Abstract
Intestinal T cells and group 3 innate lymphoid cells (ILC3) control the composition of the microbiota and gut immune responses. Within the gut there coexists ILC3 subsets which either express or lack the Natural cytoxicity receptor (NCR) NKp46. We identify here the transcriptional signature associated with the T-bet-dependent differentiation of NCR− ILC3 into NCR+ ILC3. Contrary to the prevailing view, we show by conditional deletion of the key ILC3 genes Stat3, Il22, Tbx21 and Mcl1 that NCR+ ILC3 were redundant for the control of mouse colonic infections with Citrobacter rodentium in the presence of T cells. However, NCR+ ILC3 were essential for cecum homeostasis. Our data show that interplay between intestinal ILC3 and adaptive lymphocytes results in robust complementary fail-safe mechanisms ensuring gut homeostasis.
Dysregulation of intestinal barrier function disrupts gut microbiota and results in inflammation, diarrhea and chronic disease. Mucosal immunity is essential to control the composition of gut commensal flora and maintain health in the face of continual exposure to potentially pathogenic bacteria in the gastrointestinal tract. Interleukin 22 (IL-22) plays a crucial role in this immune control of gut commensal and pathogenic bacteria and is secreted by a heterogeneous population of lymphocytes expressing the nuclear hormone receptor RORγt (encoded by the gene Rorc)1, including subsets of T helper cells (TH17, TH22) and group 3 innate lymphoid cells (ILCs) 2-7.
ILC3 are a heterogeneous population of innate lymphoid cells particularly abundant at mucosal sites. They can be divided into three main subsets, based on their role during embryogenesis and cell surface expression of the NCR, NKp46 (hereafter referred to as NCR+ or NCR− cells). NCR− ILC3 comprise lymphoid-tissue inducer (LTi) cells, which were originally found in the fetus and are required for the development of lymph nodes and Peyer’s patches 8. LTi cells do not require expression of the transcription factor PLZF9, and can be divided into CD4− and CD4+ cells. Another subset of NCR− ILC3 requires PLZF, and give rise to NCR+ ILC3 in a T-bet- and Notch-dependent manner 10, 11. NCR− ILC3 can promote colitis in a model of inflammatory bowel disease 12. By contrast, NCR+ ILC3 have been shown to be essential for regulating the balance between commensal and pathogenic bacteria through their IL-23-driven production of IL-22, particularly during Citrobacter rodentium infection 13, 14. C. rodentium is a Gram negative mouse-restricted pathogenic bacterium that can be used as a model of the human enteric pathogens enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC). It colonizes the intestinal mucosa, leading to the formation of attaching and effacing lesions that result from the effacement of the brush border microvilli. IL-22 is essential for the control of C. rodentium infection, as it stimulates the secretion of antimicrobial peptides and protects epithelial function 15. As a consequence, animals lacking this cytokine rapidly succumb to the disease 16. NCR+ ILC3 play a critical role in protection against C. rodentium in mice globally deficient for genes also expressed in B or T cells, such as Rag2, Tbx21 or Cxcr6 11, 13, 14, 17. However, these mouse models have profound or partial immunodeficiencies, confounding the interpretation of the findings and the selective contributions of NCR− and NCR+ ILC3. Overall, the analysis of ILC3 biology has been hampered by the lack of models selectively targeting these cells. Here, we explored the biology of innate producers of IL-22 in mouse conditional deletion models targeting NCR+ ILC3 but preserving B and T cells, to elucidate the specific contributions of NCR+ ILC3 to intestinal protection. Our comprehensive analysis of gut ILC3 revealed the redundant role of NCR+ ILC3 in the control of C. rodentium infection in immunocompetent hosts, and a selective role of NCR+ ILC3 in cecum homeostasis.
Results
Expression profiles of ILC3 subsets
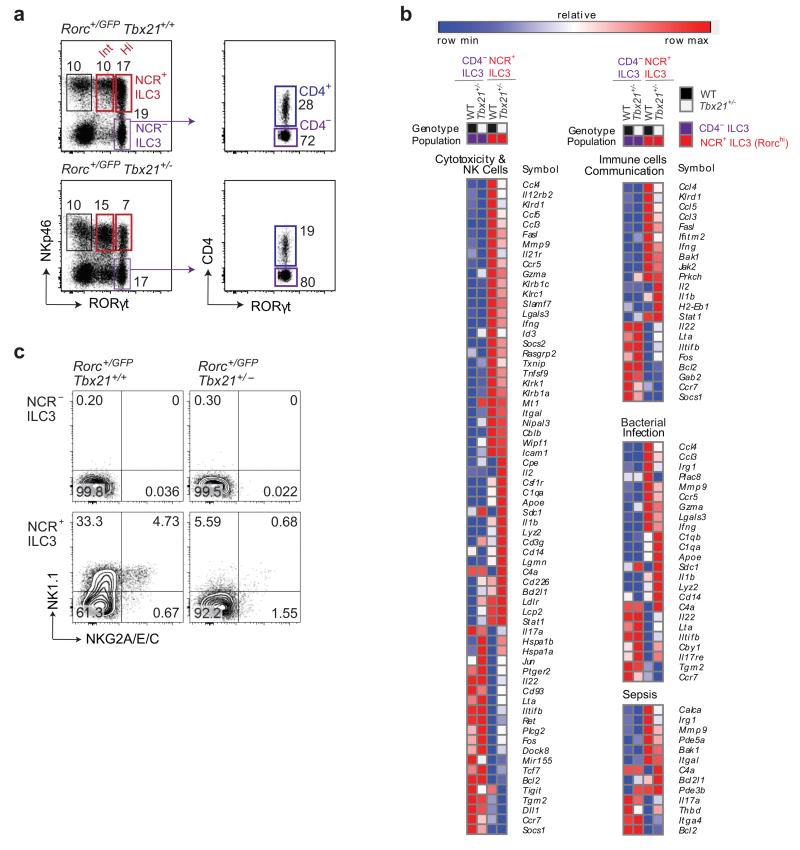
To date, RORγt+ IL-22-producing ILC3s have been divided into at least four different subsets, including T-bet-independent NCR− ILC3 comprising CD4+NKp46− and CD4−NKp46− ILC3, and T-bet-dependent NCR+ ILC3 comprising NCR+RORγtint and NCR+RORγthi ILC3 18, 19. We investigated the relationships between these four ILC3 populations, by isolating CD4+NCR− (CD4+), CD4−NCR− (DN), NCR+RORγtint and NCR+RORγthi ILC3 from the small intestine of Rorc+/GFP reporter mice (Fig. 1a) and performing a genome-wide transcriptome analysis by RNA sequencing (RNAseq). Within the NCR− ILC3 subsets, the transcriptional profiles of the CD4+NCR− and CD4−NCR− ILC3 were almost identical (Table 1), indicating that they could be considered as a single population of NCR− ILC3. Similarly, NCR+RORγtint and NCR+RORγthi NCR+ILC3 had very similar transcriptional programs (Table 1), differing only for the expression of a set of receptors guiding localization and including Ccl5, Cxcr5 Ccl4 and Ccr5 (Supplementary Table 1). Thus, NKp46+RORγtint and NKp46+RORγthi cells were also extremely similar and could be considered as a single population of NCR+ ILC3. By contrast, robust differences in transcriptional profile were found between NCR− ILC3 and NCR+ ILC3. The expression of genes encoding transcription factors, such as Idb2, Gata3 and Rorc, was similar in the different subsets, whereas the expression of Tbx21 and Prdm1 was upregulated in NCR+ ILC3. Genes encoding a number of other transcriptional regulators were also found to be differentially expressed, including interferon regulatory factor 8 (Irf8), which was upregulated, and Batf3 and Tox2, which were downregulated in NCR+ ILC3. Thus, NCR+ ILC3 and NCR− ILC3 are distinct ILC3 subsets with different gene transcription programs consistent with microarray analysis 20, validating our RNAseq approach.
Figure 1. RNAseq analysis of ILC3 subsets.
(a) Sorting strategy for RNAseq analyses of ILC3 subsets isolated from the small intestine of Rorcgfp/+Tbx21+/+ (wild-type) after gating on live CD45+ lin−(CD3−CD19−) cells (left plot). CD4+ NKp46− RORγt+ and CD4−NKp46− RORγt+ segregated among NCR− ILC3 (right plot).
(b) Analyses of gene clusters regulated by T-bet between Tbx21+/+ and Tbx21+/− NCR+ ILC3 at steady state. Ingenuity pathway analysis (IPA) was carried out for genes differentially expressed between NCR− and NCR+ ILC3 affected by the loss of a single copy of T-bet. Significant enrichment was observed for several pathways, grouped into three main categories: cytotoxicity & NK cells, immune cell communication and infectious disease. The expression values for the genes of these enriched pathways were converted into heat maps, using the mean and maximum values for each gene (Log2 reads per kilobase of transcript per million reads, RPKM).
