Summary
Background
Mitochondria form a dynamics tubular network within the cell. Proper mitochondria movement and distribution are critical for their localized function in cell metabolism, growth, and survival. In mammalian cells, mechanisms of mitochondria positioning appear dependent on the microtubule cytoskeleton, with kinesin or dynein motors carrying mitochondria as cargos and distributing them throughout the microtubule network. Interestingly, the timescale of microtubule dynamics occurs in seconds, and the timescale of mitochondria distribution occurs in minutes. How does the cell couple these two time constants?
Results
Fission yeast also relies on microtubules for mitochondria distribution. We report here a new microtubule-dependent but motor-independent mechanism for proper mitochondria positioning in fission yeast. We identify the protein mmb1p, which binds to mitochondria and microtubules. Mmb1p attaches the tubular mitochondria to the microtubule lattice at multiple discrete interaction sites. Mmb1 deletion causes mitochondria to aggregate, with the long-term consequence of defective mitochondria distribution and cell death. Mmb1p decreases microtubule dynamicity.
Conclusion
Mmb1p is a new microtubule-mitochondria binding protein. We propose that mmb1p act to couple long-term mitochondria distribution to short-term microtubule dynamics by attenuating microtubule dynamics, thus enhancing the mitochondria-microtubule interaction time.
Introduction
The mitochondria network is composed of interconnected tubular structures that undergo fusion, fission, and translocation throughout the cell [1, 2]. Proper mitochondria positioning is essential for cellular metabolism, growth and survival [3]. The actin and microtubule cytoskeleton both play key roles in mitochondria positioning. However, depending on the species or cell types, different cytoskeletal components may be employed. Despite the diversity of organisms and cell types, some general mechanisms for mitochondria distribution have emerged. For example, budding yeast S. cerevisiae, fungus Aspergillus, and plants anchor their mitochondria to the actin cytoskeleton, and uses actin-polymerization as a motive force for moving [4]. In contrast, C. elegans, Drosophila, and mammalian neuronal cells mainly attach their mitochondria to motors such as kinesin and dynein to be transported on the microtubule cytoskeleton [5-7]. Thus, microtubule- and motor-coupled mitochondria positioning appears one major mechanism. However, the majority (~70%) of mitochondria in mammalian neuronal cells are stationary and remained stably attached to microtubules [8]. In addition, for many cell types, microtubule dynamics can be very rapid, with polymer turn-over time in tens of seconds [9, 10]. In contrast, mitochondria, while clearly coupled to microtubules, do not exhibit the same fast turn-over time. How does a cell couple fast microtubule dynamics to slow mitochondria dynamics? And how does a cell statically attach the mitochondria to the microtubules?
The fission yeast Schizosaccharomyces pombe is a good model system to address mechanisms of coupling between mitochondria and microtubule dynamics. Fission yeast uses a microtubule-dependent but motor-independent mechanism for mitochondria positioning [7]. Interphase cells have several linear bundles of antiparallel microtubules organized along the cell long axis, with the plus ends interacting with the cell tips [11]. Colocalized with the microtubules are tubular strands of mitochondria [12]. Electron tomographic reconstruction showed mitochondria intertwined around microtubules [13], with typical separation distances of ~20 nm [14].
We report here a new fission yeast protein mmb1p. Mmb1p binds the mitochondria to the microtubule lattice at multiple sites. In the absence of mmb1p, mitochondria aggregate at either cell tips, leading to infrequent mitochondria mis-segregation during the cell cycle and subsequent cell death. Mmb1p attenuates microtubule dynamicity, making microtubules more stable. We propose a model where mmb1p anchors mitochondria to microtubules and acts to enhance mitochondriamicrotubule contact time, thus preventing mitochondria aggregation and promote mitochondria extension. This model can explain how cells couple long-term mitochondria distribution to short-term microtubule dynamics. Our model contrasts with a previous model which suggests that microchondria extension is driven by microtubule polymerization via their coupling to the +TIP CLASP protein peg1p [15]. Mmb1p function may represent a general mechanism of microtubule-dependent but motor-independent mitochondria distribution in cells.
Results
In a fission yeast random GFP insertional screen [16], and a genome-wide YFP tag project [17], the product of the previously uncharacterized gene SPBC25B2.07c was identified as a putative microtubule binding protein. Subsequently, in a screen for meiosis up-regulated genes, SPBC25B2.07c was identified as mug164, with no further characterization [18]. During the course of this study, we found that SPBC25B2.07c functions to bind mitochondria to microtubules (see below). Therefore, we renamed this gene mmb1+ (mitochondria microtubule binder 1) to reflect its biological function.
Mmb1p is a cytoplasmic microtubule binding protein
To investigate the localization of mmb1p throughout the cell cycle, we examined wildtype cells co-expressing mmb1-GFP and mCherry-atb2 (tubulin). Mmb1p appeared as dots along interphase cytoplasmic microtubules (Fig. 1A). In deed, mmb1p was found on all cytoplasmic microtubules throughout the cell cycle, such as the interphase, the astral, and the post-anaphase arrays of microtubules (Fig. S1A), suggesting that mmb1p may bind to microtubules directly. Interestingly, mmb1p also appeared as dim tubular structures in cytoplasmic regions where there were no apparent microtubules (Fig. 1A and Fig. S1A), suggesting that mmb1p may interact with other cytoplasmic structures.
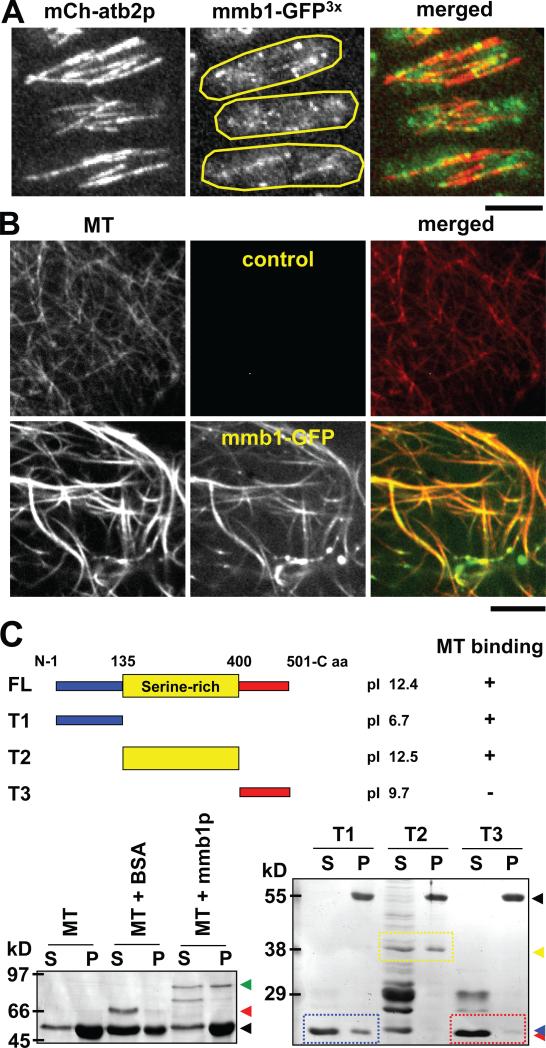
Figure 1. Mmb1p is a microtubule binding protein.
