Abstract
P2X receptors, as ATP-gated non-selective trimeric ion channels, are permeable to Na+, K+ and Ca2+. Comparing with other ligand-gated ion channel families, P2X receptors are distinct in their unique gating properties and pathophysiological roles, and have attracted attention as promising drug targets for a variety of diseases, such as neuropathic pain, multiple sclerosis, rheumatoid arthritis and thrombus. Several small molecule inhibitors for distinct P2X subtypes have entered into clinical trials. However, many questions regarding the gating mechanism of P2X remain unsolved. The structural determinations of P2X receptors at the resting and ATP-bound open states revealed that P2X receptor gating is a cooperative allosteric process involving multiple domains, which marks the beginning of the post-structure era of P2X research at atomic level. Here, we review the current knowledge on the structure-function relationship of P2X receptors, depict the whole picture of allosteric changes during the channel gating, and summarize the active sites that may contribute to new strategies for developing novel allosteric drugs targeting P2X receptors.
Keywords: P2X receptors, ATP, ligand-gated ion channel, channel gating, allosteric change, drug design
Introduction
P2X receptors, a distinct family of non-selective trimeric ligand-gated channels, are mainly permeable to Na+, K+ and Ca2+1,2,3. Binding extracellular adenosine 5′-triphosphate (ATP) from either emiocytosis or cytoclasis at the P2X receptors induces channel opening and a following ion flux4. Since the identification of ATP as a signal molecule of P2X5, seven subtypes of P2X receptors have been cloned and denoted as P2X1 to P2X7, with functional channels assembled by homo- or heterotrimers (Figure 1)6,7,8,9,10,11. P2X receptors are widely expressed in excitatory and non-excitatory cells, such as neuron, glia, platelet, epithelia and macrophage, and participate in many important physiological and pathological processes, including synaptic transmission, pain perception, inflammation, cardiovascular modulation, immunomodulation and tumorigenesis3,4,12,13,14,15,16. Heritable mutations in P2X receptors are the major causes of some disorders. For example, mutations in human P2X2 lead to hearing loss17,18,19; loss of function of the P2X4 receptor is related to increased pulse pressure20; and many non-synonymous single nucleotide polymorphisms (NS-SNPs) in the P2X7 receptor were identified as associated with chronic lymphocytic leukemia and osteoporosis21. Due to their roles in a variety of physiological and pathological processes, P2X receptors have drawn attention as promising drug targets22,23,24,25,26,27 and progress has been made toward this outcome28,29. For example, AF-219, a selective P2X3 receptor antagonist, alleviated chronic coughing in a phase II clinical trial29,30.
Figure 1.
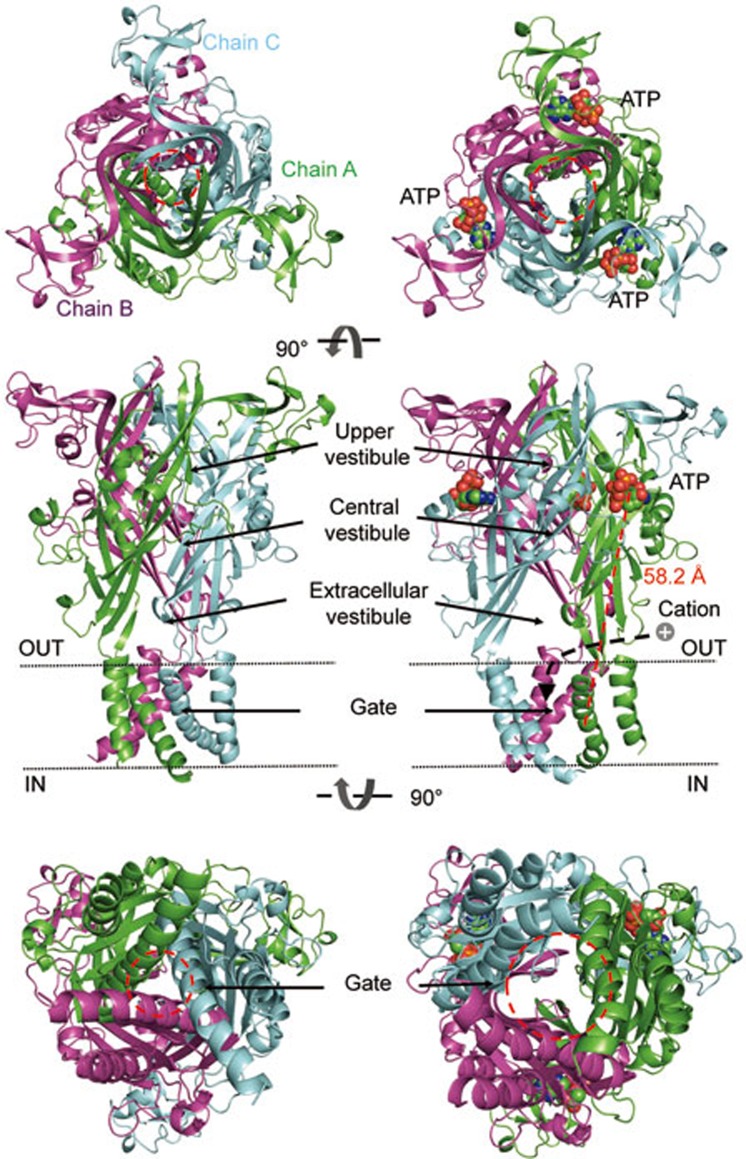
The three-dimensional architectures of the zfP2X4 receptor viewed from the extracellular side (upper), parallel to the membrane (middle) and the intracellular side (lower) at resting (left, PDB ID code: 3H9V) and ATP-bound open (right, PDB ID code: 4DW1) states. The red dashed line indicates the distance between the N9 atom of the purine ring of ATP and the Cα atom of A347. The black dashed arrow indicates the ion influx pathway. Subunits A, B and C are colored by green, magenta and cyan cartoon, respectively. ATP molecules are shown as spheres. Red dashed circles indicate the boundary of the upper vestibule or the gate. All figures were made with PyMol (http://www.pymol.org).
However, the lead compound targeting P2X receptors could only be obtained via high-throughput screening, a rather time-consuming and costly process. Rational drug design requires knowledge of channel gating and structures of P2X receptors. In 2009, the first crystal structure of the zebrafish P2X4 (zfP2X4) receptor at the closed/apo state with a resolution of 3.1 Å was reported by Kawate et al7. This structural determination marked the beginning of a new era at the atomic level for P2X researches31,32. In 2012, Hattori et al reported the open crystal structure of the zfP2X4 receptor with ATP in its binding site33, which confirmed previous studies on ATP recognition and provided structural insight into the channel gating of P2X receptors34. Despite lacking obvious similarities in primary structures between P2X receptors and acid-sensing ion channels (ASICs, another member in the trimeric ligand-gated ion channel family), those two families exhibit unanticipated similarities in their three-dimensional (3D) architecture. The transmembrane (TM) domains of those two families assemble in a similar pattern, with the three extracellular domains intertwined with each other7,33,35,36,37. The individual subunit of the two families forms different shapes, with ASIC1a resembling a human hand37 and the P2X4 receptor a dolphin rising from water (Figure 2A and 2B). Different domains of P2X are thus named as head, dorsal fin (DF), left flipper (LF), right flipper (RF), body and fluke (Figure 2A). Benefiting from those crystal structures, progress has been made in the structure-function research on P2X receptors, aiding rational drug design targeting this important ion channel family. Because these channels are ligand-gated ion channels, the gating process of P2X receptors starts with the ligand binding to the channel opening until the ultimate close of the channel, and this involves a series of step-by-step conformational changes. In this review, we focus on the roles of each domain of the P2X receptors and the stepwise domain-domain interactions during channel gating. We also summarize the binding sites of small molecules targeting P2X receptors, which provides insights into the gating mechanism of P2X receptors and the structural basis for future drug design.
Figure 2.
