Abstract
Background
Breast cancer often metastasizes into bone and leads to osteolytic lesions. The underlying mechanisms, however, are complex and not fully understood. Syndecan-1 is a proteoglycan that has various functions relevant for tumor progression including cell–cell communication and cell–matrix interactions. Moreover, its two glycosaminoglycan-binding sites suggest that it may interfere with glycoproteins such as osteoprotegerin, a potent inhibitor of osteoclastogenesis. Thus, we hypothesize that tumor-derived syndecan-1 alters osteoclast biology by modulating osteoprotegerin.
Methods
Syndecan-1 expression was down-regulated via siRNA and the cell fate of the breast cancer cell lines MCF-7, T-47D, and MDA-MB-231 was investigated. Furthermore, we determined the regulation of syndecan-1 by dexamethasone, a commonly used antiemetic in breast cancer therapy. Additionally, we analyzed the genesis and activity of osteoclasts in indirect co-culture experiments using supernatants from MCF-7 cells with deficient and sufficient levels of syndecan-1.
Results
Dexamethasone time- and dose-dependently increased syndecan-1 expression up to 4-fold but did not alter cell behavior. Syndecan-1 up-regulation did not affect the survival or migration of breast cancer cells. Depletion of syndecan-1 using siRNA led to decreased vitality of progesterone receptor-positive cell lines. In MCF-7 cells osteoprotegerin production was up-regulated 2.5-fold after syndecan-1 knock-down. The culture of osteoclast precursors with the supernatant of MCF-7 cells with reduced syndecan-1 levels suppressed osteoclast formation and activity by 21% and 23%, respectively. Adding neutralizing antibodies to osteoprotegerin to the breast cancer supernatants reversed osteoclastogenesis.
Conclusion
Thus, we identified tumor-derived syndecan-1 as a novel positive regulator of osteoclastogenesis and new player in the tumor-bone dialog.
Abbreviations: ACTB, β-actin; C, control; DEX, dexamethasone; ERBB2, v-erb-b2 erythroblastic leukemia viral oncogene homolog 2; ER, estrogen receptor; GAPDH, glyceraldehyde 3-phosphate-dehydrogenase; OPG, osteoprotegerin; PR, progesterone receptor; RANKL, receptor activator of NF-κB ligand; SDC1, syndecan-1
Keywords: Breast cancer, Osteoclast, Osteoprotegerin, Syndecan-1
1. Introduction
The proteoglycan syndecan-1 (also known as CD138) interacts with a variety of proteins via its heparin sulfate side chains or the core protein itself and therefore regulates key cellular functions such as apoptosis, proliferation, and epithelial-mesenchymal transition [1], [2], [3], [4], [5], [6], [7]. Because of its various interactions, several studies have investigated the role of syndecan-1 in tumor progression. While in multiple myeloma, high serum levels of soluble syndecan-1 correlate with a poor prognosis, the association of syndecan-1 expression and clinical outcome was ambiguous in breast cancer [8], [9], [10], [11]. One study found a correlation between stromal and epithelial syndecan-1 expression and poor prognosis, whereas another study observed a poor clinical outcome in breast cancer cases without syndecan-1 expression [9], [10]. Although the exact reasons for these different findings remain unclear, it has been suggested that they may related to the presence of two functionally different syndecan-1 isoforms [5]. While membranous syndecan-1 facilitates the proliferation of breast cancer cells, the soluble form triggers invasion [5]. Despite extensive efforts to unravel the role of syndecan-1 in breast cancer, little is known about how it is regulated. Of note, zoledronic acid, a widely used drug against osteoporosis and skeletal metastases, was found to inhibit syndecan-1 expression in breast cancer cell lines [12].
In multiple myeloma, the interaction between syndecan-1 and osteoprotegerin (OPG), the physiological antagonist of the osteoclast promoting factor receptor activator of NF-κB (RANKL), has been investigated in more detail [13]. The observed syndecan-1-mediated internalization and degradation of OPG may explain low OPG serum levels in patients with multiple myeloma [13]. Furthermore, two other studies demonstrated that tumor-derived syndecan-1 affects bone physiology [14], [15].
Here, we aimed to identify novel regulators of syndecan-1 in breast cancer. We hypothesized that changes in syndecan-1 expression affect osteoclastogenesis. Our results show that (i) dexamethasone increases syndecan-1 expression and that (ii) depletion of syndecan-1 decreases cell viability of hormone receptor-positive breast cancer cells and increases OPG expression, thus suppressing osteoclast differentiation and activation. Hence, syndecan-1 participates in the tumor-bone dialog and alters the bone microenvironment to stimulate osteoclastogenesis.
2. Methods
2.1. Cultivation and treatment of cells
All breast cancer cell lines (MCF-7, T-47D, and MDA-MB-231) were cultured in DMEM/Ham's F-12 (PAA, Pasching, Austria), 10% fetal calf serum (FCS) supreme (Lonza, Pasching, Austria) and 1% penicillin/streptomycin (PAA, Pasching, Austria). Cells were grown in a humidified atmosphere of 95% air and 5% CO2. To assess the effects of dexamethasone (DEX), 70% confluent cells were serum starved for 12 h prior to DEX exposure (Sigma-Aldrich, Darmstadt, Germany) in various concentrations (10−9–10−6 M). To antagonize the effects of DEX, RU-486 (Calbiochem, Darmstadt, Germany) was used at a concentration of 10−5 M. Cells were also treated with zoledronic acid (ZOL, provided by Novartis, Nürnberg, Germany) and the aromatase inhibitor (AI) 4-(imidazolyl)-1nitro-9H-9-xanthenone (Calbiochem, Darmstadt, Germany) in various concentrations (ZOL: 10−10–10−6 M, AI: 7.5×10−8 M and 10−7 M).
