Abstract
The bulk of models used to understand the species diversification on Madagascar have been constructed using vertebrate taxa. It is not clear how these models affect less vagile species that may interact at a variety of spatial scales. Several studies on vertebrates have divided Madagascar into east-west bioclimatic regions, suggesting there is a fundamental division between eastern wet-adapted and western dry-adapted taxa. An alternative model of ecogeographic constraints shows a north-south division. We test whether the diversification in a small arthropod with variable degrees of dispersal conform to either model of ecogeographic constraints proposed for vertebrate taxa. We employ a molecular taxonomic dataset using ~2 kilobases nuDNA (Wg, LW Rh, Abd-A, 28s) and 790 basepairs mtDNA (CO1), along with geographic and habitat data, to examine the diversification patterns of the ant genus Mystrium Roger, 1862, (Subfamily Amblyoponinae) from Madagascar. The nuclear and mitochondrial phylogenies were both congruent with morphospecies as indicated in a recent revision of the genus. Species of Mystrium practice different colony reproductive strategies (winged queens vs non-winged queens). Alternate reproductive strategies led to inequalities in female dispersal ability among species, providing an additional layer for examination of the impacts of vagility on divergence, especially when measured using a maternally inherited locus. Mystrium species distribution patterns support these models of ecogeographic constraints. Reproductive strategy effected how Mystrium mtDNA lineages were associated with large-scale habitat distinctions and various topographical features. Furthermore, in some cases we find microgeographic population structure which appears to have been impacted by localized habitat differences (tsingy limestone formations, littoral forest) on a scale much smaller than that found in vertebrates. The current system offers a finer scale look at species diversification on the island, and helps achieve a more universal understanding of the generation of biodiversity on Madagascar.
Introduction
Species have evolved in isolation on Madagascar since the breakup of Indo-Madagascar and the northwards drift of India and the Seychelles began 88 million years ago [1]. Today the entire island of Madagascar is a globally recognized biological hotspot, a region of extremely high biodiversity that contains a large number of endemic species [2]. Species radiations on the island have been difficult to adequately explain [3–5] despite the development of diversification models across multiple taxa [6–12]. Although these models have been constructed and evaluated largely using vertebrates, they are being used to understand species diversification and in conservation and management applications for all animal taxa. Many vertebrate species (birds, primates, lizards) typically have relatively high levels of vagility that allow them to disperse across many landscape features. It is not clear how these divergence models effect less vagile species that may interact at a variety of spatial scales [13,14]. Little literature exists for evaluating the species diversification and phylogeographic profiles of taxa with low vagility on Madagascar. Identification of a model taxon group that represents the wide variety of less vagile taxa is critical to evaluating the universality of the proposed models for explaining biodiversity.
Arthropods are being recognized as invaluable for highlighting the influence of fine-scale habitats [14, 15–17]. Ants are a hyperdiverse group of arthropods long utilized as indicators for conservation methods such as surveying and restoration monitoring [17,18]. Surveys and monitoring have typically used a morphospecies approach to distinguish among distinct taxonomic units; however, this approach is time-intensive and therefore limiting [15,19]. Molecular phylogenetic methods within the field of myrmecology have been broadening the ways that ants can be used to gain an understanding of the historical biogeography of a region and in applications for conservation [20,21]. Recent studies have drawn attention to the fine-scale patterns of habitat utilization among ants, demonstrating that this group is particularly well suited to illuminate patterns of endemism on a much smaller scale than vertebrates [5,16,19].
To evaluate the utility of one of these vertebrate derived species diversification models on Madagascar, we used molecular phylogenetic analyses of a genus of ants from Madagascar along with geographic and habitat data to examine the diversification patterns of the ant genus Mystrium Roger, 1862, (Subfamily Amblyoponinae). The species in this genus serve as a model for many small arthropods found on Madagascar, as the species examined have been collected in various habitats throughout the island. Furthermore, this group contains species which exhibit two forms of dispersal, colony foundation by either winged (alate) queens or a wingless reproductive caste (ergatoid queens), thus displaying variation in patterns of vagility among females.
Ecogeographic constraints diversification model
Many of the recent diversification mechanisms proposed for the fauna of Madagascar have been reviewed and the perspectives for testing them concisely outlined [22]. Prominent among these mechanisms is a model of ecogeographic constraints between eastern and western sites or northern and southern sites [6,9]. Madagascar has a north-south orientation, being notably longer than it is wide [23]. The Indian section was attached to the eastern side [24], giving the present island considerable topographic asymmetry [7]. This topography has given rise to fairly discrete habitat types across the island. The primary biomes include an eastern rainforest, western dry-deciduous forest, southern spiny forest, and central grasslands [25,26]. Several studies on vertebrate species [9,12,27,28] have divided Madagascar into east-west bioclimatic regions and much of the northern fourth of Madagascar is included in the eastern rainforest region. Yoder and Heckman (2006) term this model for species differentiation the “ecogeographic constraint.” This hypothesis can be tested by mapping a hierarchical outline of a phylogeny onto localities collected from across the island. Under this ecogeographic model, if there is a fundamental division between eastern wet-adapted and western dry-adapted taxa, we would expect to observe haplotypes from eastern localities and western localities segregating into mutually exclusive clades. Yoder and Heckman (2006) revised their hypothesis of ecogeographic constraints to include the possibility of a north-south split among clades after completing a study of mouse lemurs (genus Microcebus). Deep interspecific north-south splits also have been found among Madagascar’s reptiles [27,29]. However, these distinctions are usually associated with large-scale distances among populations separated along the north-south axis of the island rather than by discrete differences in climate.
Mystrium Reproductive Strategies
Mystrium species are considered to retain ancestral behavioral and morphological traits [30–33] and are part of a group that contains some of the oldest lineages of ants [34–39]. The workers of the genus Mystrium can be distinguished easily from other amblyoponine genera by their characteristic head shape and mandibles; however, in some species, morphological variation within castes in body size and shape, setae or body sculpture is remarkably high, while differences among species can be so slight as to be confounding for taxonomists [40]. A recent morphological revision of the genus Mystrium for the Malagasy region including descriptions of six new species was published by Yoshimura and Fisher [40]. Previous taxonomic works [30,41] were insufficient to diagnose the species boundaries of Mystrium in the Malagasy region.
One advantage of studying the genus Mystrium is that the differences in reproductive strategy among species can provide an additional layer of examination of the impacts of vagility on patterns of divergence within and among species. Different dispersal capabilities among species of Mystrium, conferred by alternative reproductive strategies, may be a secondary factor influencing biogeographic patterns of species diversification within the genus [42]. Mystrium species exhibit various reproductive forms including typical winged queens, small ergatoid queens [43,44], short-winged queens [45], and intercastes [45,46]. Two major colonial reproductive strategies have been described in ants. During independent colony foundation (ICF), a mated alate queen (winged queen) disperses away from her natal colony and becomes the sole foundress of a new nest, metabolizing her wing muscles to fuel initial brood care, either remaining with the nest (claustral) or leaving the nest untended to forage for food (non-claustral). Alate queens require considerable colony resources while being reared and experience a high mortality rate after leaving the nest [42,47]. Ant species that practice dependent colony foundation (DCF) have a permanently wingless reproductive caste (ergatoid queens) and the colony reproduces by fission or budding. An ergatoid queen that disperses to establish a new nest is accompanied by nestmate workers that provision the initial brood. The small ergatoid queens of Mystrium have been described as “multi-purpose” because while they are a reproductive caste they will if left unmated serve in the nurse role played by minor workers in Mystrium species that reproduce with winged queens. Smaller ergatoid queens require less per capita energy investment by colonies [47,48]. Therefore there is the possibility that female dispersal will be limited by dependent colony foundation relative to independent colony foundation. If dispersal limitation is the case, it would lead to different patterns of inheritance among maternally inherited loci which would be reflected by patterns of genetic divergence in a mitochondrial phylogeny.
We employed a molecular taxonomic dataset for the elucidation of inter- and intra-species patterns of diversification in the genus Mystrium, using ~2 kilobases (kb) of DNA sequence data from four independently segregating nuclear loci and a 790 bp fragment from the cytochrome c oxidase I (CO1) locus adjacent to and overlapping the standard region used in DNA barcoding [49]. We examine Mystrium species lineage distribution patterns across the island of Madagascar to determine whether 1) there are discernable differences in biogeographic patterns of female lineages between species with ergatoid versus alate queens, and 2) whether the diversification in a small arthropod with variable degrees of female dispersal conforms to either model of ecogeographic constraints proposed for vertebrate taxa.
Materials and Methods
Sampling
Samples in this study are from regions of Madagascar where Mystrium was collected during arthropod surveys from 1992 through 2003 [50]. Research, collecting, and export permits were obtained through collaboration with the Ministère de l' Environnement et des Forêts and the Madagascar National Parks. Currently understood species distinctions based on morphological characters were taken into account for sampling (Table 1). Mystrium oberthueri Forel (1897), M. voeltzkowi Forel (1897), M. shadow Yoshimura and Fisher (2014), M. janovitzi Yoshimura and Fisher (2014), M. barrybressleri Yoshimura and Fisher (2014) and M. mirror Yoshimura and Fisher (2014) are endemic to the island. M. rogeri Forel (1899) is endemic to Madagascar and Comoros. Mystrium camillae Emery (1889) is found across Southeast Asia east to the Philippines and south into Northern Australia. Mystrium silvestrii Santschi (1914) is found throughout West and Central Africa. All specimen data for material used in this study are available on AntWeb (www.antweb.org). All voucher specimens are deposited with the California Academy of Sciences, Department of Entomology Collection (CASC). Sequence data has been deposited in Genbank (Table 2). Only samples successfully producing for both primer pairs and considered unique sequences were included in the analyses. For this reason, each sequence often represents haplotypes shared by multiple individuals. Outgroup taxa were chosen based on their relationship to Mystrium in higher-level taxonomic studies [34,36,37]. Adetomyrma is a recently-described genus endemic to Madagascar [33] which appears to be sister taxa to Mystrium. Stigmatomma shares some lifestyle characteristics with Mystrium; both genera are specialized predators on chilopods, and in some species, queens perform non-destructive cannibalism by cutting holes in the integument of larvae to feed on the exuded hemolymph [51,52].
