Abstract
The prevalence of overweight and obesity in developed and developing countries has greatly increased the risk of insulin resistance and type 2 diabetes mellitus. It is evident from human and animal studies that obesity alters microRNA (miRNA) expression in metabolically important organs, and that miRNAs are involved in changes to normal physiology, acting as mediators of disease. miRNAs regulate multiple pathways including insulin signaling, immune-mediated inflammation, adipokine expression, adipogenesis, lipid metabolism, and food intake regulation. Thus, miRNA-based therapeutics represent an innovative and attractive treatment modality, with non-human primate studies showing great promise. In addition, miRNA measures in plasma or bodily fluids may be used as disease biomarkers and predictors of metabolic disease in humans. This review analyzes the role of miRNAs in obesity and insulin resistance, focusing on the miR-17/92, miR-143-145, miR-130, let-7, miR-221/222, miR-200, miR-223, miR-29 and miR-375 families, as well as miRNA changes by relevant tissue (adipose, liver and skeletal muscle). Further, the current and future applications of miRNA-based therapeutics and diagnostics in metabolic disease are discussed.
Introduction
In 2013, the American Medical Association recognized obesity as a disease, highlighting the importance in public health. Of particular concern is the incidence of overweight and obesity in children, with an estimated one-third of children and adolescents affected in the United States.1 In addition, metabolic and food intake programming may result in transgenerational metabolic dysfunction, with parental obesity potentially affecting the metabolic health of offspring and potentially future generations.2, 3, 4 Over the last decade, the nutrition–immunity theory has been hypothesized, suggesting that starvation leads to immunosuppression and that overnutrition/obesity promotes inflammation.5 During overnutrition, inflammation typically occurs in visceral adipose depots, where changes include increased immune cell infiltration, proliferation and activation, along with adipocyte hypertrophy, impaired adipogenesis, and inflammatory peptide production by immune cells and adipocytes.6, 7 Insulin-resistant adipocytes exhibit abnormal lipolysis (increasing circulating free fatty acids levels), which promotes ectopic lipid storage (liver, muscle).8 Abnormal hepatic function in metabolic disease (dysregulated gluconeogenesis and lipogenesis/fatty acid esterification) is likely a result of hepatocellular insulin resistance (IR), as well as insulin signaling independent mechanisms and circulating fatty acid (substrate)-dependent mechanisms.8 In the liver of organisms with metabolic disease, it is common to find activation of resident immune cell populations (Kupffer cells), lipid accumulation (fatty liver disease/nonalcoholic steatohepatitis, hepatic steatosis) and inflammatory chemokine production.9, 10 Skeletal muscle, however, tends to be a target of inflammatory cytokines originating from inflamed visceral adipose and liver tissue and is often characterized by ectopic lipid accumulation.11 The kidney and vasculature are similarly affected by circulating pro-inflammatory and pro-atherogenic compounds,11 providing a link between metabolic and cardiovascular disease. Finally, obesity and IR involve neurologic (hypothalamic) changes, which may increase food intake and the defense of increased body fat levels.12
Insulin resistance (IR), at a cellular level, is the result of blunted insulin-stimulated tyrosine phosphorylation of the insulin receptor (IRS-1/IRS-2)13 and associated downstream signaling events (translocation of glucose transporters to cell membranes) in glucose-metabolizing cells.14 Stress and inflammatory signaling events (JNK activation) result in serine phosphorylation (instead of tyrosine) of insulin receptor proteins, inhibiting insulin signaling (cellular IR).15 The current, prevailing theory is that inflammatory/stress signaling is the common causal event leading to cellular IR in energy homeostatic tissues.15 Briefly, cellular abnormalities that result in inflammation impaired insulin signaling. Impaired insulin signaling in turn contributes to the metabolic abnormalities specific to the cell type affected (adipocyte, hepatocyte, myocyte). Reviews on the role of inflammation in metabolic disease are available.15, 16 However, models of IR and obesity demonstrate abnormalities in a myriad of processes, often concomitantly, including inflammation and abnormal microRNA (miRNA) expression in various tissues and cell types.17, 18, 19 Determining if and how miRNA changes are causal in the development of IR is the main challenge of obesity-related miRNA research.
Mature miRNAs are small noncoding single-stranded RNAs (~21 nucleotides) that negatively regulate or ‘repress' target gene expression. The first miRNA was described in Caenorhabditis elegans, in 1993.20, 21 MiRNAs have since been identified in all multicellular organisms studied to date, demonstrating astonishing evolutionary conservation between vastly dissimilar species (humans, mice, fish and nematodes).22 miRNAs have important molecular roles in normal physiology and development as well as in disease processes such as cancer and obesity.
miRNA biogenesis, post-transcriptional editing and function
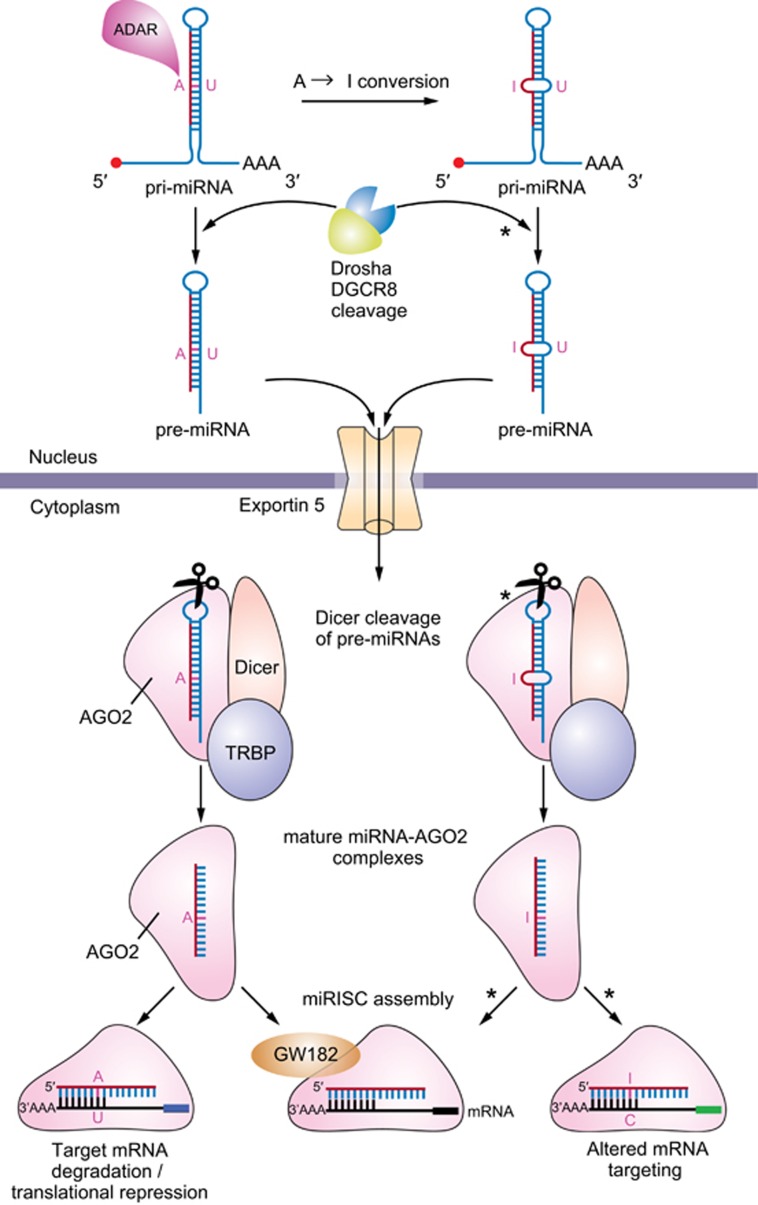
In contrast to messenger RNAs (mRNAs), miRNAs are not translated. Mature miRNAs exert biological effects by regulating the post-transcriptional regulation of protein-coding mRNAs via two recognized mechanisms: target transcript degradation/decay and inhibition of target transcript translation. There are an estimated ~45 000 miRNA-targeting sites in the human genome, affecting the expression of ~60% of genes.23 miRNA genes can be coded by the intronic regions of protein-coding genes (intronic miRNAs) or by sequences outside protein-coding genes (intergenic miRNAs).24 In addition, miRNAs genes are often found in polycistronic clusters (independent of intronic or intergenic genomic organization), with one transcriptional event (RNA polymerase II) resulting in a primary miRNA (pri-miRNA) strand with multiple stem-loop structures, which may be canonically processed into multiple miRNAs. Pri-mRNA transcripts in the nucleus, whether mono or polycistronic, undergo trimming and processing by the Drosha, DGCR8 RNase III complex into pre-miRNAs before export into the cytoplasm by exportin-5/GTP61, as shown in Figure 1.25 In the cytoplasm, the Dicer complex (Dicer, AGO2 and TRBP) cleaves the hairpin loop of the cytoplasmic pre-miRNAs resulting in an miRNA/miRNA* duplex (two distinct complementarily bound RNA strands called the guide and passenger strands (*), respectively) and assists in the loading of either strand into the miRNA-induced silencing complex.26 The miRNA-induced silencing complex is composed of Argonaute 2 (AGO2) and GW182 proteins in association with a single-stranded miRNA, most often the guide strand.27 The miRNA-induced silencing complex is capable of binding to mRNAs that are targeted by the matured miRNA guide strand. This is accomplished by the binding of the ‘seed sequence' (at the 5′ end of a mature miRNA) to the 3′ untranslated region of target mRNA transcripts. In addition, the middle and 3′ end of the miRNA can also bind mRNA and contribute to gene repression.28
Figure 1.
microRNA biogenesis and ADAR-mediated editing during miRNA maturation. ADAR, family of adenosine deaminase; miRISC, miRNA-induced silencing complex.