To obtain osteoclasts, peripheral blood mononuclear cells (PBMCs) were isolated using Biocoll (1.077 g/ml, Biochrom, Berlin, Germany) from buffy coats obtained after informed consent and following IRB approval and plated at a density of 2×106 cells/cm2 in α-MEM (Invitrogen, Darmstadt, Germany) containing 10% FCS and 1% penicillin/streptomycin. After attachment cells were cultured with 25 ng/ml M-CSF and 50 ng/ml RANKL for 21 d. For indirect cell culture, osteoclasts were cultured in 1/3 osteoclast culture media (α-MEM containing M-CSF and RANKL)+2/3 supernatant of MCF-7 cells from control cells and cells where syndecan-1 expression had been inhibited using siRNA.
2.2. Knock-down experiments
For knock-down experiments, SDC1 siRNA (ID 12527, Ambion, Applied Biosystems, Darmstadt, Germany) and for scrambled control Silencer® Select Negative Control #1 (Cat#4390844 Ambion, Applied Biosystems, Darmstadt, Germany) was introduced in cells with DharmaFECT 1 (Thermo Fisher Scientific, Schwerte, Germany). DharmaFECT1 reagent and siRNA were separately incubated in FCS-free medium (OPTI-MEM® I+GlutaMAX™—I, Invitrogen Karlsruhe, Germany) for 5 min and subsequently mixed and incubated for 20 min at room temperature. Thereafter, medium containing 10% FCS without penicillin/streptomycin was added to the siRNA and DharmaFECT-mixture to the final concentration of 50 nM. Breast cancer cells were washed with PBS and incubated with the transfection mixture for 5 h. Medium was change to stop the transfection and the cells were treated as described above.
2.3. RNA isolation and real-time PCR
RNA was isolated using HighPure RNA extraction kit (Roche, Mannheim, Germany) according to the manufacturer's protocol. Five hundred ng RNA were reverse transcribed using SuperScript II (Invitrogen, Darmstadt, Germany) and subsequently used for SYBR green-based real-time PCR using a standard protocol (Roche, Mannheim, Germany). Primers (Sigma-Aldrich, Munich, Germany) used for semi-quantitative analyses of gene expression: ACTB (β-actin) [NCBI GenBank:NM_001101]: CCAACCGCGAGAAGATGA, CCAGAGGCGTACAGGGATAG, GAPDH (glyceraldehyde 3-phosphate-dehydrogenase) [NCBI GenBank:NM_002046]: AGCCACATCGCTCAGACAC, GCCCAATACGACCAAATCC, SDC1 (syndecan-1) [NCBI GenBank:NM_001006946]: TGGACAGGAAAGAGGTGCTG, GTTTGGTGGGCTTCTGGTAG, OPG (osteoprotegerin) [NCBI GenBank:NM_002546]: GAACCCCAGAGCGAAATACAG, TAGCAGGAGACCAAAGACACTG, RANK (receptor activator of NF-κB) [NCBI GenBank:NM_001270949]: ATCTGGGACGGTGCTGTAAC, CACAGGGCAGACATACACTG, RANKL (receptor activator of NF-κB ligand) [NCBI GenBank:NM_003701]: CTGATGAAAGGAGGAAGCAC, AGTAAGGAGGGGTTGGAGAC.
PCR conditions were 95 °C for 10 min followed by 40 cycles with 95 °C for 10 s, 56 °C for 10 s and 72 °C for 30 s. The melting curve was assessed with the following program: 60 °C for 1 min and 95 °C continuously. The results were calculated applying the ∆∆CT method, and are presented in x-fold increase relative to ACTB.
2.4. Tissue qPCR array
The Tissue Scan Breast Cancer Tissue qPCR Panel II (OriGene Technologies, Inc., Rockville, USA, Cat. no. BCRT 301) was performed according to the manufacturer's protocol. For this study syndecan-1 was analyzed and normalized to ACTB (see Section 2.3).
2.5. Protein analyses
For the detection of membrane-bound proteins, the cells were cultured on cover slips and treated as described above. Subsequently cells were washed with PBS and fixed with ice-cold methanol for 1 h and −20 °C. Afterwards cells were rehydrated with PBS for 10 min and 37 °C. To avoid nonspecific binding, cells were incubated in 1% BSA for 1 h prior incubation with specific primary antibodies over night at 4 °C. Cells were than washed with PBS and treated with secondary fluorescence antibody for 1 h, washed twice and nuclei were stained with DAPI (2.5 µg/ml). Cells were washed three times with PBS and mounted with Dako Fluorescence Mounting Medium (Dako, Dako Deutschland GmbH, Hamburg, Germany).
The following primary and secondary antibodies for immunofluorescence staining were used for detection of syndecan-1: AM00592SU-N (Acris), OPG (osteoprotegerin) AF 805, (R&D), goat anti-mouse Alexa Fluor 488 (green, for SDC1) and rabbit anti-goat Alexa Fluor 594 (red, for OPG). For fluorescence microscopy the Axio M1 microscope (Carl Zeiss, Jena, Germany) was used.
For soluble proteins ELISAs for syndecan-1 (Diaclone, Besancon, France) and OPG (Immundiagnostik, Bensheim, Germany), were used according to the manufacturer's protocol.
