Abstract
Among nonhuman primates, SIV-infected Asian pigtailed macaques (PM) are relatively more susceptible to infection and disease progression than SIV-infected rhesus macaques (RM). In addition, SIV-infected African natural hosts such as the sooty mangabeys (SM) are resistant to disease. The mechanisms associated with such species-related variable clinical outcomes remain ill-defined but hold the potential to provide insights into the underlying mechanisms surrounding HIV pathogenesis. Recent findings indicate that the expression of the heterodimeric gut homing integrin α4β7 can influence both susceptibility and disease progression in RM. It was reasoned that differences in the frequencies/surface densities of α4β7-expressing lymphocytes might contribute to the differences in the clinical outcome of SIV infection among NHPs. In this article, we report that CD4+ T cells from PM constitutively express significantly higher levels of α4β7 than RM or SM. Retinoic acid, a key regulator of α4β7 expression, was paradoxically found at higher levels in the plasma of SM versus RM or PM. We also observed pairing of β7 with αE (αEβ7) on CD4+ T cells in the peripheral blood of SM, but not PM or RM. Finally, the differential mean density of expression of α4β7 in RM versus SM versus PM was predominantly dictated by species-specific sequence differences at the level of the β7 promoters, as determined by in vitro reporter/promoter construct transfection studies. We propose that differences in the regulation and expression of α4β7 may explain, in part, the differences in susceptibility and SIV disease progression in these NHP models.
The GALTs are a major target of both HIV-1 infection in humans and SIV infection in nonhuman primates (NHPs) (1–5). GALT represents the largest immune organ and contains a significant fraction of the total CD4+ T cell compartment. Because GALT is exposed, in a continuous way, to microbial challenges, the activation state of GALT CD4+ T cells is constitutively elevated (reviewed in Ref. 6). Because both HIV and SIV preferentially target activated CD4+ T cells, the increased level of activation in GALT provides an environment conducive for viral replication. As a consequence, GALT is a primary target of viral replication in the early stages of infection. This typically leads to a profound depletion of GALT CD4+ T cells and the nonspecific destruction of other cell lineages within GALT. This pathology, which is apparently irreversible, is thought to contribute to the chronic immune activation that is associated with poor prognoses (7). This putative link between viral replication in GALT and chronic immune activation prompted us and others to carry out studies aimed toward understanding the basic mechanisms by which CD4+ T cells selectively traffic to GALT, and how modulating CD4+ T cell migration into GALT might impact infection and disease progression.
We focused our initial studies on α4β7, a heterodimeric integrin receptor that mediates trafficking of cells, including the CD4+ T cells, to the GALT (8–10). Several lines of investigation highlight the potential importance of α4β7-expressing CD4+ T cells in HIV/SIV infection. These include: 1) CD4+ T cells expressing high levels of α4β7 (α4β7hi) are the preferential targets of HIV/SIV infection (11, 12); 2) certain recombinant HIV and SIV gp120s have been shown to bind to the α4β7 molecule in vitro (8, 13); 3) increased frequencies of α4β7hi-expressing CD4+ T cells within GALT at the time of infection appear to correlate with increased viral loads and rate of disease progression post SIV infection (14); 4) the i.v. administration of a novel recombinant rhesus mAb against α4β7 (α4β7 mAb) to rhesus macaques (RM) just before and during acute i.v. or intrarectal SIV infection led to marked reductions in GALT viral loads (15, 16); of note, these α4β7 mAb–treated RM did not exhibit signs of disease progression, whereas control RM succumbed to AIDS within 2 y postinfection; and 5) finally, and perhaps most intriguingly, i.v. administration of the same anti-α4β7 mAb just before and during acute intravaginal exposure to SIV prevented transmission of infection in 6 of 12 RM. When infection did occur, viremia was delayed and GALT was largely protected (17).
Although there remains much to be learned regarding the role of α4β7 in HIV pathogenesis, the earlier mentioned studies, taken together, indicate that α4β7-expressing cells are likely to play an important role in HIV disease. With these observations in mind, we set out to investigate whether differences in the frequencies of α4β7-expressing cells and/or the surface expression of this integrin could contribute to the variable susceptibility to infection, and differential rate of disease progression, that distinguish SIV infection of the three major NHP species that are routinely used to study SIV pathogenesis. It is generally recognized that pigtailed macaques (PM) are significantly more susceptible to infection and progress to disease more rapidly post SIV infection than RM using the same isolates of SIV (18, 19). Sooty mangabeys (SM), which, unlike PM or RM, are a natural host of SIV, respond to infection in a strikingly different way that suggests that they have adapted to SIV. When SM are infected (either naturally or experimentally), they typically exhibit high-level viremia, but only rarely do they experience development of immunodeficiency disease (20). Of note, extensive SIV replication occurs within the GALT of SM.
Considering these differences, we examined the frequencies and densities of α4β7-expressing cells in animals from each species. While conducting these studies, we recognized that there exists a paucity of information surrounding the manner in which α4β7 expression is regulated in NHPs. We therefore conducted studies aimed at identifying factors that differentially impact the expression of α4β7 in NHPs.
Materials and Methods
Ethics statement
We obtained heparinized blood samples and rectal biopsies from RM, SM, and PM from cohorts of animals being studied by us on various protocols or as specimen requests from the Yerkes National Primate Research Center. The housing, care, diet, and maintenance conformed with the guidelines of the Committee on the Care and Use of Laboratory Animals of the Institute of Laboratory Animal Resources, National Research Council, and the Department of Health and Human Services guidelines titled Guide for the Care and Use of Laboratory Animals. The Emory Institutional Animal Care and Use Committee approved the collection of blood and rectal biopsies before the initiation of these studies. All animals were negative for SIV, simian T cell lymphotropic virus, and simian retrovirus. PBMC samples from chimpanzees were obtained from the colonies at Yerkes National Primate Research Center, and the rest of the NHP species were obtained as a courtesy from the Texas Biomedical Research Institute, San Antonio, TX (African Green Monkeys); the Atlanta Zoo, Atlanta, GA (Drills); and the Institute for Primate Research, Karen, Kenya (the remaining African species).
Ab staining and flow-cytometric analysis
We performed polychromatic flow-cytometric analysis on aliquots of PBMCs and gut tissue isolated mononuclear cells as previously described (15). The reagents used included allophycocyanin anti-α4β7 (National Institutes of Health NHP Reagent Resource, National Institutes of Health contract HHSN272200900037C), PE-Cy5 anti-α4/CD49d (clone 9F10), PE anti-β7 integrin (clone FIB504), FITC anti-αE (#IM1856CU; Beckman Coulter), PE-Cy5 anti-β1 (clone MAR4), V450 anti-CD4 (clone L200), Alexa 700 anti-CD3 (clone SP34-2), AmCyan anti-CD8 (clone SK1), allophycocyanin-Cy7 anti-CD20 (clone L27), and PE–anti-NKG2A (clone Z199). The Abs were purchased from Beckman Coulter (Brea, CA), eBioscience (San Diego, CA), or BD Biosciences (San Jose, CA). Gated populations of CD3+ (for all T cells) and CD3− (for all other cell lineages) were analyzed for the frequency of CD4+ cells that expressed α4β7 and α4, β1, β7, and αE. Data on a minimum of 10,000 events were acquired and analyzed using FlowJo software (Treeland, Ashland, OR). Studies also included the analysis of the potential coexpression of α4β1 and α4β7. For these studies, aliquots of PBMCs from normal RM were either stained with allophycocyanin-conjugated recombinant rhesus anti-α4β7 mAb (directed at the heterodimeric form of α4β7) and a previously defined optimum concentration of biotinylated recombinant human VCAM-1 followed by PE-avidin or the same anti-α4β7 mAb followed by normal rhesus IgG and PE-avidin (for purposes of control and establishing gates). The protocol for the staining using recombinant VCAM-1 has been published elsewhere (21). One of the major caveats of these flow-cytometric studies is the concern whether the reagents used cross-react equally with the corresponding CD molecules in cells from the various NHP species. We screened multiple clones of Abs, and the earlier mentioned clones were found to provide the most satisfactory results, but caution should be noted with regard to specificity in the interpretation of the data presented in this article.
Mononuclear cell isolation from the blood and biopsy tissues
PBMCs and colorectal biopsy mononuclear cells were isolated as described previously (15). In brief, heparinized blood samples were obtained and mononuclear cells were isolated using Ficoll-Hypaque gradient centrifugation. Colorectal biopsies from select species of monkeys were subjected to a series of purification steps, and finally isolated by Percoll gradient and isolated using standard techniques as described previously (15). The yields and viability of the pooled cells extracted from the biopsy tissues varied considerably; the viability ranged from 62 to 84% as determined by trypan blue dye exclusion method.
In vitro infection of CD4+ T cells from PM, RM, and SM with SIVmac239
Enriched populations of CD4+ T cells (by negative selection; Miltenyi Biotech) from RM/PM/SM were cultured for 2 d in vitro with complete growth medium (RPMI 1640 medium + 10% heat-inactivated FCS supplemented with antibiotics and GlutaMAX) in the presence of anti-CD3/CD28–conjugated immunobeads + rHu−IL-2 (20 U/ml). The cells were then washed and cultured at 2 × 106/ml in either media (control) or infected with SIVmac239, and the supernatant fluids were collected at varying time intervals up to day 18 and assayed for levels of p27, using commercial p27 ELISA kit (ABL, Rockville, MD). Similar studies were conducted with PBMCs from PM, RM, and SM that were either treated with normal rhesus IgG (control)-coated beads or treated with a previously determined optimal concentration of the primatized recombinant anti-α4β7 mAb–coated beads, and the nonadherent population was collected and activated with Con A + IL-2 for 2 d and then infected with the same amount of SIVmac239, and the supernatant fluids collected at various time intervals and assayed for p27 as noted earlier.