(A) Images of a wildtype cell expressing mCherry-atb2 and mmb1-GFP. Shown are maximum-projection images of three interphase cells. Mmb1-GFP appears as dots or short bars decorating the microtubule lattices. Bar, 5 μm. (See also Fig. S1A)
(B) In vitro microtubule binding assay. Taxol-stabilized microtubules (MT) (red) were polymerized from porcine brain tubulin decorated with Alexa Flour® 594 dye. Recombinant His-GFP-mmb1p (green) was isolated from E. coli. Image shows mmb1-GFP decorating the microtubule lattices and bundling microtubules. Bar, 5 μm.
(C) In vitro microtubule co-sedimentation assay. Cartoon shows full length mmb1p and different truncations and their predicted isoelectric points pI. First gel shows mmb1p full length interaction with microtubules (MT). Mixtures of taxol-stabilized microtubules (MT) and BSA or His-GFP-mmb1p were centrifuged, and the supernatants (S) and pellets (P) were analyzed by gel electrophoresis and coomassie blue staining. Control MT and MT+BSA lanes show little or no co-sedimentations with the microtubule pellets (position of tubulin, black arrow head; position of BSA, red arrow head). The MT+mmb1p lane shows co-sedimentation of mmb1p and microtubules (position of mmb1p, green arrow head). Some Mmb1p and degradation products remain in the supernatant. Second gel maps the interaction domains of mmb1p with microtubules. Different truncated mmb1p versions were tested for microtubule binding (tubulin, black arrow head): the N-terminal T1 domain (position of T1, blue arrow head), middle serine-rich T2 domain (position of T2, yellow arrow head), and C-terminal T3 domain (position of T3, red arrow head). Both T1 and T2 show co-sedimentations with the microtubule pellets. T3 remains in the supernatant. (See also Fig. S1B and Fig. S2)
We next tested for direct binding of mmb1p to microtubules. Purified recombinant His-tagged mmb1-GFP was mixed with taxol-stabilized microtubules polymerized from porcine brain tubulin. In in vitro imaging assays, mmb1p appeared to decorate the microtubules (Fig. 1B), consistent with the in vivo imaging results. In co-sedimentation assays, we confirmed that a significant fraction of the soluble full-length mmb1p co-pelleted with the microtubules (Fig. 1C). We next mapped the microtubule-binding domain of mmb1p by creating recombinant truncated versions of mmb1p. Mmb1p has 501 amino acids, and a calculated isoelectric point of pI ~ 12.4. Mmb1p has a basic medial domain (T2) which is rich in serine, and flanking N- and C-terminus (T1 and T3, respectively) which are disordered (Fig. 1C). Both T1 and T2 were able to bind to microtubule in co-sedimentation assays, but T3 did not bind to microtubules (Fig. 1C and S1B). Microtubules are composed of heterodimers of αβ-tubulin, which are acidic with pI ~ 4.6. This suggests that binding of mmb1p to microtubules may be facilitated through ionic interactions. We conclude that mmb1p is a new microtubule binding protein.
Mmb1p is a mitochondria interacting protein
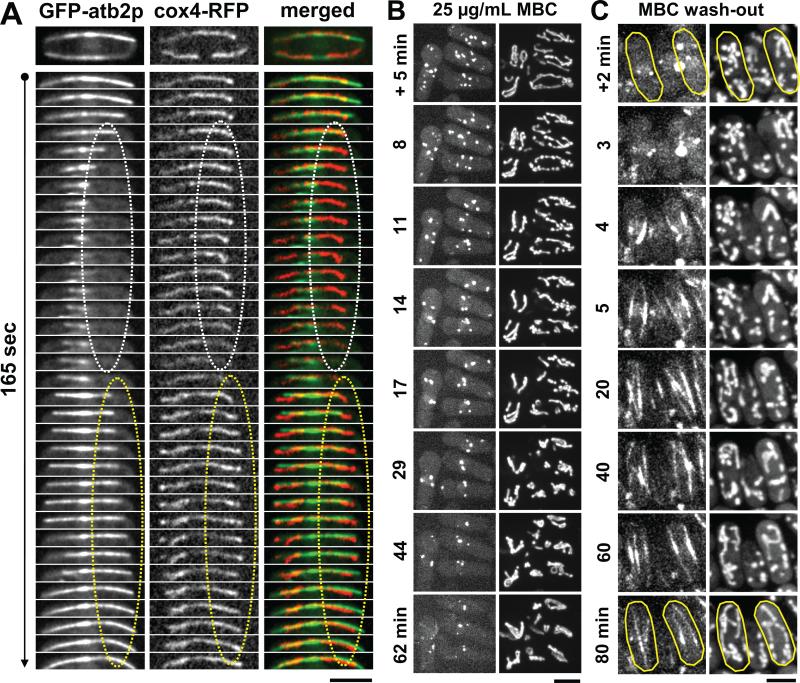
The cytoplasmic tubular structures on which mmb1p localized are reminiscent of mitochondria structures [12-14]. Therefore, we tested for colocalization of mmb1-GFP with mitochondria, which was marked with the mitochondria inner membrane protein RFP-cox4p [15]. While the fluorescent intensity of mmb1-GFP was relatively low, they were distinct from background auto-fluorescence (Fig. 2A). Mmb1p localized to mitochondria as dots and small clusters (Fig. 2A). Interestingly, in the absence of microtubules, induced by 25 μg/mL of the microtubule depolymerizing drug carbendazim (MBC), mmb1-GFP remained localized to the mitochondria in an amorphous pattern (Fig. 2B). In addition, in MBC-treated cells, which have no microtubules, the mitochondria network began to fragment and displayed defective aggregations (Fig. 2B), consistent with previous findings that mitochondria integrity and positioning are microtubule-dependent [7, 12, 15, 19].
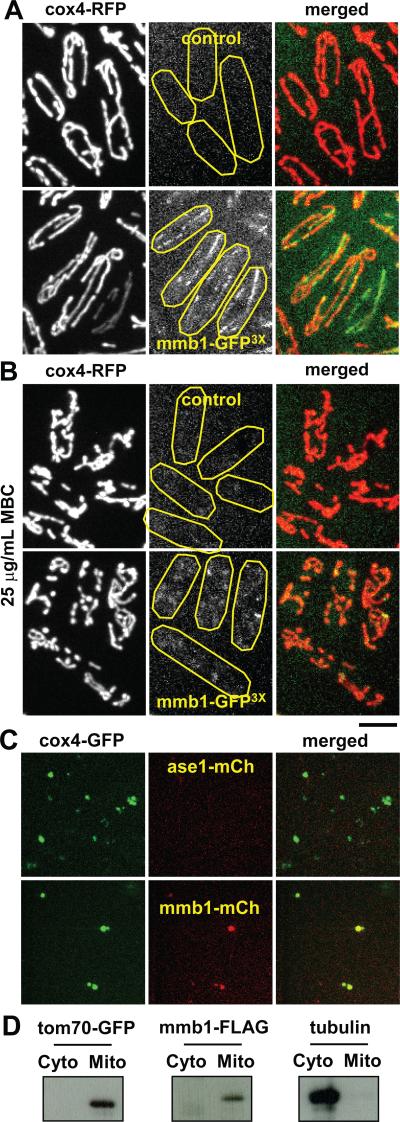
Figure 2. Mmb1p colocalizes with the mitochondria network.
