Abstract
Mismatch repair defective (MMRd) colorectal carcinoma (CRC) is a distinct molecular phenotype of colorectal cancer, including 12% of sporadic CRC and 3% of Lynch Syndrome. In order to investigate the clinicopathological characteristics of MMRd colorectal carcinoma, and to find the most effective method for preliminary screening, 296 CRC fulfilled revised Bethesda Guideline (RB) were selected from 1450 CRCs to perform both IHC staining for MLH1, MSH2, MSH6, PMS2 and MSI analysis. Sixty-eight tumors were classified as MSI-H by MSI test. Colorectal carcinomas with MSI-H were prone to be proximal located, poorly differentiated, and relatively early staged, with infrequent metastasis to lymph node as well as to distant organs, compared with MSS ones. All of the 68 MMRd CRCs presented abnormal expression of at least one mismatch repair protein (MMRP), with 48 concurrent negative of MLH1 and PMS2, 14 concurrent negative of MSH2 and MSH6, 4 isolated negative of MSH6, 1 isolated negative of PMS2, and 1 concurrent negative of 4 MMRPs. All of the MLH1 negative tumors also showed abnormal expression of PMS2. All of the MSH2 negative cases also presented negative expression of MSH6. The sensitivity and specificity of the 2-antibody IHC test contained only PMS2 and MSH6 for screening for MMRd CRC were 100% and 98.2% respectively, exactly the same as that of the 4-antibody IHC test with all of the 4 MMRPs. The diagnostic accordance rate of the 2-antibody approach and MSI analysis was 98.6%. In conclusion, MMRd CRC has characteristic clinicopathological features different from MSS CRCs. The 2-antibody IHC approach containing MSH6 and PMS2 is the most easy and effective way to detecting MMR deficiency in CRC.
Keywords: Mismatch repair, microsatellite instability, Lynch syndrome, immunohistochemistry, molecular phenotype, CRC
Introduction
Colorectal carcinoma (CRC) is the third most common cancer and the fourth leading cause of cancer death in the world, according to the GLOBOCAN 2008 estimates [1]. Cases and deaths occurred in China are increasing rapidly in recent years on account of the so called “westernization” [2,3]. The pathogenesis of colorectal cancer is still unclear, but it could be generally defined in two molecular pathways of genomic instability. One is chromosome instability, which is involved in certain oncogenes and cancer suppressor genes, such as APC, KRAS, and P53 [4]. Another way is microsatellite instability (MSI), which is due to mismatch repair system defect, and account for approximately 15% of all colorectal carcinomas. Most of the MSI colorectal carcinomas (12%) are sporadic MSI, which is attributed to acquired hypermethylation of MLH1 gene promoter, accompanied by the CpG island methylation phenotype. And 3% of the MSI CRCs are Lynch syndrome related colorectal carcinomas, once defined as HNPCC (hereditary non-polyposis colorectal cancer), a classic model of hereditary colorectal cancer [5]. The mismatch repair deficiency of Lynch syndrome is caused mostly by germline mutation of certain DNA mismatch repair gene. Thus, colorectal cancers could be classified under two molecular phenotypes, microsatellite instable and microsatellite stable (MSS) phenotype. It is more and more important to reveal the molecular phenotype of colorectal cancer, as different phenotype means different pathogenesis, prognosis, even treatment response. Mismatch repair-deficient (MMRd) colorectal cancer has distinct clinicopathological characteristics, including early onset, proximal localization, Crohn’s-like lymphocytic reaction, mucinous/signet-ring differentiation, and medullary growth pattern [6-9]. Furthermore, the clinical prognosis and treatment response of patient are closely related to the molecular mechanism underlying cancer development. Patients with MSI-H colorectal carcinoma have different outcome and chemotherapy response from those with microsatellite stable (MSS) colorectal cancer [12-14]. So, it has great significance to identify MSI in colorectal carcinoma, not only because MSI is a critical DNA marker for screening for Lynch syndrome, but also because it can help to differentiate MMRd colorectal cancers from MSS ones, which will provide valuable information for prognosis estimation and treatment individualization.
Previously, immunohistochemistry (IHC) of mismatch repair protein (MMRP) and MSI test were widely used for screening for HNPCC. And the MSI analysis based on PCR is the gold standard approach for detecting microsatellite status. American National Cancer Institute (NCI) recommended five DNA markers for MSI test. These markers are five microsatellite sequences including BAT25, BAT26, D2S123, D5S346 and D17S250. It is defined as high frequency of microsatellite instability (MSI-H) when two or more of the five markers in the tumor DNA were positive. If only one marker was positive, the tumor is termed as low frequency of MSI (MSI-L). And MSS is determined when all of the five markers were negative [7]. Another sensitive and specific method for MSI detection is IHC for MMRPs, which often involves in a traditional panel including MLH1 and MSH2. Absent expression of any one of the MMRPs indicates defective mismatch repair system in the tumor detected. It is considered that adding PMS2 can elevate the sensitivity of screening for HNPCC [15]. In the past decade, a four-antibody panel including MLH1, MSH2, MSH6, and PMS2 was becoming popular for screening for HNPCC and mismatch repair deficiency of CRC. Recent data suggest that a two-antibody panel approach using only PMS2 and MSH6 is an effective screening protocol for HNPCC related colorectal carcinoma, and even extraintestinal tumors [16-18]. In this study, the relationship between clinicopathological features and mismatch repair status was further assessed, and IHC test for mismatch repair gene proteins and MSI analysis were compared for subtyping of colorectal carcinoma by mismatch repair competency.
Materials and methods
Patients
A total of 1450 colorectal carcinomas were recruited from Fu Dan University Shanghai Cancer Center between January 2007 and March 2010. Clinical and family history of these patients was reviewed, and clinical-pathological features were examined based on the revised Bethesda Guideline [7]. Finally, 296 cases were selected for analysis, which fulfilled at least one of the following criteria: (1) Colorectal cancer diagnosed in a patient who is less than 50 years of age; (2) Presence of synchronous, metachronous colorectal, or other HNPCC-associated tumors [including colorectal, endometrial, stomach, ovarian, pancreas, ureter and renal pelvis, biliary tract, and brain (usually glioblastoma as seen in Turcot syndrome) tumors, sebaceous gland adenomas and keratoacanthoma in Muir-Torre syndrome, and carcinoma of the small bowel], regardless of age; (3) Colorectal cancer with the MSI-H histology (presence of tumor infiltrating lymphocytes, Crohn’s-like lymphocytic reaction, mucinous/signet-ring differentiation, or medullary growth pattern) diagnosed in a patient who is less than 60 years of age; (4) Colorectal cancer diagnosed in one or more first-degree relatives with an HNPCC-related tumors, with one of the cancers being diagnosed under age 50 years; (5) Colorectal cancer diagnosed in two or more first- or second-degree relatives with HNPCC-related tumors, regardless of age [7]. The informed consents of all patients and approval of Medical Ethical Committee of Shanghai Cancer Center, Fudan University were obtained at the beginning of the study.