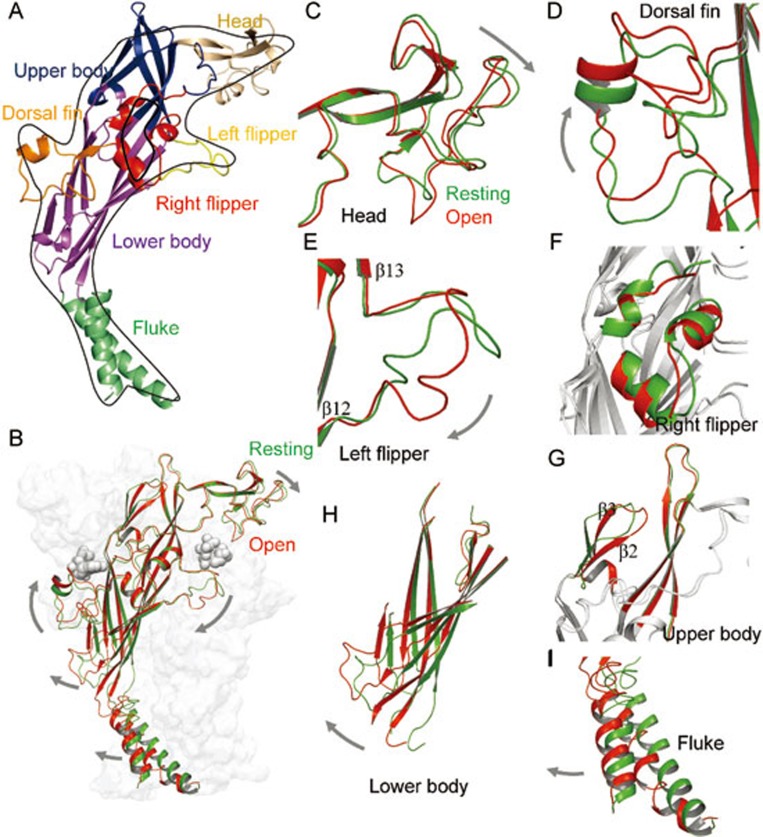
ATP-induced conformational changes of zfP2X4 receptors. (A) The P2X4 subunit has a dolphin-like shape. Distinctive body parts are shown in different colors. (B) Superposition of a single P2X4 subunit at resting (green) and open (red) states. (C–I) Superposition of head (C), dorsal fin (D), left flipper (E), right flipper (F), upper body (G), lower body (H) and fluke (I) domains at resting (green) and open (red) states. The grey arrows indicate the conformational changes after ATP binding.
Head domain
Located at the extracellular domain, the head domain of P2X subunits is composed of the residues 111–167 (zfP2X4 numbering). Although the sequence of the head domain is not highly conserved throughout the P2X family, the architecture of this domain in different subtypes shares certain similarity due to three conserved disulfide bonds that contribute to the folding of P2X receptors38,39,40,41,42 (Figure 2A and 2C). The architecture of head domain of the zfP2X4 receptor was determined by X-ray diffraction and showed a high similarity in folding pattern with rat P2X4 (rP2X4) resolved by nuclear magnetic resonance, suggesting the conservation of the P2X4 head domain in different species40. Deletion of 42 residues in the head domain of P2X1 resulted in the loss of channel function without interfering with membrane trafficking43, suggesting that the head domain is an integrant domain of channel gating. Using molecular dynamic (MD) simulations and normal mode analysis, previous studies revealed a spontaneous downward motion of the head domain, probably resulting from its inherent dynamics16,44,45 (Figure 2B and 2C). This type of motion coincides with the downward motion of the head domain demonstrated by the ATP-bound open structure and is pivotal for the channel gating of P2X receptors. Labeling L186C (rat P2X2, rP2X2, numbering) using NCS-ATP (a synthesized ATP-derived thiol-reactive compound) impedes subsequent opening of the channel by locking the channel into an ATP binding mode that is incapable of driving the downward motion of head domain46. On the contrary, ADP-ribosylation of R125 (mouse P2X7, mP2X7, numbering) (Figure 3) located in the head domain, is sufficient to activate the P2X7 receptor47, confirming the essential role of the downward motion of head domain in channel gating. It is reasonable to assume that chemicals binding to the head domain interfere with the downward motion, and therefore, alter channel gating48,49. For example, K138 in the head domain is involved in the binding of both suramin and NF449 to the P2X1 receptor (Figure 3)50,51. The data from chimeras and single-point mutations suggest that suramin and NF449 may bind to the site below the head domain of P2X1 and therefore impede the downward motion of this domain. Studies using voltage clamp flurometry and electrophysiology approaches further confirmed the pivotal role of the cysteine-rich head domain in channel activation and desensitization of the P2X1 receptor. For example, the residues N120 and G123 (rat P2X1, rP2X1, numbering) were associated with channel activation of P2X1 receptor, and P121, E122, I125 (rP2X1 numbering) were correlated with channel desensitization of P2X1 receptor52.
Figure 3.
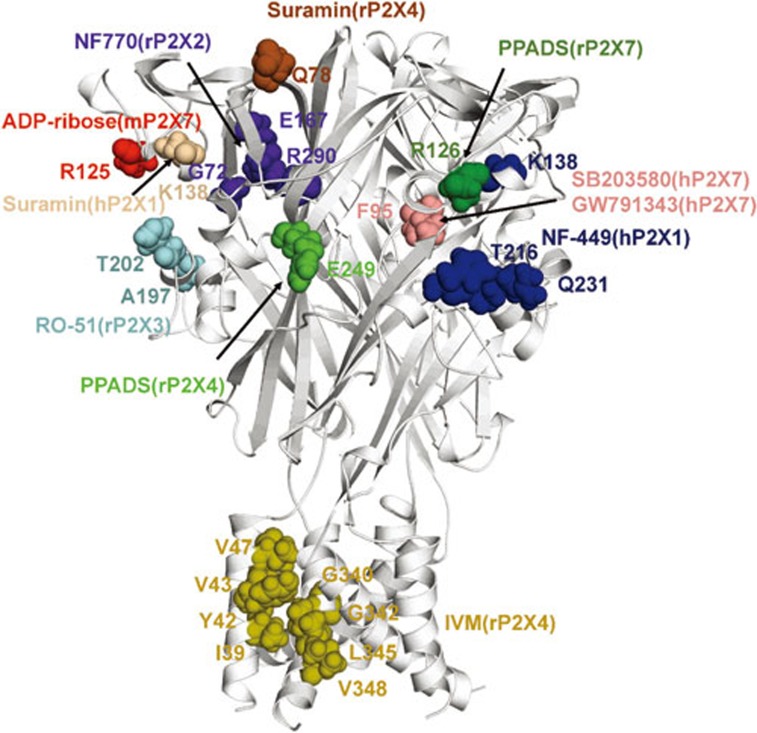
Amino residues involved in small molecule recognition of P2X receptors.
Dorsal fin domain
The dorsal fin (DF) domain (residues 206–234, zfP2X4 numbering) is a domain structurally coupled with the lower body domain. The upward motion of the DF domain is another allosteric change essential for P2X receptor activation44,45 (Figure 2A, 2B and 2D). Similar to the downward motion of the head domain, the upward motion of the DF domain is driven by its inherent dynamics. The bound-ATP directly contacts the DF domain via an interaction between its purine ring and the L217 of the DF domain (zfP2X4 numbering). This interaction induces an upward motion of the DF domain, leading to an expansion of the lower body domain and channel activation (Figures 2H and 4B). Thus, the gating mechanism of P2X receptors mimics a 'lever' system33, where the head domain and the DF domain function as the two arms of the 'lever'. TNP-ATP, a nonspecific antagonist, impedes the upward motion of the DF with a large steric bulk of trinitrophenyl moieties, and therefore inhibits channel opening33. RO-51, a bioavailable P2X3 antagonist, displays a two hundred-fold lower potency on human P2X3 (hP2X3) compared to rat P2X3 (rP2X3), due to two amino acids located in the DF domain, A197 and T202 (Figure 3)53. Thus, the upward motion of the DF domain is also essential for the channel gating of P2X receptors, and small molecules interrupting this motion effectively block P2X receptors.
Figure 4.
