Abstract
Neuroblastoma (NB) is the most common extracranial childhood tumor classified in five stages (1, 2, 3, 4 and 4S), two of which (3 and 4) identify chemotherapy-resistant, highly aggressive disease. High-risk NB frequently displays MYCN amplification, mutations in ALK and ATRX, and genomic rearrangements in TERT genes. These NB subtypes are also characterized by reduced susceptibility to programmed cell death induced by chemotherapeutic drugs. The latter feature is a major cause of failure in the treatment of advanced NB patients. Thus, proper reactivation of apoptosis or of other types of programmed cell death pathways in response to treatment is relevant for the clinical management of aggressive forms of NB. In this short review, we will discuss the most relevant genomic rearrangements that define high-risk NB and the role that destabilization of p53 and p73 can have in NB aggressiveness. In addition, we will propose a strategy to stabilize p53 and p73 by using specific inhibitors of their ubiquitin-dependent degradation. Finally, we will introduce necroptosis as an alternative strategy to kill NB cells and increase tumor immunogenicity.
Facts
High-risk NB is resistant to conventional pro-apoptotic therapies.
MYCN amplification, mutations in ALK and ATRX, and genomic rearrangements in TERT genes are frequent in high-risk NB.
Although not mutated, p53 and p73 are destabilized in NB.
Caspase 8 is often compromised in advanced NB stages.
Necroptosis is an alternative modality of programmed cell death.
Open Questions
Are there ongoing clinical trials that exploit specific apoptosis and/or necroptosis defects in NB?
Is stabilization of p53 and p73 a potentially exploitable way to induce apoptosis/differentiation in NB?
Is activation of necroptosis an alternative to kill NB cells and to increase their immunogenicity?
In neuroblastoma (NB), several genomic abnormalities have been described and the causative genes of the disease have been searched for.1, 2, 3 Some genomic defects such as deletions on chromosomes 1p and 11q or gains on 17q2, 3 have been utilized as prognostic markers although the contributing gene(s) whose alteration is responsible for the resulting phenotype, are still unknown. One of the first and doubtlessly most important genetic signature of NB is the amplification of the proto-oncogene MYCN.4, 5, 6 Amplification leading to aberrant expression of MYCN has been associated with tumor aggressiveness,7 resistance to chemotherapy1 and inability to differentiate.8 NB patients who carry MYCN amplification are classified in the high-risk group and their overall survival does not exceed 50% at 5 years from diagnosis.9 Nevertheless, there is a significant number of NB patients with poor prognosis whose DNA does not harbor MYCN amplification.1 The latter observation implies that MYCN is not the only culprit of NB aggressiveness. More recently, activating mutations of ALK were reported in both familial and sporadic cases of neuroblastoma.10, 11, 12, 13 In familiar NB, germline mutations in ALK gene have been found in ~50% of the cases.13 In addition, some sporadic NB acquire somatic mutations of ALK and ~2% display genomic amplification of the gene as reviewed in (ref. 14). ALK is a member of the insulin receptor (IR) superfamily of receptor tyrosine kinases, which shows homology with the leukocyte tyrosine kinase, the insulin-like growth factor-1 receptor kinase and the IR kinase.14 In humans, ALK is located on chromosome 2p23 and the gene encodes for a single-chain transmembrane protein.14 The mutated/amplified full-length ALK leads to cell growth and survival by the activation of the JAK–STAT, PI3K–AKT or RAS–MAPK pathways. In NB, the constitutively activated ALK is complexed with hyperphosphorylated ShcC,15 deregulating the MAPK pathway response to growth factors.16 Another relevant genetic feature in neuroblastoma is the loss-of-function mutations or deletions of the RNA-helicase ATRX.17, 18 In a study of 240 NB cases using a combination of whole-exome, genome and transcriptome sequencing Pugh et al.19 observed putative loss-of-function ATRX alterations in 9.6% of cases (6 mutations and 17 multi-exon deletions). This study confirmed that alterations of ATRX and MYCN were mutually exclusive and that ATRX alterations were enriched in older children.17
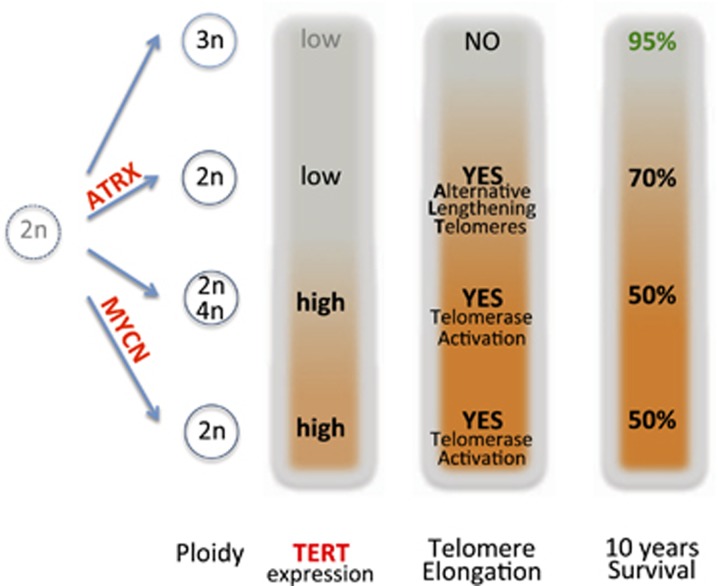
A real breakthrough in the search for genomic alterations that impact on NB aggressiveness comes from the recent observation of telomerase reverse transcriptase (TERT) activation by genetic rearrangements in high-risk NB.20 By whole-genomic sequencing of 59 NB cases the authors discovered recurrent genetic rearrangements in the chromosomal region 5p15.33 proximal of TERT. Rearrangements of this region took place only in high-risk NB (12 out of 39=31%). TERT rearrangements, ATRX mutations and MYCN amplifications occurred in a mutually exclusive manner within the high-risk group. The latter observation implies that all these alterations converge on similar effector functions. Of interest, in MYCN-amplified tumors without TERT rearrangements the expression of TERT was nevertheless increased compared with low-risk NB owing to the known function of MYCN as transcriptional activator of TERT.21 The expression of TERT was greatly increased in TERT-rearranged NBs compared with the low-risk group. Indeed, rearrangements juxtapose TERT to strong enhancers resulting in a complete epigenetic remodeling of the regulatory region without changes in the gene copy number. The whole-genomic sequencing analysis highlighted also that ATRX mutations, which define another high-risk subgroup, occur only in MYCN-non-amplified and TERT-normal NB, and are associated with alternative lengthening of telomeres (ALT) activity.20 This observation suggests that telomere lengthening is a common trait of high-risk NB (i.e., TERT-rearranged, MYCN-amplified and ATRX-mutated tumors) regardless of the mechanism that is utilized for telomere maintenance. Indeed, the most aggressive NB subtypes are characterized by telomerase activation that can derive from TERT rearrangement or MYCN amplification (which activates TERT). In light of these recent findings, we schematize in Figure 1 the different risk subgroups of NB and the genetic aberrations that define each subgroup. A question that arises from the genomic aberrations studies is as follows: which are the pathways and the genes that, following TERT rearrangements or MYCN amplification, become the executioners of the NB aggressiveness? p53, although rarely mutated in NB, is destabilized in MYCN-amplified tumors by the high expression of its ubiquitin ligase MDM2.22 Furthermore, p53 pathway is often deranged in NB cells that lack MYCN amplification but display telomere lengthening activity.23 In the next paragraph we will discuss the role of p53 family and the detrimental effect(s) that its alteration may cause in NB.
Figure 1.