(c) Flow cytometry analyses of NK1.1 and NKG2A/E/C protein levels in the indicated ILC3 subsets isolated from the intestines of RorcGFP/+Tbx21+/+ and RorcGFP/+Tbx21+/− mice. Plots are gated on live CD45+lin− RORγt+ cells and show one representative experiment. Numbers show the frequency of NCR+ ILC3 in the indicated quadrants. Plots show representative results from two independent experiments (n = 2-3 mice per genotype per experiment).
Table 1. Number of differentially expressed genes in ILC3 subsets.
| Up | NCR− ILC3 | NCR+ ILC3 | |||
|---|---|---|---|---|---|
| Down | CD4+ | DN | RORγthi | RORγtint | |
| NCR− ILC3 | CD4+ | 19 | 1011 | 744 | |
| DN | 20 | 972 | 646 | ||
| NCR+ ILC3 | RORγthi | 826 | 751 | 79 | |
| RORγtint | 560 | 536 | 84 | ||
Number of differentially expressed genes between each of the ILC3 populations sorted from Rorcgfp/+Tbx21+/+ mice. Rows show genes going up and columns show genes going down in the indicated populations.
The impact of T-bet on ILC3
T-bet is key to the differentiation of NCR− ILC3, as NCR+ ILC3 are absent from Tbx21-deficient mice 10, 11, 21, but the mechanisms by which T-bet affects ILC3 maturation are unclear. We investigated the transcriptional program of ILC3 isolated from Rorcgfp/+Tbx21+/− mice, which lack one copy of T-bet. Given that Rorcgfp/+Tbx21+/− mice have a heterogeneous phenotype, we reasoned that the loss of one allele would uncover differential regulation by T-bet without the complete loss of NCR+ ILC3, which remain present albeit at a lower frequency (Fig. 1a). We then performed two types of analysis on the RNAseq transcriptional profile dataset obtained from the following eight ILC3 populations: CD4+NCR−, CD4−NCR− (DN), NCR+RORγtint and NCR+RORγthi from Tbx21+/+ and CD4+NCR−, CD4−NCR− (DN), NCR+RORγtint and NCR+RORγthi from Tbx21+/− mice. We first compared the list of genes differentially expressed between NCR− ILC3 (CD4+and CD4−) and NCR+ ILC3 (RORγtint and RORγthi) in control mice with that for Tbx21+/− mice. In total, 674 genes displayed differences in expression by a factor of at least two between NCR− ILC3 and NCR+ ILC3 in wild-type mice; 324 of these genes did not display differential expression between these two cell types in Tbx21+/− mice (data not shown). Thus, during the transition between NCR− and NCR+ ILC3, about 50% of genes were affected by the loss of a single copy of T-bet, indicating that T-bet guides a substantial component of the NCR+ ILC3 developmental program. Second, the gene expression profiles of ILC3 from Tbx21+/− mice were compared with those from the corresponding wild-type ILC3 populations. We found no role for T-bet in CD4+ and CD4− NKp46− ILC3. Therefore, as expected, in NCR− ILC3 populations (CD4+ and CD4−), very few genes were differentially expressed between Tbx21+/+ and Tbx21+/− cells. However, significant differences in transcriptional profile were observed between Tbx21+/+ and Tbx21+/− NCR+ ILC3 (RORγtint and RORγthi): analyses of at least two-fold differences in expression between Tbx21+/+ and Tbx21+/− NCR+ ILC3 showed that 72 genes were upregulated and 86 downregulated in the wild-type cells (Supplementary Table 2). This enabled us to investigate the gene programs specifically regulated by T-bet during generation of the NCR+ ILC3 subset (Fig. 1b). Interestingly, 70% of the genes found to be down-regulated on NCR+ ILC3 in the absence of T-bet were highly expressed on NCR+ ILC3 compared to NCR− ILC3. Loss of a single allele of T-bet affected communication between innate and adaptive cells through cytokine or chemokine regulation via Il2r and interferon signaling through the Jak-Stat pathway, Il22, Ccl4 and Ccl5. T-bet also regulated a number of important gene clusters known to be preferentially associated with effector functions such as cytotoxicity and expressed by NK cells (e.g. Gzme, Gzma, Ifnγ and Il12rb). These gene clusters also included members of the killer-cell lectin-like receptor family: Klri1 (Ly49E1), Klrd1 (CD94), Klrb1c (CD161, NK1.1) and Klrc2 (NKG2C). Consistent with the genomic analyses, the T-bet-dependent regulation of NK1.1 and CD94-NKG2 heterodimers on NCR+ ILC3 was confirmed at the protein level (Fig. 1c). Thus, in addition to being required for the development of NCR+ ILC3 from NCR− ILC3, T-bet appears to play an important role in the recognition, effector function and migration of NCR+ ILC3 within tissues. T-bet is also the major driver of the ‘NK-ness’ program of NCR+ ILC3.
NCR+ ILC3 are dispensable in immunocompetent mice
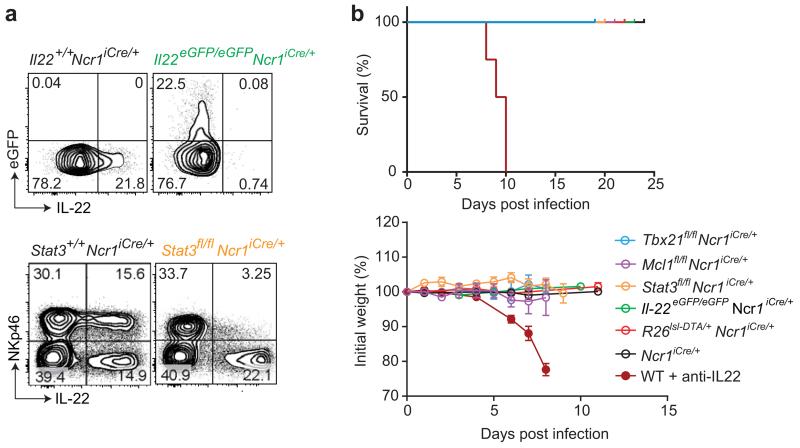
The pathway analysis of ILC3 transcriptomic data revealed that the set of genes expressed in NCR+ ILC3 and downregulated in Tbx21+/− mice was enriched in genes involved in the control of gastrointestinal diseases and bacterial infections (Fig. 1b). Consistent with these data, NCR+ ILC3 were reported pivotal in protection against C. rodentium in various models of immunodeficient mice 11, 13, 14, 17. We then investigated the role of the IL-22 produced by NCR+ ILC3 during the course of C. rodentium infection. We used a previously undescribed model of Il22eGFP/eGFPNcr1-iCre mice (Supplementary Fig. 1), in which NCR+ ILC3 were unable to produce IL-22 (Fig. 2a). We also crossed Ncr1-iCre mice with Stat3fl/fl mice, for the selective abolition of STAT3-dependent signals in NCR+ ILC3, as the transcription factor STAT3 is required for the production of IL-22 by ILC3 and Th17 cells, and the ablation of Stat3 in RORγt+ lymphocytes leads to increased morbidity and mortality after C. rodentium infection 22. NCR+ ILC3 were unable to produce IL-22 in both Il22eGFP/eGFPNcr1-iCre and Stat3fl/flNcr1-iCre mice (Fig. 2a) but both types of mice were resistant to C. rodentium infection, as no deaths, weight loss, histological abnormalities in the gut or increase in bacterial dissemination relative to controls were observed (Fig. 2b and Fig. 3). This resistance could not be accounted for by a compensatory increase in IL-22 production by NCR− ILC3, which were present in normal numbers and produced normal amounts of the cytokine (Supplementary Fig. 1c,d and Supplementary Fig. 2). By contrast, treatment with blocking anti-IL-22 led to severe colitis and the premature death of wild-type mice challenged with the same dose of C. rodentium (Fig. 2b), as previously described 16. Thus, IL-22 was essential for resistance to C. rodentium but the production of IL-22 by NCR+ ILC3 was dispensable.
Figure 2. Targeting of NCR+ ILC3 during C. rodentium infection.
(a) Upper panels show the production of IL-22 by small intestine Lin−CD45+NCR+ ILC3 from naive Il22eGFP/eGFPNcr1-iCre and control mice upon in vitro stimulation with IL-23 for 4-5 h in the presence of brefeldin A. The lower panels show the frequency of IL-22-producing cells among the NKp46+ and NKp46− cells gated on Lin−CD45.2+ small intestine lymphocytes from Stat3fl/flNcr1+/+RorcGFP/+ and Stat3fl/flNcr1-iCreRorcGFP/+ mice 8 days after C. rodentium infection. Cells were stimulated in vitro with IL-23 for 4-5 hr in the presence of brefeldin A to elicit IL-22 production. Data show representative plots from one of two independent experiments (n = 3-4 mice per genotype per experiment).