2.6. Functional analyses of breast cancer cells
Cell viability was assessed using the CellTiterBlue® Assay (Promega, Mannheim, Germany) and apoptosis using the Cell Death Detection ELISA PLUS (Roche, Mannheim, Germany) according to the manufacturer's protocol. Measurements were conducted using the FluoStar Omega (BMG Labtech, Jena, Germany).
A scratch assay was used to investigate the migration of MCF-7 cells. The cells were cultured and a scratch was done with a tip through the monolayer when they reached confluence. At time point 0 and 72 h, pictures of the scratch were taken. For analyses the scratch size was measured at 10 different regions with Image j software.
2.7. Analyses of osteoclastogenesis and activity of osteoclasts
To analyze osteoclastogenesis, cells from the indirect co-culture were fixed with an acetone/citrate buffer and afterwards stained for tartrate-resistant acid phosphatase (TRAP kit, Sigma, Munich, Germany). TRAP-positive cells with three or more nuclei were counted as osteoclasts. For measurement of osteoclast activity the OsteoLyse assay (Lonza, Pasching, Austria) was used according to the manufacturer's protocol. Briefly, osteoclast precursors were plated onto the OsteoLyse™ cell culture plate and cultured in medium that contained 33 ng/ml M-CSF and 66 ng/ml RANKL. After 7 days, medium was changed to medium consisting of 1/3 of osteoclast culture medium and 2/3 supernatant from syndecan-1 sufficient and insufficient MCF-7 cells.
2.8. Statistical analysis
Results are presented as means±standard deviation. All experiments were repeated at least three times. Outliers were determined via Grubb's test. Statistical evaluations for time- and dose-response curves were performed using a one-way analysis of variance (ANOVA) with posthoc Dunns test and single group comparisons using Student's t-test. P-values<0.05 were considered statistically significant.
3. Results
3.1. Syndecan-1 expression in breast cancer cell lines and primary breast cancer tissue
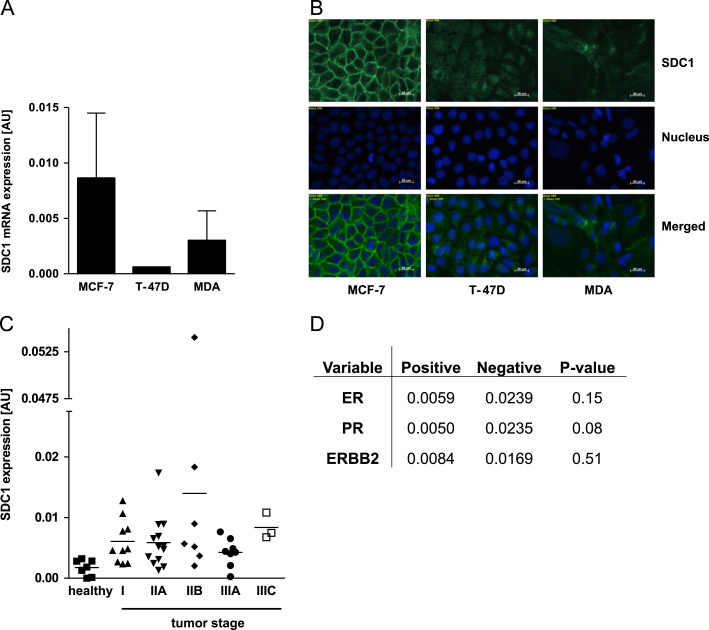
The two hormone receptor-positive cell lines MCF-7, T-47D, and the hormone receptor-negative MDA-MB-231 cells were used to comprehensively investigate the role of syndecan-1 in breast cancer cells. First, we analyzed the syndecan-1 expression in all three cell lines using real-time PCR (Fig. 1A) and immunofluorescence staining for membranous syndecan-1 (Fig. 1B). All cell lines expressed syndecan-1 with the highest levels in MCF-7 cells. While syndecan-1 was mainly located at the cell surface in MCF-7 cells, T-47D and MDA-MB-231 cells showed a lower and diffuse cytoplasmic signal for syndecan-1.
Fig. 1.
Syndecan-1 (SDC1) expression in breast cancer. SDC1 expression was analyzed using (A) real-time PCR and (B) immunofluorescence staining. Therefore, the cells were cultured in medium with 10% FCS and 1% penicillin/streptomycin. Syndecan-1 is displayed in green, the nucleus in blue. Correlation of SDC1 expression with (C) tumor stage and important (D) tumor markers were analyzed. N=3, magnification 400×, ERBB2—v-erb-b2 erythroblastic leukemia viral oncogene homolog 2, ER—estrogen receptor, MDA—MDA-MB-231, PR—progesterone receptor, and SDC1—syndecan-1.
In addition, we performed a Tissue Scan Breast Cancer Tissue qPCR Array to assess whether syndecan-1 expression is correlated with important tumor characteristics (ER, PR, ERBB2 expression or tumor stage). Syndecan-1 expression was observed in healthy and cancer tissue but no significant differences were found between these groups or within different tumor stages (Fig. 1C). Analyses of syndecan-1 with other parameters such as hormone receptor status and ERBB2 expression did not show significant correlations (Fig. 1D).