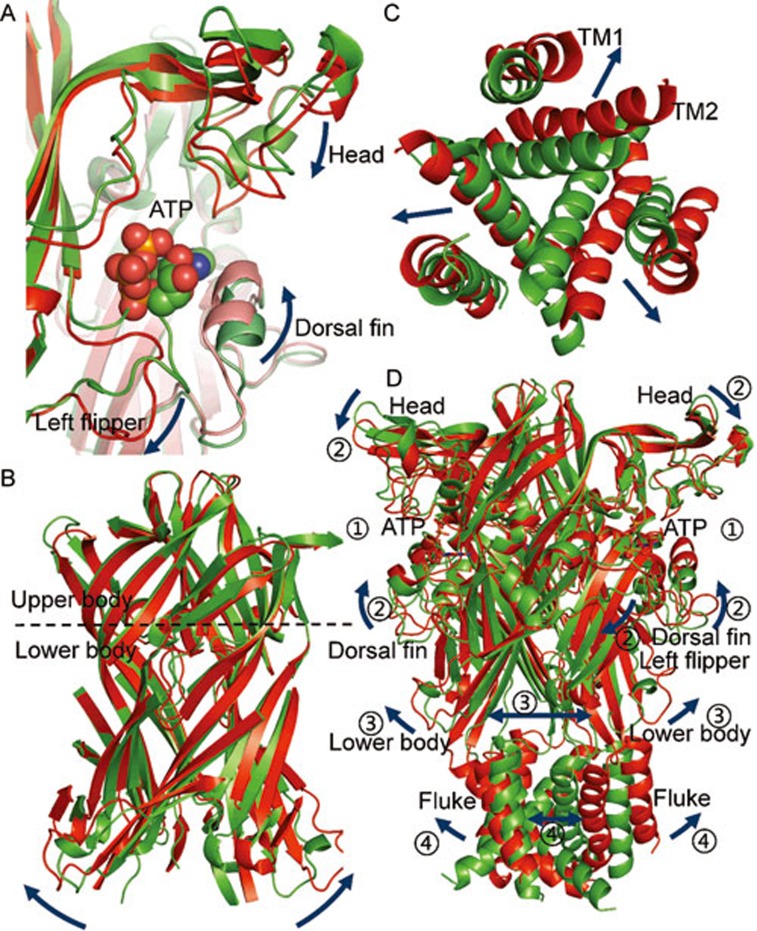
Bound ATP-evoked allosteric changes associated with channel opening in the P2X4 receptor. (A–D) Bound-ATP evoked structural rearrangements at the ATP binding site (A), body domain (B), TM region (C) and overall structure of zfP2X4 receptors (D). The dark-blue arrows indicate the conformational changes after ATP binding. Structures at the resting and open states of zfP2X4 receptors are displayed in green and red, respectively.
Left flipper domain
The left flipper (LF) domain in P2X4 is a loop structure linking β12 and β13 (Figure 2A and 2E). It is composed of the residues 281–296 (zfP2X4 numbering), with the sequences of the two ends partially conserved. Prior to the structural determination of the zfP2X4 receptor, the contribution of the LF domain to the function of P2X4 was extensively studied. One study revealed that H286 (rP2X4 numbering) is pivotal for the pH sensitivity of the P2X4 receptor in the pathophysiological range54. Mutating R278 or D280 (rP2X4 numbering) to alanine could abolish receptor function, potentially from the formation of salt bridge by R278 and D280, which are essential for channel gating the P2X4 receptor55. Those studies support an essential role of the LF in channel gating. A comparison between the resting and open structures of zfP2X4 revealed that the LF domain is driven away from the ATP binding site during the ATP bind-to-open process (Figure 2B and 2E)33, rather than approaching the binding site by the inherent dynamics of the receptors45. Alteration in interactions among I208, L217, V291 and K193 (zfP2X4 numbering), induced by ATP binding, correlates well with this movement, indicating that the proper interactions between the DF and LF domains are crucial for channel gating45, which was further confirmed by other studies56,57,58. The middle region is less conserved among P2X subtypes, which may endow subtype-specific contributions in the channel gating. For instance, an alanine substitution of S275 in the LF domain (rP2X3 numbering) revealed that this position is involved in forming the binding pocket and correlates with the recovery process of P2X3 from inactivation. In contrast, a corresponding mutation of S289 (S289A) in rP2X4 displays no significant effects on channel activation57,59. Therefore, the downward motion of the LF domain and its movement relative to the DF domain are essential for channel gating in P2X receptors, although the different roles of the LF domain in various subtypes have not been clearly delineated.
Right flipper domain
The right flipper (RF) domain (residues 178–189 and 235–254, zfP2X4 numbering) is formed by three stacked α-helices separated into two segments (Figure 2A and 2F). The superimposition of closed and open structures revealed that the RF domain does not undergo conformational changes during the channel gating of P2X receptors (Figure 2F); however, it does not imply that the RF domain is irrelevant to channel gating. One piece of evidence is that H245, a residue in the RF domain of rP2X2, is involved in zinc/copper potentiation60. E249K (rP2X4 numbering) shows sensitivity to PPADS, a chemical that has no effect on the wild-type rP2X4 receptor61, indicating that RF may act as an anchor when PPADS binds to the channel. Additionally, N187 (zfP2X4 numbering), a conserved glycosylation site located in the RF domain, is important for channel stability and membrane trafficking38,62,63,64,65. Due to the limited studies in this region, the contribution of the RF domain to channel gating, membrane trafficking and drug design requires further investigation.
Upper body domain
The upper body domain is composed of residues 75–92, 105–113 and 294–319 (zfP2X4 numbering) (Figure 2A and 2G). Using cysteine scanning mutagenesis along the upper vestibule, Damien et al found that MTS reagents had no effect on the currents of mutated P2X receptors66, indicating that the center pathway is not involved in ion permeation. The superimposition of closed and open structures revealed that the two upper bodies overlap well except for a slight right shift of β2 and β3 (Figure 2G), suggesting that the upper body might not undergo apparent conformational changes in the process of channel opening (Figure 2G), despite the direct contact between ATP and the side chains of R298 and K316 (zfP2X4 numbering). Therefore, Hattori et al proposed that the upper body behaved as a rigid body, which acted as a 'brace' in the 'lever' gating mechanism described above33. However, fast scanning atomic force microscopy revealed that the long-period application of ATP under Ca2+-free condition caused pore dilation and allowed permeation of large organic molecules, such as NMDG67,68,69. Zhao et al also reported the expansion of the upper vestibule of P2X4 during 300-ns MD simulations45. Thus, the role of the upper body domain in functional channels requires further clarification.
Lower body domain
The lower body domain is composed of residues 56–74, 93–104, 188–207, 254–281 and 320–330 (zfP2X4 numbering) and is characterized by a β-sandwich motif formed by six β-sheets (Figure 2A and 2H). Alanine/cysteine-scanning studies within the lower body had identified residues involved in ATP binding, supporting the roles of the lower body domain in agonist binding and the conformation transition pathway during channel gating70,71,72,73. Consistent with those studies, the crystal structure at the open state revealed that ATP contacts side chains and main chains of K70, K72, T189 and K193 in the lower body domain33. Thus, the residues of the lower body domain are pivotal for the agonist recognition of P2X receptors.
P2X receptors have a large extracellular domain, with ATP binding sites locating far away (58.2 Å) from channel gates (Figure 1, middle panel). Therefore, a series of conformational changes are required to translate from an extracellular domain into the TM region74. ATP binding-induced conformational changes of the LF and DF domains can trigger subsequent outward 'flexing' of the lower body domain and the expansion of the extracellular vestibule, which provides an ion influx pathway66,75 and results in an iris-like motion of TM helices to open the channel pore. Double mutations of P62C/H192C, S65C/S190C, and S65C/D315C (rP2X2 numbering) that restrain the expansion of the lower body markedly attenuated ATP-induced maximal currents of mutant receptors, which was rescued by DTT application76. Recently, the linker region between the lower body domain and TMs had also been systematically investigated77,78,79, mutations of Y54A, Q55A, F198A, W259A, F324A, and G325A (rP2X4 numbering) resulted in a loss of channel function, suggesting that these residues contribute to the conformational transition from the lower body domain to the TM region. These findings confirm the essential role of the lower body domain in conformational transitions during channel gating.
D197 (rat P2X7, rP2X7 numbering), located in the lower body domain, is pivotal for acidic pH-induced channel inhibition80, suggesting an additional function of the lower body domain that is independent to the movement of the LF and DF domains, and this was confirmed by the importance of the salt bridge E63-R274 (rP2X2 numbering) in this domain for the channel gating81. Actually, both the closure of binding site jaw and the movement of the lower body domain are required for the concomitant pore opening of P2X receptors33. Zinc is able to elicit inward currents following treatment with AM546 on the mutated (K67C) P2X2 receptor82, and the zinc-evoked currents are enhanced by lysine substitution at H319, located in the lower body domain82. Thus, the lower body domain possesses structural elements that independently affect the channel gating of P2X receptors.