MMRP immunohistochemistry
Immunohistochemistry for the four most common mismatch repair proteins were performed in all of the 296 cases using the standard Envision two-step procedure. Tumor representative blocks were carefully selected for analysis with normal-tumor junction in order to assess staining result properly. Primary monoclonal antibodies against MLH1 (clone ES05, Novocastra, Leica Biosystems Newcastle Ltd, Newcastle Upon Tyne, UK, 1:30), MSH2 (clone FE11, Calbiochem, Merck KGaA, Darmstadt, Germany, 1:50), MSH6 (clone EPR3945, Epitomics Inc, Burlingame, California, USA, 1:200), and PMS2 (clone EPR3947, Epitomics Inc, Burlingame, California, USA, 1:200) were applied to 4-μm-thick 10% formalin-fixed, paraffin-embedded tissue sections. The sections were deparaffinized in xylene for three times, every time for 10 minutes, and subsequently rehydrated through graded alcohols to distilled water. Antigen heat retrieval was performed in 1 mM EDTA (pH 9.0) for 10 minutes (PMS2 15 minutes) using a microwave oven, after that the sections were cooled down in room temperature for 1.5 hours. After rinsing in distilled water and TBS successively, sections were incubated with specific monoclonal antigen at 4°C overnight. The Dako REAL™ EnVision™ Detection System (Dako, Shanghai, China) was used as the secondary detection system according to manufacturer’s instructions. The reaction is visualized by Dako REAL™ DAB + Chromogen, and slides were counterstained with hematoxylin.
Non-neoplastic colonic mucosa, stromal cells, infiltrating lymphocytes or the centers of lymphoid follicles were used as internal positive controls. And the known MMR deficient colorectal carcinomas served as external negative controls. Two experienced pathologists evaluated the staining results independently and blindly to the MSI status. Normal expression was defined as nuclear staining within tumor cells, while negative protein expression was defined as complete absence of nuclear staining within tumor cells with concurrent internal positive controls. If internal non-neoplastic tissues showed invalid negative staining, procedure was routinely repeated.
Microsatellite instability analysis
Genomic DNA of matched tumor and non-neoplastic tissue was extracted from the paraffin-embedded tissue sections by manual microdissection on the basis of a HE-stained slide, using the QIAamp DNA Mini Kit (QIAGEN China (Shanghai) Co., Ltd., Shanghai, China). For microsatellite instability analysis, the panel of 5 markers BAT26, BAT25, D5S346, D2S123, and D17S250, recommended by NCI [19], was employed to test all of the 296 paired DNA samples. This panel composed of 2 mononucleotide repeat sequences BAT26, BAT25, and 3 dinucleotide repeat markers, D5S346, D2S123, and D17S250. The primers used for amplification were just as described previously [20]. The forward primers (Applied Biosystems, Life Technologies Corporation, California, USA) were marked with a fluorescent tag (FAM-BAT25/D2S123/D17S250, NED-BAT26, PET-D5S346) at the 5’ end. Fluorescence multiplex polymerase chain reaction (PCR) was performed with a 20 μL reaction system consisted of 10 μL 2× Taq PCR premix (containing Taq Polymerase, magnesium chloride and dNTPs) (Sangon Biotech, Shanghai, China), 2 μL of mixed primers, 1 μL genomic DNA (100 ng) and 7 μL DNASE-free H2O. After initial pre-denature at 95°C for 9 minutes, 35 cycles of: denaturation at 95°C for 30 seconds, annealing at 53°C for 30 seconds and extension at 72°C for 1 minute were performed. Final extension was at 60°C for 45 minutes. PCR reaction product (1 μL) was then mixed with 0.1 μL LIZ (internal size standard) (Applied Biosystems) and 9 μl of Hi-Di formamide (denaturant) (Applied Biosystems). Heat pre-denaturation was performed on the mixture at 95°C for 5 minutes, and the sample was kept at 4°C for 5 minutes. Then the product was put in a 96 well plate, and capillary electrophoresis was performed with an ABI 3500 Genetic Analyzer (Applied Biosystem Shanghai Division, Shanghai, China) for 45 minutes. Data was automatically collected and then analyzed with Genemapper v4.1 (Applied Biosystems). The presence of peaks in the fluorescence profile of the amplified microsatellite DNA that were absent in a corresponding profile derived from the matched normal mucosa was interpreted as microsatellite instability. If there were two or more of the 5 markers showed instability, tumors were defined as high frequency microsatellite instability (MSI-H); instability at a single locus was defined as low frequency microsatellite instability (MSI-L); and if none of the markers showed instability, the tumor was considered to be microsatellite stable (MSS).
Statistical analysis
The correlation between clinicopathological features and microsatellite status were analyzed using χ2 test. Sensitivity was defined as the IHC negative expression of mismatch repair gene products (MLH1, MSH2, MSH6, and PMS2) in screening for MMRd, with MSI test as gold standard. And specificity was the normal IHC result of MMRPs in MSI-L/MSS colorectal cancer. P values were reported correspond to two-sided test. And the statistical significance was defined as the P value <0.05. All data were processed using SPSS 16.0 (SPSS, Chicago, IL, USA).
Results
Of the 296 cases matched RB criteria, 68 (23.0%, 68/296) were classified as MSI-H, 9 (3.0%, 9/296) were MSI-L, and 219 (74.0%, 219/296) were MSS by MSI analysis. The clinicopathological characteristics of MSI colorectal carcinomas were different from MSS ones. Especially for tumor location, tumor grade, TNM stage, lymph nodes metastasis, and distant metastasis, the differences were statistically significant as showed in Table 1. Compared with MSS CRC, MSI-H CRC were more frequently located in right colon, poorly differentiated, at relatively early TNM stage, less lymph node metastasis as well as infrequent distant metastasis. Although the clinicopathological features of MSI-L tumors had no significant differences compared either to MSI-H CRC or to MSS CRC, except tumor site, the clinicopathological characteristics of MSI-L group was more close to that of MSS group.
Table 1.