3.2. Dexamethasone enhanced syndecan-1 expression in MCF-7 cells
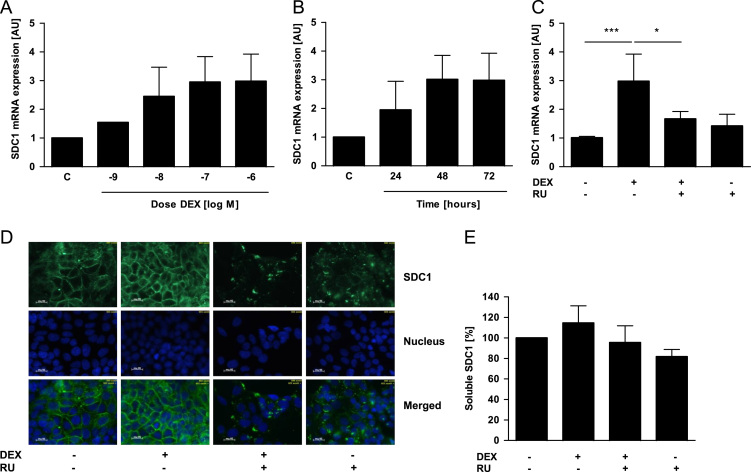
Next, we investigated syndecan-1 regulation in breast cancer cells after treatment with zoledronic acid (10−10–10−6 M); aromatase inhibitor 4-(imidazolyl)-1nitro-9H-9-xanthenone, (7.5×10−8 M and 10−7 M) and dexamethasone (10−9–10−6). Here, the most prominent effect was observed in MCF-7 cells after dexamethasone treatment (Fig. 2). Syndecan-1 mRNA levels were dose- (Fig. 2A) and time-dependently (Fig. 2B) induced by dexamethasone with the highest effect after 72 h and a concentration of 10−6 M. The dexamethasone-dependent induction of syndecan-1 was inhibited by the glucocorticoid-receptor antagonist RU-486 (Fig. 2C), which can also bind to the progesterone receptor [16]. For syndecan-1 protein analyses, we used immunofluorescence staining to detect the membrane-bound form (Fig. 2D) and ELISA (Fig. 2E) to detect the soluble form. We found that dexamethasone-treated MCF-7 cells showed a more intense immunofluorescence signal for syndecan-1 at the cell surface compared to control and RU-486-treated cells (Fig. 2D). No effect of dexamethasone was observed on the soluble syndecan-1 expression (Fig. 2E).
Fig. 2.
Dexamethasone enhances syndecan-1 (SDC1) expression in MCF-7 cells. For analyzing DEX-effects MCF-7 cells were serum starved 12 h prior to and during treatment. MCF-7 cells were treated with (A) different doses of dexamethasone (DEX) for 72 h or with (B) 10−6 M DEX for 24, 48, and 72 h. Afterwards the SDC1 expression was analyzed using real-time PCR. (C) The receptor antagonist RU-486 (10−5 M) was used to inhibit the DEX effects in MCF-7 cells. To analyze the protein expression of SDC1 in more detail we used (D) immunofluorescence staining for the membrane bound form and (E) ELISA for the soluble form. SDC1 is displayed in green, the nucleus in blue. N=3, ⁎p<0.05, ⁎⁎⁎p<0.001, magnification 400×, DEX—dexamethasone, RU—RU-486, and SDC1—syndecan-1.
In contrast to MCF-7 cells, dexamethasone did not alter the syndecan-1 expression of T-47D (Fig. S1) and MDA-MB-231 (Fig S2) cells. Furthermore, neither zoledronic acid nor aromatase-inhibitor affected syndecan-1 expression of MCF-7 and MDA-MB-231 cells (data not shown).
3.3. Functional relevance of syndecan-1 modulation
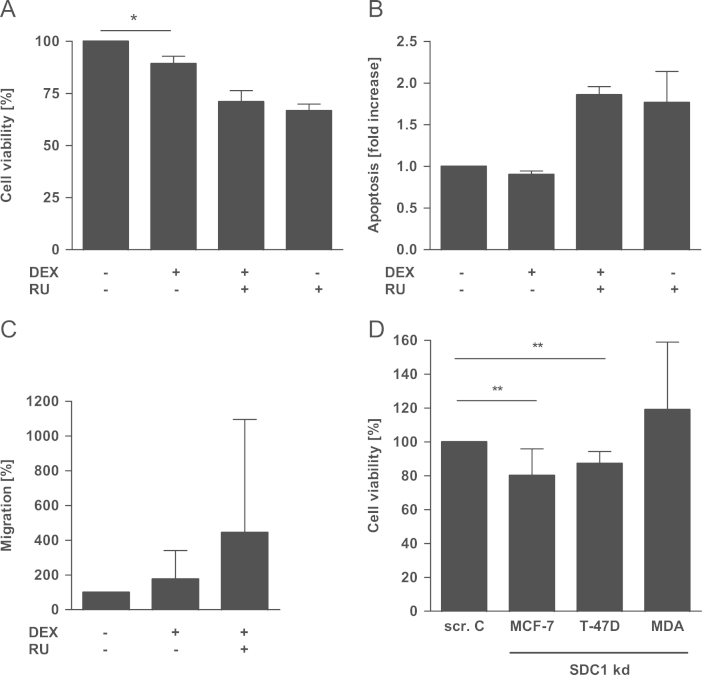
As syndecan-1 is involved in a variety of cell functions, we studied the role of dexamethasone-induced syndecan-1 expression in MCF-7. The viability of MCF-7 cells was slightly reduced (−11%) after dexamethasone treatment (Fig. 3A) but no differences were observed for apoptosis (Fig. 3B) and cell migration (Fig. 3C).