In summary, the lower body domain is not only directly involved in agonist recognition but also able to coordinate the bound-ATP induced conformational changes, conformational transitions, and the final channel pore opening.
Fluke
The TM helices (TM1 and TM2) of a single subunit delineate the 'fluke' of the 'dolphin' (Figure 2A and 2I) and are involved in many properties of P2X receptors, including unitary conductance and rectification, differential desensitization among subtypes, and voltage-dependence of P2X receptors83,84,85,86,87. Functions of the TM domains in channel gating have been deeply studied, and its roles in pore location, ion permeability and structural rearrangements were confirmed by crystal structures88,89,90,91,92,93,94,95,96,97. Under normal conditions, pore opening of P2X receptors is controlled by ATP binding. However, when cysteine was introduced at position I328 (rP2X2 numbering), the channel pore of the P2X2 receptor was directly opened by propylmethanethiosulfonate (MTSP)98, suggesting that the rearrangement of the TM domains is sufficient to initiate channel activation. Guided by this idea, azobenzene compounds, namely MEA-TMA and BMA, were linked to P2X2 receptors carrying a cysteine substitution in the TM domain by an electrophilic substitution reaction99,100, and the open or closing of the channel pore was controlled by the light-evoked isomerization. This system provides an ideal tool to control the activity of P2X receptors with high spatial and temporal accuracy.
The flukes of the three subunits form the channel pore, with TM2 lining the inner tunnel, and TM1 positioned peripherally to TM2 (Figure 4C). Both TM1 and TM2 are structurally coupled with the lower body. Outward flexing of the lower body induces the TMs to expand in an iris-like motion to open the channel pore. Due to the mismatch occurring in X-ray crystallography caused by rigorous conditions, Heymann et al proposed a model in membrane environments that stabilizes intersubunit interactions101.
N- and C-termini
The N-terminus of P2X receptors is composed of approximately 30 amino acid residues, while the C-terminus comprises approximately 30–240 amino acid residues, varying among different subtypes. To obtain a high quality crystal structure, both the N- and C-termini of zfP2X4 were truncated, which might cause the loss of intrinsic property of P2X4 to some extent. For example, the P2X4 receptor shows dynamic ionic selectivity, while the truncated P2X4 produces a constant current in response to long period exposures of ATP33. Moreover, the crystal structures made no contribution to research focusing on the intracellular domains. Nevertheless, intracellular domains were found to play important roles in membrane trafficking, channel desensitization, protein-protein interactions, and phospholipids modulation, which was unveiled by various approaches, including Western blot, co-IP, site-directed mutagenesis, and electrophysiology recording16,102,103,104,105,106. The roles of the two termini have been well summarized previously16,102,103,104,105,106, and thus, will not be further discussed in this review.
Domain-domain interactions and coordinated motions of multi-domains evoke a final channel opening of P2X receptors
So far, the location and contributions of each domain in P2X functions and channel gating that we have discussed above are at the level of the single subunit. Nevertheless, P2X receptors are intertwined trimeric membrane proteins with inter- and intrasubunit interactions present throughout the entire channel. Based on the 3D structure of the P2X4 receptor, those interactions can be divided into three compartments. (1) The binding sites of ATP are contributed by domains from different subunits, namely, the head and LF domains from the same subunit, and the DF and upper body domains from a neighbor subunit (Figure 4A). Therefore, there are three ATP binding sites in a three-fold symmetric mode, although a recent study showed that ATP binding at only two of the three sites is sufficient for channel opening107. (2) The body domains of the three subunits intertwine with each other, forming the fundamental core of the P2X receptors (Figure 4B), which is surrounded by the three ATP binding sites. This 'core' can be further divided into upper and lower sections according to the 'dolphin' body, with the upper part maintaining the stability of the P2X receptors, and the lower part translating conformational changes induced by ATP binding from the extracellular to the TM domain. (3) Flukes of three subunits constitute the TM domain of P2X (Figure 1 and 4C). Three TM2 helices compose the channel pore while TM1 embrace TM2 from outside.
These structural characteristics of P2X determine the gating process of P2X receptors (Figure 4D)33,44,45,49,108. Following ATP binding, the head domain moves downward, the DF domain moves upward and the LF domain is pushed away from the binding site. Because of the coupling between the LF, DF and lower body domains, the relative motions of the LF and DF are capable of driving the outward expansion of the lower body, followed by the movements of TMs and subsequent opening of the ion access route. In conclusion, the gating process consists of a series of complicated and coordinated motions of multiple domains, which leads to the final channel opening of P2X receptors.
Small molecules to change the channel gating of P2X receptors
As discussed above, the gating process of P2X receptors involves a cooperative system composed of multiple domains. Small molecules that interrupt this process by acting on certain sites/domains affect channel gating. According to their effects on channel function, they are classified into antagonists and modulators. Many compounds targeting P2X receptors have been developed. AF-219, a selective P2X3 receptor antagonist, has been used in the treatment of osteoarthritis pain, interstitial cystitis and respiratory disorders29. AZD9056 can selectively inhibit the P2X7 receptor and is used for treating rheumatic arthritis109. Although both compounds have entered into clinical studies, little is known regarding their binding sites or working mechanisms (Figure 3). In combination with multidisciplinary approaches, including chimera and point mutations, electrophysiology, molecular modeling and molecular docking, Evans' group discovered that NF449, a P2X1 receptor specific inhibitor, could fill up the cleft between the head and the dorsal fin domain, thus preventing the binding of ATP and the downward motion of head domain. It is known that K138 (human P2X1, hP2X1, numbering), located in the head domain, is required for the binding of suramin, a broad spectrum inhibitor of P2X receptors. However, the difference in sensitivity to suramin between human and rat homologue P2X4 was determined by Q78 (rP2X4 numbering) located in the upper body domain110. This suggests that suramin may have different binding sites in different subunits and/or have more than one binding site for a certain subunit. Similarly, the mutant E249K of rP2X4 acquires sensitivity to PPADS61; however, chimera analysis identified another domain of approximately 100 amino acids (81–183) that accounts for the higher PPADS sensitivity in the human isoform compared to the rat110. This domain is in accordance with the spatial location of R126, a residue that is responsible for the species difference in antagonists' effects of the P2X7 receptor111. NF770, a suramin derivative that competitively inhibits the P2X2 receptor at nanomolar concentrations, acts on G72, E167 and R290 (rP2X2 numbering), which are also important for ATP binding112,113. Interestingly, nearly all of those identified sites are located in or around the ATP binding pocket except for IVM, a P2X4 positive modulator, which has been identified to act on the TM domains114,115,116,117.
Questions to be answered
Although progress on the channel activation of P2X receptors has been made since the structural determination of P2X receptors, many questions remain to be addressed.
P2X receptors have seven subtypes, exhibiting different affinities for ATP, ranging from nanomolar to millimolar3, whereas the amino acid residues directly participating in ATP binding are highly conserved among different subtypes7,33. The mechanism underlying those distinct affinities remains to be further investigated.
In light of two crystal structures of P2X4, the process of ATP binding inducing channel open has been deduced. However, the recovery process from open to resting state remains to be a mystery. Furthermore, desensitization kinetics differs among the seven subtypes3. Although preliminary research indicated that TMs and intracellular domains play important roles in P2X desensitization, the mechanism requires future studies.
'Pore dilation' is one of the hottest but also toughest questions in P2X research. Two hypothetical mechanisms, namely the gating model and the Pannexin-1 model, have been proposed3. In the gating model, pore dilation resulted from long-term ATP action, which leads to additional conformational change118,119,120,121,122,123,124. While in the Pannexin-1 model, the conformational changes of the P2X receptor resulted in allosterism of an auxiliary protein coupled with the P2X receptor, such as Pannexin-1, permitting molecules to enter cells through those proteins125,126. Unfortunately, the mechanisms underlying the conformational changes from open to the dilated state of both models remain unclear.
Endogenous P2X receptors are assembled in homotrimeric as well as in heterotrimeric forms, such as P2X2/3, P2X4/6 and P2X1/58,11,127. Unlike the symmetric gating mechanism of homotrimeric P2X receptors, the gating process of heterotrimeric P2X receptors is more complicated. Limited studies have been performed on the heterotrimeric P2X receptors, mainly on subunit stoichiometry11,128,129. It remains unexplored in the field of gating mechanisms for heterotrimeric P2X receptors, including the drug-designs targeting heterotrimeric receptors.