Clearly, miRNA maturation depends on multiple pathways that regulate the biogenesis/stability/formation of miRNA-protein complexes which modulate gene expression. Though these processes have been thoroughly reviewed elsewhere,27, 29 we will briefly touch on endogenous RNA editing processes30 and their relevance to metabolic disease. A family of adenosine deaminases (ADARs) edits double-stranded RNA species, proximately responsible for the conversion of adenosine residues to inosine. Ultimately, ADARs-mediated A-to-I conversions may result in guide strand base alteration(s) and a change in targeting efficiency or complete mRNA re-targeting. Also, inhibition of Drosha (pri-miRNAs) and Dicer, TRBP, AGO2 (pre-miRNAs) cleavage may occur depending on the location of A-to-I conversion.31, 32 A-to-I conversions are best detected by next-generation sequencing, which will be discussed later in this review.33 The physiological ramifications of ADAR activity in normal and pathophysiology are still being examined, though it is likely that a majority of RNAs in a cell are edited by ADARs to some degree.34 ADAR-catalyzed conversions occur mainly in noncoding RNA sequences including the introns and untranslated regions of mRNAs and small RNA species such as miRNAs, however, the processes controlling specificity of adenosine conversion is undefined.34 It is clear, though, that ADARs affect miRNAs in humans, mice and rats and thus their role in miRNA-mediated metabolic disease should be considered when using these models. Glucose and JNK signaling regulate ADAR2 expression in the pancreatic beta cells of mice suggesting that ADARs may have a role in pancreatic adaptions in overnutrition.35
Experimental approaches to miRNA research
Systemic and organ-specific knockouts/transgenic mouse strategies are available for some miRNAs/miRNA families (http://rna.keck.ucsf.edu/miRKO-DB); this often depends on the genomic organization of the miRNA/miRNA family of interest and on embryonic lethality of miRNA knockout. Gain-of-function strategies include injection/transfection with synthetic miRNA mimics and vector-mediated miRNA overexpression by lentivirus or AAV infection. Other loss-of-function strategies include injection/transfection with anti-miRNA oligonucleotide or miR-sponges (primary cells and drosophila). miR-sponges are transgenes that harbor multiple miRNA binding sites, effectively depleting endogenous miRNA levels. In contrast, anti-miRs bind to mature miRNAs canceling out (derepressing) their effect on-target genes in a dose-dependent manner. Anti-miR approaches are thoroughly reviewed by Stenvang et al.36
There is a substantial body of literature characterizing miRNA expression in lean and obese mice and humans. However, the clarity of many publications is affected by changes in and nonstandard uses of miRNA nomenclature; miRbase.org can be used to research nomenclature. Furthermore, many publications do not cite miRNA sequences or fail to identify particular family members when reporting miRNAs with multiple sequence variants (for example, Let-7). Given their central importance in regulation of pleiotropic pathways that cut across organ systems involved in IR, miRNAs may act in concert in determining susceptibility/severity of defects associated with obesity/IR.
The Pubmed database was searched for relevant studies published between 15 September 2008 and 23 January 2015. The search terms ‘(obesity) AND microRNA' identified 344 articles, ‘(insulin resistance) AND microRNA' identified 225 articles. The literature was screened for seminal findings and for the most reported and mechanistically studied miRNAs in obesity and metabolic disease. Focus was placed on in vivo interventional (especially non-human primate) studies and human plasma biomarker research. Additional searches for specific miRNAs and ‘obesity' or ‘insulin resistance' (for example, ‘(miR-221) AND obesity' identified eight articles) were performed. Reviews, commentaries and non-original research articles were excluded.
The initial report (2009) of miRNA expression in human obesity by Kloting et al.37 examined a small cohort (n=15; 53–73 years; body mass index 25.4–38.1 kg m−2) of overweight and obese subjects with preserved glucose tolerance compared to those with type 2 diabetes mellitus (T2DM). Many of the miRNAs first identified in this study (miR-17-5p, -145, -34a, -132, -181a) have been confirmed independently in human studies and rodent models of obesity/IR.37
The miR-17/92 family
The family contains three polycistronic miRNA genes producing 15 mature miRNA species (miR-17, 18a, 18b, 20a, 20b, 93, 106a, 106b). Kloting et al.37 reported significantly lower expression of miR-17-5p in the omental adipose tissue of T2DM patients compared with normal glucose tolerance (NGT) and a negative correlation with visceral fat area (Table 1). Since these initial findings, there have been multiple reports linking plasma levels of miR-17-5p with cardiometabolic disease, suggesting usefulness as a biomarker in multiple diseases.38, 39, 40 Heneghan et al.39 confirmed lower than healthy control miR-17-5p expression in human omental adipose tissue as well as in blood from obese patients. Similar findings were also reported in human atherosclerosis by Fichtlscherer et al.,38 where miR-17 was significantly lower in the plasma of patients with coronary artery disease. Although none of these studies looked specifically at plasma exosomal miRNA expression, miR-17 is well known to be associated with the exosomal compartment (Table 2).40 Interestingly, immunologic studies by Steiner et al.41 suggest that miR-17/92 family members potentiate T helper cell proliferation. Recently, Li et al.42 reported that miR-17-5p was increased during human adipose-derived mesenchymal stem cell adipogenesis in vitro and that miR-17-5p mimic transfection resulted in enhanced adipogenesis in the same cell population via repression of bone morphogenetic protein 2 and increased CCAAT/enhancer-binding protein alpha and peroxisome proliferator-activated receptor gamma expression. Taken together, these findings suggest that the miR-17/92 family may have a role in adipogenesis and cardiometabolic disease, and be a useful positive control for plasma/exosome-based studies of obese and/or cardiovascular disease patients.
Table 1. miRNAs involved in obesity: insights from in vivo models and whole adipose.
| miRNA | Finding(s) | Intervention/phenotype | Model | Target/pathway | Author |
|---|---|---|---|---|---|
| miR-132, -181a, -17-5p, -155 | Omental, subQ fat, correlation with glucose metabolism, macrophage infiltration | NGT vs T2DM (n=15 total) | Human | — | Kloting et al.37 |
| miR-132, -17-5p, -143, -145 | Omental, subQ fat, plasma | Obese vs lean (n=50 total) | Human | — | Heneghan et al.39 |
| miR-132, -184, -338-3p | ↑, ↓, ↓ | Improved beta cell mass and activity | Mouse (db/db), Human and Rat cells | — | Nesca et al.122 |
| miR-143 | ↑ Mesenteric fat | DIO-IR, 45% E from fat | Mouse (C57BL/6J) | PPARγ, aP2, leptin pathways | Takanabe et al.46 |
| miR-143 | ↑ Insulin resistance, Protected from DIO-IR | Conditional overexpression of miR-143, miR-143 deficient | Mouse | ORP8 | Jordan et al.48 |
| miR-27a, | ↑ Omental whole adipose, Change not significant | Obese vs lean (n=15 vs 15) | Human | — | Viesti et al.123 |
| miR-143, -145 | Omental, subQ and liver difference not significant | Obese vs lean (n=15 vs 15) | Human | — | Viesti et al.123 |
| Let-7 | ↑ Insulin resistance, Protected from DIO-IR | Overexpressed—global and pancreas, whole-body inhibition | Mouse | INSR, IRS-2 | Frost and Olson59 |
| Let-7-d | ↑ Amnion | Obese pregnant women | Human | — | Nardelli et al.63 |
| miR-222, -27a | ↑ Whole adipose | Genetic IR (non-obese) | Rat (GK, WKY, BN) | — | Herrera et al.66 |
| miR-222 | ↑ Omental whole adipose | Gestational diabetes mellitus | Human | — | Shi et al.68 |
| miR-130 | ↑ Whole subcutaneous adipose | Obese vs lean (nondiabetic, normotensive) | Human (female) | — | Lee et al.52 |
| miR-130 | ↑ Whole subcutaneous adipose | HFD vs SCD | Mouse | — | Kim et al.124 |
| miR-126, -193b | ↑ SubQ adipose | Obese, IR | Human (female) | CCL2 | Arner et al.125 |
| miR-29 | ↑ Urine | Normal weight with T2DM, normo- vs albuminaria | Human | — | Peng et al.93 |
| miR-10a, -200a, -409-5p, -125-3p | ↑ Hypothalamus | Perinatal leptin blockade and DIO | Rat (Wistar) | Adiponectin pathway | Benoit et al.75 |
| miR-200a, b, -420 | ↑ Hypothalamus | Genetic obesity and IR | Mouse (ob/ob, db/db) | Insulin and leptin pathways | Crepin et al.76 |
| miR-34a, -146a, -199a-3p, -203, -210, -383 | - | Increased beta cell apoptosis | Mouse (db/db), Human and Rat cells | — | Nesca et al.122 |
| miR-375 | ↑ Pancreas | Genetic obesity | Mouse (ob/ob), KO | — | Poy et al.96 |
| miR-375, -802 | ↑ Serum, ↑ Serum, Liver, epi WAT | T2DM vs NGT, SCD vs HFD | Human plasma, Mouse | — | Higuchi et al.99 |
Abbreviations: CCL2, chemokine (C-C motif) ligand 2; DIO, diet-induced obesity; HFD, high-fat diet; IR, insulin resistance; miRNA, microRNA; NGT, normal glucose tolerance; PPARγ, peroxisome proliferator-activated receptor gamma; SCD, standard chow diet; T2DM, type 2 diabetes mellitus.
Table 2. Circulating miRNAs in human cardiometabolic disease.
| miRNA | Finding(s) | Intervention/phenotype | Model | Target/pathway | Author |
|---|---|---|---|---|---|
| miR-221, -130b, -142-3p | ↓, ↑, ↑ Plasma | Obesity | Human (children) | — | Prats-Puig et al.54 |
| miR-17-5p | ↓ Plasma | Obesity | Human | — | Heneghan et al.39 Hulsmans et al.40 |
| miR-17-5p | ↓ Plasma | Coronary artery disease | Human | — | Fichtlscherer et al.38 |
| miR-326, Let-7a, f | ↑, ↓ Plasma exosomes | Treatment naive diabetic | Human | — | Santovito et al.62 |
| Let-7b, miR-130a, | ↑, ↓ Serum | Lower glycemic index diet | Human | — | McCann et al.61 |
| miR-221, -130b, -423-5p | ↓ Plasma | Obesity | Human | — | Ortega et al.53, 69 |
| miR-221, let-7g | ↑ Plasma | Metabolic syndrome, females | Human (♀) | — | Wang et al.74 |
| miR-222, -142-3p | ↑ Plasma | Obesity | Human | — | Ortega et al.53, 69 |
| miR-222 | ↑ Plasma HDL | Familial hyperchosterolemia vs normo- | Human | — | Vickers et al.70 |
| miR-29b, -223, -126 | ↓ Plasma | T2DM vs matched controls | Human | — | Zampetaki et al.81 |
| miR-15b | ↑ Plasma | Obesity/T2DM | Human | — | Pescador et al.108 |
| miR-138, -376a, -503 | ↓ Plasma | Obesity/T2DM | Human | — | Pescador et al.108 |
| miR-34a, -375 | ↑ Plasma | T2DM vs IGT and NGT | Human | — | Kong et al.92 |
| miR-122 | ↓ Plasma | Severe NASH | Human | — | Miyaaki et al.141 |
| miR-375, -802 | ↑ Serum, | T2DM vs NGT | Human | — | Higuchi et al.99 |
Abbreviations: HDL, high-density lipoprotein; IGT, impaired glucose tolerance; miRNA, microRNA; NASH, nonalcoholic steatohepatitis; NGT, normal glucose tolerance; T2DM, type 2 diabetes mellitus.