(A) Images of wildtype cells expressing mmb1-GFP3X and cox4-RFP, a mitochondria marker. Top panel shows control cells which do not have GFP-tagged mmb1. Control cells show no detectable auto-fluorescence that can be mistaken for mmb1-GFP3X. Bottom panel shows mmb1p-GFP3X colocalizes as dots and small bars on mitochondria.
(B) Images of wildtype cells expressing mmb1-GFP3X and cox4-RFP treated for 5 min with 25 μg/mL MBC, a microtubule-depolymerization drug. In the absence of microtubules, mitochondria begin to fragment and aggregate at the cell tips and mmb1p appears as diffused signal on mitochondria. Bar, 5 μm.
(C) Mitochondria isolated from wildtype cells expressing mitochondria marker cox4-GFP and mmb1-mCherry, or cox4-GFP and the microtubule bundling protein ase1-mCherry as control. Top panel shows ase1p does not bind to isolated mitochondria. Bottom panel shows mmb1p does bind to mitochondria. Isolated mitochondria appear as small aggregates.
(D) Western blots of tom70-GFP (a bona fide mitochondria membrane protein), mmb1-FLAG, and tubulin in cellular fractions containing the cytoplasm or the isolated mitochondria. Both tom70p and mmb1p appear in the isolated mitochondria fraction. Tubulin appears in the cytoplasmic fraction.
We next tested for binding of mmb1p to mitochondria. We extracted mitochondria from cells expressing cox4-GFP and mmb1-mCherry. In in vitro imaging assays, purified mitochondria appeared as small round aggregates which showed both cox4p and mmb1p colocalization (Fig. 2C), consistent with the in vivo imaging results. As control, the microtubule binding protein ase1-mCherry [20, 21] did not co-sediment with mitochondria (Fig. 2C). For additional confirmation, we extracted mitochondria from cells expressing the FLAG-tagged mmb1p and the bona fide mitochondria membrane protein tom70p tagged with GFP, then probed for tom70-GFP and mmb1-FLAG in cell fractions containing either the cytoplasm or the purified mitochondria. Both tom70-GFP and mmb1-FLAG appeared in the mitochondria fraction (Fig. 2D). As control, tubulin remained in the cytoplasmic fraction (Fig. 2D). We conclude that mmb1p is also a mitochondria binding protein. As the primary sequence of mmb1p predicted no membrane-binding domain, we speculate that the binding of mmb1p to mitochondria may be indirect, facilitated by unknown adaptor proteins.
Further analysis of mmb1p domains responsible for mitochondria and/or microtubule binding is summarized (Fig. S2). In general, the large medial serine-rich domain of mmb1p is required for both mitochondria-microtubule binding, with the N-terminus preferentially helping microtubule-binding and the C-terminus preferentially helping mitochondria-binding (Fig. S2).
Mmb1Δ has mitochondria positioning defects
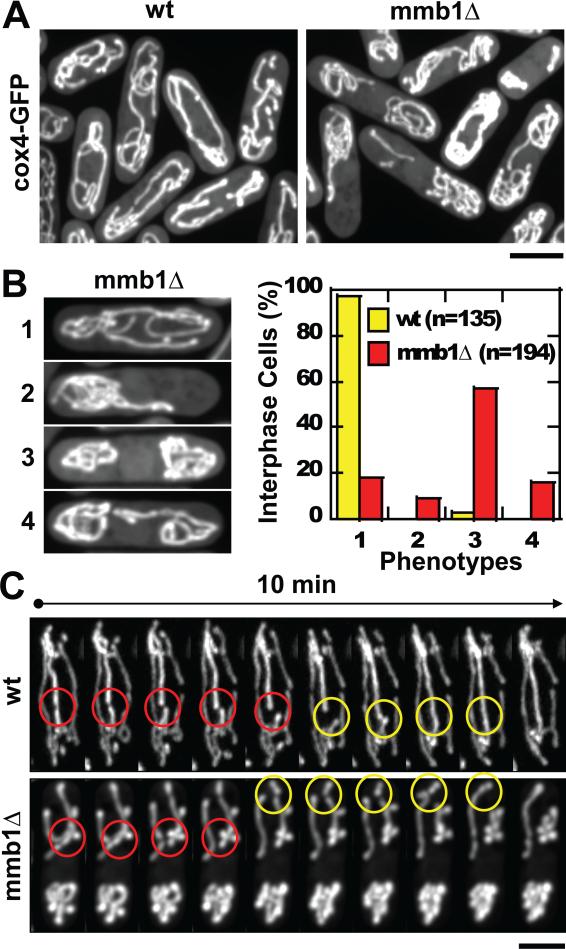
To test the function of mmb1p in mitochondria distribution, we examined mitochondria distribution in cells deleted for mmb1+ (mmb1Δ). In mmb1Δ cells expressing the mitochondria marker cox4-GFP, we observed severe mitochondria aggregation phenotypes (Fig. 3A; Movies S1A and S1B, S2A and S2B). The mitochondria aggregation phenotypes of mmb1Δ occurred at cell tips, and appeared excluded from the cell center where the nucleus is located (Fig. 3A). Whereas >95% (N=135) interphase wildtype cells showed mostly untangled mitochondria that extended continuously the length of the cells, mmb1Δ interphase cells showed several different types of aggregation, with ~70% (N=194) having mitochondria aggregates at both cell ends (phenotype 3 and 4), and ~10% having mitochondria aggregates at only one cell end (phenotype 2) (Fig. 3B). The final ~20% appeared similar to wildtype (phenotype 1).
Figure 3. Mmb1Δ cells have mitochondria positioning defects.
(A) Images of wildtype and mmb1Δ cells expressing cox4-GFP. Mmb1Δ cells show mitochondria positioning defects. Bar, 5 μm. (See also Movies S1A, S1B and S2A, S2B)
(B) Quantification of mmb1Δ mitochondria aggregation phenotypes. 1 = wildtype-like, 2 = aggregation at one cell end, 3 = aggregation at both cell ends, 4 = aggregation at both cell ends but with minor connection. Plot shows percentage of interphase wildtype and mmb1Δ cells with the different mitochondria aggregation phenotypes. (See also Fig. S3)
(C) Time-lapse images of wildtype and mmb1Δ cells expressing cox4-GFP. Both cell types exhibit apparent mitochondria fission (yellow circles) and fusion (red circles). Bar, 5 μm. (See also Movies S3A and S3B)
We next examined the cold-sensitive β-tubulin mutant nda3-311cs [22-24] expressing mCherry-atb2 and cox4-GFP, which has relatively short interphase microtubules at the permissive temperature (30 °C) and no interphase microtubules at the restrictive temperature (16 °C). In the absence of microtubules, nda3-311cs cells showed severed mitochondria aggregation phenotype (Fig. S3), reminiscent of mmb1Δ phenotype (Fig. 3A). We conclude that mmb1p binds mitochondria to microtubules, and that the absence of mmb1p or absence of microtubules lead to similar mitochondria aggregation phenotypes.