Table 1. List of current Mystrium species and species group reproductive types from the most recent revision of the genus by Yoshimura and Fisher (2014).
| camillae species group | |||
| barrybressleria | Yoshimura & Fisher, 2014 | alate queen | Madagascar |
| camillaea | Emery, 1889 | alate queen | South-East Asia, Australia |
| Labyrinth | Yoshimura & Fisher, 2014 | alate queen | Madagascar |
| Leonie | Bihn & Verhaagh, 2007 | alate queen | Indonesia |
| Maren | Bihn & Verhaagh, 2007 | alate queen | Indonesia |
| silvestriia | Santschi, 1914 | alate queen | Africa |
| mysticum species group | |||
| mysticum | Roger, 1862 | alate queen | Madagascar, Comoros |
| rogeria | Forel, 1899 | alate queen | Madagascar, Comoros |
| voeltzkowi species group | |||
| eques | Yoshimura & Fisher, 2014 | ergatoid queen | Madagascar |
| janovitzia | Yoshimura & Fisher, 2014 | ergatoid queen | Madagascar |
| mirrora | Yoshimura & Fisher, 2014 | ergatoid queen | Madagascar |
| oberthueria | Forel, 1897 | ergatoid queen | Madagascar |
| shadowa | Yoshimura & Fisher, 2014 | ergatoid queen | Madagascar |
| voeltzkowia | Forel, 1897 | ergatoid queen | Madagascar, Mayotte |
a Included in current study.
Table 2. List of all specimens, museum accession numbers, habitat types, collection locality, and GenBank accession numbers.
| Morphospecies | Casent #a | Habitat | Locality Code | Lat | Long | CO1 | Wg | Abd-A | LW Rh | 28s |
|---|---|---|---|---|---|---|---|---|---|---|
| M. voeltzkowi | 0500419* | tropical dry forest | Ankarana 210 | -12.8636 | 49.2258 | KR063453 | KP280018 | KR063375 | KR063395 | KR063415 |
| M. voeltzkowi | 0500393* | tropical dry forest | Sakaramy 325 | -12.4689 | 49.2422 | KR063468 | KP280016 | KR063368 | KR063388 | KR063408 |
| M. voeltzkowi | 0500425 | tropical dry forest | Sakaramy 325 | -12.4689 | 49.2422 | KR063479 | ||||
| M. voeltzkowi | 0500434 | tropical dry forest | Sakaramy 325 | -12.4689 | 49.2422 | KR063465 | ||||
| M. voeltzkowi | 0500436 | tropical dry forest | Sakaramy 325 | -12.4689 | 49.2422 | KR063429 | ||||
| M. voeltzkowi | 0500394 | tropical dry forest | Ankarana 210 | -12.8636 | 49.2258 | KR063478 | ||||
| M. voeltzkowi | 0500437 | tropical dry forest | Ankarana 80 | -12.9089 | 49.1097 | KR063457 | ||||
| M. voeltzkowi | 0500435* | Sambirano | Lokobe 30 | -13.4194 | 48.3311 | KR063454 | KP280019 | KR063369 | KR063389 | KR063409 |
| M. voeltzkowi | 0500397 | Sambirano | Lokobe 30 | -13.4194 | 48.3311 | KR063480 | ||||
| M. voeltzkowi | 0500431 | tropical dry forest | Ankarana 210 | -12.8636 | 49.2258 | KR063481 | ||||
| M. voeltzkowi | 0500438* | tropical dry forest | Ankarana 80 | -12.9089 | 49.1097 | KR063423 | KP280025 | KR063367 | KR063387 | KR063407 |
| M. voeltzkowi | 0500392 | tropical dry forest | Ankarana 80 | -12.9089 | 49.1097 | KR063466 | ||||
| M. voeltzkowi | 0500429 | tropical dry forest | Ankarana 80 | -12.9089 | 49.1097 | KR063452 | ||||
| M. voeltzkowi | 0500415 | tropical dry forest | Ankarana 80 | -12.9089 | 49.1097 | KR063476 | ||||
| M. voeltzkowi | 0500424 | tropical dry forest | Ankarana 80 | -12.9089 | 49.1097 | KR063435 | ||||
| M. voeltzkowi_cmplx | 0500395 | tropical dry forest | Francais 180 | -12.3228 | 49.3381 | KR063428 | ||||
| M. voeltzkowi_cmplx | 0500387 | tropical dry forest | Sakaramy 325 | -12.4689 | 49.2422 | KR063477 | ||||
| M. voeltzkowi_cmplx | 0500417* | tropical dry forest | Sakaramy 325 | -12.4689 | 49.2422 | KR063470 | KP280031 | KR063370 | KR063390 | KR063410 |
| M. mirror | 0500388 | tropical dry forest | Anabohazo 120 | -14.3089 | 47.9144 | KR063467 | ||||
| M. mirror | 0500440 | tropical dry forest | Anabohazo 120 | -14.3089 | 47.9144 | KR063427 | ||||
| M. mirror | 0501743* | TDF on tsingy | Andranopasazy 150 | -18.7094 | 44.7181 | KR063469 | KP280030 | KR063365 | KR063385 | KR063405 |
| M. mirror | 0501744 | spiny forest | Manantalinjo 150 | -24.8169 | 46.6100 | KR063430 | ||||
| M. mirror | 0501742* | spiny forest | Marie 160 | -25.5944 | 45.1469 | KR063475 | KP280027 | KR063366 | KR063386 | KR063406 |
| M. mirror | 0500412 | tropical dry forest | Anabohazo 120 | -14.3089 | 47.9143 | KR063486 | ||||
| M. shadow | 0500432* | montane rainforest | Ambre 925 | -12.5344 | 49.1794 | KR063474 | KP280023 | KR063371 | KR063391 | KR063411 |
| M. shadow | 0500439* | rainforest | Ambilanivy 600 | -13.7986 | 48.1617 | KR063449 | KP280022 | KR063374 | KR063394 | KR063414 |
| M. janovitzi | 0500088* | rainforest | Ankarana 150, 7 km | -12.9000 | 49.1167 | KP280028 | KU361295 | KU361297 | KU361296 | |
| M. janovitzi | 0746564 | rainforest | Manon 780 | -13.9767 | 48.4233 | KU361298 | ||||
| M. janovitzi | 0746565 | rainforest | Manon 400 | -13.9617 | 48.4333 | KU361299 | ||||
| M. oberthueri | 0746569 | rainforest | Marojejy 610 | -14.4358 | 49.7606 | KR063416 | ||||
| M. oberthueri | 0746566 | lowland rainforest | Amban 25 | -15.6833 | 49.9500 | KR063426 | ||||
| M. oberthueri | 0500105* | lowland rainforest | Amban 25 | -15.6833 | 49.9500 | KR063444 | KP280020 | KR063356 | KR063376 | KR063396 |
| M. oberthueri | 0746567 | rainforest | Amban 425 | -15.6667 | 49.9667 | KR063446 | ||||
| M. oberthueri | 0500091* | rainforest | Amban 425 | -15.6667 | 49.9667 | KR063439 | KP280021 | KR063357 | KR063377 | KR063397 |
| M. oberthueri | 0500136* | lowland rainforest | Cap Masoala 125 | -15.6936 | 50.1814 | KR063434 | KP280017 | KR063358 | KR063378 | KR063398 |
| M. oberthueri | 0746568 | rainforest | Sandranantitra | -18.0483 | 49.0917 | KR063425 | ||||
| M. oberthueri | 0500063 | rainforest | Sandranantitra | -18.0483 | 49.0917 | KR063455 | ||||
| M. oberthueri | 0500071* | rainforest | Andriantantely | -18.6950 | 48.8133 | KR063464 | KP280029 | KR063360 | KR063380 | KR063400 |
| M. oberthueri | 0746793 | rainforest | Andriantantely | -18.6950 | 48.8133 | KR063417 | ||||
| M. oberthueri | 0500112 | rainforest | Amban 25 | -15.6813 | 49.9580 | KR063485 | ||||
| M. oberthueri | 0500138 | lowland rainforest | Cap Masoala 125, 1 km | -15.6936 | 50.1814 | KR063484 | ||||
| M. oberthueri | 0500142 | rainforest | Marojejy 610 | -14.4358 | 49.7606 | KR063488 | ||||
| M. rogeri | 0500389 | Sambirano | Lokobe 30 | -13.4194 | 48.3311 | KR063433 | ||||
| M. rogeri | 0500391 | montane rainforest | Ambre 925 | -12.5344 | 49.1794 | KR063459 | ||||
| M. rogeri | 0500441 | montane rainforest | Ambre 925 | -12.5344 | 49.1794 | KR063463 | ||||
| M. rogeri | 0500409* | montane rainforest | Ambre 925 | -12.5344 | 49.1794 | KR063461 | KP280015 | KR063373 | KR063393 | KR063413 |
| M. rogeri | 0500390* | rainforest | Ambilanivy 600 | -13.7986 | 48.1617 | KR063472 | KP280013 | KR063372 | KR063392 | KR063412 |
| M. rogeri | 0746571 | montane rainforest | Manon 1175 | -13.9983 | 48.4283 | KR063460 | ||||
| M. rogeri | 0500097 | montane rainforest | Manon 1175 | -13.9983 | 48.4283 | KR063458 | ||||
| M. rogeri | 0500087 | lowland rainforest | Amban 25 | -15.6833 | 49.9500 | KR063424 | ||||
| M. rogeri | 0500102 | rainforest | Andri 825 | -22.2333 | 47.0000 | KR063443 | ||||
| M. rogeri | 0746570 | rainforest | Andri 785 | -22.2167 | 47.0167 | KR063431 | ||||
| M. rogeri | 0746572 | montane rainforest | Ivohibe 8.0 E | -22.4833 | 46.9683 | KR063462 | ||||
| M. rogeri | 0746573 | montane rainforest | Ivohibe 8.0 E | -22.4833 | 46.9683 | KR063471 | ||||
| M. rogeri | 0500090* | rainforest | Andri 785 | -22.2167 | 47.0167 | KR063473 | KP280014 | KR063361 | KR063381 | KR063401 |
| M. rogeri | 0746792 | montane rainforest | Ivohibe 9.0 NE | -22.4267 | 46.9383 | KR063456 | ||||
| M. rogeri | 0500119 | rainforest | Ando 330 | -24.7333 | 46.8000 | KR063432 | ||||
| M. barrybressleri | 0746560 | rainforest | Manon 400 | -13.9617 | 48.4333 | KR063441 | ||||
| M. barrybressleri | 0746559 | rainforest | Manon 780 | -13.9767 | 48.4233 | KR063440 | ||||
| M. barrybressleri | 0746558 | rainforest | Manon 780 | -13.9767 | 48.4233 | KR063442 | ||||
| M. barrybressleri | 0500082* | rainforest | Anja 875 | -14.7500 | 49.5000 | KR063451 | KP280026 | KR063359 | KR063379 | KR063399 |
| M. barrybressleri | 0746555 | rainforest | Anja 875 | -14.7500 | 49.5000 | KR063447 | ||||
| M. barrybressleri | 0746554 | rainforest | Anja 875 | -14.7500 | 49.5000 | KR063450 | ||||
| M. barrybressleri | 0746557 | rainforest | Ivohibe 7.5 ENE | -22.4700 | 46.9600 | KR063437 | ||||
| M. barrybressleri | 0746556 | rainforest | Ivohibe 7.5 ENE | -22.4700 | 46.9600 | KR063445 | ||||
| M. barrybressleri | 0500083 | littoral rainforest | Mandena | -24.9517 | 47.0017 | KR063482 | ||||
| M. barrybressleri | 0746561 | littoral rainforest | Mandena | -24.9517 | 47.0017 | KR063438 | ||||
| M. barrybressleri | 0746562 | littoral rainforest | St. Luce | -24.7717 | 47.1717 | KR063436 | ||||
| M. barrybressleri | 0500114 | rainforest | Manon 400 | -13.9617 | 48.4333 | KR063487 | ||||
| M. barrybressleri | 0500079 | rainforest | Manon 780 | -13.9767 | 48.4233 | KR063489 | ||||
| M. silvestrii | 0408192 | rainforest | Ndakan 360 | 2.3707 | 16.1725 | KR063421 | ||||
| M. camillae | 0500094* | K mixed beach forest | Brooketon Coal Mine | 5.0100 | 115.0300 | KR063448 | KP280024 | KR063362 | KR063382 | KR063402 |
| M. camillae | 0746563 | K mixed beach forest | Brooketon Coal Mine | 5.0100 | 115.0300 | KR063420 | ||||
| Adetomyrma caputleae | 0500384* | montane rainforest | Andranomay 1300 | -18.4733 | 47.9600 | KR063483 | KP280012 | KR063364 | KR063384 | KR063404 |
| Stigmatomma mg01 | 0500009 | rainforest | Ivohibe 7.5 ENE | -22.4700 | 46.9600 | KR063422 | ||||
| Stigmatomma mg01 | 0500385* | montane rainforest | Ambre 1300 | -12.5964 | 49.1594 | KR063418 | KP280011 | KR063363 | KR063383 | KR063403 |
| Stigmatomma tz06 | 0500028 | Mkomazi | 3.9667 | 37.8000 | KR063419 |
a Specimens with an asterisk (*) next to the museum accession number are included in the nuclear phylogeny.
