hs00955889_m1 | Yes |
| GJB6 | Connexin-30 | Hs00917676_m1 | Yes |
| GJC3 | Connexin-29 | Hs01384570_m1 | Yes |
| CDH1 | E-cadherin | Hs01023895_m1 | Yes |
| PVRL1 | Nectin-1 | Hs01591978_m1 | Yes |
| PVRL2 | Nectin-2 | Hs01071562_m1 | Yes |
| PVRL3 | Nectin-3 | Hs00210045_m1 | Yes |
| DSG1 | Desmoglein-1 | Hs00355084_m1 | Yes |
| DSG2 | Desmoglein-2 | Hs00170071_m1 | Yes |
| DSG3 | Desmoglein-3 | Hs00170075_m1 | Yes |
| DSG4 | Desmoglein-4 | Hs00698286_m1 | Yes |
| DSC1 | Desmocolin-1 | Hs00245189_m1 | Yes |
| DSC2 | Desmocolin-2 | Hs00951428_m1 | Yes |
| DSC3 | Desmocolin-3 | Hs00170032_m1 | Yes |
| MPDZ | MUPP1 | Hs00187106_m1 | Yes |
| TJP1 | ZO-1 | Hs01551876_m1 | Yes |
| TJP2 | ZO-2 | Hs00910541_m1 | Yes |
| TJP3 | ZO-3 | Hs00274276_m1 | Yes |
| CGN | Cingulin | Hs00430426_m1 | Yes |
| CGNL1 | Paracingulin | Hs00262671_m1 | Yes |
| MAGI1 | MAGI-1 | Hs00191026_m1 | Yes |
| MAGI3 | MAGI-2 | Hs00326365_m1 | Yes |
| INADL | PATJ | Hs00195106_m1 | Yes |
| MARVELD3 | MARVELD3 | Hs00369354_m1 | Yes |
| JUP | Plakoglobin | Hs00158408_m1 | Yes |
| DSP | Desmoplakin | Hs00189422_m1 | Yes |
| PKP1 | Plakophilin-1 | Hs00240873_m1 | Yes |
| PKP2 | Plakophilin-2 | Hs00428040_m1 | Yes |
| PKP3 | Plakophilin-3 | Hs00170887_m1 | Yes |
| PKP4 | Plakophilin-4 | Hs00269305_m1 | Yes |
| CTNNB1 | B-catenin | Hs00355049_m1 | Yes |
The gene symbols, gene names and gene expression assay IDs are provided, as well as the information whether they were expressed (Yes/No) in the present experimental model by the multi-layered gingival epithelial culture.
JAM, Junctional adhesion molecule; MAGI, membrane-associated guanylate kinase inverted; MUPP1, multi-PDZ domain containing protein 1; ZO, zonula occludens.
Statistical analysis
A two-way analysis of variance (ANOVA) was used to analyze the statistical significance of differences, using Tukey's test for multiple comparisons between groups. Differences were considered statistically significant at P < 0.05.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Mrs. Ruth Graf, Mr. Andre Meier, Mrs Verena Osterwalder and Mrs. Elpida Plattner for their excellent technical assistance.
Funding
This study was supported by the authors' Institutes, a Forschungskredit Grant by the University of Zürich (NB), and Swiss National Science Foundation Grant 320030-140772 (CA).