The miR-143-145 cluster
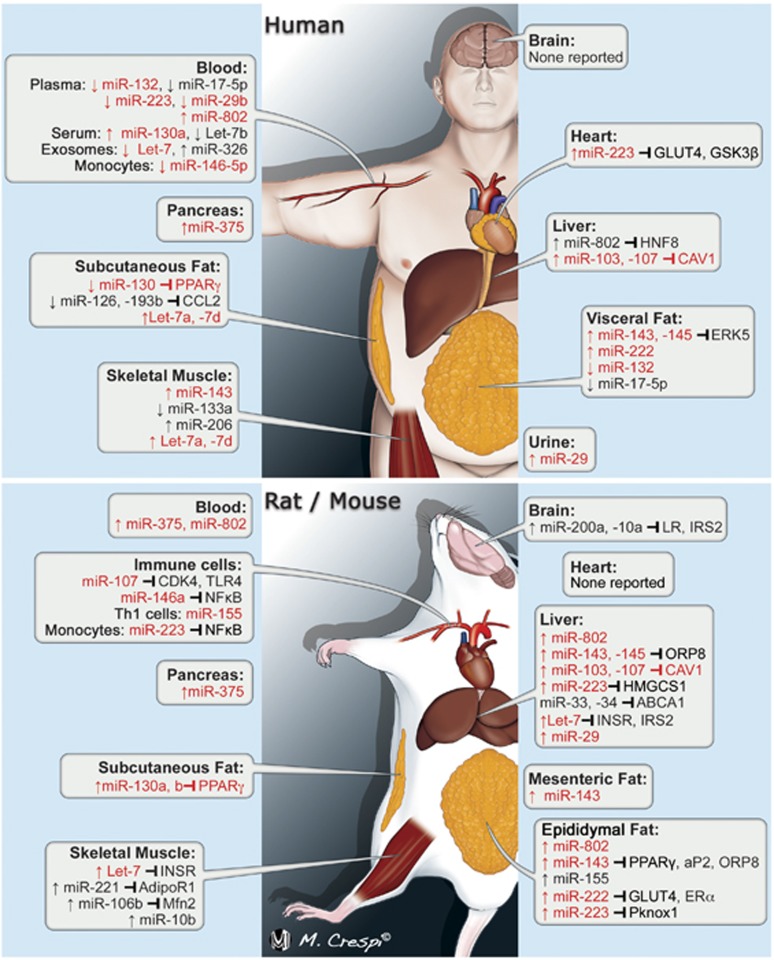
The role of miRNAs in adipocytes/adipogenesis is the most studied area of miRNA biology as it relates to obesity and IR, with 148 citations under ‘(microRNA) AND adipogenesis'. Of the adipogenesis-related miRNAs, miR-143 and miR-130 are the best studied (Table 3). miR-143 and miR-145 are located in close genomic proximity, may be transcribed in a bicistronic fashion,43 and consequently are often studied/reported concomitantly. miR-143 was first identified as a positive regulator of human adipocyte differentiation in 2004 via effects on ERK5 signaling.44 miR-143 is the only miRNA to date shown to be similarly regulated during human and mouse adipocyte differentiation (Figure 2).45 Figure 2 illustrates obesity/IR-related miRNA expression changes in humans and rodents, comparing and contrasting major findings. miR-143 expression was increased in the mesenteric adipose of high-fat diet (HFD)-fed mice,46 and tumor necrosis factor alpha treatment decreased miR-143 expression suggesting that obesity-associated inflammation may dysregulate miR-143 expression affecting adipogenesis (Table 1).47 Despite miR-143 being a positive regulator of adipogenesis, miR-143-145 cluster knockout mice were protected from obesity-induced IR, while conditional overexpression of miR-143 results in worsened IR in diet-induced obesity (DIO). miR-143 may increase IR by increasing the degradation of positive regulator of AKT signaling, oxysterol binding protein-like 8.48 Additional mechanisms for miR-143 in metabolic disease have not been reported. The role for miR-145 in obesity is less clear, though a putative role has emerged in lipolysis. Obesity increased the expression of the miR-143–145 cluster in adipose tissue/adipocytes and liver of humans and mice,39, 49, 50, 51 and overexpression of miR-145-enhanced tumor necrosis factor alpha secretion and lipolysis in human adipocytes in vitro via a nuclear factor kappa b mechanism.49 A conflicting report, however, showed that miR-145 overexpression suppressed lipolysis.50 Although significance of miR-145 in IR is under investigation, the genomic association with miR-143 suggests potential.
Table 3. Adipocytes: miRNAs involved in adipogenesis, lipogenesis and drug effects.
| miRNA | Finding(s) | Intervention/Phenotype | Model | Target/pathway | Author |
|---|---|---|---|---|---|
| miR-222, -29a, -27a | ↑ | High glucose media | 3T3-L1 cells (mouse) | — | Herrera et al.66 |
| miR-302a, -664, -1264, -378 | ↑ | Pioglitazone treatment | Human primary pre-adipocytes from subQ | PPARγ | Yu et al.126 |
| miR-338-5p, -143, 30b, -378 | ↑ | Pioglitazone treatment | Human primary pre-adipocytes from visceral | PPARγ | Yu et al.126 |
| miR-378 | ↑ Adipose, ↓ lipolysis in vitro | Cachexia, inhibited miRNA | Humans, human primary adipocytes | ATGL | Kulyte et al.127 |
| miR-375 | ↑ Adipogenesis | Overexpressed miRNA | 3T3-L1 cells (mouse) | — | Ling et al.100 |
| miR-143 | ↑, Blocked differentiation | Adipogenesis, inhibited miR-143 | Human, mouse | ERK5 | Esau et al.44 Kajimoto et al.45 |
| miR-143 | ↑, ↓ | Adipogenesis, in obesity models -TNFα treatment | Mouse primary adipocytes (ob/ob, DIO) | — | Xie et al.128 |
| miR-126, -193b, -143, Let-7d | CCL2 secretion decreased | Overexpressed miRNA | Human and mouse adipocytes | CCL2 | Arner et al.125 |
| Let-7 | ↓ Adipogenesis | Overexpressed miRNA | Human and mouse (3T3-L1) | HMGA2 | Sun et al.64 Wei et al.65 |
| miR-344 | ↓ Adipogenesis | Overexpressed miRNA | 3T3-L1 cells (mouse) | GSK3β | Chen et al.129 |
| miR-34a | ↓ Adiposity, ↑ Browning of white fat | Lentiviral-mediated downregulation of miR-34a | Mouse | FGFR1, SIRT1 | Fu et al.130 |
| miR-130a, -130b | ↓ Adipogenesis | Overexpressed miRNA | Human pre-adipocytes | PPARγ | Lee et al.52 |
| miR-130b | ↓ Fat accumulation | Microvesicles enriched in miR-130b | Porcine adipocytes | PPARγ | Pan et al.55, 56 |
| miR-130 | ↑ | TNFα treatment | 3T3-L1 cells (mouse) | PPARγ, NFκB (p65) pathway | Kim et al.124 |
| miR-181a | ↑ Lipogenesis | Overexpressed miRNA | Porcine adipocytes | TNFα, lipogenic/lipolytic pathways | Li et al.131 |
| miR-17-5p, 106a | ↑ Adipogenesis, ↓ Adipogenesis | Overexpressed, inhibited miRNA | Human adipose-derived mesenchymal stem cells | BMP2 | Li et al.42 |
| miR-145, let-7d | ↑ Lipolysis and TNFα release, ↓ lipolysis | Overexpressed miRNA | Human adipocytes | ADAM17/NF-κB and HSL pathways | Lorente-Cebrian et al.49 |
| miR-21 | ↓, Improved PI3K/Akt signaling | High glucose media, overexpressed miRNA | 3T3-L1 cells (mouse) | PTEN | Ling et al.132 |
| miR-222 | ↓ Insulin- stimulated glucose uptake | Inhibited miRNA | 3T3-L1 cells (mouse) | ERα, GLUT4 | Shi et al.68 |
| miR-103, -107 | ↑ Insulin sensitivity | Inhibited miRNA whole body | Mouse | CAV1/Insulin signaling | Trajkovski et al.133 |
| miR-145 | ↓ Lipolysis in vitro | Inhibited miRNA | Mouse primary adipocytes | FOXO1, ABHD5/KSRP pathway | Lin et al.50 |
| miR-34a | Browning of fat | Inhibited miRNA | Mouse | FGF21, SIRT1 | Fu et al.130 |
Abbreviations: BMP2, bone morphogenetic protein 2; miRNA, microRNA; NF-κB, nuclear factor kappa b; PPARγ, peroxisome proliferator-activated receptor gamma; PTEN, phosphatase and tensin homolog; TNFα, tumor necrosis factor alpha.
Figure 2.
Obesity/insulin resistance-related miRNA expression changes in humans and in murine models. miRNAs in red font were differentially expressed in human and murine obesity with arrow indicating up- or downregulation. Gene symbols or conventional gene synonyms are used to show miRNA–mRNA repression. Gene symbol in red font indicates conserved miRNA-targeting between species. The image is published with the permission of Mr Massimiliano Crespi, medical illustrator.
The miR-130 family
The miR-130 family has four members in humans: miR-130a, miR-130b, miR-301a and miR-301b (on three chromosomes). In 2011, Lee et al.52 showed lower miR-130a and -130b expression in the abdominal subcutaneous adipose of nondiabetic, obese women compared with lean subjects, although murine models have conflicting findings (Table 1). Plasma miR-130 expression was found to be lower in obese humans.53 miR-130a is highly expressed in human endothelial cells. Interestingly, patients with coronary artery disease have lower circulating miR-130a expression.38 However, these lower levels of miR-130a may be due to increased cardiovascular risk, metabolic disease or both. In contrast, it should be mentioned that miR-130b was found to be higher in the plasma of obese children (9 years of age, Table 2).54 Bioactive miR-130 in the plasma may affect distal tissue when transported by and taken up from the circulation. Microvesicular transport of plasma miRNA is a putative mechanism by which miRNAs could be mediators of local and systemic IR, similar to cytokines. Pan et al.55, 56 reported that HeLa cells overexpressing miR-130b produced microvesicles capable of decreasing adipogenesis and peroxisome proliferator-activated receptor gamma expression in cultured porcine adipocytes. Although these experiments do not provide in vivo relevance, they support the concept of microvesicle-mediated miRNA transport by the plasma.
