Abstract
Small-molecule BET inhibitors interfere with the epigenetic interactions between acetylated histones and the bromodomains of the BET family proteins, including BRD4, and they potently inhibit growth of malignant cells by targeting cancer-promoting genes. BRD4 interacts with the pause-release factor P-TEFb, and has been proposed to release Pol II from promoter-proximal pausing. We show that BRD4 occupied widespread genomic regions in mouse cells, and directly stimulated elongation of both protein-coding transcripts and non-coding enhancer RNAs (eRNAs), dependent on the function of bromodomains. BRD4 interacted physically with elongating Pol II complexes, and assisted Pol II progression through hyper-acetylated nucleosomes by interacting with acetylated histones via bromodomains. On active enhancers, the BET inhibitor JQ1 antagonized BRD4-associated eRNA synthesis. Thus, BRD4 is involved in multiple steps of the transcription hierarchy, primarily by assisting transcript elongation both at enhancers and on gene bodies.
INTRODUCTION
Epigenetic marks on histones are associated with transcriptional processes. For example, trimethylated histone H3 lysine 4 (H3K4me3) is enriched at promoters1, and monomethylated H3 lysine 4 (H3K4me1) and acetylated H3 lysine 27 (H3K27Ac) are enriched at active enhancers2,3. Also, active genes are generally associated with acetylation of H3 and H4 on gene bodies1,4. Because some epigenetically marked histones physically interact with specific protein modules, such epigenetic interactions are hypothesized to participate in the regulation of transcription. Accordingly, synthetic small molecules mimicking epigenetic marks have been developed and have provided a powerful means to investigate the fundamental roles of epigenetic interactions in physiological and pathological processes of transcription.
The bromodomain and extraterminal domain (BET) family proteins5, including BRD2, BRD3, BRD4 and BRDT, contain two bromodomains (BDs)6, which interact with acetylated histones7,8 and other acetylated proteins9,10 with varying degrees of affinity. In cells, the BDs are proposed to play a role in recruiting BET family proteins to hyperacetylated chromatin through the interaction with acetylated histones7,11,12. Small-molecule BET inhibitors such as JQ1 (ref. 13) and I-BET14 mimic the acetyl moiety and occlude the acetyl-lysine binding pocket of the BD unique to the BET family proteins. Thus, BET inhibitors are highly specific for the BET family proteins. BET inhibitors potently inhibit growth of malignant cells by reducing expression of oncogenes such as Myc15–17 and Fosl1 (ref. 18). The growing list of cancers that are effectively inhibited by BET inhibitors includes multiple myeloma15, acute myeloid leukemia16, mixed lineage leukemia17, diffuse large B cell lymphoma19,20, NUT midline carcinoma13, lung adenocarcinoma18, and prostate cancer10. Also, BET inhibitors affect immune cell functions14,21 and spermatogenesis22, and suppress cardiac hypertrophy23.
The aim of the present study was to address the role of BRD4 in epigenetic regulation of gene expression. Generally, gene transcription is regulated at the initiation24 and elongation steps25,26. Transcription initiation begins with the assembly of the preinitiation complex (PIC) on the promoter, and is followed by serine 5 phosphorylation (Ser5P) of the RNA polymerase II (Pol II) C-terminal domain (CTD) and by 5’ capping of nascent RNA27. The frequency of PIC assembly is controlled by enhancers. After initiation, a fraction of Pol II is paused within 100 bp downstream of the transcription start sites (TSS) by the actions of the negative elongation factor (NELF) and DRB-sensitivity inducing factor (DSIF)26,28. To transition to productive elongation, promoter-proximal pausing must be released by the positive elongation factor b (P-TEFb), which phosphorylates NELF and DSIF as well as serine 2 (Ser2) of Pol II CTD27,29. Pol II CTD with Ser2P and Ser5P constitutes a binding platform for proteins involved in pre-mRNA processing and histone modifications27. P-TEFb can be recruited to the promoter-proximal region by multiple factors, including the super elongation complex30,31, CDK8 (ref 32), Myc26,33, NF-κB34, and BRD4 (refs. 35–37). In addition to promoter-proximal pausing of Pol II on a DNA template, the passage of elongating Pol II along the gene body is blocked by nucleosomes in vivo38. Factors which can overcome the nucleosomal barriers include histone chaperones and ATP-dependent chromatin remodelers38. For example, the FACT (Facilitates Chromatin Transcription) complex binds a H2A–H2B dimer, and disassemble and reassemble it temporarily during the passage of Pol II without using the energy of ATP hydrolysis39,40.
Recently, developmental or tissue specific enhancers41,42 have been proposed to make strong, long-range interactions with promoters to greatly enhance PIC assembly43. Enhancer regions are also transcribed by Pol II to produce non-coding enhancer RNAs (eRNAs)44–48. Accumulating evidence suggests that eRNAs themselves may participate in enhancer function45,46,48, creating a hierarchical cascade of transcript-mediated regulation of transcription.
Among ubiquitously expressed BET family proteins, BRD4 is unique in that it interacts with P-TEFb through its C-terminal tail37. BRD4 is thought to recruit P-TEFb to hyperacetylated genomic regions. For example, during induction of interferon stimulated genes, BRD4 helps recruit P-TEFb to hyper-acetylated TSSs49. However, the TSS regions of active genes are frequently nucleosome-depleted50,51, suggesting the presence of an alternative mechanism to recruit BRD4 and P-TEFb to TSSs. BRD4 is also reported to accumulate at enhancer elements52, particularly at clustered enhancers (super-enhancers) in cancer cells53, together with Mediator complexes53, which are known to interact with BRD4 (refs. 32,35,54). Some BRD4-associated enhancers have been proposed to provide an anti-pause function for P-TEFb on promoter-proximal regions via a long-range enhancer-promoter interaction55.
In the present study, we addressed the roles of BRD4 and its bromodomains in epigenetic regulation of gene expression. We show that BRD4 acted as an elongation factor for Pol II as it traverses through acetylated chromatin. BRD4 occupied widespread genomic regions, and assisted elongation of both protein-coding transcripts and eRNAs, in a manner dependent on epigenetic interactions between the BD and acetylated histones. The BET inhibitor JQ1 antagonized BRD4-associated eRNA synthesis. Thus, the study suggests that BRD4 is involved in multiple steps of the transcription hierarchy primarily by assisting transcript elongation both at enhancers and on gene bodies.
RESULTS
JQ1 inhibited initiation and elongation of coding RNA
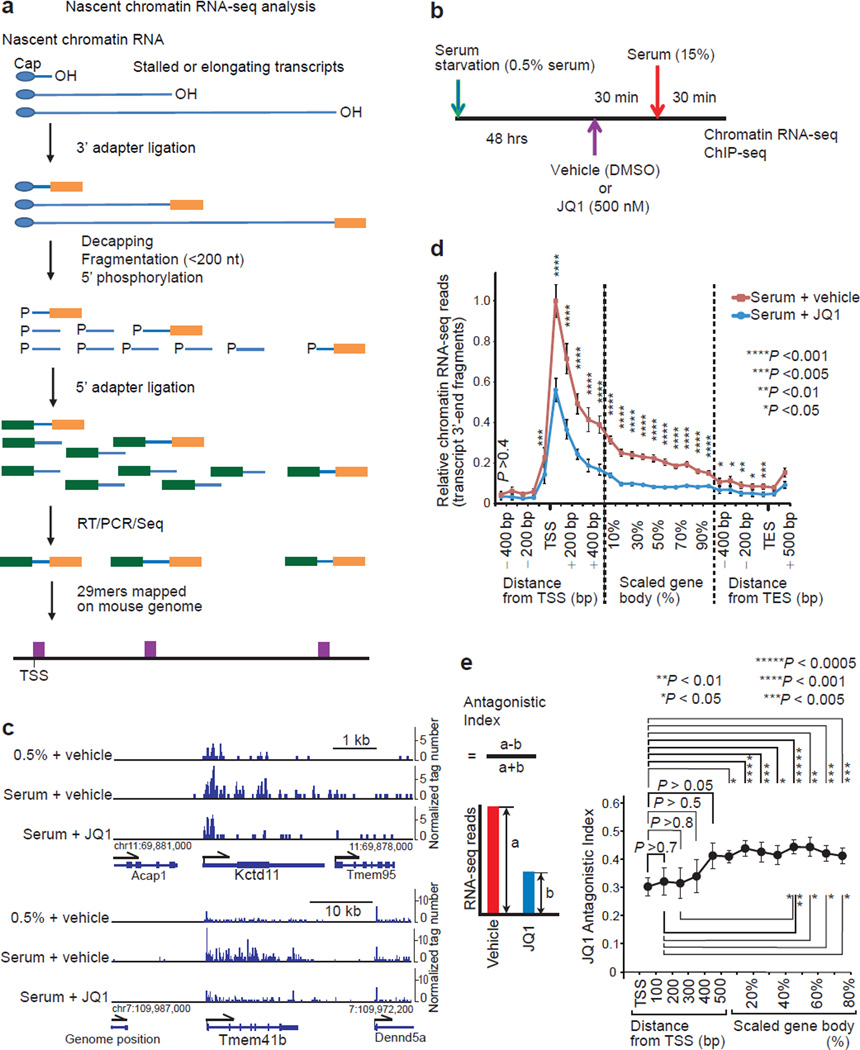
Transcription is regulated both at the Pol II recruitment and elongation steps. We employed the BET inhibitor JQ1 to analyze how BRD4 contributes to the distinct transcriptional processes. Mouse fibroblasts were synchronized by serum-starvation and then re-fed so that the transcriptional initiation of serum-response genes could be easily and uniformly captured by genome-wide sequencing of the 3’-end regions of nascent RNA (Fig. 1a, 1b, Supplementary Fig. 1). JQ1 exhibited a spectrum of inhibition, ranging from a predominant inhibition of elongation with little effect on initiation (Fig. 1c, Kctd11) to a more general inhibition (Fig. 1c, Tmem41b). Should JQ1 predominantly inhibit the pause-release action of P-TEFb without affecting the assembly of PIC, it would be expected to increase the amount of paused transcripts near the TSS. Contrary to this prediction, metagene profiles of serum-inducible and JQ-1 sensitive genes show that JQ1 reduced transcript levels throughout genes including the TSS regions (Fig. 1d). To further dissect the effect of JQ1 at different segments of a gene, the antagonistic index as defined in Fig. 1e (the difference divided by the sum; devised to avoid dividing by zero) was calculated. The index increased from 0.30 near TSS (corresponding to 46% inhibition) to 0.44 (corresponding to 61% inhibition) in the mid gene-body (Fig. 1e). Thus JQ1 antagonized both of a process regulating PIC assembly and a downstream process involved in progressive elongation rather than a typical pause-release event occurring within 100 bp downstream of TSS28.
