Abstract
Gene methylation is an important mechanism regulating gene expression and genome stability. Our previous work showed that methylation of the nitrate reductase (NR) gene NIA2 was dependent on chromomethylase 3 (CMT3). Here, we show that CMT3-mediated NIA2 methylation is regulated by ammonium in Arabidopsis thaliana. CHG sequences (where H can be A, T, or C) were methylated in NIA2 but not in NIA1, and ammonium [(NH4)2SO4] treatment completely blocked CHG methylation in NIA2. By contrast, ammonium had no effect on CMT3 methylation, indicating that ammonium negatively regulates CMT3-mediated NIA2 methylation without affecting CMT3 methylation. Ammonium upregulated NIA2 mRNA expression, which was consistent with the repression of NIA2 methylation by ammonium. Ammonium treatment also reduced the overall genome methylation level of wild-type Arabidopsis. Moreover, CMT3 bound to specific promoter and intragenic regions of NIA2. These combined results indicate that ammonium inhibits CMT3-mediated methylation of NIA2 and that of other target genes, and CMT3 selectively binds to target DNA sequences for methylation.
Keywords: ammonium, CMT3, DNA methylation, gene expression, NIA2, nitrogen assimilation, plant growth
Introduction
DNA methylation is an important epigenetic mechanism that regulates gene expression and genome stability in plants and animals (Herman and Baylin, 2003). DNA methylation in plants occurs at symmetric CG and CHG sequences and non-symmetric CHH sequences (where H can be A, C, or T). Three types of DNA methyltransferases (DNMTs) have been identified in plants, namely chromomethylase (CMT), methyltransferase (MET), and domain-rearranged methyltransferase (DRM) (Cao and Jacobsen, 2002b; Ronemus et al., 1996; Lindroth et al., 2001).
Chromomethylases contain six conserved motifs (I, IV, VI, VIII, IX, and X), a bromo adjacent homology (BAH) domain, and a chromodomain. The BAH domain is located at the N-terminal region, and the chromodomain is embedded between catalytic motifs I and IV. All DNMT1/MET1 proteins contain two BAH domains, whereas CMTs contain only one BAH domain. BAH domains are involved in protein–protein interactions, recognition of methylated histones, and nucleosome binding in animal systems (Armache et al., 2011; Kuo et al., 2012; Yang and Xu, 2013). Chromodomains function as methylated histone lysine-binding domains, facilitating recruitment to chromatin in animals (Sanchez and Zhou, 2009; Yap and Zhou, 2012). In plants, the BAH domain and chromodomain of CMT3 mediate its specific binding to histone H3 lysine 9 dimethylation (H3K9me2) (Du et al., 2012). Mutations in the BAH domain or chromodomain caused a failure of CMT3 binding to nucleosomes and a complete loss of CMT3 activity in vivo, suggesting that CMT3 associates with H3K9me2-containing nucleosomes through dual binding of its BAH and chromo domains to H3K9me2 to target DNA methylation.
Arabidopsis has three CMT genes, namely, CMT1, CMT2, and CMT3 (Henikoff and Comai, 1998; Finnegan and Kovac, 2000; McCallum et al., 2000). In the ecotype Wassilewskija (WS), CMT2 and CMT3 are predicted to encode functional proteins (Henikoff and Comai, 1998), whereas CMT1 encodes a non-functional protein (Jones et al., 2001). CMT genes have been identified in several plant species, including tobacco and maize, but they have not been identified in fungal or animal systems (Rose et al., 1998; Finnegan and Kovac, 2000). This indicates that CMT is a plant-specific DNMT. CMT2 and CMT3 have DNA methylation activity at CHG sites (Jones et al., 2001; Lindroth et al., 2001), and maintain CHH methylation at certain genomic loci (Cao and Jacobsen, 2002a; Cokus et al., 2008; Stroud et al., 2013). Zea mays methyltransferase 2 (ZMET2) and the Nicotiana benthamiana CMT3 homolog NbCMT3 were identified as CHG-specific DNMTs (Papa et al., 2001; Hou et al., 2013). MET1 is a homolog of mammalian DNMT1 that catalyzes symmetric CG methylation (Jones et al., 2001). MET1 has little effect on CHG methylation levels and it cannot substitute for CMT3 (Cokus et al., 2008; Stroud et al., 2013). The DRM DNMTs, which include DRM1, DRM2, and DRM3, have de novo DNA methylation activity at CG, CHG, and CHH sites through RNA-directed methylation pathways (Cao et al., 2003; Cokus et al., 2008; Henderson et al., 2010; Greenberg et al., 2011; Stroud et al., 2013). DRMs also maintain CHG and CHH methylation at specific genomic loci (Cao et al., 2003; Cokus et al., 2008; Henderson et al., 2010; Stroud et al., 2013). These combined results indicate that plant CHG and CHH methylation is controlled by CMT and DRM DNMTs (Chan et al., 2005; Stroud et al., 2013; Zemach et al., 2013).
The roles of plant MET1, DRM2, and CMT3 are well characterized. The expression patterns and levels of these DNMTs differ in different tissues and during different developmental stages. Mutation of these DNMTs causes severe loss of DNA methylation, which leads to abnormal development. For example, plants defective in CMT3 activity display abnormal embryo development (Pillot et al., 2010). Loss of MET1 activity delays flowering and reduces fertility, and these effects become more severe in the presence of CMT3 or DRM2 mutations (Xiao et al., 2006; Zhang and Jacobsen, 2006). A recent GUS study reported that these DNMTs are expressed in specific tissues or ubiquitously expressed at specific developmental stages (Huang et al., 2014). CMT3 is expressed only in specific organ regions that co-express DRM2 and MET1.
Nitrogen assimilation is a fundamental biological process that is essential for plant growth and development. Plants utilize nitrate as a source of environmental nitrogen, and nitrate is a potent signal that regulates nitrogen and carbon metabolism, plant growth, and development (Crawford and Forde, 2002; Forde, 2002; Stitt et al., 2002; Foyer et al., 2003). Nitrate is actively transported into cells from the soil by a nitrate transporter and then sequentially reduced to ammonia, which enters amino acid metabolic pathways primarily via the action of glutamine synthetase. NR, a key enzyme of the plant nitrogen assimilation pathway, forms homodimers that catalyze the NAD(P)H-dependent reduction of nitrate to nitrite (Campbell and Kinghorn, 1990; Solomonson and Barber, 1990). Therefore, the regulation of NR expression and NR activity is important for nitrate assimilation. A recent study reported that the Arabidopsis NRs NIA1 and NIA2 are positively regulated by sumoylation through the activity of the E3 SUMO ligase AtSIZ1 (Park et al., 2011). CMT3 sumoylation by AtSIZ1 is suggested to control NR gene expression (Kim et al., 2015a).
