Significance
Symbioses between animals and microbes are now recognized as critical to many aspects of host health. This is especially true in insects, which are associated with diverse maternally transmitted endosymbionts that can protect against parasites and pathogens. Here, we find that Spiroplasma—a defensive endosymbiont that protects Drosophila during parasitism by a virulent and common nematode—encodes a protein toxin, a ribosome-inactivating protein (RIP) related to bacterial virulence factors such as the Shiga-like toxins in Escherichia coli. We further find that nematode ribosomal RNA suffers depurination consistent with attack by a RIP when the host is protected by Spiroplasma, suggesting a mechanism through which symbiotic microbes may protect their hosts from disease.
Keywords: symbiosis, male-killing, Spiroplasma, Shiga toxin, nematode
Abstract
Vertically transmitted symbionts that protect their hosts against parasites and pathogens are well known from insects, yet the underlying mechanisms of symbiont-mediated defense are largely unclear. A striking example of an ecologically important defensive symbiosis involves the woodland fly Drosophila neotestacea, which is protected by the bacterial endosymbiont Spiroplasma when parasitized by the nematode Howardula aoronymphium. The benefit of this defense strategy has led to the rapid spread of Spiroplasma throughout the range of D. neotestacea, although the molecular basis for this protection has been unresolved. Here, we show that Spiroplasma encodes a ribosome-inactivating protein (RIP) related to Shiga-like toxins from enterohemorrhagic Escherichia coli and that Howardula ribosomal RNA (rRNA) is depurinated during Spiroplasma-mediated protection of D. neotestacea. First, we show that recombinant Spiroplasma RIP catalyzes depurination of 28S rRNAs in a cell-free assay, as well as Howardula rRNA in vitro at the canonical RIP target site within the α-sarcin/ricin loop (SRL) of 28S rRNA. We then show that Howardula parasites in Spiroplasma-infected flies show a strong signal of rRNA depurination consistent with RIP-dependent modification and large decreases in the proportion of 28S rRNA intact at the α-sarcin/ricin loop. Notably, host 28S rRNA is largely unaffected, suggesting targeted specificity. Collectively, our study identifies a novel RIP in an insect defensive symbiont and suggests an underlying RIP-dependent mechanism in Spiroplasma-mediated defense.
Symbiosis is now recognized to be a key driver of evolutionary novelty and complexity (1, 2), and symbioses between microbes and multicellular hosts are understood as essential to the health and success of diverse lineages, from plants to humans (3). Insects, in particular, have widespread associations with symbiotic bacteria, with most insect species infected by maternally transmitted endosymbionts (4, 5). Although many insect symbionts perform roles essential for host survival, such as supplementing nutrition, others are facultative and not strictly required by their hosts. These facultative symbionts have evolved diverse and intriguing strategies to maintain themselves in host populations despite loss from imperfect maternal transmission and metabolic costs to the host. These range from manipulating host reproduction to increase their own transmission (6, 7), such as by killing male hosts, to providing context-dependent fitness benefits (8). Recently, it has become clear that different insect endosymbionts have independently evolved to protect their hosts against diverse natural enemies that so far include pathogenic fungi (9), RNA viruses (10, 11), parasitoid wasps (12), parasitic nematodes (13), and predatory spiders (14, 15). This suggests that defense might be a common aspect of many insect symbioses and demonstrates that symbionts can serve as dynamic and heritable sources of protection against natural enemies (8).
Despite a growing appreciation of the importance of symbiont-mediated defense in insects, key questions remain. Most demonstrations of defense have been under laboratory conditions, and the importance of symbiont-mediated protection in natural systems is unclear in most cases (16). At the same time, the proximate causes of defense are largely unknown, although recent studies have provided some intriguing early insights: A Pseudomonas symbiont of rove beetles produces a polyketide toxin thought to deter predation by spiders (14), Streptomyces symbionts of beewolves produce antibiotics to protect the host from fungal infection (17), and bacteriophages encoding putative toxins are required for Hamiltonella defensa to protect its aphid host from parasitic wasps (18), whereas the causes of other naturally occurring defensive symbioses are unresolved. From an applied perspective, the ongoing goal of exploiting insect symbioses to arrest disease transmission to humans from insect vectors (19) makes a deeper understanding of the factors contributing to ecologically relevant and evolutionarily durable defensive symbioses urgently needed.
Here, we investigate the mechanism underlying one of the most striking examples of an ecologically important defensive symbiosis. Drosophila neotestacea is a woodland fly that is widespread across North America and is commonly parasitized by the nematode Howardula aoronymphium. Infection normally sterilizes flies (20); however, when flies harbor a strain of the inherited symbiont Spiroplasma—a Gram-positive bacterium in the class Mollicutes—they remarkably tolerate Howardula infection without loss of fecundity, and infection intensity is substantially reduced (13). The benefit conferred by this protection lends a substantial selective advantage to Spiroplasma-infected flies and has led to Spiroplasma’s recent spread across North America, with symbiont-infected flies rapidly replacing uninfected ones (21). Spiroplasma is a diverse and widespread lineage of arthropod-associated bacteria that can be commensal, pathogenic, or mutualistic (22). Maternal transmission has arisen numerous times in Spiroplasma, including strains that are well known as male-killers (22). In addition to defense against nematodes in D. neotestacea, other strains of Spiroplasma have recently been shown to protect flies and aphids against parasitic wasps and pathogenic fungi, respectively (23–25), but in no case is the mechanism of defense understood.
In theory, there are multiple avenues by which a symbiont may protect its host that include competing with parasites for limiting resources, priming host immunity, or producing factors to directly attack parasites (26). We previously assessed these possibilities in the defensive Spiroplasma from D. neotestacea (27); our findings best supported a role for toxins in defense, with Spiroplasma encoding a highly expressed putative ribosome-inactivating protein (RIP). RIPs are widespread across plants and some bacteria and include well-known plant toxins of particular human concern such as ricin, as well as important virulence factors in human toxigenic strains of Escherichia coli and Shigella (28, 29). RIPs characteristically exert their cytotoxic effects through depurination of eukaryotic 28S ribosomal RNAs (rRNAs) at a highly conserved adenine in the α-sarcin/ricin loop (SRL) of the rRNA by cleaving the N-glycosidic bond between the rRNA backbone and adenine (30, 31). The proliferation of RIPs across different lineages implies functional significance, but their ecological roles are unclear, although they often appear to have antiviral or other defensive roles (29, 32). Here, we find that Spiroplasma expresses a functional RIP distinct from previously characterized toxins that appears to specifically affect Howardula rRNA in flies coinfected with Spiroplasma and Howardula. This work suggests the mechanisms used in defensive associations to protect the host from disease as well as intriguing ecological roles for RIPs in a tripartite defensive symbiosis.
Results
Spiroplasma Strains Encode Diverse RIP-Like Sequences.
Earlier RNA-sequencing (RNA-seq) assemblies from Spiroplasma-infected D. neotestacea recovered a sequence of a putative RIP encoded in a 403 aa ORF (hereafter SpRIP) (27). Characterized RIPs typically belong to one of two classes: type I toxins are monomeric toxins that contain a single catalytically active A chain (typically ∼30 kDa). In type II toxins, this A chain is conserved but is additionally linked to a B or B5 subunit (e.g., for ricin and Shiga-like toxins, respectively) that serves as a lectin and facilitates toxin entry into target cells, typically greatly increasing cytotoxicity (28, 29, 33). The ORF of SpRIP was predicted to encode an N-terminal signal peptide, followed by a disordered region of 70 aa, and a ∼300 aa C-terminal region homologous to characterized RIP A chains. Although this is substantially longer than typical for monomeric type I toxins, we found no convincing bioinformatic evidence for the presence of a B chain homologous to those characterized from Shiga-like toxins or type II plant RIPs.