NB-risk subgroups (low and high) inferred from ploidy, ATRX mutations, MYCN amplifications, TERT activation (by genomic rearrangements) and alternative lengthening of telomeres activation
The p53 family includes three genes (p53, p63 and p73) that have a variety of roles in normal and in transformed cells.24, 25, 26 In Table 1 the prominent cellular pathways and principal regulatory circuits that involve p53 family are reported. Similarly to p63, p73 is expressed as several distinct protein isoforms.27, 28 In more detail, the usage of two alternative promoters results in the expression of two different N-terminal isoforms: the transcriptionally active p73 (TAp73) proteins, containing a complete N-terminal transactivation domain (TAD), and N-terminally truncated (ΔNp73) isoforms, which lack the TAD and might act as dominant negative molecules by inhibiting the transactivating activity of TAp73 and p53.25 Many lines of evidence have clearly demonstrated that TAp73 and ΔNp73 control several biological processes in opposite manner.29, 30 Although TAp73 is an inducer of cell cycle arrest and apoptosis, and largely mimics the tumor suppressive activities of p53,31, 32 ΔNp73 isoforms promote cancer cell survival and exhibit oncogenic properties.29 The phenotypical characterization of selectively deficient mouse models for the N-terminal p73 isoforms confirmed the role of TAp73 and ΔNp73 as tumor suppressor and pro-oncogenic factors, respectively.33, 34
Table 1. Prominent pathways and main regulatory circuits that involve p53 family.
| Pathway/regulatory circuit | References |
|---|---|
| Apoptosis | 88, 89, 90, 91, 92, 93, 94 |
| Cell growth control | 90, 95, 96, 97, 98 |
| RNA metabolism | 99, 100, 101, 102, 103, 104, 105, 106, 107, 108, 109, 110, 111, 112, 113, 114 |
| Protein degradation/stability | 67, 69, 115, 116, 117, 118, 119, 120, 121, 122, 123 |
| Autophagy | 93, 124, 125, 126, 127, 128, 129, 130, 131 |
| Splicing events | 111, 132, 133, 134, 135, 136 |
| ROS and cell metabolism | 92, 137, 138, 139, 140, 141, 142, 143, 144, 145, 146, 147 |
| Chemotherapeutic response | 56, 57, 148, 149, 150, 151, 152, 153, 154 |
| DNA damage response | 155, 156, 157, 158, 159, 160, 161, 162, 163, 164, 165, 166, 167, 168 |
| Transcription and translation | 169, 170, 171, 172, 173, 174, 175, 176, 177, 178 |
| Stemness and lineage determination | 94, 109, 179, 180, 181, 182, 183, 184, 185, 186 |
Besides their role in controlling tumor growth, p73 isoforms also contribute to the development and differentiation of neuronal tissue. TAp73 null mice show hippocampal dysgenesis with reduction of the neurogenesis in the subgranular zone of the dentate gyrus,33 while ΔNp73−/− mice show evidence of neurodegeneration, confirming thus the pro-survival role of this isoforms.34 All together, these data indicate that TAp73 and ΔNp73 are important transcription factors whose dysregulation might be an important determinant in tumorigenesis as well as in neuronal development.
p73 and NB
Alteration of the 1p chromosomal region is commonly observed in NB and the smallest region of overlapping deletions in this region has been refined within 1p36.3.35 As p73 maps at 1p36,28 it was originally hypothesized that this gene might act as tumor suppressor gene in NB. However, p73 is rarely mutated in primary NB and it is unlikely that it may function as a tumor suppressor in a classic Knudson's manner. Nevertheless, several data indicated that the altered p73 expression rather than its mutation is a determinant factor in the pathogenesis of the NB. The contribution of p73 to NB is indeed thought to depend on the TAp73 to ΔNp73 isoforms' ratio and different molecular mechanisms accounting for altered ΔN : TAp73 expression have been described in NB. Epigenetic modifications, particularly by hyper- or hypo-methylation, are crucial events in cell transformation.29 As several human malignancies, such as leukemia and Burkitt's and non-Hodgkin lymphomas, display Trp73 silencing by promoter methylation,36, 37 it has been postulated that this type of epigenetic modification could account for the decrease of the expression of the TAp73 isoform observed in NB. However, the analysis of the TAp73 promoter methylation in association with its expression level does not support the idea that the p73 gene is subjected to genome imprinting in NB.38 The idea that the TAp73 activity is associated with NB development is also supported by the role of TAp73 during the neuronal differentiation.39 Indeed, one therapeutic approach aimed to restrain NB growth is based on the pro-differentiation action of the retinoic acid.40 It has been shown that the expression of the TAp73 isoform is increased during the retinoid-driven NB differentiation and its depletion inhibits differentiation, suggesting that the TAp73 activity is functionally associated with the growth inhibition occurring during the NB differentiation.39
In contrast to TAp73, high levels of expression of ΔNp73 have been reported in primary NB.41 The increased levels of ΔNp73 observed in NB might functionally inhibit the pro-apoptotic activity of wild-type p53,42 and/or physically block the activity of TAp73 allowing the NB to escape from TAp73-driven differentiation program.39 In addition, ΔNp73 could inhibit the full activation of ATM and p53, allowing NB to be more resistant to the chemotherapic agents.34 Mechanistically, the increased levels of ΔNp73 is likely due to the epigenetic modifications as hypo-methylation of the internal P2 promoter that controls the transcription of this isoform has been observed in NB cell lines and primary tumors.38, 43
N-MYC/MDM2/p53/p73 Axis in NB
As described in the first paragraph, MYCN oncogene amplification is one of the most important biological marker of aggressive NB and it occurs in about 20% of primary tumors.44 MYCN amplification contributes to the NB development and progression by influencing many biological processes, such as cell invasion and motility, cell cycle, immune surveillance, self-renewal and apoptosis.45 TP53 mutations are rare in NB at diagnosis,46 and amplification of MYCN contributes to maintain under surveillance the p53 activity, thought to be its role in the MDM2–p53 pathway.47, 48 MDM2 is an E3 ubiquitin ligase that promotes survival by ubiquitinating and driving the degradation of p53. Several tumors, especially those expressing wild-type p53 like NB, are characterized by increased levels of MDM2 expression due to several mechanisms, such as amplification of its locus, increased transcription or increased mRNA or protein stability.49 In NB cells it has been shown that MYCN can regulate the MDM2/p53 axis by directly promoting the transcription of MDM2 thus stimulating the ubiquitin-mediated degradation of p53.22 Besides p53, MDM2 can also physically interact with TAp73 and as the affinities of MDM2 for p73 are of the same order of magnitude as those for p53, it is likely that these proteins interact in cells, as has been suggested in several studies.50, 51 However, MDM2 does not trigger TAp73 proteasome-dependent degradation but rather negatively controls the transcriptional activity of TAp73.52, 53 Therefore, by increasing the levels of MDM2, MYCN might not only stimulate p53 protein degradation but also inhibits the TAp73 transcriptional activity, enhancing thus the NB survival and chemo-resistance. It is also worth noting that some data, although controversial, suggest that MYCN can directly affect TAp73 expression levels. MYCN is indeed able to repress the transcription of TAp73 and the reduced expression of p73 correlated with the MYCN overexpression in a statistically significant manner in NB primary tumors.54 On the other hand, the overexpression of TAp73 is also able to reduce MYCN expression and thus facilitate the neuronal differentiation program, suggesting an antagonistic role of these two transcription factors on NB cell proliferation and differentiation.39, 55 Recently, it has been shown that TAp73 loss determines an increase of the vascularization of lung tumors, suggesting that TAp73 might act as a tumor suppressor by, at least in part, inhibiting tumor angiogenesis. At molecular level, TAp73 stimulates the degradation of the hypoxia-inducible factor-1 alpha (HIF-1α) in an oxygen-independent manner.56, 57 Interestingly, recent data suggest that ΔNp73 is also involved in tumor angiogenesis. Indeed, upon hypoxia ΔNp73 is stabilized and capable of inducing the expression of VEGF-A, the prototypic angiogenic gene.58 Similarly to ΔNp73, ΔNp63 is also able to increase the vascular endothelial growth factor (VEGF) secretion by leading to the stabilization of the HIF-1α protein.59 Therefore, these data suggest a cross talk between the p53 family members and the tumor angiogenesis pathways, potentially involved in the regulation of NB vascularization. Of interest, several data indicated that MYCN is functionally linked with tumor angiogenesis. Indeed, aberrant expression of MYCN had a positive effect on pro-angiogenic factors, including angiogenin and VEGF, and MYCN amplification correlates with poor survival, increased dissemination and high vascularization in NB.45 In this scenario, MYCN amplification might stimulate tumor vascularization and dissemination by also inhibiting the anti-angiogenic activity of TAp73 either directly or via MDM2.