In the present study, we investigated the effect of ammonium on CMT3-mediated methylation of NR genes. We report that ammonium inhibits CMT3-mediated NIA2 methylation without affecting on CMT3 methylation. In addition, CMT3 binds to specific regions of NIA2. Our results provide evidence that CMT3-mediated NIA2 methylation is negatively modulated by ammonium.
Materials and Methods
Plant Materials, Growth Conditions, and Ammonium Treatment
Arabidopsis thaliana Columbia-0 ecotype (wild-type, Col-0) was used in this study. For in vitro culture in artificial media, seeds were surface-sterilized for 10 min using commercial bleach containing 5% sodium hypochlorite and 0.1% Triton X-100, rinsed five times in sterilized distilled water, and then stratified for 3 days at 4°C in the dark. Seeds were then sown on agar plates containing Murashige and Skoog medium (pH 5.7), 2% sucrose, and 0.8% agar. For plants grown in a non-agar substrate, seeds were sown and grown on sterile vermiculite. All plants were grown in a growth chamber at 22°C under a 16 h light/8 h dark cycle. To examine the effect of ammonium, three different ammonium sources [(NH4)2SO4, NH4Cl, and NH4NO3] were used with similar results. Thus, (NH4)2SO4 was used for all experiments.
Immunoprecipitation of Methylated DNA
Wild-type plants were grown in vermiculite for 3 weeks, treated with 5 mM (NH4)2SO4, and grown for an additional 12 h at 22°C. Then, genomic DNA was isolated from individual plants and sonicated to produce random fragments of 200–600 bp. The fragmented DNA (4 mg) was used in a standard methylated DNA immunoprecipitation (IP) (MeDIP) assay as described previously (Pomraning et al., 2009). Methylated DNA was recovered as described previously (Kim et al., 2015a).
Illumina Genome Analyzer Sequencing
To perform second-strand synthesis of MeDIP-enriched ssDNA fragments, approximately 200 ng of MeDIP-enriched ssDNA fragments and 500 ng of random primers were mixed in a final volume of 57.9 μl, incubated at 70°C for 10 min, and then cooled gradually for 40 min. Subsequent experiments were performed as described previously (Kim et al., 2015a). End-repair of DNA fragments, addition of an adenine residue to the 3′ fragment ends, adaptor ligation, and PCR amplification using Illumina paired-end primers were performed as described previously (Pomraning et al., 2009). The products were analyzed by agarose gel electrophoresis, and bands were excised to produce libraries with 250–350 bp insert sizes, which were quantified using Quant-iT PicoGreen dsDNA Reagent and Kit (Invitrogen). Flow cells were prepared with 8 pM DNA according to the manufacturer’s recommended protocol and sequenced for 36 cycles on an Illumina Genome Analyzer II (Illumina). The obtained images were analyzed and base-called using GA pipeline software (version 1.3) with default settings (Illumina).
Mapping Reads
The reads obtained from Illumina sequencing were mapped onto the Arabidopsis genome reference sequence (Bioconductor1) using Bowtie2 as described previously (Kim et al., 2015a). The sequence reads of untreated Col-0 wild-type plants were used as controls and to categorize the methylation sequences of (NH4)2SO4-treated wild-type plants.
Bisulfite Sequencing
Genomic DNA was isolated from rosette leaves of 3-week-old wild-type plants treated with 5 mM (NH4)2SO4 or untreated plants (control). Bisulfite treatment and sample recovery were performed using the EpiTect Bisulfite Kit (QIAGEN) according to the manufacturer’s instructions. The primers were designed using MethPrimer software2. The percentage methylation (% C) was calculated as 100 × C/(C + T). Cytosine methylation in CG, CHG, and CHH contexts was analyzed and displayed using CyMATE (Hetzl et al., 2007).
Analysis of CMT3 Pull-Down of NIA2
CMT3 binding to NIA2 was examined by in vivo pull-down using a plant expression vector construct. The full-length CMT3 cDNA was amplified by PCR using FLAG4-tagged forward and reverse primers, and inserted into the plant expression vector pBA002. This generated the construct 35S-CMT3-FLAG4. Then, 3-week-old wild-type plants were infiltrated with transformed agrobacteria carrying 35S-CMT3-FLAG4 and sequentially treated with 5 mM (NH4)2SO4. Plants were incubated for 12 h at 22°C, and genomic DNA and total protein were extracted from rosette leaves of the transformed plants. CMT3-FLAG4 expression was examined by western blotting using an anti-FLAG antibody.
For the binding assay, genomic DNA was sonicated to produce random fragments of 200 - 600 bp. The fragmented DNA (4 mg) was used in the IP assay. Anti-FLAG antibody (20 μg) was added to each sample of fragmented DNA, reactions were brought to a final volume of 1 ml in IP buffer [10 mM sodium phosphate (pH 7.0), 140 mM NaCl, and 0.05% Triton X-100], and the mixture was incubated for 2 h at 4°C. Then, samples were incubated with 40 μl of protein A agarose beads for 12 h at 4°C and then washed seven times with 1 ml of IP buffer. DNA was recovered from the beads by phenol-chloroform extraction and ethanol precipitation. Purified DNA was amplified with specific primers for NIA1 and NIA2, and then the DNA levels were examined by 1.2% agarose gel electrophoresis. The primers used for quantitative PCR were as follows: NIA1, 5′-GCTAGTAAGCATAAGGAGAG-3′ (forward) and 5′-CCTTCACGTTGTAACCCATCTTCT-3′ (reverse); NIA2, 5′-TGTCTCAGTACCTAGACTCTTTGC-3′ (forward) and 5′-TGTCTCAGTACCTAGACTCTTT-3′ (reverse).