Figure 1. BET inhibitor JQ1 inhibited both initiation and elongation of protein-coding RNA.
a, Workflow for strand-specific, nascent chromatin RNA-seq sample preparation. The 29mer sequence of the most 3’ fragments of chromatin-bound transcripts was mapped on the genome sequence. b, Protocol for serum starvation and re-stimulation of NIH3T3 cells. c, Genome browser views of chromatin-bound nascent RNA-seq reads of representative serum response genes in NIH3T3 cells. Only sense-strand signals are shown. (d, e) Metagene analysis of chromatin RNA-seq reads of serum-inducible and JQ1-sensitive genes larger than 2 kb (n = 66 genes). Representative of two independent experiments. d, Metagene profiles of RNA-seq reads. Gene body regions from 500 nt downstream of transcription start site (TSS) to 500 nt upstream of transcription end site (TES) were proportionally scaled. For each gene, the RNA read-count of each metaprofile segment (100 base bin around TSS and TES, and 10% bin in the scaled gene body region) was normalized with the total read-count of the gene (from 200 nt upstream of TSS to TES) in serum+vehicle cells. For each metaprofile segment, the two-tailed paired t-test (serum+vehicle vs. serum+JQ1) was performed. Error bars, s.e.m. e, The JQ1 antagonistic index as defined in the inset was calculated on each metagene-profile segment of each gene, and the index (mean ± s.e.m.) is plotted against the segment position. The two-tailed paired t-test was performed between segments as indicated.
JQ1 antagonized BRD4 and inhibited eRNA synthesis
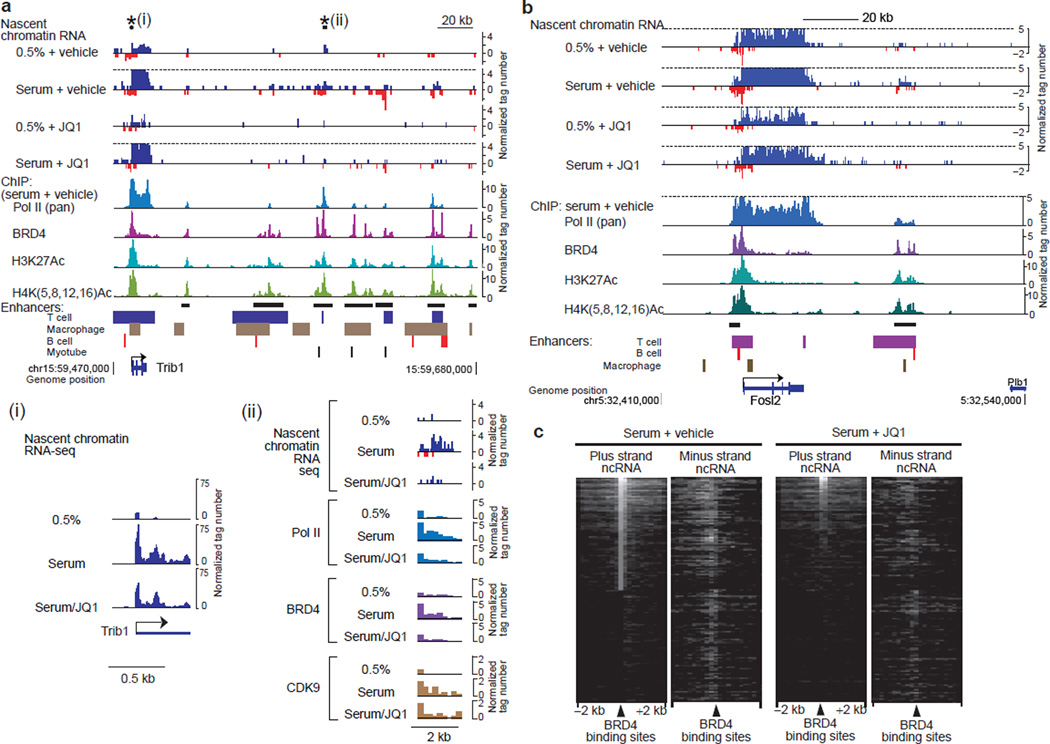
To understand the basis of the reduced transcription initiation of protein-coding genes by JQ1, we evaluated the role of BRD4 in enhancer activity. Intergenic regions flanking serum-inducible genes were found enriched for BRD4 and H3K27Ac, a marker for active enhancers, and enhancer elements characterized in multiple mouse41,42 and human53 cell types (Fig. 2a, 2b, Supplementary Fig. 1c). These enhancer regions centered around BRD4 peaks were transcribed into non-coding eRNAs, which were bidirectional, and inhibited by JQ1 in a fashion similar to the transcripts of nearby protein-coding genes (Fig. 2a-2c, Supplementary Fig. 1b, 1c). These observations indicate that, in addition to protein-coding genes, BRD4 regulates transcription also of eRNAs, which have been recently reported as crucial for enhancer activity and gene expression44–48. We further mapped and correlated the genomic distributions of Pol II (pan Pol II, as an indirect marker for eRNA synthesis) and CDK9 (the kinase component of P-TEFb) as well as BRD4 at intergenic regions (Figs. 2,3, Supplementary Fig. 1). JQ1 treatment mostly inhibited elongation of the noncoding transcripts without affecting acetylation of H3K27 and H4 (Supplementary Fig. 1b). Progressions of Pol II and BRD4 were reduced by JQ1, but that of CDK9 was less affected (Fig. 2a, ii). While JQ1 reduced the number of intergenic regions associated with BRD4, especially those associated with both BRD4 and Pol II, CDK9-associated regions was mostly unaffected (Fig. 3a). Overall, Pol II recruitment correlated significantly with BRD4 but not with CDK9 (Fig. 3b, Supplementary Fig. 2). Moreover JQ1 abolished the correlation between Pol II and BRD4 (Fig. 3b). Consistent with this, Wilcoxon’s rank score analysis indicated that the regions associated with both Pol II and BRD4 had significantly higher Pol II read-counts than those without BRD4 (Fig. 3c). These results suggest that BRD4 aids the synthesis of eRNA.
Figure 2. BET inhibitor JQ1 antagonized BRD4 and inhibits eRNA synthesis.
a, Genome browser views of chromatin-bound RNA-seq reads along the Trib1 gene and its downstream intergenic region. Sense- and antisense-strand signals are shown in upward and downward directions, respectively. Signals higher than the dotted lines are cut. ChIP-seq reads of Pol II, BRD4, H3K27Ac and H4Ac in serum+vehicle cells and known enhancer regions42 are aligned below, and potential enhancer clusters are marked by horizontal black bars. The regions marked with asterisks are shown expanded in insets (i) and (ii). Inset (i) shows rescaled views of nascent RNA-seq reads around the TSS. Inset (ii) shows expanded views of nascent RNA-seq reads and ChIP-seq reads of Pol II, BRD4 and CDK9. b, Genome browser views along the Fosl2 gene and the flanking regions. c, Heat map of chromatin-bound non-coding RNA-seq reads around intergenic BRD4 binding sites. ncRNA: non-coding RNA.
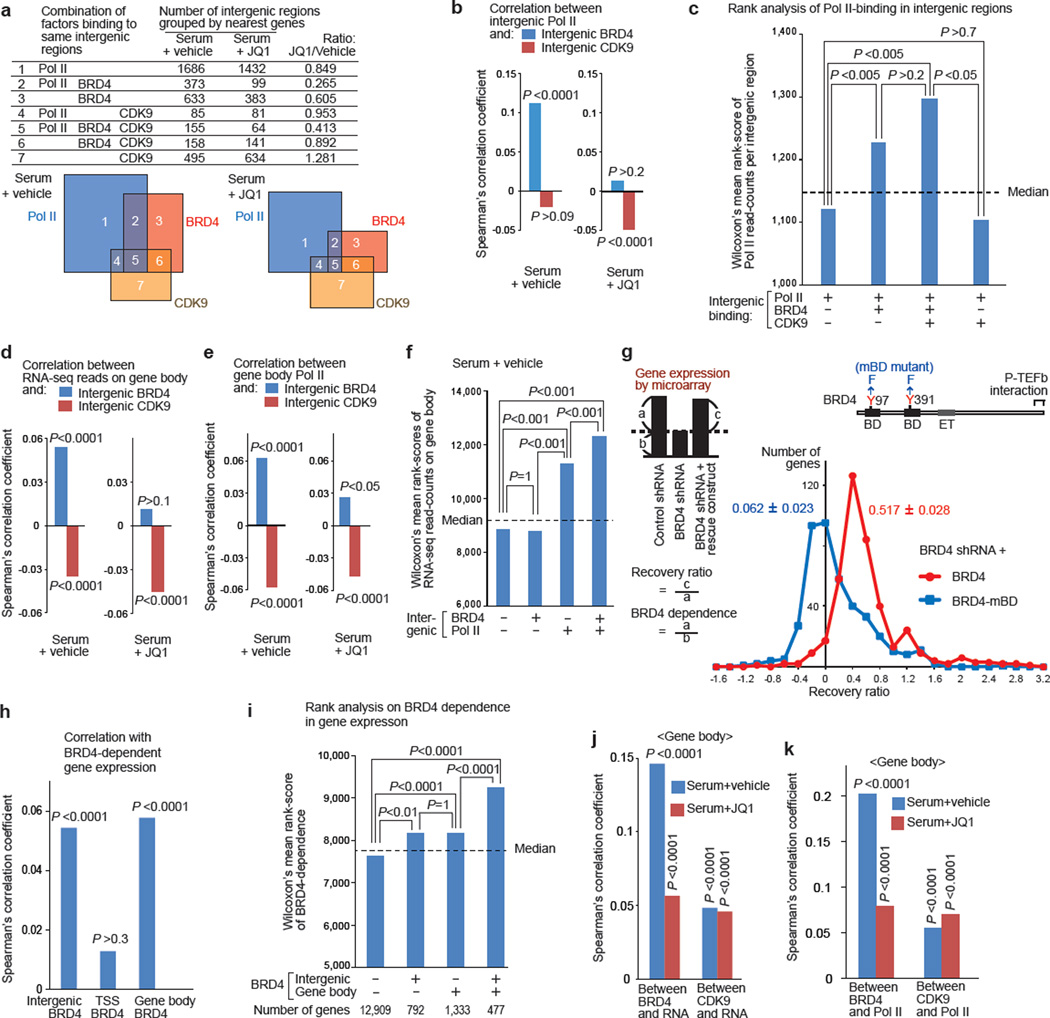
Figure 3. BRD4-dependent gene expression correlated with BRD4 enrichment in intergenic regions and on gene bodies, but not that at TSSs.
a, Combinations of Pol II (pan), BRD4, and CDK9 bindings in intergenic regions grouped by the nearest genes. The areas in Venn diagrams are proportional to the numbers listed. (b, d, e, j, k), Spearman’s rank correlation analysis: between intergenic BRD4 or CDK9 and intergenic Pol II (b); between intergenic BRD4 or CDK9 and gene-body chromatin RNA-seq reads (d) or Pol II (e); between BRD4 enrichment at distinct sites and the BRD4-dependence in gene expression as defined in g (h); and between BRD4 or CDK9 read-counts on gene bodies and chromatin RNA-seq read-counts (j) or Pol II read-counts (k). P-values represent the significance of correlation coefficients. (c, f, h, I), , Wilcoxon’s mean rank scores of Pol II read-counts in intergenic regions (c), chromatin RNA-seq read-counts on gene body (f), and BRD4-dependent gene expression (i), compared among the gene-classes as defined. Dotted line represents the median rank value, and P values were calculated using Dunn’s all-pair comparison method. g, Microarray analysis of gene expression in BRD4 knockdown cells reconstituted with a rescue construct (YFP-BRD4 or YFP-BRD4-mBD mutant). The distribution of the recovery ratio (RR) of genes repressed by BRD4 shRNA by more than 1.5 fold (n=410) is shown as the number of genes (y-axis) plotted against the RR (x-axis, binned by 0.2). Representative of two independent experiments, in which two independent cell lines per indicated transduction combination were analyzed.