Cell activation and retinoic acid treatment
PBMCs were isolated and activated in vitro using anti-CD3/CD28–conjugated immunobeads (two beads/cell, Tosyl-activated beads obtained from Life Technologies and conjugated in our laboratory with clones FN-18 for anti-CD3 and clone 9.3 for anti-CD28) in media containing human rIL-2 (20 U/ml; kind gift from Hoffman-LaRoche, Nutley, NJ) for 24 h. Aliquots of such in vitro–activated cells were then cultured for another 24 h in the same media (control) or media supplemented with 1 μM of all trans retinoic acid (RA; Sigma-Aldrich). The studies involving the use of RA were conducted in the absence of light, and the cultures were maintained in the dark until harvest. Cells were washed, fixed, and subjected to flow-cytometric analysis. Experiments involving the use of inhibitors that included FK866, a noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, were performed by their addition to the cell cultures in media as indicated. Controls included cells cultured in media without the inhibitor. The cultures were once again washed, fixed, and subjected to flow-cytometric analysis.
Measurement of RA and retinol from plasma samples
Retinoids were extracted from 150–300 μl frozen plasma using a previously described liquid–liquid extraction method (22–24). All sample preparations were performed under yellow lights. RA was quantified by liquid chromatography-tandem mass spectrometry with atmospheric pressure chemical ionization in positive-ion mode on an AB Sciex 5500 QTRAP hybrid triple-quadruple mass spectrometer (Foster City, CA) (22–24). Retinol was quantified by HPLC/UV on a Waters Aquity H-Class UPLC with a PDA detector (Milford, MA) (24). Retinoid concentrations were determined from calibration curves (for each analyte) constructed from authentic standards. The levels of plasma retinoid are expressed as moles per milliliter fluid. Results are expressed as mean ± SE based on the average of 7–12 animals.
Measurement of plasma levels of TGF-β1
Heparinized blood specimens were centrifuged at 450 × g at 4°C immediately after collection, and the plasma was removed and centrifuged one more time at 450 × g to ensure maximal depletion of platelets. The plasma was then aliquoted and cryopreserved until assay. The levels of rhesus TGF-β1 were quantitated using a commercial ELISA kit (MyBiosource, San Diego, CA). This protocol was determined to be optimal for the measurement of plasma levels of TGF-β1.
Cloning and sequencing of promoter regulatory sequences into the pGL3 vector
The full-length α4, β7, and αE promoters regulatory regions from SM, RM, and PM were amplified from genomic DNA with gene-specific primers and cloned into the pGL3 vector containing a luciferase reporter gene (Promega, Madison, WI) and sequenced using Sanger sequencing. The Emory Custom Cloning Core Facility performed the sequencing and subcloning. The sequences were deposited in GenBank with the accession numbers KP140955–KP140960 (http://www.ncbi.nlm.nih.gov/genbank/).
Phylogenetic analysis
Primate α4, β7, and αE promoter regulatory region sequences generated in this study were aligned with additional primate sequences obtained from the GenBank database through BLAST searches. In addition, we retrieved the CCR5 promoter sequences available from diverse primates. GenBank accession numbers of the retrieved primate sequences are listed in Supplemental Table I. The Integrin α4, sequence of Callimico goeldii (Goeldi’s marmoset), recently sequenced by a next-generation technique in our laboratory (M.A. Soares, unpublished results) has also been included. Alignments were generated with MEGA v.5.2 (25) using the Muscle algorithm, and the output files were subject to phylogenetic inference by maximum likelihood (ML) using PhyML v. 3.0 (26) and by Bayesian inference (BI) using Mr. Bayes v. 3.2.1 (27) in two runs with sampling at each 100 generations of a total of 1 million generations. The model of evolution was tested with the Bayesian Information Criterion on Model Generator (28), indicated Kimura 80 (29) with γ distribution for Integrin α4, and Hasegawa-Kishino-Yano (30) with γ distribution for Integrin β7, and Final trees were edited with MEGA v.2.1. Both ML bootstrap with 1.000 replicates and BI posterior probability values were determined. Only values greater than the cutoffs of 75% and 0.99, respectively, were considered significant and plotted in the final phylogenetic trees.
Transfection and reporter gene analysis
Cell culture studies
CEMX174 and HuT78 cells were grown in complete RPMI 1640 medium, and the cell lines were cultured and maintained at 37°C in a 5% CO2 humidified incubator. Cells were typically harvested during the exponential growth phase for transfection studies. The Herpes papio in vitro B lymphoblastoid cell lines (B-LCL) established from RM and SM were similarly maintained in complete RPMI 1640 medium as described earlier.
Transfection studies
CEMX174, HuT78 cells, and the monkey B-LCLs were transfected using the Amaxa nucleofection technology (Amaxa, Koln, Germany). Cells were suspended in a solution supplied with the Amaxa Human T cell nucleofector kit (VPA-1002 LONZA), following the Amaxa guidelines for cell line transfection (see Amaxa literature for further details about this kit). In brief, 100 μl of a 10 × 106 cell suspension was mixed with either 1 μg pmaxGFP only or combined with 5 μg pGL3-DNA and transferred to the provided cuvette and nucleofected using an Amaxa Nucleofector apparatus (Amaxa). Cells were transfected using the V-001 pulsing parameter and were immediately transferred into tubes containing 1 ml 37°C prewarmed culture medium, and aliquots were equally divided into individual wells of a 12-well plate with or without 1 μM RA. After transfection, cells were cultured for 24–48 h in the case of cell lines and for 6–24 h for the B-LCL, and the cultures were subjected to luciferase measurements. GFP plasmid was used as internal control in the assay to normalize for transfection efficiency. A fraction of cells from the same wells continued in culture for 72 h and were measured using flow cytometry to measure GFP.
A plasmid containing four repeats of an RA response element with luciferase reporter was used as a positive control as separate cuvette to measure RA-induced gene expression.
Statistical analysis
Statistical significance was assessed using the two-tailed, unpaired Student t test or multiple comparisons. The surface expression of CD4, CD8 was displayed as mean ± SD. Differences over time were analyzed using Tukey’s multiple comparison tests and the Mann–Whitney U test. All the reported p values were based on two-sided testing. Statistical analyses were performed using Prism 5 Statistical software packages: *p < 0.05, **p < 0.001, ***p < 0.0001.
Results
Frequencies of α4β7-expressing CD4+ T cells and other hematopoietic cell lineages from RM, PM, and SM
We first evaluated the frequencies of α4β7-expressing CD4+ T cells in PBMC samples from a number of RM, PM, and SM. A representative profile of α4β7-expressing CD4+ T cells from each of these three NHP species is displayed in Fig. 1 (with the shaded area representing the medium and the area to the right and the numbers within each panel representing the mean ± SD values of the frequencies of α4β7hi-expressing cells). As noted, there is a marked increase both in the frequencies and in the mean fluorescent intensity (MFI) of α4β7 expression by CD4+ T cells from the PM as compared with the SM and RM. When the other cell lineages were examined, the RM, by far, had the highest frequencies of CD8+ T, NK, and B cells that expressed α4β7 (see Fig. 1B). In particular, the values obtained for CD8+ T cells/NK cells and B cells in RM were clearly significantly higher than in SM (p < 0.001 and p < 0.01, respectively). We also cultured enriched populations of CD4+ T cells from each of the three species for 48 h in media containing Con A + IL-2 and then infected them with SIVmac239 as described in Materials and Methods. Supernatant fluids were collected at varying time intervals and assayed for levels of p27. As reported in Fig. 1C, the CD4+ T cells from PM not only produced the highest level of p27 but kinetically showed peak activity at day 7 as compared with day 10 for RM and SM. These data are consistent with the in vivo findings of increased susceptibility to SIV infection of PM versus RM. Furthermore, SIV infection of in vitro Con A + IL-2 activated PBMCs from all three species (n = 5 RM; n = 3 PM, and n = 5 SM) that were depleted of α4β7-expressing CD4+ T cells showed significantly lower (RM: 32 ± 17 ng/ml, PM: 28 ± 16 ng/ml, and SM: 39 ± 14 ng/ml; Fig. 1D) levels of p27 as compared with PBMCs treated with normal rhesus Ig from the same three species (Fig. 1E). Our observation that the depletion of α4β7-expressing CD4+ T cells led to significant decreases in the levels of p27 in all three species suggests that α4β7-expressing CD4+ T cells are the major source of virus in such in vitro culture conditions.
FIGURE 1.