(b) Survival curve and change in body weight of indicated mice and Ncr1iCre/+ controls following C. rodentium infection. In addition, WT mice were treated with anti-IL-22 blocking mAbs twice weekly, beginning on day -1. Pooled data from independent experiments are shown. The data shown are means ± SEM.
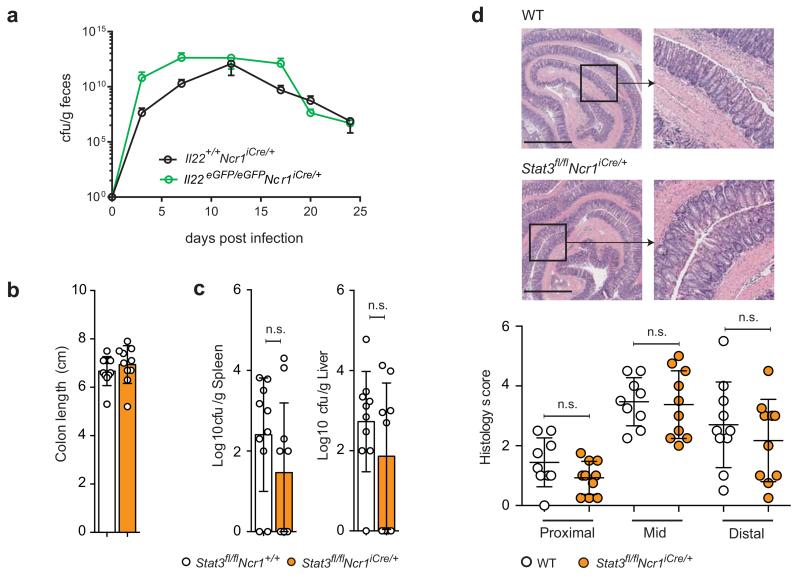
Figure 3. C. rodentium infection in the absence of IL-22 production by NCR+ ILC3.
(a) Il22eGFP/eGFPNcr1-iCre mice and controls were orally inoculated with 1010 cfu C. rodentium on day 0. Counts of C. rodentium (cfu) in the feces are shown as a function of time post-infection. Data shown are from one out of three independent experiments (n = 4-6 mice per group per experiment). Shown are means ± SD.
(b-d) Male Stat3fl/flNcr1+/+ and Stat3fl/flNcr1-iCre mice were orally inoculated with 2 × 109 cfu of C. rodentium. (b) Colon length of the indicated mice 8 days after infection. Data were pooled from three independent experiments (n = 3-4 mice per group per experiment). (c) Bacterial loads (cfu/g) were measured 8 days post-infection (liver and spleen). (d) Pathological features in the colon of the indicated mice 8 days after infection. Representative H&E staining of formalin-fixed paraffin-embedded sections (micrographs) of colons and blinded histological scoring (see Supplementary Table 3). The scale bar represents 1000 μm and the insets are × 3.4 magnifications. Data were pooled from three independent experiments (n = 3-4 mice per group per experiment). The data shown are means ± SD. Statistical analyses was performed with unpaired Student’s t-tests; n.s., not statistically significant.
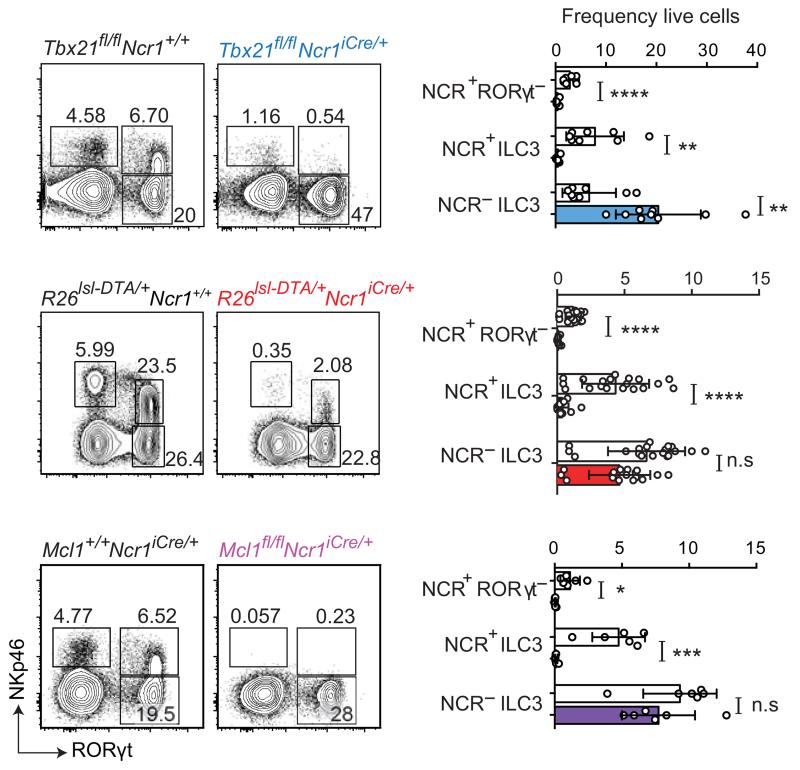
We then ablated NCR+ ILC3 to address their contribution to the control of C. rodentium infections more generally. We directly abolished Tbx21 expression in NCR+ ILC3, by crossing Ncr1iCre/+ mice with Tbx21fl/fl mice. Consistent with previous findings, the numbers of NKp46+ RORγt− cells and NCR+ ILC3 were much smaller in the small intestine and colon of Tbx21fl/flNcr1-iCre mice than in controls (Fig. 4 and Supplementary Fig. 3a). However, Tbx21fl/flNcr1-iCre mice were resistant to C. rodentium infection, as no deaths, weight loss, histological abnormalities in the gut or increase in bacterial dissemination relative to controls were observed in Tbx21fl/flNcr1-iCre mice (Fig. 2b and Supplementary Fig. 3b-d). A marked increase in the number of IL-22-producing NCR− ILC3 was observed in the Tbx21fl/flNcr1-iCre mice (Fig. 4 and Supplementary Fig. 3a), potentially compensating for the loss of NCR+ ILC3 in this model. We thus addressed the role of ILC3 during C. rodentium infection further, by generating another model of NCR+ ILC3 deficiency by crossing Ncr1iCre/+ mice with R26DTA/+ (Rosa-DTA) mice. A near-complete absence of NCR+ ILCs (both NCR+ RORγt− and NCR+ ILC3), with no increase in NCR− ILC3 numbers, was observed in the small intestine and colon of R26DTA/+Ncr1-iCre mice (Fig. 4 and Supplementary Fig. 4a, b). When R26lsl-DTA/+Ncr1iCre/+ mice were infected with C. rodentium, no morbidity or mortality was recorded (Fig. 2b). These findings are supported by the lack of difference between R26DTA/+Ncr1-iCre mice and controls in terms of colon length and histology, and bacterial dissemination, as assessed by the number of cfu in the feces, spleen and liver (Fig. 5). We further monitored the immune response to C. rodentium infection in these mice and we found no statistically significant differences in other parameters, such as the cellularity of the gut lamina propria for CD4+ T cells, CD8+ T cells, B cells, Ly6G+Ly6C+ myeloid cells and MHC class II+ CD11c+ dendritic cells, or the ability of mesenteric lymph node and splenic CD4+ T cells and CD8+ T cells to produce IL-17A or IFN-γ upon in vitro re-stimulation (Supplementary Fig. 4c-e). Cytokine (IFN-γ, IL-17A, Il-1β, IL-6, KC, TNF-α and IL-10) determinations in gut extracts revealed no differences between R26DTA/+Ncr1-iCre mice and controls (Supplementary Fig. 4f).
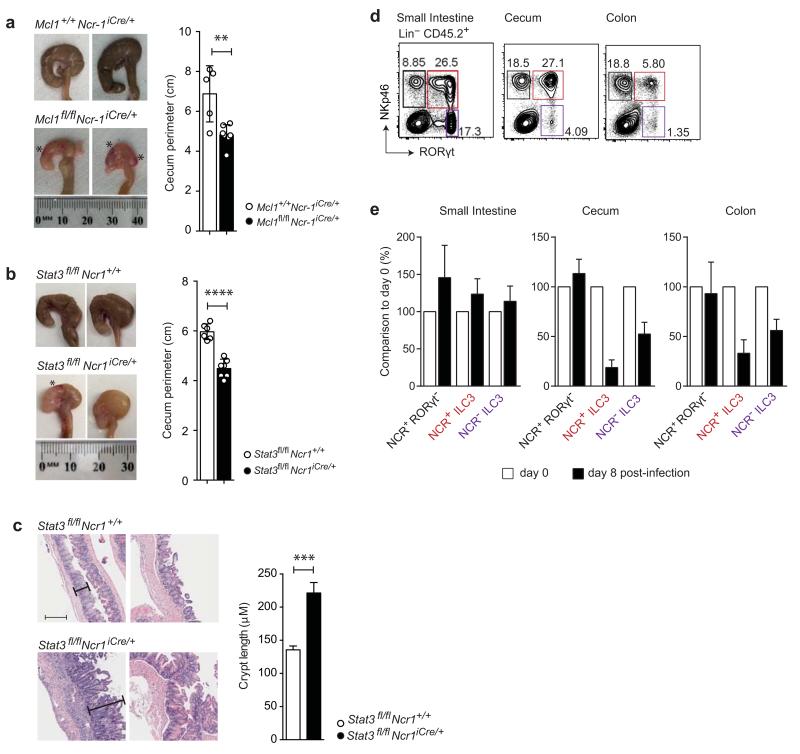
Figure 4. Impact of Tbx21 and Mcl1 deletion or DTA expression on NCR+ ILC3.