DNA isolation, Amplification, and Sequencing
Genomic DNA was extracted from specimens stored in 95% ethanol at –80°C using Qiagen Dneasy Tissues Kit (Quiagen Inc., Valencia, CA) following the protocol for animal tissues. Individual ants or portions of a specimen were placed in 1.5 ml microtubes and frozen in liquid nitrogen, then ground up thoroughly using a disposable pestle. The material was then digested overnight using 20 uL of 20 mg/mL Proteinase K at 55°C. The lysate was pipetted onto a silica-gel-membrane and purified with a series of ethanol washes using supplied Dneasy Buffers. The DNA was resuspended with 200 uL of 10 mM Tris buffer.
For the mitochondrial phylogeny, two sets of primer pairs from the 5’ end of cytochrome c oxidase I (CO1) gene were chosen from a previous study and used for polymerase chain reaction (PCR) (Table 3). For the nuclear phylogeny, the following loci were chosen from previous studies in which they provided useful resolution for ant phylogenies and were amplified according to the following protocols with some variation in annealing temperature and MgCl2 concentration: 398 bp from the wingless locus; 559 bp from the abdominal-A locus; 410 bp from the 28s rRNA locus, and 580 bp from the long-wavelength rhodopsin locus (Table 3). Reactions contained 1.5 mM MgCl2, 0.175 mM dNTPs, 0.050 U/ul Taq, 0.540 uM each primer, and 2 uL of template, for a total reaction volume of 10 uL. The amplification protocol consisted of thirty-five cycles of 30 s at 94°C, 1 min at 51°C and 2 min at 72°C, preceded by 3 min at 94°C and followed by a final extension for 10 min at 72°C. The PCR products were purified by exonuclease I and shrimp alkaline phosphatase digestion of single-stranded DNA (primers) and dNTPs (ExoSAP-IT, USB Corporation, Cleveland, Ohio, U.S.A). Samples were sequenced in 10 ul reaction volumes in both forward and reverse directions using the same primers. Dye terminator cycle sequencing was performed using one-eighth the amount of BigDye and the protocol specified by the ABI BigDye Terminator v1.1 Cycle Sequencing Kit on ABI genetic analyzer 3100 (Applied Biosystems, Foster City, CA) at the Core DNA Analysis Facility located at Sonoma State University. Sequences were assembled using the program Sequencher 4.6 (Gene Codes Corporation Inc.). All sequences were confirmed and adjusted by visual inspection of chromatograms. Sequences were relatively straightforward to align, however, LW Rh did contain an intron which always occurred at the same location, and was maintained in the dataset during analysis. The concatenated and aligned, four-gene dataset, along with the CO1 dataset, has been deposited with figshare [53].
Table 3. Primer sequences for amplification and sequencing of nuclear phylogeny with models of evolution selected by AIC in Modeltest and Mr.Modeltest.
Primer sequences used to amplify and sequence the cytochrome oxidase I (CO1) gene for the mitochondrial phylogeny and nuclear mitochondrial-like sequence (NuMts) detection and avoidance.
| Locus | Primer | Sequence 5'--->3' | Reference | Model |
|---|---|---|---|---|
| Wingless | LepWG1F | GARTGYAARTGYCAYGGYATGTCTGG | Brower and DeSalle (1998) | GTR + G |
| LepWG2R | ACTICGCRCACCARTGGAATGTRCA | Brower and DeSalle (1998) | ||
| 28s rRNA | M06F | CCCCTGAATTTAAGCATAT | Schmitz and Moritz (1994) | HKY + I |
| 28SCR | CGGTTTCACGTACTCTTGAA | Brady (2003) | ||
| LW Rh | LR-143F | GACAAAGTKCCACCRGARATGC | Ward and Downie (2005) | GTR + G |
| LR672R | CCRCAMGCVGTCATGTTRCCTTC | Ward and Downie (2005) | ||
| Abd-A | AA1172F | CACATCGGCACCGGCGATATGAG | Ward and Downie (2005) | HKY + G |
| AA1881R | GGTTGTTGGCAGGATGTCAAAGG | Ward and Downie (2005) | ||
| CO1 | M13 CI13F | ATAATTTTTTTTATAGTTATACC | Brady (2003) | |
| M13 CI14R | ATTTCTTTTTTTCCTCTTTC | Brady (2003) | ||
| CO1 | JerryF | CAACATTTATTTTGATTTTTTGG | Brady (2003) | |
| Ben3R | GCWACWACRTAATAKGTATCATG | Brady (2003) | ||
| NuMts | LF1F | ATTCAACCAATCATAAAGATATTGG | Smith et al. (2005) | |
| TRL-3382R | TYCAWTGCACTTAWTCTGCCATATTA | P.S. Ward personal communication | ||
| COII-3946R | TATTC ATANCTTCARTATCATTGRTG | P.S. Ward personal communication |
NuMts Detection and Avoidance
When relatively conserved regions of mtDNA are used to design primers that can amplify mitochondrial fragments from an unknown species, there is a risk of generating paralogous nuclear mitochondrial-like sequences (NuMts). These will cause the number of species to be overestimated if they are treated as orthologous, as a large number of these NuMts have unusually high numbers of point mutations [54]. To eliminate suspicion of pseudogenes we first checked the aligned CO1 data for known diagnostic characters such as stop codons and frameshift mutations by translating the CO1 sequences using the invertebrate genetic code in Bioedit 7.0 [55]. NuMts have been characterized in ants and will show up as an accumulation of base pair differences, generally in the third codon position, at a faster rate than the accumulation of stop codons or indels, so the above-mentioned strategies for detecting these pseudogenes may be ineffective [56]. Consequently, using additional protocols (Ward, pers. comm.), we verified the sequence of fifteen samples with the longest branches and unusually high sequence divergence values. Briefly, after amplifying a longer stretch of mitochondrial DNA with primers LF1 [19] and an ant-customized version of the Pat primer (Ward unpublished) (Table 3), the amplicons were visualized on a TAE gel, cut out, re-amplified with our CO1 primers, and sequenced again under the assumption that most NuMts are less than 1 kb in length [57].
Analyses
In the mitochondrial phylogeny a total of 72 Mystrium, three Stigmatomma, and one Adetomyrma accessions were included for analysis. The Kimuras two-parameter (K2P) model assumes equal base frequencies and two substitution types and takes into account the fact that transitions and transversions occur at different rates by adding an additional parameter “K,” thereby computing genetic distances while correcting for differences in the frequency of transition/transversion substitutions. We used the K2P model of evolution [58], with 1000 bootstrap replicates for support, to generate a neighbor-joining (NJ) tree [59] using MEGA version 6 [60]. This provided a graphic representation of the among-species divergences, allowing us to group Mystrium in MEGA according to morphospecies and calculate mtDNA divergence, both interspecific and intraspecific (within and between group means) also by using the K2P distance model.
For the mitochondrial phylogeny we ran Bayesian analysis and maximum likelihood on Cipress Science Gateway [61]. For Bayesian analysis we first partitioned the data by codon position and then used jModelTest2 [62] to estimate the best nucleotide substitution model for each codon position. For first position and third position the best model determined was GTR+I+ Γ substitution model according to Akaike’s Information Criterion (AIC). The best model determined for the second position was GTR+I. Bayesian inference (BI) analysis was carried out using Mr.Bayes 3.2.4 [63,64]. Starting from random trees, we initiated two individual runs of four Markov-chain Monte Carlo (MCMC) chains, three hot and one “cold,” with ten million iterations each, sampling every 1000 generations. Each run resulted in 10,000 trees and converged on the same topology. The first 25% of samples from the cold chain were conservatively discarded as our “burn-in” percentage. Tracer 2.0 was used to verify that stationarity had been reached (http://beast.bio.ed.ac.uk). A 50% majority-rule consensus tree was generated from the remaining trees and Bayesian posterior probability (pp) values were used as support.