P2X receptors and ASICs showed unexpected similarities in their topology, despite their unrelated primary structures. Both contain many vestibules/pockets in their extracellular domains. Multiple pockets/ligand binding sites were identified in ASICs, through which novel toxins and small molecules inhibited or activated ASICs via mechanisms distinct from the acidosis-induced channel activation130,131,132,133,134,135,136. Therefore, similar to ASICs, finding novel toxins and small molecules to activate or modulate the function of P2X receptors through interactions with those vestibules/pockets in the extracellular domain is possible.
Although the structures of P2X4 at both resting and ATP-bound open states have been determined, structures of other subtypes are required to improve our understanding of the gating process of various P2X receptors. Structures complexed with the allosteric, especially subunit specific molecules, are also in demand to provide the structural basis for rational drug designs. In addition, the structure of the full-length P2X receptor with its intracellular domains has not been developed. With the help of newly improved technology, such as cryo-EM137,138, discoveries of more P2X structures are expected.
Concluding remarks
Since ATP was identified as a signal molecule in 1975, seven subtypes of P2X have been cloned and their physiological and pathological functions recognized. As a class of trimeric ion channels, the gating mechanism of P2X receptors differs from previously identified pentameric “cys-loop” ion channel family, tetramer voltage-gated potassium ion channels, TRP channels, or glutamate receptors. The crystal structures of P2X reported in 2009 and 2012 marked the beginning of the post-structure era at the atomic level of P2X research. The gating process of P2X receptors is a complex work by multiple domains. In this work, we highlight the recent achievements in P2X structures and channel gating, aiming to illuminate the correlation between the gating process and the structural elements of P2X receptors. All those studies have paved the way for developing new drugs targeting P2X receptors, which would contribute to novel therapeutic approaches in the future.
Acknowledgments
The authors thank Drs YANG and Xiao-yang CHENG for making helpful comments on the manuscript. This work was supported by grants from the National Excellent Young Scientist Foundation of China (No 31222018), National Program on Key Basic Research Project of China (No 2014CB910302), National Natural Science Foundation of China (Nos 31570832, 31400707 and 31170787), 'Shanghai Jiao Tong University-SMC Mutual Funds' for Excellent Young Scholar, Shanghai Jiao Tong University School of Medicine 'Key Incubation Project' and Doctoral Innovation Fund Projects from Shanghai Jiao Tong University School of Medicine (No BXJ201405).
References
- 1Khakh BS. Molecular physiology of P2X receptors and ATP signalling at synapses. Nat Rev Neurosci 2001; 2: 165–74. [DOI] [PubMed] [Google Scholar]
- 2Khakh BS, North RA. P2X receptors as cell-surface ATP sensors in health and disease. Nature 2006; 442: 527–32. [DOI] [PubMed] [Google Scholar]
- 3Khakh BS, North RA. Neuromodulation by extracellular ATP and P2X receptors in the CNS. Neuron 2012; 76: 51–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4Coddou C, Yan Z, Obsil T, Huidobro-Toro JP, Stojilkovic SS. Activation and regulation of purinergic P2X receptor channels. Pharmacol Rev 2011; 63: 641–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Burnstock G. Historical review: ATP as a neurotransmitter. Trends Pharmacol Sci 2006; 27: 166–76. [DOI] [PubMed] [Google Scholar]
- 6Barrera NP, Ormond SJ, Henderson RM, Murrell-Lagnado RD, Edwardson JM. Atomic force microscopy imaging demonstrates that P2X2 receptors are trimers but that P2X6 receptor subunits do not oligomerize. J Biol Chem 2005; 280: 10759–65. [DOI] [PubMed] [Google Scholar]
- 7Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature 2009; 460: 592–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8Lewis C, Neidhart S, Holy C, North RA, Buell G, Surprenant A. Coexpression of P2X2 and P2X3 receptor subunits can account for ATP-gated currents in sensory neurons. Nature 1995; 377: 432–5. [DOI] [PubMed] [Google Scholar]
- 9Nicke A, Baumert HG, Rettinger J, Eichele A, Lambrecht G, Mutschler E, et al. P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J 1998; 17: 3016–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Torres GE, Egan TM, Voigt MM. Hetero-oligomeric assembly of P2X receptor subunits. Specificities exist with regard to possible partners. J Biol Chem 1999; 274: 6653–9. [DOI] [PubMed] [Google Scholar]
- 11Saul A, Hausmann R, Kless A, Nicke A. Heteromeric assembly of P2X subunits. Front Cell Neurosci 2013; 7: 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Surprenant A, North RA. Signaling at purinergic P2X receptors. Ann Rev Physiol 2009; 71: 333–59. [DOI] [PubMed] [Google Scholar]
- 13Burnstock G. Introduction and perspective, historical note. Frontiers Cell Neurosci 2013; 7: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Idzko M, Ferrari D, Eltzschig HK. Nucleotide signalling during inflammation. Nature 2014; 509: 310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Burnstock G. An introduction to the roles of purinergic signalling in neurodegeneration, neuroprotection and neuroregeneration. Neuropharmacology 2015. doi:10.1016/j.neuropharm.2015.05.031. [DOI] [PubMed]
- 16Habermacher C, Dunning K, Chataigneau T, Grutter T. Molecular structure and function of P2X receptors. Neuropharmacology 2015. doi:10.1016/j.neuropharm.2015.07.032. [DOI] [PubMed]
- 17Yan D, Zhu Y, Walsh T, Xie D, Yuan H, Sirmaci A, et al. Mutation of the ATP-gated P2X(2) receptor leads to progressive hearing loss and increased susceptibility to noise. Proc Natl Acad Sci U S A 2013; 110: 2228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Faletra F, Girotto G, D'Adamo AP, Vozzi D, Morgan A, Gasparini P. A novel P2RX2 mutation in an Italian family affected by autosomal dominant nonsyndromic hearing loss. Gene 2014; 534: 236–9. [DOI] [PubMed] [Google Scholar]
- 19Moteki H, Azaiez H, Booth KT, Hattori M, Sato A, Sato Y, et al. Hearing loss caused by a P2RX2 mutation identified in a MELAS family with a coexisting mitochondrial 3243AG mutation. Ann Otol Rhinol Laryngol 2015; 124 Suppl 1: 177S–83S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Stokes L, Scurrah K, Ellis JA, Cromer BA, Skarratt KK, Gu BJ, et al. A loss-of-function polymorphism in the human P2X4 receptor is associated with increased pulse pressure. Hypertension 2011; 58: 1086–U244. [DOI] [PubMed] [Google Scholar]
- 21Caseley EA, Muench SP, Roger S, Mao HJ, Baldwin SA, Jiang LH. Non-synonymous single nucleotide polymorphisms in the P2X receptor genes: association with diseases, impact on receptor functions and potential use as diagnosis biomarkers. Int J Mol Sci 2014; 15: 13344–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Evans RJ. Structural interpretation of P2X receptor mutagenesis studies on drug action. Br J Pharmacol 2010; 161: 961–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Lemoine D, Jiang RT, Taly A, Chataigneau T, Specht A, Grutter T. Ligand-gated ion channels: new insights into neurological disorders and ligand recognition. Chem Rev 2012; 112: 6285–318. [DOI] [PubMed] [Google Scholar]
- 24North RA, Jarvis MF. P2X receptors as drug targets. Mol Pharmacol 2013; 83: 759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Bartlett R, Stokes L, Sluyter R. The P2X7 receptor channel: recent developments and the use of P2X7 antagonists in models of disease. Pharmacol Rev 2014; 66: 638–75. [DOI] [PubMed] [Google Scholar]
- 26Muller CE. Medicinal chemistry of P2X receptors: allosteric modulators. Curr Med Chem 2015; 22: 929–41. [DOI] [PubMed] [Google Scholar]
- 27Dal Ben D, Adinolfi E. Purinergic P2X receptors: physiological and pathological roles and potential as therapeutic targets. Curr Med Chem 2015; 22: 782. [PubMed] [Google Scholar]
- 28Lambertucci C, Dal Ben D, Buccioni M, Marucci G, Thomas A, Volpini R. Medicinal chemistry of P2X receptors: agonists and orthosteric antagonists. Curr Med Chem 2015; 22: 915–28. [DOI] [PubMed] [Google Scholar]