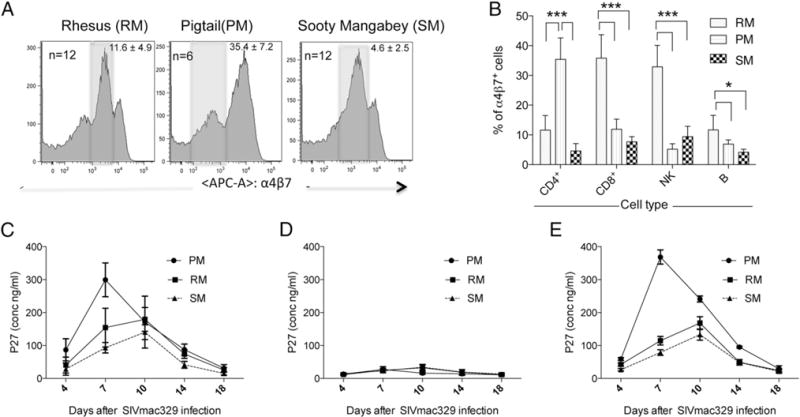
(A) Representative profiles of the surface expression of α4β7 on the gated population of CD3+ CD4+ T cells in the PBMCs from RM (n = 12), PM (n = 6), and SM (n = 12). The numbers within each profile reflect the frequency (%) of α4β7hi-expressing cells (mean ± SD); the shaded area represents the medium α4β7-expressing cells, and the area to the right reflects cells that are classified as α4β7hi. (B) Quantification of the frequency (%) of α4β7+ CD4+, CD8+, NK, and B cells (mean ± SD) from the same number of RM, PM, and SM as shown in (A). (C) Highly enriched populations of CD4+ T cells from RM, PM, and SM (n = 12 RM and SM; n = 6 PM) stimulated for 2 d in vitro with anti-CD3/28 + IL-2 were cultured in triplicate in vitro with SIVmac239, and supernatant fluid collected on days 4, 7, 10, 14, and 18 was analyzed for the levels of p27. The mean ± SD of the levels of p27 (ng/ml) for each of the three species is shown. (D) PBMCs from RM, PM, and SM (n = 5 RM, n = 3 PM, and n = 5 SM) were either depleted of α4β7-expressing CD4+ T cells or (E) treated with normal rhesus IgG and stimulated with Con A + IL-2 for 2 d and then infected with SIVmac239, and supernatant fluids were collected on days 4, 7, 10, 14, and 18 d and analyzed for the levels of p27 using commercial kits. *p < 0.05, ***p < 0.0001.
Relationship between α4β7 and other integrins expressed by CD4+ T cells
Integrins are primarily expressed as transmembrane cell adhesion molecules that bind noncovalently with their cognate ligands. They are heterodimeric proteins composed of an α- and β-chain that bind to cell-surface molecules and also to select extracellular matrix proteins (31, 32). In vertebrates, to date, at least 18 forms of the α and 8 of the β subunits have been identified that combine in a variety of combinations to generate 24 different integrin heterodimers (Fig. 2A). A detailed description of the nature of these heterodimers has been reviewed elsewhere (33). The variable levels of α4β7 expression by CD4+ T cells in the PBMCs from RM, SM, and PM prompted us to determine whether the differences noted were secondary to differential pairing of the α4 and β7 subunits with other integrin α- and β-chains, respectively, in the different species. Previous studies have documented that the α4 chain can pair with either the β7 chain or, alternatively, the β1 chain (34, 35). Detailed analysis of β7 and β1 expression in humans and mice suggests that these two β-chains effectively compete for pairing with α4 (35), and that there exists an apparent hierarchy in pairing at the molecular level such that α4 preferentially pairs with β1. Likewise, the β7 chain can alternatively pair with either α4 or αE (35). Notably, in general, neither α4, αE, β1, nor β7 remains on the cell surface in a homodimeric or an unpaired state (34). To investigate possible differences in pairing among the three species, we stained CD4+ T cells from each with mAbs specific for α4, β1, β7, and αE. We also used an mAb that recognizes the heterodimeric form of the α4β7 molecule exclusively. The gating strategy used is depicted in Fig. 2B, and a representative profile of α4β7-expressing PBMCs from RM, PM, and SM on the gated population of CD3+/CD4+ T cells is displayed in Fig. 2C (column 1, top, middle, and bottom rows, respectively). The remaining profiles reflect results obtained on the gated population of CD3+/CD4+/α4β7+ cells. As shown in the upper right quadrant of each profile in columns 2–4 of Fig. 2C, there is a distinct population of α4β7+ cells that also bind to anti-α4, -β1, and -β7, respectively. There also appears a distinct population of cells that express α4 or β1 that are nonreactive with the α4β7 mAb (Fig. 2C, columns 2, 3), which very likely reflects the pairing of these two chains to form α4β1. Of note, the CD4+ T cells from SM were unique in that they presented αE+ cells in both α4β7+ and α4β7− populations (Fig. 2C, column 5, bottom row). Detection of αE on peripheral CD4+ T cells from SM was highly reproducible and indicates a stark difference among the three NHPs analyzed (Table I). In this regard, we note RM and PM share in common with humans a near absence of αE on peripheral CD4+ T cells (data not shown). To determine whether these differences also appeared in CD4+ T cells from the gut mucosa, we obtained gut biopsies from RM (n = 6), PM (n = 2), and SM (n = 10), and isolated cells using standard procedures as described elsewhere (36) and performed flow-cytometric analysis. We found similar results as described for PBMCs, suggesting that in gut tissues α4 is also associated with either β1 or β7, forming α4β1 and α4β7 heterodimers, respectively, in all three species. In addition, αE was predominantly expressed by the central memory CD4+ T cells from SM, but none of the CD4+ T cell subsets from RM or PM (Supplemental Fig. 1A, 1B). The failure of the anti-αE reagent to react with peripheral blood–derived cells from RM and PM was not secondary to nonreactivity of the mAb because cervical biopsies from all three species showed a readily detectable population of αE-expressing CD4+ T cells in such biopsies (data not shown). Next, in efforts to determine whether α4β1 was expressed by a subset of CD4+ T cells distinct from the cells that express α4β7, PBMCs were stained with biotinylated VCAM-1 followed by PE-avidin and allophycocyanin-conjugated anti-α4β7, and the staining was performed in the presence of buffer containing Mg2+ (because VCAM-1 selectively binds to α4β1, but not α4β7, in the presence of Mg2+). As shown in Supplemental Fig. 2, a majority of the CD4+ T cells coexpress α4β1 and α4β7. Furthermore, to determine whether this pairing is influenced by age, we studied infant (3- to 8-mo-old) RM and SM and found that such pairing is present at an early age (Table I), confirming that the differences noted are not likely due to development events but are an inherent feature of the species.
FIGURE 2.
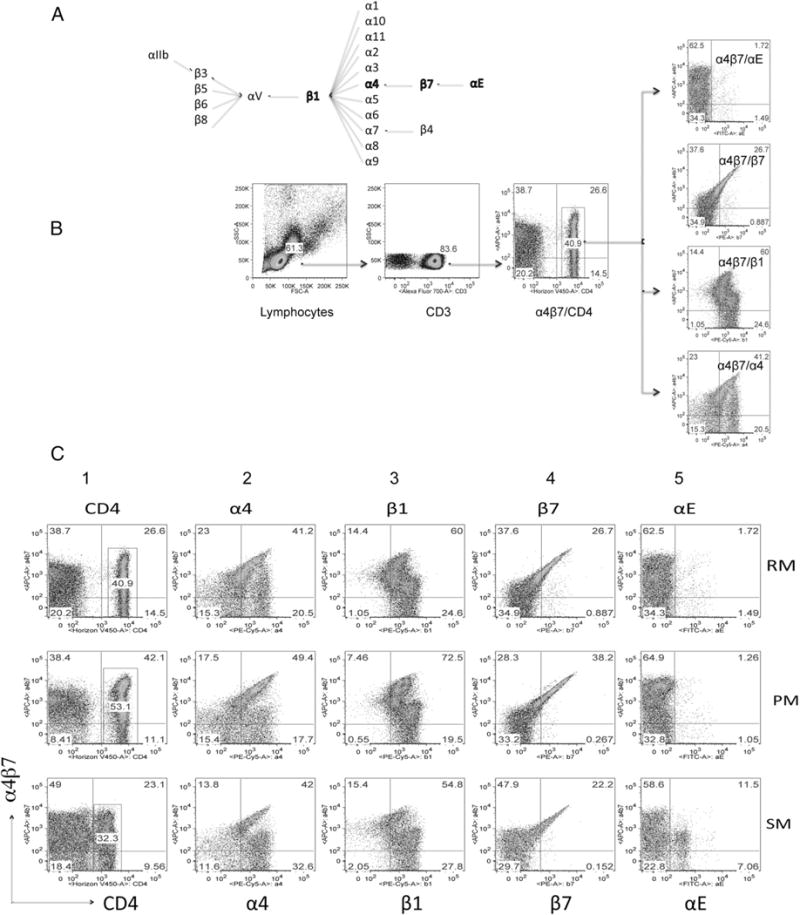
(A) Pairing patterns of various α- and β-chains of integrins and (B) a representative profile of the gating strategy that was used to define the percentages of CD4+ T cells expressing α4β7, α4, β1, β7, and αE in samples of PBMCs from the three species. (C) Representative profiles of the flow-cytometric analysis of the pairing of α4, β1, β7, αE with α4β7 integrins (columns 1–5, respectively) from RM (top row), PM (middle row), and SM (bottom row). The analysis was performed on the gated population of CD3+ CD4+ T cells (as displayed in C). This gated population was then analyzed for the expression of α4β7 by CD4+ T cells (column 1), and the gated population of CD4+ α4β7+ cells was then analyzed for their coexpression of α4 (column 2), β1 (column 3), β7 (column 4), and αE (column 5).
Table I.