For maximum likelihood analysis we used RAXML [65–67]. First, we partitioned the data by codon position and then set the models to GTR+I+Γ for all positions because of limitations in RAXML. We used the default settings for RAXML and calculated standard bootstrap values based on 1000 replicates, which are used for branch support.
We tested the relationship between geographic distance and genetic distance for M. oberthueri, M. voeltzkowi, M. mirror, M. barrybressleri and M. rogeri. First we created a pairwise genetic distance matrix using the K2P distance model in Mega 6.0 for each species [58]. We used GenAlEx 6.501 [68,69] to create pairwise geographic distance matrices and ran a Mantel test for each species using the genetic distances from Mega 6.0. Species that had a significant Mantel test, indicating there was a relationship between genetic distance and geographic distance, were included in a linear regression analysis. We used JMP® Version 11 (SAS Institute Inc., Cary, NC, 1989–2007) to run linear regressions to determine whether the slopes were significantly different among species with alternate colonial reproductive strategies.
In order to verify the species distinctions generated in the mitochondrial phylogeny and to obtain better resolution for deeper nodes in the genus, we also produced a nuclear phylogeny from a concatenated data set which includes a subsample of 19 Mystrium and two outgroup accessions and is 1948 basepairs (bp) in length. Samples for this analysis were selected as representatives of each of the species lineages identified in the mitochondrial analysis. Where possible samples representing diverse localities within each species were used to generate sequence data, although not all samples produced sequence across all loci. With a concatenated data set each gene is likely to have different sequence characteristics and rates of evolution. Therefore, we first used jModelTest2 [62] to estimate the substitution model parameters for the four genes individually, then partitioned the data in Mr.Bayes 3.2.4 [63,64], and incorporated the models of gene evolution (Table 3). Bayesian inference (BI) analysis was carried out using the best nucleotide substitution models according to both Akaike’s Information Criterion (AIC) and the likelihood-ratio test (LRT) from jModelTest2. There was no difference between the topology or Bayesian posterior probability (pp) values so only the tree based on AIC-evaluated models is presented.
Bayesian inference (BI) analysis was carried out using Mr.Bayes 3.2.4 [64,64] on CIPRES science gateway [61]. Starting from random trees, we initiated two individual runs of four Markov-chain Monte Carlo (MCMC) chains, three hot and one “cold,” with fifty million iterations each and trees were sampled every 2,000 generations, with an initial seed tree at 12,000 iterations. The analysis did not need to run longer than 50,000,000 generations, because at the end of the run the average standard deviation among topologies was below 0.000001. Each run resulted in 20,000 trees and converged on the same topology. The first 25% of samples from the cold chain were conservatively discarded as “burn-in” percentage. Tracer 2.0 was used to verify that stationarity had been reached (http://beast.bio.ed.ac.uk). A 50% majority-rule consensus tree was generated from the remaining trees. Bayesian posterior probability (pp) values, which represent the percentage of trees sampled after burn-in that recover any particular clade on the tree, were calculated as measures of support.
Maximum likelihood analysis for the nuclear phylogeny was carried out using RAxML [65–67] on the CIPRES science gateway [61]. We partitioned the data by gene and set the models to GTR+I+Γ for all positions because of limitations in RAXML. We used the default settings for RAXML and calculated standard bootstrap values based on 1000 replicates, which are used for branch support. Since RAxML is limited to incorporation of a single model of evolution, we also ran a ML bootstrap analysis using TREEFINDER [70]. We partitioned the data by gene with a data filter, then initiated a ML bootstrap analysis of 1000 replicates and incorporated the models of gene evolution suggested by jModelTest2 [62] and the AIC. A 50% majority-rule consensus tree was generated with bootstrap support. The topologies were consistent between the two methods, with little variation in the support values, and so only the tree produced using RAxML is presented.
Results
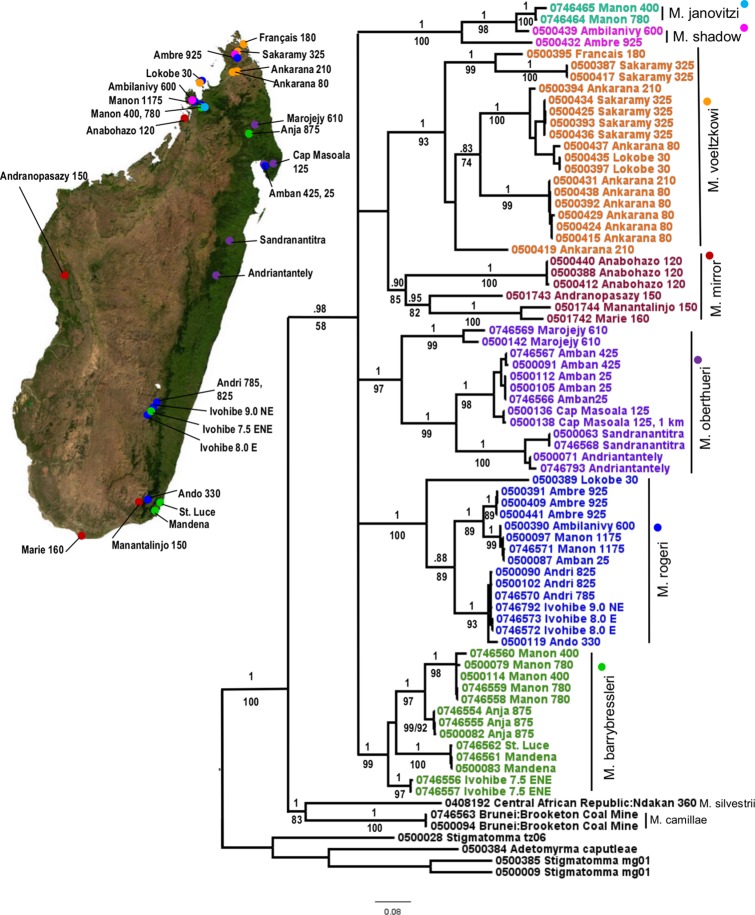
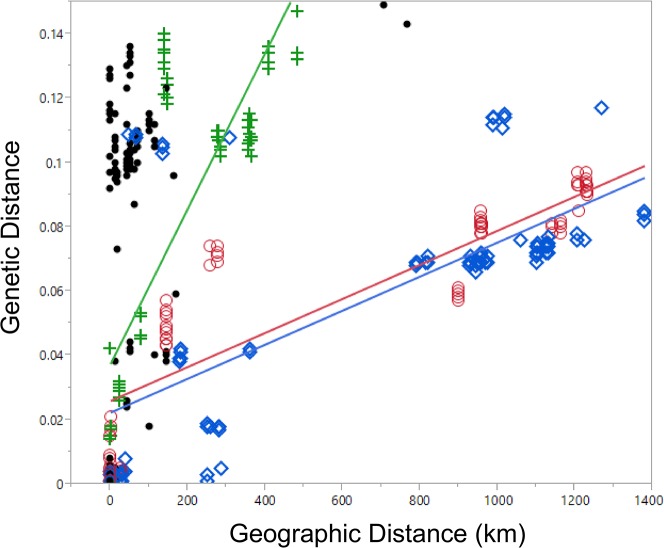
A mitochondrial phylogeny of Mystrium species from maximum likelihood analysis, with support from the Bayesian analysis, is presented (Fig 1). Phylogenetic support for published morphospecies and for mtDNA subclades within morphospecies was high across all three methods, despite a few minor differences in topology at distal branches. Average mtDNA congeneric pairwise divergence is 14.28%. Sequences are heavily AT biased; this is normally the case with insect mtDNA [71]. We found no evidence of paralogous nuclear mitochondrial-like genes in the mtDNA data set after eliminating suspect long-branch sequences using long PCR, gel extraction and re-amplification (see Methods). Comparisons of the average K2P divergence values among and within species are listed in Table 4. Average sequence divergence values between Mystrium species ranged from 13.44% to 17.67%, whereas average sequence divergence values within Mystrium species ranged from 5.23% to 11.69% and are lowest for species which use independent colony foundation. Comparisons of average K2P divergence values within and between subclades of species are listed in Table 5. The slopes of the relationship of genetic distance to geographic distance between ICF and DCF species was significantly different (F1 = 111.036, p < .0001). The slopes of the relationship of genetic distance to geographic distance were not significantly different among the ICF species M. barrybressleri and M. rogeri (F1 = 0.000, p = 0.995) (Fig 2).
Fig 1. Mitochondrial Phylogeny of Mystrium.
Maximum likelihood phylogeny based on 790 bp of CO1, summarized as a consensus tree in RaxML. Support values above branches represent Bayesian posterior probabilities (pp), those below branches ML bootstrap. Scale bar shows nucleotide changes per base pair. Taxa are labeled with specimen codes and locality codes. Symbols beside species names on the phylogeny correspond to distribution markers in the adjacent map of Madagascar.