We further investigated the contribution of intergenic BRD4(+) and CDK9(+) regions on the expression of nearby genes. Overall, the intergenic BRD4 enrichment significantly and positively correlated, in a JQ1 sensitive manner, with RNA-seq read-counts (Fig. 3d, Supplementary Fig. 2) and Pol II counts (Fig. 3e, Supplementary Fig. 2) of nearby protein-coding genes. Surprisingly, intergenic CDK9 enrichment inversely correlated with RNA-seq and Pol II counts (Fig. 3d, 3e, Supplementary Fig. 2). While the sole presence of BRD4 at intergenic regions was not associated with increased transcription of nearby genes, its co-recruitment with Pol II significantly boosted transcription (Fig. 3f). Thus we found a statistical correlation between intergenic BRD4 and nearby gene expression, which might be mediated through enhanced synthesis of eRNA.
Inter- and intragenic BRD4 correlated with gene expression
To specifically address the role of BRD4-BD interaction with acetylated histones, we constructed a mutant BRD4 (“BRD4-mBD” carrying a point mutation in each BD) which no longer binds acetylated histones effectively in vivo as tested by fluorescent resonance energy transfer (FRET) analysis7 (Supplementary Fig. 3). We first knocked down endogenous BRD4 by shRNA, and then stably reconstituted the cells with shRNA-resistant YFP-BRD4 (wild type or mBD mutant; Supplementary Fig. 4). By microarray gene expression analysis of continuously growing cells, we identified 410 BRD4-dependent annotated genes (down-regulated by at least 1.5 fold by BRD4 knockdown). Following BRD4 reconstitution, the degree of recovery in gene expression was calculated as the recovery ratio (RR) (Fig. 3g). While wild-type BRD4 restored gene expression by a mean RR of 0.517 ± 0.028 (red line), the BD-mutated BRD4 was far less efficient (RR, 0.062 ± 0.023, blue line; by a paired t-test, two-tail P = 3.4×10−35, n = 410 genes, Supplementary Fig. 4e. Representative of two sets of independently reconstituted cells.). BRD4-dependent genes included those involved in the cell cycle, transcription, DNA repair, phosphorylation and other functions, as indicated by GO annotation (Supplementary Fig. 4e).
ChIP-Seq analysis of YFP-BRD4 occupancy in continuously growing cells yielded 37,178 peaks: 28% of which resided at TSS regions, 34% on gene bodies and 38% in intergenic regions. The BRD4-dependent gene expression defined in Fig. 3g showed significant correlations with intergenic- and gene body-bound BRD4, but not with TSS-bound BRD4 (Fig. 3h, Supplementary Fig. 2). This suggests that a large fraction of BRD4 present around the TSS may not be directly engaged in transcriptional initiation or early elongation. Furthermore, the intergenic and gene-body BRD4 enrichment correlated independently with BRD4-dependent gene expression (Fig. 3i). Moreover, in serum re-stimulated cells, BRD4 on gene bodies correlated, in a JQ1-sensitive manner, with RNA-seq (Fig. 3j, Supplementary Fig. 2) or Pol II read-counts (Fig. 3k, Supplementary Fig. 2) on gene bodies. In contrast, the weaker correlation between gene-body CDK9 and RNA-seq or Pol II read-counts on gene bodies was not sensitive to JQ1 (Fig. 3j, 3k, Supplementary Fig. 2), nor did gene-body CDK9 correlate significantly with BRD4-dependent gene expression in continuously growing cells (Spearman’s correlation efficient, 0.0069; P = 0.32). Thus, BRD4 on gene bodies may play a direct role to facilitate transcript elongation in a manner dependent on its interaction with acetylated histones (and possibly also with other acetylated proteins) but not with CDK9.
BRD4 BD was not required for loading BRD4 and P-TEFb at TSS
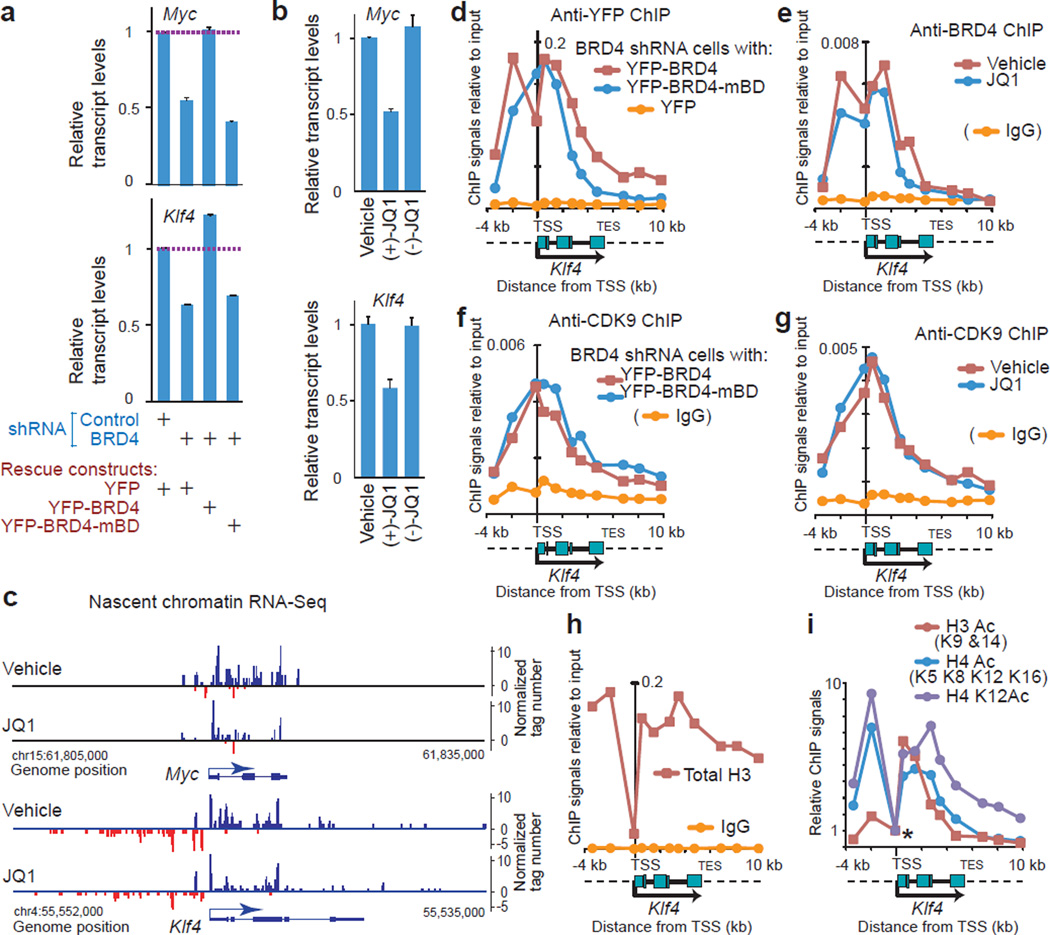
Next we selected representative BRD4-dependent genes, Myc and Klf4, and tested the role of the bromodomains in transcript elongation. BRD4 knockdown or JQ1 treatment under a continuous growth condition reduced total transcript levels of Myc and Klf4 by approximately 50% (Fig. 4a, 4b, Supplementary Fig. 5a). This suggests that the contribution of BRD4 on gene expression of Myc and Klf4 is significant, albeit not exclusive, in non-transformed cells. In BRD4 knockdown cells, wild type YFP-BRD4 effectively rescued Klf4 and Myc expression, whereas YPF-BRD4-mBD was ineffective (Fig. 5a). Transcript elongation on these genes was clearly inhibited by JQ1 (Fig. 4c; also Supplementary Fig. 5b for Pol II progression).
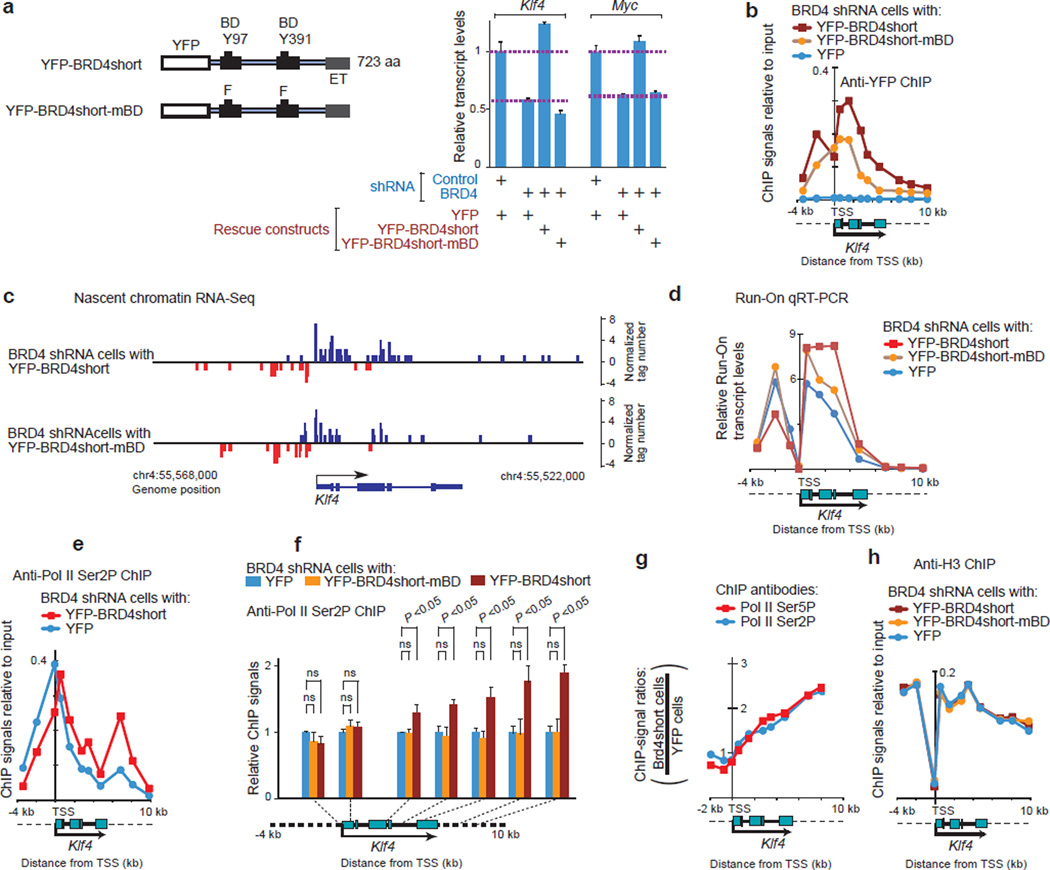
Figure 4. BD-mediated interaction with acetylated histones was required for BRD4-dependent expression of Myc and Klf4 but not for the recruitment of BRD4 and P-TEFb at the TSS.