Itch as a Potential Therapeutical Target in NB
E3 ubiquitin ligases (E3s) have been shown to have a critical role in regulating cell proliferation, differentiation or apoptosis.60, 61 For this reason, the ubiquitin system is often the target of cancer-related deregulation and is critically involved in processes such as oncogenic transformation and tumor progression. Genetic alterations, abnormal expression or dysfunction of E3s is often accompanied by the occurrence of cancer. The HECT-type E3 ubiquitin ligase Itch regulates several important biological processes, such as apoptosis, cell growth and inflammation, and several reports have demonstrated that dysregulation of Itch expression affects the apoptotic response induced by the chemotherapeutic drugs.60, 61, 62 Itch depletion by siRNA indeed increases the cytotoxic effect of anti-neoplastic drugs in cancer cell lines and in cancer stem cells.63 Furthermore, the in vivo administration of siRNA duplex targeting Itch mRNA is effective in sensitizing pancreatic cancer to gemcitabine.64 Itch exerts its biological functions mainly by controlling the proteasomal-dependent degradation of a subset of target proteins, including p73. Indeed, among several E3s controlling TAp73 protein levels,65, 66, 67 Itch is the most characterized. In detail, in unstressed cells Itch stimulates the proteasome-dependent degradation of TAp73, thereby keeping its expression levels low under normal conditions.68 In several tumor cell lines, the induction of TAp73 in response to chemotherapeutic drugs is, at least partially, accomplished through Itch downregulation. We found, in a preliminary analysis, that Itch is expressed in the majority of NB cells tested so far (data not shown). Thus, it is reasonable to hypothesize that in NB cells an Itch-dependent mechanism for negatively controlling TAp73 protein levels might occur and contribute to the chemo-resistance. Thus, targeting Itch ubiquitin ligase activity could be a feasible strategy to stabilize TAp73, enhance its pro-apoptotic activity and sensitize NB cells to the cytotoxic effects of commonly used anti-neoplastic agents. Recently, our laboratory has identified desmethyl-clomipramine (DCMI), the active metabolite of clomipramine, as inhibitor of the Itch autoubiquitylation activity and Itch-dependent ubiquitylation of p73.69 Clomipramine is an FDA-approved drug clinically used for the treatment of obsessive compulsive disorders.70 Of interest, DCMI increases the cytotoxic activity of conventional chemotherapic drugs in several cancer cell lines as well as in cancer stem cells.63, 69 Although it is still not clear whether the DCMI-mediated effect on cancer cell survival completely depends on Itch inhibition, DCMI represents the proof of principle that targeting the E3 ubiquitin ligase Itch might be a novel therapeutical approach to decrease NB cell survival and/or increase the pro-apoptotic effects of conventional anti-neoplastic agents.
Necroptosis: A Different Modality of Programmed Cell Death
Besides classical caspase-dependent apoptosis, other forms of programmed cell death exist in normal and in transformed cells, which can be activated in response to cellular stress. Necroptosis is a type of necrosis mediated by death receptors (DRs; i.e., Fas, TNFR1/2, TRAIL-R1/2, DR3 and DR6) and their ligands including CD95L (also known as FASL), TNF and TNF-related apoptosis-inducing ligand (TRAIL; also known as TNFSF10), interferons, toll-like receptors, intracellular RNA and DNA sensors, and probably other mediators.71, 72 Indeed, another receptor, the transforming growth factor-β-activated kinase 1 (TAK1), which is activated through a diverse set of intra- and extracellular stimuli, has been recently added to the list of necroptosis-inducing receptors.73
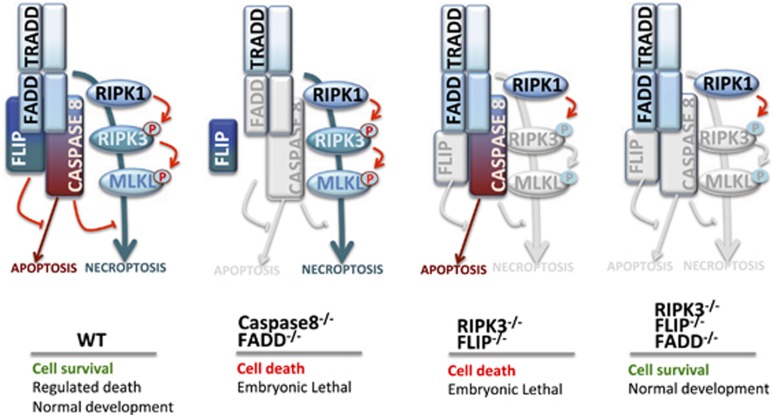
Seminal work of the laboratory of Jurg Tschopp has defined the role of the first characterized executioner of necroptosis, the receptor-interacting protein kinase 1 (RIPK1).74 This original discovery was followed by those of two other essential components of the process, RIPK375, 76, 77 and more recently MLKL.78 Necroptosis occurs in the absence of caspase activity and is regulated by the activity of a multi-protein complex called necrosome consisting of RIPK1, RIPK3 and MLKL.72, 79 In unstressed, normal conditions, FLIP (in multi-protein complex IIb) inhibits caspase 8 activity, preventing thus apoptosis. At the same time, caspase 8 (in multi-protein complex IIa) prevents the activation of RIPK1 blocking the necroptotic pathway.71 The net result of this cross-regulation is survival. Defects in this regulatory circuitry can lead to necroptosis (as in the case of the double knockout of FADD and caspase 8) or to apoptosis (in RIPK3/FLIP double knockout). Of interest, triple knockout of RIPK3, FLIP and FADD rescue the normal phenotype (cell survival). A graphic representation of normal and altered conditions in the apoptosis/necroptosis pathways is reported in Figure 2.
Figure 2.
Apoptotic and necroptotic circuitries in wild-type and knockout settings
When the necroptotic pathway is unleashed by the engagement of a death receptor, the initial activation of RIPK1 leads to that of RIPK3 by phosphorylation resulting in the recruitment and phosphorylation of MLKL, which causes a conformational change in the pseudokinase domain leading to the exposure of the four-helical bundle domain. Trimerization and movement of active MLKL to the plasma membrane initiates the final step of necroptosis, which terminates with cell rupture and dispersal of the cellular content in the interstitial space.71 Trimerization of MLKL requires both RIPK1 and RIPK3 because treating the cells with Necrostatin 1 (Nec-1), a RIPK1 inhibitor, or knocking down RIPK3 prevented the trimerization.80
Necroptosis in Inflammation and Cancer
Inflammation is a main pathologic condition in which necroptosis has an active role. Indeed, the necroptotic process causes a massive release of the so-called damage-associated molecular patterns (DAMPs) from the disintegrating cells. Some DAMP components are active promoters of the inflammatory process that exacerbate inflammation already in place.81 In sepsis, a life-threatening condition in which inflammation is a constant feature, necroptosis is associated with increased mortality during TNF-induced systemic inflammatory response syndrome.82 The detrimental effect of necroptosis in sepsis is blocked by the presence of caspase 8, which promotes RIPK1 and/or RIPK3 cleavage and inhibits necroptosis.83 On the contrary, in cancer therapy, exploitation of necroptotic cell death may open novel avenues for the treatment of apoptosis-resistant tumors. Cancer cells are known to shift from classical apoptosis to other forms of cell death such as autophagy, pyroptosis and necroptosis, some of which entail immunogenicity after anticancer treatments.84 It is also well recognized that therapy-resistant cancer stem cells (CSCs) have a higher antiapoptotic activity than that of their counterparts.85 Therefore, it would be extremely useful to exploit necroptosis induction in cancer cells for CSC-directed therapeutic application but also the resultant immunogenicity to modulate antitumor immunity.84 The latter observation is extremely important in light of the recent advances and applications of immunotherapy in cancer.86
Necroptosis Induction in NB: A Route to Novel Therapies?
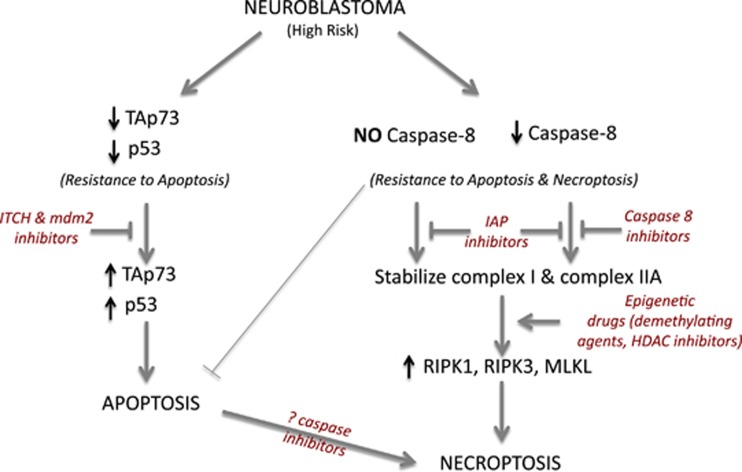
In NB the pro-apoptotic activity of caspase 8 is often compromised in advanced stages,87 nevertheless these tumors show a marked resistance to death induced by drugs that should trigger necroptosis in a context of caspase 8 deficiency. A scheme pointing out the possible points of deficiency in the necroptotic pathway in NB is depicted in Figure 3. Extensive experimental evidence is not available on the proficiency of RIPK1, RIPK3 and other necrosome components in NB. However, preliminary results from our laboratory demonstrated that caspase 8 and necroptosis-associated genes (RIPK1 and RIPK3) are expressed at significantly lower levels in NB cells compared with other tumor cell lines used as controls. Furthermore, in vitro tests suggest that several NB cell lines are resistant to necroptosis (SN and MP, unpublished results). As mutations in necroptotic genes have not been described in NB, epigenetic silencing could occur by hypermethylation of the CpG islands located in the regulatory regions of the necroptotic genes and/or by chromatin modifications. Detection of abnormalities in the activity and/or expression of different members of the necroptotic machinery may represent novel useful markers to better define NB aggressiveness and to predict its response to therapy. More importantly, reactivation of the normal function of the necroptotic pathway (e.g., by demethylating drugs and/or HDAC inhibitors) can be a strategy to rescue cell death ability in chemotherapy-resistant NB tumors defective for caspase 8. In Figure 4 are schematized our proposed approaches based on p73/p53-dependent apoptosis and on necroptosis activation with reference to the potential benefits for specific groups of NB patients.