Analysis of Lin−CD45+ small intestinal lymphocytes from naive Tbx21fl/flNcr1iCre/+, R26DTA/+Ncr1-iCre and Mcl1fl/flNcr1-iCre mice and their respective controls. Flow cytometry plots show intracellular RORγt and surface NKp46 expression. Numbers show the frequency of cells within the indicated gate. Data are pooled from at least three independent experiments (n = 2-4 mice per group per experiment) and the mean ± SD is shown. Statistical analyses were performed with unpaired Student’s t-tests. n.s., not statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001.
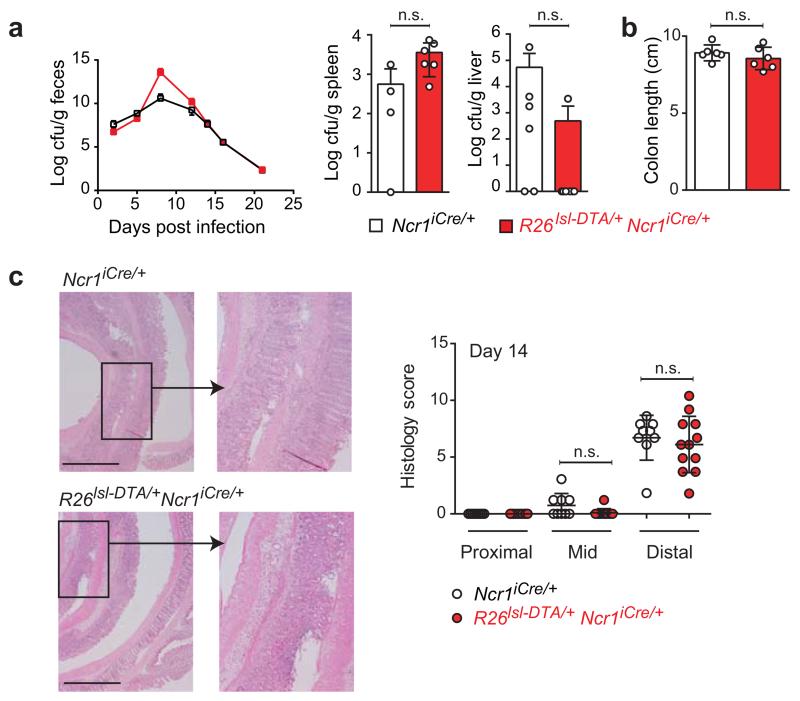
Figure 5. C. rodentium infection in the absence of NCR+ ILC3.
R26DTA/+Ncr1-iCre and Ncr1-iCre littermates were orally inoculated with 1010 cfu C. rodentium on day 0. (a) Bacterial loads (cfu/g) were determined over the course of the experiment (feces) and at 14 days post-infection (liver and spleen). The data shown are representative of three independent experiments (n = 4-8 mice per group). (b) Colon length and (c) pathological features, for R26DTA/+Ncr1-iCre and Ncr1-iCre controls, 14 days after infection. H&E staining of formalin-fixed paraffin-embedded sections (micrographs) and histological scoring (scatter plots) of colons are shown. The scale bar represents 1000 μm and the insets are x 3.4 magnifications. Histological scores for colon damage were determined on 1- 2 H&E sections per mouse. Data are pooled from two to three independent experiments (n = 4-9 mice/genotype). Statistical analyses were performed with unpaired Student’s t-tests. n.s., not statistically significant.
Mcl-1 is an anti-apoptotic protein induced by common γ chain (γc) receptor signaling and required for the maintenance of NK cells in vivo 23. We investigated whether Mcl-1 was also required for the maintenance of NCR+ ILC3 in the gut. Both NCR+ RORγt− and NCR+ ILC3 populations were absent from Mcl1fl/flNcr1-iCre mice (Fig. 4). These data demonstrate the key role of Mcl-1 in maintaining all NKp46+ ILC in vivo. Mcl1fl/flNcr1-iCre mice constitute another model of immune deficiency in which NCR+ ILC3 are absent but NCR− ILC3 and T cells are unaffected. We challenged Mcl1fl/flNcr1-iCre mice with C. rodentium. As for R26DTA/+Ncr1-iCre mice, Mcl1fl/flNcr1-iCre mice were resistant to the infection (Fig. 2b), with no signs of disease or bacterial dissemination detected by histology, or on the basis of colon length or the number of cfu in the spleen and liver (Supplementary Fig. 5). Thus, the results for five different genetic models converged to reveal that NCR+ ILC3 were dispensable for the control of C. rodentium infection in immunocompetent mice.
The lower amounts of IL-22 production by ILC3 in Ahr-deficient mice has been shown to allow the proliferation of commensal segmented filamentous bacteria (SFB), which are known to promote the development of Th17 cells 24. We therefore also investigated the consequences of a lack of NCR+ ILC3 for the gut microbiota at steady state, by deep sequencing. No difference in the presence of particular bacterial families or SFB numbers in the feces, or in Th17 counts in the small intestine was detected in comparisons of R26DTA/+Ncr1-iCre mice and control mice housed in the same conditions (Supplementary Fig. 6a-c). Similarly, the bacterial families present in the gut of Mcl1fl/flNcr1-iCre mice did not differ significantly from those present in the gut of controls (not shown). Thus NCR+ ILC3 deficiency had no impact on the gut microbiota in immunocompetent hosts at steady state.
A role for NCR+ ILC3 in immunodeficient mice
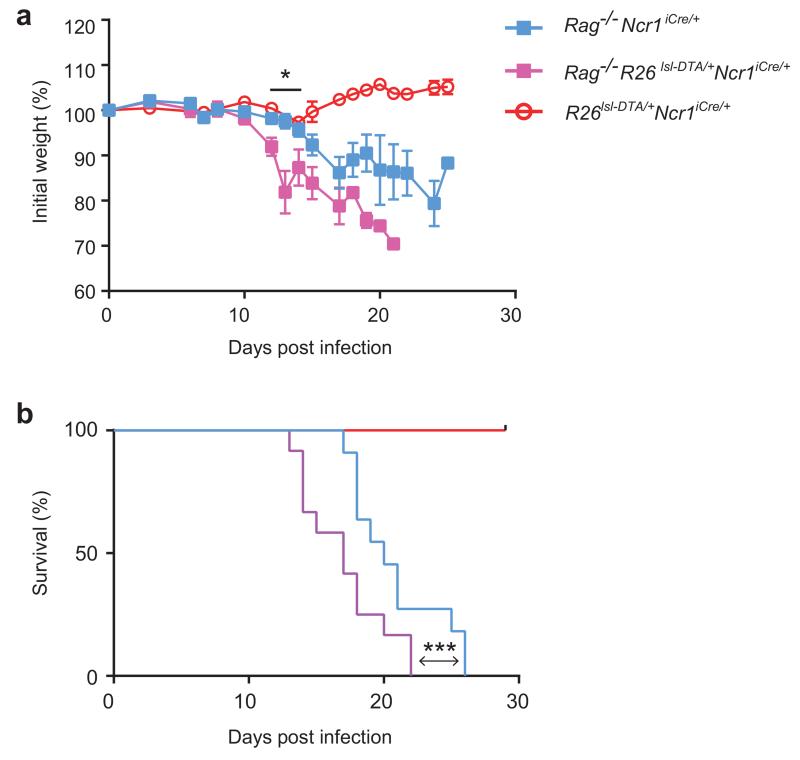
The results presented above contrast with previous reports of a critical role for NCR+ ILC3 in protection against C. rodentium in Rag2-, Tbx21-, Cxcr6- or Id2-deficient mice 11, 13, 14, 17, 25. As all these mouse models display profound or partial immunodeficiencies, we investigated the role of NCR+ ILC3 in immunodeficient and immunocompetent mice directly, by crossing R26DTA/+Ncr1-iCre mice with Rag2−/− mice, which lack T and B cells. Rag2−/−Ncr1iCre/+ mice, Rag2−/−R26DTA/+Ncr1-iCre and R26DTA/+Ncr1-iCre littermates were then infected with C. rodentium and their resistance to bacterial infection was evaluated by monitoring mouse weight and survival. Consistent with our previous findings, R26DTA/+Ncr1-iCre mice were resistant to C. rodentium (Fig. 6a and Fig. 6b). After initial control of the infection, Rag2−/−Ncr1iCre/+ mice lost weight and started to succumb to the disease on day 17 post-infection (Fig. 6a and Fig. 6b), as previously reported 26. Rag2−/−R26DTA/+Ncr1-iCre mice were even more susceptible to the infection than their Rag2−/−Ncr1-iCre littermates, as shown by their greater weight loss on days 12 to 14 post-infection, with the first mice dying on day 13 and all mice dead by day 22, as opposed to day 26 for the Rag2−/−Ncr1-iCre mice (Fig. 6a and Fig. 6b). Thus, NCR+ ILC3 cells were not essential for the control of C. rodentium in immunocompetent hosts, whereas they contributed to the control of C. rodentium in immunodeficient mice.