The site of CMT3 binding to NIA2 was determined using genomic DNA isolated from 3-week-old wild-type plants, which was sonicated to produce 200 - 600 bp fragments. The fragmented DNA (4 mg) was immunoprecipitated and analyzed as described above. Purified DNA was amplified with gene-specific primers, and the DNA levels were examined by 1.2% agarose gel electrophoresis. The following forward and reverse primers (21 oligonucleotides) were designed for PCR amplification of different regions (fragments) of NIA2: fragment - 3,000 to - 2,500 bp, 5′-TCAATAAGAGGAGGCCACAAA-3′ (forward) and 5′-ATTGTATTATATATATCAAAG-3′ (reverse); fragment - 2,499 to - 2,000 bp, 5′-CTGGCCAACATCTATTCATTA-3′ (forward) and 5′-ATATATATGATTTTTATATAC-3′ (reverse); fragment - 1,999 to - 1,500 bp, 5′-TGAAACTGCTATATGCAAGTA-3′ (forward) and 5′-CTAATTTTGGGTAACCAATAT-3′ (reverse); fragment - 1,499 to - 1,000 bp, 5′-AAGTTCACAAGAAAATCAATA-3′ (forward) and 5′-TTTCTTATTAAACGTTATTTT-3′ (reverse); fragment - 999 to - 500 bp, 5′-ATAAATATTGTATGATTATTA-3′ (forward) and 5′-TTTTCCTTTTTATTTTAGTCG-3′ (reverse); fragment - 499 to - 1 bp, 5′-TGTTTTGATCACATTTTTATA-3′ (forward) and 5′-TTGGAAAGTGTATAATCGTAA-3′ (reverse); fragment 1 to 500 bp, 5′-CATGGCGGCCTCTGTAGATAA-3′ (forward) and 5′-GTTTGACGAATCCGGTCACCT-3′ (reverse); fragment 501–1,000 bp, 5′-GGCCCATGAAATTCACCATGG-3′ (forward) and 5′-GTTGTCCTTGAAATGGTAGAA-3′ (reverse); fragment 1,001–1,500 bp, 5′-AGAGTTTTACCTTCTTTGGTA-3′ (forward) and 5′-ACCTCCACACGGGTCACTTTT-3′ (reverse); fragment 1,501–2,000 bp, 5′-CACGGTAGATGGTGGAGAGAC-3′ (forward) and 5′-ATCGTGTACAATCATAGATAT-3′ (reverse); fragment 2,001–2,500 bp, 5′-TCCTTATGGATCACCCGGGTG-3′ (forward) and 5′-TGGGTGGACACCGCCAAAGTA-3′ (reverse); fragment 2,501–3,412 bp, 5′-AGATTCCCTAACGGCGGGCTC-3′ (forward) and 5′-ATGAAAAAATGAACATATTCA-3′ (reverse).
Examination of DNA Methylation by McrBC-PCR
The methylation analysis procedure involved 5-methylcytosine-specific restriction enzyme (McrBC)-based PCR using a previously published protocol (Lippman et al., 2003). Genomic DNA (500 ng) was isolated from wild-type plants treated with 5 mM (NH4)2SO4 or untreated plants (control) and digested with 30 units of McrBC endonuclease (New England Biolabs) for 3 h at 37°C. Then, quantitative PCR analyses were performed as described previously (Kim et al., 2015a). The primers used for quantitative PCR were as follows: NIA1, 5′-GCTAGTAAGCATAAGGAGAG-3′ (forward) and 5′-CCTTCACGTTGTAACCCATCTTCT-3′ (reverse); NIA2, 5′-TGTCTCAGTACCTAGACTCTTTGC-3′ (forward) and 5′-TGTCTCAGTACCTAGACTCTTT-3′ (reverse); CMT3, 5′-GCTAGTAAGCATAAGGAGAG-3′ (forward) and 5′-CCTTCACGTTGTAACCCATCTTCT-3′ (reverse); tubulin, 5′-GTGAGCGAACAGTTCACAGC-3′ (forward) and 5′-TTATTGCTCCTCCTGCACTT-3′ (reverse).
Transcript Level Analysis for Genes Involved in NR Pathways and CMT3
Wild-type plants were grown for 3 weeks in vermiculite and treated with or without 5 mM (NH4)2SO4 for 12 h. Total RNA was extracted from rosette leaves, quantified, and divided into equal amounts. First-strand cDNA was synthesized from 5 μg of total RNA using an iScript cDNA Synthesis Kit (Bio-Rad), and cDNA was amplified by real-time qRT-PCR (MyiQ, Bio-Rad) according to the manufacturer’s instructions. PCR analysis and sequencing were performed as described previously (Kim et al., 2015b). The primers used for quantitative PCR were the same as those described in the previous section.
Results
Ammonium Reduces Genome Methylation Levels
Nitrogen is an essential element for plant growth, and plants take up nitrogen as nitrate or ammonium through roots or leaves to support biosynthetic production. Nitrate, the uptake of which is mediated by nitrate transporters, is converted to ammonia by NR and nitrite reductase, indicating that ammonia affects plant development as an end product before its incorporation into glutamine by glutamine synthase. Therefore, we first examined the effect of ammonium on genome methylation. For this experiment, wild-type plants were grown for 3 weeks in vermiculite and treated with or without 5 mM (NH4)2SO4 before being subjected to MeDIP sequencing analysis to determine the methylation level of the whole genome. The results showed that CpG and C methylation were lower in (NH4)2SO4-treated plants than in control plants (Table 1).
Table 1.
CpG and C enrichment analysis of ammonium-treated plants by MeDIP sequencing.
| No treatment | (NH4)2SO4 | |
|---|---|---|
| CG regions | 15,407,837 | 13,311,909 |
| C regions | 105,277,236 | 98,704,801 |
| Total base(bp) | 3,101,970,144 | 3,938,852,604 |
| Alignment rate(%) | 79.67 | 91.72 |
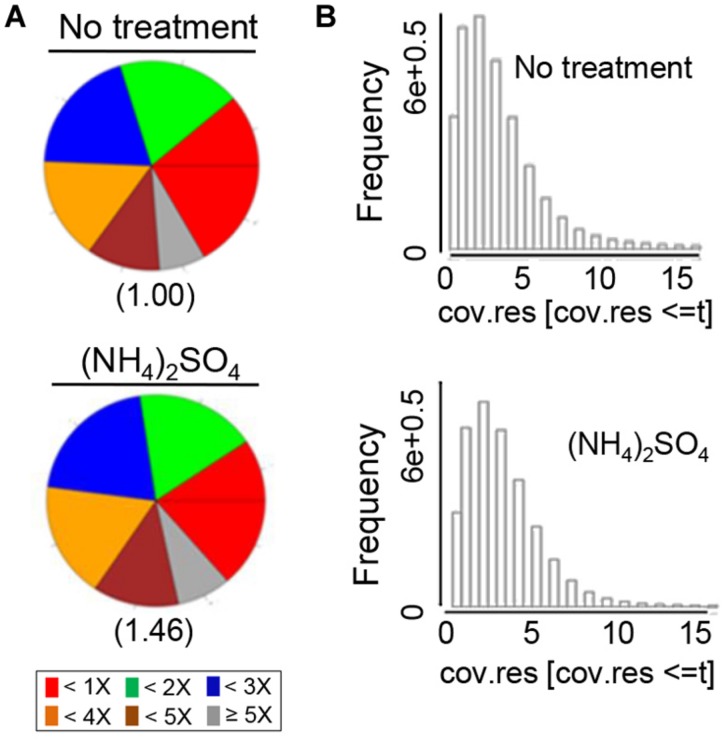
The number of reads that overlapped with CpG islands was determined to quantify the genomic coverage, which showed that the CpG coverage and sequence pattern coverage were lower in (NH4)2SO4-treated plants than in control plants (Figure 1). Therefore, the chromosomal distribution of methylated DNA was analyzed in wild-type plants treated with and without 5 mM (NH4)2SO4. Extensive DNA methylation was detected in the heterochromatic regions of each of the five chromosomes of control plants, although some noisy peaks were detected in other regions (Figure 2, left panel). This methylation pattern was similar to that reported previously (Zhang et al., 2006). In (NH4)2SO4-treated plants, methylation levels in the heterochromatic regions were lower than those in control plants (Figure 2, right panel). A comparison of whole genome methylation indicated that the methylation levels of 6,070 gene loci were downregulated in (NH4)2SO4-treated plants compared with those in control plants (Supplementary Table S1). This suggests that gene methylation status can be regulated by ammonium and lead to the upregulation of gene expression.