(a, b), Myc and Klf4 expressions in BRD4 knockdown cells reconstituted with the rescue constructs indicated (a) and NIH3T3 cells treated with JQ1 stereoisomers (1 µM, 2 h) or vehicle (DMSO) (b), detected by qRT-PCR. Values were normalized to Gapdh and represent means ± s.e.m. (n = 3 technical replicates.) c, Genome browser views of strand-specific chromatin-bound RNA-seq reads across the Myc and Klf4 gene loci in NIH3T3 cells treated with JQ1 (500 nM, 1 h) or vehicle (DMSO). (d, f), ChIP-qPCR assays demonstrating binding of BRD4 (d) and CDK9 (f) on the Klf4 gene locus (from −4 kb to +10 kb relative to the TSS) in BRD4 knockdown cells expressing YFP-BRD4 proteins (wild type or mBD mutant) or YFP alone. Signals relative to input DNA (y-axis) are plotted against genomic positions of PCR primers (x-axis). Dotted lines in the diagram of the Klf4 gene locus represent the flanking regions. Background signals (yellow lines) were assessed in cells expressing YFP-alone (d) or using normal rabbit IgG (f). (e, g) ChIP-qPCR assays demonstrating binding of BRD4 (e) and CDK9 (g) on the Klf4 gene locus in cells treated with JQ1 (1 µM, 2 h) or vehicle (DMSO). (h, i), ChIP assays using anti-histone antibodies indicated. In i, ChIP signals were normalized, relative to the signals at the TSS (marked by an asterisk).
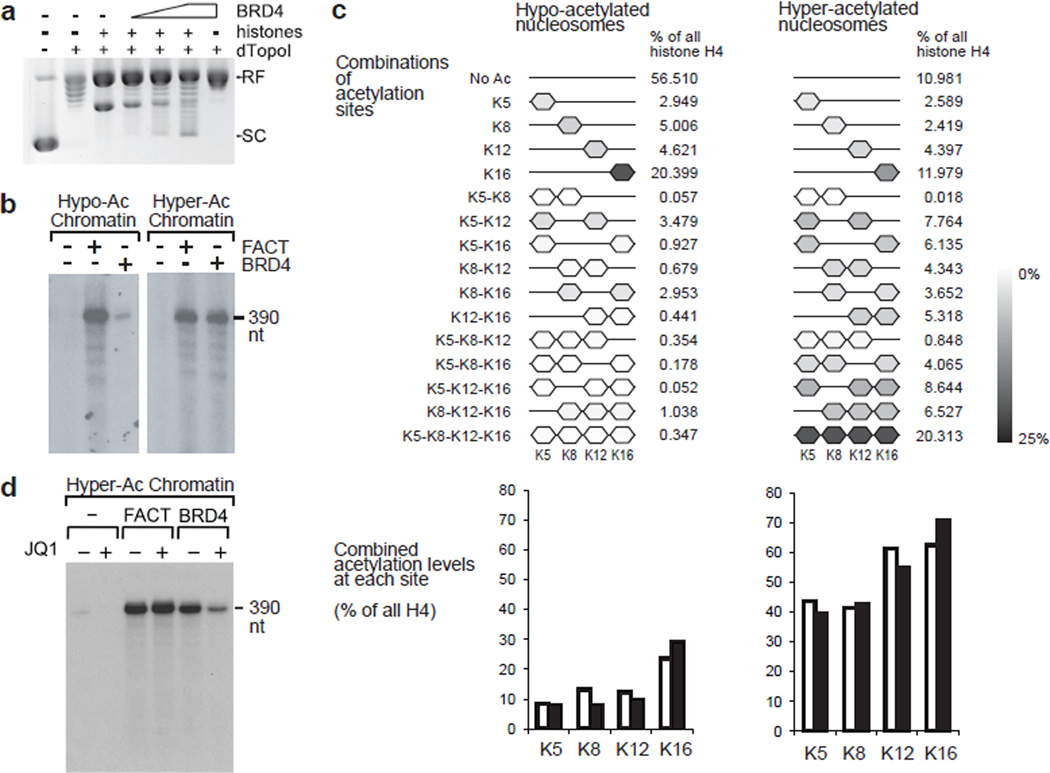
Figure 5. BRD4 possesses a histone chaperone activity and assisted elongating Pol II to transcribe through hyperacetylated nucleosomes in vitro.
a, In vitro plasmid supercoiling assays. RF indicates the position of circular plasmid DNA relaxed in the presence of Drosophila topoisomerase I fragment ND423 (dTopoI). SC indicates the position of supercoils introduced by hyperacetylated core histones (6 pmoles) and BRD4 (1, 2.5, and 5 pmoles). (b-d) The effects of BRD4 and the FACT complex on in vitro transcription of hypo- and hyper-acetylated chromatin templates (b), and the effects of JQ1 on transcription of hyper-acetylated chromatin template (d). Labeled RNA was resolved on 10% polyacrylamide urea gels. c, Relative quantification of H4 acetylations by quantitative mass spectrometry in the hypoacetylated and hyperacetylated histones used in reconstituted transcription assays. Results of two independent experiments. For the display of combinations of acetylated sites, means of two experiments are shown. For combined acetylation levels at each acetylation site, individual results are shown by open and filled bars. Uncropped images of the blots are shown in Supplementary Data Set 1.
ChIP-qPCR and ChIP-seq assays revealed that BRD4 is widely distributed over the Klf4 and Myc genes (Fig. 4d, red line, Supplementary Fig. 5c, 5d). Despite the lack of rescue activity on gene expression, noticeable amounts of BRD4-mBD were detected around the TSS and the proximal portion of gene body (Fig. 4d, blue line, Supplementary Fig. 5c). Similarly, JQ1 did not largely reduce the amount of BRD4 around the TSS (Fig. 4e). Also, the amount of P-TEFb (CDK9) was comparable in BRD4-mBD reconstituted cells or JQ1 treated cells relative to wild type BRD4 or mock-treated cells, respectively (Fig. 4f, 4g; blue lines, Supplementary Fig. 5e). These data indicate that BD-mediated interactions with acetylated nucleosomes (and possibly other proteins) are not essential for loading BRD4 or P-TEFb at the TSSs, especially at the Klf4 TSS region that is devoid of nucleosomes and acetylated histone marks (Fig. 4h, 4i). Given that BRD4 interacts with several components of Mediator32,35,54, BRD4 may be recruited by Mediator complexes, which are present around the Klf4 and Myc TSS regions (data not shown). In contrast, this JQ1-resistant recruitment of BRD4 cannot be accounted for by indirect recruitment from distant enhancers such as super-enhancers where BRD4 dissociates at low concentrations of JQ1(ref. 53).
BRD4 assisted elongation through nucleosomes in vitro
For the BRD4-dependent gene expression, therefore, the function of BDs is crucially required at a step beyond the recruitment of P-TEFb and BRD4 itself. Accordingly, we considered the possibility that BRD4 actively participates in the transcript elongation process. BRD4 possesses an intrinsic kinase activity and may complement P-TEFb for Pol II phosphorylation on Ser2 (ref. 56). However, the BRD4’s kinase activity itself does not require the function of BDs56, and thus it does not account for the BD-dependent post-recruitment function of BRD4.
It was previously reported that BRD2, another BET protein, possesses a histone chaperone activity and aids elongating Pol II in vitro11, raising the possibility that BRD4 may also possess the similar activity. Thus we first confirmed histone chaperone activity of BRD4 by plasmid supercoiling assays (Fig. 5a), and proceeded to test in vitro transcription activity. BRD4 allowed Pol II to transcribe through nucleosomes in a manner dependent on histone hyper-acetylation (Fig. 5b, 5c, Supplementary Fig. 6). Importantly, JQ1 specifically inhibited the activity of BRD4 but not that of the FACT complex (Fig. 5d). Since only nucleosomes were hyperacetylated in these experiments, that would exclude the possibility that other potential protein interactions with BRD4 bromodomains are involved in this process. Thus, BRD4 assists Pol II to transcribe through hyperacetylated nucleosomes via its histone chaperone activity, which is dependent on its BD-acetylated histone interactions.
BRD4 facilitated elongation in a BD dependent manner in vivo
Next, in order to test whether BRD4 assists the passage of elongating Pol II complexes in vivo, and whether BRD4 functions independently of its P-TEFb interaction in this process, we expressed short forms of BRD4 (BRD4short and its mBD mutant) which lack the C-terminal P-TEFb interacting domain37 in BRD4 knockdown cells (Supplementary Fig. 4). This short form5,57 essentially corresponds to the human isoform C, which lacks the DNA damage modulating activity reported for isoform B58.
Strikingly, BRD4short but not BRD4short-mBD rescued expression of Klf4, Myc, and many other genes in BRD4 knockdown cells (Fig. 6a). ChIP analysis revealed that both forms of short BRD4 proteins were bound to the nucleosome-depleted TSS region of Klf4 at similar levels, but that they were distributed differently along the highly acetylated gene body (Fig. 6b). Nascent RNA-seq analysis showed that transcripts extended more extensively toward the TES with expression of wild type BRD4short than with BRD4short-mBD (Fig. 6c; Supplementary Fig. 7a for Myc). This was further verified by quantitative reverse-transcription PCR (qRT-PCR) analysis of nuclear run-on chromatin RNA samples (Fig. 6d, Supplementary Fig. 7b for Myc). We further examined the progression of Pol II along gene bodies. Considerable levels of Ser2 phosphorylated Pol II, presumably representing an elongating form, were detected in BRD4 knockdown cells and used to monitor the progression of Pol II by ChIP assays. Consistent with the transcript analyses, the distribution of Ser2P Pol II was shifted toward and beyond the TES in cells reconstituted with YFP-BRD4short when compared with cells expressing the mBD mutant (Fig. 6e, 6f, Supplementary Fig. 7c for Myc). The distribution of serine 5 phosphorylated Pol II was also shifted by BRD4short in a similar manner to Ser2P (Fig. 6g), confirming that the observed changes were due to altered distribution of Pol II. No clear difference in nucleosome deposition was observed among these cells, as assessed by histone H3 (Fig. 6h). These results collectively support the idea that BRD4short facilitated the progression of Pol II along the gene body via its histone chaperone activity, depending on the BD-acetylated histone interaction in vivo. Although the possibility that other acetylated proteins may interact with BDs in vivo cannot be ruled out, it would be in addition to, but not instead of, the interaction with acetylated histones.