The let-7 family
Let-7 is well-conserved across species, has a substantial role in developmental processes, is considered an ‘oncogenic' miRNA in vertebrates and has a substantial role in metabolic disease. Let-7 was the first human miRNA discovered,57 and the family contains 11 members on three chromosomes in humans. Transgenic mouse experiments have shown that let-7 is a potent regulator of glucose metabolism and peripheral IR, by targeting IGF1R, insulin receptor (INSR) and insulin receptor substrate-2 (IRS-2) in skeletal muscle (Table 4) and liver tissues.58 Let-7 and the RNA binding protein Lin28 form a regulatory axis affecting insulin production and sensitivity. Lin28 overexpressing mice fed a HFD had dramatically improved glucose metabolism via inhibition of let-7 (as well as let-7-independent mechanisms). Similarly, let-7 overexpressing mice demonstrated glucose intolerance and peripheral IR on a normal or high-fat diet, despite increased insulin production and secretion. Frost and Olson59 reported complementary findings with global and pancreas-specific let-7 overexpression. Let-7 anti-miR administration partially ameliorated the effect of HFD on IR measures and liver TG accumulation in mice through anti-let-7-mediated insulin receptor derepression (Table 5). Let-7 also has a role in other tissues including skeletal muscle, where Let-7a and -7d were higher in the skeletal muscle tissue of T2DM patients compared with body mass index-matched NGT controls.60 In addition, let-7 may repress the anti-inflammatory Th2 cytokine interleukin 13 (IL-13) in myotubes, suggesting that let-7 potentially modulates muscle inflammation via IL-13 repression.60
Table 4. Skeletal muscle: miRNAs in peripheral IR.
| miRNA | Finding(s) | Intervention/phenotype | Model | Target/pathway | Author |
|---|---|---|---|---|---|
| miR-133a, -143, -144 | ↓, ↑ | Obese (NGT, IGT, T2DM) | Human (n=118 total) | — | Gallagher et al.101 |
| miR-494 | ↑, ↑ Insulin resistance | TNFα treatment | Mouse C2C12 cells | Insulin signaling | Lee et al.144 |
| miR-221 | ↑ | Genetic IR | Mouse (ob/ob) | AdipoR1/Adiponectin and PTB pathway | Lustig et al.71 |
| Let-7 | ↑ Insulin resistance, protected from DIO-IR | Overexpressed—Global and pancreas, inhibited miRNA—whole body | Mouse | INSR (muscle) | Frost et al.59 |
| Let-7a, d | ↑, ↑ Insulin resistance ↓ IL-13 secretion | Overweight (NGT vs T2DM), Overexpressed miRNA | Human, primary human myotubes | IL-13 | Jiang et al.60 |
| Let-7 | ↑ Insulin resistance | Overexpressed miRNA—muscle specific | Mouse | Lin28a, IGF1R, INSR, IRS-2 | Zhu et al.58 |
| miR-106b | ↑, ↑ Insulin resistance, | TNFα treatment, overexpressed miRNA | Mouse C2C12 myotubes | Mfn2, mitochondrial dysfunction | Zhang et al.145 |
| miR-10b | ↓ Skeletal muscle | Genetic IR (non-obese) | Rat (GK, WKY, BN) | — | Herrera et al.66 |
Abbreviations: AdipoR1, adiponectin receptor 1; DIO, diet-induced obesity; IGT, impaired glucose tolerance; INSR, insulin receptor; IR, insulin resistance; miRNA, microRNA; NGT, normal glucose tolerance; PTB, polypyrimidine tract-binding protein; T2DM, type 2 diabetes mellitus; TNFα, tumor necrosis factor alpha.
Table 5. miRNAs involved in hepatic glucose homeostasis.
| miRNA | Finding(s) | Intervention/phenotype | Model | Target/pathway | Author |
|---|---|---|---|---|---|
| miR-143 | ↑ | Genetic and DIO models | Mouse (db/db) | ORP8 | Jordan et al.48 |
| miR-145 | ↑, ↑ Insulin resistance | SCD fed, Resistin injected (6 days), overexpressed miRNA | Mouse (C57BL/6J), HepG2 cells | — | Wen et al.51 |
| miR-130a | ↓ Insulin resistance and steatosis | Adenoviral overexpressed | Mouse | Grb10 | Xiao et al.134 |
| Let-7 | ↑ Insulin resistance, protected from DIO-IR and fatty liver | Overexpressed—Global and pancreas, whole-body miRNA inhibition | Mouse | INSR, IRS-2 (liver) | Frost and Olson59 |
| miR-195, -103 | ↑ | Genetic IR (non-obese) | Rat (GK, WKY, BN) | — | Herrera et al.66 |
| miR-21 | ↑ | Unsaturated fatty acid treatment | Rat hepatocytes | PTEN | Vinciguerra et al.135 |
| miR-802 | ↑, ↑ Insulin resistance | HFD vs SCD, overexpressed miRNA | Mouse and human | HNF1B | Kornfeld et al.136 |
| miR-181a | ↑, ↑ Insulin resistance - ↓ SIRT1 | Genetic obesity/IR, overexpressed miRNA | Mouse (db/db), HepG2 cells | SIRT1 | Zhou et al.137 |
| miR-103, -107 | ↑ Insulin resistance, improved glucose tolerance | Hepatic miR-107 overexpression, inhibition of miRNAs | Mouse (C57BL/6J) | CAV1 | Trajkovski et al.133 |
| miR-29 | ↑, ↓ miRNA expression | HFD vs SCD mice, pioglitazone treatment | Mouse and Rat (fa/fa) | PPARGC1A, HMGCS2, ABHD5/FOXA2 | Kurtz et al.90 |
| miR-370 | ↑ Lipogenesis, ↓ Lipogenesis | Overexpressed miRNA, inhibited | Human HepG2 cells in vitro | miR-122, SREBP1c, DGAT2, Cpt1α | Iliopoulos et al.138 |
| miR-221 | ↓ AdipoR1 | Overexpressed miRNA | HumanHepG2 cells in vitro | AdipoR1/Adiponectin and PTB pathway | Lustig et al.71 |
| miR-126-3p, -24-3p | ↓ | Genetic obesity/IR | Mouse (ob/ob) | — | Liang et al.139 |
| miR-200s (miR-200a,b,c) | ↓ | Genetic obesity/IR | Mouse (db/db) | FOG2, IL6 | Dou et al.77 |
| miR-122 | ↓ Steatosis, ↓ Plasma cholesterol | Inhibited miRNA | Mouse SCD and HFD models | SREBP1c, AMPK pathway | Esau et al.140 |
| miR-122 | ↓ Plasma | Mild vs severe steatosis/NAFLD | Human | — | Miyaaki et al.141 |
| miR-33 | ↑ HDL, ↑ Reverse cholesterol transport | Inhibited miRNA | Mouse (LDLr−/−—western diet) | ABCA1/SREBF2 | Rayner et al.142, 143 |
Abbreviations: AdipoR1, adiponectin receptor 1; DIO, diet-induced obesity; HDL, high-density lipoprotein; HFD, high-fat diet; INSR, insulin receptor; IR, insulin resistance; miRNA, microRNA; ORP8, oxysterol binding protein-like 8; PTEN, phosphatase and tensin homolog; SCD, standard chow diet.
Let-7 has substantial potential as a biomarker of metabolic disease. In a human interventional study reducing the glycemic load in the diet of healthy premenstrual women, let-7b was the most dramatically altered microRNA, with nearly an eightfold increase of plasma let-7b after 12 months (Table 2).61 Similarly, when comparing plasma exosomes from obese diabetic patients naive to treatment and normal patients, Let-7a and -7f were found to be lower in the obese cohort.62 Interestingly, after receiving treatment, let-7 levels increased in the diabetic patients to levels not significantly different than normal controls. Let-7d was found to be more highly expressed in the amnion of obese pregnant women (Table 1).63 Finally, let-7 has been reported in adipogenesis, with overexpression of let-7 in pre-adipocytes resulting in reduced adipogenesis.64, 65 Taken together, the data strongly suggest that members of the let-7 family modulate systemic insulin sensitivity and glucose metabolism by effects on the insulin signaling/PI3K and mTOR pathways and may have significant potential as a blood biomarker of glycemic control and metabolic disease.
The miR-221/222 family
The miR-221 family in mice and humans consists of two members, miR-221 and miR-222, both located in close proximity on the X chromosome and both linked to metabolic disease. With the most references in plasma miRNA-based reports, miR-222 shows arguably the greatest promise as a clinical biomarker of metabolic disease. Functionally, miR-222 is a negative regulator of adipocyte insulin sensitivity in humans and rodents. Adipose levels of miR-222 are elevated in murine models of diabetes.66, 67 Human data from Shi et al. also reported higher miR-222 expression in the omental adipose tissue of women with gestational diabetes at the time of cesarean delivery compared with pregnant women with NGT (Table 1).68 miR-222 seems to negatively regulate adipose insulin sensitivity via repression of ERα and GLUT4.68 miR-222 was found to be significantly higher in the whole plasma of two distinct cohorts of obese human patients.53, 69 Similarly, high-density lipoproteins (HDL) isolated from patients with familial hyperchosterolemia had 8.2-fold higher levels of miR-222 than HDL from normal patients (Table 2).70 The functional significance of this differential in circulating miR-222 expression is unknown, however, plasma transport of this miRNA to adipose tissues may affect glucose metabolism.70 It should be mentioned that circulating miR-222 levels may be rapidly altered by insulin administration.69 These reports suggest a role for miR-222 in adipocyte insulin sensitivity, hormone-induced IR in white adipose tissue and as a putative circulating marker for diabetes and cardiovascular disease.
miR-221 may regulate IR via effects on adiponectin expression. In cultured human adipocytes, miR-221 mimics repressed adiponectin expression. Recently, Lustig et al.71 showed that miR-221 was selectively increased in the livers of ob/ob mice, but not in muscle, even though miR-221 repressed adiponectin receptor 1 in liver and muscle cells in vitro. In vitro miR-221 levels of human adipocytes were negatively correlated with tumor necrosis factor alpha mRNA levels and body mass index of donors.72 Conversely, subcutaneous adipose expression of miR-221 was positively correlated with body mass index in nondiabetic Pima Indians.73 Ortega et al.53 reported decreased plasma miR-221 levels in obese humans, while Wang et al.74 reported increased miR-221 in non-obese females with metabolic syndrome vs controls. The disparity in these findings may be owed to methodological differences or undefined variables in the cohorts. Despite the conflicting human findings, miR-221 may potentiate the development of IR via suppression of adiponectin signaling and shows potential as a plasma marker.
The miR-200 family (miR-8/miR-200 family)
Studies of obesity on hypothalamic leptin/food intake mechanisms have revealed a putative role for miR-200a in food intake regulation and body mass accumulation. Benoit et al.75 administered a leptin antagonist to Wistar rats shortly after birth. Leptin antagonism resulted in increased IR and increased miR-200a in the hypothalamus (interestingly, hypothalamic miR-222 was also upregulated). Rats were then fed a HFD. The leptin-antagonized group exhibited significantly higher hypothalamic miR-200a levels than control rats on the same diet. Interestingly, when fed a chow diet, leptin-antagonized rats had lower miR-200a levels than control rats. In a subsequent study using ob/ob, db/db mouse models, hypothalamic miR-200a was increased in obesity, with leptin treatment resulting in normalization/downregulation of miRNA expression.76 Importantly, miR-200a inhibition in the hypothalamus of ob/ob mice increased leptin receptor expression, reduced body weight and improved markers of whole-body insulin sensitivity. Differential miR-200 expression has also been reported in the livers of db/db mice77 and the visceral white adipose of HFD-fed mice,67 where it likely has an important role in adipogenesis via regulation of the Wnt pathway.78 These data present a strong case for miRNAs, especially miR-200a in food intake and appetite regulation making miR-200 a significant candidate for future investigation.
miR-223
miR-223 is the lone member of the family and resides on the X chromosome. Although immune cell-mediated (innate and adaptive) inflammation is known to have a significant role in obesity and IR, there are few miRNAs linked to macrophage ‘polarization' in the context of obesity. Though the most thoroughly investigated function of miR-223, outside of cancer, involves monocyte–macrophage differentiation and macrophage activation,79, 80 differential miR-223 expression has been linked to both human and murine obesity (Figure 2).17, 81 Zhuang et al.17 showed that HFD-fed miR-223 knockout mice had worsened IR and that miR-223 deficient macrophages showed increased inflammatory potential compared with miR-223 containing macrophages. The increased inflammatory stress in knockout animals was hypothesized to exacerbate obesity-related metabolic disease through derepression of PBX/knotted 1 homeobox 1 (Pknox1). miR-223 is associated in human circulation with vesicles/exosomes, lipoproteins and AGO2 complexes.70, 82, 83 In humans, plasma miR-223 was reported to be lower in T2DM patients vs normal multivariable-matched controls.81 Perhaps conversely, HDL isolated from patients with familial hyperchosterolemia had 3781-fold greater miR-223 than HDL from normal patients.70 The functional significance of differential plasma miRNA expression in metabolic disease is unknown at this time, though a repressive effect by miR-223 delivered by an exosomal fraction has been demonstrated experimientally.83 Other miRNAs implicated in adaptive and innate immune activation/polarization with the potential to have a role in obesity-mediated inflammation are: miR-155, miR-107(ref. 84) and miR-146-5p.85 miR-155-deficient macrophages had a decreased inflammatory protential,86 and miR-155 inhibited adipogenesis in adipocytes.87 Finally, it should be mentioned that miR-223 is robustly expressed in the liver and regulates liver cholesterol biosynthesis and HDL uptake through repression of various targets,88 suggesting a role in cardiovascular implications of obesity.