Fig. 3.
Functional relevance of syndecan-1 (SDC1) modulation. Cells were serum starved 12 h prior to and during treatment. After DEX-treatment (10−6 M, 72 h) the (A) viability of MCF-7 cells was analyzed. Additionally we investigated (B) apoptosis and (C) migration of DEX-treated MCF-7 cells. (D) Cell viability was assessed after SDC1 knock-down in MCF-7, T-47D and MDA-MB-231 cells. N=3, ⁎p<0.05, ⁎⁎p<0.01, DEX—dexamethasone, MDA-MDA-MB-231, RU—RU-486, SDC1 kd—syndecan-1 knock-down, and scr. C—scrambled control.
To assess the role of syndecan-1 in breast cancer cell biology more directly, syndecan-1 expression was inhibited in all three cell lines using siRNA (Fig. S3). We found that the viability of hormone receptor-positive breast cancer cells MCF-7 and T-47D was reduced by 20% (p<0.001) and 13% (p<0.001), respectively, after inhibition of syndecan-1 expression (Fig. 3D). The viability of the hormone receptor-negative MDA-MB-231 cell line was not altered after syndecan-1 knock-down (Fig. 3D).
3.4. Osteoprotegerin is up-regulated after syndecan-1 knock-down in MCF-7 cells
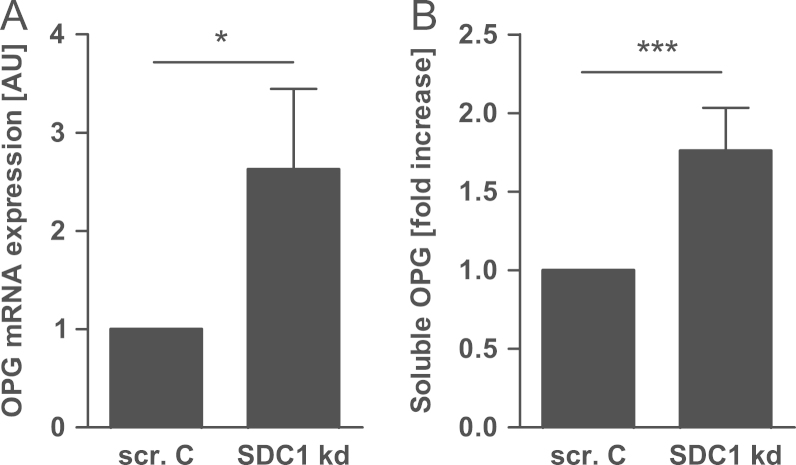
Because of the known interaction of syndecan-1 with OPG we analyzed the expression of OPG, RANK and RANKL after syndecan-1 knock-down in MCF-7 cells. The mRNA expression as well as the protein secretion of OPG was up-regulated 2.5-fold (Fig. 4A) and 1.8 fold (Fig. 4B), respectively. For T-47D (Fig. S4A) and MDA-MB-231 (Fig. S4B) cells, OPG expression after syndecan-1 knock-down was also analyzed. Compared with the OPG regulation in MCF-7 cells only minimal up-regulation of OPG was observed for MDA-231 (Fig. S4B) and no altered OPG expression for T-47D (Fig. S4A). Syndecan-1 knock-down did not alter the mRNA expression of RANK and RANKL in all three cell lines (data not shown).
Fig. 4.
OPG is up-regulated after SDC1 knock-down in MCF-7 cells. After SDC1 knock-down cells were cultured in medium without FCS for 36 h. OPG expression was analyzed using (A) real-time PCR and (B) ELISA. N=4, ⁎p<0.05, ⁎⁎⁎p<0.001, SDC1 kd—syndecan-1 knock-down, and scr. C—scrambled control.
3.5. Osteoclastogenesis is inhibited by the supernatant from MCF-7 cells with reduced syndecan-1 expression
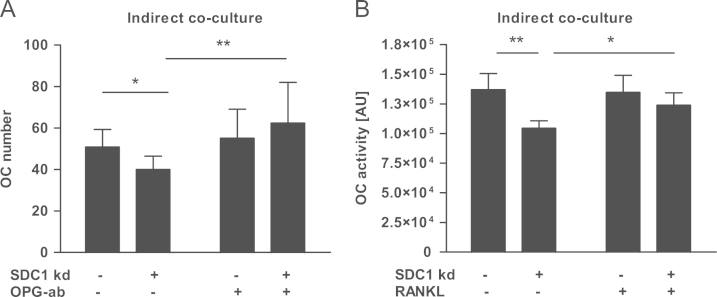
To test whether the up-regulation of OPG after syndecan-1 knock-down in MCF-7 cells functionally affects osteoclast differentiation, indirect co-culture experiments were performed. The supernatants from control and MCF-7 cells treated with siRNA against syndecan-1 were collected and transferred to osteoclasts. After 21 days the number and activity of osteoclasts were analyzed. Both parameters, osteoclast differentiation and activity, were reduced by 21% and 23%, respectively (Fig. 5A and B). The inhibition of osteoclastogenesis and activity was reversed by addition of substances which neutralize OPG (OPG antibody/RANKL).
Fig. 5.
Activity and genesis of osteoclasts is inhibited by indirect coculture with SDC1 insufficient MCF-7 supernatant. Osteoclasts were cultured in medium containing osteoclast culture media (one third) and supernatant of MCF-7 cells with and without SDC1 siRNA treatment (two thirds). Subsequently, the (A) number and (B) activity of osteoclasts were assessed. N=4, ⁎p<0.05, ⁎⁎p<0.01, ab—antibody, and SDC1 kd—syndecan-1 knock-down.