Figure 3.
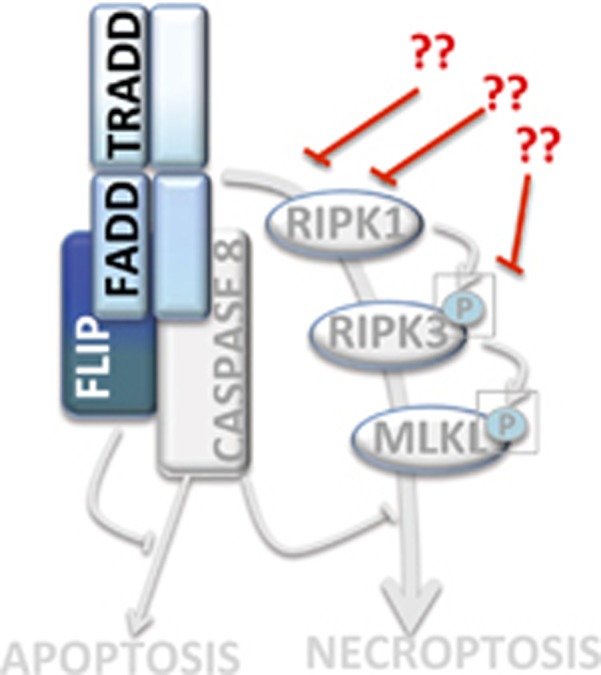
Apoptosis and necroptosis defects in NB. Caspase 8 is often defective in high-risk NB, or its pro-apoptotic activity can be blocked by FLIP. The potential points at which the necroptotic circuitry is interrupted are shown in red
Figure 4.
Proposed approaches (in red) to activate apoptotic or necroptotic response in specific subgroups of high-risk NB
Concluding Remarks
NB has been a model for geneticists and molecular biologists who classified genetic abnormalities and identified causative genes of the disease.7, 13, 17, 20 However, despite intensive research, improvements in clinical outcome of NB have been achieved mostly for low-/intermediate-risk tumors.3 Indeed, metastatic NB remains a difficult-to-treat cancer that has benefited relatively little of research advancements. A survey of the ongoing clinical trials (https://clinicaltrials.gov) highlights the coexistence of trials aimed at optimizing existing therapeutic schedules and those that utilize biological/targeted drugs alone or in combination with well-characterized chemotherapeutic drugs. A selection of current clinical trials is reported in Table 2. Few attempts are underway to exploit specific defects in apoptosis and necroptosis of NB cells. In this sense, our proposal outlined in the previous paragraphs, although not yet mature for a therapeutic application, is aimed at steering preclinical and clinical research toward the exploitation of specific pro-apoptotic and pro-necroptotic targets in NB cells minimizing harmful effects in the patients. As a further clue of the importance of genomic variations in NB, Oldridge et al.187 have recently reported genomic predisposition to NB mediated by a SNP in a super-enhancer region of the LMO1 gene.
Table 2. Current selected clinical trials on NB.
| Clinical trials | Interventions | URL |
|---|---|---|
| 124I-Metaiodobenzylguanidine (MIBG) PET/CT Diagnostic Imaging and Dosimetry for Patients With Neuroblastoma: A Pilot Study | Radiation: 124I-MIBG (no-carrier added) Radiation: 124I-MIBG (carrier added) | https://ClinicalTrials.gov/show/NCT01583842 |
| European Low and Intermediate Risk Neuroblastoma Protocol (low and intermediate pediatric NB and neonatal suprarenal masses) | Drug: chemotherapy | https://ClinicalTrials.gov/show/NCT01728155 |
| Phase II Study of Proton Radiation Therapy for Neuroblastoma | Radiation: proton beam radiation therapy | https://ClinicalTrials.gov/show/NCT02112617 |
| Immunomonitoring of Children With Neuroblastoma | Immunological analyses | https://ClinicalTrials.gov/show/NCT01295762 |
| Bivalent Vaccine With Escalating Doses of the Immunological Adjuvant OPT-821, in Combination With Oral β-glucan for High-Risk Neuroblastoma | Biological: adjuvant OPT-821 in a vaccine containing two antigens (GD2L and GD3L) covalently linked to KLH | https://ClinicalTrials.gov/show/NCT00911560 |
| Biomarkers in Tumor Tissue Samples From Patients With Newly Diagnosed Neuroblastoma or Ganglioneuroblastoma | Laboratory biomarker analysis; cytology specimen collection procedure | https://ClinicalTrials.gov/show/NCT00904241 |
| Multimodal Molecular Targeted Therapy to Treat Relapsed or Refractory High-risk Neuroblastoma | Drug: dasatinib Drug: rapamycin Drug: irinotecan Drug: temozolomide Drug: irinotecan Drug: temozolomide | https://ClinicalTrials.gov/show/NCT01467986 |
| Study of DNA in Blood Samples From Patients With Neuroblastoma | Laboratory biomarker analysis Genetic: polymerase chain reaction Genetic: polyacrylamide gel electrophoresis Genetic: DNA analysis | https://ClinicalTrials.gov/show/NCT00898391 |
| Monitor Response to Treatment in Neuroblastoma Using 3&Apos;-Deoxy-3&Apos;-Fluorothymidine-Positron Emission Tomography (FLT-PET) | Device: FLT-PET | https://ClinicalTrials.gov/show/NCT01308905 |
| Expanded Access Study of Fenretinide Lym-X-Sorb Plus Ketoconazole in Neuroblastoma | Drug: fenretinide Lym-X-Sorb oral powder Drug: ketoconazole | https://ClinicalTrials.gov/show/NCT02075177 |
| Activated T Cells Armed With GD2 Bispecific Antibody in Children and Young Adults with Neuroblastoma and Osteosarcoma | Biological: IL-2 Biological: GD2Bi-aATC Biological: GM-CSF Other: laboratory evaluations of immune responses | https://ClinicalTrials.gov/show/NCT02100930 |
| Anti-GD2 3F8 Monoclonal Antibody and GM-CSF for High-Risk Neuroblastoma | Biological: anti-GD2 3F8 monoclonal antibody Drug: GM-CSF (granulocyte-macrophage colony-stimulating factor) Drug: oral isotretinoin | https://ClinicalTrials.gov/show/NCT02100930 |
| Fenretinide Lym-X-Sorb+Ketoconazole+Vincristine for Recurrent or Resistant Neuroblastoma | Drug: fenretinide/LXS oral powder Drug: ketoconazole Drug: vincristine | https://ClinicalTrials.gov/show/NCT02163356 |
| Pilot Study of Activated T-Cell Therapy for Refractory/Relapsed Neuroblastoma | Biological: activated T lymphocyte | https://ClinicalTrials.gov/show/NCT01802138 |
| 3rd Generation GD-2 Chimeric Antigen Receptor and iCaspase Suicide Safety Switch, Neuroblastoma, GRAIN | Genetic: iC9-GD2 T-cell lymphocytes – frozen cells Genetic: iC9-GD2 T-cell lymphocytes – fresh cells Drug: cyclophosphamide Drug: fludarabine Drug: pembrolizumab | https://ClinicalTrials.gov/show/NCT01822652 |
All trials above are recruiting and no results are available yet. From www.clinicaltrials.gov
Acknowledgments
This work was supported by AIRC IG grant (2014-IG15653), AIRC 5xmille grant (2010-MCO #9979) and Fondazione Roma NCDs grant awarded to GM, and by MFAG AIRC 15523 grant awarded to AP. SN was sponsored by a post-doctoral fellowship from AIRC 5xmille grant.