The miR-29 family
The miR-29 family has four members, miR-29a, b-1, b-2 and c on two chromosomes. Since 2007, miR-29 has been known to negatively regulate insulin signaling in adipocytes,89 however, a definitive mechanism has not been elucidated. Recently, however, Kurtz et al.90 showed that miR-29 was upregulated in the livers of DIO mice and in Zucker Diabetic Fatty (fa/fa) rats. In this model, miR-29 functioned through regulation of the transcription factor FOXA2 (FOXA2-mediated regulation of PPARGC1A, HMGCS2 and ABHD5). Interestingly, pioglitazone treatment normalized miR-29 levels in both murine models. Obesity in pregnant sheep leads to increased miR-29 expression in the liver tissue of offspring lambs, along with decreased markers of insulin signaling, suggesting fetal programming of miR-29 expression.91 Interestingly, miR-29 in T cells represses both Tbet and Eomes, transcription factors involved with IFNγ expression; consequently miR-29 mimetics suppress Th1/inflammatory potential of T cells.41 miR-29 upregulation in immune cells may be protective (anti-inflammatory), whereas upregulation in metabolic tissues may impair insulin signaling.
Perhaps more importantly, miR-29b shows greatest promise as a biomarker for T2DM and atherosclerotic disease. Zampetaki et al.81 examined plasma miRNA expression from a relatively large prospective human cohort (n=822). They reported lower plasma miR-29b (and miR-223 expression) in T2DM patients vs controls matched for multiple variables in a smaller cohort (Table 2). Others have reported increased circulating miR-29a in T2DM.92 Peng et al.93 examined urinary miRNA expression in patients with T2DM, with a focus on diabetic nephropathy and atherosclerotic measures. miR-29a (but not b or c) was higher in patients with albuminuria, a measure of diabetic nephropathy, compared with normoalbuminurimic patients (Table 1). miR-29b was also positively correlated with intimal thickness in patients. miR-29 may be a useful marker for cardiometabolic disease, especially the atherogenic risk associated with obesity in humans.
miR-375
The role of miRNAs in the pancreas is well-studied, see Plaisance et al.94 for a current review. One miRNA in particular, miR-375, has an important role in multiple tissues including the pancreas during obesity. miR-375 is highly expressed in pancreatic beta cells and is important in B cell maintenance, potentiating increased insulin production during murine IR.95, 96 Mice lacking miR-375 exhibit hyperglycemia owing to decreased insulin production (lack of beta cell expansion), while ob/ob mice have increased miR-375 expression.96 Interestingly, miR-375 and miR-184 form a network with AGO2, which likely regulates pancreatic beta cell expansion in human and murine IR.97 miR-184 represses AGO2 which in turn modulated miR-375 functionality. Processes that regulate the RNA-induced silencing complex, in this case differential expression of another miRNA, have the potential to affect gene and protein expression via modulation of miRNA function. There is also evidence that pancreatic miR-375 is involved with inherited metabolic abnormalities in rats.98 miR-375 was significantly increased in the plasma of T2DM humans vs control groups in two studies (Table 2),92, 99 and miR-375 promotes adipogenesis in mouse pre-adipocytes via regulation of ERK1/2 signaling upstream of peroxisome proliferator-activated receptor gamma (Table 3).100 The role of miR-375 in obesity and IR warrants continued attention.
Skeletal muscle miRNAs
The role of miRNAs in human skeletal muscle glucose metabolism is potentially important, though not well investigated (Table 4). Gallagher et al.101 published the only comprehensive profiling study of skeletal muscle miRNA expression in IR/T2DM. The authors examined three groups with relatively robust sample sizes (T2DM n=45, IGT n=26, NGT n=47) by percutaneous needle biopsy. All the patients were characterized by oral glucose tolerance testing. Profiling identified 29 miRNAs, some of which are known to be important in obesity-related mechanism in other tissues. Significantly upregulated miRNAs include miR-143 and miR-144, and downregulated miRNAs include miR-133a and miR-126-5p. miR-133a was most consistently inversely correlated with fasting glucose, HbA1c, and 2-h glucose tolerance. It should be noted that there were no significant differences in mRNA expression. miR-126 was shown in another study to be lower in patients with T2DM vs a normal glucose tolerance group.81 It seems that multiple, muscle-specific miRNAs (‘myomiRs') are regulated in diabetes though the pathophysiological consequences of this differential are not currently understood and suggest that further investigation is warranted.
Discussion of microRNAs and microRNA-based technologies in translational medicine and human health
miRNAs have both a compensatory and pathophysiological role in metabolic tissues during obesity, the development of IR, and T2DM. In addition, circulating and biofluid-based miRNAs measures can lend insight into distal tissue miRNA changes, microvesicle and lipoprotein miRNA enrichment and can be used as biomarkers or ‘miRNA signatures' of disease.
miRNAs have a role in transgenerational obesity and metabolic dysfunction
The development of obesity in children is a complex process involving genetic and epigenetic predisposition and various external influences. The exposure of the developing fetus to abnormal metabolic conditions (hyperglycemic, hyperinsulinemic, low-nitrogen and so on) alters genomic methylation affecting gene expression in childhood, adulthood and in subsequent generations, creating a predisposition for metabolic disease and/or obesity. There is strong evidence that epigenetic mechanisms affect metabolic fitness, with certain genomic regions (imprinted loci) prone to altered methylation. It is estimated that 7% of miRNA coding regions are located in imprinted loci in humans.102 Epigenetics of obesity as it relates miRNAs is little studied, though it offers great potential to expand our understanding of transgenerational predisposition to metabolic disease. A pubmed search for ‘((microRNA) AND (epigenetic)) AND obesity' resulted in 26 publications and ‘(miRNA promoter methylation) AND obesity' resulted in only three. Kameswaran et al.103 showed that the MEG3-DLK1 locus was hypermethylated in the beta cells of T2DM patients, significantly decreasing the expression of miRNAs coded by this locus. Although Kameswaran et al.103 are the only researchers to demonstrate altered miRNA promoter methylation in T2DM, there is evidence to suggest that miRNA expression in offspring may be affected by maternal nutrition.98 HFD feeding in mouse dams resulted in altered hepatic miR-122 and miR-370 expression in offspring, though this was not proven to be because of epigenetic mechanisms.104 Nevertheless, miR-122 and miR-370 have been shown to have a causal role in hepatic lipid metabolism. Similarly, maternal obesity in ewes increased offspring hepatic expression of miRNAs well known to be associated with metabolic disease in humans and rodents (miR-29b, -103, -107, -143).91 miRNAs likely mediate the phenotypes observed in obesity-related, transgenerational metabolic disturbances.
miRNAs as biomarkers
The presence of miRNAs in human biofluids (plasma, serum, urine, tears, saliva, colostrum, amniotic, cerebrospinal and seminal fluid)105 has resulted in the pursuit of miRNA-based biomarkers (miRNA signatures) for multiple diseases including cardiometabolic disease (Table 2). With the proper development, miRNA-based biomarkers have the potential to identify metabolic problems during disease latency (preclinical), assess severity of disease, identify patients with a predisposition to metabolic disease (assess risk), address disease etiology, confirm diagnosis/reduce misdiagnosis on the basis of current clinical markers and monitor response to interventions. The relative postprandial stability of some plasma miRNAs (vs other measures such as blood glucose or insulin) as well as the unique kinetic responses to therapy are potentially attractive aspects of their implementation in a clinical setting that have not been explored. There is also potential utility in subgroups of patients not optimally served by current clinical measures (overnight fasting) such as the elderly, children, persons of a non-Caucasian background, pre-diabetic patients with normal fasting glucose who have prolonged postprandial hyperglycemia, or otherwise healthy people consuming a high glycemic diet.
Plasma miRNAs have great potential as biomarkers as they have been shown to have excellent stability106 at room temperature and during multiple freeze–thaw cycles probably owing to association with AGO2 complexes,82 lipoproteins70 and enrichment in circulating vesicles. The first step in developing a biomarker, showing a statistically relevant difference between a healthy and metabolic disease cohort, has clearly been met by multiple studies identifying numerous plasma miRNAs that are associated with metabolic disease in a range of demographics (children, women, during pregnancy, IR at low body mass). However, it is also clear that there are marked inconsistencies in the findings for specific plasma miRNAs from different groups (for example, miR-223). This is, in part, likely owing to differences in RNA isolation, detection and normalization methodologies; a recent methods publication compares platforms.107 This highlights the need for increased standardization of plasma-based miRNA measures to optimize comparison of findings by different groups and to address stability of putative biomarker over short time periods and in the presence of various stimuli. For instance, plasma miR-222 appears to be a top biomarker candidate based on multiple reports, but its use may be problematic because of rapidly changing levels of expression dependent on variables such as insulin.69
For the most part, plasma-based research has not furthered the development of plasma miRNA biomarkers of metabolic disease, including necessary risk prediction analysis such as odds ratio and risk ratio calculations or diagnostic analysis such as receiver operating characteristic curve analysis and likelihood ratios (positive/negative LHRs); only two publications performed these analyses (odds ratio,81 receiver operating characteristic curves108). Furthermore, analysis to show the value of plasma miRNA biomarkers compared with current clinical markers in patient care is absent from the literature, but is needed. The proper statistical handling and development of clinical biomarkers is vital to the advancement of this area.109 It is important to note that there are no reports comparing plasma miRNAs in humans with allelic variants associated with T2DM. Longitudinal studies in patients with and without known genomic variants and the development of metabolic disease have the potential to yield important information regarding prognostic and predictive value of plasma miRNAs compared with the penetrance of known genomic variants.
miRNAs as therapeutics
Given the role of miRNAs in multiple pathways with pathophysiologic relevance to IR/obesity, there is great interest in miRNA-based therapeutic strategies. However, at the time of writing, there were 10 registered clinical trials at clinicaltrials.gov (search: ‘(microRNA) AND obesity') none of which aimed to test miRNA-based therapeutics in humans; all studies aim to measure miRNA expression in a disease state or in response to weight or exercise. Currently, anti-miRs are the most common in vivo approach when designing miRNA-based therapeutics.110 Anti-miRs sequester endogenous mature miRs, rescuing (derepressing) target gene expression. In 2012, van Rooij et al.110 published an exhaustive review on miRNA therapeutics covering the discovery, therapeutic development, main private industry players and patent issues associated with miRNA-based therapeutics. According to van Rooij, ‘The key requirements for an anti-miR are that the chemistry must be cell permeable, cannot be rapidly excreted, must be stable in vivo and should bind to the miRNA of interest with high specificity and affinity.' Anti-miRNAs chemistry (modifications to the sugar moiety, the nucleobase or the internucleotide linkages) is a topic of much interest, with the goal of increased stability (nuclease resistance), increased binding affinity and optimized in vivo functionality.36 Stenvang et al.36 published an excellent review of the available anti-miRNA oligonucleotides that have been developed.