References
- 1.Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol 2012; 27:409-19; PMID:23134607; http://dx.doi.org/ 10.1111/j.2041-1014.2012.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G, Lamont RJ. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol 2014; 44:328-38; PMID:24338806; http://dx.doi.org/ 10.1002/eji.201344202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med 2015; 21:172-83; PMID:25498392; http://dx.doi.org/ 10.1016/j.molmed.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol 2014; 35:3-11; PMID:24269668; http://dx.doi.org/ 10.1016/j.it.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000 2006; 40:50-76; PMID:16398685; http://dx.doi.org/ 10.1111/j.1600-0757.2005.00148.x [DOI] [PubMed] [Google Scholar]
- 6.Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dental Res 2012; 91:816-20; PMID:22772362; http://dx.doi.org/ 10.1177/0022034512453589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schenkein HA. Host responses in maintaining periodontal health and determining periodontal disease. Periodontol 2000 2006; 40:77-93; PMID:16398686; http://dx.doi.org/ 10.1111/j.1600-0757.2005.00144.x [DOI] [PubMed] [Google Scholar]
- 8.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr.. Microbial complexes in subgingival plaque. J Clin Periodontol 1998; 25:134-44; PMID:9495612; http://dx.doi.org/ 10.1111/j.1600-051X.1998.tb02419.x [DOI] [PubMed] [Google Scholar]
- 9.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol 2010; 8:481-90; PMID:20514045; http://dx.doi.org/ 10.1038/nrmicro2337 [DOI] [PubMed] [Google Scholar]
- 10.Tonetti MS, Imboden MA, Lang NP. Neutrophil migration into the gingival sulcus is associated with transepithelial gradients of interleukin-8 and ICAM-1. J Periodontol 1998; 69:1139-47; PMID:9802714; http://dx.doi.org/ 10.1902/jop.1998.69.10.1139 [DOI] [PubMed] [Google Scholar]
- 11.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun 1998; 66:1660-5; PMID:9529095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukita S, Furuse M. The structure and function of claudins, cell adhesion molecules at tight junctions. Ann N Y Acad Sci 2000; 915:129-35; PMID:11193568; http://dx.doi.org/ 10.1111/j.1749-6632.2000.tb05235.x [DOI] [PubMed] [Google Scholar]
- 13.Furuse M. Molecular basis of the core structure of tight junctions. Cold Spring Harb Perspect Biol 2010; 2:a002907; PMID:20182608; http://dx.doi.org/ 10.1101/cshperspect.a002907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kast JI, Wanke K, Soyka MB, Wawrzyniak P, Akdis D, Kingo K, Rebane A, Akdis CA. The broad spectrum of interepithelial junctions in skin and lung. J Allergy Clin Immunol 2012; 130:544-7 e4; PMID:22704535; http://dx.doi.org/ 10.1016/j.jaci.2012.04.044 [DOI] [PubMed] [Google Scholar]
- 15.Fujita T, Hayashida K, Shiba H, Kishimoto A, Matsuda S, Takeda K, Kawaguchi H, Kurihara H. The expressions of claudin-1 and E-cadherin in junctional epithelium. J Periodontal Res 2010; 45:579-82; PMID:20337884; http://dx.doi.org/ 10.1111/j.1600-0765.2009.01230.x [DOI] [PubMed] [Google Scholar]
- 16.Meyle J, Gultig K, Rascher G, Wolburg H. Transepithelial electrical resistance and tight junctions of human gingival keratinocytes. J Periodontal Res 1999; 34:214-22; PMID:10444745; http://dx.doi.org/ 10.1111/j.1600-0765.1999.tb02244.x [DOI] [PubMed] [Google Scholar]
- 17.Barnett ML, Szabo G. Gap junctions in human gingival keratinized epithelium. J Periodontal Res 1973; 8:111-26; PMID:4123067 [DOI] [PubMed] [Google Scholar]
- 18.Thurnheer T, Belibasakis GN, Bostanci N. Colonisation of gingival epithelia by subgingival biofilms in vitro: role of “red complex” bacteria. Arch Oral Biol 2014; 59:977-86; PMID:24949828; http://dx.doi.org/ 10.1016/j.archoralbio.2014.05.023 [DOI] [PubMed] [Google Scholar]
- 19.Belibasakis GN, Thurnheer T, Bostanci N. Interleukin-8 responses of multi-layer gingival epithelia to subgingival biofilms: role of the “red complex” species. PloS one 2013; 8:e81581; PMID:24339946; http://dx.doi.org/ 10.1371/journal.pone.0081581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bostanci N, Bao K, Wahlander A, Grossmann J, Thurnheer T, Belibasakis GN. Secretome of gingival epithelium in response to subgingival biofilms. Mol Oral Microbiol 2015; 30(4):323-35; PMID:25787257; http://dx.doi.org/ 10.1111/omi.12096 [DOI] [PubMed] [Google Scholar]
- 21.Soyka MB, Wawrzyniak P, Eiwegger T, Holzmann D, Treis A, Wanke K, Kast JI, Akdis CA. Defective epithelial barrier in chronic rhinosinusitis: the regulation of tight junctions by IFN-gamma and IL-4. J Allergy Clin Immunol 2012; 130:1087-96 e10; PMID:22840853; http://dx.doi.org/ 10.1016/j.jaci.2012.05.052 [DOI] [PubMed] [Google Scholar]