Frequencies of CD4, α4, β1, β7, and αE that express α4β7+ in the PBMCs of RM, SM, and PM
| Species of Monkey | n | % CD4+/α4β7+ | % α4β7+/α4+ | α4β7+/β7+ | α4β7+/β1+ | α4β7+/αE+ |
|---|---|---|---|---|---|---|
| PBMCs | ||||||
| PM | 4 | 44.8 ± 6.7 | 46.6 ± 15.0 | 26.1 ± 11.5 | 46.3 ± 13.7 | 1.0 ± 0.7 |
| RM | 6 | 37.1 ± 11.8 | 62.5 ± 23.5 | 15.0 ± 6.6 | 63.8 ± 17.3 | 0.62 ± 0.4 |
| SM | 10 | 15.6 ± 8.5 | 57.37 ± 7.0 | 17.0 ± 8.2 | 64.9 ± 14.0 | 11.67 ± 7.2 |
| Infant RM | 10 | 47.4 ± 7.3 | 20.7 ± 7.5 | 7.8 ± 3.4 | 23.1 ± 5.7 | 0.15 ± 0.04 |
| Infant SM | 4 | 60.4 ± 8.4 | 18.7 ± 8.1 | 7.5 ± 4.3 | 20.3 ± 24.0 | 4.77 ± 3.5 |
| Gut | ||||||
| PM | 2 | 83.9 ± 9.3 | 6.46 ± 0.7 | 16.57 ± 6.8 | 19.3 ± 3.2 | 7.8 ± 1.6 |
| RM | 6 | 60.5 ± 7.5 | 4.35 ± 0.7 | 5.24 ± 1.0 | 5.46 ± 2.4 | 4.9 ± 1.7 |
| SM | 12 | 46.8 ± 9.3 | 20.0 ± 11.6 | 12.36 ± 6.9 | 30.8 ± 13.6 | 15.18 ± 6.5 |
Numbers in bold highlight the major differences.
Cloning, sequencing, and phylogenetic analysis of α4, β7, αE, and CCR5 promoters from RM, SM, and PM and their reporter gene analysis
We next reasoned that the differences in integrin expression among the three NHPs examined may be because of differences in the promoters. This prompted us to PCR amplify and clone the α4, β7, and αE promoters using cDNA from RM, SM, and PM into a pGAL4 vector. We sequenced several clones, obtained consensus sequences, and identified several regulatory sequences present in each of the promoters. We then performed a phylogenetic analysis that incorporated Old World and New World monkey sequences obtained from the literature or GenBank. As a point of reference, we also included CCR5 promoter sequences. As shown in Fig. 3A, the α4 sequences of new world and old world monkeys formed separate clusters. Next, in old world monkeys, the α4 sequences from the Asian RM and PM formed a close cluster and as expected, the α4 sequences from the African SM formed a different cluster, suggesting that there is divergence in the α4 promoter sequences among RM, PM, and SM. Next, we also aligned β7 promoter sequences (Fig. 3B) and found similar separate clusters between old and new world monkey species, and as with α4 promoter sequences, β7 sequences from RM and PM formed a closer cluster compared with sequences from SM. Similar results were obtained for αE and CCR5 promoters as shown in Fig. 3C and 3D. The SM promoters always formed separate clusters from PM and RM for each of the promoters analyzed, suggesting that the differences in clustering of the promoter sequences indeed are a reflection of the divergence times of the different species. This difference could potentially contribute to differential expression of each of the integrin subunits and the subsequent formation of heterodimers in cells from RM, PM, and SM.
FIGURE 3.
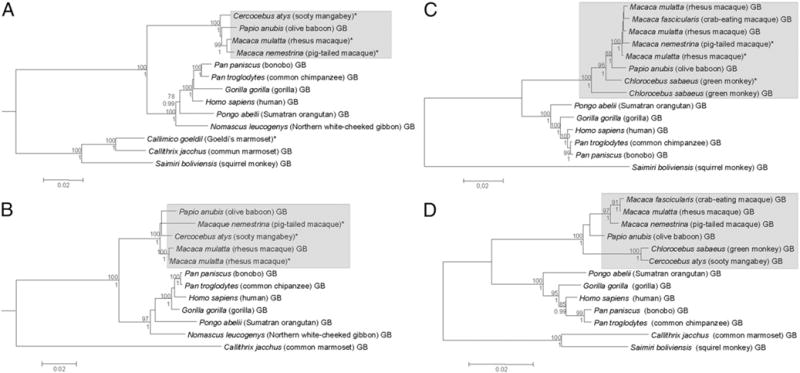
Phylogenetic inferences of Integrin α4 (α4; A), Integrin β7 (β7; B), integrin αE (αE; C), and CCR5 (D) promoters of RM, SM, and PM compared with those of other NHPs. For all four trees shown, both ML and BI analyses were carried out, and they both rendered the same tree topologies, which could be shown in single consensual trees. Sequences with asterisks represent those generated in this study, whereas those with the “GB” suffix denote sequences retrieved from the GenBank database. Numbers above and below each node of the trees are the ML bootstrap and BI posterior probability values, respectively, and only values >75% and 0.99, respectively, are shown. Scale bars at the bottom of the trees represent nucleotide substitutions per site.
Furthermore, we also examined the α4, β7, and αE promoter regulatory sequences from PM, RM, and SM for putative transcription factor binding sites (TFBSs). As shown in Fig. 4, extensive genetic variations are apparent. First, the α4 promoter (Fig. 4A) is composed of >3240 bp, and binding sites (represented by bars) for several common transcriptional factors (SRY, CdXA, AML-1aDelta E, MZF1, LYf-1) were noted within the α4 promoter sequences from the three species of NHPs; several of them were restricted at different locations within the promoters. The β7 promoter regulatory sequence (Fig. 4B) is shorter than the α4 sequence (2465-bp length), and we noted similar TFBSs as seen with the α4 promoters but also the identification of several additional TFBSs such as those for the p300, IFN-stimulated response element, among other transcription factors. Notably, whereas the CP2 promoter-binding site was restricted to the α4 promoter, the p300 and IFN-stimulated response element were restricted to the β7 promoter. Analysis of the αE promoter regulatory sequences (Fig. 4C) showed that although the RM and PM promoter regulatory region was composed of 3000 bp, there is a large insertion of ~300 bp in the SM αE promoter regulatory sequence. However, we could not identify any unique TFBSs within this 300-bp insertion noted for SM. Taken together, these TFBS analyses failed to identify any readily detectable major difference in TFBSs between these species for each of the α4, β7, and αE genes.
FIGURE 4.
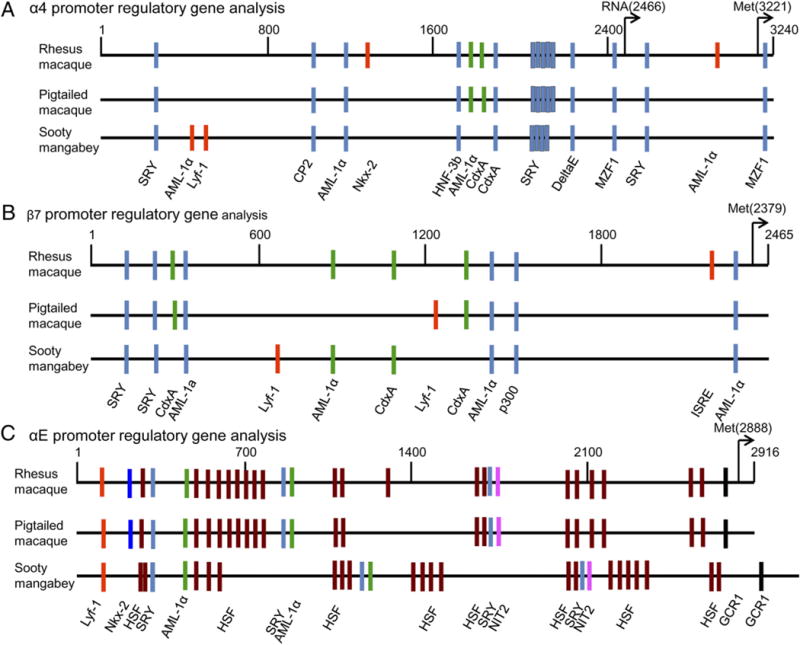
TFBS analysis of the (A) α4, (B) β7, and (C) αE promoters from RM, PM, and SM. Colored bars represent binding sites of the various transcription factors.
In parallel, we also examined the functional response of each of the promoters regulatory regions for α4 and β7 integrin chain from each of the three species using transient transfection assays. We used both established (CEMX174 and HUT78) and B-LCLs prepared from the PBMCs of PM, RM, and SM. GFP plasmids were used as internal controls during transfection studies to normalize gene expression levels for transfection efficiency. As shown in Fig. 5, the levels of α4 promoter activity from PM, RM, and SM were similar in each of the two established cell lines and in the B-LCLs tested. We also tested the β7 promoter constructs. Whereas the PM showed the highest levels of β7 promoter activity, the RM β7 promoter showed significantly lower levels, but the lowest levels of β7 promoter–driven gene expression were noted for SM as compared with PM and RM (Fig. 5). These findings were highly reproducible (a minimum of three experiments) and confirmed that the SM β7 promoter–driven expression was consistently lower as compared with the β7 promoter from RM and PM, even within species-specific cells, indicating that these findings were indeed promoter specific. The finding that CD4+ T cells from SM express significantly higher constitutive and induced levels of αE presumably in association with β7 on the cell surface prompted us to analyze gene expression levels of the αE promoter. Of interest, we found that although the promoter construct for αE gave essentially similar signals in the two established cell lines (Fig. 6A, 6B), the αE promoter gave markedly lower signals in the B-LCL from SM, but not RM (Fig. 6C, 6D). These data suggest that the preferential pairing of αE with β7 in SM is not likely secondary to differences at the promoter level but due to yet to be defined mechanisms.
FIGURE 5.