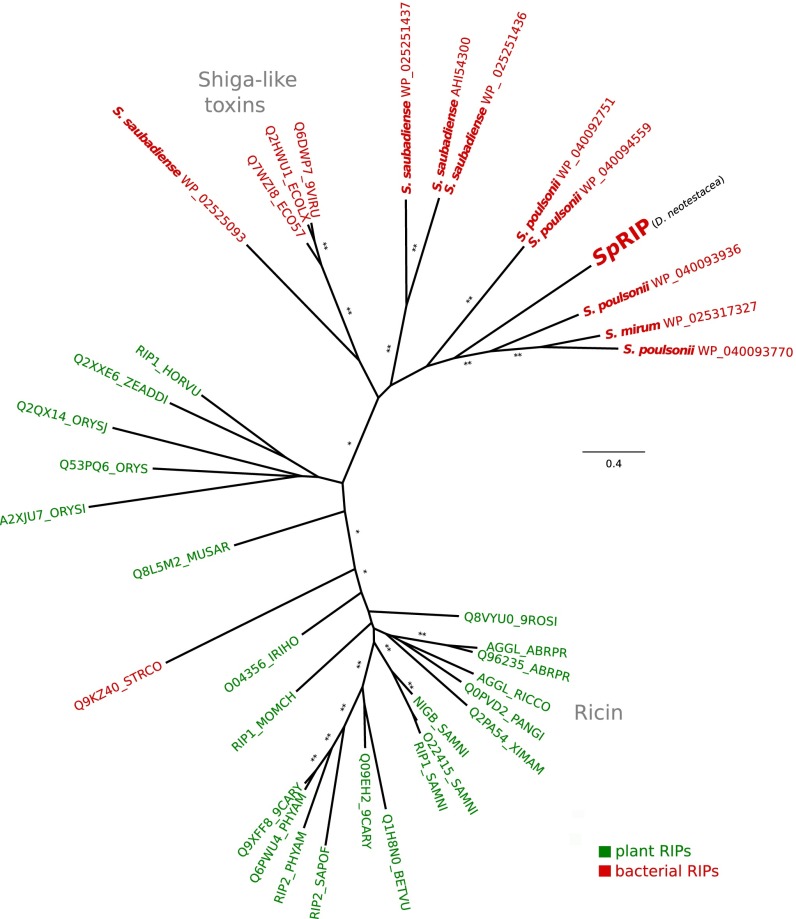
BLASTp searches recovered putative RIPs encoded by other Spiroplasma strains, including the recently sequenced defensive and male-killing MSRO strain of Spiroplasma poulsonii from Drosophila melanogaster (34). Phylogenetic analysis of these protein sequences with selected seed sequences from the Pfam protein family database (PF00161) placed RIP-like sequences from Spiroplasma strains with bacterial RIPs such as the Shiga-like toxins (Fig. 1), and alignments confirmed the presence of the known conserved catalytic residues of RIPs in SpRIP (29).
Fig. 1.
Spiroplasma strains encode divergent RIP-like sequences. Maximum-likelihood amino acid phylogeny of SpRIP from the defensive D. neotestacea strain of Spiroplasma, aligned with other putative Spiroplasma RIPs and selected RIP seed sequences from the Pfam database (PF00161). Support values are calculated based on 1,000 bootstraps using FastTree; the scale bar denotes substitutions per site. Single and double asterisks represent 70% and 90% bootstrap support, respectively. Spiroplasma-encoded sequences are shown in bold.
Spiroplasma Expresses a Functional RIP.
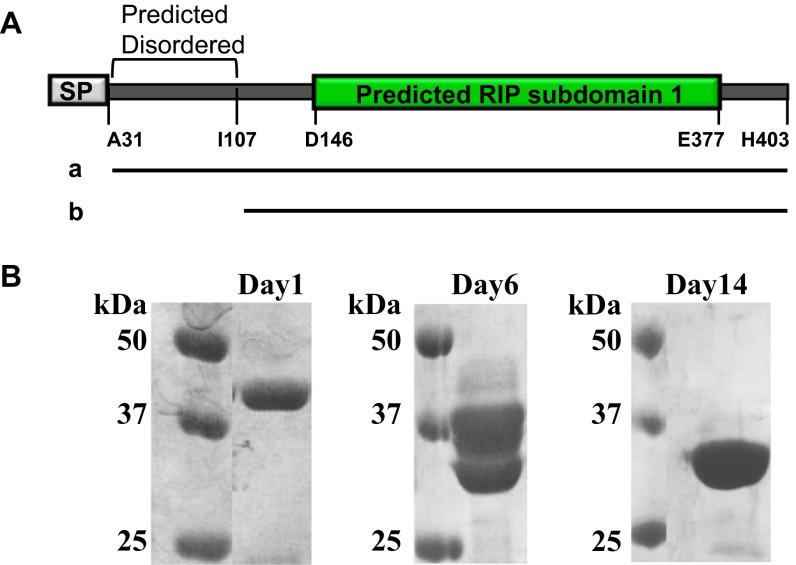
To characterize SpRIP, we first recombinantly produced a codon optimized version of the protein using E. coli (Fig. S1; signal peptide removed; Ala31 through His403). The purified 44-kDa protein degraded to a stable 34-kDa product after 2 wk in Hepes-buffered saline (HBS) at 4 °C (Fig. S1). Consistent with our expectation, mass spectrometric analysis confirmed this to be a result of proteolysis of the ∼70 residues of the N-terminal region predicted to be disordered. Subsequent assays were performed using the stable 34-kDa protein that lacked the predicted disordered region.
Fig. S1.
The purified recombinant SpRIP degraded into a stable product over 2 wk of time at 4 °C. (A) Schematic of SpRIP domain prediction. SP, signal peptide. Black horizontal line a represents the recombinant protein (Ala31 to His403) produced in this study. b represents the stable degradation product from purified recombinant SpRIP. (B) SDS/PAGE analysis of the purified recombinant SpRIP (44 kDa) incubated at 4 °C over time. The final stable degradation product (∼34 kDa on day 14 gel) was analyzed by mass spectrometry.
We used a modified, highly sensitive RT-quantitative (q)PCR–based assay to assess depurination activity (35, 36). In brief, depurination at the SRL leaves an abasic site following RIP attack but does not directly cleave the phosphodiester bond of the sugar phosphate backbone. These assays exploit the property of reverse transcriptases to incorporate deoxyadenosine monophosphate (dAMP) opposite this abasic lesion during reverse transcription, resulting in a quantifiable signature shift from T (the complement of A) to A at the site of depurination in resultant cDNA. To exploit this, we developed qPCR primers to rabbit 28S rRNA for use in cell-free rabbit reticulocyte lysate-based assays.