Table 4. species of Mystrium calculated using the K2P model of evolution in Mega version 6.0.
| M. oberthueri | M. voeltzkowi | M. rogeri | M. mirror | M. barrybressleri | M. janovitzi | M. shadow | M. camillae | M. silvestrii | |
|---|---|---|---|---|---|---|---|---|---|
| Average within | 8.19% | 8.16% | 5.23% | 11.69% | 6.92% | 1.17% | 9.27% | 0.13% | |
| M. oberthueri | |||||||||
| M. voeltzkowi | 16.01% | ||||||||
| M. rogeri | 15.70% | 15.35% | |||||||
| M. mirror | 16.61% | 15.59% | 17.10% | ||||||
| M. barrybressleri | 14.96% | 15.40% | 14.48% | 16.26% | |||||
| M. janovitzi | 15.41% | 14.70% | 13.95% | 16.25% | 14.26% | ||||
| M. shadow | 15.54% | 15.93% | 15.03% | 16.19% | 14.06% | 6.81% | |||
| M. camillae | 15.86% | 16.08% | 14.04% | 17.67% | 13.93% | 13.43% | 13.44% | ||
| M. silvestrii | 15.96% | 15.32% | 13.87% | 17.61% | 13.65% | 14.60% | 15.96% | 12.08% |
Table 5. Cytochrome c Oxidase I sequence divergence values within and between subclades of Mystrium morphospecies calculated using the K2P model of evolution in Mega version 6.0.
| M. rogeri | North | South | ||
|---|---|---|---|---|
| Within subclade | 2.47% | 0.68% | ||
| North | ||||
| South | 7.23% | |||
| M. barrybressleri | Manon | Anja | Ivohibe | Littoral |
| Within subclade | 0.99% | 0.34% | 0.51% | 0.34% |
| Manon clade | ||||
| Anja clade | 4.80% | |||
| Ivohibe | 8.09% | 5.92% | ||
| Mandena/St. Luce | 9.27% | 7.98% | 7.14% | |
| M. voeltzkowi | Clade 1 | Clade 2 | Complex | |
| Within subclade | 0.39% | 2.69% | 4.86% | |
| Clade 1 | ||||
| Clade 2 | 10.59% | |||
| Complex | 12.44% | 11.46% | ||
| M. oberthueri | Marojejy | Pennisula | So. Toam | |
| Within subclade | 4.17% | 1.92% | 3.52% | |
| Marojejy | ||||
| Pennisula | 12.90% | |||
| So. Toamasina | 13.63% | 10.80% | ||
| M. mirror | Anabohazo 120 | Manatalinjo 150 | Marie 160 | Andranopasazy150 |
| Within subclade | 0.34% | |||
| Anabohazo 120 | ||||
| Manatalinjo 150 | 16.01% | |||
| Marie 160 | 16.84% | 5.85% | ||
| Andranopasazy 150 | 16.52% | 14.88% | 14.26% |
Fig 2. Genetic Distance by Geographic Distance.
Linear regression of the relationship of genetic distance to geographic distance by species. The slopes of the lines of the ICF species M. rogeri (blue diamonds) and M. barrybressleri (red circles) are not significantly different from each other (F1 = 0.000, p = 0.995), but are significantly different from the DCF species M. oberthueri (green cross) (F1 = 111.036, p < .0001). The DCF species M. voeltzkowi and M. mirror are represented by black dots and were not included in the regression analysis.
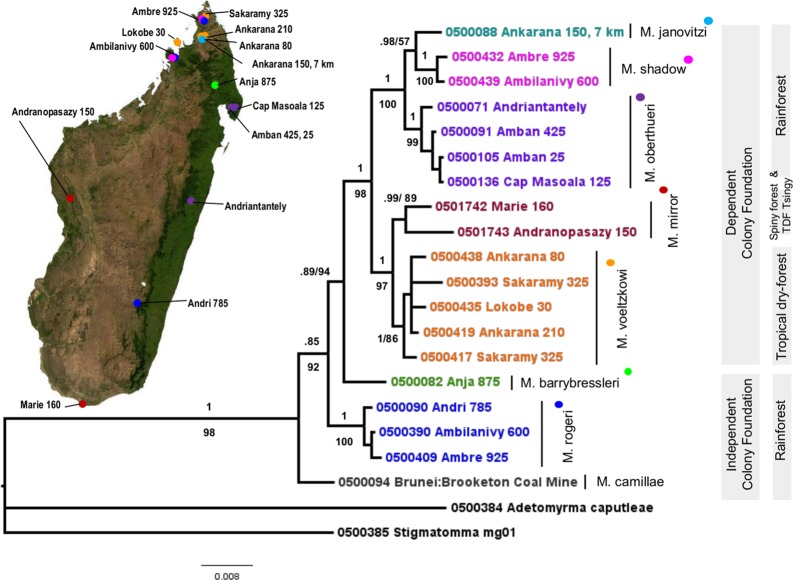
A nuclear phylogeny of Mystrium species constructed from the maximum likelihood analysis, with support from Bayesian analysis, is also presented (Fig 3). Mystrium is a monophyletic group that is well supported in all analyses, in the mitochondrial phylogeny (1.0 pp, 100% MLbs, 99% NJbs) as well as the nuclear phylogeny (1.0 pp, 98% MLbs). All species of Mystrium endemic to Madagascar form their own clade in the mitochondrial phylogeny (.98 pp, 58% MLbs, 72% NJbs) as well as the nuclear phylogeny (.85 pp, 92% MLbs) and the off-island species M. camillae (nucDNA/mtDNA) and M. silvestrii (mtDNA only) are basalmost. The species that reproduce by dependent colony foundation, M. shadow, M. janovitzi, M. oberthueri, M. mirror, and M. voeltzkowi form a distinct monophyletic group in the nuclear phylogeny (1.0 pp, 98% MLbs).
Fig 3. Nuclear Phylogeny of Mystrium.
Maximum likelihood phylogeny based on 1948 bp of the four nuclear genes Wg, Abd-A, LW Rh, 28s, summarized as a consensus tree in RaxML. Support values above branches represent Bayesian posterior probabilities (pp), those below branches ML bootstrap. Scale bar shows nucleotide changes per base pair. Colonial reproductive strategy and habitat type are labeled in adjacent bars.
Mystrium rogeri form a well-supported mtDNA clade (1.0 pp, 100% MLbs, 97% NJbs) and a monophyletic group in the nuclear phylogeny (1.0 pp, 100% MLbs). Sequence divergence within M. rogeri range from a low of 0.127% to a high of 10.759%. In the mtDNA phylogeny, M. rogeri fall into two clades: a northern clade that is spread across the mountains from the northwest to the northeast encompassing Montagne d’Ambre, R.S. Manongarivo, Foret d’Ambilanivy and down the Masoala peninsula, and a southern clade that is spread across south-central to southern Madagascar (Table 5). An additional mtDNA lineage of M. rogeri is from the island of Nosy Be (Lokobe 30).
Mystrium barrybressleri form a well-supported mtDNA clade (1.0 pp, 99% MLbs, 100% NJbs) and a distinct evolutionary lineage in the nuclear phylogeny. Sequence divergence within M. barrybressleri range from a low of 0.127% to a high of 9.022%. In the mtDNA phylogeny, M. barrybressleri contain four subclades restricted by locality: a northwest clade (R.S. Manongarivo), northeast clade (Res. Anjanaharibe-Sud), a south-central clade (R.S. Ivohibe), and a southeast clade (Mandena/St. Luce) (Table 5).
Mystrium oberthueri form a well-supported mtDNA clade (1.0 pp, 97% MLbs, 98% NJbs) and a monophyletic group in the nuclear phylogeny (1.0 pp, 99% MLbs). Sequence divergence within M. oberthueri range from a low of 0.127% to a high of 13.165%. In the mtDNA phylogeny, M. oberthueri contain three subclades: a Marojejy clade (Marojejy 610), a Masoala peninsula clade (Amban 25, Amban 425, Cap Masoala 125), and a southern Toamasina clade (Andriantantely, Sandranantitra) (Table 5).
M. shadow and M. janovitzi form a clade in the mtDNA (1.0 pp, 100% MLbs) and nuclear phylogeny (.98 pp, 57% MLbs). Mystrium oberthueri forms a well-supported clade with M. shadow and M. janovitzi in the nuclear phylogeny (1.0 pp, 100% MLbs).
Mystrium mirror form a well-supported clade in the mtDNA (.90 pp, 85% MLbs, 73% NJbs) and nuclear phylogeny (.99 pp, 89% MLbs). Sequence divergence values within M. mirror range from a low of 0.0127% to a high of 16.84%. In the mtDNA phylogeny, M. mirror form one subclade (Forêt d'Anabohazo) and several mtDNA lineages, which were collected at widely dispersed localities, including tropical dry-forest on and off tsingy limestone formations and in spiny desert localities (Table 5). Mystrium voeltzkowi and M. mirror are sister taxa in both the mitochondrial (1.0 pp, 94% MLbs, 76% NJbs) and nuclear phylogenies (1.0 pp, 97% MLbs).
Mystrium voeltzkowi form a well-supported mtDNA clade (1.0 pp, 93% MLbs, 93% NJbs) and form a monophyletic group in the nuclear phylogeny (1.0 pp, 86% MLbs). Sequence divergence values of M. voeltzkowi range from a low of 0.127% to a high of 12.674%. Mystrium voeltzkowi was collected in the northern highlands at sites within the Réserve Spéciale de l'Ankarana (Ankarana 210, Ankarana 80), or within the Réserve Spéciale d'Ambre (Sakaramy 325), with two more specimens from the coastal island of Nosy Be (Lokobe 30), and one from Montagne des Français (Français 180, mtDNA phylogeny only). Within the mtDNA phylogeny M. voeltzkowi form several clades: ‘clade 1’ contains haplotypes collected at Ankarana 80 and Ankarana 210, ‘clade 2’ contains haplotypes collected at Ankarana 80, Ankarana 210, Sakaramy 325, and Lokobe 30 (Table 5). An additional haplotype collected from Ankarana 210 forms an independent mtDNA lineage. A third mtDNA clade (Sakaramy 325 and Français 180) is made up of specimens that were morphologically keyed out to ‘M. voeltzkowi complex’ by taxonomists at the California Academy of Sciences. Some samples collected within Réserve Spéciale de l'Ankarana (Ankarana 210, Ankarana 80), or within the Réserve Spéciale d'Ambre (Sakaramy 325), have high mtDNA sequence divergence among haplotypes collected at the same site (8.9%-12%) and low divergence among haplotypes collected at sites 71–82 kilometers apart (0.4%-2.5%).
Discussion
We found unusually high levels of mtDNA sequence divergence within and among the species studied, with subclades within species showing divergence levels greater than typically found among ant species. We found that reproductive strategy appears to have had an effect on how Mystrium species female lineages are associated with large-scale habitat distinctions and various topographical features, (i.e. their phylogeography). Mystrium species female lineage distribution patterns provide support for both models of ecogeographic constraints [6,9], thus appearing to account for lineage diversification in a manner similar to what was found for vertebrate species. However, in some cases, microgeographic population structure was found to be associated with species which appear to have been impacted by localized habitat differences on a scale much smaller than that found in vertebrates.