Mitochondria fusion and fission are integral functions of the mitochondria network [3, 25]. As we could not easily quantify the frequencies of fission and fusion, particularly in mmb1Δ cells which have aggregated mitochondria, we can not rule out the possibility that mmb1p also plays a role in mitochondria fission and fusion. However, we clearly observed fission and fusion events in mmb1Δ cells (Fig. 3C; Movies S3A and S3B), suggesting that mmb1p does not have an inhibitory role in fusion and fission. We conclude that mmb1p mainly functions in mitochondria positioning.
Mmb1p binds mitochondria to microtubules
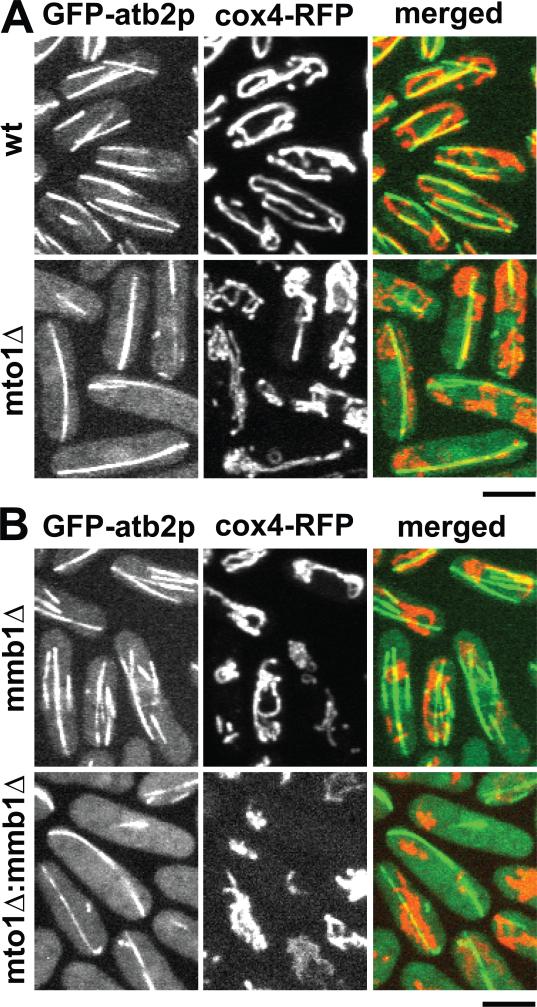
We next tested the model that mmb1p binds mitochondria to microtubules. We reasoned that if mmb1p binds mitochondria to microtubules, then the positions of mitochondria would coincide with the positions of the microtubules, regardless of where the microtubules may be. Wildtype fission yeast cells have 3-4 bundles of interphase microtubules organized by multiple iMTOCs [11]. The mutant mto1Δ fails to organize multiple iMTOCs and thus has only 1 bundle of microtubules [26]. We compared mitochondria distributions in wildtype and mto1Δ cells expressing GFP-atb2 and RFP-cox4. Wildtype cells showed long and tubular mitochondria, and most of which showed colocalization with the multiple linear microtubule bundles (Fig. 4A and Movie S2A and S4A). In mto1Δ cells, we also observed some tubular mitochondria colocalizing with the lone microtubule bundle, while other mitochondria appeared aggregated, presumably because there were not enough microtubules for all mitochondria to bind to (Fig. 4A).
Figure 4. Positive correlation between microtubule and mitochondria distribution.
(A) Images of wildtype and mto1Δ cells expressing GFP-atb2p and cox4-RFP. Mto1Δ cells typically have only one interphase microtubule bundle. The mitochondria distribution correlates with the microtubule bundles. Bar, 5 μm.
(B) Images of mmb1Δ and mto1Δ:mmb1Δ double-deletion mutant cells expressing GFP-atb2p and cox4-RFP. Mmb1Δ does not change the number or organization of the microtubule bundles. Mitochondria are aggregates in mmb1Δ mutants. The mitochondria distribution shows no apparent correlation with the microtubule bundles. Bar, 5 μm. (See also Fig. S4)
In mmb1Δ and mto1Δ:mmb1Δ double deletion cells, the number and organization of microtubule bundles appeared unchanged compared to wildtype and mto1Δ cells, respectively (Fig. 4B), indicating that mmb1p does not affect microtubule organization and architecture. However, in these mmb1Δ and mto1Δ:mmb1Δ cells, the mitochondria failed to distribute correspondingly with the microtubules, but instead aggregated at the cell ends (Fig. 4B and Movie S2B and S4B). We failed to observe long tubular mitochondria which colocalized with the microtubule bundles in mmb1Δ and mto1Δ:mmb1Δ cells, consistent with the model that mmb1p binds mitochondria to microtubules.
As further confirmation of mitochondria-microtubule interaction, we tested the microtubule-bundler mutant ase1Δ [20, 21] and the microtubule length mutant mal3Δ [27]. Ase1Δ cells have un-bundled, but still relatively long interphase microtubules, and thus the mitochondria remained stretched out along the microtubules (Fig. S4). In contrast, mal3Δ cells have short interphase microtubules, and thus the mitochondria appeared as aggregations (Fig. S4).
We note that the various mitochondria aggregation phenotypes of mmb1Δ cells appeared similar to the mitochondria aggregation phenotypes seen in nda3-311cs cells (Fig. S3), mal3Δ cells (Fig. S4), and wildtype cells treated with MBC to depolymerize microtubules (Fig. 2B and Fig. 5C), suggesting that mmb1p functions to bind mitochondria to microtubules, and that defects in microtubule dynamic organization will lead to severe defects in mitochondria positioning.
Figure 5. Microtubule and mitochondria dynamics have different intrinsic timescale.