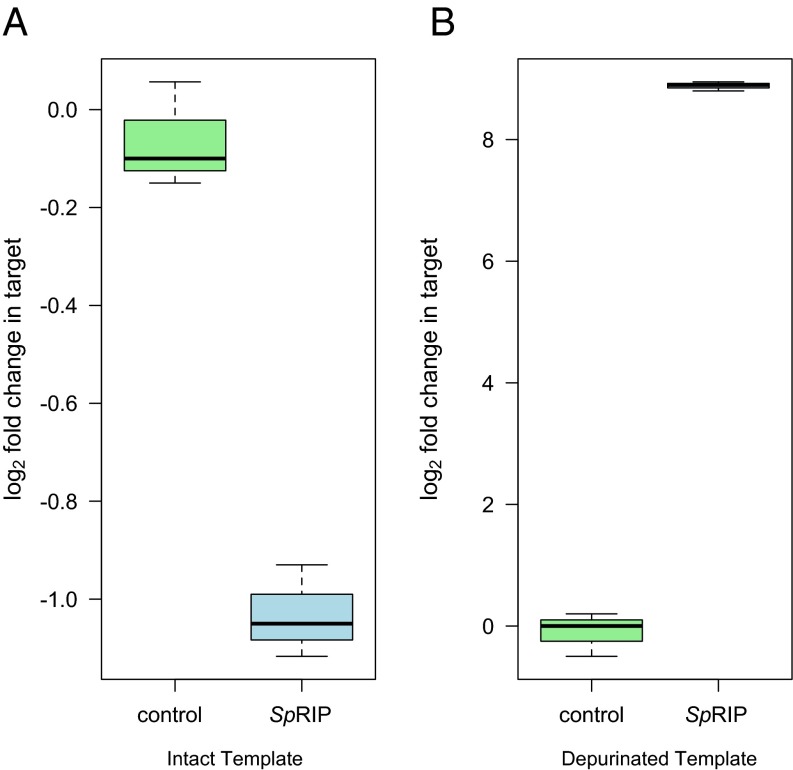
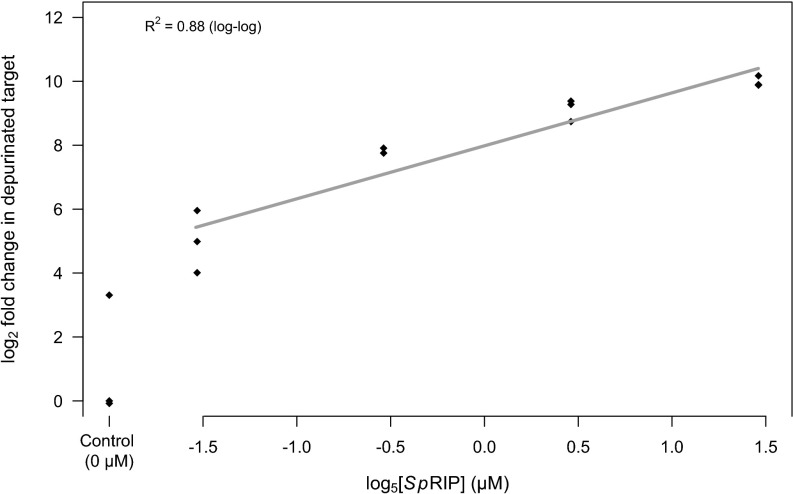
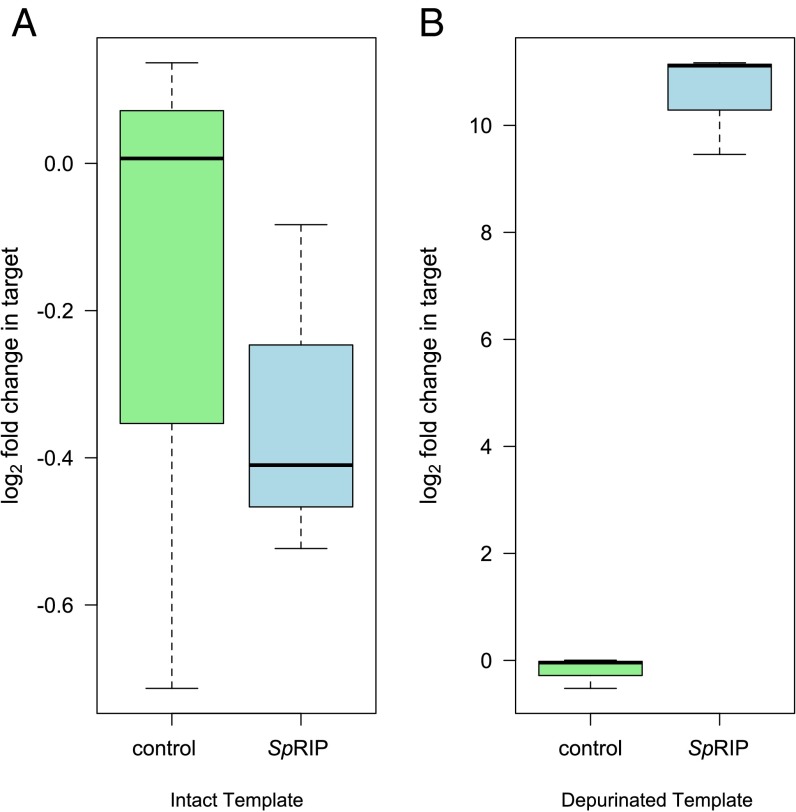
Incubating reticulocyte lysate with SpRIP led to a ∼50% decrease in the abundance of the cDNA representing intact 28S rRNA relative to negative controls (Fig. 2A; hereafter intact template; t3.94 = 11.68, P < 0.001) and, correspondingly, more than a 1,000-fold increase in cDNA representing depurinated rRNA (Fig. 2B; hereafter depurinated template; t2.18 = 42.22, P < 0.001). A 4 × fivefold serial dilution of SpRIP also confirmed depurination across a range of concentrations, with clear dose dependence to <0.1 μM [Fig. 3; linear regression of log2(depurination) vs. log5([SpRIP]); R2 = 0.88, P < 0.001]. This supports the predicted depurination function for SpRIP, with enzyme-dependent depurination likely proceeding through cleavage of the N-glycosidic bond, as is observed in other RIPs.
Fig. 2.
SpRIP depurinates rabbit 28S rRNA at the site of RIP attack in cell-free assays. Abundance of cDNA representing intact (A) and depurinated (B) rRNA template after incubation with 5.25 μM of recombinant SpRIP for 30 min at 30 °C (n = 6). SpRIP significantly decreases the abundance of intact template and increases the abundance of depurinated template (P < 0.001 for both).
Fig. 3.
SpRIP causes dose-dependent depurination of rabbit 28S rRNA in cell-free assays (n = 14). Fit represents linear regression of log2(depurination) as a function log5([SpRIP]).
SpRIP Depurinates Howardula Ribosomal RNA in Vitro.
To confirm SpRIP activity against Howardula nematode rRNA, we designed a RT-qPCR assay to measure depurination of Howardula rRNA. This assay was able to specifically differentiate Howardula rRNA from that of the fly host, with cDNA reverse-transcribed from nematode-uninfected fly negative controls yielding no amplification.
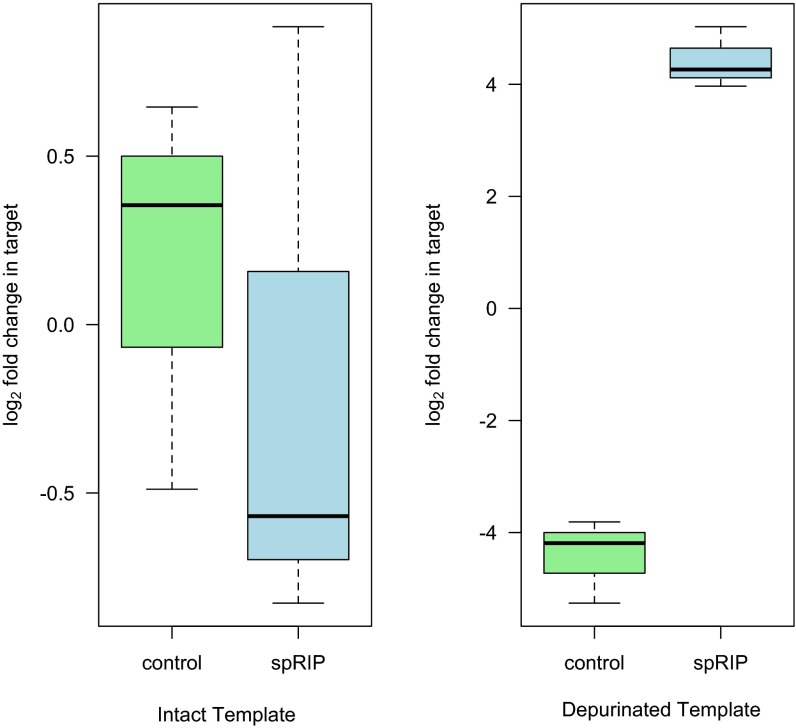
We harvested live Howardula by gently grinding infected Drosophila falleni (Spiroplasma-negative) in insect Ringer’s solution. We incubated this homogenate, containing viable juvenile nematodes, with recombinant SpRIP at 21 °C for 4 h, using a lower temperature to avoid directly killing nematodes during incubation. Incubation with the toxin again dramatically increased the abundance of depurinated template by more than 2,000-fold (Fig. 4 A and B; t2.38 = 18.34, P < 0.001). In contrast to the reticulocyte lysate assay, there was no substantial decrease in the abundance of intact template under these conditions (t2.94 = 0.51, P = 0.65). We performed a further experiment incubating single nematode motherworms to limit substrate availability, again not observing appreciable depletion of intact 28S rRNA under these conditions, despite large increases in depurinated template (Fig. S2; t3.89 = 0.31 and t2.03 = 8.68, and P = 0.77 and P = 0.01, respectively), suggesting that a proportion of Howardula ribosomes might not be accessible to the stable but truncated recombinant SpRIP used here.