FIGURE 1.
MeDIP sequencing was used to detect whole genome methylation in ammonium-treated and untreated control Arabidopsis plants. Genomic coverage was quantified by the number of DNA methylation measurements that overlapped with CpG islands. (A) CpG coverage was determined by counting the number of reads. CpG islands were identified using the program MEDIPS. The numbers below the diagrams indicate “pattern not covered.” (B) The methylation pattern is presented as sequence pattern coverage. The parameter t indicates the maximal coverage depth to be plotted (15 reads per sequence pattern).
FIGURE 2.
DNA methylation landscape in the genome of ammonium-treated plants. MeDIP sequencing was performed using whole genomic DNA isolated from wild-type and 5 mM (NH4)2SO4-treated plants. Each peak shows the distribution of sequenced read count in chromosome. Boxes indicate heterochmatic regions in each of the five chromosomes.
Similar results were obtained by performing replicate experiments using other ammonium sources including NH4Cl, indicating that changes in DNA methylation levels in response to treatment with 5 mM (NH4)2SO4 are caused by ammonium and not by sulfate.
Ammonium Treatment Reduces NIA2 Methylation Levels
MeDIP sequencing analyses indicated that the whole genome methylation level was lower in (NH4)2SO4-treated plants than in control plants. Therefore, we examined the methylation level of NR genes, which encode the first enzyme in the nitrate reduction pathway. The Arabidopsis genome database contains two NR genes, NIA1 and NIA2, and their methylation levels were analyzed by bisulfite sequencing of genomic DNA isolated from control and (NH4)2SO4-treated plants. The results showed that NIA1 and NIA2CG methylation levels were lower in (NH4)2SO4-treated plants than in control plants (Table 2; Supplementary Table S2). CHG methylation of NIA1 was not detected in control and (NH4)2SO4-treated plants, whereas CHG methylation of NIA2 was significantly reduced from 11.11 to 0% in response to (NH4)2SO4 treatment (Table 2; Supplementary Table S2). CHH methylation was slightly increased from 0 to 1.28% in NIA1 in response to (NH4)2SO4, whereas CHH methylation was not detected in NIA2 even after (NH4)2SO4 treatment (Table 2; Supplementary Table S2). The overall NIA2 methylation level was reduced by (NH4)2SO4 treatment, but this had no effect on NIA1 methylation (Table 2; Supplementary Table S2). The same results were obtained in replicate experiments with other ammonium sources.
Table 2.
Methylation analysis of NIA1 and NIA2 genes by bisulfite sequencing after ammonium treatment.
|
(A) NIA1 |
(B) NIA2 |
||||
|
No treatment |
No treatment |
||||
| Methylated (%) | Non-methylated (%) | Methylated (%) | Non-methylated (%) | ||
| CG | 10.00 | 90.00 | CG | 11.11 | 88.88 |
| CHG | 0.00 | 100.00 | CHG | 11.11 | 88.88 |
| CHH | 0.00 | 100.00 | CHH | 16.66 | 83.33 |
| All | 1.89 | 98.11 | All | 14.81 | 85.19 |
|
(NH4)2SO4 |
(NH4)2SO4 |
||||
| Methylated (%) | Non-methylated (%) | Methylated (%) | Non-methylated (%) | ||
| CG | 5.55 | 94.44 | CG | 5.88 | 94.11 |
| CHG | 0.00 | 100.00 | CHG | 0.00 | 100.00 |
| CHH | 1.28 | 98.71 | CHH | 0.00 | 100.00 |
| All | 1.90 | 98.10 | All | 1.01 | 98.99 |
The region of NIA1 corresponds to positions from 1751st to 2128th (from transcription start site), and the region of NIA2 corresponds to positions from 1552nd to 1898th (from transcription start site).
Ammonium Inhibits CMT3 Binding to NIA2
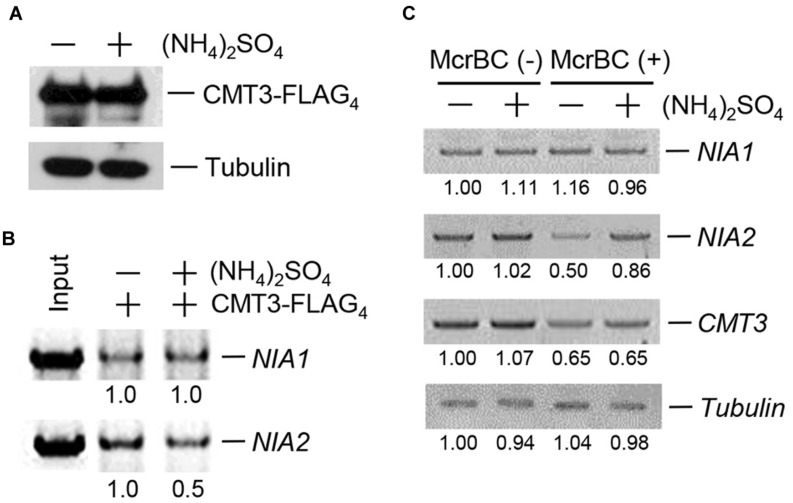
The complete loss of CHG methylation in NIA2 in response to (NH4)2SO4 led us to investigate the effect of (NH4)2SO4 on CMT3 binding to NIA2. The plant expression vector 35S-CMT3-FLAG4 was constructed and introduced into Agrobacterium, which was used to transform 3-week-old wild-type plants by infiltration. Then, CMT3-FLAG4 expression was analyzed in transformed plants after sequential treatment with 5 mM (NH4)2SO4 for 12 h and isolation of genomic DNA and total proteins. Western blot analysis showed that CMT3-FLAG4 was expressed in both untreated control and (NH4)2SO4-treated plants (Figure 3A). On the basis of this result, a DNA/protein-binding assay was performed using sonicated genomic DNA and an anti-FLAG antibody, and the pull-down DNA amount was estimated by PCR analysis with gene-specific primers for NIA1 and NIA2. The results showed that complex formation between CMT3 and NIA2 was inhibited in (NH4)2SO4-treated plants, whereas complex formation between CMT3 and NIA1 was not affected by (NH4)2SO4 treatment (Figure 3B).