Glossary
- NB
Neuroblastoma
- TERT
telomerase reverse transcriptase
- DAMPs
damage-associated molecular patterns
The authors declare no conflict of interest.
Footnotes
Edited by RA Knight
References
- 1Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3: 203–216. [DOI] [PubMed] [Google Scholar]
- 2Coco S, Theissen J, Scaruffi P, Stigliani S, Moretti S, Oberthuer A et al. Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer 2012; 131: 1591–1600. [DOI] [PubMed] [Google Scholar]
- 3Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet 2007; 369: 2106–2120. [DOI] [PubMed] [Google Scholar]
- 4Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 1983; 305: 245–248. [DOI] [PubMed] [Google Scholar]
- 5Schwab M, Varmus HE, Bishop JM, Grzeschik KH, Naylor SL, Sakaguchi AY et al. Chromosome localization in normal human cells and neuroblastomas of a gene related to c-myc. Nature 1984; 308: 288–291. [DOI] [PubMed] [Google Scholar]
- 6Schwab M, Varmus HE, Bishop JM. Human N-myc gene contributes to neoplastic transformation of mammalian cells in culture. Nature 1985; 316: 160–162. [DOI] [PubMed] [Google Scholar]
- 7Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984; 224: 1121–1124. [DOI] [PubMed] [Google Scholar]
- 8Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature 1985; 313: 404–406. [DOI] [PubMed] [Google Scholar]
- 9Suita S, Tajiri T, Kaneko M, Hirai M, Mugishima H, Sugimoto T et al. Implications of MYCN amplification in patients with stage 4 neuroblastoma who undergo intensive chemotherapy. J Pediatr Surg 2007; 42: 489–493. [DOI] [PubMed] [Google Scholar]
- 10Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 2008; 455: 971–974. [DOI] [PubMed] [Google Scholar]
- 11George RE, Sanda T, Hanna M, Frohling S, Luther W, Zhang J et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 2008; 455: 975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Janoueix-Lerosey I, Lequin D, Brugieres L, Ribeiro A, de Pontual L, Combaret V et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastomal. Nature 2008; 455: 967–970. [DOI] [PubMed] [Google Scholar]
- 13Mosse YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 2008; 455: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Azarova AM, Gautam G, George RE. Emerging importance of ALK in neuroblastoma. Semin Cancer Biol 2011; 21: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Miyake I, Hakomori Y, Shinohara A, Gamou T, Saito M, Iwamatsu A et al. Activation of anaplastic lymphoma kinase is responsible for hyperphosphorylation of ShcC in neuroblastoma cell lines. Oncogene 2002; 21: 5823–5834. [DOI] [PubMed] [Google Scholar]
- 16Osajima-Hakomori Y, Miyake I, Ohira M, Nakagawara A, Nakagawa A, Sakai R. Biological role of anaplastic lymphoma kinase in neuroblastoma. Am J Pathol 2005; 167: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Cheung NK, Zhang J, Lu C, Parker M, Bahrami A, Tickoo SK et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 2012; 307: 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Molenaar JJ, Koster J, Zwijnenburg DA, van SP, Valentijn LJ, van dP I et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 2012; 483: 589–593. [DOI] [PubMed] [Google Scholar]
- 19Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D et al. The genetic landscape of high-risk neuroblastoma. Nat Genet 2013; 45: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Peifer M, Hertwig F, Roels F, Dreidax D, Gartlgruber M, Menon R et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 2015; 526: 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Mac SM, D'Cunha CA, Farnham PJ. Direct recruitment of N-myc to target gene promoters. Mol Carcinog 2000; 29: 76–86. [PubMed] [Google Scholar]
- 22Slack A, Chen Z, Tonelli R, Pule M, Hunt L, Pession A et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proc Natl Acad Sci USA 2005; 102: 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Farooqi AS, Dagg RA, Choi LM, Shay JW, Reynolds CP, Lau LM. Alternative lengthening of telomeres in neuroblastoma cell lines is associated with a lack of MYCN genomic amplification and with p53 pathway aberrations. J Neurooncol 2014; 119: 17–26. [DOI] [PubMed] [Google Scholar]
- 24Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol 2010; 2: a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ 2006; 13: 962–972. [DOI] [PubMed] [Google Scholar]
- 26Yang A, Kaghad M, Caput D, McKeon F. On the shoulders of giants: p63, p73 and the rise of p53. Trends Genet 2002; 18: 90–95. [DOI] [PubMed] [Google Scholar]
- 27Jost CA, Marin MC, Kaelin WG Jr. p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature 1997; 389: 191–194. [DOI] [PubMed] [Google Scholar]
- 28Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997; 90: 809–819. [DOI] [PubMed] [Google Scholar]
- 29Rufini A, Agostini M, Grespi F, Tomasini R, Sayan BS, Niklison-Chirou MV et al. p73 in Cancer. Genes Cancer 2011; 2: 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Stiewe T, Putzer BM. Role of p73 in malignancy: tumor suppressor or oncogene? Cell Death Differ 2002; 9: 237–245. [DOI] [PubMed] [Google Scholar]
- 31Rossi M, Sayan AE, Terrinoni A, Melino G, Knight RA. Mechanism of induction of apoptosis by p73 and its relevance to neuroblastoma biology. Ann NY Acad Sci 2004; 1028: 143–149. [DOI] [PubMed] [Google Scholar]
- 32Ramadan S, Terrinoni A, Catani MV, Sayan AE, Knight RA, Mueller M et al. p73 induces apoptosis by different mechanisms. Biochem Biophys Res Commun 2005; 331: 713–717. [DOI] [PubMed] [Google Scholar]
- 33Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev 2008; 22: 2677–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev 2010; 24: 549–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35White PS, Maris JM, Beltinger C, Sulman E, Marshall HN, Fujimori M et al. A region of consistent deletion in neuroblastoma maps within human chromosome 1p36.2-36.3. Proc Natl Acad Sci USA 1995; 92: 5520–5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Corn PG, Kuerbitz SJ, van Noesel MM, Esteller M, Compitello N, Baylin SB et al. Transcriptional silencing of the p73 gene in acute lymphoblastic leukemia and Burkitt's lymphoma is associated with 5' CpG island methylation. Cancer Res 1999; 59: 3352–3356. [PubMed] [Google Scholar]
- 37Martinez-Delgado B, Melendez B, Cuadros M, Jose GM, Nomdedeu J, Rivas C et al. Frequent inactivation of the p73 gene by abnormal methylation or LOH in non-Hodgkin's lymphomas. Int J Cancer 2002; 102: 15–19. [DOI] [PubMed] [Google Scholar]
- 38Banelli B, Casciano I, Romani M. Methylation-independent silencing of the p73 gene in neuroblastoma. Oncogene 2000; 19: 4553–4556. [DOI] [PubMed] [Google Scholar]
- 39De Laurenzi L, Raschella V, Barcaroli G, Annicchiarico-Petruzzelli D, Ranalli M, Catani MV et al. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem 2000; 275: 15226–15231. [DOI] [PubMed] [Google Scholar]
- 40Wagner LM, Danks MK. New therapeutic targets for the treatment of high-risk neuroblastoma. J Cell Biochem 2009; 107: 46–57. [DOI] [PubMed] [Google Scholar]
- 41Casciano I, Mazzocco K, Boni L, Pagnan G, Banelli B, Allemanni G et al. Expression of DeltaNp73 is a molecular marker for adverse outcome in neuroblastoma patients. Cell Death Differ 2002; 9: 246–251. [DOI] [PubMed] [Google Scholar]
- 42Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 2000; 289: 304–306. [DOI] [PubMed] [Google Scholar]
- 43Casciano I, Banelli B, Croce M, Allemanni G, Ferrini S, Tonini GP et al. Role of methylation in the control of DeltaNp73 expression in neuroblastoma. Cell Death Differ 2002; 9: 343–345. [DOI] [PubMed] [Google Scholar]
- 44Bordow SB, Norris MD, Haber PS, Marshall GM, Haber M. Prognostic significance of MYCN oncogene expression in childhood neuroblastoma. J Clin Oncol 1998; 16: 3286–3294. [DOI] [PubMed] [Google Scholar]
- 45Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 2013; 3: a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Imamura J, Bartram CR, Berthold F, Harms D, Nakamura H, Koeffler HP. Mutation of the p53 gene in neuroblastoma and its relationship with N-myc amplification. Cancer Res 1993; 53: 4053–4058. [PubMed] [Google Scholar]
- 47Gu L, Zhang H, He J, Li J, Huang M, Zhou M. MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene 2012; 31: 1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48He J, Gu L, Zhang H, Zhou M. Crosstalk between MYCN and MDM2-p53 signal pathways regulates tumor cell growth and apoptosis in neuroblastoma. Cell Cycle 2011; 10: 2994–3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res 1998; 26: 3453–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Peirce SK, Findley HW. The MDM2 antagonist nutlin-3 sensitizes p53-null neuroblastoma cells to doxorubicin via E2F1 and TAp73. Int J Oncol 2009; 34: 1395–1402. [PubMed] [Google Scholar]