High Levels of Divergence among Mystrium Lineages
We recovered strong support in both the nuclear and mitochondrial phylogenies for the species of Mystrium included in this study, corresponding with the morphospecies described in the most recent revision of the genus [40]. However, variation between discontinuous evolutionary groupings (subclades) within some morphospecies of Mystrium was many times higher than the 2% or 3% mitochondrial sequence divergence threshold that was used to successfully identify the majority of morphospecies of ants in Madagascar [19] and the 1.9% conspecific mitochondrial sequence divergence value measured among ants of North America [19]. Thus, the mtDNA intraspecific sequence divergence values within some lineages of Mystrium are values well above those levels typically seen among species of ants. The large amount of intraspecific phenotypic variation found in Mystrium may be a reflection of this high genetic divergence. Further work using nuclear loci and expanded sampling will be necessary to confirm the extraordinarily high levels of divergence within species of Mystrium.
Within M. mirror each geographic location consists of a unique clade (Anabohazo 120) or mtDNA lineage (Manatalinjo 150, Marie 160, Andranopasazy 150) and average intraspecific sequence divergence was 11.69%. Intraspecific sequence divergence among the Anabohazo clade and other evolutionary lineages of M. mirror (16%) rivals sequence divergence levels between Mystrium species. Sequence divergence among clades within M. oberthueri was also high (10.8% to 13.6%). Comparisons between the Marojejy clade and the remaining M. oberthueri clades yielded the highest intraspecific divergence, likely due to the high mountains and sheer cliffs in the region. These formations are formed from ancient granites and gneisses, materials very resistant to weathering. Our results provide evidence of geographically segregated clades with high phylogenetic divergence within two DCF species of Mystrium.
Geographically segregated clades with high sequence divergence in species with ergatoid queens have been found in previous studies with ants [20]. Deep CO1 divergences occur between different collection localities in the ant genera Odontomachus and Anochetus (O. coquereli, A. goodmani, A. boltoni), which also have ergatoid queens [20,42]. Fisher and Smith (2008) predicted that species with ergatoid queens will have high mtDNA sequence divergence among clades at geographically localized areas because of their reduced dispersal ability. Thus, female-limited dispersal might lead to an extremely site-specific phylogeographic signal in ergatoid queen species.
Reproductive Strategy Effect on Phylogeography
Mystrium species that reproduce by dependent colony foundation form a distinct monophyletic group (Fig 3). The species that reproduce by independent colony foundation, M. barrybressleri and M. rogeri, are basal. This suggests that the colony foundation strategy of DCF has arisen once within Mystrium and is the derived condition. Dependent colony foundation has been determined to be the derived state in other ants [42] and the condition of the loss of wings is thought to be irreversible [47].
The phylogeographic patterns support the different levels of female dispersal by species that reproduce via independent colony foundation versus species that reproduce via dependent colony foundation. Intraspecific mitochondrial sequence divergence in M. rogeri and M. barrybressleri is lower than intraspecific sequence divergence of the Malagasy Mystrium reproducing by DCF (Figs 1 and 2). The greater vagility afforded by having winged queens leads to shallower mtDNA divergence within ICF species. M. oberthueri was used as a proxy for DCF species when we compared the slopes of the lines between ICF and DCF species that showed a pattern of isolation by distance. Within the DCF species M. mirror divergence values appear to reach a saturation level, making an estimation of the relationship between genetic distance and geographic distance problematic without further sampling (r2 = 0.749). We excluded M. voeltzkowi from the analysis because within M. voeltzkowi there was also no significant correlation between genetic distance and geographic distance (r2 = 0.0259). Instead, geographic barriers to gene flow leading to microendemism in the region may be generating a unique pattern among haplotypes, as discussed below.
We were not surprised to find lower intraspecific divergence values among female lineages in species that reproduce by independent colony foundation because gene flow in ICF species is facilitated by the ability of winged females to traverse greater distances and cross barriers. These same barriers would effectively reduce gene flow in species which have females that reproduce via DCF. In heterogeneous environments consisting of a set of discrete patches that persist for a finite time, movement between patches may only occur by flight, which for females only occurs in species practicing ICF. Conversely, Mystrium with ergatoid queens displayed high intraspecific sequence divergence values between conspecifics from different localities. Colonies that practice DCF are limited by terrestrial dispersal from their natal colony and the necessary accompaniment of sister workers. The short-range dispersal of ergatoid queen colonies may have had a severe effect on female gene flow [42, 44, 47, 72, 73]. In a study using mtDNA this low level of gene flow was observed across multiple spatial scales in the population genetic structure of the ant species Diaccamma cyaneiventre [72]. High divergence levels are likely a product of minimal genetic exchange among populations. As such, sequence divergence levels within DCF species are expected to be higher.
Although the influence of reproductive strategy appears to impact female dispersal, its impact on each species as a whole remains uncertain because males in these species may be able to disperse more broadly. A rigorous study of within-species variation using nuclear loci would be necessary to elucidate the impact of reproductive strategy on speciation as a whole.
Large-Scale East-West Ecogeographic Constraints
With a better understanding of the patterns of female dispersal in Mystrium, we evaluated a vertebrate-derived biogeographic model of species divergence for ants, which by comparison are likely to have reduced levels of dispersal. Among the largest-scale patterns proposed was that of ecogeographic constriants separating the more mesic rainforest on the eastern side of the island from areas of less mesic conditions of tropical dry forest and spiny desert found on the western side of the island (Figs 1 and 3). The sharp ecological distinction between the humid eastern rainforest and arid western habitat are thought to constitute a barrier to gene flow. Our results suggest that this climatological barrier has caused a basal split between eastern and western clades in the phylogeography of Mystrium species. Mystrium oberthueri and M. shadow are both sister taxa and rainforest-restricted species, suggesting that these two species prefer mesic conditions and are hylophilous. The range of M. obertheuri is on the eastern side of the island and M. shadow is restricted to rainforest habitat that is part of the eastern climatological rainforest zone which stretches across the northern part of the island. Mystrium mirror and M. voeltzkowi are sister taxa restricted to less mesic habitats, and range from spiny desert in the arid south or tropical dry forest on the western and northwestern side of the island. Mystrium mirror has multiple evolutionary lineages that fall out according to habitat (Fig 1). Climate has been correlated with ant diversity, with a combination of temperature and precipitation often representing the best two climatic predictors for diversity of litter-dwelling ants [74]. Thus, east-west phylogenetic splits among species of Mystrium support the model for speciation due to ecogeographic constraints proposed for vertebrate taxa [6,9].
Large-Scale North-South Ecogeographic Constraints
A phylogenetic split was found among haplotypes collected in the north and the south in the ICF species M. barrybressleri and M. rogeri. Although it may be an artifact of geographic distance and limited sampling in the center of the island for these species, this split may be related to the island’s mountain ranges. Mountains are thought to have acted as refugia for humid forests during periods of drier climate [75] and offered opportunities for allopatric speciation of populations that remained isolated on mountain tops [10]. The diversification of northern and southern populations of M. rogeri and M. barrybressleri during paleoclimatic oscillations may have led to an intraspecific division among haplotypes. The waning and waxing of different climatic regimes during the quaternary is thought to have led to periods when montane vegetation descended a notable distance along the flanks of mountains during warmer and wetter periods or was isolated around summits during cooler and drier periods [76]. The mountains may act as species pumps [27], supplying species that have undergone montane diversification to recolonize surrounding lowland habitat in areas where climatic conditions prevented lowland habitation for a period of geologic time. Having the mountains of the north serve as species pumps for northern lowlands and the mountains of the south serve as species pumps for southern lowlands could explain the deep phylogenetic split between northern and southern haplotypes of M. rogeri and M. barrybressleri. Thus, phylogenetic splits among north-south subclades within Mystrium potentially support both Yoder and Heckmans (2006) revised model of ecogeographic constraints along the long axis of the island and similar patterns observed in other vertebrate species [28,29]. Further sampling in the intermediate latitudinal range of the island will be essential to test where such a geographic limit between northern and southern clades in the ICF species M. barrybressleri and M. rogeri lies, and how intermediate samples will impact the phylogeographic patterns of these species.
Microendemism: Littoral Forest Habitat
One pattern found did not correspond to established diversification models found in vertebrates: cases of lineage diversification associated with small-scale habitat distinctions. In one of these cases, larger levels of genetic differentiation were found among the southern clades of M. barrybressleri than among the southern clades of M. rogeri, both ICF species. One possible explanation for this difference is that some M. barrybressleri haplotypes were collected from the Mandena/St. Luce locality, which is littoral rainforest habitat (Table 2, Fig 1). Littoral rainforests are narrow patches of coastal forest known to contain unique assemblages of plants and animals [77], and this habitat difference may be a factor in generating an isolated and distinct southeastern Madagascar M. barrybressleri clade [16]. A similar pattern has been reported previously in other species of Malagasy ants [16] and suggests that the littoral forest habitat may provide a unique habitat harboring microgeographic scales of localized endemism in less vagile organisms.
Microendemism:Tsingy Habitat
A second pattern of localized patterns of microgeographic distinction among clades occurs in the DCF species M. voeltzkowi and is associated with the presence of limestone outcrops (tsingy) throughout or surrounding the tropical dry forest in the northern region of Madagascar.
Within M. voeltzkowi, some haplotypes collected at the same locality show a great deal of mtDNA genetic divergence. The Ankarana tsingy in the northern part of Madagascar appears in patches with a total extension of about 200 km2. Here, three M. voeltzkowi haplotypes from the Réserve Spéciale de l’Ankarana (Ankarana 210) do not group together, and have high (i.e. species level 8.9%–10%) sequence divergence among them (Fig 1). Other M. voeltzkowi collected together at a different site within the Réserve Spéciale de l'Ankarana (Ankarana 80) also show high sequence divergence values (10.3%). Haplotype sequence divergence is even greater among M. voeltzkowi collected at the same locality in the Forêt d’Ambre (Sakaramy 325) (12%). In contrast, some M. voeltzkowi collected at sites 71–82 km apart have relatively low sequence divergence (0.4%–2.5%), as among haplotypes collected at Ankarana 80 and Sakaramy 325 (sequence divergence <1%). Sequence divergence values, therefore, do not relate to the isolation of populations because of distance, but rather the amount of time the populations have been separated, which may have been caused by some other isolating effect such as a physical barrier to gene flow.
Local vicariance events in the tropical dry forest of the north may have caused the long-term isolation of populations of M. voeltzkowi. Tsingy develops due to direct rainfall on limestone which is very well bedded, clean, has low porosity, and is full of joints [78]. Just as the formation of tsingy is continuous, its destruction is continuous as well; pinnacles (towers of old tsingy) many meters high may fall as side slopes crack or dissolve. Tsingy is an area of constant change, and likely the site of small-scale habitat fragmentation, as patches of deciduous forest exist throughout and around the limestone. For M. voeltzkowi, changes in the landscape likely have presented effective barriers to gene flow. Segregation of ant populations, time and time again, over the evolution of the Ankarana tsingy is likely to have been the driving factor in the diversification of this species. Likewise, it explains why haplotypes collected farther apart in continuous dry-deciduous forest habitat are more genetically similar than those found closer together in tsingy.