- 29Ford AP, Undem BJ. The therapeutic promise of ATP antagonism at P2X3 receptors in respiratory and urological disorders. Frontiers Cell Neurosci 2013; 7: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Abdulqawi R, Dockry R, Holt K, Layton G, McCarthy BG, Ford AP, et al. P2X3 receptor antagonist (AF-219) in refractory chronic cough: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 2015; 385: 1198–205. [DOI] [PubMed] [Google Scholar]
- 31Young MT. P2X receptors: dawn of the post-structure era. Trends Biochem Sci 2010; 35: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Hausmann R, Kless A, Schmalzing G. Key sites for P2X receptor function and multimerization: overview of mutagenesis studies on a structural basis. Curr Med Chem 2015; 22: 799–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Hattori M, Gouaux E. Molecular mechanism of ATP binding and ion channel activation in P2X receptors. Nature 2012; 485: 207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Chataigneau T, Lemoine D, Grutter T. Exploring the ATP-binding site of P2X receptors. Frontiers Cell Neurosci 2013; 7: 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Baconguis I, Hattori M, Gouaux E. Unanticipated parallels in architecture and mechanism between ATP-gated P2X receptors and acid sensing ion channels. Curr Opin Struc Biol 2013; 23: 277–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Kellenberger S, Grutter T. Architectural and functional similarities between trimeric ATP-gated P2X receptors and acid-sensing ion channels. J Mol Biol 2015; 427: 54–66. [DOI] [PubMed] [Google Scholar]
- 37Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature 2009; 460: 599–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Ennion SJ, Evans RJ. Conserved cysteine residues in the extracellular loop of the human P2X(1) receptor form disulfide bonds and are involved in receptor trafficking to the cell surface. Mol Pharmacol 2002; 61: 303–11. [DOI] [PubMed] [Google Scholar]
- 39Clyne JD, Wang LF, Hume RI. Mutational analysis of the conserved cysteines of the rat P2X2 purinoceptor. J Neurosci 2002; 22: 3873–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Igawa T, Abe Y, Tsuda M, Inoue K, Ueda T. Solution structure of the rat P2X4 receptor head domain involved in inhibitory metal binding. FEBS Lett 2015; 589: 680–6. [DOI] [PubMed] [Google Scholar]
- 41Rokic MB, Tvrdonova V, Vavra V, Jindrichova M, Obsil T, Stojilkovic SS, et al. Roles of conserved ectodomain cysteines of the rat P2X4 purinoreceptor in agonist binding and channel gating. Physiol Res 2010; 59: 927–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Kowalski M, Hausmann R, Dopychai A, Grohmann M, Franke H, Nieber K, et al. Conformational flexibility of the agonist binding jaw of the human P2X3 receptor is a prerequisite for channel opening. Br J Pharmacol 2014; 171: 5093–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43El-Ajouz S, Ray D, Allsopp RC, Evans RJ. Molecular basis of selective antagonism of the P2X1 receptor for ATP by NF449 and suramin: contribution of basic amino acids in the cysteine-rich loop. Br J Pharmacol 2012; 165: 390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Jiang R, Taly A, Lemoine D, Martz A, Cunrath O, Grutter T. Tightening of the ATP-binding sites induces the opening of P2X receptor channels. EMBO J 2012; 31: 2134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Zhao WS, Wang J, Ma XJ, Yang Y, Liu Y, Huang LD, et al. Relative motions between left flipper and dorsal fin domains favour P2X4 receptor activation. Nat Commun 2014; 5: 4189. [DOI] [PubMed] [Google Scholar]
- 46Jiang R, Lemoine D, Martz A, Taly A, Gonin S, Prado de Carvalho L, et al. Agonist trapped in ATP-binding sites of the P2X2 receptor. Proc Natl Acad Sci U S A 2011; 108: 9066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Adriouch S, Bannas P, Schwarz N, Fliegert R, Guse AH, Seman M, et al. ADP-ribosylation at R125 gates the P2X7 ion channel by presenting a covalent ligand to its nucleotide binding site. FASEB J 2008; 22: 861–9. [DOI] [PubMed] [Google Scholar]
- 48Huang LD, Fan YZ, Tian Y, Yang Y, Liu Y, Wang J, et al. Inherent dynamics of head domain correlates with ATP-recognition of P2X4 receptors: insights gained from molecular simulations. PloS One 2014; 9: e97528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Jiang R, Taly A, Grutter T. Moving through the gate in ATP-activated P2X receptors. Trends Biochem Sci 2013; 38: 20–9. [DOI] [PubMed] [Google Scholar]
- 50Sim JA, Broomhead HE, North RA. Ectodomain lysines and suramin block of P2X1 receptors. J Biol Chem 2008; 283: 29841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Braun K, Rettinger J, Ganso M, Kassack M, Hildebrandt C, Ullmann H, et al. NF449: a subnanomolar potency antagonist at recombinant rat P2X(1) receptors. Naunyn Schmiedebergs Arch Pharmacol 2001; 364: 285–90. [DOI] [PubMed] [Google Scholar]
- 52Lorinczi E, Bhargava Y, Marino SF, Taly A, Kaczmarek-Hajek K, Barrantes-Freer A, et al. Involvement of the cysteine-rich head domain in activation and desensitization of the P2X1 receptor. Proc Natl Acad Sci U S A 2012; 109: 11396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53Serrano A, Mo G, Grant R, Pare M, O'Donnell D, Yu XH, et al. Differential expression and pharmacology of native P2X receptors in rat and primate sensory neurons. J Neurosci 2012; 32: 11890–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Clarke CE, Benham CD, Bridges A, George AR, Meadows HJ. Mutation of histidine 286 of the human P2X4 purinoceptor removes extracellular pH sensitivity. J Physiol 2000; 523 Pt 3: 697–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Zemkova H, Yan Z, Liang Z, Jelinkova I, Tomic M, Stojilkovic SS. Role of aromatic and charged ectodomain residues in the P2X(4) receptor functions. J Neurochem 2007; 102: 1139–50. [DOI] [PubMed] [Google Scholar]
- 56Jie Y, Zhang L, Xu H, Gao C, Ma W, Li Z. Involvement of the left-flipper-to-dorsal-fin interface of the zebrafish P2X4 receptor in ATP binding and structural rearrangement. Neurosci Lett 2014; 582: 1–5. [DOI] [PubMed] [Google Scholar]
- 57Tvrdonova V, Rokic MB, Stojilkovic SS, Zemkova H. Identification of functionally important residues of the rat P2X4 receptor by alanine scanning mutagenesis of the dorsal fin and left flipper domains. PloS One 2014; 9: e112902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Zhang LM, Xu HJ, Jie YL, Gao C, Chen WJ, Yin SK, et al. Involvement of ectodomain Leu 214 in ATP binding and channel desensitization of the P2X4 receptor. Biochemistry 2014; 53: 3012–9. [DOI] [PubMed] [Google Scholar]
- 59Petrenko N, Khafizov K, Tvrdonova V, Skorinkin A, Giniatullin R. Role of the ectodomain serine 275 in shaping the binding pocket of the ATP-gated P2X3 receptor. Biochemistry 2011; 50: 8427–36. [DOI] [PubMed] [Google Scholar]
- 60Lorca RA, Coddou C, Gazitua MC, Bull P, Arredondo C, Huidobro-Toro JP. Extracellular histidine residues identify common structural determinants in the copper/zinc P2X(2) receptor modulation. J Neurochem 2005; 95: 499–512. [DOI] [PubMed] [Google Scholar]
- 61Buell G, Lewis C, Collo G, North RA, Surprenant A. An antagonist-insensitive P2X receptor expressed in epithelia and brain. EMBO J 1996; 15: 55–62. [PMC free article] [PubMed] [Google Scholar]
- 62Torres GE, Egan TM, Voigt MM. N-Linked glycosylation is essential for the functional expression of the recombinant P2X2 receptor. Biochemistry 1998; 37: 14845–51. [DOI] [PubMed] [Google Scholar]
- 63Rettinger J, Aschrafi A, Schmalzing G. Roles of individual N-glycans for ATP potency and expression of the rat P2X1 receptor. J Biol Chem 2000; 275: 33542–7. [DOI] [PubMed] [Google Scholar]
- 64Vacca F, D'Ambrosi N, Nestola V, Amadio S, Giustizieri M, Cucchiaroni ML, et al. N-Glycans mutations rule oligomeric assembly and functional expression of P2X3 receptor for extracellular ATP. Glycobiology 2011; 21: 634–43. [DOI] [PubMed] [Google Scholar]