- 51Lau LM, Nugent JK, Zhao X, Irwin MS. HDM2 antagonist Nutlin-3 disrupts p73-HDM2 binding and enhances p73 function. Oncogene 2008; 27: 997–1003. [DOI] [PubMed] [Google Scholar]
- 52Balint E, Bates S, Vousden KH. Mdm2 binds p73 alpha without targeting degradation. Oncogene 1999; 18: 3923–3929. [DOI] [PubMed] [Google Scholar]
- 53Zeng X, Chen L, Jost CA, Maya R, Keller D, Wang X et al. MDM2 suppresses p73 function without promoting p73 degradation. Mol Cell Biol 1999; 19: 3257–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Zhu X, Wimmer K, Kuick R, Lamb BJ, Motyka S, Jasty R et al. N-myc modulates expression of p73 in neuroblastoma. Neoplasia 2002; 4: 432–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Horvilleur E, Bauer M, Goldschneider D, Mergui X, de la Motte A, Benard J et al. p73alpha isoforms drive opposite transcriptional and post-transcriptional regulation of MYCN expression in neuroblastoma cells. Nucleic Acids Res 2008; 36: 4222–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56Amelio I, Inoue S, Markert EK, Levine AJ, Knight RA, Mak TW et al. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1alpha degradation. Proc Natl Acad Sci USA 2015; 112: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57Stantic M, Sakil HA, Zirath H, Fang T, Sanz G, Fernandez-Woodbridge A et al. TAp73 suppresses tumor angiogenesis through repression of proangiogenic cytokines and HIF-1alpha activity. Proc Natl Acad Sci USA 2015; 112: 220–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Dulloo I, Hooi PB, Sabapathy K. Hypoxia-induced DNp73 stabilization regulates Vegf-A expression and tumor angiogenesis similar to TAp73. Cell Cycle advance online publication 12 August 2015 [e-pub ahead of print; PMID: 26267146]. [DOI] [PMC free article] [PubMed]
- 59Bid HK, Roberts RD, Cam M, Audino A, Kurmasheva RT, Lin J et al. DeltaNp63 promotes pediatric neuroblastoma and osteosarcoma by regulating tumor angiogenesis. Cancer Res 2014; 74: 320–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60Melino G, Gallagher E, Aqeilan RI, Knight R, Peschiaroli A, Rossi M et al. Itch: a HECT-type E3 ligase regulating immunity, skin and cancer. Cell Death Differ 2008; 15: 1103–1112. [DOI] [PubMed] [Google Scholar]
- 61Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 2008; 14: 10–21. [DOI] [PubMed] [Google Scholar]
- 62Hansen TM, Rossi M, Roperch JP, Ansell K, Simpson K, Taylor D et al. Itch inhibition regulates chemosensitivity in vitro. Biochem Biophys Res Commun 2007; 361: 33–36. [DOI] [PubMed] [Google Scholar]
- 63Bongiorno-Borbone L, Giacobbe A, Compagnone M, Eramo A, De MR, Peschiaroli A et al. Anti-tumoral effect of desmethylclomipramine in lung cancer stem cells. Oncotarget 2015; 6: 16926–16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64de la Fuente M, Jones MC, Santander-Ortega MJ, Mirenska A, Marimuthu P, Uchegbu I et al. A nano-enabled cancer-specific ITCH RNAi chemotherapy booster for pancreatic cancer. Nanomedicine 2015; 11: 369–377. [DOI] [PubMed] [Google Scholar]
- 65Chaudhary N, Maddika S. WWP2-WWP1 ubiquitin ligase complex coordinated by PPM1G maintains the balance between cellular p73 and DeltaNp73 levels. Mol Cell Biol 2014; 34: 3754–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66Peschiaroli A, Scialpi F, Bernassola F, Pagano M, Melino G. The F-box protein FBXO45 promotes the proteasome-dependent degradation of p73. Oncogene 2009; 28: 3157–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67Sayan BS, Yang AL, Conforti F, Tucci P, Piro MC, Browne GJ et al. Differential control of TAp73 and DeltaNp73 protein stability by the ring finger ubiquitin ligase PIR2. Proc Natl Acad Sci USA 2010; 107: 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68Rossi M, De Laurenzi V, Munarriz E, Green DR, Liu YC, Vousden KH et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J 2005; 24: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69Rossi M, Rotblat B, Ansell K, Amelio I, Caraglia M, Misso G et al. High throughput screening for inhibitors of the HECT ubiquitin E3 ligase ITCH identifies antidepressant drugs as regulators of autophagy. Cell Death Dis 2014; 5: e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70Gillman PK. Tricyclic antidepressant pharmacology and therapeutic drug interactions updated. Br J Pharmacol 2007; 151: 737–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature 2015; 517: 311–320. [DOI] [PubMed] [Google Scholar]
- 72Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714. [DOI] [PubMed] [Google Scholar]
- 73Mihaly SR, Ninomiya-Tsuji J, Morioka S. TAK1 control of cell death. Cell Death Differ 2014; 21: 1667–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S et al. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 2000; 1: 489–495. [DOI] [PubMed] [Google Scholar]
- 75Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76He S, Wang L, Miao L, Wang T, Du F, Zhao L et al. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009; 137: 1100–1111. [DOI] [PubMed] [Google Scholar]
- 77Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC et al. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009; 325: 332–336. [DOI] [PubMed] [Google Scholar]
- 78Sun L, Wang H, Wang Z, He S, Chen S, Liao D et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012; 148: 213–227. [DOI] [PubMed] [Google Scholar]
- 79Vandenabeele P, Melino G. The flick of a switch: which death program to choose? Cell Death Differ 2012; 19: 1093–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80Cai Z, Jitkaew S, Zhao J, Chiang HC, Choksi S, Liu J et al. Plasma membrane translocation of trimerized MLKL protein is required for TNF-induced necroptosis. Nat Cell Biol 2014; 16: 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81Kaczmarek A, Vandenabeele P, Krysko DV. Necroptosis: the release of damage-associated molecular patterns and its physiological relevance. Immunity 2013; 38: 209–223. [DOI] [PubMed] [Google Scholar]
- 82Aziz M, Jacob A, Wang P. Revisiting caspases in sepsis. Cell Death Dis 2014; 5: e1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83Duprez L, Takahashi N, Van HF, Vandendriessche B, Goossens V, Vanden Berghe T et al. RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 2011; 35: 908–918. [DOI] [PubMed] [Google Scholar]
- 84Inoue H, Tani K. Multimodal immunogenic cancer cell death as a consequence of anticancer cytotoxic treatments. Cell Death Differ 2014; 21: 39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85Todaro M, Alea MP, Di Stefano AB, Cammareri P, Vermeulen L, Iovino F et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell 2007; 1: 389–402. [DOI] [PubMed] [Google Scholar]
- 86Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science 2015; 348: 69–74. [DOI] [PubMed] [Google Scholar]
- 87Casciano I, De AA, Croce M, Pagnan G, Di VA, Allemanni G et al. Expression of the caspase-8 gene in neuroblastoma cells is regulated through an essential interferon-sensitive response element (ISRE). Cell Death Differ 2004; 11: 131–134. [DOI] [PubMed] [Google Scholar]
- 88Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015; 523: 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89Fatt MP, Cancino GI, Miller FD, Kaplan DR. p63 and p73 coordinate p53 function to determine the balance between survival, cell death, and senescence in adult neural precursor cells. Cell Death Differ 2014; 21: 1546–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90Green DR, Chipuk JE. p53 and metabolism: inside the TIGAR. Cell 2006; 126: 30–32. [DOI] [PubMed] [Google Scholar]
- 91Jang CW, Shibata Y, Starmer J, Yee D, Magnuson T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev 2015; 29: 1377–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015; 520: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93Venkatanarayan A, Raulji P, Norton W, Chakravarti D, Coarfa C, Su X et al. IAPP-driven metabolic reprogramming induces regression of p53-deficient tumours in vivo. Nature 2015; 517: 626–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94Yallowitz AR, Alexandrova EM, Talos F, Xu S, Marchenko ND, Moll UM. p63 is a prosurvival factor in the adult mammary gland during post-lactational involution, affecting PI-MECs and ErbB2 tumorigenesis. Cell Death Differ 2014; 21: 645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95Aylon Y, Oren M. Living with p53, dying of p53. Cell 2007; 130: 597–600. [DOI] [PubMed] [Google Scholar]