The opposite pattern has been predicted for populations with restricted queen dispersal, especially when measured using maternally inherited mitochondrial genes (mtDNA genes) [79]. Population viscosity is the phenomenon of greater genetic similarity among colonies that are physically nearby in a continuous population than among colonies located further away [79]. This pattern has been observed in species with social insect queens performing DCF that have restricted dispersal, particularly in species without aerial dispersal [42, 72, 73]. Although limited queen dispersal may cause genetic population viscosity, especially when measured using a maternally inherited locus (mtDNA), nuclear genes inherited from both parents may be unaffected, because males can be efficient dispersers and thus mitigate the effects of restricted queen dispersal. Similarly, it would be instructive to include a series of nuclear genes in a broader study to test the patterns of divergence generated here for M. voeltzkowi and determine if the dispersal of males would degrade the remarkably high divergences among individuals collected at the same localities. However, within the Ankarana tsingy, populations of M. voelzkowi certainly do not conform to this pattern of population viscosity predicted for colonies of social insects with limited dispersal.
Conclusions
Here we showed that there were contrasting phylogeographic patterns which occurred in species with differential reproductive abilities when measured using a maternally inherited locus. These patterns are likely a reflection of the influence of colonial reproductive strategy on diversification within the genus Mystrium. Using molecular methods we confirmed Mystrium morphological species delineation, and demonstrated that the reproductive strategy of dependent colony foundation has likely arisen once. Moreover, we tested one of the prominent proposed mechanisms for vertebrate species diversification with a small colonial invertebrate model and found that similar patterns occurred in species of Mystrium ants. However, we also discovered cases of fine-scale micro-endemism in both ICF species (littoral forest clade, M. barrybressleri) and DCF species (Ankarana tsingy, M. voeltzkowi). Divergence patterns of M. voeltzkowi, in particular, suggested that the diversification of female lineages associated with the DCF strategy appear to be especially sensitive to ecological perturbations. In conclusion, this work demonstrates that both testing divergence mechanisms in species with a range of dispersal capabilities and taking into account relevant information regarding species-specific biological mechanisms and life history traits are both invaluable for building a more universal understanding of species diversification mechanisms for regions of high endemism such as Madagascar.
Acknowledgments
We thank the Sonoma State University DNA Analysis Facility and the California Academy of Sciences for providing facilities to gather these data. We thank Gary D. Ouellette and Kristy Deiner for valuable contributions to the data collection. We thank Phil Ward and Brian Lavin for contributions to the evaluation of the data. Moreover, the fieldwork on which this study is based could not have been completed without the gracious support of the Malagasy people and the Arthropod Inventory Team (Balsama Rajemison, Jean-Claude Rakotonirina, Jean-Jacques Rafanomezantsoa, Chrislain Ranaivo, Hanitriniana Rasoazanamavo, Nicole Rasoamanana, Clavier Randrianandrasana, Njaka Ravelomanana, and Manoa Ramamonjisoa).
Data Availability
Datasets and phylogenies are publically available on FigShare: http://dx.doi.org/10.6084/m9.figshare.1324480 and GenBank accession numbers for all specimens (KR063356-KR063489, KP280011-KP280031, KU3612995-KU361299) are available in Table 2.
Funding Statement
This study was supported by the National Science Foundation under Grant No. DEB-0072713 (BLF), DEB-0344731 (BLF), DEB-0842395(BLF), and DEB-9981667 (BLF and DJG). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ali JR, Aitchison JC. Gondwana to Asia: Plate tectonics, paleogeography and the biological connectivity of the Indian sub-continent from the Middle Jurassic through latest Eocene (166–35 Ma). Earth-Sci Rev. 2008;88: 145–166. 10.1016/j.earscirev.2008.01.007 [DOI] [Google Scholar]
- 2.Goodman SM, Benstead JP. Updated estimates of biotic diversity and endemism for Madagascar. Oryx. 2005;39: 73–77. 10.1017/S0030605305000128 [DOI] [Google Scholar]
- 3.Yoder AD, Olson LE, Hanley C, Heckman KL, Rasoloarison R, Russell AL, et al. A multidimensional approach for detecting species patterns in Malagasy vertebrates. Proc Natl Acad Sci U S A. 2005;102: 6587–6594. 10.1073/pnas.0502092102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu Rev Ecol Evol Syst. 2006;37: 405–431. 10.1146/annurev.ecolsys.37.091305.110239 [DOI] [Google Scholar]
- 5.Kremen C, Cameron A, Moilanen A, Phillips SJ, Thomas CD, Beentje H, et al. Aligning conservation priorities across taxa in Madagascar with high-resolution planning tools. Science. 2008;320: 222–226. 10.1126/science.1155193 [DOI] [PubMed] [Google Scholar]
- 6.Yoder AD, Rasoloarison RM, Goodman SM, Irwin JA, Atsalis S, Ravosa MJ, et al. Remarkable species diversity in Malagasy mouse lemurs (Primates, Microcebus). Proc Natl Acad Sci. 2000;97: 11325–11330. 10.1073/pnas.200121897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodman SM, Ganzhorn JU. Biogeography of lemurs in the humid forests of Madagascar: the role of elevational distribution and rivers. J Biogeogr. 2004;31: 47–55. 10.1111/j.1365-2699.2004.00953.x [DOI] [Google Scholar]
- 8.Wilmé L, Goodman SM, Ganzhorn JU. Biogeographic evolution of Madagascar’s microendemic biota. Science. 2006;312: 1063–1065. 10.1126/science.1122806 [DOI] [PubMed] [Google Scholar]
- 9.Yoder AD, Heckman KL. Mouse lemur phylogeography revises a model of ecogeographic constraint in Madagascar Primate Biogeography. Springer; 2006. pp. 255–268. [Google Scholar]
- 10.Wollenberg KC, Vieites DR, Van Der Meijden A, Glaw F, Cannatella DC, Vences M. Patterns of endemism and species richness in Malagasy cophyline frogs support a key role of mountainous areas for speciation. Evolution. 2008;62: 1890–1907. 10.1111/j.1558-5646.2008.00420.x [DOI] [PubMed] [Google Scholar]
- 11.Townsend TM, Vieites DR, Glaw F, Vences M. Testing species-level diversification hypotheses in Madagascar: The case of microendemic Brookesia leaf chameleons. Syst Biol. 2009;58: 641–656. 10.1093/sysbio/syp073 [DOI] [PubMed] [Google Scholar]
- 12.Pearson RG, Raxworthy CJ. The evolution of local endemism in Madagascar: watershed versus climatic gradient hypotheses evaluated by null biogeographic models. Evolution. 2009;63: 959–967. 10.1111/j.1558-5646.2008.00596.x [DOI] [PubMed] [Google Scholar]
- 13.Wiens JA. Spatial scaling in ecology. Funct Ecol. 1989;3: 385–397. 10.2307/2389612 [DOI] [Google Scholar]
- 14.Hortal J, Roura-Pascual N, Sanders N, Rahbek C. Understanding (insect) species distributions across spatial scales. Ecography. 2010;33: 51 10.1111/j.1600-0587.2009.06428.x [DOI] [Google Scholar]
- 15.Lawton JH, Bignell DE, Bolton B, Bloemers GF, Eggleton P, Hammond PM, et al. Biodiversity inventories, indicator taxa and effects of habitat modification in tropical forest. Nature. 1998;391: 72–76. 10.1038/34166 [DOI] [Google Scholar]
- 16.Fisher BL, Girman DJ. Biogeography of ants in eastern Madagascar. In: Lourenço WR, Goodman SM, editors. Diversité et endémisme à Madagascar. Mémoires de la Sociétéde Biogéographie, Paris; 2000. pp. 331–344.