Fig. 4.
SpRIP depurinates Howardula 28S rRNA at the site of RIP attack in assays with live Howardula. Abundance of cDNA representing intact (A) and depurinated (B) rRNA template after incubation with 5.25 μM of recombinant SpRIP for 4 h at 21 °C (n = 6). Intact template is not clearly decreased, whereas abundance of depurinated template is significantly increased (P = 0.65 and P < 0.001, respectively).
Fig. S2.
Incubation of single Howardula motherworms with SpRIP for 4 h leads to an increase in cDNA representing depurinated Howardula 28S rRNA but no decrease in intact rRNA (P = 0.01 and 0.77, respectively; n = 6).
RNA-Seq Shows Depurination of Howardula 28S rRNA at the SRL in the Presence of Spiroplasma.
To test for evidence of Howardula attack by a RIP in vivo, we revisited RNA-seq reads generated during a previous experiment, in which we sequenced RNA of D. neotestacea and Howardula in the presence and absence of Spiroplasma infection (27). We reasoned that signal of depurination should be observed in reads mapping to the SRL of Howardula 28S rRNA given the reliance of the RNA-seq on reverse transcription during library construction, causing a shift in the sequencing read at the site of depurination.
Mapping raw reads to near full-length 28S rRNA for Howardula revealed a highly significant signal of depurination with a shift from A to T (or the complement) in 3.8% of reads mapping to the adenine target of RIPs in Spiroplasma-infected flies (P < 10−180; coverage, 2,807 reads). In contrast, this signal was not present in Howardula reads from Spiroplasma-uninfected flies (P > 0.1; coverage, 15,389). The same analysis mapping a subset of raw reads to D. neotestacea 28S rRNA also revealed significant evidence of depurination, but to a much lesser extent, with a shift detectable in only 0.4% of reads (P < 10−17; coverage, 4,822). Again, there was no evidence of depurination in the absence of Spiroplasma (P > 0.1; coverage, 3,485). This near 10-fold greater depurination of Howardula versus D. neotestacea rRNA suggests substantial differences in exposure and/or susceptibility of host versus parasite ribosomes to a RIP in the presence of Spiroplasma, as we would expect. This signal of depurination is also likely conservative; rRNA depurinated by RIPs is highly susceptible to hydrolysis of the sugar-phosphate backbone at the site of depurination (31), and the freezing undergone by these samples before library construction might be expected to decrease the detectability of depurination (35). The magnitude of these effects should, however, be interpreted with caution as the polyA RNA enrichment used before sequencing would have depleted rRNA, also potentially affecting the observed signal.
qPCR Confirms That Howardula 28S rRNA Is Depurinated in Vivo During Spiroplasma Infection.
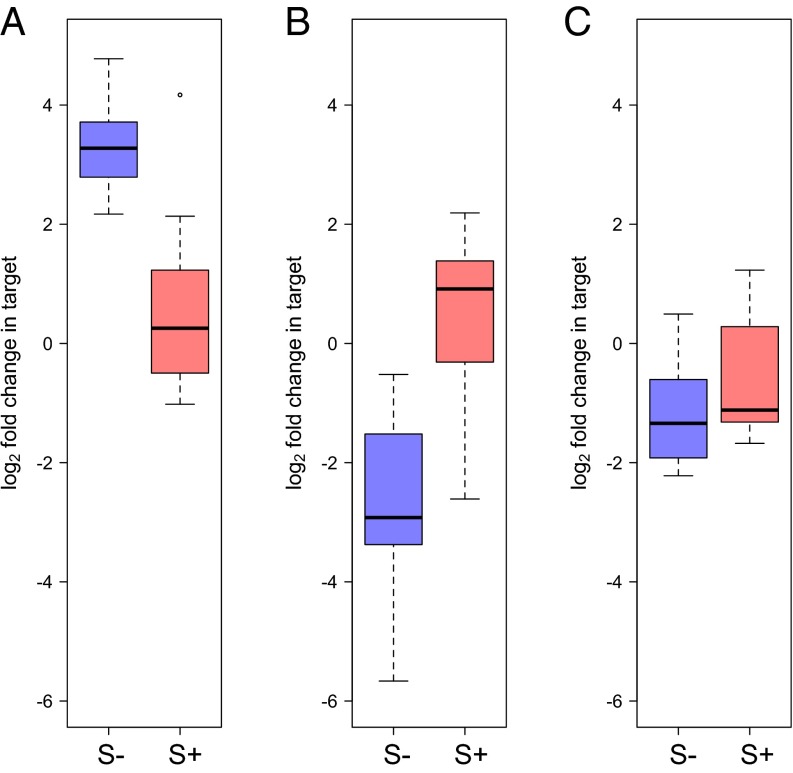
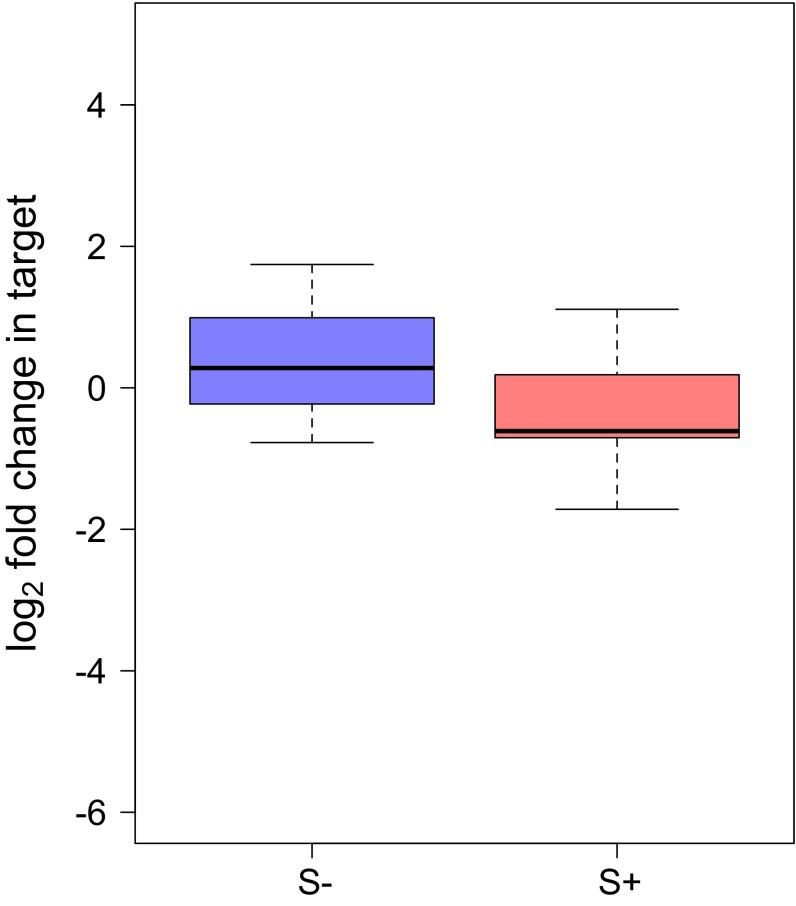
We applied the RT-qPCR assay for depurination of Howardula 28S rRNA to Howardula-infected adult flies, infected and uninfected with Spiroplasma, collected 1 d posteclosion. Howardula 28S rRNA with an intact SRL was dramatically decreased in the presence of Spiroplasma (t8.88 = 3.37, P = 0.008; Fig. 4A). Because this assay is normalized to an upstream region of rRNA not predicted to be affected by RIPs, this represents a ∼sixfold depletion of Howardula rRNA intact at the SRL in the presence of Spiroplasma, relative to the abundance of an upstream region of the 28S rRNA. Notably, the depurinated template was also ∼20-fold more detectable (Fig. 5B; t10.53 = 3.36, P = 0.007) in the presence of Spiroplasma. As fold changes are reported with respect to control samples with no expected depurination, a decrease of sixfold in intact SRL would be expected to be a much greater decrease than a 20-fold increase in depurinated template over a (negligible) background signal, although we do not directly measure absolute rRNA abundances. Depurination at the SRL is known to be a potent inducer of apoptosis (33, 37) and, as mentioned, renders the rRNA backbone highly susceptible to hydrolysis (31), likely accounting for the apparently modest accumulation of depurinated rRNA coincident with a large decrease in relative signal for the intact SRL.