Figure 6. Role of NCR+ ILC3 during C. rodentium infection in immunocompetent and immunodeficient hosts.
Change in body weight (a) and survival curves (b) of Rag2−/−Ncr1-iCre mice, Rag2−/− R26DTA/+Ncr1-iCre and R26DTA/+Ncr1-iCre littermates following C. rodentium infection. Data are pooled from 2 independent experiments (n = 3-11 mice per genotype in total). In (a), the data shown are the means ± SEM, * P < 0.04 in unpaired Student’s t-tests comparing Rag2−/− R26DTA/+Ncr1-iCre and R26DTA/+Ncr1-iCre mice at day 12, 13 and 14 post-infection. In (b), statistical analysis was performed with Log-rank Mantel-Cox test, *** P < 0.001.
Role of NCR+ ILC3 in cecum homeostasis
In contrast to the lack of impact of NCR+ ILC3-deficiency in the small intestine or the bulk of the colon, the absence of NCR+ ILC3 in Mcl1fl/flNcr1-iCre mice during C. rodentium infection resulted in a decrease in cecum size, together with ulceration and bleeding (Fig. 7a). These pathological features in the cecum were also observed when Stat3fl/flNcr1-iCre mice were infected with C. rodentium (Fig. 7b). They were accompanied by crypt hyperplasia and inflammation, as shown by the H&E staining of cecum sections (Fig. 7c). At steady state, no alteration to the cecum, small intestine or colon was observed in any of the five genetic models of NCR+ ILC3 deficiency (Mcl1fl/flNcr1-iCre, R26DTA/+Ncr1-iCre, Tbx21fl/flNcr1iCre/+, Il22eGFP/eGFPNcr1-iCre and Stat3fl/flNcr1iCre/+) (data not shown). Thus, NCR+ ILC3 play a critical role in protecting the cecum during C. rodentium infection, and this function is abolished in conditions in which NCR+ ILC3 cannot produce IL-22.
Figure 7. Role of NCR+ ILC3 in cecum homeostasis during C. rodentium infection.
(a) Representative images of wild-type and Mcl1fl/flNcr1-iCre ceca 8 days after infection with 2 × 109 cfu C. rodentium. * indicates bleeding (left). Cecum size was determined by measuring the perimeter of the organ along the antimesenteric to mesenteric borders. The histogram shows data pooled from two independent experiments (n=2-4 mice per genotype per experiment).
(b) Representative images of wild-type and Stat3fl/flNcr1-iCre ceca 8 days after infection with 2 × 109 cfu C. rodentium. * shows bleeding. Cecum size was determined by measuring the perimeter of the organ along the antimesenteric to mesenteric borders. The histogram shows data pooled from two independent experiments (n = 3-4 mice per genotype per experiment). (a, b) The data shown are the mean ± SD. Statistical analyses were performed with unpaired Student’s t-tests. **P < 0.01 and ****P < 0.0001.
(c) Histological analysis of ceca from C57BL/6 and Stat3fl/flNcr1-iCre mice 9 days after infection. Quantification of cecum crypt length from C57BL/6 and Stat3fl/flNcr1-iCre mice (n = 8 mice per genotype in total) pooled from two independent experiments. The data shown are means ± SEM. Statistical analyses were performed with unpaired Student’s t-tests, ***P < 0.001.
(d-e) Characterization of NCR+RORγt− cells and ILC3 populations in different compartments of the gastrointestinal tract. (d) Representative flow cytometry plots showing RORγt-GFP and surface NKp46 expression in Lin−CD45+ cells isolated from the lamina propria of the small intestine, cecum and colon of naive RorcGFP/+ mice. Data is representative of two independent experiments (n=3 per group per experiment) (e) WT mice were infected with 1010 c.f.u. C. rodentium on day 0. Graphs show the ratio of the frequency of the indicated populations on day 8 post-infection (black bars) over that of day 0 (white bars). Representative data from one of two independent experiments are shown (n=6 mice) as means ± SD.
We further dissected the critical role of NKp46+ cells in the control of cecum homeostasis during C. rodentium infection, by examining the regional distribution of gut ILCs. C. rodentium is a colonic infection, but it begins in the cecum. In wild-type mice the protective role of NKp46+ cells was associated with a high frequency of NCR+ ILC3 in the cecum at steady state, contrasting with the low frequency of NCR+ ILC3 in the colon (Fig. 7d). At the junction of the small intestine and the colon, the cecum was globally characterized by a distribution of ILCs with features of both the small intestine, with a high frequency of NCR+ ILC3, and the colon, with a low frequency of NCR− ILC3 (Fig. 7d). In wild-type mice this distribution of intestinal ILCs was profoundly altered over the course of C. rodentium infection, as the frequency of NCR+ ILC3 was considerably lower in the cecum and colon (Fig. 7e). The causes and consequences of these changes remain to be fully explored. A characteristic feature of cecal NCR+ ILC3 is their high density of NK1.1 cell surface expression, contrasting with the low density of NK1.1 at the surface of small intestine NCR+ ILC3 and the intermediate expression on colon NCR+ ILC3 (Supplementary Fig. 7a). The role of NK1.1 is unknown, but these data provide an explanation for the deleterious effect of anti-NK1.1 treatment on Rag2−/− mice infected with C. rodentium reported in a previous study 14. Indeed, treatment with depleting anti-NK1.1 antibodies decreased the numbers of NCR+ ILC3, particularly in the cecum, but also in the colon to some extent, whereas it had no effect on NCR+ ILC3 numbers in the small intestine (Supplementary Fig. 7b). Anti-NK1.1 antibodies also depleted NKp46+RORγt− cells, including NK cells, ILC1 and ex-ILC3 (Supplementary Fig. 7b). However, all NKp46+RORγt− cells produce interferon-γ (IFN-γ) and IFN-γ-deficient mice are not more sensitive to C. rodentium infection 26, by contrast to IL-22-deficient mice 16. Overall, these results suggest that cecal NK1.1+ NCR+ ILC3 are involved in the control of C. rodentium infection, through their production of IL-22.
Discussion
The aim of this study was to dissect the biology of NCR+ ILC3, a subset of IL-22-producing innate lymphoid cells of the gut, and their interplay with adaptive immunity in the context of a colonic infection, in the C. rodentium model.
By using Ncr1iCre/+ mice, in which NCR+ cells are selectively targeted, we were able to highlight the intrinsic role of Tbx21, Mcl1 and Stat3 in the development, maintenance and function of NCR+ ILC3. The generation of Il22eGFP/eGFPNcr1-iCre mice made it also possible to carry out functional fate mapping of IL-22-producing NCR+ ILC3, without detectable cofounding effects due to under-reporting for the Il22-Cre allele 27. Using these mice, we detected no IL-22 production by intestinal RORγt−NKp46+ cells, including NK cells, ILC1 and ‘ex-ILC3’ 28. These findings contrast with those of other studies reporting the production of IL-22 by human and mouse NK cells from various inflamed tissues 29, 30, 31, 32, 33, and they lead us to reconsider whether RORγt− ILCs could produce IL-22.