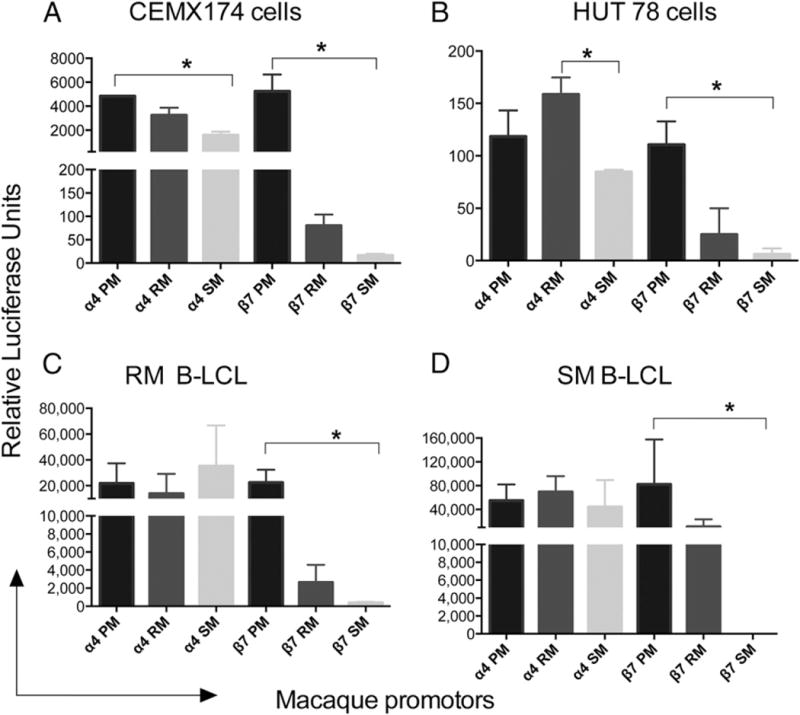
Reporter gene analysis: the various α4 and β7 integrin reporter constructs of promoters from PM, RM, and SM were assayed after transfection as shown. Transfections were performed using AMAXA, and reporter gene expression was measured at 24–48 h for cell lines and within 6–24 h for the B-LCL. Experiments were repeated three times and data calculated. RA response element was used as a positive control and a GFP plasmid as an internal control in the assay to normalize for transfection efficiency. Data shown are expressed as mean ± relative luciferase units using the (A) CEMX174 cell line, (B) HUT78 cell line, (C) rhesus B-LCL, and (D) SM B-LCL. *p < 0.05.
FIGURE 6.
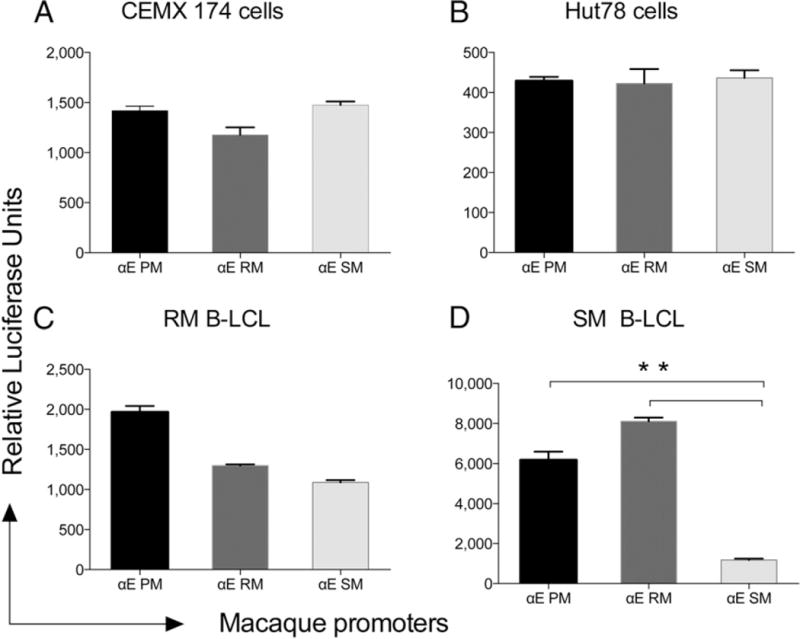
Reporter gene analysis of αE promoter–driven gene expression analysis using (A) CEMX174 cell line, (B) HUT78 cell line, (C) rhesus B-LCL, and (D) SM B-LCL. All experimental conditions were similar to those described in the legend for Fig. 5. **p < 0.001.
Biological role of RA in cell activation and expression of α4β7
It has been previously documented that select cell lineages within GALT synthesize RA from vitamin A. RA attracts cells to home to the gut tissue via the upregulation of α4β7 (37, 38). We next asked whether the species-specific differences described earlier might, in part, be due to differences in the response of CD4+ T cells from the different species to RA. We failed to identify any detectable dose or kinetic difference in the response of cells from these species to RA for optimal α4β7 expression (data not shown). Activated cells treated with RA upregulated the expression (MFI) of α4β7 >4- to 8-fold compared with unstimulated control cells in cultures from RM as shown for representative profile (Fig. 7A) with similar results obtained for RM, PM, and SM (Fig. 7B). As previously shown with humans and mice (39), preactivation of the CD4+ T cells is a requirement for RA-mediated upregulation of α4β7 expression by RA. This prerequirement for cell activation was confirmed with the use of FK866, a noncompetitive inhibitor of nicotinamide phosphoribosyltransferase. As shown in Fig. 7C and 7D, the addition of FK866 completely blocked RA induction of α4β7 on activated RM PBMCs.
FIGURE 7.
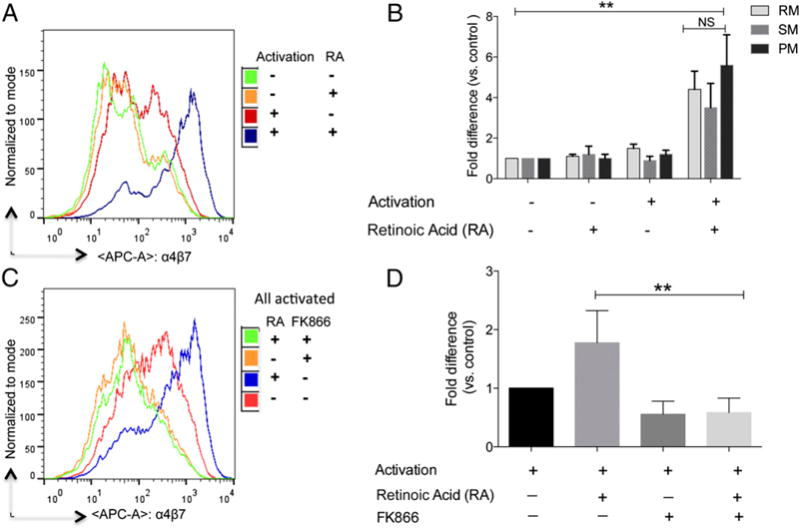
Requirements for cell activation for the upregulation of α4β7 expression by RA treatment. (A) Aliquots of PBMCs from normal healthy RM, PM, and SM were cultured for 48 h in complete RPMI 1640 media alone (green), complete media containing 1 μM RA alone for the last 24 h (orange), complete media containing anti-CD3/28 immunobeads + IL-2 alone (red), and the anti-CD3/28 immunobeads + IL-2 along with 1 μM RA for the last 24 h (blue). (C) The same experiment as outlined in (A) except an additional culture containing the immunobeads + IL-2 and 1 μM RA was set up that included the addition of the inhibitor FK866 that was added at the initiation of the culture. The cells were washed and the flow-cytometric profile of CD4+ T cells that expressed α4β7 is shown in (A) and (C). The experiment was repeated three times, and the data obtained were expressed as fold change (mean ± SD) of MFI relative to baselines (B and D). **p < 0.001.
Measurement of plasma levels of RA and retinol from the three groups of monkeys
In addition to inherent differences in the RA signaling pathway in T cells that might contribute to species-specific differences in their α4β7 expression, we reasoned that the availability of the ligand (i.e., RA) in vivo could also play a role. To assess this possibility, we measured retinol and RA levels in the plasma of PM, RM, and SM (n = 10 each). Surprisingly, we found higher plasma levels of RA in SM than that in RM or PM, as shown in Fig. 8A (p < 0.0001). In contrast, there was no significant difference in plasma retinol concentration of SM versus either of the two species, although mean retinol levels between RM and PM did show a significant difference (p < 0.05; Fig. 8B). The relatively higher levels of RA (known to induce the expression of α4β7) in SM when compared with RM was surprising in light of the relatively lower frequency of α4β7+ T cells in SM. A possible explanation for these apparently discordant observations may lie in the increased plasma levels of TGF-β1 in SM as compared with RM and PM (Fig. 9), and the manner in which αE and α4 both pair with β7. It is known that in humans and mice, β1 and β7 compete for binding to α4, and it appears that at the molecular level α4 preferentially binds to β1. A similar preference may exist for αE and α4 in competing for β7, such that increased expression of αE leads to a downregulation of α4β7. In response, SM may increase the production of RA. Yet for reasons not yet understood, increased RA levels are unable to counterbalance the high levels of TGF-β1, resulting in low levels of α4β7 on CD4+ T cells. In this regard it will be important to determine the hierarchy of pairing of αE versus α4 with β7 because it is that pairing that may establish a distinct trafficking pattern in SM. Ultimately, then, TGF-β1 may play a key role in the origin of disease resistance in SM.
FIGURE 8.