Fig. 5.
Howardula 28S rRNA is depurinated in vivo in the presence of Spiroplasma, and the relative abundance of intact 28S rRNA is dramatically reduced. Intact Howardula rRNA is reduced ∼sixfold in the presence of Spiroplasma (A) (P = 0.008), whereas the abundance of depurinated template representing RIP-induced depurination increases ∼20-fold (B) (P = 0.007). Drosophila rRNA is not significantly depurinated in the same samples (C) (P = 0.16). cDNA used in analysis was reverse-transcribed from whole 1-d-old Howardula-infected flies with (S+) and without (S–) Spiroplasma infection (n = 13 flies).
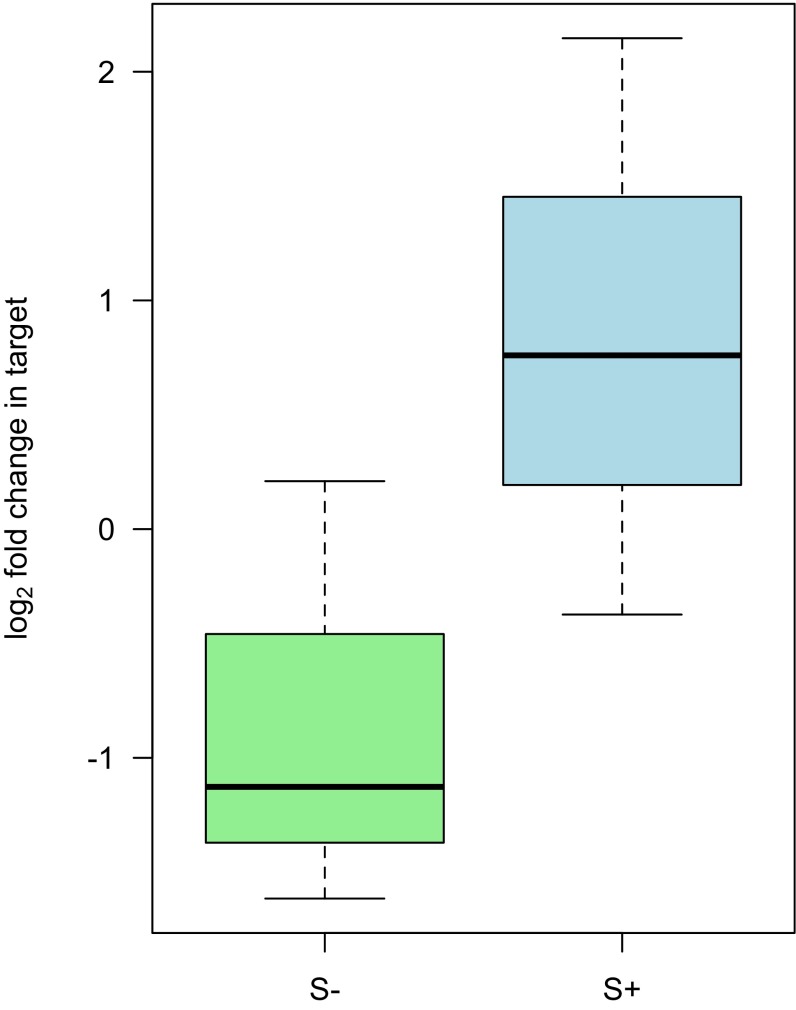
To corroborate RNA-seq results and confirm that depurination is more specific to Howardula, we designed an RT-qPCR assay for D. neotestacea rRNA (Table S1). We assayed fly ovaries—host tissue known to be high in Spiroplasma density—in gravid females with and without Spiroplasma, as well as in the whole-fly samples that showed depurination of the Howardula SRL (above). Although there appeared to be slightly elevated RIP-specific depurination in host rRNA in the presence of Spiroplasma (Fig. 5C and Fig. S3), this was not significant above controls in either assay at these sample sizes (P = 0.30 and P = 0.14 for whole flies and ovaries, respectively) and was substantially lower than that observed in Howardula in the same samples. We also assayed for a signal of depletion of intact template in flies in the presence of Spiroplasma similar to that observed for Howardula but observed no such signal (Fig. S4; P = 0.21), consistent with substantially greater effects on Howardula. This confirms the greater level of depurination in the parasite versus host, suggesting that an RIP disproportionately targets Howardula rRNA when Spiroplasma is present.
Table S1.
Efficiencies of primers used for all assays
| Primer set | Primer sequences, forward and reverse, 5′ to 3′ | R2 | Efficiency | Slope 95% confidence interval |
| Drosophila | ||||
| Intact | CGACAGCATTCCTGCGTAGTAAGA | 0.995 | 106.6% | −3.34 < s < −3.01 |
| ACAATGCAAATTGCCCCTTA | ||||
| Depurinated | CGACAGCATTCCTGCGTAGTAAGT | 0.997 | 101.4% | −3.44 < s < −3.13 |
| ACAATGCAAATTGCCCCTTA | ||||
| Normalizer | CAAGGACATTGCCAGGTAGG | 0.997 | 102.7% | −3.40 < s < −3.12 |
| AGCTTTTGCTGTCCCTGTGT | ||||
| Howardula | ||||
| Intact | TGATAGTAATCCTGCTTAGTAAGA | 0.997 | 98.0% | −3.52 < s < −3.22 |
| CACCGGAGAGCAACGATATT | ||||
| Depurinated | TGATAGTAATCCTGCTTAGTAAGT | 0.998 | 105.4% | −3.32 < s < −3.08 |
| CACCGGAGAGCAACGATATT | ||||
| Normalizer | CAAATGCCTCGTCGGATG | 0.991 | 92.1% | −3.81 < s < −3.24 |
| GCCAAAGCCTCCCACTTATAC | ||||
| Rabbit | ||||
| Intact | GGGTTTAGACCGTCGTGAGA | 0.998 | 79.6% | −4.03 < s < −3.83 |
| AGTGGAACCGCAGGTTCAGA | ||||
| Depurinated | GGGTTTAGACCGTCGTGAGA | 0.997 | 78.9% | −4.13 < s < −3.80 |
| TGTGGAACCGCAGGTTCAGA | ||||
| Normalizer | CGTTGGATTGTTCACCCACT | 0.999 | 96.4% | −3.48 < s < −3.33 |
| CATACACCAAATGTCTGAACCTG | ||||
Efficiency and specificity were validated on standard curves of 5 × 10-fold serial dilutions of synthetic DNA (IDT gBlocks) for all primer sets, except for the primer pair for the Howardula normalizing primer set, which was tested using a dilution series of cDNA reverse-transcribed from Howardula-infected flies. Bases specific to sites of depurination are in bold, and deliberate mismatches are underlined.