Clinicopathological features and microsatellite status
| Clinicopathological features | MSI-H | MSI-L | MSS | P-HvsL* | P-HvsS | P-LvsS |
|---|---|---|---|---|---|---|
| Sex | ||||||
| Male | 41 | 5 | 115 | 1.000 | 0.26 | 1.000 |
| Female | 27 | 4 | 104 | |||
| Age | ||||||
| <50 | 41 | 4 | 104 | 0.585 | 0.065 | 1.000 |
| ≥50 | 27 | 5 | 115 | |||
| Tumor site | ||||||
| Right colon | 35 | 1 | 64 | 0.037 | 0.002 | 0.228 |
| Left colon | 19 | 4 | 107 | |||
| Rectum | 14 | 4 | 48 | |||
| Tumor grade | ||||||
| I/II | 46 | 7 | 176 | 0.815 | 0.029 | 0.261 |
| III | 22 | 2 | 43 | |||
| Mucinous/signet ring differentiation | ||||||
| Yes | 23 | 4 | 61 | 0.798 | 0.345 | 0.482 |
| No | 45 | 5 | 158 | |||
| Lymphocytic infiltration† | ||||||
| Yes | 14 | 0 | 29 | 0.198 | 0.138 | 0.608 |
| No | 54 | 9 | 190 | |||
| TNM stage | ||||||
| I/II | 46 | 5 | 104 | 0.729 | 0.004 | 0.893 |
| III | 22 | 4 | 115 | |||
| Lymph nodes metastasis | ||||||
| Yes | 21 | 4 | 112 | 0.662 | 0.003 | 0.957 |
| No | 47 | 5 | 107 | |||
| Distant metastasis | ||||||
| Yes | 2 | 2 | 34 | 0.099 | 0.011 | 0.941 |
| No | 66 | 7 | 185 | |||
| N | 68 | 9 | 219 |
P-HvsL, P-HvsS, P-LvsS refers to P value of Pearson’s test or Fischer’s exact test for the comparison of clinicopathological features of CRC with different microsatellite status, MSI-H group with MSI-L group, MSI-H group with MSS group, and MSI-L group with MSS group; right colon including cecum, ascending colon, and hepatic flexure; left colon including splenic flexure, decending colon, and sigmoid colon; Tumor grade I/II/III correspond to Well/moderately/poor differentiation.
When it came to IHC test of MMRPs, there were 72 (24.3%, 72/296) tumors showed absence expression of at least one MMRP, with 68 MSI-H, 2 MSI-L, and 2 MSS. Of the 72 cases, PMS2 was negative in 52 (17.6%, 52/296) tumors, 50 of which were MSI-H, 2 were MSS. And MLH1 was negative in 51 (17.2%, 51/296) tumors, 49 of that were MSI-H, and 2 were MSS. Fifty-one tumors displayed absent co-expression of PMS2 and MLH1. All MLH1 negative tumors also showed absent expression of PMS2, with only one PMS2 negative tumor displaying intact expression of MLH1 (Figure 1). Simultaneously, 21 of 296 (7.1%) tumors showed absent expression of MSH6, of which 19 tumors were classified as MSI-H, 2 were MSI-L. And 15 of 296 (5.1%) cases displayed negative expression of MSH2; all of the 15 tumors were defined as MSI-H by MSI test. Similarly, all of the MSH2 negative tumors also showed absent expression of MSH6 protein, and 6 cases exhibited just MSH6 negative with normal expression of MSH2 (Figure 2). The two MSS tumors with abnormal MMRP expression showed exactly the same IHC pattern, which was concurrent negative of MLH1 and PMS2 with clonally absent of MSH6. The results of the 296 cases’ MSI status and IHC expression pattern of MMRPs are listed in Table 2. The sensitivity and specificity of the 2-panel IHC test including PMS2 and MSH6 for screening for mismatch repair defect were 100% and 98.2% respectively, exactly the same as that of the 4-antibody panel IHC with all of the 4 MMRPs. For the 2-antibody IHC involving MLH1 and MSH2 previously used, the sensitivity and specificity for MMRd detection were 92.6% and 99.1% separately. Detecting of PMS2 and MSH6 improved the sensitivity of the old IHC screening method, without reducing the specificity obviously, although the difference was not significant statistically (P=0.058). The diagnostic accordance rate of IHC test with PMS2 and MSH6 and MSI analysis was 98.6% (Table 3).
Figure 1.
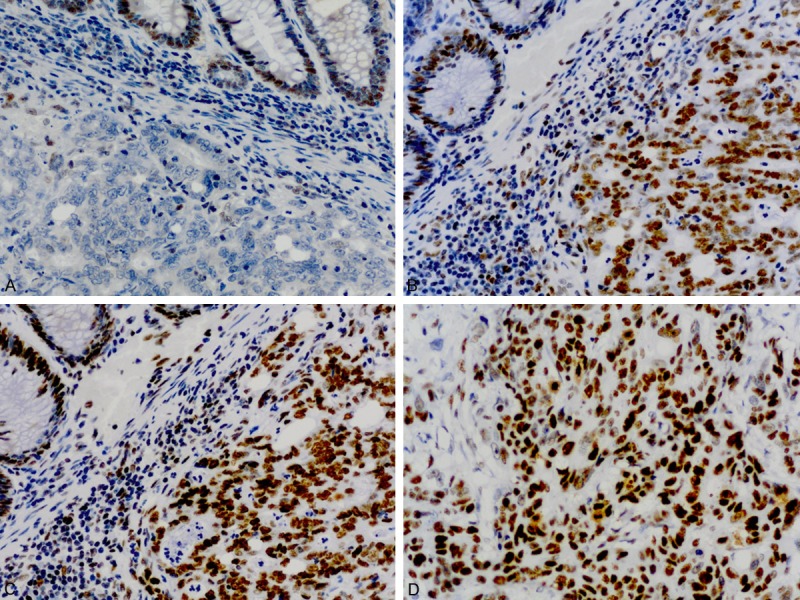
Immunohistochemical staining pattern of a MSI-H colorectal carcinoma with isolated loss of PMS2 (A), and intact staining of MLH1 (B), MSH2 (C) and MSH6 (D).
Figure 2.
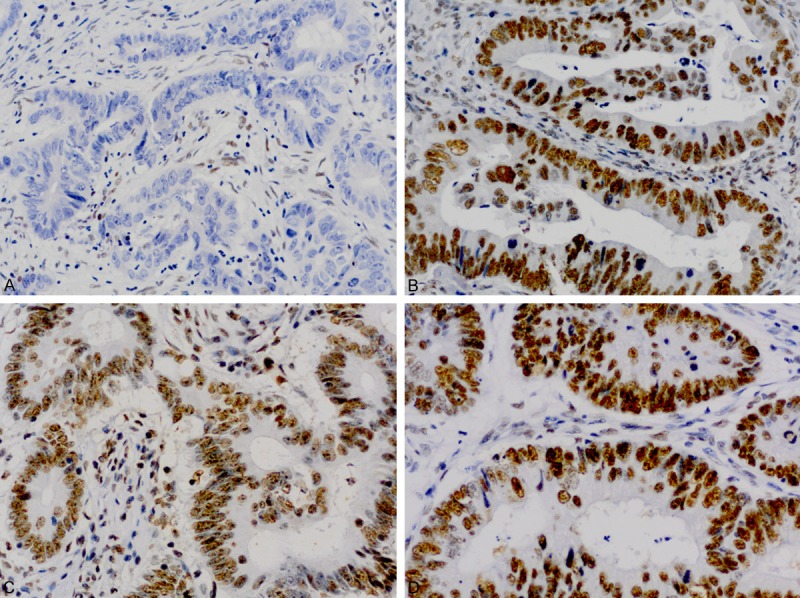
Immunohistochemical staining pattern of a MSI-H colorectal carcinoma with isolated loss of MSH6 (A) and intact staining of MSH2 (B), MLH1 (C), and PMS2 (D).