- 65Lenertz LY, Wang Z, Guadarrama A, Hill LM, Gavala ML, Bertics PJ. Mutation of putative N-linked glycosylation sites on the human nucleotide receptor P2X7 reveals a key residue important for receptor function. Biochemistry 2010; 49: 4611–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66Samways DS, Khakh BS, Dutertre S, Egan TM. Preferential use of unobstructed lateral portals as the access route to the pore of human ATP-gated ion channels (P2X receptors). Proc Natl Acad Sci U S A 2011; 108: 13800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67Shinozaki Y, Sumitomo K, Tsuda M, Koizumi S, Inoue K, Torimitsu K. Direct observation of ATP-induced conformational changes in single P2X(4) receptors. PLoS Biol 2009; 7: e1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68Khakh BS, Bao XR, Labarca C, Lester HA. Neuronal P2X transmitter-gated cation channels change their ion selectivity in seconds. Nat Neurosci 1999; 2: 322–30. [DOI] [PubMed] [Google Scholar]
- 69Virginio C, MacKenzie A, Rassendren FA, North RA, Surprenant A. Pore dilation of neuronal P2X receptor channels. Nat Neurosci 1999; 2: 315–21. [DOI] [PubMed] [Google Scholar]
- 70Allsopp RC, El Ajouz S, Schmid R, Evans RJ. Cysteine scanning mutagenesis (residues Glu52-Gly96) of the human P2X1 receptor for ATP: mapping agonist binding and channel gating. J Biol Chem 2011; 286: 29207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71Roberts JA, Valente M, Allsopp RC, Watt D, Evans RJ. Contribution of the region Glu181 to Val200 of the extracellular loop of the human P2X1 receptor to agonist binding and gating revealed using cysteine scanning mutagenesis. J Neurochem 2009; 109: 1042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72Young MT, Zhang YH, Cao L, Broomhead H, Jiang LH. Role of the domain encompassing Arg304-Ile328 in rat P2X2 receptor conformation revealed by alterations in complex glycosylation at Asn298. Biochem J 2008; 416: 137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73Yan Z, Liang Z, Obsil T, Stojilkovic SS. Participation of the Lys313-Ile333 sequence of the purinergic P2X4 receptor in agonist binding and transduction of signals to the channel gate. J Biol Chem 2006; 281: 32649–59. [DOI] [PubMed] [Google Scholar]
- 74Roberts JA, Allsopp RC, El Ajouz S, Vial C, Schmid R, Young MT, et al. Agonist binding evokes extensive conformational changes in the extracellular domain of the ATP-gated human P2X1 receptor ion channel. Proc Natl Acad Sci U S A 2012; 109: 4663–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75Kawate T, Robertson JL, Li M, Silberberg SD, Swartz KJ. Ion access pathway to the transmembrane pore in P2X receptor channels. J Gen Physiol 2011; 137: 579–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76Stelmashenko O, Compan V, Browne LE, North RA. Ectodomain movements of an ATP-gated ion channel (P2X2 receptor) probed by disulfide locking. J Biol Chem 2014; 289: 9909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77Gao C, Yu Q, Xu H, Zhang L, Liu J, Jie Y, et al. Roles of the lateral fenestration residues of the P2X(4) receptor that contribute to the channel function and the deactivation effect of ivermectin. Purinergic Signal 2015; 11: 229–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78Rokic MB, Stojilkovic SS, Vavra V, Kuzyk P, Tvrdonova V, Zemkova H. Multiple roles of the extracellular vestibule amino acid residues in the function of the rat P2X4 receptor. PloS One 2013; 8: e59411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79Rokic MB, Stojilkovic SS, Zemkova H. Structural and functional properties of the rat P2X4 purinoreceptor extracellular vestibule during gating. Frontiers Cell Neurosci 2014; 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80Liu X, Ma W, Surprenant A, Jiang LH. Identification of the amino acid residues in the extracellular domain of rat P2X(7) receptor involved in functional inhibition by acidic pH. Br J Pharmacol 2009; 156: 135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81Jiang R, Martz A, Gonin S, Taly A, de Carvalho LP, Grutter T. A putative extracellular salt bridge at the subunit interface contributes to the ion channel function of the ATP-gated P2X2 receptor. J Biol Chem 2010; 285: 15805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82Dellal SS, Hume RI. Covalent modification of mutant rat P2X2 receptors with a thiol-reactive fluorophore allows channel activation by zinc or acidic pH without ATP. PloS One 2012; 7: e47147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83Werner P, Seward EP, Buell GN, North RA. Domains of P2X receptors involved in desensitization. Proc Natl Acad Sci U S A 1996; 93: 15485–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84Fujiwara Y, Keceli B, Nakajo K, Kubo Y. Voltage- and [ATP]-dependent gating of the P2X(2) ATP receptor channel. J Gen Physiol 2009; 133: 93–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85Keceli B, Kubo Y. Functional and structural identification of amino acid residues of the P2X2 receptor channel critical for the voltage- and [ATP]-dependent gating. J Physiol 2009; 587: 5801–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86Keceli B, Kubo Y. Voltage- and ATP-dependent structural rearrangements of the P2X2 receptor associated with the gating of the pore. J Physiol 2014; 592: 4657–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87Cao L, Broomhead HE, Young MT, North RA. Polar residues in the second transmembrane domain of the rat P2X2 receptor that affect spontaneous gating, unitary conductance, and rectification. J Neurosci 2009; 29: 14257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88Samways DS, Li Z, Egan TM. Principles and properties of ion flow in P2X receptors. Frontiers Cell Neurosci 2014; 8: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89Rassendren F, Buell G, Newbolt A, North RA, Surprenant A. Identification of amino acid residues contributing to the pore of a P2X receptor. EMBO J 1997; 16: 3446–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90Kracun S, Chaptal V, Abramson J, Khakh BS. Gated access to the pore of a P2X receptor: structural implications for closed-open transitions. J Biol Chem 2010; 285: 10110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91Cao L, Young MT, Broomhead HE, Fountain SJ, North RA. Thr339-to-serine substitution in rat P2X2 receptor second transmembrane domain causes constitutive opening and indicates a gating role for Lys308. J Neurosci 2007; 27: 12916–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92Khakh BS, Egan TM. Contribution of transmembrane regions to ATP-gated P2X2 channel permeability dynamics. J Biol Chem 2005; 280: 6118–29. [DOI] [PubMed] [Google Scholar]
- 93Samways DS, Khakh BS, Egan TM. Allosteric modulation of Ca2+ flux in ligand-gated cation channel (P2X4) by actions on lateral portals. J Biol Chem 2012; 287: 7594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94Li M, Kawate T, Silberberg SD, Swartz KJ. Pore-opening mechanism in trimeric P2X receptor channels. Nat Commun 2010; 1: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95Silberberg SD, Chang TH, Swartz KJ. Secondary structure and gating rearrangements of transmembrane segments in rat P2X4 receptor channels. J Gen Physiol 2005; 125: 347–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96Browne LE, Cao L, Broomhead HE, Bragg L, Wilkinson WJ, North RA. P2X receptor channels show threefold symmetry in ionic charge selectivity and unitary conductance. Nat Neurosci 2011; 14: 17–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97Li M, Chang TH, Silberberg SD, Swartz KJ. Gating the pore of P2X receptor channels. Nat Neurosci 2008; 11: 883–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98Rothwell SW, Stansfeld PJ, Bragg L, Verkhratsky A, North RA. Direct gating of ATP-activated ion channels (P2X2 receptors) by lipophilic attachment at the outer end of the second transmembrane domain. J Biol Chem 2014; 289: 618–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99Browne LE, Nunes JP, Sim JA, Chudasama V, Bragg L, Caddick S, et al. Optical control of trimeric P2X receptors and acid-sensing ion channels. Proc Natl Acad Sci U S A 2014; 111: 521–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100Lemoine D, Habermacher C, Martz A, Mery PF, Bouquier N, Diverchy F, et al. Optical control of an ion channel gate. Proc Natl Acad Sci U S A 2013; 110: 20813–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101Heymann G, Dai J, Li M, Silberberg SD, Zhou HX, Swartz KJ. Inter- and intrasubunit interactions between transmembrane helices in the open state of P2X receptor channels. Proc Natl Acad Sci U S A 2013; 110: E4045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102Grimes L, Young MT. Purinergic P2X receptors: structural and functional features depicted by X-Ray and molecular modelling studies. Curr Med Chem 2015; 22: 783–98. [DOI] [PubMed] [Google Scholar]