- 22.Bao K, Papadimitropoulos A, Akgul B, Belibasakis GN, Bostanci N. Establishment of an oral infection model resembling the periodontal pocket in a perfusion bioreactor system. Virulence 2015:1-9; PMID:NOT_FOUND [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000 2005; 38:135-87; PMID:15853940; http://dx.doi.org/ 10.1111/j.1600-0757.2005.00107.x [DOI] [PubMed] [Google Scholar]
- 24.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, et al.. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 2011; 10:497-506; PMID:22036469; http://dx.doi.org/ 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langbein L, Grund C, Kuhn C, Praetzel S, Kartenbeck J, Brandner JM, Moll I, Franke WW. Tight junctions and compositionally related junctional structures in mammalian stratified epithelia and cell cultures derived therefrom. Eur J Cell Biol 2002; 81:419-35; PMID:12234014; http://dx.doi.org/ 10.1078/0171-9335-00270 [DOI] [PubMed] [Google Scholar]
- 26.Ye P, Yu H, Simonian M, Hunter N. Ligation of CD24 expressed by oral epithelial cells induces kinase dependent decrease in paracellular permeability mediated by tight junction proteins. Biochem Biophys Res Commun 2011; 412:165-9; PMID:21806966; http://dx.doi.org/ 10.1016/j.bbrc.2011.07.067 [DOI] [PubMed] [Google Scholar]
- 27.Ye P, Yu H, Simonian M, Hunter N. Expression patterns of tight junction components induced by CD24 in an oral epithelial cell-culture model correlated to affected periodontal tissues. J Periodontal Res 2014; 49:253-9; PMID:23713517; http://dx.doi.org/ 10.1111/jre.12102 [DOI] [PubMed] [Google Scholar]
- 28.Hatakeyama S, Yaegashi T, Oikawa Y, Fujiwara H, Mikami T, Takeda Y, Satoh M. Expression pattern of adhesion molecules in junctional epithelium differs from that in other gingival epithelia. J Periodontal Res 2006; 41:322-8; PMID:16827727; http://dx.doi.org/ 10.1111/j.1600-0765.2006.00875.x [DOI] [PubMed] [Google Scholar]
- 29.Abe D, Kubota T, Morozumi T, Shimizu T, Nakasone N, Itagaki M, Yoshie H. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. J Periodontal Res 2011; 46:345-53; PMID:21382035; http://dx.doi.org/ 10.1111/j.1600-0765.2011.01349.x [DOI] [PubMed] [Google Scholar]
- 30.Carmichael RP, McCulloch CA, Zarb GA. Quantitative immunohistochemical analysis of keratins and desmoplakins in human gingiva and peri-implant mucosa. J Dental Res 1991; 70:899-905; PMID:1708791; http://dx.doi.org/ 10.1177/00220345910700050701 [DOI] [PubMed] [Google Scholar]
- 31.Carmichael RP, McCulloch CA, Zarb GA. Immunohistochemical localization and quantification of desmoplakins I & II and keratins 1 and 19 in plastic-embedded sections of human gingiva. J Histochem Cytochem 1991; 39:519-28; PMID:1706376; http://dx.doi.org/ 10.1177/39.4.1706376 [DOI] [PubMed] [Google Scholar]
- 32.Matsuyama T, Izumi Y, Sueda T. Culture and characterization of human junctional epithelial cells. J Periodontol 1997; 68:229-39; PMID:9100198; http://dx.doi.org/ 10.1902/jop.1997.68.3.229 [DOI] [PubMed] [Google Scholar]
- 33.Troyanovsky SM, Troyanovsky RB, Eshkind LG, Krutovskikh VA, Leube RE, Franke WW. Identification of the plakoglobin-binding domain in desmoglein and its role in plaque assembly and intermediate filament anchorage. J Cell Biol 1994; 127:151-60; PMID:7929560; http://dx.doi.org/ 10.1083/jcb.127.1.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye P, Chapple CC, Kumar RK, Hunter N. Expression patterns of E-cadherin, involucrin, and connexin gap junction proteins in the lining epithelia of inflamed gingiva. J Pathol 2000; 192:58-66; PMID:10951401; http://dx.doi.org/ 10.1002/1096-9896(2000)9999:9999%3c::AID-PATH673%3e3.0.CO;2-T [DOI] [PubMed] [Google Scholar]
- 35.Ammann TW, Belibasakis GN, Thurnheer T. Impact of early colonizers on in vitro subgingival biofilm formation. PloS one 2013; 8:e83090; PMID:24340084; http://dx.doi.org/ 10.1371/journal.pone.0083090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ammann TW, Bostanci N, Belibasakis GN, Thurnheer T. Validation of a quantitative real-time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species-specific quantification of an in vitro subgingival biofilm model. J Periodontal Res 2013; 48:517-26; PMID:23278531; http://dx.doi.org/ 10.1111/jre.12034 [DOI] [PubMed] [Google Scholar]
- 37.Belibasakis GN, Guggenheim B, Bostanci N. Down-regulation of NLRP3 inflammasome in gingival fibroblasts by subgingival biofilms: involvement of Porphyromonas gingivalis. Innate Immun 2013; 19:3-9; PMID:22522430; http://dx.doi.org/ 10.1177/1753425912444767 [DOI] [PubMed] [Google Scholar]