(A) Time-lapse images of a wildtype cell expressing GFP-atb2p and cox4-RFP. As the microtubule shrinks (white dotted ovals), the attached mitochondrion slowly retracts. As the microtubule grows (yellow dotted ovals), the mitochondrion binds to the microtubule lattice. Bar, 5 μm. (See also Fig. S5 and Movie S4)
(B) Time-lapse images of wildtype cells expressing mCherry-atb2p and cox4-GFP being treated with 25 μg/mL MBC, a microtubule-depolymerizing drug. After +5 min of MBC treatment, the microtubules are mostly depolymerized, but the mitochondria remain mostly extended. Aggregation of mitochondria is strongly apparent only at >20 min after MBC treatment. Bar, 5 μm. (See also Movie S5A)
(C) Time-lapse images of wildtype cells expressing mCherry-atb2p and cox4-GFP first treated for 20 min with 25 μg/mL MBC, then MBC is washed-out at time 0 min. Within 5 min of MBC wash-out, the microtubules are re-polymerized and recover their lengths, but the mitochondria remain aggregated. Mitochondria re-extension is strongly apparent only at >40 min. Bar, 5 μm. (See also Movie S5B)
Mitochondria and microtubule dynamics have different intrinsic timescale
Thus far our results are consistent with the model that mmb1p binds mitochondria to microtubules, and that microtubule dynamics drives mitochondria distribution. Nevertheless, we noted that mitochondria dynamics and microtubule dynamics exhibited very different intrinsic timescale. High-temporal resolution imaging of cells co-expressing GFP-atb2p and cox4-RFP showed that when a microtubule grew, it appeared to attract mmb1p-coupled mitochondria to its lattice (Fig. 5A; Fig. S5 and Movie S4). Mitochondria appeared floppy in regions without microtubules; and the same floppy mitochondrion appeared to straighten out while attaching to the microtubule when a microtubule is re-polymerized next to it (Fig. 5A; Fig. S5 and Movie S4). When a microtubule shrank, it appeared to release the mmb1p-coupled mitochondrion from its lattice (Fig. 5A and Fig. S5 and Movies S3 and S4A), returning the mitochondrion to the previously more floppy state. Within the microtubule growth and shrinkage time, the interacting mitochondrion did not appear to exhibit correlated extension and retraction phases, respectively (Fig. 5A and Fig. S5 and Movies S3 and S4A), e.g., the mitochondria remained extended but floppy as the microtubule shrank. We noted that while a mitochondrion position and extension correlated with the microtubule lattice, the tips of the mitochondrion often did not correlate with the growing or shrinking tips of the microtubules (Fig. 5A; Fig. S5 and Movie S4). This has implications for possible mechanisms of microtubule-dependent mitochondria positioning (see Discussion).
We envision that the microtubules have relatively fast dynamics, growing and shrinking with microns per minute timescale. In contrast, the mitochondria have relatively slow dynamics, extending and retracting its membranous structure with microns per tens of minute timescale. We tested this hypothesis using drug-induced microtubule depolymerization and regrowth experiments. In experiments where microtubule shrinkage was induced by 25 μg/mL MBC, where as the microtubule cytoskeleton disassembled within <5 min of drug treatment, the corresponding mitochondria network took >20 min to retract into aggregates (Fig. 5B; Movie S5A). Similarly, in MBC wash-out experiments, where as the microtubule cytoskeleton re-assembled <5 min after drug removal, the aggregated mitochondria network took >30 min to re-extend fully (Fig. 5C; Movie S5B). We conclude that the intrinsic timescale of microtubule and mitochondria dynamics are different by about one order of magnitude. How does mmb1p couple the seemingly fast microtubule dynamics to the slow mitochondria dynamics?
Mmb1p attenuates microtubule dynamics
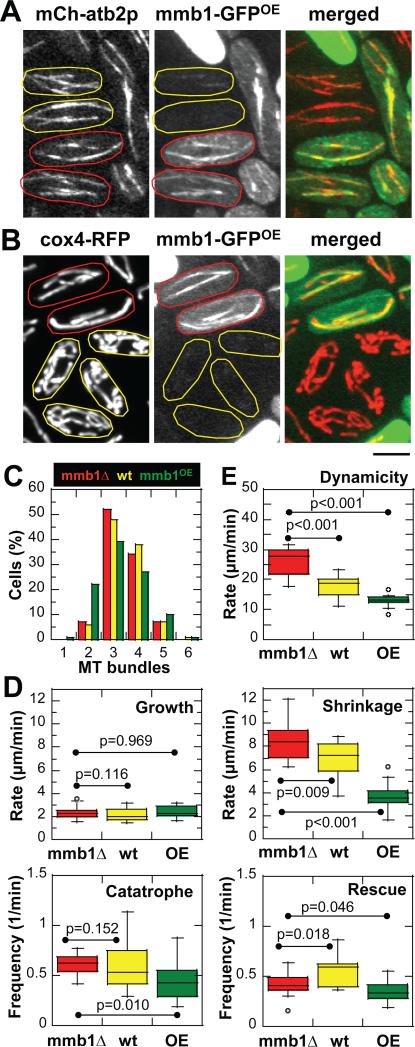
We reasoned that as a microtubule binding protein, mmb1p may alter parameters of microtubule dynamics in such manner as to better facilitate mitochondria-microtubule binding. For example, more stable microtubules would allow longer mitochondriamicrotubule contact time and thus promote mitochondria extension. We measured and compared the four microtubule dynamic parameters: growth rate Vgrowth, shrinkage rate Vshrinkage, catastrophe (switch from growth to shrinkage) frequency Fcatastrophe, and rescue (switch from shrinkage to growth) frequency Frescue [28], in mmb1Δ, wildtype, and mmb1OE (over-expression) cells. Mmb1p over-expression was achieved by transforming mmb1Δ cells with the thiamine-inducible multi-copy plasmid containing mmb1-GFP. As we could not control for plasmid copy number in each transformed cell, we limited our analysis to cells which showed ~2-folds mmb1-GFP over-expression fluorescence signal compared to wildtype cell containing one copy of mmb1-GFP at its endogenous locus. We observed that in some mmb1OE cells, there were more mmb1p binding throughout the microtubule lattice (Fig. 6A), as compared to wildtype cells (see Fig. 1A). Mmb1OE cells also showed more straightened and less floppy mitochondria structure compared to wildtype cells (Fig. 6B), or to mmb1Δ cells (see Fig. 3A). These results are consistent with the interpretation that more mmb1p induces more mitochondria binding to microtubules.
Figure 6. Mmb1p attenuates microtubule dynamics.
(A) Images of mmb1Δ cells expressing mCherry-atb2p and mmb1-GFPOE. Mmb1-GFP over-expression is variable from cell to cell: low expression (yellow), high expression (red). We chose cells with ~2-folds higher mmb1-GFP fluorescence signal compared to wildtype mmb1-GFP cells to analyze microtubule dynamics. Note that over-expressed mmb1p decorates microtubule lattices strongly (red).
(B) Images of mmb1Δ cells expressing cox4-RFP and mmb1-GFPOE. Mmb1-GFP over-expression is variable from cell to cell: low expression (yellow), high expression (red). In cells with higher mmb1-GFP signal (red), the mitochondria appear straightened and stretched, consistent with strong binding to microtubules. In cells with lower mmb1-GFP signal (yellow), the mitochondria appear less stretched, consistent with less binding to microtubules. Bar, 5 μm.