FIGURE 3.
CMT3 binding to NIA2 decreased in response to ammonium. (A) Wild-type Arabidopsis was infiltrated by Agrobacteria transformed with 35S-CMT3-FLAG4, and then treated with or without 5 mM (NH4)2SO4. CMT3-FLAG4 expression was examined by western blotting with anti-FLAG antibody. Tubulin was used as the loading control. (B) CMT3-FLAG4 was pulled down with an anti-FLAG antibody, and DNA was extracted. The purified DNA was analyzed by PCR using gene-specific primers, and NIA1 and NIA2 levels were examined by agarose gel electrophoresis. Numbers under lanes indicate relative intensities. DNA levels were normalized to a value of 1.00 for DNA level in the “-” (NH4)2SO4 lane in each panel. (C) Genomic DNA was isolated from wild-type Arabidopsis treated with or without 5 mM (NH4)2SO4 and then digested with McrBC. The remaining DNA was used for PCR amplification with specific primers for NIA1, NIA2, and CMT3. Numbers under lanes indicate relative intensities. DNA levels were normalized to a value of 1.00 for DNA level in the “-” McrBC and “-” (NH4)2SO4 lane in each panel. Tubulin was used as the loading control.
The methylation levels of NIA1, NIA2, and CMT3 in (NH4)2SO4-treated plants were examined by McrBC digestion and PCR analysis with gene-specific primers. The gels showed similar band intensities for NIA1 and CMT3 in untreated control and (NH4)2SO4-treated plants, indicating that NIA1 and CMT3 methylation levels did not change in response to (NH4)2SO4 treatment (Figure 3C). However, the NIA2 band intensity increased in (NH4)2SO4-treated plants, indicating that NIA2 methylation was reduced in response to (NH4)2SO4 treatment (Figure 3C). Similar binding patterns and methylation levels were observed in response to treatment with other ammonium sources.
Ammonium Upregulates NIA2 Gene Expression
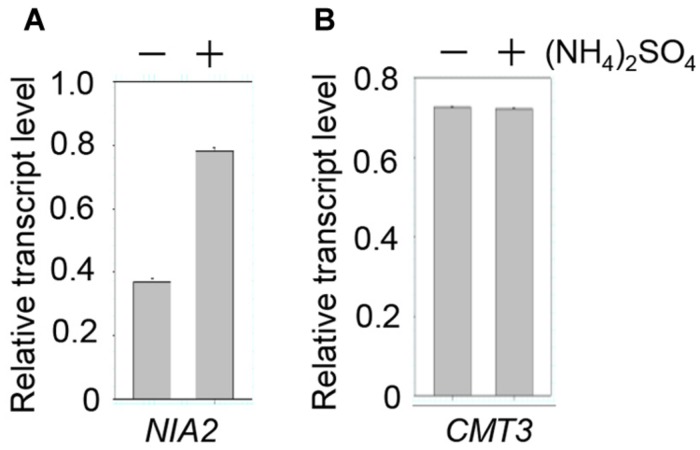
Our results showed that NIA2 methylation was inhibited by ammonium treatment, which suggests that ammonium induces NIA2 expression. NIA2 transcription was examined by qRT-PCR, which showed approximately 2-fold higher NIA2 transcript levels in (NH4)2SO4-treated plants than in control plants (Figure 4A). This indicates that NIA2 upregulation in (NH4)2SO4-treated plants resulted from low methylation levels caused by the lack of CHG methylation of NIA2. However, because low NIA2 methylation levels could also be caused by low CMT3 expression, we evaluated CMT3 expression levels and detected no differences in CMT3 transcript levels between (NH4)2SO4-treated and control plants (Figure 4B). These data indicate that low methylation levels and high transcription levels of NIA2 in response to (NH4)2SO4 were caused by impaired CHG methylation of NIA2. Similar gene expression patterns were detected with other ammonium sources.
FIGURE 4.
NIA2 expression was upregulated by ammonium. Total RNA was isolated from wild-type Arabidopsis plants treated with and without 5 mM (NH4)2SO4, and then subjected to real-time qRT-PCR analysis with specific primers for NIA2 (A) and CMT3 (B).
CMT3 Binds to Specific Regions of NIA2
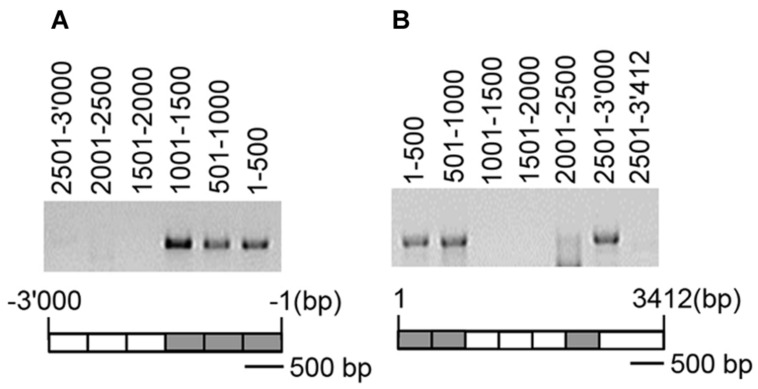
CMT3 catalyzed the CHG methylation of NIA2 but not that of NIA1. Therefore, the CMT3-binding region in NIA2 was detected using IP and PCR analysis. Wild-type Arabidopsis plants were transformed by infiltration with Agrobacteria carrying 35S-CMT3-FLAG4, and the DNA/CMT3-FLAG4 complex was pulled down with an anti-FLAG antibody. DNA was then amplified by PCR using specific primers for the promoter and coding regions of NIA2. The following three regions formed complexes with CMT3-FLAG4: the 1.5 kb promoter region from-1,500 to -1 bp (Figure 5A), the 1.0 kb coding region from 1 to 1,000 bp, and the 0.5 kb coding region from 2,501 to 3,000 bp (Figure 5B). These results indicate that CMT3 binds to specific regions in the NIA2 promoter and coding sequence, suggesting that CMT3 catalyzes NIA2 methylation at specific sequences or structures.
FIGURE 5.