Table 2.
The IHC expression patterns of MMRPs and microsatellite status in colorectal carcinomas
| IHC expression pattern | Number of cases | Microsatellite status | ||
|---|---|---|---|---|
|
| ||||
| MSI-H (%) | MSI-L (%) | MSS (%) | ||
| at least 1 MMRP (-) | 72 | 68 (94.4) | 2 (2.8) | 2 (2.8) |
| MLH1 (-) PMS2 (-) | 50 | 48 (96.0) | 0(0) | 2 (4.0)* |
| PMS2 (-) alone | 1 | 1 (100) | 0 (0) | 0 (0) |
| MSH2 (-) MSH6 (-) | 14 | 14 (100) | 0 (0) | 0 (0) |
| MSH6 (-) alone | 6 | 4 (66.7) | 2 (33.3) | 0 (0) |
| 4 MMRPs (-) | 1 | 1 (100) | 0 (0) | 0 (0) |
| 4 MMRPs (+) | 224 | 217 (96.9) | 7 (3.1) | 0 (0) |
2 MSS tumors presented concurrent loss of MLH1 and PMS2 with clonal loss of MSH6.
Table 3.
Comparison of the three IHC panels for predicting MSI in colorectal carcinomas
| MSI status | MLH1+MSH2 | MLH1+MSH2+PMS2+MSH6 | PMS2+MSH6 | |||
|---|---|---|---|---|---|---|
|
| ||||||
| At least 1 MMRP negative | Intact expression | At least 1 MMRP negative | Intact expression | At least 1 MMRP negative | Intact expression | |
| MSI-H | 63 | 5 | 68 | 0 | 68 | 0 |
| MSI-L/MSS | 2 | 226 | 4 | 224 | 4 | 224 |
| Total number | 65 | 231 | 72 | 224 | 72 | 224 |
Discussion
The molecular phenotype of colorectal carcinoma is closely related to its clinicopathological characteristics, biological behavior, prognosis and even therapy response of patient, both in sporadic and hereditary colorectal cancer [4,10,11,21]. So, it is important to recognize different CRC by pathological-molecular features. Microsatellite instability (MSI) is a distinct characteristic of mismatch repair defective CRC, which including 12% of sporadic CRC and 3% of hereditary CRC, Lynch syndrome [5,22]. Microsatellite instability was well defined in Lynch syndrome, and then recommended to be screened with Bethesda guidelines, which was revised in 2003 [7]. It was issued that colorectal carcinoma with MSI-H (MMRd CRC) has characteristic clinical and pathological features different from MSS CRC, even including prognosis and chemotherapy response [23-25]. Even recently, Le etc. reported that mismatch-repair status predicted clinical benefit of immune checkpoint blockade with pembrolizumab [26]. In order to define the clinicopathological features of MMRd colorectal carcinoma, and to find the most effective screening method for MMRd colorectal carcinoma, we recruited 296 tumors matched revised Bethesda Guidelines from 1450 CRCs. MSI analysis revealed 68 CRCs with MSI-H, 9 with MSI-L and 219 with MSS. The analysis of correlation between clinicopathological characteristics and different microsatellite status showed that MMRd CRCs truly have distinct clinical and pathological features. Colorectal carcinomas with MSI-H in this study display a predilection for right colon, with 51.5% of MMRd CRCs located in proximal colon, which is similar to the result of previous studies [27-30]. Compared to MSS CRCs, 32.3% of CRCs with MSI-H showed poor differentiation, which is significantly different from MSS CRCs (19.6%). But when it came to mucinous or signet ring differentiation, the difference between MMRd (51.1%) and MSS (38.6%) colorectal carcinomas was not so significant as the results of other studies [6,9,29,31-35]. That is partially in line with these previous researches. Lymphocytic infiltration is generally recognized as a striking characteristic of MMRd CRCs, as many studies issued [6,29,31,33,36-38]. In our study, 20.6% of MMRd CRCs presented Crohn-like lymphoid reaction, peritumoral lymphocytes, or tumor-infiltrating lymphocytes, which is relatively higher than MSS CRCs (13.2%), but with no statistic differences. Patients with MSI-H colorectal carcinoma in our series have relatively lower stage compared with patients with MSS tumor, with 32.4% of MMRd CRCs and 52.5% of MSS CRCs at Ⅲ/Ⅳ TNM stage. And MMRd tumors less prone to metastasize to lymph nodes or distant organs compared to MSS ones, with only 30.9% and 2.9% of tumors have lymph nodes and distant metastasis respectively. That is similar to the results of previous researches [39-41], and can possibly explain the relative better prognosis of MMRd colorectal cancers compared with MSS ones [42,43]. Additionally, our study suggest that colorectal carcinomas with MSI-L in this series are more close to MSS ones morphologically and biologically, as previous studies issued [44,45]. Although neither the differences between the MSI-L group and MSI-H group, nor the differences between MSI-L group and MSS group are statistically significant.
Microsatellite instability is caused by mismatch repair deficiency, presenting accumulation of insertion or deletion mutations at microsatellite across entire genome. The most reliable method for MMRd detecting is microsatellite segments analysis with fluorescent multiplex PCR-capillary electrophoresis. But it is complex and costly. Another common method for MMRd detecting is immunohistochemistry test of mismatch repair gene products. Negative expression of anyone of the MMRPs in the tumor tissue indicates to mismatch repair deficiency of the tumor. Most researches around 2000 recommended the classical panel containing MLH1 and MSH2, with considerable predictive value for a MSI-H phenotype or germline mutation of MMR gene [46-49], but always missed cases with abnormal MSH6 or PMS2, or protein intact missense mutation of MLH1 [50-52].