Fig. S3.
rRNA in D. neotestacea ovaries is not strongly depurinated at the site of RIP attack in the presence of Spiroplasma (S+) (n = 6; P = 0.14). Abundance of depurinated template normalized to upstream rRNA is presented. Ovaries were dissected from 1–2-wk-old gravid females.
Fig. S4.
D. neotestacea 28S rRNA does not show appreciable depletion of signal of 28S rRNA intact at the SRL in the presence of Spiroplasma (S– vs. S+) (P = 0.21; n = 13), in contrast to the strong signal of depletion observed in Howardula rRNA from the same flies (Fig. 5).
Discussion
Recent years have seen an increasing awareness that symbiotic associations can be critical in protecting multicellular hosts against parasites and pathogens. Many host-associated microbes produce metabolites that are known or suspected to function in defensive capacities (14, 17, 38), but the effectors that defend against specific enemies in maternally transmitted insect endosymbionts, whose success is typically intimately linked to that of the host (39), are poorly understood. This is largely due to the difficulty of working with uncultivable symbiotic lineages: Here neither Howardula nor Spiroplasma can currently be grown outside of the host, precluding many approaches to establishing function in these systems. There is great interest in exploiting insect symbioses to interrupt disease transmission from insects to humans, and a lack of understanding of the mechanisms underlying evolutionarily durable defensive symbioses impedes a full evaluation of the efficacy of these strategies.
Here, we show that the Spiroplasma defensive symbiont currently sweeping through North American populations of a common woodland Drosophila encodes a divergent RIP and that a virulent and common nematode parasite shows rRNA depurination consistent with RIP attack when Spiroplasma is present. Although some other facultative symbionts of insects are known to produce potent toxins, such as the pederin produced by Pseudomonas symbionts of Paederus rove beetles, Spiroplasma in D. neotestacea is remarkable for the extent to which the association has been selected upon due to its defensive properties, resulting in its rapid spread across North America. It is also remarkable due to the extent to which a prevalent parasite, Howardula, is affected. This association thus allows exploration not only of Spiroplasma’s defensive factors but also of ways in which Howardula might counterevolve to mitigate them. Understanding the mechanisms responsible for this defense can thereby clarify the proximate causes of ecologically relevant defensive symbioses, as well as the ways in which they might be circumvented by their targets.
In vivo, depurination of Howardula ribosomes occurs to a much greater extent than in the Drosophila host during Spiroplasma infection, demonstrating substantially greater effects on the parasite. Indeed, effective targeting of invading parasites would be expected of a toxin functioning in defense. It is unclear how this specificity is achieved here; there is substantial precedent for specificity of type I plant RIPs, which can have highly varying toxicities against different cell lines in vitro, although the molecular basis is mostly unknown (40, but see ref. 41). Similarly, the B5 subunit of Shiga-like toxins—the closest characterized relatives of SpRIP—binds specifically to the glycosphingolipid Gb3 of mammalian cells, triggering toxin endocytosis into Gb3-bearing cells and leading to heightened toxicity against specific tissues and cell types (30). In the case of SpRIP, whether the predicted disordered region might function similarly as a ligand, specifically binding to receptors of Howardula and other parasites, is unclear but is suggested by the lack of a strong decrease in intact rRNA in in vitro assays against Howardula with recombinant SpRIP lacking this region. In addition, a potential pore-forming toxin is encoded directly upstream of SpRIP (27), and it might be that such factors provide entry for RIPs into Howardula cells, potentiating toxicity.
Intriguingly, SpRIP is the first characterized of what appears to be a relatively diverse array of RIPs encoded by different Spiroplasma strains (Fig. 1), some of which are primarily known as either insect pathogens or male-killers and one of which—the MSRO strain of S. poulsonii—is also defensive against parasitoid wasps (24). Many but not all of these putative toxins have retained the essential residues of RIPs, whereas some also possess extensive modifications that include uncharacterized C-terminal domains of hundreds of amino acids. This conservation and proliferation of RIP-like sequences across Spiroplasma strains suggests functional importance in some capacity, and it is tempting to speculate that they might play roles in other defensive symbioses or in male-killing. Indeed, the apoptotic hallmarks of MSRO-induced male-killing bear similarity to those induced by RIPs in other systems (42). Putative Shiga-like toxins are also encoded in the genomes of phages that are essential to the protection against parasitoid wasps that is conferred to aphids by Hamiltonella (43), and it will be interesting to test whether these also target ribosomes.
Recent studies that transfer Spiroplasma strains to new host species have revealed interesting variation in the fitness consequences and defensive properties of novel host–Spiroplasma associations. When established in new host species, the D. neotestacea strain of Spiroplasma successfully protects against nematode infection (44). On the other hand, although other strains, including at least one predicted to encode RIPs, were able to stably persist in D. neotestacea, only the native strain protected against Howardula (45), suggesting that particular Spiroplasma-encoded RIPs might be specific to different parasites or pathogens. This is consistent with the high degree of divergence observed between Spiroplasma RIPs (Fig. 1) as well as our finding of Howardula-specific depurination. With respect to effects on the host, others have suggested that some degree of mis-targeting of toxins contributes to the virulence that is sometimes observed in novel (laboratory-initiated) Spiroplasma infections (46), and our findings are also consistent with this possibility. In D. neotestacea, we have observed no fitness costs associated with Spiroplasma (e.g., ref. 21), and population cage experiments also suggest that direct costs of Spiroplasma—toxin-induced or otherwise—are negligible or context-dependent in this symbiosis (47).
In sum, we present evidence of a novel RIP encoded by a Drosophila defensive symbiont and find that Howardula suffers a much greater degree of rRNA depurination than the Drosophila host due to Spiroplasma; whether RIPs might act in concert with other factors in this association remains to be determined. The continued goal of understanding the complex interactions that underpin ecologically important symbioses require a deeper understanding of these factors, as does the aim of exploiting defensive symbioses to limit disease transmission to humans. Our findings shed light on these factors, while also suggesting an intriguing function for RIPs in nature as players in this tripartite defensive symbiosis.
Materials and Methods
Phylogenetic Analysis.
Putative RIPs from Spiroplasma were accessed using BLASTp searches against the National Center for Biotechnology Information (NCBI) nonredundant database with SpRIP as a query and were included based on a low E-value and high degree of coverage (48). We aligned these and selected RIP sequences from the Pfam seed database using kalign (49) and constructed maximum likelihood trees [1,000 bootstraps generated in PHYLIP (50)] with FastTree (51) following model selection in MEGA (52) (Whelan and Goldman model in FastTree used).
Expression and Purification of SpRIP.
The gene encoding full-length SpRIP was codon optimized for expression in E. coli and synthesized by GenScript, with the region coding for the mature protein (Ala31 through His403—signal sequence removed) subcloned into a modified pET28a expression vector containing an N-terminal tobacco etch virus (TEV) protease-cleavable hexahistidine tag.