CMT3 bound to specific regions of NIA2. Wild-type Arabidopsis was infiltrated with agrobacteria carrying 35S-CMT3-FLAG4, and DNA/CMT3-FLAG4 complexes were pulled down with an anti-FLAG antibody. Genomic DNA was extracted from pulled-down samples and subjected to PCR analysis with gene-specific primers. (A) DNA fragments were PCR amplified with specific primers for the NIA2 promoter, and the amplified DNA levels were examined by agarose gel electrophoresis. (B) DNA fragments were PCR amplified with specific primers for the NIA2 coding region, and the amplified DNA levels were analyzed by agarose gel electrophoresis. The amplified DNA positions are illustrated under the agarose gels. Gray boxes indicate highly amplified DNA fragments; white boxes indicate very low levels of amplified DNA fragments or positions with no amplification.
Discussion
The current study aimed to define the role of CMT3 in nitrogen assimilation. We examined the effect of ammonium on whole genome methylation and observed that ammonium reduced the levels of whole genome methylation (Table 1 and Supplementary Table S1). Plants utilize ammonium for carbon metabolism, nitrogen fixation, and biosynthesis of metabolic building blocks. Therefore, the effect of ammonium on reducing genome methylation suggests that high intracellular nitrogen concentration induces gene demethylation or represses gene methylation, thereby upregulating the expression of numerous genes involved in nitrogen metabolism. The carbohydrate-to-nitrogen (CN) ratio has a central and interactive role in regulating post-germination growth because the level of nitrogen is regulated with respect to that of carbohydrate during development (Huppe and Turpin, 1994). This suggests that ammonium treatment increases the intracellular nitrogen concentration, which affects nitrogen-sensitive metabolic networks. These combined data indicate that ammonium treatment induces the expression of genes involved in nitrogen assimilation by activating DNA demethylases or inactivating DNMTs.
CMT3-mediated genome methylation is a critical epigenetic modification that regulates plant growth and development. CMT3 functions in flower and reproductive organ development, embryogenesis, seed viability (Xiao et al., 2006; Pillot et al., 2010), organogenesis, and shoot regeneration from root explants (Shemer et al., 2015). CMT3 is involved in shoot growth regulation in Arabidopsis (Lin et al., 2015) and plays a role in tobacco leaf development by modulating the jasmonate pathway (Coustham et al., 2014). In addition, CMT3 acts as a functional regulator of oxidative stress to prevent genome instability and the accumulation of mutations caused by ionizing irradiation (Sidler et al., 2015). These data indicate that CMT3 catalyzes the methylation of numerous genes and may methylate and silence transposable elements (Kato et al., 2003). Therefore, CMT3 functions as a plant-specific DNMT that modulates growth, development, and differentiation throughout the plant life cycle from germination to seed maturation.
Our previous study showed that CHG methylation occurs on NIA2 but not on NIA1 and that CMT3 catalyzes CHG methylation of NIA2 (Kim et al., 2015a). In the present study, CHG methylation of NIA2 was not detected in ammonium-treated plants (Table 2), suggesting that ammonium treatment inactivates CMT3 activity or represses CMT3 expression. The mechanism of CMT3 methylation of target genes has not been completely elucidated. Three possible mechanisms are proposed to explain how CMT3 distinguishes target DNA sequences. First, CMT3 may distinguish target genes through specific siRNAs in a siRNA/CMT3 complex, leading to CHG methylation of the specific target genes. This is similar to the recently reported DRM2 methylation of genes with target sequence homology to specific siRNAs as an AGO4/siRNA/DRM2 complex (Matzke and Mosher, 2014; Zhong et al., 2014). Since AGO4/siRNA complex formation is a prerequisite for the interaction of siRNAs with other components including methylases, identification of AGO4/siRNA complex interacting with CMT3 may provide the mechanism that CMT3 distinguishes target DNA sequences.
Second, the BAH domain and chromodomain of CMT3 recognize H3K9me2, leading to CMT3 binding to nucleosomes to methylate target DNA sequences (Du et al., 2012). This indicates that the BAH domain and chromodomain guide CMT3 to specific genomic regions by interacting with specific chromatin modifications. Therefore, examination of H3K9me2 of NIA1 and NIA2 genes in vivo may also provide clues as to why CMT3 selectively methylates the NIA2 gene.
Third, the citrullination of DNMT3A by peptidylarginine deiminase (PADI4) stimulates its DNMT activity (Deplus et al., 2014). Citrullination affects protein structure and function (Lee et al., 2005; Gyorgy et al., 2006; Guo and Fast, 2011). CMT3 can be citrullinated by a plant PADI4 homolog. Therefore, CMT3 citrullination can modulate its ability to interact with partners involved in CMT3 targeting to specific genes and DNA methylation, suggesting that citrullination can help CMT3 discriminate among potential target sequences.
CHG methylation of NIA2 in control wild-type plants but not in ammonium-treated plants (Table 2) indicated that CMT3-mediated CHG methylation of NIA2 was blocked by ammonium treatment, suggesting that ammonium treatment represses CMT3 expression. However, CMT3 transcript levels and methylation levels were not affected by ammonium treatment (Figure 4B). This indicates that repression of CMT3-mediated CHG methylation of NIA2 in response to ammonium treatment may not result from changes in CMT3 levels, but may be caused by changes in CMT3 activity due to post-translational modification. Our previous study showed that the DNMT activity of CMT3 was increased by sumoylation by the E3 SUMO ligase of AtSIZ1 (Kim et al., 2015a). This suggests that ammonium treatment may block AtSIZ1-mediated CMT3 sumoylation or other CMT3 modifications that are required for CMT3 activation.
CHG methylation correlates with H3K9m2 (Bernatavichute et al., 2008; Deleris et al., 2012), and the H3K9m2 histone MET triple mutant kyp/suvh5/suvh6 shows a reduction in CHG methylation similar to that found in the cmt3 mutant (Ebbs and Bender, 2006; Stroud et al., 2013). Some CHG sites also lost methylation in the met1 mutant, indicating that CHG methylation partly depends on CG methylation (Stroud et al., 2013). A lack of CHG methylation correlates with a lack of the H3K9m2 marker, which is maintained by the histone methylase INCREASE IN BONSAI METHYLATION1 (IBM1) (Saze and Kakutani, 2011). These results suggest that the reduction in genome methylation levels in response to ammonium treatment can be affected by other DNA and histone METs. Therefore, further investigation of these mutants is needed to discover how ammonium regulates gene methylation or demethylation, including that of NIA2.
Conclusion
The present study showed that ammonium has no effect on CMT3 methylation and expression, whereas it blocks CMT3-mediated CHG methylation of NIA2, which induces NIA2 expression. Future studies should be aimed at identifying CMT3-interacting factors, including siRNAs and proteins, and at examining the modulation of CMT3 DNMT activity by phytohormones or chemicals including ammonium. This may help elucidate specific mechanisms regulating CMT3-mediated genome methylation during specific growth and developmental stages, as well as during nitrogen assimilation.