We used all of the four MMRPs including MLH1, MSH2, MSH6 and PMS2 for IHC detecting, to select the most effective panel for MMRd screening. In the 72 tumors with at least one MMRP negative, the most frequent expression pattern of MMRPs was concurrent lost of MLH1 and PMS2, account for 69.4% (50) of all cases. And the concurrent negative expression of MSH2 and MSH6 was the second common pattern, with 19.4% (14) of tumors showed as this. Isolated MSH6 lost was presented in 6 cases (8.3%), followed by isolated PMS2 negative in 1 case (1.4%). That is similar to the result of the previous related studies list in Table 4 [14-16,51-54]. And one tumor displayed negative expression of all of the four MMRPs (1.4%). This pattern as well as some staining variants listed in Table 4 is rare, and the mechanism is still unclear. Neither isolated MLH1 nor isolated MSH2 lost was found in our series. The result is concordant to the molecular characteristics of MMRPs. As researches in vitro and in vivo proved [13,55-61], that mismatch repair gene products existing in cells are always stay as heterodimers complex. And MLH1 and MSH2 are obligatory partners, combined with their secondary partners PMS2 and MSH6 respectively. If degradation of the former partners occurs, caused by the mutation of respective MMR gene, the later partners will not exist anymore. But the opposite situation is not true. Of the 72 tumors with abnormal MMRPs expression, 68 were classified as MSI-H, 2 were MSI-L and 2 were MSS by MSI analysis. That is all of the 68 MSI-H colorectal cancers present abnormal MMRPs expression. Forty-eight of them displayed concurrent absent expression of MLH1 and PMS2. The 14 cases with concurrent negative expression of MSH2 and MSH6, and the one with all 4 MMRPs negative were all MSI-H tumors. The other MSI-H tumors were 4 isolated MSH6 negative, and 1 isolated PMS2 negative. Both of the 2 MSI-L tumors were isolated MSH6 absent, which is concordant with the fact that MSH6 mutations often cause low frequency MSI [62,63]. An interest phenomenon was observed in our study, that 2 MSS tumors with concurrent negative of MLH1 and PMS2 also displayed clonally absent expression of MSH6. And the staining heterogenicity is different from the focal, weak, or ambiguous IHC staining of MSH6 discussed in limited articles [16,61]. Shia et al. [63] described some similar IHC patterns of limited expression of MSH6, and issued that the completely loss of MLH1 and PMS2, with simultaneously reductive staining of MSH6 pattern might be attributed to neoadjuvant chemotherapy or somatic mutation of the coding region microsatellite of MSH6 gene. But it is worth noticing that, in our cases, the MSH6 negative tumor tissues in the same tumor were different from those positive morphologically. Positive tumor glands showed strong staining of MSH6, and negative tumor glands presented completely loss of MSH6, without any transition between them. Actually, the positive tumor glands and the negative glands were slightly different in differentiation. The 2 tumors with special staining pattern of MLH1, PMS2 and MSH6 are both MSS (the MMR status were confirmed for the second test). And both of the two patients haven’t received neoadjuvant chemotherapy. The molecular mechanism under the phenomenon is unclear, and further study is needed to elucidate it.
Table 4.
Immunohistochemichal staining patterns for MLH1, MSH2, MSH6 and PMS2 in colorectal carcinomas in literatures
| Reference | Patients | Total number | IHC patterns | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| ≥1 MMRP-* | MLH1-PMS2- | MSH2-MSH6- | PMS2-alone | MSH6-alone | Other abnormal patterns | Intact expression | |||
| Mojtahed et al. [16] | Selected‡ | 323 | 59 | 34 (58%) | 12 (20%) | 5 (8%) | 8 (14%) | 0 | 264 |
| Hall et al. [15] | Unselected | 344 | 104 | 89 (86%) | 13 (13%) | 1 (1%) | 1 (1%) | 1 (MLH1-PMS2-MSH6-) | 240 |
| Shia et al. [14] | RB (190), <40 y (42) | 232 | 70 | 41 (59%) | 14 (20%) | 9 (13%) | 6 (9%) | 0 | 162 |
| Hampel et al. [51] | Unselected | 483 | 71 | 45 (63%) | 12 (17%) | 2 (3%) | 9 (13%) | 1 (MLH1-), 1 (MLH1-PMS2-MSH6-), 1 (MLH1-MSH6-) | 412 |
| Watson et al. [52] | <60 y, MSI | 69 | 60 | 35 (58%) | 17 (28%) | 2 (3%) | 3 (5%) | 1 (MSH2-), 1 (PMS2-MSH6-), 1 (MLH1-PMS2-MSH6-) | 8§ |
| Truninger et al. [53] | Unselected | 1048 | 139 | 103 (74%) | 15 (11%) | 16 (12%) | 5 (4%) | 0 | 909 |
| Southey et al. [54] | <45 y | 105 | 26 | 13 (50%) | 7 (27%) | 2 (8%) | 4 (15%) | 0 | 79 |
| Our study | RB† | 296 | 72 | 50 (69%) | 14 (19%) | 1 (1%) | 6 (8%) | 1 (MLH1-MSH2-MSH6-PMS2-) | 224 |
| Total | 2900 | 601 | 410 (68%) | 104 (17%) | 38 (6%) | 42 (7%) | 2298 | ||
At least one mismatch repair protein negative.
Revised Bethesda Guideline.
Study groups consisted of 63 unselected cases and 260 cases selected based on one of the conditions: age <50 years; RB; CRC with MSI-H histology;
7/8 tumors with variable heterogeneous staining of one or more mismatch repair proteins.
Because that every tumor with absent MLH1 expression also showed negative expression of PMS2, and every tumor with absent MSH2 expression also showed negative expression of MSH6, the sensitivity and specificity of the IHC panel including PMS2 and MSH6 for screening for MSI-H tumor were the exact same as that of the traditional 4 MMRPs panel (Table 3), using MSI analysis as golden standard. To detect PMS2 and MSH6 can recognize missed cases by old panel of MLH1 and MSH2, with extremely high sensitivity reaching 100%.
Microsatellite instability test as the golden standard for MMRd screening is used in many clinical and pathological laboratory in developed contrary. But the complicated procedure and the expensive cost make it difficult to be universally implemented in small laboratories especially in developing countries. Immunohistochemistry test for MMRPs is simple and economical [64], with ideal sensitivity and specificity, compared with MSI analysis. Every clinical and pathological laboratory with IHC facilities can carry out the MMRPs detection for MMRd screening. Additionally, the MMRP detection result could direct the follow-up gene test of Lynch syndrome validation. Actually, for some cases that the tumor tissue contained scarce tumor cells, such as mucin-rich and desmoplastic adenocarcinomas, IHC has its unique advantage compare to molecular test [32]. So, as a sensitive and economical method easily implemented, IHC test for MMRPs should be carried out universally in clinical and pathological laboratories for MMRd screening and genetic counseling of Lynch syndrome, especially in developing countries. However, MSI test is a method for function detect, which can’t be replaced in certain conditions, such as MSI tumors with intact IHC expression (non-functional protein expression), or abnormal of other mismatch repair genes other than the traditional four genes [64]. The uncertainty of IHC staining and the subjectivity of result interpretation were also noticed by the writers and previous researchers [16,18,46,54]. Internal control should be emphasized, and ambiguous staining should be reconfirmed by repetition. Actually, IHC staining for MLH1, PMS2, MSH2 and MSH6 was repeated for 2-3 times in 7, 9, 1 and 3 tumors respectively to finally confirm the result. And the selection of proper antibodies of MMRPs initiatively is very important. So the quality control of IHC for MMRPs, the standardization of interpretation and the experience of interpreting pathologists are basically required. In order to obtain enough cases for study in limited time, we used the revised Bethesda Guideline as an initial screening method for MMRd validation. A random designed prospective study should be put into practice for further research.