- 96Burnley P, Rahman M, Wang H, Zhang Z, Sun X, Zhuge Q et al. Role of the p63-FoxN1 regulatory axis in thymic epithelial cell homeostasis during aging. Cell Death Dis 2013; 4: e932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97Hayashi MT, Cesare AJ, Rivera T, Karlseder J. Cell death during crisis is mediated by mitotic telomere deprotection. Nature 2015; 522: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98Wu J, Liang S, Bergholz J, He H, Walsh EM, Zhang Y et al. DeltaNp63alpha activates CD82 metastasis suppressor to inhibit cancer cell invasion. Cell Death Dis 2014; 5: e1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99Amelio I, Lena AM, Bonanno E, Melino G, Candi E. miR-24 affects hair follicle morphogenesis targeting Tcf-3. Cell Death Dis 2013; 4: e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100Antonov AV, Knight RA, Melino G, Barlev NA, Tsvetkov PO. MIRUMIR: an online tool to test microRNAs as biomarkers to predict survival in cancer using multiple clinical data sets. Cell Death Differ 2013; 20: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101Candi E, Amelio I, Agostini M, Melino G. MicroRNAs and p63 in epithelial stemness. Cell Death Differ 2015; 22: 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102Fiori ME, Barbini C, Haas TL, Marroncelli N, Patrizii M, Biffoni M et al. Antitumor effect of miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ 2014; 21: 774–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103Fortunato O, Boeri M, Moro M, Verri C, Mensah M, Conte D et al. Mir-660 is downregulated in lung cancer patients and its replacement inhibits lung tumorigenesis by targeting MDM2-p53 interaction. Cell Death Dis 2014; 5: e1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104Hoffman Y, Bublik DR, Pilpel Y, Oren M. miR-661 downregulates both Mdm2 and Mdm4 to activate p53. Cell Death Differ 2014; 21: 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M et al. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014; 5: e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106Lezina L, Purmessur N, Antonov AV, Ivanova T, Karpova E, Krishan K et al. miR-16 and miR-26a target checkpoint kinases Wee1 and Chk1 in response to p53 activation by genotoxic stress. Cell Death Dis 2013; 4: e953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107Ng WL, Chen G, Wang M, Wang H, Story M, Shay JW et al. OCT4 as a target of miR-34a stimulates p63 but inhibits p53 to promote human cell transformation. Cell Death Dis 2014; 5: e1024. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 108Ren ZJ, Nong XY, Lv YR, Sun HH, An PP, Wang F et al. Mir-509-5p joins the Mdm2/p53 feedback loop and regulates cancer cell growth. Cell Death Dis 2014; 5: e1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 2013; 495: 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110Hearn JM, Romero-Canelon I, Munro AF, Fu Y, Pizarro AM, Garnett MJ et al. Potent organo-osmium compound shifts metabolism in epithelial ovarian cancer cells. Proc Natl Acad Sci USA 2015; 112: E3800–E3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T et al. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc Natl Acad Sci USA 2004; 101: 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112Pritchard DM, Watson AJ, Potten CS, Jackman AL, Hickman JA. Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc Natl Acad Sci USA 1997; 94: 1795–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M et al. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 2011; 470: 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature 2008; 451: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115Liu J, Zhang C, Wang XL, Ly P, Belyi V, Xu-Monette ZY et al. E3 ubiquitin ligase TRIM32 negatively regulates tumor suppressor p53 to promote tumorigenesis. Cell Death Differ 2014; 21: 1792–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116Peuget S, Bonacci T, Soubeyran P, Iovanna J, Dusetti NJ. Oxidative stress-induced p53 activity is enhanced by a redox-sensitive TP53INP1 SUMOylation. Cell Death Differ 2014; 21: 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117Sane S, Abdullah A, Boudreau DA, Autenried RK, Gupta BK, Wang X et al. Ubiquitin-like (UBX)-domain-containing protein, UBXN2A, promotes cell death by interfering with the p53-Mortalin interactions in colon cancer cells. Cell Death Dis 2014; 5: e1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118Zhang HH, Li SZ, Zhang ZY, Hu XM, Hou PN, Gao L et al. Nemo-like kinase is critical for p53 stabilization and function in response to DNA damage. Cell Death Differ 2014; 21: 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119Chen C, Gorlatova N, Kelman Z, Herzberg O. Structures of p63 DNA binding domain in complexes with half-site and with spacer-containing full response elements. Proc Natl Acad Sci USA 2011; 108: 6456–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120Han X, Zhang L, Chung J, Mayca PF, Tran A, Seachrist DD et al. UbcH7 regulates 53BP1 stability and DSB repair. Proc Natl Acad Sci USA 2014; 111: 17456–17461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 2000; 408: 377–381. [DOI] [PubMed] [Google Scholar]
- 122Pearson M, Carbone R, Sebastiani C, Cioce M, Fagioli M, Saito S et al. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 2000; 406: 207–210. [DOI] [PubMed] [Google Scholar]
- 123Topisirovic I, Gutierrez GJ, Chen M, Appella E, Borden KL, Ronai ZA. Control of p53 multimerization by Ubc13 is JNK-regulated. Proc Natl Acad Sci USA 2009; 106: 12676–12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124Garufi A, Pucci D, D'Orazi V, Cirone M, Bossi G, Avantaggiati ML et al. Degradation of mutant p53H175 protein by Zn(II) through autophagy. Cell Death Dis 2014; 5: e1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125Simon HU, Yousefi S, Schmid I, Friis R. ATG5 can regulate p53 expression and activation. Cell Death Dis 2014; 5: e1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126: 121–134. [DOI] [PubMed] [Google Scholar]
- 127Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature 2009; 458: 1127–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell 2013; 155: 1216–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011; 147: 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013; 504: 296–300. [DOI] [PubMed] [Google Scholar]
- 131Viale A, Pettazzoni P, Lyssiotis CA, Ying H, Sanchez N, Marchesini M et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature 2014; 514: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132Marcel V, Fernandes K, Terrier O, Lane DP, Bourdon JC. Modulation of p53beta and p53gamma expression by regulating the alternative splicing of TP53 gene modifies cellular response. Cell Death Differ 2014; 21: 1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133Slatter TL, Hung N, Bowie S, Campbell H, Rubio C, Speidel D et al. Delta122p53, a mouse model of Delta133p53alpha, enhances the tumor-suppressor activities of an attenuated p53 mutant. Cell Death Dis 2015; 6: e1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134Solomon H, Sharon M, Rotter V. Modulation of alternative splicing contributes to cancer development: focusing on p53 isoforms, p53beta and p53gamma. Cell Death Differ 2014; 21: 1347–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 2015; 29: 63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136Chen Y, Zhang L, Jones KA. SKIP counteracts p53-mediated apoptosis via selective regulation of p21Cip1 mRNA splicing. Genes Dev 2011; 25: 701–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137Ci Y, Shi K, An J, Yang Y, Hui K, Wu P et al. ROS inhibit autophagy by downregulating ULK1 mediated by the phosphorylation of p53 in selenite-treated NB4 cells. Cell Death Dis 2014; 5: e1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138Evstafieva AG, Garaeva AA, Khutornenko AA, Klepikova AV, Logacheva MD, Penin AA et al. A sustained deficiency of mitochondrial respiratory complex III induces an apoptotic cell death through the p53-mediated inhibition of pro-survival activities of the activating transcription factor 4. Cell Death Dis 2014; 5: e1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139He Z, Liu H, Agostini M, Yousefi S, Perren A, Tschan MP et al. p73 regulates autophagy and hepatocellular lipid metabolism through a transcriptional activation of the ATG5 gene. Cell Death Differ 2013; 20: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140Kostecka A, Sznarkowska A, Meller K, Acedo P, Shi Y, Mohammad Sakil HA et al. JNK-NQO1 axis drives TAp73-mediated tumor suppression upon oxidative and proteasomal stress. Cell Death Dis 2014; 5: e1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141Xu J, Wang J, Hu Y, Qian J, Xu B, Chen H et al. Unequal prognostic potentials of p53 gain-of-function mutations in human cancers associate with drug-metabolizing activity. Cell Death Dis 2014; 5: e1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142Bertout JA, Majmundar AJ, Gordan JD, Lam JC, Ditsworth D, Keith B et al. HIF2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci USA 2009; 106: 14391–14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143Billia F, Hauck L, Grothe D, Konecny F, Rao V, Kim RH et al. Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc Natl Acad Sci USA 2013; 110: 6085–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144Cheung EC, Ludwig RL, Vousden KH. Mitochondrial localization of TIGAR under hypoxia stimulates HK2 and lowers ROS and cell death. Proc Natl Acad Sci USA 2012; 109: 20491–20496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA 2010; 107: 7455–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146Raj L, Ide T, Gurkar AU, Foley M, Schenone M, Li X et al. Selective killing of cancer cells by a small molecule targeting the stress response to ROS. Nature 2011; 475: 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 147Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S et al. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci USA 2010; 107: 7461–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148Becker MS, Schmezer P, Breuer R, Haas SF, Essers MA, Krammer PH et al. The traditional Chinese medical compound Rocaglamide protects nonmalignant primary cells from DNA damage-induced toxicity by inhibition of p53 expression. Cell Death Dis 2014; 5: e1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149Bunjobpol W, Dulloo I, Igarashi K, Concin N, Matsuo K, Sabapathy K. Suppression of acetylpolyamine oxidase by selected AP-1 members regulates DNp73 abundance: mechanistic insights for overcoming DNp73-mediated resistance to chemotherapeutic drugs. Cell Death Differ 2014; 21: 1240–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150Cheng J, Fan YH, Xu X, Zhang H, Dou J, Tang Y et al. A small-molecule inhibitor of UBE2N induces neuroblastoma cell death via activation of p53 and JNK pathways. Cell Death Dis 2014; 5: e1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151Salah Z, Bar-mag T, Kohn Y, Pichiorri F, Palumbo T, Melino G et al. Tumor suppressor WWOX binds to DeltaNp63alpha and sensitizes cancer cells to chemotherapy. Cell Death Dis 2013; 4: e480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152Weilbacher A, Gutekunst M, Oren M, Aulitzky WE, van der Kuip H. RITA can induce cell death in p53-defective cells independently of p53 function via activation of JNK/SAPK and p38. Cell Death Dis 2014; 5: e1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153Memmi EM, Sanarico AG, Giacobbe A, Peschiaroli A, Frezza V, Cicalese A et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci USA 2015; 112: 3499–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154Viticchie G, Agostini M, Lena AM, Mancini M, Zhou H, Zolla L et al. p63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci USA 2015; 112: 11577–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155Adamovich Y, Adler J, Meltser V, Reuven N, Shaul Y. AMPK couples p73 with p53 in cell fate decision. Cell Death Differ 2014; 21: 1451–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156Dashzeveg N, Taira N, Lu ZG, Kimura J, Yoshida K. Palmdelphin, a novel target of p53 with Ser46 phosphorylation, controls cell death in response to DNA damage. Cell Death Dis 2014; 5: e1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157Desantis A, Bruno T, Catena V, De NF, Goeman F, Iezzi S et al. Che-1 modulates the decision between cell cycle arrest and apoptosis by its binding to p53. Cell Death Dis 2015; 6: e1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158Lezina L, Aksenova V, Ivanova T, Purmessur N, Antonov AV, Tentler D et al. KMTase Set7/9 is a critical regulator of E2F1 activity upon genotoxic stress. Cell Death Differ 2014; 21: 1889–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159Manzl C, Fava LL, Krumschnabel G, Peintner L, Tanzer MC, Soratroi C et al. Death of p53-defective cells triggered by forced mitotic entry in the presence of DNA damage is not uniquely dependent on Caspase-2 or the PIDDosome. Cell Death Dis 2013; 4: e942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160Nair BC, Krishnan SR, Sareddy GR, Mann M, Xu B, Natarajan M et al. Proline, glutamic acid and leucine-rich protein-1 is essential for optimal p53-mediated DNA damage response. Cell Death Differ 2014; 21: 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161Phesse TJ, Myant KB, Cole AM, Ridgway RA, Pearson H, Muncan V et al. Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ 2014; 21: 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162Puzio-Kuter AM, Laddha SV, Castillo-Martin M, Sun Y, Cordon-Cardo C, Chan CS et al. Involvement of tumor suppressors PTEN and p53 in the formation of multiple subtypes of liposarcoma. Cell Death Differ 2015; 22: 1785–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163Tomasini R, Secq V, Pouyet L, Thakur AK, Wilhelm M, Nigri J et al. TAp73 is required for macrophage-mediated innate immunity and the resolution of inflammatory responses. Cell Death Differ 2013; 20: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164Deriano L, Chaumeil J, Coussens M, Multani A, Chou Y, Alekseyenko AV et al. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature 2011; 471: 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165Elyada E, Pribluda A, Goldstein RE, Morgenstern Y, Brachya G, Cojocaru G et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature 2011; 470: 409–413. [DOI] [PubMed] [Google Scholar]
- 166Poruchynsky MS, Komlodi-Pasztor E, Trostel S, Wilkerson J, Regairaz M, Pommier Y et al. Microtubule-targeting agents augment the toxicity of DNA-damaging agents by disrupting intracellular trafficking of DNA repair proteins. Proc Natl Acad Sci USA 2015; 112: 1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167Rai P, Parrish M, Tay IJ, Li N, Ackerman S, He F et al. Streptococcus pneumoniae secretes hydrogen peroxide leading to DNA damage and apoptosis in lung cells. Proc Natl Acad Sci USA 2015; 112: E3421–E3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 2002; 416: 560–564. [DOI] [PubMed] [Google Scholar]
- 169Wang B, Niu D, Lam TH, Xiao Z, Ren EC. Mapping the p53 transcriptome universe using p53 natural polymorphs. Cell Death Differ 2014; 21: 521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170Wiman KG. p53 talks to PARP: the increasing complexity of p53-induced cell death. Cell Death Differ 2013; 20: 1438–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171Zaccara S, Tebaldi T, Pederiva C, Ciribilli Y, Bisio A, Inga A. p53-directed translational control can shape and expand the universe of p53 target genes. Cell Death Differ 2014; 21: 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172Zambetti GP. Expanding the reach of the p53 tumor suppressor network. Cell Death Differ 2014; 21: 505–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature 2007; 450: 721–724. [DOI] [PubMed] [Google Scholar]
- 174Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA et al. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006; 444: 629–632. [DOI] [PubMed] [Google Scholar]
- 175Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, Liu W et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 2000; 407: 645–648. [DOI] [PubMed] [Google Scholar]
- 176Martin K, Trouche D, Hagemeier C, Sorensen TS, La Thangue NB, Kouzarides T. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature 1995; 375: 691–694. [DOI] [PubMed] [Google Scholar]
- 177Soria C, Estermann FE, Espantman KC, O'Shea CC. Heterochromatin silencing of p53 target genes by a small viral protein. Nature 2010; 466: 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K. Modulation of microRNA processing by p53. Nature 2009; 460: 529–533. [DOI] [PubMed] [Google Scholar]
- 179Aylon Y, Sarver A, Tovy A, Ainbinder E, Oren M. Lats2 is critical for the pluripotency and proper differentiation of stem cells. Cell Death Differ 2014; 21: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180Kim J, Nakasaki M, Todorova D, Lake B, Yuan CY, Jamora C et al. p53 induces skin aging by depleting Blimp1+ sebaceous gland cells. Cell Death Dis 2014; 5: e1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181Li L, Ng DS, Mah WC, Almeida FF, Rahmat SA, Rao VK et al. A unique role for p53 in the regulation of M2 macrophage polarization. Cell Death Differ 2015; 22: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009; 460: 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 1999; 398: 708–713. [DOI] [PubMed] [Google Scholar]
- 184Schnittke N, Herrick DB, Lin B, Peterson J, Coleman JH, Packard AI et al. Transcription factor p63 controls the reserve status but not the stemness of horizontal basal cells in the olfactory epithelium. Proc Natl Acad Sci USA 2015; 112: E5068–E5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature 1999; 398: 714–718. [DOI] [PubMed] [Google Scholar]
- 186Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness'. Nature 2008; 452: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187Oldridge DA, Wood AC, Weichert-Leahey N, Crimmins I, Sussman R, Winter C et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature advance online publication, 11 November 2015; doi: 10.1038/nature15540 [e-pub ahead of print]. [DOI] [PMC free article] [PubMed]