Recombinant SpRIP was produced using the E. coli BL21 codon plus strain. Chemically competent cells were transformed and grown in 2XYT media containing 50 μg/mL ampicillin and 35 μg/mL chloramphenicol at 37 °C with shaking. Overnight culture was diluted 20-fold into 1 L ZYP-5052 autoinduction media at the same antibiotic concentration and grown for 4 h at 37 °C before the temperature was reduced to 30 °C for overnight cell growth.
Bacterial cells were harvested by centrifugation and lysed by French press. We purified protein in the cell lysate by Ni-NTA batch bind. Briefly, the cell lysate was diluted in a Ni-NTA binding buffer (20 mM Hepes, pH 8.0, 1 M NaCl, 30 mM imidazole) and incubated with 2 mL Ni-NTA slurry at 4 °C for 1 h with stirring. Following the incubation, the recombinant protein was eluted from Ni-NTA resin in 5 mL fractions with 250 mM imidazole in elution buffer (20 mM Hepes, pH 8.0, 1 M NaCl). Elutions were pooled and buffer exchanged into HBS (20 mM Hepes, pH 7.5, 150 mM NaCl), with 2% (vol/vol) glycerol and 0.5 mM EDTA. The N-terminal hexahistidine tag was cleaved with TEV protease using the established protocol from Sigma-Aldrich. The TEV-treated RIP was further purified by cation exchange chromatography using 0–1.0 M gradient of NaCl in 20 mM Hepes buffer, pH 6.8, and finally by size exclusion chromatography in HBS [20 mM Hepes, pH 7.5, 300 mM NaCl, with 2% (vol/vol) glycerol]. Fractions were analyzed by SDS/PAGE, and the monomeric fractions as defined by size exclusion chromatography elution profile were pooled and concentrated.
Incubation of the purified recombinant protein (44 kDa) at 4 °C for 2 wk in HBS resulted in a stable degradation product of 34 kDa as shown by SDS/PAGE (Fig. S1). To identify the sequence of the proteolyzed fragment, the 34-kDa band was excised from the gel, reduced, alkylated, and in silico-digested with trypsin. Mass spectrometric analysis of the digested peptides was done with a Voyager DE-STR mass spectrometer (Applied Biosystems) using mass range 800–3,500. For comparison with the MS-captured peptide masses, the full-length recombinant protein sequence was submitted to Protein Prospector–MS digest server (University of California, San Francisco), which reports the predicted trypsin-digested peptide masses. The MS data showed that ∼70 residues from the N terminus were proteolyzed.
The final yield of SpRIP was 0.72 mg purified protein·L−1 cell culture. BBE31, a surface protein of Borrelia burgdorferi, was purified by the same method and was used alternatively to BSA as a negative control in incubations, showing that SpRIP’s activity did not a result from contamination from the expression system.
Bioinformatics for RNA-Seq.
RNA-seq reads originating from ref. 27 were used to test for evidence of depurination of the 28S rRNA SRL at the level of cDNA (deposited in the NCBI Sequence Read Archive, accession no. PRJNA295093). In brief, a factorial experiment was conducted in which we sequenced the metatranscriptome of D. neotestacea in the presence and absence of Spiroplasma and Howardula infection. Targeted reassemblies of Howardula and Drosophila 28s rRNA were conducted in Geneious 7 (Biomatters, Ltd.) to obtain near full-length 28S rRNAs spanning the conserved SRL for both species. Raw reads (or a random subset thereof) from Howardula-infected libraries with and without Spiroplasma were mapped to these assemblies (default low sensitivity setting) and P values for variants called in Geneious.
Design and Validation of RT-qPCR for Depurination.
For rabbit, Howardula, and Drosophila ribosomes, we designed RT-qPCR assays following the methods of refs. 35 and 36 (Table S1). In summary, primers were designed with the 3′ terminal base of either the forward or reverse primer complementary to the site of depurination, with separate primers designed to detect intact (A) versus depurinated (T) template and a secondary mismatch to increase specificity (35, 36). The reverse primer for each assay was designed in Primer3 (53) and chosen to bind to a region of divergence between Howardula and Drosophila for those assays. A second normalizing primer set for each assay was designed for upstream rRNA regions not predicted to be affected by RIPs. Because these sequences are contiguous on the rRNA, the normalizing and SRL regions are expected to occur 1:1 in controls (e.g., in the absence of a RIP).
All assays were tested for target specificity using synthetic DNA (IDT gBlocks) with and without a transversion to T at the predicted site of depurination. In all cases, no cross-amplification of the nontarget template occurred until ∼12 Ct later than target amplification, indicating primer pairs are ∼4,000× more specific to their target templates, making this the saturation limit of the assay. Because samples with no depurination will cross-amplify at this point, fold changes should be interpreted in a relative manner—changes to reaction conditions that affect specificity will affect baseline measures of depurination. Fold change in targets was calculated using the ΔΔCt method, normalized to amplification of rRNA upstream of the site of depurination, and mean Ct values for each target in each separate experiment, or a reference sample from the control treatment when standardizing control samples to 0 was desired (36). If any samples were rerun (for the in vivo experiments), ΔCts were calculated with respect to a standard of pooled cDNA for normalization across plates. Efficiencies and R2 values (Table S1) for primers for detection of intact and depurinated template were calculated using 5 × 10-fold serial dilutions of synthetic DNA or random-primed cDNA (for the Howardula normalizing primer set only).
Total RNA was extracted from samples (reticulocyte lysate, whole flies, or nematode motherworms) using TRIzol (Invitrogen). For each experiment, either 500 or 1,000 ng of RNA was reverse-transcribed using SuperScript II (Invitrogen) and random priming, following quantification with a NanoDrop spectrophotometer. We found that delays in reverse transcription or freeze-thaw cycles decreased detectability of depurinated rRNA, so RNA was reverse-transcribed immediately following RNA extraction. qPCR reactions were run at 1/10 cDNA dilutions in duplicate 10 or 20 μL reactions on a BioRad CFX96 thermal cycler with BioRad SsoFast EvaGreen Supermix. Two cDNA samples for tests of in vivo depurination that could not be reliably amplified with normalizing primer sets were excluded from analysis (2/21). Control samples with no expected depurination in which the primer set for depurinated template failed to amplify were conservatively assigned the highest reliably amplified Ct value for the primer set for the experiment during analysis. All statistical analyses were conducted in R v.3.2.1 (54), primarily using linear models or Welch's t test with log2 transformations of response variables to meet test assumptions.
For in vivo tests of depurination, Spiroplasma-infected and uninfected D. neotestacea were reared and infected with Howardula as detailed in ref. 27.
Acknowledgments
We thank Perry Howard for constructive discussions and David Stuss and Matt Ballinger for comments that improved the manuscript. P.T.H. was supported by Natural Sciences and Engineering Research Council (NSERC) and University of Victoria scholarships. This work was supported by NSERC Discovery grants (to S.J.P. and M.J.B.) and a Sinergia grant from the Swiss National Science Foundation (to S.J.P.). M.J.B. gratefully acknowledges the Canada Research Chair program for salary support. S.J.P. is a fellow of the Canadian Institute for Advanced Research’s Integrated Microbial Biodiversity Program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the NCBI Sequence Read Archive (accession no. PRJNA295093).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518648113/-/DCSupplemental.