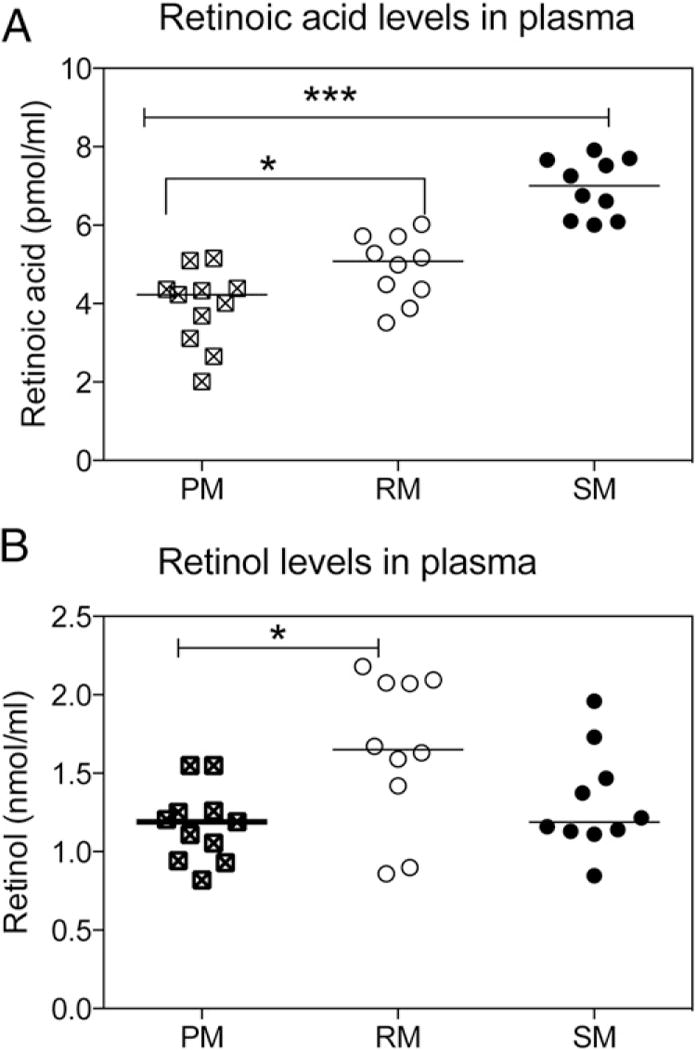
Measurements of plasma levels of RA and retinol. Levels of RA and retinol were extracted from the plasma of RM, SM, and PM (n = 10 from each species). (A) Levels of RA (pmol/ml) and (B) retinol (nmol/ml) in plasma were quantified by liquid chromatography-tandem mass spectrometry (RA) or HPLC (retinol). Data shown are means ± SD. *p < 0.05, ***p < 0.0001.
FIGURE 9.
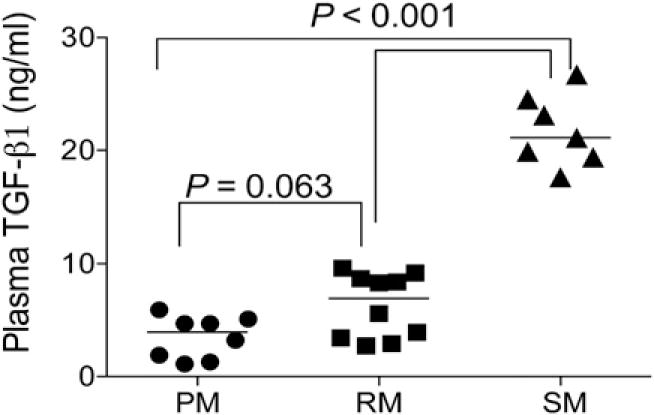
Measurement of plasma TGF-β1. Plasma levels of TGF-β1 were measured using a commercial ELISA kit and plasma from uninfected PM (n = 18), RM (n = 10), and SM (n = 7) and reported as ng/ml. The p values were presented in the figure.
Levels of α4β7 expression by hematopoietic cells from natural versus non-natural hosts of SIV
The fact that African SM that are the natural hosts of SIV showed the lowest level of α4β7-expressing CD4+ T cells and the two Asian non-natural hosts (PM and RM) showed higher levels of α4β7-expressing CD4+ T cells prompted us to determine whether the low levels noted for SM were characteristic of other African NHPs, most of which are also natural hosts of SIV (20). PBMCs from blood samples from a number of African NHP species were analyzed for the frequencies of CD4+ T, CD8+ T, NK, and B cells. As shown in Table II, although we observed differences in the frequencies of the various cell lineages that express α4β7 in the blood of the various African species, there is no apparent link between the expression of α4β7-expressing cells and natural versus non-natural hosts. In addition, because the blood samples studied were obtained from monkeys of varying ages, we carried out studies to determine whether the species-specific difference in α4β7 expression in the three species that we noted earlier was acquired as a function of age. We therefore studied infant (3- to 8-mo-old) RM and SM, and found that such differences were already apparent at an early age (Table II). Thus, the differences we noted were not due to ontogenetic differences in the species studied.
Table II.
Frequencies of CD4+ T cells, CD8+ NK cells, and B cells that express α4β7hi in the PBMCs of Asian and African NHPs
| Species of Monkey/Chimp | n | % CD4 | % CD4+ α4β7hi | % CD8+ α4β7hi | % NK α4β7hi | % CD20+ α4β7hi |
|---|---|---|---|---|---|---|
| RM | 12 | 44.5 ± 12.3 | 11.6 ± 4.9* | 35.8 ± 7.9 | 32.9 ± 7.2 | 11.68 ± 4.9 |
| Pig-tailed | 6 | 28.6 ± 7.7 | 35.4 ± 7.2* | 11.9 ± 3.4 | 5.2 ± 1.8 | 6.9 ± 1.4 |
| SM | 12 | 22.3 ± 6.1 | 4.6 ± 2.5* | 7.7 ± 1.7 | 9.4 ± 3.5 | 4.2 ± 1.0 |
| Infant RM | 10 | 34.44 ± 9.2 | 12.6 ± 4.1 | 16.1 ± 6.4 | ND | 7.1 ± 4.2 |
| Infant SM | 4 | 55.15 ± 15.9 | 5.1 ± 1.6 | 4.31 ± 1.4 | 1.45 ± 0.73 | 0.85 ± 0.5 |
| African Green | 6 | 14.9 ± 7.4 | 5.2 ± 1.5 | 12.6 ± 4.2 | 18.6 ± 7.3 | 11.4 ± 4.7 |
| Baboons | 5 | 42.5 ± 8.1 | 12.2 ± 6.1 | 2.9 ± 1.8 | 5.6 ± 4.2 | 8.9 ± 2.9 |
| Blue sykes | 3 | 18.2 ± 4.3 | 11.7 ± 4.4 | 7.2 ± 1.8 | 5.8 ± 1.4 | 4.8 ± 1.1 |
| Chimpanzee | 6 | 45.6 ± 6.1 | 2.9 ± 0.8 | 2.8 ± 1.1 | 3.3 ± 1.1 | 4.9 ± 2.3 |
| Drill | 3 | 13.4 ± 3.2 | 15.6 ± 4.7 | 3.7 ± 1.1 | 4.2 ± 1.7 | 5.6 ± 2.2 |
| Gray mangabey | 3 | 26.1 ± 1.0 | 9.6 ± 3.4 | 8.1 ± 2.4 | 6.3 ± 2.9 | 5.3 ± 1.4 |
| Stump tailed | 6 | 33.1 ± 6.5 | 10.9 ± 5.7 | 4.9 ± 2.1 | 4.8 ± 3.1 | 1.9 ± 0.4 |
| Sykes | 3 | 26.4 ± 1.7 | 7.9 ± 3.4 | 4.2 ± 1.8 | 5.6 ± 4.2 | 7.8 ± 1.9 |
Numbers in bold highlight the major differences.
p < 0.0001, differences between PM and SM; p < 0.001, differences between PM and RM.
Discussion
The α4β7 integrin is one of the major gut-homing markers expressed by a variety of hematopoietic cell lineages in both mouse and human (34, 40). Its role in facilitating the gut homing of cells with pathogenic potential has been documented in the pathology of a number of human diseases that involve GALT, including graft-versus-host disease (41), Crohn’s disease (42), ulcerative colitis (43), and inflammatory bowel disease (44). These findings have prompted studies aimed at determining the therapeutic efficacy of mAbs (such as vedolizumab) against α4β7 in patients with Crohn’s, ulcerative colitic, and inflammatory bowel disease with some very encouraging results (reviewed in Ref. 45). More recently, the role of α4β7-expressing cells has been highlighted in human HIV-1 infection and the NHP model of SIV infection (14–17, 46, 47) in which, during the early stages of disease, the GALT is the major target of infection and pathology. Thus, α4β7-expressing CD4+ T cells have been shown to be one of the major targets of HIV/SIV infection, and increased frequencies of this subset within the GALT have been shown to be associated with greater levels of infection and a more rapid disease course (14). Our laboratory has been involved in studies defining the in vivo effects of the administration of an anti-α4β7 mAb during acute SIV infection of RM infected i.v., intrarectally (15, 16), and more recently while undergoing low-dose repeated intravaginal SIV exposure (17). The central rationale for these studies was to determine whether inhibiting the interaction between HIV/SIV env and the α4β7 molecule and/or impeding the trafficking of cells to the GALT using mAbs against α4β7 could limit GALT pathology and possibly prevent mucosal transmission. Indeed, the results of such studies have shown great promise in limiting GALT pathology and even prevention of SIV transmission (17).
Considering the results of our studies noted earlier coupled with the report that SIV infection results in markedly different disease course in SM, RM, and PM, it was reasoned that a more complete characterization of the expression of α4β7 in NHPs might provide some unique insights into the basis for these distinct clinical outcomes. To this end, we carried out the studies reported in this article that show that CD4+ T cells from PM, the species that is associated with rapid disease progression, show the greatest levels of constitutive and RA-induced α4β7 expression. SM, which rarely progress to disease, showed the lowest α4β7 expression levels, whereas RM express a relatively intermediate level (rank order: PM > RM > SM). Thus, this order parallels in an inverse way the rate of disease progression associated with SIV infection of these three species of NHPs. These findings appear to remarkably parallel the differences in the expression of CCR5 by CD4+ T cells in these species (48, 49). The potential relationship(s) between the regulation of α4β7 and CCR5 expression is a subject currently under study.