Early evidence from preclinical trials in non-human primates shows encouraging pharmacokinetic properties of naked modified oligonucleotides including low toxicity. In 2008, Elmen et al.111 published findings in African green monkeys using intravenous injection of naked/unconjugated 15-mer locked nucleic acid (LNA)-anti-miR-122. The authors reported no detectible toxicity and long-lasting on-target effects. Rayner et al.112 administered 2′-fluoro/methoxyethyl modified anti-miR-33 (5 mg kg−1; Regulus Therapeutics, San Diego, CA, USA) to African green monkeys via subcutaneous injection 2 × per week for 2 weeks, then once per week for 10 weeks. After 8 weeks, they reported a 50% increase in HDL cholesterol in anti-miR treated monkeys.112 Pleiotropic effects of miRNA therapy, however, are of concern owing to the number of possible mRNA targets/pathway that may be affected by administration of a potent miRNA mimetic or inhibitor, especially with long-term administration. For instance, short-term anti-miR-33 in mice resulted in decreased atherosclerosis with no observed side effects,113 while long-term use led to increased circulating TG levels and hepatosteatosis suggesting that off-target/pleiotropic effects were significant. Thorough monitoring of multiple pathways are prudent when investigating the in vivo use of miRNA-based therapeutics.114
Though currently not investigated in non-human primate models, ‘Tiny LNA' technology, which uses 8-mer LNA-anti-miRs, shows particular promise in mice as a potential next step in in vivo anti-miR therapeutics. Obad et al.115 showed that tiny LNAs targeted the seed sequence of mature miRNAs and were able to derepress target genes as well as 15-mer LNA-anti-miRs. Theoretically full-length anti-miRs may have increased potential to bind nonspecifically in the translated portion of an mRNA, inducing an siRNA-like, knockdown effect. The authors showed that there were no significant off-target effects with tiny LNA technology when examining mRNAs with tiny LNA binding sites in the 5′ translated area, making tiny LNAs potentially superior to full-length LNA anti-miRs for in vivo studies, though additional comparisons are needed. The authors also showed that binding multiple mature miRNAs at non-seed sequences had little to no effect on miRNA activity. Interestingly, 8-mer anti-miR-21 modified by 2′-O-Me (instead of LNA technology) was not effective at blocking miRNA activity; the mechanism behind this is unknown.
Technological advancements in miRNA research
Ultra-deep RNA-Seq technology is a major advancement in studying noncoding RNA species. Though small RNA-Seq is costly and analysis heavy compared with other detection platforms, it has the potential to identify novel obesity-associated change(s) in RNA species unavailable from current microarray and PCR-based platforms, such as miRNA isomers. miRNA ‘genes' may produce miRNA transcript variants (‘isomiRs') owing to processing or post-transcriptional modifications, such as the activity of ADARs as discussed in the introduction.116 Two novel 5′-shifted isomiRs of miR-375 have been identified in mouse insulinoma cells that have distinctly different targeting than ‘canonical' miR-375.117 The authors also found that isomiRs had potential targets in genes associated with the development of T2DM.117
Software-based tools are numerous with many online and propriety sources. Valuable online resources for miRNA-target interactions such as miRWalk,118 miRTarBase119 and Targetscan, as well as databases specifically for circulating miRNAs120 that attempt to incorporate information from various online resources (PubMed, miRBase, miRo, DAVID and so on) and associate them with disease. However, annotation of these resources for obesity researches is limited. For instance, miRWalk yield no results for ‘obesity' or ‘metabolic disease'. MiRandola search under ‘Diseases and Malignant Cell Line'—term ‘obesity' yielded three miRNAs such as miR-138, miR-15b, miR-376a; while term ‘metabolic syndrome' yielded two miRNAs such as let-7 g and miR-221.120 Advancements in online miRNA analytics specifically optimized for metabolic disease, in addition to journal-mandated submission of findings to an online database of miRNAs profiling (for example, miRandola) will significantly contribute to information processing as the field expands (biomarkers, off-target effects of mimetics/anti-miRs and so on).
Conclusion
Understanding the role of miRNAs in whole-body metabolism and the pathophysiology of metabolic disease has been challenged in part by the complexity of miRNA–mRNA interactions as well as methodologic and technical limitations, especially in vivo. Future studies utilizing available miRNA profiling data from human disease, combined with miRNA detection techniques that are more sensitive, accurate and flexible (small RNA sequencing, quantitative PCR methods for blood miRNAs), and advancing bioinformatics/biostatistical analyses of complex data sets will undoubtedly result in further clarification of miRNA-mediated pathways in cardiometabolic disease. Experimentally, the development of conditional and tissue-specific miRNA knockout mice will be especially valuable in studying miRNAs that are cell/tissue specific or whose deletions are embryonically lethal (for example, miR-133a).121 Reports of miRNA–mRNA relationships based on in vitro data may have limited physiologic relevance and must be properly vetted in vivo. From a therapeutics standpoint, advancements in miRNA-based mimics and inhibitors, with increased specificity and in vivo stability, will likely potentiate efficacy and support preclinical testing of additional miRNAs in non-human primates. In the near future, we will likely see the maturation of circulating miRNAs as biomarkers of metabolic disease with their eventual clinical use in metabolic disease testing, risk assessment, and/or grading of disease. Future advances will further clarify the role of miRNAs in (1) the circulation (vesicular, nonvesicular, lipoprotein-associated) and effects in distal tissues, (2) innate and adaptive immune cell-mediated inflammation during overnutrition (for example, visceral adipose), (3) central regulation of appetite and food intake, (4) lipoprotein metabolism and fibrosis in the liver during overnutrition, (5) beta cell expansion in overnutrition and failure in T2DM and (6) cross-generational effects of obesity, when well-defined miRNA–mRNA pathways are established.
Acknowledgments
This work was supported by a K01 award (National Institute Of Diabetes And Digestive And Kidney Diseases of the National Institutes of Health under Award Number K01DK099475). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
The author declares no conflict of interest.
References
- 1Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of childhood and adult obesity in the United States, 2011-2012. JAMA 2014; 311: 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Fullston T, Ohlsson Teague EM, Palmer NO, DeBlasio MJ, Mitchell M, Corbett M et al. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J 2013; 27: 4226–4243. [DOI] [PubMed] [Google Scholar]
- 3Gonzalez-Bulnes A, Astiz S, Ovilo C, Lopez-Bote CJ, Sanchez-Sanchez R, Perez-Solana ML et al. Early-postnatal changes in adiposity and lipids profile by transgenerational developmental programming in swine with obesity/leptin resistance. J Endocrinol 2014; 223: M17–M29. [DOI] [PubMed] [Google Scholar]
- 4Multhaup ML, Seldin MM, Jaffe AE, Lei X, Kirchner H, Mondal P et al. Mouse-human experimental epigenetic analysis unmasks dietary targets and genetic liability for diabetic phenotypes. Cell Metab 2015; 21: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest 2005; 115: 1111–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6Nagareddy PR, Kraakman M, Masters SL, Stirzaker RA, Gorman DJ, Grant RW et al. Adipose tissue macrophages promote myelopoiesis and monocytosis in obesity. Cell Metab 2014; 19: 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7Feinstein R, Kanety H, Papa MZ, Lunenfeld B, Karasik A. Tumor necrosis factor-alpha suppresses insulin-induced tyrosine phosphorylation of insulin receptor and its substrates. J Biol Chem 1993; 268: 26055–26058. [PubMed] [Google Scholar]
- 8Vatner DF, Majumdar SK, Kumashiro N, Petersen MC, Rahimi Y, Gattu AK et al. Insulin-independent regulation of hepatic triglyceride synthesis by fatty acids. Proc Natl Acad Sci USA 2015; 112: 1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J et al. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med 2005; 11: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Ferreira DM, Simao AL, Rodrigues CM, Castro RE. Revisiting the Metabolic Syndrome and Paving the Way for microRNAs in Non-Alcoholic Fatty Liver Disease. FEBS J 2014; 281: 2503–2524. [DOI] [PubMed] [Google Scholar]
- 11Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 2006; 116: 1793–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Thaler JP, Yi C-X, Schur EA, Guyenet SJ, Hwang BH, Dietrich MO et al. Obesity is associated with hypothalamic injury in rodents and humans. J Clin Invest 2012; 122: 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Hotamisligil GS, Peraldi P, Budavari A, Ellis R, White MF, Spiegelman BM. IRS-1-mediated inhibition of insulin receptor tyrosine kinase activity in TNF-alpha- and obesity-induced insulin resistance. Science 1996; 271: 665–668. [DOI] [PubMed] [Google Scholar]
- 14De Alvaro C, Teruel T, Hernandez R, Lorenzo M. Tumor necrosis factor alpha produces insulin resistance in skeletal muscle by activation of inhibitor kappaB kinase in a p38 MAPK-dependent manner. J Biol Chem 2004; 279: 17070–17078. [DOI] [PubMed] [Google Scholar]
- 15Hotamisligil GS. Inflammation and metabolic disorders. Nature 2006; 444: 860–867. [DOI] [PubMed] [Google Scholar]
- 16Chawla A, Nguyen KD, Goh YPS. Macrophage-mediated inflammation in metabolic disease. Nat Rev Immunol 2011; 11: 738–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Zhuang G, Meng C, Guo X, Cheruku PS, Shi L, Xu H et al. A Novel Regulator of Macrophage Activation: miR-223 in Obesity Associated Adipose Tissue Inflammation. Circulation 2012; 125: 2892–2903. [DOI] [PubMed] [Google Scholar]
- 18Carolan E, Hogan AE, Corrigan M, Gaotswe G, O'Connell J, Foley N et al. The impact of childhood obesity on inflammation, innate immune cell frequency and metabolic microRNA expression. J Clin Endocrinol Metab 2013; 99: E474–E478. [DOI] [PubMed] [Google Scholar]
- 19Olivo-Marston SE, Hursting SD, Perkins SN, Schetter A, Khan M, Croce C et al. Effects of calorie restriction and diet-induced obesity on murine colon carcinogenesis, growth and inflammatory factors, and microRNA expression. PLoS One 2014; 9: e94765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993; 75: 855–862. [DOI] [PubMed] [Google Scholar]
- 21Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75: 843–854. [DOI] [PubMed] [Google Scholar]
- 22Lee C-T, Risom T, Strauss WM. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol 2007; 26: 209–218. [DOI] [PubMed] [Google Scholar]