4. Discussion
In this study, the expression and regulation of syndecan-1 in breast cancer cells were investigated. Furthermore, we determined the role of syndecan-1 in breast cancer pathophysiology and its impact on tumor cell–osteoclast interaction.
Analyses of basal syndecan-1 expression showed differences in syndecan-1 levels as well as staining pattern for this proteoglycan in the breast cancer cell lines that were employed. Different localization of syndecan-1 have been found in previous studies showing lysosomal syndecan-1 in poorly differentiated cells, while a membranous pattern was present in more differentiated cell lines [17], [18]. This may also be the case in our study.
Despite numerous studies addressing the role of syndecan-1 in breast cancer its function in this tumor entity remains incompletely understood and only few regulators of syndecan-1 are known [1], [3], [5], [6], [7], [15], [19], [20], [21], [22]. In this study, we identified dexamethasone as an inducer of membranous syndecan-1 expression in the MCF-7 cell line. The other tested substances zoledronic acid and the aromatase inhibitor did not alter syndecan-1 in any of the breast cancer cell lines investigated. Interestingly, a recently published study reported a significant down-regulation of syndecan-1 in MCF-7 as well as MDA-MB-231 cells by zoledronic acid when used at a concentration of 20 µM [12]. The use of a lower concentration of zoledronic acid in our study could be a reason for this discrepancy.
Because of high levels of basal syndecan-1 expression and the dexamethasone-mediated syndecan-1 up-regulation in MCF-7 cells, further experiments were focused on this cell line. Next experiments addressed the functional relevance of dexamethasone-mediated syndecan-1 up-regulation in MCF-7 cells. Since membrane-bound syndecan-1 was reported to support proliferation and inhibit invasion of breast cancer cells [5], we analyzed the cell viability and migration in MCF-7 cells after dexamethasone treatment. We found a decrease in cell viability after dexamethasone treatment whereas migration was not altered. Because of this unexpected result we performed syndecan-1 knock-down experiments in all three cell lines and subsequently measured viability. Only the hormone receptor-positive cell lines MCF-7 and T-47D responded with a decrease in cell viability after syndecan-1 knock-down, but not the triple-negative MDA-MB-231 cells. Thus, a certain threshold of syndecan-1 may be necessary for the viability of hormone-responsive breast cancer cells but increased levels do not further stimulate cell viability.
The fact that only the hormone receptor-positive cell lines responded to the syndecan-1 knock-down raised the question whether there is a link between the progesterone receptor status and syndecan-1 expression. However, the data from the tissue microarray did not support this assumption, as no correlation between the syndecan-1 expression and progesterone receptor status was found. This result is in line with two previous studies showing no correlation between syndecan-1 and the progesterone receptor status [9], [10]. Nevertheless, another study found that a high syndecan-1 expression correlated with a negative progesterone receptor status [8]. In two of these three studies lobular and other types of carcinomas were also included besides ductal carcinomas, which may be the reason for these conflicting results [8], [9]. However, studies with a greater sample size are needed to resolve this discrepancy.
The direct impact of soluble syndecan-1 on the bone compartment was previously shown in two studies [14], [15]. In a mouse model of breast cancer, where heparanase-overexpressing MDA-Met cells were used, osteoclastogenesis was enhanced and shown to be dependent on the presence of syndecan-1 expression [15]. In another study using a mouse model, syndecan-1-expressing multiple myeloma cells produced fewer osteolytic lesions compared to cells that did not express syndecan-1. Additional in vitro experiments in murine bone marrow cells showed that the syndecan-1 ectodomain inhibits osteoclast formation and promotes osteoblast development [14]. An indirect effect of syndecan-1 was also discussed for multiple myeloma where the protein interaction with OPG was proposed as a cause for lower OPG levels in patient with osteolytic lesions compared to patients without this complication [13]. In this study, we found that OPG expression was up-regulated after syndecan-1 knock-down in MCF-7 cells. Thus, showing that syndecan-1 does not only alter OPG function at the protein level, but also at transcriptional level. Moreover, the modulation of OPG by syndecan-1 knock-down was sufficient to alter osteoclast biology. Thus, our study supports the osteoclast-promoting effects of syndecan-1 and identified it as a novel direct regulator of OPG.
5. Conclusion
In summary, we identified dexamethasone as an inducer of syndecan-1 expression in breast cancer cells and show that syndecan-1 represents a novel regulator of OPG expression. This modulation of OPG inhibits osteoclastogenesis, which may be a critical mechanism in the tumor cell/bone cell dialog.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Authors' contribution
PBM and ST performed experiments. LCH, PBM and MR designed the experiments and wrote the paper. PBM, ST, TDR, MR, AG, and LCH contributed substantially to data interpretation and critical reading the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
The authors would like to thank Ms. B. Zeiler and Mr. P. Böhme for their excellent technical assistance and Ms. T. Reiche for her secretarial assistance. The work was funded by the Wilhelm Sander-Foundation (2007.00501, 2007.005.02), the DFG Research group SKELMET FOR (HO 1875/12-1 and 13-1) to LCH and MeDDriveStart 2013/2014 to PBM.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jbo.2013.11.001.
Contributor Information
Peggy Benad-Mehner, Email: Peggy.Benad@uniklinikum-dresden.de.