Figure 6. BRD4 facilitated transcript elongation in a BD dependent manner in vivo.
a, Diagrams of YFP-BRD4short and YFP-BRD4short-mBD, and Myc and Klf4 expressions detected by qRT-PCR. Values were normalized to Gapdh and represent means ± s.e.m. (n = 3 technical replicates.) Mouse BRD4short consists of the same N-terminal 720 amino acids as full length BRD4 and unique 3 amino acids added at the C-terminus. b, ChIP assays for YFP-BRD4 proteins indicated. c, Nascent transcript progression measured by chromatin RNA-seq analysis. d, Nuclear run-on RNAs, analyzed by qRT-PCR using Klf4 locus specific primers. The BrU-labeled run-on RNAs were fragmented, and immunoprecipitated with anti-BrdU antibody. (e, f), ChIP assays for Ser2-phosphorylated Pol II. In f, means ± s.e.m of three independent cell lines per indicated transduction combination are plotted after normalization with cells expressing YFP alone. Two-tailed Student’s t test. ns: not significant. g, ChIP-qPCR signal-ratios (signals in cells reconstituted with YFP-BRD4short over those with YFP alone) for Ser2P Pol II and Ser5P Pol II along the Klf4 genome locus. h, ChIP-qPCR assays demonstrating deposition of H3 (pan H3) on the Klf4 gene locus in cells transduced as indicated.
BRD4 interacted with Pol II elongation complexes in vivo
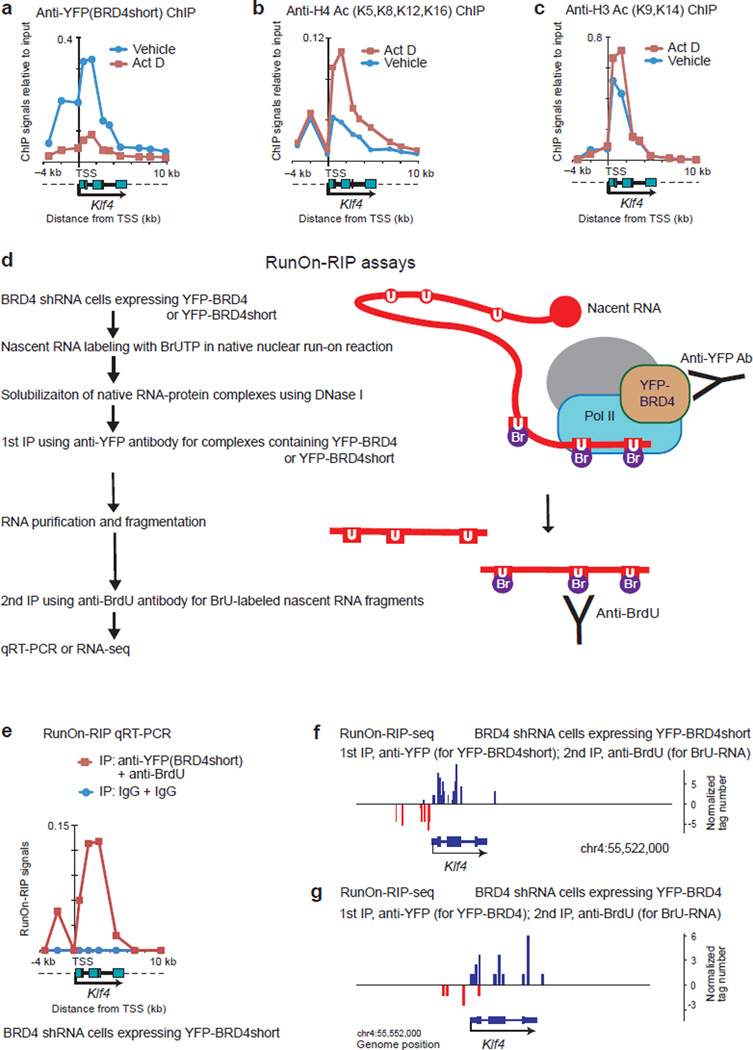
We envisioned that BRD4 may have traveled together with the Pol II elongation complexes. When transcription was blocked by actinomycin D (Act D), BRD4 distribution was dramatically reduced (Fig. 7a, red line), although acetylation marks on histones H3 and H4 were unchanged or even increased (Fig. 7b, 7c). This indicates that interactions with acetylated histones are not necessarily sufficient to recruit BRD4 on gene bodies, and that ongoing transcription may also be required. Thus, the passage of Pol II complexes and BRD4 along the gene body and the interaction of BRD4 with acetylated nucleosomes are mutually dependent to some degree.
Figure 7. BRD4 interacted physically with Pol II elongation complexes in vivo.
(a–c) ChIP assays for YFP-BRD4short (a), H4 Ac (tetra-acetylated at K5, K8, K12 and K16) (b) and H3 Ac (di-acetylated at K9 and K14) (c). BRD4-knockdown cells expressing YFP-BRD4short were treated with actinomycin D (Act D, 2 µg/ml) or vehicle (DMSO) for 2 h. d, Workflow of RunOn-RIP (run-on RNA immunoprecipitation) assays for native complexes containing BRD4 and elongating RNA. Nuclei isolated from BRD4 knockdown cells reconstituted with YFP-BRD4 or YFP-BRD4short were subjected to a nuclear run-on reaction without using sarkosyl. U, uracil; BrU, bromo-uracil. The BrU-labeled, nuclear run-on transcripts were sequentially immunoprecipitated with anti-YFP antibody (for YFP-BRD4 or YFP-BRD4short) and anti-BrdU antibody (for BrU labeled RNA fragments). e, Quantitative RT-PCR analysis of RunOn-RIP signals along the Klf4 gene locus. BRD4 knockdown cells reconstituted with YFP-BRD4short were analyzed. Background signals were assessed using normal rabbit IgG. (f, g) Genome browser views of RunOn-RIP-seq reads around the Klf4 gene locus, representing the actively elongating front of transcripts that were associated with YFP-BRD4short (f) or YFP-BRD4 (g).
To document a physical association of BRD4 with actively elongating Pol II complexes, elongating transcripts were labeled with BrUTP in a mild nuclear run-on reaction, and the transcripts associated with native protein-RNA complexes were sequentially immunoprecipitated with antibodies against YFP (for YFP-BRD4 or YFP-BRD4short) and BrdU (for BrU-labeled RNA) (Fig. 7d). BRD4 or BRD4short was associated with elongating Klf4 and Myc transcripts as shown by qRT-PCR (Fig. 7e, Supplementary Fig. 8b) or RNA-seq (Fig. 7f,7g, Supplementary Fig. 8c). These results indicate that actively transcribing Pol II complexes are associated with BRD4 in vivo.
DISCUSSION
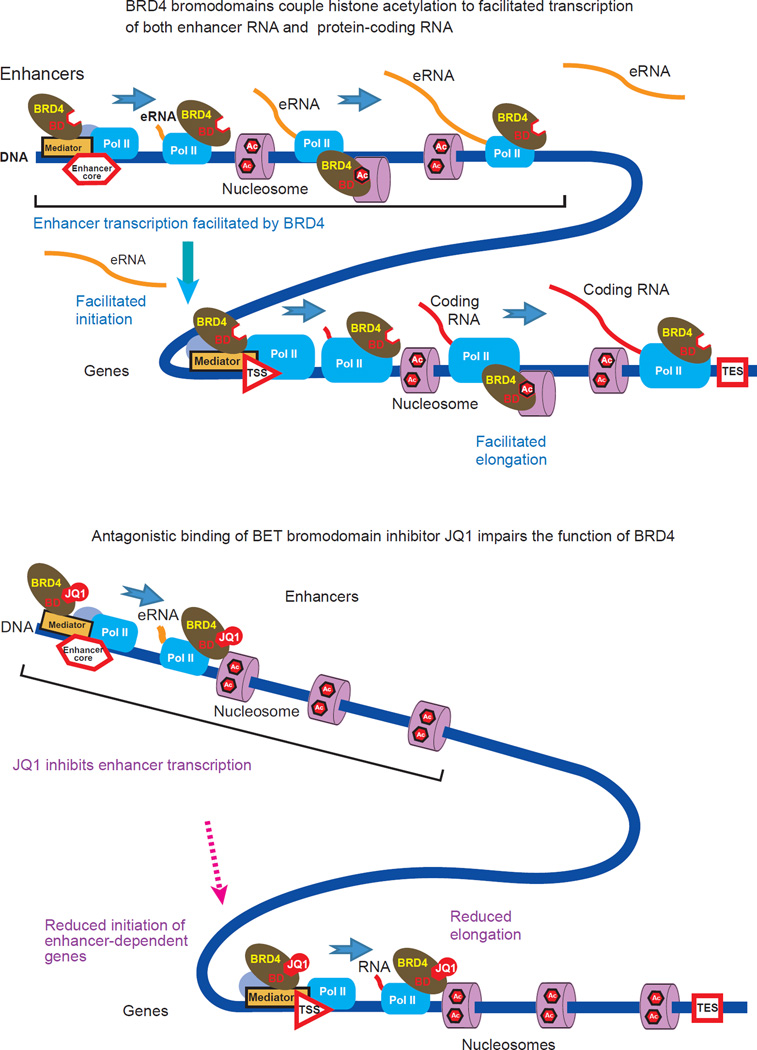
In the present study, we show that BRD4 functioned as a histone chaperone, utilizing its interaction with acetylated histones, and assisting the passage of Pol II through hyperacetylated nucleosomes (Fig. 8). This bromodomain-dependent process represents a mechanism which was directly antagonized by BET inhibitors (Fig. 8). Our results demonstrate that the chaperone-mediated activity of BRD4 assisted transcript elongation at both enhancers and within protein-coding genes. The apparent inhibition of transcription initiation by JQ1 we observed suggests that BRD4 may also be associated with the initiation of protein coding-gene transcription, possibly via an eRNA-mediated mechanism. Thus BRD4 assisted, and BET inhibitors antagonized, transcription regulatory mechanisms at multiple levels.
Figure 8. A model depicting the interdependence between the passage of Pol II elongation complexes through nucleosomes and acetyl-histone binding of BRD4 at enhancers and within protein-coding genes.
BRD4 contributes to the progression of the elongation complexes via its histone chaperone activity depending on its interaction with hyper-acetylated nucleosomes. The BET bromodomain inhibitor JQ1 occludes the acetyl-histone binding pockets of the BRD4 bromodomains, disabling the elongation-facilitating activity of BRD4.