- 17.Kaspari M, Majer JD. Using ants to monitor environmental change In: Agosti D, Majer JD, Alonso LE, Schultz TR, editors. Ants: Standard methods for measuring biodiversity. Washington, DC: Smithsonian Institution Press; 2000. pp. 89–98. [Google Scholar]
- 18.Underwood EC, Fisher BL. The role of ants in conservation monitoring: if, when, and how. Biol Conserv. 2006;132: 166–182. 10.1016/j.biocon.2006.03.022 [DOI] [Google Scholar]
- 19.Smith MA, Fisher BL, Hebert PDN. DNA barcoding for effective biodiversity assessment of a hyperdiverse arthropod group: the ants of Madagascar. Philos Trans R Soc B Biol Sci. 2005;360: 1825–1834. 10.1098/rstb.2005.1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher BL, Smith MA. A Revision of Malagasy Species of Anochetus Mayr and Odontomachus Latreille (Hymenoptera: Formicidae). PLoS ONE. 2008;3: e1787 10.1371/journal.pone.0001787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreau CS. Inferring ant evolution in the age of molecular data (Hymenoptera: Formicidae). Myrmecol News. 2009;12: 201–210. [Google Scholar]
- 22.Vences M, Wollenberg KC, Vieites DR, Lees DC. Madagascar as a model region of species diversification. Trends Ecol Evol. 2009;24: 456–465. 10.1016/j.tree.2009.03.011 [DOI] [PubMed] [Google Scholar]
- 23.Goodman SM, Benstead JP, editors. Natural history of Madagascar Chicago: University of Chicago Press; 2003. [Google Scholar]
- 24.Wells N. Some hypotheses on the Mesozoic and Cenozoic paleoenvironmental history of Madagascar In: Goodman SM, Benstead JP, editors. The natural history of Madagascar. University of Chicago Press; 2003. pp. 16–34. [Google Scholar]
- 25.Koechlin J. Flora and vegetation of Madagascar In: Battistini R, Richard-Vindard G, editors. Biogeography and ecology in Madagascar. Springer; Netherlands; 1972. pp. 145–190. [Google Scholar]
- 26.Schatz GE. Endemism in the Malagasy tree flora. Divers Endem Madag. 2000;1: 1–8. [Google Scholar]
- 27.Smith SA, De Oca ANM, Reeder TW, Wiens JJ. A phylogenetic perspective on elevational species richness patterns in Middle American treefrogs: Why so few species in lowland tropical rainforests? Evolution. 2007;61: 1188–1207. 10.1111/j.1558-5646.2007.00085.x [DOI] [PubMed] [Google Scholar]
- 28.Boumans L, Vieites DR, Glaw F, Vences M. Geographical patterns of deep mitochondrial differentiation in widespread Malagasy reptiles. Mol Phylogenet Evol. 2007;45: 822–839. 10.1016/j.ympev.2007.05.028 [DOI] [PubMed] [Google Scholar]
- 29.Raselimanana AP, Noonan B, Karanth KP, Gauthier J, Yoder AD. Phylogeny and evolution of Malagasy plated lizards. Mol Phylogenet Evol. 2009;50: 336–344. 10.1016/j.ympev.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 30.Brown WL Jr. I960. Contributions toward a reclassification of the Formicidae, ш. Tribe Amblyoponini Mus Сотр Zool Bull. 122: 145–230. [Google Scholar]
- 31.Wilson EO. The insect societies Cambridge, Mass: Harvard University Press; 1971. [Google Scholar]
- 32.Hölldobler B, Wilson EO. The Ants, 1990. Harv Belknap Camb. [Google Scholar]
- 33.Ward PS. Adetomyrma, an enigmatic new ant genus from Madagascar (Hymenoptera: Formicidae), and its implications for ant phylogeny. Syst Entomol. 1994;19: 159–175. 10.1111/j.1365-3113.1994.tb00585.x [DOI] [Google Scholar]
- 34.Saux C, Fisher BL, Spicer GS. Dracula ant phylogeny as inferred by nuclear 28S rDNA sequences and implications for ant systematics (Hymenoptera: Formicidae: Amblyoponinae). Mol Phylogenet Evol. 2004;33: 457–468. 10.1016/j.ympev.2004.06.017 [DOI] [PubMed] [Google Scholar]
- 35.Brady SG, Schultz TR, Fisher BL, Ward PS. Evaluating alternative hypotheses for the early evolution and diversification of ants. Proc Natl Acad Sci. 2006;103: 18172–18177. 10.1073/pnas.0605858103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreau CS, Bell CD, Vila R, Archibald SB, Pierce NE. Phylogeny of the ants: Diversification in the age of angiosperms. Science. 2006;312: 101–104. 10.1126/science.1124891 [DOI] [PubMed] [Google Scholar]
- 37.Ouellette GD, Fisher BL, Girman DJ. Molecular systematics of basal subfamilies of ants using 28S rRNA (Hymenoptera: Formicidae). Mol Phylogenet Evol. 2006;40: 359–369. 10.1126/science.1124891 [DOI] [PubMed] [Google Scholar]
- 38.Kück P, Hita Garcia F, Misof B, Meusemann K. Improved phylogenetic analyses corroborate a plausible position of Martialis heureka in the Ant Tree of Life. PLoS ONE. 2011;6: e21031 10.1371/journal.pone.0021031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreau CS, Bell CD. Testing the museum versus cradle tropical biological diversity hypothesis: phylogeny, diversification, and ancestral biogeographic range evolution of the ants. Evolution. 2013;67: 2240–2257. 10.1111/evo.12105 [DOI] [PubMed] [Google Scholar]
- 40.Yoshimura M, Fisher BL. A revision of the ant genus Mystrium in the Malagasy region with description of six new species and remarks on Amblyopone and Stigmatomma (Hymenoptera, Formicidae, Amblyoponinae). ZooKeys. 2014; 1 10.3897/zookeys.394.6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menozzi C. Revisione della formiche del genere Mystrium Roger. Zool Anz. 1929;82: 518–536. [Google Scholar]
- 42.Cronin AL, Molet M, Doums C, Monnin T, Peeters C. Recurrent evolution of dependent colony foundation across eusocial insects. Annu Rev Entomol. 2013;58: 37–55. 10.1146/annurev-ento-120811-153643 [DOI] [PubMed] [Google Scholar]
- 43.Molet M, Peeters C, Fisher BL. Dwarf wingless reproductives improve colony economy in Mystrium ants from Madagascar. International Union for the Study of Social Insects Congress. Washington, DC; 2006.
- 44.Molet M, Peeters C, Fisher BL. Winged queens replaced by reproductives smaller than workers in Mystrium ants. Naturwissenschaften. 2007;94: 280–287. 10.1007/s00114-006-0190-2 [DOI] [PubMed] [Google Scholar]
- 45.Molet M, Fisher BL, Ito F, Peeters C. Shift from independent to dependent colony foundation and evolution of “multi-purpose”ergatoid queens in Mystrium ants (subfamily Amblyoponinae). Biol J Linn Soc. 2009;98: 198–207. 10.1111/j.1095-8312.2009.01257.x [DOI] [Google Scholar]
- 46.Molet M, Wheeler DE, Peeters C. Evolution of novel mosaic castes in ants: modularity, phenotypic plasticity, and colonial buffering. Am Nat. 2012;180: 328–341. 10.1086/667368 [DOI] [PubMed] [Google Scholar]
- 47.Peeters C. Convergent evolution of wingless reproductives across all subfamilies of ants, and sporadic loss of winged queens (Hymenoptera: Formicidae). Myrmecol News. 2012;16: 75–91. [Google Scholar]
- 48.Molet M, Peeters C, Follin I, Fisher BL. Reproductive caste performs intranidal tasks instead of workers in the ant Mystrium oberthueri. Ethology. 2007;113: 721–729. 10.1111/j.1439-0310.2007.01376.x [DOI] [Google Scholar]
- 49.Hebert PDN, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc R Soc B Biol Sci. 2003;270: 313–321. 10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fisher BL. A model for a global inventory of ants: a case study in Madagascar. Proc-Calif Acad Sci. 2005;56: 86. [Google Scholar]
- 51.Masuko K. Larval hemolymph feeding: a nondestructive parental cannibalism in the primitive ant Amblyopone silvestrii Wheeler (Hymenoptera: Formicidae). Behav Ecol Sociobiol. 1986;19: 249–255. 10.1007/BF00300639 [DOI] [Google Scholar]
- 52.Wheeler GC, Wheeler J. An additional use for ant larvae (Hymenoptera: Formicidae). Entomol News USA. 1988;99: 23–24. [Google Scholar]
- 53.Natalie Graham. Phylogeography in response to reproductive strategies and ecogeographic isolation in ant species on Madagascar: Genus Mystrium (Formicidae: Amblyoponinae). [Internet]. 2015. Available: 10.6084/m9.figshare.1324480 [DOI] [PMC free article] [PubMed]
- 54.Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci. 2008;105: 13486–13491. 10.1073/pnas.0803076105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. 1999. pp. 95–98.10780396 [Google Scholar]
- 56.Martins J, Solomon SE, Mikheyev AS, Mueller UG, Ortiz A, Bacci M. Nuclear mitochondrial-like sequences in ants: evidence from Atta cephalotes (Formicidae: Attini). Insect Mol Biol. 2007;16: 777–784. 10.1073/pnas.0803076105 [DOI] [PubMed] [Google Scholar]
- 57.Pamilo P, Viljakainen L, Vihavainen A. Exceptionally high density of NUMTs in the honeybee genome. Mol Biol Evol. 2007;24: 1340–1346. 10.1093/molbev/msm055 [DOI] [PubMed] [Google Scholar]
- 58.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16: 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- 59.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4: 406–425. [DOI] [PubMed] [Google Scholar]
- 60.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol Biol Evol. 2013;30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010. IEEE; 2010. pp. 1–8.
- 62.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9: 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17: 754–755. [DOI] [PubMed] [Google Scholar]
- 64.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 65.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52: 696–704. 10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- 66.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 2008;57: 758–771. 10.1080/10635150802429642 [DOI] [PubMed] [Google Scholar]
- 67.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; btu033 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peakall ROD, Smouse PE. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012;28: 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jobb G, Von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4: 18 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Crozier RH, Crozier YC. The mitochondrial genome of the honeybee Apis mellifera: complete sequence and genome organization. Genetics. 1993;133: 97–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Doums C, Cabrera H, Peeters C. Population genetic structure and male-biased dispersal in the queenless ant Diacamma cyaneiventre. Mol Ecol. 2002;11: 2251–2264. 10.1046/j.1365-294X.2002.01619.x [DOI] [PubMed] [Google Scholar]
- 73.Clémencet J, Viginier B, Doums C. Hierarchical analysis of population genetic structure in the monogynous ant Cataglyphis cursor using microsatellite and mitochondrial DNA markers. Mol Ecol. 2005;14: 3735–3744. 10.1111/j.1365-294X.2005.02706.x [DOI] [PubMed] [Google Scholar]
- 74.Weiser MD, Sanders NJ, Agosti D, Andersen AN, Ellison AM, Fisher BL, et al. Canopy and litter ant assemblages share similar climate–species density relationships. Biol Lett. 2010;6: 769–772. 10.1098/rsbl.2010.0151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raxworthy CJ, Nussbaum RA. Systematics, speciation and biogeography of the dwarf chameleons (Brookesia; Reptilia, Squamata, Chamaeleontidae) of northern Madagascar. J Zool. 1995;235: 525–558. [Google Scholar]
- 76.Burney DA. Theories and facts regarding Holocene environmental change before and after human colonization In: Goodman SM, Patterson BD, editors. Natural change and human impact in Madagascar. Washington, DC: Smithsonian Institution Press; 1997. pp. 75–79. [Google Scholar]
- 77.Du Puy DJ, Moat J. Vegetation mapping and classification in Madagascar (using GIS): implications and recommendations for conservation of biodiversity In: Huxley CR, Lock JM, Cutler DF, editors. Chorology, taxonomy and ecology of the floras of Africa and Madagascar. Royal Botanic Gardens; 1998. pp. 97–117. [Google Scholar]
- 78.Veress M, Toth G, Zentai Z, Schläffer R. The Ankarana tsingy and its development. Carpathian J Earth Environ Sci. 2009;4: 95–108. [Google Scholar]
- 79.Rousset. Genetic differentiation between individuals. J Evol Biol. 2000;13: 58–62. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets and phylogenies are publically available on FigShare: http://dx.doi.org/10.6084/m9.figshare.1324480 and GenBank accession numbers for all specimens (KR063356-KR063489, KP280011-KP280031, KU3612995-KU361299) are available in Table 2.