Our initial hypothesis was that the level of α4β7 expression on CD4+ T cells would correlate with disease resistance/susceptibility distinguishing natural host species of SIV from the non-natural hosts. To address this issue, we examined PBMC samples from a variety of NHP species for the frequencies of α4β7 expression on CD4+ T cells, CD8+ T cells, NK cells, and CD20+ B cells. As shown in Table II, there did not appear to be any strict relationship between the Asian and African origin of the donor of the PBMCs and the frequencies of the α4β7-expressing cell lineages. These data need to be interpreted with caution because we are assuming that the reagents we used are equally cross-reactive between species. We did screen a large number of Ab clones and the mAbs we used were identified as the most optimal available. We submit, however, that the data from the studies reported in this article provide one important clue. The data indicate that CD4+ T cells from SM express relatively lower levels of α4β7, but ~10% of these CD4+ T cells constitutively coexpress readily detectable levels of αE most likely associated with β7 (see Fig. 2). This distinguishes SM, not only from PM and RM, but also from humans, all of which exhibit a very low frequency of αE+ CD4+ T cells circulating in blood. Thus, it appears that in the peripheral blood of SM (the natural hosts of SIV), the β7 integrin is expressed and pairs with the αE chain. The αE chain is not expressed by cells from the peripheral blood of RM or PM (the non-natural hosts). It is quite possible that this unique expression profile of integrins may promote a pattern of homing that is distinct from homing to the GALT leading to distinct disease course. In support of this hypothesis, differential localization of SIV-infected target cells to the small as compared with the large intestine has been previously shown to lead to marked differences in clinical outcome in RM (50). These findings are consistent with the report of marked differences in the composition of cells that comprise the innate and acquired immune system in the small versus the large intestine (51). In this regard, it is important to note that results from previous studies suggest that α4β7+ T cells switch to express αEβ7+ T cells once they localize to mucosal tissues and that this expression is regulated by TGF-β1 (33, 52–55). Studies of the mechanisms by which TGF-β1 regulates the levels of αEβ7 expression revealed that although this cytokine increases the expression of αE and β7 mRNA transcripts and the cell-surface expression of αEβ7, it also decreases α4 at the mRNA level (54). Furthermore, it has been suggested that there is a known axis between TGF-β1, RA, and the pairing of β1, β7, α4, and αE expressed by T cells at least in mice (56). These findings are consistent with differences in the plasma levels of TGF-β1 that we noted to be significantly higher in SM as compared with RM and PM (Fig. 9). We thus posit that the gut epithelial cells from SM synthesize significant levels of TGF-β1 that induce the upregulation of αEβ7 that results in differences in the trafficking of the SIV-susceptible cells that, in turn, contributes to the disease resistance of this species. Results of the studies reported in this article show that this difference in integrin expression patterns in SM as compared with RM and PM was not only true for blood cells but also for α4β7+ T cells isolated from gut biopsies of these species. Studies are in progress to determine whether this phenomenon is unique to SM or is a general phenomenon for other natural hosts of SIV.
Data from our transient transfection studies with reporter promoter constructs further suggest that a correlation exists between β7 promoter activity and surface expression of α4β7. We observed that β7 promoter–driven gene expression was lower in SM as compared with RM and PM, which was inversely correlated with the surface expression levels of αE in the different species; expression levels of αE were much higher in SM as compared with RM and PM. Furthermore, there was no correlation between surface expression of αE and promoter-driven expression among the three species, suggesting that regulation may be occurring through yet to be defined mechanisms. This finding suggests that there may be a feedback mechanism in these species to regulate these integrins that contributes to the susceptibility or control of SIV-induced disease. Recently, it has been shown that virion-associated or free gp120 had the capacity to reduce the activation and proliferation of naive B cells by releasing TGF-β1, and which, in turn, upregulated the expression of FcRL4 (53). These data suggest that TGF-β1 induction by infection might contribute to the pathogenesis. The data presented in this article suggest that differential levels of TGF-β1 in the NHP species being studied might contribute to differences in disease progression. The promoter gene analysis revealed the presence of extensive variations among the three species tested and, to the best of our knowledge, this is the first study to report promoter-driven gene expression studies of α4, β7, and αE in the three species of NHPs used extensively in HIV pathogenesis and vaccine studies. Phylogenetic analysis further confirms that SM α4, β7, αE, and CCR5 sequences formed separate clusters from RM and PM, suggesting that there may be evolutionary-conserved sequence patterns that exist among different species of monkeys, which helps them to recognize pathogen as friend or enemy.
In conclusion, results from the studies presented in this article appear to show differences in the expression pattern of the integrins α4β7 and αEβ7, together with differences in RA and TGF-β1 plasma levels in the NHPs analyzed. Such differences suggest that susceptible species (RM, PM) have the same pattern and this pattern is distinct from the disease-resistant species (SM). Of note, αEβ7 expression pattern in humans is similar to the susceptible NHP species. Such differences could contribute to distinct trafficking patterns in vivo and, as a consequence, impact disease progression. These observations form the foundation for future studies that may help identify which of these differences alone or in concert are the basis for the different disease outcome of SIV infection.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Emory Cloning Core Facility and Dr. O. Laur for the cloning and sequencing of the constructs in the studies outlined. We are also extremely grateful to Stephanie Ehnert and the veterinary/support staff of the Yerkes National Primate Research Center for help in maintaining and procuring all the blood samples used in the studies reported in this article. Our special thanks to the veterinarians at the Atlanta Zoo for help in obtaining samples from the drills, the Southwest National Primate Research Center for the provision of African green monkey blood samples, and the professional and support staff of the Institute for Primate Research, located in Karen, Nairobi, Kenya, for help in obtaining blood samples from the various other African species.
This work was supported by the National Institutes of Health (Grants AI078773 and AI98628 to A.A.A.), the Yerkes National Primate Research Center of Emory University (Base Grant NIH-OD 51POD1113), and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Abbreviations used in this article
- BI
Bayesian inference
- B-LCL
B lymphoblastoid cell line
- MFI
mean fluorescent intensity
- ML
maximum likelihood
- NHP
nonhuman primate
- PM
pigtailed macaque
- RA
retinoic acid
- RM
rhesus macaque
- SM
sooty mangabey
- TFBS
transcription factor binding site
Footnotes
Disclosures
The authors have no financial conflicts of interest.
The sequences presented in this article have been submitted to GenBank under accession numbers KP140955–KP140960.
The online version of this article contains supplemental material.
ORCID: 0000-0002-7423-1763 (S.N.B.).