In conclusion, MMRd CRC is a distinct molecular phenotype of colorectal carcinoma, with characteristic clinicopathological features. A two-antibody panel containing PMS2 and MSH6 is the simplest method for primary screening for MMRd CRC. And it is as effective as the traditional four-antibody panel, with halving of the expenditure. It should be universally carried out in clinical and pathological laboratories for molecular subtyping of CRC based on MMR competency, especially in developing countries.
Acknowledgements
This work has been supported by the Program of Science & Technology Commission of Shanghai Municipality (STCSM) (No. 06PJ14019).
Disclosure of conflict of interest
None.
References
- 1.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Guo P, Huang ZL, Yu P, Li K. Trends in cancer mortality in China: An update. Ann Oncol. 2012;23:2755–2762. doi: 10.1093/annonc/mds069. [DOI] [PubMed] [Google Scholar]
- 4.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 5.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shia J, Ellis NA, Paty PB, Nash GM, Qin J, Offit K, Zhang XM, Markowitz AJ, Nafa K, Guillem JG, Wong WD, Gerald WL, Klimstra DS. Value of histopathology in predicting microsatellite instability in hereditary nonpolyposis colorectal cancer and sporadic colorectal cancer. Am J Surg Pathol. 2003;27:1407–1417. doi: 10.1097/00000478-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, Lindblom A, Lynch HT, Peltomaki P, Ramsey SD, Rodriguez-Bigas MA, Vasen HF, Hawk ET, Barrett JC, Freedman AN, Srivastava S. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smyrk TC, Watson P, Kaul K, Lynch HT. Tumorinfiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–2422. [PubMed] [Google Scholar]
- 9.Greenson JK, Bonner JD, Ben-Yzhak O, Cohen HI, Miselevich I, Resnick MB, Trougouboff P, Tomsho LD, Kim E, Low M, Almog R, Rennert G, Gruber SB. Phenotype of microsatellite unstable colorectal carcinomas: Well-differentiated and focally mucinous tumors and the absence of dirty necrosis correlate with microsatellite instability. Am J Surg Pathol. 2003;27:563–570. doi: 10.1097/00000478-200305000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J. Clin. Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 11.Fallik D, Borrini F, Boige V, Viguier J, Jacob S, Miquel C, Sabourin JC, Ducreux M, Praz F. Microsatellite instability is a predictive factor of the tumor response to irinotecan in patients with advanced colorectal cancer. Cancer Res. 2003;63:5738–5744. [PubMed] [Google Scholar]
- 12.Bertagnolli MM, Niedzwiecki D, Compton CC, Hahn HP, Hall M, Damas B, Jewell SD, Mayer RJ, Goldberg RM, Saltz LB, Warren RS, Redston M. Microsatellite instability predicts improved response to adjuvant therapy with irinotecan, fluorouracil, and leucovorin in stage III colon cancer: Cancer and Leukemia Group B Protocol 89803. J. Clin. Oncol. 2009;27:1814–1821. doi: 10.1200/JCO.2008.18.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Jong AE, van Puijenbroek M, Hendriks Y, Tops C, Wijnen J, Ausems MG, Meijers-Heijboer H, Wagner A, van Os TA, Brocker-Vriends AH, Vasen HF, Morreau H. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:972–980. doi: 10.1158/1078-0432.ccr-0956-3. [DOI] [PubMed] [Google Scholar]
- 14.Shia J, Tang LH, Vakiani E, Guillem JG, Stadler ZK, Soslow RA, Katabi N, Weiser MR, Paty PB, Temple LK, Nash GM, Wong WD, Offit K, Klimstra DS. Immunohistochemistry as firstline screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: a 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol. 2009;33:1639–1645. doi: 10.1097/PAS.0b013e3181b15aa2. [DOI] [PubMed] [Google Scholar]
- 15.Hall G, Clarkson A, Shi A, Langford E, Leung H, Eckstein RP, Gill AJ. Immunohistochemistry for PMS2 and MSH6 alone can replace a four antibody panel for mismatch repair deficiency screening in colorectal adenocarcinoma. Pathology. 2010;42:409–413. doi: 10.3109/00313025.2010.493871. [DOI] [PubMed] [Google Scholar]
- 16.Mojtahed A, Schrijver I, Ford JM, Longacre TA, Pai RK. A two-antibody mismatch repair protein immunohistochemistry screening approach for colorectal carcinomas, skin sebaceous tumors, and gynecologic tract carcinomas. Mod Pathol. 2011;24:1004–1014. doi: 10.1038/modpathol.2011.55. [DOI] [PubMed] [Google Scholar]
- 17.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: Development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 18.Dietmaier W, Wallinger S, Bocker T, Kullmann F, Fishel R, Ruschoff J. Diagnostic microsatellite instability: definition and correlation with mismatch repair protein expression. Cancer Res. 1997;57:4749–4756. [PubMed] [Google Scholar]
- 19.Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–130. doi: 10.1111/j.1365-2559.2006.02549.x. [DOI] [PubMed] [Google Scholar]
- 20.Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- 21.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 22.Blake C, Tsao JL, Wu A, Shibata D. Stepwise deletions of polyA sequences in mismatch repair-deficient colorectal cancers. Am J Pathol. 2001;158:1867–1870. doi: 10.1016/S0002-9440(10)64143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 24.Ribic CM, Sargent DJ, Moore MJ, Thibodeau SN, French AJ, Goldberg RM, Hamilton SR, Laurent-Puig P, Gryfe R, Shepherd LE, Tu D, Redston M, Gallinger S. Tumor microsatelliteinstability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–257. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carethers JM, Smith EJ, Behling CA, Nguyen L, Tajima A, Doctolero RT, Cabrera BL, Goel A, Arnold CA, Miyai K, Boland CR. Use of 5-fluorouracil and survival in patients with microsatellite-unstable colorectal cancer. Gastroenterology. 2004;126:394–401. doi: 10.1053/j.gastro.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 26.