The proposed model of histone chaperone activity is distinct from the conventional model12,49,53,55, in which BRD4 functions as an adapter to recruit P-TEFb and releases Pol II from pausing near the TSS. In the present model, BRD4 is also a direct effector that functions at enhancers and on gene-bodies. The two models are not mutually exclusive, and may well cooperate. Several lines of evidence support the histone chaperone-mediated mechanism of BRD4. (1) BRD4-dependent gene expression correlated with the presence of BRD4 in intergenic regions and on gene bodies, but not at TSSs. (2) The presence of P-TEFb did not correlate well with Pol II reads or RNA-seq reads. (3) In BRD4 knockdown cells, exogenous expression of a short form of BRD4, which does not interact with P-TEFb, was able to rescue the expression of BRD4-dependent genes. Furthermore, the bromodomain mutant of full length BRD4, which maintains the ability to interact with P-TEFb, failed to rescue gene expression even when both were loaded at the TSS.
Mutagenesis of the BD in BRD4 provided a powerful tool to study the function of this domain in vivo. For a subset of BRD4-dependent genes, including Klf4 and Myc, the primary function of the BDs was not to load BRD4 at the TSS, but to facilitate transcript elongation. We monitored the elongation process by three different methods. In addition to conventional Pol II ChIP analysis, we also mapped nuclear run-on transcripts to monitor the positions of Pol II without relying on ChIP antibodies. Furthermore, we directly monitored the position of the elongating end of nascent transcripts. The results were all consistent and suggested that the essential role of BRD4 BDs in gene expression was to help elongate transcripts under our experimental condition.
In addition to BRD4, BRD2 and BRD3 also possess histone chaperone activity and have the ability to facilitate transcript elongation through nucleosomes in vitro. Furthermore, BRD2 and BRD3 are also associated with acetylated nucleosomes in vivo, and participate in gene expression7,11,59. Thus the histone chaperone activity could be the primary target of BET inhibitors. This is in line with previous reports, in which individual knockdowns of BRD2, BRD3 and BRD4 mimicked actions of BET inhibitors14,19,20.
While BRD4 is also known to possess kinase activity for Pol II Ser2, this activity is not the target of BET inhibitors56. Ser2 phosphorylation of Pol II, which can also be achieved by P-TEFb, creates a platform to recruit factors involved in RNA processing such as splicing and 3’end processing27. In contrast, the histone chaperone activity of BRD4 assists the passage of Pol II through nucleosomes. Thus BRD4 appears to have distinct functions that likely cooperate in vivo.
In order to address the passage of BRD4 along the gene body, we devised a sequential immunoprecipitation method, referred to as Run-on RIP assay, to pull-down native Pol II elongation complexes containing both nascent RNA and BRD4. Our results showed that actively elongating transcripts from Myc and Klf4 genes were co-immunoprecipitated with BRD4, suggesting that BRD4 was physically associated with elongating Pol II complexes. This assay may be helpful to further dissect Pol II elongation complexes, as many other elongation factors may associate with Pol II as it traverses gene bodies.
BRD4 can be found throughout the genome. In the present study, we noted a significant correlation between BRD4-dependent gene expression and the presence of BRD4 in intergenic regions and on gene bodies, but not at TSS regions. We envision that BRD4 is loaded at the TSS for both active and inactive genes. In BRD4-dependent active genes, the loaded BRD4 then moves downstream with Pol II complexes so that the gene body/TSS ratio of BRD4 increases in proportion to gene activity. That ongoing transcription was required for BRD4 to move along the gene body suggests that BRD4 is not sufficient to drive transcription by itself. Once transcript elongation has been initiated, BRD4 may assist the passage of Pol II through nucleosomes in a manner dependent on the state of nucleosome acetylation. However, neither the frequency of Pol II complex assembly nor nucleosome acetylation is directly regulated by BRD4. These additional requirements may account for, at least in part, the relatively small statistical correlations observed between the amount of BRD4 and transcriptional output (see Fig. 3).
At enhancers, BRD4 is associated with eRNAs, which are bidirectional and efficiently inhibited by JQ1. The similar association has recently been observed for estrogen receptor-dependent enhancers60. The present correlation analysis suggested that BRD4 assists eRNA synthesis, which may be associated with enhancer function. The present analysis further suggested that it is the eRNA, but not BRD4 itself or P-TEFb, that was statistically associated with nearby gene expression. Thus we envision that BRD4 at enhancers may influence the expression of associated target genes indirectly through its effect on eRNA synthesis. It is worth noting however that the observed inhibition of transcription initiation by JQ1 could also suggest a more direct role for BRD4 in the initiation step.
In summary, the present study reveals a role of BRD4 in transcript elongation at enhancers and gene bodies. The elongation-facilitating function of BRD4 on the gene body depends on the function of BD after the initial recruitment of BRD4 and its physical association with Pol II complexes. This function is most likely accounted for by its BD-dependent histone chaperone activity. Whereas small molecule drugs targeting epigenetic interactions are designed to interfere with factor recruitment, BET inhibitors act more directly by occluding a functional interface of BRD4. As more epigenetic drugs enter into clinical trials, the roles of each epigenetic interaction must be fully understood in order to predict and interpret the drug action precisely. Based on the present study, BET inhibitors may preferentially target hyper-acetylated genes with active enhancers.
Online Methods
Cells
NIH3T3 cells were cultured in DMEM supplemented with donor bovine serum and antibiotics. HEK293T and Platinum-E retroviral packaging cell line (Cell Biolabs) were cultured in DMEM supplemented with fetal bovine serum and antibiotics.
Expression and knockdown plasmid constructs
Retroviral backbone vectors for protein expression (pMSCVpuro; Clontech) and knockdown (pSRneo; Oligoengine) were purchased. pSRneo-shBRD4 and pSRneo-shCont are described12,35. Mouse BRD4 cDNA was described61. BRD4short cDNA was produced by PCR. Point mutations of bromodomains were created by mutagenesis using QuikChange II site-directed mutagenesis kit (Stratagene). All the BRD4 constructs described in the present study contained nucleotide substitutions around the target site of BRD4 shRNA12,35 so that they are resistant to BRD4 shRNA while keeping the amino acid coding unchanged at the corresponding region.
BRD4 knockdown and reconstitution
NIH3T3 cells were sequentially infected with shRNA retrovirus and YFP-BRD4 expressing retrovirus. Virus infection was carried out in the presence of polybrene according to a standard method12. The transduced cells were selected with G418 and puromycin.
Live cell FRET analysis
For FRET analysis, 8 × 105 HEK293T cells seeded on 6 cm dishes were transfected with expression vectors for a CFP-fusion protein (4 µg) and a YFP-histone protein (0.2 µg) using FuGene6 (Roche). Live cells were harvested 24–48 hr. after transfection, and immediately analyzed on a CyAn ADP (Beckman Coulter) as described7 with some modification. Excitation lasers and filters for detection channels were: a solid-state violet laser (405 nm) and a 450/50 nm bandpass filter for CFP; an argon laser (488 nm) and a 546/10 nm bandpass filter for YFP; and the 405 nm solid-state violet laser and a 546/10 nm bandpass filter for FRET. All three signals were detected from the same individual cells by sequentially illuminating them with the 488 nm and 405 nm lasers. Results were analyzed with Summit software v4.3.1, and compensation values for spillover signals were determined with the data from singly transfected cells. Signals from individual cells were grouped into a matrix of 28 (CFP-signals) x 32 (YFP-signals) grids, and mean FRET signals representing those grids were displayed by three-dimensional surface plots using JMP software (SAS Institute, Cary, NC).
In vitro chromatin assembly and transcription from chromatin templates
Chromatin was assembled on the pG5MLP transcription template that contained five GAL4 binding sites upstream of the adenovirus major late promoter and a 390 base pair G-less cassette. For assembly, 4 µg of pG5MLP DNA was incubated with 4 µg of the purified core histones (either hypo-acetylated or hyper-acetylated), 40 µg BSA, 500 ng GAL4-AD, 100 ng RSF, 3 mM ATP, 30 mM creatine phosphate (Sigma), 0.2 µg creatine phosphokinase (Sigma) in 10 mM Hepes-KOH (pH 7.6) containing 5 mM MgCl2, 50 mM KCl, 0.2 mM EDTA, and 5% (v/v) glycerol, as described11. Core histones (hypo-acetylated and hyper-acetylated) were purified from HeLa cells as described62. Quantification of histone post-translational modifications in the hypoacetylated and hyperacetylated histone preparations was performed by mass spectrometry as described in detail63. The purification of RSF and GAL4-AD was performed as described11,64. Transcription reactions contained 200 ng of chromatin assembled pG5ML (−/+ 200 ng of FACT or BRD4 as indicated) with 10 ng TFIIA, 10 ng TBP, 10 ng TFIIB, 10 ng TFIIF, 50 ng TFIIE, 50 ng TFIIH and 100 ng RNA polymerase II, 3 mM each of ATP and CTP, 0.03 mM UTP, 12 units of RNAsin (Promega), and 5 µCi/reaction [α32P]-UTP (800 Ci/mmol Perkin-Elmer) in 10 mM Hepes-KOH (pH7.9), containing 50 mM KCl, 6 mM MgCl2 and 1.25% PEG (w/v) (average mw 8000) and were incubated at 30°C for one hour 11. Transcription was stopped with 20 mM EDTA, containing 200 mM NaCl, 1% SDS (w/v) and 25 µg glycogen and the G-less transcripts were then digested with 50 units RNAse T1 (Roche) for 30 minutes at 30°C. Following deproteinization the transcripts were run on 6% polyacrylamide gels containing 8 M urea in 1X TBE (0.09 M Tris-borate pH 7.5, 2 mM EDTA). For JQ1 inhibition, 200 ng of FACT or BRD4 was incubated with 60 µM of JQ1 in DMSO or DMSO alone (vehicle) for 1 hour prior to adding them to the assay. The purification of FACT, BRD4, TFIIA, TFIIB, TBP, TFIIF, TFIIE, TFIIH and RNA polymerase II was performed as described11,65–67.
Supercoiling Assay
Six pmoles of hyperacetylated core histones from HeLa cells were pre-incubated with 0, 1, 2.5, or 5 pmoles of BRD4 in 10 µL Assembly Buffer (10 mM Tris-HCl pH 7.6, 100 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.05 mg/ml BSA) for 30 min at 30°C. In parallel reaction, 1 pmole of supercoiled pFASTBAC1 plasmid was relaxed with 50 ng of Drosophila topoisomerase I fragment ND423 in 10 µL of Assembly Buffer containing 5 mM KCl at 30°C for 30 min. One microliter of the topoisomerase I-relaxed circular DNA (100 fmoles) was added to each histone/BRD4 mixture and the reaction was incubated at 30°C for 2.5 hrs. The assembly reaction was stopped by adding an equal volume of Stop Buffer (10 mM EDTA pH 8.0, 0.05% SDS, 1 µg/µL glycogen) and 2.5 µg proteinase K was added. The mixture was further incubated for 30 min at 45 °C. DNA was subsequently isolated with phenol:chloroform extraction and ethanol precipitation, resolved on a 0.8% agarose gel, and visualized by ethidium bromide staining. ND423 was kindly provided by James Kadonaga (UCSD).