We also show that neither the production of IL-22 by NCR+ ILC3, nor the cells themselves were essential for the control of intestinal infection by C. rodentium in immunocompetent hosts. In these mice, NCR+ ILC3 did not control the gut commensal microbiota either. Previous studies have shown that NCR+ ILC3 play a critical role in protection against C. rodentium in mice with profound or partial immunodeficiencies 11, 13, 14, 17. Our results are consistent with previous reports 10, 34, and challenge the current dogma in revealing that NCR+ ILC3 are not essential for the control of intestinal infections in the presence of T cells. We also show that NCR+ ILC3 can confer some protection against C. rodentium if T cells are absent. The critical role of IL-22-producing T cells in the control of C. rodentium infection has already been demonstrated 26. Previous studies investigated the contributions of TH17 and TH22 cells during the late phases of C. rodentium infection 26, and suggested that the IL-22 produced by the total ILC3 population (consisting of multiple subsets) was important early in C. rodentium infection, but that once T cells were activated, ILC3 appeared to be dispensable 13, 16, 22, 26, 27, 35. Thus, NCR+ ILC3, NCR− ILC3 and TH17 or TH22 cells can display some functional redundancy. It has also been suggested that NCR− ILC3 provide an adequate barrier to C. rodentium infection 36, as they produce large amounts of IL-22 in the gut 27. However, we show here that these cells are not sufficient to confer protection against C. rodentium in T cell-compromised mice in the presence or absence of NCR+ ILC3. Nevertheless, formal elucidation of the role of NCR− ILC3 in the control of the gut commensal and pathogenic microbiota awaits the development of tools for the selective ablation of this cell population.
The interplay between T cells and ILCs is a matter of great interest. ILCs can interact with CD4 T cells via MHC class II molecules, limiting pathological adaptive immune cell responses to commensal bacteria 37. It has also been suggested that T cells function as antigen-specific sensors for the activation of ILCs, to amplify and instruct local immune responses 38. We describe here another type of relationship between T cells and ILC3, in which NCR+ ILC3 can partly overcome the lack of T cells for protection against the pathogenic bacterium C. rodentium, but become redundant if T cells are present. This is, to our knowledge, the first formal demonstration of redundancy between T cells and ILCs. T cells and ILCs have evolved similar effector functions despite their use of different recognition strategies: a germline-encoded receptor for ILCs vs. antigen receptors generated by site-specific somatic recombination for T cells. It is thus tempting to speculate that ILC3 and TH17 and TH22 cells have co-evolved, leading to the selection of robust fail-safe mechanisms for ensuring adequate control of the commensal gut microbiota and protection against intestinal infections. Selective pressure for redundancy might have led to the similarities between ILCs and T cells in terms of their distribution and effector function.
Unexpectedly, severe pathological features were observed in the cecum when NCR+ ILC3 were ablated. Similar severe cecal lesions were also observed in IL-22-deficient mice upon infection with C. rodentium 16, supporting a role for NCR+ ILC3, the only NKp46+ cells that can produce IL-22, in the cecal phenotype. The cecum is the first site in the gastrointestinal tract to be colonized by C. rodentium during infection, with invasion of the colon occurring later (~day 8) 39. We thus propose a model in which NCR+ ILC3 are redundant with T cells for protection against lethality and colon disease during intestinal infections, but critical for protecting against cecum damage at the earliest stage of infection. The cecum is a blind pouch at the junction between the small intestine and the large intestine. It is a source of bacteria and it is also linked to the appendix in humans and mice, in which it is referred to as the cecal patch. The appendix has generally been seen as an evolutionary vestige, but recent phylogenetic studies challenge the apparent lack of function of the cecum and appendix 40. In particular, it has been suggested that the cecum and appendix provide an important reservoir for maintenance of the gut flora, which is anatomically protected from the intestinal lumen and could allow seeding of the gut after dysbiosis. Our data demonstrating that NCR+ ILC3 play a critical role in the homeostasis of the cecum upon bacterial infection opens up new possibilities for delineating the role of ILC3 in the pathophysiology of the cecum and appendix and in maintaining the health of the intestinal tract. In particular, it will be interesting to determine whether the balance between NCR− ILC3 and NCR+ ILC3 is involved in the still unknown mechanisms leading to appendicitis and resistance or susceptibility to inflammatory bowel diseases, as these two conditions may be related 41.
Online Methods
Mice
All the mice used were bred and maintained in specific pathogen-free facilities at the WEHI or CIML. C57BL/6, Rag2tm1.1Flv × Il2rgtm1.1Flv (Rag2γc), Stat3fl/fl 42, RorcGFP 43, Tbx21−/− 44, Mcl1fl/fl 23, Ncr1-iCre 45, R26DTA/+ (Rosa-DTA) and Rag2−/− × R26DTA/+ mice were used. All animal experiments were performed with the approval of either the Animal Ethics Committees of the Walter and Eliza Hall Institute according to National Health and Medical Research Council (NHMRC) of Australia guidelines, or the Animal Ethics Committees of the Ciml and in accordance with European laws.
Generation of Il22fl/fl mice
A loxP site was inserted downstream from Il22 exon 1 and a reporter/selection cassette (loxP-SA-P2A-DTR-T2A-eGFP-polyA-frt-Hyg-frt) was inserted downstream from the Il22 polyA sequence (Supplementary Fig. 1a). The genomic coordinates for the floxed region are: NCBIM37: chr10:117,642,470-117,647,823. Il22eGFP/eGFPNcr1-iCre mice were obtained at Mendelian frequencies; they developed normally and were fertile.
Inoculation with Citrobacter rodentium
Overnight cultures of bacterial strains were centrifuged and the pellet was resuspended in PBS. Food was withdrawn for 8 hours before infection, and the six- to nine-week-old mice (male and female) were inoculated by oral gavage with 200 μl of a suspension of 2 × 109 or 1 × 1010 cfu of C. rodentium. The viable count of the inoculum was determined retrospectively by plating dilutions of the inoculum on plates with appropriate antibiotics, or on McConkey agar (CM0007B; Oxoid) plates supplemented with 0.68% sodium thiosulfate (Sigma) and 0.08% ammonium citrate (Sigma). Animals were allocated to experimental groups so as to ensure even distributions of age, sex and weight. Infection experiments were carried out at least twice, regardless of sample size, to ensure reproducibility, and results shown correspond to combined experimental data from all repeats. Mice were weighed daily and feces were collected at two- to four-day intervals for bacterial counts. The viable bacterial count per 100 mg of feces was determined by plating serial dilutions feces, in duplicate, on medium containing selective antibiotics. The colon and small intestine were then harvested and their length was measured. For the assessment of bacterial dissemination, the spleen and liver were collected from infected mice, weighed and homogenized. The homogenates were then cultured overnight at 37°C on LB agar plates containing 10 mM naladixic acid or McConkey agar plates, as described above.
Isolation of lymphoid cells
Intestinal lymphoid cells were isolated from the intestine by incubation in 1 mM EDTA/Ca2+ Mg2+-free Hanks medium or in PBS supplemented with 1 mM EDTA, 15 mM HEPES and 10% FCS (1-2 × 20-30 min at 37°C), with gentle shaking to remove intestinal epithelial cells. Supernatants were discarded and tissues were then incubated, with gentle shaking, for 2 × 30 min or 1 × 45 min in 1-1.5 mg/ml (w/v) collagenase type III (Worthington), 200 μg/ml DNase I (Roche), 0.4 units/mL Dispase (Gibco) in mouse tonicity RPMI-1640 + 2% (v/v) FCS, or for 45 min at 37°C in RPMI supplemented with 15 mM HEPES and 300 units/mL collagenase type VIII (Sigma). Preparations were filtered and mononuclear cells were isolated on a 40%/80% or 90/100% Percoll gradient. Lymphocytes were recovered from the interface and washed twice. Mesenteric lymph nodes and splenic lymphoid cells were isolated by mashing organs on filters with 70 μm filters and resuspending the material retained on the filter in PBS supplemented with 1 mM EDTA.
In vitro T cell restimulation
mLN or splenic cells were cultured for 4 hours in complete RPMI in the presence of PMA (200 ng/mL), ionomycin (1 μg/mL) and BD GolgiStop and GolgiPlug (1:1000) (BD Biosciences), for the determination of intracellular cytokines.
Flow cytometry
For surface molecule analyses, cells were stained with antibodies against the following markers: CD19 (clone 1D3), CD3ε (clone 1452C11), NKp46 (clone 29A1.4), CD4 (clone GK1.4), NK1.1 (clone PK136), NKG2A/E/C (clone 20d5), CD94 (clone 18d3), KLRG-1 (clone 2F1), CD11c (clone N418) (from eBioscience), CD45.2 (clone 104), CD117 (clone 2B8), CD8α (clone 53-6.7), (I-A/I-E (clone M5/114.15.2), Ly6-C (clone AL-21), Ly6-G (clone 1A8) (from BD Bioscience) and CD49b (clone DX5, generated at the WEHI hybridoma facility). Live cells were identified by propidium iodide exclusion or with a fixable blue dead cell stain kit (Invitrogen). For intracellular staining, cells were stained for surface markers and with Fixable Viability Dye (eBiosciences), then fixed and permeabilized with either the eBioscience intracellular staining kit or the Cytofix/Cytoperm™ kit (BD Biosciences). Intracellular staining was then carried out with antibodies against the transcription factors Rorγt (clone Q31-378, BD Biosciences or AFKJS-9, eBiosciences), aand T-bet (clone eBio4B10, eBioscience) or the cytokines IL-22 (clone IL22JOP, eBioscience or kindly provided by JC Renauld; antibody coupled to Alexa Fluor 647 with the antibody labeling kit from Life Technologies), IFN-γ (clone XMG1.2) and IL-17A (clone TC11-18h10) (BD Bioscience). IL-22 production was induced in cells by stimulation with IL-23 (10-40 ng/ml) for 4-5 hours in the presence of BD GolgiStop™ and BD GolgiPlug™ (1:1000) (BD Biosciences) in complete RPMI-1640 medium (containing 10% FCS, 50 mM 2-mercaptoethanol, 1 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin) at 37°C. Flow cytometry was performed with a CantoII™, LSRFortessa™ X-20 or LSR-II cytometer (BD Biosciences) and FlowJo analysis software (Freestar).