(C) Histogram of interphase microtubule bundle numbers for mmb1Δ, wildtype and mmb1OE cells. Mmb1p does not change the number or organization of interphase microtubules. (D) Box plots showing comparison of microtubule growth and shrinkage rates, and catastrophe and rescue frequencies between mmb1Δ, wildtype and mmb1OE cells. Mmb1p decreases the shrinkage rate of microtubules. P-values are from Mann-Whitney tests of nonparametric data set. (See also Fig. S6 and Table S1)
(E) Box plot showing comparison of microtubule dynamicity between mmb1Δ, wildtype and mmb1OE cells. Mmb1p decreases the dynamicity of microtubules. P-values are from Mann-Whitney tests of nonparametric data set. (see also Table S1)
Microtubule bundle number, organization, and distribution were unchanged in mmb1Δ, wildtype and mmb1OE cells (Fig. 6C), with a typical interphase cell having 3-4 bundles of microtubules. We summarize microtubule dynamic parameters measured for mmb1Δ, wildtype and mmb1OE cells here (Fig. 6D and Table S1). In general, mmb1p did not significantly affect parameters such as microtubule growth rate, catastrophe or rescue frequencies. However, mmb1p appeared to have a strong negative effect on the microtubule shrinkage rate. Microtubule shrinkage rate decreased from 8.34 ± 1.53 μm/min (mmb1Δ), to 6.90 ± 1.53 μm/min (wt), to 3.61 ± 1.00 μm/min (mmb1OE), respectively (Fig. 6D and Table S1). While it remains a challenge to interpret mmb1Δ or mmb1OE and their effects on microtubule dynamics in living cells due to possible pleiotropic effects, our data are consistent with the model that mmb1p binds microtubules and stabilizes the microtubule lattice from shrinkage. Consistent with mmb1p stabilizing microtubules from shrinkage, we observed short microtubule remnants in mmb1OE cells treated briefly with the microtubule-depolymerizing drug MBC (Fig. S6).
One consequence of reduced microtubule shrinkage rate is reduced microtubule dynamicity. Dynamicity has been defined as changes in polymer length, both during growth and shrinkage, per unit time [29]. We define dynamicity rate to take into account all the four parameters of microtubule dynamics mentioned above. Microtubule dynamicity rate decreased from 26.48 ± 6.03 μm/min (mmb1Δ), to 17.68 ± 3.69 μm/min (wt), to 13.13 ± 2.18 μm/min (mmb1OE) (Fig. 6E and Table S1), suggesting that mmb1p attenuates microtubule dynamics and stabilizes microtubules. There is a 17% decrease in microtubule shrinkage rate and a 33% decrease in microtubule dynamicity going from mmb1Δ to wildtype cells with one copy of mmb1-GFP. Mmb1p over-expression decreased microtubule shrinkage rate and dynamicity even further. The consequence of decreased microtubule dynamics and increased microtubule stabilization is an increase in the microtubule polymer lifetime, which would prolong the mitochondria-microtubule contact time and thus may favors mitochondria extension.
Mmb1p promotes proper mitochondria inheritance and cell survival
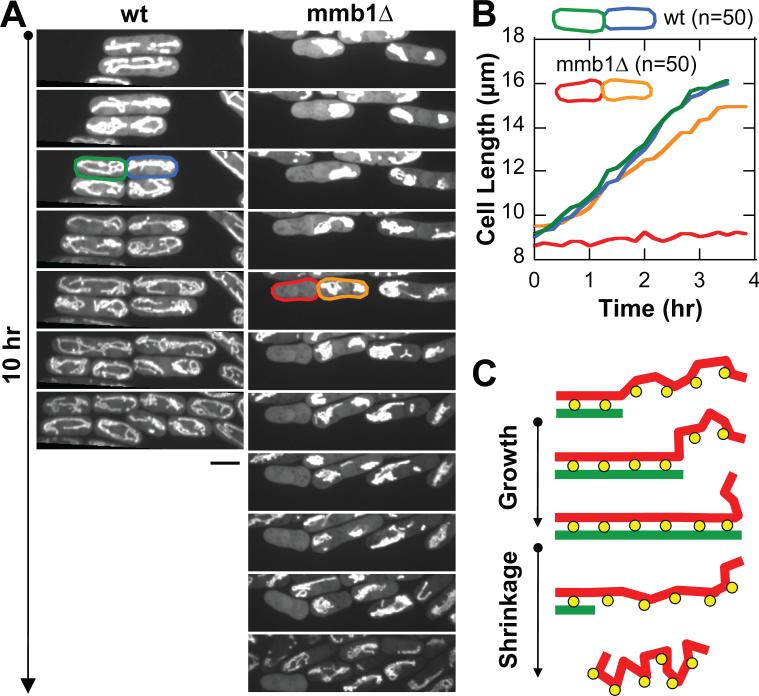
Finally, we examined the consequence of mitochondria positioning defects in mmb1Δ cells. We reasoned that mitochondria aggregates seen at one cell end in ~10% of mmb1Δ cells (Fig. 3B) would result in asymmetric inheritance of mitochondria during cell division, and this would lead to cell survival defects. We examined wildtype and mmb1Δ cells expressing the mitochondria marker cox4-GFP over several cell cycles. Wildtype cells showed symmetric inheritance of mitochondria (Fig. 7A), and symmetric and equal cell growth after successive cell divisions (Fig. 7B). In contrast, mmb1Δ cells which displayed mitochondria aggregates at one cell end exhibited asymmetric mitochondria inheritance during cell division, with one daughter cell receiving no mitochondria (Fig. 7A). Failure to inherit mitochondria occurred in ~10% (N=50) of dividing mmb1Δ cells. Cells with no mitochondria showed no growth (Fig. 7B). We conclude that mmb1p promotes proper mitochondria inheritance and cell survival.
Figure 7. Mmb1p promotes proper mitochondria inheritance and cell survival.
(A) Time-lapse images following several cell cycles of wildtype and mmb1Δ cells expressing cox4-GFP. Approximately 10% of mmb1Δ cells have mitochondria aggregation at one cell end, leading to asymmetric mitochondria inheritance during cell division – one daughter cell has no mitochondria (red) and the other has all the mitochondria (orange). Cells with no mitochondria do not grow. Bar, 5 μm.
(B) Plot of cell growth comparing the wildtype cells (green, blue) and the mmb1Δ cells (red, orange) taken from (A).
(C) A model for mmb1p-mediated (yellow) mitochondria-microtubule binding (red, green). Mmb1p associates tightly to mitochondria. The mitochondria tubular network tends to aggregate over long timescale in the absence of microtubules. The microtubule cytoskeleton, particularly microtubule growth, facilitates weak binding of Mmb1p, leading to continued mitochondria extension. (See also Fig. S7)
Discussion
We presented a new fission yeast protein mmb1p. Mmb1p localizes as dots on both the microtubule lattice and the mitochondria tubular network, and functions as a mitochondria-microtubule binder. In cells lacking mmb1p, the mitochondria no longer appear extended and tubular, but instead appear as aggregates asymmetrically distributed in the cell. The asymmetric positioning of mitochondria can lead to mitochondria inheritance failure during cell division and subsequent cell death.
A mechanism for microtubule-dependent mitochondria positioning by mmb1p
We propose that mmb1p is a mitochondria-microtubule binder (Fig. 7C). Mmb1p has binding affinity for the mitochondria tubular network and the microtubule lattices. As a predicted highly basic protein, mmb1p (pI ~ 14.2) can thus bind directly through ionic interaction with the microtubule lattice, which is highly acidic (pH ~ 4.6). Mmb1p is not predicted to have a membrane binding domain. Thus, its binding to mitochondria may be facilitated by other yet unknown proteins.