- 23Friedman RC, KK-H Farh, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009; 19: 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 2010; 11: 252–263. [DOI] [PubMed] [Google Scholar]
- 25Rottiers V, Naar AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol 2012; 13: 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009; 136: 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11: 597–610. [DOI] [PubMed] [Google Scholar]
- 28Martin HC, Wani S, Steptoe AL, Krishnan K, Nones K, Nourbakhsh E et al. Imperfect centered miRNA binding sites are common and can mediate repression of target mRNAs. Genome Biol 2014; 15: R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11: 228–234. [DOI] [PubMed] [Google Scholar]
- 30Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu Rev Genet 2000; 34: 499–531. [DOI] [PubMed] [Google Scholar]
- 31Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 2006; 13: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA 2004; 10: 1174–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 2010; 35: 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2004; 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Yang L, Huang P, Li F, Zhao L, Zhang Y, Li S et al. c-Jun amino-terminal kinase-1 mediates glucose-responsive upregulation of the RNA editing enzyme ADAR2 in pancreatic beta-cells. PLoS One 2012; 7: e48611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 2012; 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37Kloting N, Berthold S, Kovacs P, Schon MR, Fasshauer M, Ruschke K et al. MicroRNA expression in human omental and subcutaneous adipose tissue. PLoS One 2009; 4: e4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Fichtlscherer S, De Rosa S, Fox H, Schwietz T, Fischer A, Liebetrau C et al. Circulating microRNAs in patients with coronary artery disease. Circ Res 2010; 107: 677–684. [DOI] [PubMed] [Google Scholar]
- 39Heneghan HM, Miller N, McAnena OJ, O'Brien T, Kerin MJ. Differential miRNA expression in omental adipose tissue and in the circulation of obese patients identifies novel metabolic biomarkers. J Clin Endocrinol Metab 2011; 96: E846–E850. [DOI] [PubMed] [Google Scholar]
- 40Hulsmans M, Holvoet P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res 2013; 100: 7–18. [DOI] [PubMed] [Google Scholar]
- 41Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CDC et al. MicroRNA-29 regulates T-box transcription factors and interferon-γ production in helper T cells. Immunity 2011; 35: 169–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Li H, Li T, Wang S, Wei J, Fan J, Li J et al. miR-17-5p and miR-106a are involved in the balance between osteogenic and adipogenic differentiation of adipose-derived mesenchymal stem cells. Stem Cell Res 2013; 10: 313–324. [DOI] [PubMed] [Google Scholar]
- 43Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009; 460: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 2004; 279: 52361–52365. [DOI] [PubMed] [Google Scholar]
- 45Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 2006; 12: 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H et al. Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun 2008; 376: 728–732. [DOI] [PubMed] [Google Scholar]
- 47Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009; 58: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Jordan SD, Kruger M, Willmes DM, Redemann N, Wunderlich FT, Bronneke HS et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat Cell Biol 2011; 13: 434–446. [DOI] [PubMed] [Google Scholar]
- 49Lorente-Cebrián S, Mejhert N, Kulyté A, Laurencikiene J, Åström G, Hedén P et al. MicroRNAs regulate human adipocyte lipolysis: effects of miR-145 are linked to TNF-α. PLoS One 2014; 9: e86800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Lin Y-Y, Chou C-F, Giovarelli M, Briata P, Gherzi R, Chen C-Y. KSRP and microRNA 145 are negative regulators of lipolysis in white adipose tissue. Mol Cell Biol 2014; 34: 2339–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Wen F, Yang Y, Jin D, Sun J, Yu X, Yang Z. MiRNA-145 is involved in the development of resistin-induced insulin resistance in HepG2 cells. Biochem Biophys Res Commun 2014; 445: 517–523. [DOI] [PubMed] [Google Scholar]
- 52Lee EK, Lee MJ, Abdelmohsen K, Kim W, Kim MM, Srikantan S et al. miR-130 suppresses adipogenesis by inhibiting peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol 2011; 31: 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Ortega FJ, Mercader JM, Catalán V, Moreno-Navarrete JM, Pueyo N, Sabater M et al. Targeting the circulating microRNA signature of obesity. Clin Chem 2013; 59: 781–792. [DOI] [PubMed] [Google Scholar]
- 54Prats-Puig A, Ortega FJ, Mercader JM, Moreno-Navarrete JM, Moreno M, Bonet N et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab 2013; 98: E1655–E1660. [DOI] [PubMed] [Google Scholar]
- 55Pan S, Yang X, Jia Y, Li R, Zhao R. Microvesicle-shuttled miR-130b reduces fat deposition in recipient primary cultured porcine adipocytes by inhibiting PPAR-g expression. J Cell Physiol 2014; 229: 631–639. [DOI] [PubMed] [Google Scholar]
- 56Pan S, Zheng Y, Zhao R, Yang X. MicroRNA-130b and microRNA-374b mediate the effect of maternal dietary protein on offspring lipid metabolism in Meishan pigs. Br J Nutr 2013; 109: 1731–1738. [DOI] [PubMed] [Google Scholar]
- 57Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000; 408: 86–89. [DOI] [PubMed] [Google Scholar]
- 58Zhu H, Shyh-Chang N, Segre A V, Shinoda G, Shah SP, Einhorn WS et al. The Lin28/let-7 axis regulates glucose metabolism. Cell 2011; 147: 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA 2011; 108: 21075–21080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60Jiang LQ, Franck N, Egan B, Sjogren RJ, Katayama M, Duque-Guimaraes D et al. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am J Physiol Endocrinol Metab 2013; 305: E1359–E1366. [DOI] [PubMed] [Google Scholar]
- 61McCann SE, Liu S, Wang D, Shen J, Hu Q, Hong CC et al. Reduction of dietary glycaemic load modifies the expression of microRNA potentially associated with energy balance and cancer pathways in pre-menopausal women. Br J Nutr 2013; 109: 585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62Santovito D, De Nardis V, Marcantonio P, Mandolini C, Paganelli C, Vitale E et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in Diabetes: effect of glycemic control. J Clin Endocrinol Metab 2014; 99: E1681–E1685. [DOI] [PubMed] [Google Scholar]
- 63Nardelli C, Iaffaldano L, Ferrigno M, Labruna G, Maruotti GM, Quaglia F et al. Characterization and predicted role of the microRNA expression profile in amnion from obese pregnant women. Int J Obes (Lond) 2014; 38: 466–469. [DOI] [PubMed] [Google Scholar]
- 64Sun T, Fu M, Bookout AL, Kliewer SA, Mangelsdorf DJ. MicroRNA let-7 regulates 3T3-L1 adipogenesis. Mol Endocrinol 2009; 23: 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65Wei J, Li H, Wang S, Li T, Fan J, Liang X et al. let-7 enhances osteogenesis and bone formation while repressing adipogenesis of human stromal/mesenchymal stem cells by regulating HMGA2. Stem Cells Dev 2014; 23: 1452–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66Herrera BM, Lockstone HE, Taylor JM, Ria M, Barrett A, Collins S et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia 2010; 53: 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI, Kyriazopoulou VE et al. Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS One 2012; 7: e34872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68Shi Z, Zhao C, Guo X, Ding H, Cui Y, Shen R et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERalpha expression in estrogen-induced insulin resistance. Endocrinology 2014; 155: 1982–1990. [DOI] [PubMed] [Google Scholar]
- 69Ortega FJ, Mercader JM, Moreno-Navarrete JM, Rovira O, Guerra E, Esteve E et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 2014; 37: 1375–1383. [DOI] [PubMed] [Google Scholar]
- 70Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011; 13: 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71Lustig Y, Barhod E, Ashwal-Fluss R, Gordin R, Shomron N, Baruch-Umansky K et al. RNA-binding protein PTB and microRNA-221 coregulate AdipoR1 translation and adiponectin signaling. Diabetes 2014; 63: 433–445. [DOI] [PubMed] [Google Scholar]
- 72Chou W-W, Wang Y-T, Liao Y-C, Chuang S-C, Wang S-N, Juo S-HH. Decreased microRNA-221 is associated with high levels of TNF-α in human adipose tissue-derived mesenchymal stem cells from obese woman. Cell Physiol Biochem 2013; 32: 127–137. [DOI] [PubMed] [Google Scholar]
- 73Meerson A, Traurig M, Ossowski V, Fleming JM, Mullins M, Baier LJ. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-alpha. Diabetologia 2013; 56: 1971–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74Wang Y-T, Tsai P-C, Liao Y-C, Hsu C-Y, Juo S-HH. Circulating microRNAs have a sex-specific association with metabolic syndrome. J Biomed Sci 2013; 20: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75Benoit C, Ould-Hamouda H, Crepin D, Gertler A, Amar L, Taouis M. Early leptin blockade predisposes fat-fed rats to overweight and modifies hypothalamic microRNAs. J Endocrinol 2013; 218: 35–47. [DOI] [PubMed] [Google Scholar]
- 76Crepin D, Benomar Y, Riffault L, Amine H, Gertler A, Taouis M. The over-expression of miR-200a in the hypothalamus of ob/ob mice is linked to leptin and insulin signaling impairment. Mol Cell Endocrinol 2014; 384: 1–11. [DOI] [PubMed] [Google Scholar]
- 77Dou L, Zhao T, Wang L, Huang X, Jiao J, Gao D et al. miR-200 s contribute to interleukin-6 (IL-6)-induced insulin resistance in hepatocytes. J Biol Chem 2013; 288: 22596–22606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78Kennell JA, Gerin I, MacDougald OA, Cadigan KM. The microRNA miR-8 is a conserved negative regulator of Wnt signaling. Proc Natl Acad Sci USA 2008; 105: 15417–15422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB et al. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol 2012; 189: 3795–3799. [DOI] [PubMed] [Google Scholar]
- 80Li T, Morgan MJ, Choksi S, Zhang Y, Kim YS, Liu ZG. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat Immunol 2010; 11: 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res 2010; 107: 810–817. [DOI] [PubMed] [Google Scholar]
- 82Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 2011; 108: 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83Ismail N, Wang Y, Dakhlallah D, Moldovan L, Agarwal K, Batte K et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 2013; 121: 984–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84Foley NH, O'Neill LA. miR-107: a toll-like receptor-regulated miRNA dysregulated in obesity and type II diabetes. J Leukoc Biol 2012; 92: 521–527. [DOI] [PubMed] [Google Scholar]
- 85Hulsmans M, Van Dooren E, Mathieu C, Holvoet P. Decrease of miR-146b-5p in monocytes during obesity is associated with loss of the anti-inflammatory but not insulin signaling action of adiponectin. PLoS One 2012; 7: e32794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86Du F, Yu F, Wang Y, Hui Y, Carnevale K, Fu M et al. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol 2014; 34: 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87Liu S, Yang Y, Wu J. TNFα-induced up-regulation of miR-155 inhibits adipogenesis by down-regulating early adipogenic transcription factors. Biochem Biophys Res Commun 2011; 414: 618–624. [DOI] [PubMed] [Google Scholar]