Nuclear extracts
Nuclear extracts were prepared as described68.
Quantitative RT-PCR of total RNA
Expressions of genes were quantified by real-time PCR using the Fast Sybr Green Master Mix and gene-specific primers on a 7500 Fast Real-Time PCR System (Applied Biosystems).
Microarray
Amplification and labeling of cDNA and hybridization were carried out at Precision Biomarker Resources (Evanston, IL, USA). Biotinylated cDNA was generated from 150 ng total RNA. Five micrograms of fragmented cDNA were hybridized to expression microarrays (Mouse Exon 1.0 ST array, Affymetrix). Affymetrix Expression Console software was used to generate expression values from the CEL files using the RMA expression measure.
ChIP antibodies
GFP (ab290, 5 µl used; abcam, Cambridge, MA), BRD4 (10 µg; ref. 49), CDK9 (sc-8338, 10 µg; sc-484, 10 µg; both from Santa Cruz Biotechnology), H3 (ab1791, 5 µg; abcam), acetyl H3 (06–599, 5 µg; Millipore, Billerica, MA), H3 acetyl K27 (ab4729, 2 µg; abcam), acetyl H4 (06–866, 5 µg; Millipore), H4 acetyl K12 (ab1761, 5 µg; abcam), Pol II (pan) (sc-899, 5 µg; Santa Cruz Biotechnology), Ser2P Pol II (ab5095, 5 µg; abcam), Ser5P Pol 11 (ab5131, 5 µg; abcam), or normal rabbit IgG (12–370, 5 µg; Millipore).
ChIP-qPCR and ChIP-Seq
Cells (2×107 cells) were cross-linked with 1% formaldehyde and sonicated. Fraction of lysate was kept aside as input control. For ChIP-qPCR, lysates typically corresponding to 2×106 cells were immunoprecipitated using Protein A-coupled magnetic beads, and beads were washed as described69. Recovered DNA was subjected to qPCR using locus specific primer pairs. The results were normalized using the input DNA. For ChIP-Seq analysis, 2×107 cells were subjected for an IP and 3–30 ng of DNA were recovered depending on antibodies. Libraries compatible to Illumina platform were prepared according to the manufacturer’s protocol and were sequenced by Illumina GA analyzer or HiSeq 2000. For GA generated reads, reads were mapped to mouse genome (mm9), and peaks were called via CisGenome as described previously69. For HiSeq generated reads, reads of 50 bases were aligned to the mouse genome build mm9 with Bowtie 0.12.8 (ref. 70), allowing two mismatches. Uniquely mapped and non-redundant reads were used for peak calling using SICER 1.1 (ref. 71) for the epigenetic marks and MACS 1.4.2 (ref. 72) for BRD4, CDK9 and Pol II. An FDR < 0.001 and a p-value < 1.0E-05 were used with SICER and MACS, respectively. HOMER73 was used to link peaks to nearby genes and identify colocalization of peaks among samples.
Preparation of nascent chromatin RNA libraries
Chromatin-associated RNA was isolated as described24 with modifications. Briefly, cells (5×106 cells) were scraped in Swelling Buffer (10 mM TrisHCl, pH 7.5; 2 mM MgCl2; 3 mM CaCl2; RNaseOUT), and the cell pellets were lysed in Sucrose Lysis Buffer (5 mM TrisHCl, pH 7.5; 1 mM MgCl2; 3 mM CaCl2; 0.5% NP-40, 0.25 M sucrose; RNaseOUT). The nuclear pellets were resuspended in Chromatin Wash Buffer (10 mM TrisHCl, pH 7.5; 150 mM NaCl; 7.5 mM MgCl2; 0.1 mM EDTA; 0.5 M urea; 0.5% NP-40; 0.5 mM DTT; RNaseOUT), vortexed shortly, incubated on ice for 1 min, and spun. Chromatin-associated RNA was extracted from the nucleoplasmic supernatant according to the Trizol protocol, treated with DNase I (10 units), and re-extracted with acid phenol chloroform and chloroform/isoamyl alcohol.
The chromatin-associated RNA (1 to 2 µg) was depleted of rRNA using RiboMinus Eukaryote Kit for RNA-seq (Invitrogen). To degrade uncapped RNAs, the RNA preparation was sequentially treated with 20 units of RNA 5’ Polyphosphatase (Epicentre) and 1 unit of Terminator 5’-phosphate-dependent exonuclease (Epicentre), each followed by phenol chloroform, chloroform extraction and ethanol precipitation. The 3’ hydroxyl group of the nascent RNA was ligated with a pre-adenylated RNA 3’ Adapter (TruSeq Small RNA Sample Preparation kit, Illumina: TGGAATTCTCGGGTGCCAAGG) using T4 RNA ligase 2, Truncated (NEB, 200 units) for 3 hr at 25°C. Fragmentation of the RNA was performed as described74 in 200 mM NaOH on ice for 20 to 30 min (most fragments <200 nt, checked by Agilent Bioanalyzer), followed by neutralization with an equal volume of 1 M TrisHCl, pH 6.8. The RNA was purified through a BioSpin P-30, RNase-free column (BioRad). The cap structure was removed with 10 units of Tobacco Acid Pyrophosphatase (Epicentre). The 5’-ends of the hydrolyzed RNA fragments were phosphorylated with 30 units of T4 Polynucleotide Kinase, 3’ Phosphatase Minus (NEB) and 1 mM ATP. Ligation of RNA 5’-adapter (TruSeq Small RNA Sample Preparation kit, Illumina: GUUCAGAGUUCUACAGUCCGACGAUC), reverse transcription and PCR with multi-index primers were performed according to the Illumina protocol.
Run-on qRT-PCR
Nuclear run-on labeling of RNA with BrUTP, and the purification, fragmentation and immunoprecipitation (twice with anti-BrdU antibody) of the labeled RNA were performed according to the GRO-seq protocol74, using 5×106 cells per transduced NIH3T3 cell type. cDNA was made from the immunoprecipitated RNA fragments using random hexamers, and qPCR was performed with specific primers using Fast Sybr Green system.
Run-On RNA Immunoprecipitation
Immunoprecipitation of the RNA that is associated with YFP-BRD4- or YFP-BRD4short- containing complexes was performed on BrUTP-labeled nuclear run-on RNA obtained from 1×107 cells. The present protocol is an adaptation of the GRO-seq74, RIP-seq, and TruSeq Small RNA Sample Prep (Illumina) protocols with modifications.
Run-on labeling
NIH3T3 cells transduced to express YFP-BRD4 or YFP-BRD4short (5×106 cells) were swelled on ice for 5 min with Swelling Buffer (see above). Cells were scraped, centrifuged, and resuspended in 5 ml of Swelling Buffer. Five milliliters of 2x Lysis Buffer (10 mM TrisHCl pH7.5, 2 mM MgCl2, 3 mM CaCl2, 1% NP-40, 20% glycerol, RNaseOUT, protease inhibitors) was added, and cells were lysed. The nuclei were resuspended in 0.2 ml of 1X Lysis Buffer. Ten milliliters of 1x Lysis Buffer was added, and the nuclei were centrifuged and resuspended in 0.2 ml of Freezing-Storage Buffer (50 mM TrisHCl pH 8.0, 5 mM MgCl2, 0.1 mM DTT, 40% glycerol, RNaseOUT, protease inhibitors). After washing in the same buffer, the nuclei were resuspended in 0.1 ml Freezing-Storage Buffer and frozen in ethanol dry ice. The nuclei were thawed on ice, and mixed well with 0.1 ml of 2x Nuclear Run-On Master Mix without Sarkosyl (10 mM TrisHCl pH8.0, 300 mM KCl, 5 mM MgCl2, 1 mM DTT, 0.5 mM ATP, 0.5 mM GTP, 0.00233 mM CTP, 0.5 mM Br-UTP, RNaseOUT, protease inhibitors) by gentle but thorough pipetting, repeated at least 15 times. The run-on mixture was incubated in 30°C water bath for 5 min with occasional shaking, and then placed on ice.
Native RNA Immunoprecipitation
The run-on nuclei (0.2 ml) were mixed with 0.3 ml of Digestion Buffer (30 mM TrisHCl pH7.5, 7 mM MgCl2, 1.7 mM CaCl2, RNaseOUT, protease inhibitors), and digested with 200 units DNase I for 30 min on ice. The reaction was added with 8 µL of 0.5 M EDTA and 0.5 ml of RIP BindAjust Buffer (10 mM TrisHCl pH7.5, 238 mM NaCl, 0.35% NP-40, RNaseOUT, protease inhibitors). Insoluble materials were removed by centrifugation and the supernatant was combined (1×107 cells equivalent per sample). For pre-clearance of samples, four microcentrifuge tubes containing Dynabeads Protein A (1.5 mg per tube) were prepared per sample. In parallel, anti-YFP or IgG Dynabeads were prepared. For this, six tubes containing prewashed Dynabeads Protein A (1 mg each) and 2 µL of anti-GFP antibody (abcam ab290) or normal rabbit IgG were prepared per sample. Before use, these beads were washed twice in 1ml RIP Wash Buffer (20 mM TrisHCl pH7.5, 150 mM NaCl, 1 mM EDTA, 0.2% NP-40, RNaseOUT, protease inhibitors), followed by incubation in RIP Blocking Buffer (1 mg/ml BSA, 0.1% PVP, 20 mM TrisHCl pH7.5, 150 mM NaCl, 1 mM EDTA, 0.2% NP-40, RNaseOUT, protease inhibitors) for 2 hr with rotation at 4°C. The Dynabeads Protein A beads for pre-clearance were washed twice with RIP Wash Buffer, and each of the four tubes was added with 1/4 of the samples. After 2 hr of rotation at 4°C, the beads were separated on the magnet, and the supernatants were collected and combined as a pre-cleared sample. The anti-YFP Dynabeads were washed twice with RIP Wash Buffer, and each of the six tubes was added with 1/6 of the pre-cleared sample. The tubes were rotated for 16 hr at 4°C, after which the beads were washed five times with 1 ml of RIP Wash Buffer. After the final wash, the beads from three tubes were combined and resuspended in 0.1 ml 1x DNase I Buffer, to which 4 µL of DNase I was added and gently mixed. After 15 min incubation at room temperature, 1.6 µL of 0.5 M EDTA, 1 µL of 10% SDS and 1.5 µL of 20 mg/ml Proteinase K were added, and the tubes were incubated for 30 min at 55°C. After cooling to room temperature, 0.9 ml Trizol was added. The RNA pellet was resuspended in 6.5 µL of water per tube and the RNA from two tubes was combined.