One interpretation of our findings is that mmb1p passively binds mitochondria to microtubules. However, the fact that mmb1p attenuates microtubule dynamics and enhances microtubule stability suggests a model where it helps prolong the binding of mitochondria to microtubules. As a membranous tubular network, mitochondria would tend toward retraction and aggregation in the absence of forces which help extend the mitochondria. Microtubules serve to extend mitochondria. When a microtubule grows toward the cell tip, mmb1p, which is bound to a mitochondrion, attaches itself to the lattice of the growing microtubule, and thus acts as a ratchet to prevent the mitochondrion from retracting and aggregating. Further, mmb1p also reduces the microtubule shrinkage rate and attenuates microtubule dynamicity. This would directly result in a longer lifetime for the microtubule, which would further enforce and maintain the binding of mitochondria to microtubules. When a microtubule completely depolymerizes, which happens in tens of seconds, the mitochondrion begins to retract and aggregate. However, mitochondria retraction and aggregation occur at longer timescale, typically observed in tens of minutes. Thus the effect of mitochondria retraction is not immediately apparent after microtubule shrinkage. A new replacement microtubule will grow to act as a new scaffold for a mitochondrion to bind and extend, before it has time to completely retract and aggregate.
But how does an aggregated mitochondrion become extended in the first place? Mmb1p attenuates microtubule shrinkage rate and dynamicity. This, of course, enhances the mitochondria-microtubule contact time. However, mmb1p has to balance its microtubule dynamic suppression activity, without suppressing microtubule dynamics too much to the point of causing adverse pleiotropic effects on cells such as cell polarity or cell division which depend on microtubules. In the context of mitochondria-microtubule binding, by keeping growth rate, catastrophe and rescue frequencies relatively intact, and by slightly suppressing shrinkage rate, mmb1p enables many repeated events of microtubule growth over a long period of time. This helps push and extend the mitochondrion. Thus, mmb1p couples the long timescale mitochondria dynamics to the short timescale of microtubule dynamics. Our model predicts that any mutation that affects microtubule number or length or dynamics would directly lead to mitochondria positioning defects.
Different models for mitochondria positioning in fission yeast
Fission yeast uses the microtubule cytoskeleton to position its mitochondria throughout the cell cycle [7, 12]. However, unlike higher eukaryotes, which can use motor-mediated transport of mitochondria on microtubules [30-38], fission yeast has not been reported to use motors for mitochondria positioning. It was previously reported that the conserved CLASP family of microtubule +TIP protein peg1p (also called cls1p) couples the mitochondria tubular tips to the plus ends of microtubules in fission yeast [15]. Thus, plus end microtubule growth would extend the mitochondria, providing a mechanism for microtubule-dependent mitochondria positioning [15]. This model implies that a mitochondrion would be extended at the same rate as a microtubule grows.
Our current work favors a different model (Fig. 7C). We failed to observed direct interaction or coupling between the tips of mitochondria and the microtubule plus ends (Fig. 5A and Fig. S5). Instead, we observed mmb1p-dependent binding of mitochondria to the lattices of microtubules. Mmb1p attenuates microtubule dynamics, specifically decreasing the microtubule shrinkage rate, and thus maintains longer mitochondriamicrotubule interaction. Integrated over long timescale, the slow mitochondria dynamics can be coupled to the fast microtubule dynamics.
In an attempt to reconcile the differences between these two models, we reexamined to role of peg1p in coupling mitochondria to microtubule tips (Fig. S7). We failed to confirm the reported [15] colocalization of peg1p to the mitochondrion tip and the microtubule plus end (Fig. S7). Our results suggest that peg1p plays no direct role in binding mitochondria to the growing microtubule tips. However, we cannot rule out that peg1p may play an indirect role in mitochondria positioning: by modifying microtubule dynamic parameters in the short term [39], peg1p may influence mitochondria distribution in the long term.
Different mechanisms for mitochondria positioning
To date, two general mechanisms have been proposed for mitochondria positioning. Budding yeast, Aspergillus, and plants use the actin cytoskeleton and actin polymerization dynamics to position their mitochondria [4]. C. elegans, Drosophila, and mammalian neuronal cells use the microtubule cytoskeleton and motors such as kinesin or dynein to position their mitochondria [1, 2]. Interestingly, in neurons the majority of mitochondria are immobile. There, the protein syntaphilin acts to bind mitochondria to microtubules in a stationary manner [8], perhaps through inhibition of dynein-dependent mitochondria movement [40]. Thus, mitochondria positioning includes either mitochondria transport to the proper cellular position or mitochondria stably maintained at specific cellular position.
A mitochondria microtubule binder such as mmb1p would not transport mitochondria on microtubule in the same way that kinesin or dynein would. Instead, mmb1p actively attenuates microtubule dynamicity, therefore making microtubules more stable and increasing the microtubule lifetime and prolonging the mitochondriamicrotubule contact time, and thus maintain mitochondria extension. Mmb1p, when binding mitochondria to microtubules, would acts like a ratchet to prevent the mitochondria from retraction and aggregation. Fission yeast mitochondria positioning is microtubule-dependent but motor-independent [7, 12]. Thus, to the best of our knowledge, mmb1p may be the first reported protein to function in microtubule-dependent but motor-independent mitochondria positioning, and may represent another general mechanism for mitochondria positioning and inheritance. This mechanism may be important for non-neuronal cells which are relatively small in size, and therefore do not need to move their mitochondria long distances, but only need to properly extend their mitochondria.
Supplementary Material
Highlights.
We report a new protein mmb1p in fission yeast.
Mmb1p binds mitochondria to dynamic microtubules.
The absence of mmb1p leads to mitochondria aggregation and segregation defects.
Mmb1p may be motor-independent mechanism for mitochondria distribution.
Acknowledgements
C.F., D.J., J.C., and G. V-C. created tools and reagents and performed experiments. C.F. and P.T.T. analyzed the data and wrote the paper. We thank F. Chang (Columbia University), F. Chiron (UCSD), A. Paoletti (Institute Curie), M. Sato (Tokyo University), T. Toda (CRUK), and M.P. Yaffe (UCSD), for kindly providing reagents. We thank L Pon (Columbia University) and A. Paoletti (Institut Curie) for helpful discussions. We thank members of the labs of E. Bi (Penn) and P.T. Tran (Penn) for helpful discussions. J.C. is supported by a PhD fellowship from the FCT through Complexite du Vivant, UPMC. This work is supported by grants from NIH, ACS, ANR, FRM, LaLigue and HFSP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Experimental Procedures
Detail description of experimental procedures is described in the Supplemental Information section.