Stefanie Thiele, Email: Stefanie.Thiele@uniklinikum-dresden.de.
Tilman D. Rachner, Email: Tilman.Rachner@uniklinikum-dresden.de.
Andy Göbel, Email: Andy.Goebel@uniklinikum-dresden.de.
Martina Rauner, Email: Martina.Rauner@uniklinikum-dresden.de.
Lorenz C. Hofbauer, Email: Lorenz.Hofbauer@uniklinikum-dresden.de.
Appendix A. Supplementary materials
Syndecan-1 (SDC1) expression after dexamethasone treatment in T-47D cells. T-47D cells were serum starved 12 h prior to and during treatment with dexamethasone. T-47D cells were treated with (A) different doses of dexamethasone (DEX) (B) and/ or with RU-486 for 72 h and afterwards SDC1 expression was analyzed using realtime PCR. (C) To analyze the protein expression of SDC1 we used immunofluorescence staining for the membrane bound form. SDC1 is displayed in green, the nucleus in blue. N=3, magnification 400x, DEX – dexamethasone, RU – RU-486, SDC1 – syndecan-1.
Syndecan-1 (SDC1) expression after dexamethasone treatment in MDA-231 cells. MDA-231 cell line cells were serum starved 12 h prior to and during treatment with dexamethasone. MDA-231 cells were treated with (A) different doses of dexamethasone (DEX) (B) and/ or with RU-486 for 72 h and afterwards SDC1 expression was analyzed using real-time PCR. To analyze the protein expression of SDC1 in more detail we used (C) immunofluorescence staining for the membrane bound form and (D) ELISA for the soluble form. SDC1 is displayed in green, the nucleus in blue. N=3, magnification 400x, DEX – dexamethasone, RU – RU-486, SDC1 – syndecan-1.
Syndecan-1 knock-down in breast cancer cell lines. Syndecan-1 knock-down was performed in (A) MCF-7, (B) T-47D and (C) MDA-231 cells and after 24, 48 and 72 h SDC1 expression was analyzed using real-time PCR. N=3, scr. C – scrambled control, SDC1 – syndecan-1.
OPG expression after syndecan-1 knock-down in T-47D and MDA-MB-231 cells. Syndecan-1 knock-down was performed in (A) T-47D and (B) MDA-231 cells. After 36 h OPG expression was analyzed using real-time PCR. N=3, ***p<=0.001, scr. C – scrambled control, SDC1 kd – syndecan-1 knock-down.
References
- 1.Beauvais D.M., Rapraeger A.C. Syndecans in tumor cell adhesion and signaling. Reprod Biol Endocrinol. 2004;2:3. doi: 10.1186/1477-7827-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernfield M., Gotte M., Park P.W., Reizes O., Fitzgerald M.L., Lincecum J. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem. 1999;68:729–777. doi: 10.1146/annurev.biochem.68.1.729. [DOI] [PubMed] [Google Scholar]
- 3.Kato M., Saunders S., Nguyen H., Bernfield M. Loss of cell surface syndecan-1 causes epithelia to transform into anchorage-independent mesenchyme-like cells. Mol Biol Cell. 1995;6(5):559–576. doi: 10.1091/mbc.6.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manon-Jensen T., Itoh Y., Couchman J.R. Proteoglycans in health and disease: the multiple roles of syndecan shedding. FEBS J. 2010;277(19):3876–3889. doi: 10.1111/j.1742-4658.2010.07798.x. [DOI] [PubMed] [Google Scholar]
- 5.Nikolova V., Koo C.Y., Ibrahim S.A., Wang Z., Spillmann D., Dreier R. Differential roles for membrane-bound and soluble syndecan-1 (CD138) in breast cancer progression. Carcinogenesis. 2009;30(3):397–407. doi: 10.1093/carcin/bgp001. [DOI] [PubMed] [Google Scholar]
- 6.Sun D., McAlmon K.R., Davies J.A., Bernfield M., Hay E.D. Simultaneous loss of expression of syndecan-1 and E-cadherin in the embryonic palate during epithelial-mesenchymal transformation. Int J Dev Biol. 1998;42(5):733–736. [PubMed] [Google Scholar]
- 7.Sun H., Berquin I.M., Owens R.T., O‘Flaherty J.T., Edwards I.J. Peroxisome proliferator-activated receptor gamma-mediated up-regulation of syndecan-1 by n-3 fatty acids promotes apoptosis of human breast cancer cells. Cancer Res. 2008;68(8):2912–2919. doi: 10.1158/0008-5472.CAN-07-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbareschi M., Maisonneuve P., Aldovini D., Cangi M.G., Pecciarini L., Angelo Mauri F. High syndecan-1 expression in breast carcinoma is related to an aggressive phenotype and to poorer prognosis. Cancer. 2003;98(3):474–483. doi: 10.1002/cncr.11515. [DOI] [PubMed] [Google Scholar]
- 9.Leivonen M., Lundin J., Nordling S., von Boguslawski K., Haglund C. Prognostic value of syndecan-1 expression in breast cancer. Oncology. 2004;67(1):11–18. doi: 10.1159/000080280. [DOI] [PubMed] [Google Scholar]
- 10.Loussouarn D., Campion L., Sagan C., Frenel J.S., Dravet F., Classe J.M. Prognostic impact of syndecan-1 expression in invasive ductal breast carcinomas. Br J Cancer. 2008;98(12):1993–1998. doi: 10.1038/sj.bjc.6604400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seidel C., Sundan A., Hjorth M., Turesson I., Dahl I.M., Abildgaard N. Serum syndecan-1: a new independent prognostic marker in multiple myeloma. Blood. 2000;95(2):388–392. [PubMed] [Google Scholar]