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, Biedrzycki B, Donehower RC, Zaheer A, Fisher GA, Crocenzi TS, Lee JJ, Duffy SM, Goldberg RM, de la Chapelle A, Koshiji M, Bhaijee F, Huebner T, Hruban RH, Wood LD, Cuka N, Pardoll DM, Papadopoulos N, Kinzler KW, Zhou S, Cornish TC, Taube JM, Anders RA, Eshleman JR, Vogelstein B, Diaz LJ. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward R, Meagher A, Tomlinson I, O’Connor T, Norrie M, Wu R, Hawkins N. Microsatellite instability and the clinicopathological features of sporadic colorectal cancer. Gut. 2001;48:821–829. doi: 10.1136/gut.48.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA, Borten MM, Stitz R, Searle J, McKeone D, Fraser L, Purdie DR, Podger K, Price R, Buttenshaw R, Walsh MD, Barker M, Leggett BA, Jass JR. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halvarsson B, Anderson H, Domanska K, Lindmark G, Nilbert M. Clinicopathologic factors identify sporadic mismatch repair-defective colon cancers. Am J Clin Pathol. 2008;129:238–244. doi: 10.1309/0PP5GDRTXUDVKAWJ. [DOI] [PubMed] [Google Scholar]
- 30.Jover R, Paya A, Alenda C, Poveda MJ, Peiro G, Aranda FI, Perez-Mateo M. Defective mismatch-repair colorectal cancer: clinicopathologic characteristics and usefulness of immunohistochemical analysis for diagnosis. Am J Clin Pathol. 2004;122:389–394. doi: 10.1309/V9PG-K2Y2-60VF-VULR. [DOI] [PubMed] [Google Scholar]
- 31.Jass JR, Do KA, Simms LA, Iino H, Wynter C, Pillay SP, Searle J, Radford-Smith G, Young J, Leggett B. Morphology of sporadic colorectal cancer with DNA replication errors. Gut. 1998;42:673–679. doi: 10.1136/gut.42.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kakar S, Aksoy S, Burgart LJ, Smyrk TC. Mucinous carcinoma of the colon: Correlation of loss of mismatch repair enzymes with clinicopathologic features and survival. Mod Pathol. 2004;17:696–700. doi: 10.1038/modpathol.3800093. [DOI] [PubMed] [Google Scholar]
- 33.Wright CL, Stewart ID. Histopathology and mismatch repair status of 458 consecutive colorectal carcinomas. Am J Surg Pathol. 2003;27:1393–1406. doi: 10.1097/00000478-200311000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Yearsley M, Hampel H, Lehman A, Nakagawa H, de la Chapelle A, Frankel WL. Histologic features distinguish microsatellite-high from microsatellite-low and microsatellite-stable colorectal carcinomas, but do not differentiate germline mutations from methylation of the MLH1 promoter. Hum Pathol. 2006;37:831–838. doi: 10.1016/j.humpath.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenkins MA, Hayashi S, O’Shea AM, Burgart LJ, Smyrk TC, Shimizu D, Waring PM, Ruszkiewicz AR, Pollett AF, Redston M, Barker MA, Baron JA, Casey GR, Dowty JG, Giles GG, Limburg P, Newcomb P, Young JP, Walsh MD, Thibodeau SN, Lindor NM, Lemarchand L, Gallinger S, Haile RW, Potter JD, Hopper JL, Jass JR. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: A population-based study. Gastroenterology. 2007;133:48–56. doi: 10.1053/j.gastro.2007.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson P, Lin KM, Rodriguez-Bigas MA, Smyrk T, Lemon S, Shashidharan M, Franklin B, Karr B, Thorson A, Lynch HT. Colorectal carcinoma survival among hereditary nonpolyposis colorectal carcinoma family members. Cancer. 1998;83:259–266. [PubMed] [Google Scholar]
- 38.Aarnio M, Mustonen H, Mecklin JP, Jarvinen HJ. Prognosis of colorectal cancer varies in different high-risk conditions. Ann Med. 1998;30:75–80. doi: 10.3109/07853899808999387. [DOI] [PubMed] [Google Scholar]
- 39.Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, Roncalli M, Gennari L, Santoro A. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 2007;13:3831–3839. doi: 10.1158/1078-0432.CCR-07-0366. [DOI] [PubMed] [Google Scholar]
- 40.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gryfe R, Swallow C, Bapat B, Redston M, Gallinger S, Couture J. Molecular biology of colorectal cancer. Curr Probl Cancer. 1997;21:233–300. doi: 10.1016/s0147-0272(97)80003-7. [DOI] [PubMed] [Google Scholar]
- 42.Laiho P, Launonen V, Lahermo P, Esteller M, Guo M, Herman JG, Mecklin JP, Jarvinen H, Sistonen P, Kim KM, Shibata D, Houlston RS, Aaltonen LA. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002;62:1166–1170. [PubMed] [Google Scholar]
- 43.Halford SE, Sawyer EJ, Lambros MB, Gorman P, Macdonald ND, Talbot IC, Foulkes WD, Gillett CE, Barnes DM, Akslen LA, Lee K, Jacobs IJ, Hanby AM, Ganesan TS, Salvesen HB, Bodmer WF, Tomlinson IP, Roylance RR. MSI-low, a real phenomenon which varies in frequency among cancer types. J Pathol. 2003;201:389–394. doi: 10.1002/path.1453. [DOI] [PubMed] [Google Scholar]
- 44.Lindor NM, Burgart LJ, Leontovich O, Goldberg RM, Cunningham JM, Sargent DJ, Walsh-Vockley C, Petersen GM, Walsh MD, Leggett BA, Young JP, Barker MA, Jass JR, Hopper J, Gallinger S, Bapat B, Redston M, Thibodeau SN. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J. Clin. Oncol. 2002;20:1043–1048. doi: 10.1200/JCO.2002.20.4.1043. [DOI] [PubMed] [Google Scholar]
- 45.Dieumegard B, Grandjouan S, Sabourin JC, Le Bihan ML, Lefrere I, Bellefqih , Pignon JP, Rougier P, Lasser P, Benard J, Couturier D, Bressac-de PB. Extensive molecular screening for hereditary non-polyposis colorectal cancer. Br J Cancer. 2000;82:871–880. doi: 10.1054/bjoc.1999.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cawkwell L, Gray S, Murgatroyd H, Sutherland F, Haine L, Longfellow M, O’Loughlin S, Cross D, Kronborg O, Fenger C, Mapstone N, Dixon M, Quirke P. Choice of management strategy for colorectal cancer based on a diagnostic immunohistochemical test for defective mismatch repair. Gut. 1999;45:409–415. doi: 10.1136/gut.45.3.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muller W, Burgart LJ, Krause-Paulus R, Thibodeau SN, Almeida M, Edmonston TB, Boland CR, Sutter C, Jass JR, Lindblom A, Lubinski J, MacDermot K, Sanders DS, Morreau H, Muller A, Oliani C, Orntoft T, Ponz DLM, Rosty C, Rodriguez-Bigas M, Ruschoff J, Ruszkiewicz A, Sabourin J, Salovaara R, Moslein G. The reliability of immunohistochemistry as a prescreening method for the diagnosis of hereditary nonpolyposis colorectal cancer (HNPCC)--results of an international collaborative study. Fam Cancer. 2001;1:87–92. doi: 10.1023/a:1013840907881. [DOI] [PubMed] [Google Scholar]