Adapter ligation and BrU-RNA immunoprecipitation
The precipitated RNA was ligated with a pre-adenylated RNA 3’ adapter (Illumina) using T4 RNA ligase 2, Truncated (NEB, 200 units). The RNA was fragmented by base hydrolysis in 0.2 M NaOH, neutralized and immunoprecipitated with anti-BrdU agarose beads according to the GRO-seq protocol74 except that buffered phenol/chloroform (pH 8.0) was used instead of acid phenol/chloroform. The 5’ cap structure was removed with Tobacco Acid Pyrophosphatase (Epicentre, 10 unit). The 5’-ends of the hydrolyzed RNA fragments were phosphorylated with T4 Polynucleotide Kinase (NEB, 30 units) and 1 mM ATP. Ligation of RNA 5’-adapter (Illumina), reverse transcription and PCR with multi-index primers were performed according to the Illumina protocol, and the sample was subjected to massive parallel sequencing. For qRT-PCR analysis, cDNA was made using random hexamers, and qPCR was performed with specific primers using Fast Sybr Green system.
Chromatin RNA-Seq data analysis
RNA-Seq data were generated with an Illumina’s HiSeq 2000 system. Raw sequencing data were processed with CASAVA 1.8.2 to generate FastQ files. Reads of sequentially varying length were mapped to the mouse genome mm9 and the 5’ 29 bases were identified as giving the maximum number of genomic matches. The 29 base reads were thus used throughout this study. Reads aligned to ribosomal DNA were first removed. The ribo-free reads were then mapped to the mouse genome mm9 with bowtie 0.12.8 (uniquely mapped only, allowing 2 mismatches). Strand specific bed files and bigWig files were made with BEDtools75, the UCSC genome browser tool bedGraphToBigWig, and in house developed python scripts. Partek Genomic Suite 6.6 was used to calculate exon-mapped-reads-only RPKMs (Reads Per Kilobase per Million mapped reads) and all-reads (intron-mapped reads also included) RPKMs. For a list of individual genes of interest, strand specific bed files were used to generate strand specific read density profiles around TSS (±500 base, 100 base bin), gene body (10% bin), and TES (±500 base, 100 base bin), using an in-house developed python program. To study the nascent RNA-Seq data around BRD4 peaks, Homer was used to call the peaks and assign peaks as either genic or intergenic. The nascent RNA-Seq read density profiles were then generated with Homer around the BRD4 peak centers (±2k, 200 base bin). The read density heatmap was made with Partek Genomic Suite 6.6.
Metagene profiling
We first generated a list of genes, total gene-body read counts of which were upregulated by serum by 1.5 fold or more, compared with those in serum-starved cells. From this, the second list was generated, containing genes, total read counts of which were inhibited by JQ1 by 1.5 fold or more. With visual inspection on a genome browser, we excluded the genes that met the following criteria. (1) Genes smaller than 2 kb. (2) Genes overlapping with other genes including RNA genes. (3) Genes not separated from other genes including RNA genes by at least 0.5 kb. (4) Genes, over which RNA-seq reads from other genes spread. (5) Genes, RNA-seq reads of which were not detected at annotated TSS. (6) Genes with sharp RNA-seq read spikes of great intensity at regions other than TSS.
Scaled metagene profile was generated as described76. The region spanning from 500 bp downstream of TSS to 500 bp upstream of TES was defined as the “scaled gene body” region. Normalized profiles were generated for each gene by dividing subregional read counts in each experimental condition by total read counts of the gene (from 200 bp upstream of TSS to TES) obtained with the serum-starved and vehicle-treated cells (Supplementary Fig. 1a) or the serum-stimulated and vehicle-treated cells (Fig. 1d). The antagonistic index was calculated as shown in Fig. 1e in order to avoid division by zero.
Statistics
JMP 10 software (SAS Institute, Cary, NC) was used for computation of the Spearman rank-correlation coefficient, and the Kruskal-Wallis test on the joint Wilcoxon ranking, performed with Dunn’s all pair comparison procedure.
Supplementary Material
Acknowledgments
We thank L. J. Core and J. T. Lis for GRO-seq protocol; and V. Sartorelli and D. Clark for critical discussion and reading of the manuscript. This work was supported by the Intramural Research Programs of NICHD, NIAMS, and NIAID.
Footnotes
Accession codes
Microarray and deep sequencing (ChIP-seq, nascent RNA-seq, and RunOn-RIP-seq) data are available under GEO accession numbers GSE56370 and GSE58731, respectively.
Note: Supplementary information is available in the online version of the paper.
Author Contributions
T.K. conceived the study, designed experiments, performed all experiments except supercoiling assays and in vitro transcription analysis, interpreted all data and wrote the manuscript. Y.K. performed RNA-seq, GRO-seq, ChIP-seq, interpreted data and wrote the manuscript. G.L., E.C, and B.A.G. performed in vitro transcription analysis and plasmid supercoiling assays. H.-W.S, S.R.B. and G.V. performed genome-wide data processing. T.D.H. oversaw JQ1 experiment. D.R., U.S., J.J.O. and K.O. discussed the results and edited the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu SY, Chiang CM. The double bromodomain-containing chromatin adaptor Brd4 and transcriptional regulation. The Journal of biological chemistry. 2007;282:13141–13145. doi: 10.1074/jbc.R700001200. [DOI] [PubMed] [Google Scholar]
- 6.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanno T, et al. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Molecular cell. 2004;13:33–43. doi: 10.1016/s1097-2765(03)00482-9. [DOI] [PubMed] [Google Scholar]
- 8.Jung M, et al. Affinity map of bromodomain protein 4 (BRD4) interactions with the histone H4 tail and the small molecule inhibitor JQ1. The Journal of biological chemistry. 2014;289:9304–9319. doi: 10.1074/jbc.M113.523019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi J, et al. Disrupting the interaction of BRD4 with diacetylated Twist suppresses tumorigenesis in basal-like breast cancer. Cancer cell. 2014;25:210–225. doi: 10.1016/j.ccr.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeRoy G, Rickards B, Flint SJ. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Molecular cell. 2008;30:51–60. doi: 10.1016/j.molcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mochizuki K, et al. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. The Journal of biological chemistry. 2008;283:9040–9048. doi: 10.1074/jbc.M707603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ceribelli M, et al. Blockade of oncogenic IkappaB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11365–11370. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bandukwala HS, et al. Selective inhibition of CD4+ T-cell cytokine production and autoimmunity by BET protein and c-Myc inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14532–14537. doi: 10.1073/pnas.1212264109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matzuk MM, et al. Small-molecule inhibition of BRDT for male contraception. Cell. 2012;150:673–684. doi: 10.1016/j.cell.2012.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anand P, et al. BET bromodomains mediate transcriptional pause release in heart failure. Cell. 2013;154:569–582. doi: 10.1016/j.cell.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buratowski S. Progression through the RNA polymerase II CTD cycle. Molecular cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoansNature reviews. Genetics. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Molecular cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 30.Lin C, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Molecular cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He N, et al. HIV-1 Tat and host AFF4 recruit two transcription elongation factors into a bifunctional complex for coordinated activation of HIV-1 transcription. Molecular cell. 2010;38:428–438. doi: 10.1016/j.molcel.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature structural & molecular biology. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eberhardy SR, Farnham PJ. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. The Journal of biological chemistry. 2002;277:40156–40162. doi: 10.1074/jbc.M207441200. [DOI] [PubMed] [Google Scholar]
- 34.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Molecular cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 35.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Molecular cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Molecular cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 37.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petesch SJ, Lis JT. Overcoming the nucleosome barrier during transcript elongation. Trends in genetics : TIG. 2012;28:285–294. doi: 10.1016/j.tig.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belotserkovskaya R, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 40.Reinberg D, Sims RJ., 3rd de FACTo nucleosome dynamics. The Journal of biological chemistry. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- 41.Vahedi G, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kieffer-Kwon KR, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. nature. 2013;498:516–520. doi: 10.1038/nature12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lam MT, et al. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. nature. 2013;498:511–515. doi: 10.1038/nature12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mousavi K, et al. eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Molecular cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel MC, et al. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Molecular and cellular biology. 2013;33:2497–2507. doi: 10.1128/MCB.01180-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valouev A, et al. Determinants of nucleosome organization in primary human cells. nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramirez-Carrozzi VR, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W, et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. The Journal of biological chemistry. 2012;287:43137–43155. doi: 10.1074/jbc.M112.413047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang YW, et al. Mammalian mediator of transcriptional regulation and its possible role as an end-point of signal transduction pathways. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8538–8543. doi: 10.1073/pnas.95.15.8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–1595. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Devaiah BN, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ottinger M, et al. Kaposi’s sarcoma-associated herpesvirus LANA-1 interacts with the short variant of BRD4 and releases cells from a BRD4- and BRD2/RING3-induced G1 cell cycle arrest. Journal of virology. 2006;80:10772–10786. doi: 10.1128/JVI.00804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Floyd SR, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leroy G, et al. Proteogenomic characterization and mapping of nucleosomes decoded by Brd and HP1 proteins. Genome biology. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagarajan S, et al. Bromodomain protein BRD4 is required for estrogen receptor-dependent enhancer activation and gene transcription. Cell reports. 2014;8:460–469. doi: 10.1016/j.celrep.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online Method
- 61.Dey A, et al. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Molecular and cellular biology. 2000;20:6537–6549. doi: 10.1128/mcb.20.17.6537-6549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein A, Mitchell M. Generation of different nucleosome spacing periodicities in vitro. Possible origin of cell type specificity. Journal of molecular biology. 1988;203:1029–1043. doi: 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- 63.Leroy G, et al. A quantitative atlas of histone modification signatures from human cancer cells. Epigenetics & chromatin. 2013;6:20. doi: 10.1186/1756-8935-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.LeRoy G, Orphanides G, Lane WS, Reinberg D. Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science. 1998;282:1900–1904. doi: 10.1126/science.282.5395.1900. [DOI] [PubMed] [Google Scholar]
- 65.LeRoy G, Drapkin R, Weis L, Reinberg D. Immunoaffinity purification of the human multisubunit transcription factor IIH. The Journal of biological chemistry. 1998;273:7134–7140. doi: 10.1074/jbc.273.12.7134. [DOI] [PubMed] [Google Scholar]
- 66.Maldonado E, Drapkin R, Reinberg D. Purification of human RNA polymerase II and general transcription factors. Methods in enzymology. 1996;274:72–100. doi: 10.1016/s0076-6879(96)74009-0. [DOI] [PubMed] [Google Scholar]
- 67.Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- 68.Schnitzler GR. Isolation of histones nucleosome cores from mammalian cells. Current protocols in molecular biology / edited by Frederick M. Ausubel… [et al.] 2001;21:25. doi: 10.1002/0471142727.mb2105s50. Unit 21 Chapter. [DOI] [PubMed] [Google Scholar]
- 69.Wei L, et al. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 2010;32:840–851. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larschan E, et al. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.