- 12.Dedes P.G., Gialeli C., Tsonis A.I., Kanakis I., Theocharis A.D., Kletsas D. Expression of matrix macromolecules and functional properties of breast cancer cells are modulated by the bisphosphonate zoledronic acid. Biochim Biophys Acta. 2012;1820(12):1926–1939. doi: 10.1016/j.bbagen.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Standal T., Seidel C., Hjertner O., Plesner T., Sanderson R.D., Waage A. Osteoprotegerin is bound, internalized, and degraded by multiple myeloma cells. Blood. 2002;100(8):3002–3007. doi: 10.1182/blood-2002-04-1190. [DOI] [PubMed] [Google Scholar]
- 14.Dhodapkar M.V., Abe E., Theus A., Lacy M., Langford J.K., Barlogie B. Syndecan-1 is a multifunctional regulator of myeloma pathobiology: control of tumor cell survival, growth, and bone cell differentiation. Blood. 1998;91(8):2679–2688. [PubMed] [Google Scholar]
- 15.Kelly T., Suva L.J., Nicks K.M., MacLeod V., Sanderson R.D. Tumor-derived syndecan-1 mediates distal cross-talk with bone that enhances osteoclastogenesis. J Bone Miner Res. 2010;25(6):1295–1304. doi: 10.1002/jbmr.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollow K., Grill H.J., Elger W., Christmann P., Manz B., Juchem M. Vergleichende Untersuchungen der synthetischen Antigestagene RU 38 486, ZK 98734 und ZK 98299 auf der Rezeptorebene. Arch Gynecol Obstet. 1989;245:929–930. [Google Scholar]
- 17.Burbach B.J., Friedl A., Mundhenke C., Rapraeger A.C. Syndecan-1 accumulates in lysosomes of poorly differentiated breast carcinoma cells. Matrix Biol. 2003;22(2):163–177. doi: 10.1016/s0945-053x(03)00009-x. [DOI] [PubMed] [Google Scholar]
- 18.Gotte M., Kersting C., Radke I., Kiesel L., Wulfing P. An expression signature of syndecan-1 (CD138), E-cadherin and c-met is associated with factors of angiogenesis and lymphangiogenesis in ductal breast carcinoma in situ. Breast Cancer Res. 2007;9(1):R8. doi: 10.1186/bcr1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexander C.M., Reichsman F., Hinkes M.T., Lincecum J., Becker K.A., Cumberledge S. Syndecan-1 is required for Wnt-1-induced mammary tumorigenesis in mice. Nat Genet. 2000;25(3):329–332. doi: 10.1038/77108. [DOI] [PubMed] [Google Scholar]
- 20.Beauvais D.M., Burbach B.J., Rapraeger A.C. The syndecan-1 ectodomain regulates alphavbeta3 integrin activity in human mammary carcinoma cells. J Cell Biol. 2004;167(1):171–181. doi: 10.1083/jcb.200404171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maeda T., Desouky J., Friedl A. Syndecan-1 expression by stromal fibroblasts promots breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene. 2006;25(9):1408–1412. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 22.Purushothaman A., Chen L., Yang Y., Sanderson R.D. Heparanase stimulation of protease expression implicates it as a master regulator of the aggressive tumor phenotype in myeloma. J Biol Chem. 2008;283(47):32628–32636. doi: 10.1074/jbc.M806266200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Syndecan-1 (SDC1) expression after dexamethasone treatment in T-47D cells. T-47D cells were serum starved 12 h prior to and during treatment with dexamethasone. T-47D cells were treated with (A) different doses of dexamethasone (DEX) (B) and/ or with RU-486 for 72 h and afterwards SDC1 expression was analyzed using realtime PCR. (C) To analyze the protein expression of SDC1 we used immunofluorescence staining for the membrane bound form. SDC1 is displayed in green, the nucleus in blue. N=3, magnification 400x, DEX – dexamethasone, RU – RU-486, SDC1 – syndecan-1.
Syndecan-1 (SDC1) expression after dexamethasone treatment in MDA-231 cells. MDA-231 cell line cells were serum starved 12 h prior to and during treatment with dexamethasone. MDA-231 cells were treated with (A) different doses of dexamethasone (DEX) (B) and/ or with RU-486 for 72 h and afterwards SDC1 expression was analyzed using real-time PCR. To analyze the protein expression of SDC1 in more detail we used (C) immunofluorescence staining for the membrane bound form and (D) ELISA for the soluble form. SDC1 is displayed in green, the nucleus in blue. N=3, magnification 400x, DEX – dexamethasone, RU – RU-486, SDC1 – syndecan-1.
Syndecan-1 knock-down in breast cancer cell lines. Syndecan-1 knock-down was performed in (A) MCF-7, (B) T-47D and (C) MDA-231 cells and after 24, 48 and 72 h SDC1 expression was analyzed using real-time PCR. N=3, scr. C – scrambled control, SDC1 – syndecan-1.
OPG expression after syndecan-1 knock-down in T-47D and MDA-MB-231 cells. Syndecan-1 knock-down was performed in (A) T-47D and (B) MDA-231 cells. After 36 h OPG expression was analyzed using real-time PCR. N=3, ***p<=0.001, scr. C – scrambled control, SDC1 kd – syndecan-1 knock-down.