- 48.Christensen M, Katballe N, Wikman F, Primdahl H, Sorensen FB, Laurberg S, Orntoft TF. Antibody-based screening for hereditary nonpolyposis colorectal carcinoma compared with microsatellite analysis and sequencing. Cancer. 2002;95:2422–2430. doi: 10.1002/cncr.10979. [DOI] [PubMed] [Google Scholar]
- 49.Mangold E, Pagenstecher C, Friedl W, Fischer HP, Merkelbach-Bruse S, Ohlendorf M, Friedrichs N, Aretz S, Buettner R, Propping P, Mathiak M. Tumours from MSH2 mutation carriers show loss of MSH2 expression but many tumours from MLH1 mutation carriers exhibit weak positive MLH1 staining. J Pathol. 2005;207:385–395. doi: 10.1002/path.1858. [DOI] [PubMed] [Google Scholar]
- 50.Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox E. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- 51.Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Clendenning M, Sotamaa K, Prior T, Westman JA, Panescu J, Fix D, Lockman J, LaJeunesse J, Comeras I, de la Chapelle A. Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J. Clin. Oncol. 2008;26:5783–5788. doi: 10.1200/JCO.2008.17.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watson N, Grieu F, Morris M, Harvey J, Stewart C, Schofield L, Goldblatt J, Iacopetta B. Heterogeneous staining for mismatch repair proteins during population-based prescreening for hereditary nonpolyposis colorectal cancer. J Mol Diagn. 2007;9:472–478. doi: 10.2353/jmoldx.2007.060162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truninger K, Menigatti M, Luz J, Russell A, Haider R, Gebbers JO, Bannwart F, Yurtsever H, Neuweiler J, Riehle HM, Cattaruzza MS, Heinimann K, Schar P, Jiricny J, Marra G. Immunohistochemical analysis reveals high frequency of PMS2 defects in colorectal cancer. Gastroenterology. 2005;128:1160–1171. doi: 10.1053/j.gastro.2005.01.056. [DOI] [PubMed] [Google Scholar]
- 54.Southey MC, Jenkins MA, Mead L, Whitty J, Trivett M, Tesoriero AA, Smith LD, Jennings K, Grubb G, Royce SG, Walsh MD, Barker MA, Young JP, Jass JR, St JD, Macrae FA, Giles GG, Hopper JL. Use of molecular tumor characteristics to prioritize mismatch repair gene testing in early-onset colorectal cancer. J. Clin. Oncol. 2005;23:6524–6532. doi: 10.1200/JCO.2005.04.671. [DOI] [PubMed] [Google Scholar]
- 55.Koi M, Umar A, Chauhan DP, Cherian SP, Carethers JM, Kunkel TA, Boland CR. Human chromosome 3 corrects mismatch repair deficiency and microsatellite instability and reduces N-methyl-N’-nitro-N-nitrosoguanidine tolerance in colon tumor cells with homozygous hMLH1 mutation. Cancer Res. 1994;54:4308–4312. [PubMed] [Google Scholar]
- 56.Umar A, Koi M, Risinger JI, Glaab WE, Tindall KR, Kolodner RD, Boland CR, Barrett JC, Kunkel TA. Correction of hypermutability, Nmethyl-N’-nitro-N-nitrosoguanidine resistance, and defective DNA mismatch repair by introducing chromosome 2 into human tumor cells with mutations in MSH2 and MSH6. Cancer Res. 1997;57:3949–3955. [PubMed] [Google Scholar]
- 57.Watanabe Y, Haugen-Strano A, Umar A, Yamada K, Hemmi H, Kikuchi Y, Takano S, Shibata Y, Barrett JC, Kunkel TA, Koi M. Complementation of an hMSH2 defect in human colorectal carcinoma cells by human chromosome 2 transfer. Mol Carcinog. 2000;29:37–49. [PubMed] [Google Scholar]
- 58.Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci U S A. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boland CR, Koi M, Chang DK, Carethers JM. The biochemical basis of microsatellite instability and abnormal immunohistochemistry and clinical behavior in Lynch syndrome: from bench to bedside. Fam Cancer. 2008;7:41–52. doi: 10.1007/s10689-007-9145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berends MJ, Wu Y, Sijmons RH, Mensink RG, van der Sluis T, Hordijk-Hos JM, de Vries EG, Hollema H, Karrenbeld A, Buys CH, van der Zee AG, Hofstra RM, Kleibeuker JH. Molecular and clinical characteristics of MSH6 variants: An analysis of 25 index carriers of a germline variant. Am J Hum Genet. 2002;70:26–37. doi: 10.1086/337944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buttin BM, Powell MA, Mutch DG, Babb SA, Huettner PC, Edmonston TB, Herzog TJ, Rader JS, Gibb RK, Whelan AJ, Goodfellow PJ. Penetrance and expressivity of MSH6 germline mutations in seven kindreds not ascertained by family history. Am J Hum Genet. 2004;74:1262–1269. doi: 10.1086/421332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shia J, Klimstra DS, Nafa K, Offit K, Guillem JG, Markowitz AJ, Gerald WL, Ellis NA. Value of immunohistochemical detection of DNA mismatch repair proteins in predicting germline mutation in hereditary colorectal neoplasms. Am J Surg Pathol. 2005;29:96–104. doi: 10.1097/01.pas.0000146009.85309.3b. [DOI] [PubMed] [Google Scholar]
- 63.Shia J, Zhang L, Shike M, Guo M, Stadler Z, Xiong X, Tang LH, Vakiani E, Katabi N, Wang H, Bacares R, Ruggeri J, Boland CR, Ladanyi M, Klimstra DS. Secondary mutation in a coding mononucleotide tract in MSH6 causes loss of immunoexpression of MSH6 in colorectal carcinomas with MLH1/PMS2 deficiency. Mod Pathol. 2013;26:131–138. doi: 10.1038/modpathol.2012.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Debniak T, Kurzawski G, Gorski B, Kladny J, Domagala W, Lubinski J. Value of pedigree/clinical data, immunohistochemistry and microsatellite instability analyses in reducing the cost of determining hMLH1 and hMSH2 gene mutations in patients with colorectal cancer. Eur J Cancer. 2000;36:49–54. doi: 10.1016/s0959-8049(99)00208-7. [DOI] [PubMed] [Google Scholar]