Author Contributions
HSS designed the studies. JYK, and DYK performed experiments. JYK, SIK, YJK, JTS, and HSS interpreted data. JYK and HSS wrote the manuscript. All authors commented on the results and the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by a grant from the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center no. PJ01108701), Rural Development Administration, Republic of Korea.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01161
References
- Armache K. J., Garlick J. D., Canzio D., Narlikar G. J., Kingston R. E. (2011). Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science 334 977–982. 10.1126/science.1210915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatavichute Y. V., Zhang X., Cokus S., Pellegrini M., Jacobsen S. E. (2008). Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3:e3156 10.1371/journal.pone.0003156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell W. H., Kinghorn K. R. (1990). Functional domains of assimilatory nitrate reductases and nitrite reductases. Trends Biochem. Sci. 15 315–319. 10.1016/0968-0004(90)90021-3 [DOI] [PubMed] [Google Scholar]
- Cao X., Aufsatz W., Zilberman D., Mette M. F., Huang M. S., Matzke M., et al. (2003). Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr. Biol. 13 2212–2217. 10.1016/j.cub.2003.11.052 [DOI] [PubMed] [Google Scholar]
- Cao X., Jacobsen S. E. (2002a). Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc. Natl. Acad. Sci. U.S.A. 99 16491–16498. 10.1073/pnas.162371599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S. E. (2002b) Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 9 1138–1144. 10.1016/S0960-9822(02)00925-9 [DOI] [PubMed] [Google Scholar]
- Chan S. W., Henderson I. R., Jacobsen S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Genetics 6 351–360. [DOI] [PubMed] [Google Scholar]
- Cokus S. J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C. D., et al. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215–219. 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustham V., Vlad D., Deremetz A., Gy I., Cubillos F. A., Kerdaffrec E., et al. (2014). SHOOT GROWTH1 maintains Arabidopsis epigenomes by regulating IBM1. PLoS ONE 9:e84687 10.1371/journal.pone.0084687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford N. M., Forde B. G. (2002). “Molecular and developmental biology of inorganic nitrogen nutrition,” in The Arabidopsis Book Vol. 1 eds Meyerowitz E., Somerville C. (Rockville, MD: American Society of Plant Biologists Press; ), e0011 10.1199/tab.0011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A., Stroud H., Bernatavichute Y., Johnson E., Klein G., Schubert D., et al. (2012) Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 8:e1003062 10.1371/journal.pgen.1003062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R., Denis H., Putmans P., Calonne E., Fourrez M., Yamamoto K., et al. (2014). Citrullination of DNMT3A by PADI4 regulates its stability and controls DNA methylation. Nucleic Acids Res. 42 8285–8296. 10.1093/nar/gku522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Zhong X., Bernatavichute Y. V., Stroud H., Feng S., Caro E., et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151 167–180. 10.1016/j.cell.2012.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebbs M. L., Bender J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18 1166–1176. 10.1105/tpc.106.041400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan E. J., Kovac K. A. (2000). Plant DNA methyltransferases. Plant Mol. Biol. 43 189–201. 10.1023/A:1006427226972 [DOI] [PubMed] [Google Scholar]
- Forde B. G. (2002). Local and long-range signaling pathways regulating plant responses to nitrate. Annu. Rev. Plant Biol. 53 203–224. 10.1146/annurev.arplant.53.100301.135256 [DOI] [PubMed] [Google Scholar]
- Foyer C. H., Parry M., Noctor G. (2003). Markers and signals associated with nitrogen assimilation in higher plants. J. Exp. Bot. 54 585–593. 10.1093/jxb/erg053 [DOI] [PubMed] [Google Scholar]
- Greenberg M. V., Ausin I., Chan S. W., Cokus S. J., Cuperus J. T., Feng S., et al. (2011). Identification of genes required for de novo DNA methylation in Arabidopsis. Epigenetics 6 344–354. 10.4161/epi.6.3.14242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Fast W. (2011). Citrullination of inhibitor of growth 4 (ING4) by peptidylarginine deminase 4 (PAD4) disrupts the interaction between ING4 and p53. J. Biol. Chem. 286 17069–17078. 10.1074/jbc.M111.230961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyorgy B., Toth E., Tarcsa E., Falus A., Buzas E. I. (2006). Citrullination: a posttranslational modification in health and disease. Int. J. Biochem. Cell Biol. 38 1662–1677. 10.1016/j.biocel.2006.03.008 [DOI] [PubMed] [Google Scholar]
- Henderson I. R., Deleris A., Wong W., Zhong X., Chin H. G., Horwitz G. A., et al. (2010). The de novo cytosine methyltransferase DRM2 requires intact UBA domains and a catalytically mutated paralog DRM3 during RNA-directed DNA methylation in Arabidopsis thaliana. PLoS Genet. 6:e1001182 10.1371/journal.pgen.1001182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Comai L. (1998). A DNA methyltransferase homolog with a chromodomain exists in multiple polymorphic forms in Arabidopsis. Genetics 149 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J. G., Baylin S. B. (2003). Gene silencing in cancer in association with promoter hypermethylation. N. Engl. J. Med. 349 2042–2054. 10.1056/NEJMra023075 [DOI] [PubMed] [Google Scholar]
- Hetzl J., Foerster A. M., Raidl G., Mittelsten Scheid O. (2007). CyMATE : a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 51 526–536. 10.1111/j.1365-313X.2007.03152.x [DOI] [PubMed] [Google Scholar]
- Hou P. Q., Lee Y. I., Hsu K. T., Lin Y. T., Wu W. Z., Lin J. Y., et al. (2013). Functional characterization of Nicotiana benthamiana chromomethylase 3 in developmental programs by virus-induced gene silencing. Physiol. Plant. 150 119–132. 10.1111/ppl.12071 [DOI] [PubMed] [Google Scholar]
- Huang J. J., Wang H. H., Liang W. H., Xie X. J., Guo G. Q. (2014). Developmental expression of Arabidopsis methyltransferase genes MET1, DRM2 and CMT3. Mol. Biol. 48 782–789. 10.1134/S0026893314050057 [DOI] [PubMed] [Google Scholar]
- Huppe H. C., Turpin D. H. (1994). Integration of carbon and nitrogen metabolism in plant and algal cells. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 577–607. 10.1146/annurev.pp.45.060194.003045 [DOI] [Google Scholar]
- Jones L., Ratcliff F., Baulcombe D. C. (2001). RNA-directed transcriptional gene silencing in plants can be inherited independently of the RNA trigger and requires Met1 for maintenance. Curr. Biol. 11 747–757. 10.1016/S0960-9822(01)00226-3 [DOI] [PubMed] [Google Scholar]