Cell purification
Small intestinal lamina propria lymphocytes (pooled from 3-6 mice) were stained with surface antibodies and cells were purified with a BD FACSAria™ cell sorter (BD Biosciences). Cells were sorted into 50% v/v FCS/PBS or the appropriate cell culture medium in tubes previously coated with heat-inactivated FCS.
mRNAseq analysis
Total RNA for RT-quantitative PCR was prepared from purified ILC populations with an RNeasy mini kit (Qiagen). We generated cDNA from total RNA with OligodT primers and the Thermoscript reverse transcriptase (Invitrogen). Quantitative PCR was then performed with the SensiMix SYBR no-Rox kit (Bioline). Approximately 5 × 105 lin− (CD3−CD19−) CD45.2+ cells from each ILC subset were sorted from the intestines of RorcGFP/+ and RorcGFP/+ Tbx21+/− mice and two biological replicates were generated and subjected to 50 bp single-end sequencing on an Illumina HiSeq2000 at the Australian Genome Research Facility (Melbourne, Australia). More than 30 million reads were generated for each replicate and aligned with the GRCm38/mm10 build of the Mus musculus genome, using the Subread aligner 46. Genewise counts were obtained with featureCounts 47. Reads overlapping exons in annotation build 38.1 of the NCBI RefSeq database were included. Genes were filtered out and excluded from downstream analysis if they failed to achieve a CPM (counts per million mapped reads) value of at least 0.5 in at least two libraries. Counts were converted to log2 counts per million, quantile-normalized and precision-weighted with the voom function of the limma package 48. A linear model was fitted to each gene 49 and empirical Bayes moderated t-statistics were used to assess differences in expression 50. Genes were called differentially expressed if they achieved a false discovery rate of <= 0.1 and an average RPKM (reads per kilobase per million) of >= 8 in at least one of the two groups being compared.
Histology
Colons were gently flushed clean and cut into three sections, corresponding to the proximal, mid and distal colon. Alternatively, colons were cut longitudinally, cleaned and formed into a ‘Swiss’ roll, to visualize the full length of the colon on sectioning. The cecum was opened along the mesenteric side and its contents cleaned. Tissues were fixed in 10% neutral buffered formalin and processed for wax embedding and routine hematoxylin and eosin (H&E) staining. The ceca were sectioned longitudinally, parallel to the mesenteric axis. Images were collected and analyzed with Aperio™ ImageScope v11.2.0.780 software. Colitis severity was assessed blind, by determining a combined score of immune cell infiltration and epithelial damage based on hyperproliferation, crypt loss and the presence of ulcers (Supplementary Table 3).
Colon homogenates
Explants of distal colon were isolated, weighed and cut. They were then homogenized in 2 mL of complete RPMI 1640 medium (as previously described) and incubated for 24 h at 37°C. The supernatants were collected for cytokine measurement.
Cytokine measurement
Levels of IFN-γ, IL-17A, TNFα, IL-6, IL-1β, KC and IL-10 were assessed with a cytometric bead array (BD Bioscience), according to the manufacturer’s protocol.
Microbiota sequencing
Fresh mouse stool samples were collected and immediately stored at −80°C. DNA was extracted from stools, and its quality and quantity were checked with NanoDrop and Qubit, together with the BR assay. Barcoding PCR was carried out with indexed primers targeting the V3-V5 of the 16S rRNA gene. AccuPrime™ Pfx SuperMix (Invitrogen - 12344-040) was used for PCR. The PCR mix consisted of 18 μL of AccuPrime™ Pfx SuperMix, 0.5 μL of both the V3-340F and V5-926R primers (0.2 μM) and 1 μL of DNA (10 ng). PCR was carried out as follows: 95°C for 2 min, 30 cycles of 95°C for 20 s, 55°C for 15 s, 72°C for 5 min and a final extension step at 72°C for 10 min. PCR products were then quantified with the Qubit BR DNA assay kit. Equal amounts of each PCR mixture were pooled and thoroughly mixed. The amplicon pool was then loaded on an agarose gel (1.5%) and subjected to electrophoresis for 45 min at 100 V. The band corresponding to V3-V5 of the 16S rDNA amplicon was extracted from the gel and purified with the Qiaquick gel extraction kit (Qiagen - 28704). Amplicon quantity and integrity were checked by agarose gel electrophoresis and with the Qubit BR DNA assay kit. The amplicon was stored at −20°C until use. The 16S rDNA amplicon library was sequenced at the Institut Curie NGS platform, on a MiSeq machine, with the 2*300 bp V3 kit. Reads was merged with FLASH and trimmed with an in-house analysis script (length ≥ 500 bp, quality ≥ 25). Reads were clustered at 97% identity with the vsearch pipeline (http://zenodo.org/record/15524#.VPIujy4f2J8). Chimeric OTU were identified with UCHIME and discarded from subsequent analysis. The taxonomy of representative OTU sequences was determined with RDP classifier. OTU sequences were aligned, using ssu-align. The phylogenetic tree was inferred from multiple alignments of the OTUs generated with Fattree2. Statistical analyses were performed with R software (R Development Core Team 2012) and various related packages (Ade4,Vegan, ape, phyloseq). Weighted unifrac distances were calculated from a microbiota abundance table and phylogenetic tree. Statistical tests were performed with the Wilcoxon test (p-value). The false discovery rate method was used to correct for multiple testing (q-value).
SFB quantification
Mice were housed by genotype, and feces were collected at the ages of 10 and 15 weeks. The protocol for DNA extraction was adapted from the RBB+C method described by Zhongtang Yu and Mark Morrison, 2004. In brief, feces were homogenized in lysis buffer, using FASTprep. DNA was precipitated in ammonium acetate and washed with 70% ethanol. For the removal of RNA and protein, samples were further purified with the DNeasy Blood and Tissue Kit from Qiagen. Standard SYBRGreen qPCR was performed. Total bacteria detection was carried out with primers 1369f (5’-CGG TGA ATA CGT TCC CGG-3’) and 1492r (5’-TAC GGC TAC CTT GTT ACG ACT T-3’); SFB were detected with primers 779f (5’-TGTGGGTTGTGAATAACAAT-3’) and 1008r (5’- GCGGGCTTCCCTCATTACAAGG-3’). DNA from the feces of SFB single-colonized mice was used as a control for calculation of the relative amount of SFB by the Pfaffl-Method: Ratio=〖(E_target)〗^(ΔCT,target (calibrator-test))/〖(E_ref)〗^(ΔCT,ref (calibrator-test)).
Statistical analysis
All statistical analyses were performed with GraphPad Prism version 6.0d (Graphpad Software, San Diego, California, USA). We used unpaired two-tailed Student’s t-tests for pairwise comparisons. Unpaired and paired Student’s t-tests, nonparametric Mann-Whitney or Log-rank Mantel-Cox tests were used, as indicated. The data shown are the mean ± standard deviation (SD) or mean ± standard error of mean (SEM) as indicated in figure legends. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 throughout.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants and fellowships from the National Health and Medical Research Council (NHMRC) of Australia (G.T.B., S.C., N.H.) a NHMRC Dora Lush Postgraduate Research Scholarship (L.R); ARC Future Fellowships (J.R.G. and G.T.B.); We thank M. Camilleri, J. Vella, E. Loza and L. Inglis and the staff of the animal and flow cytometry facilities of WEHI and CIML for technical assistance; and Dr Susan Kaech (Yale), who kindly provided T-bet floxed mice. The E.V. laboratory members also thank L. Chasson and C. Laprie for providing expertise in histology and for the blind scoring of sections. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIIS. The EV laboratory is supported by the European Research Council (THINK Advanced Grant), the Ligue Nationale contre le Cancer (Equipe Labellisée) and by institutional grants from INSERM, CNRS and Aix-Marseille University to CIML. E.V. is a scholar of the Institut Universitaire de France.
Footnotes
Accession codes. GEO: mRNA-seq, GSE72909.