- 88Vickers KC, Landstreet SR, Levin MG, Shoucri BM, Toth CL, Taylor RC et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc Natl Acad Sci USA 2014; 111: 14518–14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89He A, Zhu L, Gupta N, Chang Y, Fang F. Overexpression of micro ribonucleic acid 29, highly up-regulated in diabetic rats, leads to insulin resistance in 3T3-L1 adipocytes. Mol Endocrinol 2007; 21: 2785–2794. [DOI] [PubMed] [Google Scholar]
- 90Kurtz CL, Peck BC, Fannin EE, Beysen C, Miao J, Landstreet SR et al. microRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes 2014; 63: 3141–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91Nicholas LM, Rattanatray L, MacLaughlin SM, Ozanne SE, Kleemann DO, Walker SK et al. Differential effects of maternal obesity and weight loss in the periconceptional period on the epigenetic regulation of hepatic insulin-signaling pathways in the offspring. FASEB J 2013; 27: 3786–3796. [DOI] [PubMed] [Google Scholar]
- 92Kong L, Zhu J, Han W, Jiang X, Xu M, Zhao Y et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol 2011; 48: 61–69. [DOI] [PubMed] [Google Scholar]
- 93Peng H, Zhong M, Zhao W, Wang C, Zhang J, Liu X et al. Urinary miR-29 correlates with albuminuria and carotid intima-media thickness in type 2 diabetes patients. PLoS One 2013; 8: e82607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94Plaisance V, Waeber G, Regazzi R, Abderrahmani A. Role of microRNAs in islet beta-cell compensation and failure during diabetes. J Diabetes Res 2014; 2014: 618652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004; 432: 226–230. [DOI] [PubMed] [Google Scholar]
- 96Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA 2009; 106: 5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97Tattikota SG, Rathjen T, McAnulty SJ, Wessels H-H, Akerman I, van de Bunt M et al. Argonaute2 mediates compensatory expansion of the pancreatic β cell. Cell Metab 2014; 19: 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98Dumortier O, Hinault C, Gautier N, Patouraux S, Casamento V, Van Obberghen E. Maternal protein restriction leads to pancreatic failure in offspring: role of misexpressed microRNA-375. Diabetes 2014; 63: 3416–3427. [DOI] [PubMed] [Google Scholar]
- 99Higuchi C, Nakatsuka A, Eguchi J, Teshigawara S, Kanzaki M, Katayama A et al. Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism 2015; 64: 489–497. [DOI] [PubMed] [Google Scholar]
- 100Ling H-Y, Wen G-B, Feng S-D, Tuo Q-H, Ou H-S, Yao CH et al. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clin Exp Pharmacol Physiol 2011; 38: 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP et al. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med 2010; 2: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102Labialle S, Cavaillé J. Do repeated arrays of regulatory small-RNA genes elicit genomic imprinting?: Concurrent emergence of large clusters of small non-coding RNAs and genomic imprinting at four evolutionarily distinct eutherian chromosomal loci. Bioessays 2011; 33: 565–573. [DOI] [PubMed] [Google Scholar]
- 103Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab 2014; 19: 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104Benatti RO, Melo AM, Borges FO, Ignacio-Souza LM, Simino LA, Milanski M et al. Maternal high-fat diet consumption modulates hepatic lipid metabolism and microRNA-122 (miR-122) and microRNA-370 (miR-370) expression in offspring. Br J Nutr 2014; 111: 2112–2122. [DOI] [PubMed] [Google Scholar]
- 105Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ et al. The microRNA spectrum in 12 body fluids. Clin Chem 2010; 56: 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008; 105: 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, Chen C et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods 2014; 11: 809–815. [DOI] [PubMed] [Google Scholar]
- 108Pescador N, Perez-Barba M, Ibarra JM, Corbaton A, Martinez-Larrad MT, Serrano-Rios M. Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS One 2013; 8: e77251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109Ray P, Le Manach Y, Riou B, Houle TT. Statistical evaluation of a biomarker. Anesthesiology 2010; 112: 1023–1040. [DOI] [PubMed] [Google Scholar]
- 110Van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res 2012; 110: 496–507. [DOI] [PubMed] [Google Scholar]
- 111Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S et al. LNA-mediated microRNA silencing in non-human primates. Nature 2008; 452: 896–899. [DOI] [PubMed] [Google Scholar]
- 112Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011; 478: 404–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113Distel E, Barrett TJ, Chung K, Girgis NM, Parathath S, Essau CC et al. miR33 inhibition overcomes deleterious effects of diabetes mellitus on atherosclerosis plaque regression in mice. Circ Res 2014; 115: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114Goedeke L, Salerno A, Ramírez CM, Guo L, Allen RM, Yin X et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol Med 2014; 6: 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 2011; 43: 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu A-L et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008; 18: 610–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117Baran-Gale J, Fannin EE, Kurtz CL, Sethupathy P. Beta cell 5'-shifted isomiRs are candidate regulatory hubs in type 2 diabetes. PLoS One 2013; 8: e73240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by ‘walking' the genes of three genomes. J Biomed Inform 2011; 44: 839–847. [DOI] [PubMed] [Google Scholar]
- 119Hsu S-D, Tseng Y-T, Shrestha S, Lin Y-L, Khaleel A, Chou C-H et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014 42: D78–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120Russo F, Di Bella S, Nigita G, Macca V, Laganà A, Giugno R et al. miRandola: extracellular circulating microRNAs database. PLoS One 2012; 7: e47786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep 2012; 1: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122Nesca V, Guay C, Jacovetti C, Menoud V, Peyot ML, Laybutt DR et al. Identification of particular groups of microRNAs that positively or negatively impact on beta cell function in obese models of type 2 diabetes. Diabetologia 2013; 56: 2203–2212. [DOI] [PubMed] [Google Scholar]
- 123Viesti A, Collares R, Salgado W, Pretti da Cunha Tirapelli D, dos Santos JS. The expression of LEP, LEPR, IGF1 and IL10 in obesity and the relationship with microRNAs. PLoS One 2014; 9: e93512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124Kim C, Lee H, Cho YM, Kwon O-J, Kim W, Lee EK. TNFα-induced miR-130 resulted in adipocyte dysfunction during obesity-related inflammation. FEBS Lett 2013; 587: 3853–3858. [PubMed] [Google Scholar]
- 125Arner E, Mejhert N, Kulyte A, Balwierz PJ, Pachkov M, Cormont M et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes 2012; 61: 1986–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126Yu J, Kong X, Liu J, Lv Y, Sheng Y, Lv S et al. Expression profiling of PPARγ-regulated microRNAs in human subcutaneous and visceral adipogenesis in both genders. Endocrinology 2014; 155: 2155–2165. [DOI] [PubMed] [Google Scholar]
- 127Kulyté A, Lorente-Cebrián S, Gao H, Mejhert N, Agustsson T, Arner P et al. MicroRNA profiling links miR-378 to enhanced adipocyte lipolysis in human cancer cachexia. Am J Physiol Endocrinol Metab 2014; 306: E267–E274. [DOI] [PubMed] [Google Scholar]
- 128Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009; 58: 1050–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129Chen H, Wang S, Chen L, Chen Y, Wu M, Zhang Y et al. MicroRNA-344 inhibits 3T3-L1 cell differentiation via targeting GSK3β of Wnt/β-catenin signaling pathway. FEBS Lett 2014; 588: 429–435. [DOI] [PubMed] [Google Scholar]
- 130Fu T, Seok S, Choi S, Huang Z, Suino-Powell K, Xu HE et al. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Mol Cell Biol 2014; 34: 4130–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131Li H, Chen X, Guan L, Qi Q, Shu G, Jiang Q et al. MiRNA-181a regulates adipogenesis by targeting tumor necrosis factor-α (TNF-α) in the porcine model. PLoS One 2013; 8: e71568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132Ling HY, Hu B, Hu XB, Zhong J, Feng SD, Qin L et al. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp Clin Endocrinol Diabetes 2012; 120: 553–559. [DOI] [PubMed] [Google Scholar]
- 133Trajkovski M, Hausser J, Soutschek J, Bhat B, Akin A, Zavolan M et al. MicroRNAs 103 and 107 regulate insulin sensitivity. Nature 2011; 474: 649–653. [DOI] [PubMed] [Google Scholar]
- 134Xiao F, Yu J, Liu B, Guo Y, Li K, Deng J et al. A novel function of microRNA 130a-3p in hepatic insulin sensitivity and liver steatosis. Diabetes 2014; 63: 2631–2642. [DOI] [PubMed] [Google Scholar]
- 135Vinciguerra M, Sgroi A, Veyrat-Durebex C, Rubbia-Brandt L, Buhler LH, Foti M. Unsaturated fatty acids inhibit the expression of tumor suppressor phosphatase and tensin homolog (PTEN) via microRNA-21 up-regulation in hepatocytes. Hepatology 2009; 49: 1176–1184. [DOI] [PubMed] [Google Scholar]
- 136Kornfeld JW, Baitzel C, Konner AC, Nicholls HT, Vogt MC, Herrmanns K et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 2013; 494: 111–115. [DOI] [PubMed] [Google Scholar]
- 137Zhou B, Li C, Qi W, Zhang Y, Zhang F, Wu JX et al. Downregulation of miR-181a upregulates sirtuin-1 (SIRT1) and improves hepatic insulin sensitivity. Diabetologia 2012; 55: 2032–2043. [DOI] [PubMed] [Google Scholar]
- 138Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res 2010; 51: 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139Liang T, Liu C, Ye Z. Deep sequencing of small RNA repertoires in mice reveals metabolic disorders-associated hepatic miRNAs. PLoS One 2013; 8: e80774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006; 3: 87–98. [DOI] [PubMed] [Google Scholar]
- 141Miyaaki H, Ichikawa T, Kamo Y, Taura N, Honda T, Shibata H et al. Significance of serum and hepatic microRNA-122 levels in patients with non-alcoholic fatty liver disease. Liver Int 2014; 34: e302–e307. [DOI] [PubMed] [Google Scholar]
- 142Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest 2011; 121: 2921–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010; 328: 1570–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144Lee H, Jee Y, Hong K, Hwang GS, Chun K-HH. MicroRNA-494, upregulated by tumor necrosis factor-alpha, desensitizes insulin effect in C2C12 muscle cells. PLoS One 2013; 8: e83471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145Zhang Y, Yang L, Gao YF, Fan ZM, Cai XY, Liu MY et al. MicroRNA-106b induces mitochondrial dysfunction and insulin resistance in C2C12 myotubes by targeting mitofusin-2. Mol Cell Endocrinol 2013; 381: 230–240. [DOI] [PubMed] [Google Scholar]