- Kato M., Miura A., Bender J., Jacobsen S. E., Kakutani T. (2003). Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr. Biol. 13 421–426. 10.1016/S0960-9822(03)00106-4 [DOI] [PubMed] [Google Scholar]
- Kim D. Y., Han Y. J., Kim S., Song J. T., Seo H. S. (2015a). Arabidopsis CMT3 activity is positively regulated by AtSIZ1-mediatedsumoylation. Plant Sci. 239 209–215. 10.1016/j.plantsci.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Kim S. I., Park B. S., Kim D. Y., Yeu S. Y., Ong S. I., Song J. T., et al. (2015b). E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signaling and plant development. Biochem. J. 469 299–314. 10.1042/BJ20141302 [DOI] [PubMed] [Google Scholar]
- Kuo A. J., Song J., Cheung P., Ishibe-Murakami S., Yamazoe S., Chen J. K., et al. (2012). The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484 115–119. 10.1038/nature10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. H., Coonrod S. A., Kraus W. L., Jelinek M. A., Stallcup M. R. (2005). Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U.S.A. 102 3611–3616. 10.1073/pnas.0407159102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. T., Wei H. M., Lu H. Y., Lee Y. I., Fu S. F. (2015). Developmental– and tissue-specific expression of NbCMT3-2 encoding a chromomethylase in Nicotiana benthamiana. Plant Cell Physiol. 56 1124–1143. 10.1093/pcp/pcv036 [DOI] [PubMed] [Google Scholar]
- Lindroth A. M., Cao X., Jackson J. P., Zilberman D., McCallum C. M., Henikoff S., et al. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080. 10.1126/science.1059745 [DOI] [PubMed] [Google Scholar]
- Lippman Z., May B., Yordan C., Singer T., Martienssen R. (2003). Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 1:e67 10.1371/journal.pbio.0000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M. A., Mosher R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15 394–408. 10.1038/nrg3683 [DOI] [PubMed] [Google Scholar]
- McCallum C. M., Comai L., Greene E. A., Henikoff S. (2000). Targeted screening for induced mutations. Nat. Biotechnol. 18 455–457. 10.1038/74542 [DOI] [PubMed] [Google Scholar]
- Papa C. M., Springer N. M., Muszynski M. G., Meeley R., Kaeppler S. M. (2001). Maize chromomethylase Zea Methyltransferase2 is required for CpNpG methylation. Plant Cell 13 1919–1928. 10.2307/3871328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. S., Song J. T., Seo H. S. (2011). Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat. Commun. 2 400 10.1038/ncomms1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillot M., Baroux C., Vazquez M. A., Autran D., Leblanc O., Vielle-Calzada J. P., et al. (2010). Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22 307–320. 10.1105/tpc.109.071647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning K. R., Smith K. M., Freitag M. (2009). Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods 47 142–150. 10.1016/j.ymeth.2008.09.022 [DOI] [PubMed] [Google Scholar]
- Ronemus M. J., Galbiati M., Ticknor C., Chen J., Dellaporta S. L. (1996). Demethylation-induced developmental pleiotropy in Arabidopsis. Science 273 654–657. 10.1126/science.273.5275.654 [DOI] [PubMed] [Google Scholar]
- Rose T. M., Schultz E. R., Henikoff J. G., Pietrokovski S., McCallum C. M., Henikoff S. (1998). Consensus-degenerate hybrid oligonucleotide primers for amplification of distantly related sequences. Nucleic Acids Res. 26 1628–1635. 10.1093/nar/26.7.1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R., Zhou M. M. (2009). The role of human bromodomains in chromatin biology and gene transcription. Curr. Opin. Drug Discov. Devel. 12 659–665. [PMC free article] [PubMed] [Google Scholar]
- Saze H., Kakutani T. (2011). Differentiation of epigenetic modifications between transposons and genes. Curr. Opin. Plant Biol. 14 81–87. 10.1016/j.pbi.2010.08.017 [DOI] [PubMed] [Google Scholar]
- Shemer O., Landau U., Candela H., Zemach A., Eshed Williams L. (2015). Competency for shoot regeneration from Arabidopsis root explants is regulated by DNA methylation. Plant Sci. 238 251–261. 10.1016/j.plantsci.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Sidler C., Li D., Kovalchuk O., Kovalchuk I. (2015). Development-dependent expression of DNA repair genes and epigenetic regulators in Arabidopsis plants exposed to ionizing radiation. Radiat. Res. 183 219–232. 10.1667/RR13840.1 [DOI] [PubMed] [Google Scholar]
- Solomonson L. P., Barber M. J. (1990). Assimilatory nitrate reductase-functional properties and regulation. Annu. Rev. Plant Physiol. Plant Mol. Biol. 41 225–253. 10.1146/annurev.pp.41.060190.001301 [DOI] [Google Scholar]
- Stitt M., Müller C., Matt P., Gibon Y., Carillo P., Morcuende R., et al. (2002). Steps towards an integrated view of nitrogen metabolism. J. Exp. Bot. 53 959–970. 10.1093/jexbot/53.370.959 [DOI] [PubMed] [Google Scholar]
- Stroud H., Greenberg M. V., Feng S., Bernatavichute Y. V., Jacobsen S. E. (2013). Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152 352–364. 10.1016/j.cell.2012.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W., Custard K. D., Brown R. C., Lemmon B. E., Harada J. J., Goldberg R. B., et al. (2006). DNA methylation is critical for Arabidopsis embryogenesis and seed viability. Plant Cell 18 805–814. 10.1105/tpc.105.038836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N., Xu R. M. (2013). Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Crit. Rev. Biochem. Mol. Biol. 48 211–221. 10.1021/bi101885m [DOI] [PubMed] [Google Scholar]
- Yap K. L., Zhou M. M. (2012). Structure and mechanisms of lysine methylation recognition by the chromodomain in gene transcription. Biochemistry 50 1966–1980. 10.1021/bi101885m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., Kim M. Y., Hsieh P. H., Coleman-Derr D., Eshed-Williams L., Thao K., et al. (2013). The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153 193–205. 10.1016/j.cell.2013.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Jacobsen S. E. (2006). Genetic analyses of DNA methyltransferase in Arabidopsis thaliana. Cold Spring Harb. Symp. Quant. Biol. 71 439–447. 10.1101/sqb.2006.71.047 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S. W.-L., Chen H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201. 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Zhong X., Du J., Hale C. J., Gallego-Bartolome J., Feng S., Vashisht A. A., et al. (2014). Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157 1050–1060. 10.1016/j.cell.2014.03.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.