References
- 1.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCune JM. The dynamics of CD4+ T-cell depletion in HIV disease. Nature. 2001;410:974–979. doi: 10.1038/35073648. [DOI] [PubMed] [Google Scholar]
- 3.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smit-McBride Z, Mattapallil JJ, McChesney M, Ferrick D, Dandekar S. Gastrointestinal T lymphocytes retain high potential for cytokine responses but have severe CD4(+) T-cell depletion at all stages of simian immunodeficiency virus infection compared to peripheral lymphocytes. J Virol. 1998;72:6646–6656. doi: 10.1128/jvi.72.8.6646-6656.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Veazey RS, DeMaria M, Chalifoux LV, Shvetz DE, Pauley DR, Knight HL, Rosenzweig M, Johnson RP, Desrosiers RC, Lackner AA. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]
- 6.Smith PM, Garrett WS. The gut microbiota and mucosal T cells. Front Microbiol. 2011;2:111. doi: 10.3389/fmicb.2011.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley JM, Douek DC. The mucosal barrier and immune activation in HIV pathogenesis. Curr Opin HIV AIDS. 2008;3:356–361. doi: 10.1097/COH.0b013e3282f9ae9c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, et al. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 9.Cicala C, Arthos J, Fauci AS. HIV-1 envelope, integrins and co-receptor use in mucosal transmission of HIV. J Transl Med. 2011;9(Suppl. 1):S2. doi: 10.1186/1479-5876-9-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, Jelicic K, Kottilil S, Macleod K, O’Shea A, et al. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci USA. 2009;106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kader M, Wang X, Piatak M, Lifson J, Roederer M, Veazey R, Mattapallil JJ. Alpha4(+)beta7(hi)CD4(+) memory T cells harbor most Th-17 cells and are preferentially infected during acute SIV infection. Mucosal Immunol. 2009;2:439–449. doi: 10.1038/mi.2009.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Xu H, Gill AF, Pahar B, Kempf D, Rasmussen T, Lackner AA, Veazey RS. Monitoring alpha4beta7 integrin expression on circulating CD4+ T cells as a surrogate marker for tracking intestinal CD4+ T-cell loss in SIV infection. Mucosal Immunol. 2009;2:518–526. doi: 10.1038/mi.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, Ogundare O, Pascuccio M, Patel N, Wei D, et al. The genotype of early-transmitting HIV gp120s promotes α (4) β(7)-reactivity, revealing α (4) β(7) +/CD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2011;7:e1001301. doi: 10.1371/journal.ppat.1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinelli E, Veglia F, Goode D, Guerra-Perez N, Aravantinou M, Arthos J, Piatak M, Jr, Lifson JD, Blanchard J, Gettie A, Robbiani M. The frequency of α4β7(high) memory CD4+ T cells correlates with susceptibility to rectal simian immunodeficiency virus infection. J Acquir Immune Defic Syndr. 2013;64:325–331. doi: 10.1097/QAI.0b013e31829f6e1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ansari AA, Reimann KA, Mayne AE, Takahashi Y, Stephenson ST, Wang R, Wang X, Li J, Price AA, Little DM, et al. Blocking of α4β7 gut-homing integrin during acute infection leads to decreased plasma and gastrointestinal tissue viral loads in simian immunodeficiency virus-infected rhesus macaques. J Immunol. 2011;186:1044–1059. doi: 10.4049/jimmunol.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwa S, Kannanganat S, Nigam P, Siddiqui M, Shetty RD, Armstrong W, Ansari A, Bosinger SE, Silvestri G, Amara RR. Plasmacytoid dendritic cells are recruited to the colorectum and contribute to immune activation during pathogenic SIV infection in rhesus macaques. Blood. 2011;118:2763–2773. doi: 10.1182/blood-2011-02-339515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byrareddy SN, Kallam B, Arthos J, Cicala C, Nawaz F, Hiatt J, Kersh EN, McNicholl JM, Hanson D, Reimann KA, et al. Targeting α4β7 integrin reduces mucosal transmission of simian immunodeficiency virus and protects gut-associated lymphoid tissue from infection. Nat Med. 2014;20:1397–1400. doi: 10.1038/nm.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klatt NR, Harris LD, Vinton CL, Sung H, Briant JA, Tabb B, Morcock D, McGinty JW, Lifson JD, Lafont BA, et al. Compromised gastrointestinal integrity in pigtail macaques is associated with increased microbial translocation, immune activation, and IL-17 production in the absence of SIV infection. Mucosal Immunol. 2010;3:387–398. doi: 10.1038/mi.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canary LA, Vinton CL, Morcock DR, Pierce JB, Estes JD, Brenchley JM, Klatt NR. Rate of AIDS progression is associated with gastrointestinal dysfunction in simian immunodeficiency virus-infected pigtail macaques. J Immunol. 2013;190:2959–2965. doi: 10.4049/jimmunol.1202319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ansari AA, Silvestri G. Comparative studies of natural and non-natural hosts of SIV—an overview. In: Ansari AA, Silvestri G, editors. Natural Hosts of SIV: Implication in Aids. Elsevier; San Diego, CA: 2014. pp. 1–18. [Google Scholar]
- 21.Cicala C, Arthos J. Virion attachment and entry: HIV gp120 Env biotinylation, gp120 Env, or integrin ligand-binding assay. Methods Mol Biol. 2014;1087:3–12. doi: 10.1007/978-1-62703-670-2_1. [DOI] [PubMed] [Google Scholar]
- 22.Kane MA, Folias AE, Napoli JL. HPLC/UV quantitation of retinal, retinol, and retinyl esters in serum and tissues. Anal Biochem. 2008;378:71–79. doi: 10.1016/j.ab.2008.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kane MA, Folias AE, Wang C, Napoli JL. Quantitative profiling of endogenous retinoic acid in vivo and in vitro by tandem mass spectrometry. Anal Chem. 2008;80:1702–1708. doi: 10.1021/ac702030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane MA, Napoli JL. Quantification of endogenous retinoids. Methods Mol Biol. 2010;652:1–54. doi: 10.1007/978-1-60327-325-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 27.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 28.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, Mclnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 30.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 31.Humphries MJ. Integrin structure. Biochem Soc Trans. 2000;28:311–339. [PubMed] [Google Scholar]
- 32.Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119:3901–3903. doi: 10.1242/jcs.03098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gorfu G, Rivera-Nieves J, Ley K. Role of beta7 integrins in intestinal lymphocyte homing and retention. Curr Mol Med. 2009;9:836–850. doi: 10.2174/156652409789105525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell ID, Humphries MJ. Integrin structure, activation, and interactions. Cold Spring Harb Perspect Biol. 2011;3:3. doi: 10.1101/cshperspect.a004994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowell CA, Mayadas TN. Overview: studying integrins in vivo. Methods Mol Biol. 2012;757:369–397. doi: 10.1007/978-1-61779-166-6_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahashi Y, Byrareddy SN, Albrecht C, Brameier M, Walter L, Mayne AE, Dunbar P, Russo R, Little DM, Villinger T, et al. In vivo administration of a JAK3 inhibitor during acute SIV infection leads to significant increases in viral load during chronic infection. PLoS Pathog. 2014;10:e1003929. doi: 10.1371/journal.ppat.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agace W. Generation of gut-homing T cells and their localization to the small intestinal mucosa. Immunol Lett. 2010;128:21–23. doi: 10.1016/j.imlet.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 38.Zeng R, Oderup C, Yuan R, Lee M, Habtezion A, Hadeiba H, Butcher EC. Retinoic acid regulates the development of a gut-homing precursor for intestinal dendritic cells. Mucosal Immunol. 2013;6:847–856. doi: 10.1038/mi.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Beilhack A, Schulz S, Baker J, Beilhack GF, Wieland CB, Herman EI, Baker EM, Cao YA, Contag CH, Negrin RS. In vivo analyses of early events in acute graft-versus-host disease reveal sequential infiltration of T-cell subsets. Blood. 2005;106:1113–1122. doi: 10.1182/blood-2005-02-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Chang CH, Stein R, Cardillo TM, Gold DV, Goldenberg DM. Prevention of acute graft-versus-host disease in a xenogeneic SCID mouse model by the humanized anti-CD74 antagonistic antibody milatuzumab. Biol Blood Marrow Transplant. 2013;19:28–39. doi: 10.1016/j.bbmt.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 42.Sandborn WJ, Feagan BG, Rutgeerts P, Hanauer S, Colombel JF, Sands BE, Lukas M, Fedorak RN, Lee S, Bressler B, et al. GEMINI 2 Study Group. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2013;369:711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 43.Bansal GP, Malaspina A, Flores J. Future paths for HIV vaccine research: exploiting results from recent clinical trials and current scientific advances. Curr Opin Mol Ther. 2010;12:39–46. [PubMed] [Google Scholar]
- 44.Villablanca EJ, Cassani B, von Andrian UH, Mora JR. Blocking lymphocyte localization to the gastrointestinal mucosa as a therapeutic strategy for inflammatory bowel diseases. Gastroenterology. 2011;140:1776–1784. doi: 10.1053/j.gastro.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jovani M, Danese S. Vedolizumab for the treatment of IBD: a selective therapeutic approach targeting pathogenic a4b7 cells. Curr Drug Targets. 2013;14:1433–1443. doi: 10.2174/13894501113146660206. [DOI] [PubMed] [Google Scholar]
- 46.McKinnon LR, Nyanga B, Chege D, Izulla P, Kimani M, Huibner S, Gelmon L, Block KE, Cicala C, Anzala AO, et al. Characterization of a human cervical CD4+ T cell subset coexpressing multiple markers of HIV susceptibility. J Immunol. 2011;187:6032–6042. doi: 10.4049/jimmunol.1101836. [DOI] [PubMed] [Google Scholar]
- 47.Richardson SI, Mkhize N, Abdool Karim S, Gray E, Morris L. Role of integrin α4β7 in HIV transmission and pathogenesis. AIDS Vaccine 2013. 2013 OA07.02 (Abstr.) [Google Scholar]
- 48.Chahroudi A, Cartwright E, Lee ST, Mavigner M, Carnathan DG, Lawson B, Carnathan PM, Hashempoor T, Murphy MK, Meeker T, et al. Target cell availability, rather than breast milk factors, dictates mother-to-infant transmission of SIV in sooty mangabeys and rhesus macaques. PLoS Pathog. 2014;10:e1003958. doi: 10.1371/journal.ppat.1003958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chakrabarti LA. Natural Hosts of SIV: Implication in AIDS. Elsevier Academic Press; San Diego, CA: 2014. The different modes of resistance to AIDS: lessons from HIV/SIV controllers and SIV natural hosts. [Google Scholar]
- 50.Mori K, Sugimoto C, Ohgimoto S, Nakayama EE, Shioda T, Kusagawa S, Takebe Y, Kano M, Matano T, Yuasa T, et al. Influence of glycosylation on the efficacy of an Env-based vaccine against simian immunodeficiency virus SIVmac239 in a macaque AIDS model. J Virol. 2005;79:10386–10396. doi: 10.1128/JVI.79.16.10386-10396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14:667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 52.Cummins JE, Jr, Bunn WJ, Hall SD, Donze HH, Mestecky J, Jackson S. In vitro exposure to highly cytopathic HIV-1 X4 strains increases expression of mucosa-associated integrins on CD4(+) T cells. Virology. 2001;280:262–272. doi: 10.1006/viro.2000.0781. [DOI] [PubMed] [Google Scholar]
- 53.Jelicic K, Cimbro R, Nawaz F, Huang W, Zheng X, Yang J, Lempicki RA, Pascuccio M, Van Ryk D, Schwing C, et al. The HIV-1 envelope protein gp120 impairs B cell proliferation by inducing TGF-β1 production and FcRL4 expression. Nat Immunol. 2013;14:1256–1265. doi: 10.1038/ni.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kilshaw PJ, Murant SJ. Expression and regulation of beta 7(beta p) integrins on mouse lymphocytes: relevance to the mucosal immune system. Eur J Immunol. 1991;21:2591–2597. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- 55.El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, Drachenberg CB, Hadley GA. TGF-beta-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med. 2005;201:1647–1657. doi: 10.1084/jem.20041044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang SG, Park J, Cho JY, Ulrich B, Kim CH. Complementary roles of retinoic acid and TGF-β1 in coordinated expression of mucosal integrins by T cells. Mucosal Immunol. 2011;4:66–82. doi: 10.1038/mi.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