- 103Hausmann R, Kless A, Schmalzing G. Key sites for P2X receptor function and multimerization: overview of mutagenesis studies on a structural basis. Curr Med Chem 2015; 22: 799–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104North RA. Molecular physiology of P2X receptors. Physiol Rev 2002; 82: 1013–67. [DOI] [PubMed] [Google Scholar]
- 105Surprenant A, North RA. Signaling at purinergic P2X receptors. Ann Rev Physiol 2009; 71: 333–59. [DOI] [PubMed] [Google Scholar]
- 106Bernier LP, Ase AR, Seguela P. Post-translational regulation of P2X receptor channels: modulation by phospholipids. Frontiers Cell Neurosci 2013; 7: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107Stelmashenko O, Lalo U, Yang Y, Bragg L, North RA, Compan V. Activation of trimeric P2X2 receptors by fewer than three ATP molecules. Mol Pharmacol 2012; 82: 760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108Du J, Dong H, Zhou HX. Gating mechanism of a P2X4 receptor developed from normal mode analysis and molecular dynamics simulations. Proc Natl Acad Sci U S A 2012; 109: 4140–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109Keystone EC, Wang MM, Layton M, Hollis S, McInnes IB, Team DCS. Clinical evaluation of the efficacy of the P2X7 purinergic receptor antagonist AZD9056 on the signs and symptoms of rheumatoid arthritis in patients with active disease despite treatment with methotrexate or sulphasalazine. Ann Rheum Dis 2012; 71: 1630–5. [DOI] [PubMed] [Google Scholar]
- 110Garcia-Guzman M, Soto F, Gomez-Hernandez JM, Lund PE, Stuhmer W. Characterization of recombinant human P2X4 receptor reveals pharmacological differences to the rat homologue. Mol Pharmacol 1997; 51: 109–18. [DOI] [PubMed] [Google Scholar]
- 111Michel AD, Clay WC, Ng SW, Roman S, Thompson K, Condreay JP, et al. Identification of regions of the P2X(7) receptor that contribute to human and rat species differences in antagonist effects. Br J Pharmacol 2008; 155: 738–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112Wolf C, Rosefort C, Fallah G, Kassack MU, Hamacher A, Bodnar M, et al. Molecular determinants of potent P2X2 antagonism identified by functional analysis, mutagenesis, and homology docking. Mol Pharmacol 2011; 79: 649–61. [DOI] [PubMed] [Google Scholar]
- 113Hausmann R, Gunther J, Kless A, Kuhlmann D, Kassack MU, Bahrenberg G, et al. Salt bridge switching from Arg290/Glu167 to Arg290/ATP promotes the closed-to-open transition of the P2X2 receptor. Mol Pharmacol 2013; 83: 73–84. [DOI] [PubMed] [Google Scholar]
- 114Khakh BS, Proctor WR, Dunwiddie TV, Labarca C, Lester HA. Allosteric control of gating and kinetics at P2X(4) receptor channels. J Neurosci 1999; 19: 7289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115Priel A, Silberberg SD. Mechanism of ivermectin facilitation of human P2X4 receptor channels. J Gen Physiol 2004; 123: 281–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116Silberberg SD, Li M, Swartz KJ. Ivermectin interaction with transmembrane helices reveals widespread rearrangements during opening of P2X receptor channels. Neuron 2007; 54: 263–74. [DOI] [PubMed] [Google Scholar]
- 117Popova M, Trudell J, Li KX, Alkana R, Davies D, Asatryan L. Tryptophan 46 is a site for ethanol and ivermectin action in P2X4 receptors. Purinergic Signal 2013; 9: 621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118Chaumont S, Khakh BS. Patch-clamp coordinated spectroscopy shows P2X2 receptor permeability dynamics require cytosolic domain rearrangements but not Panx-1 channels. Proc Natl Acad Sci U S A 2008; 105: 12063–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119Yan Z, Li S, Liang Z, Tomic M, Stojilkovic SS. The P2X7 receptor channel pore dilates under physiological ion conditions. J Gen Physiol 2008; 132: 563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120Yan ZH, Khadra A, Li S, Tomic M, Sherman A, Stojilkovic SS. Experimental characterization and mathematical modeling of P2X7 receptor channel gating. J Neurosci 2010; 30: 14213–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121Yan Z, Khadra A, Sherman A, Stojilkovic SS. Calcium-dependent block of P2X7 receptor channel function is allosteric. J Gen Physiol 2011; 138: 437–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122Khadra A, Yan Z, Coddou C, Tomic M, Sherman A, Stojilkovic SS. Gating properties of the P2X2a and P2X2b receptor channels: experiments and mathematical modeling. J Gen Physiol 2012; 139: 333–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123Allsopp RC, Evans RJ. Contribution of the juxtatransmembrane intracellular regions to the time course and permeation of ATP-gated P2X7 receptor ion channels. J Biol Chem 2015; 290: 14556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124Kido Y, Kawahara C, Terai Y, Ohishi A, Kobayashi S, Hayakawa M, et al. Regulation of activity of P2X7 receptor by its splice variants in cultured mouse astrocytes. Glia 2014; 62: 440–51. [DOI] [PubMed] [Google Scholar]
- 125Pelegrin P, Surprenant A. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J 2006; 25: 5071–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126Pelegrin P, Surprenant A. Pannexin-1 couples to maitotoxin- and nigericin-induced interleukin-1beta release through a dye uptake-independent pathway. J Biol Chem 2007; 282: 2386–94. [DOI] [PubMed] [Google Scholar]
- 127Torres GE, Haines WR, Egan TM, Voigt MM. Co-expression of P2X1 and P2X5 receptor subunits reveals a novel ATP-gated ion channel. Mol Pharmacol 1998; 54: 989–93. [DOI] [PubMed] [Google Scholar]
- 128Kowalski M, Hausmann R, Schmid J, Dopychai A, Stephan G, Tang Y, et al. Flexible subunit stoichiometry of functional human P2X2/3 heteromeric receptors. Neuropharmacology 2015; 99: 115–30. [DOI] [PubMed] [Google Scholar]
- 129Jiang LH, Kim M, Spelta V, Bo X, Surprenant A, North RA. Subunit arrangement in P2X receptors. J Neurosci 2003; 23: 8903–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130Dawson RJ, Benz J, Stohler P, Tetaz T, Joseph C, Huber S, et al. Structure of the acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nat Commun 2012; 3: 936. [DOI] [PubMed] [Google Scholar]
- 131Baconguis I, Gouaux E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 2012; 489: 400–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132Yu Y, Chen Z, Li WG, Cao H, Feng EG, Yu F, et al. A nonproton ligand sensor in the acid-sensing ion channel. Neuron 2010; 68: 61–72. [DOI] [PubMed] [Google Scholar]
- 133Bohlen CJ, Chesler AT, Sharif-Naeini R, Medzihradszky KF, Zhou S, King D, et al. A heteromeric Texas coral snake toxin targets acid-sensing ion channels to produce pain. Nature 2011; 479: 410–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134Diochot S, Baron A, Salinas M, Douguet D, Scarzello S, Dabert-Gay AS, et al. Black mamba venom peptides target acid-sensing ion channels to abolish pain. Nature 2012; 490: 552–5. [DOI] [PubMed] [Google Scholar]
- 135Wemmie JA, Taugher RJ, Kreple CJ. Acid-sensing ion channels in pain and disease. Nat Rev Neurosci 2013; 14: 461–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136Grunder S, Pusch M. Biophysical properties of acid-sensing ion channels (ASICs). Neuropharmacology 2015; 94: 9–18. [DOI] [PubMed] [Google Scholar]
- 137Nogales E, Scheres SHW. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Mol Cell 2015; 58: 677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138Cheng YF. Single-Particle Cryo-EM at crystallographic resolution. Cell 2015; 161: 450–7. [DOI] [PMC free article] [PubMed] [Google Scholar]