References
- 1.Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moran NA. Symbiosis. Curr Biol. 2006;16(20):R866–R871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 3.McFall-Ngai M, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110(9):3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zug R, Hammerstein P. Still a host of hosts for Wolbachia: Analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One. 2012;7(6):e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell JA, et al. A veritable menagerie of heritable bacteria from ants, butterflies, and beyond: Broad molecular surveys and a systematic review. PLoS One. 2012;7(12):e51027. doi: 10.1371/journal.pone.0051027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Werren JH, Baldo L, Clark ME. Wolbachia: Master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6(10):741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- 7.Hunter MS, Perlman SJ, Kelly SE. A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. Proc Biol Sci. 2003;270(1529):2185–2190. doi: 10.1098/rspb.2003.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaenike J. Population genetics of beneficial heritable symbionts. Trends Ecol Evol. 2012;27(4):226–232. doi: 10.1016/j.tree.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Scarborough CL, Ferrari J, Godfray HC. Aphid protected from pathogen by endosymbiont. Science. 2005;310(5755):1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 10.Teixeira L, Ferreira A, Ashburner M. The bacterial symbiont Wolbachia induces resistance to RNA viral infections in Drosophila melanogaster. PLoS Biol. 2008;6(12):e2. doi: 10.1371/journal.pbio.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedges LM, Brownlie JC, O’Neill SL, Johnson KN. Wolbachia and virus protection in insects. Science. 2008;322(5902):702. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- 12.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 2003;100(4):1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: Recent spread of a Drosophila defensive symbiont. Science. 2010;329(5988):212–215. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 14.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99(22):14002–14007. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellner RLL, Dettner K. Differential efficacy of toxic pederin in deterring potential arthropod predators of Paederus (Coleoptera: Staphylinidae) offspring. Oecologia. 1996;107(3):293–300. doi: 10.1007/BF00328445. [DOI] [PubMed] [Google Scholar]
- 16.Oliver KM, Smith AH, Russell JA. Defensive symbiosis in the real world—Advancing ecological studies of heritable, protective bacteria in aphids and beyond. Funct Ecol. 2013;28(2):341–355. [Google Scholar]
- 17.Kroiss J, et al. Symbiotic Streptomycetes provide antibiotic combination prophylaxis for wasp offspring. Nat Chem Biol. 2010;6(4):261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 18.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325(5943):992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoffmann AA, et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature. 2011;476(7361):454–457. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 20.Jaenike J. Interactions between and their mycophagous nematode Drosophila to community parasites: From physiological to community ecology. Oikos. 1995;72(2):235–244. [Google Scholar]
- 21.Cockburn SN, et al. Dynamics of the continent-wide spread of a Drosophila defensive symbiont. Ecol Lett. 2013;16(5):609–616. doi: 10.1111/ele.12087. [DOI] [PubMed] [Google Scholar]
- 22.Anbutsu H, Fukatsu T. Spiroplasma as a model insect endosymbiont. Environ Microbiol Rep. 2011;3(2):144–153. doi: 10.1111/j.1758-2229.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 23.Xie J, Vilchez I, Mateos M. Spiroplasma bacteria enhance survival of Drosophila hydei attacked by the parasitic wasp Leptopilina heterotoma. PLoS One. 2010;5(8):e12149. doi: 10.1371/journal.pone.0012149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie J, Butler S, Sanchez G, Mateos M. Male killing Spiroplasma protects Drosophila melanogaster against two parasitoid wasps. Heredity (Edinb) 2014;112(4):399–408. doi: 10.1038/hdy.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett. 2013;16(2):214–218. doi: 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 26.Haine ER. Symbiont-mediated protection. Proc R Soc B Biol Sci. 2008;275(1633):353–361. doi: 10.1098/rspb.2007.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamilton PT, Leong JS, Koop BF, Perlman SJ. Transcriptional responses in a Drosophila defensive symbiosis. Mol Ecol. 2014;23(6):1558–1570. doi: 10.1111/mec.12603. [DOI] [PubMed] [Google Scholar]
- 28.Bergan J, Dyve Lingelem AB, Simm R, Skotland T, Sandvig K. Shiga toxins. Toxicon. 2012;60(6):1085–1107. doi: 10.1016/j.toxicon.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 29.de Virgilio M, Lombardi A, Caliandro R, Fabbrini MS. Ribosome-inactivating proteins: From plant defense to tumor attack. Toxins (Basel) 2010;2(11):2699–2737. doi: 10.3390/toxins2112699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stirpe F. Ribosome-inactivating proteins. Toxicon. 2004;44(4):371–383. doi: 10.1016/j.toxicon.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Endo Y, Tsurugi K. Mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. Nucleic Acids Symp Ser. 1986;262(17):187–190. [PubMed] [Google Scholar]
- 32.Lainhart W, Stolfa G, Koudelka GB. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J Bacteriol. 2009;191(16):5116–5122. doi: 10.1128/JB.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johannes L, Römer W. Shiga toxins--From cell biology to biomedical applications. Nat Rev Microbiol. 2010;8(2):105–116. doi: 10.1038/nrmicro2279. [DOI] [PubMed] [Google Scholar]
- 34.Paredes JC, et al. Genome sequence of the Drosophila melanogaster male-killing Spiroplasma strain MSRO endosymbiont. MBio. 2015;6(2):1–12. doi: 10.1128/mBio.02437-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melchior WB, Jr, Tolleson WH. A functional quantitative polymerase chain reaction assay for ricin, Shiga toxin, and related ribosome-inactivating proteins. Anal Biochem. 2010;396(2):204–211. doi: 10.1016/j.ab.2009.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Pierce M, Kahn JN, Chiou J, Tumer NE. Development of a quantitative RT-PCR assay to examine the kinetics of ribosome depurination by ribosome inactivating proteins using Saccharomyces cerevisiae as a model. RNA. 2011;17(1):201–210. doi: 10.1261/rna.2375411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iordanov MS, et al. Ribotoxic stress response: Activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol. 1997;17(6):3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakabachi A, et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 2013;23(15):1478–1484. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 39.Hamilton PT, Perlman SJ. Host defense via symbiosis in Drosophila. PLoS Pathog. 2013;9(12):e1003808. doi: 10.1371/journal.ppat.1003808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandvig K, van Deurs B. Delivery into cells: Lessons learned from plant and bacterial toxins. Gene Ther. 2005;12(11):865–872. doi: 10.1038/sj.gt.3302525. [DOI] [PubMed] [Google Scholar]
- 41.Chan WY, Huang H, Tam SC. Receptor-mediated endocytosis of trichosanthin in choriocarcinoma cells. Toxicology. 2003;186(3):191–203. doi: 10.1016/s0300-483x(02)00746-1. [DOI] [PubMed] [Google Scholar]
- 42.Harumoto T, Anbutsu H, Fukatsu T. Male-killing Spiroplasma induces sex-specific cell death via host apoptotic pathway. PLoS Pathog. 2014;10(2):e1003956. doi: 10.1371/journal.ppat.1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Degnan PH, Moran NA. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol. 2008;74(21):6782–6791. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haselkorn TS, Cockburn SN, Hamilton PT, Perlman SJ, Jaenike J. Infectious adaptation: Potential host range of a defensive endosymbiont in Drosophila. Evolution. 2013;67(4):934–945. doi: 10.1111/evo.12020. [DOI] [PubMed] [Google Scholar]
- 45.Haselkorn TS, Jaenike J. Macroevolutionary persistence of heritable endosymbionts: Acquisition, retention and expression of adaptive phenotypes in Spiroplasma. Mol Ecol. 2015;24(14):3752–3765. doi: 10.1111/mec.13261. [DOI] [PubMed] [Google Scholar]
- 46.Nakayama S, et al. Can maternally inherited endosymbionts adapt to a novel host? Direct costs of Spiroplasma infection, but not vertical transmission efficiency, evolve rapidly after horizontal transfer into D. melanogaster. Heredity (Edinb) 2015;114(6):539–543. doi: 10.1038/hdy.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaenike J, Brekke TD. Defensive endosymbionts: A cryptic trophic level in community ecology. Ecol Lett. 2011;14(2):150–155. doi: 10.1111/j.1461-0248.2010.01564.x. [DOI] [PubMed] [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassmann T, Sonnhammer ELL. Kalign--An accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 51.Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura K, et al. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23(10):1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 54.R Core Team 2014 R: A Language and Environment for Statistical Computing. Available at www.r-project.org/