COMPETING FINANCIAL INTERESTS
E.V. is the cofounder and a shareholder of Innate Pharma. The other authors have no conflicting financial interest to declare.
REFERENCES
- 1.Dudakov JA, Hanash AM, van den Brink MR. Interleukin-22: Immunobiology and Pathology. Annu Rev Immunol. 2015;33:747–85. doi: 10.1146/annurev-immunol-032414-112123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 3.McKenzie AN, Spits H, Eberl G. Innate lymphoid cells in inflammation and immunity. Immunity. 2014;41:366–374. doi: 10.1016/j.immuni.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Diefenbach A, Colonna M, Koyasu S. Development, differentiation, and diversity of innate lymphoid cells. Immunity. 2014;41:354–365. doi: 10.1016/j.immuni.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells (ILC3s): Origin, differentiation, and plasticity in humans and mice. Eur J Immunol. 2015;45:2171–2182. doi: 10.1002/eji.201545598. [DOI] [PubMed] [Google Scholar]
- 6.Eberl G, Di Santo JP, Vivier E. The brave new world of innate lymphoid cells. Nat Immunol. 2015;16:1–5. doi: 10.1038/ni.3059. [DOI] [PubMed] [Google Scholar]
- 7.Eberl G, Colonna M, Di Santo JP, McKenzie AN. Innate lymphoid cells: a new paradigm in immunology. Science. 2015;348:aaa6566. doi: 10.1126/science.aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mebius RE, Rennert P, Weissman IL. Developing lymph nodes collect CD4+CD3− LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- 9.Constantinides MG, Gudjonson H, McDonald BD, Ishizuka IE, Verhoef PA, Dinner AR, et al. PLZF expression maps the early stages of ILC1 lineage development. Proc Natl Acad Sci USA. 2015;112:5123–5128. doi: 10.1073/pnas.1423244112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- 11.Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol. 2013;14:389–395. doi: 10.1038/ni.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, et al. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med. 2013;210:917–931. doi: 10.1084/jem.20122308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 2009;457:722–725. doi: 10.1038/nature07537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonnenberg GF, Fouser LA, Artis D. Functional Biology of the IL-22-IL-22R Pathway in Regulating Immunity and Inflammation at Barrier Surfaces. Adv Immunol. 2010;107:1–29. doi: 10.1016/B978-0-12-381300-8.00001-0. [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 17.Satoh-Takayama N, Serafini N, Verrier T, Rekiki A, Renauld JC, Frankel G, et al. The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity. 2014;41:776–788. doi: 10.1016/j.immuni.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 18.Artis D, Spits H. The biology of innate lymphoid cells. Nature. 2015;517:293–301. doi: 10.1038/nature14189. [DOI] [PubMed] [Google Scholar]
- 19.van de Pavert SA, Vivier E. Differentiation and function of group 3 innate lymphoid cells, from embryo to adult. Int Immunol. 2015:pii: dxv052. doi: 10.1093/intimm/dxv052. [DOI] [PubMed] [Google Scholar]
- 20.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol. 2015;16:306–317. doi: 10.1038/ni.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sciume G, Hirahara K, Takahashi H, Laurence A, Villarino AV, Singleton KL, et al. Distinct requirements for T-bet in gut innate lymphoid cells. J Exp Med. 2012;209:2331–2338. doi: 10.1084/jem.20122097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, Qiu J, Tu T, Yang X, Deng L, Anders RA, et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity. 2014;40:25–39. doi: 10.1016/j.immuni.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sathe P, Delconte RB, Souza-Fonseca-Guimaraes F, Seillet C, Chopin M, Vandenberg CJ, et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nature communications. 2014;5:4539. doi: 10.1038/ncomms5539. [DOI] [PubMed] [Google Scholar]
- 24.Qiu J, Guo X, Chen ZM, He L, Sonnenberg GF, Artis D, et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity. 2013;39:386–399. doi: 10.1016/j.immuni.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X, Liang Y, Zhang Y, Lasorella A, Kee BL, Fu YX. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity. 2015;42:731–743. doi: 10.1016/j.immuni.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu R, O’Quinn DB, Silberger DJ, Schoeb TR, Fouser L, Ouyang W, et al. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 2012;37:1061–1075. doi: 10.1016/j.immuni.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahlfors H, Morrison PJ, Duarte JH, Li Y, Biro J, Tolaini M, et al. IL-22 fate reporter reveals origin and control of IL-22 production in homeostasis and infection. J Immunol. 2014;193:4602–4613. doi: 10.4049/jimmunol.1401244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, et al. Regulated Expression of Nuclear Receptor RORgammat Confers Distinct Functional Fates to NK Cell Receptor-Expressing RORgammat(+) Innate Lymphocytes. Immunity. 2010;33:736–751. doi: 10.1016/j.immuni.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dhiman R, Indramohan M, Barnes PF, Nayak RC, Paidipally P, Rao LV, et al. IL-22 produced by human NK cells inhibits growth of Mycobacterium tuberculosis by enhancing phagolysosomal fusion. J Immunol. 2009;183:6639–6645. doi: 10.4049/jimmunol.0902587. [DOI] [PubMed] [Google Scholar]
- 30.Guo H, Topham DJ. Interleukin-22 (IL-22) production by pulmonary Natural Killer cells and the potential role of IL-22 during primary influenza virus infection. J Virol. 2010;84:7750–7759. doi: 10.1128/JVI.00187-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Male V, Hughes T, McClory S, Colucci F, Caligiuri MA, Moffett A. Immature NK cells, capable of producing IL-22, are present in human uterine mucosa. J Immunol. 2010;185:3913–3918. doi: 10.4049/jimmunol.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang Q, Ahn YO, Southern P, Blazar BR, Miller JS, Verneris MR. Development of IL-22-producing NK lineage cells from umbilical cord blood hematopoietic stem cells in the absence of secondary lymphoid tissue. Blood. 2011;117:4052–4055. doi: 10.1182/blood-2010-09-303081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu X, Weiss ID, Zhang HH, Singh SP, Wynn TA, Wilson MS, et al. Conventional NK cells can produce IL-22 and promote host defense in Klebsiella pneumoniae pneumonia. J Immunol. 2014;192:1778–1786. doi: 10.4049/jimmunol.1300039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macho-Fernandez E, Koroleva EP, Spencer CM, Tighe M, Torrado E, Cooper AM, et al. Lymphotoxin beta receptor signaling limits mucosal damage through driving IL-23 production by epithelial cells. Mucosal Immunol. 2015;8:403–413. doi: 10.1038/mi.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–1325. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gasteiger G, Rudensky AY. Interactions between innate and adaptive lymphocytes. Nat Rev Immunol. 2014;14:631–639. doi: 10.1038/nri3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiles S, Clare S, Harker J, Huett A, Young D, Dougan G, et al. Organ specificity, colonization and clearance dynamics in vivo following oral challenges with the murine pathogen Citrobacter rodentium. Cell Microbiol. 2004;6:963–972. doi: 10.1111/j.1462-5822.2004.00414.x. [DOI] [PubMed] [Google Scholar]
- 40.Smith HF, Fisher RE, Everett ML, Thomas AD, Bollinger RR, Parker W. Comparative anatomy and phylogenetic distribution of the mammalian cecal appendix. J. Evol. Biol. 2009;22:1984–1999. doi: 10.1111/j.1420-9101.2009.01809.x. [DOI] [PubMed] [Google Scholar]
- 41.Frisch M, Pedersen BV, Andersson RE. Appendicitis, mesenteric lymphadenitis, and subsequent risk of ulcerative colitis: cohort studies in Sweden and Denmark. Bmj. 2009;338:b716. doi: 10.1136/bmj.b716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol Cell Biol. 2001;21:1621–1632. doi: 10.1128/MCB.21.5.1621-1632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eberl M, Jomaa H, Hayday AC. Integrated immune responses to infection - cross-talk between human gammadelta T cells and dendritic cells. Immunology. 2004;112:364–368. doi: 10.1111/j.1365-2567.2004.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nieuwenhuis EE, Neurath MF, Corazza N, Iijima H, Trgovcich J, Wirtz S, et al. Disruption of T helper 2-immune responses in Epstein-Barr virus-induced gene 3-deficient mice. Proc Natl Acad Sci USA. 2002;99:16951–16956. doi: 10.1073/pnas.252648899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc Natl Acad Sci USA. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thakur C, Lu Y, Sun J, Yu M, Chen B, Chen F. Increased expression of mdig predicts poorer survival of the breast cancer patients. Gene. 2014;535:218–224. doi: 10.1016/j.gene.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 48.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 50.McCarthy DJ, Smyth GK. Testing significance relative to a fold-change threshold is a TREAT. Bioinformatics. 2009;25:765–771. doi: 10.1093/bioinformatics/btp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.