References
- 1.Boldogh IR, Pon LA. Mitochondria on the move. Trends Cell Biol. 2007;17:502–510. doi: 10.1016/j.tcb.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Frederick RL, Shaw JM. Moving mitochondria: establishing distribution of an essential organelle. Traffic. 2007;8:1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 4.Boldogh IR, Pon LA. Interactions of mitochondria with the actin cytoskeleton. Biochim Biophys Acta. 2006;1763:450–462. doi: 10.1016/j.bbamcr.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Hollenbeck PJ, Saxton WM. The axonal transport of mitochondria. J Cell Sci. 2005;118:5411–5419. doi: 10.1242/jcs.02745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo GJ, Louie K, Wellington A, Macleod GT, Hu F, Panchumarthi S, Zinsmaier KE. Drosophila Miro is required for both anterograde and retrograde axonal mitochondrial transport. J Neurosci. 2009;29:5443–5455. doi: 10.1523/JNEUROSCI.5417-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaffe MP, Stuurman N, Vale RD. Mitochondrial positioning in fission yeast is driven by association with dynamic microtubules and mitotic spindle poles. Proc Natl Acad Sci U S A. 2003;100:11424–11428. doi: 10.1073/pnas.1534703100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang JS, Tian JH, Pan PY, Zald P, Li C, Deng C, Sheng ZH. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner MK, Hunt AJ, Goodson HV, Odde DJ. Microtubule assembly dynamics: new insights at the nanoscale. Curr Opin Cell Biol. 2008;20:64–70. doi: 10.1016/j.ceb.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 11.Sawin KE, Tran PT. Cytoplasmic microtubule organization in fission yeast. Yeast. 2006;23:1001–1014. doi: 10.1002/yea.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaffe MP, Harata D, Verde F, Eddison M, Toda T, Nurse P. Microtubules mediate mitochondrial distribution in fission yeast. Proc Natl Acad Sci U S A. 1996;93:11664–11668. doi: 10.1073/pnas.93.21.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanbe T, Kobayashi I, Tanaka K. Dynamics of cytoplasmic organelles in the cell cycle of the fission yeast Schizosaccharomyces pombe: three-dimensional reconstruction from serial sections. J Cell Sci. 1989;94(Pt 4):647–656. doi: 10.1242/jcs.94.4.647. [DOI] [PubMed] [Google Scholar]
- 14.Hoog JL, Schwartz C, Noon AT, O'Toole ET, Mastronarde DN, McIntosh JR, Antony C. Organization of interphase microtubules in fission yeast analyzed by electron tomography. Dev Cell. 2007;12:349–361. doi: 10.1016/j.devcel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 15.Chiron S, Bobkova A, Zhou H, Yaffe MP. CLASP regulates mitochondrial distribution in Schizosaccharomyces pombe. J Cell Biol. 2008;182:41–49. doi: 10.1083/jcb.200712147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding DQ, Tomita Y, Yamamoto A, Chikashige Y, Haraguchi T, Hiraoka Y. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells. 2000;5:169–190. doi: 10.1046/j.1365-2443.2000.00317.x. [DOI] [PubMed] [Google Scholar]
- 17.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 18.Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, Smith GR, Moreno S. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol. 2005;15:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jourdain I, Gachet Y, Hyams JS. The dynamin related protein Dnm1 fragments mitochondria in a microtubule-dependent manner during the fission yeast cell cycle. Cell Motil Cytoskeleton. 2009;66:509–523. doi: 10.1002/cm.20351. [DOI] [PubMed] [Google Scholar]
- 20.Loiodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita A, Sato M, Fujita A, Yamamoto M, Toda T. The roles of fission yeast ase1 in mitotic cell division, meiotic nuclear oscillation, and cytokinesis checkpoint signaling. Mol Biol Cell. 2005;16:1378–1395. doi: 10.1091/mbc.E04-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- 23.Toda T, Umesono K, Hirata A, Yanagida M. Cold-sensitive nuclear division arrest mutants of the fission yeast Schizosaccharomyces pombe. J Mol Biol. 1983;168:251–270. doi: 10.1016/s0022-2836(83)80017-5. [DOI] [PubMed] [Google Scholar]
- 24.Umesono K, Toda T, Hayashi S, Yanagida M. Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J Mol Biol. 1983;168:271–284. doi: 10.1016/s0022-2836(83)80018-7. [DOI] [PubMed] [Google Scholar]
- 25.Shaw JM, Nunnari J. Mitochondrial dynamics and division in budding yeast. Trends Cell Biol. 2002;12:178–184. doi: 10.1016/s0962-8924(01)02246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawin KE, Lourenco PC, Snaith HA. Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr Biol. 2004;14:763–775. doi: 10.1016/j.cub.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 27.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Toso RJ, Jordan MA, Farrell KW, Matsumoto B, Wilson L. Kinetic stabilization of microtubule dynamic instability in vitro by vinblastine. Biochemistry. 1993;32:1285–1293. doi: 10.1021/bi00056a013. [DOI] [PubMed] [Google Scholar]
- 30.Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end-directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 31.Khodjakov A, Lizunova EM, Minin AA, Koonce MP, Gyoeva FK. A specific light chain of kinesin associates with mitochondria in cultured cells. Mol Biol Cell. 1998;9:333–343. doi: 10.1091/mbc.9.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tanaka Y, Kanai Y, Okada Y, Nonaka S, Takeda S, Harada A, Hirokawa N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- 33.Gorska-Andrzejak J, Stowers RS, Borycz J, Kostyleva R, Schwarz TL, Meinertzhagen IA. Mitochondria are redistributed in Drosophila photoreceptors lacking milton, a kinesin-associated protein. J Comp Neurol. 2003;463:372–388. doi: 10.1002/cne.10750. [DOI] [PubMed] [Google Scholar]
- 34.Ni CZ, Wang HQ, Xu T, Qu Z, Liu GQ. AtKP1, a kinesin-like protein, mainly localizes to mitochondria in Arabidopsis thaliana. Cell Res. 2005;15:725–733. doi: 10.1038/sj.cr.7290342. [DOI] [PubMed] [Google Scholar]
- 35.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pilling AD, Horiuchi D, Lively CM, Saxton WM. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol Biol Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horiuchi D, Barkus RV, Pilling AD, Gassman A, Saxton WM. APLIP1, a kinesin binding JIP-1/JNK scaffold protein, influences the axonal transport of both vesicles and mitochondria in Drosophila. Curr Biol. 2005;15:2137–2141. doi: 10.1016/j.cub.2005.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita T, Maturana AD, Ikuta J, Hamada J, Walchli S, Suzuki T, Sawa H, Wooten MW, Okajima T, Tatematsu K, Tanizawa K, Kuroda S. Axonal guidance protein FEZ1 associates with tubulin and kinesin motor protein to transport mitochondria in neurites of NGF-stimulated PC12 cells. Biochem Biophys Res Commun. 2007;361:605–610. doi: 10.1016/j.bbrc.2007.07.050. [DOI] [PubMed] [Google Scholar]
- 39.Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen YM, Gerwin C, Sheng ZH. Dynein light chain LC8 regulates syntaphilin-mediated mitochondrial docking in axons. J Neurosci. 2009;29:9429–9438. doi: 10.1523/JNEUROSCI.1472-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.